Abstract
Gene activation by steroid hormone receptors involves the recruitment of the steroid receptor coactivator (SRC)/p160 coactivator LXXLL motifs to activation function 2 (AF2) in the ligand binding domain. For the androgen receptor (AR), AF2 also serves as the interaction site for the AR NH2-terminal FXXLF motif in the androgen-dependent NH2-terminal and carboxyl-terminal (N/C) interaction. The relative importance of the AR AF2 site has been unclear, since the AR FXXLF motif interferes with coactivator recruitment by competitive inhibition of LXXLL motif binding. In this report, we identified the X chromosome-linked melanoma antigen gene product MAGE-11 as an AR coregulator that specifically binds the AR NH2-terminal FXXLF motif. Binding of MAGE-11 to the AR FXXLF α-helical region stabilizes the ligand-free AR and, in the presence of an agonist, increases exposure of AF2 to the recruitment and activation by the SRC/p160 coactivators. Intracellular association between AR and MAGE-11 is supported by their coimmunoprecipitation and colocalization in the absence and presence of hormone and by competitive inhibition of the N/C interaction. AR transactivation increases in response to MAGE-11 and the SRC/p160 coactivators through mechanisms that include but are not limited to the AF2 site. MAGE-11 is expressed in androgen-dependent tissues and in prostate cancer cell lines. The results suggest MAGE-11 is a unique AR coregulator that increases AR activity by modulating the AR interdomain interaction.
The androgen receptor (AR) is a member of the steroid receptor subfamily of nuclear receptors. Like other steroid receptors, AR has multiple domains involved in ligand and DNA binding and transcriptional activation. Recently, several unique properties of AR that distinguish it from other steroid receptors have gained attention. High-affinity androgen binding stabilizes AR (26), which is in contrast to most steroid receptors that are down regulated by agonist binding. Agonist-induced AR stabilization results in part from the NH2-terminal and carboxyl-terminal (N/C) interdomain interaction mediated by the androgen-dependent interaction between the AR NH2-terminal FXXLF motif and activation function 2 (AF2) in the ligand binding domain (16, 17). The FXXLF motif 23FQNLF27 is part of an amphipathic α-helical region that is similar in structure to the LXXLL motifs of the steroid receptor coactivator (SRC)/p160 family of coactivators. The AR FXXLF motif is highly conserved among vertebrates, supporting its functional importance across species (13, 18). Recent cocrystal structures and binding studies have confirmed preferential binding of the FXXLF motif to the AR AF2 site and adaptability of AF2 to coactivator LXXLL motif binding through an induced-fit mechanism (15, 22). FXXLF motif binding to AF2 requires binding of ligands that display agonist activity in vivo (27). In transient transfection reporter gene assays, the N/C interaction is required for the activation of some, but not all, androgen-regulated genes (2, 18). One consequence of the AR interdomain interaction is inhibition of recruitment of the SRC/p160 family of coactivators by competitive binding of FXXLF at the coactivator LXXLL motif binding site in AF2 (14). AR activation by the SRC/p160 family of coactivators is reduced by the interdomain interaction and by sequence changes in AF2 during evolution that favor FXXLF over LXXLL motif binding (15, 20).
Preference for FXXLF binding by AF2 but adaptability to coactivator LXXLL motif binding brings into question the role of the SRC/p160 family of coactivators in AR functional activity. The level of transcriptional intermediary factor 2 (TIF2) (also known as SRC2 or glucocorticoid receptor [GR] interacting protein 1 [GRIP1]) is low in normal prostate epithelial cells (9). In contrast, a majority of advanced prostate cancers that recur after androgen deprivation therapy can have increased levels of SRC1 and TIF2 (SRC2 or GRIP1) (10), suggesting that these coactivators have an important role in AR action that contributes to prostate tumor development and progression (1, 8). Increased expression of the SRC1 and TIF2 coactivators increases AR transactivation at gene promoters in transient transfection assays, even when the promoter depends on the AR N/C interaction for maximal activation (18). This is attributed to overall conservation of the AR AF2 binding site that allows coactivator LXXLL motif binding, albeit with lower affinity than for the FXXLF motif (15, 20).
These observations led us to postulate the existence of an AR coregulator that binds the AR FXXLF motif that would expose the AF2 site for coactivator binding. In this report, we made use of the AR NH2-terminal FXXLF peptide as bait in a Saccharomyces cerevisiae two-hybrid screen of a human testis library to identify the melanoma antigen gene product MAGE-11 as a novel AR coregulator. MAGE-11 competes for the androgen-induced AR N/C interaction by specifically binding the AR FXXLF motif and relieves inhibition at AF2.
MATERIALS AND METHODS
Plasmids.
Expression vectors previously described include pCMVhAR (31); pCMVhAR1-660 (42); pCMVhAR-FXXAA (14); GAL-AR1-503 (30); VP-AR1-660; VP-AR1-660FXXAA (17); GAL-AR20-30, GAL-AR20-36, GAL-AR16-30 and GAL-AR16-36 (20); pcDNA3HA-AR624-919 (17, 18); pCMVhAR-C576A; pCMVhAR-R617M-K618M-K632M-R633M (ARm4) (52); pSG5-TIF2 (47); and the thyroid hormone receptor activator molecule expression vector pSG5-TRAM1 (44). pCMVhAR1-660FXXAA was prepared by digestion of pCMVhAR-FXXAA with TthIII and XbaI and blunt ending and religating the plasmid. GAL-AR16-36 mutants were described previously (20) or were prepared by cloning annealed complementary oligonucleotides into pGAL0, which expresses S. cerevisiae GAL4 DNA binding domain residues 1 to 147. pCMV5-Flag-hAR was constructed by cloning a PCR-amplified BglII- and AflII-digested AR NH2-terminal fragment containing the Flag tag sequence DYKDDDDK, following the first methionine into pCMVhAR digested with the same enzymes. GR-FXXLF and GR-FXXAA contain human AR residues 1 to 132 with the wild-type and mutant FXXLF motif expressed as a fusion protein with human GR residues 132 to 777 in pCMV5. They were constructed by cloning the PCR-amplified AR NH2-terminal region into the KpnI and SalI sites of pCMV5hGR. pCMVhGR1-550 was prepared by inserting a KpnI- and BamHI-digested human GR fragment into the same sites of pCMV5. p5mPRB-1-688 was prepared by digestion of p5mPRB with BclI and BglII, religation of the vector, and expansion of the isolated clone in SCS110 cells to avoid methylation that interferes with the BclI restriction site. GAL-AR4-52 and GAL-AR4-52FXXAA were constructed by digestion of pGBT8-AR4-52 with EcoRI and BamHI and ligating the fragment into pGAL0 digested with the same enzymes. GST-AR4-52 and GST-AR4-52FXXAA were constructed by PCR amplification of pGBT8-AR4-52 with primers with EcoRI and XhoI ends and cloning of the fragment into these sites in pGEX-4T-1.
The parent yeast bait vector pGBT8 was kindly provided by Yue Xiong, University of North Carolina at Chapel Hill. Wild-type bait vector pGBT8-AR4-52 and the FXXAA mutant containing the AR NH2-terminal sequences 23FQNLF27 and 23FQNAA27 were constructed by PCR amplification of the region spanning amino acids 4 to 52 of pCMVhAR and pCMVhAR-FXXAA and cloning the fragments into the EcoRI and BamHI sites of pGBT8. pACT2-MAGE-11 was rescued from a positive yeast clone identified by two-hybrid screening of an amplified human testis library. Sequencing revealed a TGA stop codon between the GAL4 activation domain and the MAGE-11 coding regions. Translation read-through can occur in yeast (45) to produce the GAL-AD-MAGE-11 fusion protein. For expression in mammalian assays, MAGE-11 clones were constructed to begin at the first ATG of the coding region for the 429-amino-acid full-length protein or at the second codon for the fusion proteins.
GAL-MAGE-11 containing full-length MAGE-11 residues 2 to 429 and partial form GAL-MAGE-11-112-429 (GAL-MAGE-11-112-429) were constructed by PCR amplification of pACT2-MAGE-11 with primers with EcoRI and XhoI ends. The fragment was cloned into the EcoRI and SalI sites of pGAL0. VP-MAGE-11 containing full-length MAGE-11 residues 2 to 429 and VP-MAGE-11-112-429 (VP-MAGE-11-112-429) were constructed by the same strategy. GAL-MAGE-11 fragments were constructed by PCR amplification of GAL-MAGE-11 and cloning the fragments into pGAL0. MAGE-11 fragments were amplified with EcoRI- and SalI-ended primers (for GAL-MAGE-11-2-252, -2-362, -112-252, -112-362, and -222-362) or EcoRI- and XhoI-ended primers (for GAL-MAGE-11-222-429) and cloned into pGAL0 digested with EcoRI and SalI. For GAL-MAGE-11-332-429, primer ends and cloning sites were EcoRI and SacI. For GAL-MAGE-11-2-121, GAL-MAGE-11 was digested with Bsu36I and ClaI, blunt ended with the Klenow fragment of DNA polymerase I, and self ligated.
pSG5-MAGE-11 encoding full-length MAGE-11 amino acid residues 1 to 429 was constructed by digestion of pACT2-MAGE-11 with EcoRI and BglII. The fragment was cloned into pSG5 digested with the same enzymes. pSG5-MAGE-11-111-429 was constructed by digestion of pSG5-MAGE-11 with EcoRI and PshAI. The vector was blunt ended with the Klenow fragment of DNA polymerase I and self ligated. pCMV-Flag-MAGE-11 encoding full-length MAGE-11 was constructed by digestion of GAL-MAGE-11 with EcoRI and ClaI and ligation of the fragment into pCMV-Flagb (provided by Yi Zhang and Qin Feng, University of North Carolina at Chapel Hill) digested with the same enzymes. DNA sequences were verified for all PCR-amplified regions.
Yeast two-hybrid screen.
A two-hybrid screen of an amplified human testis library was performed with YEASTMAKER Transformation System 2 (Clontech) as previously described (13). Yeast strain HF7c was transformed with pGBT8-AR4-52 and plated on synthetic medium lacking Trp. The yeast clone expressing the pGBT8-AR4-52 bait vector was transformed with 50 μg of amplified human testis MATCHMAKER cDNA library (BD Clontech) and plated on 25 15-cm dishes containing synthetic medium minus Leu, Trp, and His. Over a period of 9 days, colonies were scored according to their time of appearance and were transferred to plates containing synthetic medium lacking Leu, Trp, and His. Clones were retested by a two-hybrid interaction β-galactosidase filter assay (Clontech) by transference to a Whatman no. 5 filter, followed by immersion in liquid N2 and thawing at room temperature for 5 min. Freshly prepared Z buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 (pH 7.0), 50 mM 2-mercaptoethanol and 33 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per ml was added to the filters and incubated at room temperature to score blue colonies after 2 h to overnight. Plasmids from the positive yeast colonies in the β-galactosidase filter assay were rescued. Colonies that grew in synthetic medium lacking Leu gradually lost the bait vector but not the library plasmids. The library vector was purified with a YEASTMAKER yeast plasmid isolation kit (catalogue number K1611-1).
Cell transfections.
Human hepatocellular carcinoma HepG2 cells (American Type Culture Collection) were cultured and transfected with Effectene reagent (QIAGEN) as previously described (13). Typically, transfections were performed in 12-well tissue culture plates containing 2 × 105 HepG2 cells/well, 0.1 μg of 5XGAL4Luc3 reporter vector, and 50 ng of the GAL4 and VP16 expression vectors or 5 ng of pCMVhAR. To determine the effect of MAGE-11 on AR transactivation, monkey kidney CV1 cells (4.2 × 105 cells/6-cm dish) were transfected with calcium phosphate (14). The (prostate-specific antigen) PSA-Enh-Luc (PSA-Luc) reporter, provided by Michael Carey (University of California at Los Angeles), contains the PSA upstream enhancer region (21). Probasin (−244 to −96)3-Luc was provided by Robert Matusik (Vanderbilt University) and contains three copies of the rat probasin promoter region. Luciferase activity was determined 48 h after the addition of hormone by harvesting cells in lysis buffer containing 1% Triton X-100, 2 mM EDTA, and 25 mM Tris phosphate (pH 7.8) (14). Luciferase light units were assayed with a LumiStar Galaxy multiplate reader luminometer (BMG Labtechnologies). Transfection data shown are representative of at least three independent experiments.
Ligand dissociation assays.
Ligand dissociation rate studies were performed with COS cells (4 × 105 cells/well of 6-well culture plates) transfected with DEAE-dextran (19) with 1 μg of pCMVhAR/well with or without 1 μg of pSG5-MAGE-11 or pSG5-MAGE-11-111-429/well. At 48 h after transfection, cells were incubated with 5 or 10 nM 3H-labeled R1881 (82 Ci/mmol; PerkinElmer Life Sciences) for 2.5 h at 37°C, followed by the addition of 50 μM unlabeled R1881. At increasing time intervals at 37°C, cells were washed and harvested in sodium dodecyl sulfate (SDS)-containing buffer, and radioactivity remaining with the cells was determined by scintillation counting. At least three independent experiments were performed to determine the mean and standard deviation of the half-time of dissociation.
GST affinity matrix binding assays.
In vitro protein interaction assays were performed by expressing glutathione S-transferase (GST)-AR4-52 and GST-AR4-52-FXXAA in BL21 Escherichia coli cells treated with 1 mM isopropyl-1-thio-β-d-galactoside as previously described (13, 17). GST-AR fusion proteins were extracted in 0.1 M NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl floride, 1 mM dithiothrietol, and 20 mM Tris-HCl (pH 8.0) and incubated with glutathione agarose beads (Amersham Biosciences). In vitro translation was performed in the presence of 4 μCi of [35S]methionine (Perkin-Elmer Life Sciences) for pcDNA3HA-AR624-919 and 8 μCi of [35S]methionine for pSG5-MAGE-11 with the TNT T7 Quick Coupled transcription-translation system (Promega). Washed beads were extracted with SDS buffer. Input lanes contained 5 or 10% of the binding reaction mixtures as indicated. The data shown are representative of three independent experiments.
Immunochemical methods.
For coimmunoprecipitation and immunoblot experiments, COS cells (2 × 106 cells/10-cm dish) were transfected with DEAE-dextran (14) with the indicated amounts of wild-type and mutant pCMVhAR, pCMV-Flag-AR, pCMV-Flag-MAGE-11, pSG5-MAGE-11, pSG5-TIF2, or GAL-AR peptide vectors. Cells were incubated with and without hormone for 24 h in medium containing 10% charcoal-stripped fetal calf serum (Gemini or HyClone). Cells were harvested in phosphate-buffered saline. For immunoprecipitations, cells were incubated for 30 min at 4°C in, per 10-cm dish, 0.5 to 1 ml of lysis buffer containing 0.15 M NaCl, 0.5% NP-40, 50 mM NaF, and 50 mM Tris-HCl (pH 7.5) plus 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche) in the absence and presence of hormone. For immunoblot analysis, the radioimmunoprecipitation assay extraction buffer contained 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.5 mM EDTA, 50 mM Tris-HCl (pH 7.4), and 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche). After centrifugation for 30 min at 14,000 × g, protein concentration was determined (Bio-Rad). For coimmunoprecipitation, samples were precleared with 100 μl of agarose (Sigma) and incubated for 1 h at 4°C. Supernatants of a 15-min 13,000 rpm centrifugation were incubated with 15 μl of anti-Flag M2 affinity gel (catalogue number A2220; Sigma) for 1 h at 4°C. Agarose pellets were washed four times with 1 ml of lysis buffer in the absence and presence of hormone. Adsorbed proteins were released in 50 μl of 4% SDS, 20% glycerol, 0.2% 2-mercaptoethanol, and 20 mM Tris-HCl (pH 6.8) and analyzed by immunoblotting. For immunoblots of cell extracts, samples (10 μg of protein/lane) were separated on 10% acrylamide gels containing SDS. After electrophoresis, gel proteins were electrophoretically transferred to nitrocellulose membranes, and the blots were incubated with anti-Flag M2 monoclonal antibody (1:2,000 dilution, catalogue number F-3165; Sigma), anti-TIF2 antibody (1:1,000 dilution; Transduction Laboratories), rabbit AR32 immunoglobulin G (IgG) polyclonal antibody (0.42 μg/ml), an anti-MAGE-11 peptide antibody described below (2.9 μg/ml), β-actin antibody AC-15 (1:5,000 dilution; Abcam, Inc.), or anti-GAL4 DNA binding domain antibody (1:500 dilution; Santa Cruz). Immunoreactive bands were visualized by chemiluminescence (SuperSignal Western Dura Extended Duration substrate; Pierce Biotechnology, Inc., Rockford, Ill.).
Immunocytochemistry was performed in COS cells (1.25 × 106 cells/well of 12 plates with cover glass) or the indicated human foreskin fibroblast or prostate cancer cell lines (13). Cells were transfected with Effectene with 0.2 μg of wild-type and mutant pCMVhAR with and without 0.2 μg of pCMVFlag-MAGE-11 and treated for 24 h at 37°C with or without 10 nM dihydrotestosterone (DHT). Prostate cancer cell lines were transfected with 0.2 μg of pCMVFlag-MAGE-11 with Effectene and treated with 10 nM DHT. Cells were fixed with paraformaldehyde, permeabilized with Triton X-100, blocked with bovine serum albumin (13), and incubated for 1 h with primary antibodies anti-Flag M2 monoclonal antibody (1:1,000 dilution; Sigma) and anti-AR polyclonal antibody ab3510 (1:250 dilution; Abcam, Inc.). Cells were incubated with secondary antibodies Rhodamine (tetramethyl rhodamine isocyanate)-conjugated AffiniPure donkey anti-mouse IgG (1:75 dilution; Jackson ImmunoResearch Laboratories, Inc.) and fluorescein isothiocyanate-conjugated AffiniPure donkey anti-rabbit IgG (1:50 dilution; Jackson ImmunoResearch Laboratories, Inc.). Slides were viewed with a Zeiss LSM 210 confocal microscope at an original magnification of ×63.
MAGE-11 NH2-terminal 13 to 26 amino acid residue peptide C13SPASIKRKKKREDS26, where C was added to the NH2 terminus, was predicted to be antigenic (Princeton Biomolecules) and was used to raise a rabbit polyclonal antibody (Pocono Rabbit Farm & Laboratory, Inc., Canadensis, Pa.). Immunoreactivity for MAGE-11 was verified on immunoblots of extracts from COS cells transfected with pSG5-MAGE-11 and pCMV-Flag-MAGE-11. The antibody was purified by antigen affinity chromatography with activated immunoaffinity Affi-Gel 10 gel (Bio-Rad). The peptide antigen was coupled to the column in 0.2 M ethanolamine (pH 8.0). Antiserum was incubated for 2 h at 4°C and the antibody was eluted with 0.1 M glycine (pH 3.0) in 0.1 volume of 1 M Tris-HCl (pH 8.0) and used for immunoblotting at a concentration of 2.9 μg/ml.
RNA amplification.
In vitro reverse transcription was performed according to the manufacturer of SuperScript II reverse transcriptase (Invitrogen Life Technologies) in 20-μl reaction mixtures with 2 μg of total RNA as a template, extracted from cells and tissues with Trizol reagent (Invitrogen Life Technologies). The first PCR amplification of the template was performed with 1 μl of the 20-μl reverse transcription product in reaction buffer containing 1.5 mM MgCl2 and 0.1 μM forward (5′-GGAGACTCAGTTCCGCAGAG-3′) and reverse (5′-TGGGACCACTGTAGTTGTGG-3′) primers that amplify the coding region spanning amino acids 2 to 57 of MAGE-11. PCRs were performed for 5 min at 95°C, followed by 35 cycles (each cycle consisting of 50 s at 95°C, 50 s at 55°C, and 50 s at 72°C), followed by 10 min at 72°C. A second PCR was performed using 2 μl of the initial PCR product as a template with the same primers and the amplification conditions. Due to a genomic intron, the resulting 168-zbp fragment cannot be obtained from genomic DNA.
Nucleotide sequence accession number.
DNA and the protein sequence for full-length MAGE-11 were deposited under accession number AY747607 in GenBank.
RESULTS
Identification of MAGE-11 as an AR NH2-terminal FXXLF motif interacting protein.
We showed previously that the androgen-dependent AR N/C interaction between the AR NH2-terminal 23FQNLF27 sequence and AF2 in the ligand binding domain is required for optimal activation of androgen-responsive promoter-enhancer regions (18). The AR N/C interaction appears to promote selective gene activation through AF1 in the AR NH2-terminal domain. On the other hand, the androgen-induced AR N/C interaction competitively inhibits SRC/p160 coactivator binding to AF2 (14). We therefore proposed that a putative coregulator may exist that could bind the AR NH2-terminal FXXLF motif, relieve inhibition at AF2, and increase AR transactivation by the SRC/p160 coactivators.
We performed a yeast two-hybrid screen of a human testis library using the AR NH2-terminal FXXLF motif region as bait. The strategy was based on our previous observations that screening with short α-helical regions from the intrinsically unstructured AR NH2-terminal transcriptional activation domain is a useful approach to identify interacting proteins (13). Approximately 5 × 105 yeast library clones were screened with pGBT8-AR4-52 as bait, which contains the AR NH2-terminal FXXLF motif sequence. We identified 119 positive clones that grew on synthetic medium lacking Leu, Trp, and His. Two of the clones were positive by β-galactosidase filter assays. One clone encoded RanBPM, a previously reported AR-interacting protein (36). However, interaction between RanBPM and AR was not diminished by mutation of the FXXLF motif, indicating that binding to AR was not FXXLF motif specific (data not shown). The second positive clone encoded the melanoma antigen gene family protein, MAGE-11. Binding of MAGE-11 to AR4-52 was abolished by an AR4-52-FXXAA mutant in a yeast two-hybrid assay (data not shown), suggesting a specific interaction between MAGE-11 and the AR FXXLF motif.
A BLAST search of the human genome database confirmed that the isolated clone was coded from the Xq28 gene locus of the MAGE gene family (23, 39) (Fig. 1A). The five exons predict a 1,837-nucleotide MAGE-11 mRNA. Four of the exons encode the 429-amino-acid full-length MAGE-11 protein. MAGE-11 has a calculated molecular mass of 48 kDa but, as we will show, an apparent molecular mass of 70 kDa on SDS gels. The cloning results confirm a previous report of the additional upstream coding regions for the 110 NH2-terminal amino acids unique to MAGE-11 (23).
FIG. 1.
Human MAGE-11 gene, mRNA, and protein, and AR NH2-terminal fragments. (A) The MAGE-11 gene is carried on the long arm of the X chromosome at Xq28 and includes five exons that transcribe into a 1,837-nucleotide residue mRNA. The four-exon open reading frame (shown in black) translates to a 429-amino-acid-residue MAGE-11 protein (shown in white). The AR interaction region is based on data shown in Fig. 9. (B) AR NH2-terminal fragments of full-length human AR1-919 were expressed as GAL and VP16 fusion proteins in two-hybrid assays. Each fragment contains the region spanning amino acids 16 to 36 with the FXXLF interaction motif sequence 23FQNLF27 (underlined).
Specific interaction between MAGE-11 and the AR NH2-terminal FXXLF motif.
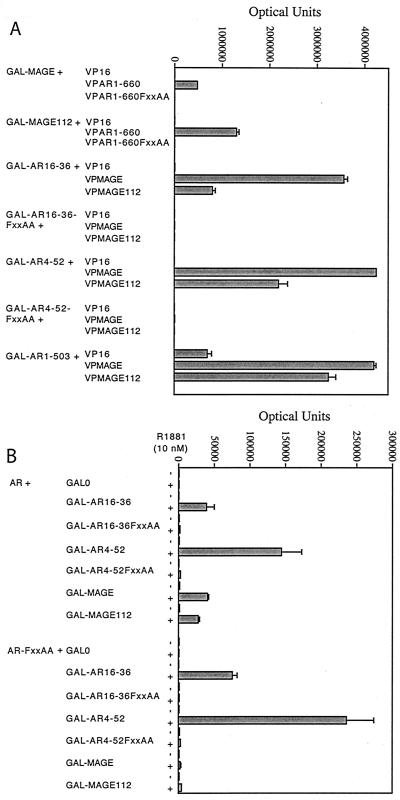
We made use of mammalian two-hybrid assays to demonstrate a dependence of MAGE-11 binding to AR on the AR FXXLF motif. The GAL DNA binding domain and VP16 transactivation domain fusion proteins were expressed that contained full-length MAGE-11 or a previously reported short form (24, 39) that corresponds to MAGE-11-112-429. GAL and VP fusion proteins were created to contain the 23FQNLF27 amphipathic α-helical region within increasing lengths of the AR NH2-terminal region (Fig. 1B). A GAL-responsive luciferase reporter vector was cotransfected into HepG2 cells, and assays were performed in the absence and presence of 10 nM R1881, a synthetic androgen.
In the absence of androgen, GAL-MAGE-11 and GAL-MAGE-11-112-429 interacted with VP-AR1-660 (Fig. 2A). The interaction depended on the AR FXXLF motif, since no interaction was observed with VP-AR1-660-FXXAA containing the 23FQNAA27 mutation. We also observed strong interaction between VP-MAGE-11 or VP-MAGE-11-112-429 with GAL-AR16-36, GAL-AR4-52, and GAL-AR1-503.
FIG. 2.
FXXLF motif dependence of the AR and MAGE-11 interaction. Two-hybrid interaction assays were performed with HepG2 cells transfected with Effectene with 0.1 μg of 5XGAL4Luc3/well in 12-well plates with (A) 50-ng/well GAL-MAGE-11, GAL-MAGE-11-112-429 (GAL-MAGE112), or the indicated GAL-AR fusion proteins, with the VP16 empty vector control, wild-type or mutant VP-AR1-660, VP-MAGE-11, or VP-MAGE-11-112-429 (VP-MAGE112), and (B) 5 ng of pCMVhAR or pCMVhAR-FXXAA/well with 50 ng of the indicated GAL vectors/well. Cells were incubated for 24 h in the absence (A) or absence and presence (B) of 10 nM R1881. The FXXAA mutation is AR sequence 23FQNLF27 changed to 23FQNAA27.
In the presence of androgen, we observed an FXXLF motif-dependent interaction between full-length AR and the same GAL-AR FXXLF motif peptides and GAL-MAGE-11 (Fig. 2B). The interaction of GAL-AR16-36 or GAL-AR4-52 with AR in the presence of androgen (Fig. 2B) reflects the androgen-dependent N/C interaction between the AR NH2-terminal FXXLF motif in the GAL-AR fragment and AF2 in the ligand binding domain of full-length AR (17). Note that in this experimental design, luciferase activity in the two-hybrid assay requires androgen for the nuclear transport of full-length AR (20). This is in contrast to the androgen-independent interactions shown in Fig. 2A that made use of GAL4 and VP16 fusion proteins of AR NH2-terminal fragments that lack the ligand binding domain and are in the nucleus independent of androgen (52). Expression of the fusion proteins was confirmed by immunoblot analysis (Fig. 3B and data not shown).
FIG. 3.
Differences in flanking sequence requirements for FXXLF motif binding to AR and MAGE-11. (A) Two-hybrid interaction assays were performed with HepG2 cells as described in the legend to Fig. 2, except that 10 ng of pCMVhAR (AR1-919)/well (left) or 50 ng of VP-MAGE-11/well (right) with 50 ng of the indicated GAL-AR vectors/well was used. GAL-AR16-36FXXAA indicates the 23FQNAA27 mutation. Each of the indicated amino acid changes was introduced into the GAL-DNA binding domain fusion vector GAL-AR16-36. (B) Immunoblots of GAL-AR peptides were performed using extracts from COS cells (2 × 106/10-cm dish) transfected with 10 μg of GAL-AR peptide DNA with DEAE-dextran. Cells were treated with 1 μM MG132 (Sigma), a proteasome inhibitor, 24 and 1 h prior to harvest. The GAL4 DNA binding domain antibody (1:500 dilution; Santa Cruz) was used to probe 25 μg of total protein in each lane. (C) Diagram of the predicted amphipathic α-helical AR NH2-terminal FXXLF motif regions required to interact with AF2 in the AR ligand binding domain and MAGE-11. The predicted ideal α-helix may be disrupted to the left at G21. Hydrophobic residues F23, L26, and F27 contact the hydrophobic surface at AR AF2 in the N/C interaction (15). These residues plus V30, V33, and I34 in the extended helix are predicted to contact the binding surface of MAGE-11.
The results demonstrate that the same AR NH2-terminal FXXLF motif region that mediates the androgen-dependent AR N/C interaction also interacted with MAGE-11. The data confirm the yeast two-hybrid results that MAGE-11 binding to AR depended on the AR FXXLF motif contained within a 21-amino-acid residue region of the AR NH2-terminal domain.
Sequence requirements for FXXLF motif binding to MAGE-11.
The flanking sequence requirements for the AR FXXLF motif to bind MAGE-11 and AF2 in AR were found to have similarities but also striking differences. Wild-type and mutant GAL-AR peptides were coexpressed in two-hybrid assays in HepG2 cells with full-length AR (Fig. 3A, left) and with VP-MAGE-11 (Fig. 3A, right) in the presence of a GAL-responsive luciferase reporter. In agreement with the data in Fig. 2A, interaction of GAL-AR16-36 with AR or VP-MAGE-11 was eliminated by the FXXAA mutation in GAL-AR16-36.
We tested several shorter fragments of the FXXLF motif region, since we showed previously that AR20-30 binds AF2 in AR more effectively than AR16-36. At that time, we speculated that R31 flanking FXXLF, which is deleted in AR20-30, undergoes electrostatic repulsion at the positive-charge cluster-containing charge clamp residue K720 (20). We found that MAGE-11 did not bind GAL-AR20-30, GAL-AR20-36, or GAL-AR16-30, even though GAL-AR20-30 and GAL-AR16-30 (Fig. 3A, top) interacted to a greater extent with AR than does GAL-AR16-36, as previously reported (20).
We also noted a number of differences in the FXXLF motif-flanking sequence in GAL-AR16-36 required to bind AR and MAGE-11 (Fig. 3A, bottom). Mutations S29D, V30E, R31A, R31D, V33E, and I34A in GAL-AR16-36 each increased binding to AR AF2, whereas these mutations eliminated binding to MAGE-11. T18A and E32A increased binding of GAL-AR16-36 to both MAGE-11 and AR. S29A increased MAGE-11 binding but decreased AR binding; S16D, K17A, T18D, and V30A each increased MAGE-11 binding, with little effect on AR binding. S16A increased binding to AR but not MAGE-11, and R20A and R20D were detrimental to both interactions. Expression levels of the GAL-AR fusion proteins varied somewhat, but the differences did not correlate with the observed interactions (Fig. 3).
The data show that the flanking sequence of the AR FXXLF motif has different requirements for binding MAGE-11 and the AR AF2 site. Optimal binding to MAGE-11 required the extended 21-amino-acid FXXLF α-helical region, which is nearly twice the length of the minimal 11-amino-acid FXXLF region that optimally bound AF2 in the AR N/C interaction (Fig. 3C). The data also suggest that the MAGE-11 binding site differs substantially from AF2 in the AR ligand binding domain (15, 22).
Specificity of MAGE-11 binding to the AR FXXLF motif.
We found that MAGE-11 binding to the AR NH2-terminal FXXLF motif is highly specific. FXXLF motif sequences previously reported in several putative AR coregulators and LXXLL motif regions from the SRC/p160 coactivators did not interact with MAGE-11 in two-hybrid assays performed with HepG2 cells. Included in the comparison were ARA70/RFG (49, 50), ARA54 (25), and ARA55/Hic5 (7, 48), whose FXXLF motifs are required for the androgen-dependent interaction between these coregulators and the AR AF2 site (19). No interaction was found between VP-MAGE-11 and GAL-FXXLF fusion proteins containing ARA70-321-340, ARA70-321-499, ARA54-447-465, ARA54-361-474, ARA55-314-333, ARA55-427-444, or ARA55-251-444 (data not shown). Little or no interaction was detected between GAL-MAGE-11 and the LXXLL motif regions from the SRC/p160 coactivator family. These included VP-LXXLL motif fusion proteins containing SRC1-1139-1441, SRC1-568-1441, TIF2-624-1287, TRAM1-604-1297, and p300-1-133 (data not shown).
The results indicate a specific interaction between MAGE-11 and the AR FXXLF motif. The lack of an interaction between MAGE-11 and other FXXLF motifs from several potentially relevant AR coregulators supports our findings that residues within the extended α-helical region flanking FXXLF establish specificity for MAGE-11binding.
In vitro interaction between AR and MAGE-11.
A direct interaction between AR and MAGE-11 was supported by in vitro GST affinity matrix binding assays. An interaction between GST-AR4-52 and 35S-labeled MAGE-11 was observed in the absence of androgen, which was eliminated with GST-AR4-52FXXAA (Fig. 4). Similarly, an FXXLF motif-dependent interaction was observed in the presence of DHT between GST-AR4-52 and 35S-labeled AR624-919 containing the AR ligand binding domain. 35S-labeled MAGE-11 migrates a 70 kDa, and 35S-labeled AR ligand binding domain migrates a 41 kDa.
FIG. 4.
In vitro binding of AR and MAGE-11. GST affinity matrix assays were performed with GST-AR NH2-terminal fusion peptides and 35S-labeled MAGE-11 expressed from pSG5-MAGE-11 (left) and 35S-labeled AR624-919 expressed from pcDNA3HA-AR624-919 containing the AR ligand binding domain (right). GST empty vector pGEX-4T-1 (GST-AR-0), GST-AR4-52, and GST-AR4-52FXXAA were expressed in E. coli and incubated in the presence of 35S-labeled MAGE-11 (lanes 2 to 4), and 35S-labeled AR624-919 in the presence of 0.2 μM DHT (lanes 6 to 8). Input lanes contained 5% of the total reaction mixture for MAGE-11 (lane 1) and 10% of the reaction mixture for AR624-919 (lane 5). 35S-labeled MAGE-11 (70 kDa) and AR ligand binding domain fragment 624-919 (41 kDa) are indicated by arrows.
Coimmunoprecipitation of AR, MAGE-11 and TIF2.
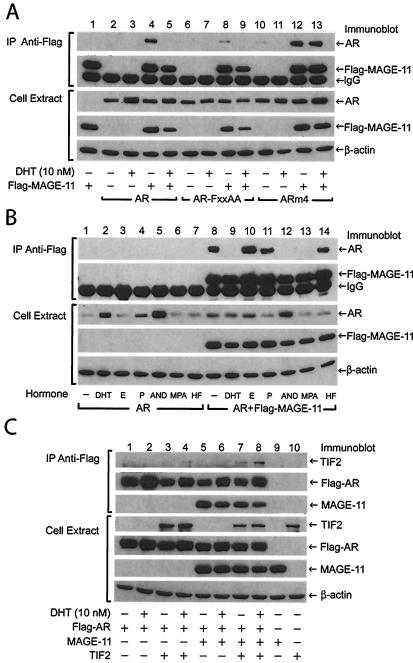
We found that coimmunoprecipitation of AR and MAGE-11 is selectively modulated by ligand binding and AR subcellular localization (Fig. 5A and B, top). AR transiently expressed in COS cells coimmunoprecipitates with Flag-MAGE-11 with a Flag antibody when assayed in the absence of DHT (Fig. 5A, lane 4). However, the interaction between AR and MAGE-11 was barely detectable in the presence of 10 nM DHT (Fig. 5A, lane 5, and Fig. 5B, lane 9). In the absence of DHT, the interaction between AR and MAGE-11 was greatly diminished by the AR-FXXAA mutant (Fig. 5A, lane 8). AR was coimmunoprecipitated with the Flag antibody only when Flag-MAGE-11 was expressed, suggesting the absence of nonspecific binding (Fig. 5A, top, lanes 2, 3, 6, and 7).
FIG. 5.
Coimmunoprecipitation of AR, MAGE-11, and TIF2. (A and B) COS cells were transfected with 2 μg of pCMVhAR, pCMVhAR-FXXAA, and pCMVhARm4/10-cm dish in the absence and presence of 5 μg of pCMV-Flag-MAGE-11 with DEAE-dextran. Cells were incubated for 24 h in media containing 10% charcoal-stripped serum (Gemini) in the absence and presence of 10 nM DHT, 50 nM estradiol (E), 50 nM progesterone (P), 50 nM androstenedione (AND), 50 nM MPA, or 1 μM hydroxyflutamide (HF) as indicated. Cells were extracted in 0.5% NP-40 lysis buffer and immunoprecipitated with anti-Flag antibody as described in Materials and Methods. Immunoprecipitated proteins (A and B, top two panels) and a portion of the whole cell protein lysates (A and B, lower three panels) (10 μg/lane) were separated by electrophoresis on 10% acrylamide gels containing SDS. Immunoblots of immunoprecipitated proteins and cell extracts (10 μg) were exposed to X-ray film for 10 s. (C) COS cells were transfected with 2 μg of pCMV-Flag-hAR, 4 μg of pSG5-TIF2, and 2 μg of pSG5-MAGE-11; treated with and without 10 nM DHT; and extracted as above. Anti-Flag antibody immunoprecipitates of Flag-AR (top three panels) and whole cell extracts (bottom four panels; 20 μg protein/lane) were separated by electrophoresis on 10% acrylamide gels containing SDS. Immunoblots of the immunoprecipitates were exposed to X-ray film for 2 s for Flag-AR and MAGE-11 protein and 4 h for TIF2, and the cell lysates were exposed for 5 s for TIF2, 2 s for Flag-AR, and 1 s for MAGE-11 and β-actin. Immunoblots were probed as indicated with AR32 rabbit polyclonal antibody for AR (A and B) and Flag-AR (C), anti-Flag mouse monoclonal antibody for Flag-MAGE-11 (A and B), anti-TIF2 antibody (BD Bioscience) (C), peptide affinity-purified rabbit polyclonal anti-MAGE-11 antibody for MAGE-11 (C), and anti-β-actin antibody (A to C). The immunoblots are representative of more than three independent experiments.
We tested a previously described AR nuclear transport mutant, ARm4, that contains mutations in the AR bipartite nuclear transport signal. ARm4 retains high-affinity androgen binding but does not translocate to the nucleus in the presence of androgen (52). In contrast to wild-type AR which is nuclear in the presence of DHT, this constitutively cytoplasmic mutant coimmunoprecipitated with MAGE-11 in the absence and presence of DHT (Fig. 5A, lanes 12 and 13).
We also found that the stable association between AR and MAGE-11 persists in the presence of ligands that promote AR nuclear localization but are AR antagonists or poor agonists (26). These included 50 nM estradiol, 50 nM progesterone, and 1 μM hydroxyflutamide (Fig. 5B, lanes 10, 11, and 14). However, like DHT (10 nM), the stable association between AR and MAGE-11 was reduced in the presence of 50 nM androstenedione and 50 nM medroxyprogesterone acetate (MPA) (Fig. 5B, lanes 12 and 13). Coimmunoprecipitation of AR with Flag-MAGE-11 was indicative of a specific interaction, since AR was not detected when MAGE-11 was not expressed (Fig. 5B, lanes 1 to 7).
The coimmunoprecipitation results suggest that MAGE-11 stably interacts in the cytoplasm with the ligand-free AR and in the nucleus when AR is bound to an antagonist or poor agonist. In the presence of stronger agonists, the AR and MAGE-11 interaction becomes more transient. The effectiveness of androstenedione and MPA in disrupting the interaction with MAGE-11 was attributed in part to the high ligand concentrations. The persistent interaction of MAGE-11 with the constitutively cytoplasmic AR raised the possibility that both agonist binding and AR nuclear transport are required to destabilize the AR and MAGE-11 interaction.
To investigate whether MAGE-11 influences the interaction between AR and the p160 coactivator TIF2 and whether MAGE-11, AR, and TIF2 form an intracellular complex, we performed coimmunoprecipitation studies using Flag-AR (Fig. 5C). The 70-kDa MAGE-11 protein was detected with an antibody raised against the MAGE-11 NH2-terminal peptide C13SPASIKRKKKREDS26, a sequence specific to MAGE-11. We noted an increase in TIF2 levels in cell extracts with the coexpression of AR in the presence of 10 nM DHT. In the Flag antibody immunoprecipitates, an association between AR and TIF2 was evident in the presence of DHT (Fig. 5C, lanes 3 and 4). We found that the AR and TIF2 interaction increased with coexpression of MAGE-11 in the absence and presence of androgen (Fig. 5C, lanes 7 and 8). Neither MAGE-11 nor TIF2 was detected in the immunoprecipitates in the absence of coexpressed Flag-AR (Fig. 5C, lanes 9 and 10). The results suggest that binding of MAGE-11 to AR increases the interaction between AR and TIF2.
MAGE-11 influences AR steady-state levels.
Immunoblots of cell lysates from the immunoprecipitation studies shown in Fig. 5 show the expected androgen-induced increase in AR in the presence of 10 nM DHT. AR stabilization by DHT results from the AR N/C interaction (14), as evidenced by the lack of an increase with AR-FXXAA (Fig. 5A, lanes 6 and 7). We found that coexpression of MAGE-11 interferes with DHT-induced AR stabilization, since AR levels declined (Fig. 5B, lanes 8 and 9). MAGE-11 levels also decreased in the presence of 10 nM DHT, in association with the reduced interaction between AR and MAGE-11 (Fig. 5A, lanes 4 and 5). However, no decrease in MAGE-11 was observed in the presence of DHT and the nuclear transport mutant, ARm4 (Fig. 5A, lanes 12 and 13). A decrease in MAGE-11 was also observed with AR-FXXAA, suggesting an effect of MAGE-11 independent of the AR FXXLF motif.
The effect of MAGE-11 on steady-state levels of AR transiently expressed in COS cells was further investigated at increasing concentrations of androgen. DHT concentrations of 2 and 10 nM are below and near saturating levels, respectively, based on 3H-labeled R1881 binding and AR nuclear transport studies in COS cells transiently expressing AR, and 50 nM DHT is saturating (references 17 and 52 and unpublished results). Coexpression of MAGE-11 in the presence of 2 and 10 nM DHT reduced AR levels in association with decreased levels of MAGE-11 (Fig. 6A, lanes 1 to 7). The level of AR-FXXAA also decreased with MAGE-11 expression but to a lesser extent. This result again raised the possibility that MAGE-11 has effects on AR that are FXXLF motif independent. In contrast, addition of 50 nM DHT apparently overcame the destabilizing effect of MAGE-11 on AR but not AR-FXXAA, and androgen-induced AR stabilization was restored (Fig. 6A, lanes 8 and 16). The AR-FXXAA mutant lacks an N/C interaction and was not stabilized by androgen (14). In [35S]methionine-labeled AR pulse-chase experiments performed with COS cells in the presence of 10 nM DHT, the destabilizing effect of MAGE-11 on AR degradation could not be detected over the stabilizing influence of DHT on AR (t1/2 = 4.7 ± 0.5 h) and the AR-FXXAA mutant (t1/2 = 3.3 ± 0.4 h).
FIG. 6.
Effect of MAGE-11 on AR steady-state levels. (A) Immunoblots are shown of whole-cell protein lysates of COS cells transfected with 2 μg of pCMVhAR or pCMVhAR-FXXAA without and with 5 μg of pCMV-Flag-MAGE-11 in the absence and presence of 2, 10, and 50 nM DHT. Whole-cell extracts (10 μg) were analyzed with the antibodies indicated on the right. The blot was exposed to X-ray film for 50 s. (B) Immunoblots of pCMVhAR1-660 and pCMVhAR1-660-FXXAA (2 μg) expressed in COS cells (2 × 106 cells/10-cm dish) using DEAE-dextran in the absence and presence of 5 μg of pCMV-Flag-MAGE-11 are shown. Each lane contains 15 μg of total protein, and the blots were exposed to X-ray film for 3 s.
Paradoxically, we found that in the absence of androgen, MAGE-11 increases AR levels (Fig. 6A, lanes 1 and 5). Pulse-chase studies with [35S]methionine-labeled AR in COS cells in the absence of DHT confirmed the half-time of AR degradation (t1/2 = 1.3 ± 0.1 h) increased twofold with MAGE-11 expression (t1/2 = 2.4 ± 0.3 h). Although we observed some increase in AR-FXXAA in the absence of androgen compared to wild-type AR as previously reported (14), the level of AR-FXXAA did not increase with MAGE-11 expression (Fig. 6A, lanes 9 and 13). In agreement with these results, the half-time of AR-FXXAA degradation (t1/2 = 1.5 ± 0.1 h) in the absence of androgen was less affected by MAGE-11 (t1/2 = 1.8 ± 0.1 h). MAGE-11 also increased the level of AR1-660, a constitutively active AR NH2-terminal fragment that lacks the ligand binding domain, but not the level of AR1-660-FXXAA (Fig. 6B).
The results support a stable and specific interaction between MAGE-11 and AR in the absence of androgen that increases AR steady-state levels. The stabilizing effect of MAGE-11 in the absence of DHT is mediated at least in part through its interaction at the AR FXXLF site. In the presence of androgen, the interaction between AR and MAGE-11 is more transient and AR steady-state levels decline. The decrease in AR-FXXAA levels with MAGE-11 expression in the presence of androgen suggests that FXXLF motif-independent mechanisms contribute to the effects of MAGE-11 on AR.
AR and MAGE-11 colocalize in the cytoplasm and nucleus.
Immunocytochemical staining of AR and MAGE-11 was determined to further establish their intracellular association. Immunostaining of transiently expressed AR in COS cells resulted in a typical cytoplasmic staining pattern in the absence of androgen and nuclear staining in the presence of DHT (Fig. 7A and B) (52). Flag-tagged MAGE-11 localized predominantly in nuclei with some cytoplasmic staining (Fig. 7C). We found that Flag-MAGE-11 and AR colocalize in the cytoplasm in the absence of androgen with persistent nuclear staining of MAGE-11 (Fig. 7D to F). With the addition of androgen, AR and MAGE-11 colocalized to the nucleus (Fig. 7G to I). Cytoplasmic colocalization of MAGE-11 and AR was also seen in the presence of 10 or 50 nM estradiol, and we observed both cytoplasmic and nuclear AR with 10 and 50 progesterone or 50 nM and 1 μM hydroxyflutamide. AR was nuclear with 10 and 50 nM androstenedione or MPA (data not shown).
FIG.7.
Immunocytochemistry of AR and MAGE-11. Cells were transiently transfected with pCMV-Flag-MAGE-11 or pCMVhAR alone or together with Effectene. Transiently expressed AR (A, B, and D to I) and endogenous AR (J to O) are represented by green fluorescence and were detected with rabbit anti-AR polyclonal antibody ab3510 (Abcam, Inc.). Transiently expressed Flag-MAGE-11 (C to O), represented by red fluorescence, was detected with anti-Flag M2 mouse monoclonal antibody. Yellow is indicative of AR and MAGE-11 colocalization. Representative illustrations are shown for pCMVhAR in COS cells without DHT (A), pCMVhAR in COS cells without DHT (B), pCMV-Flag-MAGE-11 in COS cells without DHT (C), pCMVhAR and pCMV-Flag-MAGE-11 in COS cells without DHT (D to F), and pCMVhAR and pCMV-Flag-MAGE-11 in COS cells with 50 nM DHT (G to I). pCMV-Flag-MAGE-11 was transfected alone into human foreskin fibroblasts (J to L), and human prostate cancer cell lines LNCaP (M), CWR22-RV1 (N), and LAPC4 (O), each incubated with 10 nM DHT. Original magnification, ×63.
Transient expression of Flag-MAGE-11 in human foreskin fibroblasts showed colocalization of MAGE-11 with endogenous AR in nuclei in the presence of androgen (Fig. 7J to L). Similarly, MAGE-11 colocalized with endogenous AR in human prostate cancer cell lines, LNCaP (Fig. 7M), CWR22-RV1 (Fig. 7N), and LAPC4 (Fig. 7O) assayed in the presence of androgen. Persistent cytoplasmic MAGE-11 was also evident in both fibroblasts and prostate cancer cells.
In contrast, the addition of androgen changed the immunostaining pattern of Flag-MAGE-11 and the nuclear transport mutant ARm4 from typical cytoplasmic staining (Fig. 8A to C) to colocalization in prominent cytoplasmic spots (Fig. 8D to F). For DNA binding mutant AR-C576A, addition of DHT changed cytoplasmic staining (Fig. 8G to I) to striking colocalization in subnuclear bodies (Fig. 8J to L). The results support an intracellular interaction between AR and MAGE-11 in the cytoplasm in the absence of androgen and in the nucleus in the presence of androgen.
FIG. 8.
Immunocytochemistry of AR mutants and MAGE-11. COS cells were transiently transfected with pCMV-Flag-MAGE-11 together with nuclear transport mutant pCMVhAR-R617M-K618M-K632M-R633M (ARm4) in the absence (A to C) and presence (D to F) of 50 nM DHT or with DNA binding domain mutant pCMVhAR-C576A in the absence (G to I) or presence (J to L) of 50 nM DHT. AR (green fluorescence), Flag-MAGE-11 (red fluorescence), and the merge (yellow fluorescence) are shown. Original magnification, ×63.
Inherent transcriptional activity and FXXLF-interacting domain of MAGE-11.
Expression of GAL-MAGE-11 fusion proteins in mammalian two-hybrid assays demonstrated that MAGE-11 lacks a major intrinsic transactivation domain. Of the fusion proteins created to span the MAGE-11 sequence, only GAL-MAGE-11-2-252 had a low level of transcriptional activity (Fig. 9, top).
FIG. 9.
Inherent transcriptional activity of MAGE-11 and the interaction domain for the AR FXXLF motif. Two-hybrid interaction assays were performed in HepG2 cells by transient transfection in 12-well plates with Effectene with 0.1 μg of 5XGAL4Luc3 reporter and 50 ng of the indicated GAL-MAGE-11 fusion protein vectors with either VP16 activation domain empty vector control (50 ng/well) (VP16-AD, top) or VP-AR1-660 (50 ng/well) (bottom). Luciferase activity and error were determined in the absence of hormone.
We also found that like AR, MAGE-11 binds the AR NH2-terminal FXXLF motif through its carboxyl-terminal region (Fig. 9, bottom). GAL-MAGE-11-2-429 and GAL-MAGE-11-112-429 interacted to a similar extent with VP-AR1-660, which contains the AR FXXLF motif. Both of these larger MAGE-11 fragments interacted to a greater extent than GAL-MAGE-11-222-429. Shorter carboxyl-terminal fragments of MAGE-11 did not interact with AR. We also found that a MAGE-11 LXXLL motif sequence 229LVHLL233 interacted only weakly with AR (data not shown). Thus, the carboxyl-terminal 112 to 429 residues of MAGE-11 represent the AR-interacting domain (Fig. 1A).
Competition for the N/C interaction.
We performed two-hybrid assays and ligand dissociation rate studies to demonstrate that binding of MAGE-11 to the AR NH2-terminal FXXLF motif competes for the androgen-induced N/C interaction. Using the assay that first demonstrated the AR N/C interaction (30), we found that coexpression of MAGE-11 inhibited the interaction between VP-AR1-660 and GAL-AR624-919 in the presence of androgen (Fig. 10A).
FIG. 10.
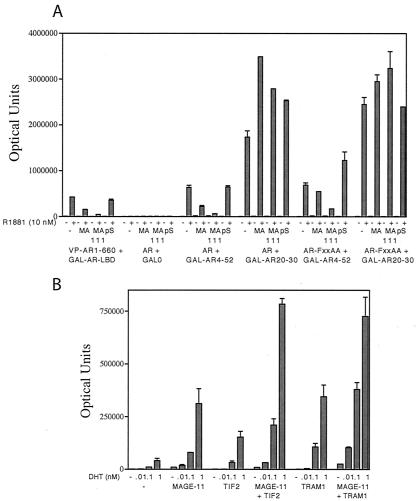
Competition for the AR N/C interaction by MAGE-11 and a synergistic increase in AR transactivation. (A) Two-hybrid interaction assays were performed with HepG2 cells in 12-well plates with VP-AR1-660 and GAL-AR624-919 (GAL-AR-LBD) (50 ng/well) or 5 ng of pCMVhAR and pCMVhAR-FXXAA expressed together with 50 ng of pGAL0 empty vector control (GAL0), GAL-AR4-52, or GAL-AR20-30, without further vector addition (−), or with 50 ng pSG5-MAGE-11 (MA), pSG5-MAGE-11-111-429 (MA 111), or the pSG5 empty vector (pS). Assays were performed in the absence and presence of 10 nM R1881. (B) A total of 100 ng of pCMVhAR was expressed in CV1 cells in 6-cm dishes with calcium phosphate precipitation of DNA in the absence and presence of 2 μg of pSG5-MAGE-11, pSG5-TIF2, or pSG5-TRAM1 or in combination as indicated, together with 5 μg of PSA-Luc. Transfected cells were incubated for 40 h in the absence and presence of 0.01, 0.1, and 1 nM DHT as indicated. Parallel transfections of pCMVhAR and the pSG5 empty vector control showed no increase in activity (data not shown).
In a recent study, we showed that GAL-AR4-52 and GAL-AR20-30 compete for the AR N/C interaction by binding AF2 in AR (20). In this assay, the N/C interaction of full-length AR itself is not directly measured. Luciferase activity results from the interaction between the GAL-FXXLF peptide and AR that contains the strong AF1 NH2-terminal transactivation domain. MAGE-11, on the other hand, lacks a major activation domain (Fig. 9). We found that coexpression of MAGE-11 inhibited the interaction between AR and GAL-AR4-52. We observed similar but slightly less inhibition of AR-FXXAA by MAGE-11. The decrease in luciferase activity appeared to be attributable largely to MAGE-11 binding to GAL-AR4-52. Increased exposure of AF2 resulting from the AR-FXXAA mutation would be expected to increase binding of GAL-AR4-52 to AF2 (14), which could account for the reduced inhibition of AR-FXXAA by MAGE-11.
To investigate this further, we tested GAL-AR20-30, since this short FXXLF motif peptide strongly engages the AR AF2 site indicative of the N/C interaction but does not bind MAGE-11 (Fig. 3). In this assay, the effect of MAGE-11 was mediated only by binding the NH2-terminal FXXLF motif of full-length AR. We found that MAGE-11 increased the interaction between GAL-AR20-30 and AR (Fig. 10A). When AR-FXXAA was tested, binding of GAL-AR20-30 increased and was less dependent on MAGE-11. The results suggest that binding of MAGE-11 to the AR NH2-terminal FXXLF motif competes for the AR N/C interaction and exposes AF2 to increased binding, in this case, of GAL-AR20-30.
Competition by MAGE-11 for the agonist-induced AR N/C interaction was also evident from the dissociation rate of bound androgen. These studies were based on previous observations that the N/C interaction slows the dissociation rate of bound androgen (14, 17, 51). COS cells expressing AR in the absence and presence of MAGE-11 were incubated with 5 nM [3H]R1881. The half-time (t1/2) of [3H]R1881 dissociation from AR (t1/2 = 109 ± 11 min) decreased with coexpression of MAGE-11 (t1/2 = 71 ± 6 min) or MAGE-11-111-429 (t1/2 = 71 ± 7 min). In contrast, dissociation of [3H]R1881 from AR-FXXAA (t1/2 = 40 ± 5 min) was unaffected by coexpression of MAGE-11 (t1/2 = 40 ± 4 min) or MAGE-11-111-429 (t1/2 = 44 ± 3 min).
The data provide evidence that MAGE-11 binding to the AR NH2-terminal FXXLF motif competes for the AR N/C interaction and exposes the AF2 site in the ligand binding domain. Increased dissociation of androgen from the ligand binding pocket is indicative of this competitive interaction (14, 17, 51).
Effect of MAGE-11 on AR functional activity.
To further address the possibility that binding of MAGE-11 to the AR NH2-terminal FXXLF increases the accessibility of AF2, we determined the effect of MAGE-11 on AR transactivation in the absence and presence of increased SRC/p160 coactivator expression. We found that MAGE-11 increased AR transactivation of a PSA-Luc reporter to an extent similar to that seen with the coexpression of TIF2 and TRAM1 (Fig. 10B). Furthermore, the effects of MAGE-11 and the SRC/p160 coactivators on AR transactivation appeared to be synergistic. We also observed a small, biphasic increase in ligand-independent AR transactivation with increasing MAGE-11 expression that contributed to a persistent effect of MAGE-11 on the transcriptional activity of AR-FXXAA (Fig. 11A). Transactivation by AR-FXXAA in the absence of MAGE-11 was almost undetectable (Fig. 11A) due to the dependence of the PSA-Luc reporter activation on the AR N/C interaction, a dependence that is overcome by increased expression of the SRC/p160 coactivators (18). The small but noticeable increase in AR transactivation with MAGE-11 expression in the absence of androgen raised the possibility that MAGE-11 can influence transcriptional activity of the ligand free AR. The dose dependent increase in AR-FXXAA activity with MAGE-11 expression suggested that the transcriptional effects of MAGE-11 are not entirely FXXLF motif dependent.
FIG. 11.
FXXLF motif-dependent effects of MAGE-11 on receptor mediated transactivation. (A) The effect of MAGE-11 on AR transactivation was tested by transfecting 100 ng of pCMVhAR or pCMVhAR-FXXAA into CV1 cells with 5 μg of PSA-Luc reporter in the absence and presence of 0.2, 0.5, and 2 μg of pSG5-MAGE-11. Luciferase activity was determined in the absence and presence of 1 nM DHT. (B) The effect of MAGE-11 on GR transactivation was tested by transfecting 100 ng of pCMVhGR, pCMVhAR1-132-GR132-777 chimera (GR-FXXLF), and pCMVhAR1-132-FXXAA-GR132-777 (GR-FXXAA) into CV1 cells with 5 μg of PSA-Luc reporter in the absence and presence of 0.2, 0.5, and 2 μg of pSG5-MAGE-11, as indicated. Luciferase activity was determined in the absence and presence of 10 nM dexamethasone (DEX). (C) The effect of MAGE-11 on AR transactivation of the MMTV and probasin enhancer-promoters was tested in CV1 cells transfected with 25 ng of pCMVhAR and 5 μg of MMTV-Luc or with 10 ng of pCMVhAR and 5 μg of probasin (−244 to −96)3-Luc in the absence and presence of 0.1, 0.5, and 2 μg of pSG5-MAGE-11. (D) The FXXLF motif dependence of the MAGE-11-induced increase in constitutive activity of NH2-terminal and DNA binding domain fragments of AR, GR, and PR was tested with CV1 cells transfected with 50 ng of pCMVhAR1-660, pCMVhAR1-660FXXAA, pCMVhGR1-550, p5mPRB-1-688, or pCMV5 empty vector (p5)/6-cm dish with 5 μg of PSA-Luc in the absence and presence of 0.2 and 0.5 μg of pSG5-MAGE-11.
To explore this further, the specificity of the stimulatory effects of MAGE-11 was tested with GR and AR-GR chimeras and the PSA-Luc reporter (Fig. 11B). The transcriptional activity of wild-type GR was only moderately increased with the coexpression of MAGE-11. In contrast, the AR-GR chimera AR1-132-GR132-777 (GR-FXXLF), which contains the AR NH2-terminal FXXLF motif region, showed a striking increase in activity with the coexpression of MAGE-11. The increase in activity depended on the FXXLF motif, since it was not observed with a GR-FXXAA mutant.
The data suggest that the AR NH2-terminal FXXLF motif region is sufficient to mediate the strong stimulatory effect of MAGE-11. However, the effect of MAGE-11 was not entirely specific to the AR FXXLF motif, since we also observed moderate increases in GR and PR-B activity on the PSA and mouse mammary tumor virus (MMTV)-Luc reporter vectors with higher expression levels of MAGE-11 (Fig. 11B and data not shown). MAGE-11 also increased AR transactivation of the MMTV-Luc and probasin promoters (Fig. 11C). However, MAGE-11 did not increase the activity of the constitutive viral reporters, pSV2-Luc, pSG5-Luc, and CMV-Luc (data not shown), suggesting that the effects of MAGE-11 on transcriptional activation are receptor mediated.
MAGE-11 also strongly increased the constitutive transcriptional activity of AR1-660, an AR NH2-terminal and DNA binding domain fragment that has only weak activity on the PSA-Luc reporter in the absence of MAGE-11 (Fig. 11D). The striking increase in AR1-660 transcriptional activity in response to MAGE-11 was FXXLF motif dependent, since the activity of the AR1-660-FXXAA mutant only marginally increased with MAGE-11 expression. Similar small increases were seen with GR1-550 and PRB-1-688, which are corresponding NH2-terminal and DNA binding domain fragments but lack an FXXLF motif. However, higher amounts of MAGE-11 expression vector DNA (2 μg) increased the activity of AR-FXXAA and the other receptor NH2-terminal fragments (data not shown). The data suggest that the presence of the AR FXXLF motif increases the sensitivity of AR to the stimulatory effects of MAGE-11.
Taken together, MAGE-11 appears to increase AR transactivation through mechanisms that include, but may not be limited to, its interaction with the FXXLF motif. The data support that MAGE-11 binding to the AR NH2-terminal FXXLF motif exposes AF2 in the ligand binding domain to activation by the SRC/p160 family of coactivators. The data also show a striking stimulatory effect of MAGE-11 on the AR NH2-terminal region that is independent of AF2.
In vivo expression of MAGE-11.
Northern blot analysis of RNA from several prostate cancer cell lines and human testis failed to reveal MAGE-11 mRNA (data not shown), suggesting that MAGE-11 expression is low. RT-PCR amplification of total RNA revealed the 168-bp fragment indicative of MAGE-11 mRNA in LNCaP, LNCaP-C4-2, CWR-R1, CWR22-RV1, and LAPC4 prostate cancer cell lines (Fig. 12A, top). However, detection of MAGE-11 mRNA in PC-3 and DU-145, two prostate cancer cell lines that do not express AR, required a second amplification using the initial PCR product as template (Fig. 12A, bottom). MAGE-11 mRNA was also detected in HeLa cells, a HeLaAR1C cell line stably expressing Flag-AR, was weakly detected in a human foreskin fibroblast cell line, HepG2, and COS cells, but was not detected in CV1 cells. MAGE-11 mRNA was also expressed in samples from testis, ovary, prostate, cancerous prostate, breast, and adrenal tissue, but was weak to undetectable in liver and lung tissue (Fig. 12B).
FIG. 12.
Endogenous MAGE-11 expression. (A) MAGE-11 mRNA was analyzed by RT-PCR. Total RNA (2 μg) was reverse transcribed, and the resulting cDNAs were amplified by 35 PCR cycles (top). In addition, 2 μl of the first PCRs was used as a template for a second 35-cycle PCR amplification with the same primers (bottom). Amplified MAGE-11 DNA fragments are shown for pSG5-MAGE-11 (lane 1), human foreskin fibroblasts (HFF, lane 2) and prostate cancer cell lines LNCaP (lane 3), LNCaP-C4-2 (lane 4), CWR-R1 (lane 5), CWR22-RV1 (lane 6), LAPC4 (lane 7), PC-3 (lane 8), and DU-145 (lane 9), as well as from HeLa cells (lane 10), HeLaAR1C cell line stably expressing human AR (lane 11), HepG2 (lane 12), COS cells (lane 13), CV1 cells (lane 14), human testis tissue (lane 15), and the no-RNA control (lane 16). The products were analyzed on 2% agarose gels. (B) Total RNA was isolated from adult human tissues with Trizol, reverse transcribed, and PCR amplified as described in Materials and Methods. Shown are the results of 35 PCR cycles with cDNA from human testis (lane 1), prostate (lane 2), a prostate tumor (lane 3), breast (lane 4), ovary (lane 5), lung (lane 6), adrenal gland (lane 7), and liver (lane 8). (C) Endogenous MAGE-11 protein (70 kDa) was detected in immunoblots with the anti-MAGE-11 peptide antibody described in Materials and Methods. The indicated cell lines were cultured to near confluence, and total protein was extracted with radioimmunoprecipitation assay buffer as described in Materials and Methods. Total protein (100 μg) was analyzed in each lane, and immunoblot analyses were performed with the peptide affinity-purified MAGE-11 antibody (2.9 μg/ml) raised in a rabbit. The MAGE-11 antibody was untreated (lanes 1 to 8) or preadsorbed overnight at 4°C by adding 5 μl of 0.5 mg of peptide immunogen/ml to 28 μl of 0.52-mg/ml antibody (lanes 9 to 16). Transiently expressed pSG5-MAGE-11 from COS cells was analyzed with 1 μg (lanes 1 and 9) and 2 μg (lanes 8 and 16) of total protein. The HeLaAR1C cell line expresses a stably integrated Flag-tagged human AR (P. D. Reynolds and E. M. Wilson, unpublished studies). The MAGE-11 antibody was relatively specific to MAGE-11 but also interacted on immunoblots with a ∼80-kDa DNA helicase (data not shown).
Endogenous MAGE-11 protein was evident on immunoblots of extracts from several cancer cell lines cultured in the absence of androgen and probed with the MAGE-11 antibody raised against the MAGE-11 NH2-terminal peptide. Endogenous MAGE-11 in HeLa cells and in the CWR-R1, LNCaP, PC-3, and DU-145 prostate cancer cell lines comigrated with 70-kDa recombinant MAGE-11 expressed in COS cells (Fig. 12C). Only faint comigrating bands persisted after preincubating the MAGE-11 antibody with the MAGE-11 peptide immunogen (Fig. 12C, right), and there was no evidence for an NH2-terminally truncated form of MAGE-11. It was interesting that HeLaAR1C cells stably expressing Flag-tagged full-length human AR had higher MAGE-11 protein levels than HeLa cells lacking AR (Fig. 12C, lanes 2 and 3). The similar MAGE-11 mRNA levels evident in HeLa and HeLaAR1C cells (Fig. 12A) raise the possibility that the stabilizing effect of MAGE-11 on AR observed above in the absence of androgen contributes to the stabilization of MAGE-11.
DISCUSSION
Identification of MAGE-11 as an AR coregulator.
We have identified the X-linked protein MAGE-11 as a novel AR coregulator. MAGE-11 specifically binds the AR NH2-terminal FXXLF motif that mediates the androgen-dependent N/C interaction with AF2 in the AR ligand binding domain. The molecular interaction between AR and MAGE-11 in the absence and presence of androgen is supported by their intracellular colocalization, coimmunoprecipitation, GST affinity matrix binding, and the MAGE-11-induced increases in AR transcriptional activity and ligand dissociation.
MAGE-11 forms a stable complex with the ligand-free AR that results in increased AR stability. Increased AR stabilization in the absence of androgen raises AR protein levels, which could contribute to the increase in AR transcriptional activity observed in the presence of MAGE-11. In contrast, binding of a strong agonist such as DHT decreases the association between AR and MAGE-11. This is consistent with competition by MAGE-11 for the agonist-induced N/C interaction between the AR FXXLF motif and AF2. At subsaturating concentrations of DHT, increased turnover of AR and MAGE-11 appears to result from this competition. Higher levels of DHT apparently overcome this effect and restore AR stabilization mediated by the N/C interaction.
We observed a persistent association between MAGE-11 and an AR nuclear transport mutant in the presence of DHT that suggests that the N/C interaction alone is not sufficient to destabilize MAGE-11 binding. Our earlier studies with this ARm4 mutant indicate binding kinetics consistent with an N/C interaction (51). Furthermore, persistence of a stable MAGE-11 and AR interaction in the presence of ligands such as estradiol and progesterone that induce AR nuclear transport but do not induce the AR N/C interaction suggests that nuclear targeting of AR is also not sufficient to destabilize the AR and MAGE-11 interaction. The data indicate that both AR nuclear transport and binding of an agonist are required for MAGE-11 and AR AF2 to competitively interact with the AR NH2-terminal FXXLF motif. We have shown that competition between MAGE-11 binding and the N/C interaction increases AR transactivation of androgen-responsive enhancer-promoters in a manner that is synergistic with the SRC/p160 family of coactivators. MAGE-11 binding and the N/C interaction appear to act in concert with coactivators to modulate AR activity. Binding of MAGE-11 to the AR NH2-terminal FXXLF motif mimics the effect of an AR-FXXAA mutant (14). Both expose the AF2 site in the AR ligand binding domain that is otherwise inhibited in the presence of androgen by the N/C interaction and allow increased recruitment and activation by the SRC/p160 family of coactivators.
The MAGE gene family.
The MAGE gene family consists of 12 homologous proteins coded by a 3.5-Mb segment on the long arm of the X chromosome at human Xq28. MAGE-11 is in a region encoding MAGE-7 and -9 (39). Six other MAGE genes are expressed exclusively in tumors, such as melanomas (hence the family name); lung, colon, and breast cancers; laryngeal tumors; sarcomas; and leukemias (5, 39). Tumor-selective expression of some MAGE family members is attributed to genome-wide demethylation of CpG dinucleotides at promoters (6), a common feature of cancer cells (4). MAGE-11 was previously reported in a variety of tumor cell lines, including HeLa, monkey kidney COS cells, 3T6 mouse fibroblasts, rat glial tumor cells, hamster CHO cells, and human 293 cells (24). MAGE-11 was also detected in testis and placenta (5), suggesting its expression is not limited to cancer cells.
However, there are discrepancies regarding the size of the MAGE-11 protein. Previous studies indicate an apparent molecular mass of 48 kDa (24) that was predicted from a single coding exon (GenBank accession number NM005366) (24, 39). This exon corresponds to the last exon of MAGE-11 shown in Fig. 1A. The previously predicted protein lacked the first 110 NH2-terminal residues reported here for MAGE-11. This putative truncated form of MAGE-11 would initiate at the third methionine, residue 111 of full-length MAGE-11. We observed a 70-kDa MAGE-11 protein, which agrees with a recent study (23) that identified three additional upstream exons encoding the NH2-terminal region unique to full-length MAGE-11 (Fig. 1A). When we created the truncated form of MAGE-11 to initiate at the third methionine, it was 52 kDa after in vitro translation and analysis with SDS gels (data not shown). Previous reports of a similarly sized endogenous MAGE-11 made use of an antibody raised against the carboxyl-terminal region of MAGE-11. However, this region is homologous with other members of the MAGE family, raising the possibility of cross-reaction with other family members. Our data nevertheless support the expression of full-length MAGE-11 in human testis, human foreskin fibroblasts, and several human cancer cell lines.
Mechanism of interaction between MAGE-11 and the AR FXXLF motif.
The AR FXXLF motif that binds MAGE-11 is identical to the motif that binds AF2 in the AR ligand binding domain mediating the androgen-dependent AR N/C interaction. However, detailed deletion mapping and point mutagenesis studies revealed overlapping yet distinct binding regions. AR binding to MAGE-11 requires an extended 21-amino-acid α-helical region between AR residues 16 to 36 that contains 23FQNLF27. This minimal region is nearly twice as long as the 11-amino-acid AR20-30 α-helical region required to bind AF2 in the AR ligand binding domain. The more-extended AR FXXLF binding region may allow MAGE-11 to compete for the N/C interaction in the presence of an AR agonist. Other regions outside but flanking the AR NH2-terminal domain FXXLF motif region required for MAGE-11 binding have also been suggested to modulate AR N/C interactions (43).
The entire carboxyl-terminal region of MAGE-11 is required to bind the AR FXXLF motif. This is reminiscent of the carboxyl-terminal region of AR that forms the ligand binding domain and the AF2 hydrophobic cleft that binds FXXLF. Cocrystal structures of the AR ligand binding domain have revealed the hydrophobic AF2 surface that accommodates the amphipathic α-helical regions of the FXXLF and LXXLL motifs (15, 22). The structure of MAGE-11 has not been determined, but analysis using the PONDR Protein Disorder Predictor (http://www.pondr.com/PONDR/pondr.cgi) suggests a highly ordered arrangement of α-helical regions between MAGE-11 residues 220 to 429. It is possible that a similar but unique binding surface on MAGE-11 interacts with the extended α-helical region of the AR FXXLF motif. On the other hand, binding of LXXLL peptides to protein domains other than nuclear receptor ligand binding domains has been reported (37). Binding of the FXXLF motif to MAGE-11 may occur through a mechanism similar to the interaction of the Stat6 transactivation domain LXXLL peptide binding to the PAS-B domain of SRC1 (37).
The stabilizing influence of MAGE-11 on AR in the absence of androgen and androgen-induced AR stabilization that results from the AR N/C interaction raises the possibility that the amphipathic α-helical FXXLF motif region also serves as an AR degradation signal (40). MAGE-11 binding to the FXXLF motif region could mask the putative signal and increase AR stabilization as observed in the absence of hormone. In the presence of DHT, FXXLF motif binding to AF2 could similarly protect against degradation resulting in the well-documented AR stabilization that results from the N/C interaction.
The intracellular association between AR and MAGE-11 is supported by their colocalization in the cytoplasm in the absence of androgen and in the nucleus in the presence of androgen. We also observed that MAGE-11 colocalizes in a prominent punctate cytoplasmic staining pattern, previously reported for this AR nuclear transport mutant (52). Dependence of the punctate pattern on androgen binding argues against nonspecific aggregation and suggests that the AR transport mutant becomes trapped in an as-yet-uncharacterized cytoplasmic substructure on route to the nucleus. For a DNA binding mutant, AR colocalizes with MAGE-11 in prominent subnuclear structures that resemble promyelocytic leukemia (PML) bodies. It was previously reported that agonist, but not antagonist-bound, AR triggers the subnuclear movement of AR and SRC1 out of the PML body into more filamentous nuclear speckles (38). Entrapment of an AR DNA binding domain mutant together with MAGE-11 in PML bodies would reinforce the idea that MAGE-11 facilitates the interaction between AR and the SRC/p160 coactivators.
A competition model.
Our search for an FXXLF motif binding protein was based on the premise that a coregulator that binds the AR NH2-terminal FXXLF motif with high specificity might relieve inhibition of AF2 imposed by the androgen-induced N/C interaction (14). The data presented in this report have led us to propose the model in Fig. 13. DHT binding to AR establishes an equilibrium between the AR N/C interaction (Fig. 13, top right) and AR AF2 binding of the SRC/p160 coactivators (Fig. 13, bottom right). In each instance, MAGE-11 is released in association with AR-mediated gene activation. In the presence of DHT, an AR, MAGE-11, and TIF2 intermediate (Fig. 13, bottom left) is supported by the persistent but reduced association of MAGE-11. The decrease in AR-associated MAGE-11 in the presence of DHT suggests MAGE-11 is released from the AR FXXLF binding site in parallel with formation of the AR N/C interaction or exposure of AF2 to SRC/p160 coactivator binding. This androgen-dependent interrelationship is supported by the synergistic increase in transcriptional activity that occurs in the presence of MAGE-11 and TIF2. It is also supported by the increased association of TIF2 and decreased association of MAGE-11 with the DHT-bound AR. By competing for FXXLF motif binding, MAGE-11 appears to function as an AR coregulator, in part by increasing accessibility of AF2 to SRC/p160 coactivator recruitment. There is also evidence that MAGE-11 acts through additional mechanisms independent of its interaction with the FXXLF motif.
FIG. 13.
Model for the effect of MAGE-11 on AR transactivation. In the absence of androgen, MAGE-11 binds the AR NH2-terminal domain (NTD) FXXLF motif (Fx) and stabilizes the ligand-free AR. Binding of DHT induces AR nuclear transport and a conformational change in AF2 in the ligand binding domain (LBD) that enables FXXLF motif binding in the N/C interaction and release of MAGE-11. Although not depicted, the AR FXXLF motif-AF2 N/C interaction is thought to occur between monomers in an antiparallel AR dimer. The androgen-dependent AR N/C interaction inhibits the activity of AF2, making AF1 the dominant activation domain. In the nucleus, MAGE-11 competes for and relieves the inhibitory effect of the N/C interaction, resulting in increased activation by AF2 binding of the LXXLL motifs (Lx) of the SRC/p160 family of coactivators. The model suggests that MAGE-11 functions as an AR coregulator by occupying the AR FXXLF site and facilitates intermittent binding of coactivators reported to occur at enhancer-promoter regions of androgen-regulated genes.
It is well recognized that the AR ligand binding domain expressed as a fragment or fusion protein has little or no transcriptional activity when assayed in mammalian cells (16). This reflects the relatively inefficient recruitment of coactivator LXXLL motifs by AR AF2, even in transformed cell lines where endogenous coactivator levels are typically higher than for normal cells (9). The inherent low activity of the AR ligand binding domain results from the 5- to 10-fold-lower binding affinity of AF2 for the SRC/p160 coactivator LXXLL motifs than the AR NH2-terminal FXXLF motif (15, 20). Reduced binding affinity for the LXXLL motifs results from sequence changes in AR AF2 that apparently evolved to favor FXXLF motif binding (15). Under conditions of increased SRC/p160 coactivator expression (18) as observed in recurrent prostate cancer (10), AR AF2 can recruit and be activated by this family of endogenous SRC/p160 coactivators (9, 41). The increase in AR transactivation that we observe in response to MAGE-11 without transiently overexpression of the SRC/p160 coactivators suggests that MAGE-11 binding to AR increases the recruitment of endogenous coactivators. Coactivator recruitment by the AR AF2 site in full-length AR is probably more efficient than coactivator recruitment by the AR ligand binding domain fragment alone, since additional interactions are reported to occur between the coactivator and the AR NH2-terminal region (16).
In the absence of androgen, the predominant binding of MAGE-11 to AR was not surprising, since AR FXXLF motif binding to AF2 that characterizes the AR N/C interaction requires the presence of a high-affinity androgen bound in the ligand binding pocket (51). Isolation of a stable complex between AR and MAGE-11 in the absence of androgen and our observations that MAGE-11 colocalizes with AR in the cytoplasm strongly indicate that binding of MAGE-11 to AR predominates in the absence of androgen. Moreover, in the presence of ligands such as estradiol, progesterone, and hydroxyflutamide that can target AR to the nucleus but do not induce the N/C interaction, a stable interaction with MAGE-11 persists in the nucleus.
On the other hand, MPA at high concentrations does not promote the AR N/C interaction between VP-AR1-660 and GAL-AR624-919 and instead acts as an inhibitor in the presence of DHT (27). In agreement with this, MPA at high concentrations does not stabilize AR (Fig. 5B, lane 6) and is not a strong androgen in vivo (27). However, recently we found that MPA at relatively low concentrations does promote the interaction of GAL-AR20-30 and GAL-AR4-52 with full-length AR (unpublished studies). The discrepancy between the assays is currently not understood. We reported previously that an AR NH2-terminal WXXLF motif contributes to the N/C interaction, but the influence of specific ligands is not known. Like the AR NH2-terminal FXXLF motif, the WXXLF motif interacts with AF2 in an androgen-dependent manner (17). The effectiveness of MPA to displace MAGE-11 may contribute to its strong transcriptional effects observed in transient transfection assays (27).
In the presence of DHT, competition between MAGE-11 binding and the AR N/C interaction depends on the DHT concentration and subcellular location of AR. At subsaturating concentrations of androgen (≤10 nM DHT for AR expressed in COS cells), steady-state levels of AR protein were reduced, suggesting that MAGE-11 interferes with the N/C interaction that mediates androgen-induced AR stabilization. However, the AR N/C interaction prevailed at higher androgen levels, and stabilization was restored for wild-type AR but not for the AR-FXXAA mutant defective in the N/C interaction. At androgen levels that fully saturate the AR, the N/C interaction apparently prevailed over MAGE-11 binding.
Even more compelling evidence for the intracellular competition between MAGE-11 and the N/C interaction is that MAGE-11 increases the dissociation rate of bound androgen from AR. Slow androgen dissociation is a hallmark of the AR N/C interaction (14, 17, 51). However, we also found that the nuclear transport mutant that localizes in the cytoplasm in the presence of DHT stably associates with MAGE-11 in the absence and presence of androgen. In the presence of androgen, ARm4 engages in an N/C interaction, based on its stabilization by androgen and by the slow rate of dissociation of bound ligand (51).
Thus, high-affinity agonist binding and AR nuclear transport both appear to be necessary to establish the dynamic relationship between MAGE-11 and AF2 binding at the AR NH2-terminal FXXLF site. By binding the FXXLF motif, MAGE-11 competes for the androgen dependent N/C interaction, exposing AF2 to activation by coactivators. Increased coactivator binding likely accounts in part for the MAGE-11-induced increase in AR transcriptional activity of androgen-responsive gene promoters, an increase that is synergistic with coactivator expression. Moreover, the studies support multiple protein interactions at a common FXXLF binding site whereby MAGE-11 influences AR turnover, the N/C interaction, and transcriptional activity in the absence and presence of agonist binding. Binding of MAGE-11 may contribute to the intermittent binding of coactivators that has been reported during AR mediated gene activation (41).
In vivo function of MAGE-11 and its role in prostate cancer.
Unlike most of the other MAGE family members that are cell surface tumor-associated antigens, MAGE-11 is confirmed by our studies as a cytoplasmic and nuclear protein that is excluded from nucleoli (24). A putative nuclear targeting signal in MAGE-11 was recognized previously (23). Localization of the MAGE family to the X chromosome and homology to a group of proteins in the sex-determining region of the X chromosome support a role for MAGE-11 in AR function during phenotypic sex determination. MAGE-11 is highly conserved in primates, although a cDNA and genomic DNA database homology search indicates MAGE-11 may not exist in lower species. However, a related MAGEB family of genes was recently reported that is ∼60% homologous to the MAGE (MAGEA) family (32). The MAGEB gene family maps to the Xp21 region of the X chromosome and is associated with a dose-sensitive sex reversal phenotype (29, 32, 35) and with the DAX-1 gene, where mutations cause X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism (34).
From an evolutionary perspective, the AR AF2 region of the ligand binding domain is more highly conserved among all species that express AR than is the FXXLF motif. This suggests that the AR AF2 region evolved prior to the FXXLF motif. Exclusive expression of MAGE-11 in primates may continue this evolutionary progression in AR regulation, suggesting that mechanisms that control the function of AR, one of the youngest members of the nuclear hormone receptor superfamily, continue to evolve.
It remains to be established in which cell types and under what physiological conditions MAGE-11 influences AR activity. MAGE-11 mRNA was not detectable by Northern blotting and required RT-PCR methods. Endogenous MAGE-11 protein was detected in prostate cancer cells and at higher levels in HeLa cells when the AR was stably expressed. Since MAGE-11 mRNA levels were not higher in HeLaAR1C cells stably expressing AR, the increase in MAGE-11 protein suggests that AR stabilizes MAGE-11 in a reciprocal relationship with the stabilizing effect of MAGE-11 on AR in the absence of androgen. The melanoma gene family of proteins is characterized by expression in cancer cells, but MAGE-11 is also expressed in normal tissues including testis, placenta (5), and (as shown here) ovary and prostate. The presence of MAGE-11 mRNA in a human foreskin fibroblast cell line also supports a role for MAGE-11 in AR transcriptional activity in normal cells.
Our studies further suggest that MAGE-11 may be important in prostate cancer. We found higher levels of expression of MAGE-11 mRNA in prostate cancer cells that express AR and that have been shown to have increased levels of the SRC/p160 family of coactivators (9). The MAGE-11-induced stabilization of AR in the absence of hormone could account for the increased levels of AR frequently observed with recurrent prostate cancer (3, 28, 46) and may contribute to the increased ligand-independent AR transcriptional activity. This, together with increased levels of the coactivators SRC1 and TIF2 (10), could enhance the sensitivity of AR to low androgen levels (11, 12, 33) and to the effects of mitogen signaling (9). The data support that MAGE-11 is a new player in the cellular pathways that mediate AR transactivation in normal and cancer cells.
Acknowledgments
We thank Frank S. French for reviewing the manuscript, Susan H. Hall for providing human tissues, and Rebecca I. Kalman, John T. Minges, Emily B. Askew, Andrew T. Hnat, Jonathan L. Faggart, K. Michelle Cobb, and De-Ying Zang for excellent technical assistance.
This work was supported by Public Health Service grant HD16910 from the National Institutes of Child Health and Development, by cooperative agreement (U54-HD35041) as part of the Specialized Cooperative Centers Program in Reproductive Research of the National Institutes of Health, and by Fogarty International Center Training and Research in Population and Health grant R03TW001234 awarded to Frank S. French by the National Institute of Health, supporting S.B. and B.H.
REFERENCES
- 1.Balk, S. P. 2002. Androgen receptor as a target in androgen-independent prostate cancer. Urology 60:132-138. [DOI] [PubMed] [Google Scholar]
- 2.Callewaert, L., G. Verrijdt, V. Christiaens, A. Haelens, and F. Claessens. 2003. Dual function of an amino-terminal amphipathic helix in androgen receptor-mediated transactivation through specific and nonspecific response elements. J. Biol. Chem. 278:8212-8218. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. D., D. S. Welsbie, C. Tran, S. H. Baek, R. Chen, R. Vessella, M. G. Rosenfeld, and C. L. Sawyers. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10:33-39. [DOI] [PubMed] [Google Scholar]
- 4.Counts, J. L., and J. I. Goodman. 1995. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell 83:13-15. [DOI] [PubMed] [Google Scholar]
- 5.De Plaen, E., K. Arden, C. Traversari, J. J. Gaforio, J. P. Szikora, C. De Smet, F. Brasseur, P. van der Bruggen, B. Lethé, C. Lurquin, R. Brasseur, P. Chomez, O. De Backer, W. Cavenee, and T. Boon. 1994. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics 40:360-369. [DOI] [PubMed] [Google Scholar]
- 6.De Smet, C., O. De Backer, I. Faraoni, C. Lurquin, F. Brasseur, and T. Boon. 1996. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc. Natl. Acad. Sci. USA 93:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto, N., S. Yeh, H. Y. Kang, S. Inui, H. C. Chang, A. Mizokami, and C. Chang. 1999. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J. Biol. Chem. 274:8316-8321. [DOI] [PubMed] [Google Scholar]
- 8.Gelmann, E. P. 2002. Molecular biology of the androgen receptor. J. Clin. Oncol. 20:3001-3015. [DOI] [PubMed] [Google Scholar]
- 9.Gregory, C. W., X. Fei, L. A. Ponguta, B. He, H. M. Bill, F. S. French, and E. M. Wilson. 2004. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J. Biol. Chem. 279:7119-7130. [DOI] [PubMed] [Google Scholar]
- 10.Gregory, C. W., B. He, R. T. Johnson, O. H. Ford, J. L. Mohler, F. S. French, and E. M. Wilson. 2001. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 61:4315-4319. [PubMed] [Google Scholar]
- 11.Gregory, C. W., R. T. Johnson, J. L. Mohler, F. S. French, and E. M. Wilson. 2001. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 61:2892-2898. [PubMed] [Google Scholar]
- 12.Grossmann, M. E., H. Huang, and D. J. Tindall. 2001. Androgen receptor signaling in androgen-refractory prostate cancer. J. Natl. Cancer Inst. 93:1687-1697. [DOI] [PubMed] [Google Scholar]
- 13.He, B., S. Bai, A. T. Hnat, R. I. Kalman, J. T. Minges, C. Patterson, and E. M. Wilson. 2004. An androgen receptor NH2-terminal conserved motif interacts with carboxyl terminus of Hsp70-interacting protein CHIP. J. Biol. Chem. 279:30643-30653. [DOI] [PubMed] [Google Scholar]
- 14.He, B., N. T. Bowen, J. T. Minges, and E. M. Wilson. 2001. Androgen-induced NH2- and COOH-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 276:42293-42301. [DOI] [PubMed] [Google Scholar]
- 15.He, B., R. T. Gampe, A. J. Kole, A. T. Hnat, T. B. Stanley, G. An, E. L. Stewart, R. I. Kalman, J. T. Minges, and E. M. Wilson. 2004. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 16:425-438. [DOI] [PubMed] [Google Scholar]
- 16.He, B., J. A. Kemppainen, J. J. Voegel, H. Gronemeyer, and E. M. Wilson. 1999. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 274:37219-37225. [DOI] [PubMed] [Google Scholar]
- 17.He, B., J. A. Kemppainen, and E. M. Wilson. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275:22986-22994. [DOI] [PubMed] [Google Scholar]
- 18.He, B., L. W. Lee, J. T. Minges, and E. M. Wilson. 2002. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J. Biol. Chem. 277:25631-25639. [DOI] [PubMed] [Google Scholar]
- 19.He, B., J. T. Minges, L. W. Lee, and E. M. Wilson. 2002. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem. 277:10226-10235. [DOI] [PubMed] [Google Scholar]
- 20.He, B., and E. M. Wilson. 2003. Electrostatic modulation in steroid receptor recruitment of LXXLL and FXXLF motifs. Mol. Cell. Biol. 23:2135-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, W., Y. Shostak, P. Tarr, C. Sawyers, and M. Carey. 1999. Cooperative assembly of androgen receptor into a nucleoprotein complex that regulates the prostate-specific antigen enhancer. J. Biol. Chem. 274:25756-25768. [DOI] [PubMed] [Google Scholar]
- 22.Hur, E., S. J. Pfaff, E. S. Payne, H. Gron, B. M. Buehrer, and R. J. Fletterick. 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2:e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irvine, R. A., and G. A. Coetzee. 1999. Additional upstream coding sequences of MAGE-11. Immunogenetics 49:585. [DOI] [PubMed] [Google Scholar]
- 24.Jurk, M., E. Kremmer, U. Schwarz, R. Forster, and E. L. Winnacker. 1998. MAGE-11 protein is highly conserved in higher organisms and located predominantly in the nucleus. Int. J. Cancer 75:762-766. [DOI] [PubMed] [Google Scholar]
- 25.Kang, H. Y., S. Yeh, N. Fujimoto, and C. Chang. 1999. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J. Biol. Chem. 274:8570-8576. [DOI] [PubMed] [Google Scholar]
- 26.Kemppainen, J. A., M. V. Lane, M. Sar, and E. M. Wilson. 1992. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones.> J. Biol. Chem. 267:968-974. [PubMed] [Google Scholar]
- 27.Kemppainen, J. A., E. Langley, C. I. Wong, K. Bobseine, W. R. Kelce, and E. M. Wilson. 1999. Distinguishing androgen receptor agonists and antagonists: distinct mechanisms of activation by medroxyprogesterone acetate and dihydrotestosterone. Mol. Endocrinol. 13:440-454. [DOI] [PubMed] [Google Scholar]
- 28.Koivisto, P., J. Kononen, C. Palmberg, T. Tammela, E. Hyytinen, J. Isola, J. Trapman, K. Cleutjens, A. Noordzij, T. Visakorpi, and O. P. Kallioniemi. 1997. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res. 57:314-319. [PubMed] [Google Scholar]
- 29.Lalli, E., and P. Sassone-Corsi. 2003. DAX-1, an unusual orphan receptor at the crossroads of steroidogenic function and sexual differentiation. Mol. Endocrinol. 17:1445-1453. [DOI] [PubMed] [Google Scholar]
- 30.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 31.Lubahn, D. B., D. R. Joseph, M. Sar, J. Tan, H. N. Higgs, R. E. Larson, F. S. French, and E. M. Wilson. 1988. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol. Endocrinol. 2:1265-1275. [DOI] [PubMed] [Google Scholar]
- 32.Lurquin, C., C. De Smet, F. Brasseur, F. Muscatelli, V. Martelange, E. De Plaen, R. Brasseur, A. P. Monaco, and T. Boon. 1997. Two members of the human MAGEB gene family located in Xp21.3 are expressed in tumors of various histological origins. Genomics 46:397-408. [DOI] [PubMed] [Google Scholar]
- 33.Mohler, J. L., C. W. Gregory, O. H. Ford, D. Kim, C. M. Weaver, P. Petrusz, E. M. Wilson, and F. S. French. 2004. The androgen axis in recurrent prostate cancer. Clin. Cancer Res. 10:440-448. [DOI] [PubMed] [Google Scholar]
- 34.Muscatelli, F., T. M. Strom, A. P. Walker, E. Zanaria, D. Recan, A. Meindl, B. Bardoni, S. Guioli, G. Zehetner, W. Rabl, H. P. Schwarz, J. C. Kaplan, G. Camerino, T. Meitinger, and A. P. Monaco. 1994. Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372:672-676. [DOI] [PubMed] [Google Scholar]
- 35.Muscatelli, F., A. P. Walker, E. De Plaen, A. N. Stafford, and A. P. Monaco. 1995. Isolation and characterization of a MAGE gene family in the Xp21.3 region. Proc. Natl. Acad. Sci. USA 92:4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao, M. A., H. Cheng, A. N. Quayle, H. Nishitani, C. C. Nelson, and P. S. Rennie. 2002. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem. 277:48020-48027. [DOI] [PubMed] [Google Scholar]
- 37.Razeto, A., V. Ramakrishnan, C. M. Litterst, K. Giller, C. Griesinger, T. Carlomagno, N. Lakomek, T. Heimburg, M. Lodrini, E. Pfitzner, and S. Becker. 2004. Structure of the NCoA-1/SRC-1 PAS-B domain bound to the LXXLL motif of the STAT6 transactivation domain. J. Mol. Biol. 336:319-329. [DOI] [PubMed] [Google Scholar]
- 38.Rivera, O. J., C. S. Song, V. E. Centonze, J. D. Lechleiter, B. Chatterjee, and A. K. Roy. 2003. Role of the promyelocytic leukemia body in the dynamic interaction between the androgen receptor and steroid receptor coactivator-1 in living cells. Mol. Endocrinol. 17:128-140. [DOI] [PubMed] [Google Scholar]
- 39.Rogner, U. C., K. Wilke, E. Steck, B. Korn, and A. Poustka. 1995. The melanoma antigen gene (MAGE) family is clustered in the chromosomal band Xq28. Genomics 29:725-731. [DOI] [PubMed] [Google Scholar]
- 40.Sadis, S., C. Atienza, and D. Finley. 1995. Synthetic signals for ubiquitin-dependent proteolysis. Mol. Cell. Biol. 15:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 42.Simental, J. A., M. Sar, M. V. Lane, F. S. French, and E. M. Wilson. 1991. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 266:510-518. [PubMed] [Google Scholar]
- 43.Steketee, K., C. A. Berrevoets, H. J. Dubbink, P. Doesburg, R. Hersmus, A. O. Brinkmann, and J. Trapman. 2002. Amino acids 3-13 and amino acids in and flanking the 23FxxLF27 motif modulate the interaction between the N-terminal and ligand-binding domain of the androgen receptor. Eur. J. Biochem. 269:5780-5791. [DOI] [PubMed] [Google Scholar]
- 44.Tan, J. A., S. H. Hall, P. Petrusz, and F. S. French. 2000. Thyroid receptor activator molecule, TRAM-1, is an androgen receptor coactivator. Endocrinology 141:3440-3450. [DOI] [PubMed] [Google Scholar]
- 45.Tork, S., I. Hatin, J. P. Rousset, and C. Fabret. 2004. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 32:415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visakorpi, T., E. Hyytinen, P. Koivisto, M. Tanner, R. Keinanen, C. Palmberg, A. Palotie, T. Tammela, J. Isola, and O. P. Kallioniemi. 1995. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 9:401-406. [DOI] [PubMed] [Google Scholar]
- 47.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, L., J. Guerrero, H. Hong, D. B. DeFranco, and M. R. Stallcup. 2000. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell 11:2007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. USA 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, Z. X., B. He, S. H. Hall, E. M. Wilson, and F. S. French. 2002. Domain interactions between coregulator ARA(70) and the androgen receptor (AR). Mol. Endocrinol. 16:287-300. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, Z. X., M. V. Lane, J. A. Kemppainen, F. S. French, and E. M. Wilson. 1995. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol. Endocrinol. 9:208-218. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, Z. X., M. Sar, J. A. Simental, M. V. Lane, and E. M. Wilson. 1994. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J. Biol. Chem. 269:13115-13123. [PubMed] [Google Scholar]