Abstract
Helicobacter pylori strains possessing the cag pathogenicity island (PaI) are associated with the development of gastroduodenal diseases, including gastric cancer. cag PaI products induce the secretion of interleukin-8 (IL-8) from epithelial cells and facilitate the translocation of CagA into the cell cytosol. In East Asia, where the incidence of gastric cancer is high, most strains possess the cag PaI. To date, however, no cag PaI phenotypic data have been provided for strains isolated in mainland China. Here we used 31 Chinese strains to determine the genotypic and phenotypic status of the cag PaI. All strains possessed cagA and cagE, and we observed a variation in the length of cagA variable regions. Nucleotide sequencing of the cagA variable region revealed that CagA was of two types, a short “Western” form with two tyrosine phosphorylation sites and a longer “East Asian” form with three tyrosine phosphorylation sites. Coculture of strains with AGS epithelial cells showed that strains could induce IL-8 secretion from the cells and that CagA with three phosphorylation sites became more phosphorylated than that with two and could induce significantly (P < 0.001) more cells to elongate. We hypothesize that the preponderance of the more active East Asian form of cagA may underlie the high rate of gastric cancer in China.
Helicobacter pylori is a spiral, gram-negative, microaerophilic bacterium that is present in the stomachs of approximately half of the world's population. H. pylori has been identified as a causative agent of chronic gastritis and of gastric and duodenal ulceration, and infection with this organism is an important risk factor for the development of gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma (12, 23). Several putative H. pylori virulence factors have been identified as additional risk factors involved in ulceration and gastric cancer, including the cytotoxin-associated gene (cag) pathogenicity island (PaI).
The cag PaI comprises a 40-kb DNA segment integrated into the H. pylori chromosome in some strains (9) and encodes a type IV secretory system, which is associated with enhanced virulence and increased mucosal inflammation. The cagA gene encodes the CagA protein and has been used as a marker for the presence of the cag PaI. The type IV secretory system facilitates the translocation of CagA into the cytosol of epithelial cells; CagA localizes to the inner surface of the plasma membrane (21) and subsequently undergoes tyrosine phosphorylation in the host cells (2, 7, 16, 19, 21) by Src family protein tyrosine kinases (2, 20, 22). The CagA tyrosine phosphorylation motifs (TPMs) have been mapped to the Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs present in the C-terminal region of the protein (3, 13, 15, 22). CagA proteins generally possess two or more EPIYA motifs within the variable region, which accounts for the size variation observed between CagA proteins isolated from different H. pylori strains. CagA proteins with greater numbers of TPMs become more phosphorylated and lead to enhanced formation of the “hummingbird” phenotype (1, 13). Analysis of cagA in H. pylori strains isolated from patients in East Asia, where the incidence of stomach cancer is high, has shown differences between the cagA variable region of those isolates and that of “Western” isolates (24) and also an association between the number of CagA TPMs and the development of atrophic gastritis and gastric cancer in Japan (5, 24).
In this study we looked at the CagA status of H. pylori strains from Chinese patients. Previously, Zhou et al. (25) showed that all 18 H. pylori strains isolated from the Hangzhou region of China possessed cagA and that 17 of 18 were of the “East Asian” type, although no sequence data were provided to show differences between cagA genes from different strains. Zhou et al. (26) showed that 79 of 82 Chinese H. pylori strains possessed cagA genes which, as determined by PCR amplification of the 3′ variable regions, were of three types, although no data were provided to show the composition of each of these variable regions. Neither study presented phenotypic data regarding the expression and tyrosine phosphorylation of CagA from these strains. We therefore decided to look at the cag PaI status and cagA variable region in H. pylori strains isolated from eastern China in terms of the numbers of CagA TPMs, tyrosine phosphorylation, and hummingbird phenotype formation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori strains were isolated from 31 Chinese patients, living in Jiangsu Province of mainland China, with chronic gastritis alone (12 cases), duodenal ulcers (11 cases), gastric ulcers (5 cases), both duodenal and gastric ulcers (2 cases), and gastric cancer (1 case), and were grown on horse blood agar plates (Oxoid, Basingstoke, United Kingdom) at 37°C in a microaerobic environment.
PCR amplification of cagA and cagE genes.
H. pylori genomic DNA was extracted from minimally passaged strains as previously described (4). PCR amplification of the cagA 3′ variable region with primers cag2 and cag4 as previously described for South African and European strains (18) was unsuccessful. The reverse primer was therefore redesigned as either cagA31 (5′-CGGCTATGCTCAACCTGGTGGAAAACTTGAACG) or cagA26R (5′-GCTTTAGCTTCTGATACC), and the PCR was performed under otherwise identical conditions. For nucleotide sequencing of cagA variable regions, PCR-amplified products (with primers cag2 and cagA26R) were purified with a QIAquick purification kit (Qiagen, Crawley, United Kingdom) before sequence analysis by the Biopolymer Synthesis and Analysis Unit, Queen's Medical Centre, University of Nottingham, United Kingdom. PCR amplification of cagE was carried out with primers PBRTF (5′-AAGGGTAAAGAAATGGGACTGAAT) and PBRTR (5′-GGAAGCGTGATAAAAGAGCAATGT). A reaction mixture containing a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.4 nM concentration of each primer, 0.05 U of Taq DNA polymerase (Roche Diagnostics, Penzberg, Germany) per μl, and 1 μl of genomic DNA, in buffer (10 mM Tris-HCl [pH 8.3], 1.5 mM magnesium chloride, 50 mM potassium chloride), was incubated at 95°C for 90 s; followed by 35 cycles of 95°C for 30 s, 56°C for 60 s, and 72°C for 90 s; and a final extension at 72°C for 5 min.
Infection of AGS cells with H. pylori strains and preparation of cell lysates.
AGS human gastric epithelial cells were seeded into 25-cm2 flasks (106 cells/flask) in Ham F-12 nutrient mixture and incubated at 37°C in a 5% CO2-air humidified atmosphere until almost confluent. H. pylori strains were resuspended in Ham F-12 nutrient mixture and diluted to an optical density at 550 nm of 0.1 (multiplicity of infection, ∼100) before addition to AGS cells for 6 h at 37°C in a 5% CO2-air humidified atmosphere. Infected AGS cells were washed twice with phosphate-buffered saline (PBS), and the cells were scraped from the flasks in 5 ml of PBS containing 1 mM sodium vanadate, harvested by centrifugation at 1,000 × g for 10 min, and then resuspended in 100 μl of PBS-vanadate and 50 μl of 4× sample loading buffer (0.2 M Tris-HCl [pH 6.8], 0.4 M dithiothreitol, 8% [wt/vol] sodium dodecyl sulfate, 40% [vol/vol] glycerol, 0.4% [wt/vol] bromophenol blue). Samples were heated at 100°C for 5 min before analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with antiphosphotyrosine monoclonal antibodies (clone PY99; Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-CagA polyclonal antibodies (Austral Biologicals, San Ramon, Calif.). Quantification of the degree of CagA phosphorylation was performed by densitometry with a Bio-Rad (Hemel Hempstead, United Kingdom) GS-800 calibrated densitometer and Quantity One software and was expressed as a ratio of phospho-CagA to total CagA.
Analysis of AGS cell hummingbird phenotype formation.
AGS cells were seeded into six-well dishes at a density of 2 × 105 cells/well and incubated at 37°C in a 5% CO2-air humidified atmosphere for 24 h. H. pylori strains were cocultured with AGS cells as described above, except that cells were incubated for 1 to 2 days, before the cells were examined for hummingbird phenotype formation (1) by microscopy of randomly chosen fields with a Nikon Eclipse TE200 microscope. Protrusions characteristic of the hummingbird phenotype (at least 100) were measured by using Lucia G (version 3.5) software. Statistical analysis was performed with a two-tailed Student's t test.
Nucleotide sequence accession numbers.
The nucleotide sequences of the cagA variable regions have been entered into the EMBL nucleotide sequence database under accession numbers AJ832140 to AJ832149.
RESULTS
PCR amplification of cagA and cagE genes.
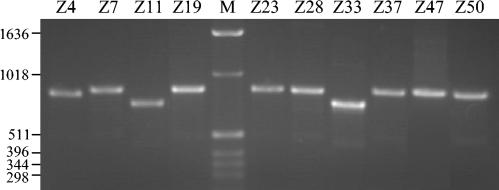
PCR amplification of cagA from Chinese H. pylori strains, using primers cag2 and cagA31, revealed that all (31 of 31) strains possessed this gene, although there was size variation in the lengths of products generated. Most strains possessed a cagA fragment of ∼890 bp, whereas three of the strains possessed smaller fragments of ∼840 bp (strain Z4) and ∼770 bp (strains Z11 and Z33) (Fig. 1). Amplification of cagE revealed that all (31 of 31) strains also possessed this gene.
FIG. 1.
cagA variable regions display size variation by PCR. PCR amplification of the 3′ cagA variable regions from Chinese H. pylori strains with primers cag2 and cagA31 is shown. Lane M, size markers (in base pairs).
Sequencing of the cagA variable region.
As we found size variation within cagA between strains, we decided to perform nucleotide sequence analysis of the cagA variable regions from 10 strains (the three with shorter cagA genes and seven of those with longer cagA genes). The cagA variable region was amplified by PCR with primers cag2 and cagA26R prior to nucleotide sequencing. We found that CagA variable regions from these Chinese isolates fell into two types: a short one possessing two EPIYA motifs and a longer one possessing three EPIYA motifs (Fig. 2). Of the three shorter cagA genes, we found that one (strain Z4) possessed three variable-region TPMs but had a deletion between the second and third EPIYA motifs, whereas the other two (strains Z11 and Z33) possessed only two EPIYA motifs. We also found that the variable region possessing two TPMs had a Western-type sequence, whereas the variable region with three TPMs had an East Asian-type sequence, characterized by the EATSAINRKIDRINKIASAGKGVGGFSGA pattern following the second EPIYA motif (Fig. 2).
FIG. 2.
Deduced amino acid sequences of 10 CagA variable regions. cagA variable regions were amplified by PCR with primers cag2 and cagA26R, and the products were purified and subjected to nucleotide sequencing. The deduced amino acid sequences are shown.
Expression and tyrosine phosphorylation of CagA.
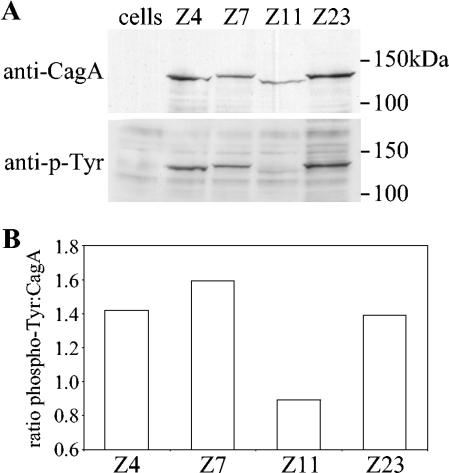
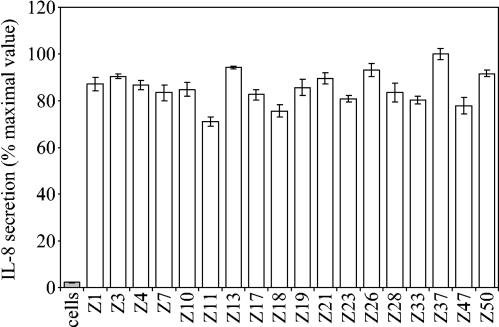
We next went on to look at the expression of CagA by Chinese H. pylori strains and whether they became phosphorylated within AGS epithelial cells by coculture. We tested 18 of the strains and found that all of them expressed CagA, which became tyrosine phosphorylated within AGS cells (Fig. 3), and all of these strains induced interleukin-8 secretion from AGS cells (Fig. 4). As expected, CagA proteins with two TPMs were of lower molecular weight than those with three TPMs, and densitometric analysis revealed that CagA proteins possessing three EPIYA motifs became more phosphorylated than those with only two EPIYA motifs (Fig. 3).
FIG. 3.
CagA with two variable-region TPMs is less phosphorylated than CagA with three variable-region TPMs. (A) AGS cells were cocultured with H. pylori strains possessing two (strain Z11) or three (strains Z4, Z7, and Z23) variable-region EPIYA motifs for 6 h at 37°C before the cells were lysed and the samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with antiphosphotyrosine monoclonal antibodies (lower panel). The blots were then stripped and reprobed with anti-CagA polyclonal antibodies (upper panel). (B) Densitometric analysis of the degree of CagA phosphorylation, expressed as a ratio of phospho-Tyr intensity to CagA protein intensity. The data shown are representative of those from three separate experiments.
FIG. 4.
Interleukin-8 (IL-8) secretion induced by H. pylori strains. Chinese H. pylori strains were cocultured with AGS cells for 6 h before the amount of secreted interleukin-8 was determined by enzyme-linked immunosorbent assay. The results are expressed as percentages of the maximal value (for strain Z37). Errors bars represent standard errors of the means.
Induction of AGS cell hummingbird phenotype formation by CagA.
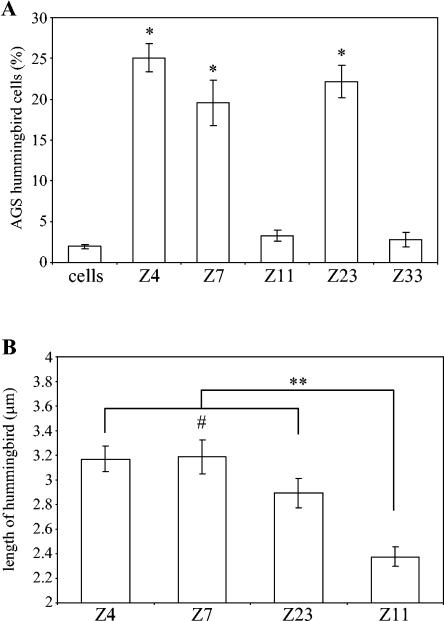
CagA phosphorylation within epithelial cells leads to hummingbird phenotype formation, the extent of which appears to be increased in proportion to the number of EPIYA motifs (1). We found, following 24-h coculture of AGS cells with H. pylori strains, that hummingbird phenotype formation was significantly (P < 0.001) more pronounced in AGS cells incubated with strains expressing CagA with three variable-region EPIYA motifs (Fig. 5A). Only 3% of AGS cells displayed the hummingbird phenotype when cocultured with strains expressing CagA with two variable-region EPIYA motifs, compared to an average of 16% for those with three TPMs. We also found that the average lengths of protrusions characteristic of the hummingbird phenotype induced were significantly (P < 0.0001) shorter for the CagA proteins with two variable-region EPIYA motifs than for those with three EPIYA motifs (Fig. 5B).
FIG. 5.
AGS cell hummingbird phenotype formation is reduced by CagA with two variable-region TPMs. AGS cells were cocultured with H. pylori strains Z4, Z7, and Z23 (three variable-region EPIYA motifs) and with strains Z11 and Z33 (two variable-region EPIYA motifs) for 1 day before cells were visualized by microscopy. (A) Percentages of AGS cells displaying the hummingbird phenotype in randomly chosen microscopic fields. *, P < 0.001 (Z4, Z7, or Z23, compared to Z11, Z33, or cells alone). (B) Average lengths of protrusions characteristic of hummingbird phenotype of at least 100 randomly chosen hummingbird phenotype AGS cells cocultured with each strain. **, P < 0.0001; #, P > 0.05. Errors bars represent standard errors of the mean.
DISCUSSION
H. pylori is recognized as a major causative agent of chronic gastritis and peptic ulcer disease and has been identified as a major risk factor for gastric cancer. In most studies, strains possessing cagA have been associated with an increased risk for the development of gastric cancer (8, 11), although some studies found no significant association (14, 17). This is especially true in East Asia, where more than 90% of strains possess cagA. As CagA shows size differences due to variations in the number of TPMs within the variable region of the protein, the degree of CagA phosphorylation may be a more important determinant of virulence than possession of cagA (1, 5, 24). Indeed, studies using H. pylori strains isolated from Japanese subjects have shown that strains expressing CagA with higher numbers of TPMs are more prevalent in patients with atrophic gastritis and gastric cancer (5, 24), and a study using H. pylori isolated from South Africans found that six of seven strains expressing CagA proteins with four or more variable-region EPIYA motifs were from patients with gastric cancer (1).
In this study we looked at the CagA status of H. pylori strains from Chinese patients. Most (28 of 31; 90%) of the strains possessed a longer form of cagA, but 3 of 31 strains possessed cagA genes with shorter variable regions, of which one was found to possess a deletion between the second and third EPIYA motifs. Sequencing analysis revealed that strains with longer cagA genes possessed three TPMs which were of the East Asian type, as they possessed the motif EATSAINRKIDRINKIASAGKGVGGFSGA (5), whereas the two cagA alleles with only two EPIYA motifs were of the Western type. The longer type of cagA variable region corresponded to the type I cagA described by Zhou et al. (26), which was found to occur in 67 of 71 Chinese strains (94%). Analysis of the degree of CagA phosphorylation by AGS cell coculture showed that CagA proteins with three TPMs became more phosphorylated than those with two TPMs. This also shows that CagA with only two EPIYA motifs can become phosphorylated, in agreement with other studies showing that all EPIYA motifs can become phosphorylated (3, 13, 15, 22).
Phosphorylation of CagA within the epithelial cell cytosol leads to hummingbird phenotype formation, which is dependent upon the degree of CagA phosphorylation. CagA proteins with greater numbers of EPIYA motifs become more phosphorylated and induce more AGS cells to display the hummingbird phenotype and increase the length of cellular protrusions (1, 13). Higashi et al. (13) showed, by mutagenesis of CagA from strain NCTC 11637, which has five EPIYA motifs (in an ABCCC pattern), that the C type of motif was responsible for binding SHP-2 and promoting hummingbird phenotype formation and that CagA with more C-type motifs caused more cellular rearrangements. It has also been shown that CagA proteins from East Asian H. pylori strains bind to SHP-2 phosphatase more strongly than those from Western strains, due to the presence of the pY-(S/T/A/V/I)-X-(V/I/L)-X-(W/F) SHP-2 phosphatase binding motif (10) in the C-type motif, EPIYATIDFD (6, 13). CagA proteins lacking this motif (or the EPIYATIDDL motif present in CagA proteins from Western strains) do not appear to bind the phosphatase (6). Coculture of AGS cells with the Chinese H. pylori strains used in this study revealed that the CagA proteins with three EPIYA motifs induce significantly more cells to undergo transformation into the hummingbird phenotype and induce longer protrusions than CagA proteins with only two EPIYA motifs, due presumably to lack of the third (C-type) TPM.
In summary, we show that, in H. pylori strains from Jiangsu Province in China, there are two allelic forms of cagA: the more commonly occurring form of East Asian origin and the infrequently occurring form of Western origin. Strains with these different forms of cagA display marked differences in CagA phosphorylation and induction of the hummingbird phenotype. As all strains possess cagA, the differences in Cag phenotypes may represent important differences in the pathogenic potentials of strains infecting different patients. Large studies are now needed to examine the association between the number of tyrosine phosphorylation sites and disease, especially among populations where cagA is virtually ubiquitous, such as that in China.
Acknowledgments
This work was funded by a grant from Cancer Research UK. Youli Zhang is funded by a scholarship from the Jiangsu Provincial Department of Education of China to conduct the research in the United Kingdom. John Atherton is funded by a Senior Clinical Fellowship from the Medical Research Council, United Kingdom.
We thank Joanne Rhead and Geraldine Flaujac-Lafontaine for technical assistance.
REFERENCES
- 1.Argent, R. H., M. Kidd, R. J. Owen, R. J. Thomas, M. C. Limb, and J. C. Atherton. 2004. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127:514-523. [DOI] [PubMed] [Google Scholar]
- 2.Asahi, M., T. Azuma, S. Ito, Y. Ito, H. Suto, Y. Nagai, M. Tsubokawa, Y. Tohyama, S. Maeda, M. Omata, T. Suzuki, and C. Sasakawa. 2000. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi, M., Y. Tanaka, T. Izumi, Y. Ito, H. Naiki, D. Kersulyte, K. Tsujikawa, M. Saito, K. Sada, S. Yanagi, A. Fujikawa, M. Noda, and Y. Itokawa. 2003. Helicobacter pylori CagA containing ITAM-like sequences localized to lipid rafts negatively regulates VacA-induced signalling in vivo. Helicobacter 8:1-14. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C. 1997. Molecular methods for detecting ulcerogenic strains of H. pylori, p. 133-143. In C. L. Clayton and H. L. T. Mobley (ed.), Methods in molecular medicine, Helicobacter pylori protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 5.Azuma, T., A. Yamakawa, S. Yamazaki, K. Fukuta, M. Ohtani, Y. Ito, M. Dojo, Y. Yamazaki, and M. Kuriyama. 2002. Correlation between variation of the 3′ region of the cagA gene in Helicobacter pylori and disease outcome in Japan. J. Infect. Dis. 186:1621-1630. [DOI] [PubMed] [Google Scholar]
- 6.Azuma, T., S. Yamazaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, Y. Ito, M. Dojo, Y. Yamazaki, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189:820-827. [DOI] [PubMed] [Google Scholar]
- 7.Backert, S., S. Moese, M. Selbach, V. Brinkmann, and T. F. Meyer. 2001. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol. 42:631-644. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 9.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza, D., L. J. Fabri, A. Nash, D. J. Hilton, N. A. Nicola, and M. Baca. 2002. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry 41:9229-9236. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo, C., J. C. Machado, P. Pharoah, R. Carvalho, A. F. Capelinha, W. Quint, C. Caldas, L.-J. van Doorn, F. Carneiro, and M. Sobrinho-Simões. 2002. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J. Natl. Cancer Inst. 94:1680-1687. [DOI] [PubMed] [Google Scholar]
- 12.Helicobacter and Cancer Collaborative Group. 2001. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case controlled studies nested within prospective cohorts. Gut 49:347-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miehlke, S., K. Kibler, J. G. Kim, N. Figura, S. M. Small, D. Y. Graham, and M. F. Go. 1996. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am. J. Gastroenterol. 91:1322-1325. [PubMed] [Google Scholar]
- 15.Mimuro, H., T. Suzuki, J. Tanaka, M. Asahi, R. Haas, and C. Sasakawa. 2002. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 10:745-755. [DOI] [PubMed] [Google Scholar]
- 16.Odenbreit, S., J. Püls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 17.Pan, Z. J., R. W. van der Hulst, M. Feller, S. D. Xiao, G. N. Tytgat, J. Dankert, and A. van der Ende. 1997. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J. Clin. Microbiol. 35:1344-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudi, J., C. Kolb, M. Maiwald, D. Kuck, A. Sieg, P. R. Galle, and W. Stremmel. 1998. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J. Clin. Microbiol. 36:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal, E. D., S. Falkow, and L. S. Tompkins. 1996. Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. USA 93:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyer, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277:6775-6778. [DOI] [PubMed] [Google Scholar]
- 21.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 23.Stolte, M., E. Bayerdörffer, A. Morgner, B. Alpen, T. Wündisch, C. Thiede, and A. Neubauer. 2002. Helicobacter and gastric MALT lymphoma. Gut 50(Suppl. III):19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, J., J. Zhang, C. Xu, and L. He. 2004. cagA genotype and variants in Chinese Helicobacter pylori strains and relationship to gastroduodenal diseases. J. Med. Microbiol. 53:231-235. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, W., S. Yamazaki, A. Yamakawa, M. Ohtani, Y. Ito, Y. Keida, H. Higashi, M. Hatakeyama, J. Si, and T. Azuma. 2004. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol. Med. Microbiol. 40:81-87. [DOI] [PubMed] [Google Scholar]