Abstract
Metal ions play pivotal roles in protein structure, function and stability. The functional and structural diversity of proteins in nature expanded with the incorporation of metal ions or clusters in proteins. Approximately one-third of these proteins in the databases contain metal ions. Many biological and chemical processes in nature involve metal ion-binding proteins, aka metalloproteins. Many cellular reactions that underpin life require metalloproteins. Most of the remarkable, complex chemical transformations are catalysed by metalloenzymes. Realization of the importance of metal-binding sites in a variety of cellular events led to the advancement of various computational methods for their prediction and characterization. Furthermore, as structural and functional knowledgebase about metalloproteins is expanding with advances in computational and experimental fields, the focus of the research is now shifting towards de novo design and redesign of metalloproteins to extend nature’s own diversity beyond its limits. In this review, we will focus on the computational toolbox for prediction of metal ion-binding sites, de novo metalloprotein design and redesign. We will also give examples of tailor-made artificial metalloproteins designed with the computational toolbox.
Keywords: artificial metalloproteins, computational protein design, computational protein re-design, de novo design, metal binding sites
Introduction
Function of a protein and its interactions with other molecules such as proteins, nucleic acids are determined by protein’s 3D structure. There are over 124000 protein structures deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Databank (PDB) by June 2016 [1]. In 2012, Yu and colleagues estimated that approximately one-third of the proteins deposited in the PDB database contained metal ions essential for function [2]. In cells, 30–40% of the proteins depend upon at least one metal ion to carry out their biological functions [3,4]. It has been well known that regions or residues of proteins that interact with the metal ligands are very well conserved both in sequence and in structure [5,6].
In many biological systems, proteins that require a metal ion to carry out their physiological function, aka metalloproteins, are very widespread and they perform a variety of functions as storage and transport proteins, enzymes, regulators of gene expression and signal transduction cascade proteins [7–10]. Metalloproteins have been in the focus of the biological research for many years. This intense focus could be traced back to the early X-ray crystallography studies that led to the discovery of presence of a metal atom (iron) along with 3D structure of myoglobin [11,12] and haemoglobin proteins [13].
Metal ions can be crucial for protein structure and function. Proteins are involved in a variety of cellular events and catalytic reactions. Chemical nature of the side chains of amino acids that made up proteins provides them with limited proportion of the chemical functionality seen in nature. On the other hand, association of proteins with cofactors such as small organic molecules, single metal ions or clusters with metal and non-metal atoms granted these proteins with a diversity of functions [12]. Metal ions are involved in nucleophilic catalysis events, in induction of the conformational changes in proteins, in electron transport, in folding and stabilization of protein structures [14]. Metalloproteins play crucial roles in development and progress of a number of diseases including brain diseases such as prion, amyotrophic lateral sclerosis (ALS), Parkinson’s and Alzheimer’s where an effective treatment and cure is still missing [15]. They are also implicated in apoptosis and aging [16,17]. In plants and other microorganisms, they improve metal-adsorption capacities [18]. Well-characterized plant metalloproteins such as metallothioneins and phytochelatins are involved in the uptake of the essential micronutrients required for plant metabolism and detoxification of heavy metal ions [19]. Many metalloproteins in plants are involved in the electron transport chain of the photosynthetic machinery [20,21]. Furthermore, metal ions play a crucial role in protein folding and stability [22–24]. Realization of the importance and involvement of metal-binding sites in a diversity of cellular processes and functions, led to the advancement of various computational tools and algorithms for prediction, identification and engineering of these sites. These advances present us new means to understand the biological function of a protein; to decipher the underlying mechanisms of protein folding and stability and thus improve protein function and stability. An active research area was already established with studies involving de novo design, redesign, prediction, optimization and stabilization of protein structure. It has been almost 30 years since the introduction of a helical protein designed from the first principles [25]. In this era, focus is now shifting towards introduction of novel functions to proteins; protein stabilization by de novo protein design or redesign; use of metal cofactors. Subsequently, this approach extends the nature’s repository of protein structure and function beyond its natural limits.
In this review, we will focus on the computational tools for the prediction of metal-binding sites on a protein and emphasize the computational tools that are used in de novo design and redesign of metal ion-binding sites. We will also highlight some noteworthy artificial metalloproteins that were designed using the developed computational tools.
Computational tools for de novo design and redesign of metalloproteins
Unprecedented increase in the number of protein sequences and structures deposited in the public databases due to advances in genome-sequencing technologies and experimental methods for structure determination, poses a great challenge for researchers in terms of prediction of the biological functions of the deposited proteins. Since metal ions play crucial roles in many biological processes, their presence in protein structures reveals essential information about protein’s inherent function(s). Owing to the fact that determination of structural and functional features of a protein such as metal ion-binding sites using experimental methods is still challenging because of the problems related to cost, time and automation of processes, there is an increasing demand for prediction of those features using computational methods. Use of computational methods for the prediction of metal ion-binding sites not only contributes to expansion of the existing knowledgebase, but also aids de novo metalloprotein design and redesign by providing necessary structural information about the metal co-ordination environment. This information is used in the design of tailored and stable metalloproteins with improved functions. Therefore, prosperity of a metalloprotein design largely depends on the quantity and quality of available information on metal ion-binding sites, protein scaffolds and physiochemical rules that direct the folding of a polypeptide into a functional protein [26,27].
Computational prediction of metal ion-binding sites
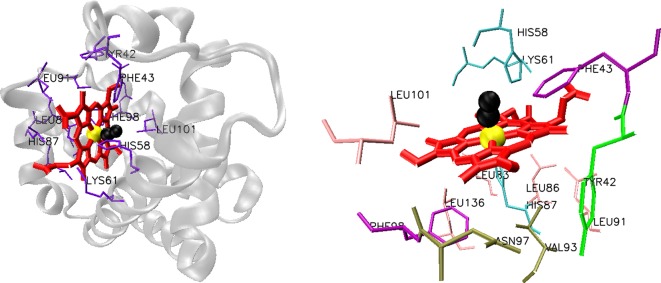
Metal ion-binding sites in proteins exhibit a wide range of diversity. In some proteins, backbone oxygen and nitrogen atoms are involved in metal ion binding, whereas in some proteins side chain oxygen, nitrogen and sulfur atoms are involved. Metal ions were also found to be selective in their binding to their respective ligands. In a 2012 paper, Yu and colleagues used 1109 metal ion-binding polypeptides and predicted the metal ion-binding sites and verified that metal ions preferentially bind to certain residues on the protein [2]. For Ca+2, favoured amino acids were predicted to be D, E, N and G; for Cu+2 H; for Mg+2 D and E; for Fe+3 H, D, E, C and Y. Residues that reside partially within 3.5 Å of the metal ion were considered as metal ion-binding residues. As the metal ion-binding sites diversify, correct identification of the ligands in metal ion-binding sites with experimental methods becomes more problematic without prior knowledge of the identity of the bound metal ion since current high-throughput methods based on X-ray absorption spectroscopy can only identify its presence but cannot identify the residues involved in metal ion binding [28,29]. Besides, most of the computational tools for metal ion-binding site prediction use the information mostly derived from crystals of the metal ion-bound form of the proteins, holo (metal ion-bound) forms whereas a majority of the recently resolved structures are from the apo (metal ion-free) forms [30]. Moreover, metal ion binding is a dynamic process that often results in structural rearrangements of the residues in binding pockets [31,32]. Additionally, in some cases, a metal ion is buried in a prosthetic group, tightly bound, specific non-polypeptide unit required for the biological function of some proteins, such as haem that consists of a central Fe (II/III) atom in a protoporphyrin ring [13] as in haemoglobin structure (Figure 1). Consequently, prediction and identification of metal ion-binding sites pose a much more complex problem that cannot be solved solely based on simple geometrical criteria or sequence information. Therefore, different computational methods and algorithms have been introduced for the prediction of metal ion-binding sites and the residues involved in this process. Moreover, prediction of most probable sites of metal ion-binding motifs in a newly identified protein would facilitate redesign of that site to better accommodate a metal to modify its structural and functional properties. After prediction and comparison with known motifs, this knowledge would enable establishment of a metal ion-binding site library that could be utilized for protein engineering purposes.
Figure 1. Structure of chain C of cross-linked carbonmonoxy haemoglobin A from Homo sapiens (left).
Haem-coordinating residues are shown on the right. Haem group (red) with bound Fe (yellow) and carbonmonoxide (black). PDB ID: 1SDK.
Most computational methods rely on the sequence information and they target the identification of metal-binding motif or motifs in well characterized metal ion-binding proteins. These motifs are then used to search for similar patterns in newly identified proteins. Machine learning methods have also been introduced to the problem only recently [7,33–37]. Programs such as MetalPredator [38], MetalDetector v2.0 [39], SeqCHED Server [34], ZincFinder [35], SVMProt [36,37] employ sequence based approaches for the prediction of the metal ion-binding sites. For instance MetalDetector v2.0 [39] uses cysteine and histidine residues and therefore focuses only on transition metals, haem and Fe/S clusters for the prediction, by predicting the metal-bonding state of these residues and number of bound metal ions. On the other hand, MetalPredator [38] utilizes two libraries of Hidden Markov Model profiles representative of Pfam domains and structural motifs that bind Fe/S clusters. Some prediction software such as FINDSITE-metal [40], mFASD [41], TEMSP [42], FEATURE metal scanning [43–45] utilize structural information for prediction of the metal ion-binding sites. A few of them such as MetSite [46], 3DLigandSite [47], Fragment Transformation Method [2] combine information from both protein sequence and structure. Use of ab initio methods as in IonCom [48] server has also been recently realized. Several helper tools such as MetalS2 [49], MetalS3 [50], FindGeo [51], CheckMyMetal (CMM) [52] were developed for pairwise structural alignment, database mining of metal-binding sites, determination of metal co-ordination geometry, validation of metal-binding sites in PDB structures respectively. Software developed for the prediction of metal ion-binding sites, their rationale and web links are summarized in Table 1. Although these servers provide a large selection of different algorithms for the prediction, some of them are only limited to the prediction of a single metal (i.e. MetalPredictor and MetalDetectorv2.0 only predict Fe–S cluster-binding sites whereas FEATURE metal scanning tool, TEMSP, ZincFinder predict Zn2+-binding sites, the rest of the servers provide a diverse range of metals that are associated with metalloproteins). Databases such as MetalPDB [4], MetalMine [53], Metal Interactions in Protein Structures (MIPS) [54], MESPEUS_10 [55], COMe [56], MetLigDB [57] as listed in Table 2, encompass information on both sequence motifs that define metal ion-binding sites and structure of metal ions and their corresponding residues that are deposited in various sequence and structural databases.
Table 1.
Software designed for metal ion-binding site prediction
Table 2.
List of databases for metalloproteins, metal ion-binding site motifs and structural information on metal ion-binding sites
| Databases | ReferenceLink | |
|---|---|---|
| MetalPDB | [4] | http://metalweb.cerm.unifi.it/ |
| MetalMine | [53] | http://metalmine.naist.jp/metalmine009/index.html |
| MIPS | [54] | http://dicsoft2.physics.iisc.ernet.in/mips/ |
| MESPEUS_10 | [55] | http://mespeus.bch.ed.ac.uk/MESPEUS_10/ |
| COMe | [55,56] | http://www.flymine.org/come/ |
| MetLigDB | [57] | http://silver.sejong.ac.kr/MetLigDB |
Computational design tools, strategies for de novo design and redesign of metalloproteins
Many of the biological and chemical reactions that establish the foundations of life such as water oxidation, carbon dioxide reduction, nitrogen fixation, photosynthesis require involvement of metalloproteins or metalloenzymes [58]. Chemical transformations achieved by metalloproteins are diverse [59]. Despite the diversity of these reactions, we are still far from completely understanding the principles that govern these metalloproteins when performing their functions. Much effort has been put to fully mimic or exploit these processes. As de novo protein design and redesign methods have been advancing for the past decades, application of these methods to artificial metalloprotein and metalloenzyme design and redesign is inevitable. Previous research has created fully functional de novo designed metalloproteins that exhibit activity for a variety of reactions ranging from ester and organophosphate hydrolysis [60,26] to nitric oxide reduction [60,61].
In metalloproteins, metal ions either interact with amino acid side chains or accompanying residues. Most of the metal-binding sites are known to be promiscuous and can accommodate non-native metals with similar properties [62]. Redesign of metalloproteins usually involves modification of a well-characterized and well-known native protein through engineering or introduction of metal-binding ligands to protein structure to accommodate non-native metals. For de novo design of metalloproteins, protein structures should be designed from scratch starting from the amino acid sequences. Residues for metal ion binding should also be introduced to the structure [63]. Both methods require correctly folded and functional final protein structures with introduced metal ions or cofactors. De novo design approach is more complex since physiochemical rules that drive the protein folding [64] have not yet been deciphered completely. Moreover, “inverse folding problem” adds another layer to this complexity. De novo protein design uses the “inverse folding problem”, which states that different combinations of amino acid sequences can fold into the same 3D structure and resulting proteins can perform the same function [65,66]. In protein design, we start with a rigid or flexible backbone structure and try to determine a sequence that would fold into this structure. As a variety of sequences fold into the same structure, there is degeneracy. Therefore, the accuracy and availability of the template structures for protein design has a huge affect in the success of the final design.
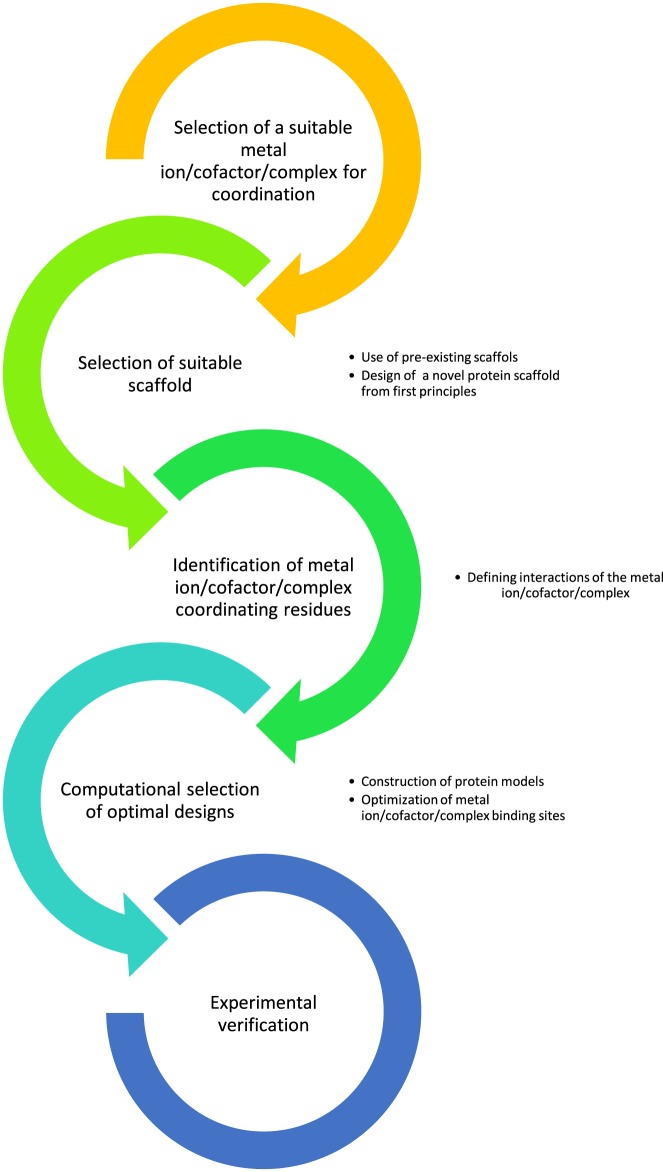
The current collection of metalloproteins is largely coming from nature’s own repository. This repository is now being diversified by de novo design and redesign approaches with the aid of the knowledgebase of metal ion-binding sites. A typical workflow for this procedure is shown in Figure 2. Successful application of de novo design and redesign approaches create a diversity of advanced proteins with either new or modified functions. Additionally, through the application of these methods, an excellent platform is formed to study structure–function relationship and protein folding problem further.
Figure 2. A typical workflow for de novo metalloprotein design and redesign.
Construction of artificial or redesigned metalloproteins requires detailed information on the nature of the metal ion-binding motifs, how the metal ions are co-ordinated (ligand–metal interactions and binding pocket geometry, geometrical arrangements, redox states) and finally a detailed information on the structure–function of the scaffold protein that would accommodate the metal ion. This, in turn, requires the use of state of the art computational tools for design and experimental techniques for analysis and characterization. A variety of deterministic, stochastic and probabilistic methods have been applied for de novo protein design [67]. With the recent advances in the computational field (high performance computing, better algorithms, faster processors), backbone flexibility is also incorporated into the calculations [68].
A compelling number of programs such as Protein WISDOM [68], HostDesigner [69], METALSEARCH [70], CORE [71], RosettaDesign [72], EvoDesign [73], RosettaBackrub [74], IPRO (OptGraft procedure) [75], PyRosetta [76] has been in use for de novo protein design (Table 3). These software provide in silico workbenches for the design of novel, stable and functional proteins. Most of these programs use force fields to represent molecular interactions and to search for sequences with lowest energy in the target fold since the minimum free energy conformation of a protein is considered as the native fold of the protein.
Table 3.
Programs for de novo protein design
| Software | Ref.Link | |
|---|---|---|
| METALSEARCH | [70] | Not available |
| Protein WISDOM | [68] | http://www.proteinwisdom.org |
|
HostDesigner |
[69] | https://sourceforge.net/projects/hostdesigner-v3-0/ |
|
CORE |
[71] | Not available |
| RosettaDesign | [72] | http://rosettadesign.med.unc.edu |
|
RosettaBackrub |
[74] | https://kortemmelab.ucsf.edu/backrub/cgi-bin/rosettaweb.py?query=index |
| EvoDesign | [73] | http://zhanglab.ccmb.med.umich.edu/EvoDesign |
|
IPRO (OptGraft) |
[75] | http://www.maranasgroup.com/submission/ipro2014.htm |
| PyRosetta | [76] | http://www.pyrosetta.org/ |
Although all of them have been used for de novo design of metalloproteins, only three of them, HostDesigner, METALSEARCH and IPRO (OptGraft) are specific for de novo design or redesign of metalloproteins. HostDesigner is a combinatorial chemistry-based program that uses two complementary algorithms LINKER and OVERLAY for creation of a number of candidate structures and to discover metal ion receptors in these structures [69]. Two user-defined complex fragments are connected with linking fragments deposited in the library and structures are built by LINKER. Next, a single user-defined complex structure is built by OVERLAY through the superimposition of the linking fragments on to this structure.
METALSEARCH, being one of the earliest design programs, creates lists of four residues that might form tetrahedral metal ion-binding sites if amino acids in the native protein were replaced by cysteine or histidine residues. Program also provides dihedral angles of the amino acids and the co-ordinates of the predicted metal ion as output [70]. OptGraft is a computational program that is present in IPRO suite of programs and it is specifically designed for transferring a binding site on to an existing protein scaffold. For this purpose, possible binding pocket placement combinations are exhaustively explored using mixed-integer linear optimization and a ranked list of possible designs that fit the geometric criteria and orientation of the native binding pocket is generated. Moreover, this procedure also assesses the impact of the new pocket on the protein structure and if there is a potential distortion in the structure, small mutations in the neighbouring residues are introduced to counteract their probable distortional effects [75].
Applications
Computational design of novel redox centres
METALSEARCH was used for de novo design of a rubredoxin-like Fe site [ 77], for the introduction of zinc-binding sites to the designed four helix bundle protein α4 and to the Ig-binding B1 domain of the Streptococcal protein G [ 78, 79]. It has been long known that iron ion in rubredoxin proteins is co-ordinated by four cysteine residues. Farinas et al. [ 77] designed a redox centre that mimics rubredoxin’s into the Ig-binding B1 domain of the Streptococcal protein G by taking into account the backbone movements while maintaining the structural integrity and stability of the B1 protein. For this purpose, they first used METALSEARCH to identify potential metal ion-binding sites that are capable of tetrahedral co-ordination in the B1 protein structure. Contrary to previous work of the same group [ 78, 79], they made an attempt to use a more realistic backbone flexibility description in the computational part. As an input to the METALSEARCH, rather than using the averaged NMR structure, researchers selected 6 randomly calculated NMR structures out of 60 that reflected a more realistic backbone flexibility. Co(II) and Cd(II) bound designed variants’ stability and integrity were confirmed with spectroscopic methods. Fe(II) complex of the designed variant was produced and analysis and characterization of the rubredoxin centre revealed that the mutant rubredoxin centre mimicked oxidized rubredoxin. Nanda et al. [ 80] also managed to design a redox active minimal β protein scaffold (RM1) that exhibit rubredoxin activity through formation of a stable, redox-active 4-Cys thiolate Fe(II/III) site. The designed complex was further shown to be stable through repeated cycles of oxidation and reduction even in the presence of an oxygen containing environment.
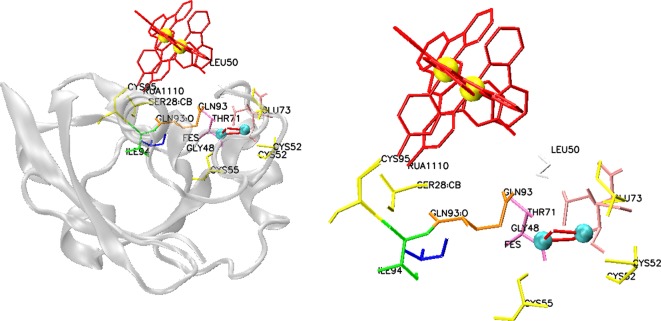
In another design approach, computationally designed metalloprotein using an in-house developed CORE software [71] that functions as an artificial redox centre was produced with the ability to mimic photosynthesis [ 81]. An antiparallel four helix bundle containing two helix–loop–helix peptides connected through a disulfide bridge was used as the scaffold. This scaffold is functionalized through histidine-coordinated Ruthenium(II) Bipyridine (Ru(bpy)2) and haem cofactors by engineering of the binding sites. The exterior hydrophilicity of the bundle was provided by the salt bridges formed primarily between glutamic acid and lysine residues, whereas identity of the hydrophobic core residues inside the helices were determined using the CORE software. An exemplary structure of Ru(bpy)2(mbpy)-modified bovine adrenodoxin protein was shown in Figure 3 to show the metal co-ordination. The backbone structure that was used as an input to the software contains alanine in all positions with the exception of the exterior hydrophilic residues and cofactor-coordinating residues. Software predicts the optimum hydrophobic core residues based on the protein thermal stability and co-operativity. Consequently, the resulting protein mimics photosynthesis following photo-excitation participating in multiple oxidation and reduction cycles with exogenous electron acceptors and donors.
Figure 3. Structure of Ru(bpy)2(mbpy)-modified bovine adrenodoxin protein (left).
Ru(bpy)2 complex (red) is covalently bound to adrenodoxin via Cys95. An iron–sulfur cluster (cyan-red) is also present in the protein. Ruthenium is shown in yellow (PDB ID: 2BT6). Ru(bpy)2 complex (red) and iron–sulfur cluster (red-cyan) co-ordinating residues are shown on the right.
Computational design of artificial metalloproteins and metalloenzymes
Computational protein design software Rosetta Design, is a highly versatile tool that can be exploited for analysis of protein–metal interactions in the design of artificial metalloproteins. Recently, this software was used to improve the catalytic performance of an artificially designed metalloenzyme and to design a promiscuous metalloprotein where an incorporated unnatural amino acid (2,2′-bipyridin-5yl) alanine (Bpy–Ala) residue co-ordinates the metal ion with octahedral co-ordination geometry [80,81]. Heinisch et al. [82] adopted a computational design approach for the genetic optimization of a first-generation artificial metalloenzyme using this software. Previous research produced an assembly of Noyori-type pianostool complex [(5-Cp*)Ir(pico)Cl] in a native human carbonic anhydrase II enzyme that is capable of asymmetric transfer hydrogenation of a salsolidine precursor (a cyclic imine) with moderate enantioselectivity [83] and combinations of these first-generation artificial metalloenzymes with naturally occurring enzymes were also successfully utilized in redox cascade reactions [84]. The crystal structure of the complex revealed that the metal-coordinating moiety was not fully occupied, which led to a modest activity and selectivity. Therefore, RosettaDesign [72] was used to improve cofactor-binding sites and the second co-ordination sphere around cofactor in the first generation artificial metalloenzymes. The software produced four scaffold variants that have mutations for protein backbone stabilization and that enable hydrophobic cofactor burial. The produced designs not only exhibited increased affinity towards the cofactor but also increased activity, turnover number and enantioselectivity with respect to the first-generation artificial metalloenzyme. Mills et al. [85] employed a similar design approach with the use of RosettaMatch algorithm [86] and RosettaDesign [72] for the co-ordination of metal ions by a metalloprotein through the use of unnatural amino acids. For this purpose, an active site capable of catalysing oxidative ring opening of catechol substrates was designed. Four theoretical enzyme active sites, also referred as “theozymes” were constructed including unnatural amino acid Bpy–Ala, catechol containing dopamine molecule, a tyrosine and a histidine ligand for metal co-ordination. RosettaMatch was adopted for the identification of a set of backbone positions in native scaffolds that could accommodate theozyme geometrical restraints. RosettaDesign was then employed to introduce additional stabilizing interactions for the Bpy–Ala complex to the RosettaMatch output. An additional filtering step was applied to find the optimum structures. Although 13 genes that encode artificial metalloproteins are designed and synthesized, only five of the designed proteins yielded soluble, full length proteins in the presence of the unnatural amino acid. These proteins were shown to be promiscuous and bind to divalent cations including Co2+, Zn2+, Fe2+ and Ni2+. Moreover, structural analysis via X-ray crystallography revealed that the designed proteins exhibit only slight deviations from the designed models.
Additionally, the OptGraft procedure in IPRO software suite was used by Fazelinia et al. [75] to graft a calcium-binding pocket of thermitase protein to the first domain of CD2 protein (a rat cell adhesion protein that does not bind calcium). Minimally perturbed structures were created by identification of suitable locations for the grafting using OptGraft procedure. Candidate residues that would ensure the desired spatial restraints for the binding pocket were determined using CHARMM energy calculations. Designs with higher stability were further selected. Novel CD2 variants were shown to exhibit higher affinities for terbium and selective binding to calcium.
Researchers also developed in-house programs for the de novo design of metal ion-binding sites. Zhu et al. [ 87] managed to incorporate computationally engineered zinc-binding sites to de novo designed DS119 protein with βαβ fold with 1:1 stoichiometry using an in-house developed program. Likewise, Yeung et al. [88] used myoglobin as scaffold to introduce nitric oxide reductase activity by grafting three histidines and one glutamate residues that would accommodate a haem and non-haem FeB centre. They followed a molecular dynamics (MD)-based modelling approach for the in silico introduction of mutations to myoglobin scaffold and to evaluate the ability of the designed metal ion-binding site to bind an iron ion. To achieve this three in silico mutations (L29H, F43H and V68E) were introduced into sperm whale myoglobin structure using the VMD software suite. Additionally, molecular dynamics simulations (NVT ensemble, 310 K, 10000 steps, 1 fs/step) with NAMD software were performed to estimate the ability of the mutant protein to bind Fe atom through the engineered metal ion-binding site. The triple mutant of the sperm whale myoglobin was constructed and mutant protein was expressed in Escherichia coli. The mutant protein was further purified and characterized. Finally, the tailor-made protein was shown to exhibit nitric oxide reductase activity.
Computational design of metal-templated interfaces
In 2010, Salgado et al. used Metal Templated Interface Redesign (MeTIR) approach to gain insights about the role played by metal co-ordination via metal co-ordinated nucleation process in the formation of early protein folds and complexes throughout evolution [89]. For this purpose, they have redesigned a monomeric protein, namely cytochrome cb562 with non-self-associating surface and haem groups in its structure to obtain a variant that exhibits Zn co-ordinated self-association properties. A second redesign step was introduced to Zn co-ordinated D2-symmetrical tetramer to engineer the complex further into a complex that exhibits self-assembly in the presence or absence of metals. Mutations were introduced only into the residues on the surface of cytochrome cb562 to allow easy tracing of the structure with crystallographic methods and to preserve the overall fold. Residues inside the protein (with low solvent accessible surface are (SASA) values), residues contacting heme groups or Zn atoms, residues that were involved in side chain-main chain H-bonding interactions remained and they were not selected for mutations. A variant of RosettaDesign algorithm was employed for the rotamer optimization of the selected residues to find minimal number of mutations that make a maximum impact towards the self-assembly of the monomeric protein. The energy and packing scores obtained, ranked and position of candidate residues with respect to the undesirable residues were evaluated to find optimal residues at the interface for redesign.
Knowledge-driven metalloprotein designs
In some cases, computational programs were not used for the design. Instead, designs were based on previous information derived from sequence, structure and function of proteins, previous literary knowledgebase and intuition of the researchers [90–92]. In most of the cases, previously designed protein scaffolds are modified to engineer the binding pockets [63, 88, 91, 93–96]. In one recent case, noble metals were introduced to iron-binding sites of the haem proteins such as myoglobin and the designed protein was subjected to directed evolution [97]. First, apo-form of haem proteins were expressed in E. coli using minimal media devoid of Fe for the minimization of haemin biosynthesis, at low temperature to alleviate the instability of the apo-form. Myoglobin from Physeter macrocephalus and cytochrome P450 BM3h from Bacillus megaterium were found to be overexpressed with or without mOCR stability tags in relatively high yields. These haem lacking apo-forms were purified and holo-forms of the proteins were reconstituted with noble metal-porphyrin IX cofactors. Next, eight myoglobin variants with mutations in the axial ligand position (His93) were expressed in apo-form and then reconstituted with various noble metal-porphyrin IX cofactors. Consequently, the resulting proteins had the capability to catalyse the functionalization of C–H bonds to form C–C bonds by carbene insertion and add carbenes to both β-substituted vinylarenes and unactivated aliphatic α-olefins.
Another approach involves the use of biotin–streptavidin (Sav) system to anchor metal complexes to a specific location in a protein [59 ,98–100]. This methodology was applied to artificial cupredoxin synthesis recently [59]. A systematic modulation of the primary and secondary co-ordination spheres of artificial copper proteins was performed. Biotinylated copper complexes within a cysteine containing variant of Sav have exhibited properties similar to native cupredoxins. Fine tuning of the position of the Cu centres inside the modified Sav was achieved by changing length of the linker between biotin and the copper complex. A similar approach was employed by Heinisch et al. [82] and Quinto et al. [98] in the design of an artificial metalloenzyme that exhibits activity as an NAD(P)H dependent transfer hydrogenase recently. To this end, a biotinylated iridium cofactor was introduced to variants of Sav scaffold that resulted in the successful activity of the artificial metalloenzymes.
Conclusions and future perspectives
Metalloproteins advanced through millions of years of protein evolution. Considering this huge evolutionary history, we are still far from perfecting our de novo design attempts. Computational design offers an alternative but complementary route to directed evolution experiments for the introduction of novel functionalities. Learning from nature’s toolbox and rules that drive protein evolution, we can improve our designs [95]. Considerable efforts have been put for the design and production of new artificial metalloproteins [85]. However, there are still challenges such as achieving accuracy at the atomic level since flexible polar amino acid side chains form metal-binding sites through alternative ways. Computational design principles can be used to design a library of variants that could co-ordinate a metal cofactor. Additionally, it can be used to reduce number of the variants for experimental characterization [82]. Initially, the metalloprotein design was focused on the design and formation of mono-metal centres on the designed proteins. This horizon was expanded by the introduction of di-metal centres [91, 101] and current research is focused on building multi-metal cluster containing artificial metalloproteins [58] and specifically metalloenzymes since most remarkable and complex chemical transformations are achieved through the use of multi-metal centres [59]. As these horizons expand, design of different scaffolds from first principles that would host different metal centres is also needed. Moreover, directed evolution should complement de novo protein design efforts to produce artificial metalloproteins with improved properties. Although an increasing number of graphical user interfaces for protein design and metal ion-binding site prediction are being developed, use of computational programs for the design of artificial metalloproteins does not follow the same pace. Therefore, there is an emerging need to develop user-friendly tools specific for metalloprotein design and engineering.
Abbreviations
- BIM
bioinorganic motif
- bpy
bipyridine
- Bpy–Ala
(2,2′-bipyridin-5yl) alanine
- CHARMM
Chemistry at Harvard Macromolecular Mechanics
- CMM
Check My Metal
- fs
femtosecond
- MIPS
metal interactions in protein structures
- PDB
Protein Databank
- Ru(bpy)2
ruthenium(II) bpy
- Sav
Streptavidin
- SASA
Solvent Accessible Surface Area
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Rose P.W., Bi C., Bluhm W.F., Christie C.H., Dimitropoulos D., Dutta S. et al. (2013) The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 41, D475–D482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu C.-H., Lin Y.-F., Lin J.-J. and Yu C.-S. (2012) Prediction of metal ion-binding sites in proteins using the fragment transformation method. Plos ONE 7, e39252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreini C., Bertini I. and Rosato A. (2009) Metalloproteomes: a bioinformatic approach. Acc. Chem. Res. 42, 1471–1479 [DOI] [PubMed] [Google Scholar]
- 4.Andreini C., Cavallaro G., Lorenzini S. and Rosato A. (2013) MetalPDB: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 41, D312–D319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasampalidis I.N., Pitas I. and Lyroudia K. (2007) Conservation of metal-coordinating residues. Proteins 68, 123–130 [DOI] [PubMed] [Google Scholar]
- 6.Opella S.J., DeSilva T.M. and Veglia G. (2002) Structural biology of metal-binding sequences. Curr. Opin. Chem. Biol. 6, 217–223 [DOI] [PubMed] [Google Scholar]
- 7.Brylinski M. and Skolnick J. (2011) FINDSITE-metal: Integrating evolutionary information and machine learning for structure-based metal binding site prediction at the proteome level. Proteins 79, 735–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barondeau D.P. and Getzoff E.D. (2004) Structural insights into protein-metal ion partnerships. Curr. Opin. Struct. Biol. 14, 765–774 [DOI] [PubMed] [Google Scholar]
- 9.Yamashita M.M., Wesson L., Eisenman G. and Eisenberg D. (1990) Where metal ions bind in proteins. Proc. Natl. Acad. Sci. U.S.A. 87, 5648–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm R.H., Kennepohl P. and Solomon E.I. (1996) Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 [DOI] [PubMed] [Google Scholar]
- 11.Kendrew J.C., Bodo G., Dintzis H.M., Parrish R.G., Wyckoff H. and Phillips D.C. (1958) A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 181, 662–666 [DOI] [PubMed] [Google Scholar]
- 12.Finkelstein J. (2009) Metalloproteins. Nature 460, 813. [DOI] [PubMed] [Google Scholar]
- 13.Perutz M.F., Muirhead H., Cox J.M. and Goaman L.C.G. (1968) Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 Å resolution: the atomic model. Nature 219, 131–139 [DOI] [PubMed] [Google Scholar]
- 14.Regan L. (1995) Protein design: novel metal-binding sites. Trends Biochem. Sci. 20, 280–285 [DOI] [PubMed] [Google Scholar]
- 15.Gaggelli E., Kozlowski H., Valensin D. and Valensin G. (2006) Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis). Chem. Rev. 106, 1995–2044 [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Chakraborty S., Hosseinzadeh P., Yu Y., Tian S., Petrik I. et al. (2014) Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 114, 4366–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lothian A., Hare D.J., Grimm R., Ryan T.M., Masters C.L. and Roberts B.R. (2013) Metalloproteomics: principles, challenges and applications to neurodegeneration. Front. Aging Neurosci. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fosso-Kankeu E. and Mulaba-Bafubiandi A.F. (2014) Implication of plants and microbial metalloproteins in the bioremediation of polluted waters: a review. Phys. Chem. Earth Parts A/B/C 67–69, 242–252 [Google Scholar]
- 19.Cobbett C. and Goldsbrough P. (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182 [DOI] [PubMed] [Google Scholar]
- 20.Yruela I. (2013) Transition metals in plant photosynthesis. Metallomics 5, 1090–1109 [DOI] [PubMed] [Google Scholar]
- 21.Merchant S. and Dreyfuss B.W. (1998) Posttranslational assembly of photosynthetic metalloproteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 25–51 [DOI] [PubMed] [Google Scholar]
- 22.Botelho H.M., Koch M., Fritz G. and Gomes C.M. (2009) Metal ions modulate the folding and stability of the tumor suppressor protein S100A2. FEBS J. 276, 1776–1786 [DOI] [PubMed] [Google Scholar]
- 23.Sujak A., Sanghamitra N.J.M., Maneg O., Ludwig B. and Mazumdar S. (2007) Thermostability of proteins: role of metal binding and pH on the stability of the dinuclear Cu(A) site of Thermus thermophilus. Biophys. J. 93, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palm-Espling M.E., Niemiec M.S. and Wittung-Stafshede P. (2012) Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 1823, 1594–1603 [DOI] [PubMed] [Google Scholar]
- 25.Regan L. and DeGrado W. (1988) Characterization of a helical protein designed from first principles. Science 241, 976–978 [DOI] [PubMed] [Google Scholar]
- 26.Lu Y., Yeung N., Sieracki N. and Marshall N.M. (2009) Design of functional metalloproteins. Nature 460, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Zhang Z. and Song J. (2012) Computational enzyme design approaches with significant biological outcomes: progress and challenges. Comput. Struct. Biotechnol. J. 2, e201209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passerini A., Lippi M. and Frasconi P. (2012) Predicting metal-binding sites from protein sequence. IEEE/ACM Trans. Comput. Biol. Bioinform. 9, 203–213 [DOI] [PubMed] [Google Scholar]
- 29.Shi W., Punta M., Bohon J., Sauder J.M., D’Mello R., Sullivan M. et al. (2011) Characterization of metalloproteins by high-throughput X-ray absorption spectroscopy. Genome Res. 21, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babor M., Gerzon S., Raveh B., Sobolev V. and Edelman M. (2008) Prediction of transition metal-binding sites from apo protein structures. Proteins 70, 208–217 [DOI] [PubMed] [Google Scholar]
- 31.Asante-Appiah E., Seeholzer S.H. and Skalka A.M. (1998) Structural determinants of metal-induced conformational changes in HIV-1 integrase. J. Biol. Chem. 273, 35078–35087 [DOI] [PubMed] [Google Scholar]
- 32.de Peredo A.G., Saint-Pierre C., Latour J.-M., Michaud-Soret I. and Forest E. (2001) Conformational changes of the ferric uptake regulation protein upon metal activation and DNA binding; first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310, 83–91 [DOI] [PubMed] [Google Scholar]
- 33.Liu T. and Altman R.B. (2009) Prediction of calcium-binding sites by combining loop-modeling with machine learning. BMC Struct. Biol. 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy R., Edelman M. and Sobolev V. (2009) Prediction of 3D metal binding sites from translated gene sequences based on remote-homology templates. Proteins 76, 365–374 [DOI] [PubMed] [Google Scholar]
- 35.Passerini A., Andreini C., Menchetti S., Rosato A. and Frasconi P. (2007) Predicting zinc binding at the proteome level. BMC Bioinformatics 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai C.Z., Han L.Y., Ji Z.L., Chen X. and Chen Y.Z. (2003) SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 31, 3692–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H., Han L., Zhang H., Zheng C., Xie B., Cao Z. et al. (2006) Prediction of the functional class of metal-binding proteins from sequence derived physicochemical properties by support vector machine approach. BMC Bioinformatics 7, S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valasatava Y., Rosato A., Banci L. and Andreini C. (2016) MetalPredator: a web server to predict iron-sulfur cluster binding proteomes. Bioinformatics 32, 2850–2852 [DOI] [PubMed] [Google Scholar]
- 39.Passerini A., Lippi M. and Frasconi P. (2011) MetalDetector v2.0: predicting the geometry of metal binding sites from protein sequence. Nucleic Acids Res. 39, W288–W292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brylinski M. and Skolnick J. (2011) FINDSITE-metal: integrating evolutionary information and machine learning for structure-based metal-binding site prediction at the proteome level. Proteins 79, 735–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He W., Liang Z., Teng M. and Niu L. (2015) mFASD: a structure-based algorithm for discriminating different types of metal-binding sites. Bioinformatics 31, 1938–1944 [DOI] [PubMed] [Google Scholar]
- 42.Zhao W., Xu M., Liang Z., Ding B., Niu L., Liu H. et al. (2011) Structure-based de novo prediction of zinc-binding sites in proteins of unknown function. Bioinformatics 27, 1262–1268 [DOI] [PubMed] [Google Scholar]
- 43.Liang M.P., Banatao D.R., Klein T.E., Brutlag D.L. and Altman R.B. (2003) WebFEATURE: an interactive web tool for identifying and visualizing functional sites on macromolecular structures. Nucleic Acids Res. 31 3324–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebert J.C. and Altman R.B. (2008) Robust recognition of zinc binding sites in proteins. Protein Sci. 17, 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S., Liang M.P. and Altman R.B. (2008) The SeqFEATURE library of 3D functional site models: comparison to existing methods and applications to protein function annotation. Genome Biol. 9, R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodhi J.S., Bryson K., McGuffin L.J., Ward J.J., Wernisch L. and Jones D.T. (2004) Predicting metal-binding site residues in low-resolution structural models. J. Mol. Biol. 342, 307–320 [DOI] [PubMed] [Google Scholar]
- 47.Wass M.N., Kelley L.A. and Sternberg M.J.E. (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38, W469–W473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X., Dong Q., Yang J. and Zhang Y. (2016) Recognizing metal and acid radical ion-binding sites by integrating ab initio modeling with template-based transferals. Bioinformatics 32, 3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreini C., Cavallaro G., Rosato A. and Valasatava Y. (2013) MetalS2: a tool for the structural alignment of minimal functional sites in metal-binding proteins and nucleic acids. J. Chem. Inf. Model 53, 3064–3075 [DOI] [PubMed] [Google Scholar]
- 50.Valasatava Y., Rosato A., Cavallaro G. and Andreini C. (2014) MetalS(3), a database-mining tool for the identification of structurally similar metal sites. J. Biol. Inorg. Chem. 19, 937–945 [DOI] [PubMed] [Google Scholar]
- 51.Andreini C., Cavallaro G. and Lorenzini S. (2012) FindGeo: a tool for determining metal coordination geometry. Bioinformatics 28, 1658–1660 [DOI] [PubMed] [Google Scholar]
- 52.Zheng H., Chordia M.D., Cooper D.R., Chruszcz M., Müller P., Sheldrick G.M. et al. (2013) Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc. 9, 156–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi H Metalmine [Internet] (2009) MetalMine: a database of functional metal-binding sites in proteins. [cited 2016 Aug 2]. Available from: http://metalmine.naist.jp/metalmine009/about.html [Google Scholar]
- 54.Hemavathi K., Kalaivani Kaani M., Udayakumar A., Sowmiya G., Jeyakanthan J. and Sekar K. (2010) MIPS: metal interactions in protein structures. J. Appl. Cryst. 43, 196–199 [Google Scholar]
- 55.Hsin K.-Y., Sheng Y., Harding M.M., Taylor P. and Walkinshaw M.D. (2008) MESPEUS: a database of the geometry of metal sites in proteins. J. Appl. Cryst. 41, 963–968 [Google Scholar]
- 56.Degtyarenko K. and Contrino S. (2004) COM e: the ontology of bioinorganic proteins. BMC Struct. Biol. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi H., Kang H. and Park H. (2011) MetLigDB: a web-based database for the identification of chemical groups to design metalloprotein inhibitors. J. Appl. Cryst. 44, 878–881 [Google Scholar]
- 58.Fehl C. and Davis B.G. (2016) Proteins as templates for complex synthetic metalloclusters: towards biologically programmed heterogeneous catalysis. Proc. Math. Phys. Eng. Sci. 472, 20160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mann S.I., Heinisch T., Weitz A.C., Hendrich M.P., Ward T.R. and Borovik A.S. (2016) Modular artificial cupredoxins. J. Am. Chem. Soc. 138, 9073–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu F., Cangelosi V.M., Zastrow M.L., Tegoni M., Plegaria J.S., Tebo A.G. et al. (2014) Protein design: toward functional metalloenzymes. Chem. Rev. 114, 3495–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nastri F., Chino M., Maglio O., Bhagi-Damodaran A., Lu Y. and Lombardi A. (2016) Design and engineering of artificial oxygen-activating metalloenzymes. Chem. Soc. Rev. 45, 5020–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pordea A. (2015) Metal-binding promiscuity in artificial metalloenzyme design. Curr. Opin. Chem. Biol. 25, 124–132 [DOI] [PubMed] [Google Scholar]
- 63.Cangelosi V.M., Deb A., Penner-Hahn J.E. and Pecoraro V.L. (2014) A de novo designed metalloenzyme for the hydration of CO2. Angew. Chem. Int. Ed. Engl. 53, 7900–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dill K.A. and MacCallum J.L. (2012) The protein-folding problem, 50 years on. Science 338, 1042–1046 [DOI] [PubMed] [Google Scholar]
- 65.Drexler K.E. (1981) Molecular engineering: an approach to the development of general capabilities for molecular manipulation. Proc. Natl. Acad. Sci. U.S.A. 78, 5275–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pabo C. (1983) Molecular technology: designing proteins and peptides. Nature 301, 200. [DOI] [PubMed] [Google Scholar]
- 67.Fung H.K., Welsh W.J. and Floudas C.A. (2008) Computational de novo peptide and protein design: rigid templates versus flexible templates. Ind. Eng. Chem. Res. 47, 993–1001 [Google Scholar]
- 68.Smadbeck J., Peterson M.B., Khoury G.A., Taylor M.S. and Floudas C.A. (2013) Protein WISDOM: a workbench for in silico de novo design of biomolecules. J. Vis. Exp. 25, 10.3791/50476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hay B.P. and Firman T.K. (2002) HostDesigner: a program for the de novo structure-based design of molecular receptors with binding sites that complement metal ion guests. Inorg. Chem. 41, 5502–5512 [DOI] [PubMed] [Google Scholar]
- 70.Clarke N.D. and Yuan S.-M. (1995) Metal search: a computer program that helps design tetrahedral metal-binding sites. Proteins 23, 256–263 [DOI] [PubMed] [Google Scholar]
- 71.Jiang X., Farid H., Pistor E. and Farid R.S. (2000) A new approach to the design of uniquely folded thermally stable proteins. Protein Sci. 9, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y. and Kuhlman B. (2006) RosettaDesign server for protein design. Nucleic Acids Res. 34, W235–W238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitra P., Shultis D. and Zhang Y. (2013) EvoDesign: de novo protein design based on structural and evolutionary profiles. Nucleic Acids Res. 41, W273–W280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lauck F., Smith C.A., Friedland G.F., Humphris E.L. and Kortemme T. (2010) RosettaBackrub–a web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 38, W569–W575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fazelinia H., Cirino P.C. and Maranas C.D. (2009) OptGraft: A computational procedure for transferring a binding site onto an existing protein scaffold. Protein Sci. 18, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhury S., Lyskov S. and Gray J.J. (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farinas E. and Regan L. (1998) The de novo design of a rubredoxin-like Fe site. Protein Sci. 7, 1939–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klemba M., Gardner K.H., Marino S., Clarke N.D. and Regan L. (1995) Novel metal-binding proteins by design. Nat. Struct. Mol. Biol. 2, 368–373 [DOI] [PubMed] [Google Scholar]
- 79.Regan L. and Clarke N.D. (1990) A tetrahedral zinc(II)-binding site introduced into a designed protein. Biochemistry 29, 10878–10883 [DOI] [PubMed] [Google Scholar]
- 80.Nanda V., Rosenblatt M.M., Osyczka A., Kono H., Getahun Z., Dutton P.L. et al. (2005) De novo design of a redox-active minimal rubredoxin mimic. J. Am. Chem. Soc. 127, 5804–5805 [DOI] [PubMed] [Google Scholar]
- 81.Cristian L., Piotrowiak P. and Farid R.S. (2003) Mimicking photosynthesis in a computationally designed synthetic metalloprotein. J. Am. Chem. Soc. 125, 11814–11815 [DOI] [PubMed] [Google Scholar]
- 82.Heinisch T., Pellizzoni M., Dürrenberger M., Tinberg C.E., Köhler V., Klehr J. et al. (2015) Improving the catalytic performance of an artificial metalloenzyme by computational design. J. Am. Chem. Soc. 137, 10414–10419 [DOI] [PubMed] [Google Scholar]
- 83.Monnard F.W., Heinisch T., Nogueira E.S., Schirmer T. and Ward T.R. (2011) Human carbonic anhydrase II as a host for piano-stool complexes bearing a sulfonamide anchor. Chem. Commun. (Camb.) 47, 8238–8240 [DOI] [PubMed] [Google Scholar]
- 84.Gamenara D. and Dominguez de Maria P. (2014) Enantioselective imine reduction catalyzed by imine reductases and artificial metalloenzymes. Org. Biomol. Chem. 12, 2989–2992 [DOI] [PubMed] [Google Scholar]
- 85.Mills J.H., Khare S.D., Bolduc J.M., Forouhar F., Mulligan V.K., Lew S. et al. (2013) Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy. J. Am. Chem. Soc. 135, 13393–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zanghellini A., Jiang L., Wollacott A.M., Cheng G., Meiler J., Althoff E.A. et al. (2006) New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 15, 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu C., Zhang C., Liang H. and Lai L. (2011) Engineering a zinc binding site into the de novo designed protein DS119 with a βαβ structure. Protein Cell 2, 1006–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeung N., Lin Y.-W., Gao Y.-G., Zhao X., Russell B.S., Lei L. et al. (2009) Rational design of a structural and functional nitric oxide reductase. Nature 462, 1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salgado E.N., Ambroggio X.I., Brodin J.D., Lewis R.A., Kuhlman B. and Tezcan F.A. (2010) Metal templated design of protein interfaces. Proc. Natl. Acad. Sci. U.S.A. 107, 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rufo C.M., Moroz Y.S., Moroz O.V., Stöhr J., Smith T.A., Hu X. et al. (2014) Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 6, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanaka T., Mizuno T., Fukui S., Hiroaki H., Oku J., Kanaori K. et al. (2004) Two-metal ion, Ni(II) and Cu(II), binding alpha-helical coiled coil peptide. J. Am. Chem. Soc. 126, 14023–14028 [DOI] [PubMed] [Google Scholar]
- 92.Nanda V. and Koder R.L. (2010) Designing artificial enzymes by intuition and computation. Nat. Chem. 2, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zastrow M.L., Peacock A.F., Stuckey J.A. and Pecoraro V.L. (2012) Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat. Chem. 4, 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zastrow M.L. and Pecoraro V.L. (2013) The influence of active site location on catalytic activity in de novo designed zinc metalloenzymes. J. Am. Chem. Soc. 135, 5895–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tebo A.G. and Pecoraro V.L. (2015) Artificial metalloenzymes derived from three-helix bundles. Curr. Opin. Chem. Biol. 25, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Plegaria J.S., Herrero C., Quaranta A. and Pecoraro V.L. (2016) Electron transfer activity of a de novo designed copper center in a three-helix bundle fold. Biochim. Biophys. Acta 1857, 522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Key H.M., Dydio P., Clark D.S. and Hartwig J.F. (2016) Abiological catalysis by artificial haem proteins containing noble metals in place of iron. Nature 534, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quinto T., Haussinger D., Kohler V. and Ward T.R. (2015) Artificial metalloenzymes for the diastereoselective reduction of NAD(+) to NAD(2)H. Org. Biomol. Chem. 13, 357–360 [DOI] [PubMed] [Google Scholar]
- 99.Lo C., Ringenberg M.R., Gnandt D., Wilson Y. and Ward T.R. (2011) Artificial metalloenzymes for olefin metathesis based on the biotin-(strept)avidin technology. Chem. Commun. (Camb.) 47, 12065–12067 [DOI] [PubMed] [Google Scholar]
- 100.Okamoto Y., Köhler V. and Ward T.R. (2016) An NAD(P)H-dependent artificial transfer hydrogenase for multienzymatic cascades. J. Am. Chem. Soc. 138, 5781–5784 [DOI] [PubMed] [Google Scholar]
- 101.Shiga D., Funahashi Y., Masuda H., Kikuchi A., Noda M., Uchiyama S. et al. (2012) Creation of a binuclear purple copper site within a de novo coiled-coil protein. Biochemistry 51, 7901–7907 [DOI] [PubMed] [Google Scholar]