Abstract
We performed pulsed-field gel electrophoresis on Escherichia coli O157 isolates (n = 318) from 199 healthy animals in a longitudinal study carried out on nine farms. Investigation of the restriction types proved that at the farm level, the same clones can be detected on sampling occasions separated by as much as 17 months. The cohort animals were repeatedly sampled, and for some of these, the same clones were obtained on sampling occasions separated by as much as 8 months.
Escherichia coli strains that produce Shiga-like toxins, also known as verocytotoxins, have been implicated as the cause for human disease (hemorrhagic colitis and hemolytic-uremic syndrome). One particular serovar, E. coli O157, represents a major public health concern worldwide. Healthy asymptomatic cattle are the best-recognized animal reservoir for these organisms, and shedding via feces occurs only intermittently, typically lasting 3 to 4 weeks (5, 10, 16). Several molecular techniques have been used to investigate the epidemiology of E. coli O157, and pulsed-field gel electrophoresis (PFGE) is currently considered the “gold standard” for DNA fingerprinting of these strains (17). The ecology and epidemiology of this organism in cattle appear to be very complex, often involving multiple clones on a single farm (9, 14). There is some previously published information about on-farm persistence in different cattle production systems (8, 15, 16). However, the information about persistence in individual animals is very scarce. A longitudinal study was conducted to provide information on the persistence and maintenance of colonization with E. coli O157 at the farm level as well as the individual-animal level in the context of a typical farm in the United Kingdom.
Bacterial strains.
E. coli O157 isolates (n = 648) were cultured from bovine fecal samples from nine epidemiologically unrelated farms dispersed across England and Wales (9, 12). All the isolates were confirmed as serotype O157 by serum agglutination (11); the first confirmed colony from a positive sample was selected as representative for that animal and was stored at −80°C. The first sampling visit occurred during the months of August to November 1999 and used previously described epidemiological methods (12). The longitudinal study started in April 2000; during the first visit, as many as 90 animals under the age of 24 months (“original cohort”) were randomly selected for sampling on each farm selected. Subsequently, monthly sampling visits were carried out until February 2001. Samples of rectal feces (one per animal) were collected from individuals in the original cohort, as well as from any cattle born since the previous visit and any new acquisitions to the herd.
A total of 318 E. coli O157 isolates from 199 healthy bovines on nine farms were characterized by PFGE (Table 1). This selection covered all isolates from animals that yielded two or more positive results on several sampling occasions, as well as a random representation of isolates from animals that were found to be positive only on one occasion. These comprised 29 to 100% of the available isolates for each of the farms. Preparation and XbaI digestion of DNA for PFGE and analysis of results were conducted as described previously (2).
TABLE 1.
E. coli O157 isolations by farm and sampling date
| Site | No. of animals sampled (no. positive for E. coli O157) at the following timea:
|
No. (%)b of isolates typed by PFGE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 08/99 to 11/99 | 04/00 | 05/00 | 06/00 | 07/00 | 08/00 | 09/00 | 10/00 | 11/00 | 12/00 | 01/01 | 02/01 | ||
| Farm 2 | 80 (38) | NS | 90 (8) | 92 (4) | 90 (4) | 90 (5) | NS | 90 (1) | 91 (12) | 90 (53) | 90 (30) | 86 (28) | 111 (60) |
| Farm 3 | 46 (12) | NS | 56 (23) | 53 (14) | 61 (1) | 46 (0) | 54 (7) | 57 (0) | 61 (0) | 60 (0) | 60 (0) | 60 (0) | 35 (61) |
| Farm 4 | 57 (9) | NS | 72 (5) | 26 (1) | 23 (4) | 23 (7) | 42 (8) | 74 (13) | 92 (11) | NS | 92 (15) | 92 (7) | 27 (34) |
| Farm 5 | 64 (1) | NS | 89 (7) | 75 (8) | 83 (2) | 98 (4) | 135 (4) | 87 (1) | NS | NS | NS | NS | 8 (29) |
| Farm 6 | 87 (11) | 90 (2) | 48 (0) | 55 (0) | NS | 58 (0) | 63 (1) | 88 (0) | 90 (1) | 91 (1) | 88 (0) | 87 (2) | 10 (55) |
| Farm 8 | 46 (5) | 90 (11) | 83 (43) | 82 (17) | 80 (6) | 53 (14) | 54 (8) | 58 (1) | 62 (3) | 90 (3) | 112 (12) | NS | 37 (30) |
| Farm 9 | 72 (37) | 90 (0) | 93 (0) | 90 (1) | 91 (35) | 92 (23) | 92 (23) | 101 (9) | 102 (9) | 101 (8) | 101 (0) | 90 (0) | 78 (54) |
| Farm 10 | 46 (5) | 31 (0) | 40 (0) | 41 (0) | 45 (0) | 40 (0) | 36 (0) | 33 (0) | 32 (0) | NS | 33 (1) | 33 (1) | 4 (57) |
| Farm 11 | 61 (4) | NS | NS | 37 (0) | NS | 35 (0) | 37 (0) | 38 (0) | 41 (0) | 40 (2) | 41 (1) | 41 (1) | 8 (100) |
Sampling dates are given as month/year. NS, no sampling was conducted.
Percentage of the total isolates available for a given farm that was typed.
Table 2 details the persistence of PFGE subtypes at the farm level. The most persistent types (X49 and X25) were isolated from samples from farms 4 and 8, taken 17 and 16 months apart, respectively. Types X8 and X24 were found in isolates from farms 8 and 9, respectively, on six consecutive sampling occasions. On every farm, a variety of types were found on particular sampling occasions and never found again. This situation does not seem to be unique to the United Kingdom; previous work from the United States indicates that individual strains can be isolated from some dairy herds for as long as 2 years (16), for as long as 10 months in cattle ranges (15), and over the entire feeding period on cattle feedlots (8).
TABLE 2.
On-farm persistence of PFGE types
| Site | PFGE type isolated at the following time (mo/yr)a:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 08/99 to 11/99 | 04/00 | 05/00 | 06/00 | 07/00 | 08/00 | 09/00 | 10/00 | 11/00 | 12/00 | 01/01 | 02/01 | |
| Farm 2 | NS | NS | X39 | X39 | X39 | X39 | ||||||
| NS | NS | X44 | X44 | X44 | X44 | X44 | ||||||
| NS | NS | X45 | X45 | X45 | ||||||||
| Farm 3 | X24 | NS | X24 | X24 | X24 | − | X24 | − | − | − | − | − |
| NS | X26 | X26 | ||||||||||
| Farm 4 | X49 | NS | X46 | NS | X46 | X46 | ||||||
| X49 | NS | X49 | X49 | X49 | X49 | NS | X49 | |||||
| NS | − | X50 | X50 | NS | ||||||||
| NS | X52 | X52 | NS | |||||||||
| NS | X54 | X54 | NS | X54 | ||||||||
| Farm 5 | − | NS | X31 | X31 | − | NS | NS | NS | NS | |||
| NS | X56 | X56 | NS | NS | NS | NS | ||||||
| Farm 6 | X22 | X22 | − | − | NS | − | − | − | − | |||
| NS | X42 | X42 | ||||||||||
| Farm 8 | X8 | X8 | X8 | X8 | X8 | X8 | NS | |||||
| X25 | X25 | X25 | X25 | NS | ||||||||
| X30 | X30 | X30 | X30 | X30 | X30 | NS | ||||||
| Farm 9 | X24 | X24 | X24 | X24 | X24 | X24 | ||||||
| − | − | X26 | X26 | X26 | − | − | ||||||
| X27 | X27 | X27 | X27 | |||||||||
| Farm 10 | − | − | − | − | − | − | − | − | NS | X24 | X24 | |
| Farm 11 | NS | NS | − | NS | − | − | − | − | X8 | X8 | ||
−, E. coli O157 was not isolated from the premises. NS, the farm was not sampled on that occasion. An empty cell indicates that E. coli O157 isolates were obtained, but they presented PFGE types that were never found again on that farm at any sampling time.
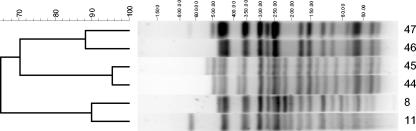
Table 3 shows a summary of the types persisting in animals that were positive for E. coli O157 on two or more sampling occasions. In most cases, several different PFGE types were found persisting in different animals from the same farm. However, in one case (farm 3), the same clone persisted in all the animals for which persistence could be demonstrated. The most persistent clone at the individual-animal level was X24, which was found in isolates recovered from samples from animals 13 and 17 on farm 3, taken 7 months apart. On several occasions (animal 156, farm 2; animal 56, farm 8; animal 9, farm 9) the same clones were found on four consecutive sampling occasions. Types X44 and X45, X46 and X47, and X8 and X11 were very similar; each of these pairs differed only in ≤2 restriction fragments (Fig. 1). For this reason, they have been included in Table 3. One suspects that these variations may be due to clonal turnover in the animal. Similar small variations in PFGE profiles have been reported before in experimental and natural infections (1, 6).
TABLE 3.
Persistence of PFGE types in individual animals that were positive for E. coli 0157 on two or more occasions and from which a PFGE result was available
| Farm | Animal | PFGE type isolated at the following time (mo/yr)a:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 08/99 to 11/99 | 04/00 | 05/00 | 06/00 | 07/00 | 08/00 | 09/00 | 10/00 | 11/00 | 12/00 | 01/01 | 02/01 | ||
| Farm 2 | A133 | NS | NS | NS | NS | NS | − | NS | − | X44 | X44 | NS | NS |
| A151 | NS | NS | NS | NS | NS | − | NS | − | − | − | NS | ||
| A156 | NS | NS | NS | NS | NS | − | NS | − | X39 | X39 | X39 | X39 | |
| A162 | NS | NS | NS | NS | NS | − | NS | − | − | X39 | X39 | − | |
| A166 | NS | NS | NS | NS | NS | − | NS | − | − | X39 | X39 | NS | |
| A179 | NS | NS | NS | NS | NS | NS | NS | − | − | X44 | X44 | − | |
| A196 | NS | NS | NS | NS | NS | NS | NS | − | − | X44 | X45 | − | |
| A206 | NS | NS | NS | NS | NS | NS | NS | NS | − | − | |||
| A207 | NS | NS | NS | NS | NS | NS | NS | NS | − | X44 | X44 | ||
| A208 | NS | NS | NS | NS | NS | NS | NS | NS | − | − | |||
| A210 | NS | NS | NS | NS | NS | NS | NS | NS | − | X44 | X45 | ||
| A212 | NS | NS | NS | NS | NS | NS | NS | NS | X44 | X44 | − | ||
| A215 | NS | NS | NS | NS | NS | NS | NS | NS | − | X44 | X44 | ||
| A217 | NS | NS | NS | NS | NS | NS | NS | NS | − | X44 | − | X45 | |
| A219 | NS | NS | NS | NS | NS | NS | NS | NS | − | X44 | X44 | ||
| A225 | NS | NS | NS | NS | NS | NS | NS | NS | − | − | |||
| A230 | NS | NS | NS | NS | NS | NS | NS | NS | − | X39 | X39 | − | |
| A247 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | X39 | X39 | |
| A259 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | X44 | X45 | |
| A260 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | X44 | X45 | |
| A265 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | X44 | X44 | |
| A268 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | X44 | X44 | |
| Farm 3 | A4 | NS | NS | NS | − | NS | NS | NS | NS | NS | |||
| A10 | NS | NS | NS | − | − | NS | NS | NS | NS | NS | |||
| A13 | X24 | NS | − | X24 | NS | NS | NS | NS | NS | NS | NS | NS | |
| A17 | X24 | NS | X24 | X24 | NS | − | − | NS | NS | NS | NS | NS | |
| A19 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | |||
| A21 | NS | NS | X24 | X24 | NS | NS | NS | NS | NS | NS | NS | NS | |
| A26 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | |||
| A28 | NS | NS | X24 | − | − | − | X24 | NS | NS | NS | NS | NS | |
| A32 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | |||
| A35 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | |||
| A36 | NS | NS | X24 | X24 | NS | NS | NS | NS | NS | NS | NS | NS | |
| A40 | NS | − | − | − | − | NS | NS | NS | NS | NS | |||
| A45 | NS | − | − | − | NS | NS | NS | NS | NS | NS | |||
| A53 | NS | − | − | − | NS | NS | NS | NS | NS | NS | |||
| A55 | NS | − | − | − | − | NS | NS | NS | NS | NS | |||
| Farm 4 | A2 | NS | NS | − | − | − | X52 | − | X52 | NS | − | − | |
| A5 | NS | NS | − | − | − | X49 | − | − | X49 | NS | − | − | |
| A6 | NS | NS | − | − | X54 | − | X54 | − | NS | − | − | ||
| A8 | NS | NS | − | − | − | − | − | − | NS | ||||
| A18 | NS | NS | − | NS | − | − | X49 | X49 | − | NS | − | ||
| A21 | NS | NS | − | − | − | − | X49 | − | − | NS | X49 | − | |
| A79 | NS | NS | NS | NS | NS | NS | NS | NS | X46 | NS | − | X46 | |
| A82 | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||||
| A92 | NS | NS | NS | NS | NS | NS | NS | NS | − | NS | X46 | X47 | |
| Farm 5 | A19 | NS | NS | − | − | X56 | NS | X56 | − | NS | NS | NS | NS |
| A41 | NS | NS | X31 | X31 | − | NS | NS | NS | NS | NS | NS | NS | |
| A43 | NS | NS | NS | NS | NS | NS | NS | NS | NS | NS | |||
| Farm 8 | A17 | NS | X8 | X8 | − | X8 | NS | NS | NS | NS | NS | NS | NS |
| A22 | NS | X8 | X11 | X8 | − | NS | NS | NS | NS | NS | NS | NS | |
| A23 | NS | − | X8 | − | X8 | NS | NS | NS | NS | NS | NS | NS | |
| A24 | NS | X8 | X8 | X8 | − | NS | NS | NS | NS | NS | NS | NS | |
| A48 | NS | X30 | X30 | − | NS | NS | NS | NS | NS | NS | NS | NS | |
| A56 | NS | X30 | X30 | X30 | X30 | NS | NS | NS | NS | NS | NS | NS | |
| A78 | NS | − | − | X30 | X30 | NS | NS | NS | NS | NS | NS | NS | |
| A93 | NS | NS | − | X8 | − | X8 | NS | NS | NS | NS | NS | NS | |
| A102 | NS | NS | NS | X8 | − | − | X8 | − | NS | NS | NS | NS | |
| A104 | NS | NS | NS | − | − | NS | NS | NS | NS | NS | |||
| A138 | NS | NS | NS | NS | NS | − | X30 | X30 | NS | NS | NS | NS | |
| A193 | NS | NS | NS | NS | NS | NS | NS | NS | X25 | − | X25 | NS | |
| Farm 9 | A3 | NS | − | − | − | − | − | − | − | − | NS | ||
| A7 | − | − | − | − | − | − | − | − | − | NS | |||
| A9 | NS | − | − | − | − | X24 | X24 | X24 | X24 | − | − | NS | |
| A16 | NS | − | − | − | − | − | NS | NS | NS | NS | |||
| A29 | NS | − | − | − | X24 | X24 | − | X24 | − | NS | NS | NS | |
| A31 | NS | − | − | − | X24 | − | − | − | X24 | X24 | − | NS | |
| A47 | − | − | − | − | − | − | − | − | − | − | |||
| A53 | − | − | − | − | − | − | − | − | − | − | |||
| A56 | NS | − | − | − | X27 | X27 | X27 | − | − | − | − | − | |
| A59 | − | − | − | X24 | X24 | − | − | − | − | − | − | ||
| A74 | NS | − | − | − | − | X24 | X24 | − | − | − | − | − | |
| A77 | NS | − | − | − | − | − | − | − | − | − | |||
| A81 | NS | − | − | − | X26 | X26 | − | − | − | − | |||
| A82 | NS | − | − | − | − | X25 | X26 | − | − | − | − | ||
| A92 | NS | NS | − | − | − | − | − | − | − | − | |||
| A93 | NS | NS | − | − | − | − | X24 | − | − | X24 | − | − | |
| Farm 10 | A39 | NS | NS | NS | − | − | − | NS | − | − | NS | X24 | X24 |
| Farm 11 | A5 | NS | NS | NS | − | NS | − | − | − | − | X8 | X8 | |
| A13 | NS | NS | − | NS | − | − | − | − | − | − | |||
−, the sample from the animal was negative for E. coli O157. NS, the animal was not sampled on that occasion. Empty cells indicate that the sample from the animal was positive for E. coli O157, but the isolates showed fingerprints that were never found again in isolates from the same animal.
FIG. 1.
Dendrogram generated with Bionumerics software showing the PFGE XbaI types for three pairs of E. coli O157 isolates. The bands generated were analyzed by using the Jaccard coefficient and the unweighted-pair group method with arithmetic averages. Molecular weights (in kilobase pairs) are given above the gel.
A study from Australia showed that indistinguishable PFGE types could be consistently isolated from cattle feces for as long as 49 days (10). However, it also seems true that individual animals may shed multiple strains simultaneously, and the spectrum of types changes over time (7). Some studies have indicated that E. coli O157 strains seem to be continually replaced by new and distinct types in cattle (4, 13). This agrees with the results in our study, where on several occasions, animals appeared to shed different types at different times. All of this seems to indicate that E. coli O157 strains cycle with a high turnover through individual cattle, though predominant strains may persist collectively on a farm (3).
We are aware that the selection of only a fraction of our isolates for PFGE, as well as the fact that only one colony per positive animal was stored, may have influenced our results. However, in spite of these limitations, we have successfully demonstrated that some clones can be isolated from the same farms over a period of at least 17 months and from the same animal over a period of at least 7 months. We cannot be sure if these results are a reflection of persistent colonization of the animal by a particular clone or if they are due instead to residual contamination of the farm environment and recycling through the host. In any case, we have demonstrated that defined PFGE clones persist on farms in the United Kingdom for a significant period. In most cases, the persisting types coincided with the most prevalent clones for the farm (9), indicating that less prevalent clones may represent only transient episodes of contamination.
Acknowledgments
We thank Tom Cheasty and Geraldine Willshaw (PHLS, Colindale, United Kingdom) for phage typing of the strains.
This work was supported by DEFRA grant OZ0138.
REFERENCES
- 1.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 2.Avery, S. M., E. Liebana, C. A. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting mathods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cobbold, R., and P. Desmarchelier. 2001. Characterisation and clonal relationships of Shiga-toxigenic Escherichia coli (STEC) isolated from Australian dairy cattle. Vet. Microbiol. 79:323-335. [DOI] [PubMed] [Google Scholar]
- 4.Galland, J. C., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karch, H., H. Russmann, H. Schmidt, A. Schwarzkopf, and J. Heesemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1997. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J. Clin. Microbiol. 35:892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebana, E., R. P. Smith, E. Lindsay, I. McLaren, C. Cassar, F. A. Clifton-Hadley, and G. A. Paiba. 2003. Genetic diversity among Escherichia coli O157:H7 isolates from bovines living on farms in England and Wales. J. Clin. Microbiol. 41:3857-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Midgley, J., and P. Desmarchelier. 2001. Pre-slaughter handling of cattle and Shiga toxin-producing Escherichia coli (STEC). Lett. Appl. Microbiol. 32:307-311. [DOI] [PubMed] [Google Scholar]
- 11.Paiba, G. A., J. C. Gibbens, S. J. Pascoe, J. W. Wilesmith, S. A. Kidd, C. Byrne, J. B. Ryan, R. P. Smith, M. McLaren, R. J. Futter, A. C. Kay, Y. E. Jones, S. A. Chappell, G. A. Willshaw, and T. Cheasty. 2002. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Vet. Rec. 150:593-598. [DOI] [PubMed] [Google Scholar]
- 12.Paiba, G. A., J. W. Wilesmith, S. J. Evans, S. J. Pascoe, R. P. Smith, S. A. Kidd, J. B. Ryan, I. M. McLaren, S. A. Chappell, G. A. Willshaw, T. Cheasty, N. P. French, T. W. Jones, H. F. Buchanan, D. J. Challoner, A. D. Colloff, M. P. Cranwell, R. G. Daniel, I. H. Davies, J. P. Duff, R. A. Hogg, F. D. Kirby, M. F. Millar, R. J. Monies, M. J. Nicholls, and J. H. Payne. 2003. Prevalence of faecal excretion of verocytotoxigenic Escherichia coli O157 in cattle in England and Wales. Vet. Rec. 153:347-353. [DOI] [PubMed] [Google Scholar]
- 13.Rahn, K., S. A. Renwick, R. P. Johnson, J. B. Wilson, R. C. Clarke, D. Alves, S. McEwen, H. Lior, and J. Spika. 1997. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol. Infect. 119:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renter, D. G., and J. M. Sargeant. 2002. Enterohemorrhagic Escherichia coli O157: epidemiology and ecology in bovine production environments. Anim. Health Res. Rev. 3:83-94. [PubMed] [Google Scholar]
- 15.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]