Abstract
Background
Insulin receptor substrate (Irs) proteins are essential for insulin signaling as they allow downstream effectors to dock with, and be activated by, the insulin receptor. A family of four Irs proteins have been identified in mice, however the gene for one of these, IRS3, has been pseudogenized in humans. While it is known that the Irs gene family originated in vertebrates, it is not known when it originated and which members are most closely related to each other. A better understanding of the evolution of Irs genes and proteins should provide insight into the regulation of metabolism by insulin.
Results
Multiple genes for Irs proteins were identified in a wide variety of vertebrate species. Phylogenetic and genomic neighborhood analyses indicate that this gene family originated very early in vertebrae evolution. Most Irs genes were duplicated and retained in fish after the fish-specific genome duplication. Irs genes have been lost of various lineages, including Irs3 in primates and birds and Irs1 in most fish. Irs3 and Irs4 experienced an episode of more rapid protein sequence evolution on the ancestral mammalian lineage. Comparisons of the conservation of the proteins sequences among Irs paralogs show that domains involved in binding to the plasma membrane and insulin receptors are most strongly conserved, while divergence has occurred in sequences involved in interacting with downstream effector proteins.
Conclusions
The Irs gene family originated very early in vertebrate evolution, likely through genome duplications, and in parallel with duplications of other components of the insulin signaling pathway, including insulin and the insulin receptor. While the N-terminal sequences of these proteins are conserved among the paralogs, changes in the C-terminal sequences likely allowed changes in biological function.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-0994-z) contains supplementary material, which is available to authorized users.
Keywords: Insulin receptor substrate, Gene duplication, Protein evolution, Episodic evolution, Phylogeny, Vertebrate, Pseudogene
Background
The intracellular actions of insulin are initiated by the binding of the hormone insulin to its specific cell surface receptor, the insulin receptor [1, 2]. The insulin receptor is a heterotetrameric protein consisting of two extracellular alpha subunits and two transmembrane beta subunits that are connected by disulfide bridges [3, 4]. The binding of insulin to the extracellular alpha subunits of the receptor induces a conformational change that activates the intracellular tyrosine kinase domain found in the beta subunits [5, 6]. Once the tyrosine kinase activity is triggered, the insulin receptor autophosphorylates key tyrosine residues (Tyr-1158, Tyr-1162, and Tyr1163, in the human sequence) in the intracellular portion of the beta subunit [7]. Phosphorylation of these sites then allows interactions with docking proteins, which are also subsequently tyrosine phosphorylated by the insulin receptor tyrosine kinase activity [8], and downstream signaling via SH-2 domain-containing proteins to yield physiological responses [2]. Insulin can initiate several different signaling pathways that regulate metabolic responses, cell survival, growth, and differentiation [1, 2, 9].
Docking proteins are key molecules as they allow the aggregation of components of signaling cascades [7]. The first insulin receptor docking protein identified in mammalian cells was Insulin receptor substrate (Irs1) [10], with three additional docking proteins (Irs2, Irs3, and Irs4) subsequently characterized and found to share similarity in their sequences with Irs1 [11–13]. The four characterized members of the Irs protein family share similar protein architectures, with fairly well conserved N-terminal pleckstrin homology (PH) and phosphotyrosine binding (PTB) domains located near their N-termini and having relatively long C-terminal extensions [14–18]. The C-terminal extensions, which show lower levels of similarity than the N-terminal region, contain multiple tyrosine phosphorylation motifs (as well as serine/threonine phosphorylation motifs) that interact with multiple signaling proteins [14–18]. The PH and PTB domains aid in targeting Irs proteins to the plasma membrane and insulin receptor, respectively [19, 20], while differences in the tyrosine phosphorylation motifs in the C-terminal sequences of the Irs proteins allow interactions with distinct downstream signaling pathways [15, 18]. Only three of the four Irs proteins found in the mouse are functional in humans, as the IRS3 gene sequence has been pseudogenized [21]. Intriguingly, Irs3, at only 494 amino acids in length, is less than half the size of the other three characterized Irs proteins, which are about 1200–1300 amino acids in length [10–13]. Compared to the other Irs proteins, Irs3 has a shorter C-terminal domain but retains similar-sized PH and PTB domains [12, 18]. Additional proteins containing both the PH and PTB domains have been identified (i.e., Dok4 and Dok5) that interact with the insulin receptor, however these proteins lack C-terminal extension with multiple phosphotyrosine motifs [22]. While Irs proteins were initially identified due to their interaction with the insulin receptor, they also interact, as docking proteins, with receptors for other growth factors, such as the insulin growth factor 1 receptor (IGF1R) and the insulin-related receptor (IRR), that also contain intracellular tyrosine domains [23, 24].
Irs proteins exert their unique functions through a combination of tissue-specific expression and differential binding of downstream signaling proteins [14–18, 25]. Irs1 is found in many classical targets of insulin action and is important for insulin sensitivity and embryonic and post-natal body growth [26]. Irs2 is found in an overlapping set of tissues with Irs1, however appears to have a more important role in mediating the neuronal effects of insulin [27] and the growth and survival of pancreatic beta-cells [28]. On the other hand, the function of Irs4 has been difficult to identify as genetic knockouts of this gene have little physiological effect [29]. However, when these knockouts are combined with a brain-specific Irs2 knockout, unique changes in energy regulation and glucose homeostasis are observed [30]. Irs3 is not essential for growth or glucose metabolism [31] and its expression is restricted to white adipocyte tissue in mice [12, 32] (and is absent in humans [21]), suggesting a possible, but non-essential, role for this protein in adipose tissue in rodents. In contrast to other Irs proteins, the PH domain of Irs3 has an additional role in targeting Irs3 to the nucleus, in addition to the plasma membrane, a localization necessary for Irs3 induced glucose uptake [33]. Loss of the Irs3 gene on the human lineage indicates that the function of this gene is not essential in some mammals, and raises questions about the necessity of multiple Irs proteins.
A single Irs-like protein, named Chico, has been found in Drosophila melanogaster that also interacts with the Drosophila insulin receptor [34]. Like the mammalian Irs proteins, Chico is a large protein of about 1000 amino acids in length that contains PH and PTB domains near its N-termini and multiple phosphotyrosine motifs in its C-terminal region [34]. Only a few studies have examined the origin and evolution of the vertebrate Irs gene family, where it has been concluded that these genes diverged on the vertebrate lineage but these studies have reached differing conclusions concerning the relationships among the 4 Irs proteins [17, 35–37]. A number of questions remain to be answered. While it appears that the Irs genes duplicated and diverged from each other on the vertebrate lineage, before the mouse-human divergence, how early in vertebrate evolution this occurred is currently unknown. Did the duplications occur very early in vertebrate evolution in parallel with the duplications of other members of the insulin signaling pathway such as insulin [38] and the insulin receptor [39, 40]? Irs3 was lost on the human (primate) lineage [21]. Was this loss a unique event, or has this gene been lost on other lineages? Have other Irs genes been lost on other vertebrate lineages? Which gene(s) are best conserved (i.e., potentially most essential), both in terms of retention in genomes and in conservation of their sequences within vertebrates? Why is the Irs3 protein sequence much shorter than for other Irs proteins? When did the protein become smaller? Here we show that the Irs genes duplicated very early in vertebrate evolution, likely at a similar time as the origin of the insulin and insulin receptor gene families [38–40] and as a consequence of the two rounds of genome duplications that occurred in the vertebrate ancestor [41, 42]. Our analyses also show that the Irs3 has been lost on multiple independent lineages, and that the genes for other Irs proteins, including Irs1 and Irs2, have occasionally been lost. The length of the Irs3 protein was reduced on the early tetrapod lineage, after divergence for fish, and was followed by a period of rapid sequence evolution in an early mammalian ancestor. Intriguingly, Irs4 also experienced an episode of rapid evolution, in parallel with Irs3, early in mammalian evolution.
Results
Number of insulin receptor substrate (Irs) genes in vertebrate genomes
To estimate the number of insulin receptor substrate (Irs) genes in the genomes of diverse vertebrate species, we conducted BLAST searches [43] of 64 diverse vertebrate genomes in the Ensembl database [44]. As the Ensembl database does not include a species repressing the class Chondrichthyes (cartilaginous fish), we also searched the Elephant shark genome [45], thus the genomes of a total of 65 vertebrate species, representing all vertebrate classes, were examined. Genes were named (see Additional file 1: Table S1) based on orthology-paralogy relationships derived from sequence similarity as well as our phylogenetic and genomic location analyses described below. The numbers of species searched and the numbers of each type of Irs gene found in the different groups of vertebrates is summarized in Table 1. Sequences belonging to all four types of Irs genes were found in diverse representatives of mammals, reptiles, amphibians, lobe-finned fish, and bony fish (Table 1 and Additional file 1: Table S1). Within the bony fish, a single copy of each Irs gene was identified in the spotted gar (Additional file 1: Table S1), a species that diverged before the fish-specific genome duplication [46]. Among the remaining species of bony fish, all of which experienced the fish-specific genome duplication, most had two copies of the Irs2, Irs3, and Irs4-like genes, but only a few had Irs1 genes (Table 1 and Additional file 1: Table S1). Of the fish species that are descendants of the fish-specific genome duplication, only two (zebrafish and cavefish/Mexican tetra) had an Irs1-like gene, and both of these species had only a single copy of this gene, in contrast to the duplicate copies of the other Irs genes found in these (and other) fish species (Additional file 1: Table S1). No Irs3 genes were found in birds or the single representative of cartilaginous fish, although Irs1, Irs2, and Irs4 were identified in both groups (Table 1 and Additional file 1: Table S1). A single genomic sequence encoding an incomplete Irs-like coding region was found in the lamprey (jawless fish), which showed some affinity to Irs2 sequences, but its orthology could not be confidently assessed (Table 1 and Additional file 1: Table S1).
Table 1.
Numbers of Irs-like genes found in diverse vertebrates in the genome and coding sequence databases
| Speciesa | Irs1 b | Irs2 b | Irs3 b | Irs4 b | |
|---|---|---|---|---|---|
| Mammals | 43 | 81 | 42 | 81 (73) | 39 | 73 (28) | 31 | 38 (29) | 42 | 78 (51) |
| Birds | 5 | 54 | 5 | 51 (4) | 5 | 32 (5) | 0 | 0 (0) | 5 | 54 (4) |
| Reptiles | 2 | 5 | 2 | 5 (4) | 2 | 5 (1) | 2 | 5 (4) | 2 | 5 (4) |
| Amphibians | 1 | 2 | 1 | 2 (1) | 1 | 1 (1) | 1 | 1 (1) | 1 | 2 (2) |
| Lobe-finned fish | 1 | 1 | 1 | 1 (1) | 1 | 1 (1) | 1 | 1 (1) | 1 | 1 (0) |
| Bony fish | 11 | 25 | 3 | 5 (5) | 21 | 44 (43) | 21 | 49 (46) | 21 | 47 (29) |
| Cartilaginous fish | 1 | 1 | 1 | 1 (1) | 1 | 1 (1) | 0 | 0 (0) | 1 | 1 (1) |
| Jawless fish | 1 | 0 | 0 | 0 (0) | 1 | 0 (0) | 0 | 0 (0) | 0 | 0 (0) |
| Total | 65 | 167 | 55 | 146 (89) | 71 | 157 (80) | 56 | 94 (81) | 73 | 188 (91) |
aNumber of species with identified genes (or searched if no genes were found): Number of species with genome sequences searched | Number of species searched only for coding sequences
bNumber of unique genes or coding sequences found for each gene: Number of genomic sequence | Number of coding sequences (Number of full-length or near full-length sequences)
Many of the Irs genes identified in our searches of the Ensembl database were incomplete (i.e., did not predict complete open reading frames). Some of the incomplete coding sequences contained unsequenced gaps (Ns) in the genome assemblies, while others could have been due to sequencing errors or pseudogenization. To complement the sequences identified from the Ensembl database, a BLAST search [42] was conducted of the NCBI database [47] to identify Irs coding sequences (Table 1 and Additional file 2: Table S2). Searches of the NCBI database identified a larger number (167) of vertebrate species with Irs coding sequences than the Ensembl database, but many of these are from species do not contain near complete genome sequences (e.g., Xenopus laevis), thus the full complement of Irs genes in these species might not have been found. A second limitation of our NCBI searches was that we only identified Irs-like sequences that had been annotated as coding sequences (i.e., if the gene was not annotated or was a pseudogene it would not be found) (see Additional file 1: Table S1 and Additional file 2: Table S2). The total number of vertebrate species with identified Irs-like genes was 172 (59 in both Ensembl and NCBI, 1 in both the Elephant Shark Genome project and NCBI, 107 only in NCBI, and 5 only in Ensembl, see Table 1 and Additional file 1: Table S1 and Additional file 2: Table S2). The distribution of the Irs-like gene paralogs among vertebrate classes identified in the NCBI database was similar to that seen with the Ensembl database (Table 1 and Additional file 2: Table S2).
Phylogeny of vertebrate insulin receptor substrate (Irs) genes
To better establish the orthology-paralogy relationships among the identified Irs genes, and determine when duplications of the Irs genes occurred, phylogenetic relationships of the sequences were established using maximum likelihood [48, 49] and Bayesian approaches [50, 51]. A total of 341 full-length, or near-full length (those missing only short portions of sequence at the N- or C-termini of their predicted proteins), Irs-like coding sequences from 172 vertebrate species (including 89 Irs1, 80 Irs2, 81 Irs3, and 91 Irs4 sequences (Table 1 and Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Figure S1) were used in this analysis. Maximum likelihood phylogenetic analysis of putative Irs orthologs yielded topologies consistent with the expected species topologies (Additional file 4: Figure S2, Additional file 5: Figure S3, Additional file 6: Figure S4 and Additional file 7: Figure S5; similar results were obtained using Bayesian methods, results not shown), suggesting that the analyzed genes were orthologous.
The relationship among the Irs paralogs was established using these full-length Irs sequences and rooted using Irs-like sequences from three non-vertebrate outgroup species (see Additional file 2: Table S2). Both the Maximum liklihood (Fig. 1) and Bayesian (Additional file 8: Figure S6) analyses demonstrated that each of the four Irs ortholog groups form strongly supported monophyletic clades that diverged from each other prior to the divergence of jawed and jawless vertebrates. Both analyses displayed the same relationships among the paralogs, with Irs2 and Irs4 being most closely related, then both grouping with Irs1, and Irs3 genes being the most distantly related group (Fig. 1 and Additional file 8: Figure S6). Like our finding of distinct Irs paralogs in most classes of vertebrates (see Table 1), these results indicate that the Irs gene family originated early in vertebrate evolution. An intriguing feature of both analyses was that the mammalian Irs3 and Irs4 sequences have longer ancestral mammalian lineages, suggesting episodes of more rapid sequence evolution for these genes in the early mammal.
Fig. 1.
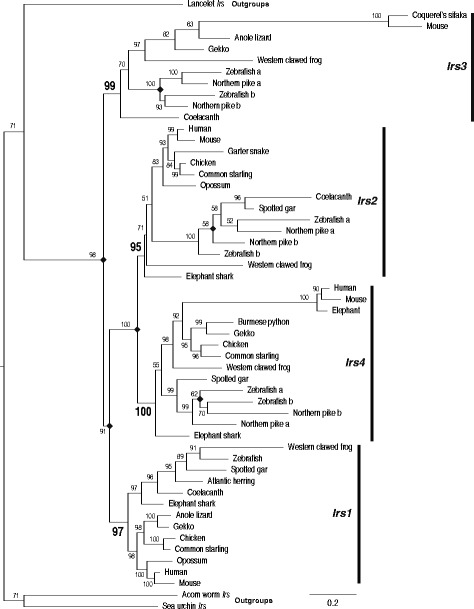
Phylogeny of vertebrate Insulin receptor substrate (Irs) gene family sequences. Phylogeny of Irs sequences from diverse vertebrate species generated by Maximum likelihood using IQ-tree [49]. A similar phylogeny was generated by Bayesian methods [50, 51] (see Additional file 8: Figure S6). Phylogeny was rooted with sequences from acorn worm, purple sea urchin, and Florida lancelet (labeled as outgroups). Selected Irs sequences were chosen to represent the diversity of vertebrates, with different vertebrate Irs genes identified on the right. Similar results were obtained if other Irs sequences were used. Numbers at the nodes bootstrap support. Branch lengths are proportional to the inferred amount of change, with the scale bar at the bottom right. Diamonds indicate gene duplication events
Origin of vertebrate insulin receptor substrate (Irs) genes
Our phylogenetic analysis indicates that the Irs gene family originated early in vertebrate evolution; however, due to the absence of full-length gene sequences from the lamprey, we were unable to show whether any of the duplications preceded the earliest divergence within this group. Genome duplications occurred prior to the divergence of jawed and jawless vertebrates and explain the presence of multiple gene families in vertebrate genomes [41, 42, 52]. With genome duplications, paralogous genome segments are created where different chromosome share sets of paralogous genes [41, 52]. To determine whether the Irs genes originated through genome duplications we examined the genomic neighborhoods surrounding the four mouse Irs genes. As shown in Fig. 2a, the mouse Irs1, Irs2, and Irs4 genes are each found adjacent to a pair of collagen type IV genes (Col4a4 and Col4a23, Col4a1 and Col4a2, and Col4a5 and Col4a6, respectively) on different chromosomes. The same arrangement was found for the human IRS1, IRS2, and IRS4 genes (results not shown). The Irs3 gene, on the other hand, is not located near any collagen gene (Fig. 2a). Whether this difference seen in the genomic neighborhood for Irs3 reflects changes in genomic organization after genomic duplications, or whether Irs3 originated via a different mechanism cannot be determined at this time. However, these results do suggest that the Irs1, Irs2, and Irs4 genes originated via genome duplications in an early vertebrate, and as Irs3 diverged earlier from the other Irs genes, this supports origin of this gene family at or before the genome duplications on the early vertebrate lineage.
Fig. 2.
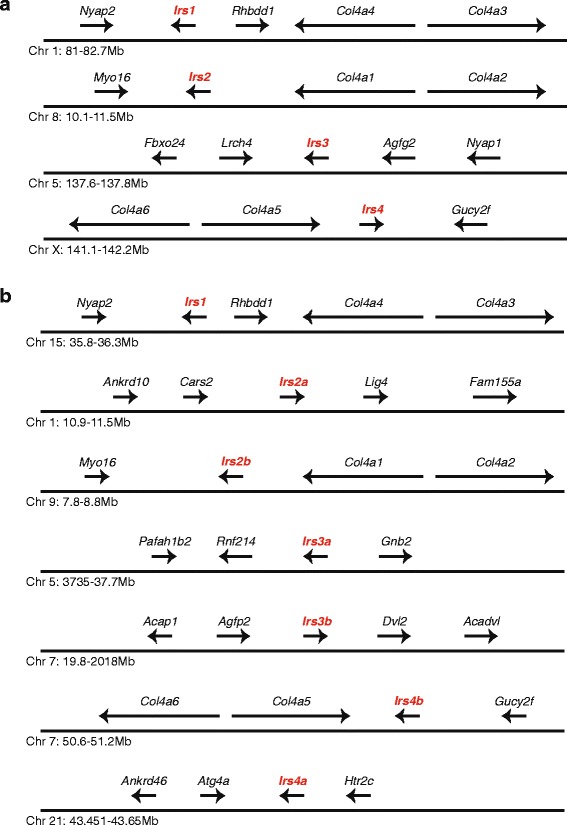
Genomic organization of genes near Irs genes in the mouse and zebrafish genomes. The relative organization and orientation of genes near insulin receptor substrate (Irs) genes in (a) mouse and (b) zebrafish. Chromosomes and genomic locations are from Ensembl [44] (see Additional file 1: Table S1). Irs genes are labeled in red. Gene sizes and distances between genes are not to scale. Arrowheads indicate direction of transcription. Gene symbols are: Irs1–4, insulin receptors substrates 1–4; Col4a1–6, collagen, type IV, alpha1–6; Rhbdd1, rhomboid domain containing 1; Nyap2, Neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2; Myo16, Myosin XVI; Fbxo24, F-box protein 24; Lrch4, Leucine-rich repeats and calponin homology (CH) domain containing 4; Agfp2, ArfGAP with FG repeats 2; Nyap1, Neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1; Gucy2f, Guanylate cyclase 2f; Ankrd10, Ankyrin repeat domain 10; Ankrd46, Ankyrin repeat domain 46; Cars2, Cysteinyl-tRNA synthetase 2; Lig4, Ligase IV, DNA, ATP-dependent; Fam155a, Family with sequence similarity 155, member A; Pafah1b2, Platelet-activating factor acetylhydrolase, isoform 1b, subunit 2; Rnf214, Ring finger protein 214; Gnb2, Guanine nucleotide binding protein (G protein), beta 2; Acap1, ArfGAP with coiled-coil, ankyrin repeat and PH domains 1; Dvl2, Dishevelled segment polarity protein 2; Acadvl, Acyl-Coenzyme A dehydrogenase, very long chain; Atg4a, Autophagy related 4A, cysteine peptidase; Htr2c, 5-hydroxytryptamine (serotonin) receptor 2C
Duplication of Irs genes in Bony fish
Duplicate copies of Irs2, Irs3, and Irs4 were found in most species of bony fish examined (Table 1 and Additional file 1: Table S1 and Additional file 2: Table S2). Bony fish experienced an additional genome duplication not shared by other vertebrates [53, 54], thus duplicated Irs genes would be expected. Duplicated Irs genes were not found in the genome of the spotted gar, a species that diverged from other bony fish prior to the fish-specific genome duplication [46]. Phylogenetic analysis of the Irs2, Irs3, and Irs4 sequences (Additional file 5: Figure S3, Additional file 6: Figure S4 and Additional file 7: Figure S5) demonstrated that the duplications of these genes occurred early in bony fish evolution consistent with the fish-specific genome duplication. When the genomic neighborhoods surrounding the zebrafish Irs genes were examined, only one of the fish duplicates (Irs1, Irs2b, Irs3b, and Irs4b) was located in a genomic neighborhood orthologous to those seen in mice (Fig. 2b), while the second paralogous gene (Irs2a, Irs3a, and Irs4a) resided in genomic regions with no similarity in gene composition to the genomic region found in mice.
Loss of the Irs3 Gene on the primate lineage
While mice have 4 Irs genes, only 3 functional Irs genes are found in humans, as Irs3 contains mutations that introduce a stop codon and delete part of the coding sequence [21]. Genomic sequences similar to Irs3 were identified in a number of primate genomes in the Ensembl database; however, intact coding sequences could only be predicted for the Tree shrew and the Mouse lemur (Additional file 1: Table S1). Similarly, searches of the NCBI database for coding sequences similar to Irs3 only identified potentially functional Irs3 coding sequences in three primate species, the Mouse lemur, Coquerel’s sifaka, and Sunda flying lemur (Additional file 2: Table S2). Complete coding sequences could be predicted for the Mouse lemur and Coquerel’s sifaka but the sequences from the other two primates contained unsequenced gaps. Importantly, all four of these species with potentially intact Irs3 gene sequences are early branching lineages within primates [55]. Alignment of the Irs3 genomic sequences from diverse primates (see Additional file 9: Figure S7) using MultiPipMaker [56, 57] demonstrated that the sequences were not well conserved as a large number of frameshift mutations were identified along with large deletions, including those previously identified in the human IRS3 pseudogene sequence [21]. These results suggest that Irs3 was inactivated early in primate evolution, but after divergence of the Mouse lemur and Coquerel’s sifaka. When MultiPipMaker alignments were generated using the human sequence as the master sequence (results not shown), an Alu repetitive element that disrupts the human IRS3 coding region [21] was found to be shared by Irs3 sequences from primates that lack an intact coding sequence, suggesting that the insertion of this element into the gene occurred at about the same time as the pseudogenization of the gene.
Loss of the Irs3 Gene in birds
In addition to the absence of Irs1 in most bony fish and Irs3 in most primates, another notable group of animals that lack a specific Irs gene is birds, where no Irs3 coding or gene sequences were identified (Table 1 and Additional file 1: Table S1 and Additional file 2: Table S2). In contrast to primates, where genomic sequences similar to Irs3 were found containing mutations that disrupt the coding sequences (see above), genomic sequences similar to Irs3 were not found in any of the bird genomes examined (Additional file 1: Tables S1). To exclude the possibility that the avian Irs3 sequences had rapidly evolved, and thus were not detectable in the BLAST searches [38], we attempted to use genomic neighborhoods to identify these genes. However, searches for the genes that flank the mammalian Irs3 gene (i.e., Lrch4 and Agfg2, see Fig. 2) also failed to find orthologs of these genes (results not shown). These results suggest that the Irs3 genomic region, including adjacent genes, had been deleted from the genomes of birds.
Episodic evolution of vertebrate insulin receptor substrate (Irs) genes
Visual inspection of the phylogenies generated from the Irs coding sequences, using both single gene (Additional file 4: Figure S2, Additional file 5: Figure S3, Additional file 6: Figure S4 and Additional file 7: Figure S5) and gene family (Fig. 1 and Additional file 8: Figure S6) phylogenies, suggested accelerated evolution on the mammalian ancestral lineages for Irs3 and Irs4. Branch lengths displayed in our phylogenetic analysis are proportional to the number of inferred nucleotide substitutions. For both Irs3 and Irs4, mammals have accumulated more changes than sequences from the other vertebrate classes, suggesting that these genes experienced accelerated evolution early in mammalian evolution. To determine whether the longer branches are due to increased numbers of amino acid substitutions in the Irs3 and Irs4 protein sequences we conducted relative rate tests [58] with protein sequences encoded by Irs genes from four different mammalian species (if available) and 6 non-mammalian species (Additional file 10: Table S3). For all relative rate comparisons, the mammalian Irs3 and Irs4 protein sequences accumulated significantly higher numbers of amino acid substitutions compared to protein sequences from a diverse array of non-mammalian species. In contrast, only a small number of the comparisons with Irs1 displayed significantly higher numbers of amino acid substitution on the mammalian lineage, with none being significantly higher on the mammalian lineage for Irs2, although there were a few cases of significantly higher numbers on the non-mammalian lineage for this protein (Additional file 10: Table S3). These results show that the proteins encoded by Irs3 and Irs4, but not Irs1 or Irs2, have accumulated increased numbers of amino acid substitutions on the mammalian lineage.
Changes in the lengths of vertebrate insulin receptor substrate (Irs) proteins
We then examined whether changes in the rate of amino acid sequence evolution resulted in changes in the structure of the Irs proteins. Previously, it had been reported that mouse Irs3 is much shorter than any other mouse Irs proteins, or Irs1, Irs2, and Irs4 proteins from other species [10–13]. To determine whether this was a general feature of Irs3 proteins or was specific to a subgroup of species we calculated the lengths of Irs proteins from species representing diverse groups of vertebrates (Table 2). Lengths of Irs proteins from species not listed in this table were generally similar to their most closely related representative shown in the table (results not shown). Most Irs protein sequences have a length of about 1000–1300 amino acids, except Irs3 from tetrapods (amphibians, reptiles, and mammals, Table 2). Irs3 proteins from zebrafish and coelacanth (and other fish) have length similar to those of the other Irs proteins. These observations suggest that the length of Irs3 progressively shortened from a full-length sequence of 1000–1300 amino acids, which was retained in fish, to one of ~800 residues that is found today in amphibians (Xenopus), to ~600 residues and found in reptiles (garter snake), to ~500 residues found in mammals (mouse) (Table 2). Most of the reduction in Irs3 protein length occurred on the lineages leading to tetrapods (ancestor of amphibians and mammals) and amniotes (ancestor of reptiles and mammals), and not on the lineage leading to mammals. This suggests that the reduction in the length of Irs3 is not associated with the accelerated protein sequence evolution observed for this sequence in the early mammalian lineage. Irs4, which also experienced increased rates of amino acid sequence evolution on the lineage leading to mammals, does not show any major changes in protein length among vertebrate classes, nor do Irs1 or Irs2 (Table 2).
Table 2.
Lengths of Irs proteins from representative vertebrate speceis
| Protein | Human | Mouse | Snakea | Chicken | Xenopus | Coelacanth | Gara | Zebrafish a | Zebrafish b | Sharkb |
|---|---|---|---|---|---|---|---|---|---|---|
| Irs1 | 1242 | 1231 | 1186 | 1178 | 1091 | 1076 | >1085c | 1099 | NAb | 1099 |
| Irs2 | 1338 | 1321 | 1105 | 1148 | 1006 | 1069 | 1069 | 1032 | 1062 | >1082 |
| Irs3 | NAb | 495 | 662 | NAb | 809 | 1034 | NAb | 1181 | 1245 | NAb |
| Irs4 | 1257 | 1216 | 1191 | 1164 | 1077 | NAb | 1120 | 1158 | 1051 | 1200 |
aSnake is garter snake; Gar is spotted gar; Shark is elephant shark
bNA, not applicable, gene was not found, incomplete, or absent
cSpotted gar Irs1 is missing part of its C-terminus; Elephant shark Irs2 is missing part of its N-terminus
Conservation of Irs protein sequences
Since each of the Irs proteins have differing roles in insulin signaling [14–18, 25], we examined whether these roles generated differences in the constraints acting across the Irs protein sequences. To avoid lineage-specific effects, we only examined Irs protein sequences from species where full-length sequences for all four Irs genes had been identified (see Additional file 1: Table S1 and Additional file 2: Tables S2). As such, the four genes would then have existed in parallel in the same genomes for their entire evolutionary history and therefore have likely experienced similar evolutionary pressures at the genomic level. A total of 10 species, 9 mammals and one amphibian (mouse, rat, golden hamster, prairie vole, prairie deer mouse, Coquerel’s sifaka, mouse lemur, Mouflon sheep, killer whale, and Xenopus tropicalis), were found to have complete coding sequences for all 4 Irs proteins (Additional file 1: Table S1 and Additional file 2: Tables S2). If we included sequences from zebrafish, where the single Irs1 sequence was used and one of the two paralogs for Irs2, Irs3, and Irs4 were selected we obtained similar results for the following analyses (results not shown). Conservation of sequences was assessed using Jenson-Shannon Divergence (JS) scores [59] (Additional file 11: Table S4) and was plotted for each Irs protein and the complete set of four Irs proteins in Fig. 3. Irs1 has the highest average conservations score (0.75) followed by Irs2 (0.73), Irs4 (0.67), and Irs3 (0.45). These results suggest that the episodes of more rapid sequence evolution seen on the early mammalian lineages for Irs3 and Irs4 (see above) resulted in an acceleration relative to all Irs proteins, and that the non-mammalian Irs3 and Irs4 sequences might be evolving at rates similar to those for Irs1 and Irs2. As expected, the PH and PTB domains show strong conservation in all 4 Irs proteins, with the protein sequences between and flanking these domains generally showing lower levels of conservation (Fig. 3 and Additional file 11: Table S4). Strong conservation of the PH and PTB domains might be expected as all Irs proteins interact with plasma membranes and insulin receptors, the functions of these domains [19, 20]. Many locations in the C-terminal extensions of the 4 Irs proteins also display high levels of conservation, however it appears that Irs3, and to some extent Irs4, have lower levels than the other two proteins. In comparisons of all 4 Irs proteins, 9 short regions show high levels of conservation, as indicated by having JS score in the top 10% (Fig. 3e). This suggests that only limited parts of the C-terminal region have functions that are conserved across all Irs gene family members, while regions that are not conserved across all family members, but conserved within orthologs might have ortholog-specific functions.
Fig. 3.
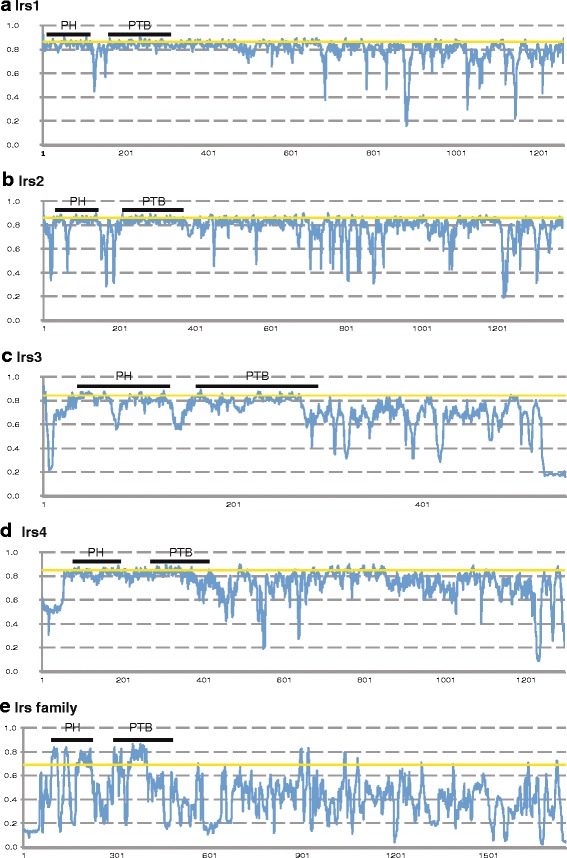
Conservation of Irs protein sequences. JS divergence scores for aligned Irs protein sequences from 10 vertebrate species. (a) Irs1, (b) Irs2, (c) Irs3, (d) Irs4, (e) all Irs family members. JS scores are presented in Additional file 11: Table S4. Position in alignment is shown at the bottom of each graph. The locations of the PH and PTB are shown as bars near the top of each graph. JS scores above the yellow horizontal line are in the top 10% of JS scores for that alignment
Tyrosine phosphorylation of Irs protein sequences
Phosphorylation of tyrosine residues in Irs proteins, especially those in the C-terminal extension, is important for signaling [2, 8, 9]. Potential tyrosine phosphorylation sites were predicted [60] for the Irs protein sequences from the 10 vertebrates that had complete sequences for all 4 family members (Table 3). Approximately 60% of the tyrosine residues in any Irs protein sequence were predicted to be phosphorylation sites. As expected, being the shortest Irs protein, Irs3 has least number of tyrosine residues (average 17.2 residues per sequence) and putative tyrosine phosphorylation sites (average of 10.1), compared to the other Irs proteins (Irs1: 33.8 and 20.4, Irs2: 36.6 and 20.1, Irs4: 31.2 and 15.8, tyrosine and putative tyrosine phosphorylation sites, respectively). Irs3 also showed the lowest conservation of tyrosine residues (10/17.2 = 58%) and putative tyrosine phosphorylation sites (5/10.1 = 50%) in protein alignments. Lower levels of conservation were also seen for Irs4 (tyrosine residues: 17/ 31.2 = 54%, and tyrosine phosphorylation sites: 9/15.8 = 57%). In contrast, conservation of both tyrosine residues (Irs1: 27/33.8 = 80%, Irs2: 25/36.6 = 68%) and putative tyrosine phosphorylation sites (Irs1: 16/20.4 = 78%, and Irs2: 13/20.1 = 65%) were higher for Irs1 and Irs2 (see Table 3 and Additional file 12: Figure S8). Only 4 tyrosine residues were conserved across all Irs sequences in all 10 species (see Additional file 12: Figure S8), with two of these being predicted tyrosine phosphorylation sites for all sequences. Of these four conserved residues, the two sites that are not conserved putative tyrosine phosphorylation sites are located in the PH domain, while the two putative tyrosine phosphorylation sites that are conserved are in the C-terminal extension (residues 608 and 628, 649 and 671, 350 and 361, and 672 and 689 in mouse Irs1, Irs2, Irs3, and Irs4, respectively) (see Additional file 12: Figure S8). The two putative tyrosine phosphorylation sites located in the C-terminal extension are located in regions that have strong conservation among the four Irs protein sequences (Additional file 12: Figure S8).
Table 3.
Tyrosine phosphoryation of Irs proteins
| Irs1 | Irs2 | Irs3 | Irs4 | |||||
|---|---|---|---|---|---|---|---|---|
| Species | Ya | pYb | Y | pY | Y | pY | Y | pY |
| Mouse | 34 | 20 | 36 | 21 | 17 | 10 | 29 | 13 |
| Rat | 35 | 22 | 36 | 21 | 18 | 13 | 30 | 17 |
| Golden hamster | 34 | 20 | 37 | 20 | 14 | 8 | 31 | 15 |
| Prairie vole | 34 | 21 | 37 | 20 | 17 | 11 | 34 | 13 |
| Prarie deer mouse | 34 | 20 | 37 | 20 | 18 | 12 | 27 | 15 |
| Coqurel’s sifka | 34 | 20 | 38 | 21 | 16 | 7 | 33 | 15 |
| Mouse lemur | 34 | 20 | 38 | 20 | 16 | 8 | 31 | 16 |
| Mouflon sheep | 34 | 19 | 38 | 20 | 14 | 8 | 30 | 15 |
| Killer whale | 33 | 20 | 38 | 21 | 17 | 8 | 28 | 15 |
| Xenopus tropicalis | 32 | 22 | 31 | 17 | 25 | 16 | 39 | 24 |
| Average | 33.8 | 20.4 | 36.6 | 20.1 | 17.2 | 10.1 | 31.2 | 15.8 |
| Conservedc | 27 | 16 | 25 | 13 | 10 | 5 | 17 | 9 |
aNumber of tyrosine residues in the sequence
bNumber of tyrosine residues predicted to be phosphorylated
cNumber of residues conserved across the 10 sequences
Discussion
Origin of the Irs gene family
While multiple Irs-like genes have been previously characterized in several mammalian species [10–13, 17, 36], only a few non-mammalian Irs-like genes have been identified, which limited the ability to resolve when this gene family originated and how the different genes are related to each other [17, 35–37]. Here, our searches have identified a large number of Irs-like genes from a diverse array of vertebrate classes, which should allow better estimation of the time when this gene family originated and how the different genes are related to each other. Searches of vertebrate genomes identified multiple Irs-like sequences in the genomes of representative species for all vertebrate classes except Agnatha (Jawless fish) (Table 1 and Additional file 1: Table S1 and Additional file 2: Tables S2). However, given the low coverage of the sea lamprey somatic genome [61] and the loss of DNA in this species due to genomic remodeling in somatic tissue [62], Irs-like sequences may have been missed in this jawless fish. These observations suggest that the Irs gene family originated early in vertebrate evolution, and possibly before the earliest divergence of extant vertebrate species.
Phylogenetic analyses of the sequences (Fig. 1 and Additional file 8: Figure S6) strengthened this conclusion, demonstrating that the multiple genes originated early in vertebrate evolution and were not due to parallel duplications on diverse lineages. Analysis of genomic neighborhoods is a powerful tool for identifying orthologs [63], especially in gene families where multiple sequences have similar levels of similarity to a putative ortholog, where only the true othologs share genomic neighborhoods [64]. In this context, we used genomic neighborhoods to confirm the orthology of many of the diverse Irs genes found in vertebrates. When the genomic locations of Irs-like genes were examined (Fig. 2), three of the 4 Irs genes were found to be in genomic neighborhoods that shared similar gene contents. The sharing of paralogus genes among genomic neighborhoods is consistent with these genes originating through genome duplications [65], which suggests that at least 3 of the 4 Irs genes originated via the two rounds of genome duplication that occurred in the common ancestral vertebrate lineage [41, 42]. Interestingly, both the insulin [38] and the insulin receptor [39, 40] gene families originated very early in vertebrate evolution, and potentially via the same genome duplications. Irs proteins not only interact with the insulin receptor, but also with other receptors, including the Insulin growth factor I (IGF-1) receptor and the Insulin-related receptor (Irr) [23, 24]. These observations suggest that duplications of the genes for the ligands, receptors, and docking proteins could lead to increased specialization in these signaling pathways, and the possibility to evolve new functions.
Change in number of Irs genes
While Irs gene originated very early in vertebrate evolution, the number of Irs genes is found to vary between species. Similar variations in the numbers of genes within gene families involved in insulin signaling in vertebrates have previously been reported [38, 66, 67]. Early studies demonstrated that the Irs3 gene was lost on the human lineage [21], and our analysis indicates that it was possibly inactivated by the insertion of a repetitive DNA element early in primate evolution (results not shown). Irs3 genes were also lost on the lineage leading to birds. A number of genes involved in insulin-regulated metabolism have been lost in the chicken [68], some of which have been shown to be missing in wide variety of birds (e.g., Resistin [64]), suggesting that the loss of Irs3 might have been part of an adaptation by birds to their new locomotive style. Teleost fish experienced a genome duplication [53], however rapid loss of many of the duplicates occurred [54]. Here we found duplicated copies of Irs2, Irs3, and Irs4 in most teleost fish genomes, but most of these species have lost both copies of Irs1 (see Table 1 and Additional file 1: Table S1 and Additional file 2: Tables S2). The presence of multiple Irs genes, and the overlap in the functions of the Irs proteins [14–18] suggests a degree of redundancy among these genes allowing species to adapt to the loss of one (or more) of these genes.
Evolution of Irs proteins
Duplication of genes should allow the specialization of distinct proteins to unique biological roles [69, 70], thus duplication of the Irs genes might have allowed the evolution of novel regulatory roles for the insulin signaling pathway. While all Irs proteins are involved in insulin signaling, they each appear to have unique, but to some extent overlapping, biological roles [14–18]. Changes in the numbers of Irs genes also shows that the genes have retained a degree of redundancy and have not completely sub-functionalized since their origin. Despite the overlap in function, differences in evolutionary patters can be seen among the Irs genes. Irs3 and Irs4 both experienced episodes of more rapid protein sequence evolution on the common ancestral lineage leading to mammals (Additional file 10: Table S3), which suggests either a temporary relaxation of evolutionary constraints on these sequences on this lineage or that the rapid evolution was driven by positive selection. Both patterns of evolution could have resulted in changed biological functions for these proteins, and might explain why Irs3 and Irs4 might have functions that are less essential than Irs1 or Irs2. Irs3 is non-essential as loss of this gene in humans is tolerated [21], and our data shows that a number of primates, birds and potentially other vertebrates can survive without this gene. Knockout of Irs4 has little physiological effect [29], while Irs1 or Irs2 knockout mice have much more pronounced physiological defects [26, 27, 71, 72].
Further evidence for the diversification of the function of the Irs proteins is derived from the conservation plots. When each Irs protein is individually examined, areas of strong sequence conservation are seen across the entire protein sequence, although to a lower extent for Irs3, which might be due to the rapid evolution on the early mammalian lineage (Fig. 3a-d). However, when conservation is examined across the family of Irs proteins (Fig. 3e), most of the conservation is concentrated in the regions encoding the PH and PTB domains, sequences that are important for localizing these proteins to the plasma membrane [19] and insulin receptors [20], respectively. The plasma membrane localization, and insulin receptor interactions of these proteins have been conserved, but the C-terminal extension, which allow interaction with downstream signaling partners [15, 18], show greater levels of divergence to account for changes in downstream functions. However, there are a few areas of the C-terminal extension that are strongly conserved among all Irs, including two putative tyrosine phosphorylation sites that have been shown to be important in Irs1 and Irs2 for interactions with phosphatidylinositol 3-kinase (PI3K) [73–76], a key downstream signaling protein of insulin receptors [76]. Thus, interaction with PI3K appears to be conserved among all Irs proteins, but changes in interactions with other signaling proteins might explain the differences in biological function of the different Irs proteins.
Conclusions
Here we have shown that the Irs gene family originated early in vertebrate evolution, with at least three of the genes likely generated during the two rounds of genome duplication that occurred in the vertebrate ancestor. Most groups of vertebrates have retained all 4 Irs genes, although some groups have lost genes, including primates and birds that have lost Irs3 and most fish that have lost Irs1. Duplication of Irs genes is only seen in fish that have experienced the fish-specific genome duplication, leading to duplicated Irs2, Irs3, and Irs4 genes. This suggests that while there are redundancies in the function of Irs gene, thus can tolerate the loss of a gene, gain of Irs genes is likely harmful, except when other genes in the insulin signaling pathway are duplicated. This conclusion is agreement with the finding of an increased number of retained duplicated genes involved in signal transduction pathways found in fish after the fish-specific genome duplications [77]. The protein sequences of Irs1 and Irs2 are strongly conserved across vertebrates while Irs3 and Irs4 show lower levels of conservation. In addition to lower sequence conservation, the length of Irs3 progressively shorted along the lineage leading to mammals. Comparisons among the paralogous Irs sequences shows that most of the sequence is well conserved within a paralog, but only the PH and TTB domains, those responsible for binding to plasma membranes and the insulin receptor, are conserved between paralogs. Only a few regions within the C-terminal extensions of these proteins are conserved among Irs paralogs, suggesting that divergence in these sequences has allowed divergence in function.
Methods
Database searches
Molecular sequence databases maintained by Ensembl [44] and the National Center for Biotechnology Information (NCBI) [47] were searched in January 2016 for insulin receptor substrate (Irs1, Irs2, Irs3, and Irs4)-like coding sequences. We initially searched the databases using the tBLASTn algorithm [43] using previously characterized mouse Irs1, Irs2, Irs3, and Irs4 protein sequences as queries. Putative Irs-like protein sequences identified were then used in subsequent tBLASTn searches. We also investigated the elephant shark (the sole representative of cartilaginous fish with a near-complete genome sequence) genome generated by the Elephant Shark Genome Project [45, 78]. All sequences that had E-scores below 0.01 were examined. Sequences identified by BLAST were used in reciprocal BLASTx searches of the mouse proteomes to ensure that their best matches were Irs-like sequences.
To examine genomic neighborhoods near Irs-like genes genomic comparisons were conducted using PipMaker and MultiPipMaker [56, 57]. Genes neighboring the Irs-like genes were identified from the genome assemblies in Ensembl [44] and the Elephant Shark Genome Project [78]. The organization of genes adjacent to the Irs-like genes was used to determine whether the genes of interest reside in conserved genomic neighborhoods.
Phylogenetic analysis
Phylogenies of vertebrate Irs-like gene coding sequences were generated using full-length, or near full-length (i.e., missing a short part of their N- or C-termini), Irs1, Irs2, Irs3, and Irs4 coding sequences from diverse vertebrate and outgroups (see Additional file 1: Table S1 and Additional file 2: Tables S2) and outgroups. Irs-like coding sequences were aligned using MAFFT [79] as implemented at the Guidance web site [80, 81], using default parameters. Similar results were obtained if Clustal Omega [82] was used as the alignment program. DNA sequence alignments were based on codons to retain protein alignments. The reliability of the alignments was examined using Guidance [80, 81] and trimmed alignments using sites that had values above the default cut-off of 0.93 were generated.
Phylogenetic trees of the sequences were generated using Bayesian methods with MrBayes 3.2 [50, 51, 83], maximum likelihood with IQ-tree [49, 84], and neighbor-joining distance approaches with MEGA6.06 [85]. Bayesian trees were generated from coding sequences with MrBayes 3.2 using parameters selected by ModelFinder [86], whose results are presented in Additional file 13: Figure S9. MrBayes was run for 2,000,000 generations with four simultaneous Metropolis-coupled Monte Carlo Markov chains sampled every 100 generations. The average standard deviation of split frequencies dropped to less than 0.02 for all analyses. The first 25% of the trees were discarded as burn-in with the remaining samples used to generate the consensus trees. Trace files generated by MrBayes were examined by Tracer [87] to verify if they had converged. Maximum likelihood trees, constructed with 1000 replications by the ultrafast approximation [88], were generated with IQ-tree [49] on the IQ-tree webserver [84] using parameters for the substitution model suggested by ModelFinder [86]. The maximum likelihood search was initiated from a tree generated by BIONJ and the best tree was identified after heuristic searches using the nearest neighbor interchange (NNI) algorithm. MEGA6.06 [85] was used to construct bootstrapped (1000 replications) neighbor-joining distance trees, using either Maximum Composite Likelihood distances for the DNA sequences or JTT distances for the proteins sequences. Similar results were obtained, but with lower confidence (bootstrap or posterior probabilities) intervals if alternative outgroups were used (results not shown).
With respect to orthology-paralogy issues, choice of outgroup, alignment method (MAFFT [79] or Clustal [82]), or the use of full-length or trimmed (based on Guidance scores [81]) alignments had little influence on the key findings of these analyses. Methods that relied on shorter sequences (i.e., trimmed alignments or protein sequences) or simpler models of sequence evolution (i.e., neighbor-joining or parsimony) tended to yield weaker support for the earlier diverging lineages, but none of our analyses were in significant conflict with the key inferences of the phylogeny presented in Fig. 2 or Additional file 11: Figure S8.
Analysis of protein sequence conservation
Conservation of proteins sequences was assessed using Jenson-Shannon (JS) divergence scores [62] on the JS Divergence web server [89], using a window size of 3 and the BLOSUM62 matrix as background. Putative tyrosine phosphorylation sites in the protein sequences were predicted using NetPhos [63, 90].
Additional files
This file is in Excel format. Genomic locations of Irs-like genes in sequenced vertebrate genomes identified from the Ensembl database. (XLSX 40 kb)
This file is in Excel format. Accession numbers of Irs-like coding sequences from vertebrates and outroup species identified from the NCBI database. (XLSX 218 kb)
This file is in Word format. Coding sequences for full-length and near full-length Irs genes from diverse vertebrates and outgroups. (PDF 200 kb)
This file is in PDF format. Phylogeny of vertebrate Irs1 sequences. (PDF 172 kb)
This file is in PDF format. Phylogeny of vertebrate Irs2 sequences. (XLSX 58 kb)
This file is in PDF format. Phylogeny of vertebrate Irs3 sequences. (XLSX 53 kb)
This file is in PDF format. Phylogeny of vertebrate Irs4 sequences. (DOCX 627 kb)
This file is in PDF format. Phylogeny of the vertebrate Irs gene family rooted with non-vertebrate Irs-like genes. (PDF 525 kb)
This file is in PDF format. Alignment of primate Irs3 genomic sequences. (PDF 518 kb)
This file is in Excel format. Relative rates of evolution of Irs genes in mammals and non-mammals. (PDF 518 kb)
This file is in Excel format. JS Divergence Scores from alignments of Irs proteins from 10 vertebrates. (PDF 499 kb)
This file is in PDF format. Alignment of Irs protein sequences with conserved residues highlighted. (PDF 450 kb)
This file is in PDF format. ModelFinder results for the coding sequences used in the phylogenetic analyses. (PDF 224 kb)
Acknowledgements
This work has been supported by a grant from the Canadian Institutes of Health Research CCI-109605 (to DMI). The funding body did not have any role in the design, analysis, or interpretation of data or in the writing of the manuscript and the decision to submit the manuscript for publication.
Availability of data and materials
The data set supporting the results of this article is included within the article’s additional files (see Additional file 3: Figure S1).
Authors’ contributions
AA and DMI designed the research and outlined the manuscript, obtained and analyzed the data, and drafted the manuscript. The authors have read, edited, and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-0994-z) contains supplementary material, which is available to authorized users.
References
- 1.Pirola L, Johnston AM, Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47:170–184. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 2.Myers MG, Jr, White MF. Insulin signal transduction and the IRS proteins. Annu Rev Pharmacol Toxicol. 1996;36:615–658. doi: 10.1146/annurev.pa.36.040196.003151. [DOI] [PubMed] [Google Scholar]
- 3.De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 4.Hubbard SR. The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb Perspect Biol. 2013;5:a008946. doi: 10.1101/cshperspect.a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward CW, Lawrence MC. Ligand-induced activation of the insulin receptor: a multi-step process involving structural changes in both the ligand and the receptor. BioEssays. 2009;31:422–434. doi: 10.1002/bies.200800210. [DOI] [PubMed] [Google Scholar]
- 6.Du Y, Wei T. Inputs and outputs of insulin receptor. Protein Cell. 2014;5:203–213. doi: 10.1007/s13238-014-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei L, Hubbard SR, Hendrickson WA, Ellis L. Expression, characterization, and crystallization of the catalytic core of the human insulin receptor protein-tyrosine kinase domain. J Biol Chem. 1995;270:8122–8130. doi: 10.1074/jbc.270.14.8122. [DOI] [PubMed] [Google Scholar]
- 8.Brummer T, Schmitz-Peiffer C, Daly RJ. Docking proteins. FEBS J. 2010;277:4356–4369. doi: 10.1111/j.1742-4658.2010.07865.x. [DOI] [PubMed] [Google Scholar]
- 9.Jensen M, De Meyts P. Molecular mechanisms of differential intracellular signaling from the insulin receptor. Vitam Horm. 2009;80:51–75. doi: 10.1016/S0083-6729(08)00603-1. [DOI] [PubMed] [Google Scholar]
- 10.Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, et al. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 11.Sun XJ, Wang LM, Zhang Y, Yenush L, Myers MG, Jr, Glasheen E, et al. Role of IRS-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 12.Lavan BE, Lane WS, Lienhard GE. The 60-kDa phosphotyrosine protein in insulin-treated adipocytes is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:11439–11443. doi: 10.1074/jbc.272.17.11439. [DOI] [PubMed] [Google Scholar]
- 13.Lavan BE, Fantin VR, Chang ET, Lane WS, Keller SR, Lienhard GE. A novel 160-kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:21403–21407. doi: 10.1074/jbc.272.34.21403. [DOI] [PubMed] [Google Scholar]
- 14.White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182:3–11. doi: 10.1023/A:1006806722619. [DOI] [PubMed] [Google Scholar]
- 15.Giovannone B, Scaldaferri ML, Federici M, Porzio O, Lauro D, Fusco A, et al. Insulin receptor substrate (IRS) transduction system: distinct and overlapping signaling potential. Diabetes Metab Res Rev. 2000;16:434–441. doi: 10.1002/1520-7560(2000)9999:9999<::AID-DMRR159>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Withers DJ. Insulin receptor substrate proteins and neuroendocrine function. Biochem Soc Trans. 2001;29:525–529. doi: 10.1042/bst0290525. [DOI] [PubMed] [Google Scholar]
- 17.White MF. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab. 2002;283:E413–E422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 18.Thirone AC, Huang C, Klip A. Tissue-specific roles of IRS proteins in insulin signaling and glucose transport. Trends Endocrinol Metab. 2006;17:72–78. doi: 10.1016/j.tem.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs AR, LeRoith D, Taylor J. Insulin receptor substrate-1 pleckstrin homology and phosphotyrosine-binding domains are both involved in plasma membrane targeting. J Biol Chem. 2001;276:40795–40802. doi: 10.1074/jbc.M105194200. [DOI] [PubMed] [Google Scholar]
- 20.Wolf G, Trüb T, Ottinger E, Groninga L, Lynch A, White MF, et al. PTB domains of IRS-1 and Shc have distinct but overlapping binding specificities. J Biol Chem. 1995;270:27407–27410. doi: 10.1074/jbc.270.46.27407. [DOI] [PubMed] [Google Scholar]
- 21.Björnholm M, He AR, Attersand A, Lake S, Liu SC, Lienhard GE, et al. Absence of functional insulin receptor substrate-3 (IRS-3) gene in humans. Diabetologia. 2002;45:1697–1702. doi: 10.1007/s00125-002-0945-z. [DOI] [PubMed] [Google Scholar]
- 22.Cai D, Dhe-Paganon S, Melendez PA, Lee J, Shoelson SE. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J Biol Chem. 2003;278:25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- 23.De Meyts P. Insulin and its receptor: structure, function and evolution. BioEssays. 2004;26:1351–1362. doi: 10.1002/bies.20151. [DOI] [PubMed] [Google Scholar]
- 24.Marino-Buslje C, Martin-Martinez M, Mizuguchi K, Siddle K, Blundell TL. The insulin receptor: from protein sequence to structure. Biochem Soc Trans. 1999;27:715–726. doi: 10.1042/bst0270715. [DOI] [PubMed] [Google Scholar]
- 25.Lavin DP, White MF, Brazil DP. IRS proteins and diabetic complications. Diabetologia. 2016;59:2280–2291. doi: 10.1007/s00125-016-4072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araki E, Lipes MA, Patti ME, Brüning JC, Haag B, 3rd, Johnson RS, et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 27.Schubert M, Brazil DP, Burks DJ, Kushner JA, Ye J, Flint CL, et al. Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J Neurosci. 2003;23:7084–7092. doi: 10.1523/JNEUROSCI.23-18-07084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Withers DJ, Burks DJ, Towery HH, Altamuro SL, Flint CL, White MF. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nature Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 29.Fantin VR, Wang Q, Lienhard GE, Keller SR. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and glucose homeostasis. Am J Physiol Endocrinol Metab. 2000;278:E127–E133. doi: 10.1152/ajpendo.2000.278.1.E127. [DOI] [PubMed] [Google Scholar]
- 30.Sadagurski M, Dong XC, Myers MG, Jr, White MF. Irs2 and Irs4 synergize in non-LepRb neurons to control energy balance and glucose homeostasis. Mol Metab. 2013;3:55–63. doi: 10.1016/j.molmet.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu SC1, Wang Q, Lienhard GE, Keller SR: Insulin receptor substrate 3 is not essential for growth or glucose homeostasis. J Biol Chem 1999, 274:18093–18099. [DOI] [PubMed]
- 32.Sciacchitano S, Taylor SI. Cloning, tissue expression, and chromosomal localization of the mouse IRS-3 gene. Endocrinology. 1997;138:4931–4940. doi: 10.1210/endo.138.11.5518. [DOI] [PubMed] [Google Scholar]
- 33.Maffucci T, Razzini G, Ingrosso A, Chen H, Iacobelli S, Sciacchitano S, et al. Role of pleckstrin homology domain in regulating membrane targeting and metabolic function of insulin receptor substrate 3. Mol Endocrinol. 2003;17:1568–1579. doi: 10.1210/me.2001-0211. [DOI] [PubMed] [Google Scholar]
- 34.Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, et al. Autonomous control of cell and organ size by CHICO, a drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/S0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 35.Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty C, Agoramoorthy G, Hsu MJ. Exploring the evolutionary relationship of insulin receptor substrate family using computational biology. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGaugh SE, Bronikowski AM, Kuo CH, Reding DM, Addis EA, Flagel LE, et al. Rapid molecular evolution across amniotes of the IIS/TOR network. Proc Natl Acad Sci U S A. 2015;112:7055–7060. doi: 10.1073/pnas.1419659112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olinski RP, Lundin LG, Hallböök F. Genome duplication-driven evolution of gene families: insights from the formation of the insulin family. Ann N Y Acad Sci. 2005;1040:426–428. doi: 10.1196/annals.1327.080. [DOI] [PubMed] [Google Scholar]
- 39.Hernández-Sánchez C, Mansilla A, de Pablo F, Zardoya R. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol. 2008;25:1043–1053. doi: 10.1093/molbev/msn036. [DOI] [PubMed] [Google Scholar]
- 40.Rentería ME, Gandhi NS, Vinuesa P, Helmerhorst E, Mancera RL. A comparative structural bioinformatics analysis of the insulin receptor family ectodomain based on phylogenetic information. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huminiecki L, Heldin CH. 2R and remodeling of vertebrate signal transduction engine. BMC Biol. 2010;8:146. doi: 10.1186/1741-7007-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hokamp K, McLysaght A, Wolfe KH. The 2R hypothesis and the human genome sequence. J Struct Funct Genom. 2003;3:95–110. doi: 10.1023/A:1022661917301. [DOI] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ensembl Genome Browser [http://www.ensembl.org/index.html].
- 45.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH: The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 2016, 48:427–37. [DOI] [PMC free article] [PubMed]
- 47.National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/].
- 48.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cañestro C, Albalat R, Irimia M, Garcia-Fernàndez J. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Semin Cell Dev Biol. 2013;24:83–94. doi: 10.1016/j.semcdb.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Gen Genomics. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 54.Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci U S A. 2015;112:14918–14923. doi: 10.1073/pnas.1507669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. [DOI] [PMC free article] [PubMed]
- 56.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, et al. PipMaker--a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A. NISC comparative sequencing program, green ED, Hardison RC, Miller W: MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tajima F. Simple methods for testing molecular clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 60.Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 61.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SM, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson YW, Nanlohy KG, Libants SV, Yeh CY, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W: Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013, 45:415–21. [DOI] [PMC free article] [PubMed]
- 62.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci U S A. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurokawa T, Uji S, Suzuki T. Identification of cDNA coding for a homologue to mammalian leptin from pufferfish, Takifugu rubripes. Peptides. 2005;26:745–750. doi: 10.1016/j.peptides.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 64.Hu Q, Tan H, Irwin DM. Evolution of the vertebrate Resistin Gene family. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuraku S, Meyer A. Detection and phylogenetic assessment of conserved synteny derived from whole genome duplications. Methods Mol Biol. 2012;855:385–395. doi: 10.1007/978-1-61779-582-4_14. [DOI] [PubMed] [Google Scholar]
- 66.Arroyo JI, Hoffmann FG, Opazo JC. Gene turnover and differential retention in the relaxin/insulin-like gene family in primates. Mol Phylogenet Evol. 2012;63:768–776. doi: 10.1016/j.ympev.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 67.Hoffmann FG, Opazo JC. Evolution of the relaxin/insulin-like gene family in placental mammals: implications for its early evolution. J Mol Evol. 2011;72:72–79. doi: 10.1007/s00239-010-9403-6. [DOI] [PubMed] [Google Scholar]
- 68.Daković N, Térézol M, Pitel F, Maillard V, Elis S, Leroux S, et al. The loss of adipokine genes in the chicken genome and implications for insulin metabolism. Mol Biol Evol. 2014;31:2637–2646. doi: 10.1093/molbev/msu208. [DOI] [PubMed] [Google Scholar]
- 69.Massingham T, Davies LJ, Liò P. Analysing gene function after duplication. Bioessays. 2001;23:873–876. doi: 10.1002/bies.1128. [DOI] [PubMed] [Google Scholar]
- 70.Freeling M, Scanlon MJ, Fowler JE. Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr Opin Genet Dev. 2015;35:110–118. doi: 10.1016/j.gde.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 71.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S: Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–6. [DOI] [PubMed]
- 72.Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–903. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 73.Sun XJ, Crimmins DL, Myers MG, Jr, Miralpeix M, White MF. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol. 1993;13:7418–7428. doi: 10.1128/MCB.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Esposito DL, Li Y, Vanni C, Mammarella S, Veschi S, Della Loggia F, et al. A novel T608R missense mutation in insulin receptor substrate-1 identified in a subject with type 2 diabetes impairs metabolic insulin signaling. J Clin Endocrinol Metab. 2003;88:1468–1475. doi: 10.1210/jc.2002-020933. [DOI] [PubMed] [Google Scholar]
- 75.Landis J, Shaw LM. Insulin receptor substrate 2-mediated phosphatidylinositol 3-kinase signaling selectively inhibits glycogen synthase kinase 3β to regulate aerobic glycolysis. J Biol Chem. 2014;289:18603–18613. doi: 10.1074/jbc.M114.564070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asano T, Fujishiro M, Kushiyama A, Nakatsu Y, Yoneda M, Kamata H, et al. Role of phosphatidylinositol 3-kinase activation on insulin action and its alteration in diabetic conditions. Biol Pharm Bull. 2007;30:1610–1616. doi: 10.1248/bpb.30.1610. [DOI] [PubMed] [Google Scholar]
- 77.Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009;9:127. doi: 10.1186/1471-2148-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elephant Shark Genome Project [http://esharkgenome.imcb.a-star.edu.sg/].
- 79.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucl Acids Res. 2010;38:W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guidance2 Web Server [http://guidance.tau.ac.il/ver2/].
- 82.Clustal Omega Web Server [http://www.ebi.ac.uk/Tools/msa/clustalo/].
- 83.MrBayes 3.2.2 Web Site [http://mrbayes.sourceforge.net/].
- 84.IQ-tree Web Server [http://iqtree.cibiv.univie.ac.at/].
- 85.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;2017 in press [DOI] [PMC free article] [PubMed]
- 87.Tracer v1.6 Web Site [http://tree.bio.ed.ac.uk/software/tracer/].
- 88.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.JS Distance Web Server [http://compbio.cs.princeton.edu/conservation/].
- 90.NetPhos 2.0 Web Server [http://www.cbs.dtu.dk/services/NetPhos/].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file is in Excel format. Genomic locations of Irs-like genes in sequenced vertebrate genomes identified from the Ensembl database. (XLSX 40 kb)
This file is in Excel format. Accession numbers of Irs-like coding sequences from vertebrates and outroup species identified from the NCBI database. (XLSX 218 kb)
This file is in Word format. Coding sequences for full-length and near full-length Irs genes from diverse vertebrates and outgroups. (PDF 200 kb)
This file is in PDF format. Phylogeny of vertebrate Irs1 sequences. (PDF 172 kb)
This file is in PDF format. Phylogeny of vertebrate Irs2 sequences. (XLSX 58 kb)
This file is in PDF format. Phylogeny of vertebrate Irs3 sequences. (XLSX 53 kb)
This file is in PDF format. Phylogeny of vertebrate Irs4 sequences. (DOCX 627 kb)
This file is in PDF format. Phylogeny of the vertebrate Irs gene family rooted with non-vertebrate Irs-like genes. (PDF 525 kb)
This file is in PDF format. Alignment of primate Irs3 genomic sequences. (PDF 518 kb)
This file is in Excel format. Relative rates of evolution of Irs genes in mammals and non-mammals. (PDF 518 kb)
This file is in Excel format. JS Divergence Scores from alignments of Irs proteins from 10 vertebrates. (PDF 499 kb)
This file is in PDF format. Alignment of Irs protein sequences with conserved residues highlighted. (PDF 450 kb)
This file is in PDF format. ModelFinder results for the coding sequences used in the phylogenetic analyses. (PDF 224 kb)
Data Availability Statement
The data set supporting the results of this article is included within the article’s additional files (see Additional file 3: Figure S1).


