Summary
Background
The incidence of HIV infection in young women in Africa is very high. We did a large-scale community-wide phylogenetic study to examine the underlying HIV transmission dynamics and the source and consequences of high rates of HIV infection in young women in South Africa.
Methods
We did a cross-sectional household survey of randomly selected individuals aged 15–49 years in two neighbouring subdistricts (one urban and one rural) with a high burden of HIV infection in KwaZulu-Natal, South Africa. Participants completed structured questionnaires that captured general demographic, socioeconomic, psychosocial, and behavioural data. Peripheral blood samples were obtained for HIV antibody testing. Samples with HIV RNA viral load greater than 1000 copies per mL were selected for genotyping. We constructed a phylogenetic tree to identify clusters of linked infections (defined as two or more sequences with bootstrap or posterior support ≥90% and genetic distance ≤4.5%).
Findings
From June 11, 2014, to June 22, 2015, we enrolled 9812 participants, 3969 of whom tested HIV positive. HIV prevalence (weighted) was 59.8% in 2835 women aged 25–40 years, 40.3% in 1548 men aged 25–40 years, 22.3% in 2224 women younger than 25 years, and 7.6% in 1472 men younger than 25 years. HIV genotyping was done in 1589 individuals with a viral load of more than 1000 copies per mL. In 90 transmission clusters, 123 women were linked to 103 men. Of 60 possible phylogenetically linked pairings with the 43 women younger than 25 years, 18 (30.0%) probable male partners were younger than 25 years, 37 (61.7%) were aged 25–40 years, and five (8.3%) were aged 41–49 years: mean age difference 8.7 years (95% CI 6.8–10.6; p<0.0001). For the 92 possible phylogenetically linked pairings with the 56 women aged 25–40 years, the age difference dropped to 1.1 years (95% CI –0.6 to 2.8; p=0.111). 16 (39.0%) of 41 probable male partners linked to women younger than 25 years were also linked to women aged 25–40 years. Of 79 men (mean age 31.5 years) linked to women younger than 40 years, 62 (78.5%) were unaware of their HIV-positive status, 76 (96.2%) were not on antiretroviral therapy, and 29 (36.7%) had viral loads of more than 50 000 copies per mL.
Interpretation
Sexual partnering between young women and older men, who might have acquired HIV from women of similar age, is a key feature of the sexual networks driving transmission. Expansion of treatment and combination prevention strategies that include interventions to address age-disparate sexual partnering is crucial to reducing HIV incidence and enabling Africa to reach the goal of ending AIDS as a public health threat.
Funding
President’s Emergency Program for AIDS Relief, US Centers for Disease Control and Prevention, South African Medical Research Council, and MAC AIDS Fund.
Introduction
In southern and eastern Africa, the incidence of HIV in young women (aged <25 years) remains high, despite extensive prevention campaigns and scale-up of antiretroviral therapy (ART).1 The Joint UN Programme on HIV/AIDS (UNAIDS) has identified young women in these regions as the highest priority group for HIV prevention to help achieve the global goal of ending AIDS as a public health threat by 2030.1
A substantial imbalance in HIV prevalence exists between men and women throughout southern and eastern Africa, and women acquire HIV at a much earlier age than their male peers (appendix p 4). Although the reasons for these differences are not fully understood, the sexual network structure and dynamics might have prevalence2 suggest that older men are probably the main source of infection for young women, this suggestion has been disputed in two recent studies.3,4 However, findings from these studies were based on reported ages of male partners presumed to be the source of, or at least related to, HIV infections in women. The lack of clarity and definitive data on whether age-disparate sexual partnering is important for HIV transmission has hampered HIV prevention in southern and eastern Africa, which hosts more than 50% of the global HIV burden.1 Current prevention programmes in South Africa and several other countries have steered clear of including this sociobehavioural factor in their prevention messages and education programmes.
Current HIV prevention programmes have had limited impact on reducing HIV incidence in young women. Prevention strategies targeting the key underlying drivers of HIV transmission could provide a more focused approach to HIV prevention in this high-risk group. Phylogenetic analysis can be used to identify drivers of HIV transmission, as seen in the ATHENA cohort study of men who have sex with men in the Netherlands.5 Phylodynamic models can assess how transmission dynamics change over time, as used in a study in men who have sex with men in Detroit, USA.6 However, longitudinal cohorts with detailed clinical and behavioural data are rare in Africa.7
The purpose of our large-scale community-wide phylogenetic study was to examine the underlying HIV transmission dynamics and the source and consequences of high rates of HIV infection in young women in a district with a high burden of HIV in KwaZulu-Natal, South Africa. In this community, the reported incidence of HIV in women younger than 25 years is 10.2 per 100 person-years (36 seroconversions during 353 person-years of follow-up in the placebo group of a microbicide trial).8
Methods
Study population
We did a household-based HIV survey in two neighbouring subdistricts (one urban and one rural) of the uMgungundlovu district of KwaZulu-Natal, South Africa. The household survey is described in detail in the study protocol.9 In summary, this was a cross-sectional survey of randomly selected individuals aged 15–49 years (appendix p 2). We used multistage sampling to randomly select households and recruit a household-representative sample of men and women. The primary sampling unit was the enumeration area, the secondary sampling unit was household, and the tertiary sampling unit was the individual within a household. 221 of 591 enumeration areas were drawn randomly, and households within each enumeration area were identified by a global positioning system. In the selected households, one individual was selected at random to be included in the study. Eligible participants received study information and provided written informed consent (or parental consent and assent if aged <18 years) in the preferred language of English or isiZulu before enrolment in the study.
The study was approved by the Biomedical Research Ethics Committee, University of KwaZulu-Natal; the US Centers for Disease Control and Prevention; and the KwaZulu-Natal Provincial Department of Health.
Procedures
Each participant was assigned a unique study number linked to the structured questionnaires that captured general demographic, socioeconomic, psychosocial, and behavioural data. This information included, but was not limited to, self-reported HIV testing history, ART use, sexually transmitted infections, and male circumcision status.
Peripheral blood samples were collected by trained staff. HIV antibody testing was done with the HIV enzyme Vironostika HIV Uniform II Antigen/Antibody microELISA system (BioMérieux, Marcy l’Étoile, France) and Elecsys HIV 1/2 combi PT assay (Roche Diagnostics, Penzberg, Germany). Samples that produced an indeterminate result were resolved using the ADVIA Centaur HIV Antigen/Antibody Combo (CHIV) assay (Siemens, Tarrytown, NY, USA). HIV antibody-positive samples were confirmed with the HIV-1 Western Blot assay (Bio-Rad Laboratories, Redmond, WA, USA). HIV antibody-negative samples were also tested with a pooled nucleic acid amplification test for detection of HIV-1 RNA using the COBAS AmpliPrep/COBAS TaqMan HIV-1 version 2.0 assay (Roche), in pools of ten samples. Pooled samples testing positive were disaggregated and retested individually. For all HIV-1-positive samples, CD4 cell counts were measured by BD FACSCalibur flow cytometry (BD Biosciences, San Jose, CA, USA). Viral loads were determined for all HIV-positive samples with the COBAS AmpliPrep/COBAS TaqMan HIV-1 version 2.0 assay.
Samples with HIV RNA viral loads greater than 1000 copies per mL were selected for genotyping (1589 sequences generated), which was done in three accredited laboratories in South Africa—namely, the National Institute of Communicable Diseases (NICD) HIV Genotyping Laboratory in Johannesburg (194 sequences), the Wellcome Trust Africa Centre for Population Health Virology Laboratory in Durban (685), and the Global Clinical Viral Laboratory in Durban (710). The inter-laboratory quality control procedure between the Global Clinical Viral Laboratory and the NICD generated nearly identical (mean pairwise distance at nucleotide level 0.000 [range 0.000–0.002] and aminoacid level 0.001 [range 0.000–0.006]) sequences for the random sample of 14 (0.9%) plasma specimens tested in both laboratories. Overall, the analysis showed sufficient nucleotide and aminoacid similarity between pairs of sequences to pass the inter-laboratory validation process. The Africa Centre for Population Health Virology Laboratory participates in an external proficiency testing programme of the French National Agency for Research on AIDS and Viral Hepatitis (ANRS).
All three laboratories generated HIV-1 sequences using the SATuRN/Life Technologies genotyping system,10 with standard operating procedures common to all three laboratories. In short, this protocol uses four sequencing primers for the generation of the 1197 bp pol sequence covering all 99 HIV-1 protease codons and the first 300 codons of the reverse transcriptase gene. Geneious software was used for sequence assembly of the Sanger electropherogram reads. The viral sequences were analysed using the HIV-1 Quality Analysis Tool (For more on the HIV-1 Quality Analysis Tool see http://www.bioafrica.net/software.php) and the Calibrated Population Resistance (CPR) tool11 to identify sequencing problems such as frameshifts, stop codons, and unusual polymorphisms. To further identify potential contamination, the generated sequences were compared with a local BLAST database, which included 11 500 unique sequences.12 Sequences with more than 98% identity to sequences in the database were regarded as potential contaminants. For all potential contaminants, the laboratory where they were produced as well as the RNA extraction and sequencing dates were identified. If the RNA extraction or sequencing happened on the same day, the RNA was re-extracted and resequenced at different dates or different laboratories, or both. Sequences that did not pass this quality step were not used for the construction of the phylogenetic tree. HIV subtyping was done with the REGA HIV-1 Subtyping Tool (version 3.0) and by maximum likelihood phylo- genetic confirmation. All but five of the 1589 sequences were subtype C.
Phylogenetic analysis
HIV-1 sequences were aligned using MAFFT13 and visually inspected in AliView.14 The alignment was edited manually until a perfect codon-based alignment was produced. A maximum likelihood tree was reconstructed using FastTree15 through a computer cluster of the Centre for High Performance Computing at the University of KwaZulu-Natal. The model of evolution was estimated from the dataset with ModelTest. The selected model was the general time reversible substitution model16 with γdistributed rate variation among sites.17 To assess the clade support, we applied the Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) with 1000 pseudo-replicates. To refine the topology, we re-estimated the maximum likelihood tree with 100 extra rounds of branch moves. This process was done with both nearest-neighbour interchanges and subtree-prune- regraft tree topology operators, as applied in FastTree.
HIV-1 transmission clusters were identified from the maximum likelihood tree with the ClusterPicker software application.18 We defined a transmission cluster as any clade with a support higher than 90% (SH-aLRT) and whose sequences had an intraclade genetic distance of less than or equal to 4.5%, parameters that are commonly used for the identification of transmission clusters.5,18 Phylogenetic linkage was regarded as the relation between two or more individuals within a cluster and, hence, were probable nodes within a transmission network. Every sequence within each cluster had to be connected to another sequence by less than or equal to 4.5% genetic distance; the mean genetic diversity was 2.8% and the mean branch support was 99.6%. All sequences that were identified in phylogenetic clusters were extracted and a confirmatory phylogenetic analysis done. This analysis involved the construction of a maximum likelihood phylogenetic tree with 1000 bootstraps using PhyML, with the same substitution model (general time reversible with γ-distributed rate variation among sites) as used for the initial FastTree-constructed phylogenetic tree. Additionally, we used MrBayes to construct a second phylogenetic tree with Bayesian methods. Because this Bayesian tree did not differ from the maximum likelihood tree, only the latter was analysed further. The maximum likelihood tree was analysed with ClusterPicker with the previously described parameters (ie, branch support >90% and genetic distance ≤4.5%) and compared with epidemiological data by use of ClusterMatcher.18 Lastly, trees were visualised and annotated with key demographic and epidemiological data in FigTree.
Directionality of transmission was assessed through a stratified age and sex prevalence analysis based on the probability of one sampled individual transmitting to another within a cluster being a function of the sequencing coverage and the prevalence in the population.6 Within each cluster, all possible pairings were identified and stratified by age of women and male partner’s age into three categories (<25 years, 25–40 years, and >40 years), on the basis of previous HIV prevalence rates in this community, thereby generating nine groups of age–sex pairings. We calculated the number of pairings in each of the nine groups and the groups that constituted most pairings were analysed for directionality by determining whether men or women had higher HIV prevalence.
Statistical analysis
Taking into account the complex multilevel sampling approach and adjusting for non-response, weighted data were analysed with SAS version 9.4 survey procedures (see appendix for details of sample size calculation and weighting procedures). Two-sample t tests or Kruskal-Wallis non-parametric tests, and χ² or Fisher’s exact tests, were used to compare continuous and categorical descriptive outcomes, respectively.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. ABMK, TdO, TG, SSAK, and AG had full access to all the data in the study. ABMK, TdO, and SSAK had final responsibility for the decision to submit for publication.
Results
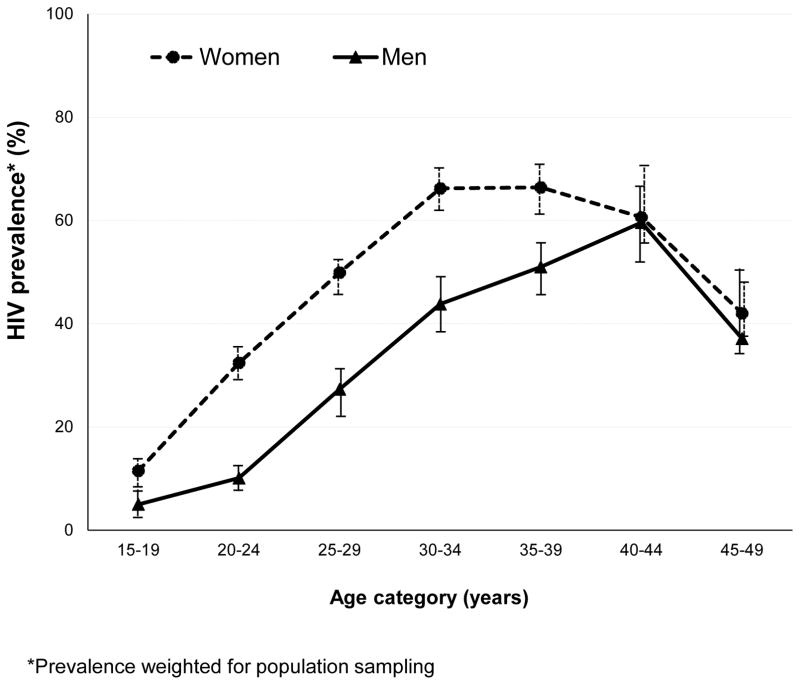
Enrolment for the study began on June 11, 2014, and ended on June 22, 2015. 14 618 households were eligible; of these, 2144 (14.7%) heads of households refused to take part in the study, 753 (5.2%) households had no one at home after three visits, and 432 (3.0%) households had no one home after the first visit, but were never revisited because the protocol-specified sample size was met before they needed to be. 11 289 (77.2%) households were enrolled, 398 of whom did not have an eligible person living in the household. 10 891 potentially eligible individuals were approached for participation in the study; 577 (5.3%) refused to take part (246 men and 331 women), 488 (4.5%) were still in the process of completing enrolment when the study ended, and 14 (0.1%) had no HIV results available. 9812 individuals were enrolled and tested for HIV, of whom 3969 (40.5%) were HIV positive (weighted 36.3%, 95% CI 34.8–37.8; table 1). 2955 (47.2%) of 6265 women (44.1%, 42.3–45.9) had HIV compared with 1014 (28.6%) of 3547 men (28.0%, 25.9–30.1; p<0.0001; table 2). HIV prevalence was 62.0% (1758 of 2835; weighted 59.8%) in women aged 25–40 years and was highest in those aged 35–39 years (517 [68.0%] of 760; weighted 66.4%; figure 1). In individuals younger than 25 years, HIV prevalence was about three times higher in women than in men (p<0.0001; table 2).
Table 1.
Demographic characteristics and HIV disease parameters on the entire sample and the progressive sub-populations in a KwaZulu-Natal community in South Africa
| All individuals sampled (N=9812)* | HIV positive individuals (N=3969)* | Individuals with Viral Load >1000 copies per mL (N=1847)* | Individuals with sequenced HIV (N=1589)*# | Individuals in transmission clusters (N=469) | Individuals in heterosexual transmission clusters
|
||
|---|---|---|---|---|---|---|---|
| Female (N=123) | Male (N=103) | ||||||
| Number of women (%) | 6265 (51.8%) | 2955 (62.8%) | 1264 (55.6%) | 1102 (55.7%) | 318 (67.8%) | - | - |
| Age in categories | |||||||
| <25 years | 3696 (39.0%) | 690 (16.1%) | 443 (22.1%) | 372 (21.7%) | 128 (27.3%) | 43 (35.0%) | 21 (20.4%) |
| 25–40 years | 4383 (46.2%) | 2402 (64.0%) | 1118 (64.1%) | 956 (63.7%) | 263 (56.1%) | 56 (45.5%) | 64 (62.1%) |
| ≥40 years | 1733 (14.8%) | 877 (20.0%) | 286 (13.8%) | 261 (14.6%) | 78 (16.6%) | 24 (19.5%) | 18 (17.5%) |
| Age mean (SE) | 29.0 (0.2) | 33.0 (0.2) | 31.0 (0.2) | 31.2 (0.3) | 30.9 (0.4) | 30.7 (0.8) | 32.1 (0.8) |
| Education | |||||||
| No schooling | 418 (2.8%) | 185 (3.1%) | 85 (2.9%) | 73 (3.2%) | 29 (6.2%) | 9 (7.3%) | 6 (5.8%) |
| Primary / Incomplete secondary | 4828 (52.0%) | 2112 (56.3%) | 940 (54.3%) | 811 (55.5%) | 217 (46.3%) | 59 (48.0%) | 52 (50.5%) |
| Secondary | 4009 (39.8%) | 1525 (37.1%) | 753 (38.9%) | 645 (37.7%) | 199 (42.4%) | 54 (43.9%) | 38 (36.9%) |
| Tertiary | 552 (5.3%) | 146 (3.5%) | 69 (3.9%) | 60 (3.6%) | 24 (5.1%) | 1 (0.8%) | 7 (6.8%) |
| Unknown | 5 (<0.1%) | 1 (<0.1%) | - | - | - | - | - |
| Marital status | |||||||
| Married | 879 (9.1%) | 335 (8.8%) | 103 (5.7%) | 83 (5.2%) | 15 (3.2%) | 3 (2.4%) | 3 (2.9%) |
| Single | 8223 (84.9%) | 3220 (82.0%) | 1565 (85.8%) | 1440 (91.0%) | 441 (94.0%) | 117 (95.1%) | 97 (94.2%) |
| Stable relationship but not married | 236 (2.3%) | 136 (3.8%) | 51 (2.9%) | 43 (2.6%) | 8 (1.7%) | 2 (1.6%) | 2 (1.9%) |
| Other | 474 (3.7%) | 278 (5.4%) | 128 (5.6%) | 23 (1.3%) | 5 (1.1%) | 1 (0.8%) | 1 (1.0%) |
| Condom use in last 12 months | |||||||
| Reported always using condoms in last12 months | 1587 (16.9%) | 789 (21.1%) | 300 (16.6%) | 253 (16.5%) | 77 (16.4%) | 17 (13.8%) | 18 (17.5%) |
| Reported never using a condom in last12 months | 1501 (15.6%) | 583 (14.9%) | 304 (16.4%) | 265 (15.0%) | 72 (15.4%) | 21 (17.1%) | 17 (16.5%) |
| Mean age at sexual debut (SE) | 17.9 (0.1) | 18.1 (0.1) | 17.9 (0.1) | 17.9 (0.1) | 17.9 (0.2) | 18.3 (0.3) | 17.8 (0.4) |
| Number of lifetime partners | |||||||
| One partner only | 1967 (19.8%) | 626 (16.3%) | 299 (16.8%) | 249 (16%) | 78 (14.9%) | 27 (23.3%) | 9 (6.8%) |
| Between 2 and 5 partners | 4050 (41.6%) | 2011 (51.1%) | 881 (47.2%) | 769 (47.9%) | 219 (46.4%) | 61 (55.9%) | 37 (38.4%) |
| More than 5 partners | 1040 (13.5%) | 518 (17.6%) | 262 (19.3%) | 229 (19.6%) | 65 (20.8%) | 8 (6.7%) | 30 (34.9%) |
| Not reported | 2755 (25.2%) | 814 (15%) | 405 (16.6%) | 342 (16.5%) | 107 (17.8%) | 27 (14.2%) | 27 (19.9%) |
| Number of partners in the last 12 months | |||||||
| Zero partners | 3113 (30.9%) | 956 (21.8%) | 428 (21.3%) | 375 (22.3%) | 113 (21.8%) | 35 (24.9%) | 25 (19.6%) |
| One partner only | 5473 (55.7%) | 2482 (63.9%) | 1139 (61.1%) | 976 (59.7%) | 280 (55.8%) | 77 (67.6%) | 50 (47.3%) |
| Between 2 and 5 partners | 610 (8.7%) | 236 (8.9%) | 129 (11.4%) | 112 (11.9%) | 34 (14.6%) | 3 (4.1%) | 14 (20.2%) |
| More than 5 partners | 55 (0.8%) | 23 (0.7%) | 20 (1.2%) | 16 (1.2%) | 7 (2.5%) | 1 (0.3%) | 3 (5.4%) |
| Not reported | 556 (3.8%) | 269 (4.6%) | 131 (5.0%) | 110 (5.0%) | 35 (5.3%) | 7 (3.1%) | 11 (7.5%) |
| Transactional sex† | 1325 (10.8%) | 649 (13.3%) | 303 (12.8%) | 255 (12.6%) | 84 (13.9%) | 29 (17.3%) | 17 (15.2%) |
| Income per month (R15 = US$1) | |||||||
| No income | 1290 (10.7%) | 527 (10.7%) | 265 (11.6%) | 225 (11.4%) | 70 (14.9%) | 18 (14.6%) | 14 (13.6%) |
| R1 – R500 | 907 (6.2%) | 450 (8.3%) | 219 (9.7%) | 183 (9.8%) | 72 (15.4%) | 21 (17.1%) | 15 (14.6%) |
| R501 – R2,500 | 4156 (41.6%) | 1729 (43.0%) | 805 (42.0%) | 706 (43.0%) | 192 (40.9%) | 57 (46.3%) | 36 (35.0%) |
| R2,501 – R6,000 | 1902 (24.3%) | 732 (23.2%) | 332 (22.9%) | 276 (22.1%) | 83 (17.7%) | 20 (16.3%) | 19 (18.4%) |
| >R6,000 | 674 (9.0%) | 220 (7.5%) | 87 (6.4%) | 80 (6.4%) | 24 (5.1%) | 5 (4.1%) | 5 (4.9%) |
| No Response | 883 (8.2%) | 311 (7.4%) | 139 (7.3%) | 119 (7.4%) | 28 (6.0%) | 2 (1.6%) | 14 (13.6%) |
| HIV status (self-reported) $ | |||||||
| Positive | 2367 (22.1%) | 2337 (59.8%) | 718 (37.8%) | 629 (39.1%) | 164 (35.0%) | 49 (39.8%) | 27 (26.2%) |
| Negative | 4708 (51.7%) | 823 (22.1%) | 588 (34.7%) | 493 (33.0%) | 149 (31.8%) | 39 (31.7%) | 23 (22.3%) |
| Don't know | 2673 (25.8%) | 772 (17.4%) | 520 (26.6%) | 449 (27.1%) | 149 (31.8%) | 32 (26.0%) | 51 (49.5%) |
| No response | 64 (0.4%) | 37 (0.6%) | 21 (0.8%) | 18 (0.9%) | 7 (1.5%) | 3 (2.4%) | 2 (1.9%) |
| Currently on antiretroviral therapy | 1598 (15.4%) | 1592 (42.3%) | 178 (10.5%) | 161 (11.8%) | 19 (4.1%) | 5 (4.1%) | 5 (4.9%) |
| Median Viral Load (copies per mL) (Inter-Quartile Range) | - | 402 (0 – 23656) | 26846 (9424 – 76148) | 24289 (7485– 72403) | 24222 (6738 – 68000) | 16629 (5769 – 38133) | 31000 (11906 – 85412) |
| Mean CD4 cell count (cells per μL) (SE) | - | 526 (6.2) | 439 (9.8) | 450 (9.9) | 459 (12.6) | 493 (26.5) | 424 (26.3) |
Analyses are weighted to adjust for the multi-level sampling
Only individuals with viral load above 1000 could be included in this sequencing analysis
Self-reported information on HIV status was not available in 3 men and 2 women
SE: Standard error
Assessed in most recent sexual partner only
Table 2.
Community-based HIV prevalence and proportion of individuals with viral sequences by sex in each of the three age categories
| HIV prevalence: weighted % (n/N=unweighted %) | Total | |||
|---|---|---|---|---|
|
|
||||
| Age category | < 25 years | 25–40 years | 41–49 years | |
| Women | 22.3% (567/2224= 25.5%) | 59.8% (1758/2835= 62.0%) | 50.1% (630/1206=52.2%) | 44.1% (2955/6265=47.2%) |
| Men | 7.6% (123/1472=8.4%) | 40.3% (644/1548=41.6%) | 47.2% (247/527=46.9%) | 28.0% (1014/3547=28.6%) |
|
| ||||
| Proportion of individuals with viral sequences (n/N) | Total | |||
|
|
||||
| Age category | < 25 years | 25–40 years | 41–49 years | |
| Women | 52.7% (299/567) | 35.7% (627/1758) | 27.9% (176/630) | 37.3% (1102/2955) |
| Men | 59.3% (73/123) | 51.1% (329/644) | 34.4% (85/247) | 48.0% (487/1014) |
Figure 1.
HIV prevalence rates in a KwaZulu-Natal community in South Africa by age and gender
1592 (40.1%; weighted 42.3%) of the 3969 HIV-positive individuals reported that they were on ART (table 1), coverage that was similar to national treatment coverage in ART-eligible individuals at the time of the study.1 1412 (88.8%; weighted 88.3%) of 1590 participants who reported using ART and whose viral loads were tested had HIV RNA viral loads of less than 1000 copies per mL, whereas 697 (29.5%; weighted 27.1%) of 2366 individuals who reported that they were not on ART had viral loads less than 1000 copies per mL. 13 HIV-positive individuals had samples insuffcient for viral load testing. 2292 (57.9%; weighted 58.0%) of the 3956 HIV-positive individuals whose viral loads were tested had detectable viral loads of at least 20 copies per mL; 1847 (46.5%) had more than 1000 copies per mL and were selected for sequencing (table 1).
Blood samples were available from 1688 individuals whose viruses were selected for sequencing. The viruses of 95 of these individuals could not be sequenced and four sequences did not pass the quality control step, resulting in 1589 pol sequences of 1200 bp for 86.0% of the individuals with viral loads of more than 1000 copies per mL. Applying the criteria of more than 90% SH-aLRT clade support and genetic distance of less than or equal to 4.5%, 469 (29.5%) sequences grouped into 202 transmission clusters (see appendix p 3 for cluster size distribution). 151 (32.2%) of the 469 sequences were from men and 318 (67.8%) were from women, reflecting the sex distribution in the whole sequenced dataset (1102 women and 487 men).
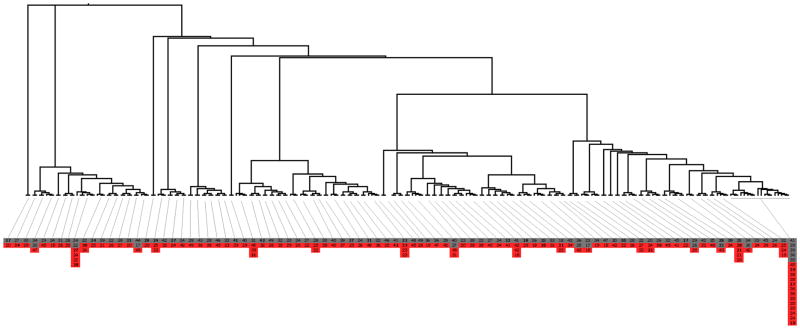
90 of the 202 clusters had at least one woman and one man (figure 2). 123 women were linked to 103 men in the 90 clusters. Within these clusters, 43 (35.0%) women were younger than 25 years, 56 (45.5%) were aged 25–40 years, and 24 (19.5%) were older than 40 years.
Figure 2. Maximum Likelihood tree for 90 heterosexual transmission clusters.
Clusters were with a bootstrap support higher than 90% and whose sequences had an intra-clade genetic distance <= 4.5%. In total, 123 women were linked to 103 men in the 90 heterosexual clusters. For better visualization of the clusters, tree is represented using proportional branch lengths transformation. The age of the individuals in each transmission cluster is presented inside boxes. Boxes are coloured based on gender, where grey represents males and red represents females.
49 women (39.8%) were aware of their serostatus compared with 27 men (26.2%; p=0.035; table 1). Because only individuals with viral loads of more than 1000 copies per mL are included in this analysis, the small numbers of people in transmission clusters who reported being on ART were expected (table 1). Median viral load was significantly higher in men than in women (31 000 copies per mL in men vs 16 629 copies per mL in women; p=0.002); 41 (33.3%) women and 37 (35.9%) men had viral loads of more than 50 000 copies per mL.
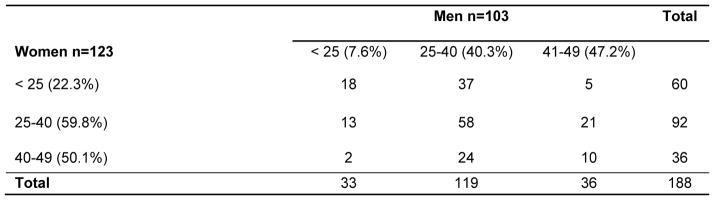
In the 90 clusters, there were 188 possible pairings between the 123 women and 103 men. A comparison of all possible pairings across age and sex categories (figure 3) identified two more frequent sets of linkages: between women aged 25–40 years and men aged 25–40 years (58 [30.9%] pairings) and between women younger than 25 years and men aged 25–40 years (37 [19.7%] pairings]), which constituted just over 50% of the linkages. In the 60 possible pairings between the 43 women younger than 25 years who were linked to 41 probable male partners, the mean age difference was 8.7 years (95% CI 6.8–10.6; p<0.0001). In these 60 possible pairings, 18 (30.0%) of the probable male partners were younger than 25 years, 37 (61.7%, 95% CI 49.7–74.3) were aged 25–40 years, and five (8.3%) were aged 41–49 years. Of the 41 probable male partners linked to a woman younger than 25 years, 16 (39.0%) were also linked to a 25–40-year-old woman (figure 4); the mean age of these male partners was 29.6 years (SD 7.1) and they were linked to both a younger woman (mean age 22.1 years) and an older woman (mean age 32.6 years). In the 92 possible pairings between the 56 women aged 25–40 years who were linked to 36 probable male partners, the mean age difference was 1.1 years (95% CI–0.6 to 2.8; p=0.111). Of 79 men (mean age 31.5 years) linked to women younger than 40 years, 62 (78.5%) were unaware of their HIV-positive status, 76 (96.2%) were not on ART, and 29 (36.7%) had viral loads of more than 50 000 copies per mL.
Figure 3.
All possible pairings from 90 male-female clusters stratified by the age group. The community-based HIV prevalence in each age group is shown in parentheses.
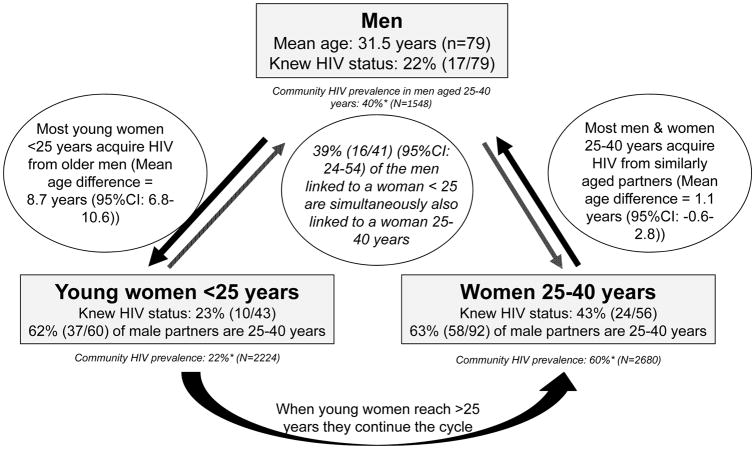
Figure 4.
Schematic presentation of the sexual networks of HIV positive men and women in phylogenetically identified heterosexual transmission clusters in a KwaZulu-Natal community in South Africa *weighted HIV prevalence
The mean age of male partners of HIV-positive women aged 20 years or younger was 30.5 years; the mean age difference was 11.5 years (SD 7.8; p<0.0001). The age gap decreased to a mean difference of 7.0 years (SD 6.7) in HIV-positive women aged 21–25 years and to a mean difference of 1.5 years (SD 9.1) in women aged 26–30 years (table 3).
Table 3.
Mean ages of male partners for each age category of women in a KwaZulu-Natal community in South Africa.
| HIV positive women in male-female clusters Ages of men linked in clusters* | HIV positive women from the community survey Self-reported age of last 3 partners | HIV negative women from the community survey Self-reported age of last 3 male partners* | p-value$ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Age of women (years) | No. women | No. men | Mean age HIV +ve male partners (years) | Mean difference in age (years) | No. women | No. men | Mean age male partners (years) Weighted (Unweighted) | Mean difference in age (years) Weighted (Unweighted) | No. women | No. men | Mean age male partners (years) Weighted (Unweighted) | Mean difference in age (years) Weighted (Unweighted) | |
| 16–20 | 18 | 25 | 30.5 | 11.5 | 164 | 140 | 23.4 (23.8) | 4.4 (4.7) | 501 | 565 | 22.3 (22.4) | 3.8 (3.9) | 0.171 |
| 21–25 | 31 | 41 | 30.4 | 7.0 | 553 | 436 | 29.8 (31.0) | 6.7 (7.8) | 612 | 692 | 26.1 (26.5) | 3.3 (3.6) | 0.226 |
| 26–30 | 18 | 19 | 29.2 | 1.5 | 739 | 566 | 32.8 (34.7) | 4.8 (6.7) | 436 | 537 | 31.2 (31.5) | 3.5 (3.7) | 0.178 |
| 31–35 | 14 | 27 | 35.6 | 1.7 | 664 | 523 | 39.2 (43.0) | 6.3 (10.1) | 221 | 268 | 36.5 (36.7) | 3.5 (3.9) | 0.166 |
| 36–40 | 18 | 42 | 37.0 | −0.5 | 593 | 497 | 40.4 (40.8) | 2.7 (2.9) | 227 | 255 | 41.8 (42.3) | 4.0 (4.3) | 0.005 |
| >40# | 24 | 34 | - | - | 734 | 602 | 46.0 (47.7) | 2.8 (3.0) | 532 | 590 | 57.0 (57.3) | 11.6 (11.5) | 0.074 |
The ages of male partners of HIV positive women are derived from transmission clusters while the ages of male partners of HIV negative women are self-reported ages of their last 3 male partners
The mean age and age difference of male partners for HIV positive women >40 years cannot be correctly calculated as men above age 50 years were not enrolled in this study and are therefore not included in the cluster analysis. The 34 male partners listed are only those partners <50 years.
p-value comparing the self-reported ages of partners for HIV positive and negative women
The age gap between HIV-negative women and their self-reported male partners (not linked through clustering) was smaller (plus or minus 3.8 years) and more consistent across the age groups. Educational levels, age of sexual debut, number of lifetime and current sexual partners, sexual frequency, and condom use were similar in the various subpopulations, irrespective of HIV status and linkage in clusters (table 1). Overall, the sample included 4% of the census-defined population. Sequencing coverage of HIV-positive individuals in the sample was higher than 25% in all age categories, being highest in men (402 [52.4%; weighted 51.3%] of 767) and women (926 [39.8%; weighted 36.5%] of 2325) aged 40 years and younger (table 2). Since sequencing coverage was not skewed and there is no evidence of super-spreaders driving HIV transmission in the opposite direction to the prevalence gradient, the dominant directionality of transmission was determined to occur from the highest prevalence of 59.8% (women aged 25–40 years) to 40.3% (probable male partners aged 25–40 years) to 22.3% (women aged <25 years; table 2, figures 3 and 4).
Discussion
Phylogenetic linkage, together with prevalence gradients, suggest that men aged 25–40 years were the primary source of the high rates of HIV acquisition in adolescent girls and young women (15–25 years). Many of these men had acquired HIV infection from women aged 25–40 years, which is the group with the highest prevalence of HIV. Over time, when the current group of adolescent girls and young women reach their 30s, we expect that they will then constitute the next group of women with high HIV prevalence, thereby perpetuating the cycle of HIV transmission to men in their 30s who will infect the next cohort of adolescent girls and young women (figure 4). The risks of HIV transmission within this cycle are enhanced at a community level by two key factors. First, two-fifths of HIV-positive individuals in this community were not aware of their HIV-positive status, and second, only two-thirds of individuals who knew they had HIV had initiated ART, resulting in suboptimal viral suppression. In particular, many of the men linked to women in the dual age groups (ie, <25 years and 25–40 years) were likely to have recently acquired HIV, were unaware of their HIV status, and had viral loads associated with high rates of transmission.19
Our results differ from those reported elsewhere; age disparity was not a risk factor for HIV acquisition in either a cohort study in which women reported the age of their most recent sexual partner on a yearly basis3 or in a clinical trial in which the age of a single primary partner was reported by women at baseline for infections occurring up to a year later.4 The major shortcoming in both studies is that the men whose ages were analysed might not have been the source of the women’s HIV exposure or infection. Investigators of both studies were unable to determine accurately which men transmitted HIV to the women enrolled.
Rapidly advancing bioinformatics methods such as phylogenetics20 and phylodynamics12,21 overcome this limitation and provide information about clearly linked individuals within a transmission cluster. A strength of our approach is that the phylogenetic clusters minimise the assumptions required to identify the chains of transmission. However, an important assumption remains; that viral transmission occurred among linked individuals within a cluster, acknowledging that not all partners in a chain of transmission may have been included in a cluster.
Our study also has several weaknesses. First, although this was a large cross-sectional study, our sample of 9812 individuals consisted of only 4% of the population, limiting the extent to which we were able to uncover linkages. Since the 4% sample was representative, we have extrapolated its findings to the community, in recognition of the limitations of this sample size.
Second, we did not have detailed longitudinal, clinical, and behavioural data to assist in establishing directionality. We therefore inferred directionality on the basis of the prevalence gradients, which is a crude approach and might not apply uniformly across the 188 possible pairings in the 90 male–female clusters. Although it is a reasonable assumption that most transmission will probably occur from high to low prevalence, we were not able to quantify the exact proportion of transmissions occurring against the gradient. However, there was no evidence of super-spreaders to drive transmission against the prevalence gradient.
Third, our study sampled an urban and a neighbouring rural community from one province in one country and is therefore not representative of all southern and eastern African countries. However, phylogenetic studies require a large sample size from a single community to maximise the opportunity to identify transmission clusters. Although several similar large-scale studies will be required in other settings to assess the generalisability of our results, the differences in HIV prevalence in young men and women in South Africa are also present in many southern and eastern African countries (appendix p 4). Findings from a phylogenetic study in Rakai, Uganda, showed that a substantial proportion of HIV infections were transmitted within married couples.22 A bioinformatics study in Mochudi, Botswana, identified acute subepidemics introduced from outside the region between 1997 and 2007.23 However, both studies did not specifically address the issue of the source of infection in young women.
Fourth, our analysis excluded individuals on successful therapy and is therefore skewed to more recent infections because only individuals with viral loads greater than 1000 copies per mL could be included. Alternatively, this limitation can be viewed as a strength, because the data describe the current and recent epidemic with limited contamination from long- established infections and people on treatment who pose minimal risk of spreading HIV. The high rate of sequencing failure with current sequencing technology in individuals with low viral loads will make these kinds of studies much more difficult to do as treatment coverage increases. In this community with 42% ART coverage, our large study of almost 10 000 individuals and more than 1500 sequences identified only 90 male–female clusters. Although this number is fewer than expected, it reflects what is possible from large studies at this stage of the HIV epidemic when ART is being rapidly rolled out and clusters becoming increasingly diffcult to identify. Finally, a potential bias could be the non-response rate. In this study, non-response at household and individual levels were within the range factored into the design.
Age disparity in sexual relationships has long been postulated as an important contributor to HIV transmission in Africa.24 Empirically, age disparity has been difficult to quantify but an analysis of a 1998–2000 community survey in Zimbabwe reported a small but significant increase in HIV risk in men caused by age-disparate relationships.25 Our phylogenetic analysis, which was made possible by the rapidly decreasing cost of gene sequencing, provides an empiric demonstration of the key role of the high HIV incidence in adolescent girls and young women transmitted by men (on average 8 years older) as a key component of HIV transmission in South Africa.
In several countries, including South Africa, men have been more challenging than women to reach with HIV testing and ART because men are often highly mobile while seeking job opportunities.26,27 However, other options to reduce HIV transmission are available. Prevention efforts need to include clear messages about age-disparate relationships between young women and men in their 30s, as part of an overall longer-term effort to change community norms in this regard. Over and above existing prevention and treatment programmes, three targeted interventions could help to break the cycle of HIV transmission in this setting: rapid scale-up of universal testing and treatment of men and women aged 25–40 years to reduce the risk of these high HIV prevalence groups transmitting HIV; scale-up of circumcision in men younger than 25 years to decrease their risk of acquiring HIV in their highest risk years; and provision of pre-exposure prophylaxis for young women to empower them to reduce the high risk of acquiring HIV. This combination prevention approach targeting three points in the transmission cycle could help to reduce HIV incidence in young women, an essential initial step to set Africa on the path to the UNAIDS 2030 goal of ending AIDS.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for articles published on or before Aug 24, 2016, with the search terms “HIV” and “age-disparate” and “Africa”. We applied no date or language restrictions. Despite extensive prevention campaigns and antiretroviral therapy (ART) scale-up, HIV incidence in adolescent girls and young women in southern and eastern Africa remains high. Although age and sex differences in several HIV prevalence studies suggested that older men were likely to be an important source of infection for adolescent girls and young women, this finding has been disputed in recent studies. Thus, whether age disparate sexual partnering is important for HIV transmission in Africa is unclear.
Added value of this study
In this community-wide survey of 9812 individuals in a setting with a high burden of HIV, viral sequencing in 1589 HIV-positive individuals who had viral loads greater than 1000 copies per mL enabled a phylogenetic analysis of clusters of men and women that showed a cycle of HIV transmission driven by high rates of HIV acquisition in adolescent girls and young women (15–25 years), principally from men close to or in their 30s (an average of 8 years older). These men are likely to have acquired HIV from women aged 25–40 years, who have the highest HIV prevalence. When the current group of adolescent girls and young women reach their 30s, they will then constitute the next group of women with high HIV prevalence, thereby perpetuating the cycle of HIV transmission to men in their 30s who will infect the next cohort of adolescent girls and young women. Our findings highlight the importance of age-disparate relationships in driving HIV transmission in southern and eastern Africa.
Implications of all the available evidence
The paucity of definitive data about the link between age-disparate sexual partnerships and HIV transmission has hampered HIV prevention in southern and eastern Africa. Current prevention programmes in South Africa and several other countries have steered clear of including this sociobehavioural factor in their prevention messages and education programmes. Our results clarify that sexual partnering between adolescent girls or young women and older men is a key feature of the sexual networks driving transmission of HIV. Our study provides the necessary evidence to guide changes in HIV programmes by identifying a combination prevention-and-treatment approach targeting three points in the cycle of HIV transmission that could help to reduce HIV incidence in young women, an essential initial step to set Africa on the path to the UNAIDS goal of ending AIDS by 2030.
Acknowledgments
This study was funded by the US President’s Emergency Plan for AIDS Relief (PEPFAR) through the US Centers for Disease Control and Prevention (CDC) under terms of the cooperative agreement 3U2GGH000372-02 W1. Community outreach activities were supported by the MAC AIDS Fund. Partial funding for the gene sequencing and bioinformatics was provided by the South African Medical Research Council. The study received support from the South African Government’s Department of Science and Technology (DST) and National Research Foundation (NRF) for the CAPRISA repository through the DST-NRF Centre of Excellence in HIV Prevention. ABMK is supported by a joint South African Medical Research Council/National Institutes of Health grant (R01HD083343). TdO and TG are supported by a research Flagship grant from the South African Medical Research Council (MRC-RFA-UFSP-01-2013/UKZN HIVEPI), a Royal Society Newton Advanced Fellowship (TdO), and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no 634650. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funders of this study. We acknowledge the support of the uMgungundlovu Health District, Provincial Department of Health, and the uMgungundlovu district municipality, local traditional leadership, and community members for all their support throughout the study. We give sincere thanks to Natasha Samsunder, Phumzile Khumalo, Gillian Hunt, and Lara Lewis. We also thank Sureshnee Pillay, Theresa Smith, and Hloniphile Mthiyane for assisting with the sequencing and to Rachael Dellar for previous discussions. We also thank our study collaborators Gavin George and Kaymarlin Govender from Health Economics and HIV/AIDS Research Division (HEARD) and Alex Welte from the South African Centre for Epidemiological Modelling and Analysis (SACEMA). A special thanks to all the study field staff and to all the participants who contributed their time and biological samples.
Footnotes
Contributors
ABMK is the principal investigator of the survey. ABMK, CC, and DK were responsible for the field work and quality assurance; AG, TdO, and TG for statistical analysis; SM, AP, and TdO for laboratory testing and quality assurance; SSAK, TdO, TG, ABMK, QAK, and CB contributed to analysis and interpretation of the data and writing of the first draft of the report. All authors critically reviewed and approved the final version of the report.
Declaration of interests
We declare no competing interests.
References
- 1.UNAIDS. Global AIDS update - 2016. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS (UNAIDS); 2016. [Google Scholar]
- 2.Abdool Karim SS, Churchyard GJ, Abdool Karim Q, Lawn SD. HIV infection and tuberculosis in South Africa: an urgent need to escalate the public health response. Lancet. 2009;374(9693):921–33. doi: 10.1016/S0140-6736(09)60916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harling G, Newell ML, Tanser F, Kawachi I, Subramanian SV, Barnighausen T. Do Age-Disparate Relationships Drive HIV Incidence in Young Women? Evidence from a Population Cohort in Rural KwaZulu-Natal, South Africa. J Acquir Immune Defic Syndr. 2014;66(4):443–51. doi: 10.1097/QAI.0000000000000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkus JE, Nair G, Montgomery ET, Mishra A, Palanee-Phillips T, Ramjee G, et al. Age-Disparate Partnerships and Risk of HIV-1 Acquisition Among South African Women Participating in the VOICE Trial. J Acquir Immune Defic Syndr. 2015;70(2):212–7. doi: 10.1097/QAI.0000000000000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratmann O, van Sighem A, Bezemer D, Gavryushkina A, Jurriaans S, Wensing A, et al. Sources of HIV infection among men having sex with men and implications for prevention. Sci Transl Med. 2016;8(320):320ra2. doi: 10.1126/scitranslmed.aad1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volz EM, Frost SDW. Inferring the Source of Transmission with Phylogenetic Data. PLoS Comput Biol. 2013;9(12):e1003397. doi: 10.1371/journal.pcbi.1003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis AM, Herbeck JT, Brown AL, Kellam P, de Oliveira T, Pillay D, et al. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest? J Acquir Immune Defic Syndr. 2014;67(2):181–95. doi: 10.1097/QAI.0000000000000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and Safety of Tenofovir Gel, an Antiretroviral Microbicide, for the Prevention of HIV Infection in Women. Science. 2010;329:1168–74. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kharsany AB, Cawood C, Khanyile D, Grobler A, McKinnon LR, Samsunder N, et al. Strengthening HIV surveillance in the antiretroviral therapy era: rationale and design of a longitudinal study to monitor HIV prevalence and incidence in the uMgungundlovu District, KwaZulu-Natal, South Africa. BMC public health. 2015;15:1149. doi: 10.1186/s12889-015-2179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manasa J, Lessells R, Rossouw T, Naidu K, Van Vuuren C, Goedhals D, et al. Southern African Treatment Resistance Network (SATuRN) RegaDB HIV drug resistance and clinical management database: supporting patient management, surveillance and research in southern Africa. Database (Oxford) 2014;2014:bat082. doi: 10.1093/database/bat082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gifford RJ, Liu TF, Rhee SY, Kiuchi M, Hue S, Pillay D, et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics. 2009;25(9):1197–8. doi: 10.1093/bioinformatics/btp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadler T, Bonhoeffer S. Uncovering epidemiological dynamics in heterogeneous host populations using phylogenetic methods. Philos Trans R Soc Lond B Biol Sci. 2013;368(1614):20120198. doi: 10.1098/rstb.2012.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22):3276–8. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavaré S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. In: Miura RM, editor. Some Mathematical Questions in Biology-DNA Sequence Analysis. Providence, RI, USA: American Mathematical Society; 1986. pp. 57–86. [Google Scholar]
- 17.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39(3):306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 18.Ragonnet-Cronin M, Hodcroft E, Hue S, Fearnhill E, Delpech V, Brown AJ, et al. Automated analysis of phylogenetic clusters. BMC Bioinformatics. 2013;14:317. doi: 10.1186/1471-2105-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, et al. Viral Load and Heterosexual Transmission of Human Immunodeficiency Virus Type 1. N Eng J Med. 2000;342(13):921. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 20.Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, et al. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci U S A. 2012;109(37):15066–71. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volz EM, Koelle K, Bedford T. Viral phylodynamics. PLoS Comput Biol. 2013;9(3):e1002947. doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabowski MK, Lessler J, Redd AD, Kagaayi J, Laeyendecker O, Ndyanabo A, et al. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med. 2014;11(3):e1001610. doi: 10.1371/journal.pmed.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novitsky V, Kuhnert D, Moyo S, Widenfelt E, Okui L, Essex M. Phylodynamic analysis of HIV sub-epidemics in Mochudi, Botswana. Epidemics. 2015;13:44–55. doi: 10.1016/j.epidem.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson RM, May RM, Boily MC, Garnett GP, Rowley JT. The spread of HIV-1 in Africa: sexual contact patterns and the predicted demographic impact of AIDS. Nature. 1991;352(6336):581–9. doi: 10.1038/352581a0. [DOI] [PubMed] [Google Scholar]
- 25.Gregson S, Nyamukapa CA, Garnett GP, Mason PR, Zhuwau T, Carael M, et al. Sexual mixing patterns and sex-differentials in teenage exposure to HIV infection in rural Zimbabwe. Lancet. 2002;359(9321):1896–903. doi: 10.1016/S0140-6736(02)08780-9. [DOI] [PubMed] [Google Scholar]
- 26.Auld AF, Shiraishi RW, Mbofana F, Couto A, Fetogang EB, El-Halabi S, et al. Lower Levels of Antiretroviral Therapy Enrollment Among Men with HIV Compared with Women - 12 Countries, 2002–2013. MMWR Morb Mortal Wkly Rep. 2015;64(46):1281–6. doi: 10.15585/mmwr.mm6446a2. [DOI] [PubMed] [Google Scholar]
- 27.Houlihan CF, Bland RM, Mutevedzi PC, Lessells RJ, Ndirangu J, Thulare H, et al. Cohort Profile: Hlabisa HIV Treatment and Care Programme. Int J Epidemiol. 2011;40(2):318–26. doi: 10.1093/ije/dyp402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.