Abstract
Metastatic disease is a significant contributor to cancer patient mortality. We previously reported that the Kinase Suppressor of Ras1 (KSR1) scaffold protein for the Erk mitogen-activated protein kinase pathway coimmunoprecipitated the metastasis suppressor protein Nm23-H1. We now hypothesize that altered expression levels of Nm23-H1 influence the binding properties, stability, and function of the KSR1 scaffold. Increased coimmunoprecipitation of Hsp90 with KSR1 was observed in either stable or transient transfectants of nm23-H1 in MDA-MB-435 human breast carcinoma cells. Similar trends were also observed in the cytoplasmic and nuclear fractions of cells. Cells expressing high levels of Nm23-H1 exhibited increased KSR1 degradation in the presence of either cycloheximide or an Hsp90-directed drug currently in clinical trial, 17-allylamino-17-demethoxygeldanamycin (17-AAG). In agreement with KSR1 degradation data, high-Nm23-H1-expression cells were preferentially inhibited in anchorage-independent colonization assays by 17-AAG. KSR1 scaffold binding patterns are dynamic in both the cytoplasmic and nuclear compartments, modulated by metastasis suppressor expression. Metastasis suppressor expression levels can impact traditional signaling pathways, such as the Erk pathway, resulting in altered tumor cell sensitivity to cancer therapeutics.
Kinase Suppressor of Ras1 (KSR1) was hypothesized to be a scaffold for the Erk mitogen-activated protein (MAP) kinase pathway (reviewed in references 39 and 56). Scaffold proteins bind two or more components of a signaling pathway and may regulate the amplitude, threshold, spatiotemporal characteristics, and specificity of response. Discovered in Ras-related genetic screens with Drosophila and Caenorhabditis elegans, KSR1 functioned downstream of Ras (24, 67, 70) and was required for MAP kinase activation (2, 45, 56). Mammalian KSR1 binds Mek1 and -2, Erk1 and -2, G-protein β and γ subunits, and accessory proteins, such as Hsp90, c-Tak1, PP2A, CNK, and 14-3-3 (2, 6, 40, 42, 46, 65), supporting the scaffold hypothesis. Translocation of the protein complex from the cytoplasm to the plasma membrane to assemble a functional Raf/Mek complex was observed upon growth factor stimulation, controlled by KSR1 serine 392 phosphorylation and 14-3-3 binding status (40, 46) (56). KSR1 transfection has either stimulated or inhibited Erk signaling in cell lines, depending on the level of KSR1 protein expressed (reviewed in reference 39).
KSR1 null mice exhibited normal development (29, 42). Crosses of KSR1 null mice to MMTV-polyoma middle T mice (wild-type ras) resulted in delayed tumor formation (42), while crosses to Tg.AC mice (v-Ha-ras mutated at codons 12 and 59) inhibited skin papilloma formation (29). Antisense inhibition of KSR1 also inhibited mutant Ras-dependent pancreatic cancer xenograft growth (76).
Other aspects of KSR1 function remain incompletely understood. KSR1 has also been localized to the nucleus, which may be important for the subcellular routing of Mek (9). KSR1−/− mouse embryo fibroblasts exhibited reduced Erk activation in response to 12-O-tetradecanoylphorbol-13-acetate, but Raf activation of Mek remained normal (29). A kinase function for KSR1 has been debated (77, 80). KSR1 phosphorylation has been reported to be influenced by zinc ion concentrations in C. elegans (81).
We recently reported complex interactions between KSR1 and the Nm23-H1 metastasis suppressor (19). Metastatic disease represents one of the most difficult challenges in cancer therapy. Both positive and negative signaling pathways regulate tumor metastasis, including multiple metastasis suppressor genes (64). When reexpressed in a metastatic cell line, these genes suppress in vivo metastasis without significant effects on the size of the primary tumor. The nm23 gene family has been demonstrated to have suppressive activity for the development of lymph node, lung and/or liver metastases in 11 independent studies (3, 7, 17, 25, 26, 36, 38, 52, 59, 68, 69). The biochemical mechanism of metastasis suppression is thought to involve attenuation of signaling for tumor cell motility, invasion, and colonization. At least four classes of Nm23 biochemical activities may contribute to altered signaling, including protein-protein interactions (4, 12, 16, 28, 44, 47, 51, 55, 57, 58, 66), regulation of GTP-binding protein function (15, 48, 50, 72, 79, 82), DNA-associated activities (14, 53, 54, 63), and histidine-dependent protein phosphotransferase activity (11, 73, 74). A role for KSR1 in Nm23-H1 attenuation of Erk activation was investigated. Transfection of wild-type nm23-H1, but not a control vector or a mutant nm23-H1 incapable of inhibiting motility, was associated with low Erk activation (19). Nm23-H1 coimmunoprecipitated KSR1 from 293T cells and human MDA-MB-435 breast carcinoma cells, and Nm23-H1 phosphorylated KSR1 on serines 392 and 434 of KSR1 in vitro (19). The data suggested the hypothesis that Nm23-H1 overexpression and/or kinase activity altered the scaffold properties of KSR1 and attenuated Erk signaling needed for metastatic invasion and colonization.
In this report, the steady-state scaffold function of KSR1 was investigated in control and Nm23-H1-transfected breast carcinoma cells in response to the milieu of factors present in serum, mimicking conditions in which metastases thrive in vivo. We observed increased binding of Hsp90 to KSR1 in Nm23-H1-transfected cells. To date, Hsp90 has been shown to promote either the folding or the proteasome-mediated degradation of multiple client proteins depending on the complex of bound cochaperone proteins (reviewed in references 21 and 33). We report the downstream consequences of an increased Hsp90-KSR1 interaction in Nm23-H1-overexpressing breast carcinoma cells, including decreased KSR1 stability and increased sensitivity to a Hsp90-directed therapeutic agent.
MATERIALS AND METHODS
Plasmids.
The hemagglutinin (HA)-tagged KSR1 plasmid (HA-KSR1) was constructed by amplifying the murine wild-type ksr1 cDNA (kindly provided by Deborah K. Morrison) via PCR and subcloning into KpnI-NotI sites of pcDNA3 (Invitrogen). The green fluorescent protein (GFP)-KSR1 plasmid was created by amplifying the EcoRI-KpnI fragment encoding the murine wt-ksr1 cDNA via PCR and subcloning into EcoRI-KpnI sites of pEGFP-C1 (Clontech). To construct Nm23-H1-Flag, a 459-bp EcoRV-BamHI fragment encoding the human nm23-H1 cDNA from pSVnm23-H1-Flag was ligated into a pcDNA3 (Invitrogen) expression vector previously digested with BamHI and blunted with Klenow polymerase. All clones were verified by double-stranded sequencing.
Cell lines, transfections, and drug treatments.
Human MD-MBA-435 breast carcinoma cells or stable transfectants were previously described (26, 31) and were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were plated at a density of 2 × 106 cells/100-mm plate and transfected with HA-KSR1, Nm23-H1-Flag, or control vectors using Effectene transfection reagent (QIAGEN). The cell culture medium was changed 24 h later, and the cells were harvested 48 h posttransfection.
Reagents.
17-Allylamino-17-demethoxygeldanamycin (17-AAG) was kindly provided by the Cancer Therapy Evaluation Program, National Cancer Institute. A 50 mM stock solution in dimethyl sulfoxide (DMSO) was frozen at −80°C in aliquots and protected from light. Cycloheximide (CHX) was obtained from Sigma. MG-132 proteasome inhibitor and the MEK inhibitor PD90859 were purchased from Calbiochem.
Cell fractionation.
Cells were scraped in ice-cold phosphate-buffered saline, collected by centrifugation, and incubated in hypotonic buffer (5 mM sodium phosphate [pH 7.5], 2 mM MgCl2, complete mini EDTA-free protease inhibitor cocktail [Roche Molecular Biochemicals], dual phosphatase inhibitor cocktails 1 and 2 [Sigma], 1 mM phenylmethylsulfonyl fluoride) for 30 min on ice. Cells were sheared by repeated passage through a 25-gauge needle, and the lysate was precleared by low-speed centrifugation (12,000 × g for 5 min). The pellet was washed with hypotonic buffer, centrifuged at low speed, and lysed with NE-PER extraction reagent (Pierce) including protease and phosphatase inhibitors to obtain nuclear proteins. The supernatant was centrifuged at 100,000 × g for 1 h. The resulting supernatant was used as the cytoplasmic extract. The pellet was washed, recentrifuged, and lysed in hypotonic buffer containing 1% (vol/vol) Triton X-100 as the membrane extract. To verify the purity of each fraction, 30 μg of protein were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis, and a Western blot was probed with the following antibodies: anti-Na, K-ATPase (membrane marker; Novus Biological), anti-α-tubulin (cytoplasmic marker; Oncogene), and anti-c-Jun (nuclear marker; Santa Cruz).
Drug treatments.
MDA-MB-435 cells were transfected as described above. Twenty-four hours posttransfection, cells were trypsinized, and 3 × 105 cells were plated in six-well plates. Twelve hours later, 100 μM CHX was added. A similar procedure was used for 17-AAG treatments with the exception that cells were plated in 150-mm dishes, trypsinized, and replated after 16 h into four 100-mm dishes. The reversible proteasome inhibitor MG-132 (Calbiochem) was used at a 15 μM concentration and added to the cells 1 h before 17-AAG.
Coimmunoprecipitations and Western blot analysis.
For Western blots, cells were lysed in RIPA buffer (20 mM Tris-HCl [pH 8], 137 mM NaCl, 10% glycerol, 1% Nonidet P-40 [NP-40], 2 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS), containing complete mini EDTA-free protease inhibitor cocktail (Roche Molecular Biochemicals) and phosphatase inhibitor cocktails 1 and 2 (Sigma). Lysates were normalized to equivalent total protein. The phospho-MAP kinase (P-p44/P-p42) and the total MAP kinase (p44/p42) antibodies were obtained from Cell Signaling. After incubation with horseradish peroxidase-conjugated secondary antibody, proteins were visualized by autoradiography with LumiGlo reagents (Cell Signaling).
For coimmunoprecipitations, cells were lysed in NP-40 buffer (20 mM Tris-HCl [pH 8], 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA), containing complete mini EDTA-free protease inhibitor cocktail (Roche Molecular Biochemicals), phosphatase inhibitor cocktails 1 and 2 (Sigma), and 1 mM phenylmethylsulfonyl fluoride, and the lysates were precleared with protein G-agarose (Roche Molecular Biochemicals) for 1 h at 4°C. Eight hundred micrograms of lysate was incubated overnight at 4°C with 5 μg of mouse anti-HA antibody (Covance, Princeton, N.J.) or the same amount of control immunoglobulin G antibody (Santa Cruz Biotechnology). The immunocomplexes were precipitated after incubation with 30 μl of protein-G-agarose for 1 h at 4°C, washed three times with NP-40 buffer and twice with 1× phosphate-buffered saline and resuspended with 30 μl of Laemmli buffer. Proteins were separated by SDS-polyacrylamide electrophoresis and transferred to a nitrocellulose membrane. The membrane was then immunoblotted with anti-HA (Covance), anti-14-3-3β (K-19; Santa Cruz Biotechnology), anti-Mek-1/2 (Upstate), anti-Hsp90 (H-114; Santa Cruz Biotechnology), or anti-Nm23 (clone 56; BD Transduction Laboratories) (Ab 11; Cymbus Biotechnology). Western blots were processed as described above.
Soft agar assay.
Soft agar colonization assays were performed as previously described (49). Briefly, a suspension of 3 × 104 cells, containing 0.3% (wt/vol) soft agar, was mixed with various concentrations of 17-AAG or PD90859 prior to setting. The cell layer was overlaid onto a layer of culture medium containing 0.7% (wt/vol) agar in 24-well plates. Colonies were counted after 3 weeks by using an ocular with a grid. Colonies were defined as having a diameter of 0.125 mm as assessed by the ocular grid. Each point is the mean ± standard error of the mean (SEM) of triplicate determinations.
Statistical analyses.
Results shown are representative of three experiments conducted, unless otherwise noted. Differences were tested by a two-tailed Student t test using Instat (GraphPad Software, Inc.).
RESULTS
Elevated Nm23-H1 expression results in greater Hsp90 binding to cytoplasmic KSR1.
Previous studies indicated that the Nm23-H1 metastasis suppressor interacted with the KSR1 Erk scaffold protein and that stable Nm23-H1-overexpressing MDA-MB-435 breast carcinoma cell lines exhibited reduced Erk activation (19). We hypothesized that high Nm23-H1 levels attenuated Erk activation by altering the scaffold properties of KSR1. The data in Fig. 1 show the coimmunoprecipitation of Nm23-H1, KSR1, and one scaffold binding protein, Hsp90, in a panel of stable control and nm23-H1-transfected cell lines. MDA-MB-435 human breast carcinoma cell lines stably expressing a vector construct (C-100), wild-type nm23-H1 (H1-177), or the P96S mutant of nm23-H1 (S-22) were previously described (26, 31). The H1-177 cell line was less metastatic in vivo and exhibited less in vitro motility and anchorage-independent colonization activity than the C-100 control line (23, 26). The S-22 cell line exhibited in vitro motility at or above that of the control line (31). In contrast, the anchorage-dependent proliferative rates of the cell lines were comparable (31). Each cell line was transiently transfected with HA-KSR1, since endogenous KSR1 was not detectable using commercially available antibodies, and cell lysates were fractionated to assess the scaffold binding properties in specific cell compartments.
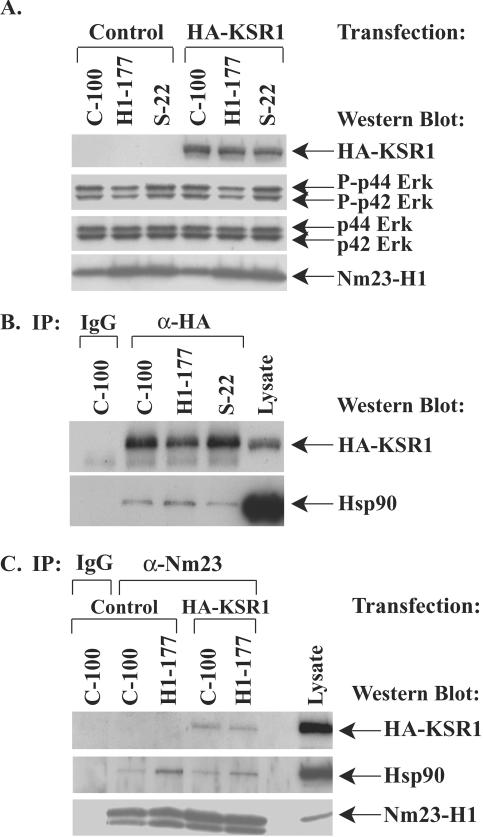
FIG. 1.
Nm23-H1 inhibition of motility is correlated with greater Hsp90 binding to KSR1 among a panel of stable Nm23-H1 transfectants. MDA-MB-435 breast carcinoma cells were previously stably transfected with a control vector (C-100), wild-type nm23-H1 metastasis suppressor (H1-177) or a P96S mutation of nm23-H1 (S-22); in vitro motility experiments demonstrated that the H1-177 transfectants exhibited reduced motility responses to either serum or autotaxin compared to C-100 or S-22 cells (31). Each cell line was transiently transfected with a HA-tagged KSR1 (HA-KSR1) construct, and cytoplasmic lysates were prepared. (A) Transfection of exogenous HA-KSR1 does not affect the level of phospho-Erk (P-Erk). C-100, H1-177, and S-22 cell lines were transfected with a control vector or HA-KSR1, cytoplasmic lysates were analyzed by Western blotting with anti-HA, anti-P-Erk, anti-Erk, and anti-Nm23-H1 antibodies. (B) KSR1 was immunoprecipitated from equivalent amounts of cytoplasmic fractions of cell lysates using anti-HA (α-HA), and binding of Hsp90 was determined by Western blotting. (C) Nm23-H1, HA-KSR1, and Hsp90 are in the same complex. Cytoplasmic lysates from C-100 and H1-177 cell lines transfected with a control vector or HA-KSR1 were immunoprecipitated with anti Nm23-H1 antibody (α-Nm23), and binding of HA-KSR1 and Hsp90 was determined by Western blotting. IgG, immunoglobulin G.
Western blot analysis of cytoplasmic extracts of stable Nm23-H1 cell lines are shown in Fig. 1A. Lysates from each cell line transiently transfected with a control vector or HA-KSR1 were compared to assess the effect of elevated KSR1 expression on Erk activation. Activated Erk was reduced in the Nm23-H1-overexpressing H1-177 cell line compared to the control (C-100) or mutant (S-22) cell lines regardless of the HA-KSR1 transfection status. This trend recapitulated that observed previously in the same cell lines without transient ectopic expression of HA-KSR1 (19) and established that the level of KSR1 transfection used did not alter the Erk phosphorylation status. Nm23-H1 expression was approximately eightfold higher in the H1-177 and S-22 lines than in the control C-100 cell line.
HA-KSR1 was immunoprecipitated from equivalent amounts of total protein in cytoplasmic lysates of the C-100, H1-177, and S-22 cell lines (Fig. 1B), and the relative amount of Hsp90 bound to KSR1 was determined by Western blotting of the immunoprecipitates. KSR1 was expressed at comparable levels in each cell line. Nm23-H1-overexpressing H1-177 cells exhibited greater Hsp90 binding to KSR1 than the control C-100 or S-22 mutant Nm23-H1-expressing cells. A control immunoglobulin G did not immunoprecipitate KSR1 or coimmunoprecipitate Hsp90. In a separate experiment, the Nm23-H1-overexpressing H1-177 cell line also exhibited higher levels of Hsp90 binding to cytoplasmic KSR1 than a stable transfectant expressing a S120G mutant of nm23-H1, the I-205 cell line, which was also not motility inhibited (data not shown). In the experiment shown in Fig. 1C, control C-100 and Nm23-H1-overexpressing H1-177 cells were first immunoprecipitated with anti-Nm23-H1 and then assessed for binding of KSR1 and Hsp90. A trend similar to that seen in Fig. 1B, increased binding of Hsp90 to KSR1, was demonstrated with H1-177 cells with HA-KSR1 transiently transfected. The data also demonstrate three-way complex formation between Nm23-H1, KSR1, and Hsp90 with either anti-HA-KSR1 or anti-Nm23 as immunoprecipitating antibodies (Fig. 1B and C). Thus, a direct correlation of motility-suppressed behavior and quantitatively higher binding of Hsp90 to the cytoplasmic KSR1 scaffold was observed.
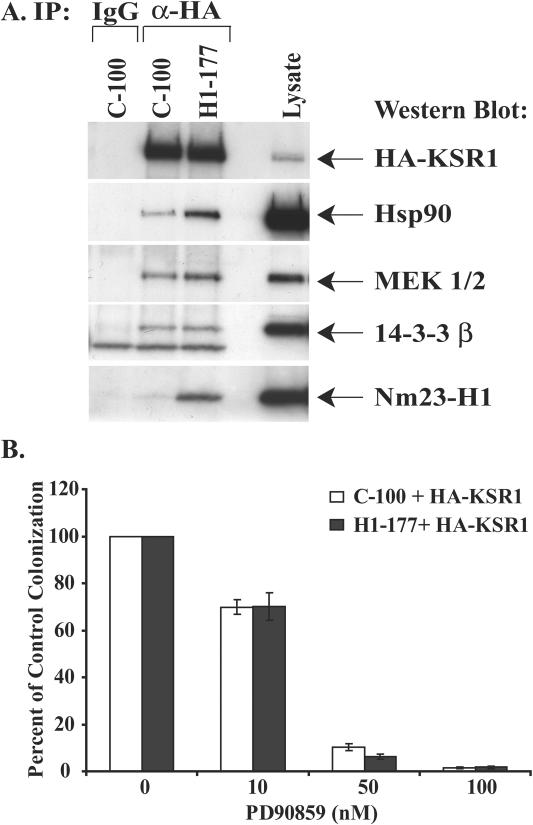
Not all of the known KSR1-associated proteins exhibited altered binding levels between the control C-100 and Nm23-H1-overexpressing H1-177 cell lines. In the experiment shown in Fig. 2A, the cells were transiently transfected with HA-KSR1, and KSR1 was immunoprecipitated from cytoplasmic lysates. Western blot analysis of the KSR1 immunoprecipitations was performed to ascertain relative binding levels of other KSR1 scaffold proteins. Increased binding of Hsp90 to KSR1 was again observed with the H1-177 cell line. In addition, a low level of increased Mek1-2 binding but equivalent 14-3-3β binding were observed with the H1-177 cells. Binding of Erk proteins to KSR1 was at the lower limit of detection but comparable (data not shown). The data demonstrate specificity in altered KSR1 binding protein patterns. To analyze the potential biological implications of the increased association of Mek with KSR1 in the H1-177 cell line, we performed a soft agar colonization assay. Although nm23-H1 transfection had no effect on anchorage-dependent proliferation in vitro or primary tumor size in vivo, nm23-H1 transfectants exhibited decreased colonization in soft agar, thought to represent the ability to grow in a foreign site (26). C-100 and H1-177 cells transiently transfected with HA-KSR1 were exposed to different concentrations of the Mek inhibitor PD90859, and the results from two independent experiments were plotted in Fig. 2B as a percentage of control colonization (vehicle control). Colonization of both cell lines was inhibited by PD90859 with the same sensitivity, indicating that Mek contributions to colonization were equivalent.
FIG. 2.
Increased KSR1 scaffold protein binding is specific to Hsp90 and Mek but not 14-3-3β in Nm23-H1-overexpressing cells. (A) Binding of Hsp90 and to a lesser extent Mek is increased in Nm23-H1-overexpressing cells compared to that in control cells. The control C-100 and Nm23-H1-overexpressing H1-177 cell lines were transiently transfected with HA-KSR1. KSR1 was immunoprecipitated (IP) from cytoplasmic lysates, and the coimmunoprecipitation of Hsp90, Mek1/2, 14-3-3β, and Nm23-H1 was determined by Western blotting. (B) The effect of the Mek inhibitor PD90859 on soft agar colonization of C-100 and H1-177 breast carcinoma cells. Equivalent numbers of cells were plated in soft agar transiently transfected with HA-KSR1. Colonies were counted 3 weeks later. Data represent the means ± SEM for triplicate cultures from two experiments, normalized to the percentage of vehicle control colonization. IgG, immunoglobulin G; α-HA, anti-HA.
Because long-term-established cell lines could accumulate alterations, it was possible that increased binding of Hsp90 to KSR1 was an artifact of long-term culture of the H1-177 cell line. The relative level of Hsp90 binding to KSR1 was determined with MDA-MB-435 cells transiently transfected with HA-KSR1 and either a control Flag vector or Nm23-H1-Flag (Fig. 3). Figure 3A shows the relative purity of the cytoplasmic and nuclear subcellular fractions as well as exogenous and endogenous Nm23-H1 expression levels. In the experiment shown in Fig. 3B, comparable amounts of KSR1 were immunoprecipitated from cytoplasmic lysates of Nm23-H1-Flag/HA-KSR1- and control Flag/HA-KSR1-transfected cells. Coimmunoprecipitation of KSR1 and endogenous and Nm23-H1-Flag was observed and was more pronounced in the Nm23-H1-Flag/HA-KSR1 transfectants. MDA-MB-435 cells transiently transfected with Nm23-H1-Flag/HA-KSR1 exhibited greater binding of Hsp90 to cytoplasmic KSR1 than control Flag/HA-KSR1-transfected cells. Densitometric analysis of three experiments showed a 2.5- to 3.2-fold increase in binding of Hsp90 to KSR1 in the Nm23-H1 transient transfectants. These data indicate that the trend of Nm23-H1 overexpression and increased Hsp90 binding to KSR1 is not an artifact of long-term culture of stably transfected cell lines.
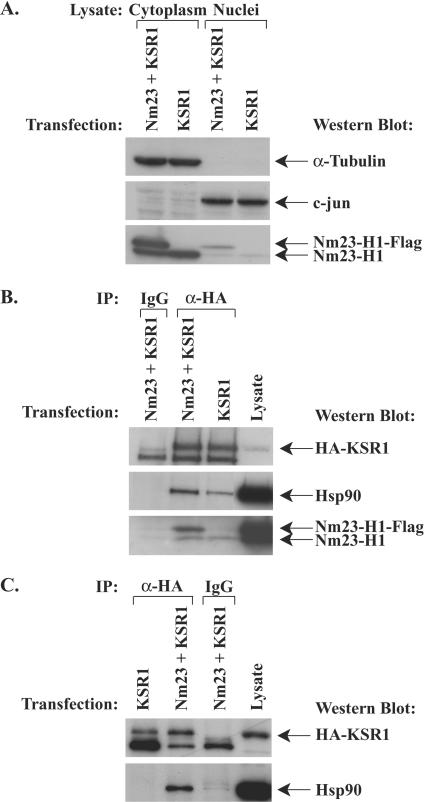
FIG. 3.
MDA-MB-435 cells transiently transfected with Nm23-H1 and KSR1 also show increased Hsp90 binding to KSR1. MDA-MB-435 cells were transiently transfected with combinations of HA-KSR1 and either a control Flag vector or Nm23-H1-Flag, and subcellular fractions were prepared. (A) Western blot of lysates for cytoplasmic (α-tubulin) and nuclear (c-jun) markers, as well as endogenous and transiently transfected Nm23-H1. (B) Immunoprecipitation of HA-KSR1 from equivalent amounts of cytoplasmic lysates; coimmunoprecipitation of Hsp90 and Nm23-H1 was determined by Western blotting. (C) Immunoprecipitation of HA-KSR1 from equivalent amounts of nuclear lysates; coimmunoprecipitation of Hsp90 and HA-KSR1 was determined by Western blotting.
The nuclear KSR1 scaffold is also dynamic.
While most reports have focused on KSR1 in the cytoplasmic and membrane compartments, KSR1 has been reported in the cell nucleus and has been proposed to regulate nucleocytoplasmic partitioning of Mek (9). It is unclear if nuclear KSR1 binds the same scaffold proteins as cytoplasmic-membranous KSR1. Nuclear expression of a transiently transfected GFP-KSR1 construct was observed in both the control C-100 and Nm23-H1-overexpressing H1-177 cell lines by both Western blot analysis and immunofluorescence (data not shown). Incubation of MDA-MB-435 cells with the nuclear export inhibitor leptomycin B caused nuclear accumulation of GFP-KSR1, in agreement with previously published findings with fibroblasts (data not shown) (9). We then examined whether the nuclear KSR1 scaffold bound similar proteins and whether the relative levels of binding observed in the cytoplasm were maintained in the nucleus (Fig. 3C). HA-KSR1 immunoprecipitated from nuclear lysates of MDA-MB-435 cells transiently transfected with Nm23-H1-Flag/HA-KSR1 bound more Hsp90 than nuclear KSR1 immunoprecipitated from control Flag/HA-KSR1-transfected cells. Binding of Hsp90 to KSR1 was approximately threefold higher in Nm23-H1-transfected cells than in control transfectants as assessed by densitometry for three independent experiments. A similar trend was observed with nuclear lysates of stable control C-100 and Nm23-H1-overexpressing H1-177 cell lines (data not shown). Thus, Nm23-H1 overexpression resulted in coordinate increased Hsp90 binding to KSR1 in the cytoplasmic and nuclear cell compartments.
Although membrane fractions were obtained from each cellular lysate, levels of HA-KSR1 protein were at the limit of detection. Transfection of greater HA-KSR1 was avoided, since it has been proposed to alter the stoichiometry of the KSR1 scaffold to its binding proteins (39). Western blots of total cellular lysates from control C-100 and Nm23-H1-overexpressing H1-177 cell lines contained comparable amounts of total Hsp90 (data not shown). Thus, increased binding of Hsp90 to KSR1 in cells expressing high levels of Nm23-H1 did not reflect overall protein trends but an increased affinity.
Nm23-H1 overexpression promotes KSR1 degradation.
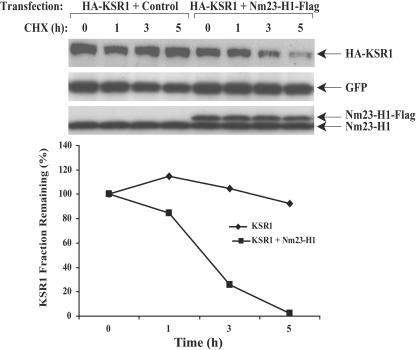
To date, Hsp90 has been shown to promote either the folding or the proteasome-mediated degradation of client proteins depending on the complex of bound proteins and cochaperones. Geldanamycin and its derivative form, 17-AAG, are benzoquinone ansamycin antibiotics that stabilize Hsp90 in its ADP-bound conformation with a class of cochaperones that promote client protein degradation. Hsp90 binding to KSR1 in fibroblasts has been previously reported; treatment of cells with 17-AAG accelerated KSR1 degradation (65). These data suggested that KSR1 is a client protein of Hsp90. If elevated Nm23-H1 expression resulted in enhanced binding of Hsp90 to KSR1, it was hypothesized that this interaction may exert functional consequences on KSR1 stability. MDA-MB-435 cells were transiently transfected with HA-KSR1, pEGFP as a transfection control, and either a control Flag or Nm23-H1-Flag construct. Twenty-four hours posttransfection, the cells were trypsinized, and equivalent cell numbers were plated in six-well plates. The cells were permitted to attach for 12 h and then were exposed to 100 μM CHX to inhibit protein synthesis. HA-KSR1 protein levels were determined by Western blotting and normalized to GFP levels (Fig. 4). With protein synthesis blocked, only 25 and 2.5% of KSR1 remained after 3 and 5 h of CHX treatment, respectively, in the Nm23-H1-Flag/HA-KSR1-transfected cells. In comparison, KSR1 was more stable in cells expressing low, endogenous levels of Nm23-H1, with 105 and 92.5% of protein remaining at the same time points. Similar trends were observed using stable transfectants (data not shown).
FIG. 4.
KSR1 stability is decreased in Nm23-H1-overexpressing breast carcinoma cells exposed to cycloheximide. MDA-MB-435 cells were transiently transfected with pEGFP as a control for transfection efficiency, a HA-KSR1 vector, and either a control Flag or an Nm23-H1-Flag vector. Cells were then treated with 100 μM cycloheximide (a protein synthesis inhibitor) for various amounts of time. Lysates were analyzed by Western blotting with anti-HA, anti-Nm23-H1, and anti-GFP as a transfection control. Densitometric analysis of KSR1 expression normalized to GFP levels is graphed below.
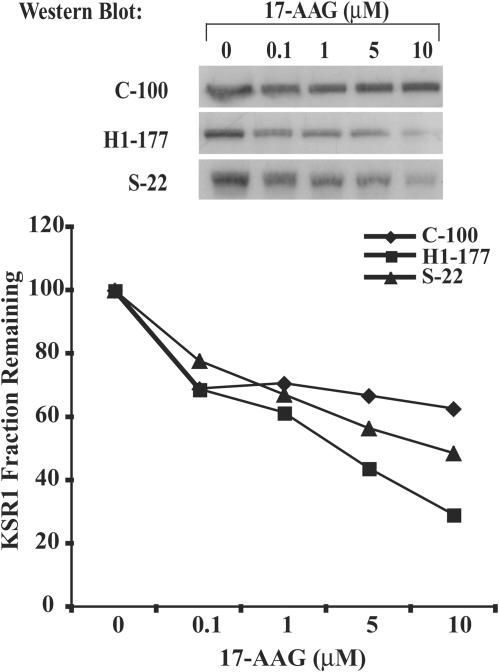
To examine the impact of Hsp90 on KSR1 stability in a molecularly targeted fashion, the 17-AAG form of geldanamycin was used. Stable MDA-MB-435 control C-100, P96S Nm23-H1-overexpressing S-22, and wild-type Nm23-H1-overexpressing H1-177 cell lines were transiently transfected with HA-KSR1 and pEGFP and exposed to 0 to 10 μM of 17-AAG for 6 h. In Fig. 5, KSR1 protein levels were determined by Western blotting, and the fraction of KSR1 remaining was normalized to GFP levels. KSR1 protein levels declined 30% on average in H1-177 cells exposed to 10 μM 17-AAG compared to untreated levels, while similarly treated control C-100 cells and mutant Nm23-H1 S-22 cells retained on average 60 and 50% of untreated KSR1, respectively. Similar trends were observed with 1 and 5 μM 17-AAG, where KSR1 in H1-177 cells showed a greater extent of degradation. In experiments not shown, time course data confirmed this trend. C-100 and H1-177 cells were cultured with or without 1 μM 17-AAG for 0 to 24 h, and KSR1 expression was determined as described above. Control C-100 and Nm23-H1-overexpressing H1-177 KSR levels (as a percentage of those at the zero time point) were 67 and 45%, respectively, at 12 h and 49 and 16%, respectively, at 24 h of culture. The data provide additional evidence that KSR1 is a client protein of Hsp90 and that relative levels of Nm23-H1 metastasis suppressor expression influence Hsp90-dependent degradation of KSR1.
FIG. 5.
Degradation of KSR1 is increased in cells overexpressing Nm23-H1 treated with the Hsp90 inhibitor 17-AAG. Control C-100, mutant P96S Nm23-H1 S-22, and wild-type Nm23-H1 H1-177 cell lines transiently cotransfected with HA-KSR1 and pEGFP were treated with various amounts of 17-AAG for 6 h. Equivalent amounts of protein from total cell lysate were analyzed by Western blotting with anti-HA. Western blots shown are a representative experiment; densitometric analysis of KSR1, normalized to GFP expression, for three independent experiments is graphed below.
17-AAG affects Nm23-H1, Hsp90, and KSR1 complex formation.
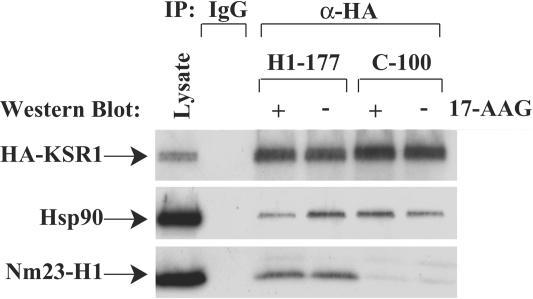
To assess if 17-AAG treatment of cells expressing different levels of Nm23-H1 would affect Nm23-H1, Hsp90, and KSR1 complex formation, C-100 and H1-177 cells transiently transfected with HA-KSR1 were incubated with 1 μM 17-AAG or DMSO for 1 h. HA-KSR1 was immunoprecipitated from equivalent amounts of total protein in cytoplasmic lysates of the C-100 and H1-177 cell lines, and the relative amounts of Hsp90 and Nm23-H1 bound to KSR1 were determined by Western blotting (Fig. 6). Untreated H1-177 cells again showed increased binding of Hsp90 to KSR1 compared to untreated control C-100 cells. In the presence of 17-AAG, binding of Hsp90 to KSR1 was reduced approximately 50% in H1-177 cells but not in control C-100 cells. The release of client proteins from Hsp90 in response to blockade of ADP/ATP recycling has been reported to be associated with their degradation (8, 30). In experiments not shown, KSR1 degradation in H1-177 cells was only partially sensitive to the MG132 proteasome inhibitor, suggesting that proteasomal and other degradative pathways may be involved.
FIG. 6.
17-AAG disrupts Nm23-H1-KSR1-Hsp90 complex formation in H1-177 cells. Control C-100 and Nm23-H1-overexpressing H1-177 cells were transiently transfected with HA-KSR1 and exposed to 1 μM 17-AAG or DMSO (vehicle control) for 1 h. Cytoplasmic lysates were immunoprecipitated with anti-HA, and the binding of Hsp90 and Nm23-H1 proteins was determined by Western blotting.
Nm23-H1 overexpression confers increased sensitivity to 17-AAG in soft agar colonization.
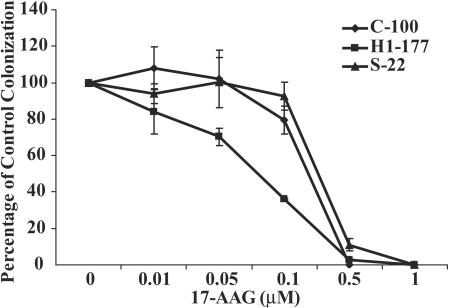
Given the correlation of increased Nm23-H1 expression, increased Hsp90 complex binding to KSR1, and enhanced degradation of KSR1, we examined whether this pathway exerted biological consequences. Nm23-H1 overexpression has been reported to decrease in vitro motility and soft agar colonization (26). The control C-100, the Nm23-H1-overexpressing H1-177, and the P96S Nm23-H1-overexpressing S-22 cell lines were grown in soft agar with or without 17-AAG. Colonization by H1-177 cells was two- to threefold less than that of the control C-100 or mutant Nm23-H1 S-22 cells, in agreement with previously reported data (25). The relative effect of 17-AAG on colonization of the cell lines was quantitated as a percentage of control colonization, without 17-AAG (Fig. 7). Colonization of all three cell lines was inhibited by 17-AAG but with altered sensitivities. Nm23-H1-overexpressing H1-177 cells were preferentially sensitive to 17-AAG inhibition of colonization, particularly in the lower 17-AAG concentration range thought to be therapeutically relevant. The 50% inhibitory concentration for H1-177 cells was ∼0.07 μM, compared to ∼0.25 μM for the C-100 and S-22 cells, an approximately threefold difference. In experiments not shown, H1-177 cells transiently transfected with a control or HA-KSR1 construct exhibited virtually identical sensitivity to 17-AAG in soft agar colonization, indicating that the experimental procedure of transient HA-KSR1 transfection did not alter the biological sensitivity in this assay. The data indicate that Nm23-H1 overexpression conferred increased sensitivity to an Hsp90-specific agent in anchorage-independent growth assays.
FIG. 7.
17-AAG preferentially inhibits the colonization of Nm23-H1 overexpressing MDA-MB-435 breast carcinoma cells. Equivalent cell numbers of control C-100, P96S mutant Nm23-H1 S-22 and wild type Nm23-H1 overexpressing H1-177 cell lines were plated in soft agar with semilog dilutions of 17-AAG. Colonies were counted three weeks later. Data represent the means ± SEM for triplicate cultures (for a representative experiment of n = 3), expressed as a percentage of vehicle control colonization for each line. C-100 and H1-177 colonization was significantly different: with 0.05 μM 17-AAG, P = 0.0046 (Student's t test).
DISCUSSION
Signal transduction inhibitors have attracted widespread interest in breast cancer therapy (reviewed in references 41 and 60). A new aspect of signaling concerns the scaffolds which assemble the participating signaling proteins, determine their subcellular localization, and influence the amplitude, duration, and specificity of response. We have investigated KSR1 function in metastatic breast carcinoma cells. The MDA-MB-435 cell line was derived from a breast metastasis and retains in vivo metastatic competency, although it exhibits gene expression profiles of breast and melanoma lines (62). This is consistent with findings with other aggressive cell lines, where metastatic tumor cells can take on the phenotypes of endothelial cells and other cells (20, 34). Our experiments examined steady-state conditions in serum, which mimic the growth factor milieu encountered by primary tumors and metastases. The cell line chosen has wild-type Ras, similar to the majority of breast cancers (27, 37, 61). Under these conditions, we present evidence indicating that scaffold function constitutes a dynamic part of the signaling machinery and may influence cancer progression and treatment.
The metastasis suppressor Nm23-H1 is shown herein to modulate the stability of the KSR1 Erk MAP kinase scaffold protein via its differential binding of Hsp90 complexes. This new pathway may underlie in part the observed inhibition of Erk MAP kinase pathway activity in cells expressing high levels of Nm23-H1 (19), which is hypothesized to contribute to its metastasis-suppressive activity. Erk activity may be required for outgrowth of metastases in a distant site. Transfection of nm23-H1 into MDA-MB-435 breast carcinoma cells in either a transient or stable fashion resulted in two- to threefold greater binding of Hsp90 to KSR1. Other KSR1 scaffold proteins, such as 14-3-3β, did not exhibit alterations in binding levels, providing evidence of specificity. The mutant Nm23-H1 transfectants S-22 and I-205, which failed to inhibit in vitro motility upon transfection (31), also exhibited lower levels of Hsp90 binding to KSR1. Hsp90 is a chaperone protein, involved in the maturation and/or degradation of numerous client proteins. Limited evidence has been previously reported indicating that KSR1 is an Hsp90 client protein in 293T cells. The geldanamycins (17-AAG), which stabilize Hsp90 in the ADP-bound state favoring protein degradation, accelerated the degradation of KSR1 in fibroblasts (65). Based on these observations, we asked if elevated binding of Hsp90 to the KSR1 scaffold in Nm23-H1-overexpressing cells resulted in its enhanced degradation. In agreement with this hypothesis, enhanced degradation of KSR1 was observed in either cycloheximide- or 17-AAG-treated H1-177 cells. The degradation of KSR1 in high-Nm23-H1-expression breast carcinoma cells correlated with reduced phospho-Erk expression and increased sensitivity to 17-AAG in soft agar colonization assays. These data support the hypothesis that KSR1 scaffold function can be modulated based on Hsp90 binding and, in turn, metastasis suppressor expression. Several caveats are important. (i) These studies are based on two- to eightfold increases in Nm23-H1 expression among the transient and stable transfectants, levels within the range observed in tumor cohorts where quantitated (10, 18, 71). (ii) Complex formation between Nm23-H1, KSR1, and Hsp90 represented a minor proportion of total protein levels. For Nm23-H1, developmental studies have indicated that only 4% of its protein level can exert phenotypic effects (78). Other functions for these abundant proteins exist and may contribute to the observed colonization phenotype. (iii) It is noteworthy that multiple regulatory points may exist for important signaling proteins, such as KSR1. A Ras-responsive E3 ubiquitin ligase protein, Impedes Mitogenic Signal Propagation protein, was recently reported to be degraded in response to Ras activation, permitting Raf-Mek-KSR1 complex formation (35).
Current experiments are investigating the mechanism of increased Hsp90 binding to KSR1 in Nm23-H1-overexpressing breast carcinoma cells. Our data fail to suggest that the Nm23-H1-induced phosphorylation of KSR1 on serines 392 and 434 accounts for the altered Hsp90 binding, based on site-directed mutagenesis experiments (data not shown). Rather, overall Nm23-H1 protein levels appear to be important. The fact that Nm23-H1 and Hsp90 coimmunoprecipitate regardless of the presence of exogenous KSR1 led us to hypothesize that Nm23-H1 could work as a cochaperone itself to confer specificity to the KSR1-Hsp90 complex and promote the degradation of KSR1. This would explain the fact that we did not find any significant differences in the amount of tested cochaperones (Hsp70/Hsc70, Cdc37, Hop, p23, and BAG-1) bound to KSR1 in C-100 and H1-177 cells after treatment with 17-AAG (data not shown).
Our data also suggest that the KSR1 scaffold may have important functions in the nucleus. Little is known of KSR1 nuclear function. Brennan et al. (9) reported that leptomycin B-treated fibroblasts accumulated nuclear KSR1, consistent with a Crm1-dependent nuclear shuttling process. Mutations in KSR1 affecting its interaction with Mek blocked nuclear import of KSR1, and microinjection of Mek or KSR1 promoted cytoplasmic localization of the other protein. These data suggested the hypothesis that KSR1 serves to regulate the subcellular distribution of Mek. Data from Saccharomyces cerevisiae are also consistent with an important role for scaffolds in the nucleus. Ste5 is the MAP kinase scaffold protein required for mating (13) and continuously shuttles from the cytoplasm to the nucleus but translocates to the plasma membrane upon pheromone stimulation. Nuclear shuttling of Ste5 controls the amount of Ste5 available to bind membrane receptors and enhances pathway activation (32). In our experiments, KSR1 immunoprecipitated from nuclear extracts of MDA-MB-435 breast carcinoma cells bound Erk pathway members and was subject to the same changes in Hsp90 binding between low- and high-Nm23-H1-expression cell lines that were observed in the cytoplasm. Thus, nuclear KSR1 is not merely binding Mek but exists as a more complete scaffold with similar regulatory influences, where a function to directly bind membranous Raf-1 appears unlikely. KSR1 complexes were chromatographed in the 250- to 500-kDa range (42), and it is unknown whether this complex can move in toto from the cytoplasm to the nucleus or has to reform in each compartment. We hypothesize that membrane-associated KSR1 may have a distinct protein binding pattern, since the KSR1 coimmunoprecipitation pattern from total versus cytoplasmic or nuclear lysates of breast carcinoma cells differs (data not shown). This hypothesis could not be tested, however, since the transfection of greater KSR1 to enable detection is thought to upset the stoichiometry of binding proteins, thereby altering the function of the Erk pathway.
Finally, the data speak to the potential importance of metastatic competence as a contributor to therapeutic efficacy. The geldanamycins affect degradation of numerous client proteins; the 17-AAG form is in phase I-II clinical trials and the water-soluble 17-DMAG form has entered phase I trial. Several Hsp90 clients have been identified in breast cancer, including Her-2, Raf-1, mutant p53, Akt, and hormone receptors (1, 5). Our data showing that increased binding of Hsp90 to KSR1 leads to its more-rapid degradation identifies KSR1 as another client in breast cancer. The concentrations of 17-AAG used herein are consistent with KSR1 being a clinically tractable target. Using 17-AAG, we demonstrated that the trend of increased Nm23-H1 expression, increased Hsp90 binding to KSR1, and more-rapid KSR1 degradation has functional consequences; high-Nm23-H1-expression breast cells were approximately threefold more sensitive to 17-AAG inhibition of soft agar colonization. These data suggest that 17-AAG may not be optimally active against low-Nm23-H1-expression, highly metastatic tumor cells and that drug combinations may need to be investigated in the metastatic setting. The 17-AAG sensitivity of tumor cells dichotomized by the expression of other metastasis-associated genes will be of interest to further develop this hypothesis. It should be noted that most drugs have never been tested on low- versus high-metastatic-potential tumor cells before clinical trial, and many others may unfortunately exhibit similar trends. 17-AAG has been previously reported to modulate several metastasis-related proteins and to inhibit the motility of tumor cells (43, 75); to our knowledge the present data are the first comparison of function in cells with various degrees of metastatic competency. Current experiments are addressing 17-AAG combinations with standard and new therapeutic agents for in vitro and in vivo efficacy against highly metastatic breast carcinoma lines. Such an approach may ultimately improve our clinical trial efficacy, in contrast to current situations where drugs are developed on primary xenograft data but then asked to work in a metastatic clinical setting (22).
Acknowledgments
We thank Len Neckers, Urologic Oncology Branch, CCR, NCI, for multiple helpful discussions. We also thank Deborah Morrison, NCI, and the Cancer Therapy Evaluation Program, NCI, for reagents.
REFERENCES
- 1.An, W., R. Schnur, L. Neckers, and M. Blagosklonny. 1997. Depletion of p185erbB2, Raf-1 and mutant p53 proteins by geldanamycin derivatives correlates with antiproliferative activity. Cancer Chemother. Pharmacol. 40:60-64. [DOI] [PubMed] [Google Scholar]
- 2.Anselmo, A., R. Bumeister, J. Thomas, and M. White. 2002. Critical contribution of linker proteins to Raf kinase activation. J. Biol. Chem. 277:5940-5943. [DOI] [PubMed] [Google Scholar]
- 3.Baba, H., T. Urano, K. Okada, K. Furukawa, E. Nakayama, H. Tanaka, K. Iwasaki, and H. Shiku. 1995. Two isotypes of murine nm23/nucleoside diphosphate kinase, nm23-M1 and nm23-M2, are involved in metastatic suppression of a murine melanoma line. Cancer Res. 55:1977-1981. [PubMed] [Google Scholar]
- 4.Baillat, G., S. Gaillard, F. Castets, and A. Monneron. 2002. Interactions of phocein with nucleoside diphosphate kinase, Eps15, and dynamin I. J. Biol. Chem. 277:18961-18966. [DOI] [PubMed] [Google Scholar]
- 5.Basso, A., D. Solit, P. Munster, and N. Rosen. 2002. Ansamycin antibiotics inhibit Akt activation and cyclin D expression in breast cancer cells that overexpress HER2. Oncogene 21:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, B., H. Xing, K. Yan, N. Gautam, and A. Muslin. 1999. KSR-1 binds to G-protein bg subunits and inhibits bg-induced mitogen activated protein kinase activation. J. Biol. Chem. 274:7982-7986. [DOI] [PubMed] [Google Scholar]
- 7.Bhujwalla, Z., E. Aboagye, R. Gilles, V. Chack, C. Mendola, and J. Backer. 1999. Nm23-transfected MDA-MB-435 human breast carcinoma cells form tumors with altered phospholipid metabolism and pH: a 31P nuclear magnetic resonance study in vivo and in vitro. Magn. Reson. Med. 41:897-903. [DOI] [PubMed] [Google Scholar]
- 8.Bonvini, P., T. Gastaldi, B. Falini, and A. Rosolen. 2002. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK+ CD30+ lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 62:1559-1566. [PubMed] [Google Scholar]
- 9.Brennan, J., D. Volle, O. V. Chaika, and R. Lewis. 2002. Phosphorylation regulates the nucleocytoplasmic distribution of kinase suppressor of Ras. J. Biol. Chem. 277:5369-5377. [DOI] [PubMed] [Google Scholar]
- 10.Caligo, M., G. Cipollini, A. Berti, P. Viacava, P. Collecchi, and G. Bevilacqua. 1997. NM23 gene-expression in human breast carcinomas—loss of correlation with cell-proliferation in the advanced phase of tumor progression. Int. J. Cancer 74:102-111. [DOI] [PubMed] [Google Scholar]
- 11.Cuello, F., R. Schulze, F. Heemeyer, H. Meyer, S. Lutz, K. Jakobs, F. Niroomand, and T. Wieland. 2003. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gβ subunits. J. Biol. Chem. 278:7220-7226. [DOI] [PubMed] [Google Scholar]
- 12.Du, J., and G. Hannon. 2002. The centrosomal kinase Aurora-A/STK15 interacts with putative tumor suppressor NM23-H1. Nucleic Acids Res. 30:5465-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elion, E. 2001. The Ste5 scaffold. J. Cell Sci. 114:3967-3978. [DOI] [PubMed] [Google Scholar]
- 14.Fan, Z., P. Beresford, D. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659-672. [DOI] [PubMed] [Google Scholar]
- 15.Fishbach, M., and J. Settleman. 2003. Specific biochemical inactivation of oncogenic ras proteins by nucleoside diphosphate kinase. Cancer Res. 63:4089-4094. [PubMed] [Google Scholar]
- 16.Fournier, H., S. Dupe-Manet, D. Bouvard, M. Laconbe, C. Marie, M. Block, and C. Albiges-Rizo. 2002. Integrin cytoplasmic domain-associated protein 1a (ICAP-1a) interacts directly with the metastasis suppressor nm23-H2, and both proteins are targeted to newly formed cell adhesion sites upon integrin engagement. J. Biol. Chem. 277:20895-20902. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, M., A. Ishii, Y. Yasutomo, N. Shimada, N. Ishikawa, N. Hanai, N. Nagara, T. Irimura, G. Nicolson, and N. Kimura. 1996. Metastatic potential of rat mammary adenocarcinoma cells associated with decreased expression of nucleoside diphosphate kinase/nm23: reduction by transfection of NDP kinase a isoform, an nm23-H2 gene homolog. Int. J. Cancer 65:531-537. [DOI] [PubMed] [Google Scholar]
- 18.Goodall, R., H. Dawkins, P. Robbins, E. Hahnel, M. Sarna, R. Hahnel, J. Papadimitriou, J. Harvey, and G. Sterrett. 1994. Evaluation of the expression levels of nm23-H1 RNA in primary breast cancer, benign breast disease, axillary lymph nodes and normal breast tissue. Pathology 26:423-428. [DOI] [PubMed] [Google Scholar]
- 19.Hartsough, M., D. Morrison, M. Salerno, D. Palmieri, T. Ouatas, M. Mair, J. Patrick, and P. Steeg. 2002. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of ras (KSR), via a histidine protein kinase pathway. J. Biol. Chem. 277:32389-32399. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix, M., E. Seftor, D. Kirschmann, and R. Seftor. 2000. Can breast cancer cells undergo vasculogenic mimicry? Br. Cancer Res. 2:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs, J., W. Xu, and L. Neckers. 2003. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 3:213-217. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J., S. Decker, D. Zaharevitz, L. Rubenstein, J. Vendetti, S. Schepartz, S. Kalyandrug, M. Christian, S. Arbuck, M. Hollingshead, and E. Sausville. 2001. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer 84:1424-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantor, J. D., B. McCormick, P. S. Steeg, and B. R. Zetter. 1993. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer Res. 53:1971-1973. [PubMed] [Google Scholar]
- 24.Kornfield, K., D. Hom, and H. Horvitz. 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83:903-913. [DOI] [PubMed] [Google Scholar]
- 25.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. K. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65:25-35. [DOI] [PubMed] [Google Scholar]
- 26.Leone, A., U. Flatow, K. VanHoutte, and P. S. Steeg. 1993. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization, and enzymatic activity. Oncogene 8:2325-2333. [PubMed] [Google Scholar]
- 27.Lintig, F. V., A. Dreilinger, N. Varki, A. Wallace, D. Casteel, and G. Boss. 2000. Ras activation in human breast cancer. Breast Cancer Res. Treat. 62:51-62. [DOI] [PubMed] [Google Scholar]
- 28.Lombardi, D., A. Sacchi, G. D'Agostino, and G. Tibursi. 1995. The association of the Nm23-M1 protein and beta-tubulin correlates with cell differentiation. Exp. Cell Res. 217:267-271. [DOI] [PubMed] [Google Scholar]
- 29.Lozano, J., R. Xing, Z. Cai, H. Jensen, C. Trempus, W. Mark, R. Cannon, and R. Kolesnick. 2003. Deficiency of kinase suppressor of ras1 prevents oncogenic ras signaling in mice. Cancer Res. 63:4232-4238. [PubMed] [Google Scholar]
- 30.Luo, J., and J. Benovic. 2003. G protein-coupled receptor kinase interaction with Hsp90 mediates kinase maturation. J. Biol. Chem. 278:50908-50914. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald, N., J. Freije, M. Stracke, R. Manrow, and P. Steeg. 1996. Site directed mutagenesis of nm23-H1: mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J. Biol. Chem. 271:25107-25116. [DOI] [PubMed] [Google Scholar]
- 32.Mahanty, S., Y. Wang, F. Farley, and E. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98:501-512. [DOI] [PubMed] [Google Scholar]
- 33.Maloney, A., and P. Workman. 2002. HSP90 as a new therapeutic target for cancer therapy: the story unfolds. Expert Opin. Biol. Ther. 2:3-24. [DOI] [PubMed] [Google Scholar]
- 34.Maniotis, A., R. Folberg, A. Hess, E. Seftor, L. Gardner, J. Pe'er, J. Trent, P. Meltzer, and M. Hendrix. 1999. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am. J. Pathol. 155:739-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matheny, S., C. Chen, R. Kortum, G. Razidlo, R. Lewis, and M. White. 2004. Ras regulates assembly of mitogenic signalling complexes throughout the effector protein IMP. Nature 427:256-260. [DOI] [PubMed] [Google Scholar]
- 36.Miele, M. E., A. D. L. Rosa, J. H. Lee, D. J. Hicks, J. U. Dennis, P. S. Steeg, and D. R. Welch. 1997. Suppression of human melanoma metastasis following introduction of chromosome 6 is independent of NME1 (nm23). Clin. Exp. Metastasis 15:259-265. [DOI] [PubMed] [Google Scholar]
- 37.Miyakis, S., G. Sourvinos, and D. Spandidos. 1998. Differential expression and mutation of the ras family genes in human breast cancer. Biochem. Biophys. Res. Commun. 251:609-612. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki, H., M. Fukuda, Y. Ishijima, A. Negishi, R. Hirayama, N. Ishikawa, T. Amagasa, and N. Kimura. 1999. Overexpression of nm23-H2/NDP kinase B in a human oral squamous cell carcinoma cell line results in reduced metastasis, differentiated phenotype in the metastatic site, and growth factor-independent proliferative activity in culture. Clin. Cancer Res. 5:4301-4307. [PubMed] [Google Scholar]
- 39.Morrison, D. 2001. KSR: a MAPK scaffold of the ras pathway? J. Cell Sci. 114:1609-1612. [DOI] [PubMed] [Google Scholar]
- 40.Muller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. Morrison. 2001. C-TAK1 regulates ras signaling by phosphorylating the MAPK scaffold, KSR. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 41.Nahta, R., G. Hortobagyi, and F. Esteva. 2003. Novel pharmacological approaches in the treatment of breast cancer. Expert Opin. Investig. Drugs 12:909-921. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen, A., W. Burack, J. Stock, R. Kortum, O. Chaika, M. Afkarian, W. Muller, K. Murphy, D. Morrison, R. Lewis, J. McNeish, and A. Shaw. 2002. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 22:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen, D., S. Desai, A. Chen, T. Weiser, and D. Schrump. 2000. Modulation of metastasis phenotypes of non-small cell lung cancer cells by 17-allylamino 17-demethoxy geldanamycin. Ann. Thorac. Surg. 70:1853-1860. [DOI] [PubMed] [Google Scholar]
- 44.Nosaka, K., M. Kawahara, M. Masuda, Y. Satomi, and H. Nishino. 1998. Association of nucleoside diphosphate kinase nm23-H2 with human telomeres. Biochem. Biophys. Res. Comm. 243:342-348. [DOI] [PubMed] [Google Scholar]
- 45.Ohmachi, M., C. Rocheleau, D. Church, E. Lambie, T. Schedl, and M. Sundaram. 2002. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 12:427-433. [DOI] [PubMed] [Google Scholar]
- 46.Ory, S., M. Zhou, T. Conrads, T. Veenstra, and D. Morrison. 2003. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr. Biol. 13:1356-1364. [DOI] [PubMed] [Google Scholar]
- 47.Otero, A. 1997. Copurification of vimentin, energy metabolism enzymes and a MER5 homolog with nucleoside diphosphate kinase. Identification of tissue-specific interactions. J. Biol. Chem. 272:14690-14694. [DOI] [PubMed] [Google Scholar]
- 48.Otsuki, Y., M. Tanaka, S. Yoshii, N. Kawazoe, K. Nakaya, and H. Sugimura. 2001. Tumor metastasis suppressor nm23H1 regulates Rac GTPase by interaction with Tiam1. Proc. Natl. Acad. Sci. USA 98:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ouatas, T., D. Halverson, and P. Steeg. 2003. Dexamethasone and medroxyprogesterone acetate elevate Nm23-H1 metastasis suppressor expression in metastatic human breast carcinoma cells: new uses for old compounds. Clin. Cancer Res. 9:3763-3772. [PubMed] [Google Scholar]
- 50.Palacios, F., J. Schweitzer, R. Boshans, and C. D'Souza-Schorey. 2002. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat. Cell Biol. 4:929-936. [DOI] [PubMed] [Google Scholar]
- 51.Paravicini, G., M. Steinmayr, E. André, and M. Becker-André. 1996. The metastasis suppressor candidate nucleoside diphosphate kinase Nm23 specifically interacts with members of the ROR/RZR nuclear orphan receptor subfamily. Biochem. Biophys. Res. Commun. 227:82-87. [DOI] [PubMed] [Google Scholar]
- 52.Parhar, R. S., Y. Shi, M. Zou, N. R. Farid, P. Ernst, and S. Al-Sedairy. 1995. Effects of cytokine mediated modulation of Nm23 expression on the invasion and metastatic behavior of B16F10 melanoma cells. Int. J. Cancer 60:204-210. [DOI] [PubMed] [Google Scholar]
- 53.Postel, E. 2003. Multiple biochemical activities of NM23/NDP kinase in gene regulation. J. Bioenerg. Biomembr. 35:31-40. [DOI] [PubMed] [Google Scholar]
- 54.Postel, E., and B. Abramczyk. 2003. Escherichia coli nucleoside diphosphate kinase is a uracil-processing DNA repair nuclease. Proc. Natl. Acad. Sci. USA 100:13247-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reymond, A., S. Volorio, G. Merla, M. Al-Maghtheh, O. Zuffardi, A. Bulfone, A. Ballabio, and M. Zollo. 1999. Evidence for interaction between human PRUNE and nm23-H1 NDPKinase. Oncogene 18:7244-7252. [DOI] [PubMed] [Google Scholar]
- 56.Roy, F., G. Laberge, M. Douziech, D. Ferland-McCollough, and M. Therrien. 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roymans, D., K. Vissenberg, C. DeJonghe, R. W. G. Engler, N. Kimura, B. Grobben, P. Claes, J. Verbelen, C. VanBroeckhoven, and H. Slegers. 2001. Identification of the tumor metastasis suppressor Nm23-H1/Nm23-R1 as a constitutent of the centrosome. Exp. Cell Res. 262:145-153. [DOI] [PubMed] [Google Scholar]
- 58.Roymans, D., R. Willems, K. Vissenberg, C. DeJonghe, B. Groben, P. Claes, I. Lascu, D. V. Bockstaele, J. Verbelen, C. VanBroeckhoven, and H. Slegers. 2000. Nucleoside diphosphate kinase b (Nm23-R1/NDPKb) is associated with intermediate filaments and becomes upregulated upon cAMP-induced differentiation of rat C6 glioma. Exp. Cell Res. 261:127-138. [DOI] [PubMed] [Google Scholar]
- 59.Russell, R., A. Pedersen, J. Kantor, K. Geisinger, R. Long, N. Zbieranski, A. Townsend, B. Shelton, N. Brunner, and T. Kute. 1998. Relationship of nm23 to proteolytic factors, proliferation and motility in breast cancer tissues and cell lines. Br. J. Cancer 78:710-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sausville, E., Y. Elsayed, M. Monga, and G. Kim. 2003. Signal transduction-directed cancer treatments. Annu. Rev. Pharmacol. Toxicol. 43:199-231. [DOI] [PubMed] [Google Scholar]
- 61.Schondorf, T., A. Andrack, D. Niederacher, A. Scharl, M. Becker, H. Engel, and U. Gohring. 1999. H-ras gene amplification or mutation is not common in human primary breast cancer. Oncol. Rep. 6:1029-1033. [DOI] [PubMed] [Google Scholar]
- 62.Sellappan, S., R. Grijalva, X. Zhou, W. Yang, M. B. Eli, G. Mills, and D. Yu. 2004. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 64:3479-3485. [DOI] [PubMed] [Google Scholar]
- 63.Siddiqui-Jain, A., C. Grand, D. Bearss, and L. Hurley. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a samll molecule to repress c-MTC transcription. Proc. Natl. Acad. Sci. USA 99:11593-11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steeg, P. 2003. Metastasis suppressors alter the signal transduction of cancer cells. Nat. Cancer Rev. 3:55-63. [DOI] [PubMed] [Google Scholar]
- 65.Stewart, S., M. Sundaram, Y. Zhang, J. Lee, M. Han, and K. Guan. 1999. Kinase suppressor of ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 19:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian, C., M. Cotter, and E. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 67.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki, E., T. Ota, K. Tsukuda, A. Okita, K. Matsuoka, M. Murakami, H. Doihara, and N. Shimizu. 2004. nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int. J. Cancer 108:207-211. [DOI] [PubMed] [Google Scholar]
- 69.Tagashira, H., K. Hamazaki, N. Tanaka, C. Gao, and M. Namba. 1998. Reduced metastatic potential and c-myc overexpression of colon adenocarcinoma cells (Colon 26 line) transfected with nm23-R2 rat nucleoside diphosphate kinase a isoform. Int. J. Mol. Med. 2:65-68. [DOI] [PubMed] [Google Scholar]
- 70.Therrien, M., H. Chang, N. Solomon, F. Karim, D. Wassarman, and G. Rubin. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83:879-888. [DOI] [PubMed] [Google Scholar]
- 71.Toulas, C., J. Mihura, C. de Balincourt, B. Marques, E. Marek, G. Soula, H. Roche, and G. Favre. 1996. Potential prognostic value in human breast cancer of cytosolic Nme1 protein detection using an original hen specific antibody. Br. J. Cancer 73:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tseng, Y., D. Vincent, J. Zhu, A. Adeyinka, J. Moyers, P. Watson, and C. Kahn. 2001. Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and Nm23. Cancer Res. 61:2071-2079. [PubMed] [Google Scholar]
- 73.Wagner, P., and N.-D. Vu. 2000. Histidine to aspartate phosphotransferase activity of nm23 protein: phosphorylation of aldolase C on Asp 319. Biochem. J. 346:623-630. [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner, P., and N.-D. Vu. 1995. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J. Biol. Chem. 270:21758-21764. [DOI] [PubMed] [Google Scholar]
- 75.Webb, C., C. Hose, S. Koochekpour, M. Jeffers, M. Oskarsson, E. Sausville, A. Monks, and G. VandWoude. 2000. The geldanamycins are potent inhibitors of the hepatocyte growth factor/scatter factor-Met-urokinase plasminogen activator-pasmin proteolytic network. Cancer Res. 60:342-349. [PubMed] [Google Scholar]
- 76.Xing, H., C. Cordon-Cardo, X. Deng, W. Tong, L. Campodonico, Z. Fuks, and R. Kolesnick. 2003. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates ras-mediated pancreatic cancer. Nat. Med. 9:1267-1268. [DOI] [PubMed] [Google Scholar]
- 77.Xing, H., and R. Kolesnick. 2001. Kinase suppressor of ras signals through Thr269 of c-Raf-1. J. Biol. Chem. 276:9733-9741. [DOI] [PubMed] [Google Scholar]
- 78.Xu, J., L. Liu, F. Deng, L. Timmons, E. Hersperger, P. Steeg, M. Veron, and A. Shearn. 1996. Rescue of the awd mutant phenotype by expression of human Nm23/NDP kinase in Drosophila. Dev. Biol. 177:544-557. [Google Scholar]
- 79.Yaguchi, H., N. Ohkura, T. Tsukada, and K. Yamaguchi. 2002. Menin, the multiple endocrine neoplasia type 1 gene product, exhibits GTP-hydrolyzing activity in the presence of the tumor metastasis suppressor nm23. J. Biol. Chem. 277:38197-38204. [DOI] [PubMed] [Google Scholar]
- 80.Yan, F., and B. Polk. 2001. Kinase suppressor of ras is necessary for tumor necrosis factor a activation of extracellular signal-regulated kinase/mitogen-activated protein kinase in intestinal epithelial cells. Cancer Res. 61:963-989. [PubMed] [Google Scholar]
- 81.Yoder, J., H. Chong, K.-L. Guan, and M. Han. 2004. Modulation of KSR activity in Caenorhabditis elegans by Zn ions, PAR-1 kinase and PP2A phosphatase. EMBO J. 23:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu, J., Y.-H. Tseng, J. Kantor, C. Rhodes, B. Zetter, J. Moyers, and C. Kahn. 1999. Interaction of the Ras-related protein associated with diabetes Rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc. Natl. Acad. Sci. USA 96:14911-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]