Summary
Recognition between sperm and the egg surface marks the beginning of life in all sexually reproducing organisms. This fundamental biological event depends on the species-specific interaction between rapidly evolving counterpart molecules on the gametes. We report biochemical, crystallographic, and mutational studies of domain repeats 1–3 of invertebrate egg coat protein VERL and their interaction with cognate sperm protein lysin. VERL repeats fold like the functionally essential N-terminal repeat of mammalian sperm receptor ZP2, whose structure is also described here. Whereas sequence-divergent repeat 1 does not bind lysin, repeat 3 binds it non-species specifically via a high-affinity, largely hydrophobic interface. Due to its intermediate binding affinity, repeat 2 selectively interacts with lysin from the same species. Exposure of a highly positively charged surface of VERL-bound lysin suggests that complex formation both disrupts the organization of egg coat filaments and triggers their electrostatic repulsion, thereby opening a hole for sperm penetration and fusion.
Keywords: biological evolution, fertilization, protein interaction domains and motifs, sperm-ovum interactions, X-ray crystallography, zona pellucida, zona pellucida glycoproteins
Graphical Abstract
Highlights
-
•
Sperm-binding repeats of mollusk VERL and mouse ZP2 egg coat proteins fold similarly
-
•
Structures of VERL/lysin complexes reveal the atomic basis of egg coat-sperm recognition
-
•
A medium-affinity VERL repeat selectively binds lysin from the same species
-
•
VERL/lysin recognition suggests a mechanism for sperm penetration through the egg coat
Molecular insights into egg-sperm recognition point to the basis of species-specific fertilization.
Introduction
The first contact between germ cells during fertilization is mediated by the surface of the egg coat. This specialized extracellular matrix, called zona pellucida (ZP) in mammals and vitelline envelope (VE) in non-mammals, must be recognized and penetrated by sperm in order for gamete plasma membrane fusion to occur (Wassarman, 1999). ZP3 and ZP2, the two major subunits of the ZP, have long been implicated in the initial interaction between mammalian egg and sperm (Bleil et al., 1981, Wassarman, 1999). In particular, recent studies suggested that the N-terminal region of ZP2 crucially regulates gamete recognition in mouse and human (Avella et al., 2014, Gahlay et al., 2010). However, although significant progress was recently made in elucidating plasma membrane contacts essential for the fusion of mammalian gametes (Aydin et al., 2016, Ohto et al., 2016, Bianchi et al., 2014, Inoue et al., 2005), the identity of possible counterpart molecules of ZP2 and ZP3 on sperm remains elusive.
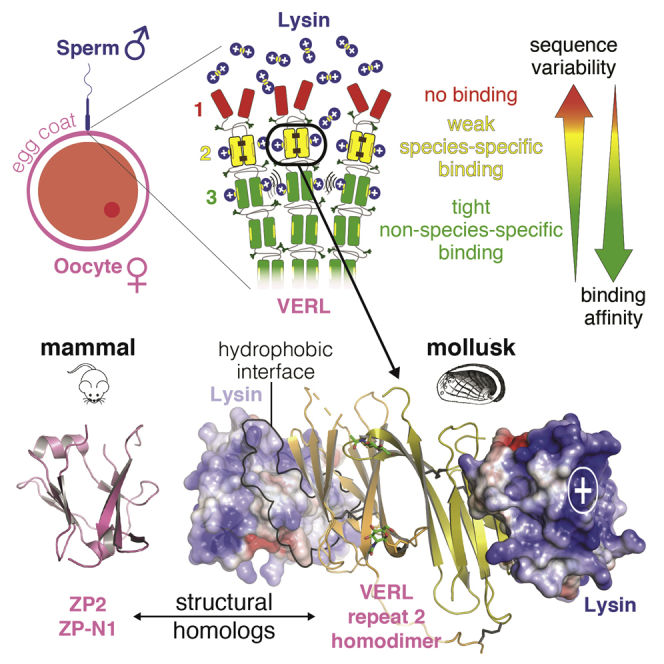
Seminal studies of fertilization in the marine mollusk abalone, a classic invertebrate model system, identified sperm acrosomal protein lysin and egg coat subunit VE receptor for lysin (VERL) as the molecular partners responsible for the initial recognition between gametes (Lewis et al., 1982). Despite the overlapping habitats and breeding seasons, hybrids of free-spawning abalone species rarely occur, and lysin and VERL are thought to play a major role in this phenomenon by ensuring the species-specificity of fertilization (Swanson and Vacquier, 1997). Non-glycosylated 16 kDa lysin, which is released by sperm upon acrosome reaction and dissolves the VE non-enzymatically, was the first fertilization protein to be crystallographically determined (Shaw et al., 1993). In contrast, heavy glycosylation of 2 MDa VERL—whose mass is 50% carbohydrate (Swanson and Vacquier, 1997)—prevented structural investigations of its large N-terminal sperm-binding region, which consists of an array of 22 sequence repeats (VR1–22) spaced by linkers (Galindo et al., 2002) (Figure 1A, top). As a result of rapid evolution, lysin and VERL repeats 1 and 2 are under strong positive selection, while the remaining repeats 3–22 evolve neutrally (Galindo et al., 2003). How sequence variability affects VE dissolution as a result of the interaction between lysin and individual VERL repeats is unknown.
Figure 1.
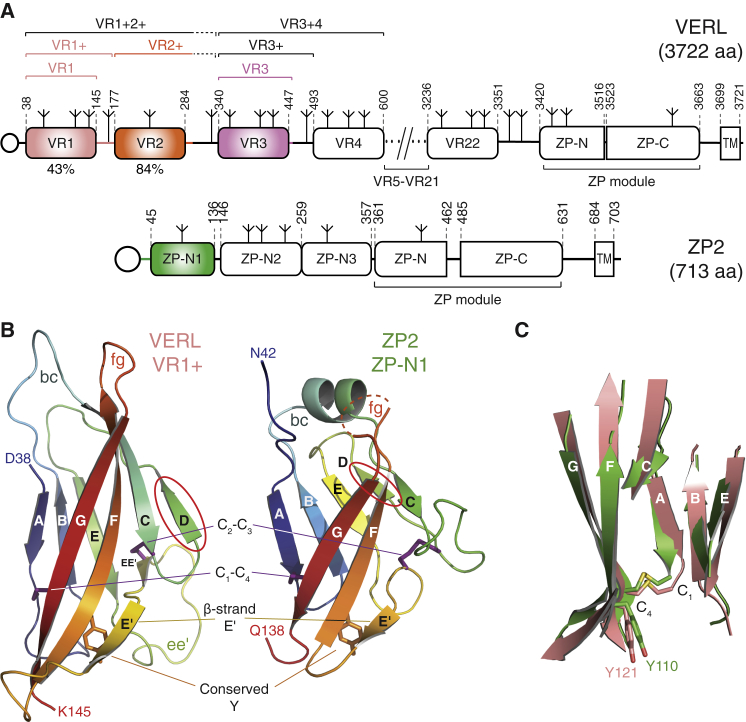
The N-Terminal Repeats of Mollusk VERL and Mammalian ZP2 Adopt a Common ZP-N Fold
(A) Domain organization of red abalone VERL and mouse ZP2. Construct boundaries are marked by brackets and detailed in the Key Resources Table; regions corresponding to constructs whose structure was determined in this work are colored. Open circles and inverted tripods represent signal peptides and N-glycans, respectively. TM, transmembrane domain. Percentage sequence identity between VR1 or VR2 and VR3 is indicated below the respective domains; repeats 3–22 are ∼99% identical.
(B) Crystal structures of red abalone VERL VR1+ and mouse ZP2 ZP-N1, shown in cartoon representation and rainbow-colored from blue (N terminus) to red (C terminus). ZP-N domain features are highlighted, with the two invariant disulfides with 1-4, 2-3 connectivity and the conserved Tyr residue in β strand F shown as dark magenta and orange sticks, respectively. The partially disordered fg loop of ZP2 is represented by a dashed line. A red ellipse marks β strand D, which belongs to different sheets in the two structures. N- and C-terminal residues defined in the electron density are indicated.
(C) Structural comparison of VR1+ (salmon) and ZP-N1 (green) identifies a core of 52 residues that can be superimposed with a Cα RMSD of 1.9 Å.
Interestingly, the functional N-terminal region of ZP2 also evolves under positive Darwinian selection (Swanson and Vacquier, 2002, Swanson et al., 2001) and was suggested to consist of three repeated domains that fold like the ZP-N moiety of the “ZP domain”—a polymerization module conserved at the C terminus of all egg coat proteins, including ZP2 and VERL (Callebaut et al., 2007, Han et al., 2010, Monné et al., 2008) (Figure 1A, bottom). More recently, VERL repeats were also hypothesized to adopt a ZP-N fold (Swanson et al., 2011); however, because VERL and ZP2 repeats are only 10% identical in sequence, this possibility remained highly speculative.
Here, we provide definitive experimental proof that the functional regions of ZP2 and VERL are structurally homologous, and, by determining crystallographic structures of VERL/lysin complexes, we reveal the first atomic details of how sperm recognizes the egg coat. Together with biochemical and mutational studies, our findings bring insights into the molecular basis of species-specific gamete interaction and sperm penetration through the egg coat.
Results
A Common Fold Links the Sperm-Interacting Regions of Invertebrate and Vertebrate Egg Coat Proteins
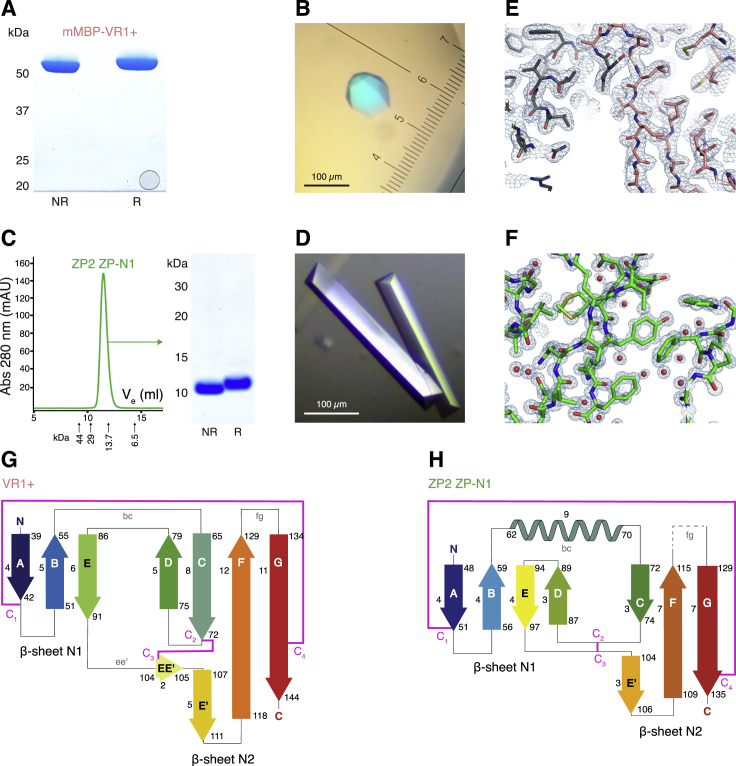
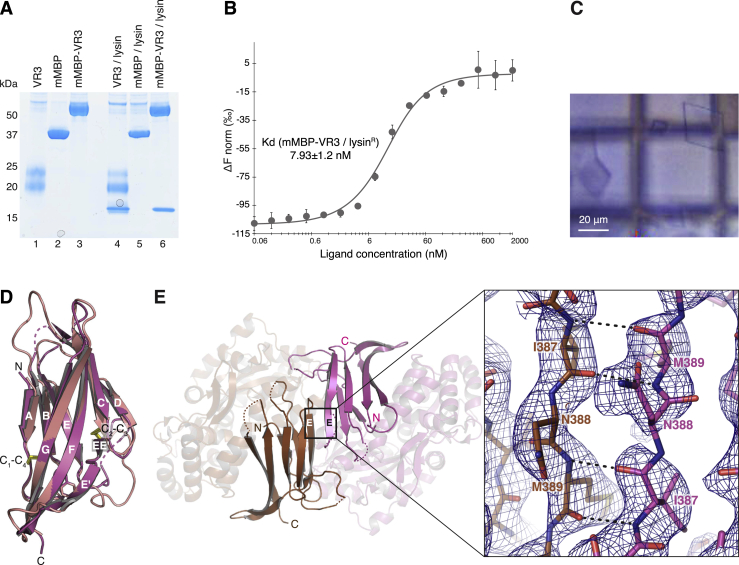
To investigate whether VERL and ZP2 are structurally related, we expressed in mammalian cells VR1+, a construct including red abalone VERL repeat 1 and the 30-residue linker between repeats 1 and 2 (residues D38–I176; Figure 1A). Although deglycosylated VR1+ did not crystallize, mammalianized maltose-binding protein (mMBP) fusions of an equivalent fragment carrying N-glycosylation site mutations (Bokhove et al., 2016) and a variant lacking most of the linker region (VR1; residues D38–A151) yielded crystals diffracting to 2.0 and 1.8 Å resolution, respectively (Figures S1A and S1B; Table S1). In parallel, crystals of an unfused construct including the first putative ZP-N domain of mouse ZP2 (residues V35–Q138; Figure 1A) were obtained, that diffracted beyond 1 Å resolution (Figures S1C and S1D; Table S2).
Figure S1.
Structure Determination of mMBP-VR1+ and ZP2 ZP-N1, Related to Figure 1
(A) Coomassie-stained SDS-PAGE analysis of purified mMBP-VR1+. The relative shift in migration between samples run in non-reducing (NR) and reducing (R) conditions indicates presence of disulfide bonds.
(B) Crystal of mMBP-VR1+.
(C) SEC suggests that purified ZP2 ZP-N1 is a monomer. The elution volume (Ve) of standard markers is indicated. Right panel, SDS-PAGE analysis of the SEC peak.
(D) Crystals of ZP2 ZP-N1.
(E and F) 2mFo-DFc electron density maps contoured at 1 σ of mMBP-VR1+ (gray, mMBP; salmon, VR1+) and ZP2 ZP-N1 (green).
(G and H) Topology diagrams of VR1+ and ZP2 ZP-N1. Secondary structure elements are colored as in Figure 1B and their boundaries and length are indicated; conserved ZP-N domain disulfides C1-C4 and C2-C3 are represented by magenta lines.
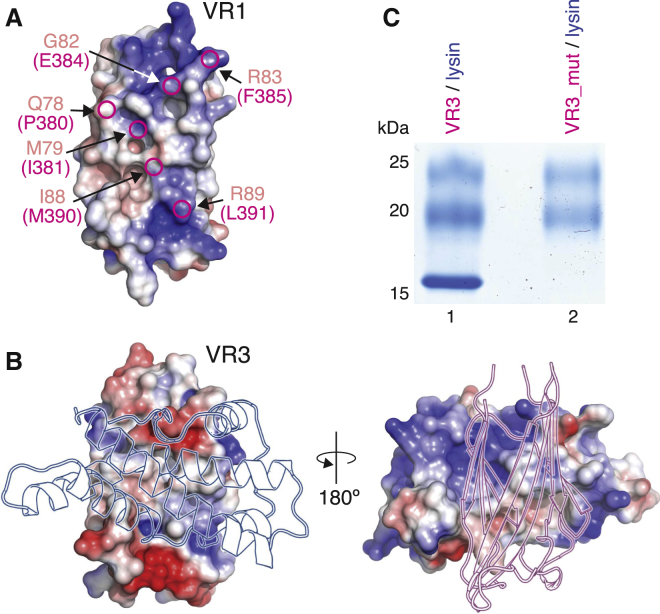
The structures of VR1+ and ZP-N1 were independently solved by molecular replacement (MR) with MBP and sulfur-single-wavelength anomalous dispersion (S-SAD), respectively (Figures S1E and S1F), revealing a common immunoglobulin-like fold with all the hallmarks of the ZP-N domain (Han et al., 2010, Monné et al., 2008) (Figure 1B). The two structures are highly similar, and their β sandwich cores can be superimposed with a root-mean-square deviation (RMSD) of 1.9 Å (Figure 1C). Notable differences are an α helix in the bc loop of ZP2 and the position of β strand D. The latter belongs to sheet N1 in ZP2 but is displaced to sheet N2 in VR1+, with an almost 90-degree rotation of disulfide C2-C3 compared to the one in ZP2 (Figures 1B, S1G, and S1H). VR1+ also has a much shorter fg loop than ZP3 ZP-N, where this loop mediates protein homodimerization by interacting with the ZP-C moiety of a second molecule (Han et al., 2010). Moreover, as a result of structural flexibility, VERL repeat 1/repeat 2 linker residues S146–L175 are not defined in the electron density, so that the structures of VR1+ and VR1 are essentially identical.
VERL Repeats 2 and 3, but Not Highly Sequence-Divergent Repeat 1, Form Gamete Recognition Complexes with Lysin
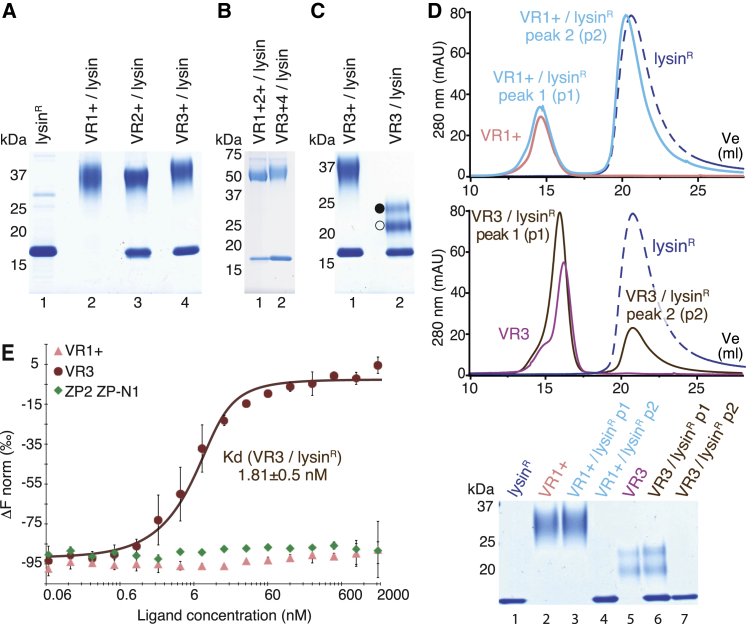
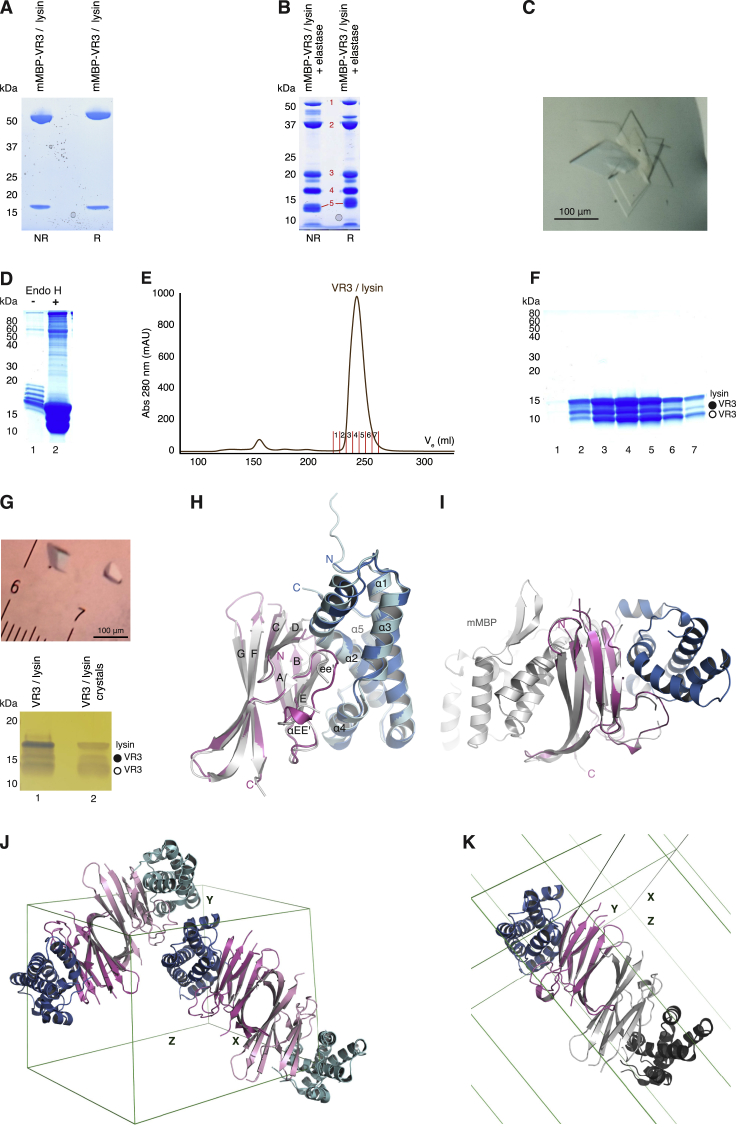
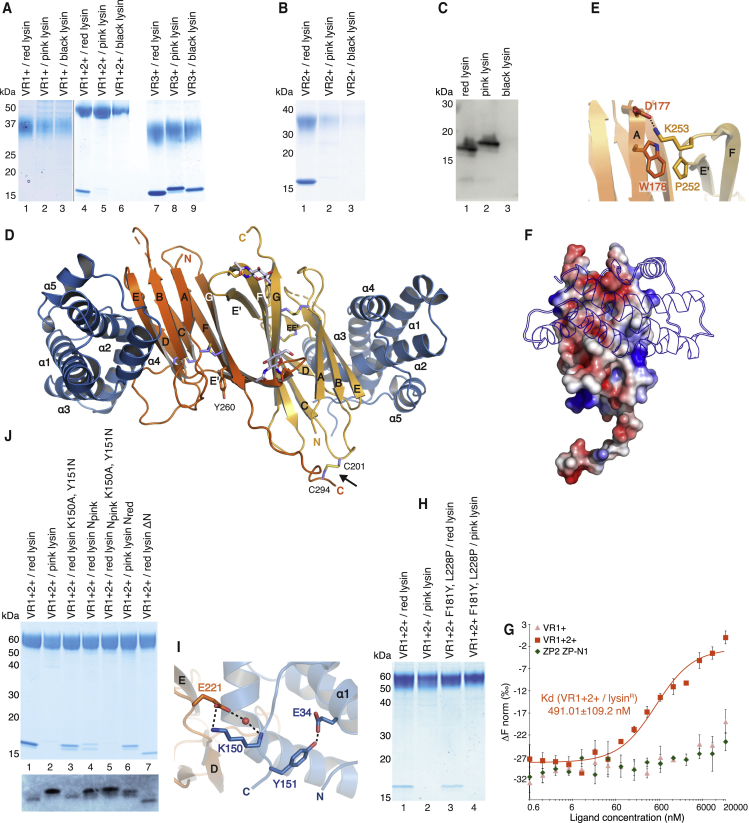
Previous studies suggested that abalone VE dissolution depends on direct interaction between lysin and VERL (Swanson and Vacquier, 1997). However, it is unknown if lysin binds equally to positively selected VERL repeats 1 and 2, which are 43% and 84% identical to repeat 3, and repeats 3–22, which are almost identical in sequence because of concerted evolution (Galindo et al., 2002, Galindo et al., 2003). To clarify this point, His-tagged VR1+ and equivalent constructs encompassing VERL repeats 2 and 3 were co-expressed with untagged lysin in mammalian cells. His-pull-down of conditioned media shows that VR2+ and VR3+, but not VR1+, bind lysin (Figure 2A). Accordingly, co-expression of double-repeat constructs show that VR1+2+ binds roughly half the amount of lysin as VR3+4 (Figure 2B). Moreover, a VR3 protein lacking the interdomain linker region that is disordered in the VR1+ structure pulls down lysin as efficiently as VR3+ (Figure 2C), suggesting that the linker is not involved in complex formation.
Figure 2.
Identification of a Minimal, High-Affinity VERL/Lysin Complex
(A–C) Coomassie-stained SDS-PAGE analysis of pull-down experiments of lysin co-transfected with different His-tagged VERL constructs. Lane 1 in (A) is a refolded lysin control. Open and closed circles in (C) indicate VR3 isoforms resulting from alternative signal peptide cleavages (see STAR Methods).
(D) SEC analysis of purified VR1+ (top) and VR3 (middle) by themselves and mixed with lysinR in a 1:2 ratio. Peaks are normalized to 80 mAU. Bottom: SDS-PAGE analysis of SEC peak fractions.
(E) MST analysis of lysinR interaction with VR1+, VR3, and negative control ZP2 ZP-N1. Results are shown as mean ± SD.
See also Figure S2.
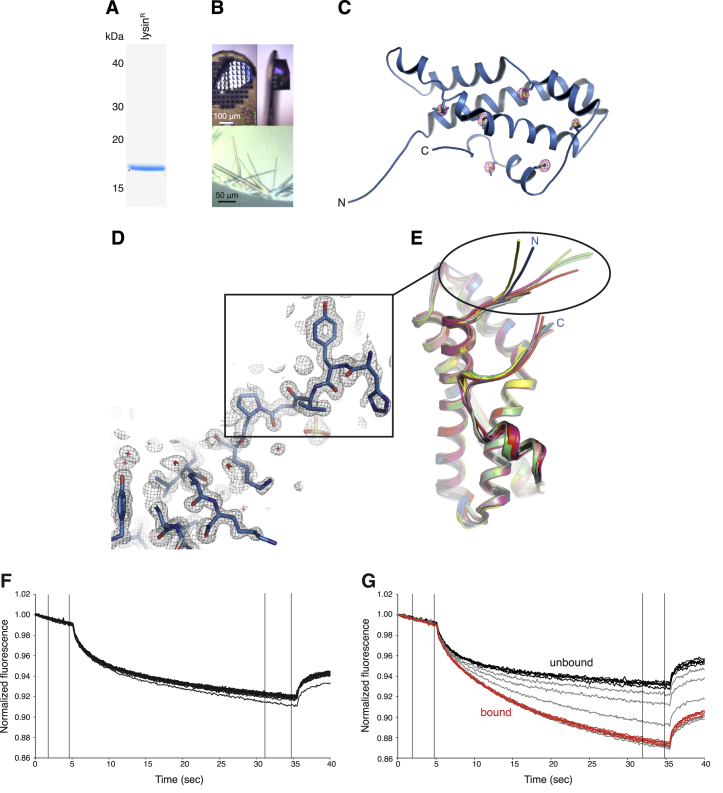
To biochemically confirm pull-down experiments, we used bacterially expressed lysin (lysinR). After refolding, this crystallizes as native lysin (Lyon and Vacquier, 1999, Shaw et al., 1993), and de novo determination of its structure by in-house S-SAD yielded a model that was refined to 0.99 Å using synchrotron data collected from dehydrated crystals (Figures S2A–S2D; Table S3). As also shown by a previously unreported 2.11 Å monoclinic crystal form (Figure S2B, bottom; Table S3), the five-helical bundle structure of lysinR is essentially identical to that of its native counterpart (Kresge et al., 2000, Shaw et al., 1993) (Figure S2E). Size-exclusion chromatography (SEC) shows that lysinR co-elutes with VR3 but not VR1+ (Figure 2D). Moreover, microscale thermophoresis (MST) (Wienken et al., 2010) indicates that lysinR binds VR3 with nanomolar affinity, whereas no interaction is detected with VR1+ even at 0.1 mM concentration (Figures 2E, S2F, and S2G).
Figure S2.
Structure Determination and Functional Analysis of LysinR, Related to Figure 2
(A) Coomassie-stained SDS-PAGE analysis of purified lysinR.
(B) Orthorhombic (top panels) and monoclinic (bottom panel) crystals of lysinR.
(C) The sulfur atoms of 6 Met residues are identified by a 5 σ anomalous difference map of lysinR (red mesh), which is shown superimposed onto the refined model of the protein.
(D) Detail of the hypervariable N-terminal region of lysinR in the refined 0.99 Å resolution 2mFo-DFc electron density map of the protein, contoured at 1 σ.
(E) Comparison of chains from different structures highlights the flexibility of lysin N terminus. Blue, orthorhombic lysinR; shades of green, chains of monoclinic lysinR; shades of gray, chains of PDB: 1LYN; yellow, PDB: 2LIS; shades of red, chains of PDB: 2LYN.
(F and G) MST analysis of the interaction of lysinR with different VERL repeats. MST time traces obtained by titrating 10 nM labeled lysin with increasing concentrations of VR1+ up to 0.1 mM (F) and VR3 up to 2 μM (G). Traces corresponding to bound and unbound states are colored red and black, respectively, whereas traces corresponding to partially bound intermediates are shown in gray. VR3 binds to lysin with high affinity, but no binding is detected to VR1+.
Together, these results show that lysin binds both VR2+ and VR3+, but not VR1+, and identify VR3 (and possibly VR2) as a minimal VERL fragment that forms a high-affinity complex with lysin in vitro. Considering that VR3 is ∼99% identical in sequence to repeats 4–22 (Galindo et al., 2002), we pursued this repeat as a general example of VERL/lysin interaction.
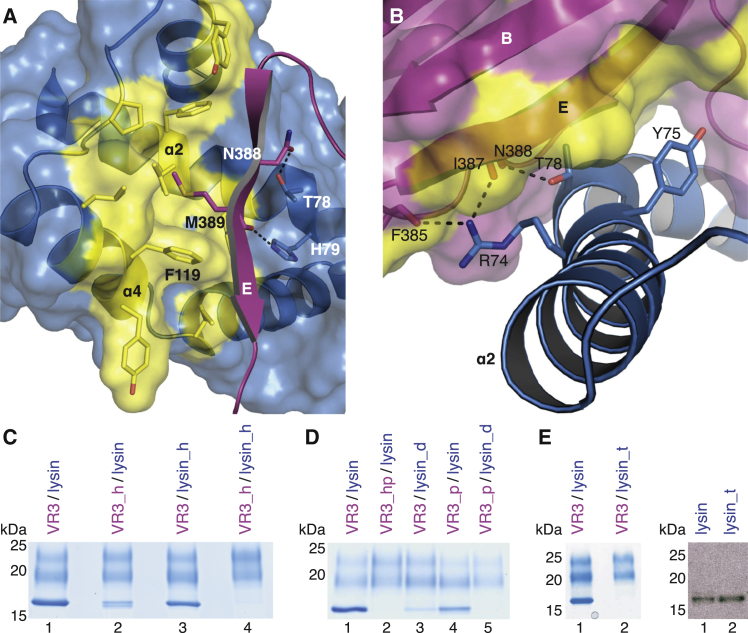
Crystal Structures of VR3 and the VR3/Lysin Complex
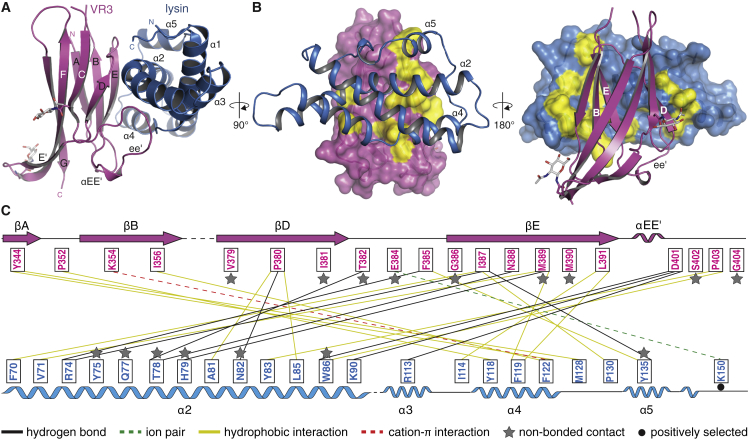
To reveal the molecular basis of gamete interaction and understand how lysin discriminates between VERL repeats 1 and 3, we first crystallized an mMBP-VR3 fusion that binds lysin with the same affinity as unfused VR3 (Figures S3A–S3C). The resulting 2.9 Å resolution structure of unbound VR3, solved by MR with mMBP (Table S1), is highly similar to that of VR1 (RMSD 0.9 Å; Figure S3D). Subsequently, we obtained triclinic crystals of a VR3/lysin complex generated by limited proteolysis of mMBP-VR3 co-expressed with lysin (Figures S4A–S4C) as well as trigonal crystals of unfused VR3 bound to lysin (Figures S4D–S4G). The two complex structures, determined at 1.7 Å and 1.64 Å resolution by MR with VR3 and lysin (Table S4), are essentially identical despite different crystal growth conditions and packing. The proteins form a 1:1 complex (Figure 3A), held together by an extensive contiguous interface burying an average accessible surface area of 1148 Å2. This involves about 20 residues from each binding partner, mostly located in βB, βD-βE, loop ee′ of VR3 and α2, α4-α5 of lysin (Figures 3 and S5; Table S5). Except for VR3 loop ee′, which is partially disordered in the free protein but leans against lysin α2 in the complex, the proteins do not change structure upon binding (Figure S4H). Thus, abalone gamete recognition essentially consists in rigid-body docking of lysin onto VERL.
Figure S3.
mMBP-Fused VR3 Binds Lysin and Crystallizes with Antiparallel Pairing of the E Strands of Two Molecules, Related to Figures 3 and 7
(A) Coomassie-stained SDS-PAGE analysis of His-pull-down experiments of individually expressed VR3, mMBP and mMBP-VR3 (lanes 1-3), as well as of the same constructs co-expressed with lysin (lanes 4-6). mMBP-VR3 and unfused VR3 bind lysin comparably. mMBP was used as negative control.
(B) MST determination of mMBP-VR3/lysinR Kd shows that unfused and mMBP-fused VR3 bind to lysinR with similar affinity (compare with Figure 2E). Results are shown as mean ± SD.
(C) Microcrystals of mMBP-VR3 on the mesh used for diffraction data collection.
(D) VR3 (dark pink) and VR1 (salmon) can be superimposed over 81 residues with a Cα RMSD of 0.9 Å. Conserved ZP-N disulfide bonds are shown as yellow sticks; disordered loops are represented by dashed lines.
(E) The asymmetric unit of the mMBP-VR3 crystal contains an antiparallel VR3 homodimer held together by interactions involving the β sheets E of opposite chains. The resulting interface is scored as highly significant by PISA (Krissinel and Henrick, 2007) and buries an average accessible surface area of 646 Å2. The overall arrangement of the two mMBP-VR3 chains is depicted in the left panel, and details of the 1 σ 2mFo-DFc electron density map of the interface region are shown in the right panel. Dashed lines indicate intermolecular hydrogen bonds.
Figure S4.
Purification and Crystallization of Elastase-Treated (mMBP-)VR3/Lysin and Unfused VR3/Lysin, Related to Figures 3 and 4
(A) Coomassie-stained SDS-PAGE analysis of purified mMBP-VR3/lysin complex material.
(B) Treatment of mMBP-VR3 lysin with elastase produces five major protein bands, indicated by red numbers. Based on immunoblot analysis (data not shown), digestion products are intact mMBP-VR3 (1), mMBP (2), partially degraded mMBP (3), lysin (4) and VR3 (5).
(C) Crystals obtained using elastase-treated mMBP-VR3/lysin material.
(D) Coomassie-stained SDS-PAGE of IMAC-purified VR3/lysin complex from HEK293S cells, before and after Endo H treatment.
(E) SEC of the Endo H-treated VR3/lysin complex shows that it elutes as a single peak, which was collected into 7 fractions.
(F) Coomassie-stained SDS-PAGE analysis of the fractions of the VR3/lysin complex SEC peak. Lane numbers match the peak fraction numbers in (E). Open and close circles indicate VR3 isoforms resulting from alternative signal peptide cleavages (see STAR Methods).
(G) Trigonal crystals of the VR3/lysin complex (top) and silver-stained SDS-PAGE analysis of dissolved crystals (bottom). In lane 1, 2 μg of purified VR3/lysin were loaded; lane 2 shows material from 5 crystals of the complex.
(H) The structure of mMBP-fused VR3 (gray) and orthorhombic lysinR (cyan) can be superimposed on the corresponding moieties of the VR3/lysin complex (dark pink and blue) with a Cα RMSD of 0.7 Å and 0.3 Å, respectively. This comparison indicates that no significant structural change occurs during complex formation. The mMBP moiety of mMBP-VR3 is not shown for simplicity.
(I) Superimposition of the VR3 moieties of mMBP-VR3 (gray) and the VR3/lysin complex (dark pink) shows that mMBP (light gray) lies on the opposite side of VR3 as lysin (blue). This explains why mMBP fusion does not interfere with complex formation between VR3 and lysin.
(J) The asymmetric unit of the triclinic crystal form contains two homodimers of VR3 (dark and light pink), each of which is bound to two molecules of lysin (blue and cyan). The unit cell is shown in green and crystal axes are indicated.
(K) The asymmetric unit of the trigonal crystal form contains a single copy of the VR3/lysin complex (dark pink/blue), whose VR3 moiety is engaged in the same homodimer observed in the triclinic crystals by interacting with a symmetry-related VR3/lysin complex (light and dark gray). Unit cells and crystal axes are depicted as in (J).
Figure 3.
Molecular Basis of VR3/Lysin Interaction
(A) Cartoon representation of the VR3 (dark pink)/lysin (blue) complex, with N-glycans shown as sticks. The trigonal structure is shown here and subsequent figures.
(B) Surface representations of VR3 (left) and lysin (right), highlighting hydrophobic regions (yellow) at the interface with the respective partners (cartoon). Main secondary structure elements involved in the interaction are marked.
(C) Scheme of the interface. Interactions are represented by lines connecting residues and color coded by property as indicated below the panel.
See also Figure S3, Figure S4, Figure S5, Figure S6 and Tables S4 and S5.
Figure S5.
Sequence Comparison of VERL Repeats and Lysin from Different Haliotis Species, Related to Figures 3 and 6
(A) Multiple sequence alignment, showing the intra- and interspecies similarity of VR1, VR2 and VR3. Sequences are listed in order of decreasing similarity to red abalone VERL. Secondary structure, based on the crystal structures of red abalone VR1+, VR2+ (from chain B of the VR2+/lysin complex) and VR3 (from the trigonal VR3/lysin complex), is depicted on top; consensus sequences are shown at the bottom (cons., brown). Conserved ZP-N Cys are marked and their connectivity is indicated (magenta). Interface residues of the VR3/lysin and VR2+/lysin complexes are indicated by red boxes; corresponding positions in VR1 are outlined by dashed red boxes. Hydrophobic interface residues are highlighted in yellow. Positively selected residues (Galindo et al., 2003, Kresge et al., 2001) are marked by black circles above the alignment. Abalone species abbreviations are: RUF, H. rufescens (red); SOR, H. sorenseni (white); WAL, H. walallensis (flat); KAM, H. kamtschatkana (pinto); GIG, H. gigantea (giant); DIS, H. discus hannai (Japanese); COR, H. corrugata (pink); CRA, H. cracherodii (black); FUL, H. fulgens (green); DIV, H. diversicolor (variously colored); AUS, H. australis (Australian); VAR, H. varia (variable); OVI, H. ovina (sheep’s ear); ROE, H. roei (Roe’s); RUB, H. rubra (blacklip); CON, H. conicopora; SCA, H. scalaris (staircase); LAE, H. laevigata (greenlip); PUS, H. pustulata; MID, H. midae (perlemoen); CYC, H. cyclobates (whirling); IRI, H. iris (paua); TUB, H. tuberculata (green ormer). Californian species (Lee and Vacquier, 1995) are marked by an asterisk next to their name abbreviation. Consensus symbols: uppercase, high consensus (> 90%); lowercase, low consensus (> 50%); !, I or V; $, L or M; %, F or Y; #, N or D or Q or E.
(B) Multiple sequence alignment of lysin. Conventions are as in (A), with interface residues of the VR3/lysin and VR2+/lysin complexes indicated by open red boxes in the alignment and closed red boxes below the consensus line, respectively. The secondary structure of red abalone lysin (from the trigonal VR3/lysin complex) is displayed, and solvent-exposed basic residues that do not interact with VR3 or VR2+ in the complex structures are highlighted in blue.
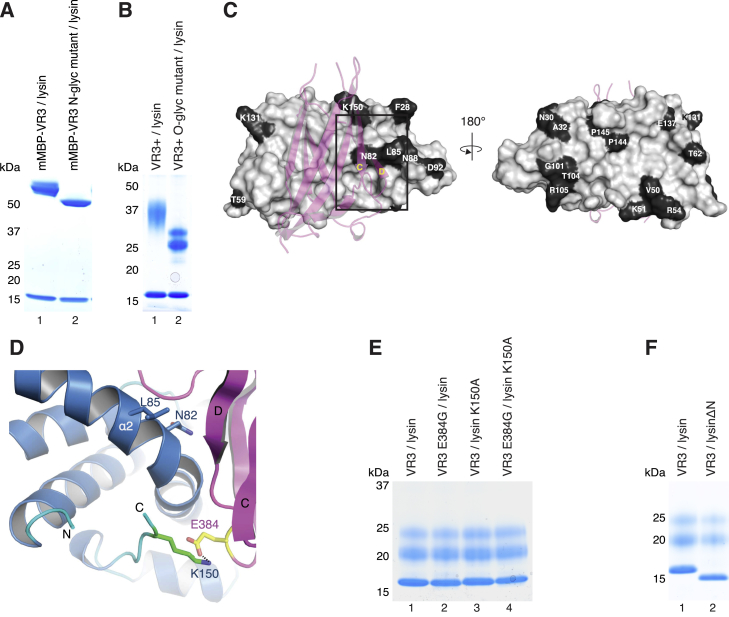
The mode of interaction explains why mMBP fusion of VR3 does not interfere with its ability to bind lysin (Figure S4I). Moreover, since lysin is not glycosylated (Lewis et al., 1982, Shaw et al., 1993) and all VR3 N-glycans are on the opposite side of the interface (Figure 3A), the structure indicates that complex formation does not depend on N-glycosylation of VERL. This is confirmed by the functional characterization of a VR3 construct where all N-glycosylation sites are mutated, which still binds lysin (Figure S6A). Although the electron density maps of free and complexed VERL repeats do not show evidence of O-glycosylation, this post-translational modification is likely to contribute significantly to the carbohydrate composition of VERL (Swanson and Vacquier, 1997) and was hypothesized to play a role in the interaction with lysin (Gallo and Costantini, 2012). Consistent with the high sequence conservation of VERL interdomain linkers (Swanson et al., 2011) and the aforementioned structural flexibility of the C-terminal region of VR1+, mutational analysis suggests that the linker of VR3+ is heavily O-glycosylated. However, in agreement with the observation that VR3+ and VR3 bind lysin with comparable efficiency (Figure 2C), inactivation of linker O-glycosylation sites does not affect VR3+/lysin interaction (Figure S6B).
Figure S6.
Glycans and Positively Selected Residues Do Not Influence VR3/Lysin Complex Formation, Related to Figure 3
(A and B) VR3 or VR3+ constructs carrying mutations of N-glycosylation sites (N373Q, N417Q, N438Q) or interdomain linker O-glycosylation sites (T456G, T459G, S460G, T467A, T470G, S471G, S472G, S478G, S479G, S485G, S489G) are significantly less glycosylated than counterpart wild-type proteins, but bind lysin equally well.
(C) Surface representation of lysin bound to VR3 (depicted as dark pink cartoon), with positively selected residues highlighted in black. Only N82, L85 and K150 lie at the complex interface.
(D) Close-up of the trigonal VR3/lysin complex, with the three positively selected residues of lysin at the interface with VR3 shown as sticks. Whereas N82 and L85 are conserved among all Californian species of abalone, K150 - which makes an ion pair with VR3 E384 - is variable. Note how the N-terminal region of lysin, which is only defined in the electron density map from F28 onward, is positioned away from interface with VR3.
(E) Disruption of the lysin K150/VR3 E384 ion pair by individual or combined mutation of the interacting residues does not hinder complex formation.
(F) A lysin mutant lacking hypervariable N-terminal residues R19-L29 is pulled down by VR3 as efficiently as wild-type lysin.
The VERL/lysin interface is largely hydrophobic in nature (Figures 3B and 3C) but also involves hydrogen bonds between VR3 βE and lysin α2 (Figure 3C). Consistent with a high-affinity interaction (Figure 2E), complex formation is not abolished by individual replacement of VR3 M389 or lysin F119—two residues that interact hydrophobically (Figure 4A)—with polar counterparts (Figure 4C, lanes 2 and 3). However, binding is lost when these mutations are combined (Figure 4C, lane 4). Similarly, whereas VR3 N388A or lysin T78A, H79A mutations reduce binding to different extents (Figure 4D, lanes 4 and 3), and their combination totally abolishes VERL/lysin interaction (Figure 4D, lane 5) by breaking the hydrogen bond network (Figure 4A). Complex formation is also disrupted by the combination of VR3 mutations M389K and N388A (Figures 4A and 4D, lane 2) or a triple mutant of lysin impairing the α2/VR3 βE interface (Figures 4B and 4E, lane 2). Together, these results support a lock-and-key recognition mechanism involving an extended surface area.
Figure 4.
VR3/Lysin Complex Formation Depends on Both Hydrophobic and H-Bonding Interactions
(A) Close up of the VR3/lysin interface, oriented as in the right panel of Figure 3B. Functionally important residues and surrounding hydrophobic amino acids are in stick representation. Only β strand E of VR3 is shown for clarity. Dashed black lines indicate hydrogen bonds.
(B) Lysin α2 interface residues mutated in construct lysin_t (R74A, Y75A, T78A).
(C–E) Functional analysis of VR3 and lysin interface mutants by His-pull-down. VR3_h, M389K; VR3_p, N388A; VR3_hp, N388A, M389K; lysin_h, F119S; lysin_d, T78A, H79A. Right panel of (E), anti-lysin immunoblot shows that individually expressed lysin_t is secreted as efficiently as wild-type lysin.
See also Figures S4 and S5 and Tables S4 and S5.
Molecular Basis of VERL Repeat Discrimination by Lysin
Conservation of VR3 interface residues (Figure S5A) explains why positively selected VR2 also binds lysin (Figure 2A). On the contrary, six interface residues of VR3 are different in VR1 (Figure 5A and Figure S5A), which is also less electrostatically complementary to basic lysin than relatively acidic VR3 (Figure 5B). This rationalizes why VR1 does not bind lysin (Figure 2), as confirmed by loss of complex formation upon introduction of the VR1 substitutions into VR3 (Figure 5C).
Figure 5.
Substitution of VR3 Interface Residues Explains Why VR1 Does Not Bind Lysin
(A) VR1 surface colored by electrostatic potential. Circles mark non-positively selected amino acids of VR1 (salmon) that differ from VR3 residues (magenta) at the interface with lysin.
(B) Unlike the corresponding region in VR1 (A), the interacting surface of VR3 (left) is electrostatically complementary to that of lysin (right).
(C) A VR3 mutant where the residues highlighted in (A) are replaced by the corresponding residues of VR1 (VR3_mut, lane 2) does not pull down lysin.
VR2 Is Important for the Species-Specificity of VERL/Lysin Interaction
In vitro studies of VE dissolution by lysin show a significant degree of species-specificity (Lyon and Vacquier, 1999, Swanson and Vacquier, 1997), and rapid evolution of lysin and VERL by positive selection was suggested to play an important role in this process by affecting gamete recognition (Lyon and Vacquier, 1999, Swanson and Vacquier, 2002). Interestingly, the aforementioned differences between VR1 and VR3 exclusively involve non-positively selected residues (Figures 5A and S5A) (Galindo et al., 2003). Moreover, only 3 of the 23 positively selected amino acids in lysin (Kresge et al., 2001) contribute to the VR3/lysin interface (Figures S5B and S6C). Of these, only K150, which makes a peripheral ion pair with VR3 E384 (Figure S6D), differs in three of the seven Californian abalone species spawning in the same area as red abalone (Figure S5B). VR3 mutation E384G, lysin mutation K150A, or their combination have no effect on binding (Figure S6E). Furthermore, the structure of the complex shows that the hypervariable N-terminal region of lysin, which adopts different conformations in structures of the free protein (Figure S2E) and was implicated in species-specific VE dissolution (Lyon and Vacquier, 1999), is largely disordered and makes no contacts with VR3. Accordingly, deletion of N-terminal lysin residues R19–L29 has no effect on VR3 binding (Figure S6F). Collectively, these results suggest that positive selection does not affect the high-affinity interaction between lysin and VR3 (or, by inference, VR4–VR22).
To understand the basis of species-specificity, we co-expressed different repeats of red abalone VERL with lysins from red, pink, and black abalone (Figures 6A and 6B). Whereas VR1+ does not bind lysin from any species—excluding a possible decoy function for this repeat—VR3+ binds comparably to red and pink lysins. Moreover, VR3+ rescues secretion of black lysin, which, unlike the other lysins, is not secreted by mammalian cells when expressed by itself (Figure 6C). Remarkably, parallel experiments performed with VR2+ or VR1+2+ (a protein that was used for subsequent functional studies because, unlike VR2+, it is highly secreted also when expressed individually), showed that these constructs bind red lysin much more efficiently than pink lysin; moreover, neither VR2+ nor VR1+2+ rescue secretion of black lysin (Figures 6B and 6A, respectively). Together, these findings suggest that VR2+ plays a major role in determining the species-specificity of red abalone VERL/lysin interaction.
Figure 6.
VR2/Lysin Recognition Contributes to the Species-Specificity of Gamete Interaction
(A–B) His-pull-down analysis of lysins from different species of abalone co-expressed with (A) VR1+, VR1+2+, VR3+ or (B) VR2+ constructs of red abalone VERL. Like VR1+2+ (A, lane 4), VR2+ binds efficiently only to red abalone lysin (B, lane 1); however, different from VR1+2+, VR2+ is not well secreted by mammalian cells when it does not form a complex (B, lanes 2 and 3).
(C) Polyclonal anti-lysin immunoblot analysis of the secretion levels of individually expressed lysins.
(D) Cartoon representation of the crystal structure of the VR2+/lysin complex, with the two moieties of the VR2+ homodimer and lysin colored orange/yellow and blue, respectively. The intermolecular disulfide stabilizing the VR2+ homodimer is indicated by an arrow.
(E) Details of non-covalent interactions mediating VR2+ homodimerization.
(F) As observed in the case of VR3 (Figure 5B), the surface of VR2+ is electrostatically complementary to lysin.
(G) MST analysis of red lysinR interaction with red VR1+2+ and VR1+, as well as negative control ZP2 ZP-N1. Results are shown as mean ± SD.
(H) Amino acid substitutions converting the interface of red VR2 to that of pink do not change the species-specificity of the interaction between VR1+2+ and lysin.
(I) Close-up of the VR2+/lysin complex structure, showing how the C-terminal region of red lysin interacts with the de loop of VR2+.
(J) His-pull-down analysis of VR1+2+ co-transfected with N- and C-terminal variants of red and pink lysin. Bottom: An immunoblot with anti-lysin, showing the secretion levels of the same lysin constructs expressed individually.
See also Figures S5 and S7, and Tables S4 and S6.
Crystal Structures of VR2+ and Its Species-Specific Complex with Lysin
VR2+ contains two additional Cys residues not found in other repeats, C201 and C294, the latter of which lies within the linker region. To maintain this possibly structurally relevant characteristic, we engineered a crystallization construct that included VERL residues I176–P298 and carried Ala mutations of linker O-glycosylation sites S293, S296, and S297. This allowed us to determine a 2.5 Å resolution structure of free VR2+ (Figures S7A–S7C; Table S1), as well as a 1.8 Å structure of the complex between VR2+ and lysin (Figures 6D, S7C, and S7D; Table S4). In both structures, pairs of VR2+ molecules form antiparallel dimers stabilized by contacts between conserved residues in β strand A of one copy and the e′f loop of the other (Figures 6D, 6E, S5, and S7C). Interestingly, the same interactions and quaternary structure are observed in both crystal forms of VR3/lysin (Figures S4J and S4K), but not in the structures of free VR1+, VR1, and VR3, whose A strands pack against mMBP. In addition to these non-covalent contacts, the VR2+ homodimer is secured by intermolecular disulfide bonding of the two additional Cys, C201 in the bc loop and C294 in the interdomain linker (Figure 6D). However, this connection must be very flexible because it is only resolved in one of the two asymmetric unit copies of VR2+/lysin (Figure S7C). The visible VR2+ linker does not contact lysin in the structure, and, as previously observed in the case of VR3+ (Figure 2C), its deletion does not affect lysin binding (data not shown).
Figure S7.
Crystal Structures of VR2+ and the VR2+/Lysin Complex, Related to Figures 6 and 7
(A) Crystal of free VR2+ grown in drops set up with the VR2+/lysin complex.
(B) Silver-stained non-reducing SDS-PAGE analysis of a crystal like the one shown in (A) indicates that this exclusively contains a homodimeric, intermolecularly disulfide-bonded form of VR2+ (orange arrow). Due to Endo H treatment of N-glycans and a shorter linker with O-glycosylation site mutations, the VR2+ construct used for X-ray crystallography has a significantly smaller mass than the corresponding protein used for functional studies (Figures 2A and 6B).
(C) Crystal structure of free VR2+ in cartoon representation (left panel), with the two moieties of the homodimer colored as in Figure 6D. The structure is essentially identical to that of lysin-bound VR2+ (right panel; RMSD 0.7 Å over 156 Cα atoms). As a consequence of the structural disorder of the C-terminal interdomain linker, the two intermolecular disulfide bonds involving C201 and C204 are not resolved in the electron density map of free VR2+ (left panel, thin ovals); similarly, only one of the disulfides is visible in the case of the VR2+/lysin complex (right panel, thick oval), where the linker packs against a symmetry-related copy of lysin (not shown). Disordered linkers and C201/C294 are represented by dotted lines and closed circles, colored according to the respective molecules.
(D) Crystal of the VR2+/lysin complex.
VR2+ is highly similar to VR3 (RMSD 0.6 Å over 83 Cα atoms) and has a comparable surface charge distribution (Figures 6F and 5B); moreover, its complex with lysin is also essentially superimposable on VR3/lysin (average RMSD 1.6 Å over 428–442 Cα atoms). However, the VR2+/lysin interface involves seven residues fewer than VR3/lysin (Figure S5; Tables S5 and S6). Accordingly, computational comparison of the structures indicates that the VR2+/lysin interface has an interaction energy that is ∼20% lower than that of VR3+/lysin. This was confirmed by MST analysis, which showed that—in agreement with the observation that VR1+2+ pulls down lysin less efficiently than VR3+ (Figure 6A, compare lanes 4 and 7)—the affinity of lysinR for VR1+2+ is two orders of magnitude lower than for VR3 (Figures 6G and 2E).
Although red VR2 discriminates its cognate lysin from equally well-expressed pink lysin (Figures 6A–6C), only two of the amino acid differences between red and pink VERL (F181Y, L228P) fall within the VR2 interface with lysin; notably, the former change affects a positively selected site (Figure S5A). Introduction of both substitutions into red VR1+2+ does not hinder red lysin binding nor does it allow VR1+2+ to interact with pink lysin (Figure 6H), suggesting that, in addition to VR2, the species-specificity of VERL/lysin interaction must also involve amino acid differences in lysin.
The Suboptimal Interface of VR2 Amplifies the Effect of Lysin Substitutions, Generating Species-Specificity
Out of 17 lysin residues at the interface with VR2, only positively selected K150 differs between red and pink lysin (Figure S5B). As also observed in the VR3/lysin complex (Figure S6D), K150 makes an ion pair with an invariant Glu residue of VERL (E221 in VR2; Figure 6I). This interaction is reinforced by a water-mediated hydrogen bond between E221 and the main chain nitrogen of K150 itself and facilitated by an intramolecular hydrogen bond between Y151 and E34 that stabilizes the conformation of the C-terminal loop of lysin. Interestingly, Y151 also differs in pink lysin. In agreement with the prediction that the K150A Y151N substitutions found in pink lysin would impair the interaction of the C terminus of lysin with VR2, introduction of both changes into red lysin lowers its binding to VR1+2+ (Figure 6J, lane 3). Moreover, a major reduction in binding is observed upon replacement of the positively selected N-terminal region of red lysin with that of its pink homolog, which differs in 10 positions out of 11 (Figures S5B and 6J, lane 4). A red lysin mutant combining both types of changes is indistinguishable from pink lysin (Figure 6J, lane 5), and, conversely, swapping the N-terminal region of pink lysin with that of red allows it to bind red VR1+2+ (Figure 6J, lane 6). Taken together, these results support the aforementioned VE dissolution assays (Lyon and Vacquier, 1999) and suggest that the hypervariable N-terminal region of lysin contributes significantly to the species-specificity of VR2/lysin interaction. However, in agreement with the observation that deletion of red lysin residues R19–L29 does not disrupt its binding to VR1+2+ (Figure 6J, lane 7), the N termini of the lysin molecules in the asymmetric unit of the VR2+/lysin complex are either disordered or not in contact with VR2+. This suggests that N-terminal sequence of pink lysin interferes with its binding to red VR2.
Remarkably, equivalent mutations of the N- and C-terminal regions of red lysin do not affect its high-affinity interaction with VR3 (Figures S6E and S6F) but impair binding to VR2 (Figure 6J). Considering that changes in the VR2 interface have no effect on lysin binding (Figure 6H), these observations suggest that VR2 contributes to the species-specificity of VERL/lysin interaction by amplifying the effect of amino acid substitutions in lysin (Figure S5) as a consequence of its relatively low binding affinity compared to VR3 (Figures 6G and 2E).
Discussion
More than 100 years ago, it was proposed that fertilization depends on a lock-and-key mechanism between egg and sperm (Lillie, 1914). Our work provides a firm structural bridge between invertebrate and vertebrate fertilization, reveals the first details of how sperm and egg coat interact at the atomic level, and gives insights into the species-specificity of gamete recognition in an experimental model system. At the same time, the data has implications for the higher-order organization of VERL repeats, suggests a mechanism for egg coat penetration by sperm, and has evolutionary and functional relevance for mammalian fertilization.
VERL Repeat Array Architecture Shapes VE Recognition and Dissolution by Lysin
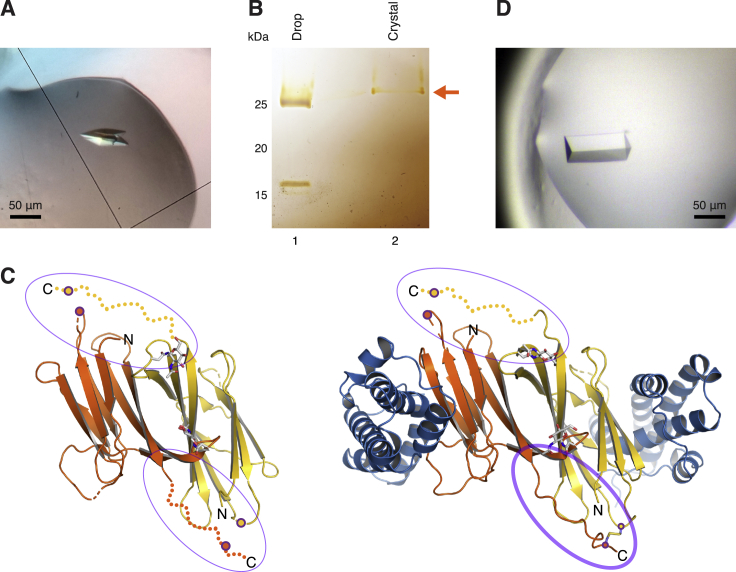
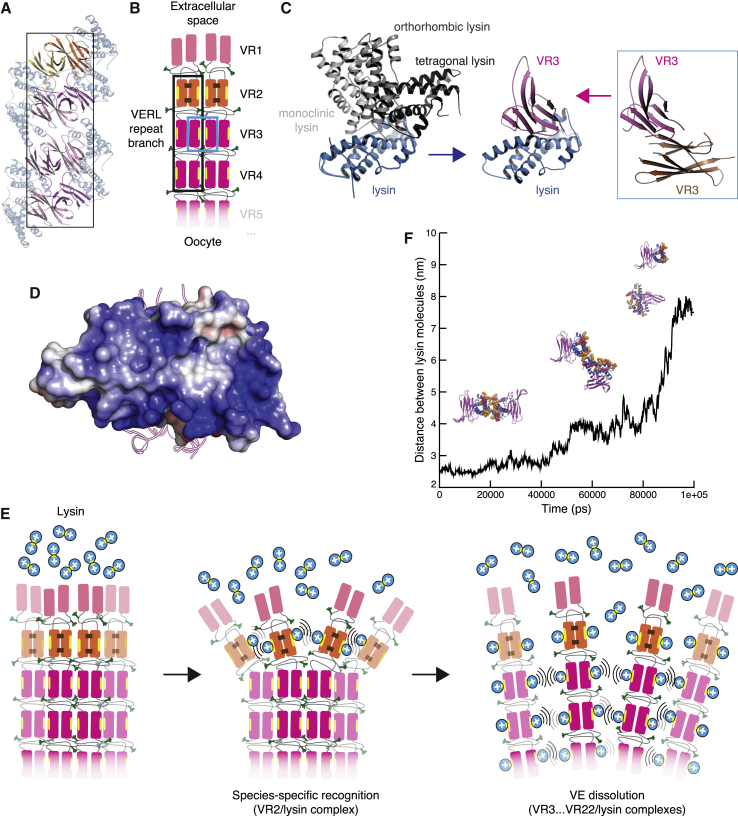
Although O-glycosylation and flexibility of the interdomain linker regions of VERL (Figures S6B and S7C) precluded structural investigation of fragments including more than one repeat, all our structures of non-mMBP-fused constructs show the same antiparallel homodimeric arrangement of repeats (Figures 6D, S4J, S4K, and S7C). Moreover, repeat homodimers recurrently stack to generate filament-like structures running through the crystals (Figure 7A, and data not shown). This suggests that a similar organization may be found in the VE, where an additional structural constraint would be introduced by the VR2/VR2 pair (Figure 6D) whose intermolecular disulfides are consistent with the observation that red abalone VERL is a dimer under non-reducing conditions (Swanson and Vacquier, 1997). Thus, VERL may for example consist of two identical intertwined chains, whose corresponding repeats pair with each other as a result of the register imposed by covalent VR2/VR2. Although alternative models are possible due to the extensive length of the interdomain linkers, stacking of paired repeats would invariantly result in the formation of branches that protrude from the polymeric core of VE filaments toward the extracellular space. Together with the lateral juxtaposition of branches originating from other VERL molecules, this would position VR1 at the surface of the VE, followed by a thin layer of VR2/VR2 repeats and finally a thick layer consisting of stacks of paired near-identical repeats 3–22 (Figure 7B). When combined with the finding that VR2 and lysin interact species specifically with relatively low affinity (Figure 6) whereas VR3 and lysin form a tight non-species-specific complex (Figures 2 and 3 and 6A), this model both rationalizes the different sequence conservation of VERL repeats (Galindo et al., 2003) and provides a structural explanation for the observation that species-specific recognition of the VE precedes its general dissolution (Lyon and Vacquier, 1999).
Figure 7.
The Repeated Structure of VERL Filaments Suggests a Mechanism for Egg Coat Recognition and Penetration by Sperm
(A) Stacked VR2+ homodimers adopt a filament-like arrangement in the VR2+/lysin complex crystal. The boxed area suggests the repeat organization outlined in (B).
(B) Possible architecture of VERL repeat branches. Filled rectangles represent individual repeats, colored as in the previous figures and connected by flexible interdomain linkers (thin gray lines). Brown staples represent VR2+ intermolecular disulfide bonds. Yellow ellipses and green tripods indicate hydrophobic lysin-binding sites and O-glycans, respectively. Stacking of homodimeric VERL repeats within each branch is based on the packing shown in (A) (black rectangle); lateral interaction between repeats of adjacent branches is supported by the VR3/VR3 contacts depicted in (C) (blue rectangle).
(C) Surfaces mediating VR3/lysin interaction (center) make homomeric contacts in crystals of isolated lysin (left) and VR3 (right).
(D) As exemplified by the VR3/lysin complex structure, VERL repeat-bound lysin exposes a highly basic surface to the solvent.
(E) Proposed mechanism of VE recognition and non-enzymatic dissolution by lysin. Positively charged lysin molecules are represented by blue circles, with the hydrophobic VERL repeat-binding site shown as a yellow ellipse.
(F) 100 ns molecular dynamics simulation of two facing lysin molecules bound to VR3 repeats representing adjacent branches of VERL. Strong electrostatic repulsion between the exposed basic surfaces of the lysin molecules causes the complexes to be pushed apart over time. Basic surface Lys and Arg residues are in sphere representation and colored red and orange, respectively.
See also Figures S3 and S7, Tables S1, S3, and S4, and Movie S1.
One question that remains open is the biological function of repeat VR1, which does not bind lysin from the same or other species (Figures 2 and 6A) and is most likely monomeric due to lack of conserved residues mediating homodimerization of VR2 and VR3 (such as the Trp in β strand A and Lys in the e′f loop; Figures S5A and 6E). Considering that VR1 is found in all homologs of VERL so far characterized, possible exposure of this rapidly evolving repeat on the surface of the VE may allow it to play a role in pathogen resistance (Messier and Stewart, 1994)—a process that could then be evolutionarily coupled to the generation of species-specificity at the level of VR2. However, given the biochemical complexity of the abalone VE (Aagaard et al., 2010, Swanson and Vacquier, 1997), a yet-to-be-identified function of VR1 in fertilization cannot be excluded.
Mechanism of Sperm Penetration through the Egg Coat
How does lysin create a hole in the VE for sperm passage? Analysis of tetragonal crystals of the free protein led to the suggestion that the hydrophobic patch of lysin mediates its homodimerization before it contacts the VE (Kresge et al., 2000). Consistent with a weak homodimeric interface, the same region of lysin makes alternative contacts in different crystal forms of the unbound molecule (Figure 7C, left).
Similarly, in crystals of free mMBP-fused VR3, the complementary hydrophobic region of VR3 interacts with another copy of itself via antiparallel pairing of the respective E strands (Figure 7C, right; Figure S3E). Considering the architecture of VERL discussed above, in the VE this kind of interaction could mediate the lateral adhesion of repeats belonging to adjacent VERL branches (Figure 7B).
Most importantly, the crystal structures of lysin in complex with VR2+ and VR3 reveal that the hydrophobic regions of the two counterparts directly mediate their interaction (Figures 3, 4, and 6D). Together with the finding that VERL repeat-bound lysin exposes a highly positively charged surface to the solvent (Figure 7D), these observations support a simple mechanism of non-enzymatic sperm penetration through the egg coat (Figure 7E). Upon sperm acrosome reaction and release of lysin onto the egg coat, the aforementioned homomeric interactions of lysin and VERL are swapped for the more extensive, higher-affinity heteromeric interface observed in our complex structures (Figures 3 and 6D). Consistent with early electron microscopy observations (Lewis et al., 1982), this would disrupt VERL fiber organization—first slowly, at the level of the species-specific VR2 repeat layer, and then rapidly through the stacks of repeats 3–22. At the same time, it would cause adjacent VERL branches to electrostatically repel each other, due to close juxtaposition of the highly basic surface of VERL repeat-bound lysins. In agreement with this possibility, molecular dynamics simulations performed in seawater conditions suggest that repulsion between the basic surfaces of two lysin molecules bound to facing VERL repeats would effectively push them apart (Figure 7F; Movie S1). Moreover, the asymmetric unit contents of the VR2+/lysin complex (Figure 7A), which is computationally scored as highly significant and could mimic a lysin-bound VERL branch, hints that contacts between lysin molecules attached to successive homodimeric VERL repeats may contribute to the cooperativity of VE dissolution (Swanson and Vacquier, 1997). As a result of progressive VERL architecture disruption and electrostatic repulsion between VERL repeat-bound lysins, a hole could form into the egg coat, allowing sperm to penetrate and fuse with the plasma membrane.
Implications for the Evolution of Fertilization and Mammalian Gamete Interaction
Despite being separated by 0.6 billion years of evolution, mollusk and human use a common C-terminal ZP module as a building block for the assembly of their egg coats (Bork and Sander, 1992, Galindo et al., 2002, Han et al., 2010). By showing that the functional N-terminal repeats of VERL and ZP2 also share a common fold (Figures 1B and 1C), our study brings conclusive experimental evidence that the similarity between the invertebrate VE and the mammalian ZP extends well beyond the core of their filaments. This implies that the egg extracellular structure that is involved in the initial interaction with sperm has a much more ancient evolutionary origin than Juno, the mammalian-specific protein that interacts with sperm Izumo1 to trigger the fusion of gamete plasma membranes (Bianchi et al., 2014, Inoue et al., 2005).
At the same time, our findings immediately raise the question of whether a lysin-like molecule exists in mammalian sperm, that binds ZP2. This is because post-fertilization cleavage of the latter has long been known to regulate mammalian sperm binding to the oocyte (Bleil et al., 1981, Gahlay et al., 2010), and its N-terminal region—essentially corresponding to the ZP2 ZP-N1 construct that we crystallized (Figures 1A and 1B)—was suggested to directly interact with sperm (Avella et al., 2014). Considering the insignificant sequence similarity between VERL and ZP2 repeats, as well as the fact that lysin lacks distinctive features such as Cys residues involved in structurally crucial disulfide bonds, it may be very difficult to recognize bioinformatically a putative structural homolog of lysin in mammals. However, comparison of the ZP2 ZP-N1 structure (Figure 1B) with that of the VERL/lysin complexes (Figures 3A and 6D) indicates that a VERL/lysin-like interface would not be compatible with the different location of β strand D observed in ZP-N1 (Figures 1B and S1H). This suggests that either ZP2 evolved a different way to bind sperm or that it regulates gamete interaction only indirectly by altering the overall structure of the egg coat (Hoodbhoy and Dean, 2004). Of relevance to the first possibility is the observation that ZP2 residues recently reported to be important for human gamete recognition (Avella et al., 2014) largely correspond to ZP-N1 α helix bc, β strand C, and loop cd (Figure 1B). As a result of the different position of β strand D, these structural elements are exposed on the face of ZP2 that corresponds to the lysin-interacting region of VR2 and VR3. Thus ZP2 may use the same surface as VERL repeats to bind sperm but not necessarily interact with a structural homolog of lysin.
Lysin is among the two most abundant proteins in abalone sperm (Lewis et al., 1982), a requirement for an external fertilization system where opening of a hole into the egg coat is achieved via the essentially irreversible binding of a soluble sperm molecule that is released extracellularly to titrate a large number of VERL repeats. Considering that the human ZP is ∼30-fold thicker than abalone VE and that human sperm lacks a secreted acrosomal protein as abundant as lysin, a putative ZP2 counterpart is more likely to be a sperm-surface-associated protein that makes transient, low-affinity interactions with ZP2 while sperm penetrates through the ZP. Akin to the case of Juno/Izumo1 (Aydin et al., 2016, Ohto et al., 2016), recognition of ZP2 by human sperm may therefore depend on establishing a relatively small interface with the same egg ZP-N fold that mediates gamete interaction in mollusk.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-lysin rabbit polyclonal antibody | Vacquier et al., 1990 | N/A |
| Penta·His Antibody, BSA-free | QIAGEN | Cat. # 34660; RRID: AB_2619735 |
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat. # 115-035-003; RRID: AB_10015289 |
| Peroxidase AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat. # 111-035-144; RRID: AB_2307391 |
| Bacterial and Virus Strains | ||
| E. coli BL21 Star (DE3) | Thermo Fisher Scientific | Cat. # C601003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Modified Eagle Medium (DMEM), high glucose, NEAA, no glutamine | Thermo Fisher Scientific | Cat. # 10938025 |
| Elastase from porcine pancreas | Sigma-Aldrich | Cat. # E0127 |
| Fetal Bovine Serum (FBS) European Grade | Biological Industries | Cat. # 04-007-1A |
| L-glutamine | Thermo Fisher Scientific | Cat. # 25030024 |
| Polyethyleneimine (PEI), branched | Sigma-Aldrich | Cat. # 408727 |
| Critical Commercial Assays | ||
| Monolith Protein Labeling Kit BLUE-NHS (Amine Reactive) | NanoTemper Technologies GmbH | Cat. # MO-L003 |
| Deposited Data | ||
| mMBP-VR1+ coordinates and structure factors | This study | PDB: 5II4 |
| mMBP-VR1 coordinates and structure factors | This study | PDB: 5II5 |
| ZP2 ZP-N1 coordinates and structure factors | This study | PDB: 5II6 |
| Orthorhombic lysin (S-SAD) coordinates and structure factors | This study | PDB: 5II7 |
| Orthorhombic lysin (high resolution) coordinates and structure factors | This study | PDB: 5II8 |
| Monoclinic lysin coordinates and structure factors | This study | PDB: 5II9 |
| mMBP-VR3 coordinates and structure factors | This study | PDB: 5IIC |
| Triclinic VR3/lysin complex coordinates and structure factors | This study | PDB: 5IIA |
| Trigonal VR3/lysin complex coordinates and structure factors | This study | PDB: 5IIB |
| VR2+ coordinates and structure factors | This study | PDB: 5MR2 |
| VR2+/lysin complex coordinates and structure factors | This study | PDB: 5MR3 |
| Experimental Models: Cell Lines | ||
| Human Embryonic Kidney (HEK) 293T | Aricescu et al., 2006 | N/A |
| Human Embryonic Kidney (HEK) 293S GnTI- | ATCC | Cat. # CRL-3022 |
| Recombinant DNA | ||
| Mammalian expression vector for VR1+ (H. rufescens VERL D38-I176) {pHLsec3-VR1+} | This study | N/A |
| Mammalian expression vector for mMBP-VR1+ (H. rufescens VERL D38-L175 [N115Q, N122T, N142Y, N171Q]) {pHLmMBP2-VR1+} | This study | N/A |
| Mammalian expression vector for mMBP-VR1 (H. rufescens VERL D38-A151 [N115Q, N122T, N142Y]) {pHLmMBP1-VR1} | This study | N/A |
| Mammalian expression vector for ZP2 ZP-N1 (M. musculus ZP2 M1-Q138 [N83S]) {pHLsec3-ZP2 ZP-N1} | This study | N/A |
| Mammalian expression vector for VR2+ (H. rufescens VERL D177-I339) {pHLsec3-VR2+} | This study | N/A |
| Mammalian expression vector for VR3+ (H. rufescens VERL D340-I492) {pHLsec3-VR3+} | This study | N/A |
| Mammalian expression vector for lysin (H. rufescens lysin M1-K154) {pHLsec3-Lysin} | This study | N/A |
| Mammalian expression vector for VR1+2+ used in Figure 2B (H. rufescens VERL D38-S304) {pHLsec3-VR1+2+(304)} | This study | N/A |
| Mammalian expression vector for VR3+4 (H. rufescens VERL D340-S601) {pHLsec3-VR3+4} | This study | N/A |
| Mammalian expression vector for VR3 (H. rufescens VERL D340-K453) {pHLsec3-VR3} | This study | N/A |
| Bacterial expression vector for lysinR (H. rufescens lysin R19-M152) {pJexpress411-LYSIN-02} | This study | N/A |
| Mammalian expression vector for mMBP-VR3 (H. rufescens VERL D340-K453) {pHLmMBP1-VR3} | This study | N/A |
| Mammalian expression vector for mMBP-VR3 N-glyc mutant (H. rufescens VERL D340-K453 [N373Q, N417Q, N438Q]) {pHLmMBP2-VR3 Nglyc mutant} | This study | N/A |
| Mammalian expression vector for VR3+ O-glyc mutant (H. rufescens VERL D340-I492 [T456G, T459G, S460G, T467A, T470G, S471G, S472G, S478G, S479G, S485G, S489G]) {pHLsec3-VR3+ O-glyc mutant} | This study | N/A |
| Mammalian expression vector for VR3_h (H. rufescens VERL D340-K453 [M389K]) {pHLsec3-VR3_h} | This study | N/A |
| Mammalian expression vector for lysin_h (H. rufescens lysin M1-K154 [F119S]) {pHLsec3-Lysin_h} | This study | N/A |
| Mammalian expression vector for VR3_p (H. rufescens VERL D340-K453 [N388A]) {pHLsec3-VR3_p} | This study | N/A |
| Mammalian expression vector for lysin_d (H. rufescens lysin M1-K154 [T78A, H79A]) {pHLsec3-Lysin_d} | This study | N/A |
| Mammalian expression vector for VR3_hp (H. rufescens VERL D340-K453 [N388A, M389K]) {pHLsec3-VR3_hp} | This study | N/A |
| Mammalian expression vector for lysin_t (H. rufescens lysin M1-K154 [R74A, Y75A, T78A]) {pHLsec3-Lysin_t} | This study | N/A |
| Mammalian expression vector for VR3_mut (H. rufescens VERL D340-K453 [P380Q, I381M, E384G, F385R, M390I, L391R]) {pHLsec3-VR3_mut} | This study | N/A |
| Mammalian expression vector for VR3 E384G (VERL D340-K453 [E384G]) {pHLsec3-VR3 E384G} | This study | N/A |
| Mammalian expression vector for lysin K150A (H. rufescens lysin M1-K154 [K150A]) {pHLsec3-Lysin K150A} | This study | N/A |
| Mammalian expression vector for lysinΔN (H. rufescens lysin M1-K154 [ΔR19-L29]) {pHLsec3-LysinΔN} | This study | N/A |
| Mammalian expression vector for pink abalone lysin (H. corrugata lysin M1-K155) {pHLsec3-Pink Lysin} | This study | N/A |
| Mammalian expression vector for black abalone lysin (H. cracherodii lysin M1-K156) {pHLsec3-Black Lysin} | This study | N/A |
| Mammalian expression vector for VR1+2+ used in Figure 6 (H. rufescens VERL D38-I339) {pHLsec3-VR1+2+(339)} | This study | N/A |
| Mammalian expression vector for VR2+ crystallization construct (H. rufescens VERL I176-P298 [S293A, S296A, S297A]) {pHLsec3-VR2+XR} | This study | N/A |
| Mammalian expression vector for VR1+2+ F181Y, L228P (H. rufescens VERL D38-I339 [F181Y, L228P]) {pHLsec3-VR1+2+ F181Y, L228P} | This study | N/A |
| Mammalian expression vector for lysin K150A, Y151N (H. rufescens lysin M1-K154 [K150A, Y151N]) {pHLsec3-Lysin K150A, Y151N} | This study | N/A |
| Mammalian expression vector for lysin Npink (H. corrugata lysin M1-I30 + H. rufescens lysin N30-K154) {pHLsec3-Lysin Npink} | This study | N/A |
| Mammalian expression vector for lysin Npink K150A, Y151N (H. corrugata lysin M1-I30 + H. rufescens lysin N30-K154 [K150A, Y151N]) {pHLsec3-Lysin Npink K150A, Y151N} | This study | N/A |
| Mammalian expression vector for pink abalone lysin Nred (H. rufescens lysin M1- L29 + H. corrugata lysin R31-K155) {pHLsec3-Pink Lysin Nred} | This study | N/A |
| pHLmMBP1 | Bokhove et al., 2016 | N/A |
| pHLmMBP2 | Bokhove et al., 2016 | N/A |
| pHLsec | Aricescu et al., 2006 | N/A |
| pHLsec3 | This study | N/A |
| pJexpress411 | DNA2.0/ATUM | https://www.atum.bio |
| pProEX HT-endoglycosidase H | Dan Leahy | N/A |
| pRARE | Novagen/Merck Millipore | Cat. # 70954 |
| Software and Algorithms | ||
| aimless | Evans and Murshudov, 2013 | http://www.ccp4.ac.uk/html/aimless.html |
| APBS | Baker et al., 2001 | http://www.poissonboltzmann.org |
| ARP/wARP | Langer et al., 2008 | http://www.embl-hamburg.de/ARP |
| CHARMM27 force field | Mackerell et al., 2004 | https://www.charmm.org |
| Coot | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| DIALS | Gildea et al., 2014 | http://dials.github.io |
| ESPript | Robert and Gouet, 2014 | http://espript.ibcp.fr |
| FoldX | Schymkowitz et al., 2005 | http://foldxsuite.crg.eu |
| GROMACS | Pronk et al., 2013 | http://www.gromacs.org |
| HKL-3000R | Minor et al., 2006 | http://www.hkl-xray.com/hkl-3000 |
| LINCS algorithm | Hess, 2008 | N/A |
| MO.Affinity Analysis | NanoTemper Technologies GmbH | http://www.nanotemper-technologies.com/products/monolith-series/software |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| multAlin | Corpet, 1988 | http://multalin.toulouse.inra.fr/multalin |
| PDB2PQR | Dolinsky et al., 2007 | http://www.poissonboltzmann.org |
| PDBsum | N/A | http://www.ebi.ac.uk/pdbsum |
| Phaser | McCoy et al., 2007 | http://www.phaser.cimr.cam.ac.uk |
| PHENIX | Adams et al., 2010 | http://www.phenix-online.org |
| PIC | Tina et al., 2007 | http://pic.mbu.iisc.ernet.in |
| PISA | Krissinel and Henrick, 2007 | http://www.ebi.ac.uk/pdbe/pisa |
| Privateer | Agirre et al., 2015 | http://www.ccp4.ac.uk/html/privateer.html |
| PyMOL | Schrödinger, LLC | http://www.pymol.org |
| SETTLE algorithm | Miyamoto and Kollman, 1992 | N/A |
| SHARP | Bricogne et al., 2003 | http://www.globalphasing.com/sharp |
| SHELX | Sheldrick, 2008 | http://shelx.uni-ac.gwdg.de/SHELX |
| SOLOMON | Abrahams and Leslie, 1996 | http://www.ccp4.ac.uk/html/solomon.html |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera |
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de |
| xia2 | Winter, 2010 | http://xia2.github.io |
| Other | ||
| Monolith NT.115 Premium Coated Capillaries | NanoTemper Technologies GmbH | Cat. # MO-K005 |
| Ni-NTA Agarose | QIAGEN | Cat. # 30230 |
| Ni Sepharose excel | GE Healthcare | Cat. # 17-3712-02 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Luca Jovine (luca.jovine@ki.se).
Experimental Model and Subject Details
Cell Lines
Mammalian cell expression was performed in HEK293T cells (a kind gift of R. Aricescu and Y. Zhao, University of Oxford) or HEK293S GnTI- cells (ATCC). Cells were cultivated at 37°C, 5% CO2 in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Biological Industries) and 4 mM L-glutamine (Thermo Fisher Scientific).
Bacterial expression was carried out using E. coli BL21 Star (DE3) (Thermo Fisher Scientific) co-transformed with plasmid pRARE (extracted from E. coli Rosetta (DE3) cells; Novagen/Merck Millipore). A single colony of freshly transformed cells was used to inoculate 50 mL liquid Luria-Bertani broth (Sigma-Aldrich) supplemented with 34 μg/mL kanamycin (Sigma-Aldrich). After overnight culture at 37°C, 200 rpm, 10 mL cell suspension were seeded into 1 L of the same medium. When the absorbance of the culture reached an OD600 of 0.5, protein expression was induced for 3 hr at 37°C with 0.5 mM IPTG (Thermo Fisher Scientific).
Method Details
Constructs
For mammalian expression of 6His-tagged VERL and ZP2 ZP-N1 constructs, as well as untagged lysin proteins, synthetic genes (DNA2.0/ATUM, Invitrogen GeneArt/Thermo Fisher Scientific or GeneScript) or cDNA fragments amplified by PCR with PfuTurbo DNA polymerase (Agilent Technologies) were subcloned into pHLsec3, a vector derived from pHLsec (Aricescu et al., 2006), or pHLmMBP vectors (Bokhove et al., 2016). Whereas ZP2 ZP-N1 and lysin proteins included native signal peptides, all VERL constructs were secreted using a Crypα signal sequence. Glycosylation site mutants of ZP2 ZP-N1 (N83S) as well as VERL VR1+ (N115Q, N122T, N142Y, N171Q), VR1 (N115Q, N122T, N142Y) and VR2+ (S293A, S296A, S297A) were used for structure determination. With the exception of mMBP-VR3 N-glyc mutant and VR3+ O-glyc mutant (Figures S6A and S6B), VERL constructs used for binding assays contained all glycosylation sites. Expression of lysinR in E. coli was performed using pJexpress411 (DNA2.0/ATUM).
Mutations were introduced by synthesis, using a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) or by overlap extension PCR. All constructs were verified by DNA sequencing (Eurofins Genomics).
Details of all expression constructs are provided in the Key Resources Table, and boundaries of VERL and ZP2 recombinant proteins are marked by brackets in Figure 1A.
Protein Expression
mMBP-VR1+, mMBP-VR1 and ZP2 ZP-N1 were expressed in HEK293T cells. For structural studies, mMBP-VR3, mMBP-VR3/lysin, VR3/lysin and VR2+/lysin were expressed in HEK293S GnTI- cells, which secrete proteins carrying Endoglycosidase H (Endo H)-sensitive Man5GlcNAc2 N-glycans (Chang et al., 2007, Reeves et al., 2002). Transient transfections were performed essentially as described (Aricescu et al., 2006, Bokhove et al., 2016), using 25 kDa branched PEI (Sigma-Aldrich) and a 1:1 DNA ratio in the case of VERL and lysin co-expression experiments. Large-scale production of lysinR for crystallographic studies and biochemical assays was performed using E. coli BL21 Star (DE3) co-transformed with pRARE, as detailed above.
Protein Purification
For purification of proteins expressed in mammalian cells, conditioned media was harvested 3 days post-transfection, 0.22 μm-filtered (Sarstedt) and adjusted to 20 mM Na-HEPES pH 7.8, 150 mM NaCl and 5-20 mM imidazole (immobilized metal affinity chromatography (IMAC) binding buffer). 10 mL Ni-NTA agarose (QIAGEN) or Ni Sepharose excel (GE Healthcare) slurry were added per liter of medium and incubated either for 1 hr at room temperature or overnight at 4°C. After collecting IMAC beads into a column and washing them with binding buffer, protein was batch-eluted with IMAC elution buffer (20 mM Na-HEPES pH 7.8, 150 mM NaCl, 500 mM imidazole). HEK293S cell-expressed mMBP-VR3, mMBP-VR3/lysin, VR3/lysin and VR2+/lysin were deglycosylated with Endo H (1:10 mass ratio) for 1 hr at 37°C in 120 mM Na/K phosphate pH 6.0, prior to elution. Proteins were then concentrated using centrifugal filtration devices (Amicon) with appropriate molecular weight cut-offs and further purified by SEC at 4°C, using an ÄKTAFPLC chromatography system (GE Healthcare). A HiLoad 26/600 Superdex 200 pg column (GE Healthcare) and a 20 mM Na-HEPES pH 7.8, 150 mM NaCl running buffer were used for unfused VERL proteins and the VR3/lysin complex; for mMBP-fused material, the buffer was supplemented with 10 mM D-maltose. A HiLoad 26/60 Superdex 75 pg column (GE Healthcare) and a 10 mM Tris-HCl, 50 mM NaCl buffer was used for ZP2 ZP-N1. Fractions containing protein peaks were pooled, concentrated and used for crystallization trials.
To obtain crystals of the VR2+/lysin complex, it was essential to supplement SEC-purified material with lysinR, obtained as described below. After dialysis against 10 mM MES pH 6.0, 250 mM NaCl, the sample was applied to a 5 mL HiTrap CM Sepharose FF ion-exchange chromatography (IEX) column (GE Healthcare) equilibrated with the same buffer. Pure complex was eluted with a salt gradient to 1.5 M NaCl, dialyzed against 20 mM Na-HEPES pH 8.0, 200 mM NaCl and finally concentrated for crystallization.
LysinR was purified as previously described (Lyon and Vacquier, 1999), with modifications. Pelleted cells from 1 L E. coli culture were resuspended in 40 mL lysis buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA) containing 60 ku rLysozyme (Merck Millipore), 750 u Benzonase nuclease (Sigma-Aldrich) and 1 tablet cOmplete EDTA-free protease inhibitor cocktail (Roche), incubated for 30 min at room temperature and disrupted by sonication on ice (6 bursts of 10 s, with 10 s cooling in between). Inclusion bodies were collected by 12,000 g centrifugation for 20 min at 4°C, dissolved in 100 mL 5 M guanidine-HCl for 2-3 hr at 37°C on a magnetic stirrer and clarified by 12,000 g centrifugation for 15 min at 4°C. The supernatant was dialyzed for 12 hr at 4°C against 4 L carboxymethyl (CM) buffer (250 mM NaCl, 2 mM EDTA, 10 mM MES pH 6.0) using 10 kDa cut-off SnakeSkin dialysis tubing (Thermo Fisher Scientific). After 4 dialysis buffer changes at 12 hr intervals, the solution was clarified by 12,000 g centrifugation for 30 min at 4°C and subjected to IEX, using a 5 mL HiTrap CM Sepharose FF column (GE Healthcare) and a linear gradient of 0.25 to 1.25 M NaCl in CM buffer. Concentrated peak fractions were further purified by SEC at 4°C, using a HiLoad 26/60 Superdex 75 pg column (GE Healthcare) and a seawater-like solution (468 mM NaCl, 10 mM KCl, 10 mM CaCl2, 28 mM MgSO4, 25 mM MgCl2, 20 mM Na-HEPES pH 7.8) as running buffer.
Protein Analysis
Samples separated on SDS-PAGE gels were detected with SimplyBlue SafeStain (Thermo Fisher Scientific) or transferred to nitrocellulose membranes (GE Healthcare) for immunoblotting with either Penta·His mouse monoclonal antibody (1:1,000; QIAGEN) or anti-lysin rabbit polyclonal antibody (1:10,000) (Vacquier et al., 1990). Secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse IgG (1:10,000) or goat anti-rabbit IgG (1:5,000) (Jackson ImmunoResearch Laboratories). Chemiluminescence detection was performed with Western Lightning ECL Plus (Perkin Elmer). Unless otherwise specified, samples were analyzed in reducing conditions.
N-terminal sequencing (Cambridge Peptides) showed that the two isoforms of HEK293-produced VR3 (Figure 2C, lane 2) begin with the sequences ETGAA and AADWD, originating from alternative cleavage of the Crypα signal peptide (with the underlined sequence corresponding to VERL residues D340-D342).
Protein Crystallization
Crystallization trials by hanging or sitting drop vapor diffusion were set up at room temperature using a mosquito crystallization robot (TTP Labtech). Crystals were harvested using MicroMounts, MicroLoops or MicroMeshes (MiTeGen).
mMBP-VR1+ (16.8 mg/mL) and mMBP-VR1 (12.8 mg/mL) crystallized in 40% (v/v) PEG 600, 0.1 M CHES pH 9.5, whereas mMBP-VR3 (18.5 mg/mL) formed crystals in 20% (w/v) PEG 4000, 20% (v/v) isopropanol, 0.1 M trisodium citrate pH 5.5. For data collection, crystals were cryoprotected using mother liquor supplemented with 20% (v/v) glycerol (mMBP-VR1+, mMBP-VR1) or fished directly from the drops in which they grew (mMBP-VR3) and flash frozen in liquid nitrogen.
The ZP2 ZP-N1 crystal that yielded that data used for experimental phasing by S-SAD was grown in 14% (w/v) PEG 6000, 0.1 M MES pH 5.5, using a protein concentration of 14.6 mg/mL. The crystal was cryoprotected by stepwise addition of PEG 200 to a final concentration of 20% (v/v) in mother liquor solution, followed by 3 min soaks in equivalent solutions supplemented with 0.05 M 5-amino-2,4,6-triiodoisophthalic acid (I3C; Jena Bioscience), 0.16 M I3C and 0.27 M I3C. Atomic resolution data was obtained from a crystal grown with 2.5 mg/mL ZP2 ZP-N1 in 30% (v/v) PEG 200, 5% (w/v) PEG 3000, 0.1 M MES pH 6.0, which was cryoprotected for 1 min using a mother liquor solution containing 40% (v/v) PEG 200.
Purified lysinR, dialyzed against sodium acetate pH 5.5, was concentrated to 10 mg/mL and crystallized in 0.65 M ammonium sulfate, 0.2 M NaCl, 0.1 M CHES pH 9.5 (orthorhombic crystal form) or 5% (w/v) PEG 4000, 0.2 M sodium acetate, 0.1 M sodium citrate pH 5.5 (monoclinic crystal form). For in-house S-SAD data collection, an orthorhombic crystal was dehydrated by stepwise transfer to a mother liquor solution containing 4.0 M ammonium sulfate, 10% (v/v) glycerol and finally transferred to a gaseous nitrogen stream. To collect atomic resolution data from the same crystal form, an HC1 dehydration device (Sanchez-Weatherby et al., 2009) was used to progressively decrease the relative humidity of a specimen in mother liquor supplemented with 5% (v/v) glycerol. Upon reaching a 36% change in relative humidity over a total time of 18 min, the crystal was flash frozen in liquid nitrogen by retrieval into the beamline sample changer. Monoclinic crystals were cryoprotected using a mother liquor solution supplemented with 20% (v/v) glycerol.
Extensive crystallization trials of mMBP-VR3/lysin (23 mg/mL) did not produce any crystals. However, triclinic complex crystals were readily obtained in 20% (w/v) PEG 3350, 0.2 M diammonium citrate pH 5.0 after setting up drops with mMBP-VR3/lysin premixed with porcine elastase (1:1000 molar ratio; Sigma-Aldrich). Trigonal crystals of unfused VR3/lysin (10 mg/mL) appeared in 1.6 M ammonium sulfate, 0.1 M citric acid pH 5.0. Triclinic and trigonal complex crystals were cryoprotected for data collection using mother liquor solutions supplemented with 20% (v/v) glycerol and 20% (v/v) saturated sucrose, respectively.
Crystals of free VR2+ were obtained in drops set up with VR2+/lysin complex material not supplemented with lysinR (10 mg/mL) and 0.2 M ammonium formate pH 6.6, 20% (w/v) PEG 3350. These crystals were cryoprotected using mother liquor solution containing 15% (v/v) ethylene glycol.
VR2+/lysin complex crystals were obtained in 0.1 M Tris-HCl pH 8.5, 5% (w/v) PEG 8000, 20% (v/v) PEG 300, 10% (v/v) glycerol using lysinR-supplemented material (10 mg/mL). Specimens were directly harvested for cryocooling and data collection.
X-Ray Diffraction Data Collection
All datasets were collected from single crystals at 100 K.
Lysin S-SAD data was collected in house with a Compact HomeLab system (Rigaku) equipped with a PILATUS 200K detector (DECTRIS). Data for the trigonal crystal form of VR3/lysin was collected at beamline ID23-1 of the European Synchrotron Radiation Facility (ESRF, Grenoble) (Nurizzo et al., 2006), equipped with a PILATUS 6M-F detector (DECTRIS); data for VR2+ was collected at ESRF beamline ID23-2 (Flot et al., 2010) using a PILATUS 2M detector (DECTRIS); all other datasets were collected at ESRF beamline ID29 (de Sanctis et al., 2012), equipped with a PILATUS 6M-F detector.
Data Processing and Structure Determination
X-ray diffraction data was processed using XDS (Kabsch, 2010) (mMBP-VR1+; mMBP-VR1; mMBP-VR3; orthorhombic lysin (high resolution); VR3/lysin complexes; VR2+ and VR2+/lysin complex), DIALS (Gildea et al., 2014) and aimless (Evans and Murshudov, 2013) (ZP2 ZP-N1), HKL-3000R (Minor et al., 2006) (orthorhombic lysin (S-SAD)) or xia2 (Winter, 2010) (monoclinic lysin).
The structures of mMBP-VR1+ and mMBP-VR3 were determined by MR with Phaser (McCoy et al., 2007), using as search model an ensemble of MBP structures extracted from PDB: 3SET, 3SEX, and 4WRN. Structures were autotraced with PHENIX AutoBuild (Terwilliger et al., 2008), manually rebuilt with Coot (Emsley et al., 2010) and refined with phenix.refine (Afonine et al., 2012). Protein geometry was validated with MolProbity (Chen et al., 2010), carbohydrate structure validation was carried out using Privateer (Agirre et al., 2015). The structure of mMBP-VR1 was phased using an intermediate, rigid body-refined model of mMBP-VR1+. Rebuilding, refinement and validation were performed as above.
For experimental phasing of ZP2 ZP-N1 by S-SAD, anomalous differences in four 360-degree datasets collected at different positions of a single crystal at 7 keV (1.7712 Å) were evaluated with SHELXC followed by SHELXD (Sheldrick, 2008). This located 6 sulfur atoms (corresponding to 4 Cys, forming 2 disulfide bridges, and 2 Met). Despite of soaking in I3C, no iodine atom was identified. Phases were calculated and refined with SHARP (Bricogne et al., 2003) (phasing power 0.921, overall figure of merit 0.28), and improved with SOLOMON (Abrahams and Leslie, 1996), using an optimized solvent content of 45.2%. An initial model was built with ARP/wARP (Langer et al., 2008) that placed and docked in sequence 85 residues, which were then rigid body-refined against the atomic resolution dataset. Rebuilding, refinement and validation were performed as above.
S-SAD phasing of the structure of orthorhombic lysin was carried out with PHENIX AutoSol (Terwilliger et al., 2009), which located 15 sites (corresponding to 6 of the 7 Met of the protein, the sulfur atom of an ordered MES buffer molecule and sulfate ions) and built 127 out of 136 residues (Bayesian correlation coefficient 0.307, overall figure of merit 0.306, model correlation coefficient 0.81). The resulting refined model of lysin was rigid body-refined against the high resolution synchrotron data of the same crystal form, as well as used to solve the monoclinic structure of lysin by MR with Phaser. Rebuilding, refinement and validation were performed as above.
The triclinic structure of the VR3/lysin complex was solved by MR with Phaser, using the refined structures of VR3 and orthorhombic lysin as search models. A partially refined structure of the complex was then used to solve the trigonal crystal form of VR3/lysin by MR. Both structures were rebuilt, refined and validated as described above.
The structure of VR2+ was solved by MR with Phaser, using an ensemble of VR3 structures. Ensembles of VR2+ and lysin structures were then used as search models to solve the structure of the VR2+/lysin complex. Rebuilding, refinement and validation of both structures were performed as above.
Refinement and validation statistics calculated using phenix.table_one (Adams et al., 2010) are reported in Tables S1-S4.
Sequence and Structure Analysis
Multiple sequence alignments and consensus sequences were calculated with multAlin (Corpet, 1988) and presented with ESPript (Robert and Gouet, 2014). Structural alignments were performed using UCSF Chimera (Pettersen et al., 2004), Coot and PyMOL (Schrödinger, LLC).
Protein-protein interfaces and oligomeric states were analyzed using PDBsum (de Beer et al., 2014), PIC (Tina et al., 2007), FoldX (Schymkowitz et al., 2005) and PISA (Krissinel and Henrick, 2007) (excluding the mMBP moiety in the case of mMBP-VR3).
Electrostatic surface potential calculations were performed in seawater conditions with PDB2PQR (Dolinsky et al., 2007) and APBS (Baker et al., 2001), via the APBS Tools plugin of PyMOL; results were mapped onto the molecular surfaces by coloring them from red (−2.5 kT/e) to blue (+2.5 kT/e) through white (0 kT/e).
All structural figures were created with PyMOL.
Molecular Dynamics Simulations
Simulations were performed using GROMACS (Pronk et al., 2013) and the CHARMM27 force field (Mackerell et al., 2004).
Charges of ionizable groups were appropriate for pH 7.0 (Arg and Lys protonated; Asp and Glu unprotonated; His neutral). Tautomeric forms were based on local interactions; for lysin H56 and H79 the N3-H tautomer was used. Proteins were placed in a dodecahedric box of 13 nm, which was subsequently filled with TIP3P water molecules (Jorgensen and Chandrasekhar, 1983); Na+ and Cl- ions were randomly added to neutralize the systems and mimic the seawater environment (0.6 M ion concentrations) where VERL and lysin interact.
The particle mesh Ewald method (Darden et al., 1993) was employed to treat Coulomb interactions using a switching distance of 1.2 nm and a grid of 0.12 nm; Lennard-Jones interactions were switched off between 0.8 and 1.2 nm. Constant pressure p and temperature T were maintained by weakly coupling the system to an external bath at 1 bar and 298 K, using the Parrinello-Rahman barostat (Parrinello and Rahman, 1981) and velocity-rescaling thermostat (Bussi et al., 2007), respectively. The system was coupled to the temperature bath with a coupling time of 0.5 ps. Pressure coupling time was 5.0 ps and the isothermal compressibility 4.5·10−5 bar−1. Bond distances and angles of water were constrained using the SETTLE algorithm (Miyamoto and Kollman, 1992); other bond distances were constrained with the LINCS algorithm (Hess, 2008). A leap-frog integrator with an integration time step of 2 fs was used.
Starting structures were generated by locating the Arg/Lys-rich interface of the complexes at a minimum distance of 2 Å. Two different relative orientations of the interface (parallel and antiparallel) were used for the starting structures. During the simulation, the distance between the center of mass of the two VR3 was constrained to mimic the reorientation restriction imposed by the architecture of VERL repeat arrays. 20 ns position-restrained simulations were performed to relax ion positions. After ion equilibration, 100 ns distance-restrained simulations were performed for the different starting structures, for a total of 4 independent simulations.
Pull-Down Analysis of Protein-Protein Interaction
All pull-down experiments were performed using C-terminally His-tagged VERL constructs. 2 mL conditioned medium harvested three days post-transfection was centrifuged for 5 min at 500 x g and 0.22 μm-filtered. After adjustment to 20 mM Na-HEPES pH 7.8, 150 mM NaCl, 5 mM imidazole (binding buffer), medium was incubated with 50 μl IMAC beads for 1 hr at room temperature or overnight at 4°C. Beads were collected by centrifugation at 100 x g and washed with 10 mL binding buffer; bound material was then eluted using 100 μl 20 mM Na-HEPES pH 7.8, 150 mM NaCl, 500 mM imidazole.
Size-Exclusion Chromatography Analysis of Protein-Protein Interaction
Analysis of complex formation between VR1+ or VR3 and lysin was carried out using a Superdex 200 10/300 GL column (GE Healthcare). To avoid nonspecific binding of lysin to the column material, seawater solution supplemented with 0.0005% Triton X was used as running buffer. Analysis of individual proteins was carried out by injecting 150 μg material; complex formation was assessed using a 2:1 molar ratio of lysin to VR1+/VR3.
Binding Affinity Determination by Microscale Thermophoresis
MST analysis was performed using a NanoTemper Monolith NT.115 instrument (NanoTemper Technologies GmbH). Lysin was fluorescently labeled with a Blue-NHS labeling kit (NanoTemper Technologies GmbH), according to the manufacturer’s instructions and using seawater solution supplemented with 0.05% Tween. Lysin was labeled with a dye:protein molar ratio of 0.67. Varying concentrations of VR1+, VR3, mMBP-VR3 and VR1+2+ were titrated against labeled lysin (10 nM) in MST buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl) supplemented with 0.05% Tween. ZP2 ZP-N1 was used as negative control. Samples were loaded into Premium Coated Capillaries (NanoTemper Technologies GmbH) and MST measurements were performed using 20% MST power and 90% LED power. Laser-on and -off times were 30 s and 5 s, respectively. For each set of binding experiments, three independent MST measurements were carried out at 495 nm. Datasets were processed with the MO.Affinity Analysis software (NanoTemper Technologies GmbH), using the signal from the initial fluorescence.
Quantification and Statistical Analysis
X-Ray Crystallography
High resolution data cutoff was chosen based on statistical indicators CC1/2 and CC∗ (Evans and Murshudov, 2013, Karplus and Diederichs, 2015).
Microscale Thermophoresis
Error bars in Figures 2E, 6G, and S3B represent the standard deviation for three independent experiments.
Data and Software Availability
Data Resources
Structure factors and atomic coordinates have been deposited in the Protein Data Bank under accession numbers PDB: 5II4 (mMBP-VR1+), PDB: 5II5 (mMBP-VR1), PDB: 5II6 (ZP2 ZP-N1), PDB: 5II7 (orthorhombic lysinR, S-SAD), PDB: 5II8 (orthorhombic lysinR, high resolution), PDB: 5II9 (monoclinic lysinR), PDB: 5IIC (mMBP-VR3), PDB: 5IIA (triclinic VR3/lysin complex), PDB: 5IIB (trigonal VR3/lysin complex), PDB: 5MR2 (VR2+), and PDB: 5MR3 (VR2+/lysin complex).
Author Contributions
I.R., H.S.A.H., E.D., K.N., and L.H. performed DNA work and protein expression, analysis, and purification. I.R. performed MST experiments. H.S.A.H., I.R., E.D., K.N., and L.H. crystallized proteins. L.J., D.d.S., H.S.A.H., and L.H. collected X-ray diffraction data. L.J. solved and refined the VERL, lysin, and VERL/lysin complex structures. D.d.S. phased the ZP2 ZP-N data, which was refined by L.J. and D.d.S. K.N. was involved in structure determination of the VR2/lysin complex structure. A.V. performed MD simulations. L.J. conceived the study, coordinated the research, and wrote the manuscript together with I.R. and H.S.A.H.
Acknowledgments
We are grateful to R. Aricescu and Y. Zhao (University of Oxford) for mammalian expression vector pHLsec and HEK293T cells; D. Leahy (Johns Hopkins University School of Medicine) for E. coli expression vector pProEX HT-endoglycosidase H; V. Vacquier (University of California San Diego) and W. Swanson (University of Washington) for anti-lysin polyclonal antibodies; A. Stsiapanava (Karolinska Institutet) for data collection at ESRF ID23-1 and comments; ESRF for access to structural biology beamlines. This work was supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement 260759; Karolinska Institutet; the Center for Biosciences; the Center for Innovative Medicine; Swedish Research Council grant 2012-5093; the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine; the Sven and Ebba-Christina Hagberg Foundation; and an EMBO Young Investigator award.
Published: June 15, 2017
Footnotes
Supplemental Information includes seven figures, six tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.05.033.
Supplemental Information
References
- Aagaard J.E., Vacquier V.D., MacCoss M.J., Swanson W.J. ZP domain proteins in the abalone egg coat include a paralog of VERL under positive selection that binds lysin and 18-kDa sperm proteins. Mol. Biol. Evol. 2010;27:193–203. doi: 10.1093/molbev/msp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams J.P., Leslie A.G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre J., Iglesias-Fernández J., Rovira C., Davies G.J., Wilson K.S., Cowtan K.D. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Avella M.A., Baibakov B., Dean J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 2014;205:801–809. doi: 10.1083/jcb.201404025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H., Sultana A., Li S., Thavalingam A., Lee J.E. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature. 2016;534:562–565. doi: 10.1038/nature18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T.A.P., Berka K., Thornton J.M., Laskowski R.A. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Doe B., Goulding D., Wright G.J. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleil J.D., Beall C.F., Wassarman P.M. Mammalian sperm-egg interaction: fertilization of mouse eggs triggers modification of the major zona pellucida glycoprotein, ZP2. Dev. Biol. 1981;86:189–197. doi: 10.1016/0012-1606(81)90329-8. [DOI] [PubMed] [Google Scholar]
- Bokhove M., Sadat Al Hosseini H., Saito T., Dioguardi E., Gegenschatz-Schmid K., Nishimura K., Raj I., de Sanctis D., Han L., Jovine L. Easy mammalian expression and crystallography of maltose-binding protein-fused human proteins. J. Struct. Biol. 2016;194:1–7. doi: 10.1016/j.jsb.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Mornon J.-P., Monget P. Isolated ZP-N domains constitute the N-terminal extensions of Zona Pellucida proteins. Bioinformatics. 2007;23:1871–1874. doi: 10.1093/bioinformatics/btm265. [DOI] [PubMed] [Google Scholar]
- Chang V.T., Crispin M., Aricescu A.R., Harvey D.J., Nettleship J.E., Fennelly J.A., Yu C., Boles K.S., Evans E.J., Stuart D.I. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot D., Mairs T., Giraud T., Guijarro M., Lesourd M., Rey V., van Brussel D., Morawe C., Borel C., Hignette O. The ID23-2 structural biology microfocus beamline at the ESRF. J. Synchrotron Radiat. 2010;17:107–118. doi: 10.1107/S0909049509041168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlay G., Gauthier L., Baibakov B., Epifano O., Dean J. Gamete recognition in mice depends on the cleavage status of an egg’s zona pellucida protein. Science. 2010;329:216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo B.E., Moy G.W., Swanson W.J., Vacquier V.D. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. [DOI] [PubMed] [Google Scholar]
- Galindo B.E., Vacquier V.D., Swanson W.J. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl. Acad. Sci. USA. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A., Costantini M. Glycobiology of reproductive processes in marine animals: the state of the art. Mar. Drugs. 2012;10:2861–2892. doi: 10.3390/md10122861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea R.J., Waterman D.G., Parkhurst J.M., Axford D., Sutton G., Stuart D.I., Sauter N.K., Evans G., Winter G. New methods for indexing multi-lattice diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2652–2666. doi: 10.1107/S1399004714017039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Monné M., Okumura H., Schwend T., Cherry A.L., Flot D., Matsuda T., Jovine L. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Hess B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Hoodbhoy T., Dean J. Insights into the molecular basis of sperm-egg recognition in mammals. Reproduction. 2004;127:417–422. doi: 10.1530/rep.1.00181. [DOI] [PubMed] [Google Scholar]
- Inoue N., Ikawa M., Isotani A., Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P.A., Diederichs K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 2015;34:60–68. doi: 10.1016/j.sbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N., Vacquier V.D., Stout C.D. 1.35 and 2.07 Å resolution structures of the red abalone sperm lysin monomer and dimer reveal features involved in receptor binding. Acta Crystallogr. D Biol. Crystallogr. 2000;56:34–41. doi: 10.1107/s0907444999014626. [DOI] [PubMed] [Google Scholar]
- Kresge N., Vacquier V.D., Stout C.D. Abalone lysin: the dissolving and evolving sperm protein. BioEssays. 2001;23:95–103. doi: 10.1002/1521-1878(200101)23:1<95::AID-BIES1012>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-H., Vacquier V.D. Evolution and systematics in Haliotidae (Mollusca: Gastropoda): inferences from DNA sequences of sperm lysin. Mar. Biol. 1995;124:267–278. [Google Scholar]
- Lewis C.A., Talbot C.F., Vacquier V.D. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev. Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- Lillie F.R. Studies of fertilization. VI. The mechanism of fertilization in arbacia. J. Exp. Zool. 1914;16:523–590. [Google Scholar]
- Lyon J.D., Vacquier V.D. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev. Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier W., Stewart C.B. Molecular evolution. Dissolving the barriers. Curr. Biol. 1994;4:911–913. doi: 10.1016/s0960-9822(00)00202-5. [DOI] [PubMed] [Google Scholar]
- Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Kollman P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- Monné M., Han L., Schwend T., Burendahl S., Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- Nurizzo D., Mairs T., Guijarro M., Rey V., Meyer J., Fajardo P., Chavanne J., Biasci J.C., McSweeney S., Mitchell E. The ID23-1 structural biology beamline at the ESRF. J. Synchrotron Radiat. 2006;13:227–238. doi: 10.1107/S0909049506004341. [DOI] [PubMed] [Google Scholar]
- Ohto U., Ishida H., Krayukhina E., Uchiyama S., Inoue N., Shimizu T. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature. 2016;534:566–569. doi: 10.1038/nature18596. [DOI] [PubMed] [Google Scholar]
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.J., Callewaert N., Contreras R., Khorana H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Weatherby J., Bowler M.W., Huet J., Gobbo A., Felisaz F., Lavault B., Moya R., Kadlec J., Ravelli R.B.G., Cipriani F. Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1237–1246. doi: 10.1107/S0907444909037822. [DOI] [PubMed] [Google Scholar]
- de Sanctis D., Beteva A., Caserotto H., Dobias F., Gabadinho J., Giraud T., Gobbo A., Guijarro M., Lentini M., Lavault B. ID29: a high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J. Synchrotron Radiat. 2012;19:455–461. doi: 10.1107/S0909049512009715. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A., McRee D.E., Vacquier V.D., Stout C.D. The crystal structure of lysin, a fertilization protein. Science. 1993;262:1864–1867. doi: 10.1126/science.8266073. [DOI] [PubMed] [Google Scholar]
- Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc. Natl. Acad. Sci. USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Yang Z., Wolfner M.F., Aquadro C.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J., Aagaard J.E., Vacquier V.D., Monné M., Sadat Al Hosseini H., Jovine L. The molecular basis of sex: linking yeast to human. Mol. Biol. Evol. 2011;28:1963–1966. doi: 10.1093/molbev/msr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tina K.G., Bhadra R., Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier V.D., Carner K.R., Stout C.D. Species-specific sequences of abalone lysin, the sperm protein that creates a hole in the egg envelope. Proc. Natl. Acad. Sci. USA. 1990;87:5792–5796. doi: 10.1073/pnas.87.15.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman P.M. Mammalian fertilization: molecular aspects of gamete adhesion, exocytosis, and fusion. Cell. 1999;96:175–183. doi: 10.1016/s0092-8674(00)80558-9. [DOI] [PubMed] [Google Scholar]
- Wienken C.J., Baaske P., Rothbauer U., Braun D., Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 2010;43:186–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.