Abstract
The K-homology (KH) domain is a nucleic acid-binding domain present in many proteins but has not been reported in helicases. DDX43, also known as HAGE (helicase antigen gene), is a member of the DEAD-box protein family. It contains a helicase core domain in its C terminus and a potential KH domain in its N terminus. DDX43 is highly expressed in many tumors and is, therefore, considered a potential target for immunotherapy. Despite its potential as a therapeutic target, little is known about its activities. Here, we purified recombinant DDX43 protein to near homogeneity and found that it exists as a monomer in solution. Biochemical assays demonstrated that it is an ATP-dependent RNA and DNA helicase. Although DDX43 was active on duplex RNA regardless of the orientation of the single-stranded RNA tail, it preferred a 5′ to 3′ polarity on RNA and a 3′ to 5′ direction on DNA. Truncation mutations and site-directed mutagenesis confirmed that the KH domain in DDX43 is responsible for nucleic acid binding. Compared with the activity of the full-length protein, the C-terminal helicase domain had no unwinding activity on RNA substrates and had significantly reduced unwinding activity on DNA. Moreover, the full-length DDX43 protein, with single amino acid change in the KH domain, had reduced unwinding and binding activates on RNA and DNA substrates. Our results demonstrate that DDX43 is a dual helicase and the KH domain is required for its full unwinding activity.
Keywords: DNA, DNA enzyme, DNA helicase, DNA-protein interaction, RNA, RNA-binding protein, RNA helicase, RNA metabolism
Introduction
Helicases are molecular motors that transduce the chemical energy generated by ATP hydrolysis into an oligonucleotide strand separation and protein displacement. They are involved in virtually all aspects of nucleic acid metabolism, including replication, repair, recombination, transcription, chromosome segregation, and telomere maintenance (1–3). They move directionally along one nucleic acid strand; thus helicases are classified as 5′ → 3′ or 3′ → 5′ with respect to the strand they bind to and move along. Based on substrates, helicases can be classified as DNA or RNA helicases, although some can function on both DNA and RNA molecules (4). According to their conserved motifs, helicases are also classified into six superfamilies (5), among which SF2 is the largest superfamily that includes DEAD-box helicases.
Several subfamilies of DEAD-box helicases have generated particularly high interest because individual enzymes from these subfamilies are involved in multiple processes of RNA metabolism (2), such as transcription and pre-mRNA splicing (6, 7), ribosomes biogenesis (8), nuclear export (9), translation initiation (10), RNA degradation (11, 12), and organelle gene expression (13). Altogether, DEAD-box helicases are the largest group of enzymes involved in eukaryotic RNA metabolism. DEAD-box helicases possess a nonprocessive separation activity, and most RNA helicases examined to date do not efficiently unwind duplexes with more than one-and-a-half-helical turns in vitro, but they efficiently separate shorter duplexes (2). This activity is particularly used in many RNA metabolic processes in eukaryotic cells, where DEAD-box helicases act as important placeholders or checkpoint proteins, allowing processes to proceed in order and connect steps in the RNA metabolism machinery (15).
DDX43,2 (DEAD-box polypeptide 43; also known as HAGE (helicase antigen gene), was first identified as a cancer/testis antigen gene in a human sarcoma cell line (16). The gene is present on chromosome 6 (6q12-q13) as determined by radiation hybrid analysis and encodes a putative 73-kDa protein that belongs to the DEAD-box family of ATP-dependent RNA helicases. With the exception of testis that shows a high level of expression, DDX43 mRNA is found to be expressed by a wide range of tumor tissues at least 100-fold that of normal tissues (16). At the protein level, DDX43 is also detected at different levels in a variety of tumor tissues including bladder, brain, breast, colon, esophagus, kidney, liver, lung, stomach, and small intestine among others but not in normal tissues or at very low levels (17). DDX43 is also overexpressed in >50% of chronic myeloid leukemia, 20% of acute myeloid leukemia (18), and >40% of multiple myelomas. Therefore, considering the expression pattern and the diversity of DDX43 expression in different tumors, it may represent a suitable target for immunotherapy.
DDX43 protein contains the signature motifs of SF2 RNA helicases, including Q, I, Ia, Ib, II, III, IV, V, and VI (supplemental Fig. S1A). In addition, DDX43 also has conserved motifs Ic, Va, and Vb that are present in certain DEAD-box proteins. Besides the helicase core domain located in its C terminus, DDX43 also possesses potential K-homology (KH) domains at its N terminus (supplemental Fig. S1B) with its hallmark sequence, GXXG (19). The KH domain was first identified in the human heterogeneous nuclear ribonucleoprotein K (hnRNP K) more than two decades ago (20, 21) and subsequently has been found in a wide diversity of organisms spanning archaea, bacteria, and eukaryotes (22). KH domains exist in two different versions: type 1 KH-fold (KH1) is found in eukaryotes and often in multiple copies and type 2 KH-fold (KH2) is found in prokaryotes and often in a single copy (23, 24). The KH domain is ∼70 amino acids long with the most conserved consensus sequence VIGXXGXXI mapping to the middle of the domain (25). The KH domain is composed of three α-helices pack onto the surface of a central antiparallel β-sheet. KH1 and KH2 domains share a minimal βααβ core, with two additional α and β elements positioned at either the C terminus (KH1) or the N terminus (KH2) of this core motif (26, 27). The highly conserved GXXG loop has a specific sequence element that establishes contact with the nucleic acid, which is essential for the biochemical function of the proteins (19). KH domain-containing proteins perform a wide range of cellular functions, and several diseases, including paraneoplastic syndromes and some cancers, are associated with the loss of function of specific KH domains (19). Although KH domains have been found in a variety of proteins, no KH domain has been described in helicases.
Here, we found that DDX43 protein could function as both an RNA and DNA helicase in an ATP-dependent manner. Intriguingly, DDX43 unwound duplex RNA regardless of single-strand tail orientation, but it preferred a 5′ to 3′ polarity on RNA substrates and unwound DNA only in the 3′ to 5′ direction. Site-directed mutagenesis and biochemical assays revealed that there is a KH domain in the N terminus that is responsible for nucleic acid binding. The C-terminal helicase core domain displayed very weak unwinding on DNA substrates and no unwinding on RNA substrates. These results suggest that the KH domain is required for the unwinding activity of DDX43 helicase.
Results
Expression and purification of DDX43 protein
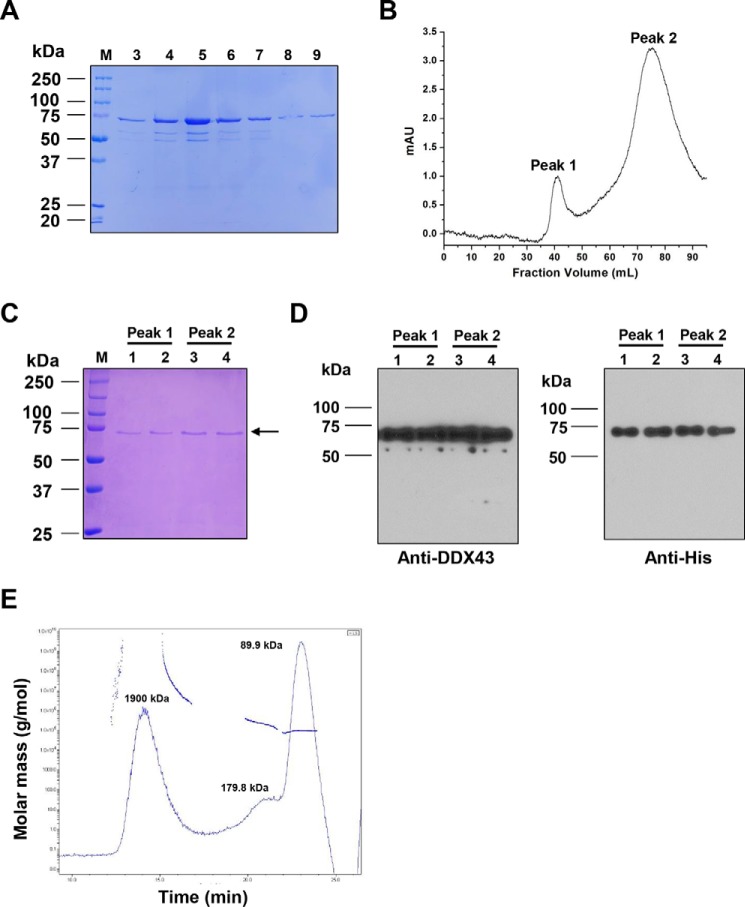
To characterize the DDX43 protein, we cloned the human DDX43 gene into a pET28a vector, overexpressed in Escherichia coli and purified through a two-step chromatography. First, the His-tagged DDX43 protein was passed through Ni-NTA affinity chromatography, and the retained proteins were eluted with imidazole. The recombinant DDX43 proteins were purified to near homogeneity as judged by their appearance on Coomassie-stained a SDS-polyacrylamide gel (Fig. 1A). The fractions with high protein concentration were pooled and applied to a Sephacryl S-300 size exclusion column. DDX43 protein eluted in two peaks, namely peak 1 and peak 2 (Fig. 1B). Identical migration bands were observed on the Coomassie stained SDS-PAGE gel for both peaks (Fig. 1C). The identity of DDX43 protein was confirmed by Western blot using anti-DDX43 and anti-His antibodies (Fig. 1D), indicating both fractions are indeed recombinant DDX43 proteins. According to the molecular mass standards used to calibrate the size exclusion column (supplemental Fig. S2), peak 1 was in the void volume of the column, likely representing aggregated DDX43 proteins, whereas the mass of peak 2 protein was 67.4 ± 5.1 kDa, which is close to the predicted monomeric form of DDX43 (72.4 kDa). Additionally, we also performed multiangle laser light scattering (MALS) and refractive index measurements to further confirm the oligomeric state of DDX43 protein. The MALS analysis revealed that the majority of the protein (86.4%) existed with a molecular mass of 89.9 kDa (Fig. 1E), in line with the previous observations of monomers from size exclusion column. A minor fraction (13.6%) was present as a dimer (179.8 kDa), which might be due to the protein concentration step using membrane cut-off. Because of the overlap of monomer and dimer peaks, the light-scattering-of-dimer fraction might have influenced the monomer, causing apparent higher molecular mass for the monomer fraction. Therefore, we collected the monomer fractions of DDX43 for the following biochemical assays.
Figure 1.
Purification and identification of DDX43 protein. A, SDS-PAGE analysis of the eluted DDX43 fractions from a Ni-NTA column. M, marker. Fractions 3–9 are shown. B, chromatographic profile of recombinant DDX43 proteins eluting from a Sephacryl S-300 HR column. Two peaks are indicated. mAU, milliabsorbance units. C, SDS-PAGE analysis of the peaks shown in B. D, Western blot analyses of the proteins shown in C with antibody against DDX43 (left) or His (right). E, SEC-MALS analysis of DDX43 protein shows the calculated molecular mass of the peak and the variation in the molecular weight across the peak for DDX43. The molecular mass for each peak was indicated.
DDX43 is an ATP-dependent RNA helicase
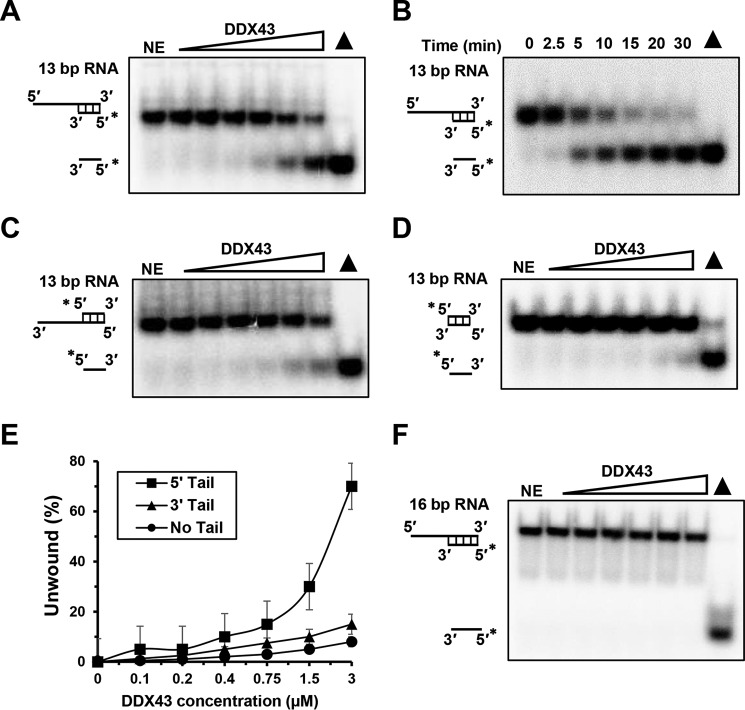
According to its primary protein sequence, DDX43 belongs to the DEAD-box protein family of RNA helicases (16); thus we started to characterize its RNA unwinding activity. Using a 5′-tailed 13-bp duplex RNA that is commonly used with the DEAD-box proteins (28), we found that DDX43 could efficiently unwind this substrate in the presence of ATP in both a concentration (0–3 μm) and a time-dependent manner (0–30 min) (Fig. 2, A and B). DDX43 also exhibited some unwinding activity on a 3′-tailed 13-bp duplex RNA (Fig. 2C) and a blunt-end 13-bp duplex RNA substrate (Fig. 2D). For example, at the highest concentration (3 μm), DDX43 unwound ∼60% of 5′-tailed dsRNA compared with ∼15% of 3′-tailed dsRNA and ∼10% of blunt-end dsRNA (Fig. 2E). However, when we increased the length from 13 to 16 bp for the 5′-tailed duplex RNA substrate, DDX43 failed to unwind this substrate under the same reaction conditions (Fig. 2F) even at high protein concentration (9 μm), suggesting that DDX43 has low processivity on duplex RNA.
Figure 2.
RNA helicase activity of DDX43 protein. A, a representative image of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate with increasing protein concentration (0–3 μm) at 37 °C for 15 min. NE, no enzyme. B, a representative image of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate and 150 nm DDX43 protein with increasing time (0–30 min) at 37 °C. C and D, representative images of helicase reactions performed by incubating 0.5 nm 3′-tailed 13-bp duplex RNA substrate (C) and 13-bp blunt end duplex RNA substrate (D) with increasing protein concentration (0–3 μm) at 37 °C for 15 min. E, quantitative analyses of RNA unwinding of DDX43 in panels A, C, and D. Data represent the mean of at least three independent experiments with S.D. indicated by error bars. F, a representative image of helicase reactions performed by incubating a 0.5 nm 5′-tailed 16-bp duplex RNA substrate with increasing protein concentrations (0–3 μm) at 37 °C for 15 min. RNA is in black.
To confirm that the unwinding activity detected was truly dependent on DDX43 and not because of contaminants in the preparation of the DDX43 protein, we changed the conserved lysine 292 to alanine (K292A) in motif I, which is essential for ATP hydrolysis, and aspartic acid 396 to alanine in motif II's DEAD-box (D396A), which helps to bind cations for enzyme activity (supplemental Fig. S1). Using the identical methods and conditions as wild-type DDX43, mutant proteins were purified to near homogeneity (supplemental Fig. S3A); however, neither K292A nor D396A displayed any unwinding activity on the 5′-tailed 13-bp duplex RNA substrate (supplemental Fig. S3, B and C).
DDX43 is an ATP-dependent 3′ → 5′ DNA helicase
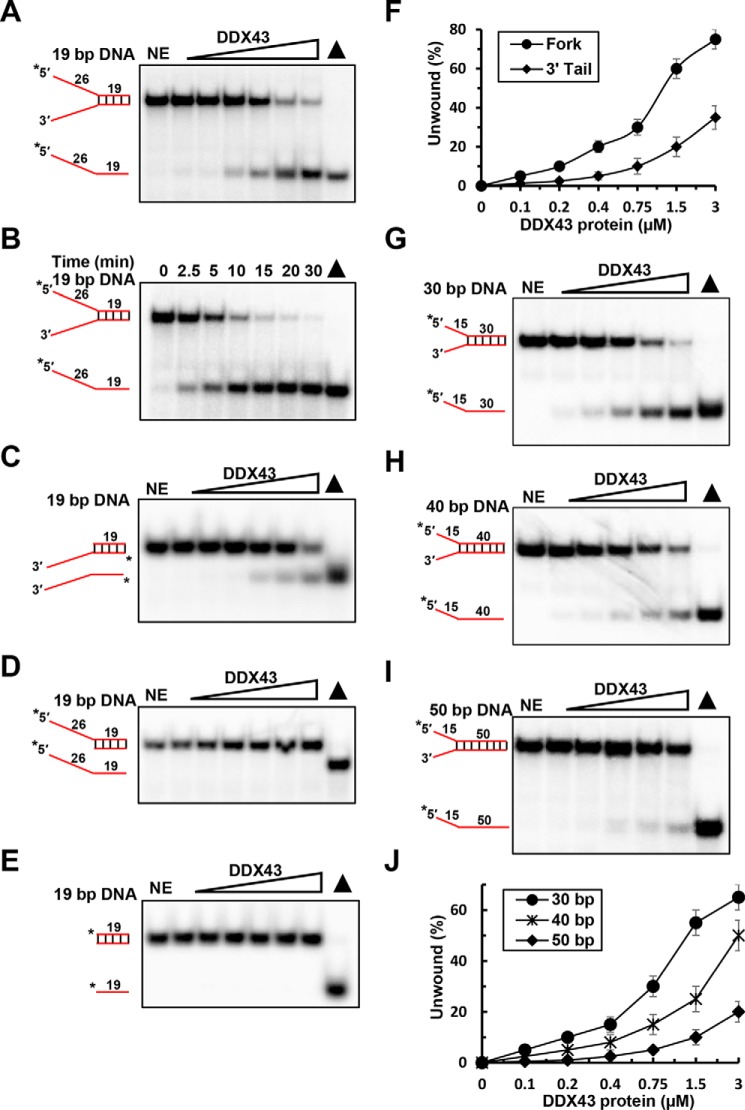
Many helicases have been found to possess dual unwinding activity, acting on both DNA and RNA substrates (4). Thus, we asked whether DDX43 can work on DNA substrates as well. Using a 19-bp forked duplex DNA substrate, we detected that DDX43 can efficiently unwind the DNA substrate in the presence of ATP in both a concentration (0–3 μm)- and a time (0–30 min)-dependent manner (Fig. 3, A and B). In contrast, it was less active on a 3′-tailed 19-bp duplex DNA (Fig. 3C) and totally inactive on a 5′-tailed 19-bp duplex DNA substrate (Fig. 3D) and a 19-bp blunt end duplex DNA substrate (Fig. 3E), indicating that DDX43 unwinds duplex DNA with defined directionality by the conventional mechanism, namely translocating in the 3′ → 5′ direction coupled to unwinding. At the highest concentration (3 μm), DDX43 unwound ∼76% of forked dsDNA and ∼25% of 3′-tailed dsDNA (Fig. 3F). Furthermore, we found that DDX43 could unwind 30-, 40-, and 50-bp forked duplex substrates; however, it had significantly reduced unwinding activity as the duplex length increased (Fig. 3, G–I). For example, at the highest concentration, DDX43 unwound ∼65% of 30-bp dsDNA, ∼50% of 40-bp dsDNA, and ∼15% of 50-bp dsDNA (Fig. 3J), suggesting that the processivity of DDX43 is moderate on DNA substrates. Also, we observed that DDX43 could unwind DNA/RNA hybrid substrates (supplemental Fig. S4A); however, it displayed higher efficiency when the tailed strand was RNA (supplemental Fig. S4, B–D).
Figure 3.
DNA helicase activity of DDX43 protein. A, a representative image of helicase reactions performed by incubating 0.5 nm 19-bp forked duplex DNA substrate with increasing protein concentrations (0–3 μm) at 37 °C for 15 min. NE, no enzyme. B, a representative image of helicase reactions performed by incubating 0.5 nm 19-bp forked duplex DNA substrate and 150 nm DDX43 protein with increasing time (0–30 min) at 37 °C. C–E, representative images of helicase reactions performed by incubating 0.5 nm 3′-tailed 19-bp duplex DNA substrate (C), 5′-tailed 19-bp duplex DNA substrate (D), or 19 bp blunt end duplex DNA substrate (E) with increasing protein concentrations (0–3 μm) at 37 °C for 15 min. F, quantitative analyses of DNA unwinding of DDX43 in panel A and C. Data represent the mean of at least three independent experiments with S.D. indicated by error bars. G–I, representative images of helicase reactions performed by incubating 0.5 nm 30-bp (G), 40-bp (H), or 50-bp (I) forked duplex DNA substrate with increasing protein concentrations (0–3 μm) at 37 °C for 15 min. J, quantitative analyses of DNA unwinding of DDX43 in panels G–I. Data represent the mean of at least three independent experiments with S.D. indicated by error bars. DNA is in red.
Again, to confirm that the DNA unwinding activity detected was truly dependent on DDX43 protein, we examined the unwinding activity of two engineered mutants, K292A and D396A, and found both mutations abolished unwinding activity of DDX43 on the DNA substrate (supplemental Fig. S3, D and E). Because many helicases have annealing activity, we also examined the annealing activity of DDX43 in the absence and presence of ATP and found that the DDX43 protein did not exhibit any annealing activity for either RNA (supplemental Fig. S5A) or DNA (supplemental Fig. S5B).
Because DDX43 can separate both dsRNA and dsDNA substrates, we sought to determine whether the single-stranded nucleic acid tail in substrates might influence the ability of DDX43 to unwind the flanking duplex. For RNA substrates, we used 8- and 18-nt 5′-tailed 13-bp duplex RNA substrate and found a slight increase in unwinding with increasing tail length (supplemental Fig. S6, A–C). For DNA substrates, increasing unwinding activity was observed when we increased the ssDNA tail from 15 to 25 nt (supplemental Fig. S6, D–F). Again, these results suggested that DDX43 unwinds RNA by a non-canonical/local destabilization mechanism while unwinding DNA by the canonical translocation mechanism, and the single-stranded nucleic acid's tail may serve as loading dock for DDX43 protein in both dsRNA and dsDNA.
ATP hydrolysis and Mg2+ are required for DDX43 unwinding
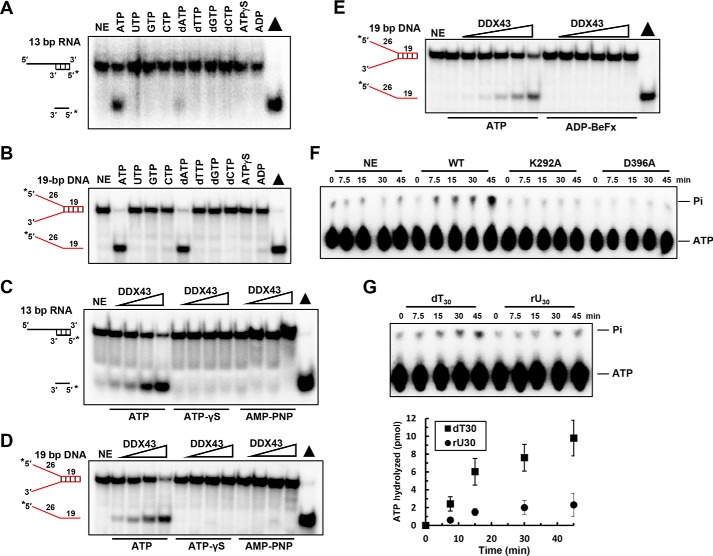
Helicases utilize the energy derived from ATP hydrolysis to separate base-paired DNA or RNA. First we aimed to identify the best NTP for the unwinding activity of DDX43 and found that efficient unwinding was observed with ATP and dATP (to a small extent) on the 5′-tailed 13-bp duplex RNA substrate (Fig. 4A), which is consistent with the previous findings for DEAD-box helicases, whereas both ATP and dATP were equally effective on the forked duplex DNA (Fig. 4B).
Figure 4.
ATP hydrolysis is required for DDX43 unwinding. A and B, representative images of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate (A) or forked 19-bp duplex DNA substrate (B) with 150 nm DDX43 protein at 37 °C for 15 min with different nucleoside triphosphates. NE, no enzyme. C and D, representative images of helicase reactions performed by incubating 0.5 nm 5′-tailed 13-bp duplex RNA substrate (C) or forked 19-bp duplex DNA substrate (D) with ATP or ATP analogs with increasing protein concentrations (0–3 μm). E, representative images of helicase reactions performed by incubating 0.5 nm 19 bp forked dsDNA substrate with increasing protein concentrations of DDX43 (0–3 μm) and 2 mm ATP or ADP-BeFx at 37 °C for 15 min. F, a representative image of ATP hydrolysis detected by TLC with DDX43 protein (2.5 μm) with M13 ssDNA effector (50 μm). WT, wild type; K292A and D396A are two engineered mutants. G, a representative image (top) of DDX43 protein ATP hydrolysis detected by TLC with indicated nucleic acid cofactor dT30 or rU30 (30 μm) and quantitative analyses of ATP hydrolysis (bottom). Data represent the mean of at least three independent experiments with S.D. indicated by error bars.
The conformational change caused by ATP binding with the DEAD-box helicases might lead to unwinding activity (29, 30); therefore, we used non-hydrolyzable ATP-analogs, ATPγS and AMP-PNP, and found no unwinding activity on DNA and RNA substrates (Fig. 4, C and D), indicating that ATP hydrolysis is essential for the unwinding activity of DDX43. So far the unwinding reactions were under multiple turnover conditions; thus, to mimic single turnover conditions, we used non-hydrolyzable ATP analog ADP-BeFx (30) that mimics the ATP pre-hydrolysis state and found that DDX43 could not unwind the DNA substrate (Fig. 4E). Thus, we concluded that ATP hydrolysis is required for DDX43 helicase activity on the substrates.
Next, we asked whether DDX43 could function as an ATPase. In the absence of DNA or RNA stimulator, DDX43 displayed no detectable ATP hydrolysis activity (data not shown). In the presence of circular single-stranded M13 DNA, the hydrolysis activity of DDX43 was greatly stimulated, whereas the engineered mutants DDX43-K292A and -D396A displayed no ATPase activity (Fig. 4F). Using dT30 or rU30 as a stimulator (both at 0.03 mm), DDX43 had greater ATP hydrolysis with dT30 compared with rU30 (Fig. 4G); for example, at the time point of 45 min, DDX43 hydrolyzed 9.8 ± 1.7 pmol of ATP in the presence of dT30 and 2.1 ± 0.9 pmol ATP in the presence of rU30. These results indicate that DDX43 possesses an intrinsic DNA- or RNA-dependent ATP hydrolysis activity. Also, we found that Mg2+ was the best cation for the helicase activity of DDX43 (supplemental Fig. S7, A and B), and ATP:Mg2+ ratios of 1:1 and 1:2 were the best for its unwinding activity (supplemental Fig. S7, C and D).
DDX43 has weak translocase activity but no RNP remodeling activity
DDX43 unwinds dsDNA from 3′ to 5′ but not from 5′ to 3′, indicating it might be a 3′ to 5′ translocase. Triplex displacement experiments have been utilized to monitor the translocase activity of helicases, such as AddAB (31), FANCM (32), and ChlR1 (33). In this assay a triple helix is formed when a third strand forms Hoogsteen base pairs with DNA duplex. If a translocase proceeds through the triplex, it will displace the third strand. Using a 5′-tailed triplex structure, we found that DDX43 could not displace the third strand, whereas ChlR1 helicase could (Fig. 5A); however, although it was weak, DDX43 could displace the third strand in a 3′-tailed triplex (Fig. 5B), indicating its 3′ to 5′ directional translocation activity.
Figure 5.
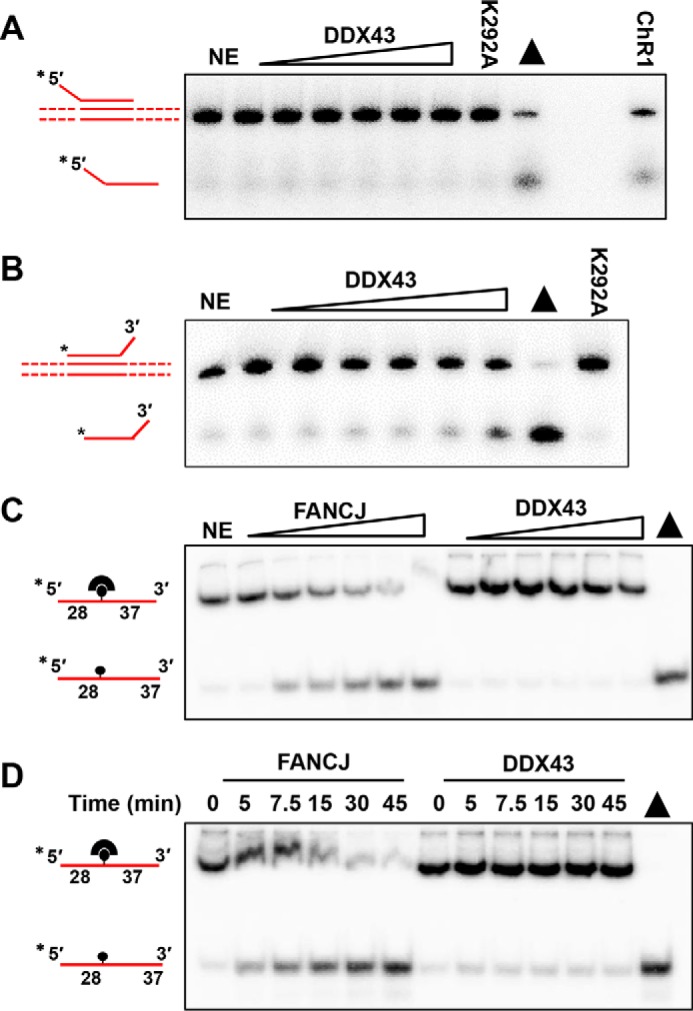
Translocase activity and protein displacement activity of DDX43 protein. A and B, translocation reactions (20 μl) were performed by incubating increasing concentrations of DDX43 protein (0–3 μm) with 0.5 nm 5′-tailed plasmid-triplex substrate (A) or 3′-tailed plasmid-triplex substrate (B) at 37 °C for 15 min under standard translocase assay conditions. ChlR1 (2.4 nm) was used as a positive control, and K292A was an engineered DDX43 mutant. Triangle, heat-denatured DNA substrate control. NE, no enzyme. C and D, representative images of protein displacement reactions performed by incubating 0.5 nm DNA substrate (X12-1-28-BioT37; supplemental Table S1) that had streptavidin bound with increasing protein concentrations of FANCJ (0–2.4 nm) and DDX43 (0–3 μm, C) at 37 °C for 15 min or increasing times (0–45 min, D) at 37 °C with a consistent protein concentration of FANCJ (1.2 nm) or DDX43 (1.5 μm).
Next, we asked whether DDX43 coupled its translocase activity to nucleoprotein remodeling activity. We performed a streptavidin displacement assay in which we monitored the ability of DDX43 to displace a streptavidin molecule bound to a biotinylated DNA. Because DDX43 behaves as a 3′ → 5′ DNA helicase, we designed a 65-mer oligonucleotide that has the biotin-conjugated 28 nt from the 5′-end and the 37 nt from the 3′-end (supplemental Table S1). Briefly, DDX43 protein and the substrate were preincubated before initiation of the reaction by adding ATP/Mg2+ mixture and an excess of biotin to trap free streptavidin molecules. As shown in Fig. 5, C and D, DDX43 failed to displace streptavidin in either a protein concentration or a time-dependent fashion. Under the same reaction conditions, FANCJ helicase was able to remove streptavidin from DNA as we have previously observed (34).
DDX43 has a KH domain in its N terminus
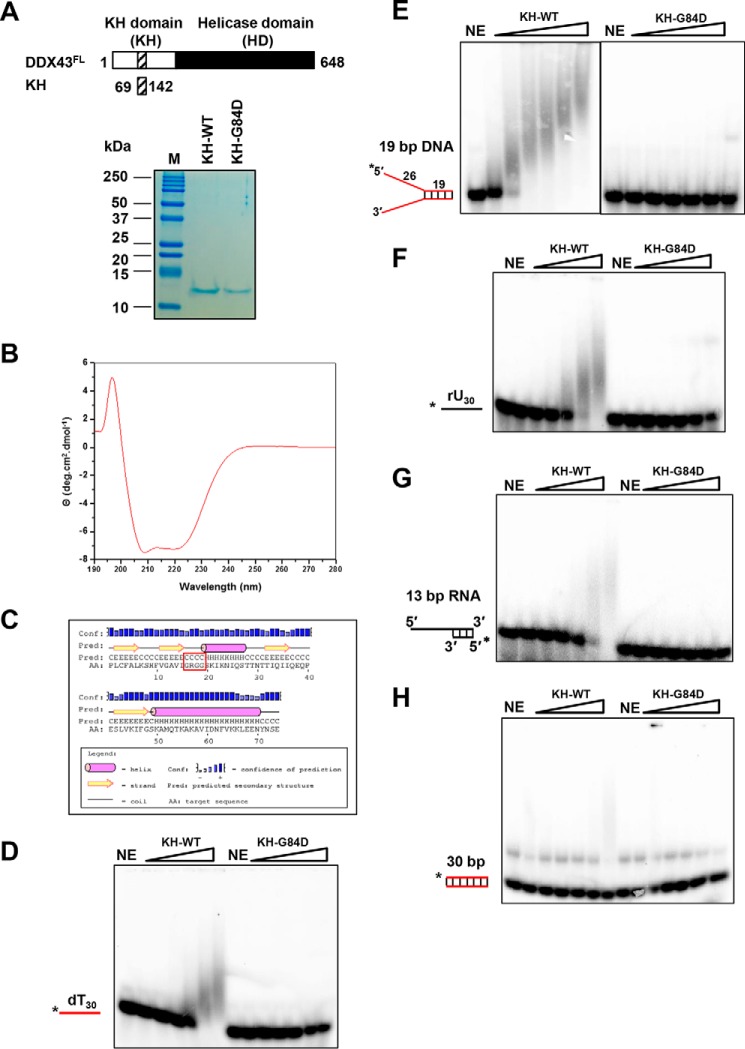
From its primary sequence, DDX43 contains three potential KH domains with three signature GXXG sequences (supplemental Fig. S1). Close inspection of the second GXXG sequence revealed that it has additional nearby conserved amino acid sequences shared with other well known KH domain-containing proteins (supplemental Fig. S8A), which was absent in the first and third sequence (data not shown). Moreover, these amino acids are conserved in DDX43 orthologues across species (supplemental Fig. S8B). To confirm experimentally that the second GXXG sequence-containing region is a KH domain, we cloned these conserved 74 amino acids (69–142 aa, named DDX43KH) into a pET28 vector and purified this protein to near homogeneity (Fig. 6A). Previous studies have shown that the first glycine residue in the GXXG sequence is essential for RNA binding function of the KH domain (35–37); thus, we changed the first glycine to aspartic acid in the GXXG sequence (DDX43KH-G84D). The far UV-wavelength scan of the protein revealed that DDX43KH is typically well folded with predominant α-helical structures, having a dip at 222 and 208 nm (Fig. 6B). This result is concordant with the previous reports of KH domains of the P-element somatic inhibitor protein (38) and Fragile X mental retardation protein (39, 40). Further analysis of the CD data with Selcon 3 (41) revealed that the KH domain of DDX43 has 41% α-helices, 35% β-sheets, and 24% unordered. This random coil might be due to the N-terminal and C-terminal His tag obtained from the vector. The secondary structure prediction by PSIPRED (42) also revealed similar results: 40% α-helices, 30% β-sheets, and random coil showing that the GRGG motif falls within the predicted βα-folded domain (Fig. 6C). Similar results were obtained for the DDX43KH-G84D proteins (data not shown). Electrophoretic mobility shift assay (EMSA) revealed that the wild-type KH domain bound the ssDNA and forked dsDNA; in contrast, the mutant, DDX43KH-G84D, showed no binding (Fig. 6, D and E). Similar results were obtained with ssRNA rU30 and dsRNA (Fig. 6, F and G). None of them bound blunt-end dsDNA (Fig. 6H), which is consistent with the structural evidence (43).
Figure 6.
Purification and characterization of DDX43 KH domain. A, schematic representation of full-length DDX43 and its KH domain (top) and purified KH domain proteins (wild type and mutant, 1 μg of protein each loaded) shown on Coomassie-stained SDS-PAGE gel (bottom). NE, no enzyme. B, circular dichroism spectrum of KH domain protein. C, secondary structure prediction of KH domain protein. The motif GRGG is highlighted in red square. D–H, representative EMSA images for KH domain proteins binding with 0.5 nm dT30 ssDNA (D), 19-bp forked duplex DNA (E, two images merged), ssRNA rU30 (F), 5′-tailed 13 bp dsRNA (G), and blunt-end dsDNA (H).
To elucidate potential sequence specificity binding for the KH domain, we labeled a dC30 (oligodeoxycytosine 30 residues), a dA30 (oligodeoxyadenine 30 residues), and a random DNA oligo (DNA 30-mer, supplemental Table S1) under the same reaction conditions as dT30 and rU30; EMSA showed that the KH domain exhibits binding affinity in the order of random DNA > rU > dT > dC > dA (Fig. 6, D and F; supplemental Fig. S9).
To eliminate the possibility that the two other GXXG sequences were the KH domain motifs, we also cloned the N-terminal region (1–253 aa, named DDX43NT) that contains all three GXXG sequence into a pET28a vector for protein expression. We changed the first glycine-to-aspartic acid in each conserved GXXG sequence, namely G46D, G84D, and G154D (named DDX43NT-G46D, NT-G84D, and NT-G154D, respectively). Three mutant proteins were purified along with wild type to near homogeneity (supplemental Fig. S10A). Using the 19-bp forked duplex DNA substrate, the EMSA result revealed that G46D and G154D bound forked dsDNA comparable with or even better than wild-type protein (supplemental Fig. S10B); however, the G84D mutation abolished its binding activity. Similar results were obtained with ssDNA (supplemental Fig. S10C) and RNA substrates (supplemental Fig. S10D). None of them bound blunt-end dsDNA (supplemental Fig. S10E). Collectively, we concluded that the second 84GXXG87 sequence-containing region is a functional KH domain responsible for nucleic acid binding.
KH domain is required for efficient helicase activity of DDX43 protein
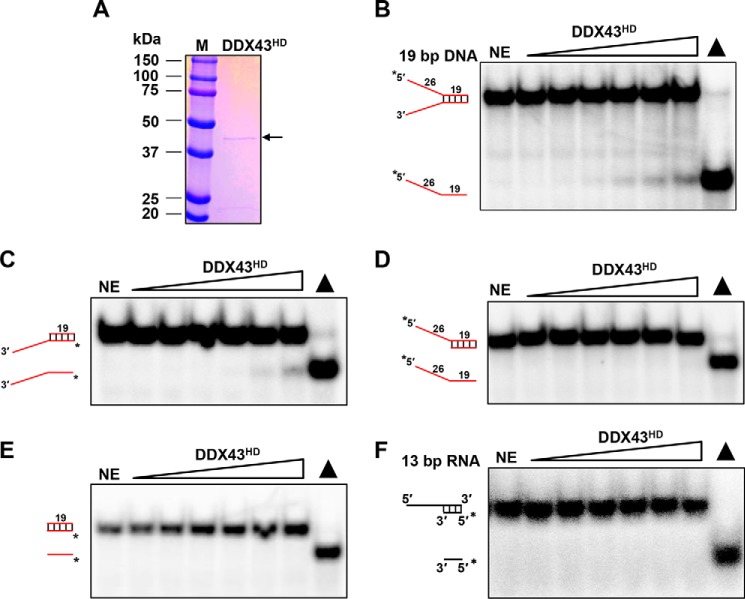
As we observed that DDX43 unwinds RNA duplex with a non-processive mechanism and DNA duplex with a processive mechanism, we suspected that the helicase core domain may be responsible for this phenomenon (5). To address this, we purified the C-terminal helicase core domain (254–648 aa, named DDX43HD) to near homogeneity (Fig. 7A). Using a 19-bp forked duplex DNA substrate, we detected unwinding activity for the helicase domain protein (Fig. 7B), although it was weaker than the full-length DDX43 protein. Moreover, it was active on a 3′-tailed 19-bp duplex DNA substrate (Fig. 7C) but not active on 5′-tailed (Fig. 7D) or blunt end 19-bp duplex DNA substrates (Fig. 7E), indicating that the helicase domain, like the full-length protein, unwinds duplex DNA with defined directionality, namely a 3′ to 5′ direction. Unexpectedly, unwinding activity was not detected for the RNA substrate at our regular reaction conditions (Fig. 7F), even at increased enzyme (9 μm) or reaction time (45 min, data not shown). These results suggest that the helicase core domain is responsible for the processivity on DNA molecules; however, it is not as active as the full-length protein.
Figure 7.
Purification and characterization of DDX43 helicase core domain. A, SDS-PAGE analysis of the DDX43 helicase domain protein eluted from a Sephacryl S-300 HR column. 1 μg of protein was loaded. NE, no enzyme. B–F, representative images of helicase reactions by incubating increasing DDX43HD protein (0–3 μm) with 0.5 nm 19-bp forked dsDNA (B), 3′-tailed 19-bp dsDNA (C), 5′-tailed 19-bp dsDNA (D), 19-bp blunt-end dsDNA (E), and 13-bp 5′-tailed dsRNA (F).
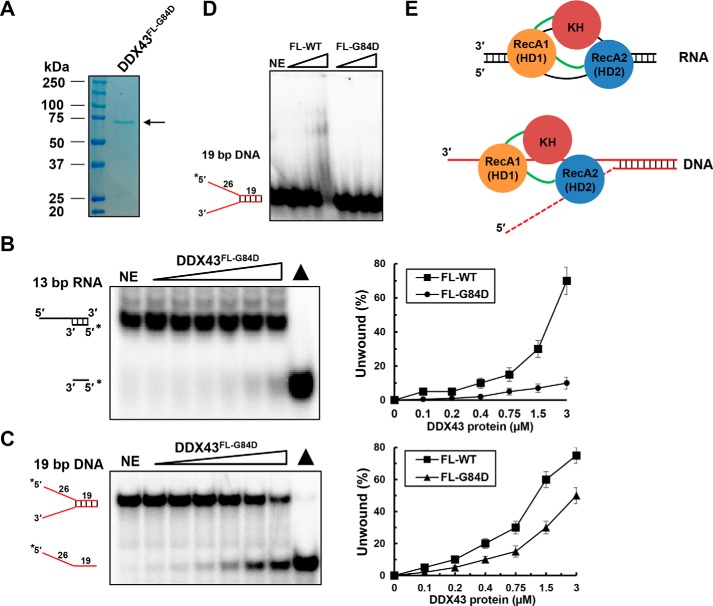
Because the helicase core domain does not exhibit efficient unwinding activity as compared with the full-length protein, we suspected that the KH domain is essential for its full functionality. To address this, we mutated the conserved glycine at position 84 to aspartic acid (named DDX43FL-G84D) in the context of full-length DDX43 protein. The mutant protein was purified to near homogeneity (Fig. 8A). Helicase assays revealed that DDX43FL-G84D had reduced unwinding activity on both RNA (Fig. 8B) and DNA substrates (Fig. 8C), suggesting that the KH domain is required for the full unwinding activity of DDX43 protein. EMSA showed that the DDX43FL-G84D had significant reduced binding ability to the forked dsDNA substrates (Fig. 8D), suggesting that the reduced unwinding activity might be due to poor binding between DDX43 protein and substrates. Taken together, these results suggest that the DDX43 protein's N-terminal KH domain binds to nucleic acids independent of its C-terminal helicase domain, whereas the C-terminal helicase domain requires N-terminal KH domain for its full activity (Fig. 8E). Our EMSA results also revealed that the N-terminal region has higher affinity to DNA and RNA molecules than the helicase core (supplemental Fig. S11). These results suggest that the presence of an ancillary KH domain is essential for the efficient biochemical activities of the C-terminal helicase domain, likely tightening the binding between helicase core and nucleic acid substrates.
Figure 8.
KH domain mutation affects unwinding activity of DDX43. A, SDS-PAGE analysis of the DDX43 full-length G84D mutant protein. NE, no enzyme. B and C, representative images (left) and quantitative analyses (right) of helicase reactions by incubating increasing DDX43FL-G84D protein (0–3 μm) with 0.5 nm 13-bp 5′-tailed dsRNA (B) or 19-bp forked dsDNA (C). D, a representative EMSA image for DDX43 full-length wild type and G84D mutant proteins binding with 0.5 nm 19-bp forked duplex DNA. E, proposed models for the role of KH domain in DDX43 helicase unwinding for dsRNA (upper) and dsDNA (bottom). RecA1 (HD1), RecA2 (HD2), and KH domains are shown in orange, blue, and dark red, respectively, and linkers are in green. The dashed red line in dsDNA representing the 5′ tail may or may not be there. For the visible purpose, the RNA:RNA bubble is larger than the reality.
Discussion
Although KH domains have been reported in many RNA-binding proteins, the role of a KH domain in a helicase has not been studied. In this study we have demonstrated that DDX43 is an ATP-dependent dual helicase that unwinds both RNA and DNA substrates. It catalyzes the unwinding reaction most efficiently in the presence of Mg2+ and ATP. Intriguingly, DDX43 does not follow a strict translocation mechanism to unwind RNA substrates, but it unwinds DNA in a unidirectional manner. We also discovered that the KH domain in the N-terminal region is involved in nucleic acid binding and is essential for the efficient unwinding activity of DDX43 helicase.
Superfamily 2 (SF2) helicases tend to be strictly specific for RNA or DNA, but it is not uncommon that a helicase can unwind both DNA and RNA substrates. Most of the DEAD-box proteins display clear specificity for RNA, whereas very few can unwind both DNA and RNA substrates, e.g. p68 (DDX5) (44) and Dbp9p (DDX56) (45). Some can unwind DNA-RNA chimeric substrates as well, e.g. Ded1 (46) and DDX1 (47). DDX43 is classified as a DEAD-box RNA helicase based on sequence homology to SF2 superfamily. Indeed, it has been reported that DDX43 unwinds duplex RNA (48). Our biochemical characterization of DDX43 indicated that both DNA and RNA can stimulate its ATP hydrolysis activity, and it can unwind both the substrates; therefore, DDX43 is an addition to the dual helicase family.
Generally, RNA helicases have low processivity on the substrate. For example, eIF4A (DDX2), a prototypic member of the DEAD box family, acts in a non-processive manner to unwind 10–15-bp duplex RNA (49). In fact, almost all members of the human DEAD-box family studied so far appear similar to eIF4A and have limited processivity; for example, 10 bp for DDX1 (50), 20 bp for DDX25 (51), 15 bp for DP103 (DDX20) (52), and 25 bp for RH-II (DDX21) (53). Unlike most helicases, the efficiency of strand separation for DEAD-box proteins is strongly dependent on the stability of the helix, thus, unwinding efficiency decreases with increase in the length and stability of the duplex. A few exceptions to this trend exist, such as DDX3 (50 bp) (54), p68 (162 bp) (55), and p72 (DDX17, 41 bp) (56) were shown to unwind moderately long RNA duplexes. However, it should be noted that reaction conditions, such as the amount of substrate and protein concentration, duration of reaction, amount of ATP, and so on may cause the difference reported. Even in the case of DDX43, it has been reported it could unwind 40-bp duplex RNA in certain conditions (48, 57). Nevertheless, most of the DEAD-box helicases have limited processivity.
Many DEAD-box proteins unwind RNA substrates without any preference for the polarity of single-stranded overhangs and unwind duplexes by interacting with an internal region of an RNA duplex and causing local strand separation; as a result, unwinding occurs without a defined polarity (46). Using a 16-bp duplex RNA substrate, Ded1 and Mss116p have been shown to unwind RNA duplexes without strict polarity, including blunt-end duplexes (46, 58). Further additions to this list are p68 (59), eIF4A (60), DDX25 (51), Has1p (61), and SRMB (62). This distinct unwinding mode of DEAD-box proteins appears uniquely suited for the localized separation of short duplexes in the cell. First, usually short duplex RNAs (no longer than 10 bp) are present in cells. Second, the structure of Mss116p with RNA revealed that a single protein monomer could bind along most of the length of U10 RNA, raising the possibility that strand separation by DEAD-box proteins does not involve any translocation at all (63).
In our study we found that DDX43 could efficiently unwind 13-bp dsRNA but not 16-bp dsRNA, suggesting that DDX43 is a low processivity DEAD-box helicase. It should also be noted that exclusive RNA duplex structures exist mainly during viral replication and transcription, and helical elements within structured RNAs are rarely longer than 10 base pairs (64). Thus, there appears to be little need for highly processive RNA helicases. We also found that DDX43 can unwind the blunt-end RNA substrate, suggesting that the unwinding could begin internally. However, DDX43 is more active on 5′-tailed than 3′-tailed dsRNA substrates, the reason for which is unknown. A very recent study reported that the Ski2-like RNA helicase Mtr4p unwinds RNA duplexes by 3′ to 5′ translocation (65), suggesting some RNA helicases may unwind RNA duplexes by translocation-based mechanisms.
Translocation polarity is a predefined and inherent feature for a particular enzyme and reflects helicase movement either in the 3′ → 5′ or 5′→3′ direction (5). Helicase domain 1 (Rec A1) interacts with the 3′-end, and helicase domain 2 (Rec A2) interacts with the 5′-end of bound single strands, resulting in an interface with a well defined asymmetry that is likely to differentiate the two possible strand orientations within the binding cleft (66). Intriguingly, in our study we found that DDX43 unwinds the DNA substrate by canonical mechanism, that is, in a defined 3′ → 5′ direction. It has been found that the helicase core domains HD1 and HD2 in the Mss116p DEAD-box helicase have modular functions enabling a novel mechanism for RNA-duplex recognition and unwinding where HD1 binds ATP and HD2 contains a nucleic acid-binding pocket, which accommodates A-form but not B-form duplexes (67). Also, the crystal structure of Msp116 with ssDNA and ssRNA revealed that the protein forms a closed-state complex with both nucleic acids. Although the interactions are similar, the closed-state complex with ssRNA contains protein contacts to RNA's 2′-OH groups that are absent in the closed-state complex with ssDNA (68). To probe the effects of RNA in various parts of the loading strand, a set of chimeric substrates was synthesized according to previous studies on NPH-II helicase (69) (supplemental Table S1). For the same duplex, such as the RNA duplex (supplemental Fig. S12, A and B) and DNA:RNA hybrid (supplemental Fig. S12, C and D), RNA is the preferred loading strand for DDX43, and even the few nucleic acids that are immediately adjacent to the junction with duplex made a difference (supplemental Fig. S12, E–G). This provides a basis for substrate specificity and might explain the differential unwinding mechanisms on RNA and DNA substrates. However, additional biochemical and structural studies are required to elucidate the unique polarity of DDX43 helicase on RNA and DNA substrates: for example, site-directed mutagenesis of helicase domain 1 in DDX43 to determine whether its unwinding is directly regulated by helicase domain 1 as reported in SF2 helicase (70). Also co-crystallization of DDX43 protein with nucleic acids may provide evidence of directional translocation.
Most DNA helicases use translocase mechanisms to unwind its substrate. A high degree of processivity is crucial for helicases involved in DNA replication, where millions of base pairs must be replicated quickly. In our study we found that DDX43 is more processive on the DNA substrates compared with the RNA substrates; however, the rate of duplex unwinding is significantly decreased with increasing length of the duplex, which is different from classic DNA helicases. A key property of DNA helicases is their ability to unwind long duplexes. Certain DNA helicases (e.g. MCM (71), RecBCD (72), TraI (73)) can processively unwind DNA tracts with longer than 500 base pairs. Even on a preferred forked duplex DNA substrate, DDX43 acts inefficiently to unwind a 50-bp duplex, suggesting that DDX43 is specifically tailored to act on short duplex substrates; however, its exact role in DNA metabolism needs further investigation.
Many helicases contain accessory domain(s) in their N-terminal or C-terminal regions that play critical roles in helicase function. It has been reported that DEAD-box helicases bind to helix extensions with low or no specificity, but the ancillary domains help in target recognition and thus unwinding. For example, Thermus thermophilus protein Hera binds structured RNAs through a RNA recognition motif (RRM)-like domain (74); another DEAD-box helicase DbpA uses its C-terminal ancillary domain to recognize a specific hairpin within the 23S rRNA and plays an important role in biogenesis of the large ribosomal subunit (75). Furthermore, deletion of the C-terminal tail, which is involved in RNA recognition in CYT-19 and Ms116p, has been found to affect the RNA chaperon activities of both proteins (76, 77). In contrast, negative regulation has been reported in DDX19 helicase, where the N-terminal extension prevents its ATP hydrolysis activity (78).
In our study we found that the KH domain in the N terminus of DDX43 is responsible for substrate binding. Consistent with literature of DEAD-box helicases, our results showed that only the full-length DDX43 protein could unwind RNA and DNA duplex substrates. The truncated protein, the C-terminal helicase domain, did not show any unwinding activity on RNA and had very weak unwinding activity on DNA, indicating that the KH domain is crucial for DDX43 to perform unwinding activity on RNA substrates, which supports the notion that the KH domain is an RNA-binding domain (79). More studies are required to determine the structural and functional role of the KH domain in DDX43 helicase; for example, whether the KH domain in DDX43 forms a βααβ core with the two additional α and β elements positioned in its C terminus and whether the KH domain dictates the sequence-specific recognition of substrates for DDX43 helicase and/or stimulates the unwinding activity of the helicase core domain by binding ssRNA/ssDNA to prevent them reannealing (Fig. 8E).
DDX43 is overexpressed in a number of cancers and thus can serve as a biomarker and immunotherapy target. Recently, it was shown that the expression of DDX43 is a potential prognostic marker and a predictor of response to anthracycline treatment in breast cancer (80, 81). Frequent expression of DDX43 was also found in chronic myeloid leukemia and acute myeloid leukemia (18, 82, 83), and its expression is associated with advanced disease and poor prognosis in chronic myeloid leukemia (83). Therefore, DDX43 is considered as an ideal target for vaccine therapy. Vaccines, dendritic cell, peptide, and whole cell-based immunotherapy can be used to target DDX43 in various cancers. Although our current biochemical evidences suggest its KH domain is essential for DDX43's enzymatic activity, the KH domain, a short functional domain, can be a candidate epitope for the development of peptide vaccines against tumor immunotherapy.
Conclusively, our biochemical results demonstrate that DDX43 is a unique dual function DEAD-box helicase, indicating its potential distinctive cellular functions that might correlate with its overexpression in cancers.
Experimental procedures
Plasmid DNA
Human DDX43 cDNA clone was purchased from the SPARC BioCentre, the Hospital for Sick Children, Toronto, Canada. DDX43 gene (full-length, N-terminal region, C-terminal helicase domain, and KH domain) was PCR-amplified and cloned into the NdeI and XhoI sites of a pET28a vector (Novagen). The lysine mutation in motif I K292A, the aspartic acid mutation in DEAD-box motif II D396A, and first glycine in GXXG sequence mutations G46D, G84D, and G154D were generated with the QuikChange site-directed mutagenesis kit (Agilent Technologies) using primers listed in supplemental Table S2. All plasmids were verified by DNA sequencing.
Recombinant protein
The plasmid pET28a-DDX43 was transformed into E. coli Rosetta 2 cells (EMD Millipore). The Rosetta 2 cells were grown at 37 °C in LB medium containing 30 μg/ml kanamycin and 34 μg/ml of chloramphenicol until the A600 reached 0.6 and then induced by the addition of 0.3 mm IPTG overnight at 15 °C. The cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C. The periplasmic material was removed from the cells as described (83). Briefly, the cells were suspended in 5 ml/g of cell mass of hypertonic buffer solution (50 mm HEPES, pH 7.4, 20% sucrose, 1 mm EDTA) and centrifuged at 8000 rpm for 10 min at 4 °C. The cells were resuspended in 5 ml/g of cell mass of hypotonic solution (5 mm MgSO4) and incubated for 10 min on ice. Cells were then pelleted by centrifugation at 4000 rpm for 10 min at 4 °C and stored at −80 °C until used. The cell suspension was lysed by sonication in buffer A (25 mm Tris, pH 8.0, 0.15 m NaCl, 100 μm Tween 20, and 10% glycerol) having a final concentration of 1 mm phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor (Roche Applied Science) at 4 °C with 5 short bursts of 10 s at intervals of 5 min. The cell debris and inclusion bodies were removed by centrifugation at 45,000 × g for 30 min at 4 °C. Recombinant His-tagged proteins were subjected to a two-step purification using nickel affinity beads (Sigma) and a Sephacryl S-300 HR 16/60 gel filtration column (GE Healthcare). The supernatant was applied to the Ni-NTA beads equilibrated with buffer A and washed with 10 column volume (CV) of buffer B (25 mm Tris, pH 8.0, 0.5 m NaCl, 100 μm Tween 20, and 10% glycerol) containing 25 mm imidazole and eluted with 5 CV of buffer B containing 250 mm imidazole. The protein fractions were confirmed with SDS-PAGE; the fractions with high protein yield were pooled and subjected to size-exclusion chromatography on a Sephacryl S-300 HR 16/60 (GE Healthcare) equilibrated with buffer A. The fractions were collected at a flow rate of 0.5 ml/min with the same buffer. The proteins in peaks were confirmed with SDS-PAGE, and the ideal fractions were pooled and concentrated. The N-terminal region, C-terminal helicase domain, and KH domain proteins were purified similar to the full-length DDX43. FANCJ protein was purified as described previously (34). All proteins were snap-frozen in liquid nitrogen and stored at −80 °C. Protein concentration was determined by the Bradford method using bovine serum albumin (BSA) as the standard.
Size exclusion chromatography and multiangle light scattering
Size-exclusion chromatography with SEC-MALS was performed using the Waters HPLC systems coupled to the refractive index and static laser light-scattering instruments (Wyatt MiniDAWN Treos and Wyatt OptiLab rEX, respectively). The DDX43 protein, purified by Ni-NTA chromatography, was concentrated using a 30-kDa centrifugal cut-off spin column (Millipore) and was injected (0.8 mg/ml; 500 μl) onto the Wyatt WTC030S5 size exclusion column (300 Å pore size for the elution of 5–1250-kDa proteins) equilibrated with buffer A (without Tween 20). The light scattering and refractive index data were used to calculate the weight-averaged molar mass and the mass fraction of each peak using the AstraTM package v 6.1.2 (Wyatt Technologies).
Circular dichroism (CD) spectroscopy
The proteins were eluted with buffer C (10 mm phosphate buffer, pH 7.4, 100 mm NaCl) in size-exclusion chromatography. The CD spectra of the KH domain proteins (0.5 mg/ml) were acquired using Chirascan plus (Applied Photophysics) in 0.5-mm cuvettes, and the spectrum was recorded between 190 and 280 nm. Baselines were adjusted with buffer, scanned 10 times, and averaged. The protein samples were also scanned 10 times and averaged. The averaged baseline was subtracted from the averaged sample spectrum. The secondary structure content was analyzed using Selcon 3 (41) with respective spectrum input.
Nucleic acid substrates
PAGE-purified oligonucleotides used for RNA or DNA substrates were purchased from IDT and listed in supplemental Table S1. The DNA substrates were prepared as described previously (84), and RNA substrates were prepared according to Jankowsky and Putnam (28). Briefly, a single oligonucleotide was 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England BioLabs) at 37 °C for 1 h. Unincorporated radionucleotides were removed by a G25 chromatography column (GE Healthcare). Single-stranded DNA or RNA substrates were kept at 4 °C and ready to use. For the double-stranded DNA substrates, a [γ-32P]ATP-labeled oligonucleotide was annealed to a 2.5-fold excess of the unlabeled complementary strands in annealing buffer (10 mm Tris-HCl, pH 7.5, 50 mm NaCl) by heating at 95 °C for 6 min and then cooling slowly to room temperature. For double-stranded RNA, a [γ-32P]ATP-labeled oligonucleotide was annealed to a 2.5-fold excess of the unlabeled complementary strands in annealing buffer (10 mm MOPS, pH 6.5, 1 mm EDTA, 50 mm KCl) by heating at 95 °C for 6 min and then cooling slowly to room temperature. All double-stranded substrates were purified by PAGE isolation, and their concentrations were determined by liquid scintillation counting before use.
Western blot
An equal amount of proteins (150 ng) were denatured at 100 °C for 5 min, then resolved on 10% polyacrylamide Tris-glycine SDS gels and transferred to PVDF membranes. The membrane was blocked in PBS containing 5% powdered milk at room temperature for 1 h followed by probe with a rabbit polyclonal anti-DDX43 antibody (1:1000, catalog #HPA031381, Sigma) or a mouse anti-His monoclonal antibody (1:5000, catalog #SAB1400618, Sigma), respectively. Goat anti-rabbit or goat anti-mouse IgG-horseradish peroxidase conjugate (cat#sc-2004, 2005, Santa Cruz Biotechnology) was used as a secondary antibodies at a 1:10,000 dilution and detected using ECL Plus (GE Healthcare).
Helicase assays
Helicase assay reaction mixtures (20 μl) contained 40 mm Tris, pH 8.0, 0.5 mm MgCl2, 15 mm NaCl, 0.01% Nonidet P-40, 0.1 mm DTT, 1 mg/ml bovine serum albumin, equimolar mixture of 2 mm ATP and MgCl2, 0.5 nm concentrations of duplex RNA or DNA substrate, and the indicated concentrations of DDX43 protein. ADP-BeFx was prepared as a mixture of 20 mm ADP (Sigma) with 10 mm of metal fluoride (BeF2, Alfa Aesar) and 50 mm of NaF (BDH Chemicals) and incubated on ice for 4 h (14). Helicase reactions were initiated by the addition of DDX43 and then incubated at 37 °C for 15 min unless otherwise indicated. Reactions were quenched with the addition of 20 μl of 2× stop buffer (17.5 mm EDTA, 0.3% SDS, 12.5% glycerol, 0.02% bromphenol blue, 0.02% xylene cyanol). For duplex RNA and DNA substrates, a 10-fold excess of unlabeled oligonucleotide (cold oligo) with the same sequence as the labeled strand was included in the quench to prevent reannealing. The products of the helicase reactions for duplex RNA substrates were resolved on nondenaturing 15% (19:1 acrylamide:bisacrylamide) polyacrylamide gels, and products of DNA unwinding reactions were resolved on nondenaturing 12% (19:1 acrylamide:bisacrylamide) polyacrylamide gels. Radiolabeled DNA or RNA species in polyacrylamide gels were visualized using a PharosFX Imager and quantitated using the Quantity One software (Bio-Rad). The percent helicase substrate unwound was calculated by using the formula, % unwinding = 100 × (P/(S + P)), where P is the product and S is the substrate. The values of P and S have been corrected after subtracting background values in the no-enzyme and heat-denatured substrate controls, respectively.
Strand annealing assays
Annealed fork 19-bp duplex DNA or 5′-tailed 13-bp duplex RNA was denatured at 95 °C for 5 min and kept on ice, then followed by the helicase assays described above in the presence or absence of ATP.
Streptavidin displacement assays
X12–1-28-BioT37 (supplemental Table S1, biotinylated dT was designed at the position of 28 nt from 5′-end and 37 nt from the 3′-end) was 5′-32P-end-labeled. DNA substrate (0.5 nm) was preincubated with 100 nm streptavidin (Sigma) for 10 min at 37 °C in helicase reaction buffer (above). Reactions were initiated by adding DDX43 or FANCJ before the addition of biotin (1 μm) and incubated at 37 °C for 15 min followed by a quench with the addition of 10 μl of stop buffer (50 mm EDTA, 40% glycerol, 0.9% SDS, 0.05% bromphenol blue, and 0.05% xylene cyanol). Products were resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) polyacrylamide gels. Radiolabeled DNA species in polyacrylamide gels were visualized using a PharosFX Imager (Bio-Rad).
EMSA
Protein/DNA or RNA binding mixtures (20 μl) contained the indicated concentrations of DDX43 and 0.5 nm specified 32P-end-labeled DNA substrate in the same reaction buffer as that used for helicase assays (see above) without ATP. The binding mixtures were incubated at room temperature for 30 min after the addition of DDX43 protein. After incubation, 3 μl of loading dye (74% glycerol, 0.01% xylene cyanol, 0.01% bromphenol blue) was added to each mixture, and samples were loaded onto native 5% (19:1 acrylamide/bisacrylamide) polyacrylamide gels and electrophoresed at 200 V for 2 h at 4 °C using 1×Tris borate-EDTA as the running buffer. The resolved radiolabeled species were visualized using a PharosFX Imager (Bio-Rad).
ATP hydrolysis assays
ATP hydrolysis was measured using [γ-32P]ATP (PerkinElmer Life Sciences) and analyzed by thin layer chromatography (TLC) on polyethyleneimine-cellulose plates (Mallinckrodt Baker). The reaction mixture (30 μl total volume) contained 40 mm Tris, pH 8.0, 0.5 mm MgCl2, 150 mm NaCl, 0.01% Nonidet P-40, 0.1 mm DTT, 1 mg/ml BSA, 850 μm ATP, 250 μm [γ-32P]ATP, 2.5 μm DDX43 protein, and ssDNA M13 (50 μm), and dT30 or rU30 (30 μm) stimulator was incubated at 37 °C for 0, 7.5, 15, 30, and 45 min. Reactions were quenched with 5 μl of 50 mm EDTA final concentration. The reaction mixture was spotted onto a PEI-cellulose TLC plate and resolved by using 0.5 m LiCl, 1 m formic acid as the carrier solvent. The TLC plate was exposed to a phosphorimaging cassette for 30 min and visualized using a PharosFX Imager (Bio-Rad).
Author contributions
T. T., V. V., and Y. W. conceived and coordinated the study and wrote the paper. J. Q. helped in the initial protein purification. M. G. prepared triplex DNA. A. K. provided technical assistance in protein purification. Y. L. and R. S. assisted in manuscript preparation. K. E. K. contributed the KH domain characterization. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Michal Boniecki (Protein Characterization and Crystallization Facility (PCCF), Department of Biochemistry, University of Saskatchewan) for assistance with multiangle light scattering and CD spectroscopy and Drs. Stanley Moore and Jeremy Lee (Department of Biochemistry, University of Saskatchewan) for suggestions for this work.
This work was supported by the Canadian Breast Cancer Foundation (to Y. W. and K. E. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Figs. S1–S12.
- DDX43
- DEAD-box polypeptide 43
- HAGE
- helicase antigen gene
- AMP-PNP
- adenylyl-imidodiphosphate
- ATPγS
- adenosine 5′-[γ-thio] triphosphate
- dT30
- oligodeoxythymine 30 residues
- rU30
- ribonucleic acid uracil 30
- HD
- helicase domain
- KH
- K-homology
- SEC
- size-exclusion chromatography
- MALS
- multiangle laser light scattering
- Ni-NTA
- nickel-nitrilotriacetic acid
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- dsRNA
- double-stranded RNA
- aa
- amino acids
- SF2
- Superfamily 2
- nt
- nucleotide(s).
References
- 1. Jarmoskaite I., and Russell R. (2014) RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 83, 697–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Linder P., and Jankowsky E. (2011) From unwinding to clamping: the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 [DOI] [PubMed] [Google Scholar]
- 3. Lohman T. M., and Bjornson K. P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 4. Pyle A. M. (2008) Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 5. Singleton M. R., Dillingham M. S., and Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 6. Liu Z. R. (2002) p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22, 5443–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hönig A., Auboeuf D., Parker M. M., O'Malley B. W., and Berget S. M. (2002) Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 22, 5698–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venema J., and Tollervey D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311 [DOI] [PubMed] [Google Scholar]
- 9. Gatfield D., Le Hir H., Schmitt C., Braun I. C., Köcher T., Wilm M., and Izaurralde E. (2001) The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11, 1716–1721 [DOI] [PubMed] [Google Scholar]
- 10. Chuang R. Y., Weaver P. L., Liu Z., and Chang T. H. (1997) Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 11. Py B., Higgins C. F., Krisch H. M., and Carpousis A. J. (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169–172 [DOI] [PubMed] [Google Scholar]
- 12. Anderson J. S., and Parker R. P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Missel A., Souza A. E., Nörskau G., and Göringer H. U. (1997) Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17, 4895–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Combeau C., and Carlier M. F. (1989) Characterization of the aluminum and beryllium fluoride species bound to F-actin and microtubules at the site of the γ-phosphate of the nucleotide. J. Biol. Chem. 264, 19017–19021 [PubMed] [Google Scholar]
- 15. Tanner N. K., and Linder P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8, 251–262 [DOI] [PubMed] [Google Scholar]
- 16. Martelange V., De Smet C., De Plaen E., Lurquin C., and Boon T. (2000) Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 60, 3848–3855 [PubMed] [Google Scholar]
- 17. Mathieu M. G., Linley A. J., Reeder S. P., Badoual C., Tartour E., Rees R. C., and McArdle S. E. (2010) HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 10, 2. [PMC free article] [PubMed] [Google Scholar]
- 18. Adams S. P., Sahota S. S., Mijovic A., Czepulkowski B., Padua R. A., Mufti G. J., and Guinn B. A. (2002) Frequent expression of HAGE in presentation chronic myeloid leukaemias. Leukemia 16, 2238–2242 [DOI] [PubMed] [Google Scholar]
- 19. Valverde R., Edwards L., and Regan L. (2008) Structure and function of KH domains. FEBS J. 275, 2712–2726 [DOI] [PubMed] [Google Scholar]
- 20. Matunis M. J., Michael W. M., and Dreyfuss G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burd C. G., and Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 22. Grishin N. V. (2001) KH domain: one motif, two folds. Nucleic Acids Res. 29, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spingola M., Armisen J., and Ares M. Jr. (2004) Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res. 32, 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashley C. T. Jr, Wilkinson K. D., Reines D., and Warren S. T. (1993) FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262, 563–566 [DOI] [PubMed] [Google Scholar]
- 25. Gibson T. J., Thompson J. D., and Heringa J. (1993) The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324, 361–366 [DOI] [PubMed] [Google Scholar]
- 26. Musco G., Stier G., Joseph C., Castiglione Morelli M. A., Nilges M., Gibson T. J., and Pastore A. (1996) Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell 85, 237–245 [DOI] [PubMed] [Google Scholar]
- 27. Siomi H., Matunis M. J., Michael W. M., and Dreyfuss G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jankowsky E., and Putnam A. (2010) Duplex unwinding with DEAD-box proteins. Methods Mol. Biol. 587, 245–264 [DOI] [PubMed] [Google Scholar]
- 29. Chen Y., Potratz J. P., Tijerina P., Del Campo M., Lambowitz A. M., and Russell R. (2008) DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. U.S.A. 105, 20203–20208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu F., Putnam A., and Jankowsky E. (2008) ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U.S.A. 105, 20209–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gilhooly N. S., and Dillingham M. S. (2014) Recombination hotspots attenuate the coupled ATPase and translocase activities of an AddAB-type helicase-nuclease. Nucleic Acids Res. 42, 5633–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meetei A. R., Medhurst A. L., Ling C., Xue Y., Singh T. R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C. G., Hoatlin M., Joenje H., de Winter J. P., and Wang W. (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37, 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ding H., Guo M., Vidhyasagar V., Talwar T., and Wu Y. (2015) The Q motif is involved in DNA binding but not ATP binding in ChlR1 helicase. PLoS ONE 10, e0140755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo M., Vidhyasagar V., Ding H., and Wu Y. (2014) Insight into the roles of helicase motif Ia by characterizing fanconi anemia group J protein (FANCJ) patient mutations. J. Biol. Chem. 289, 10551–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis H. A., Musunuru K., Jensen K. B., Edo C., Chen H., Darnell R. B., and Burley S. K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332 [DOI] [PubMed] [Google Scholar]
- 36. Barnes M., van Rensburg G., Li W. M., Mehmood K., Mackedenski S., Chan C. M., King D. T., Miller A. L., and Lee C. H. (2015) Molecular insights into the coding region determinant-binding protein-RNA interaction through site-directed mutagenesis in the heterogeneous nuclear ribonucleoprotein-K-homology domains. J. Biol. Chem. 290, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y., Mah T. F., Greenblatt J., and Friedman D. I. (2002) Evidence that the KH RNA-binding domains influence the action of the E. coli NusA protein. J. Mol. Biol. 318, 1175–1188 [DOI] [PubMed] [Google Scholar]
- 38. Chmiel N. H., Rio D. C., and Doudna J. A. (2006) Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA 12, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adinolfi S., Bagni C., Musco G., Gibson T., Mazzarella L., and Pastore A. (1999) Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 5, 1248–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pozdnyakova I., and Regan L. (2005) New insights into Fragile X syndrome: relating genotype to phenotype at the molecular level. FEBS J. 272, 872–878 [DOI] [PubMed] [Google Scholar]
- 41. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 42. McGuffin L. J., Bryson K., and Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 43. Backe P. H., Messias A. C., Ravelli R. B., Sattler M., and Cusack S. (2005) X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 44. Tuteja N., Tarique M., Banu M. S., Ahmad M., and Tuteja R. (2014) Pisum sativum p68 DEAD-box protein is ATP-dependent RNA helicase and unique bipolar DNA helicase. Plant Mol. Biol. 85, 639–651 [DOI] [PubMed] [Google Scholar]
- 45. Kikuma T., Ohtsu M., Utsugi T., Koga S., Okuhara K., Eki T., Fujimori F., and Murakami Y. (2004) Dbp9p, a member of the DEAD box protein family, exhibits DNA helicase activity. J. Biol. Chem. 279, 20692–20698 [DOI] [PubMed] [Google Scholar]
- 46. Yang Q., and Jankowsky E. (2006) The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 13, 981–986 [DOI] [PubMed] [Google Scholar]
- 47. Li L., Monckton E. A., and Godbout R. (2008) A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 28, 6413–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Linley A. J., Mathieu M. G., Miles A. K., Rees R. C., McArdle S. E., and Regad T. (2012) The helicase HAGE expressed by malignant melanoma-initiating cells is required for tumor cell proliferation in vivo. J. Biol. Chem. 287, 13633–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rogers G. W. Jr, Richter N. J., and Merrick W. C. (1999) Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274, 12236–12244 [DOI] [PubMed] [Google Scholar]
- 50. Kellner J. N., Reinstein J., and Meinhart A. (2015) Synergistic effects of ATP and RNA binding to human DEAD-box protein DDX1. Nucleic Acids Res. 43, 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang P. Z., Tsai-Morris C. H., and Dufau M. L. (1999) A novel gonadotropin-regulated testicular RNA helicase: a new member of the dead-box family. J. Biol. Chem. 274, 37932–37940 [DOI] [PubMed] [Google Scholar]
- 52. Yan X., Mouillet J. F., Ou Q., and Sadovsky Y. (2003) A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell. Biol. 23, 414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flores-Rozas H., and Hurwitz J. (1993) Characterization of a new RNA helicase from nuclear extracts of HeLa cells which translocates in the 5′ to 3′ direction. J. Biol. Chem. 268, 21372–21383 [PubMed] [Google Scholar]
- 54. Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., and Jeang K. T. (2004) Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119, 381–392 [DOI] [PubMed] [Google Scholar]
- 55. Hirling H., Scheffner M., Restle T., and Stahl H. (1989) RNA helicase activity associated with the human p68 protein. Nature 339, 562–564 [DOI] [PubMed] [Google Scholar]
- 56. Rössler O. G., Straka A., and Stahl H. (2001) Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 29, 2088–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mathieu M. G., Miles A. K., Ahmad M., Buczek M. E., Pockley A. G., Rees R. C., and Regad T. (2014) The helicase HAGE prevents interferon-α-induced PML expression in ABCB5+ malignant melanoma-initiating cells by promoting the expression of SOCS1. Cell Death Dis. 5, e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Halls C., Mohr S., Del Campo M., Yang Q., Jankowsky E., and Lambowitz A. M. (2007) Involvement of DEAD-box proteins in group I and group II intron splicing: biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365, 835–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang Y., and Liu Z. R. (2002) The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277, 12810–12815 [DOI] [PubMed] [Google Scholar]
- 60. Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., and Sonenberg N. (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rocak S., Emery B., Tanner N. K., and Linder P. (2005) Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 33, 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bizebard T., Ferlenghi I., Iost I., and Dreyfus M. (2004) Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43, 7857–7866 [DOI] [PubMed] [Google Scholar]
- 63. Del Campo M., and Lambowitz A. M. (2009) Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell. 35, 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jankowsky E., and Fairman M. E. (2007) RNA helicases: one fold for many functions. Curr. Opin. Struct. Biol. 17, 316–324 [DOI] [PubMed] [Google Scholar]
- 65. Patrick E. M., Srinivasan S., Jankowsky E., and Comstock M. J. (2017) The RNA helicase Mtr4p is a duplex-sensing translocase. Nat. Chem. Biol. 13, 99–104 [DOI] [PubMed] [Google Scholar]
- 66. Hopfner K. P., and Michaelis J. (2007) Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr. Opin. Struct. Biol. 17, 87–95 [DOI] [PubMed] [Google Scholar]
- 67. Mallam A. L., Del Campo M., Gilman B., Sidote D. J., and Lambowitz A. M. (2012) Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature 490, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mallam A. L., Sidote D. J., and Lambowitz A. M. (2014) Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 3, e04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kawaoka J., and Pyle A. M. (2005) Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 33, 644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pugh R. A., Wu C. G., and Spies M. (2012) Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J. 31, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee J. K., and Hurwitz J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. U.S.A. 98, 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bianco P. R., Brewer L. R., Corzett M., Balhorn R., Yeh Y., Kowalczykowski S. C., and Baskin R. J. (2001) Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 [DOI] [PubMed] [Google Scholar]
- 73. Lahue E. E., and Matson S. W. (1988) Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem. 263, 3208–3215 [PubMed] [Google Scholar]
- 74. Steimer L., Wurm J. P., Linden M. H., Rudolph M. G., Wöhnert J., and Klostermeier D. (2013) Recognition of two distinct elements in the RNA substrate by the RNA-binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 41, 6259–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Diges C. M., and Uhlenbeck O. C. (2001) Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 20, 5503–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mohr G., Del Campo M., Mohr S., Yang Q., Jia H., Jankowsky E., and Lambowitz A. M. (2008) Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 375, 1344–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grohman J. K., Del Campo M., Bhaskaran H., Tijerina P., Lambowitz A. M., and Russell R. (2007) Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry 46, 3013–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Collins R., Karlberg T., Lehtiö L., Schütz P., van den Berg S., Dahlgren L. G., Hammarström M., Weigelt J., and Schüler H. (2009) The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J. Biol. Chem. 284, 10296–10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nicastro G., Taylor I. A., and Ramos A. (2015) KH-RNA interactions: back in the groove. Curr. Opin. Struct. Biol. 30, 63–70 [DOI] [PubMed] [Google Scholar]
- 80. Abdel-Fatah T. M., McArdle S. E., Johnson C., Moseley P. M., Ball G. R., Pockley A. G., Ellis I. O., Rees R. C., and Chan S. Y. (2014) HAGE (DDX43) is a biomarker for poor prognosis and a predictor of chemotherapy response in breast cancer. Br. J. Cancer 110, 2450–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Abdel-Fatah T. M., McArdle S. E., Agarwal D., Moseley P. M., Green A. R., Ball G. R., Pockley A. G., Ellis I. O., Rees R. C., and Chan S. Y. (2016) HAGE in Triple-negative breast cancer is a novel prognostic, predictive, and actionable biomarker: a transcriptomic and protein expression analysis. Clin. Cancer Res. 22, 905–914 [DOI] [PubMed] [Google Scholar]
- 82. Chen Q., Lin J., Qian J., Yao D. M., Qian W., Li Y., Chai H. Y., Yang J., Wang C. Z., Zhang M., and Xiao G. F. (2011) Gene expression of helicase antigen in patients with acute and chronic myeloid leukemia. Zhongguo Shi Yan. Xue. Ye. Xue. Za Zhi. 19, 1171–1175 [PubMed] [Google Scholar]
- 83. Magnusdottir A., Johansson I., Dahlgren L. G., Nordlund P., and Berglund H. (2009) Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates. Nat. Methods 6, 477–478 [DOI] [PubMed] [Google Scholar]
- 84. Guo M., Vidhyasagar V., Talwar T., Kariem A., and Wu Y. (2016) Mutational analysis of FANCJ helicase. Methods 108, 118–129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.