Abstract
Guinea pig cytomegalovirus (GPCMV) immediate early proteins, IE1 and IE2, demonstrated structural and functional homologies with human cytomegalovirus (HCMV). GPCMV IE1 and IE2 co-localized in the nucleus with each other, the viral polymerase and guinea pig ND10 components (gpPML, gpDaxx, gpSp100, gpATRX). IE1 showed direct interaction with ND10 components by immunoprecipitation unlike IE2. Additionally, IE1 protein disrupted ND10 bodies. IE1 mutagenesis mapped the nuclear localization signal to the C-terminus and identified the core domain for gpPML interaction. Individual knockout of GPCMV GP122 or GP123 (IE2 and IE1 unique exons respectively) was lethal to the virus. However, an IE1 mutant (codons 234–474 deleted), was viable with attenuated viral growth kinetics and increased susceptibility to type I interferon (IFN-I). In HCMV, the IE proteins are important T cell target antigens. Consequently, characterization of the homologs in GPCMV provides a basis for their evaluation in candidate vaccines against congenital infection.
Keywords: congenital HCMV, guinea pig CMV, immediate-early 1, HSV-1, innate immunity, nuclear domain 10, PML, Daxx, interferon, JAK-STAT
Introduction
Human cytomegalovirus (HCMV, HHV-5) is a member of the Betaherpesvirinae subfamily and a ubiquitous pathogen. In healthy individuals, the virus is asymptomatic but establishes a lifelong mainly latent infection in the human host. HCMV can cause morbidity and mortality in immune suppressed transplant and AIDS patients. Additionally, congenital HCMV is a leading cause of birth defects in newborns, which include mental retardation, hearing loss, microcephaly, impaired vision and seizures. Globally, approximately 0.5–2.2% of live newborns have congenital CMV infection. Ultimately, HCMV affects more live births than Down Syndrome or Fetal Alcohol Syndrome, making the development of an effective vaccine or intervention strategy a high priority.
Primary HCMV infection triggers one of the highest T-cell responses observed for human viral infections directed to various viral proteins including the tegument protein pp65 and nonstructural IE1 protein. Although pp65 elicits the majority of CD8+ T cell responses, HCMV IE-72 kDa (IE1) is an important target for both CD4+ and CD8+ T cells with IE1-specific CD8+ T cell responses exceeding those of pp65-specific CD8+ T cells in some patients. In one study, 25% of HCMV infected people developed T cell responses only to IE1, suggesting that IE1 is at least equally important as, if not more than, pp65 in generating protective T cell immune responses. In contrast to pp65, which is dispensable for growth of HCMV in vitro, IE1 is essential for viral replication and expressed throughout the lytic cycle. Vaccine studies targeting IE1 eliciting a T cell response may be important in protecting against congenital CMV but this aspect has not been evaluated. However, in the two available animal models for congenital CMV (rhesus macaque and guinea pig) a T cell response against CMV has been shown to be important for protection against congenital infection. Therefore, the development of an effective vaccine strategy should include important T cell target antigens including IE1 as a priority candidate.
HCMV IE1 and IE2 gene expression is regulated by the major immediate early (MIE) promoter and generated through alternative splicing. IE1 and IE2 are encoded by shared exons but each also have a unique exon: UL123 (IE1); UL122 (IE2). IE-72 kDa (IE1) and IE-86 kDa (IE2) are considered the main, functionally active proteins that play essential roles in transcriptional activation, innate cellular immune evasion, and cell cycle regulation during viral infection. While IE1 is required at low MOI, IE2 is indispensable to viral replication. Homologs to IE1 and IE2 have been identified in various species of animal CMV.
One way by which the cellular innate immune system interferes with viral infection involves nuclear domain 10 (ND10) or promyelocytic leukemia (PML) protein bodies. These nucleoplasmic foci are involved in diverse cellular functions such as replication, stress response, senescence, and epigenetic silencing. ND10 structures constitute an interferon (IFN)-dependent antiviral mechanism against lytic replication of different viruses. Activation of IFN-stimulated response elements and PML promoter trigger a significant increase in cellular PML and Sp100 expression, which leads to a death associated protein (Daxx)- and PML-mediated antiviral response through the action of histone deacetylases (HDACs), cytokines and IFN-stimulated genes (ISGs).
The major scaffolding component of ND10 is the PML protein. While more than 150 proteins have been identified to associate with ND10 structures, in silico analysis using BIOGRID, predicts 520 proteins to physically interact with PML. Other primary components include Sp100, Daxx and ATRX. Composition, stability, and function of ND10 are tightly regulated by small ubiquitin-like modifier (SUMO). SUMO modification is a crucial posttranslational mechanism that controls chromatin structure, protein stability, function and interaction. SUMO-1 interacts, co-localizes and covalently modifies human PML, Daxx and Sp100, which subsequently regulates the recruitment as well as dispersal of ND10 components.
Viruses belonging to all three herpesvirus subfamilies target and disrupt ND10 structures to escape the host innate immune response. The alphaherpesvirus herpes simplex type 1 (HSV-1) is the best studied virus for interaction with PML. In HSV-1, the immediate early infected cell protein 0 (ICP0) overcomes ND10-mediated responses by targeting, interacting with, and disrupting ND10 components. The SUMO-targeted E3 ubiquitin ligase activity of ICP0 induces PML, Sp100, and Daxx degradation followed by ND10 dispersal and antagonization of ND10-medicated gene silencing. Analogous to ICP0, HCMV IE1 and the tegument protein pp71 have been shown to counteract ND10-mediated responses. IE1 directly interacts with PML via its core domain, which shares secondary structures with the coiled-coil domain of E3-ligase TRIM factors, leading to PML modification and disruption of ND10 bodies. Newly expressed HCMV IE1 interacts with cellular factors to regulate viral and cellular gene expression by antagonizing HDACs.
Candidate vaccine studies require evaluation in a preclinical animal model. Species-specificity of HCMV prevents direct studies in animal models and requires the use of species-specific virus and animal models. The guinea pig is perhaps the only practical animal model for high-throughput congenital CMV studies due to the lack of RhCMV negative macaques and costs related to this non-human animal model. The guinea pig is the only small animal model for congenital CMV. Both human and guinea pig placentas are hemomonochorial containing a homogenous layer of trophoblast cells separating maternal and fetal circulation unlike mice, which have three trophoblast layers. Additionally, the guinea pig has a long gestation period (65 days) that can be divided into three trimesters and congenitally infected pups have disease symptoms similar to human disease such as hearing loss.
However, guinea pig CMV (GPCMV) lacks in depth functional characterization of potential homolog viral proteins. Ultimately, understanding the role of viral proteins in infection and immunity will aid the development of a CMV vaccine. The GPCMV genome is predicted to encode over > 200 open reading frames (ORFs). A number of HCMV co-linear homolog genes have been evaluated for encoded conserved protein function. These include homologs of pp65, pp71, UL84, viral kinase UL97, viral glycoproteins gB, gM, gN, gH, gL, gO and pentameric complex. Analogous to HCMV, the GPCMV MIE locus encodes IE1 and IE2 genes with shared exons and a unique fourth exon GP123 (IE1) and GP122 (IE2). However, functional properties of GPCMV IE1 and IE2 encoded proteins have not yet been investigated. A series of studies were conducted to characterize the role of GPCMV IE1 and IE2 proteins in the virus life cycle. These studies included essential nature, interaction with ND10 bodies and IE1 mutant susceptibility to type I interferon (IFN-I). Overall, these studies demonstrated functional homology of GPCMV IE1 and IE2 to HCMV and merit the continued development of this animal model of CMV.
Results
GPCMV major immediate early (MIE) locus encodes the two major proteins IE1 and IE2 essential for viral replication
The GPCMV major MIE locus was predicted to encode two proteins, IE1 (474 amino acids,aa) and IE2 (676 aa), which are potential HCMV IE protein homologs (Fig 1(iii)). While IE2 homologs are highly conserved between different CMV species, IE1 homologs are less conserved, which is also observed for GPCMV IE1. Amino acid sequence alignment showed only 16% sequence identity between HCMV and GPCMV IE1 compared to 30% for IE2 (Table S1). To verify the sequence and exclude alternative transcripts, IE1-specific 5′ and 3′ RACE experiments were conducted on total RNA isolated from GPCMV infected GPL cells treated with cycloheximide. DNA sequence analysis of cloned RACE products were aligned to GPCMV IE1 cDNA sequence (accession #AB592928) (Fig S1) and confirmed that the IE1 coding sequence was similar to that previously described.
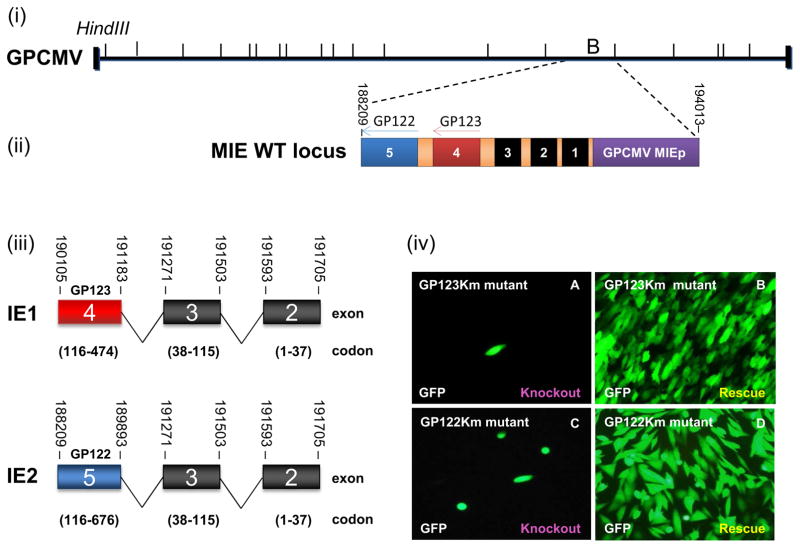
Figure 1. GPCMV major immediate early (MIE) locus encodes IE1 and IE2.
(i) Schematic representation of the GPCMV BAC genome and major immediate-early region with HindIII sites indicated. (ii) The GPCMV MIE WT locus is located within the HindIII ‘B’ fragment and encodes exons 1 to 5 on the antisense strand under the control of the MIE promoter (GPCMV MIEp) (purple). (iii) GPCMV IE1 and IE2 are splice variants that share exons 2 and 3, but are distinct in unique exon 4 (IE1, GP123) and 5 (IE2, GP122). Exon 1 is not translated. (iv) GP123Km (A–B) and GP122Km (C–D) mutant BACs were separately transfected onto GPL cells to generate virus. Each BAC was transfected individually (A and C) or in combination with a rescue plasmid encoding a WT IE1 or IE2 locus (B and D). Images taken 20 days post transfection.
GPCMV IE1 and IE2 unique exons, GP123 and GP122 respectively, were targeted for individual knockout mutagenesis to determine the essential nature of the genes. GPCMV IE1 (GP123) and IE2 (GP122) knockout BAC mutants were generated as described in materials and methods and designated GP123Km for IE1 and GP122Km for IE2. Separate transfection of knockout mutant GPCMV BAC DNA (GP123Km or GP122Km) onto GPL cells failed to produce virus unless co-transfected with a rescue locus as described in materials and methods (Fig 1(iv)). This demonstrated the essential nature of both IE1 and IE2 homolog proteins to GPCMV.
Expression and nuclear localization of GPCMV IE1 and its unique exon GP123
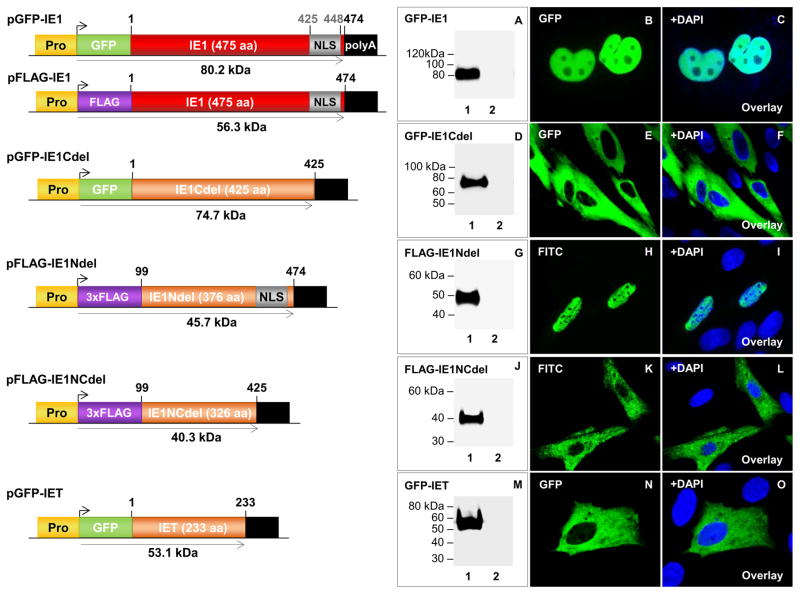
GPCMV IE1 protein was characterized by generating a full-length IE1 cDNA (474 codons) N-terminal tagged with either GFP (GFP-IE1) or FLAG epitope (FLAG-IE1) in separate plasmid expression constructs as described in materials and methods (Fig 2). Transient expression and western blot analysis identified GFP-IE1 as a protein of 80 kDa molecular weight (Fig 2; panel A), which approximated to the expected size of 80.2 kDa (Table S1). In transfected cells, IE1 demonstrated uniform, nuclear localization of GFP-IE1 (Fig 2; B–C) and FLAG-IE1 (Fig 3(iii)). Transient expression of the IE1 unique exon GP123 (GFP-GP123) produced a protein of 60 kDa (Table S1) (Fig S2(ii), panel A), which also targeted the nucleus in a uniform pattern (Fig S2(ii), B–C). The GFP-IE1 and GFP-GP123 expression pattern was similar in that both dispersed evenly throughout the nucleus. In the presence of GPCMV, no significant change was observed in the localization pattern of plasmid expressed IE1 protein (Fig 3(iv); M–O).
Figure 2. Transient expression and cellular localization of GPCMV IE1 and IE1 truncation mutants.
Schematic representation of epitope tagged ORFs of full length and truncated GPCMV IE1 GFP- and FLAG-tagged expression constructs. All expression constructs are under the control of the immediate early CMV promoter (Pro) and contain a SV40 polyadenylation signal (polyA). Nuclear localization signal (NLS, grey box). Protein expression confirmed by western blot analysis and immunofluorescence. A–C, transient expression and localization of full-length GFP-IE1. D–F, transient expression and localization of GFP-IE1Cdel. G–I, transient expression and localization of FLAG-IE1Ndel. J–L, transient expression and localization of FLAG-IE1NCdel. M–O, transient expression and localization of GFP-IET. FLAG-IE1Ndel and FLAG-IE1NCdel detected by primary anti-epitope antibody and secondary anti-mouse IgG-FITC (immunofluorescence) or anti-mouse IgG-HRP (western blot). GFP-IE1, GFP-IE1Cdel, and GFP-IET were detected by fluorescence and specific epitope antibody (western blot). Lane 1 in A, D, G, J, and M are individual transfected constructs also visualized in B, E, H, K, and N. Lane 2 are mock transfected GPL cells. Panels C, F, I, L, and O are DAPI overlays.
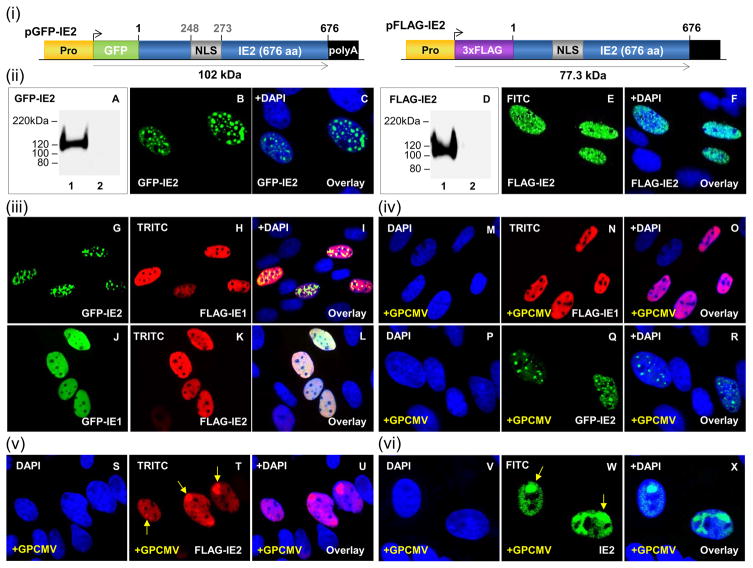
Figure 3. Transient expression and nuclear localization of GPCMV IE2 with IE1.
(i) Schematic representation of GFP- and FLAG-tagged IE2 fusion protein expression constructs. Expression constructs are under the control of the CMV immediate early promoter (Pro) and contain a SV40 polyadenylation signal (polyA). Nuclear localization signal (NLS, grey box). (ii) A–C, transient expression and localization of GFP-IE2. Lane 1, individually transfected GFP-IE2 visualized in B. Lane 2, mock transfected cell lysate. D–F, transient expression and localization of FLAG-IE2. Lane 1, individually transfected FLAG-IE2 visualized in E. Lane 2, mock transfected cell lysate. (iii) G–I, co-localization of FLAG-IE1 and GFP-IE2. G and H represent individually transfected GFP-IE2 (G) and FLAG-IE1 (H) respectively. I is the merge of G and H overlaid with DAPI. J–L, co-localization of GFP-IE1 (J) and FLAG-IE2 (K). L represents the merge of J and K overlaid with DAPI. (iv) M–O, FLAG-IE1 (N) in the presence of GPCMV (M). O, merge of M and N overlaid with DAPI. P–R, GFP-IE2 (Q) in the presence of GPCMV (P). R, overlay with DAPI. (v) S–U, FLAG-IE2 (T) in the presence of GPCMV (S). U is overlay of S and T. (vi) V–X, GPCMV IE2 (W). X, overlay with DAPI. FLAG-IE1 and FLAG-IE2 detected by primary anti-FLAG Ab and secondary anti-mouse IgG-TRITC (immunofluorescence) or anti-mouse IgG-HRP (western blot). GFP-IE1 and GFP-IE2 were detected by fluorescence and specific epitope Ab (western blot). GPCMV IE2 was detected via IE2-specific antisera. Arrows indicate accumulation of IE2.
In silico analysis of GPCMV IE1 predicted protein sequence by nuclear localization signal (NLS) mapper indicated a potential monopartite NLS (aa 425 to 438) and bipartite NLS (aa 425 and 448) within the C-terminal domain of IE1 (Fig S2(iii)). N- and C-terminus truncated versions of IE1 were generated, epitope tagged, and expression and size of each IE1 mutant confirmed by western blot analysis (Fig 2). Each mutant protein migrated with a molecular weight similar to predicted size (Table S1). Immunofluorescence analysis of transiently expressed IE1 mutants demonstrated that the C-terminus of IE1 was necessary for cellular nuclear localization, as the proteins were located in the cytoplasm (Fig 2). In contrast, the IE1Ndel was detected in the nucleus (Fig 2; H–I). Overall, this confirmed that the C-terminus of GPCMV IE1 (codons 426–474) harbors a functional NLS.
Nuclear localization of GPCMV IE2 (GP122) and co-localization with IE1
Full-length GPCMV IE2 cDNA (676 codons) was generated and N-terminal tagged with either GFP (GFP-IE2) or FLAG epitope (FLAG-IE2) in separate plasmid expression constructs (Fig 3(i)). The unique GP122 exon was similarly cloned and GFP epitope tagged (Fig S2(i)). Transient expression and western blot analysis revealed that the molecular weight for GFP-IE2 (Fig 3(ii); panel A) and GFP-GP122 (Fig S2(ii); panel D) was greater than their expected calculated sizes of 101.2 kDa and 88.8 kDa respectively (Table S1). A larger than expected molecular weight was also observed for FLAG-IE2 (Fig 3(ii); panel D), which suggested possible post-translational modification. Analysis of IE2 predicted amino acid sequence identified six potential sumoylation sites within the unique exon GP122 coding sequence (Table S1). Immunofluorescent microscopy of transfected cells demonstrated that IE2 targeted the nucleus with a punctate pattern (Fig 3(ii); B–C and E–F) compared to the uniform pattern observed for the IE1 protein and a similar result was observed for GFP-GP122 ((ii); E–F).
Next, the interaction between IE1 and IE2 proteins was evaluated in transient expression assays. Both proteins showed weak nuclear co-localization (Fig 3(iii); G–I and J–L). IE1 maintained a uniform distribution throughout the nucleus. Interestingly, while FLAG-IE2 adopted a diffuse nuclear pattern (Fig 3(iii); panel K), GFP-IE2 nuclear localization was more irregular and punctate (Fig 3(iii); panel G). In virus-infected cells, FLAG-IE1 and GFP-IE2 retained their distinct uniform and irregular distribution pattern respectively (Fig 3(iv); M–O and P–R). However, in the presence of GPCMV, FLAG-IE2 accumulated in larger punctate compartments (Fig 3(v); S–U; arrow). This pattern was identical to IE2 distribution during GPCMV infection determined via IE2-specific antisera (Fig 3(vi); panel W; arrows). During HCMV infection, IE2 accumulates in viral DNA replication compartments. Similarly, GPCMV IE1 and IE2 co-localized with the viral polymerase subunit protein (GP44), when GP44 was expressed with either IE1 or IE2 in separate experiments (data not shown). Since both IE1 and IE2 had potential for sumoylation (Table S1) both proteins were analyzed for their ability to co-localize with SUMO-1 (Fig S5). Although nuclear co-localization was observed, the lack of specific guinea pig sumoylation reagents prevented any definitive experiment to conclude that sumoylation occurred.
Transient expression and co-localizations of guinea pig ND10 components
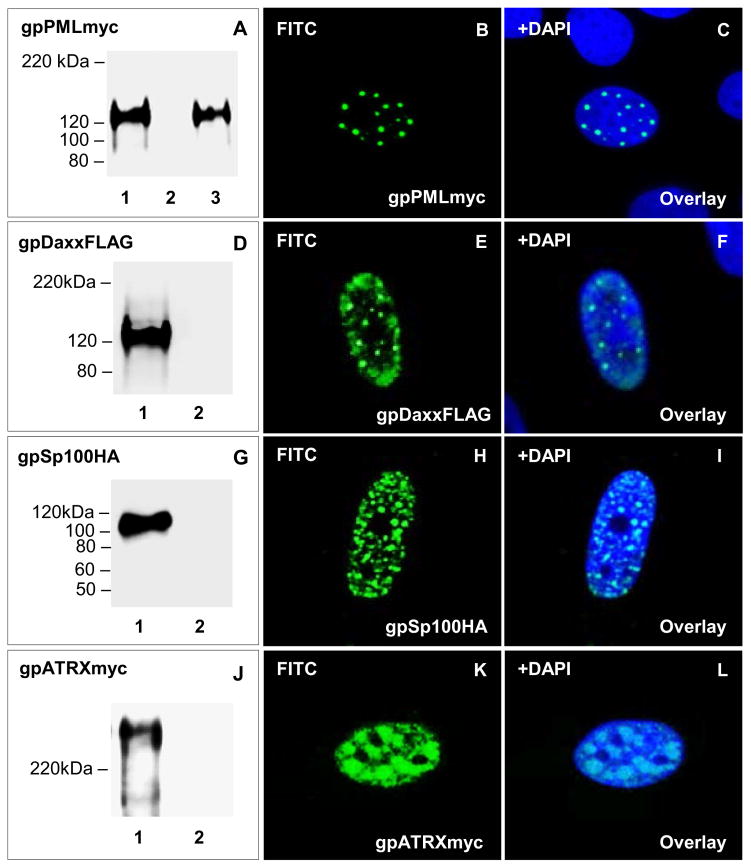
ND10 structures have been studied in different organisms, but have not been evaluated in the guinea pig. Based on the sequenced genome, the guinea pig encodes homologs to ND10 components (PML, Daxx, Sp100, and ATRX). Clustal Omega pairwise protein:protein sequence alignment of guinea pig ND10 components (gpPML, gpDaxx, gpSp100, and gpATRX) demonstrated higher sequence identity to human ND10 proteins: (1) PML (70%); (2) Daxx (77%); (3) Sp100 (45%) and (4) ATRX (90%) (Table S1) than between human and murine ND10 components (PML 64%, Daxx 71%, Sp100 30%, and ATRX 83%). While protein sequence analysis predicted guinea pig PML to contain a zinc-binding RBCC/TRIM motif that harbors a RING, B-box, and α-helical coiled-coil domains (data not shown) similar to human PML, there was insufficient conservation in commercial antibodies to PML and other ND10 components (Daxx, Sp100, and ATRX) to enable studies with cross reactive antibodies (data not shown). Consequently, epitope-tagged fusion protein expression constructs for gpPML (myc), gpDaxx (FLAG), gpSp100 (HA) and gpATRX (myc) were synthetically generated (DNA2.0) for ND10 studies as described in materials and methods. Immunofluorescence assay of separately expressed ND10 components demonstrated distinct nuclear pattern in GPL cells (Fig 4). gpPML (Fig 4; B–C) and gpSp100 (Fig 4; H–I) showed a distinct punctate nuclear distribution. The nuclear expression pattern of gpDaxx (Fig 4; E–F) and gpATRX (Fig 4; K–L) was also punctate but less defined with more protein detected within the nucleoplasm. Overall, the nuclear distribution pattern of guinea pig ND10 components (gpPML, gpDaxx, gpSp100, and gpATRX) was consistent with the pattern observed for human ND10 components.
Figure 4. Transient expression and nuclear localization of guinea pig ND10 components.
Transient expression of ND10 fusion proteins in GPL cells confirmed by western blot analysis and immunofluorescence. Lane 1, individually transfected plasmids. Lane 2, mock transfected GPL cells. Lane 3, GPCMV infected cells plus gpPML. A–C, transient expression and localization of gpPMLmyc. A shows levels of gpPMLmyc in transiently transfected GPLs in the absence or presence of GPCMV. Mock infected cells or cells infected with GPCMV (1.5 MOI) were transiently transfected with gpPMLmyc expression plasmid. B shows expression for gpPMLmyc, with C being the overlay of B with DAPI. D–F, transient expression and localization of gpDaxxFLAG. D shows levels of gpDaxxFLAG in transiently transfected GPLs. E shows localization of gpDaxxFLAG, with F being the overlay of E with DAPI. G–I, transient expression and localization of gpSp100HA. G shows levels of gpSp100HA in transiently transfected GPLs. H shows localization of gpSp100HA, with I being the overlay of H with DAPI. J–L, transient expression and localization of gpATRXmyc. J shows levels of gpATRXmyc in transiently transfected GPLs. K shows localization of gpATRXmyc, with L being the overlay of K with DAPI. gpPMLmyc, gpDaxxFLAG, gpSp100HA, and gpATRXmyc detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG-HRP (western blot) and anti-mouse or anti-rabbit IgG FITC (immunofluorescence).
Western blot analysis showed gpSp100 (113.4 kDa) (Fig 4; panel G) and gpATRX (285.8 kDa) (Fig 4; panel J) had molecular weights to predicted size. However, gpPML migrated at a higher molecular weight than its predicted value of 98 kDa to approximately 120 kDa (Fig 4; panel A). Similarly, gpDaxx was 120 kDa in molecular weight (Fig 4; panel D), which was significantly larger than its predicted size of 75.3 kDa (Table S1). In silico analysis of each ND10 protein identified possible sumoylation sites, which potentially accounted for the difference between predicted and observed protein sizes (Table S1). To determine whether guinea pig ND10 components co-localize with SUMO-1, gpPML and gpDaxx were transiently expressed in GPL cells in the presence of human HA-epitope tagged SUMO-1 (Fig S3). Both, gpPML (Fig S3; I–L) and gpDaxx (Fig S3; M–P) co-localized with SUMO-1 in the nucleus, which suggested sumoylation as the potential cause for the increase in molecular weight seen in western blot analysis compared to predicted size. However, sumoylation could not be validated because of a lack of guinea pig specific or cross-reacting antibody for the study of sumoylation.
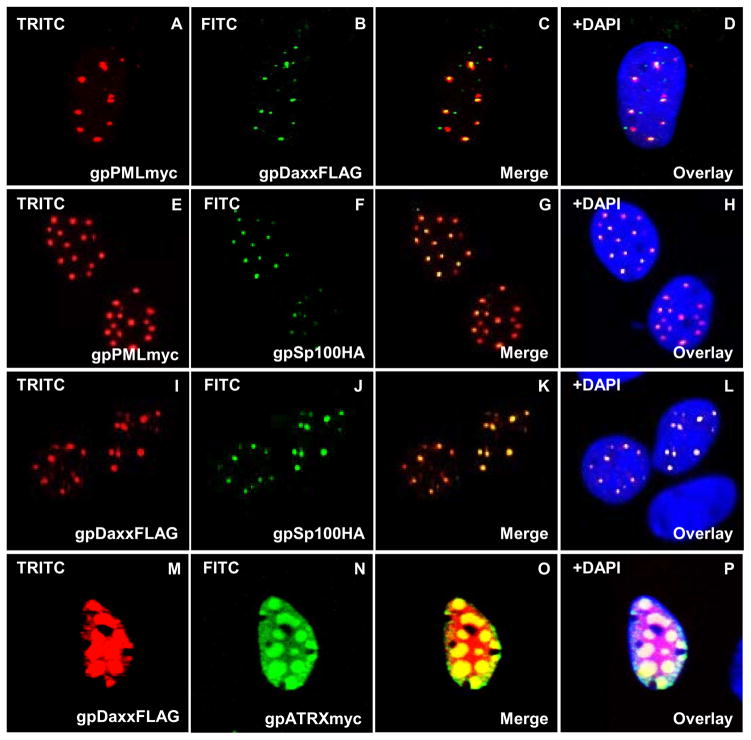
To demonstrate nuclear co-localization of these guinea pig ND10 proteins, different combination of ND10 components were transiently overexpressed in GPL cells (Fig 5). A fine, punctate, nuclear co-localization pattern was observed for combinations of: gpPML and gpDaxx (Fig 5; AD); gpPML and gpSp100 (Fig 5; E–H); gpDaxx and gpSp100 (Fig 5; I–L). While gpDaxx alone was also detected within the nucleoplasm (Fig 4; E–F), gpDaxx precisely co-localized to gpPML-and gpSp100-positive sites. The precise co-localization of gpDaxx with gpPML and gpSp100 may be explained by their transient overexpression, resulting in the sequestering of gpDaxx into an exclusively punctate pattern. Although gpDaxx and gpATRX co-localized in the nucleus (Fig 5; M–P), gpDaxx accumulated in aggregates distinct from the nuclear pattern of gpDaxx alone (Fig 4; E–F). Overall, these data indicated that transiently expressed guinea pig ND10 components have the potential to aggregate and form ND10 structures similar to that seen in human cells.
Figure 5. Nuclear co-localization of guinea pig ND10 components.
Different combinations of epitope tagged ND10 components were co-expressed in GPL cells and cellular localization investigated by immunofluorescence. A–D, gpPMLmyc and gpDaxxFLAG co-localization. gpPMLmyc (A) and gpDaxxFLAG (B) separately within the same cell. C is the merged image for A and B. D is the overlay of C with DAPI. E–H, co-localization of gpPMLmyc (E) and gpSp100HA (F), G merged image for E and F. H overlay with DAPI. I–L, colocalization of gpDaxxFLAG (I) and gpSp100HA (J), K merged image. L DAPI overlay. M–P, co-localization of gpDaxxFLAG (M) with gpATRXmyc (N). O merged image, P DAPI overlay. All immunofluorescent images were detected with primary anti-epitope Ab and secondary anti-mouse IgG-FITC or TRITC as described in materials and methods. Detection of GPCMV IE2 with anti-GPCMV IE2 as previously described.
GPCMV IE1 and IE2 interaction with guinea pig ND10 components
HCMV IE1 targets, destabilizes, and disrupts ND10 structures via its core domain. In silico sequence analysis of GPCMV IE1 revealed a conserved, stably folded globular core domain (codons 104–411) (Fig S4(i)) similar to HCMV IE1 (Fig S4(ii)) as well as a hydrophilic C-terminal domain (Fig S4). The ability of GPCMV IE1 to co-localize and interact with different guinea pig ND10 components was investigated. GPL cells were transiently co-transfected to express IE1 in the presence of gpPML, gpDaxx or gpSp100 (Fig 6(i)). Co-expression of IE1 and gpPML showed distinct nuclear co-localization (Fig 6; A–D). While individual IE1 expression showed uniform distribution within the nucleus (Fig 2; B–C), co-expression of IE1 and gpPML caused redistribution of IE1 within the nucleus into distinct compartments (Fig 6(i); A–D). IE1 also co-localized with gpSp100 (Fig 6(i); I–L) and gpATRX (Fig 6(i); M–P) in a similar punctate manner. However, IE1 failed to re-localize into an exclusively punctate pattern as seen with gpPML and showed minimal co-localization with gpDaxx (Fig 6(i); E–H).
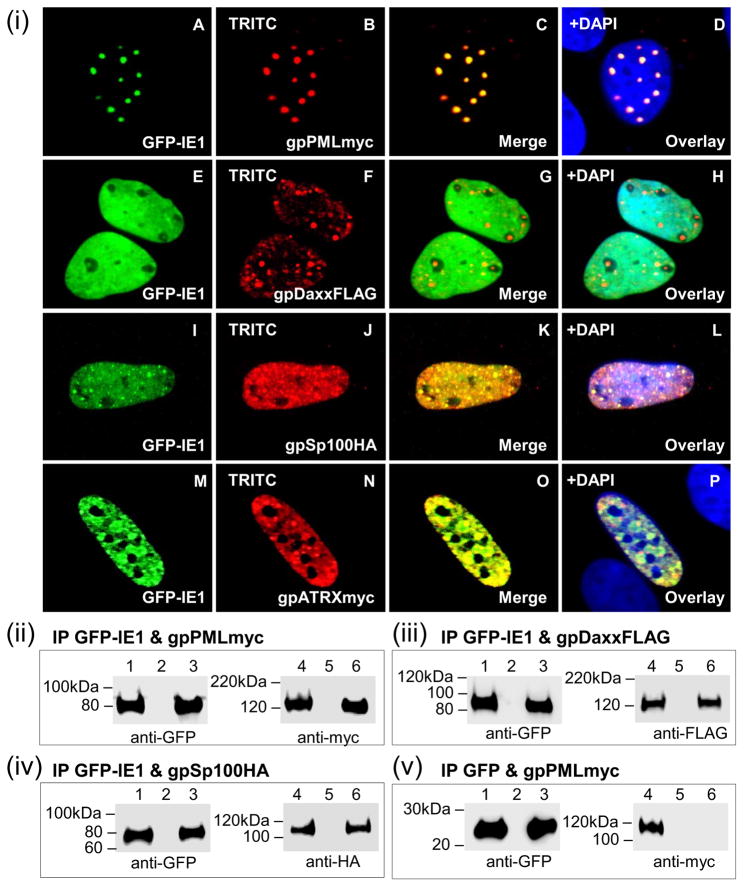
Figure 6. Nuclear co-localization of GPCMV IE1 with guinea pig ND10 components.
gpPML, gpDaxx, gpSp100, and gpATRX were evaluated for its ability to interact with IE1 by cellular co-localization (i) or GFP trap immunoprecipitation (ii-v). (i) A–D, localization of GFPIE1 (A) and gpPMLmyc (B) shown separately in the same cell. C is the merged image for A and B. D is the overlay of C with DAPI. E–H, GFP-IE1 (E) and gpDaxxFLAG (F). G merged image, H DAPI overlay. I–L, GFP-IE1 (I) and gpSp100HA (J). K merged image, L DAPI overlay. M–P, GFP-IE1 (M) and gpATRXmyc (N). O merged image, P DAPI overlay. gpPMLmyc, gpDaxxFLAG, gpSp100HA, and gpATRXmyc detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG TRITC. GFP-IE1 detected by fluorescence. (ii) GFPIE1 and gpPMLmyc co-expression and IP. (iii) GFP-IE1 and gpDaxxFLAG co-expression and IP. (iv) GFP-IE1 and gpSp100HA co-expression and IP. (v) GFP and gpPMLmyc co-expression and IP. GFP-IE1, gpPMLmyc, gpDaxxFLAG, and gpSp100HA detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG-HRP. Lanes 1 and 4, total cell lysate of transfected GPL cells. Lanes 3 and 6, IP reactions. Lanes 2 and 5, mock transfected GPL cells.
Subsequent IP studies of transiently expressed IE1 and ND10 components confirmed specific protein:protein interactions of IE1 with gpPML, gpDaxx, and gpSp100 (Fig 6(ii)–(iv)). Interaction between IE1 and gpATRX was not evaluated. Each immunoprecipitate was probed by western blot for the presence of GFP-IE1 (lanes 1 and 3) and (ii) gpPMLmyc (lanes 4 and 6), (iii) gpDaxxFLAG (lanes 4 and 6), or (iv) gpSp100HA (lanes 4 and 6) using primary anti-epitope antibodies. Analysis showed that GFP-IE1 was able to immunoprecipitate gpPMLmyc (Fig 6(ii); lane 6)) and gpSp100HA (Fig 6(iv); lane 6). Although no distinct co-localization was observed for IE1 and gpDaxx by immunofluorescence, both proteins interacted in the IP study (Fig 6(iii); lane 6). Potentially, this suggested that there might be some baseline interaction between IE1 and gpDaxx as previously reported for HCMV or secondary interaction via endogenous PML. The specificity of IE1-mediated IP was verified by GFP alone, which failed to immunoprecipitate gpPMLmyc (Fig 6(v); lane 6).
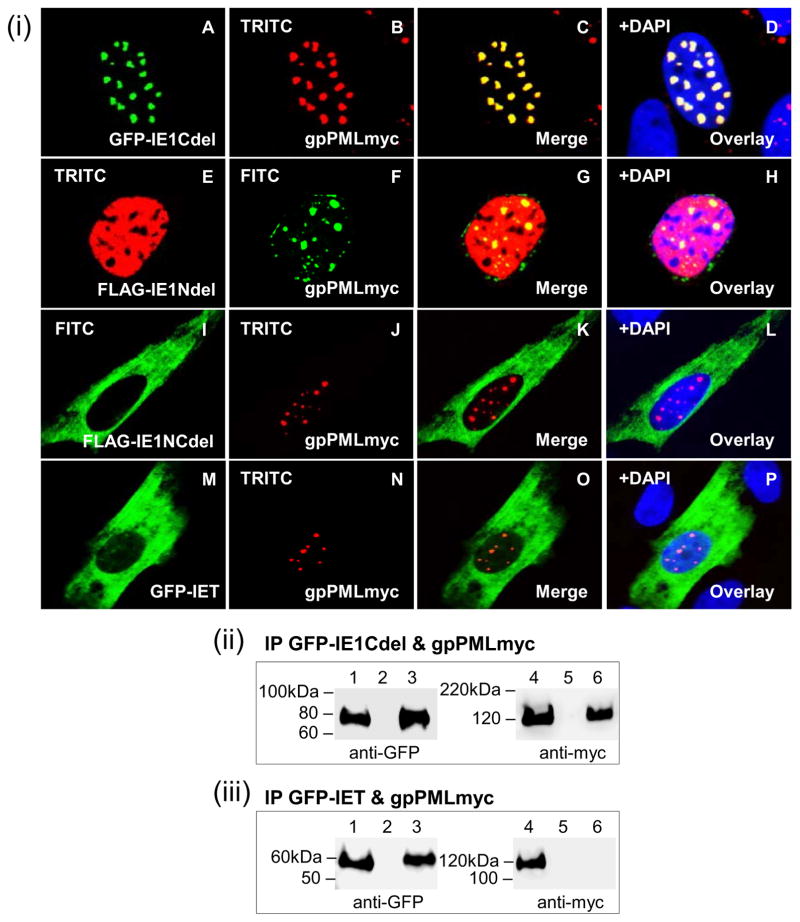
The functional domain of GPCMV IE1 required for the co-localization and interaction with gpPML was investigated with each of the four IE1 mutants (Fig 7(i)). Although the IE1Cdel mutant alone was unable to localize to the nucleus (Fig 2), co-expression with gpPML led to nuclear co-localization in distinct punctate compartments (Fig 7; A–D) similar to full-length IE1 (Fig 6; A–D). The IE1Ndel mutant was able to co-localize with gpPML but failed to adopt the characteristic punctate pattern (Fig 7; E–H). Both, the IE1NCdel (Fig 7; I–L) and IET (Fig 7; MP) mutants failed to localize to the nucleus and were unable to co-localize with gpPML. Furthermore, IP studies demonstrated the ability of IE1Cdel to interact with gpPML (Fig 7(ii); lane 6). However, IET failed to immunoprecipitate gpPML indicating a lack of interaction (Fig 7(iii); lane 6). Overall, these results demonstrated that GPCMV IE1 was able to target and directly interact with ND10 components and that both, the N- and C-termini of GPCMV IE1 play a critical role in the co-localization and interaction with gpPML.
Figure 7. Localization and interaction of gpPML with GPCMV IE1 mutants.
(i) Co-localization of different IE1 truncation mutants and gpPMLmyc in GPL cells analyzed by immunofluorescence. A–D, GFP-IE1Cdel (A) and gpPMLmyc (B) shown separately in the same cell. C merged image for A and B. D overlay of C with DAPI. E–H, FLAG-IE1Ndel (E) and gpPMLmyc (F). G merged image, with H being DAPI overlay. I–L, FLAG-IE1NCdel (I) and gpPMLmyc (J). K merged image, with L being DAPI overlay. M–P, GFP-IET (M) and gpPMLmyc (N). O merged image, with P being DAPI overlay. (ii) GFP-IE1Cdel and gpPMLmyc co-expression and IP. (iii) GFP-IET and gpPMLmyc co-expression and IP. Lanes 1 and 4, total cell lysate of transfected GPL cells. Lanes 3 and 6, IP reactions. Lanes 2 and 5, mock transfected GPL cells. gpPMLmyc, FLAG-IENdel, and FLAG-IE1NCdel detected by primary anti-epitope Ab and secondary anti-mouse IgG FITC or TRITC (immunofluorescence) and anti-mouse IgG-HRP (western blot). GFP-IE1Cdel and GFP-IET detected by fluorescence (cell localization) and specific epitope Ab (western blot).
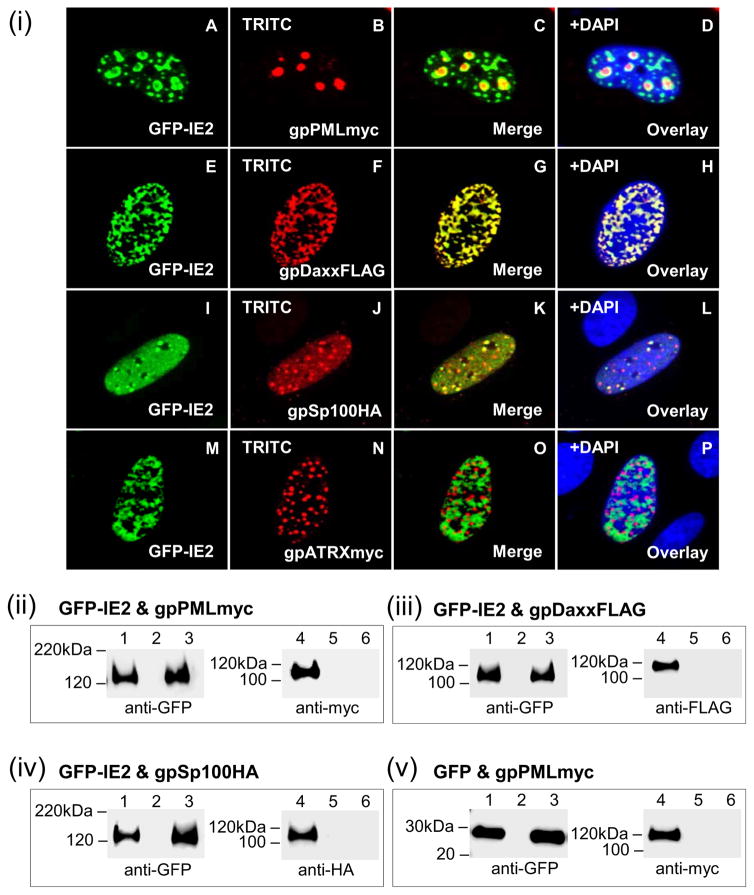
Similar to IE1, IE2 was investigated for its ability to co-localize and interact with guinea pig ND10 components (Fig 8). While GFP-IE2 alone adopted an uneven nuclear pattern (Fig 3), subnuclear localization of IE2 was partially altered in the context of ND10 components (Fig 8(i)). Co-expression of IE2 and gpPML resulted in nuclear co-localization, with IE2 adopting a punctate pattern in the periphery of individual gpPML dots (Fig 8(i); A–D). Direct co-localization of IE2 was also seen with gpDaxx (Fig 8(i); E–H). Although IE2 could be detected evenly throughout the nucleoplasm, IE2 co-localized with gpSp100 in distinct foci (Fig 8(i); IL). Despite nuclear localization, IE2 and ATRX failed to co-localize (Fig 8(i); M–P).
Figure 8. Nuclear co-localization of GPCMV IE2 and guinea pig ND10 components.
gpPML, gpDaxx, gpSp100, and gpATRX were evaluated for its ability to interact with IE2 by cellular co-localization (i) or by GFP trap immunoprecipitation (ii-v). (i) A–D, co-localization of GFP-IE2 (A) and gpPMLmyc (B) shown separately in the same cell. C is the merged image for A and B. D is the overlay of C with DAPI. E–H, GFP-IE2 (E) and gpDaxxFLAG (F). G merged image, H DAPI overlay. I–L, GFP-IE2 (I) and gpSp100HA (J). K merged image, L overlay with DAPI. M–P, GFP-IE2 (M) and gpATRXmyc (N). O merged image, P overlay with DAPI. gpPMLmyc, gpDaxxFLAG, gpSp100HA, and gpATRXmyc detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG TRITC. GFP-IE2 detected by fluorescence. (ii) GFP-IE2 and gpPMLmyc co-expression and IP. (iii) GFP-IE2 and gpDaxxFLAG co-expression and IP. (iv) GFP-IE2 and gpSp100HA co-expression and IP. (v) GFP and gpPMLmyc co-expression and IP. GFP-IE2, gpPMLmyc, gpDaxxFLAG, gpSp100HA detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG-HRP. Lanes 1 and 4, total cell lysate of transfected GPL cells. Lanes 3 and 6, IP reactions. Lanes 2 and 5, mock transfected GPL cells.
Immunoprecipitation studies were carried out on transiently expressed GFP-IE2 and each of the ND10 components (gpPML, gpDaxx, or gpSp100) in separate experiments to demonstrate specific protein:protein interactions (Fig 8(ii)–(iv)). Despite the co-localization results seen by immunofluorescence, specific protein:protein interactions between IE2 and gpPML, gpDaxx, or gpSp100 (Fig 8(ii)–(iv)) were not observed. The prediction was that IE2 most likely does not interact with ATRX, and therefore their interaction was not evaluated. A control GFP trap IP of cells transduced with AdGFP resulted in successful IP of GFP but not gpPML (Fig 8(v)). Failure of GPCMV IE2 to interact with the ND10 components suggested that IE1 and IE2 are functionally distinct with only IE1 targeting and disrupting ND10 structures.
vIET and GPCMV target ND10 cellular localization pattern
To characterize the role of GPCMV IE1 in infection, an IE1 C-terminus truncated viral mutant (vIET) was generated and confirmed as described in materials and methods. The vIET mutant carried a partial deletion (codonsΔ 234–474) within the unique exon GP123 coding sequence (Fig S5(i)) identical to the IET construct studied in transient expression assays (Fig 2).
Transfection of mutant IET GPCMV BAC DNA onto cells generated viable virus that was designated vIET. A multi growth curve of the vIET mutant showed a delay in viral replication and reduced viral titers compared to wild type (WT) virus (Fig 9(i)).
Figure 9. Dispersal of transiently expressed ND10 by WT HSV-1 and GPCMV.
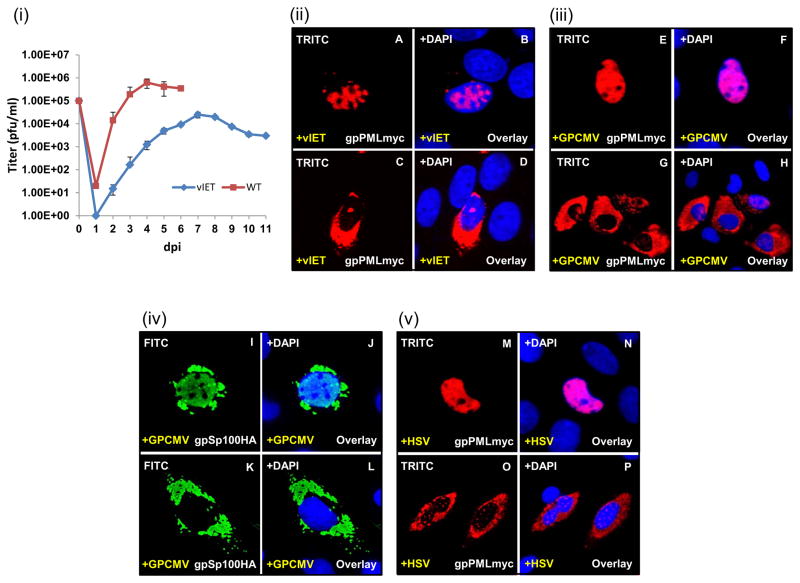
(i) GPL cells were infected with either IET or WT virus (0.1 MOI). Samples taken daily for 6 or 11 days and titrated in duplicate on GPL cells. Results plotted as virus titer versus days post infection (p.i.). (ii-v) GPL cells were infected with HSV-1 or GPCMV (0.1 MOI) prior to transient transfection with the gpPMLmyc expression construct and analyzed for gpPML localization by immunofluorescence. (ii) A–D, expression of gpPMLmyc in the presence of vIET. gpPML redistributes throughout the nucleus (A) and into the cytoplasm (C). Panels B and D are overlays for A and C with DAPI respectively. (iii) E–H, expression of gpPMLmyc in the presence of GPCMV. gpPML redistributes throughout the nucleus (E) and into the cytoplasm (G). Panels F and H are overlays for E and G with DAPI. (iv) I–L, expression of gpSp100HA in the presence of GPCMV causes redistribution of gpSP100 in the nucleus (I) and into the cytoplasm (K). Panels J and L are overlays for I and K with DAPI. (v) M–P, expression of gpPMLmyc in the presence of HSV-1 seen in the nucleus (M) and into the cytoplasm (O). Panels N and P are overlays for M and O with DAPI respectively. gpPMLmyc and gpSp100HA detected by primary anti-epitope Ab and secondary anti-mouse IgG TRITC or anti-rabbit IgG FITC respectively.
To determine the ability of vIET to target and disrupt exogenous ND10 structures, gpPML was transiently expressed in GPL cells prior to viral infection. The vIET mutant was able to target gpPML, causing its dispersal within the nucleus and re-localization to the cytoplasm (Fig 9(ii)). These findings propose an essential role for IE1 in targeting PML. Infection of GPL cells with GPCMV led to the disruption and dispersal of nuclear punctate gpPML (Fig 9(iii)) and gSp100 (Fig 9(iv)). WT HSV-1 was also shown to target transiently expressed gpPML (Fig 9(v)). HSV-1 and GPCMV both gave rise to two phenotypes that were marked by uniform, nuclear dispersal of gpPML as well as its re-localization into the cytoplasm. Since it is HSV-1 ICP0 that is required to disrupt ND10 structures during HSV-1 infection, a ICP0 double mutant virus was generated (Fig S6) and tested for its ability to target PML. However, the HSV-1 ICP0 double mutant had no impact on the localization pattern of gpPML (Fig S7(i)), emphasizing the known role of ICP0 in targeting ND10. These data confirm the ability of GPCMV to target PML bodies, and more specifically propose a role for IE1 in the dispersal of ND10 structures.
Transient gpPML expression impairs viral replication of vIET and HSV-1 ICP0Δ mutants
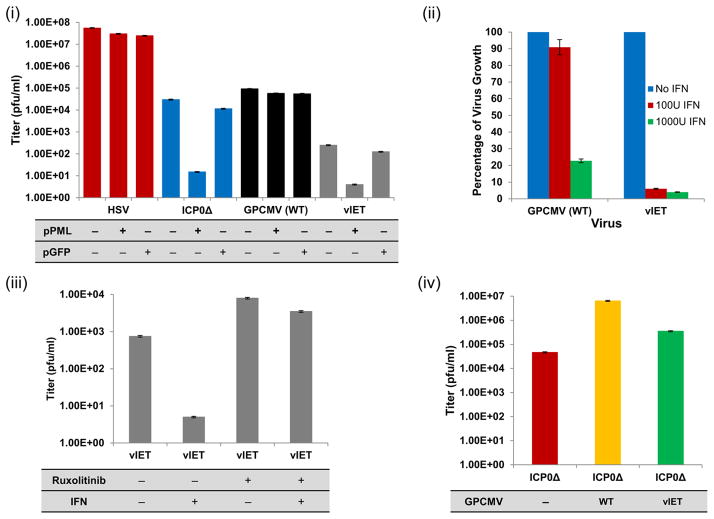
Since PML is the main ND10 component, the ability of transiently expressed gpPML to block viral replication of HSV-1 ICP0Δ or GPCMV vIET mutants was investigated (Fig 10(i)). Transient expression of gpPML decreased the production of ICP0Δ virus particles by approximately 3 logs compared to ICP0Δ alone (Fig 10(i); blue bars). Transient expression of gpPML did not decrease the viral titer of WT HSV-1 (Fig 10(i); red bars). Infection with vIET mutant resulted in a decrease in viral titer of about 2 logs (Fig 10(i); grey bars) compared to vIET alone. WT GPCMV virus was unaffected by transiently expressed gpPML (Fig 10(i); black bars). A GFP expression plasmid was used as control to confirm the antiviral specificity of gpPML. To confirm the antiviral effect of ND10 components on viral replication, GPL cells were treated with siRNA specific for gpPML and gpDaxx prior to infection with vIET mutant. In the presence of siRNA, vIET replication increased by 1.5 log compared to vIET control alone (data not shown). Overall, these data potentially indicate that gpPML negatively impacts HSV-1 ICP0Δ and GPCMV vIET virus replication in GPL cells but requires more definitive assays of siRNA target before concluding that HSV-1 ICP0 and GPCMV IE1 play similar important roles in counteracting gpPML-mediated antiviral responses.
Figure 10. ICP0Δ and vIET mutant virus replication. (i).
GPL cells transiently transfected with gpPML (+) before being infected with ICP0Δ or IET mutant virus, WT HSV, or GPCMV (0.1 MOI). (−) indicates GPL cells with intrinsic gpPML only. Cell monolayers and supernatant were harvested 3 days p.i. and titrated on GPL cells. Results plotted as virus titer versus experimental condition. (ii) IFN-I susceptibility of vIET mutant compared to WT. Cells were incubated overnight in the absence or presence of 100 IU/ml (red) or 1000 IU/ml (green) human IFN-I prior to virus infection (0.1 MOI). After 72 hrs, supernatant and monolayer were harvested and titrated in duplicate on GPL cells. Results blotted as percentage of virus growth. (iii) GPL cells were grown in the absence (−) or presence (+) of ruxolitinib (10 μM) and IFN-I (100 IU/ml). Cells were pretreated 6 hrs prior to infection with vIET (0.1 MOI). Cells and supernatants were harvested 4 days p.i. and titrated on GPL cells. Results plotted as virus titer versus experimental condition. (iv) Complementation of ICP0Δ mutant virus by GPCMV. GPL cells were infected with either WT GPCMV (yellow) or vIET (green) (3×104 pfu) prior to being superinfected with ICP0Δ mutant virus (102 pfu). ICP0Δ mutant virus alone (red) served as control. Cells and supernatants were harvested after 3 days and titrated on U2OS cells. Results plotted as virus titer versus experimental condition.
GPCMV IE1 C-terminus mutant virus susceptibility to IFN-I is restored by JAK/STAT inhibitor
GPCMV IE1 function in counteracting IFN-I responses was evaluated. Cells were pretreated with 100 IU/ml or 1000 IU/ml universal type I alpha interferon (PBL Assay Science) or untreated as control cells and subsequently infected at 0.1 pfu/cell (Fig 10(ii)). 100 IU/ml IFN moderately impaired WT GPCMV compared to untreated control (decreased to 90%). In contrast, the vIET mutant demonstrated significant replication impairment (decreased to below 10%) compared to untreated control. This indicated increased susceptibility to IFN-I as a result of IE1 mutation. Increasing the IFN-I pretreatment to 1000 IU/ml blocked WT GPCMV replication by 80% and vIET mutant by > 95% compared to untreated controls. The increased sensitivity of vIET to IFN treatment emphasized the importance of IE1 to overcome IFN-based immunity.
In mammals, the JAK-STAT signaling cascade regulates numerous cellular functions including innate immune responses, where it mediates cellular transcriptional responses to cytokines including IFN. To characterize the mechanism by which GPCMV IE1 counteracts the innate immune system, vIET infected GPL cells were grown in the absence or presence of IFN-I and/or the Janus kinase (JAK) inhibitor ruxolitinib, which has a selectivity for subtypes 1 (JAK1) and 2 (JAK2) of the JAK-STAT signaling pathway. vIET mutant replication was significantly impaired in the presence of 100 IU/ml IFN compared to untreated control (Fig 10(iii)). In the presence of ruxolitinib alone, there was a 1 log increase in vIET yield compared to vIET alone (Fig 10(iii)), which indicated that the JAK-STAT pathway was involved in the restriction of GPCMV replication. Simultaneous addition of IFN-I and ruxolitinib resulted in almost a log higher titer than vIET only infected cells (Fig 10(iii)). Overall, these data demonstrated that IFN-I based innate immunity mediates through the JAK-STAT pathway in guinea pig cells and that GPCMV IE1 is required for counteracting IFN-I based innate immune response.
Complementation of HSV-1 ICP0Δ mutant by GPCMV
Functional homology between HSV-1 ICP0 and GPCMV IE1 was established by initially analyzing HSV-1 ICP0 for its expression and localization pattern, as well as ability to co-localize with guinea pig ND10 components (Fig S7(ii)). Transient expression of only ICP0 in plasmid transfected GPL cells showed a nuclear punctate pattern (Fig S7(ii); E–G). Co-expression of ICP0 with different guinea pig ND10 components showed distinct punctate nuclear colocalization (Fig S7(ii)). Based on ICP0 co-localization with guinea pig ND10 components, the ability of GPCMV to complement a HSV ICP0 null mutant virus was investigated (Fig 10(iv)). GPL cells were co-infected with ICP0Δ virus and GPCMV or vIET. The ICP0Δ mutant was impaired for replication compared to WT HSV-1 but WT GPCMV complemented ICP0Δ virus by 2 log, while the vIET mutant promoted an increase in viral titer by 1 log. These results showed that GPCMV IE1 can complement HSV-1 ICP0Δ mutant demonstrating potential similar function and a partial deletion of IE1 reduced the complementation by 1 log.
Discussion
The observed positional and functional homology between HCMV and GPCMV tegument protein homologs, viral kinase, and glycoproteins raised the question of functional homology in other positional homolog viral genes and proteins. GPCMV MIE locus potentially encoded homologs of HCMV IE1 and IE2. Characterization of GPCMV IE1 and IE2 demonstrated that each encoded separate NLS within their unique exons GP123 and GP122 respectively. In contrast, HCMV IE1 and IE2 have a shared NLS within their N-terminus (exon 2) and distinct NLS sequences within their unique exons (UL123 or UL122). Distinct NLS sequences were also present in unique IE1 and IE2 coding exons for rat CMV (RCMV) and mouse CMV (MCMV), which stress the importance of IE1 and IE2 homologs as nuclear targeting proteins. GPCMV knockout mutagenesis demonstrated the separate essential nature of IE1 (GP123) and IE2 (GP122) in GPCMV infection, which potentially indicated conservation of essential function between HCMV and GPCMV MIE proteins. In contrast, the MCMV and RCMV homologs of IE1 are non-essential in tissue culture. Unlike HCMV and GPCMV, MCMV IE1 was predicted to express two globular domains and an extended hydrophilic C-terminus (Fig S4(iii)). Therefore, structural and functional homologies between HCMV and GPCMV IE1 and IE2 possibly indicate a higher conservation between viruses compared to MCMV.
As part of an immune-evasive strategy, a range of herpesviruses has evolved mechanisms to target and disrupt ND10 structures, leading to morphological changes and cellular redistribution. HSV-1 was the first herpesvirus described to co-localize with ND10 domains and mediate their disruption via ICP0. In HCMV, the IE1 protein functions as key antagonist against ND10-mediated immunity through direct interaction with ND10 components after infection. Protein:protein sequence alignment of guinea pig ND10 components showed a higher sequence identity with human ND10 proteins compared to mouse. To determine the impact of GPCMV IE1 and IE2 on guinea pig ND10 structures, epitope-tagged expression constructs of major ND10 components were initially tested for cellular expression (Fig 4) and co-localization (Fig 5). As an experimental approach, ND10 components were overexpressed, as there are currently no antibodies available to detect guinea pig ND10 proteins (Hornig, unpublished data). Guinea pig ND10 components adopted a punctate nuclear form and co-localized with each other as previously shown for human ND10 components. While GPCMV IE1 co-localized with gpPML, gpSp100 and gpATRX similar to HCMV IE1, it failed to co-localize with gpDaxx in a punctate pattern (Fig 6) but interaction could be demonstrated by IP assay. Data have previously shown that the viral tegument protein pp71 targets ND10 structures by associating with human Daxx (hDaxx) leading to its proteasomal degradation and removal from ND10. HCMV pp71 contains a Daxx-binding domain motif, which may also be present in the GPCMV pp71 homolog (GP82). Transient expression studies of GPCMV pp71 homolog with gpDaxx demonstrated colocalization similar to HCMV pp71 and hDaxx (data not shown).
Studies with N- and C-terminal truncated GPCMV IE1 mutants revealed that the N-terminus (1–98 aa) has a role in IE1, and most likely IE2, co-localization with PML and its re-organization into a punctate pattern (Fig 7), while the IE1 core domain (IE1CORE) (104–411 aa) was required for PML interaction (Fig S4). These observations are consistent with studies on the crystal structure of HCMV IE1, which demonstrated that it is the IE1CORE domain (exons 2 and 3) that binds PML, induces PML desumoylation and ND10 disruption. While this core domain is intact in the IE1Cdel mutant (codons 1–425), nearly half of it is missing in the IET mutant (codons 1–233), which could explain an inability to interact with gpPML. Further studies are needed to determine the ability of the IET mutant to interact with ND10 components such as gpDaxx, gpSp100, or gpATRX. Additionally, extensive C-terminal deletion of IE1 may impact on the stably folded globular core domain and protein structure impacting on the ability of IE1 to target and interact with ND10 structures. HCMV IE1 studies have shown that HCMV carrying mutations within the IE1CORE have impaired growth kinetics and fail to interact with PML.
In HCMV, the IE2 protein demonstrates a dynamic interaction with ND10 components, ranging from juxtaposition of IE2 to precise co-localization and separation, which contrasts with IE1. Transiently expressed GPCMV IE2 aggregated in the periphery of gpPML and co-localized with gpSp100 and gpDaxx (Fig 8(i)) similar to HCMV IE2-86, which co-localizes with ND10 bodies but fails to disrupt them. ATRX is known to interact with Daxx, while HCMV IE2 recruitment ND10 sites has been attributed to interactions with histone acetyltransferases and viral DNA. Protein:protein studies failed to demonstrate a direct interaction between GPCMV IE2 and gpPML, gpDaxx, or gpSp100 (Fig 8(ii)–(iv)), leading to the conclusion that GPCMV IE2 targets ND10 structures indirectly by means of alternative adopter protein or DNA interactions and recruitment mechanisms. Similar to HCMV IE2, GPCMV IE2 may be recruited to ND10 via interactions with various proteins including retinoblastoma proteins and p53, which are predicted to interact with PML and localize to ND10 sites. However, this awaits further evaluation.
Interestingly, western blot analysis of transiently expressed proteins indicated larger than expected molecular weights for gpPML and gpDaxx. In silico amino acid sequence analysis revealed the presence of several potential sumoylation sites within each ND10 component (Table S1). Studies in human and murine cells have shown that modification of the SUMO Interacting Motif (SIM) and SUMO-mediated modification of PML aid in ND10 formation, stability and function. hPML and hDaxx have higher than predicted molecular weights, which is most likely due to posttranslational modification by SUMO but a lack of specific antibodies to guinea pig SUMO prevents evaluation of this hypothesis for gpPML and gpDaxx.
The ability of HSV-1 ICP0 to target PML and thus evade ND10-mediated innate immune responses has been well documented. Since HSV-1 has a wide host range in tissue culture, WT HSV-1 and a HSV-1 ICP0Δ mutant viruses were used as controls and compared to WT GPCMV and the C-terminus truncated vIET mutant (codonsΔ 234–474) for their ability to counter the antiviral effect of gpPML (Fig 10). The trans-dominant effect of gpPML significantly decreased ICP0Δ and vIET mutant replication. For the HSV-1 ICP0Δ mutant, this is based on the fact that ND10 disruption is mediated by the E3 ubiquitin ligase activity of ICP0. Our data suggested that GPCMV IE1 is essential for viral replication and required for counteracting PML function and the IFN response analogous to HSV-1 ICP0, HCMV IE1 and Ad E4 ORF3. Knockdown of endogenous gpPML and gpDaxx by siRNA indicated a potential antiviral role for gpPML and gpDaxx but awaits more definitive assays on siRNA target to reach any conclusion.
Infection studies revealed the ability of HSV-1 to target gpPML, while the HSV-1 ICP0Δ mutant failed to disperse gpPML (Fig S98(ii)). The vIET mutant retained the ability to disrupt gpPML similar to GPCMV unlike transient expression of IET which failed to disrupt PML (Fig 9 and 7). Potentially, the vIET mutant may exert its function through alternative interactions with cellular or viral proteins, such as GP82 (pp71 homolog) to interact with gpDaxx. HCMV IE1 has a structural sequence analogous to the coiled-coil domain of E3 ligase TRIM proteins, which raised the question of whether GPCMV IE1 carries E3 ligase activity. However, in silico analysis of GPCMV IE1 failed to predict B-box and RING-finger domains characteristically seen with TRIM domains and ICP0. The vIET mutant showed a delay in viral replication and increased susceptibility to IFN-I (Fig 10), which suggested that GPCMV IE1 is directly or indirectly involved in overcoming the IFN-I response by regulating expression of evasion proteins similar to HCMV. Furthermore, these data suggest that the C-terminus and core domain play an essential role in mediating these processes. The C-terminus of HCMV IE1 is thought to mediate binding to STAT2 in order to disrupt IFN signaling. Hydrophilicity analysis of GPCMV IE1 protein (Fig S4(i); red box) revealed the presence of acidic residues within its C-terminal region similar to HCMV IE1 (Fig S4(ii); red box), which suggests a potential binding domain for STAT2 similar to HCMV IE1. However, specific interactions await further study complicated by lack of dedicated reagents. During infection, PML complexes with HDAC1 and 2, and STAT1 and 2 to induce ISG expression and mediate an antiviral response. However, HCMV IE1 suppresses ISGs by targeting STATs, HDACs and PML. Direct involvement of the JAK-STAT pathway in counteracting GPCMV was demonstrated by blocking JAK1 and JAK2, which resulted in enhanced viral replication (Fig 10).
The data presented in this paper need to be viewed in light of ND10 overexpression, since GPL cells are intrinsically positive for ND10. Additionally, in humans, several PML isoforms (PMLI-VIIb) have been identified, which are thought to play distinct as well as overlapping functions during viral infection. Although not part of this study, NCBI database analysis identified eight different guinea pig PML variants (data not shown), which suggests that the guinea pig has the potential to encode various PML isoforms. An important area for future evaluation is viral tropism to guinea pig trophoblast cells of the placenta. PML expression in human placenta is selective to trophoblasts, which may present a mechanism by which CMV transmission across the placenta can be prevented, or reduced, but this awaits further study in the guinea pig. The vIET mutant was generated on the backdrop of a virus lacking a pentameric complex (PC), necessary for infection of epithelial cells. Future studies will include the engineering of a vIET mutant that expresses the full pentameric complex (UL128-131 homolog), to evaluate GPCMV in guinea pig epithelial cells in vitro and in vivo and impact on viral dissemination.
Overall, GPCMV IE1 and IE2 hold an essential function in viral replication similar to HCMV. GPCMV IE1 and IE2 seem to play different roles in targeting and counteracting the innate immune response. This paper identifies a functional NLS within the IE1 C-terminus (GP123) and proposes a role for the N-terminus in nuclear redistribution of IE1 to co-localize with gpPML. The GPCMV IE1 (IE1CORE) was identified to target and interact with guinea pig ND10 components and counteract the IFN-I response through the STAT-JAK pathway in guinea pig fibroblast cells. Its role as an innate immune antagonist and immunogenic protein proposes GPCMV IE1 to be an ideal candidate for live subunit vaccine development. Overall, these studies further strengthen the guinea pig model for CMV studies and continued development of intervention strategies in this model.
Materials and Methods
Cells, viruses and oligonucleotides
GPCMV (strain 22122, ATCC VR682), first and second-generation GPCMV BAC derived viruses were propagated on guinea pig fibroblast lung cells (GPL; ATCC CCL-158™) as previously described. High titer stock was generated as previously described. Vero cells (ATCC; CCL-81™) and human osteosarcoma U2OS cells (ATCC; HTB-96™) were cultured in high glucose DMEM medium supplemented with 10% fetal calf serum (FCS). WT HSV-1 (17+strain) virus and ICP0Δ mutant virus stocks were generated on U2OS cells. Oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, TX) (Table S2).
GPCMV IE Transcript 5′ and 3′ RACE
GPL cells were pre-treated with cycloheximide (SIGMA) (200 μg/ml) prior to infection with WT GPCMV at a multiplicity of infection (MOI) of 10 pfu/cell. Total cellular RNA was extracted with the RNeasy Mini kit (Qiagen). 5′ and 3′ RACE was performed using the SMARTer® RACE 5′/3′ kit (Clontech) according to the manufacturer’s protocol. Individual 5′ and 3′RACE reactions with GPCMV IE1 gene-specific F5′ and R3′ RACE primers (Table S2) gave rise to DNA products of 1.5 kb (data not shown). Each fragment was PCR amplified, gel purified and subcloned into the pRACE plasmid according to the manufacturer’s protocol (Gentech®). Positive clones were confirmed by colony PCR and HindIII and EcoRI restriction enzyme digestion (data not shown).
Cloning and generation of expression vectors of GPCMV IE1, IE2 and IE1 mutants
The predicted GPCMV coding sequences were based on the 22122 viral genome sequence (Genbank accession #AB592928). GPCMV IE2: 191593–191705 (exon 2), 191271–191503 (exon 3), and 188209–189893 (exon 4, GP122). GPCMV IE1: 191593–191705 (exon 2), 191271–191503 (exon 3), and 190105–191183 (exon 4, GP123) (Fig 1). Full-length cDNAs for IE1 and IE2 ORFs were generated using the GeneArt® Seamless Cloning and Assembly approach (Invitrogen) according to the manufacture’s protocol. The vector backbone and individual exons were PCR-amplified using IE1 and IE2 specific primers. For the seamless cloning of IE1 cDNA, forward and reverse primers for pUC19(IE1), Exon2and3(IE1), and Exon4(IE1) were used (Table S2). IE2 cDNA was assembled using forward and reverse primers for pUC19(IE2), Exon2and3(IE2), and Exon4(IE2) (Table S2). One Shot® competent cells were transformed according to the manufacturer’s protocol. Positive ampicillin resistant clones were screened by BamH1 and EcoR1 double digest to confirm correct fragment assembly. GPCMV IE1 and IE2 cDNAs were confirmed by sequencing. Full-length GPCMV IE1 cDNA was subsequently PCR amplified as previous described to add Pst1 restriction sites to the 5′ and 3′ ends using primers FIE1Pst1 and RIE1Pst1 (Table S2) generating IE1Pst1. Full-length GPCMV IE2 cDNA was PCR amplified introducing 5′ and 3′ HindIII restriction sites using primers FIE2HindIII and RIE2HindIII (Table S2) generating IE2HindIII.
IE1 and IE2 ORFs were individually cloned into the pAcGFP1-C3 expression plasmid (Clontech) to generate N-terminus GFP-tagged fusion proteins. The IE1Pst1 cassette was cloned into the Pst1 restriction site of pAcGFP1-C3 to give rise to pGFP-IE1 (Fig 2). The IE2HindIII fragment was subcloned into the HindIII site of pAcGFP1-C3 to generate pGFP-IE2 (Fig 3(i)). N-terminus FLAG-epitope tagged expression plasmids for IE1 and IE2 were generated by in frame cloning of each ORF into the pCMV-3Tag-1A expression plasmid (Agilent Technologies). IE1Pst1 was cloned into the Pst1 restriction site to create pFLAG-IE1 (Fig 2). IE2HindIII was inserted into the HindIII site to generate pFLAG-IE2 (Fig 3(i)).
GPCMV IE1 mutants were generated by restriction enzyme-mediated collapse of either the N- or C-terminus, or both. The pGFP-IE1Cdel mutant was generated through AccI mediated C-terminus collapse (codonsΔ 426–474). The pFLAG-IE1Ndel mutant was created by BamHI N-terminus collapse (codonsΔ 1–98) of pFLAG-IE1. BamHI and AccI double digestion of pFLAG-IE1 generated the pFLAG-IE1NCdel mutant (codonsΔ 1–98 and 426–474). The IET mutant (codonsΔ 234–474) was synthetically generated (DNA 2.0) before being subcloned as a BglII fragment into the pEGFP-C1 expression vector (Clontech) to give rise to pGFP-IET (Fig 2).
Generation of unique exon GP122 and GP123 epitope- or GFP-tagged mammalian expression constructs
Unique exon GP123 (codons 116–474) was initially PCR amplified using primers FGP123exBam and RGP123exBam (Table S2) and cloned into the pCMV2a expression vector (Clontech) as a BamHI fragment. GP123 was subsequently isolated from pCMVGP123ex as a BamH1 fragment and sub-cloned into the pAcGFP-C3 (Clontech) expression vector to generate a N-terminus GFP-GP123 fusion protein expression construct (pGFP-GP123). Similarly, GP122 was PCR amplified using primers FGP122exBam and RGP122exBam (Table S2) and cloned into the pCMV2a expression vector as a BamHI fragment. Unique exon GP122 BamHI fragment was inserted into the pAcGFP-C3 (Clontech) expression vector to generate plasmid pGFP-GP122. Constructs were confirmed by sequencing.
Generation of GP123KmFRT vector, GP123Km intermediate and IET BAC mutants
As previously described, an inducible ET recombination system (Gene Bridges) was used to generate GPCMV BAC mutants. The second-generation GPCMV BAC was used to generate the GP123Km intermediate and IET BAC mutants. The GP123 ORF was partially deleted (GPCMV nucleotide positions 190621–190830) (Fig S5(i)) by targeted mutagenesis of the GPCMV WT BAC to generate a C-terminus truncated IE1 (codonsΔ 233–474). A GP123 shuttle vector was synthetically generated to allow sequence specific deletion of GP123 and a KmFRT drug resistant cassette was introduced into the BamH1 site between flanking 5′ and 3′ sequences to enable selection of GP123 mutants (pGP123KmFRT). The 5′ flanking sequence ranged from nucleotide 856–564 within GP123. The 3′ flanking sequence encoded nucleotides 354–84 within GP123. The pGP123KmFRT shuttle vector was linearized by EcoRI digestion, purified, and used in homologous recombination reaction with a second-generation GPCMV BAC. This gave rise to the GP123Km GPCMV BAC intermediate. Positive clones were identified by HindIII digestion, as insertion of the Km cassette into the GP123 locus introduced an additional HindIII site. HindIII digestion cut the original 25.7 kb fragment (Fig S5(ii); yellow, lane 2) into a 2.6 kb and 24 kb band (Fig S5(ii); blue, lane 1). Next, the Km cassette was removed by Flp-FRT recombinase system which generated the IET GPCMV BAC mutant. In restriction profile analysis, removal of the Km cassette resulted in partial deletion of GP123 that decreased the original 25.7 kb fragment (Fig S5(ii); yellow, lane 4) by 200 bp in the IET BAC mutant (Fig S5(ii); blue, lane 3). Primers FGP123exBm and RGP123exBm (Table S2) were used to verify correct genome modification of the GP123 locus in the IET mutant by PCR (Fig S5(iii)). The GP123 WT locus was 1.1 kb in size (lane 2) and the IET mutant was 0.9 kb (lane 1). The intermediate IETKm mutant BAC was 2.1 kb in size (lane 3) (Fig S5(iii)).
Generation of individual GPCMV IE1 and IE2 knockout BAC mutants, analysis, and rescue
The GP123 knockout BAC was generated by Km cassette insertion and a 496 bp deletion of the GP123 ORF (codons 62–226) within a first-generation GPCMV BAC. The FGP123Km and RGP123Km primers (Table S2) were used to PCR amplify the Km cassette with homolog sequence to GP123 exon. The product was band isolated and used in homologous recombination with WT GPCMV BAC. Mutant GPCMV BAC clones were confirmed by restriction enzyme profile analysis with EcoRI and also by PCR analysis of the modified GP123 locus (data not shown).
The GP122 knockout shuttle vector was generated by insertion of a kanamycin (Km) cassette into the GP122 ORF at a unique EcoRV site (GPCMV nucleotide position 188745) to disrupt the GP122 ORF at codon 382 (IE2 at codon 498) in a pUC construct encoding GP122 as an EcoRI fragment. This generated plasmid pNEBGP122Km. Using primers FGP122Ec and RGP122Ec (Table S2), the GP122Km cassette was PCR amplified, band isolated and used in knockout mutagenesis of the first-generation GPCMV WT BAC to generate GP122Km BAC. Mutant clones were confirmed by restriction enzyme profile analysis with HindIII and PCR analysis of the GP122 locus (data not shown).
To generate recombinant viruses, large-scale GPCMV BAC DNA MAXI preps of IE1 and IE2 mutants were prepared and transfected onto GPL cells using Lipofectamine 2000 (Invitrogen) as previously described. GPCMV BACs encoded a GFP reporter gene to enable development of virus plaques from single transfected cells. For IE2 rescue virus the WT unique exon GP122 plasmid was co-transfected with GP122Km BAC DNA onto GPL cells. For IE1 rescue virus, the WT GP123 locus and flanking sequence was PCR amplified using FIEexonRes and RIEexonRes primers from WT GPCMV DNA (Table S2) and co-transfected with GP123Km BAC DNA onto GPL cells. Transfections were followed for 3–4 weeks and green fluorescent protein (GFP)-positive cells or plaques identified by microscopy.
Epitope-tagged ND10 component mammalian expression constructs
Epitope-tagged expression constructs for gpPML protein (accession #XP_003462251), gpDaxx ( #XP_013012418), gpSp100 (#XM_013157411) protein, and gpATRX (#XM_003471056) were synthetically generated (DNA 2.0). The gpPML and gpATRX ORFs were N-terminal myc epitope-tagged. The gpSp100 ORF was N-terminal tagged with a HA epitope tag. The gpDaxx ORF was C-terminally tagged with a FLAG epitope. Each ORF was placed under the control of the HCMV MIE promoter 5′ end and a SV40 polyadenylation sequence at the 3′ end.
Generation of HSV-1 ICP0 BAC mutant
The predicted HSV-1 ICP0 coding sequence was based on the complete (17+) viral genome sequence (accession #NC_001806). In the HSV-1 genome, there are two copies of ICP0 located in the inverted repeats b and b’ (Fig S6(ii)). The ICP0 gene is multi-spliced. The coding sequence co-ordinates for ICP0 are 2262–2318 for exon 1, 3084–3750 for exon 2, and 3887–5490 for exon 3. The second ICP0 ORF extends from exon 1 (120884–122487) through exon 2 (122624–1232900 to exon 3 (124056–124112). Both ICP0 loci were knockout out by targeted mutagenesis of the HSV-1 BAC using a Km drug resistance donor cassette flanked by ICP0 target regions (Fig S6(ii)) to introduce partial deletion of exon 2 and exon 3 (Fig S6(i)). Primers FHSVKOO and RHSVKOO (Table S2) were designed to amplify a Km cassette thereby introducing 5′ and 3′ sequences homologous to the ICP0 target region. The PCR product was band isolated, purified and used in homologous recombination with WT HSV-1 BAC. Individual clones of HSV-1 ICP0Δ BAC DNA were analyzed by HindIII restriction digestions to verify correct HSV-1 genome modification (Fig S6(iii)). Insertion of a Km cassette introduced additional HindIII sites. For the first locus, HindIII digestion resulted in the loss of the 26.5 kb band to generate two new bands of 8 kb and 18 kb. For the second locus, HindIII digestion resulted in the loss of the 34.6 kb band and gave rise to two bands of 10.5 kb and 23.7 kb. Knockout of ICP0 was confirmed by western blot (Fig S6(iv)).
Immunofluorescence and Immunoprecipitation assays
Immunofluorescence, immunoprecipitation, and western blot assays were performed as previously described. For western blots, anti-epitope tag primary antibodies were used at 1/1000. For immunofluorescence assays, primary antibodies were used at 1/500: mouse anti-HSV-1 ICP0 (Santa Cruz); rabbit anti-HA (Novus Biologicals); mouse anti-FLAG (Novus Biological); mouse anti-GFP (Santa Cruz); mouse anti-Myc-c (Novus Biologicals); rabbit anti-GPCMV IE2. Secondary antibodies anti-rabbit or anti-mouse IgG HRP-linked secondary antibodies for western blot (Cell Signaling) were used at 1/5000 and anti-mouse/rabbit IgG FITC or TRITC were used at 1/1000 for immunofluorescent staining. High titer recombinant defective adenovirus expressing GFP (AdGFP) was generated by Welgen Inc..
IFN or JAK/STAT inhibitor treatment of vIET mutant
On day 0, GPL cells were pre-treated with 100 IU/ml or 1000 IU/ml universal human IFN-I (PBL Assay Science). The next day, IFN pre-treated and control untreated cells were infected separately with either WT GPCMV or vIET mutant virus (0.1 MOI) and incubated at 37°C with 5% CO2. After 72 hrs, supernatants and monolayers were harvested and titrated in duplicate on GPL cells as previously described. IFN-I IU/ml were confirmed by VSV growth experiments in the presence or absence of different concentrations of IFN-I on GPL cells prior to reported studies (data not shown). GPL cells were pre-treated with 10 μM of the JAK1/JAK2 inhibitor ruxolitinib (Invivogen) for 1–6 hrs. Depending on the experimental condition, some cells were treated with 100 IU/ml of universal human IFN-I (PBL Assay Science). Cells were subsequently infected with vIET (0.1 MOI) for 1 hr. Cells were washed and fresh F-12 medium supplemented with 10% FCS plus 10 μM ruxolitinib were added then incubated at 37°C with 5% CO2 for 4 days. Cells and supernatant were harvested and virus titrated on GPL cells.
gpPML-mediated inhibition assay
On day 1, GPL cells in 6 well dishes were transfected with gpPML (3μg) expression plasmid or GFP expression vector (pEGFPC-1). The following day, cells were infected with either HSV-1 ICP0Δ or GPCMV IET mutant viruses at 0.1 MOI for 1hr. The inoculum was washed and fresh media added to the cells. Plates were incubated at 37°C with 5% CO2 for 3 days before cell monolayer and supernatant were harvested and titrated on GPL cells as previous described.
GPCMV complementation assay of HSV-1 ICP0 mutant virus
On day 0, GPL cells in 6 well dishes were infected with 3×104 pfu of GPCMV vIET or GPCMV WT viruses. On day 1, cells were infected with GFP-labeled HSV ICP0Δ virus (102 pfu/cell). Cells and supernatant were harvested 5 days p.i. and titrated on U2OS cells for HSV-1.
Small interfering RNA (siRNA) knockdown assay against gpPML and gpDaxx
On day 0, 100nM of each siRNA against gpPML and gpDaxx were transfected overnight onto GPL cells as previously described. On day 1, cells were infected with vIET (0.01 MOI) for 1hr. Cells were washed and fresh F-12 medium supplemented with 10% FCS added. New siRNA (100nM each) was transfected overnight onto vIET infected GPL cells. Cells were incubated at 37°C with 5% CO2. Cells and supernatant were harvested 3 days p.i. and virus titrated on GPL cells.
Supplementary Material
Table S1. Summary of calculated and observed protein sizes with predicted sumoylation sites and HCMV homology.
Table S2. Oligonucleotides used for PCR reactions.
Fig S1. GPCMV IE1 specific 5′ and 3′ RACE analysis. IE1 specific 5′ and 3′ RACE analysis was performed as described in materials and methods. Positive clones were confirmed by PCR and restriction profile analysis before being sequenced. Sequence analysis of duplicate 5′ and 3′ RACE clones confirmed the published GPCMV IE1 cDNA genome sequence (accession #AB592928).
Fig S2. Nuclear expression of unique exons GP123 (IE1) and GP122 (IE2). (i) Schematic representation of GFP-tagged GP123 and GP122 fusion protein expression constructs. (ii) GFP-GP123 and GFP-GP122 protein expression by western blot analysis and immunofluorescence. A–C, expression and localization of GFP-GP123. D–F, expression and localization of GFP-GP122. Lane 1, plasmid transfect GPL cells. Lane 2, mock transfected GPL cells. GFP-GP123 and GFP-GP122 detected by fluorescence (cell localization) or primary anti-epitope Ab and anti-mouse IgG-HRP secondary (western blot). (iii) Predicted nuclear localization signals for GPCMV IE1 and IE2 as described in material and methods. cNLS Mapper was used to predict nuclear localization signal sequences for IE1 and IE2 using a cutoff score of 7.
Fig S3. Co-localization of SUMO-1 with gpPML, gpDaxx, GPCMV IE1 and IE2. A–D, co-localization of SUMO-HA (A) and GFP-IE1 (B). C is the merged image of A and B. D represents the overlay with DAPI. E–H, co-localization of SUMO-HA (E) with GFP-IE2 (F). G is the merged image for E and F, with H being the overlay of G with DAPI. I–L, co-localization of SUMO-HA (I) and gpPMLmyc (J). K is the merge of I and J. L represents the overlay with DAPI. M–P, co-localization of SUMO-HA (M) and gpDaxxFLAG (N). O is the merged image for M and N, with P being the overlay with DAPI. Detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG FITC or TRITC.
Fig S4. Prediction of intrinsically unstructured regions and hydrophilicity of GPCMV, HCMV, and MCMV IE1 protein. The IUPred program (“structured domains”) was used to predict and plot the tendency of intrinsic disorder in the GPCMV, HCMV, and MCMV IE1 proteins. Scores ≥ 0.5 (blue line) indicate disorder, while scores < 0.5 represent structured regions. Kyte-Doolittle hydrophilicity analysis (MacVector Inc.) of GPCMV, HCMV, and MCMV IE1 protein. (i) Disorder score for GPCMV IE1 predicts the presence of a central globular core domain (aa 104–411). The C-terminus contains a hydrophilic stretch (aa 415–474) (red box). (ii) The HCMV IE1 (accession #GQ244523.1) globular core domain ranges from aa 77–378. The C-terminus contains a hydrophilic stretch (aa 458–491) (red box). (iii) Disorder score for MCMV IE1 (accession #CCE57451) predicts two globular core domains (aa 41–72 and 87–372). The C-terminus contains a hydrophilic stretch (aa 393–595) (red box).
Fig S5. Generation of the vIET knockout virus. (i) Genome sequence of GPCMV unique exon GP123 (yellow). The 200 bp deletion introduced within the unique exon is underlined. Primer sequences used for confirmation of GP123 locus modification are bolded (Table S2). (ii) Restriction profile analysis of the GPCMV IETKm BAC mutant intermediate (lane 1) and IET mutant (lane 3) compared to WT GPCMV BAC (lanes 2 and 4). HindIII digestion of GPCMV IETKm intermediate BAC DNA cut the original 25.7 kb fragment (yellow, lane 2) into a 2.6 kb and 24 kb band (blue, lane 1). HindIII restriction profile analysis of GPCMV IET BAC DNA decreased the original 25.7 kb fragment (yellow, lane 4) by 200bp (blue, lane 3). (iii) PCR analysis of IETKm intermediate and IET compared to WT GPCMV. Lane 1, IET BAC (0.9 kb). Lane 2, WT BAC (1.1 kb). Lane 3, IETKm BAC intermediate (2.1 kb).
Fig S6. Generation of HSV-1 ICP0Δ double knockout virus. (i) Homologous recombination introduced a partial deletion within exons 2 (yellow) and 3 (grey) of both ICP0 loci. Primer pair sequences (Table S2) used to amplify the Km cassette used in homologous recombination are in bold. Deleted sequence within exons 2 and 3 is underlined. (ii) Schematic representation of the HSV-1 genome and insertion of a Km cassette into ICP0 loci within inverted repeats b and b’ by homologous recombination with WT HSV BAC. (iii) HindIII restriction profile analysis confirmed modification of both loci by insertion of a Km cassette. HindIII digestion resulted in the loss of the 26.5 kb band (lane 1, blue) to generate two new bands of 8 kb and 18 kb (lane 2, yellow). For the second locus, HindIII digestion resulted in the loss of the 34.6 kb band (lane 1, red) and gave rise to two bands of 10.5 kb and 23.7 kb (lane 2, green). (iv) Western blot analysis of HSV-I ICP0 knockout virus detected by primary anti-HSV ICP0 Ab and secondary anti-mouse IgG-HGP. Lane 3, ICP0Δ virus. Lane 4, ICP0 expression plasmid. Lane 5, mock infected GPL cells. Lane 6, WT HSV-1 virus.
Fig S7. HSV-1 ICP0 co-localization with guinea pig ND10 components. (i) A–D, ICP0Δ mutant virus fails to disrupt gpPMLmyc (A and B), showing a phenotype that is identical to transiently expressed gpPMLmyc only (C and D). (ii) GPL cells were transfected with expression plasmids encoding gpPMLmyc, gpDaxxFLAG, or gpSp100HA in the presence of ICP0 to evaluate cellular co-localization. E–G, ICP0 transient expression alone. G represents E merged with F. H–J, co-localization of ICP0 and gpPMLmyc. ICP0 (H) and gpPMLmyc (I) individually expressed in the same cell. J is the merged image overlaid with DAPI. K–M, colocalization of ICP0 (K) and gpDaxxFLAG (L). M being merged image overlaid with DAPI. NP, co-localization of ICP0 (N) with gpSp100HA (O). P is merged image overlaid with DAPI. Imunofluorescence detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG FITC or TRITC.
Acknowledgments
The ICP0 expression plasmid was a kind gift from Dr. R. Everett (University of Glasgow, UK). The SUMO-1 expression plasmid was generously provided by Dr. V. G. Wilson (Texas A&M University, Health Science Center). The second-generation GPCMV BAC N13R10 was a gift from Dr. M. McVoy (Virginia Commonwealth University). This research was funded by National Institute of Health (NIH/NIAID); R01AI100933; R01AI098984; R21AI090156.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.La Rosa C, Diamond DJ. The immune response to human CMV. Future Virol. 2012;7(3):279–293. doi: 10.2217/fvl.12.8. http://dx.doi.org/10.2217/fvl.12.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wreghitt TG, Teare EL, Sule O, Devi R, Rice P. Cytomegalovirus infection in immunocompetent patients. Clin Infect Dis. 2003;37(12):1603–1606. doi: 10.1086/379711. http://dx.doi.org/10.1086/379711. [DOI] [PubMed] [Google Scholar]
- 3.Ross SA, Boppana SB. Congenital cytomegalovirus infection: outcome and diagnosis. Semin Pediatr Infect Dis. 2005;16(1):44–49. doi: 10.1053/j.spid.2004.09.011. http://dx.doi.org/10.1053/j.spid.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Cannon MJ. Congenital cytomegalovirus (CMV) epidemiology and awareness. J Clin Virol. 2009;46(4):S6–S10. doi: 10.1016/j.jcv.2009.09.002. http://dx.doi.org/10.1016/j.jcv.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Peckham CS. Cytomegalovirus in the neonate. J Antimicrob Chemother. 1989;23(Suppl E):17–21. doi: 10.1093/jac/23.suppl_e.17. http://dx.doi.org/10.1093/jac/23.suppl_E.17. [DOI] [PubMed] [Google Scholar]
- 6.Cannon MJ, Davis KF. Washing our hands of the congenital cytomegalovirus disease epidemic. BMC Public Health. 2005;5:70. doi: 10.1186/1471-2458-5-70. http://dx.doi.org/10.1186/1471-2458-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Berg PJ, van Stijn A, Ten Berge IJ, van Lier RA. A fingerprint left by cytomegalovirus infection in the human T cell compartment. J Clin Virol. 2008;41(3):213–217. doi: 10.1016/j.jcv.2007.10.016. http://dx.doi.org/10.1016/j.jcv.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Khan N, Cobbold M, Keenan R, Moss PA. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J Infect Dis. 2002;185(8):1025–1034. doi: 10.1086/339963. http://dx.doi.org/10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 9.Kern F, Surel IP, Faulhaber N, Frömmel C, Schneider-Mergener J, Schönemann C, Reinke P, Volk HD. Target structures of the CD8(+)-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73(10):8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. http://www.ncbi.nlm.nih.gov/pubmed/10482568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson L, Piccinini G, Lilleri D, Revello MG, Wang Z, Markel S, Diamond DJ, Luzuriaga K. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection. J Immunol. 2004;172(4):2256–2264. doi: 10.4049/jimmunol.172.4.2256. http://www.ncbi.nlm.nih.gov/pubmed/14764694. [DOI] [PubMed] [Google Scholar]
- 11.Alp NJ, Allport TD, Van Zanten J, Rodgers B, Sissons JG, Borysiewicz LK. Fine specificity of cellular immune responses in humans to human cytomegalovirus immediate-early 1 protein. J Virol. 1991;65(9):4812–4820. doi: 10.1128/jvi.65.9.4812-4820.1991. http://www.ncbi.nlm.nih.gov/pubmed/1714519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pahl-Seibert M-F, Juelch M, Podlech J, Thomas D, Deegen P, Reddehase MJ, Holtappels R. Highly Protective In Vivo Function of Cytomegalovirus IE1 Epitope-Specific Memory CD8 T Cells Purified by T-Cell Receptor-Based Cell Sorting. J Virol. 2005;79(9):5400–5413. doi: 10.1128/JVI.79.9.5400-5413.2005. http://dx.doi.org/10.1128/JVI.79.9.5400-5413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slezak SL, Bettinotti M, Selleri S, Adams S, Marincola FM, Stroncek DF. CMV pp65 and IE-1 T cell epitopes recognized by healthy subjects. J Transl Med. 2007;5:17. doi: 10.1186/1479-5876-5-17. https://dx.doi.org/10.1186/1479-5876-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunde T, Kichner A, Hoffmeister B, Habedank D, Hetzer R, Cherepnew G, Proesch S, Reineke P, Volk H-D, Lehmkuhl H, Kern F. Protection from cytomegalovirus after transplantation is correlated with immediate early 1–specific CD8 T cells. JEM. 2005;201(7):1031–1036. doi: 10.1084/jem.20042384. http://dx.doi.org/10.1084/jem.20042384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Roy E, Baron M, Faigle W, Clément D, Lewinsohn DM, Streblow DN, Nelson JA, Amigorena S, Davignon JL. Infection of APC by human cytomegalovirus controlled through recognition of endogenous nuclear immediate early protein 1 by specific CD4(+) T lymphocytes. J Immunol. 2002;169(3):1293–1301. doi: 10.4049/jimmunol.169.3.1293. http://dx.doi.org/10.4049/jimmunol.169.3.1293. [DOI] [PubMed] [Google Scholar]
- 16.Schmolke S, Kern HF, Drescher P, Jahn G, Plachter B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol. 1995;69(10):5959–5968. doi: 10.1128/jvi.69.10.5959-5968.1995. http://www.ncbi.nlm.nih.gov/pubmed/7666500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93(21):11321–11326. doi: 10.1073/pnas.93.21.11321. http://www.ncbi.nlm.nih.gov/pubmed/8876134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bialas KM, Tanaka T, Tran D, Varner V, Cisneros De La Rosa E, Chiuppesi F, Wussow F, Kattenhorn L, Macri S, Kunz EL, Estroff JA, Kirchherr J, Yue Y, Fan Q, Lauck M, O'Connor DH, Hall AH, Xavier A, Diamond D, Barry PA, Kaur A, Permar SR. Maternal CD4+ T cells protect against severe congenital cytomegalovirus disease in a novel nonhuman primate model of placental cytomegalovirus transmission. Proc Natl Acad Sci U S A. 2015;112(44):13645–13650. doi: 10.1073/pnas.1511526112. http://dx.doi.org/10.1073/pnas.1511526112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi KY, Root M, McGregor A. A Novel Non-Replication-Competent Cytomegalovirus Capsid Mutant Vaccine Strategy Is Effective in Reducing Congenital Infection. J Virol. 2016;90(17):7902–7919. doi: 10.1128/JVI.00283-16. http://dx.doi.org/10.1128/JVI.00283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleiss MR, Lacayo JC, Belkaid Y, McGregor A, Stroup G, Rayner J, Alterson K, Chulay JD, Smith JF. Preconceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J Infect Dis. 2007;195(6):789–798. doi: 10.1086/511982. http://dx.doi.org/10.1086/511982. [DOI] [PubMed] [Google Scholar]
- 21.Paulus C, Nevels M. The human cytomegalovirus major immediate-early proteins as antagonists of intrinsic and innate antiviral host responses. Viruses. 2009;1(3):760–779. doi: 10.3390/v1030760. http://dx.doi.org/10.3390/v1030760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenberg RM, Witte PR, Stinski MF. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56(3):665–675. doi: 10.1128/jvi.56.3.665-675.1985. http://www.ncbi.nlm.nih.gov/pubmed/2999423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim ET, Kim Y-E, Kim YJ, Lee MK, Hayward GS, Ahn J-H. Analysis of human cytomegalovirus-encoded SUMO targets and temporal regulation of SUMOylation of the immediate-early proteins IE1 and IE2 during infection. PLoS One. 2014;9(7):e103308. doi: 10.1371/journal.pone.0103308. http://dx.doi.org/10.1371/journal.pone.0103308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arend KC, Ziehr B, Vincent HA, Moorman NJ. Multiple Transcripts Encode Full-Length Human Cytomegalovirus IE1 and IE2 Proteins during Lytic Infection. J Virol. 2016;90(19):8855–8865. doi: 10.1128/JVI.00741-16. http://dx.doi.org/10.1128/JVI.00741-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaves RF, Mocarski ES. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72(1):366–379. doi: 10.1128/jvi.72.1.366-379.1998. http://www.ncbi.nlm.nih.gov/pubmed/9420235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229(1):155–158. doi: 10.1006/excr.1996.0353. http://dx.doi.org/10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 27.Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growth control. Gene. 2002;290(1–2):19–34. doi: 10.1016/s0378-1119(02)00566-8. http://dx.doi.org/10.1016/S0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez V, Spector DH. Subversion of cell cycle regulatory pathways. Curr Top Microbiol Immunol. 2008;325:243–262. doi: 10.1007/978-3-540-77349-8_14. http://www.ncbi.nlm.nih.gov/pubmed/18637510. [DOI] [PubMed] [Google Scholar]
- 29.Marchini A, Liu H, Zhu H. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol. 2001;75(4):1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. http://dx.doi.org/10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang YN, Jeang KT, Lietman T, Hayward GS. Structural Organization of the Spliced Immediate-Early Gene Complex that Encodes the Major Acidic Nuclear (IE1) and Transactivator (IE2) Proteins of African Green Monkey Cytomegalovirus. J Biomed Sci. 1995;2(2):105–130. doi: 10.1007/BF02253062. http://www.ncbi.nlm.nih.gov/pubmed/11725047. [DOI] [PubMed] [Google Scholar]
- 31.Sandford GR, Ho K, Burns WH. Characterization of the major locus of immediateearly genes of rat cytomegalovirus. J Virol. 1993;67(7):4093–4103. doi: 10.1128/jvi.67.7.4093-4103.1993. http://www.ncbi.nlm.nih.gov/pubmed/8389919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keil GM, Ebeling-Keil A, Koszinowski UH. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61(6):1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC254196/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maul GG. Nuclear domain 10, the site of DNA virus transcription and replication. Bioassays. 1998;20(8):660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. http://dx.doi.org/10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80(16):8006–8018. doi: 10.1128/JVI.00743-06. http://dx.doi.org/10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. doi: 10.1038/nrm2277. http://dx.doi.org/10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 36.Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. 2011;31(1):145–158. doi: 10.1089/jir.2010.0111. http://dx.doi.org/10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 37.Tavalai N, Stamminger T. New insights into the role of the subnuclear structure ND10 for viral infection. BBA. 2008;1783(11):2207–2221. doi: 10.1016/j.bbamcr.2008.08.004. http://dx.doi.org/10.1016/j.bbamcr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Stadler M, Chelbi-Alix MK, Koken MH, Venturini L, Lee C, Saïb A, Quignon F, Pelicano L, Guillemin MC, Schindler C, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11(12):2565–2573. http://www.ncbi.nlm.nih.gov/pubmed/8545113. [PubMed] [Google Scholar]
- 39.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. http://dx.doi.org/10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 40.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115(Pt 16):3319–3330. doi: 10.1242/jcs.115.16.3319. http://www.ncbi.nlm.nih.gov/pubmed/12140263. [DOI] [PubMed] [Google Scholar]
- 41.Preston CM, Nicholl MJ. Role of the cellular protein hDaxx in human cytomegalvirus immediate-early gene expression. J Gen Virol. 2006;87(Pt 5):1113–1121. doi: 10.1099/vir.0.81566-0. http://dx.doi.org/10.1099/vir.0.81566-0. [DOI] [PubMed] [Google Scholar]
- 42.Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80(8):3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. http://dx.doi.org/10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavalai N, Stamminger T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses. 2009;1(3):1240–1264. doi: 10.3390/v1031240. http://dx.doi.org/10.3390/v1031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20(49):7223–7233. doi: 10.1038/sj.onc.1204765. http://dx.doi.org/10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 45.Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. 2010;6(1):51–67. doi: 10.7150/ijbs.6.51. http://www.ncbi.nlm.nih.gov/pubmed/20087442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.BioGRID3.4 interaction database. 2016 http://thebiogrid.org/. [Online]
- 47.Lallemand-Breitenbach V, de Thé H. PML Nuclear Bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. http://dx.doi.org/10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiner S, Wodrich H. Virion factors that target Daxx to overcome intrinsic immunity. J Virol. 2013;87(19):10412–10422. doi: 10.1128/JVI.00425-13. http://dx.doi.org/10.1128/JVI.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129(2–3):278–287. doi: 10.1006/jsbi.2000.4239. https://dx.doi.org/10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 50.Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20(49):7243–7249. doi: 10.1038/sj.onc.1204758. http://dx.doi.org/10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 51.Wilson VG, Rangasamy D. Intracellular targeting of proteins by sumoylation. Exp Cell Res. 2001;271(1):57–65. doi: 10.1006/excr.2001.5366. http://dx.doi.org/10.1006/excr.2001.5366. [DOI] [PubMed] [Google Scholar]
- 52.Cubeñas-Potts C, Matunis MJ. SUMO: A Multifaceted Modifier of Chromatin Structure and Function. Developmental Cell. 2013;24(1):1–12. doi: 10.1016/j.devcel.2012.11.020. http://dx.doi.org/10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. JCB. 1997;139(7):1621–1634. doi: 10.1083/jcb.139.7.1621. http://www.ncbi.nlm.nih.gov/pubmed/9412458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang MS, Ryu SW, Kim E. Modification of Daxx by small ubiquitin-related modifier-1. Biochem Biophys Res Commun. 2002;295(2):495–500. doi: 10.1016/s0006-291x(02)00699-x. http://www.ncbi.nlm.nih.gov/pubmed/12150977. [DOI] [PubMed] [Google Scholar]
- 55.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ETH, Strauss JF, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147(2):221–234. doi: 10.1083/jcb.147.2.221. http://www.ncbi.nlm.nih.gov/pubmed/10525530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10(5):538–546. doi: 10.1038/ncb1716. http://dx.doi.org/10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 57.Rivera-Molina YA, Martínez FP, Tang Q. Nuclear domain 10 of the viral aspect. World J Virol. 2013;2(3):110–122. doi: 10.5501/wjv.v2.i3.110. http://dx.doi.org/10.5501/wjv.v2.i3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74(Pt12):2679–2690. doi: 10.1099/0022-1317-74-12-2679. https://dx.doi.org/10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 59.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72(8):6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. http://www.ncbi.nlm.nih.gov/pubmed/9658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chelbi-Alix MK, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18(4):935–941. doi: 10.1038/sj.onc.1202366. http://dx.doi.org/10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 61.Everett RD, Bell AJ, Lu Y, Orr A. The replication defect of ICP0-null mutant herpes simplex virus 1 can be largely complemented by the combined activities of human cytomegalovirus proteins IE1 and pp71. J Virol. 2013;87(2):987–990. doi: 10.1128/JVI.01103-12. http://dx.doi.org/10.1128/JVI.01103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn J-H, Hayward GS. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. Virol. 1997;71(6):4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. http://www.ncbi.nlm.nih.gov/pubmed/9151854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn J-H, Brignole EJ, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18(8):4899–4913. doi: 10.1128/mcb.18.8.4899. http://www.ncbi.nlm.nih.gov/pubmed/9671498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scherer M, Klingl S, Sevvana M, Otto V, Schilling E-M, Stump JD, Müller R, Reuter N, Sticht H, Muller YA, Stamminger T. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog. 2014;10(11):e1004512. doi: 10.1371/journal.ppat.1004512. http://dx.doi.org/10.1371/journal.ppat.1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang ES, Zhang Z, Cai H, Huang DY, Huong SM, Cha CY, Huang ES. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J Virol. 2009;83(23):12388–12398. doi: 10.1128/JVI.00304-09. http://dx.doi.org/10.1128/JVI.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poma EE, Kowalik TF, Zhu L, Sinclair JH, Huang ES. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70(11):7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. http://www.ncbi.nlm.nih.gov/pubmed/8892909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci USA. 2004;101(49):17234–17239. doi: 10.1073/pnas.0407933101. http://dx.doi.org/10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barry PA, Strelow L. Development of Breeding Populations of Rhesus Macaques (Macaca mulatta) That Are Specific Pathogen-free for Rhesus Cytomegalovirus. Comp Med. 2008;58(1):43–46. http://www.ncbi.nlm.nih.gov/pubmed/19793455. [PMC free article] [PubMed] [Google Scholar]
- 69.Mess A. The guinea pig placenta: model of placental growth dynamics. Placenta. 2007;28(8–9):812–815. doi: 10.1016/j.placenta.2007.02.005. http://dx.doi.org/10.1016/j.placenta.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, Mysorekar IU, Diamond MS. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell. 2016;165(5):1081–1091. doi: 10.1016/j.cell.2016.05.008. http://dx.doi.org/10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson KP. Mouse Cytomegalovirus: Placental Infection. J Infect Dis. 1969;120(4):445–450. doi: 10.1093/infdis/120.4.445. http://www.ncbi.nlm.nih.gov/pubmed/4309994. [DOI] [PubMed] [Google Scholar]
- 72.Rossant J, Cross JC. Placental development: Lessons from mouse mutants. Nat Rev Genet. 2001;2(7):538–548. doi: 10.1038/35080570. http://dx.doi.org/10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 73.Schleiss MR, McVoy MA. Guinea Pig Cytomegalovirus (GPCMV): A Model for the Study of the Prevention and Treatment of Maternal-Fetal Transmission. Future Virol. 2010;5(2):207–217. doi: 10.2217/fvl.10.8. https://dx.doi.org/10.2217/fvl.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schleiss MR, McGregor A, Choi Y, Date SV, Cui X, McVoy M. Analysis of the nucleotide sequence of the guinea pig cytomegalovirus (GPCMV) genome. Virol J. 2008;5:139. doi: 10.1186/1743-422X-5-139. http://dx.doi.org/10.1186/1743-422X-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamada S, Nozawa N, Katano H, Fukui Y, Tsuda M, Tsutsui Y, Kurane I, Inoue N. Characterization of the guinea pig cytomegalovirus genome locus that encodes homologs of human cytomegalovirus major immediate-early genes, UL128, and UL130. Virol. 2009;391(1):99–106. doi: 10.1016/j.virol.2009.05.034. http://dx.doi.org/10.1016/j.virol.2009.05.034. [DOI] [PubMed] [Google Scholar]
- 76.McGregor A, Liu F, Schleiss MR. Molecular, biological, and in vivo characterization of the guinea pig cytomegalovirus (CMV) homologs of the human CMV matrix proteins pp71 (UL82) and pp65 (UL83) J Virol. 2004;78(18):9872–9889. doi: 10.1128/JVI.78.18.9872-9889.2004. http://dx.doi.org/10.1128/JVI.78.18.9872-9889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGregor A, Choi KY, Schleiss MR. Guinea pig cytomegalovirus GP84 is a functional homolog of the human cytomegalovirus (HCMV) UL84 gene that can complement for the loss of UL84 in a chimeric HCMV. Virol. 2011;410(1):76–87. doi: 10.1016/j.virol.2010.10.028. http://dx.doi.org/10.1016/j.virol.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGregor A, Choi KY, Cui X, McVoy MA, Schleiss MR. Expression of the human cytomegalovirus UL97 gene in a chimeric guinea pig cytomegalovirus (GPCMV) results in viable virus with increased susceptibility to ganciclovir and maribavir. Antiviral Res. 2008;78(3):250–259. doi: 10.1016/j.antiviral.2008.01.008. http://dx.doi.org/10.1016/j.antiviral.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coleman S, Hornig J, Maddux S, Choi KY, McGregor A. Viral Glycoprotein Complex Formation, Essential Function and Immunogenicity in the Guinea Pig Model for Cytomegalovirus. PLOS ONE. 2015;10(8):e0135567. doi: 10.1371/journal.pone.0135567. http://dx.doi.org/10.1371/journal.pone.0135567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coleman S, Choi KY, Root M, McGregor A. A homolog pentameric complex dictates viral epithelial tropism, pathogenicity and congenital infection rate in guinea pig cytomegalovirus. PLoS. 2016;12(7):e1005755. doi: 10.1371/journal.ppat.1005755. http://dx.doi.org/10.1371/journal.ppat.1005755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barry PA, Alcendor DJ, Power MD, Kerr H, Luciw PA. Nucleotide sequence and molecular analysis of the rhesus cytomegalovirus immediate-early gene and the UL121-117 open reading frames. J Virol. 1996;215(1):61–72. doi: 10.1006/viro.1996.0007. http://dx.doi.org/10.1006/viro.1996.0007. [DOI] [PubMed] [Google Scholar]
- 82.cNLS Mapper. [Online] 2016 http://nls-mapper.iab.keio.ac.jp/cgibin/NLS_Mapper_form.cgi.
- 83.Ahn J-H, Jang W-J, Hayward GS. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73(12):10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. http://www.ncbi.nlm.nih.gov/pubmed/10559364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.EMBL-EBI. Clustal Omega. Pairwise Sequence Alignment. 2016 [Online] http://www.ebi.ac.uk/Tools/psa/
- 85.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. http://dx.doi.org/10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 86.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. http://www.ncbi.nlm.nih.gov/pubmed/9600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100(19):10635–10640. doi: 10.1073/pnas.1937626100. http://dx.doi.org/10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sourvinos G, Tavalai N, Berndt A, Spandidos DA, Stamminger T. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J Virol. 2007;81(18):10123–10136. doi: 10.1128/JVI.01009-07. http://dx.doi.org/10.1128/JVI.01009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reeves M, Woodhall D, Compton T, Sinclair J. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J Virol. 2010;84(14):7185–7194. doi: 10.1128/JVI.02231-09. http://dx.doi.org/10.1128/JVI.02231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martínez FP, Cruz Cosme RS, Tang Q. Murine cytomegalovirus major immediateearly protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J Gen Virol. 2010;91(Pt 11):2664–2676. doi: 10.1099/vir.0.022301-0. http://dx.doi.org/10.1099/vir.0.022301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shuai K, Liu B. Regulation of JAK–STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–911. doi: 10.1038/nri1226. http://dx.doi.org/10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 92.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–2473. doi: 10.1038/sj.onc.1203476. http://dx.doi.org/10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 93.Mesa RA. Ruxolitinib, a selective JAK1 and JAK2 inhibitor for the treatment of myeloproliferative neoplasms and psoriasis. IDrugs. 2010;13(6):394–403. http://www.ncbi.nlm.nih.gov/pubmed/20506062. [PubMed] [Google Scholar]
- 94.Lee HR, Huh YH, Kim YE, Lee K, Kim S, Ahn JH. N-terminal determinants of human cytomegalovirus IE1 protein in nuclear targeting and disrupting PML-associated subnuclear structures. Biochem Biophys Res Commun. 2007;356(2):499–504. doi: 10.1016/j.bbrc.2007.03.007. http://dx.doi.org/10.1016/j.bbrc.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 95.Delmas S, Martin L, Baron M, Nelson JA, Streblow DN, Davignon JL. Optimization of CD4+ T lymphocyte response to human cytomegalovirus nuclear IE1 protein through modifications of both size and cellular localization. J Immunol. 2005;175(10):6812–6819. doi: 10.4049/jimmunol.175.10.6812. http://dx.doi.org/10.4049/jimmunol.175.10.6812. [DOI] [PubMed] [Google Scholar]
- 96.Pizzorno MC, Mullen MA, Chang YN, Hayward GS. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65(7):3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. http://www.ncbi.nlm.nih.gov/pubmed/1645794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghazal P, Visser AE, Gustems M, García R, Borst EM, Sullivan K, Messerle M, Angulo A. Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J Virol. 2005;79(11):7182–7194. doi: 10.1128/JVI.79.11.7182-7194.2005. http://dx.doi.org/10.1128/JVI.79.11.7182-7194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sandford GR, Schumacher U, Ettinger J, Brune W, Hayward GS, Burns WH, Voigt S. Deletion of the rat cytomegalovirus immediate-early 1 gene results in a virus capable of establishing latency, but with lower levels of acute virus replication and latency that compromise reactivation efficiency. J Gen Virol. 2010;91(Pt 3):616–621. doi: 10.1099/vir.0.016022-0. http://dx.doi.org/10.1099/vir.0.016022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20(49):7266–7273. doi: 10.1038/sj.onc.1204759. http://dx.doi.org/10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 100.Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13(21):5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC395452/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lang M, Jegou T, Chung I, Richter K, Münch S, Udvarhelyi A, Cremer C, Hemmerich P, Engelhardt J, Hell SW, Rippe K. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123(Pt 3):392–400. doi: 10.1242/jcs.053496. http://dx.doi.org/10.1242/jcs.053496. [DOI] [PubMed] [Google Scholar]
- 102.Hofmann H, Sindre H, Stamminger T. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol. 2002;76(11):5769–5783. doi: 10.1128/JVI.76.11.5769-5783.2002. http://www.ncbi.nlm.nih.gov/pubmed/11992005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ishov AM, Vladimirova OV, Maul GG. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J Virol. 2002;76(15):7705–7712. doi: 10.1128/JVI.76.15.7705-7712.2002. http://www.ncbi.nlm.nih.gov/pubmed/12097584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lukashchuk V, McFarlane S, Everett RD, Preston CM. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J Virol. 2008;82(24):12543–12554. doi: 10.1128/JVI.01215-08. http://dx.doi.org/10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scherer M, Otto V, Stump JD, Klingl S, Müller R, Reuter N, Muller YA, Sticht H, Stamminger T. Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1–382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses. J Virol. 2015;90(3):1190–1205. doi: 10.1128/JVI.01973-15. https://dx.doi.org/10.1128/JVI.01973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ishov AM, Stenberg RM, Maul GG. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. JCB. 1997;138(1):5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84(8):4026–4040. doi: 10.1128/JVI.02597-09. http://dx.doi.org/10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bryant LA, Mixon P, Davidson M, Bannister AJ, Kouzarides T, Sinclair JH. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J Virol. 2000;74(16):7230–7237. doi: 10.1128/jvi.74.16.7230-7237.2000. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC112244/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fortunato EA, Sommer MH, Yoder K, Spector DH. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71(11):8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC192274/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pampin M, Simonin Y, Blondel B, Percherancier Y, Chelbi-Alix MK. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J Virol. 2006;80(17):8582–8592. doi: 10.1128/JVI.00031-06. https://dx.doi.org/10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24(3):331–339. doi: 10.1016/j.molcel.2006.09.013. http://dx.doi.org/10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hwang J, Kalejta RF. Human Cytomegalovirus Protein pp71 Induces Daxx SUMOylation. J Virol. 2009;83(13):6591–6598. doi: 10.1128/JVI.02639-08. http://dx.doi.org/10.1128/JVI.02639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim ET, Kim YE, Huh YH, Ahn JH. Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. J Virol. 2010;84(16):8111–8123. doi: 10.1128/JVI.00459-10. https://dx.doi.org/10.1128/JVI.00459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11(6):400–411. doi: 10.1038/nrmicro3015. http://dx.doi.org/10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 115.Maul GG, Everett RD. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75(Pt 6):1223–1233. doi: 10.1099/0022-1317-75-6-1223. http://dx.doi.org/10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 116.Smith MC, Boutell C, David J. HSV-1 ICP0: paving the way for viral replication. Future Virol. 2011;6(4):421–429. doi: 10.2217/fvl.11.24. http://dx.doi.org/10.2217/fvl.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ullman AJ, Hearing P. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J Virol. 2008;82(15):7325–7335. doi: 10.1128/JVI.00723-08. https://dx.doi.org/10.1128/JVI.00723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raghavan B, Cook CH, Trgovcich J. The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling. Viruses. 2014;6(4):1502–1524. doi: 10.3390/v6041502. http://dx.doi.org/10.3390/v6041502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krauss S, Kaps J, Czech N, Paulus C, Nevels M. Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J Virol. 2009;83(24):12854–12870. doi: 10.1128/JVI.01164-09. http://dx.doi.org/10.1128/JVI.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim YE, Ahn JH. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 2015;11(3):e1004785. doi: 10.1371/journal.ppat.1004785. https://dx.doi.org/10.1371/journal.ppat.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. PNAS. 2006;103(10):3840–3845. doi: 10.1073/pnas.0600007103. http://dx.doi.org/10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential Roles of PML Isoforms. Front Oncol. 2013;3:125. doi: 10.3389/fonc.2013.00125. http://dx.doi.org/10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121(Pt 16):2731–274310. doi: 10.1242/jcs.031922. http://dx.doi.org/10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 124.Chen Y, Wright J, Meng X, Leppard KN. Promyelocytic Leukemia Protein Isoform II Promotes Transcription Factor Recruitment To Activate Interferon Beta and Interferon-Responsive Gene Expression. Mol Cell Biol. 2015;35(10):1660–1672. doi: 10.1128/MCB.01478-14. https://dx.doi.org/10.1128/MCB.01478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Delorme-Axford E, Sadovsky Y, Coyne CB. The Placenta as a Barrier to Viral Infections. Annu Rev Virol. 2014;1(1):133–146. doi: 10.1146/annurev-virology-031413-085524. http://dx.doi.org/10.1146/annurevvirology-031413-085524. [DOI] [PubMed] [Google Scholar]
- 126.Kim CJ, Yoon BH, Jun JK, Park JO, Cho SY, Romero R, Kim YM, Yu ES. Promyelocytic leukaemia (PML) protein expression in human placenta and choriocarcinoma. J Pathol. 2003;1(1):83–89. doi: 10.1002/path.1382. http://dx.doi.org/10.1002/path.1382. [DOI] [PubMed] [Google Scholar]
- 127.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131–128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78(18):10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. http://dx.doi.org/10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, Johnson DC. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol. 2008;82(1):60–70. doi: 10.1128/JVI.01910-07. http://dx.doi.org/10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cui X, McGregor A, Schleiss MR, McVoy MA. Cloning the complete guinea pig cytomegalovirus genome as an infectious bacterial artificial chromosome with excisable origin of replication. J Virol Methods. 2008;149(2):231–239. doi: 10.1016/j.jviromet.2008.01.031. http://dx.doi.org/10.1016/j.jviromet.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGregor A, Schleiss MR. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol Genet Metab. 2001;72(1):15–26. doi: 10.1006/mgme.2000.3102. http://dx.doi.org/10.1006/mgme.2000.3102. [DOI] [PubMed] [Google Scholar]
- 131.Paredes AM, Yu D. Human Cytomegalovirus: Bacterial Artificial Chromosome (BAC) Cloning and Genetic Manipulation. Curr Protoc Microbiol. 2012;(Unit14E.4) doi: 10.1002/9780471729259.mc14e04s24. CHAPTER. http://dx.doi.org/10.1002/9780471729259.mc14e04s24. [DOI] [PMC free article] [PubMed]
- 132.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106(25):10171–10176. doi: 10.1073/pnas.0900604106. https://dx.doi.org/10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. http://dx.doi.org/10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 134.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347(4):827–839. doi: 10.1016/j.jmb.2005.01.071. http://dx.doi.org/10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of calculated and observed protein sizes with predicted sumoylation sites and HCMV homology.
Table S2. Oligonucleotides used for PCR reactions.
Fig S1. GPCMV IE1 specific 5′ and 3′ RACE analysis. IE1 specific 5′ and 3′ RACE analysis was performed as described in materials and methods. Positive clones were confirmed by PCR and restriction profile analysis before being sequenced. Sequence analysis of duplicate 5′ and 3′ RACE clones confirmed the published GPCMV IE1 cDNA genome sequence (accession #AB592928).
Fig S2. Nuclear expression of unique exons GP123 (IE1) and GP122 (IE2). (i) Schematic representation of GFP-tagged GP123 and GP122 fusion protein expression constructs. (ii) GFP-GP123 and GFP-GP122 protein expression by western blot analysis and immunofluorescence. A–C, expression and localization of GFP-GP123. D–F, expression and localization of GFP-GP122. Lane 1, plasmid transfect GPL cells. Lane 2, mock transfected GPL cells. GFP-GP123 and GFP-GP122 detected by fluorescence (cell localization) or primary anti-epitope Ab and anti-mouse IgG-HRP secondary (western blot). (iii) Predicted nuclear localization signals for GPCMV IE1 and IE2 as described in material and methods. cNLS Mapper was used to predict nuclear localization signal sequences for IE1 and IE2 using a cutoff score of 7.
Fig S3. Co-localization of SUMO-1 with gpPML, gpDaxx, GPCMV IE1 and IE2. A–D, co-localization of SUMO-HA (A) and GFP-IE1 (B). C is the merged image of A and B. D represents the overlay with DAPI. E–H, co-localization of SUMO-HA (E) with GFP-IE2 (F). G is the merged image for E and F, with H being the overlay of G with DAPI. I–L, co-localization of SUMO-HA (I) and gpPMLmyc (J). K is the merge of I and J. L represents the overlay with DAPI. M–P, co-localization of SUMO-HA (M) and gpDaxxFLAG (N). O is the merged image for M and N, with P being the overlay with DAPI. Detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG FITC or TRITC.
Fig S4. Prediction of intrinsically unstructured regions and hydrophilicity of GPCMV, HCMV, and MCMV IE1 protein. The IUPred program (“structured domains”) was used to predict and plot the tendency of intrinsic disorder in the GPCMV, HCMV, and MCMV IE1 proteins. Scores ≥ 0.5 (blue line) indicate disorder, while scores < 0.5 represent structured regions. Kyte-Doolittle hydrophilicity analysis (MacVector Inc.) of GPCMV, HCMV, and MCMV IE1 protein. (i) Disorder score for GPCMV IE1 predicts the presence of a central globular core domain (aa 104–411). The C-terminus contains a hydrophilic stretch (aa 415–474) (red box). (ii) The HCMV IE1 (accession #GQ244523.1) globular core domain ranges from aa 77–378. The C-terminus contains a hydrophilic stretch (aa 458–491) (red box). (iii) Disorder score for MCMV IE1 (accession #CCE57451) predicts two globular core domains (aa 41–72 and 87–372). The C-terminus contains a hydrophilic stretch (aa 393–595) (red box).
Fig S5. Generation of the vIET knockout virus. (i) Genome sequence of GPCMV unique exon GP123 (yellow). The 200 bp deletion introduced within the unique exon is underlined. Primer sequences used for confirmation of GP123 locus modification are bolded (Table S2). (ii) Restriction profile analysis of the GPCMV IETKm BAC mutant intermediate (lane 1) and IET mutant (lane 3) compared to WT GPCMV BAC (lanes 2 and 4). HindIII digestion of GPCMV IETKm intermediate BAC DNA cut the original 25.7 kb fragment (yellow, lane 2) into a 2.6 kb and 24 kb band (blue, lane 1). HindIII restriction profile analysis of GPCMV IET BAC DNA decreased the original 25.7 kb fragment (yellow, lane 4) by 200bp (blue, lane 3). (iii) PCR analysis of IETKm intermediate and IET compared to WT GPCMV. Lane 1, IET BAC (0.9 kb). Lane 2, WT BAC (1.1 kb). Lane 3, IETKm BAC intermediate (2.1 kb).
Fig S6. Generation of HSV-1 ICP0Δ double knockout virus. (i) Homologous recombination introduced a partial deletion within exons 2 (yellow) and 3 (grey) of both ICP0 loci. Primer pair sequences (Table S2) used to amplify the Km cassette used in homologous recombination are in bold. Deleted sequence within exons 2 and 3 is underlined. (ii) Schematic representation of the HSV-1 genome and insertion of a Km cassette into ICP0 loci within inverted repeats b and b’ by homologous recombination with WT HSV BAC. (iii) HindIII restriction profile analysis confirmed modification of both loci by insertion of a Km cassette. HindIII digestion resulted in the loss of the 26.5 kb band (lane 1, blue) to generate two new bands of 8 kb and 18 kb (lane 2, yellow). For the second locus, HindIII digestion resulted in the loss of the 34.6 kb band (lane 1, red) and gave rise to two bands of 10.5 kb and 23.7 kb (lane 2, green). (iv) Western blot analysis of HSV-I ICP0 knockout virus detected by primary anti-HSV ICP0 Ab and secondary anti-mouse IgG-HGP. Lane 3, ICP0Δ virus. Lane 4, ICP0 expression plasmid. Lane 5, mock infected GPL cells. Lane 6, WT HSV-1 virus.
Fig S7. HSV-1 ICP0 co-localization with guinea pig ND10 components. (i) A–D, ICP0Δ mutant virus fails to disrupt gpPMLmyc (A and B), showing a phenotype that is identical to transiently expressed gpPMLmyc only (C and D). (ii) GPL cells were transfected with expression plasmids encoding gpPMLmyc, gpDaxxFLAG, or gpSp100HA in the presence of ICP0 to evaluate cellular co-localization. E–G, ICP0 transient expression alone. G represents E merged with F. H–J, co-localization of ICP0 and gpPMLmyc. ICP0 (H) and gpPMLmyc (I) individually expressed in the same cell. J is the merged image overlaid with DAPI. K–M, colocalization of ICP0 (K) and gpDaxxFLAG (L). M being merged image overlaid with DAPI. NP, co-localization of ICP0 (N) with gpSp100HA (O). P is merged image overlaid with DAPI. Imunofluorescence detected by primary anti-epitope Ab and secondary anti-mouse or anti-rabbit IgG FITC or TRITC.