Abstract
Inflammation induced by innate immunity influences the development of T cell-mediated autoimmunity in multiple sclerosis (MS) and its animal model, experimental autoimmune encephalomyelitis (EAE). We found that strong activation of innate immunity induced NLRP3 inflammasome-independent and interferon-β (IFNβ)-resistant EAE (termed Type-B EAE), whereas EAE induced by weak activation of innate immunity requires NLRP3 inflammasome and is sensitive to IFNβ treatment. Instead, an alternative inflammatory mechanism, including membrane-bound lymphotoxin-β receptor (LTβR) and CXCR2, is involved in Type-B EAE development; and Type-B EAE is ameliorated by antagonizing the receptors. Relative expression of Ltbr and Cxcr2 genes was indeed enhanced in IFNβ-resistant MS patients. Importantly, remission is minimal in Type-B EAE due to neuronal damages induced by semaphorin 6B upregulation on CD4+ T cells. Our data reveal a novel inflammatory mechanism by which IFNβ-resistant EAE subtype develops.
Introduction
Multiple sclerosis (MS) is a chronic inflammatory autoimmune disease accompanied by demyelination of the central nervous system (CNS). As a multifactorial heterogeneous disease, MS is difficult to treat. Some MS patients show CNS axonal damage, which may occur independently of chronic demyelination1. Disease heterogeneity also extends to drug response. For example, IFNβ, a first-line treatment to MS patients for over 15 years in relapsing-remitting MS (RRMS), is not effective for 7–49% of RRMS patients2. Mechanisms underlying the heterogeneity in the disease etiology and patients’ responses to treatments are largely unknown.
Experimental autoimmune encephalomyelitis (EAE), an animal model of MS, has provided valuable insight to some of the pathological mechanisms of MS and has led to the development of glatiramer acetate and natalizumab as therapeutics3–5. However, there is an inherent challenge to using EAE as a model for studying disease heterogeneity without introducing confounders such as an alternative mouse strain(s), manipulating gene expression, or using unconventional reagent(s)6.
The NLRP3 inflammasome is a crucial target of IFNβ treatment in EAE. We have previously reported that IFNβ suppresses NLRP3 inflammasome activity by reducing active Rac-1 and mitochondrial ROS generation7. The NLRP3 inflammasome plays a critical role in EAE development, as demonstrated that Nlrp3−/− mice are resistant to EAE8, 9. The lack of the NLRP3 inflammasome in antigen presenting cells (APCs) dampens expression of chemokines and chemokine receptors on T helper cells and APCs, and fails their migration to the CNS8. Nevertheless, it is also known that EAE can develop without the NLRP3 inflammasome7, 10. It is, however, elusive what is an alternative mechanistic pathway to induce EAE without the NLRP3 inflammasome, and whether the NLRP3 inflammasome-independent EAE is functionally and phenotypically distinct from typical NLRP3 inflammasome-dependent EAE. Understanding these mechanisms may represent a pivotal step toward understanding heterogeneity in EAE, and possibly in MS too.
In this study, we sought to reveal the molecular and cellular mechanisms that govern the pathophysiology of Type-B EAE by dissecting mechanisms to induce an NLRP3 inflammasome-independent EAE subtype, which is induced by aggressive immunization using high doses of heat-killed Mycobacteria (Mtb) without introducing confounding factors. For Type-B EAE development, our results suggested the critical involvement and enhanced expression of lymphotoxin-β receptor (LTβR) and a chemokine receptor CXCR2, which detect membrane-bound lymphotoxin (mLT) and CXCL1, respectively. The pathway including LTβR and CXCR2 allows mice to bypass NLRP3 inflammasome-mediated inflammation for EAE induction. Although Type-B EAE does not respond to IFNβ treatment, blockade of LTβR or CXCR2 successfully ameliorates the disease. Furthermore, relative levels of Ltbr and Cxcr2 were found to be increased in IFNβ non-responder MS patients. Type-B EAE undergoes irreversible disease phenotype due to the neuronal damage mediated by semaphorin 6B (Sema6B) upregulation in CD4+ T cells in the CNS.
RESULTS
NLRP3 is dispensable for EAE high adjuvant doses
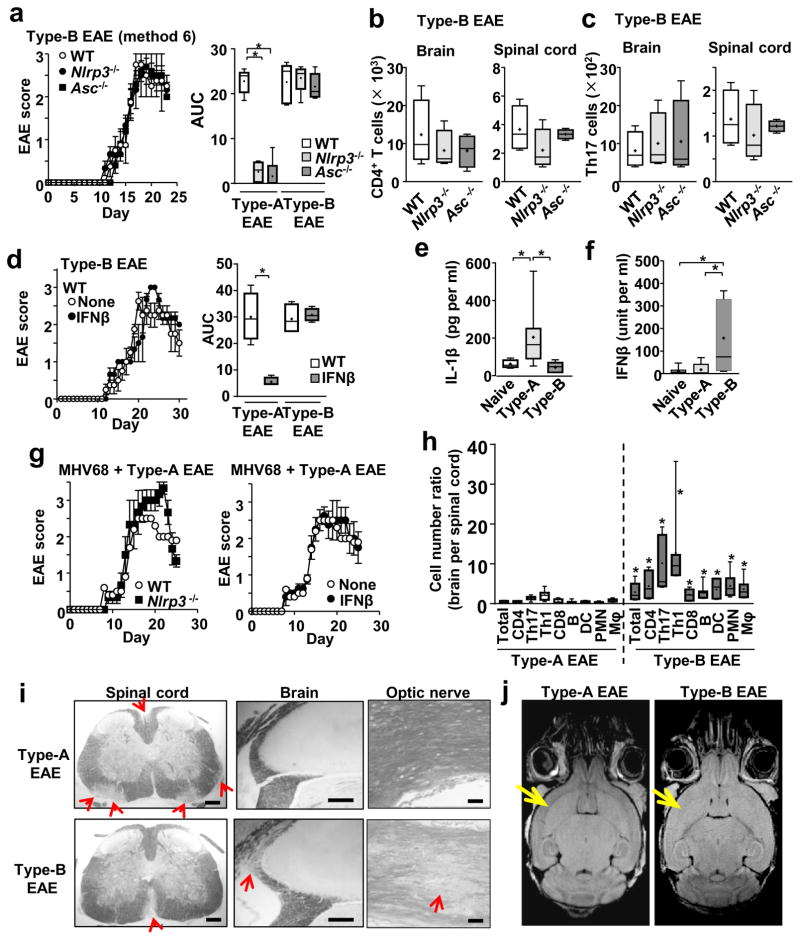
We titrated the amounts of Mtb (i.e, adjuvant) used in complete Freund’s adjuvant (CFA) to induce EAE, and evaluated disease severity in Nlrp3−/− mice (in the C57BL6 background). Nlrp3−/− mice were used, because they hardly developed EAE using the standard EAE induction method with 200 μg Mtb in CFA (Method 1 in Supplementary Figure 1a, b), as previously reported8. However, either repeated immunization or an increased Mtb dosage (400 μg) was sufficient to induce severe EAE in Nlrp3−/− mice (Method 2, 3 in Supplementary Figure 1a, b). In contrast, 25 % increase in myelin oligodendrocyte glycoprotein (MOG) peptide did not change EAE severity (Method 3 vs. 4 and 5 vs. 6 in Supplementary Figure 1a, b). Using the most aggressive EAE induction method (Method 6 in Supplementary Figure 1a, b), Asc−/− and Nlrp3−/− mice developed comparable disease severity to wild-type (WT) mice (Figure 1a), as well as similar numbers of total leukocytes, CD4+ T cells, and Th17 cells in spinal cords and brains (Fig. 1b, c; Supplementary Figure 1c). Importantly, EAE induced by the Method 6 completely abrogated the efficacy of IFNβ both in WT and Nlrp3−/− mice (Figure 1d; Supplementary Figure 1d). Here, we tentatively termed the EAE subtype, which develops without the NLRP3 inflammasome and does not respond to IFNβ, as “Type-B EAE,” in contrast to another EAE subtype, which is NLRP3 inflammasome-dependent and responds to IFNβ treatment (Figure 1d, right panel)(induced by the Method 1, and termed “Type-A EAE”). Type-B EAE mice could not clinically tell apart from Type-A EAE mice for the first 30 days in comparable body weights (Supplementary Figure 1e) and the lack of obvious difference in ataxia (Supplementary video 1). The results here also suggested that there are intermediate phenotypes between Type-A and Type-B EAE, which represent a continuum of the EAE subtypes.
Figure 1. Type-B EAE develops independent of the NLRP3 inflammasome and is resistant to IFNβ treatment.
(a) EAE scores after Type-B EAE induction (n=5). AUC was calculated from Type-A and Type-B EAE data between day 0–23 dpi. Type-A EAE: P(WT vs Nlrp3−/−) <0.0001, t(8)=12.78, P(WT vs Asc−/−) <0.0001, t(8)=10.57. Type-B EAE: P(WT vs Nlrp3−/−)=0.7236, t(8)=0.3634, P(WT vs Asc−/−)=0.6869, t (8)=0.4181. (b, c) Numbers of total CD4+ T cells (b), and Th17 cells (c) in the brains and spinal cords of mice with Type-B EAE at 17-dpi. n=4. CD4+ T in Brain: P(WT vs Nlrp3−/−)=0.4507, t(6)=0.8065, P(WT vs Asc−/−)=0.4256, t(6)=0.85456. CD4+ T in Spinal cord: P(WT vs Nlrp3−/−)=0.2260, t(6)=1.349, P(WT vs Asc−/−)=0.6887, t(6)=0.4206. Th17 in Brain: P(WT vs Nlrp3−/−)=0.6880, t(6)=0.4216, P(WT vs Asc−/−)=0.6953, t(6)=0.4111. Th17 in Spinal cord: P(WT vs Nlrp3−/−)=0.4683, t(6)=0.77416, P(WT vs Asc−/−)=0.6429, t(6)=0.4879. (d) EAE scores of Type-B EAE mice with or without IFNβ treatment. n=5. AUC between 0–30 dpi. P(Type-A)=0.0002, t(5)=6.668, P(Type-B)=0.6514, t(6)=0.4753. Treatment condition is described in Experimental Procedures. (e, f) Serum levels of IL-1β (e) and IFNβ (f) in naïve mice, Type-A and Type-B EAE mice at 9-dpi. n=7. IL-1β: P(Naïve vs Type-A) 0.0327, t(13)=2.389, P(Naïve vs Type-B)=0.1697, t(14)=1.448, P(Type-A vs Type-B)=0.0284, t(13)=2.4912. Naïve (n=8), Type-A (n=7), Type-B (n=8). IFNβ: P(Naïve vs Type-A)=0.7024, t(12)=0.3914, P(Naïve vs Type-B)=0.0339, t(12)=2.393, P(Type-A vs Type-B)=0.0409, t(12)=2.29. (g) EAE scores of WT and Nlrp3−/− mice immunized with a Type-A EAE induction method with MHV68 infection (left panel); and WT Type-A EAE mice infected with MHV68 with or without IFNβ treatment (right panel). MHV68 (105 pfu/mouse) was i.v. administered at the same time as Type-A EAE immunization. n=5. (h) Cell number ratios shown as brain-infiltrated cells divided by spinal-cord infiltrated cells (n=7). P(Total) =0.0166, t(12)=2.78, P(CD4)<0.0074, t(12)=3.216, P(Th17)=0.049, t(12)=3.436, P(Th1)=0.0193, t(12)=2.701, P(CD8)=0.098, t(12)=3.067, P(B)=0.0062, t(12)=3.311, P(DC)=0.0049, t(12)=3.436, P(PMN)=0.0051, t(12)=3.413, P(Mac)=0.0112, t(12)=2.992. (i) Representative LFB-stained images with red arrows indicating regions of demyelination. All the CNS samples were harvested at 17-dpi.. (j) T2 FLAIR MRI analysis of brain obtained from mice at 18-dpi. Yellow arrows indicate cortical region and external capsule. Image from Type B EAE mice exhibits the potential loss of myelin. *; p<0.05. All scale bars, 200 μm. All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. All the experimental data and images are representatives from at least 2 similar experiments for each.
Type-B EAE mice were characterized with low and basal levels of serum IL-1β (Figure 1e). Splenocytes from Type-B EAE mice showed significantly decreased ability to produce extracellular IL-1β, despite high expression of Il1b mRNA, and decrease in caspase-1 activity Supplementary Figure 1f–h). The results strongly suggest remarkably low levels of NLRP3 inflammasome activation in Type-B EAE. In contrast, high levels of serum IFNβ were observed in Type-B EAE mice (Figure 1f). As IFNβ suppresses NLRP3 inflammasome activity7, endogenous IFNβ may contribute to inhibit NLRP3 inflammasome activation in Type-B EAE mice.
Because Type-B EAE is induced with high dosages of adjuvant (Mtb), we sought to identify if acute infection converts the phenotype of mice, immunized with the Type-A EAE-induction, to develop Type-B EAE responses. Here, we intentionally used an agent, which is not Mtb, to test our hypothesis -- strong stimulation of innate immunity enhances Type-B EAE responses – is not specific for the Mtb adjuvant. Murine gamma herpesvirus-68 (MHV-68), equivalent to Epstein-Barr virus (EBV) in humans, was used simply because of the relevance of EBV infection to MS11. Type-A EAE induction accompanied with a high inoculum of MHV-68 made mice develop NLRP3 inflammasome-independent and IFNβ-resistant EAE (Figure 1g). Taken together, it is possible that EAE is skewed towards Type-B by strong stimulation of innate immunity with high dosages of adjuvant or acute infection.
Distinct phenotypes between Type-A and Type-B EAE
Type-B EAE mice showed significantly higher numbers of various leukocytes than Type-A EAE mice on 17-day post immunization (dpi) in brains (peak time of disease)(Figure 1h, Supplementary Figure 1i, 1j). In contrast, Type-A EAE mice showed higher numbers of these cells than Type-B EAE mice in spinal cords (Figure 1h, Supplementary Figure 1i). Type-B EAE mice suffered mild demyelination in the spinal cord, whereas demyelination in Type-A EAE mice was severe (Figure 1i). Alternatively, Type-B EAE mice, but not Type-A EAE mice, suffered severe demyelination in the brain (proximal to choroid plexus, an entry zone of immune cells into the brain12) and in the optic nerve (Figure 1i, Supplementary Figure 1k). Magnetic resonance imaging (MRI) T2 FLAIR images showed hyperintensity, reflecting demyelination and cell density changes, in the external capsule and adjacent cortical regions in the brains of Type-B EAE mice at 18-dpi and to a lesser extent in the spinal cord (Figure 1j, Supplementary Figure 1l). Type-B EAE mice also showed thermal hyperalgesia at 9-dpi (before or around EAE onset), but mice with Type-A EAE did not (Supplementary Figure 1m). As shown in thermal hyperplasia, a pain syndrome is a manifestation of some MS patients. Although EAE scores are comparable between Type-A and Type-B EAE, mice with Type-B EAE showed distinct phenotypes, which are not reflected in the EAE score.
mLT on DCs plays a critical role in Type-B EAE development
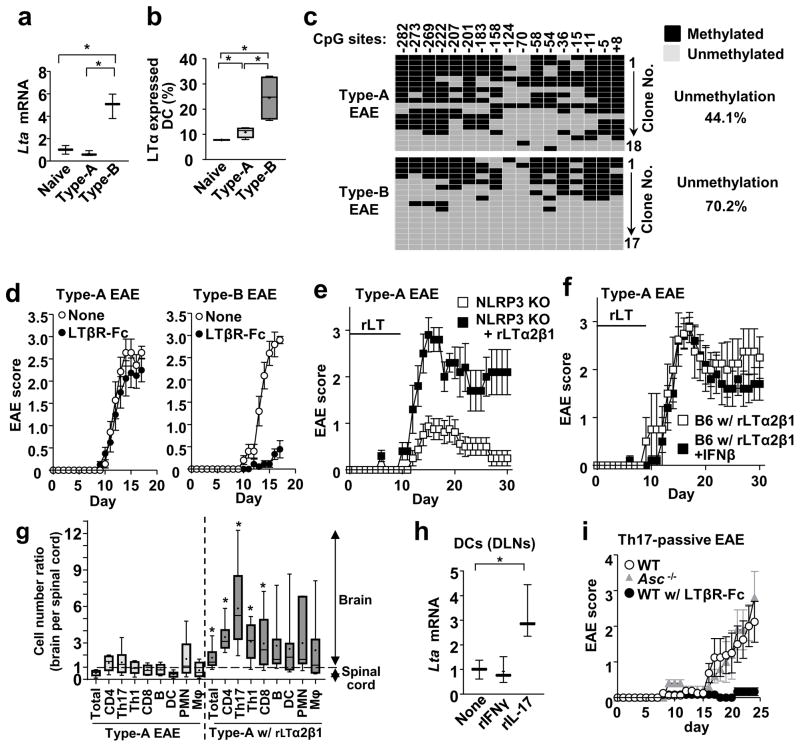
To identify molecules involved in Type-B EAE, we compared gene expression between Type-A and Type-B EAE mice. PCR array analyses indicated high expression of Lta mRNAs in dendritic cells (DCs) isolated from draining lymph nodes (DLNs) of Type-B EAE mice (Supplementary Table 1). Lta encodes lymphotoxin α (LTα, also termed TNFβ), which forms secreted homo-trimer or membrane-bound hetero-trimer with LTβ (i.e., mLT). Lymphotoxin (LT), at least, is known to be critical in the development of MS and EAE13, 14, and was observed in demyelinated regions in the brain of some MS patient15, and serum16. Blockade of LTβR, an mLT receptor, ameliorates EAE induced by repetitive immunizations17, a condition that might approximate Type-B EAE. Although Lta mRNA is widely known to be expressed in lymphocytes, DCs and macrophages expressed enhanced levels of Lta mRNA and mLT in Type-B EAE (Figure 2a, b; Supplementary Figure 2a, b). Antigen was not required to induce mLT expression on DCs because a high dose of Mtb in CFA was sufficient (Supplementary Figure 2c), although the MOG antigen helped to further induce mLT expression in Type-B EAE (Figure 2b). These results suggest that strong stimulation of innate immunity with high dose of adjuvant alone is sufficient to induce mLTc expression on DCs.
Figure 2. mLT is a key initiator of Type-B EAE.
(a) Expression levels of Lta mRNA in DCs obtained from DLNs of naïve mice or mice at 9-dpi. (n=3). P(Naive vs Type-A)=0.2646, t(4)=1.296, P(Naive vs Type-B)=0.0042, t(4)=5.88, P(Type-A vs Type-B) =0.0027, t(4)=6.61. (b) Percentages of DCs, positive for cell surface LTα expression, in DLNs of naïve mice or mice with EAE at 9-dpi, determined by flow cytometry. (n=4). P(Naive vs Type-A)=0.0253, t(6)=2.96, P(Naive vs Type-B)=0.0109, t(6)=3.637, P(Type-A vs Type-B)=0.0277, t(6)=2.891. (c) Methylation analysis by bisulfite conversion with DCs obtained from DLNs, pooled from eight WT mice, Type-A and Type-B EAE at 9-dpi. Methylated and unmethylated CpG were shown with black and gray boxes, respectively. (d) EAE scores of WT mice after Type-A and Type-B induction with or without LTβR-Fc treatment. (n=5). (e) Nlrp3−/− mice were immunized with the Type-A EAE induction method and treated with or without rLT. rLT (10 μg/kg) was i.p. administrated daily between 0–9 dpi. n=5. (f) rLT-treated WT B6 mice induced for Type-A EAE were treated with or without IFNβ (n=5). (g) Ratio of infiltrated cell numbers in the brain versus spinal cord at 17-dpi. Type-A EAE group (n=6), rLT-treated Type-A EAE group (n=7). P(Total)=0.0082, t(11)=3.22, P(CD4)=0.0024, t(11)=3.909, P(Th17)=0.0131, t(11)=2.953, P(Th1)=0.0109, t(11)=3.058, P(CD8) =0.0480, t(11)=2.225, P(B)=0.1762, t(11)=1.445, P(DC)=0.1020, t(11)=1.784, P(PMN)=0.3260, t(11)=1.027, P(Mac)=0.1737, t(11)=1.455. (h) Lta mRNA expression in DCs treated with rIL-17 (10 ng/ml) or rIFNγ (10 ng/ml) for 3h. DCs were obtained from naïve WT mice. (n=3). P(rIFNγ)=0.8433, t(4)=0.2108, P(rIL-17)=0.0295, t(4)=3.317. (i) Th17-mediated passive EAE in WT (with or without LTβR-Fc treatment) and Asc−/− mice (n=5). All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. *; p<0.05. All the experimental data sets, except for (c), are representatives from at least 2 similar experiments for each.
Induction of Lta mRNA levels in DCs was, at least in part, attributed to epigenetic alteration in the Lta gene locus. We found that the Lta promoter in DCs from Type-B EAE mice was hypomethylated, compared to naïve and Type-A EAE mice (Figure 2c; Supplementary Figure 2b). Importantly, LTβR signaling is essential for development of Type-B EAE, because an LTβR antagonist, LTβR-Fc, ameliorated Type-B EAE, but not Type-A EAE (Figure 2d). We then hypothesized that recombinant LT (rLT; LTα2β1), a soluble LTβR agonist, could “convert” the EAE phenotype from Type-A to the Type-B. With rLT treatment with Type-A EAE induction (Method 1 in Supplementary Figure 1a), Nlrp3−/− mice developed severe EAE (Figure 2e) and WT mice lost responsiveness to IFNβ treatment (Figure 2f). Moreover, rLT treatment increased the immune cell migration into the brain (Figure 2g) as observed in Type-B EAE mice (Figure 1h). Taken together, these findings strongly suggest mLT and LTβR are key factors for Type-B EAE development.
Type-B EAE and Th17 cell-mediated passive EAE
A previous report showed that IFNβ treatment did not ameliorate passive EAE induced by Th17 cell adoptive transfer18. We sought a connection between Type-B EAE and Th17-mediated passive EAE. We found that rIL-17, but not rIFNγ, upregulated Lta expression in DCs (Figure 2h), suggesting that Th17 cells may induce Type-B EAE disease through upregulation of Lta. Indeed, LTβR blockade strongly ameliorated Th17-mediated passive EAE (Figure 2i). In addition, Th17 cells were potent enough to make Asc−/− recipients develop severe EAE as well (Figure 2i). As we discuss in a later section, Type-A and Type-B EAE mice showed comparable numbers of Th17 cells in DLN. Thus, Type-B EAE responses do not appear to induce Th17 cells; rather, Th17 cell transfer induces Type-B EAE phenotypes through the mLT-LTβR axis.
CD4+ T cell gene expression profile in Type-B EAE
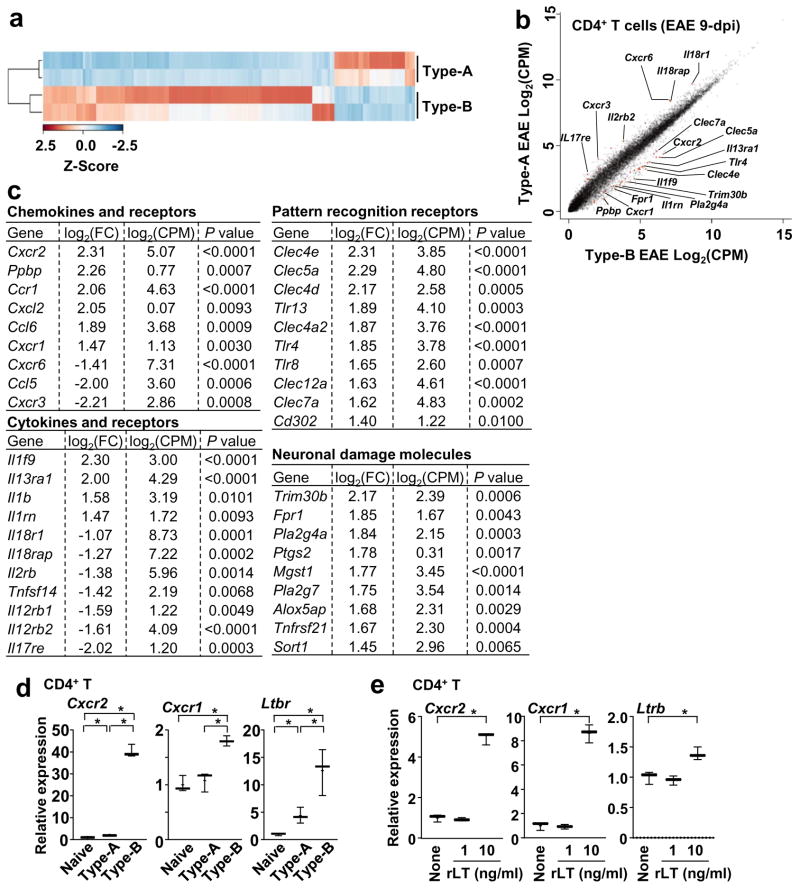
We next examined phenotypes of splenic CD4+ T cells in Type-B EAE. PCR array analyses indicated high expression of Cxcr2 and Cxcr1 mRNAs in CD4+ T cells isolated from splenocytes of Type-B EAE mice (Supplementary Table 2). To further address the broad changes of gene expression profile in CD4+ T cells, we performed high-throughput RNA-Seq analysis (Figure 3 and Supplementary Figure 3), and obtained distinct gene expression profile between Type-A EAE and Type-B EAE (Figure 3a). Six hundred sixty eight genes were upregulated more than 2-fold (P<0.05) in CD4+ T cells from Type-B EAE mice compared to those from Type-A EAE mice, while 600 genes were downregulated more than 2-fold in CD4+ T cells from Type-B EAE mice. Several genes previously detected by PCR array, such as Cxcr2 and Cxcr1 (Supplementary Table 2), were consistently detected by RNA-Seq as highly expressed genes in Type-B EAE. Interestingly, highly expressed genes in Type-B EAE include those which encode pattern-recognition receptors and neuronal damage-related molecules (Figure 3c), suggesting enhanced inflammation and neuronal damages in Type-B EAE.
Figure 3. Gene expression profile of CD4+ T cells between Type-A and Type-B EAE.
CD4+ T cells were FACS-purified from spleens at 9-dpi and submitted for the RNA-seq analysis. (a) Heatmap of of 1,846 differentially expressed genes (P<0.05; log2FC >1 or <−1) between CD4+ T cells from Type-A and Type-B EAE mice. (b) Scatter plot comparing transcriptome. (c) Selected genes from pools with P<0.01, log2FC >1 or <−1, and a log2CPM>1. (d) mRNA levels of Cxcr2, Cxcr1 and Ltbr in splenic CD4+ T cells obtained from naïve mice and mice with Type-A or Type-B EAE induction at 9-dpi determined by qPCR. (n=3). Cxcr2: P(Naive vs Type-A)=0.0231, t(4)=3.583, P(Naive vs Type-B) <0.0001, t(4)=23.22, P(Type-A vs Type-B) <0.0001, t(4)=22.75. Cxcr1: P(Naive vs Type-A)=0.5999, t(4)=0.5689, P(Naive vs Type-B)=0.0015, t(4)=7.806, P(Type-A vs Type-B)=0.0036, t(4)=6.119. Ltbr: P(Naive vs Type-A)=0.0170, t(4)=3.937, P(Naive vs Type-B)=0.0091, t(4)=4.733, P(Type-A vs Type-B)=0.0333, t(4)=3.188. (e) Levels of Cxcr2, Cxcr1 and Ltbr mRNA in CD4+ T 6h after rLT1 treatment at indicated concentrations. (n=3). P(Cxcr2) <0.0001, t(4)=19.36, P(Cxcr1) <0.0001, t(4)=16.09, P(Ltbr)=0.0116, t(4)=4.413. All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. Data sets (d, e) are representatives from at least 2 similar experiments for each.
On CD4+ T cells in spleens from Type-B EAE mice, standard qPCR confirmed the results from qPCR array and RNA-Seq analyses on abundant expression of Cxcr2, Cxcr1, and Ltbr mRNA (Figure 3d). Expression of the genes in CD4+ T cells was induced by ex vivo rLT treatment (Figure 3e), suggesting the upregulation of the mRNA in Type-B EAE was mediated by enhanced LTβR stimulation.
Comparison between Type-A and Type-B EAE
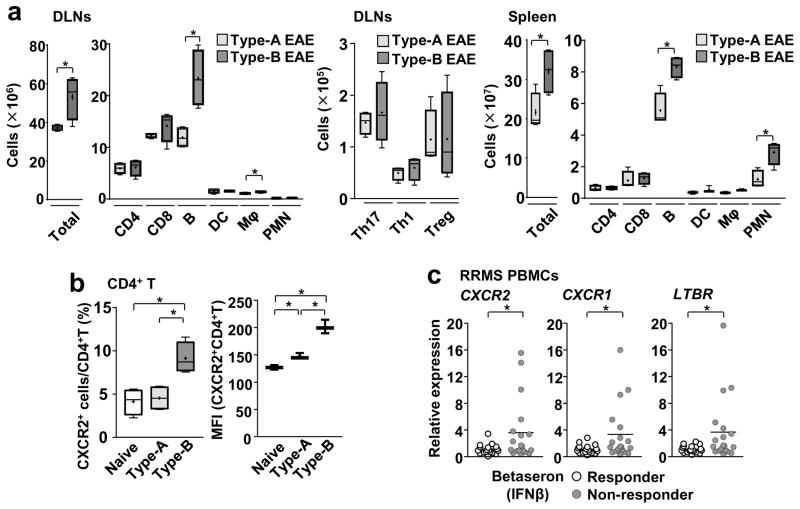
We next sought to identify differences between the two EAE subtypes at the cellular and protein levels. At the cellular level in peripheral lymphoid organs, numbers of T cells (total T, total CD4+ T, Th1, Th17, and Treg cells), DCs, macrophages, and neurtrophils/polymorphonuclear leukocytes (PMNs) were similar between two EAE subtypes, except for total leukocytes and B cells in DLNs and spleens, on day 9 (Figure 4a). However, around the disease peak time, no cell types showed significant differences in their numbers (Supplementary Figure 4a). We also examined proportions of GM-CSF-producing Th1 and Th17 cells in the spleen at 9-dpi because they are encephalitogenic19, 20. But no difference was found (Supplementary Figure 4b). Other Th17 cell subsets expressing IFNγ, GM-CSF, IL-22, or CD5L in the spinal cord at 17-dpi, also showed no difference in their population sizes between the two EAE subsets (Supplementary Figure 4d). These results suggested no marked alteration in leukocyte numbers in Type -B EAE.
Figure 4. Expression of Cxcr2, Cxcr1, and Ltbr in IFNβ-resistant EAE and MS.
(a) Cell numbers for indicated cell populations in DLNs and spleen at 9-dpi (n=4) from mice with either Type-A or Type-B EAE. DLNs: P(Total)=0.0314, t(6)=2.795, P(CD4)=0.8376, t(6)=0.2141, P(CD8)=0.2798, t(6)=1.188, P(B)=0.0078, t(6)=3.921, P(DC)=0.9606, t(6)=0.05144, P(Mac)=0.0469, t(6)=2.495, P(PMN)= 0.1138, t(6)=1.851, P(Th17)=0.5671, t(6)=0.6053, P(Th1)=0.4813, t(6)=0.7506, P(Treg)=0.9888, t(6)=0.01458. Spleen: P(Total)=0.0307, t(6)=2.813, P(CD4)=0.7759, t(6)=0.2978, P(CD8)=0.0729, t(6)=0.4435, P(B)=0.0046, t(6)=4.403, P(DC)=0.2555, t(6)=1.326, P(Mac)=0.0540, t(6)=2.703, P(PMN)=0.0104, t(6)=3.673, two-tailed unpaired Student’s t-test. (b) Proportions of CXCR2-positive cells and MFI of CXCR2 staining in splenic CD4+ T cells obtained from naïve mice and mice with Type-A or Type-B EAE induction at 9-dpi determined by flow cytometry. (n=4). %: P(Naive vs Type-A)=0.7071, t(6)=0.3942, P(Naive vs Type-B)=0.0053, t(6)=4.255, P(Type-A vs Type-B)=0.0067, t(6)=4.051. MFI: P(Naive vs Type-A)=0.0067, t(4)=5.151, P(Naive vs Type-B)=0.0006, t(4)=9.794, P(Type-A vs Type-B)=0.0024, t(4)=6.843, two-tailed unpaired Student’s t-test. (c) Relative expression of CXCR2, CXCR1 and LTBR normalized by VCAM1 in total PBMCs from IFNβ-responder and non-responder MS patients. One circle denotes one patient. (n=18). P(CXCR2)=0.0435, P(CXCR1)=0.0224, P(LTBR) =0.0129, Mann-Whitney U test. *; p<0.05. All the experimental data sets, except for (c), are representatives from at least 2 similar experiments for each.
We next focused on cell surface protein expression on leukocytes in secondary lymphoid organs. CXCR2 is known as a major chemokine receptor mainly on neutrophils. As expected, a majority of neutrophils expressed CXCR2 in both EAE subtypes, but mean fluorescent intensity (MFI) levels were significantly increased in Type-B EAE (Supplementary Figure 4e). In contrast, CD4+ T cells are not largely known to express CXCR2. Nevertheless, more than 10 % of CD4+ T cells from Type-B EAE mice expressed CXCR2 on their cell surface (Figure 4b, Supplementary Figure 4f). In addition, CXCR2+ macrophages were also more frequent and expressed higher levels of CXCR2 in Type-B EAE (Supplementary Figure 4e). Thus, the data suggested that Type-B EAE induces CXCR2 both in lymphoid and myeloid cells.
Gene expression in PBMCs from IFNβ non-responder patients
We next compared gene expression of CXCR2, CXCR1, and LTBR between IFNβ-responder and non-responder patients by using peripheral blood mononuclear cells (PBMCs), which are mixtures of lymphocytes, monocytes, and others. When relative expression levels to VCAM1 were calculated (VCAM1 is expressed in monocytes (Expression Atlas by EMBL-EBI) and is not differentially expressed between IFNβ-responder and nonresponder RRMS patients21), the data showed that relative mRNA expression of CXCR2, CXCR1, and LTBR were significantly higher in IFNβ-responders than nonresponders (Figure 4c). Here, the two-thirds of non-responders showed comparable gene expression levels to responders. It is possible that using total PBMCs evened out the changes in CD4+ T cells; thus, further analyses using purified CD4+ T cells are required to directly evaluate the gene expression. Coefficient of determination (R2) values between relative expression levels of CXCR1 and LTBR were particularly higher in non-responder than responder (Supplementary Figure 4g). Taken together, the data suggested that relative expression of CXCR2, CXCR1 and LTBR might be used to distinguish IFNβ-resistant MS.
Critical role of CXCR2 in Type-B EAE
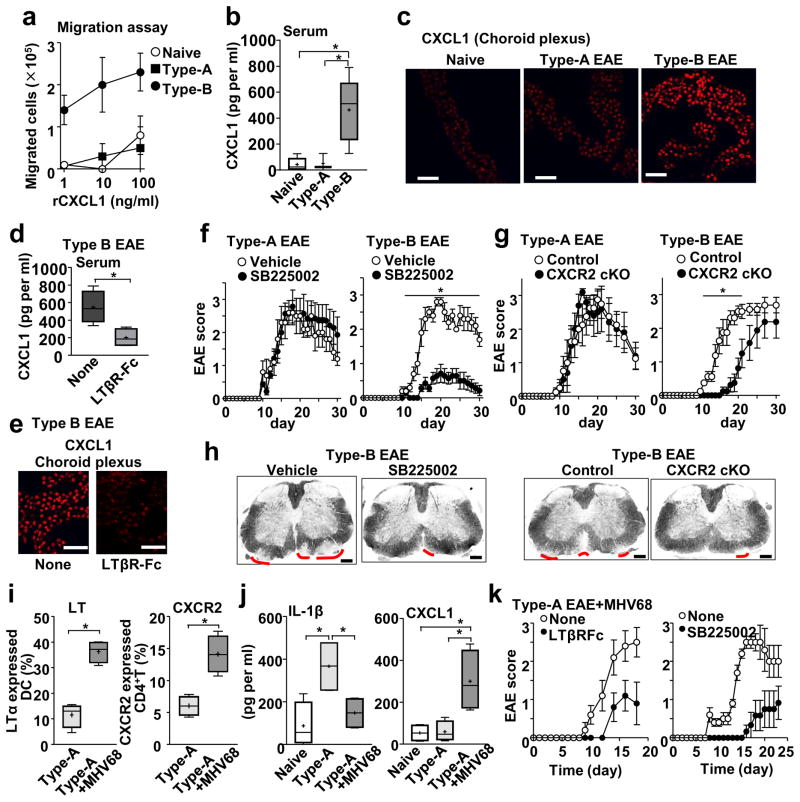
As CD4+ T cells from Type-B EAE mice express high levels of CXCR2 (Figure 4b; Supplementary Figure 4e), the cells showed strong chemotaxis toward CXCL1, a ligand of CXCR2 (Figure 5a). Type-B EAE actually showed high levels of CXCL1 in serum, choroid plexus, and the spinal cord (Figure 5b, c; Supplementary Figure 5). The CXCL1 levels in serum and choroid plexus in Type-B EAE mice were reduced by blocking LTβR in vivo (Figure 5d,e), suggesting mLT enhancing the CXCL1-CXCR2 axis in the development of Type-B EAE.
Figure 5. Involvement of the CXCL1 and CXCR2 in development of Type-B EAE and Type-A EAE with MHV68 infection.
(a) Transwell migration assay towards rCXCL1 using splenic CD4+ T cells harvested from indicated mice. n=4. (b) Levels of CXCL1 in serum from naïve mice or mice with Type-A or Type-B EAE mice on 9-dpi determined by ELISA. Naïve (n=5), Type-A (n=3), Type-B (n=5). P(Naive vs Type-A)=0.8676, t(6)=0.174, P(Naive vs Type-B)=0.0059, t(8)=3.722, P(Type-A vs Type-B)=0.0334, t(6)=2.749. (c) CXCL1 staining in the choroid plexus on 9-dpi. Scale bars, 20 μm. (d, e) Levels of CXCL1 in serum from Type-B EAE with or without LTβR-Fc treatment on 9-dpi. n=4. P=0.0174, t(6)=3.252. Scale bars, 20 μm. (f) Score of Type-A or Type-B EAE treated with or without SB225002. Vehicle-treated group (n=5), SB225002-treated group (n=7). P=0.002, t(10)=5.634. (g) EAE score in Lck-Cre Cxcr2fl/fl mice (CXCR2-cKO) and Cxcr2fl/fl mice (control). *; p<0.05. (n=8). P=0.0192, t(14)=2.646. (h) Representative LFB-stained images with red lines indicating regions of demyelination at 17-dpi. Scale bars, 200 μm. (i) Percentages LTα+ DCs in DLNs and CXCR2+CD4+T cells in spleens of mice at 9-dpi immunized with a Type-A EAE induction method with or without MHV68 infection. (n=4). P=0.0002, t(6)=7.72, P=0.0026, t(6)=4.931. (j) Serum levels of IL-1β and IFNβ at 9-dpi in naïve mice and mice immunized with a Type-A EAE induction method with or without MHV68 infection. n=4. IL-1β: P(Naive vs Type-A)=0.0152, t(6)=3.361, P(Naive vs Type-A+MHV68)=0.3925, t(6)=0.9212, P(Type-A vs Type-A+MHV68)=0.0259, t(6)=2.942. CXCL1: P(Naive vs Type-A)=0.8498, t(6)=0.1978, P(Naive vs Type-A+MHV68)=0.0174, t(6)=3.252, P(Type-A vs Type-A+MHV68) 0.0205, t(6)=3.124. (k) EAE scores of mice Type-A-induced mice with MHV68 infection. Treatment was with either LTβR-Fc or SB225002. Negative control groups did not receive the treatments (n=4). All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. All the experimental data and images are representatives from at least 2 similar experiments for each.
CXCR2 indeed was a key molecule specifically in Type-B EAE, because blockade of CXCR2 with SB225002 ameliorated Type-B EAE, but not Type-A EAE (Figure 5f). We confirmed the involvement of CXCR2 on T cells in vivo by using Cxcr2fl/flLckCre+ (CXCR2 cKO) mice, which lack CXCR2 in T cells. The lack of CXCR2 in T cells did not alter disease score in Type-A EAE. However, although CXCR2 deficiency in T cells eventually allowed mice to develop severe Type-B EAE, it significantly slowed the Type-B EAE onset (Figure 5g). SB225002-treated mice and CXCR2 cKO mice showed mild demyelination in the spinal cord at 17-dpi (Figure 5h). Taken together, we demonstrated the critical role of T cell CXCR2 in Type-B EAE.
Virus infection enhanced the Type-B EAE phenotype
We have shown that a high inoculum of MHV-68 made mice, despite of Type-A induction, develop NLRP3 inflammasome-independent and IFNβ-resistant EAE (Figure 1g). MHV68 infection also increased CXCR2+CD4+ T cells together with mLT-expressing DCs (Figure 5i). Furthermore, MHV68 infection converted Type-A-induced mice to develop Type-B EAE responses, such as reduced serum IL-1β levels and induced CXCL1 levels (Figure 5j), allowing infected mice, despite of Type-A EAE induction, to respond treatments that block LTβR or CXCR2 (Figure 5k). These results suggest strong stimulation of innate immunity triggers LTβR-CXCR2 axis to induce the Type-B EAE phenotype.
Minimum remission with neuronal damage in Type-B EAE mice
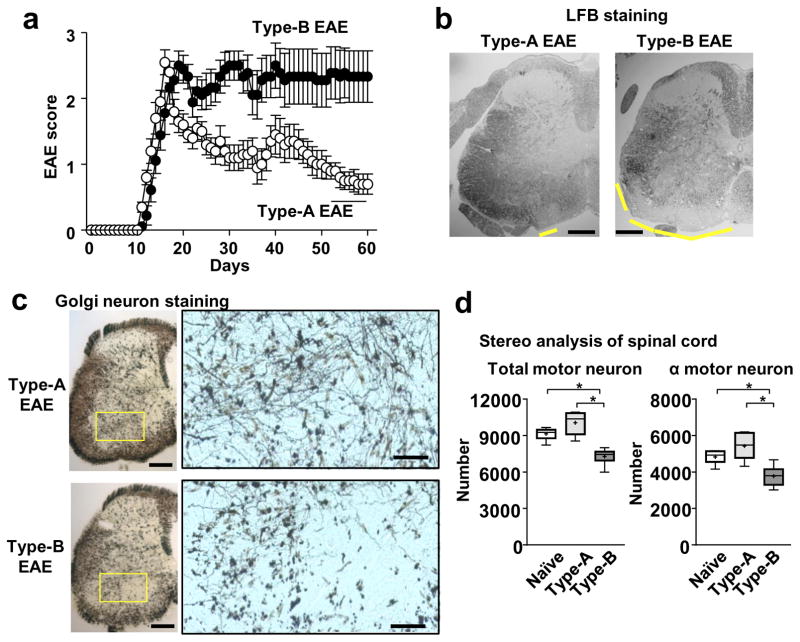
Type-B EAE mice showed persistent EAE severity with little remission (Figure 6a). To investigate the long-term disease severity, we focused the spinal cord rather than the brain, because the spinal cord is responsible for overt motor disability in EAE. Regardless of significantly severe demyelination in spinal cord (Figure 6b), leukocyte infiltration was not clearly observed in the spinal cords in Type-B EAE mice (Supplementary Figure 6a) at a later stage of EAE on 60~70-dpi. Then, rather than sustained inflammation triggered by infiltrated leukocytes, we considered the possibility of poor remyelination to explain the sustained EAE phenotype. However, Type-B EAE mice at least successfully expressed insulin-like growth factor (IGF)-1, a critical molecule in remyelination22, 23 (Supplementary Figure 6b).
Figure 6. Persistent disease severity and spinal transected neurites in Type-B EAE mice.
(a) EAE scores in WT mice after Type-A and Type-B EAE induction (n=9). (b) Representative LFB-stained images of spinal cord sections. Yellow lines indicate regions of demyelination. (c) Representative silver staining images to show neurons in spinal cord sections. Areas within the yellow rectangles were enlarged. Scale bars in (b) and (c, original size picture), 200 μm, and in (c, magnified pictures), 50 μm. (d) Motor neuron stereology. Total Nissl+ and α-motor neuron in lumber region of the spinal ventral horn were evaluated. *; p<0.05. Total motor neuron number: P=0.0001, F20.61. α motor neuron number: P=0.0032, F=11.65, by Bonferroni’s Method (n=6). All the CNS samples were harvested on 70-dpi. All the experimental data and images are representatives from at least 2 similar experiments for each.
We next evaluated morphology of neurons in spinal cord sections from EAE mice, because a previous study demonstrated the loss of neurites in some MS patients24. We performed Bielschowsky neuronal staining, and found that Type-B EAE mice showed fainter staining particularly in outer areas of the white matter, which may include the descending tracts (Supplementary Figure 6c, Arrows). Furthermore, Golgi silver staining, which stains random subsets of neurons, revealed that Type-B EAE mice at 70-dpi showed low-density, transected neurites in the spinal ventral horn containing motor neurons (Figure 6c). Stereological analysis confirmed the significant reduction of α-motor neurons and interneurons surrounding α-motor neurons in the spinal ventral horn of Type-B EAE mice (Figure 6d). In contrast, Type-A EAE mice showed similar neuronal morphology and numbers of α-motor neurons to naïve mice (Figure 6c, d; Supplementary Figure 6d). At an earlier stage of EAE (22-dpi), the difference between Type-A and Type-B EAE Golgi staining was not identified (Supplemental Figure 6e). These result suggested that the minimal remission in Type-B EAE may be attributed to irreversibly loss of axons in Type-B EAE.
Type-B EAE CD4 T cells induce neuronal retraction by Sema6B
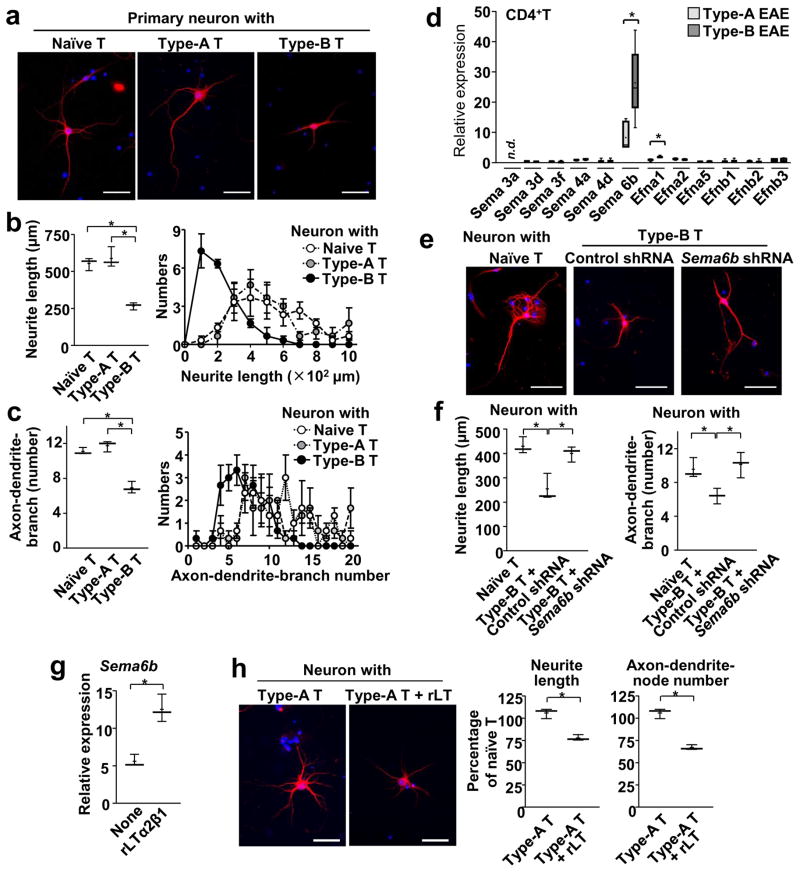
To further investigate the altered neuronal morphology in Type-B EAE mice, we examined primary hippocampal neurons co-cultured with CD4+ T cells from EAE mice. In neuron-T cell co-culture, significant suppression of neurite outgrowth and reduction in the number of axon, dendrites, and branches were observed when CD4+ T cells were from Type-B EAE mice. However, the phenomenon was not seen with T cells from Type-A EAE mice (Figure 7a). Quantitative analyses confirmed decreased lengths of neurites and numbers of axon/dendrite branches with CD4+ T cells from Type-B EAE mice (Figure 7b, c).
Figure 7. Type-B EAE-derived T cells cause neuronal retraction.
(a–c) CD4+ T cells were obtained from spleens of either naïve, Type-A EAE, or Type-B EAE mice; then, co-cultured with neurons. Representative images of hippocampal neuron stained with MAP2 (red) and DAPI (blue) one day after co-culture with indicated CD4+ T cells (a). (b) Average neurite lengths (left panel) and distributions of neurite lengths (right panel) in T cell-neuron co-culture (n=3, measured at least 18 neurons per condition). P(Naive vs Type-A)=0.5044, t(4)=0.7324, P(Naive vs Type-B)=0.0005, t(4)=10.11, P(Type-A vs Type-B)=0.0017, t(4)=7.54. Average numbers of axon and dendrite branches per neuron (left panel) and distributions of branch numbers (right panel)(c). (n=3, measured at least 18 neurons per condition). P(Naive vs Type-A)= 0.1966, t(4)=1.548, P(Naive vs Type-B)=0.0007, t(4)=9.387, P(Type-A vs Type-B)=0.0008, t(4)=9.22. (d) mRNA expression levels of genes encoding proteins in semaphorin and ephrin families in CD4+ T cells from Type-A and Type-B EAE mice at 9-dpi. Data denote as ratio of gene expression in CD4+ T cells from naïve mice. (n=3) P(Sema3d)=0.0599, t(2)=3.899, P(Sema3f)=0.8657, t(2)=0.1916, P(Sema4a)=0.4043, t(2)=1.049, P(Sema4d)=0.0792, t(2)=3.339, P(Sema6b)=0.0175, t(5)=34.915, P(Efna1)=0.0318, t(2)=5.472, P(Efna2)=0.4476, t(2)=0.9373, P(Efna5)=0.2419, t(2)=1.644, P(Efnb1)=0.0658, t(2)=3.704, P(Efnb2)=0.8169, t(2)=0.2634, P(Efnb3)=0.6110, t(2)=0.5059. “n.d.” means “not detectable”. (e, f) CD4+ T cells were obtained from either naïve or Type-B EAE mice, and Sema6b mRNA was knocked down by shRNA in the indicated group. T cells were then co-cultured with neurons. Representative images of neurons stained with MAP2 (red) and DAPI (blue)(e). P(Naive T vs Type-B T+Control shRNA)=0.0096, t(4)=4.657, P(Naive T vs Type-B T+Sema6b shRNA)=0.3377, t(4)=1.088, P(Type-B T+Control shRNA vs Type-B T+Sema6b shRNA) 0.0169, t(4)=3.944. Average neurite lengths (f, left panel) and average numbers of axon and dendrite branches per neuron (f, right panel). (n=3, measured at least 18 neurons per condition). P(Naive T vs Type-B T+Control shRNA)=0.0233, t(4)=3.573, P(Naive T vs Type-B T+Sema6b shRNA)=0.6259, t(4)=0.5273, P(Type-B T+Control shRNA vs Type-B T+Sema6b shRNA)=0.0215, t(4)=3.662. (g) Expression levels of Sema6b mRNA in CD4+ T cells isolated from Type-A EAE mice with or without rLT treatment for 6h. (n=3) P =0.0039, t(4)=5.981. (h) Representative images of MAP2-stained neurons in co-culture with rLT-treated T cell. Average neurite lengths and numbers of axon and dendrites branches per neuron were analyzed and shown. One-hundred % denotes the values obtained from neurons co-cultured with naive T cells. n=3. All scale bars in (a, e, h), 50 μm. P (length)= 0.0211, t(4)=3.686. P (number) =0.017, t(4)=7.544. All statistical analyses in this figure were performed by two-tailed unpaired Student’s t-test. All the experimental data and images are representatives from at least 2 similar experiments for each.
To identify molecules responsible for transected neurites specific for Type-B EAE, we examined expression levels of axon guidance molecules in the semaphorin and ephrin families. Among several molecules, which potently inhibit neurite outgrowth, semaphorin 6B (Sema6B; a class 6 transmembrane semaphoring)25 gene expression was particularly high in CD4+ T cells from Type-B EAE mice (Figure 7d). Knocking-down Sema6b mRNA by shRNA (62 % reduction of mRNA, Supplementary Figure 7) in CD4+ T cells from Type-B EAE mice almost completely rescued neurite outgrowth (Figure 7e, f). Importantly, rLT treatment induced Sema6b mRNA expression in CD4+ T cells isolated from Type-A EAE mice (Figure 7g). rLT-treated CD4+ T cells potently shortened neurite lengths and numbers of neurites in ex vivo co-culture (Figure 7h). In sum, these results suggested that Sema6B upregulation by the LTβR signaling in CD4+ T cells enhances neurite retraction in Type-B EAE mice.
DISCUSSION
In this study, we demonstrated that strong activation of the innate immune system substantially impacts the development of EAE and disease phenotype, i.e., high dosages of adjuvant (heat-killed Mycobacteria; Mtb) were sufficient to induce distinct subtype of EAE (Summarized in Supplementary Figure 8). This simple method enabled us to study the heterogeneity of EAE without using confounding factors such as; special reagents, genomic mutations, mRNA knock-down, and mouse strains other than commonly used C57BL/6. As an environmental cue of “strong activation of the innate immune system,” we demonstrated that acute virus infection could mimic the effect of increased Mtb and converts Type-A EAE to Type-B EAE, leading to the failure of IFNβ treatment. The result suggests that intensity of infections could modulate autoimmune responses and possibly alters treatment options, at least in part, from the mechanism we demonstrate in this study.
Type-B EAE is distinct from Type-A EAE in: (1) increased recruitment of inflammatory cells in the brain, (2) prolonged disease with more neuronal damage, and (3) gene and protein expression profiles. Yet, Type-B EAE pathology did not alter composition of major leukocyte subpopulations. Based on the gene expression patterns between two EAE subtypes, development of Type-B EAE may inhibit development of Type-A EAE. For example, high levels of endogenous IFNβ in Type-B EAE condition are considered to suppress NLRP3 inflammasome activity7, which is required for Type-A EAE. Importantly, Type-B EAE development depends on the mLT-LTβR axis. Indeed, Type-B EAE cannot be treated by IFNβ, but ameliorated by an LTβR antagonist. Furthermore, rLT treatment switches Type-A EAE to Type-B EAE. We do not call Type-B EAE in this study as “atypical EAE”, which was previously reported by a number of laboratories because Type-B EAE mice do not show phenotypes of “atypical EAE” such as severe gait ataxia, hemiparesis, and cellular infiltration in the cerebellum26–29. It is of note that Liu et al. also demonstrated that CXCR2 blockade ameliorated atypical EAE, and the CXCR2 blockade mechanism of atypical EAE was shown to work through neutrophils27. Alternatively, our data suggested blocking CXCR2 on CD4+ T cells is critical to ameliorate Type-B EAE.
We have shown that rIL-17 induces Lta expression (Lta is highly expressed in Type-B EAE). In addition, LTβR blockade ameliorates Th17-mediated EAE (and Type-B EAE). These results suggest that Th17-mediated EAE and Type-B EAE share some common pathology. Indeed, previous reports showed that Th17-mediated EAE was characterized with induced neuronal damage30 and resistance to IFNβ treatment18. We also demonstrated that Type-B EAE is characterized with neuronal damage and resistance to IFNβ treatment.
Our results suggested that mLT expressed on DCs increases CXCR2 expression on CD4+ T cells, neutrophils, and macrophages to induce Type-B EAE. In addition, mLT-mediated induction of CXCL1, as well as its receptor CXCR2, was shown to be critical for Type-B EAE induction. Although CXCR2 is widely known as a neutrophil receptor, we found that CD4+ T cells induce expression of CXCR2 in Type-B EAE; and CXCR2 on T cells are essential for early phase Type-B EAE development. CXCR2 on CD4+ T cells was also reported in other conditions, such as aging31, Alzheimer’s disease32, and tuberculosis33. Here, we found that rLT treatment induces CXCR2 expression on CD4+ T cells in vivo, as well as serum CXCL1. A previous study showed that LTβR signaling is required for upregulation of CXCL1 expression in intestinal epithelial cells for neutrophil recruitment to the bacterial infection site34. The interplay between LTβR and the CXCL1-CXCR2 axis appears to be critical to induce inflammation and is involved in the pathogenesis of Type-B EAE.
We demonstrated that, by using PBMCs from RRMS patients, IFNβ non-responder showed elevated relative expression of CXCR2, CXCR1, and LTBR than IFNβ-responders. Although only PBMCs were available to this study, further analyses using purified CD4+ T cells from patients will be required to delineate pathogenic mechanisms involving CXCR1/2 and LTβR in MS, and to evaluate whether Type-B EAE recapitulates phenotype of IFNβ-resistant MS.
Type-A and Type-B EAE have distinct phenotypes in the CNS. In particular, Type-B EAE shows minimal remission because of neuronal damage, which is irreversible and reminiscent of progressive MS that shows no disease remission with neuronal damage35, 36. Regarding neuronal damage in Type-B EAE, we demonstrated that Sema6B generated by CD4+ T cells causes neurite retraction. Previous reports demonstrated that Sema6B, detected by its receptors (plexin-A2 and plexin-A4), induces neuronal growth cone collapse and repulsion25, 37–39. The finding in the previous reports may share its mechanism to Sema6B-dependent neurite retraction in Type-B EAE. Although there is no report on Sema 6B in MS, other semaphorins were reported in the light of MS. The report demonstrated that Sema 3A and 3F transcripts are detected around and within demyelinating white matter lesions in MS patients40.
Our RNA-Seq analysis identified genes highly expressed in CD4+ T cells from Type-B EAE mice. Some genes shown in Figure 3c are considered to be involved in neuronal damage. In particular, phospholipase A2, encoded by Pla2g4a and Pla3g7 genes, induces neuronal damage as a secreted form41, 42. MGST1, encoded by Mgst1, also damages neurons by activating prostaglandin E synthase43. Alox5ap gene encodes the arachidonate 5-lipoxygenase (ALOX5) activating protein, which facilitates leukotriene biosynthesis: ALOX5 and leukotriene are detected in MS lesions and cerebrospinal fluid from MS patients, respectively44, 45, and contribute to axonal damage46. The RNA-Seq analysis did not pick up Sema6b, because its expression levels were not abundant enough for drawing a conclusion from the RNA-Seq approach. Yet, induction of Sema6b mRNA was detected with increased expression in CD4+ T cells from Type-B EAE mice by standard qPCR.
Here, we report that strong activation of innate immunity makes the EAE pathological mechanism bypass the NLRP3 inflammasome; and the EAE subtype developed without the NLRP3 inflammasome (i.e., Type-B EAE) showed distinct mechanisms in disease development and responses to drug treatment. Although further studies are required to answer whether the Type-A/Type-B EAE spectrum recapitulates the heterogeneity in MS at least to some extent, elucidating mechanisms to generate the heterogeneity in EAE may provide new molecular insights in developing prognosis and treatments of MS as a heterogeneous disease.
Online Methods
Animals
Healthy male mice of the C57BL/6 background of 6–8 weeks old were randomly selected and used in this study. The Asc−/− and Nlrp3−/− mice were gifts from Genentech under material transfer agreement (MTA), and were rederived in our facility. Cxcr2fl/fl mice were a gift from Dr. Richard M. Ransohoff (Cleveland Clinic)47. C57BL/6 and Lck-Cre transgenic mice were purchased from Jackson Laboratories. All the mice were kept group-housed (2–5 mice per cage) in a specific pathogen free facility with 12h each of a light-dark cycle. One cage housed up to five mice. Mice in one experimental group were collected from at least 2 litters. This study was approved by the Duke University Institutional Animal Care and Use Committee.
Reagents
SB225002, a specific CXCR2 inhibitor, was purchased from Tocris. LTβR-Fc (PRO154527) is a gift from Genentech under MTA. Antibodies against LT (ab100844), CXCR2 (129101), and CXCL1 (LS-B2513) were purchased from Abcam, BioLegend, and Life Span Bioscience, respectively. The rIFNβ used in EAE treatment was human rIFNβ-1b (Betaseron, Bayer). rLT (rLTα2β1; 1008-LY-010/CF) was purchased from R & D systems. Mouse rCXCL1 (573702), rIL-17 (576002) and rIFNγ (575306) were purchased from BioLegend. MOG35-55 peptide was synthesized by United Peptides. Enzyme-linked immunosorbent assay (ELISA) kits for the detection of IL-1β and CXCL1 were purchased from BD PharMingen, and Promocell, respectively.
Induction of active EAE
Type-A EAE was induced as previously described48. MOG35-55 peptide was emulsified with CFA (including Mtb), and subcutaneously injected in the flanks of mice on day 0 for one-time immunization, or day 0 and 7 for two-time immunization. Pertussis toxin (PTx; 200 ng/mouse) was administered on day 0 and 2, if the MOG immunization was performed once on day 0. If the immunization was performed twice on day 0 and 7, 200 ng/mouse PTx was administered on day 0, 2, and 7. EAE was induced by 6 different methods. Method 1: Mtb 200 μg, MOG peptide 100 μg, 1 time of immunization, Method 2: Mtb 200 μg, MOG peptide 100 μg, 2 time of immunization, Method 3: Mtb 400 μg, MOG 100 μg, 1 time of immunization, Method 4: Mtb 400 μg, MOG peptide 125 μg, 1 time of immunization, Method 5: Mtb 400 μg, MOG peptide 100 μg, 2 time of immunization, and Method 6: Mtb 400 μg, MOG peptide 125 μg, 2 time of immunization. EAE scores were assessed daily for clinical signs of EAE in a blinded fashion as previously described24. EAE severity was also evaluated as an area under the curve (AUC) from time course experiments. EAE score was evaluated as; 0.5, partial tail limpness, 1, tail limpness, 1.5, reversible impaired righting reflex, 2, impaired righting reflex, 2.5, one hind limb paralysis, 3, both hind limb paralysis, 3.5, both hind and one fore limb paralysis, 4, hind and fore limb paralysis, 5, death. We provided water gel and powder feed when score 2 to avoid body weight reduction because of no reaching to feed and water. Disease score analysis and video-recording was performed in the middle of the day during the light cycle.
Induction of passive EAE
To polarize Th17 cells, total splenocytes were obtained from naïve MOG-specific 2D2 CD4+ TCR Tg mice, and cultured with MOG35-55 peptide (3 μg/ml), IL-6 (20 ng/ml), TGFβ (4 ng/ml), and IL-23 (50 ng/ml) for 6 days. Cells (3×107) were transferred i.v. into sublethally irradiated (450 rad) B6 and Asc−/− mice.
Pharmaceutical treatment of mice wtih EAE
IFNβ (3×104 unit/mouse) were i.p. injected every other day from day 0 to 8 as previously performed24. rLT (rLTα2β1)(1008-LY-010/CF, R&D; 10 μg/kg mouse) was i.p. administered every day from day 0 to 9. CXCR2 inhibitor (SB225002, Tocris: 0.1 mg/kg mouse) was i.p. injected every day from day 0 to day 20. LTβR-Fc (PRO154527, Genentech)(150 ng/mouse) was i.p. injected every other day from day 0.
MHV68 infection
MHV68 was purchased from ATCC Cell Lines (VR-1465). MHV68 strains were amplified in BALB/3T12-3 cells (ATCC, CCL-164™), and virus-infected cells were treated with three freeze–thaw cycles, and then supernatant was isolated. Titers of purified virus were determined using the median tissue culture infective dose (TCID50) method, and convert TCID50 to plaque forming units (PFU). MHV68 (105 pfu/mouse) was inoculated through i.v. at the time of Type-A EAE immunization.
Confocal microscopy
Transverse sections (30 μm) of brains and spinal cords were stained with CXCL1 antibody (Life Span Bioscience) and DAPI (Invitrogen) then analyzed with Zeiss LSM750 laser scanning confocal microscope through the Plan-Apochromat 63×/1.40 oil DIC M27 (n=4). Images were captured with AxioCam (Zeiss) and analyzed by ZEN software (Zeiss).
Western blot analysis
To detect active caspase-1, we analyzed culture supernatant of spleen (5×106 cells) isolated from EAE at 9-dpi by Western blotting with antibody against active caspase-1 (1:500), which was gift from Genentech. Horseradish peroxidase (HRP)-conjugated secondary antibody (1:1,000, BD Pharmingen) was used to detect signals, which were visualized by enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech).
RNA and cDNA preparation for standard qPCR and PCR array analyses
For standard qPCR and PCR array analysis, splenic CD4+ T cells and CD11c+MHCII+ cells (as DCs from DLNs) were isolated by FACS-sorting on 9-dpi. Total RNA was extracted from cells with RNeasy Kit (Qiagen). cDNA synthesis was performed with qScript cDNA SuperMix (Quanta). qPCR analysis was performed with KAPA-SYBR-FAST (KAPA Bio Systems) with an initial denaturing step at 95°C for 3 min, followed by 35 cycles of a denaturation step at 94°C for 3 s and an annealing and extension step at 60°C for 30 s. The relative amounts of qPCR products were determined with the ΔΔCt method to compare the relative expression of target genes and housekeeping genes. The expression of the gene encoding β-actin was used as an internal control49. Primer sequences were indicated in Supplementary Table 3. PCR array was performed by using Chemokines & Receptors PCR Array (PAMM-022Z, Qiagen) and Cytokines & Chemokines PCR Array (PAMM-150Z, Qiagen) with CD4+ T cells DCs, respectively.
RNA-seq analysis
CD4+ T cells were purified by FACS-sorting from spleens of two mice at 9-dpi. Total RNA was extracted from cells with RNeasy Kit (Qiagen). Stranded mRNA libraries were made with the Illumina TruSeq Stranded mRNA kit (catalog number RS-122-2101). The quality of RNA libraries was checked by running them on a 2100 Bioanalyzer instrument (Agilent), pooled and sequenced on 2 lanes of Illumina HiSeq 50bp SR. RNA-seq data was processed using the TrimGalore toolkit (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore), which employs Cutadapt50 to trim low quality bases and Illumina sequencing adapters from the 3′ end of the reads. Only pairs where both reads were 20-nt or longer were kept for further analysis. Reads were mapped to the NCBIM38r73 version of the mouse genome and transcriptome51 using the STAR RNA-seq alignment tool52. Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool (http://www-huber.embl.de/users/anders/HTSeq/). Only genes that had at least 10 reads in any given library were used in subsequent analysis. Normalization and differential expression was carried out using the EdgeR53 and Bioconductor54 package with the R statistical programming environment (www.r-project.org). The exact test method55 was used to identify differentially expressed genes between the different conditions. Enriched pathways were determined by GSEA56.
RRMS patient’s information and qPCR analysis
PBMC samples from RRMS patients were obtained from the MURDOCK study cohorts (IRB# Pro00023791) with the informed consent (http://www.murdock-study.com/). Each participant self-reported their drug history, which includes the drug, start and end dates, and comments. Comments on discontinued drugs typically include the reason for discontinuation, and we selected IFNβ non-responders as those that self-reported stopping an interferon drug due to it not working. Relapses and more lesions via MRI were often cited, and some referred to positive neutralizing antibody results, but sometimes they just reported that it was not working without further description. IFNβ responders were currently taking an interferon drug with no reports of disease progression, and we weighted selection to those that explicitly reported that the drug was working for them and that they had not had relapses or developed more lesions while on it. We excluded patients who used drugs other than IFNβ. Three male patients are included in each group. We used samples from 18 patients (15 female and 3 male) per group. Ages of patients vary from 23 years old to 64 years old in the responder group (the mean of 47.0 years old, SD of 11.429; the age of one patient was unknown), and from 26 to 68 years old in the non-responder group (the mean of 47.8 years old, SD of 10.837). RNA from samples responder and non-responder groups were quantified via Ribogreen, normalized and converted to cDNA using the SuperScript® VILO™ cDNA Synthesis Kit. Gene expression assays utilized the TaqMan® Gene Expression protocol and run on the ViiA™ 7 Real-Time PCR System.
DNA methylation assay
DCs were FACS-sorted as CD11c+CD11b− from DLNs of naïve and EAE mice at 9-dpi. Genomic DNA was obtained with the GenEluteTM Mammalian Genomic DNA Miniprep kit (Sigma). Bisulfite conversion was performed with the MethylDetector kit (Active Motif), and the Lta promoter (between −363-nt and +65-nt from the transcription start site) was cloned into the pCR4-TOPO-TA vector (Life Technologies) and transformed into E. coli. Methylation status was evaluated by sequencing 17–22 clones per group. Percentages of unmethylated CpG in all the tested CpG sites were calculated.
Analyzing demyelination
For Luxol fast blue (LFB) staining, transverse sections (5 μm) of spinal cord and the brain from mice at 17-dpi were stained. MRI analysis was performed with mice with either Type-A or Type-B EAE at 18-dpi. As a procedure for MRI, 10% buffered formalin-perfused mouse brains were kept within the cranium to prevent any potential damage to the brain caused by surgical removal. All images were acquired using a 9.4 T (400 MHz) 89-mm vertical bore Oxford magnet with shielded coil providing gradients of 2200 mT/m. The system is controlled by the Agilent imaging console. The specimens were scanned using a 2D fluid attenuated inversion recovery (FLAIR) sequence with the following parameters: FOV = 22×11 mm2, matrix = 256×128, 0.5-mm slice thickness with zero gap, TR/TI/TE = 5000/925/30.8 ms, echo train length (ETL) = 8, bandwidth = 100 kHz and 12 averages.
Neuronal staining and motor neuron analysis by stereology
Neuronal staining was performed by Golgi’s silver staining with the FD Rapid Golgi staining kit (FD Neurotechonologies, Inc.) and Bielschowsly staining with Hito Bielschowsky OptimStain™ Kit (Hito Biotec) according manufacture’s protocol. For stereological analysis, 4% paraformaldehyde-fixed spinal cords were removed from EAE mice at 70 dpi. Tissue sections (30 μm) were analyzed by stereology using Stereo Investigator v11 (MicroBrightfield, Williston, VT) using a Zeiss AxioImager M2 microscope. Contours were drawn around the ventral horn with a 5× objective and neuronal counting was completed using a 63×/1.40 oil objective (Zeiss). Beginning at L6 and moving caudally, we sampled 10 lumbar spinal sections at a section interval of 5. Using the resample oversample tool in Stereo Investigator, we first determined that at a section interval of 5, a systematic random sampling (SRS) grid size of 164×90 would allow precise cell estimates in the ventral horn of the lumbar region. The mounted thickness was 22 μm after tissue shrinkage and we used 2μm guard zones. The dissector height was 18 μm and the counting frame size was 75×75μm. Stereology parameters were indicated in Supplementary Table 4. The total numbers of bilateral Nissl-stained ventral horn neurons were counted stereologically with the Stereo Investigator software by using an optical fractionator. α-motor neurons were defined by having a maximal diameter greater than 30 μm. The average number of α-motor neurons (>30μm) was 162. The average number of total Nissl-stained neurons counted per animal was 337. Our analysis produced a CE (Gunderson, m=1) of 0.078.
CD4+ T cell-neuron co-culture, and neurite length measurement
Primary neurons were obtained from the hippocampus of P2 mice, as previously described57. After papain treatment to hippocampus tissue, 1×105 cells were cultured for 8 days. Splenic CD4+ T cells were isolated with CD4 microbeads (Myltenyi) and cultured with neuron for 24h. Neuron and nuclear staining were performed with MAP2 and DAPI, respectively. Neurite lengths and numbers of total axons, dendrites and branches were analyzed with the Neurolucida software (MBF bioscience) using at least 18 neurons per condition.
Hot-plate test
To evaluate thermal sensitivity, a “hot plate test”58 (52 °C and 40-sec cut-off) was used for mice at 9-dpi during the light cycle (i.e., day time)
Chemotaxis assay
As previously described8, splenic CD4+ T cells (1×106 cells/well) obtained from naïve mice or EAE mice at 9-dpi were plated in upper chambers of Transwell (5 μm pore, Corning Costar). rCXCL1 was added to lower chambers, and cells were incubated for 3 hr at 37 °C. Numbers of migrated cell were counted, and the data are displayed after subtracting cell numbers in lower chambers in untreated group (as control).
Statistical analysis
Statistical analysis in all results, except for human sample analysis and for motor neuron stereology analysis, was evaluated with two-tailed unpaired Student’s t tests and t-values. The criterion of significance was set as P < 0.05. Statistical analysis for human samples was carried out with the Mann-Whitney U test. Differences among means in motor neuron stereology analysis were analyzed by one-way ANOVA followed by Bonferroni post hoc testing for pairwise comparison unless otherwise stated. Animals were randomly used for experiments under the criteria aforementioned in the section of “Animals.” All behavior experiments were performed in a blinded fashion. No animals or data points were excluded. Quantifications were performed from at least two experimental groups in a blinded fashion. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. Data distribution was assumed to be normal, but this was not formally tested. Blinding and randomization was performed in all experiments.
A supplementary methods checklist is available
Data Availability
The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE85946. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank A. Uezu for his help in primary hippocampal neuron culture and Golgi staining, S. McKinnon for his help in isolating optic nerves, and M. Luftig for MHV68 experiments, M. Skeen for discussions on IFNβ treatments on MS patients. We are also grateful to S. Cote, S. Arvai, and all participants in the MURDOCK Study Community Registry and Biorepository and the MURDOCK Multiple Sclerosis Cohort Study. We thank MURDOCK Study leadership and staff, including K. Newby (PI), for providing samples and support. This study was funded by the National Multiple Sclerosis Society (RG4536 to M.L.S.; RG4723 to C.L.) and NIH (R01MH096979 and P41EB015897 to C.L.).
Footnotes
Author contributions
M.I. and M.L.S. designed the study, analyzed data, and wrote the manuscript. M.I. performed most of the experiments. P.C. performed methylation assay. S.G.G. selected RRMS patient samples from the MURDOCK study cohorts. S.S. performed qPCR analysis for RRMS patient samples. C.L. performed MRI analysis. E.B. assisted neuron imaging analyses. C.L., Q.L., L.S., S.G. and E.B. contributed critical discussion for data and analysis.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Haines JD, Inglese M, Casaccia P. Axonal damage in multiple sclerosis. Mt Sinai J Med. 2011;78:231–243. doi: 10.1002/msj.20246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rio J, et al. Defining the response to interferon-beta in relapsing-remitting multiple sclerosis patients. Ann Neurol. 2006;59:344–352. doi: 10.1002/ana.20740. [DOI] [PubMed] [Google Scholar]
- 3.Rivers TM, Sprunt DH, Berry GP. Observations on Attempts to Produce Acute Disseminated Encephalomyelitis in Monkeys. J Exp Med. 1933;58:39–53. doi: 10.1084/jem.58.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnon R, Sela M. Immunomodulation by the copolymer glatiramer acetate. J Mol Recognit. 2003;16:412–421. doi: 10.1002/jmr.628. [DOI] [PubMed] [Google Scholar]
- 5.Steinman L. The discovery of natalizumab, a potent therapeutic for multiple sclerosis. J Cell Biol. 2012;199:413–416. doi: 10.1083/jcb.201207175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons SB, Pierson ER, Lee SY, Goverman JM. Modeling the heterogeneity of multiple sclerosis in animals. Trends Immunol. 2013;34:410–422. doi: 10.1016/j.it.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue M, et al. Interferon-beta therapy against EAE is effective only when development of the disease depends on the NLRP3 inflammasome. Sci Signal. 2012;5:ra38. doi: 10.1126/scisignal.2002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue M, Williams KL, Gunn MD, Shinohara ML. NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2012;109:10480–10485. doi: 10.1073/pnas.1201836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gris D, et al. NLRP3 plays a critical role in the development of experimental autoimmune encephalomyelitis by mediating Th1 and Th17 responses. J Immunol. 2010;185:974–981. doi: 10.4049/jimmunol.0904145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw PJ, et al. Cutting edge: critical role for PYCARD/ASC in the development of experimental autoimmune encephalomyelitis. J Immunol. 2010;184:4610–4614. doi: 10.4049/jimmunol.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyk DS, Alexander AK, Walker M. Acute disseminated encephalomyelitis progressing to multiple sclerosis: Are infectious triggers involved? Immunol Res. 2014 doi: 10.1007/s12026-014-8499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meeker RB, Williams K, Killebrew DA, Hudson LC. Cell trafficking through the choroid plexus. Cell Adh Migr. 2012;6:390–396. doi: 10.4161/cam.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinman L. Some misconceptions about understanding autoimmunity through experiments with knockouts. J Exp Med. 1997;185:2039–2041. doi: 10.1084/jem.185.12.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lock C, Oksenberg J, Steinman L. The role of TNFalpha and lymphotoxin in demyelinating disease. Ann Rheum Dis. 1999;58(Suppl 1):I121–128. doi: 10.1136/ard.58.2008.i121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodroofe MN, Cuzner ML. Cytokine mRNA expression in inflammatory multiple sclerosis lesions: detection by non-radioactive in situ hybridization. Cytokine. 1993;5:583–588. doi: 10.1016/s1043-4666(05)80008-0. [DOI] [PubMed] [Google Scholar]
- 16.Kraus J, et al. Serum cytokine levels do not correlate with disease activity and severity assessed by brain MRI in multiple sclerosis. Acta Neurol Scand. 2002;105:300–308. doi: 10.1034/j.1600-0404.2002.1o199.x. [DOI] [PubMed] [Google Scholar]
- 17.Columba-Cabezas S, et al. Suppression of established experimental autoimmune encephalomyelitis and formation of meningeal lymphoid follicles by lymphotoxin beta receptor-Ig fusion protein. J Neuroimmunol. 2006;179:76–86. doi: 10.1016/j.jneuroim.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Axtell RC, et al. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat Med. 2010;16:406–412. doi: 10.1038/nm.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, et al. CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell. 2015;163:1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustamante MF, Nurtdinov RN, Rio J, Montalban X, Comabella M. Baseline gene expression signatures in monocytes from multiple sclerosis patients treated with interferon-beta. PLoS One. 2013;8:e60994. doi: 10.1371/journal.pone.0060994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth GA, Spada V, Hamill K, Bornstein MB. Insulin-like growth factor I increases myelination and inhibits demyelination in cultured organotypic nerve tissue. Brain Res Dev Brain Res. 1995;88:102–108. doi: 10.1016/0165-3806(95)00088-u. [DOI] [PubMed] [Google Scholar]
- 23.McMorris FA, Smith TM, DeSalvo S, Furlanetto RW. Insulin-like growth factor I/somatomedin C: a potent inducer of oligodendrocyte development. Proc Natl Acad Sci U S A. 1986;83:822–826. doi: 10.1073/pnas.83.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magliozzi R, et al. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain. 2007;130:1089–1104. doi: 10.1093/brain/awm038. [DOI] [PubMed] [Google Scholar]
- 25.Tawarayama H, Yoshida Y, Suto F, Mitchell KJ, Fujisawa H. Roles of semaphorin-6B and plexin-A2 in lamina-restricted projection of hippocampal mossy fibers. J Neurosci. 2010;30:7049–7060. doi: 10.1523/JNEUROSCI.0073-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothhammer V, et al. Th17 lymphocytes traffic to the central nervous system independently of alpha4 integrin expression during EAE. J Exp Med. 2011;208:2465–2476. doi: 10.1084/jem.20110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. Preferential Recruitment of Neutrophils into the Cerebellum and Brainstem Contributes to the Atypical Experimental Autoimmune Encephalomyelitis Phenotype. J Immunol. 2015;195:841–852. doi: 10.4049/jimmunol.1403063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H, et al. Signal transducer and activator of transcription-3/suppressor of cytokine signaling-3 (STAT3/SOCS3) axis in myeloid cells regulates neuroinflammation. Proc Natl Acad Sci U S A. 2012;109:5004–5009. doi: 10.1073/pnas.1117218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukens JR, et al. The NLRP12 Sensor Negatively Regulates Autoinflammatory Disease by Modulating Interleukin-4 Production in T Cells. Immunity. 2015;42:654–664. doi: 10.1016/j.immuni.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siffrin V, et al. In vivo imaging of partially reversible th17 cell-induced neuronal dysfunction in the course of encephalomyelitis. Immunity. 2010;33:424–436. doi: 10.1016/j.immuni.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Mo R, et al. T cell chemokine receptor expression in aging. J Immunol. 2003;170:895–904. doi: 10.4049/jimmunol.170.2.895. [DOI] [PubMed] [Google Scholar]
- 32.Liu YJ, et al. Peripheral T cells derived from Alzheimer’s disease patients overexpress CXCR2 contributing to its transendothelial migration, which is microglial TNF-alpha-dependent. Neurobiol Aging. 2010;31:175–188. doi: 10.1016/j.neurobiolaging.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 33.Pokkali S, Das SD, RL Expression of CXC and CC type of chemokines and its receptors in tuberculous and non-tuberculous effusions. Cytokine. 2008;41:307–314. doi: 10.1016/j.cyto.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, et al. Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity. 2010;32:403–413. doi: 10.1016/j.immuni.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revesz T, Kidd D, Thompson AJ, Barnard RO, McDonald WI. A comparison of the pathology of primary and secondary progressive multiple sclerosis. Brain. 1994;117( Pt 4):759–765. doi: 10.1093/brain/117.4.759. [DOI] [PubMed] [Google Scholar]
- 36.Thompson AJ, et al. Major differences in the dynamics of primary and secondary progressive multiple sclerosis. Ann Neurol. 1991;29:53–62. doi: 10.1002/ana.410290111. [DOI] [PubMed] [Google Scholar]
- 37.Suto F, et al. Plexin-a4 mediates axon-repulsive activities of both secreted and transmembrane semaphorins and plays roles in nerve fiber guidance. J Neurosci. 2005;25:3628–3637. doi: 10.1523/JNEUROSCI.4480-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang B, Su YS, Sockanathan S. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 2009;61:359–372. doi: 10.1016/j.neuron.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suto F, et al. Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron. 2007;53:535–547. doi: 10.1016/j.neuron.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Williams A, et al. Semaphorin 3A and 3F: key players in myelin repair in multiple sclerosis? Brain. 2007;130:2554–2565. doi: 10.1093/brain/awm202. [DOI] [PubMed] [Google Scholar]
- 41.Liu NK, et al. Cytosolic phospholipase A2 protein as a novel therapeutic target for spinal cord injury. Ann Neurol. 2014;75:644–658. doi: 10.1002/ana.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolko M, Rodriguez de Turco EB, Diemer NH, Bazan NG. Neuronal damage by secretory phospholipase A2: modulation by cytosolic phospholipase A2, platelet-activating factor, and cyclooxygenase-2 in neuronal cells in culture. Neurosci Lett. 2003;338:164–168. doi: 10.1016/s0304-3940(02)01385-x. [DOI] [PubMed] [Google Scholar]
- 43.Takemiya T, et al. Endothelial microsomal prostaglandin E synthase-1 facilitates neurotoxicity by elevating astrocytic Ca2+ levels. Neurochem Int. 2011;58:489–496. doi: 10.1016/j.neuint.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Whitney LW, Ludwin SK, McFarland HF, Biddison WE. Microarray analysis of gene expression in multiple sclerosis and EAE identifies 5-lipoxygenase as a component of inflammatory lesions. J Neuroimmunol. 2001;121:40–48. doi: 10.1016/s0165-5728(01)00438-6. [DOI] [PubMed] [Google Scholar]
- 45.Neu I, Mallinger J, Wildfeuer A, Mehlber L. Leukotrienes in the cerebrospinal fluid of multiple sclerosis patients. Acta Neurol Scand. 1992;86:586–587. doi: 10.1111/j.1600-0404.1992.tb05491.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa K, Palumbo S, Toscano CD, Bosetti F. Inhibition of 5-lipoxygenase activity in mice during cuprizone-induced demyelination attenuates neuroinflammation, motor dysfunction and axonal damage. Prostaglandins Leukot Essent Fatty Acids. 2011;85:43–52. doi: 10.1016/j.plefa.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L, et al. Functional defect of peripheral neutrophils in mice with induced deletion of CXCR2. Genesis. 2013;51:587–595. doi: 10.1002/dvg.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinohara ML, Kim JH, Garcia VA, Cantor H. Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: role of intracellular osteopontin. Immunity. 2008;29:68–78. doi: 10.1016/j.immuni.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinohara ML, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Alvarez J, et al. Balloon dilation of an imperforate cor triatriatum dexter in a Golden Retriever with concurrent double-chambered right ventricle and subsequent evaluation by cardiac magnetic resonance imaging. J Vet Cardiol. 2011;13:211–218. doi: 10.1016/j.jvc.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Kersey PJ, et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 56.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 57.Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- 58.Eddy NB, Leimbach D. Synthetic analgesics. II. Dithienylbutenyl- and dithienylbutylamines. J Pharmacol Exp Ther. 1953;107:385–393. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE85946. The data that support the findings of this study are available from the corresponding author upon reasonable request.