Abstract
In recent years, exciting developments in instrument technology and experimental methodology have advanced the field of magic angle spinning (MAS) NMR to new heights. Contemporary MAS NMR yields atomic-level insights into structure and dynamics of an astounding range of biological systems, many of which cannot be studied by other methods. With the advent of fast magic angle spinning, proton detection, and novel pulse sequences, large supramolecular assemblies, such as cytoskeletal proteins and intact viruses, are now accessible for detailed analysis. In this review, we will discuss the current MAS NMR methodologies that enable characterization of complex biomolecular systems and will present examples of applications to several classes of assemblies comprising bacterial and mammalian cytoskeleton as well as HIV-1 and bacteriophage viruses. The body of work reviewed herein is representative of the recent advancements in the field, with respect to the complexity of the systems studied, the quality of the data, and the significance to the biology.
1. Introduction
In the past decade, the field of magic angle spinning (MAS) nuclear magnetic resonance (NMR) has made significant strides. This technique has advanced to the level where we can now determine structures and characterize dynamics of complex systems, including large protein assemblies, at atomic resolution. A decade ago, this effort was in its infancy with the demonstration of the proof of principle that structures of small proteins can be solved de novo. Now we are tackling a wide range of biologically pressing problems, where traditional techniques yield only limited insights or are powerless. Recent instrument technology and methodological advancements have been conducive to the study of increasingly complex biological systems. Such advancements include the development of fast magic angle spinning capabilities (up to ~ 110 kHz at present) and very high magnetic fields (up to 1 GHz at present with 1.2 GHz magnets currently in production) that yield unprecedented gains in sensitivity and resolution and enable proton detection (Holland et al., 2010; Lewandowski et al., 2011a; Zhou et al., 2007b).
As a biophysical method, MAS NMR offers many advantages over other techniques. There are no theoretical size limitations (though challenges with respect to sensitivity and resolution arise with increasing molecular weight), no solubility limitations, and no requirements for well-formed crystals or long-range order. MAS NMR can achieve atomic level resolution and also tackle very large systems such as whole cells and intact viral particles. MAS NMR can probe both structure and dynamics at or close to physiologically relevant experimental conditions including temperature and pH. These advantages allow for the characterization of highly complex biological systems to address compelling questions in biology. MAS NMR can provide unique insights into an astounding range of biological systems, including proteins embedded in native membrane environments (Brown & Ladizhansky, 2015; Naito et al., 2015), aggregates of misfolded or disordered proteins (Comellas & Rienstra, 2013; Tycko, 2011), biomaterials (Goobes, 2014), and metalloproteins (Jaroniec, 2012; Knight et al., 2013; Knight et al., 2012). MAS NMR is also well suited for the study of biological assemblies comprised of multiple components or multiple copies of the same molecule, including entire viruses and cells (Goldbourt, 2013; Loquet et al., 2013b; Weingarth & Baldus, 2013; Yan et al., 2013b). In this review, we will discuss the current MAS NMR methodology for structural and dynamics studies of biological systems with specific focus on applications to supramolecular assemblies represented by proteins associated with the cytoskeleton and viral assemblies.
2. Current Methodology for Structural and Dynamics Analysis of Biological Assemblies by MAS NMR
The general work flow for MAS NMR studies of biological systems (Fig. 1) first entails preparation of samples isotopically enriched with NMR active nuclei (namely, 13C and 15N). Proteins are subsequently prepared for MAS NMR studies by crystallization (Martin & Zilm, 2003), assembly, sedimentation (Bertini et al., 2013; Bertini et al., 2011b), or similar approaches, and packed into rotors. Optimization is key as sample conditions can have a significant impact on spectral quality. Before advanced structural or dynamics studies can be executed, site-specific resonance assignments must be obtained. This is accomplished by acquiring a suite of multidimensional spectra (typically 2D and 3D), and establishing through-space (dipolar) and/or through-bond (scalar) intra- and inter-residue homo- and heteronuclear correlations.
Fig. 1.
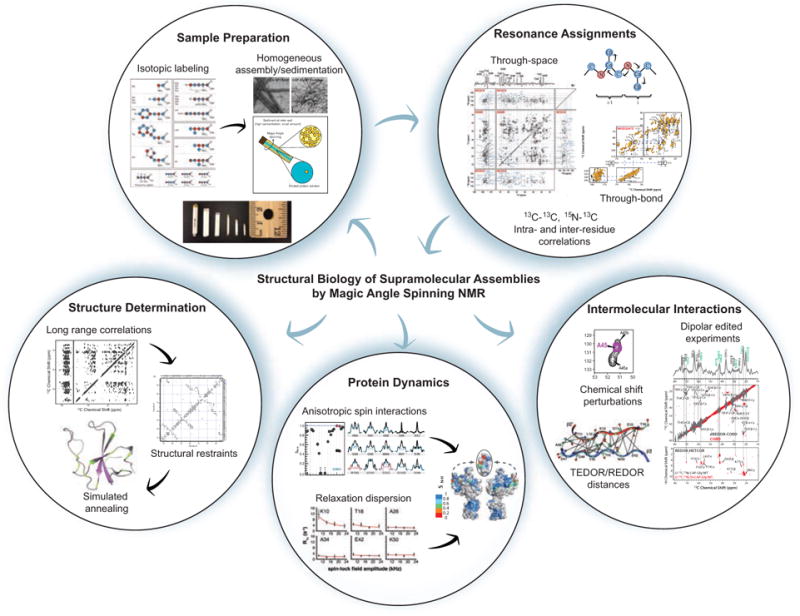
Workflow for studies of biological supramolecular assemblies by magic angle spinning NMR. Preparation of homogeneous, isotopically labeled samples and resonance assignments are the first steps of any structural biology study by MAS NMR. Resonance assignments and other experiments exploit two types of inter-nuclear correlations: through-space (dipolar-based), which selects for rigid residues, and through-bond (scalar or J coupling based), which selects for dynamic residues. Biological questions that can be addressed by MAS NMR include structure determination, protein dynamics, and intermolecular interactions. Protein structure determination generally entails first obtaining long-range, inter-nuclear distance correlations, often combined with other structural restraints, and subsequently input into simulated annealing protocols for structure calculation. Two approaches commonly used for the determination of site-specific millisecond to nanosecond protein dynamics are relaxation dispersion and measurement of reduced anisotropic interactions (e.g. chemical shift anisotropy or dipolar interactions). Finally, MAS NMR can characterize protein-protein and protein-ligand intermolecular interactions. Methods for observing these intermolecular interfaces include chemical shift perturbations, dipolar filtered experiments such as dREDOR, and quantitative distance measurements with REDOR/TEDOR based experiments. Isotopic labeling schematic reprinted with permission from Higman et al., J. Biomol. NMR, 2009, 44, 245–260. Copyright 2009 Springer (Higman et al., 2009). Sedimented solute NMR (SedNMR) figure adapted with permission from Bertini et al., Acc. Chem. Res., 2013, 46 (9), 2059–2069. Copyright 2013 American Chemical Society (Bertini et al., 2013). CA-SP1 A92E TEM image and through-space and through-bond correlation experiments reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135 (47), 17793–17803. Copyright 2013 American Chemical Society (Han et al., 2013). Structure determination and chemical shift perturbation figures and adapted with permission Yan et al., J. Mol. Biol., 2013, 425 (22), 4249–4266. Copyright 2013 Elsevier (Yan et al., 2013a). Anisotropic spin interactions and protein dynamics/structure figures adapted with permission from Lu et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14617–14622. Copyright 2015 National Academy of Sciences (Lu et al., 2015a). dREDOR figure and CAP-Gly/MT complex TEM adapted with permission from Yan et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14611–14616. Copyright 2015 National Academy of Sciences (Yan et al., 2015a). TEDOR/REDOR distances figure reprinted with permission from Nieuwkoop et al., J. Am. Chem. Soc., 2010, 132 (22), 7570–7571. Copyright 2010 American Chemical Society (Nieuwkoop & Rienstra, 2010). Relaxation dispersion figure reprinted with permission from Lewandowski et al., J. Am. Chem. Soc., 2011, 133, 16762–16765. Copyright 2011 American Chemical Society (Lewandowski et al., 2011b).
MAS NMR can access information of great interest in biology, including protein structure and dynamics, as well as protein-protein and protein-ligand interactions. Protein structure determination by MAS NMR requires the quantification of structural restraints, such as short- and long-range (separated by more than 4 residues) internuclear distances, as well as backbone torsion angles. Structural restraints are integrated into simulated annealing protocols for structure calculation. MAS NMR has been increasingly combined with other biophysical methods such as cryo-electron microscopy (cryo-EM) for structure determination of supramolecular assemblies. Anisotropic interactions, such as magnitudes and orientations of dipolar and chemical shift tensors are highly sensitive to both structure and dynamics. A wide range of methods exists to study protein dynamics with MAS NMR over timescales from picoseconds to seconds. Chemical shift perturbations and dipolar-edited correlation methods can be used to characterize protein-protein and protein-ligand interactions.
Two essential considerations for successful MAS NMR experiments are sensitivity and resolution, which can be affected by numerous factors such as protein size and dynamics, sample homogeneity, and nuclear spin interactions including dipolar and J (scalar) couplings. Challenges related to resolution and sensitivity can be alleviated or overcome using advanced hardware (i.e. faster spinning probes and higher magnetic fields) and appropriate choice of isotopic labeling schemes. Spectroscopic methods employed are a fundamental factor to maximize sensitivity and resolution, including choice of magnetization transfer method (i.e. through-space vs through-bond) and detection method (1H vs heteronuclear detection). In the following sections, we provide an overview of methods commonly employed for the study of biological systems including supramolecular assemblies by magic angle spinning NMR.
2.1 Isotopic labeling
Isotopic enrichment with magnetically active 13C and 15N is essential for the study of proteins by nuclear magnetic resonance. Beyond uniform isotopic labeling with 13C-glucose and 15NHCl4, there are many alternative labeling schemes for selective incorporation of isotopes into desired sites. Spectral crowding is a substantial challenge in MAS NMR, and beyond 3- and 4-dimensional spectra, higher magnetic fields, and fast magic angle spinning, isotope editing is used to alleviate the congestion. Sparse as well as selective isotopic labeling methods are often employed for the determination of long-range 13C-13C distance restraints and torsion angles. The common protocols include preparation of recombinant proteins from minimal media containing [2-13C]glycerol, [1,3-13C]glycerol, [1,6-13C]glucose, and [2-13C]glucose as the sole carbon source (Higman et al., 2009; Hong, 1999; LeMaster & Kushlan, 1996). Selective labeling with [2-13C]glycerol and [1,3-13C]glycerol was essential to the first protein structure determination by MAS NMR (Castellani et al., 2002). These labeling schemes exploit bacterial metabolic pathways to achieve known patterns of amino acid labeling (Goldbourt et al., 2007a). These selective labeling schemes also serve to reduce line broadening by reducing strong 13C-13C dipolar couplings and J-couplings. For further spectral simplification, amino acid specific labels can be incorporated (Mcintosh & Dahlquist, 1990), which also allow for the study of critical protein properties, such as amino acid protonation state, as well as the determination of select distance restraints with fewer ambiguities. With His-to-Gln mutations and selective labeling of His37 of M2(21-97), Hong and co-workers showed that the protonation state of this transmembrane domain residue is perturbed by the presence of the cytoplasmic domain, suggesting a mechanism of 1H conduction (Liao et al., 2015). Perdeuteration with back exchange of amide protons enables the acquisition of high-resolution proton-detected spectra and the determination of 1H-1H distance restraints by reducing the very strong 1H-1H dipolar couplings (Chevelkov et al., 2006; Reif et al., 2003; Zhou et al., 2007b) and is further discussed below. Additional selective 13C and 2H labeling schemes for aliphatic groups of Ala, Val, Leu, and Ile, developed by Kay and co-workers (Rosen et al., 1996) and first applied in the solid state by Reif and co-workers (Agarwal et al., 2006), can also be used for structural restraints as demonstrated for structure determination of ubiquitin (Agarwal et al., 2014). To characterize intermolecular interfaces and distances, differential labeling schemes have been developed. In this family of labeling schemes one region of the protein, monomer in an assembly, or binding partner contains one set of labels (e.g., 13C or 13C,15N) while its interaction partner has different labeling (e.g, 15N). In these differentially labeled samples, intermolecular interactions are then measured by experiments where magnetization is selectively transferred across the intermolecular interface, demonstrated for distance determination of select 13C-15N spin pairs in gramicidin A in early work (Fu et al., 2000), and later applied to protein studies by Baldus (Etzkorn et al., 2004) and Polenova (Marulanda et al., 2004; Yang et al., 2008). Generally, there are many isotopic labeling approaches available to an experimentalist, and an appropriate combination of isotopic labeling schemes is selected to address specific questions.
2.2 Resonance Assignments and Structure Determination
Performing resonance assignments entails obtaining homo- and heteronuclear intra-residue and sequential inter-residue correlations and is the necessary first step to any study of protein structure or dynamics by MAS NMR. Early work of note includes complete or near complete resonance assignments of BPTI (58 residues, (McDermott et al., 2000)), SH3 (62 residues, (Pauli et al., 2001)), ubiquitin (76 residues, (Igumenova et al., 2004a; Igumenova et al., 2004b), and Crh (85 residues, (Bockmann et al., 2003)). Isotropic chemical shifts yield information on secondary structure, protonation states, and dynamics (Williamson, 1990; Wishart & Sykes, 1994). For structure determination, long-range distance restraints must also be obtained. Determining resonance assignments and distance restraints require collecting a suite of multidimensional spectra using dipolar and/or scalar based correlations. From homo- and heteronuclear correlation experiments, the spin system belonging to a given amino acid is first identified from experiments including 2D 13C-13C experiments and 2D/3D 15N-13C NCACX experiments. Inter-residue correlation experiments such as 2D/3D NCOCX are then used to establish sequential, residue specific assignments. These experiments are further detailed in section 2.2.1. In large systems, assignments could be challenging due to spectral congestion and typically require a large number of experiments in conjunction with sparse isotopic labeling discussed above, as demonstrated for assignment of the 189 residue protein DsbA by Rienstra and co-workers (Sperling et al., 2010). Modern technological advancements including fast magic angle spinning (MAS frequencies of 40-110 kHz), which provide both sensitivity and resolution enhancement (Barbet-Massin et al., 2014b; Bertini et al., 2010; Laage et al., 2009; Parthasarathy et al., 2013; Samoson et al., 2005), and proton detection (Chevelkov et al., 2006; Paulson et al., 2003; Reif & Griffin, 2003; Zhou et al., 2007a) enabled the development of new experiments for time-efficient resonance assignments and recording distance restraints. Fast MAS and proton detection are further discussed in section 2.2.3.
2.2.1 Through-Space Multidimensional Correlation Spectroscopy
Through-space correlation experiments rely on distance-dependent internuclear dipolar couplings (DIS ∝ γIγS/r3). Observed correlations can be short or long range, depending on the chosen pulse sequence and experimental parameters (e.g. mixing time). Early through-space correlation experiments were optimized for MAS frequencies of 10-30 kHz. With advances in probe technology and faster spinning speeds, methods have been developed to achieve efficient polarization transfer at higher MAS rates. Common through-space homonuclear correlation experiments optimized for the slower spinning regime (10–30 kHz) include DARR (dipolar-assisted rotational resonance (Takegoshi et al., 2001)), RAD (RF-assisted diffusion (Morcombe et al., 2004)), PDSD (proton-driven spin diffusion (Szeverenyi et al., 1982)), and DREAM (dipolar recoupling enhanced by amplitude modulation (Verel et al., 2001)) to obtain 13C-13C correlations. Some applications of note on biological assemblies include PDSD for resonance assignments and structure determination of the type III secretion system (T3SS) needle (Demers et al., 2014; Loquet et al., 2011), BacA filament (Shi et al., 2015; Vasa et al., 2015), and HET-s amyloid (Wasmer et al., 2008), and DARR for detection of the Pf1 bacteriophage DNA signals (Sergeyev et al., 2011) and characterization of the HIV-1 capsid and CA-SP1 maturation intermediate (Han et al., 2010; Han et al., 2013). For determination of heteronuclear NCA, NCO, NCACX, and NCOCX correlations, 15N-13C double cross polarization (DCP), first presented by Stejskal and co-workers (Schaefer et al., 1979), is commonly employed. Baldus et al. developed frequency-selective DCP (known as SPECIFIC-CP) (Baldus et al., 1998) for selective NCA or NCO excitation, as demonstrated for resonance assignments of SH3 (Pauli et al., 2001). SPECIFIC-CP has been shown to be broadly applicable (Luca et al., 2003). Other recoupling sequences such as dipolar INEPT (insensitive nuclei enhanced by polarization transfer) for selective C-H excitation at both moderate (De Vita & Frydman, 2001; Wickramasinghe et al., 2008) and fast (Holland et al., 2010) spinning speeds have been also reported.
At MAS frequencies faster than 30 kHz, the conventional spin diffusion based experiments for recording homonuclear correlations are no longer efficient. Under these conditions, DREAM and fpRFDR (finite pulse rf driven recoupling (Ishii, 2001)) are efficient for recording one- and two-bond correlations. Another family of experiments that is particularly useful for recording long-range 13C-13C distance restraints is COmbined R2nν-Driven (CORD) dipolar recoupling sequences, where the magnetization transfer is driven by rotor-synchronized R2nν symmetry based recoupling (Hou et al., 2011a; Hou et al., 2013a; Lu et al., 2015b). The R2nν and CNnν symmetry recoupling schemes were originally presented by Levitt and co-workers (Carravetta et al., 2000). CORD utilizes a super-cycled R2nν recoupling to achieve broadband homonuclear correlations with high polarization transfer efficiency at both moderate and fast MAS rates while not suffering from dipolar truncation effects. Proton-assisted recoupling (PAR) is another method that performs well at fast MAS to obtain long distance 13C-13C (De Paepe et al., 2008; Lewandowski et al., 2009b) or 15N-15N (Lewandowski et al., 2009a) correlations. PAR is based on third spin assisted recoupling (TSAR), in which two spins are connected via dipolar couplings with a third spin leading to zero quantum (ZQ) polarization transfer. Distances of ~6–7 Å can be observed with PAR and CORD.
Beyond double cross polarization (DCP), several methods have been developed for the acquisition of long range 15N-13C correlations. PAIN-CP (proton assisted insensitive nuclei cross polarization) is a third-spin-assisted heteronuclear polarization transfer (Agarwal et al., 2013; De Paepe et al., 2011; Lewandowski et al., 2007) first presented by Griffin and co-workers, which like its homonuclear counterpart discussed above, utilizes neighboring proton spins to enhance magnetization transfer efficiency with appropriate choice of 13C, 15N, and 1H rf fields. Transferred echo double resonance (TEDOR) (Hing et al., 1992) is a REDOR (rotational echo double resonance (Gullion & Schaefer, 1989)) derived scheme that can also be used to detect 15N-13C distances up to ~8 Å. In REDOR-based pulse sequences, the dipolar coupling between two spins is reintroduced by a train of rotor-synchronized 180° pulses (Gullion & Schaefer, 1989). The resulting dephasing of magnetization is proportional to the magnitude of the dipolar coupling (and hence distance between the two spins). A variation of the TEDOR pulse sequence developed by Jaroniec et al. (Jaroniec et al., 2002), z-filtered TEDOR, is shown in Fig. 2(e). The inclusion of a z-filter is needed to eliminate artifacts due to 13C-13C J couplings in uniformly labeled systems. TEDOR-derived distance restraints have been applied to a range of systems including structure determination of microcrystalline GB1 by Rienstra and co-workers (Nieuwkoop et al., 2009) and L7Ae-bound Box C/D RNA by Carlomagno and co-workers (Marchanka et al., 2015). Pulse sequences, schematics, and model compound spectra for several through-space correlation methods are presented in Fig. 2.
Fig. 2.
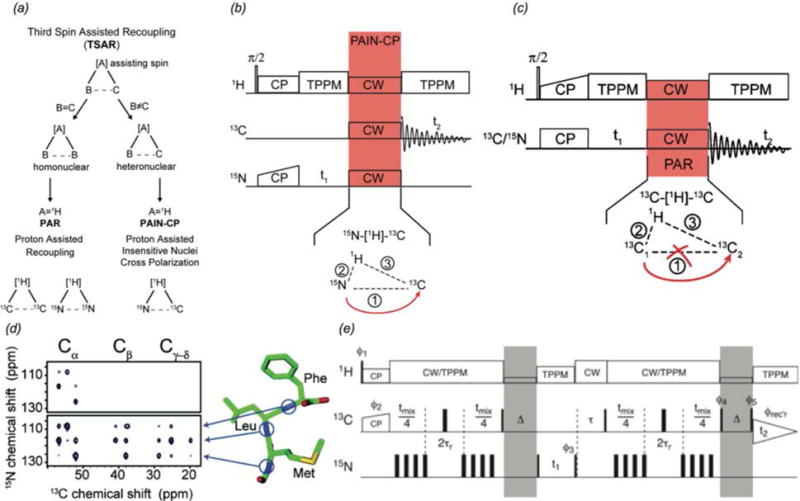
(a) Schematic for homonuclear and heteronuclear third spin assisted recoupling, a second-order mechanism which uses the dipolar couplings with a third spin to achieve magnetization transfer (De Paepe et al., 2011). (b) Pulse sequence for 2D 15N-13C PAIN-CP heteronuclear correlation experiment (De Paepe et al., 2011). (c) 2D homonuclear PAR pulse sequence (De Paepe et al., 2008). (d) 15N-13C correlation spectra of MLF: (top) DCP, (bottom) PAIN-CP, demonstrating the more efficient magnetization transfer of PAIN-CP (Lewandowski et al., 2007). (e) Pulse sequence for 15N-13C heteronuclear z-filtered TEDOR correlations (Jaroniec et al., 2002). Shaded portions indicate z-filters incorporated to eliminate artifacts arising 13C-13C J couplings in uniformly labeled samples. (a) and (b) Reprinted with permission from de Paepe et al., J. Chem. Phys., 2011, 139 (9). Copyright 2011 AIP Publishing. (c) Reprinted with permission from de Paepe et al., J. Chem. Phys., 2008, 129 (24). Copyright 2008 AIP Publishing. (d) Reprinted with permission from Lewandowski et al., J. Am. Chem. Soc., 2007, 129 (4), 728–729. Copyright 2007 American Chemical Society. (e) Reprinted with permission from Jaroniec et al., J. Am. Chem. Soc., 2002, 124 (36), 10728–10742. Copyright 2002 American Chemical Society.
2.2.2 Through-Bond Multidimensional Correlation Spectroscopy
Complementary to through-space, dipolar-based correlation experiments, scalar-based through-bond transfer mechanisms can be exploited to obtain inter-nuclear correlations. Through-bond experiments utilize the electron-mediated J coupling between neighboring residues. This transfer mechanism can be especially valuable in cases of dynamics (Heise et al., 2005) and at fast MAS frequencies, situations where dipolar couplings are partially or fully averaged. J-based experiments are also ideal at faster spinning frequencies due to the lower required decoupling power, and can allow for the necessary longer coherence evolution times (Bertini et al., 2011a). Experiments such as heteronuclear (Elena et al., 2005) or homonuclear (Linser et al., 2008) INEPT (insensitive nuclei enhanced polarization transfer, (Morris & Freeman, 1979)), homonuclear TOBSY (total through-bond correlation spectroscopy (Hardy et al., 2001)), homonuclear CTUC-COSY (constant-time uniform sign cross-peak COSY, (Chen et al., 2006)), as well as solid state INADEQUATE (Lesage et al., 1997) and refocused INADEQUATE (Lesage et al., 1999) have been used to complement dipolar-based correlation spectroscopy in the study of protein assemblies (Fig. 3). With these methods, sufficient sensitivity is attained despite the relatively small size of the J-couplings (e.g. 50 Hz 13C-13C J coupling vs 2 kHz dipolar coupling). TOBSY experiments utilize the POST-C7 symmetry sequence (Hohwy et al., 1998) to achieve efficient, scalar based polarization transfer. CTUC-COSY offers excellent sensitivity by converting both zero-quantum and double-quantum magnetization, and has been applied to detect dynamic regions of the Y145Stop human prion protein (Helmus et al., 2010) and α-Synuclein fibrils (Comellas et al., 2011), as well as to obtain pure 1-bond correlations in 13C-13C spectra of CAP-Gly (Sun et al., 2009). INADEQUATE experiments in the solid state use double quantum coherence transfer identical to solution NMR. More recent modifications of solid state INADEQUATE have included the addition of a z-filter (Cadars et al., 2007) and FSLG (frequency-switched Lee-Goldberg) homonuclear 1H-1H decoupling (Baltisberger et al., 2011) to reduce artifacts, and development of band selective INADEQUATE using the spin state selective excitation (S3E) scheme, which has been demonstrated at 60 kHz MAS (Bertini et al., 2011a). The use of scalar transfers in proton-detected experiments at fast MAS has recently been demonstrated in the solid state as well including 13C-13C INEPT transfer for resonance assignments of superoxide dismutase (SOD) (Knight et al., 2011). Pintacuda and co-workers reported the application of ‘out-and-back’ 13C-13C scalar-based transfer for resonance assignments with fast MAS (frequencies of 60 kHz and higher, (Barbet-Massin et al., 2013)). These proton-detected 3D experiments can be applied to both fully protonated samples as well as perdeuterated samples with 100% HN back exchange, and were demonstrated on AP205 bacteriophage as well as numerous other diverse classes of proteins (Barbet-Massin et al., 2014b).
Fig. 3.
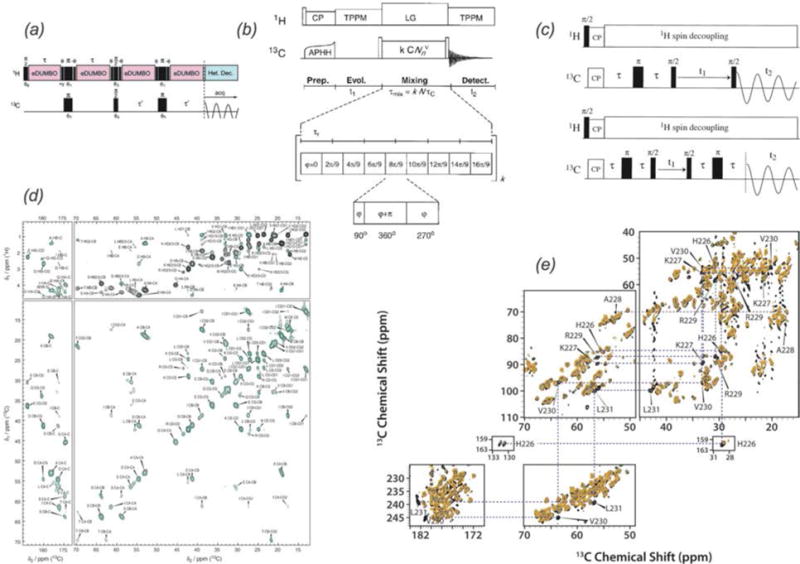
Scalar-based correlation experiments frequently used in the solid state. (a) Heteronuclear 1H-13C INEPT pulse sequence (Elena et al., 2005), (b) homonuclear 13C-13C TOBSY pulse sequence (Hardy et al., 2001), (c) homonuclear 13C-13C INADEQUATE pulse sequences, (top) solid-state INADEQUATE, (bottom) refocused INADEQUATE (Lesage et al., 1999). (d) 1H-13C INEPT (black) and 13C-13C INEPT-TOBSY spectra (green) of HET-s amyloids (Wasmer et al., 2009). (e) Direct (black) and CP (orange) INADEQUATE spectra of CA-SP1 tubular assemblies (Han et al., 2013). (a) Adapted with permission from Elena et al., J. Am. Chem. Soc., 2005, 127 (49), 17296–17302. Copyright 2005 American Chemical Society. (b) Adapted with permission from Hardy et al., J. Magn. Reson., 2001, 148 (2), 459–464. Copyright 2001 Elsevier. (c) Reprinted with permission from Lesage et al., J. Am. Chem. Soc., 1999, 121 (47), 10987–10993. Copyright 1999 American Chemical Society. (d) Reprinted with permission from Wasmer et al., J. Mol. Biol., 2009, 394 (1), 119–127. Copyright 2009 Elsevier. (e) Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135 (47), 17793–17803. Copyright 2013 American Chemical Society.
2.2.3 Proton Detection and Fast MAS
In contrast to solution NMR where dipolar couplings are averaged out by molecular tumbling, the strong 1H-1H dipolar couplings present in solid-state NMR samples lead to very broad 1H lines. As a consequence, MAS NMR experiments have customarily been acquired with direct detection of low γ nuclei such as 13C and 15N, which greatly limits sensitivity. Proton detection takes advantage of the high gyromagnetic ratio of protons for increased sensitivity and with advances in hardware is increasingly applied in solid-state NMR. Early work by Reif, Griffin, and Zilm demonstrated that with perdeuteration to reduce 1H-1H dipolar couplings and 100% amide 1H-back exchange, proton-detected heteronuclear correlation experiments could be applied in the solid state and that the anticipated sensitivity gains are realized while dipolar truncation is avoided (Paulson et al., 2003; Reif & Griffin, 2003; Reif et al., 2001). Subsequent work demonstrated that increased 1H resolution can be achieved with higher levels of deuteration (i.e., only 10–40% 1H back exchange) (Akbey et al., 2010) and/or faster MAS frequencies (Chevelkov et al., 2006; Samoson et al., 2001). Linser et al. first demonstrated the application of 1H detection to amyloids and membrane proteins (Linser et al., 2011b). With the advent of fast magic angle spinning (≥ 40 kHz), proton-detection even on fully protonated proteins, first demonstrated by Rienstra and co-workers (Zhou et al., 2007a) has become feasible with improvements in resolution and sensitivity scaling with the MAS rate (Agarwal et al., 2014; Lewandowski et al., 2011a; Marchetti et al., 2012). Further, the sensitivity gains of 1H detection enable the use of very small sample amounts (Agarwal et al., 2014; Dannatt et al., 2015). Recent works of note in the application of 1H proton detection include studies of RNA-protein interfaces by Asami et al. (Asami et al., 2013), structure determination of superoxide dismutase (SOD) by Knight et al. (Knight et al., 2012), and measurements of heteronuclear dipolar couplings in Pf1 bacteriophage by Mueller and co-workers (Park et al., 2013). Additional capabilities of 1H detection include obtaining direct information on hydrogen-bond length from 1H chemical shifts (Zhou & Rienstra, 2008). Pintacuda and co-workers have recently demonstrated a suite of 3-dimensional proton-detected experiments to enable rapid data acquisition and assignment, with data sets of sufficient quality for the automated assignment routines to be applicable (Barbet-Massin et al., 2014b). They demonstrated the use of these sequences on several challenging systems, including assemblies of AP205 bacteriophage and Measles virus (MeV) nucleocapsid (Barbet-Massin et al., 2014a; Barbet-Massin et al., 2014b).
2.2.4 Protein Structure Determination by MAS NMR
Structure determination by solid state NMR was first demonstrated on small peptides oriented in lipid bilayers with static methods by Cross (Ketchem et al., 1996; Wang et al., 2001) and Opella (Opella et al., 1999). MAS NMR structure determination of a protein was first reported by Oschkinat and co-workers for the α-spectrin SH3 domain (Castellani et al., 2002). Technical and methodological advances have enabled the application of MAS NMR to structure determination of increasingly complex systems (Fig. 4(a)). MAS NMR is particularly valuable for the high-resolution structure determination of supramolecular assemblies, which are often insoluble or noncrystalline. Protein structure determination by MAS NMR requires determination of a sufficient number of quantitative or semi-quantitative structural restraints including distance restraints obtained from homo- and heteronuclear correlation spectra, using long-range magnetization transfer techniques described above, which can probe interatomic distances of up to ~ 7Å (Fig. 4(b)). 13C-13C distances are the most frequently utilized and often make use of selectively labeled samples for spectral simplification and semi-quantitative crosspeak intensity analysis. Additional distance restraints that have been utilized include 15N-13C (Nieuwkoop et al., 2009), 15N-15N (Hu et al., 2012; Lewandowski et al., 2009a), and increasingly 1H-1H (Andreas et al., 2016; Linser et al., 2011a; Zhou et al., 2007b) distances. Very recently, Pintacuda and co-workers presented the first protein structures determined on fully protonated samples with 1H detection (Andreas et al., 2016). They acquired RFDR-based 1H-1H distance restraints with ≥ 100 kHz MAS to determine the structures of 2 proteins: GB1 and the AP205 nucleocapsid assembly. Less than 0.5 mg of U-15N, 13C protein and 2 weeks of experiment time and ‘unsupervised’ structure determination were sufficient to derive the protein structures.
Fig. 4.
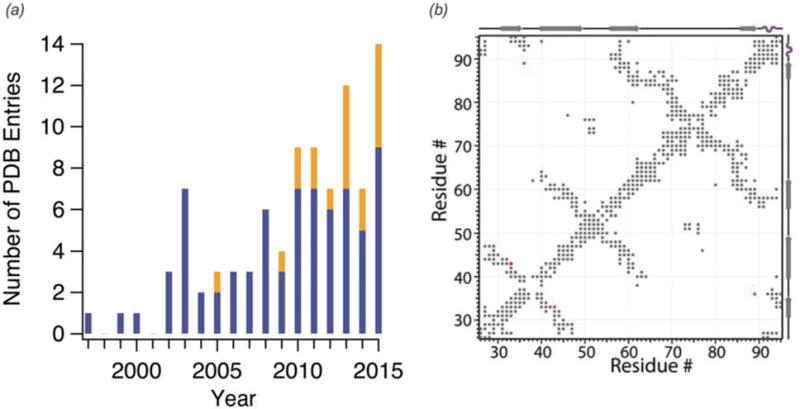
(a) PDB structures determined by solid-state NMR each year. Blue indicates structures determined by MAS NMR alone while orange indicates structures determined with an integrated approach, including methods such as electron microscopy or solution NMR in addition to SSNMR data. Year 2015 includes structures deposited through February 2016. (b) Contact map of microtubule-associated CAP-Gly illustrating all intra- and inter-residue correlations observed from MAS NMR distance restraints used in the structure calculation (Yan et al., 2015a). (b) Adapted with permission from Yan et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14611–14616. Copyright 2015 National Academy of Sciences.
In addition to inter-atomic distance restraints, anisotropic spin interactions including dipolar and chemical shift tensor magnitudes and orientations are a powerful tool for protein structure determination. These interactions exhibit secondary structure, orientation, and amino acid type dependence that can be exploited in structure determination, demonstrated extensively by Rienstra and co-workers on GB1 (Franks et al., 2008; Wylie et al., 2009; Wylie et al., 2011). In structure calculations, dipolar couplings and CSA tensors can constrain backbone torsion angles (Ladizhansky et al., 2003; Rienstra et al., 2002) as demonstrated on Aβ11–25 with ROCSA (Recoupling Of Chemical Shift Anisotropy) measurements (Chan & Tycko, 2003). Recent applications include the use of 1H-15N and 1H-13Cα dipolar couplings determined from separated local field (SLF) measurements (Das et al., 2013) as orientation restraints (with dihedral angles derived from isotropic chemical shifts) in determining the structure of CXCR1, a chemokine receptor involved in inflammatory response ((Park et al., 2012), Fig. 5). Anisotropic spin interactions can also provide valuable dynamics information, as further discussed in section 2.3. Additional structural restraints that may be incorporated into a structure calculation include predicted torsion angles based on backbone chemical shifts from TALOS (Shen & Bax, 2015; Shen et al., 2009), hydrogen bonding, and paramagnetic relaxation enhancements (PRE, (Nadaud et al., 2007)). PREs utilize the enhanced R1 relaxation of residues in close proximity to a paramagnetic center as a structural restraint and were applied to structure determination of SOD (Knight et al., 2012) and the membrane protein Anabaena sensory rhodopsin (Wang et al., 2013). Beyond intra-subunit restraints, long-range distances, such as those acquired in zf-TEDOR experiments, can contribute inter-subunit restraints for structure determination of supramolecular assemblies (Nieuwkoop & Rienstra, 2010).
Fig. 5.
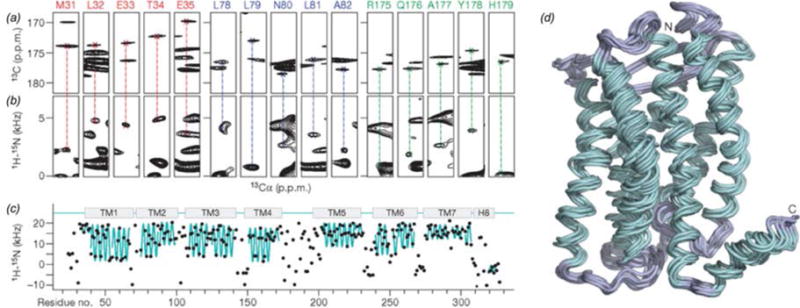
Structure determination of CXCR1 with dipolar couplings as a structural restraint (Park et al., 2012). (a) CO-Cα correlations from NCACX 3D. (b) Strip plots from SLF measurements, indicating the 1H-15N dipolar coupling strength at a given 13Cα chemical shift, corresponding to the residues indicated. (c) 1H-15N dipolar coupling vs residue number. The ‘wave’ pattern (cyan) is a feature of the transmembrane helices. (d) 10 lowest energy structures of CXCR1. Adapted with permission from Park et al., Nature, 2012, 491, 779–784. Copyright 2012 Nature Publishing Group.
Structural restraints are incorporated in simulated annealing calculations in a program such as Xplor-NIH (Schwieters et al., 2006; Schwieters et al., 2003) or CYANA (Guntert, 2004), with optimization protocols, which include molecular dynamics and Monte Carlo simulations. Recently, multiple labs have demonstrated the use of de novo structure prediction based on isotropic chemical shifts and amino acid sequence with CS-ROSETTA (Das et al., 2009; Shen et al., 2008), without requiring distance restraints. CS-ROSETTA has been incorporated into structure determination of the biological assemblies T3SS (Demers et al., 2014; Loquet et al., 2012) and the M13 bacteriophage (Morag et al., 2015). (Whether this approach is generally applicable to a wide range of systems remains to be investigated.) Rosetta enables the modeling of symmetric macromolecular assemblies and has been a key development for the atomic-resolution structure determination of these large and complex systems (DiMaio et al., 2011).
The capabilities of MAS NMR for structure determination have been further expanded in recent years by the application of integrated structure determination, wherein MAS NMR restraints are combined with other biophysical methods, such as electron microscopy, solution NMR, and molecular dynamics (MD) simulations. While exact approaches may differ, in general, the secondary structure as determined from secondary chemical shifts or high-resolution monomeric structure is mapped into the lower resolution electron density map (typically by rigid body modeling), and this structure is further refined with simulated annealing, using structure restraints such as cryo-EM structure factors and NMR distance restraints. Fig. 6(e) illustrates the iterative protocol. Lower-resolution microscopy can provide information on the symmetry and macromolecular organization, while MAS NMR data provide atomic-level structural details including inter-subunit contacts. This approach is proving to be particularly auspicious for the study of macromolecular assemblies including structure determination ofαB-crystallin (with SAXS, MD (Jehle et al., 2010)), T3SS (with cryo-EM (Demers et al., 2014; Loquet et al., 2012)), DsbB (with x-ray crystallography, MD (Tang et al., 2013)), FimA (with solution NMR, STEM (Habenstein et al., 2015)), and the mouse ASC inflammasome (with cryo-EM, Fig. 6 (Sborgi et al., 2015)).
Fig. 6.
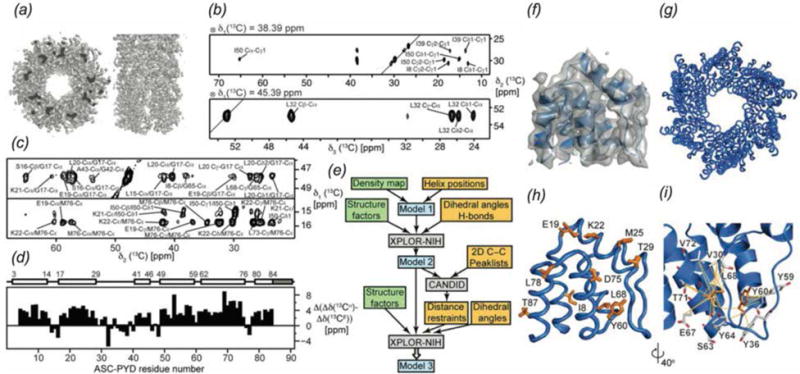
Combined use of MAS NMR and cryo-EM to determine the structure of the mouse ASC inflammasome (ASC-PYD) (Sborgi et al., 2015). (a) Electron density map determined by cryo-EM. (b) Strips from 13C-13C-13C 3D. (c) Strips from 13C-13C 2D (top) and CHHC 2D (bottom). (d) Secondary chemical shift plot, indicating the predominantly α-helical content of the protein. (e) Flow chart illustrating the protocol for structure refinement. MAS NMR data contributions are shaded yellow and cryo-EM data are shaded green. (f) Cryo-EM density reconstruction superimposed with a monomer of ASC-PYD. (g and h) Superposition of the 20 lowest energy structures of the filament and monomer. Positions of 10 arbitrary residues as determined by structure refinement are shown in orange. (i) Inter-residue interactions in a monomer of ASC-PYD. Orange lines indicate ambiguous distance restraints between Tyr60, Leu68 (orange) and neighboring residues (gray). Reprinted with permission from Sborgi et al., Proc. Nat. Acad. Sci., 2015, 112 (43), 13237–13242. Copyright 2015 National Academy of Sciences.
2.3 MAS NMR for the Study of Protein Dynamics
Protein dynamics are an essential attribute of biological function including intra-cellular transport (Desai & Mitchison, 1997) and inter-cellular signaling (Alenghat & Golan, 2013), as well as detrimental pathologies, such as in the case of amyloids (Chiti & Dobson, 2006). Relevant motions include both faster, small-amplitude motions such as backbone fluctuations and larger amplitude motions such as whole domain reorganization (Tzeng & Kalodimos, 2012). In contrast to other techniques that are used for characterization of biomolecular dynamics, such as SAXS, FRET, and AFM, NMR (both solution and MAS) yields information on multiple sites within a protein simultaneously. Furthermore, nuclear spin interactions, including the chemical shift, dipolar, and quadrupolar tensors, are sensitive probes of dynamics over many decades of motional timescales, from picoseconds to seconds, making NMR a unique technique for probing motions over the entire range of functionally relevant time scales, often in a single sample as demonstrated for GB1 (Lewandowski et al., 2015) and thioredoxin (Yang et al., 2009). MAS NMR is particularly well suited for probing protein dynamics in large biological assemblies and has shed light on a number of intriguing biological questions, such as gating of membrane proteins (Hu et al., 2010; Wang & Ladizhansky, 2014; Weingarth et al., 2014; Wylie et al., 2014), mechanisms of enzyme catalysis (Caulkins et al., 2015; Rozovsky & McDermott, 2001; Schanda et al., 2014; Ullrich & Glaubitz, 2013), and the regulation of protein-protein interactions in supramolecular assemblies (Hoop et al., 2014; Opella et al., 2008; Yan et al., 2015b). Unlike in solution NMR, the anisotropic tensorial spin interactions are recorded in MAS NMR rather than the motionally averaged residual interactions. Dipolar, CSA, and quadrupolar tensors contain orientational information and thus bear a wealth of information on the motional symmetry and amplitudes, which can be inferred only indirectly from the isotropic chemical shifts or residual dipolar interactions. Nuclear spin relaxation is also extensively used as a probe of dynamics over a wide range of conditions, and yields unprecedented insights into hierarchical protein dynamics, as was recently demonstrated (Lewandowski et al., 2015). For more extensive review of MAS NMR for the study of protein dynamics see the following review articles: (Krushelnitsky et al., 2013; Watt & Rienstra, 2014).
Recently, several groups have presented comprehensive studies of protein dynamics for systems of interest using a combination of the methods described below to gain insight into protein dynamics across multiple timescales. These works include dipolar order parameter (DOP) and 15N R1ρ studies of Anabaena sensory rhodopsin (ASR) (Good et al., 2014), R1 and R1ρ studies of GB1 (Lewandowski et al., 2015; Lewandowski et al., 2011b; Lewandowski et al., 2010), SH3 (Zinkevich et al., 2013) and superoxide dismutase (SOD) (Knight et al., 2012), 1H-15N DOP, 15N R1, and 15N CSA measurements of thioredoxin (Yang et al., 2009), DOP, 15N CSA, and peak intensity experiments on HIV-1 capsid (Byeon et al., 2012; Lu et al., 2015a), DOP and peak intensity measurements of CAP-Gly (Yan et al., 2015b), and DOP, R1, R1ρ, and R2 studies of ubiquitin (Haller & Schanda, 2013; Schanda et al., 2010).
2.3.1 Microsecond to Nanosecond Timescale Dynamics
Dynamic processes on the microsecond-to-nanosecond timescale include backbone fluctuations and rotation of side chain methyl groups. These motions can be observed with 1H-15N and 1H-13C dipolar as well as 13C, 15N, and 1H chemical shift anisotropy (CSA) tensor measurements, and R1 relaxation experiments. Dynamic processes on this timescale result in narrowing of the tensors below their rigid-limit value (Torchia & Szabo, 1985). The rigid limit 1H-15NH and 1H-13Cα dipolar coupling constants are 11.34 kHz (Yao et al., 2008) and 22.7 kHz (Alkaraghouli & Koetzle, 1975) respectively. Reduced 1H-13C and 1H-15N dipolar coupling strengths due to dynamics are also reflected in peak intensities in 1H-13C or 1H-15N cross polarization experiments as demonstrated by the differential 1H-15N CP buildup for the soluble and transmembrane domains of the N-terminal FMN binding domain of NADPH-cytochrome P450 oxidoreductase, a redox partner of cytochrome P450 (Huang et al., 2014). For further characterization of microsecond-to-nanosecond dynamics, several methods that rely on quantitative measurement of T1 relaxation, dipolar couplings, and CSA tensors have been developed. Reducing interference from strong 1H-1H couplings and spin diffusion have been important components in the development of methods to study microsecond to nanosecond dynamics.
Longitudinal spin-lattice relaxation (R1) is used to probe protein backbone mobility on the nanosecond timescale. Pseudo-quantitative R1 measurements were first conducted on Crh by Emsley and co-workers (Giraud et al., 2004). Quantitative R1 measurements have subsequently been applied to several microcrystalline systems including GB1 (Lewandowski et al., 2010), SH3 (Zinkevich et al., 2013), and ubiquitin (Haller & Schanda, 2013; Schanda et al., 2010), as well as the transmembrane protein ASR (Good et al., 2014), the metalloprotein SOD (Knight et al., 2012), and an amyloid-like fragment of the yeast prion protein Sup35p (Lewandowski et al., 2011c). While 15N R1 measurements are relatively straightforward, 13C R1 measurements require fast magic angle spinning (>60 kHz, (Lewandowski et al., 2010)) or selective labeling (Asami et al., 2015), in order to reduce 13C-13C proton driven spin diffusion.
Dipolar chemical shift correlation (DIPSHIFT), first presented by Griffin and co-workers (Munowitz et al., 1982; Munowitz et al., 1981) and extended to slower dynamics by DeAzevedo et al (Deazevedo et al., 2008) is a common technique for the measurement of 1H-15N and 1H-13C dipolar couplings and characterization of microsecond-to-nanosecond timescale dynamics. In traditional DIPSHIFT experiments, the magnetization evolves under the influence of the heteronuclear dipolar coupling, while 1H-1H couplings are suppressed with phase-modulated Lee-Goldburg decoupling (PMLG) (Vinogradov et al., 1999). Alternatively, DISPSHIFT-based RN-symmetry recoupling experiments can be used for the measurement of 1H-15N and 1H-13C dipolar couplings (Fig. 7, (Hou et al., 2011b)). These sequences selectively reintroduce heteronuclear dipolar couplings while reducing interference from homonuclear dipolar couplings (Carravetta et al., 2000). In addition, these pulse sequences are suitable for fast MAS frequencies and can be used in fully protonated systems (Hou et al., 2011b). In these R symmetry sequences, the heteronuclear dipolar coupling is reintroduced with a rotor-synchronized RNnν radio frequency pulse train applied on the proton channel. An NCA, NCO, or 13C-13C correlation dimension is incorporated for site-specific determination of dynamics. Recently a modification of the RN-DIPSHIFT experiment, Phase-Alternating R-Symmetry (PARS), was developed (Hou et al., 2014). This sequence incorporates a phase-shifted RN symmetry block applied on 1H, with π pulses applied on the X channel and efficiently suppresses effects from the 1H chemical shift anisotropy. Further, the series of X channel pulses refocuses the chemical shift, eliminating the need for a Hahn echo and giving the experiment inherently higher sensitivity than RN-DIPSHIFT. Under fast MAS conditions (≥ 60 kHz), cross polarization with variable contact time (CPVC) is another promising approach for characterization of motions on these timescales (Paluch et al., 2013; Paluch et al., 2015b; Zhang et al., 2015). This approach has been recently demonstrated for recording motions in aromatic groups in GB1 and dynein light chain LC8 proteins (Paluch et al., 2015a).
Fig. 7.
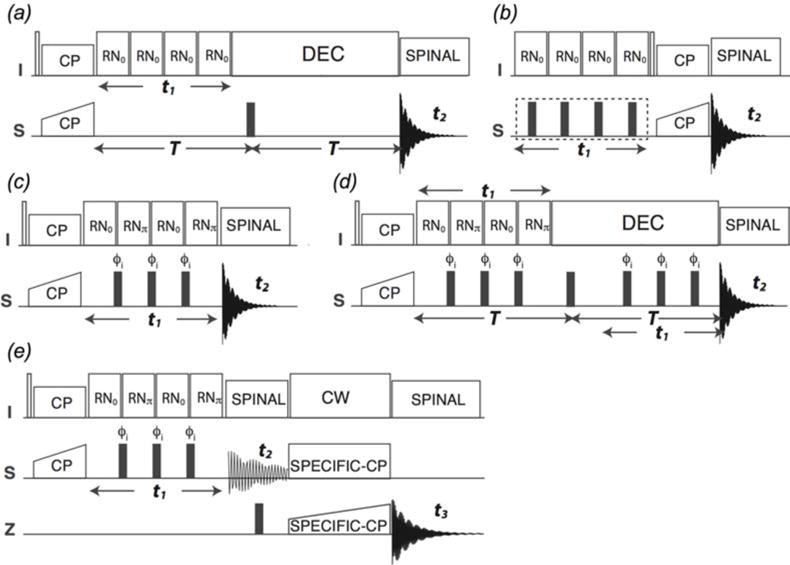
RN-symmetry based sequences for the measurement of dipolar and chemical shift anisotropy lineshapes. (Hou et al., 2014) (a) conventional RN-based DIPSHIFT, (b) 1H CSA recoupling with or without heteronuclear decoupling, (c) PARS, (d) constant time PARS, (e) 3D PARS for dipolar lineshapes measurements. Reproduced with permission from Hou et al., J. Chem. Phys., 2014, 141 (10). Copyright 2014 AIP Publishing.
CN (Chan & Tycko, 2003) and RN (Hou et al., 2012; Hou et al., 2010; Hou et al., 2013b) symmetry sequences for measurement of chemical shift tensors have also been established (dubbed ROCSA and RNCSA respectively). Like their dipolar counterparts, these experiments can be used under fast magic angle spinning and in fully protonated, uniformly 13C enriched systems, with effective suppression of homonuclear dipolar couplings. Several variations of these RN symmetry pulse sequences for the study of microsecond-to-nanosecond dynamics are presented in Fig. 7. RN-DIPSHIFT, PARS, and RNCSA experiments have been successfully applied to a range of supramolecular assemblies, with several studies highlighted below.
2.3.2 Millisecond to Microsecond Timescale Dynamics
Biological processes on the millisecond-to-microsecond timescale include domain motions and enzyme catalysis. Rotating frame (R1ρ) and transverse (R2*) spin-lattice relaxation and 15N-13C dipolar couplings are sensitive to dynamics on this timescale. Quantitative spin-lattice relaxation methods can measure exchange rates, population distributions, and chemical shift differences among exchanging sites. An important consideration for relaxation-based dynamics studies is interference from relaxation mechanisms unrelated to dynamics. These issues can be overcome by the use of deuteration and/or fast magic angle spinning (Lewandowski et al., 2011b; Quinn & McDermott, 2012; Tollinger et al., 2012).
Rotating frame based experiments measure relaxation resulting from spatial reorientation of a CSA or dipolar tensor and are sensitive to microsecond dynamics. Lewandowski et al. quantified site-specific backbone dynamics of GB1 with 15N and 13C R1ρ relaxation measurements (Fig. 8(a,b), (Lamley et al., 2015; Lewandowski et al., 2011b)). The method has been recently applied to the membrane protein Anabaena sensory rhodopsin by Ladizhansky and co-workers (Good et al., 2014). Krushelnitsky and co-workers presented a suite of relaxation studies on SH3 over a range of timescales that included R1ρ relaxation, as well as 1H-15N dipolar couplings and R1 relaxation (Zinkevich et al., 2013). As technology and methodology in the field of MAS NMR continue to advance, these experiments are being applied to increasingly complex systems.
Fig. 8.
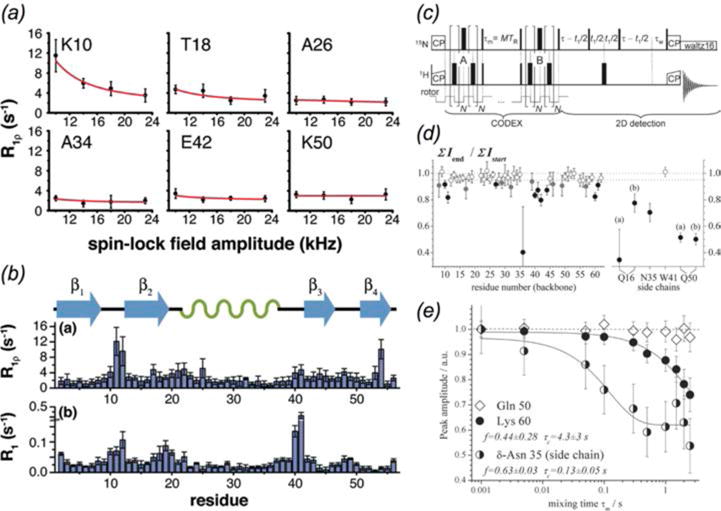
Methods for millisecond to microsecond timescale dynamics measurements. (a) Backbone amide 15N R1ρ relaxation dispersion curves for select GB1 residues (Lewandowski et al., 2011b). (b) Residue-specific 15N R1 and R1ρ relaxation rates for GB1. (c) Dipolar CODEX pulse sequence (Krushelnitsky et al., 2009). (d) Residue-specific intensity ratios for dipolar CODEX measurements of SH3. (e) Peak intensity ratio as a function of CODEX mixing time for select SH3 residues. Residues that lack slow dynamics (e.g. Gln 50) exhibit no mixing time dependence. (a,b) Reprinted with permission from Lewandowski et al., J. Am. Chem. Soc., 2011, 133 (42), 16762–16765. Copyright 2011 American Chemical Society. (c-e) Reprinted with permission from Krushelnitsky et al., J. Am. Chem. Soc., 2009, 131 (34), 12097–12099. Copyright 2009 American Chemical Society.
Carr-Purcell-Meiboom-Gill (CPMG) measurements, sensitive to sub-millisecond motions, have been well established for the study of protein dynamics in solution state NMR (Epstein et al., 1995; Farrow et al., 1994; Lorieau et al., 2010; Mandel et al., 1995; Meiboom & Gill, 1958; Volkman et al., 2001; Zhang et al., 2006). These experiments have recently been extended to applications in the solid state, as demonstrated by Schanda and co-workers on microcrystalline ubiquitin (Tollinger et al., 2012). In this study, differential line broadening of zero- and double-quantum coherences as an initial indicator of dynamics was also utilized (Dittmer & Bodenhausen, 2004). Dipolar evolution time from REDOR dephasing curves can also serve as a measure of millisecond timescale dynamics. In this case, reduced effective 15N-13C dipolar couplings at dynamic sites result in the absence of REDOR dephasing, as observed for the important linker residue Tyr145 in HIV-1 capsid assemblies (Byeon et al., 2012). Reduced 15N-13C dipolar couplings and hence the presence of millisecond timescale dynamics can also be reflected in reduced N-C cross peak intensities (Helmus et al., 2008).
Exchange experiments such as CODEX (center-band only detection of exchange) can monitor slow dynamics on the timescale of milliseconds-to-seconds. In a CODEX experiment, dipolar (Krushelnitsky et al., 2009; Li & McDermott, 2009) or CSA (deAzevedo et al., 1999) recoupling is applied before and after a mixing period. Dynamic residues will undergo only partial refocusing after the mixing period and exhibit reduced peak intensities. CODEX has been applied to characterize site-specific backbone and side chain dynamics of α-spectrin SH3 (Fig. 8(c–e), (Krushelnitsky et al., 2009)), as well as small organic molecules (Li & McDermott, 2012).
2.4 Intermolecular Interactions
Protein-protein and protein-ligand interactions are integral in the regulation of cellular processes. Understanding these interactions can be very advantageous for guided design of therapeutics. Protein-protein interactions are a hallmark of biological assemblies. MAS NMR is a powerful method for the study of intermolecular interactions as many protein-protein and protein-ligand complexes, including membrane proteins, amyloids, and viruses exist in non-crystalline or insoluble environments.
2.4.1 Chemical Shift Perturbations
Analysis of chemical shift perturbations, including intensity changes and line broadening, between free and bound states of a system of interest can indicate sites affected by the presence of a binding partner as demonstrated in early work of ligand binding to Bcl-xL with MAS NMR from Zech and McDermott (Zech et al., 2004). Chemical shift perturbations indicate not only direct protein-protein (Liu et al., 2016) or protein-ligand interactions (Schutz et al., 2011) but also allosteric structural changes that occur as a result of binding. An extension of chemical shift perturbations, changes in the CSA tensor magnitude can also serve as a probe of intermolecular interactions, such as applied to studies of cytb(5) in complex with cytP4502B4 (Pandey et al., 2013). Further MAS NMR methods exist to characterize the bona fide intermolecular binding interface. For example, paramagnetic relaxation enhancement in sites distal to the spin label can also be used as an indicator of intermolecular interactions (Wang et al., 2012). Dipolar edited correlation techniques have been particularly productive for the study of direct intermolecular interactions in biological macromolecular assemblies.
2.4.2 Dipolar-Edited Correlation Spectroscopy
One approach to identifying intermolecular interfaces is the use of differential isotopic labeling of the binding partners, where one binding partner is 13C or 13C,15N labeled and the other is 15N labeled, first demonstrated by Polenova (Marulanda et al., 2004; Yang et al., 2008) and Baldus (Etzkorn et al., 2004; Weingarth & Baldus, 2013). Long range polarization transfer across the intermolecular interface is achieved using heteronuclear mixing sequences such as REDOR (Gullion & Schaefer, 1989) or TEDOR (Hing et al., 1992), NHHC (Lange et al., 2002), or PAIN (Lewandowski et al., 2007), or a combination of these methods such as REDOR-PAINCP (Fig. 9, Panel 2, (Yang et al., 2008)). These techniques have been applied for the observation of inter-monomer interactions in microcrystalline Crh (Etzkorn et al., 2004) and structural characterization of α-synuclein amyloid fibrils (Lv et al., 2012) with NHHC correlation experiments, PAIN-derived intermolecular correlations in thioredoxin reassemblies (Yang et al., 2008), and quaternary structure of GB1 crystals with TEDOR (Nieuwkoop & Rienstra, 2010). An alternate approach is to use complementary, selective 13C labeling such as a mixture of [1-13C]glucose and [2-13C]glucose labeled protein to observe intermolecular 13C-13C correlations. This scheme has been applied to α-synuclein fibrils (Loquet et al., 2010).
Fig. 9.
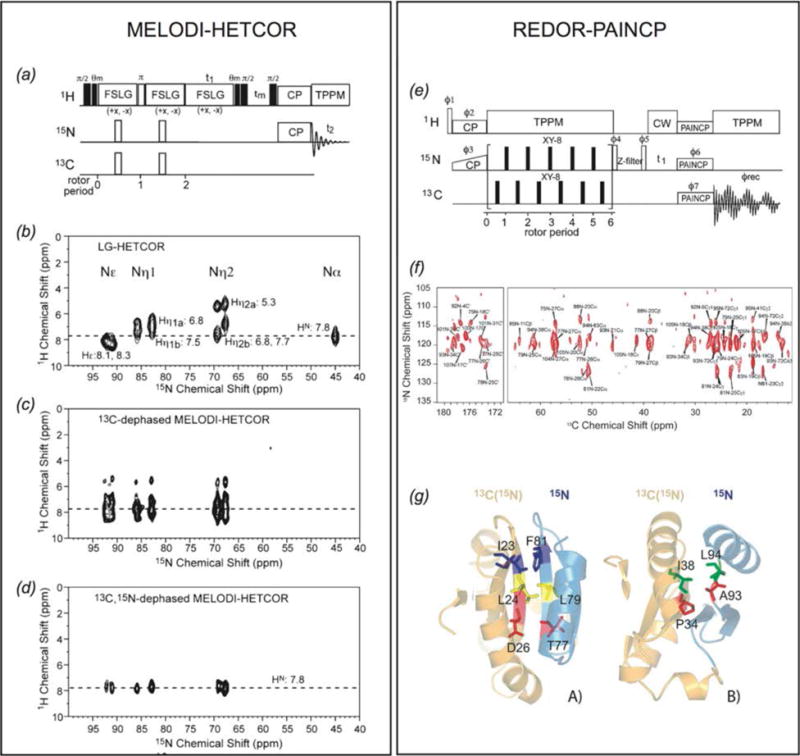
Two methods for the study of intermolecular interactions in protein assemblies. (Panel 1) MELODI-HETCOR (a) MELODI-HETCOR pulse sequence. (b–d) LG-HETCOR 1H-15N spectra of an Arg-rich membrane embedded peptide (b) no REDOR dephasing, (c) only 1H-13C REDOR dephasing, (d) both 1H-13C and 1H-15N REDOR dephasing. (Li et al., 2010) (Panel 2) REDOR-PAINCP (e) pulse sequence for REDOR-PAINCP experiment. (f) 2D 15N-13C REDOR-PAINCP spectra of thioredoxin. (g) Observed intermolecular correlations plotted onto the structure of thioredoxin. (Yang et al., 2008) (a–d) Adapted with permission fron Li et al., J. Phys. Chem. B, 2010, 114 (11), 4063–4069. Copyright 2010 American Chemical Society. (e–g) Adapted with permission from Yang et al., J. Am. Chem. Soc., 2008, 130 (17), 5798–5807. Copyright 2008 American Chemical Society.
A considerable drawback to the approach described above is the requirement that both species are isotopically labeled. Many systems of interest cannot be readily prepared with isotopic labels. Thus methods to observe binding interfaces where one binding partner is natural abundance are essential. These interface correlations can be achieved in REDOR-filter based experiments. In these experiments, REDOR dephasing is applied to 1H-13C and/or 1H-15N dipolar couplings to eliminate magnetization arising from directly bonded protons. Subsequent 1H polarization transfer arises from the source without 13C/15N labels and magnetization is transferred to the magnetically active nuclei of the isotopically labeled protein at the binding interface. This approach has been demonstrated for the study of natural abundance rhodopsin in complex with 13C-labeled 11-cis-retinal (Kiihne et al., 2005) and assemblies of U-2H,13C,15N-CAP-Gly with natural abundance polymerized microtubules assembles (Yan et al., 2015a). A significant drawback of this approach in the latter study is the presence of residual deuteration at ca. 1%, resulting in unwanted intramolecular transfers. To overcome this limitation, double-REDOR filter approach (dREDOR) was demonstrated on assemblies of U-13C,15N-CAP-Gly with natural abundance polymerized microtubules (Yan et al., 2015a). In this method, a double 1H-13C/1H-15N REDOR filter is applied to dephase the 1H signals from the U-13C,15N-CAP-Gly, with the subsequent transfer of 1H magnetization from the microtubules back to U-13C,15N-CAP-Gly across the intermolecular interface.
In addition to the observation of protein-protein or protein-ligand interactions, REDOR-filter based experiments, such as MELODI-HETCOR (Fig. 9, Panel 1, (Yao et al., 2001)) have also been applied to the study of water-protein interactions, including the identification of a hydrated arginine for an antimicrobial peptide in a lipid bilayer (Li et al., 2010) and the characterization of Pf1 bacteriophage hydration (Purusottam et al., 2013; Sergeyev et al., 2014).
In a variation of these dipolar edited methods, Asami et al. identified residues of the protein L7Ae that form the binding interface with box C/D RNA in the protein-RNA complex using 2H, 15N-labeled protein and 1H, 13C, 15N- or 2H, 13C, 15N-labeled RNA. In this experiment, protein residues interacting with 1H-RNA exhibited line broadening and weaker peak intensities relative to the corresponding peaks in the 2H-RNA complex due to 1H-dipolar interactions (Asami et al., 2013).
Beyond qualitative identification of intermolecular interfaces, the distance dependence of dipolar couplings (1/r3) can be used to quantify protein-protein and protein-ligand intermolecular distances. In addition to applications in structure calculation and dynamics studies, REDOR dephasing or TEDOR build-up curves can be used to quantify intermolecular distances. Protein-protein intermolecular distances can lend insight into higher-order structural features, as has been demonstrated for amyloid fibrils by Tycko, Dobson, and others (Fitzpatrick et al., 2013; Petkova et al., 2006; van der Wel et al., 2010) (Fig. 10) and membrane protein architecture by Griffin and co-workers (Andreas et al., 2015). REDOR distance measurements are also used for the studies of ligand-protein interactions, including binding of amantadine and its derivatives in the M2 channel by Cross (Wright et al., 2016) and Hong (Cady et al., 2010) and retinal binding in bacteriorhodopsin (Helmle et al., 2000). REDOR-derived distances can also indicate changes in protein or nucleic acid conformation upon ligand binding, demonstrated for tat peptide-bound TAR RNA of HIV-1 (Olsen et al., 2005).
Fig. 10.
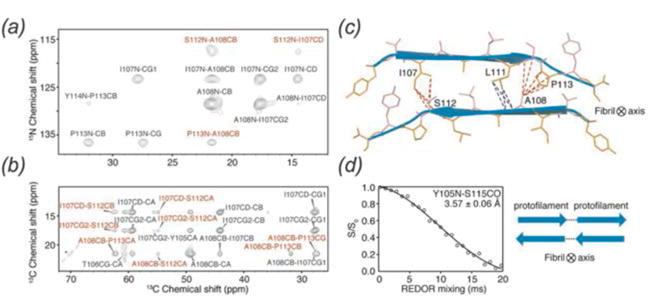
Application of REDOR distance measurements to a selectively labeled amyloid protofilament revealed anti-parallel stacking of the β-sheets. (a) 2D 15N-13C ZF-TEDOR spectrum, (b) 2D 13C-13C PDSD spectrum, (c) cross section of anti-parallel β-sheets, red and blue lines indicate intermolecular distances measured, (d) REDOR dephasing curve of residues Y105 and S115, indicating head-to-tail arrangement of the protofilament. (Fitzpatrick et al., 2013) Reproduced with permission from Fitzpatrick et al., Proc. Nat. Acad. Sci., 2013, 110 (14), 5468–5473. Copyright 2013 National Academy of Sciences.
Various classes of supramolecular assemblies studied by MAS NMR include amyloid systems (reviewed in (Comellas & Rienstra, 2013; Tycko, 2011)), the Shigella type-III secretion system (TSS3) (Demers et al., 2014; Loquet et al., 2013a; Loquet et al., 2012), the E. coli pilus protein FimA (Habenstein et al., 2015), and the mitochondrial antiviral signaling protein (MAVS) (He et al., 2016; He et al., 2015). This review focuses on two particular classes of supramolecular assemblies: cytoskeletal proteins and viruses.
3. MAS NMR of Cytoskeleton-Associated Proteins
The cytoskeleton is an essential cellular component in all domains of life. Functions of the cytoskeleton include maintenance of cell shape, motility, intracellular transport, endocytosis, and cell signaling (Fischer & Fowler, 2015). In eukaryotes, the cellular cytoskeleton is composed of three main filament types: microfilaments (actin filaments), microtubules (Nogales, 2000), and intermediate filaments. While most filaments of the prokaryotic cytoskeleton have eukaryotic analogues, there are filament types that are unique to prokaryotes (Lowe et al., 2004). Function of the cytoskeleton is crucially dependent on interactions with binding partners, including the motor proteins dynein, kinesin, and myosin (Vale, 2003). The disruption of these interactions by small molecules is a key mechanism in therapeutics for the treatment of cancers (Wood & Bergnes, 2004) and neurodegenerative diseases (Gunawardena, 2013).
3.1 Microtubules and Microtubule-Associated Proteins
Microtubules (MTs) and their associated proteins (MAPs) perform several vital physiological functions in the cell including mitosis and transport of signaling molecules (Vale, 2003). Microtubules are an important target of chemotheraputics due to their essential roles in cell division. MTs are highly dynamic and continually polymerizing and de-polymerizing in the cellular matrix (Howard & Hyman, 2003). Despite extensive structural and biochemical characterization, many open questions remain with respect to the function of microtubules and their associated proteins, including the atomic level understanding of protein-protein interactions, of the role of protein dynamics in different conformational states, and of how protein-protein interactions and dynamics come together to orchestrate cellular processes. MAS NMR can lend insight into the structure and dynamics of microtubule-MAP complexes at atomic resolution. To date, in-depth MAS NMR studies have been performed on only a handful of systems, including dynactin’s CAP-Gly domain assembled with microtubules, bactofilin, and microtubules interacting with small molecules, such as paclitaxel (Taxol) (Li et al., 2000; Paik et al., 2007), epothilone B (Kumar et al., 2010), and their derivatives. In the following we review the work on the first two cytoskeletal assemblies.
3.1.1 Structure of CAP-Gly Domain of Dynactin
Dynactin, an activator of the dynein motor assembly, is a protein complex involved in intracellular transport (Caviston & Holzbaur, 2006). Dynactin regulates dynein transport along microtubules, and mutations within its p150Glued subunit lead to neurodegenerative disorders, such as Huntington’s disease, Charcot-Marie Tooth disease, amyotropic lateral sclerosis (ALS), distal spinal bulbar muscular atrophy, and Perry syndrome (Chen et al., 2014). Within the p150Glued subunit of dynactin, the CAP-Gly (cytoskeleton-associated protein glycine-rich) is an 89 residue microtubule-binding domain (Vaughan et al., 2002; Waterman-Storer et al., 1995). Dynactin CAP-Gly is the first MAP assembled with MTs, whose structure and dynamics have been investigated in depth by MAS NMR, yielding atomic-resolution insights unavailable from other techniques and establishing a proof of principle for investigations of other cytoskeleton-associated assemblies (Ahmed et al., 2010; Sun et al., 2009; Yan et al., 2015a; Yan et al., 2013a; Yan et al., 2015b). Recently, the atomic-level resolution structure of CAP-Gly bound to polymeric microtubules was reported (Yan et al., 2015a) (PDB ID code 2MPX), the first structure of any cytoskeleton-associated protein assembled with cytoskeleton.
To determine the structure of CAP-Gly in complex with MTs, three different isotopic labeling schemes were used: U-15N,13C; U-15N, [2-13C]glucose; and U-15N, [1,6-13C]glucose. The structure was determined using hundreds of medium- and long-range distance restraints collected from 13C-13C CORD and 15N-13C PAIN-CP experiments, as well as hydrogen bonding restraints and torsion angles from TALOS+. The equivalent resolution in the structure was 1.9–2.5 Å with a tightly constrained ensemble of lowest-energy conformers. A very similar approach was previously used to determine the structure of free CAP-Gly (PDB ID code 2M02) (Yan et al., 2013a). The structure of CAP-Gly assembled on MTs indicates that, while the overall secondary structure is retained, the flexible loops of CAP-Gly have remarkably different conformations when associated with microtubules (Fig. 12). Loop β3/β4 adopts a more open conformation in the free state of CAP-Gly, and rearranges to a more closed conformation when bound to MTs. The different sidechain orientations of residues in this loop may play a role in CAP-Gly’s structural plasticity and ability to interact with different binding partners.
Fig. 12.
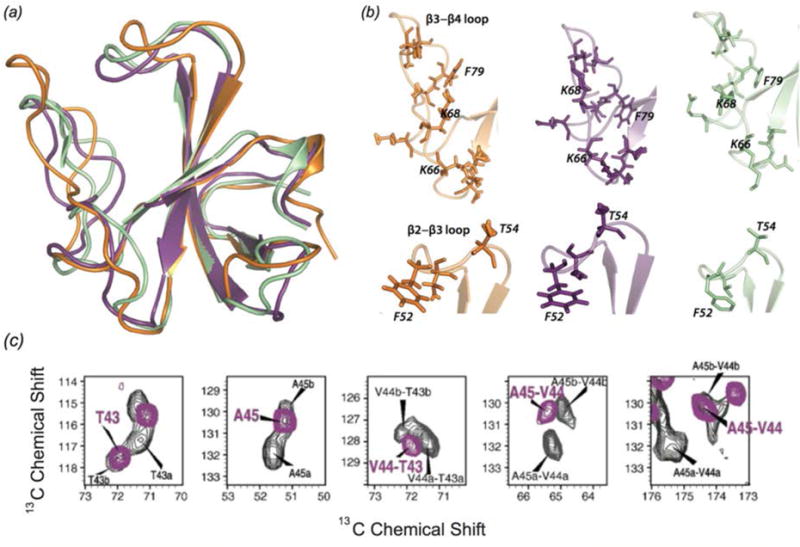
(a) Structure of CAP-Gly bound to polymerized microtubules (purple, 2MPX) and free CAP-Gly (orange, 2M02), both determined with MAS NMR, and CAP-Gly bound to EB1 (green, 2HKQ). (b) Expansion of loop regions of CAP-Gly in the three systems, indicating the differences in loop position and sidechain orientation for CAP-Gly in its three different states. (Yan et al., 2015a) (c) Chemical shift perturbations for several residues in CAP-Gly indicating multiple conformers of free CAP-Gly (black) that collapse to a single conformer in complex with EB1. (Yan et al., 2013a) (a) and (b) Adapted with permission from Yan et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14611–14616. Copyright 2015 National Academy of Sciences. (c) Adapted with permission from Yan et al., J. Mol. Biol., 2013, 425 (22), 4249–4266. Copyright 2013 Elsevier.
EB1 is another MAP that, like dynactin, localizes at the plus end of the growing microtubule. EB1 is thought to have a role in MT dynamics; specifically it may promote MT elongation (Rogers et al., 2002). EB1 interacts with the p150Glued subunit of dynactin and it is hypothesized that the two proteins form a plus end complex to regulate microtubule dynamics (Ligon et al., 2003). The p150Glued subunit may play a role in recruiting EB1 to the microtubules. Residues of CAP-Gly that are perturbed by binding of EB1 were identified by chemical shift changes. Chemical shift perturbations indicate that free CAP-Gly exists in multiple conformers, but is conformationally homogeneous when bound to MTs (Yan et al., 2013a).
3.1.2 Interface of CAP-Gly with Microtubules
The main challenge for NMR characterization of microtubules and their assemblies with associated proteins is that to date there have been no efficient isotopic labeling protocols established for tubulin. This precludes in-depth structural characterization of microtubules and limits the approaches for determination of intermolecular interfaces formed by microtubules and their binding partners. To overcome this challenge, the application of double REDOR filters (dREDOR) was explored to characterize the intermolecular interfaces that dynactin’s CAP-Gly forms with MTs and EB1 (Yan et al., 2015a). In these experiments, 1H-13C and 1H-15N REDOR filters were simultaneously applied to dephase all 1H magnetization arising from U-13C,15N CAP-Gly. Subsequently, polarization was transferred from 1H of natural abundance microtubules or EB1 to their binding interface on the surface of CAP-Gly. A 13C-13C CORD dimension was included to enable site-specific assignment of the binding interface. Fig. 13 shows dREDOR-CORD and dREDOR-HETCOR spectra of CAP-Gly in complex with microtubules (b), EB1 (c), and the intermolecular interfaces as determined by dREDOR (a left, green) and chemical shift perturbations (a right, yellow/purple). The results confirmed that loop β3/β4 including the GKNDG motif comprises the primary binding interface with MTs. CAP-Gly interacts with its binding partners on the flat side of the protein, where most of the surface-exposed hydrophobic residues are located. dREDOR experiments of the CAP-Gly/EB1 complex were consistent with the known binding interface for this complex, which has been determined previously (Hayashi et al., 2005; Honnappa et al., 2006; Yan et al., 2013a), validating the approach for characterizing the CAP-Gly/MT interface.
Fig. 13.
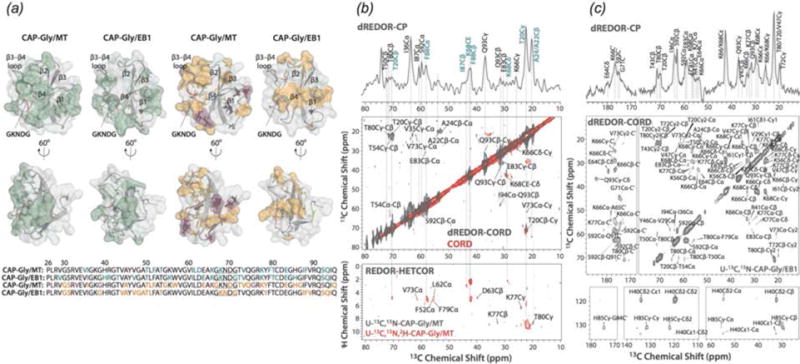
(a) Intermolecular interfaces of CAP-Gly with MT and EB1 determined with dREDOR (left, green), and observed chemical shift perturbations (right, purple/orange). For chemical shift perturbations, purple residues indicate large shifts >1 ppm, orange indicates shifts between 0.5 and 1 ppm. (b) dREDOR-HETCOR and dREDOR-CORD spectra of U-13C,15N CAP-Gly bound to MT and (c) in complex with EB1. (Yan et al., 2015a) Reproduced with permission from Yan et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14611–14616. Copyright 2015 National Academy of Sciences.
3.1.3 Dynamics of CAP-Gly
Microtubule-associated motors and their activators possess conformational plasticity, which is essential for their ability to bind to and slide along the microtubules (Howard, 2001; Vale, 2003). Conformational plasticity is directly related to internal flexibility, and therefore, knowledge of dynamics is essential for understanding the biological function of microtubule-associated protein assemblies. The dynamics of CAP-Gly free, in complex with EB1, and bound to polymeric microtubules have been characterized using MAS NMR, over a range of functionally relevant timescales from nanoseconds to milliseconds (Yan et al., 2015b). Global dynamics were probed through a temperature series of 1- and 2-dimensional 13C spectra (Fig. 14(a–c)). Site-specific dynamics were characterized using 1H-15N and 1H-13C dipolar order parameters (Fig. 14(d)) (Hou et al., 2011b). As indicated by the temperature series in Fig. 14(a–c), free and microtubule-bound CAP-Gly are dynamic across the entire range of timescales under investigation, and the motions are strongly temperature dependent. In contrast, the CAP-Gly in complex with EB1 is largely rigid on these timescales and its spectra are not temperature dependent. From the measurement of dipolar order parameters, it was found that the loops of CAP-Gly are mobile in the free protein as well as in complex with microtubules. However, consistent with 1- and 2-dimensional spectra, the dynamics of CAP-Gly are notably attenuated when in complex with EB1. Remarkably, the loops of CAP-Gly show an increase in fast timescale backbone fluctuations (nanosecond-to-microsecond), but a decrease in slower dynamics (microsecond-to-millisecond) upon binding to MTs (Fig. 14(d)). It was proposed that these observed changes in dynamics have a critical function in CAP-Gly/MT interactions. The combined structural and dynamics studies of CAP-Gly highlight the structural plasticity of this protein and the essential role this flexibility plays in CAP-Gly’s ability to adopt different conformations and interact with different binding partners.
Fig. 14.
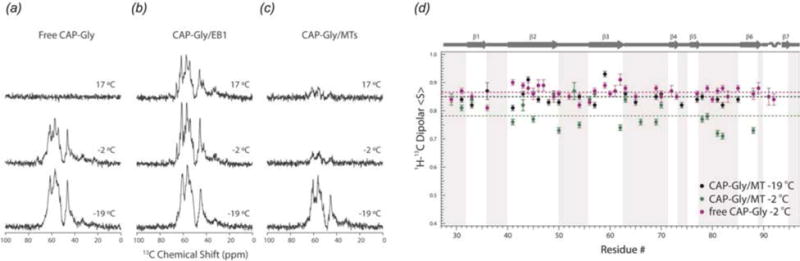
15N-13C SPECIFIC CP NCA 1D spectra indicating temperature dependence of global conformational dynamics of (a) free CAP-Gly, (b) CAP-Gly/EB1 complex, (c) CAP-Gly bound to microtubules. (d) Dipolar order parameters of free CAP-Gly at −2°C (purple), and microtubule-bound CAP-Gly at −2°C (green) and −19°C (black). The micro-to-nanosecond timescale dynamics of MT-bound CAP-Gly are enhanced at −2° in comparison to the free protein. (Yan et al., 2015b) Adapted with permission from Yan et al., J. Biol. Chem., 2015, 290 (3), 1607–1622. Copyright 2015 The American Society for Biochemistry and Molecular Biology.
3.2 Bactofilins
Bactofilins are a recently discovered class of bacterial cytoskeletal proteins. Similar to eukaryotic cystoskeletal proteins, these proteins have diverse functions, such as cellular mobility, cell shape, and attachment. Bactofilins assemble rapidly and spontaneously, making them not amenable for characterization by many biophysical techniques (Kuhn et al., 2010). Bactofilins contain a conserved central rigid DUF583 domain, and terminal regions that are more dynamic. Lange and co-workers have recently reported resonance assignments and structure for BacA from Caulobacter crescentus (Shi et al., 2015; Vasa et al., 2015). Only the core DUF583 domain (residues 39–137) was observed in dipolar based 13C-13C and 15N-13C spectra, supporting the hypothesis that while the core is rigid, the termini are dynamic and believed to have a role in protein-protein or protein-membrane interactions. Secondary structure analysis revealed at least 10 distinct β-sheet segments (Fig. 15). To observe dynamic residues, through-bond INEPT (Morris & Freeman, 1979) 13C-13C correlation spectra were acquired. Chemical shifts for residues in the INEPT spectra indicated random coil secondary structure. Interestingly, fewer resonances were observed in the INEPT spectrum than would be expected to arise from the N-and C-termini, indicating that not all residues in these regions are dynamic. While the secondary structure and dynamic behavior of BacA have similar features to amyloids (Daebel et al., 2012; Heise et al., 2005; Helmus et al., 2010), BacA has a distinct β-helical tertiary structure, as indicated by mass-per-length measurements by scanning transmission electron microscopy.
Fig. 15.
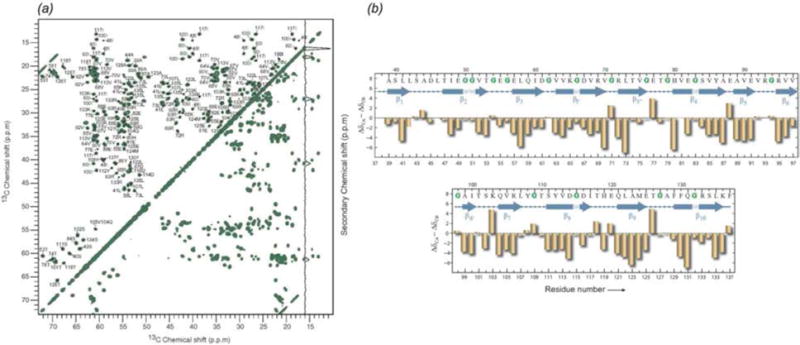
(a) 13C-13C proton-driven spin diffusion (PDSD) correlation spectrum of U-13C, 15N BacA. (b) Secondary structure of the core domain DUF583 of BacA determined from secondary chemical shift analysis. (Vasa et al., 2015) Adapted with permission from Vasa et al., Proc. Nat. Acad. Sci., 2015, 112 (2), E127–E136. Copyright 2015 National Academy of Sciences.
The atomic resolution structure determined by Shi et al. revealed that BacA is a right handed β-helix with a triangular hydrophobic core and 6 windings (Shi et al., 2015). With sparsely labeled BacA samples (1,3-13C and 2-13C glycerol), medium- and long-range distance restraints were obtained, as well as torsion angle restraints from TALOS+ and β-sheet hydrogen bond restraints. Additional 1H-1H distance restraints from a 4-dimensional HN(H)(H)NH spectrum (acquired with sine weighted Poisson-gap non-uniform sampling (Hyberts et al., 2012; Hyberts et al., 2010)) and a 2-dimensional NHHC spectrum were essential for determination of the handedness of the β helical structure. The presence of a right-handed β-helix had not been previously reported for any cytoskeletal protein. The hydrophobic core is triangular with highly conserved glycines at many of the corners and three parallel β-sheets per winding (Fig. 16). It is believed that hydrophobic interactions mediate polymerization/folding of bactofilins (Kuhn et al., 2010). Hydrogen bonds between adjacent β-strands helps to stabilize the overall structure. Windings 1 and 6 were not as well restrained due to a lack of intermolecular distance restraints, which is attributed to increased dynamics in these regions of the protein. Mutations of hydrophobic residues in winding 6 affect BacA assembly in vivo (Kuhn et al., 2010). It is likely that dynamics in this region of the protein has a role in BacA assembly.
Fig. 16.
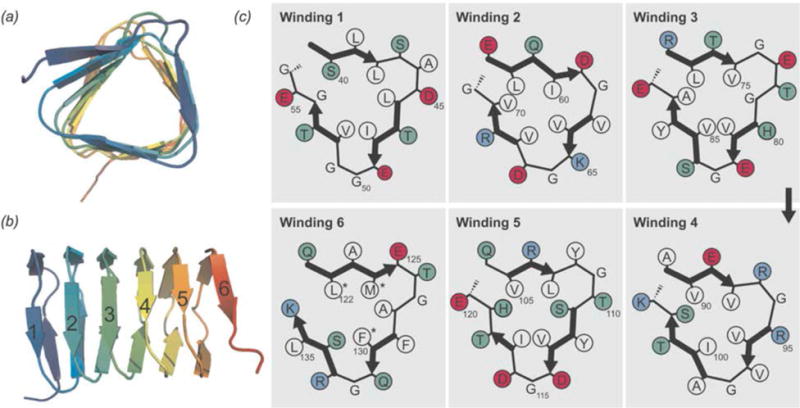
Top view (a) and side view (b) of BacA structure determined by MAS NMR. (c) Schematic representation of the 6 windings. Colors are as follows: white-hydrophobic residues, red-acidic residues, blue-basic residues, green-other residues. Mutations of asterisked residues in winding 6 affect in vivo assembly. (Shi et al., 2015) Reprinted with permission from Shi et al., Sci. Adv. 2015, 1 (11). Copyright 2015 American Association for the Advancement of Science.
4. MAS NMR of Viral Assemblies and Intact Viral Particles
Viruses are small pathogens that can impact all forms of life from bacteria, to plants and animals (Brussaard et al., 2004; Nelson & Citovsky, 2005; Pearson et al., 2009; Prangishvili, 2013; Smith & Helenius, 2004). The general virion structure is a single or double stranded DNA or RNA encapsulated by a protein coat or capsid. Some viruses include a lipid envelope surrounding the capsid as well. Viruses invade target cells and seize the host cell’s machinery to reproduce, while evading cellular defense mechanisms (Kaminskyy & Zhivotovsky, 2010). These key aspects of viral replication are of great interest as targets for the treatment of viral infections, but can also be exploited for nanotechnology and drug development. MAS NMR is an excellent tool to probe structure and dynamics of viral protein assemblies, and particularly promising is an integrated approach, where this technique is combined with other experimental (e.g., cryo-EM, X-ray crystallography, and solution NMR) and/or computational (e.g., MD simulations) methods to yield atomic-level understanding of structure and dynamics of viral assemblies. Below we discuss two important classes of viruses, HIV-1 and bacteriophages, which to date have been the most thoroughly characterized by MAS NMR (Fig. 17).
Fig. 17.
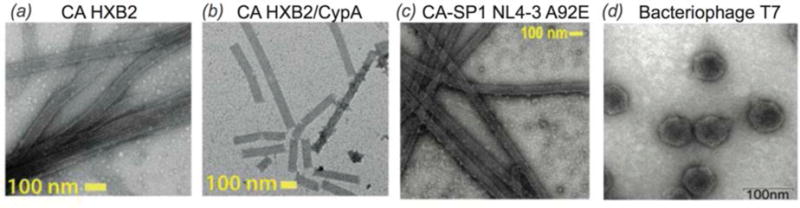
Transmission electron microscopy images of viral assemblies and viruses for MAS NMR studies: (a) tubular assemblies of HIV-1 capsid (CA), HXB2 strain, (b) CA in complex with CypA (Lu et al., 2015a), (c) tubular assemblies of CA-SP1 maturation intermediate, NL4-3 strain, A92E mutant. (Han et al., 2013), (d) T7 bacteriophage (Abramov & Goldbourt, 2014). (a, b) Adapted with permission from Lu et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14617-14622. Copyright 2015 National Academy of Sciences. (c) Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135 (47), 17793–17803. Copyright 2013 American Chemical Society. (d) Adapted with permission from Abramov et al., J. Biomol. NMR, 2014, 59 (4), 219–230. Copyright 2014 Springer.
4.1 HIV-1 Capsid and Maturation Intermediates
Acquired Immunodeficiency Syndrome (AIDS), caused by the Human Immunodeficiency Virus (HIV) is a global pandemic and affects approximately 37 million people globally (World Health Organization, 2015). A key step in the HIV lifecycle is maturation, where an immature viral particle is converted into a mature, infectious virion through a cascade of cleavage steps of the Gag polyprotein (Fig. 18 (a), Fig. 22 Panel 1). The final step of maturation is the cleavage of the SP1 tail from CA (capsid protein) and the reorganization of CA into the mature conical capsid, which encapsulates the retroviral RNA (Briggs & Krausslich, 2011; Engelman & Cherepanov, 2012). Viral maturation is of poignant interest as the target of a novel class of therapeutics termed maturation inhibitors (Adamson et al., 2009; Salzwedel et al., 2007). Maturation inhibitors such as Bevirimat (BVM) inhibit HIV-1 maturation by binding to the CA-SP1 cleavage site and preventing cleavage. Tubular assemblies of CA mimic the native capsid lattice (Byeon et al., 2009; Zhao et al., 2013). Obtaining stable, morphologically homogeneous samples for MAS NMR typically requires high ionic strength (~1–2 M NaCl). With the use of low-E and Efree probes designed to minimize heating due to radiofrequency irradiation, outstanding quality MAS NMR spectra can be obtained (Bayro et al., 2014; Byeon et al., 2012; Chen & Tycko, 2010; Han et al., 2010; Han et al., 2013; Lu et al., 2015a).
Fig. 18.
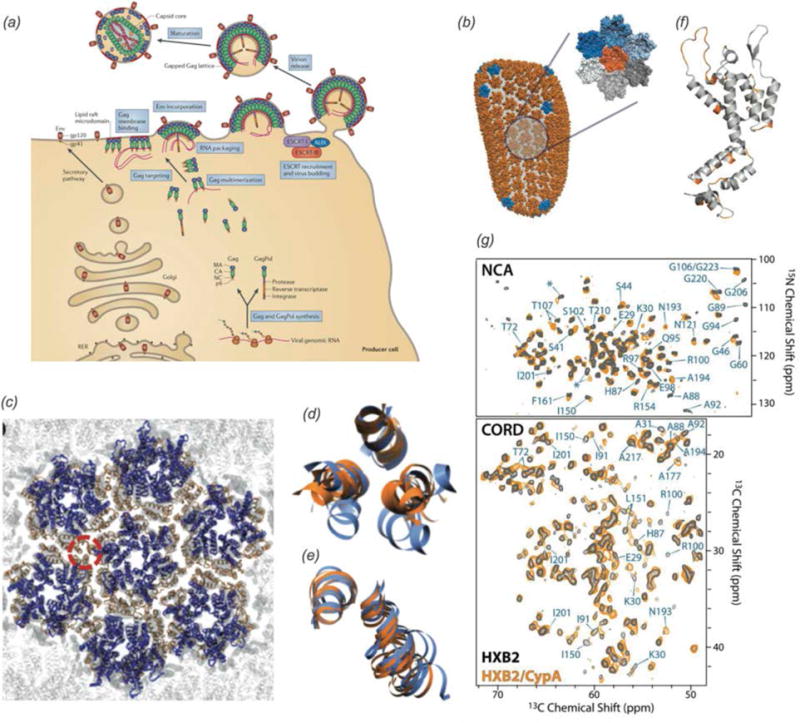
(a) Late stages of HIV-1 viral life cycle from assembly at the host cell membrane to budding and virion maturation. (Freed, 2015) (b) All-atom MD derived model of the HIV-1 capsid based on cryo-ET and solution NMR, with a hexamer of hexamers subunit shown in the expansion (Lu et al., 2015a). (c) Hexmer of hexamers of HIV-1 capsid assembly. The interhexameric trimer interface is circled. (d,e) Helix 10 trimer interface and helix 9 dimer interface respectively. Blue: hexamer of hexamers, orange: pentamer of hexamers (Zhao et al., 2013). (f) CA monomer. Residues for which chemical shift perturbations are observed upon binding of CypA are highlighted orange. (g) 15N-13C and 13C-13C correlation spectra of free CA tubular assemblies (black) and CA tubular assemblies in complex with CypA (Lu et al., 2015a) (a) Adapted with permission from Freed, Nat. Rev. Microbiol., 2015, 13. Copyright 2015 Macmillan Publishers. (b,f,g) Adapted with permission from Lu et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14617–14622. Copyright 2015 National Academy of Sciences. (c–e) Adapted with permission from Zhao et al., Nature, 2013, 497, 643–646. Copyright 2013 Nature Publishing Group.
Fig. 22.
(Panel 1) Sequence of Gag polyprotein cleavage during maturation. (Panel 2) (a–d) Direct-DARR, (e–j) INADEQUATE, and (k) CP-DARR spectra of CA (orange) and CA-SP1 (black) NL4-3 strain assembled into tubes. Selected regions show the presence of SP1 peaks not observed in the CA spectra. (Han et al., 2013) Reprinted with permission from Han et al., J. Am. Chem. Soc., 2013, 135 (47), 17793–17803. Copyright 2013 American Chemical Society.
4.1.1 Structural Characterization of HIV-1 Capsid Assemblies
CA is a 231 residue, predominantly α-helical protein with independently folding N-terminal (NTD, residues 1–143) and C-terminal (CTD, residues 148–231) domains, connected by an inter-domain linker (Deshmukh et al., 2013; Gres et al., 2015). Early studies (Chen & Tycko, 2010; Han et al., 2010) revealed that most residues in both NTD and CTD are relatively rigid, and CA assemblies retain the secondary and tertiary structure determined from solution NMR and x-ray crystallography studies. Upon maturation, the viral capsid morphology changes from spherical to conical. Mature conical HIV-1 capsids are built from hexameric and pentameric CA assemblies, whose stoichiometry in the final capsid is variable, and so is the capsid’s cone shape. Four kinds of intermolecular contacts are critical for capsid morphology and stability both in vivo and in vitro: intra-hexameric NTD-NTD and NTD-CTD, and inter-hexameric CTD-CTD interfaces around pseudo-two fold and pseudo-three fold axes (Fig. 18 c–e) (Byeon et al., 2009; Pornillos et al., 2009). The capsid’s structural polymorphism is also observed in vitro, where the morphology can be controlled by assembly conditions. CA can assemble into cones, tubes, and spheres (Barklis et al., 1998; Ehrlich et al., 2001; Ganser-Pornillos et al., 2007; Gross et al., 1997; Han et al., 2010; Han et al., 2013). MAS NMR studies of spheres, cones, and tubes indicated no major differences in secondary or tertiary structure among the three morphologies (Bayro et al., 2014; Chen & Tycko, 2010; Han et al., 2010; Han et al., 2013) but no 3D structure is available so atomic-level details of structural organization of these in vitro assemblies remain poorly understood.
HIV-1 exhibits significant sequence variability and sequence-dependent viral infectivity (Price et al., 2012). To date, two HIV-1 strains have been studied by MAS NMR: HXB2 and NL4-3 (Bayro et al., 2014; Byeon et al., 2012; Chen & Tycko, 2010; Han et al., 2010; Han et al., 2013; Lu et al., 2015a). The wild type sequences of these proteins differ by only 4 amino acids, but this variation causes considerable conformational changes across the CA sequence (Han et al., 2013). These structural perturbations may play a role in the differing viral infectivity of the two strains. Viral infectivity is also regulated through the interactions with host factors. Cyclophilin A (CypA) is one such host factor protein that modulates HIV-1 infectivity and uncoating through direct interactions with the HIV-1 capsid’s CypA-binding loop located in the NTD. The mechanism of CypA is complex and poorly understood (Colgan et al., 1996; Luban et al., 1993). A recent study yielded the structure of CypA in complex with the assembled HIV-1 capsid at 8 Å resolution (Liu et al., 2016). It was discovered, using cryo-EM-guided all-atom MD simulations and MAS NMR spectroscopy that CypA simultaneously interacts with two CA subunits in different hexamers. This binding mechanism established through the integrated cryo-EM/MAS NMR/MD approach provided insights into the mechanism of HIV-1 capsid stabilization by and recruitment of CypA to promote the viral infectivity.
Additional mutations in the various regions of CA sequence have been shown to affect the assembly morphology and efficiency, capsid stability, and interactions with host factors (Bocanegra et al., 2012; Forshey et al., 2002; Gres et al., 2015; Jiang et al., 2011; Manocheewa et al., 2013; McCarthy et al., 2013; Qi et al., 2008; Yang & Aiken, 2007). For example, mutations in the linker region of CA render the capsid assembly inefficient or abolish it completely (Jiang et al., 2011). On the contrary, the E45A mutation makes a “hyperstable” capsid, which does not disassemble, thus dramatically reducing the viral infectivity (Forshey et al., 2002; Hulme et al., 2015; Yang et al., 2012). Mutations in the CypA binding loop, such as A92E and G94 exhibits drastically reduced viral infectivity in the presence of the normally required CypA. Full infectivity can be restored by inhibition of CypA by cyclosporin. MAS NMR analysis of these mutants revealed that the mutations induce conformational changes, as well as changes in the dynamics, vide infra (Lu et al., 2015a).
To get insights into the structural changes occurring upon formation of the hexagonal CA lattice (predominant symmetry in the cones), Bayro et al. used MAS NMR-derived structural restraints in tubular CA assemblies for refinement against solution NMR and x-ray crystallography structures of monomeric CA (Bayro et al., 2014). 15N-15N backbone distances, determined by 15N-15N dipolar recoupling (15N-BARE), have been used by Tycko and coworkers to derive backbone torsion angles and secondary structure (Hu et al., 2012). In α-helices and tight turns, 15N-15N distances are short, leading to rapid signal decay, while the decay is slower in loop regions (Fig. 19(a)). Deviations of experimental 15N-BARE curves of CA tubular assemblies from simulated curves derived from crystal or solution NMR structures indicated differences in secondary structure. Using the 15N-15N distances and TALOS torsion angle predictions as structural restraints, structure refinement was carried out against CA coordinates derived from solution NMR (monomer, 2LF4 (Shin et al., 2011)) and crystal structures (hexamer, 3MGE (Pornillos et al., 2010)). Fig. 19(b–d) shows regions of the CA structure that adopt a more extended conformation in tubular assemblies than in their respective initial structures. Among these regions that undergo changes are the 310 helix near the start of the CTD, as well as the loops between helices 3 and 4, and helices 10 and 11. These perturbations were attributed to conformational changes arising from higher order oligomerization and/or lattice curvature.
Fig. 19.
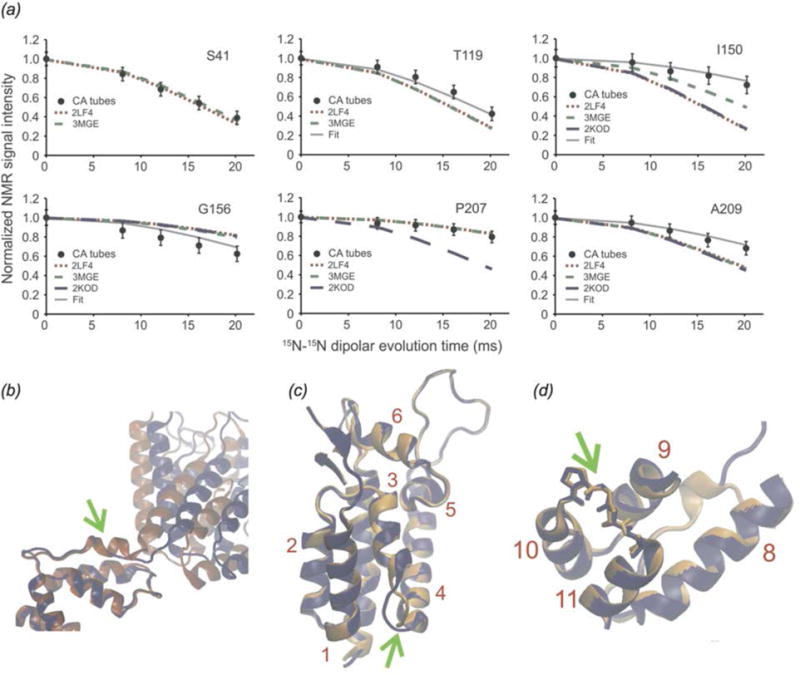
(a) 15N-15N BARE curves for selected CA residues. Circles indicate experimental curves for CA tubular assemblies. Simulated curves correspond to the following structures: (− −) 2LF4, (−) 3MGE, (−) 2KOD. (b) Initial (red) and final (blue) structure refinement against 2LF4, indicating the change in the conformation of the 310 helix. (c and d) Initial (orange) and final (blue) structure refinement against 3MGE, indicating the change in conformation of loop 3/4 and loop 10/11 respectively. (Bayro et al., 2014) Adapted with permission from Bayro et al., J. Mol. Biol., 2014, 426 (5), 1109–1127. Copyright 2014 Elsevier.
Very recently Bayro and Tycko characterized the structure of tubular assemblies of the capsid pseudo-two fold inter-hexameric interface (Bayro & Tycko, 2016) using mixed and selective labeling schemes and dipolar-based distance measurements, with additional restraints derived from the cryo-EM structure determined by Zhang and co-workers (Zhao et al., 2013). In this study, quantitative W184 to M185 intermolecular distances across the dimerization interface were obtained with 15N-13C REDOR dephasing experiments. A series of experiments were performed to characterize intra-residue distances of W184 and M185 in order to constrain the sidechain conformations including 15N-13C REDOR distances, 13C-13C BroBaRR (BroadBand Rotational Resonance] distances for M185 (Chan & Tycko, 2004), and tensor correlation experiments (Dabbagh et al., 1994) of W184 to determine the angle between the backbone Cα-N bond vector and sidechain Cδ1-Nε1 bond vector. The distances and angles determined differed somewhat from prior solution NMR (Byeon et al., 2009) and X-ray crystallography studies (Gres et al., 2015; Worthylake et al., 1999), suggesting the structure of the dimerization interface in tubular assemblies differs from that in crystals or CA in solution. Further intra-CA monomer distance constraints were obtained from 13C-13C correlations with long mixing (700 ms). With an integrated structure calculation approach described above, they were able to determine the structure of the inter-hexameric dimer interface in CA tubular assemblies, and further demonstrated that this interface is well ordered in this morphology.
4.1.2 Conformational Dynamics of HIV-1 Capsid Assemblies by MAS NMR
Protein dynamics have indispensible functions in many stages of the HIV-1 lifecycle, including assembly, release, and maturation (Freed, 2015). While CA is relatively rigid in the individual α-helices, residues in loops and the flexible linker, which are key functional regions of the protein, are dynamic, giving rise to conformational plasticity, which is directly connected to capsid morphology, interactions with host factors, and viral infectivity.
Lu et al. have characterized the site-specific dynamics of several capsid constructs, including the HXB2 and NL4-3 strains discussed above, as well as CypA-bound CA and the A92E and G94D ‘escape mutants’ (Lu et al., 2015a). These mutants have approximately 10% the infectivity of the wild type virus, however infectivity can be restored upon CypA inhibition (Ylinen et al., 2009). The CypA loop of CA (residues 84-100) undergos conformational changes upon binding of CypA as well as in the A92E and G94D “escape” mutants. 1H-15N and 1H-13C dipolar tensors and resonance intensities reported on CA dynamics on the nano-through millisecond timescales. The CypA loop was observed to exhibit an unprecedented degree of mobility over the entire range of timescales under study. As expected, the motions are attenuated upon CypA binding. A completely unexpected finding was that the dynamic profiles of the escape mutants A92E and G94D closely resembled that of CA in complex with CypA: mobility of CypA loop is dramatically attenuated due to the mutations. Furthermore, NMR parameters derived from all-atom molecular dynamics (MD) trajectories were in remarkable agreement with experimental results, corroborating that the dynamics are modulated by the mutations in the CypA loop. Fig. 20 demonstrates (a) dipolar order parameters and (b) 1H-15N dipolar lineshapes of residues in the Cyclophilin A loop with (c) peak intensities and (d) DOPs mapped to the CA structure. Taken together, these results indicate that that the capsid escapes the CypA dependence by a dynamic allostery mechanism and highlight the key role of conformational dynamics in the HIV-1 CA function.
Fig. 20.
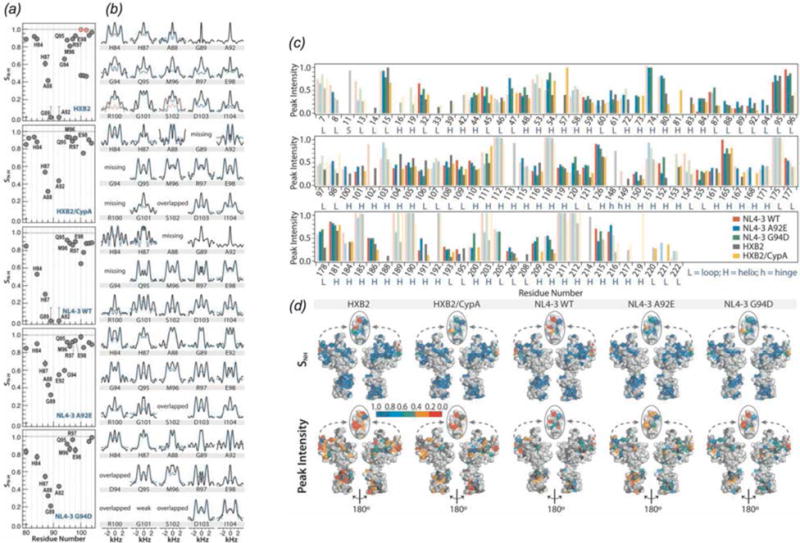
(a) 1H-15N dipolar order parameters and (b) lineshapes for residues in the CypA loop in WT CA (HXB2), cyclophilinA-bound CA (HXB2), WT CA (NL4-3), CA A92E (NL4-3), and CA G94D (NL4-3), listed from top to bottom. (c) Peak intensities observed in an NCACX correlation spectrum for each of the 5 constructs. (d) Dipolar order parameters (top) and NCA peak intensities (bottom) mapped onto the structure of CA. (Lu et al., 2015a) Adapted with permission from Lu et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14617–14622. Copyright 2015 National Academy of Sciences.
Conformational plasticity of HIV-1 CA is essential for its assembly into the viral capsid core. As discussed above, the hinge that links the NTD and CTD has been shown to play a key role in HIV-1 core stability (Byeon et al., 2009; Jiang et al., 2011). The dynamic origin and mechanism of CA’s pleiomorphic assembly was investigated by an integrated MAS and solution NMR approach, using conical assemblies of U-13C,15N-Tyr labeled samples of full length CA, of individual CTD constructs, and solutions of U-13C,15N-CA and CTD (Byeon et al., 2012). Tyrosine is an ideal probe of linker dynamics as functionally important Y145 is located in the linker; CA has only 3 additional Tyr residues located in rigid regions of the protein, which provide internal controls in the NMR characterization. To observe dynamics on the millisecond timescale, 13C-15N REDOR dephasing experiments were performed. The dephasing profiles for the three Tyr residues located in the helical regions of CA are consistent with them being rigid. On the contrary, the Y145 signals could not be dephased due to the complete averaging of the 13C-15N dipolar couplings, which is a clear manifestation of dynamics occurring on the timescales on the order of ~10 ms (Gullion & Schaefer, 1989). To probe the dynamics of the interdomain linker on the microsecond to nanosecond timescale, 1H-15N dipolar and 15N chemical shift anisotropy (CSA) lineshapes were acquired. All Tyr residues, including Y145 showed close to rigid limit dipolar and CSA parameters, establishing that Y145 is rigid on the corresponding timescales. On the basis of these results it was concluded that millisecond timescale conformational dynamics of the interdomain linker is essential for the CA assembly. Solution NMR experiments revealed that, while these millisecond timescale motions of the linker are essential for opening up the conformational space, the number of accessible conformers is finite, and their populations are controlled through electrostatic intermolecular interactions between side chains of W184 and E175. This integrated approach established a molecular switch mechanism by which dynamics and electrostatic interactions work together to permit the formation of varied capsid morphologies in the mature HIV (Byeon et al., 2012).
In another study, Tycko and co-workers observed that while CA is generally rigid, there are regions of crucial biological function that exhibit static or dynamic disorder (Bayro et al., 2014). Using scalar-based 13C-13C TOBSY (Hardy et al., 2001) experiments, dynamically disordered regions of the protein were identified. Residues attributed to the interdomain linker, loop 8/9, and the C-terminal tail were observed in this data set, indicating that these residues are dynamically disordered. To distinguish residues that are mobile on the sub-millisecond timescale, 1H T2 filtered NCA data sets were acquired. In this experiment, a 1H T2 filter is incorporated and signals decay according to the local 1H-1H dipolar coupling strength. Thus peak intensities of rigid residues are weakened to 20–30% of their peak intensity, while dynamic residues retain more than 40% of their peak intensity. 1H T2 filtered experiments indicated that many dynamic residues are in loops and/or solvent exposed, with the most mobile residues being in the CypA loop (Fig. 21), in agreement with the dipolar order parameter measurements by Lu et al. (Lu et al., 2015a). The N-terminal segment was also observed to be mobile.
Fig. 21.
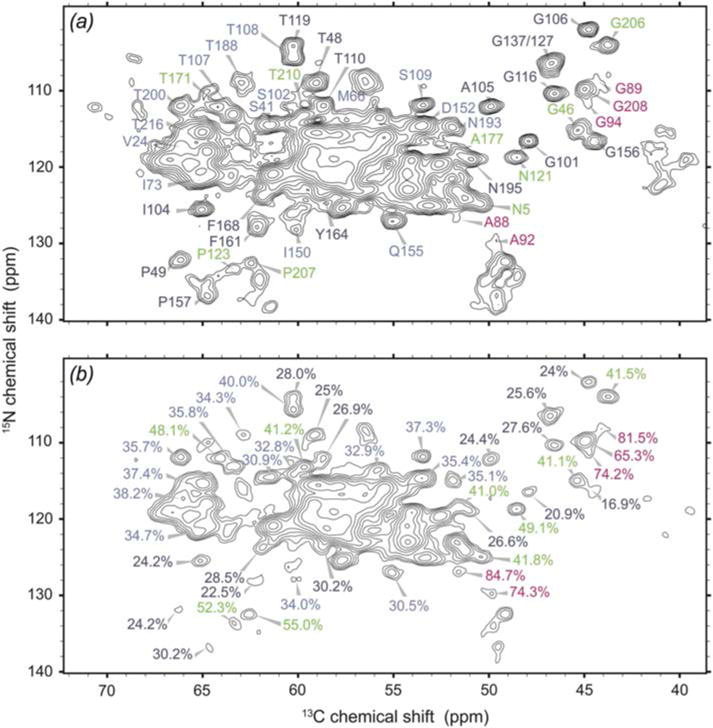
(A) NCA spectrum of CA tubular assemblies, with sufficiently resolved peaks labeled. (B) 1H T2 filtered NCA spectrum with 168 μs spin echo. Label colors correspond to peak intensity from 1H T2 filtered NCA experiment: 15–30%, dark blue; 31–40%, light blue; 41–55%, green; 56–85%, magneta. (Bayro et al., 2014) Reprinted with permission from Bayro et al., J. Mol. Biol., 2014, 426 (5), 1109–1127. Copyright 2014 Elsevier.
4.1.3 MAS NMR of HIV-1 Maturation Intermediates
Understanding the viral maturation process has been of interest both from the fundamental science standpoint and for the development of small molecule maturation inhibitors as a venue for HIV-1 treatment. Despite intense research into the maturation mechanism, many key questions remain open, including how the conical CA capsids form from the immature Gag lattice. Two hypotheses have emerged to explain how capsid reorganization takes place upon maturation: i) during maturation, the capsid gradually changes shape from spherical to conical, and ii) cleavage of SP1 triggers disassembly of the immature lattice and subsequent de novo reassembly of the mature capsid core. Recently, a small molecule Bevirimat (BVM) was discovered, which was shown to inhibit viral maturation by abolishing the final step, the cleavage of the SP1 peptide from the CA-SP1 maturation intermediate (Fig. 22, panel 1) (Adamson et al., 2010; de Marco et al., 2010; Fontana et al., 2016; Nguyen et al., 2011). Studies on the BVM interaction with the Gag and maturation intermediates by electron microscopy and biochemical methods suggest that the second hypothesis is correct, i.e., capsid formation takes place through de novo assembly rather than by the gradual lattice remodeling (Keller et al., 2013). These EM studies are limited by resolution, and to gain atomic-level insights on the final step of HIV-1 maturation, MAS NMR spectroscopy was pursued. In a recent investigation, the SP1 peptide was characterized in tubular assemblies of two strains of the CA-SP1 maturation intermediate: wild type HXB2 and NL4-3 A92E mutant (Han et al., 2013). Dipolar and scalar-based correlation experiments (Fig. 22, panel 2) revealed that the SP1 peptide in the assembled state is dynamically disordered and does not adopt a stable helical structure proposed on the basis of early solution NMR experiments on isolated SP1 peptide in organic solvents (Datta et al., 2011). The presence of the SP1 peptide affects the conformation and dynamics of CA protein: residues 226-231 of the CTD directly preceding the SP1 peptide, and which are mobile on microsecond timescale in CA assemblies, become more rigid, with the motions occurring on millisecond timescale in the CA-SP1 tubes. Most surprisingly, the dynamics of the CypA loop is modulated by the presence of the SP1 peptide: the loop becomes more flexible. Taken together, the results of this investigation support the de novo reassembly hypothesis (Han et al., 2013; Keller et al., 2013; Lu et al., 2015a).
To summarize, MAS NMR is a powerful emerging method for atomic-level characterization of HIV-1 viral assemblies. Recent work has demonstrated the significance of sequence-dependent conformational changes and dynamics of the HIV-1 capsid assemblies, as well as brought to light the importance of dynamic allostery in HIV maturation, infectivity, and interactions with host factor proteins.
4.2 Bacteriophages
Bacteriophages are a diverse class of viruses that target and infect bacteria. Bacteriophages are have applications in many areas, including molecular biology (Bax & Grishaev, 2005; Messing, 2001; Smith, 1985), nanotechnology (Huang et al., 2005; Nam et al., 2006), and drug development (Clark & March, 2006; Omidfar & Daneshpour, 2015). For review see: (Henry & Debarbieux, 2012; Salmond & Fineran, 2015). Two classes of bacteriophages have been studied by MAS NMR: filamentous and icosahedral bacteriophages. Filamentous phages, which include Pf1, fd, and M13 are long rod-like structures with a single-stranded DNA encapsulated in a capsid, composed of symmetric repeats of several thousand copies of an α-helical coat protein (Marvin, 1998). A second class is represented by tailed icosahedral viruses, such as T4 and T7, which are composed of nucleic acid encased in an icosahedral capsid, with a tail to attach to the target host cell (Ackermann, 1999). In addition to structural studies of bacteriophage capsids, protein-nucleic acid interactions in bacteriophages have been addressed by MAS NMR.
4.2.1 Structural Characterization of Bacteriophage Capsid Proteins
McDermott and co-workers first characterized the intact Pf1 bacteriophage by MAS NMR, where resonance assignments and secondary structure determination were performed and the feasibility of this approach for atomic-level analysis was established (Goldbourt et al., 2007b). Pf1 undergoes a cooperative phase transition at 10°C (Thiriot et al., 2005), with the high-temperature and low-temperature forms having slightly different helical symmetries. Goldbourt et al. mapped chemical shift perturbations observed in 13C-13C correlation spectra to the structure of Pf1 (Goldbourt et al., 2010). Most chemical shift perturbations appeared in the side chains of residues in the hydrophobic region of the protein, at the interface between subunits. They postulated that adjustments in these hydrophobic regions enabled the temperature transition of what is an overall rigid structure.
Hydration waters play critical roles in biological function, including maintenance of structure and facilitating protein dynamics. McDermott and co-workers (Sergeyev et al., 2014), and Sinha and co-workers (Purusottam et al., 2013) used 1H-15N and 1H-13C heteronuclear correlation (HETCOR) spectroscopy, including MELODI (MEdium-to-LOng DIstance) HETCOR (Yao & Hong, 2001) to probe the hydration state of Pf1. REDOR dephasing pulses (Gullion & Schaefer, 1989) were applied during HETCOR experiments to dephase magnetization arising from directly bonded protons, such that observed magnetization is known to come from direct contact with water. McDermott and co-workers also utilized water-selective T2’-filtering for spectral simplification in some HETCOR experiments and a 3-dimensional MELODI HETCOR experiment which incorporates a train of 13C and 15N REDOR dephasing pulses on both 13C and 15N with a DARR 13C-13C mixing component which allowed for the assignment of numerous protein residues in contact with water, as shown in Fig. 23. SD-HETCOR experiments (HETCOR with 1H spin diffusion (Kumashiro et al., 1998)) were also included to allow for the observation of hydration waters on the encapsulated DNA. Hydration sites include surface exposed residues at the N-terminus, as well as C-terminal residues, which contact the interior cavity of the phage, including Arg-44 and Lys-45, which participate in protein-DNA interactions, as discussed below. These results indicate that hydration waters are essential in the protein-DNA interactions, which stabilize the capsid structure. The data also revealed hydrophilic groves in the coat protein, which allow for water exchange between the interior of the phage and the exterior surface.
Fig. 23.
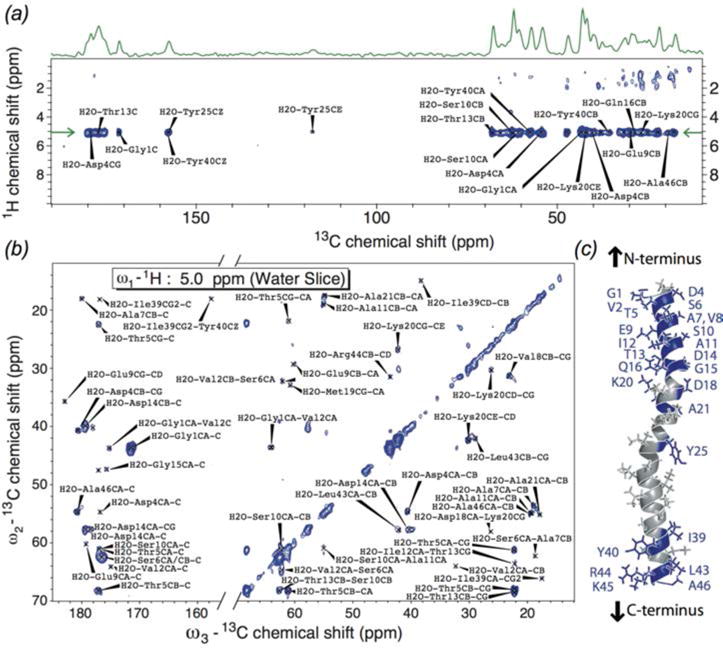
(a) 1H-13C MELODI-HETCOR of Pf1. (b) 13C-13C slice of a 1H-13C-13C 3D spectrum at the water frequency. (c) Pf1 subunit; residues interacting with water are shaded purple. Hydrated residues are concentrated at the N- and C-termini. (Sergeyev et al., 2014) Reprinted with permission from Sergeyev et al., J. Chem. Phys., 2014, 141 (22). Copyright 2014 AIP Publishing.
Goldbourt and co-workers reported the structure of the capsid protein from intact M13 bacteriophage (Morag et al., 2015). This is the first structure of an intact filamentous virus capsid solved by MAS NMR. The intra- and inter-subunit distance restraints were established using selectively labeled samples (Fig. 24 (a)) (Morag et al., 2011). The ‘fold-and-dock’ protocol of CS-Rosetta (Das et al., 2009; Shen et al., 2008) containing 35 M13 subunits to capture all unique interactions was used (DiMaio et al., 2011). This protocol is designed to determine optimum structures of higher order oligomers. M13 and other Ff class bacteriophages have 5-fold subunit symmetry around the virion axis with largely α-helical secondary structure. Many inter-subunit interactions were observed in hydrophobic regions, highlighting the importance of the hydrophobic pockets for subunit packing. Each monomer was found to participate in 4 hydrophobic pockets, with participating residues distributed across the monomer (Fig. 24(c)). Many of these key aromatic/hydrophobic residues are conserved among filamentous bacteriophages. The structure also reveals hydrogen bonding and electrostatic interactions that further contribute to capsid stability.
Fig. 24.
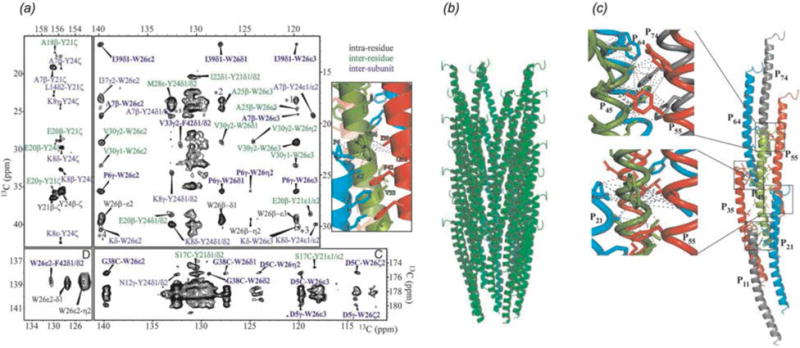
(a) 13C-13C CORD spectrum of [1,3-13C]glycerol, U-15N M13 bacteriophage with 500 ms mixing time. Intra-residue contacts are labeled in black. Inter-residue contacts within the same subunit are labeled in green. Inter-residue contacts between residues in different subunits are labeled in blue. Select inter-subunit correlations are shown on the structure. (b) Sideview of the NMR-ROSETTA model of M13 containing 35 subunits, the minimum number of subunits required to contain all unique interactions. (c) Hydrophobic pockets formed by several subunits. (Morag et al., 2015) Adapted with permission from Morag et al., Proc. Nat. Acad. Sci., 2015, 112 (4), 971–976. Copyright 2015 National Academy of Sciences.
4.2.2 Characterization of Bacteriophage Capsid Dynamics
Lorieau et al. probed backbone dynamics in Pf1 by Lee-Goldburg Cross Polarization (LGCP) experiments (Lorieau et al., 2008). 1H-13C dipolar order parameters were measured for several Cα, Cβ, Cγ, and Cδ sites. The backbone of Pf1 was found to be remarkably rigid. As would be expected, many solvent-exposed side chains exhibited reduced order parameters. Significantly, Arg 44 and Lys 45 side chains, which are directed into the phage cavity, were also observed to be highly mobile. It was postulated that the dynamics of these two residues is essential for mediating DNA-protein interactions. Opella and co-workers also measured 1H-15N dipolar lineshapes of this system with 1H-detected experiments at 50 kHz MAS (Park et al., 2013).
4.2.3 Characterization of Protein-Nucleic Acid Interactions in Bacteriophages
Nucleic acids are an integral component of viruses (Speir & Johnson, 2012). Recently several labs have demonstrated that MAS NMR can characterize nucleic acids (Cherepanov et al., 2010; Huang et al., 2012) and protein-nucleic acid interactions (Asami et al., 2013; Asami & Reif, 2013; Marchanka et al., 2015; Marchanka et al., 2013) including studies of protein-DNA interactions in intact bacteriophages (Morag et al., 2014; Sergeyev et al., 2011; Yu & Schaefer, 2008).
In contrast to other bacteriophages, Pf1 has an unusually high nucleotide-to-capsid subunit ratio of 1:1 (Day et al., 1988) and a highly extended conformation. (Other inoviruses have ratios of between 2–2.5 to 1.) Using dynamic nuclear polarization (DNP, (Ni et al., 2013)) for sensitivity enhancement, Sergeyev et al. were able to detect nucleotide resonances in the intact Pf1 bacteriophage (Sergeyev et al., 2011). The reported chemical shifts for the deoxyribose moieties capture the unusual DNA conformation of Pf1. As compared to average B-form DNA chemical shifts, C2′, C3′, C4′, and C5′ shifts in Pf1 appeared downfield, indicating a C2′-endo/gauche conformation with an anti glycosidic bond orientation. Nucleotide base chemical shifts, such as those of TC7 and CC5 fall outside the averages reported in the BMRB (Ulrich et al., 2008) and indicating that Pf1 DNA does not form typical base-pairing interactions.
Goldbourt, Morag, and Abramov used 13C-13C and 1H-mediated 31P-13C correlation experiments to characterize capsid-ssDNA interactions in the fd bacteriophage (Abramov et al., 2011; Morag et al., 2014). Initial studies demonstrated that well-resolved spectra with narrow peaks could be attained for the wild type coat protein, enabling resonance assignments and further structural characterization (Abramov et al., 2011). Prior structural studies with static solid-state NMR (SSNMR), cryo-EM, and fiber diffraction relied on the rigid Y21M mutant (Zeri et al., 2003), as the wild type protein yielded poor data quality (i.e., broad lines) in SSNMR (Tan et al., 1999) or lack of structural convergence in the case of cryo-EM (Wang et al., 2006). For the study of DNA within fd, the choice of labeling scheme was key (Morag et al., 2014). By including unlabeled aromatic amino acids in the expression media, the nucleotide resonances were not obscured by protein peaks. Broadband and efficient CORD 13C-13C correlation experiments (Hou et al., 2011a; Hou et al., 2013a), described above in section 2.2.1, were found to be ideal for the observation of DNA resonances, particularly for quaternary carbons with no directly attached protons. Nucleotide-specific assignments were achieved using a nucleotide walk, as shown in Fig. 25. In the CORD spectra acquired with long mixing times, sugar-to-capsid and base-to-capsid DNA-protein interactions were also observed. PHHC proton-mediated 31P-13C correlation spectra detected capsid-to-phosphate backbone interactions. Altogether, 56 capsid-ssDNA interactions could be assigned. These protein-DNA interactions occur primarily between positively charged Lys side chains near the C-terminus and the DNA phosphate backbone. Fd was found to have a similar sugar conformation to Pf1 (Sergeyev et al., 2011) but greater tendency for base stacking and a different protein-DNA interface.
Fig. 25.
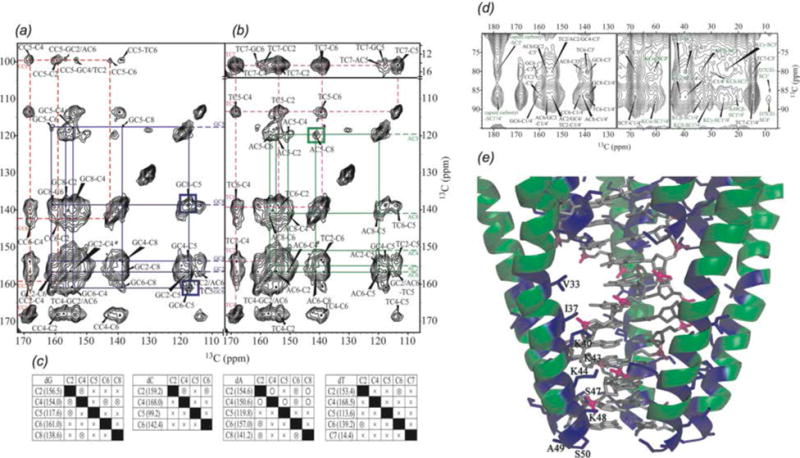
Spin system assignments of DNA in fd bacteriophage. Nucleotide walks are shown for (a) dG, blue, and dC, red and (b) dA, green, and dT, pink. (c) Assignment grid indicating all observed DNA correlations. (d) Expansion of 13C-13C CORD spectrum. Capsid-to-sugar correlations are labeled green, while intra-nucleotide resonances are labeled black. (e) Model of protein-DNA interactions in fd. (Morag et al., 2014) Adapted with permission from Morag et al., J. Am. Chem. Soc., 2014, 136 (6), 2292–2301. Copyright 2014 American Chemical Society.
Due to challenges with respect to spectral overlap and sample preparation, MAS NMR of nucleic acids has been much more limited than protein NMR. Abramov and Goldbourt were able to characterize the 40 kbp double-stranded DNA packaged within bacteriophage T7, a 50 MDa icosahedral phage (Abramov & Goldbourt, 2014). Using 13C-13C DARR and 15N-13C TEDOR correlations, all carbons and most nitrogens of the nucleic acid could be detected. The DNA packaged inside the phage was determined to be B-form with expected Watson-Crick base pairing. This study, along with the studies of nucleic acids in fd and Pf1, adds to the growing database of nucleic acid chemical shifts (particularly for large, native systems) and the relationship of chemical shifts to structure.
Yu and Schaefer used 15N-31P REDOR experiments to characterize DNA packaging in bacteriophage T4 with U-15N and [ε-15N]Lys labeled T4 (Yu & Schaefer, 2008). The dependence of REDOR dephasing on the 15N-31P distance enabled the determination that the DNA packaged inside T4 is B-form by determining the distance between the phosphate backbone and nitrogen atoms of the nucleic acid bases. T4 is packed into the phage capsid with over 1000 molecules of three lysine-rich proteins (Karam, 1994). The very close phosphate-lysine distances (as close as 3.5 Å) indicate that side chain amine groups have an important role in charge balance in the phage. Further charge balance of DNA inside the phage comes from polyamines and ammonium cations.
Bacteriophages including Pf1, fd, and M13 have also been studied in membrane-associated states, as well as by static SSNMR techniques by Opella and others. These topics are beyond the scope of the current review. We direct readers to the following reviews and key publications: (Bechinger, 1997; Cross et al., 1983; Glaubitz et al., 2000; Marassi & Opella, 2003; Opella et al., 2008; Park et al., 2010; Shon et al., 1991; Tan et al., 1999; Thiriot et al., 2005; Thiriot et al., 2004; Zeri et al., 2003).
4.3 Other Viral Assemblies
Pintacuda and co-workers reported a study of the Measles virus (MeV) nucleocapsid (Barbet-Massin et al., 2014a). Prior to this work, no atomic level information was available for assembled nucleocapsid. The MeV nucleocapsid is comprised of a relatively rigid NCORE domain and a more dynamic NTAIL domain. Proton detection was applied to both intact and cleaved assemblies with magic angle spinning at 60 kHz. Dipolar 15N-1H correlations revealed rigid residues of NCORE while scalar-based correlations detected mobile residues of NTAIL. Capsid hydration levels were also evaluated by observing T1ρ relaxation of 15N-1H peak intensities. The reduced intensities of the cleaved nucleocapsid relative to intact capsid indicate that the presence of the NTAIL domain leads to a less ordered and hence more hydrated conformation.
5. Conclusions and Future Perspectives
Magic angle spinning NMR has developed into a compelling technique for the characterization of biological assemblies at atomic level. Integrated approaches to protein structure determination utilizing MAS NMR structural restraints in conjunction with information from other methods, such as cryo-EM and molecular dynamics, are very promising for structural and dynamics analysis of high molecular weight assemblies. Continued advancements in sensitivity, resolution, and methodology will enable the detailed characterization of increasingly complex systems, such as whole cells, where substantial progress has already been made (Curtis-Fisk et al., 2008; Janssen et al., 2010; Pius et al., 2012; Reichhardt & Cegelski, 2014).
Fig. 11.
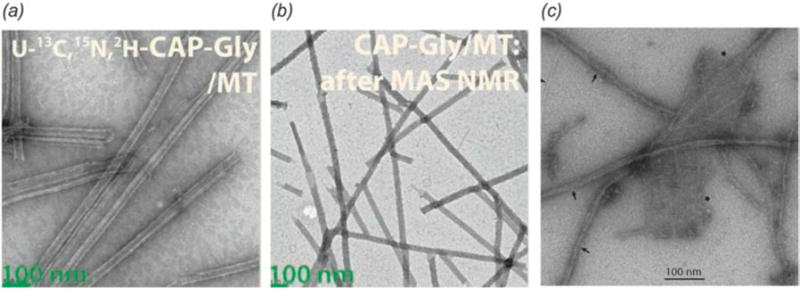
Transmission electron microscopy of cytoskeleton-associated proteins for MAS NMR experiments. (a–b) 2H,13C,15N CAP-Gly/microtubule complex before and after magic angle spinning. (Yan et al., 2015a) Adapted with permission from Yan et al., Proc. Nat. Acad. Sci., 2015, 112 (47), 14611–14616. Copyright 2015 National Academy of Sciences. (c) 13C, 15N BacA. Filament bundles are indicated by arrows, sheets are indicated by asterisks, and single filaments are indicated by arrowheads. (Vasa et al., 2015) Adapted with permission from Vasa et al., Proc. Nat. Acad. Sci., 2015, 112 (2), E127–E136. Copyright 2015 National Academy of Sciences.
Acknowledgments
This work is supported by the National Institutes of Health NIGMS grants P50GM082251 and F32GM113452 and is a contribution from the Pittsburgh Center for HIV Protein Interactions. CMQ acknowledges the support of the National Institutes of Health Postdoctoral Fellowship grant F32GM113452.
References
- World Health Organization. HIV/AIDS (Fact sheet No 360) [Google Scholar]
- ABRAMOV G, GOLDBOURT A. Nucleotide-type chemical shift assignment of the encapsulated 40 kbp dsDNA in intact bacteriophage T7 by MAS solid-state NMR. Journal of Biomolecular NMR. 2014;59(4):219–230. doi: 10.1007/s10858-014-9840-4. [DOI] [PubMed] [Google Scholar]
- ABRAMOV G, MORAG O, GOLDBOURT A. Magic-angle spinning NMR of a class I filamentous bacteriophage virus. Journal of Physical Chemistry B. 2011;115(31):9671–9680. doi: 10.1021/jp2040955. [DOI] [PubMed] [Google Scholar]
- ACKERMANN HW. Tailed bacteriophages: The order Caudovirales. Advances in Virus Research. 1999;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADAMSON CS, SAKALIAN M, SALZWEDEL K, FREED EO. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology. 2010;7 doi: 10.1186/1742-4690-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADAMSON CS, SALZWEDEL K, FREED EO. Virus maturation as a new HIV-1 therapeutic target. Expert Opinion on Therapeutic Targets. 2009;13(8):895–908. doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AGARWAL V, DIEHL A, SKRYNNIKOV N, REIF B. High resolution 1H detected 1H,13C correlation spectra in MAS solid-state NMR using deuterated proteins with selective 1H,, 2H, isotopic labeling of methyl groups. Journal of the American Chemical Society. 2006;128(39):12620–12621. doi: 10.1021/ja064379m. [DOI] [PubMed] [Google Scholar]
- AGARWAL V, PENZEL S, SZEKELY K, CADALBERT R, TESTORI E, OSS A, PAST J, SAMOSON A, ERNST M, BOCKMANN A, MEIER BH. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angewandte Chemie-International Edition. 2014;53(45):12253–12256. doi: 10.1002/anie.201405730. [DOI] [PubMed] [Google Scholar]
- AGARWAL V, SARDO M, SCHOLZ I, BOCKMANN A, ERNST M, MEIER BH. PAIN with and without PAR: variants for third-spin assisted heteronuclear polarization transfer. Journal of Biomolecular NMR. 2013;56(4):365–377. doi: 10.1007/s10858-013-9756-4. [DOI] [PubMed] [Google Scholar]
- AHMED S, SUN S, SIGLIN AE, POLENOVA T, WILLIAMS JC. Disease-associated mutations in the p150Glued subunit destabilize the CAP-Gly domain. Biochemistry. 2010;49(25):5083–5085. doi: 10.1021/bi100235z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKBEY U, LANGE S, FRANKS WT, LINSER R, REHBEIN K, DIEHL A, VAN ROSSUM BJ, REIF B, OSCHKINAT H. Optimum levels of exchangeable protons in perdeuterated proteins for proton detection in MAS solid-state NMR spectroscopy. Journal of Biomolecular NMR. 2010;46(1):67–73. doi: 10.1007/s10858-009-9369-0. [DOI] [PubMed] [Google Scholar]
- ALENGHAT FJ, GOLAN DE. Membrane protein dynamics and functional implications in mammalian cells. Functional Organization of Vertebrate Plasma Membrane. 2013;72:89–120. doi: 10.1016/B978-0-12-417027-8.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALKARAGHOULI AR, KOETZLE TF. Neutron-diffraction study of L-phenylalanine hydrochloride. Acta Crystallographica Section B-Structural Science. 1975;31:2461–2465. [Google Scholar]
- ANDREAS LB, JAUDZEMS K, STANEK J, LALLI D, BERTARELLO A, LE MARCHAND T, CALA-DE PAEPE D, KOTELOVICA S, AKOPJANA I, KNOTT B, WEGNER S, ENGELKE F, LESAGE A, EMSLEY L, TARS K, HERRMANN T, PINTACUDA G. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1602248113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDREAS LB, REESE M, EDDY MT, GELEV V, NI QZ, MILLER EA, EMSLEY L, PINTACUDA G, CHOU JJ, GRIFFIN RG. Structure and mechanism of the influenza A M2(18–60) dimer of dimers. Journal of the American Chemical Society. 2015;137(47):14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASAMI S, PORTER JR, LANGE OF, REIF B. Access to Cα backbone dynamics of biological solids by 13C T1 relaxation and molecular dynamics simulation. Journal of the American Chemical Society. 2015;137(3):1094–1100. doi: 10.1021/ja509367q. [DOI] [PubMed] [Google Scholar]
- ASAMI S, RAKWALSKA-BANGE M, CARLOMAGNO T, REIF B. Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angewandte Chemie-International Edition. 2013;52(8):2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- ASAMI S, REIF B. Proton-detected solid-state NMR spectroscopy at aliphatic sites: application to crystalline systems. Accounts of Chemical Research. 2013;46(9):2089–2097. doi: 10.1021/ar400063y. [DOI] [PubMed] [Google Scholar]
- BALDUS M, PETKOVA AT, HERZFELD J, GRIFFIN RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Molecular Physics. 1998;95(6):1197–1207. [Google Scholar]
- BALTISBERGER JH, MUSAPELO T, SUTTON B, REYNOLDS A, GURUNG L. Reduction of spin diffusion artifacts from 2D zfr-INADEQUATE MAS NMR spectra. Journal of Magnetic Resonance. 2011;208(1):70–75. doi: 10.1016/j.jmr.2010.10.006. [DOI] [PubMed] [Google Scholar]
- BARBET-MASSIN E, FELLETTI M, SCHNEIDER R, JEHLE S, COMMUNIE G, MARTINEZ N, JENSEN MR, RUIGROK RWH, EMSLEY L, LESAGE A, BLACKLEDGE M, PINTACUDA G. Insights into the structure and dynamics of measles virus nucleocapsids by 1H-detected solid-state NMR. Biophysical Journal. 2014a;107(4):941–946. doi: 10.1016/j.bpj.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARBET-MASSIN E, PELL AJ, JAUDZEMS K, FRANKS WT, RETEL JS, KOTELOVICA S, AKOPJANA I, TARS K, EMSLEY L, OSCHKINAT H, LESAGE A, PINTACUDA G. Out-and-back 13C-13C scalar transfers in protein resonance assignment by proton-detected solid-state NMR under ultra-fast MAS. Journal of Biomolecular NMR. 2013;56(4):379–386. doi: 10.1007/s10858-013-9757-3. [DOI] [PubMed] [Google Scholar]
- BARBET-MASSIN E, PELL AJ, RETEL JS, ANDREAS LB, JAUDZEMS K, FRANKS WT, NIEUWKOOP AJ, HILLER M, HIGMAN V, GUERRY P, BERTARELLO A, KNIGHT MJ, FELLETTI M, LE MARCHAND T, KOTELOVICA S, AKOPJANA I, TARS K, STOPPINI M, BELLOTTI V, BOLOGNESI M, RICAGNO S, CHOU JJ, GRIFFIN RG, OSCHKINAT H, LESAGE A, EMSLEY L, HERRMANN T, PINTACUDA G. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. Journal of the American Chemical Society. 2014b;136(35):12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARKLIS E, MCDERMOTT J, WILKENS S, FULLER S, THOMPSON D. Organization of HIV-1 capsid proteins on a lipid monolayer. Journal of Biological Chemistry. 1998;273(13):7177–7180. doi: 10.1074/jbc.273.13.7177. [DOI] [PubMed] [Google Scholar]
- BAX A, GRISHAEV A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Current Opinion in Structural Biology. 2005;15(5):563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- BAYRO MJ, CHEN B, YAU WM, TYCKO R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. Journal of Molecular Biology. 2014;426(5):1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAYRO MJ, TYCKO R. Structure of the dimerization interface in the mature HIV-1 capsid protein lattice from solid state NMR of tubular assemblies. Journal of the American Chemical Society. 2016;138(27):8538–8546. doi: 10.1021/jacs.6b03983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECHINGER B. Structure and dynamics of the M13 coat signal sequence in membranes by multidimensional high-resolution and solid-state NMR spectroscopy. Proteins-Structure Function and Genetics. 1997;27(4):481–492. doi: 10.1002/(sici)1097-0134(199704)27:4<481::aid-prot2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- BERTINI I, EMSLEY L, FELLI IC, LAAGE S, LESAGE A, LEWANDOWSKI JR, MARCHETTI A, PIERATTELLI R, PINTACUDA G. High-resolution and sensitivity through-bond correlations in ultra-fast magic angle spinning (MAS) solid-state NMR. Chemical Science. 2011a;2(2):345–348. [Google Scholar]
- BERTINI I, EMSLEY L, LELLI M, LUCHINAT C, MAO J, PINTACUDA G. Ultrafast MAS solid-state NMR permits extensive 13C and 1H detection in paramagnetic metalloproteins. Journal of the American Chemical Society. 2010;132(16):5558–5559. doi: 10.1021/ja100398q. [DOI] [PubMed] [Google Scholar]
- BERTINI I, LUCHINAT C, PARIGI G, RAVERA E. SedNMR: on the edge between solution and solid-state NMR. Accounts of Chemical Research. 2013;46(9):2059–2069. doi: 10.1021/ar300342f. [DOI] [PubMed] [Google Scholar]
- BERTINI I, LUCHINAT C, PARIGI G, RAVERA E, REIF B, TURANO P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108(26):10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOCANEGRA R, RODRIGUEZ-HUETE A, FUERTES MA, DEL ALAMO M, MATEU MG. Molecular recognition in the human immunodeficiency virus capsid and antiviral design. Virus Research. 2012;169(2):388–410. doi: 10.1016/j.virusres.2012.06.016. [DOI] [PubMed] [Google Scholar]
- BOCKMANN A, LANGE A, GALINIER A, LUCA S, GIRAUD N, JUY M, HEISE H, MONTSERRET R, PENIN F, BALDUS M. Solid state NMR sequential resonance assignments and conformational analysis of the 2 × 10.4 kDa dimeric form of the Bacillus subtilis protein Crh. Journal of Biomolecular NMR. 2003;27(4):323–339. doi: 10.1023/a:1025820611009. [DOI] [PubMed] [Google Scholar]
- BRIGGS JA, KRAUSSLICH HG. The molecular architecture of HIV. Journal of Molecular Biology. 2011;410(4):491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- BROWN LS, LADIZHANSKY V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Science. 2015;24(9):1333–1346. doi: 10.1002/pro.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUSSAARD CPD, NOORDELOOS AAM, SANDAA RA, HELDAL M, BRATBAK G. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology. 2004;319(2):280–291. doi: 10.1016/j.virol.2003.10.033. [DOI] [PubMed] [Google Scholar]
- BYEON IJ, MENG X, JUNG J, ZHAO G, YANG R, AHN J, SHI J, CONCEL J, AIKEN C, ZHANG P, GRONENBORN AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139(4):780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BYEON IJL, HOU GJ, HAN Y, SUITER CL, AHN J, JUNG J, BYEON CH, GRONENBORN AM, POLENOVA T. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. Journal of the American Chemical Society. 2012;134(14):6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CADARS S, SEIN J, DUMA L, LESAGE A, PHAM TN, BALTISBERGER JH, BROWN SP, EMSLEY L. The refocused INADEQUATE MAS NMR experiment in multiple spin-systems: interpreting observed correlation peaks and optimising lineshapes. Journal of Magnetic Resonance. 2007;188(1):24–34. doi: 10.1016/j.jmr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- CADY SD, SCHMIDT-ROHR K, WANG J, SOTO CS, DEGRADO WF, HONG M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463(7281):689–693. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARRAVETTA M, EDEN M, ZHAO X, BRINKMANN A, LEVITT MH. Symmetry principles for the design of radiofrequency pulse sequences in the nuclear magnetic resonance of rotating solids. Chemical Physics Letters. 2000;321(3–4):205–215. [Google Scholar]
- CASTELLANI F, VAN ROSSUM B, DIEHL A, SCHUBERT M, REHBEIN K, OSCHKINAT H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420(6911):98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- CAULKINS BG, YANG C, HILARIO E, FAN L, DUNN MF, MUELLER LJ. Catalytic roles of beta Lys87 in tryptophan synthase: 15N solid state NMR studies. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2015;1854(9):1194–1199. doi: 10.1016/j.bbapap.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVISTON JP, HOLZBAUR EL. Microtubule motors at the intersection of trafficking and transport. Trends in Cell Biology. 2006;16(10):530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- CHAN JCC, TYCKO R. Recoupling of chemical shift anisotropies in solid-state NMR under high-speed magic-angle spinning and in uniformly 13C-labeled systems. Journal of Chemical Physics. 2003;118(18):8378–8389. [Google Scholar]
- CHAN JCC, TYCKO R. Broadband rotational resonance in solid state NMR spectroscopy. Journal of Chemical Physics. 2004;120(18):8349–8352. doi: 10.1063/1.1737369. [DOI] [PubMed] [Google Scholar]
- CHEN B, TYCKO R. Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Science. 2010;19(4):716–730. doi: 10.1002/pro.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN LL, OLSEN RA, ELLIOTT DW, BOETTCHER JM, ZHOU DHH, RIENSTRA CM, MUELLER LJ. Constant-time through-bond 13C correlation spectroscopy for assigning protein resonances with solid-state NMR spectroscopy. Journal of the American Chemical Society. 2006;128(31):9992–9993. doi: 10.1021/ja062347t. [DOI] [PubMed] [Google Scholar]
- CHEN XJ, XU H, COOPER HM, LIU YB. Cytoplasmic dynein: a key player in neurodegenerative and neurodevelopmental diseases. Science China-Life Sciences. 2014;57(4):372–377. doi: 10.1007/s11427-014-4639-9. [DOI] [PubMed] [Google Scholar]
- CHEREPANOV AV, GLAUBITZ C, SCHWALBE H. High-resolution studies of uniformly 13C,15N-labeled RNA by solid-state NMR spectroscopy. Angewandte Chemie-International Edition. 2010;49(28):4747–4750. doi: 10.1002/anie.200906885. [DOI] [PubMed] [Google Scholar]
- CHEVELKOV V, REHBEIN K, DIEHL A, REIF B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angewandte Chemie-International Edition. 2006;45(23):3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- CHITI F, DOBSON CM. Protein misfolding, functional amyloid, and human disease. Annual Review of Biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- CLARK JR, MARCH JB. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends in Biotechnology. 2006;24(5):212–218. doi: 10.1016/j.tibtech.2006.03.003. [DOI] [PubMed] [Google Scholar]
- COLGAN J, YUAN HEH, FRANKE EK, LUBAN J. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. Journal of Virology. 1996;70(7):4299–4310. doi: 10.1128/jvi.70.7.4299-4310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COMELLAS G, LEMKAU LR, NIEUWKOOP AJ, KLOEPPER KD, LADROR DT, EBISU R, WOODS WS, LIPTON AS, GEORGE JM, RIENSTRA CM. Structured regions of α-synuclein fibrils include the early-onset Parkinson’s Disease mutation sites. Journal of Molecular Biology. 2011;411(4):881–895. doi: 10.1016/j.jmb.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COMELLAS G, RIENSTRA CM. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annual Review of Biophysics. 2013;42:515–536. doi: 10.1146/annurev-biophys-083012-130356. [DOI] [PubMed] [Google Scholar]
- CROSS TA, TSANG P, OPELLA SJ. Comparison of protein and deoxyribonucleic acid backbone structures in fd and Pf1 bacteriophages. Biochemistry. 1983;22(4):721–726. doi: 10.1021/bi00273a002. [DOI] [PubMed] [Google Scholar]
- CURTIS-FISK J, SPENCER RM, WELIKY DP. Native conformation at specific residues in recombinant inclusion body protein in whole cells determined with solid-state NMR spectroscopy. Journal of the American Chemical Society. 2008;130(38):12568–12569. doi: 10.1021/ja8039426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DABBAGH G, WELIKY DP, TYCKO R. Determination of monomer conformations in noncrystalline solid polymers by 2-dimensional NMR exchange spectroscopy. Macromolecules. 1994;27(21):6183–6191. [Google Scholar]
- DAEBEL V, CHINNATHAMBI S, BIERNAT J, SCHWALBE M, HABENSTEIN B, LOQUET A, AKOURY E, TEPPER K, MULLER H, BALDUS M, GRIESINGER C, ZWECKSTETTER M, MANDELKOW E, VIJAYAN V, LANGE A. β-Sheet core of tau paired helical filaments revealed by solid-state NMR. Journal of the American Chemical Society. 2012;134(34):13982–13989. doi: 10.1021/ja305470p. [DOI] [PubMed] [Google Scholar]
- DANNATT HRW, TAYLOR GF, VARGA K, HIGMAN VA, PFEIL MP, ASILMOVSKA L, JUDGE PJ, WATTS A. 13C and 1H-detection under fast MAS for the study of poorly available proteins: application to sub-milligram quantities of a 7 transmembrane protein. Journal of Biomolecular NMR. 2015;62(1):17–23. doi: 10.1007/s10858-015-9911-1. [DOI] [PubMed] [Google Scholar]
- DAS BB, LIN EC, OPELLA SJ. Experiments optimized for magic angle spinning and oriented sample solid-state NMR of proteins. Journal of Physical Chemistry B. 2013;117(41):12422–12431. doi: 10.1021/jp407154h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAS R, ANDRE I, SHEN Y, WU Y, LEMAK A, BANSAL S, ARROWSMITH CH, SZYPERSKI T, BAKER D. Simultaneous prediction of protein folding and docking at high resolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DATTA SAK, TEMESELEW LG, CRIST RM, SOHEILIAN F, KAMATA A, MIRRO J, HARVIN D, NAGASHIMA K, CACHAU RE, REIN A. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? Journal of Virology. 2011;85(9):4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAY LA, MARZEC CJ, REISBERG SA, CASADEVALL A. DNA packing in filamentous bacteriophages. Annual Review of Biophysics and Biophysical Chemistry. 1988;17:509–539. doi: 10.1146/annurev.bb.17.060188.002453. [DOI] [PubMed] [Google Scholar]
- DE MARCO A, MULLER B, GLASS B, RICHES JD, KRAUSSLICH HG, BRIGGS JAG. Structural analysis of HIV-1 maturation using cryo-electron tomography. Plos Pathogens. 2010;6(11) doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE PAEPE G, LEWANDOWSKI JR, LOQUET A, BOCKMANN A, GRIFFIN RG. Proton assisted recoupling and protein structure determination. Journal of Chemical Physics. 2008;129(24) doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE PAEPE G, LEWANDOWSKI JR, LOQUET A, EDDY M, MEGY S, BOCKMANN A, GRIFFIN RG. Heteronuclear proton assisted recoupling. Journal of Chemical Physics. 2011;134(9) doi: 10.1063/1.3541251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE VITA E, FRYDMAN L. Spectral editing in 13C MAS NMR under moderately fast spinning conditions. Journal of Magnetic Resonance. 2001;148(2):327–337. doi: 10.1006/jmre.2000.2255. [DOI] [PubMed] [Google Scholar]
- DEAZEVEDO ER, HU WG, BONAGAMBA TJ, SCHMIDT-ROHR K. Centerband-only detection of exchange: Efficient analysis of dynamics in solids by NMR. Journal of the American Chemical Society. 1999;121(36):8411–8412. [Google Scholar]
- DEAZEVEDO ER, SAALWACHTER K, PASCUI O, DE SOUZA AA, BONAGAMBA TJ, REICHERT D. Intermediate motions as studied by solid-state separated local field NMR experiments. Journal of Chemical Physics. 2008;128(10) doi: 10.1063/1.2831798. [DOI] [PubMed] [Google Scholar]
- DEMERS JP, HABENSTEIN B, LOQUET A, VASA SK, GILLER K, BECKER S, BAKER D, LANGE A, SGOURAKIS NG. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nature Communications. 2014;5 doi: 10.1038/ncomms5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DESAI A, MITCHISON TJ. Microtubule polymerization dynamics. Annual Review of Cell and Developmental Biology. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- DESHMUKH L, SCHWIETERS CD, GRISHAEV A, GHIRLANDO R, BABER JL, CLORE GM. Structure and dynamics of full-length HIV-1 capsid protein in solution. Journal of the American Chemical Society. 2013;135(43):16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIMAIO F, LEAVER-FAY A, BRADLEY P, BAKER D, ANDRE I. Modeling symmetric macromolecular structures in Rosetta3. PLoS One. 2011;6(6):e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DITTMER J, BODENHAUSEN G. Evidence for slow motion in proteins by multiple refocusing of heteronuclear nitrogen/proton multiple quantum coherences in NMR. Journal of the American Chemical Society. 2004;126(5):1314–1315. doi: 10.1021/ja0386243. [DOI] [PubMed] [Google Scholar]
- EHRLICH LS, LIU TB, SCARLATA S, CHU B, CARTER CA. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: Structures switching by pH-induced conformational changes. Biophysical Journal. 2001;81(5):2992–2992. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELENA B, LESAGE A, STEUERNAGEL S, BOCKMANN A, EMSLEY L. Proton to carbon-13 INEPT in solid-state NMR spectroscopy. Journal of the American Chemical Society. 2005;127(49):17296–17302. doi: 10.1021/ja054411x. [DOI] [PubMed] [Google Scholar]
- ENGELMAN A, CHEREPANOV P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nature Reviews Microbiology. 2012;10(4):279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPSTEIN DM, BENKOVIC SJ, WRIGHT PE. Dynamics of the dihydrofolate-reductase folate complex-catalytic sites and regions known to undergo conformational change exhibit diverse dynamical features. Biochemistry. 1995;34(35):11037–11048. doi: 10.1021/bi00035a009. [DOI] [PubMed] [Google Scholar]
- ETZKORN M, BOCKMANN A, LANGE A, BALDUS M. Probing molecular interfaces using 2D magic-angle-spinning NMR on protein mixtures with different uniform labeling. Journal of the American Chemical Society. 2004;126(45):14746–14751. doi: 10.1021/ja0479181. [DOI] [PubMed] [Google Scholar]
- FARROW NA, MUHANDIRAM R, SINGER AU, PASCAL SM, KAY CM, GISH G, SHOELSON SE, PAWSON T, FORMANKAY JD, KAY LE. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- FISCHER RS, FOWLER VM. Thematic minireview series: the state of the cytoskeleton in 2015. Journal of Biological Chemistry. 2015;290(28):17133–17136. doi: 10.1074/jbc.R115.663716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FITZPATRICK AWP, DEBELOUCHINA GT, BAYRO MJ, CLARE DK, CAPORINI MA, BAJAJ VS, JARONIEC CP, WANG LC, LADIZHANSKY V, MULLER SA, MACPHEE CE, WAUDBY CA, MOTT HR, DE SIMONE A, KNOWLES TPJ, SAIBIL HR, VENDRUSCOLO M, ORLOVA EV, GRIFFIN RG, DOBSON CM. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(14):5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FONTANA J, KELLER PW, URANO E, ABLAN SD, STEVEN AC, FREED EO. Identification of an HIV-1 mutation in spacer peptide 1 that stabilizes the immature CA-SP1 lattice. Journal of Virology. 2016;90(2):972–978. doi: 10.1128/JVI.02204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORSHEY BM, VON SCHWEDLER U, SUNDQUIST WI, AIKEN C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. Journal of Virology. 2002;76(11):5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKS WT, WYLIE BJ, SCHMIDT HL, NIEUWKOOP AJ, MAYRHOFER RM, SHAH GJ, GRAESSER DT, RIENSTRA CM. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(12):4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREED EO. HIV-1 assembly, release and maturation. Nature Reviews Microbiology. 2015;13(8):484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FU RQ, COTTEN M, CROSS TA. Inter- and intramolecular distance measurements by solid-state MAS NMR: Determination of gramicidin A channel dimer structure in hydrated phospholipid bilayers. Journal of Biomolecular NMR. 2000;16(3):261–268. doi: 10.1023/a:1008372508024. [DOI] [PubMed] [Google Scholar]
- GANSER-PORNILLOS BK, CHENG A, YEAGER M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131(1):70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- GIRAUD N, BOCKMANN A, LESAGE A, PENIN F, BLACKLEDGE M, EMSLEY L. Site-specific backbone dynamics from a crystalline protein by solid-state NMR spectroscopy. Journal of the American Chemical Society. 2004;126(37):11422–11423. doi: 10.1021/ja046578g. [DOI] [PubMed] [Google Scholar]
- GLAUBITZ C, GROBNER G, WATTS A. Structural and orientational information of the membrane embedded M13 coat protein by 13C-MAS NMR spectroscopy. Biochimica Et Biophysica Acta. 2000;1463(1):151–161. doi: 10.1016/s0005-2736(99)00195-9. [DOI] [PubMed] [Google Scholar]
- GOLDBOURT A. Biomolecular magic-angle spinning solid-state NMR: recent methods and applications. Current Opinion in Biotechnology. 2013;24(4):705–715. doi: 10.1016/j.copbio.2013.02.010. [DOI] [PubMed] [Google Scholar]
- GOLDBOURT A, DAY LA, MCDERMOTT AE. Assignment of congested NMR spectra: Carbonyl backbone enrichment via the Entner-Doudoroff pathway. Journal of Magnetic Resonance. 2007a;189(2):157–165. doi: 10.1016/j.jmr.2007.07.011. [DOI] [PubMed] [Google Scholar]
- GOLDBOURT A, DAY LA, MCDERMOTT AE. Intersubunit hydrophobic interactions in Pf1 filamentous phage. Journal of Biological Chemistry. 2010;285(47):37051–37059. doi: 10.1074/jbc.M110.119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDBOURT A, GROSS BJ, DAY LA, MCDERMOTT AE. Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. Journal of the American Chemical Society. 2007b;129(8):2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- GOOBES G. Past and future solid-state NMR spectroscopic studies at the convergence point between biology and materials research. Israel Journal of Chemistry. 2014;54:113–124. [Google Scholar]
- GOOD DB, WANG S, WARD ME, STRUPPE J, BROWN LS, LEWANDOWSKI JR, LADIZHANSKY V. Conformational dynamics of a seven transmembrane helical protein Anabaena Sensory Rhodopsin probed by solid-state NMR. Journal of the American Chemical Society. 2014;136(7):2833–2842. doi: 10.1021/ja411633w. [DOI] [PubMed] [Google Scholar]
- GRES AT, KIRBY KA, KEWALRAMANI VN, TANNER JJ, PORNILLOS O, SARAFIANOS SG. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349(6243):99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSS I, HOHENBERG H, KRAUSSLICH HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. European Journal of Biochemistry. 1997;249(2):592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- GULLION T, SCHAEFER J. Rotational-echo double-resonance NMR. Journal of Magnetic Resonance. 1989;81(1):196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- GUNAWARDENA S. Nanoparticles in the brain: A potential therapeutic system targeted to an early defect observed in many neurodegenerative diseases. Pharmaceutical Research. 2013;30(10):2459–2474. doi: 10.1007/s11095-013-1037-0. [DOI] [PubMed] [Google Scholar]
- GUNTERT P. Automated NMR structure calculation with CYANA. Protein NMR Techniques. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- HABENSTEIN B, LOQUET A, HWANG S, GILLER K, VASA S, BECKER S, HABECK M, LANGE A. Hybrid structure of the type 1 pilus of uropathogenic Escherichia coli. Angewandte Chemie-International Edition. 2015;54:11691–11695. doi: 10.1002/anie.201505065. [DOI] [PubMed] [Google Scholar]
- HALLER JD, SCHANDA P. Amplitudes and time scales of picosecond-to-microsecond motion in proteins studied by solid-state NMR: a critical evaluation of experimental approaches and application to crystalline ubiquitin. Journal of Biomolecular NMR. 2013;57(3):263–280. doi: 10.1007/s10858-013-9787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN Y, AHN J, CONCEL J, BYEON IJL, GRONENBORN AM, YANG J, POLENOVA T. Solid-state NMR studies of HIV-1 capsid protein assemblies. Journal of the American Chemical Society. 2010;132(6):1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN Y, HOU GJ, SUITER CL, AHN J, BYEON IJL, LIPTON AS, BURTON S, HUNG I, GOR’KOV PL, GAN ZH, BREY W, RICE D, GRONENBORN AM, POLENOVA T. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. Journal of the American Chemical Society. 2013;135(47):17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDY EH, VEREL R, MEIER BH. Fast MAS total through-bond correlation spectroscopy. Journal of Magnetic Resonance. 2001;148(2):459–464. doi: 10.1006/jmre.2000.2258. [DOI] [PubMed] [Google Scholar]
- HAYASHI I, WILDE A, MAL TK, IKURA M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Molecular Cell. 2005;19(4):449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- HE LC, BARDIAUX B, AHMED M, SPEHR J, KONIG R, LUNSDORF H, RAND U, LUHRS T, RITTER C. Structure determination of helical filaments by solid-state NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(3):E272–E281. doi: 10.1073/pnas.1513119113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE LC, LUHRS T, RITTER C. Solid-state NMR resonance assignments of the filament-forming CARD domain of the innate immunity signaling protein MAVS. Biomolecular NMR Assignments. 2015;9(2):223–227. doi: 10.1007/s12104-014-9579-6. [DOI] [PubMed] [Google Scholar]
- HEISE H, HOYER W, BECKER S, ANDRONESI OC, RIEDEL D, BALDUS M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELMLE M, PATZELT H, OCKENFELS A, GARTNER W, OESTERHELT D, BECHINGER B. Refinement of the geometry of the retinal binding pocket in dark-adapted bacteriorhodopsin by heteronuclear solid-state NMR distance measurements. Biochemistry. 2000;39(33):10066–10071. doi: 10.1021/bi0006666. [DOI] [PubMed] [Google Scholar]
- HELMUS JJ, SUREWICZ K, NADAUD PS, SUREWICZ WK, JARONIEC CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELMUS JJ, SUREWICZ K, SUREWICZ WK, JARONIEC CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. Journal of the American Chemical Society. 2010;132(7):2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENRY M, DEBARBIEUX L. Tools from viruses: bacteriophage successes and beyond. Virology. 2012;434(2):151–161. doi: 10.1016/j.virol.2012.09.017. [DOI] [PubMed] [Google Scholar]
- HIGMAN VA, FLINDERS J, HILLER M, JEHLE S, MARKOVIC S, FIEDLER S, VAN ROSSUM BJ, OSCHKINAT H. Assigning large proteins in the solid state: a MAS NMR resonance assignment strategy using selectively and extensively 13C-labelled proteins. Journal of Biomolecular NMR. 2009;44(4):245–260. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- HING AW, VEGA S, SCHAEFER J. Transferred-echo double-resonance NMR. Journal of Magnetic Resonance. 1992;96(1):205–209. [Google Scholar]
- HOHWY M, JAKOBSEN HJ, EDEN M, LEVITT MH, NIELSEN NC. Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: A compensated C7 pulse sequence. Journal of Chemical Physics. 1998;108(7):2686–2694. [Google Scholar]
- HOLLAND GP, CHERRY BR, JENKINS JE, YARGER JL. Proton-detected heteronuclear single quantum correlation NMR spectroscopy in rigid solids with ultra-fast MAS. Journal of Magnetic Resonance. 2010;202(1):64–71. doi: 10.1016/j.jmr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG M. Determination of multiple φ-torsion angles in proteins by selective and extensive 13C labeling and two-dimensional solid-state NMR. Journal of Magnetic Resonance. 1999;139(2):389–401. doi: 10.1006/jmre.1999.1805. [DOI] [PubMed] [Google Scholar]
- HONNAPPA S, OKHRIMENKO O, JAUSSI R, JAWHARI H, JELESAROV I, WINKLER FK, STEINMETZ MO. Key interaction modes of dynamic +TIP networks. Molecular Cell. 2006;23(5):663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- HOOP CL, LIN HK, KAR K, HOU ZP, POIRIER MA, WETZEL R, VAN DER WEL PCA. Polyglutamine amyloid core boundaries and flanking domain dynamics in Huntingtin fragment fibrils determined by solid-state nuclear magnetic resonance. Biochemistry. 2014;53(42):6653–6666. doi: 10.1021/bi501010q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU G, BYEON IJ, AHN J, GRONENBORN AM, POLENOVA T. Recoupling of chemical shift anisotropy by R-symmetry sequences in magic angle spinning NMR spectroscopy. Journal of Chemical Physics. 2012;137(13):134201. doi: 10.1063/1.4754149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU G, PARAMASIVAM S, BYEON IJ, GRONENBORN AM, POLENOVA T. Determination of relative tensor orientations by γ-encoded chemical shift anisotropy/heteronuclear dipolar coupling 3D NMR spectroscopy in biological solids. Physical Chemistry Chemical Physics. 2010;12(45):14873–14883. doi: 10.1039/c0cp00795a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU G, YAN S, SUN S, HAN Y, BYEON IJ, AHN J, CONCEL J, SAMOSON A, GRONENBORN AM, POLENOVA T. Spin diffusion driven by R-symmetry sequences: applications to homonuclear correlation spectroscopy in MAS NMR of biological and organic solids. Journal of the American Chemical Society. 2011a;133(11):3943–3953. doi: 10.1021/ja108650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU G, YAN S, TREBOSC J, AMOUREUX JP, POLENOVA T. Broadband homonuclear correlation spectroscopy driven by combined R2nv sequences under fast magic angle spinning for NMR structural analysis of organic and biological solids. Journal of Magnetic Resonance. 2013a;232:18–30. doi: 10.1016/j.jmr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU GJ, BYEON IJL, AHN J, GRONENBORN AM, POLENOVA T. 1H-13C/1H-15N heteronuclear dipolar recoupling by R-symmetry sequences under fast magic angle spinning for dynamics analysis of biological and organic solids. Journal of the American Chemical Society. 2011b;133(46):18646–18655. doi: 10.1021/ja203771a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU GJ, LU XY, VEGA AJ, POLENOVA T. Accurate measurement of heteronuclear dipolar couplings by phase-alternating R-symmetry (PARS) sequences in magic angle spinning NMR spectroscopy. Journal of Chemical Physics. 2014;141(10) doi: 10.1063/1.4894226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOU GJ, PARAMASIVAM S, YAN S, POLENOVA T, VEGA AJ. Multidimensional magic angle spinning NMR spectroscopy for site-resolved measurement of proton chemical shift anisotropy in biological solids. Journal of the American Chemical Society. 2013b;135(4):1358–1368. doi: 10.1021/ja3084972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- HOWARD J, HYMAN AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422(6933):753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- HU FH, LUO WB, HONG M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330(6003):505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU KN, QIANG W, BERMEJO GA, SCHWIETERS CD, TYCKO R. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N-15N and carbonyl 13C-13C dipolar recoupling data. Journal of Magnetic Resonance. 2012;218:115–127. doi: 10.1016/j.jmr.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG R, YAMAMOTO K, ZHANG M, POPOVYCH N, HUNG I, IM SC, GAN Z, WASKELL L, RAMAMOORTHY A. Probing the transmembrane structure and dynamics of microsomal NADPH-cytochrome P450 oxidoreductase by solid-state NMR. Biophysical Journal. 2014;106(10):2126–2133. doi: 10.1016/j.bpj.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG W, BARDARO MF, VARANI G, DROBNY GP. Preparation of RNA samples with narrow line widths for solid state NMR investigations. Journal of Magnetic Resonance. 2012;223:51–54. doi: 10.1016/j.jmr.2012.07.018. [DOI] [PubMed] [Google Scholar]
- HUANG Y, CHIANG CY, LEE SK, GAO Y, HU EL, DE YOREO J, BELCHER AM. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Letters. 2005;5(7):1429–1434. doi: 10.1021/nl050795d. [DOI] [PubMed] [Google Scholar]
- HULME AE, KELLEY Z, OKOCHA EA, HOPE TJ. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. Journal of Virology. 2015;89(1):643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYBERTS SG, MILBRADT AG, WAGNER AB, ARTHANARI H, WAGNER G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. Journal of Biomolecular NMR. 2012;52(4):315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYBERTS SG, TAKEUCHI K, WAGNER G. Poisson-Gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. Journal of the American Chemical Society. 2010;132(7):2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IGUMENOVA TI, MCDERMOTT AE, ZILM KW, MARTIN RW, PAULSON EK, WAND AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. Journal of the American Chemical Society. 2004a;126(21):6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- IGUMENOVA TI, WAND AJ, MCDERMOTT AE. Assignment of the backbone resonances for microcrystalline ubiquitin. Journal of the American Chemical Society. 2004b;126(16):5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- ISHII Y. C-13-C-13 dipolar recoupling under very fast magic angle spinning in solid-state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. Journal of Chemical Physics. 2001;114(19):8473–8483. [Google Scholar]
- JANSSEN GJ, DAVISO E, VAN SON M, DE GROOT HJM, ALIA A, MATYSIK J. Observation of the solid-state photo-CIDNP effect in entire cells of cyanobacteria Synechocystis. Photosynthesis Research. 2010;104(2–3):275–282. doi: 10.1007/s11120-009-9508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JARONIEC CP. Solid-state nuclear magnetic resonance structural studies of proteins using paramagnetic probes. Solid State Nuclear Magnetic Resonance. 2012:43–44. doi: 10.1016/j.ssnmr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- JARONIEC CP, FILIP C, GRIFFIN RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C, 15N-labeled solids. Journal of the American Chemical Society. 2002;124(36):10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- JEHLE S, RAJAGOPAL P, BARDIAUX B, MARKOVIC S, KUHNE R, STOUT JR, HIGMAN VA, KLEVIT RE, VAN ROSSUM BJ, OSCHKINAT H. Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nature Structural & Molecular Biology. 2010;17(9):1037–U1031. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG JY, ABLAN SD, DEREBAIL S, HERCIK K, SOHEILIAN F, THOMAS JA, TANG SX, HEWLETT I, NAGASHIMA K, GORELICK RJ, FREED EO, LEVIN JG. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology. 2011;421(2):253–265. doi: 10.1016/j.virol.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAMINSKYY V, ZHIVOTOVSKY B. To kill or be killed: how viruses interact with the cell death machinery. Journal of Internal Medicine. 2010;267(5):473–482. doi: 10.1111/j.1365-2796.2010.02222.x. [DOI] [PubMed] [Google Scholar]
- KARAM JD. Molecular Biology of Bacteriophage T4. Washington, DC: American Society for Microbiology Press; 1994. [Google Scholar]
- KELLER PW, HUANG RK, ENGLAND MR, WAKI K, CHENG NQ, HEYMANN JB, CRAVEN RC, FREED EO, STEVEN AC. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. Journal of Virology. 2013;87(24):13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KETCHEM RR, LEE KC, HUO S, CROSS TA. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. Journal of Biomolecular NMR. 1996;8(1):1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- KIIHNE SR, CREEMERS AFL, DE GRIP WJ, BOVEE-GEURTS PHM, LUGTENBURG J, DE GROOT HJM. Selective interface detection: Mapping binding site contacts in membrane proteins by NMR spectroscopy. Journal of the American Chemical Society. 2005;127(16):5734–5735. doi: 10.1021/ja045677r. [DOI] [PubMed] [Google Scholar]
- KNIGHT MJ, FELLI IC, PIERATTELLI R, EMSLEY L, PINTACUDA G. Magic angle spinning NMR of paramagnetic proteins. Accounts of Chemical Research. 2013;46(9):2108–2116. doi: 10.1021/ar300349y. [DOI] [PubMed] [Google Scholar]
- KNIGHT MJ, PELL AJ, BERTINI I, FELLI IC, GONNELLI L, PIERATTELLI R, HERRMANN T, EMSLEY L, PINTACUDA G. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(28):11095–11100. doi: 10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIGHT MJ, WEBBER AL, PELL AJ, GUERRY P, BARBET-MASSIN E, BERTINI I, FELLI IC, GONNELLI L, PIERATTELLI R, EMSLEY L, LESAGE A, HERRMANN T, PINTACUDA G. Fast resonance assignment and fold determination of human superoxide dismutase by high-resolution proton-detected solid-state MAS NMR spectroscopy. Angewandte Chemie-International Edition. 2011;50(49):11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- KRUSHELNITSKY A, DEAZEVEDO E, LINSER R, REIF B, SAALWACHTER K, REICHERT D. Direct observation of millisecond to second motions in proteins by dipolar CODEX NMR spectroscopy. Journal of the American Chemical Society. 2009;131(34):12097–12099. doi: 10.1021/ja9038888. [DOI] [PubMed] [Google Scholar]
- KRUSHELNITSKY A, REICHERT D, SAALWACHTER K. Solid-state NMR approaches to internal dynamics of proteins: from picoseconds to microseconds and seconds. Accounts of Chemical Research. 2013;46(9):2028–2036. doi: 10.1021/ar300292p. [DOI] [PubMed] [Google Scholar]
- KUHN J, BRIEGEL A, MORSCHEL E, KAHNT J, LESER K, WICK S, JENSEN GJ, THANBICHLER M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29(2):327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUMAR A, HEISE H, BLOMMERS MJJ, KRASTEL P, SCHMITT E, PETERSEN F, JEGANATHAN S, MANDELKOW EM, CARLOMAGNO T, GRIESINGER C, BALDUS M. Interaction of epothilone B (patupilone) with microtubules as detected by two-dimensional solid-state NMR spectroscopy. Angewandte Chemie-International Edition. 2010;49(41):7504–7507. doi: 10.1002/anie.201001946. [DOI] [PubMed] [Google Scholar]
- KUMASHIRO KK, SCHMIDT-ROHR K, MURPHY OJ, OUELLETTE KL, CRAMER WA, THOMPSON LK. A novel tool for probing membrane protein structure: Solid-state NMR with proton spin diffusion and X-nucleus detection. Journal of the American Chemical Society. 1998;120(20):5043–5051. [Google Scholar]
- LAAGE S, SACHLEBEN JR, STEUERNAGEL S, PIERATTELLI R, PINTACUDA G, EMSLEY L. Fast acquisition of multi-dimensional spectra in solid-state NMR enabled by ultra-fast MAS. Journal of Magnetic Resonance. 2009;196(2):133–141. doi: 10.1016/j.jmr.2008.10.019. [DOI] [PubMed] [Google Scholar]
- LADIZHANSKY V, JARONIEC CP, DIEHL A, OSCHKINAT H, GRIFFIN RG. Measurement of multiple psi torsion angles in uniformly 13C,15N-labeled alpha-spectrin SH3 domain using 3D 15N-13C-13C-15N MAS dipolar-chemical shift correlation spectroscopy. Journal of the American Chemical Society. 2003;125(22):6827–6833. doi: 10.1021/ja029082c. [DOI] [PubMed] [Google Scholar]
- LAMLEY JM, LOUGHER MJ, SASS HJ, ROGOWSKI M, GRZESIEK S, LEWANDOWSKI JR. Unraveling the complexity of protein backbone dynamics with combined 13C and 15N solid-state NMR relaxation measurements. Physical Chemistry Chemical Physics. 2015;17(34):21997–22008. doi: 10.1039/c5cp03484a. [DOI] [PubMed] [Google Scholar]
- LANGE A, LUCA S, BALDUS M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. Journal of the American Chemical Society. 2002;124(33):9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- LEMASTER DM, KUSHLAN DM. Dynamical mapping of E-coli thioredoxin via 13C NMR relaxation analysis. Journal of the American Chemical Society. 1996;118(39):9255–9264. [Google Scholar]
- LESAGE A, AUGER C, CALDARELLI S, EMSLEY L. Determination of through-bond carbon-carbon connectivities in solid-state NMR using the INADEQUATE experiment. Journal of the American Chemical Society. 1997;119(33):7867–7868. [Google Scholar]
- LESAGE A, BARDET M, EMSLEY L. Through-bond carbon-carbon connectivities in disordered solids by NMR. Journal of the American Chemical Society. 1999;121(47):10987–10993. [Google Scholar]
- LEWANDOWSKI JR, DE PAEPE G, EDDY MT, GRIFFIN RG. 15N-15N proton assisted recoupling in magic angle spinning NMR. Journal of the American Chemical Society. 2009a;131(16):5769–5776. doi: 10.1021/ja806578y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, DE PAEPE G, EDDY MT, STRUPPE J, MAAS W, GRIFFIN RG. Proton assisted recoupling at high spinning frequencies. Journal of Physical Chemistry B. 2009b;113(27):9062–9069. doi: 10.1021/jp810280t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, DE PAEPE G, GRIFFIN RG. Proton assisted insensitive nuclei cross polarization. Journal of the American Chemical Society. 2007;129(4):728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, DUMEZ JN, AKBEY U, LANGE S, EMSLEY L, OSCHKINAT H. Enhanced resolution and coherence lifetimes in the solid-state NMR spectroscopy of perdeuterated proteins under ultrafast magic-angle spinning. Journal of Physical Chemistry Letters. 2011a;2(17):2205–2211. [Google Scholar]
- LEWANDOWSKI JR, HALSE ME, BLACKLEDGE M, EMSLEY L. Direct observation of hierarchical protein dynamics. Science. 2015;348(6234):578–581. doi: 10.1126/science.aaa6111. [DOI] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, SASS HJ, GRZESIEK S, BLACKLEDGE M, EMSLEY L. Site-specific measurement of slow motions in proteins. Journal of the American Chemical Society. 2011b;133(42):16762–16765. doi: 10.1021/ja206815h. [DOI] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, SEIN J, SASS HJ, GRZESIEK S, BLACKLEDGE M, EMSLEY L. Measurement of site-specific 13C spin-lattice relaxation in a crystalline protein. Journal of the American Chemical Society. 2010;132(24):8252–8254. doi: 10.1021/ja102744b. [DOI] [PubMed] [Google Scholar]
- LEWANDOWSKI JR, VAN DER WEL PCA, RIGNEY M, GRIGORIEFF N, GRIFFIN RG. Structural complexity of a composite amyloid fibril. Journal of the American Chemical Society. 2011c;133(37):14686–14698. doi: 10.1021/ja203736z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI SH, SU YC, LUO WB, HONG M. Water-protein interactions of an arginine-rich membrane peptide in lipid bilayers investigated by solid-state nuclear magnetic resonance spectroscopy. Journal of Physical Chemistry B. 2010;114(11):4063–4069. doi: 10.1021/jp912283r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI WB, MCDERMOTT A. Investigation of slow molecular dynamics using R-CODEX. Journal of Magnetic Resonance. 2012;222:74–80. doi: 10.1016/j.jmr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI WB, MCDERMOTT AE. Characterization of slow conformational dynamics in solids: dipolar CODEX. Journal of Biomolecular NMR. 2009;45(1–2):227–232. doi: 10.1007/s10858-009-9353-8. [DOI] [PubMed] [Google Scholar]
- LI YK, POLIKS B, CEGELSKI L, POLIKS M, GRYCZYNSKI Z, PISZCZEK G, JAGTAP PG, STUDELSKA DR, KINGSTON DGI, SCHAEFER J, BANE S. Conformation of microtubule-bound paclitaxel determined by fluorescence spectroscopy and REDOR NMR. Biochemistry. 2000;39(2):281–291. doi: 10.1021/bi991936r. [DOI] [PubMed] [Google Scholar]
- LIAO SY, YANG Y, TIETZE D, HONG M. The influenza M2 cytoplasmic tail changes the proton-exchange equilibria and the backbone conformation of the transmembrane histidine residue to facilitate proton conduction. Journal of the American Chemical Society. 2015;137(18):6067–6077. doi: 10.1021/jacs.5b02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIGON LA, SHELLY SS, TOKITO M, HOLZBAUR ELF. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Molecular Biology of the Cell. 2003;14(4):1405–1417. doi: 10.1091/mbc.E02-03-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINSER R, BARDIAUX B, HIGMAN V, FINK U, REIF B. Structure calculation from unambiguous long-range amide and methyl 1H-1H distance restraints for a microcrystalline protein with MAS solid-state NMR spectroscopy. Journal of the American Chemical Society. 2011a;133(15):5905–5912. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- LINSER R, DASARI M, HILLER M, HIGMAN V, FINK U, DEL AMO JML, MARKOVIC S, HANDEL L, KESSLER B, SCHMIEDER P, OESTERHELT D, OSCHKINAT H, REIF B. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angewandte Chemie-International Edition. 2011b;50(19):4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- LINSER R, FINK U, REIF B. Proton-detected scalar coupling based assignment strategies in MAS solid-state NMR spectroscopy applied to perdeuterated proteins. Journal of Magnetic Resonance. 2008;193(1):89–93. doi: 10.1016/j.jmr.2008.04.021. [DOI] [PubMed] [Google Scholar]
- LIU C, PERILLA JR, NING J, LU M, HOU G, RAMALHO R, HIMES BA, ZHAO G, BEDWELL G, BYEON IJ, AHN J, GRONENBORN AM, PREVELIGE PE, ROUSSO I, AIKEN C, POLENOVA T, SCHULTEN K, ZHANG P. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nature Communications. 2016;7(10714) doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOQUET A, GILLER K, BECKER S, LANGE A. Supramolecular interactions probed by 13C-13C solid-state NMR spectroscopy. Journal of the American Chemical Society. 2010;132(43):15164–15166. doi: 10.1021/ja107460j. [DOI] [PubMed] [Google Scholar]
- LOQUET A, HABENSTEIN B, CHEVELKOV V, VASA SK, GILLER K, BECKER S, LANGE A. Atomic structure and handedness of the building block of a biological assembly. Journal of the American Chemical Society. 2013a;135(51):19135–19138. doi: 10.1021/ja411362q. [DOI] [PubMed] [Google Scholar]
- LOQUET A, HABENSTEIN B, LANGE A. Structural investigations of molecular machines by solid-state NMR. Accounts of Chemical Research. 2013b;46(9):2070–2079. doi: 10.1021/ar300320p. [DOI] [PubMed] [Google Scholar]
- LOQUET A, LV G, GILLER K, BECKER S, LANGE A. C-13 spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. Journal of the American Chemical Society. 2011;133(13):4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- LOQUET A, SGOURAKIS NG, GUPTA R, GILLER K, RIEDEL D, GOOSMANN C, GRIESINGER C, KOLBE M, BAKER D, BECKER S, LANGE A. Atomic model of the type III secretion system needle. Nature. 2012;486(7402):276–281. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LORIEAU JL, DAY LA, MCDERMOTT AE. Conformational dynamics of an intact virus: order parameters for the coat protein of Pf1 bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LORIEAU JL, LOUIS JM, BAX A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(25):11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWE J, VAN DEN ENT F, AMOS LA. Molecules of the bacterial cytoskeleton. Annual Review of Biophysics and Biomolecular Structure. 2004;33:177–198. doi: 10.1146/annurev.biophys.33.110502.132647. [DOI] [PubMed] [Google Scholar]
- LU M, HOU G, ZHANG H, SUITER CL, AHN J, BYEON IJ, PERILLA JR, LANGMEAD CJ, HUNG I, GOR’KOV PL, GAN Z, BREY W, AIKEN C, ZHANG P, SCHULTEN K, GRONENBORN AM, POLENOVA T. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proceedings of the National Academy of Sciences of the United States of America. 2015a;112(47):14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU X, GUO C, HOU G, POLENOVA T. Combined zero-quantum and spin-diffusion mixing for efficient homonuclear correlation spectroscopy under fast MAS: broadband recoupling and detection of long-range correlations. Journal of Biomolecular NMR. 2015b;61(1):7–20. doi: 10.1007/s10858-014-9875-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUBAN J, BOSSOLT KL, FRANKE EK, KALPANA GV, GOFF SP. Human-immunodeficiency-virus type-1 Gag protein binds to cyclophilin-A and cyclophilin-B. Cell. 1993;73(6):1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- LUCA S, HEISE H, BALDUS M. High-resolution solid-state NMR applied to polypeptides and membrane proteins. Accounts of Chemical Research. 2003;36(11):858–865. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- LV GH, KUMAR A, GILLER K, ORCELLET ML, RIEDEL D, FERNANDEZ CO, BECKER S, LANGE A. Structural comparison of couse and human α-synuclein amyloid fibrils by solid-state NMR. Journal of Molecular Biology. 2012;420(1–2):99–111. doi: 10.1016/j.jmb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- MANDEL AM, AKKE M, PALMER AG. Backbone dynamics of Escherichia coli ribonuclease H – Correlations with structure and function in an active enzyme. Journal of Molecular Biology. 1995;246(1):144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- MANOCHEEWA S, SWAIN JV, LANXON-COOKSON E, ROLLAND M, MULLINS JI. Fitness costs of mutations at the HIV-1 capsid hexamerization interface. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARASSI FM, OPELLA SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Science. 2003;12(3):403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCHANKA A, SIMON B, ALTHOFF-OSPELT G, CARLOMAGNO T. RNA structure determination by solid-state NMR spectroscopy. Nature Communications. 2015:6. doi: 10.1038/ncomms8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCHANKA A, SIMON B, CARLOMAGNO T. A suite of solid-state NMR experiments for RNA intranucleotide resonance assignment in a 21 kDa protein-RNA complex. Angewandte Chemie-International Edition. 2013;52(38):9996–10001. doi: 10.1002/anie.201304779. [DOI] [PubMed] [Google Scholar]
- MARCHETTI A, JEHLE S, FELLETTI M, KNIGHT MJ, WANG Y, XU ZQ, PARK AY, OTTING G, LESAGE A, EMSLEY L, DIXON NE, PINTACUDA G. Backbone assignment of fully protonated solid proteins by 1H detection and ultrafast magic-angle-spinning NMR spectroscopy. Angewandte Chemie-International Edition. 2012;51(43):10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- MARTIN RW, ZILM KW. Preparation of protein nanocrystals and their characterization by solid state NMR. Journal of Magnetic Resonance. 2003;165(1):162–174. doi: 10.1016/s1090-7807(03)00253-2. [DOI] [PubMed] [Google Scholar]
- MARULANDA D, TASAYCO ML, MCDERMOTT A, CATALDI M, ARRIARAN V, POLENOVA T. Magic angle spinning solid-state NMR spectroscopy for structural studies of protein interfaces. resonance assignments of differentially enriched Escherichia coli thioredoxin reassembled by fragment complementation. Journal of the American Chemical Society. 2004;126(50):16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- MARVIN DA. Filamentous phage structure, infection and assembly. Current Opinion in Structural Biology. 1998;8(2):150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]
- MCCARTHY KR, SCHMIDT AG, KIRMAIER A, WYAND AL, NEWMAN RM, JOHNSON WE. Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two Iindependent conserved determinants of Trim5 alpha specificity. Plos Pathogens. 2013;9(5) doi: 10.1371/journal.ppat.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCDERMOTT A, POLENOVA T, BOCKMANN A, ZILM KW, PAULSON EK, MARTIN RW, MONTELIONE GT. Partial NMR assignments for uniformly (13C, 15N)-enriched BPTI in the solid state. Journal of Biomolecular NMR. 2000;16(3):209–219. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- MCINTOSH LP, DAHLQUIST FW. Biosynthetic incorporation of 15N and 13C for assignment and interpretation of nuclear-magnetic-resonance spectra of proteins. Quarterly Reviews of Biophysics. 1990;23(1):1–38. doi: 10.1017/s0033583500005400. [DOI] [PubMed] [Google Scholar]
- MEIBOOM S, GILL D. Modified spin-echo method for measuring nuclear relaxation times. Review of Scientific Instruments. 1958;29(8):688–691. [Google Scholar]
- MESSING J. The universal primers and the shotgun DNA sequencing method. Protein NMR Techniques. 2001;167:13–31. doi: 10.1385/1-59259-113-2:013. [DOI] [PubMed] [Google Scholar]
- MORAG O, ABRAMOV G, GOLDBOURT A. Similarities and differences within members of the Ff family of filamentous bacteriophage viruses. Journal of Physical Chemistry B. 2011;115(51):15370–15379. doi: 10.1021/jp2079742. [DOI] [PubMed] [Google Scholar]
- MORAG O, ABRAMOV G, GOLDBOURT A. Complete chemical shift assignment of the ssDNA in the filamentous bacteriophage fd reports on its conformation and on its interface with the capsid shell. Journal of the American Chemical Society. 2014;136(6):2292–2301. doi: 10.1021/ja412178n. [DOI] [PubMed] [Google Scholar]
- MORAG O, SGOURAKIS NG, BAKER D, GOLDBOURT A. The NMR-Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(4):971–976. doi: 10.1073/pnas.1415393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORCOMBE CR, GAPONENKO V, BYRD RA, ZILM KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. Journal of the American Chemical Society. 2004;126(23):7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- MORRIS GA, FREEMAN R. Enhancement of nuclear magnetic-resonance signals by polarization transfer. Journal of the American Chemical Society. 1979;101(3):760–762. [Google Scholar]
- MUNOWITZ M, AUE WP, GRIFFIN RG. Two-dimensional separation of dipolar and scaled isotropic chemical-shift interactions in magic angle NMR spectra. Journal of Chemical Physics. 1982;77(4):1686–1689. [Google Scholar]
- MUNOWITZ MG, GRIFFIN RG, BODENHAUSEN G, HUANG TH. Two-dimensional rotational spin-echo nuclear magnetic-resonance in solids – Correlation of chemical-shift and dipolar interactions. Journal of the American Chemical Society. 1981;103(10):2529–2533. [Google Scholar]
- NADAUD PS, HELMUS JJ, HOFER N, JARONIEC CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. Journal of the American Chemical Society. 2007;129(24):7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- NAITO A, KAWAMURA I, JAVKHLANTUGS N. Recent solid-state NMR studies of membrane-bound peptides and proteins. Annual Reports on NMR Spectroscopy. 2015;86:333–411. [Google Scholar]
- NAM KT, KIM DW, YOO PJ, CHIANG CY, MEETHONG N, HAMMOND PT, CHIANG YM, BELCHER AM. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312(5775):885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- NELSON RS, CITOVSKY V. Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiology. 2005;138(4):1809–1814. doi: 10.1104/pp.104.900167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NGUYEN AT, FEASLEY CL, JACKSON KW, NITZ TJ, SALZWEDEL K, AIR GM, SAKALIAN M. The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles. Retrovirology. 2011;8 doi: 10.1186/1742-4690-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI QZ, DAVISO E, CAN TV, MARKHASIN E, JAWLA SK, SWAGER TM, TEMKIN RJ, HERZFELD J, GRIFFIN RG. High frequency dynamic nuclear polarization. Accounts of Chemical Research. 2013;46(9):1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIEUWKOOP AJ, RIENSTRA CM. Supramolecular protein structure determination by site-specific long-range intermolecular solid state NMR spectroscopy. Journal of the American Chemical Society. 2010;132(22):7570–7571. doi: 10.1021/ja100992y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIEUWKOOP AJ, WYLIE BJ, FRANKS WT, SHAH GJ, RIENSTRA CM. Atomic resolution protein structure determination by three-dimensional transferred echo double resonance solid-state nuclear magnetic resonance spectroscopy. Journal of Chemical Physics. 2009;131(9) doi: 10.1063/1.3211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOGALES E. Structural insights into microtubule function. Annual Review of Biochemistry. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- OLSEN GL, EDWARDS TE, DEKA P, VARANI G, SIGURDSSON ST, DROBNY GP. Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR. Nucleic Acids Research. 2005;33(11):3447–3454. doi: 10.1093/nar/gki626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIDFAR K, DANESHPOUR M. Advances in phage display technology for drug discovery. Expert Opinion on Drug Discovery. 2015;10(6):651–669. doi: 10.1517/17460441.2015.1037738. [DOI] [PubMed] [Google Scholar]
- OPELLA SJ, MARASSI FM, GESELL JJ, VALENTE AP, KIM Y, OBLATT-MONTAL M, MONTAL M. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nature Structural Biology. 1999;6(4):374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPELLA SJ, ZERI AC, PARK SH. Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy. Annual Review of Physical Chemistry. 2008;59:635–657. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- PAIK Y, YANG C, METAFERIA B, TANG SB, BANE S, RAVINDRA R, SHANKER N, ALCARAZ AA, JOHNSON SA, SCHAEFER J, O’CONNOR RD, CEGELSKI L, SNYDER JP, KINGSTON DGI. Rotational-echo double-resonance NMR distance measurements for the tubulin-bound paclitaxel conformation. Journal of the American Chemical Society. 2007;129(2):361–370. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALUCH P, PAWLAK T, AMOUREUX JP, POTRZEBOWSKI MJ. Simple and accurate determination of X-H distances under ultra-fast MAS NMR. Journal of Magnetic Resonance. 2013;233:56–63. doi: 10.1016/j.jmr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- PALUCH P, PAWLAK T, JEZIORNA A, TREBOSC J, HOU G, VEGA AJ, AMOUREUX JP, DRACINSKY M, POLENOVA T, POTRZEBOWSKI MJ. Analysis of local molecular motions of aromatic sidechains in proteins by 2D and 3D fast MAS NMR spectroscopy and quantum mechanical calculations. Physical Chemistry Chemical Physics. 2015a;17(43):28789–28801. doi: 10.1039/c5cp04475h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALUCH P, TREBOSC J, NISHIYAMA Y, POTRZEBOWSKI MJ, MALON M, AMOUREUX JP. Theoretical study of CP-VC: a simple, robust and accurate MAS NMR method for analysis of dipolar C-H interactions under rotation speeds faster than ca. 60 kHz. Journal of Magnetic Resonance. 2015b;252:67–77. doi: 10.1016/j.jmr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- PANDEY MK, VIVEKANANDAN S, AHUJA S, HUANG R, IM SC, WASKELL L, RAMAMOORTHY A. Cytochrome-P450-cytochrome-b(5) interaction in a membrane environment changes 15N chemical shift anisotropy tensors. Journal of Physical Chemistry B. 2013;117(44):13851–13860. doi: 10.1021/jp4086206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, DAS BB, CASAGRANDE F, TIAN Y, NOTHNAGEL HJ, CHU M, KIEFER H, MAIER K, DE ANGELIS AA, MARASSI FM, OPELLA SJ. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491(7426):779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, MARASSI FM, BLACK D, OPELLA SJ. Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly. Biophysical Journal. 2010;99(5):1465–1474. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, YANG C, OPELLA SJ, MUELLER LJ. Resolution and measurement of heteronuclear dipolar couplings of a noncrystalline protein immobilized in a biological supramolecular assembly by proton-detected MAS solid-state NMR spectroscopy. Journal of Magnetic Resonance. 2013;237:164–168. doi: 10.1016/j.jmr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARTHASARATHY S, NISHIYAMA Y, ISHII Y. Sensitivity and resolution enhanced solid-state NMR for paramagnetic systems and biomolecules under very fast magic angle spinning. Accounts of Chemical Research. 2013;46(9):2127–2135. doi: 10.1021/ar4000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAULI J, BALDUS M, VAN ROSSUM B, DE GROOT H, OSCHKINAT H. Backbone and side-chain 13C and 15N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 tesla. Chembiochem. 2001;2(4):272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- PAULSON EK, MORCOMBE CR, GAPONENKO V, DANCHECK B, BYRD RA, ZILM KW. Sensitive high resolution inverse detection NMR spectroscopy of proteins in the solid state. Journal of the American Chemical Society. 2003;125(51):15831–15836. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- PEARSON MN, BEEVER RE, BOINE B, ARTHUR K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Molecular Plant Pathology. 2009;10(1):115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETKOVA AT, YAU WM, TYCKO R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIUS J, MORROW MR, BOOTH V. 2H solid-state nuclear magnetic resonance investigation of whole Escherichia coli interacting with antimicrobial peptide MSI-78. Biochemistry. 2012;51(1):118–125. doi: 10.1021/bi201569t. [DOI] [PubMed] [Google Scholar]
- PORNILLOS O, GANSER-PORNILLOS BK, BANUMATHI S, HUA YZ, YEAGER M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. Journal of Molecular Biology. 2010;401(5):985–995. doi: 10.1016/j.jmb.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PORNILLOS O, GANSER-PORNILLOS BK, KELLY BN, HUA YZ, WHITBY FG, STOUT CD, SUNDQUIST WI, HILL CP, YEAGER M. X-Ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137(7):1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRANGISHVILI D. The wonderful world of archaeal viruses. Annual Review of Microbiology. 2013;67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- PRICE AJ, FLETCHER AJ, SCHALLER T, ELLIOTT T, LEE K, KEWALRAMANI VN, CHIN JW, TOWERS GJ, JAMES LC. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. Plos Pathogens. 2012;8(8) doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PURUSOTTAM RN, RAI RK, SINHA N. Mechanistic insights into water-protein interactions of filamentous bacteriophage. Journal of Physical Chemistry B. 2013;117(10):2837–2840. doi: 10.1021/jp310921n. [DOI] [PubMed] [Google Scholar]
- QI ML, YANG RF, AIKEN C. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. Journal of Virology. 2008;82(24):12001–12008. doi: 10.1128/JVI.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUINN CM, MCDERMOTT AE. Quantifying conformational dynamics using solid-state R1ρ experiments. Journal of Magnetic Resonance. 2012;222:1–7. doi: 10.1016/j.jmr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REICHHARDT C, CEGELSKI L. Solid-state NMR for bacterial biofilms. Molecular Physics. 2014;112(7):887–894. doi: 10.1080/00268976.2013.837983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REIF B, GRIFFIN RG. 1H detected 1H, 15N correlation spectroscopy in rotating solids. Journal of Magnetic Resonance. 2003;160(1):78–83. doi: 10.1016/s1090-7807(02)00035-6. [DOI] [PubMed] [Google Scholar]
- REIF B, JARONIEC CP, RIENSTRA CM, HOHWY M, GRIFFIN RG. 1H-1H MAS correlation spectroscopy and distance measurements in a deuterated peptide. Journal of Magnetic Resonance. 2001;151(2):320–327. doi: 10.1006/jmre.2001.2354. [DOI] [PubMed] [Google Scholar]
- REIF B, VAN ROSSUM BJ, CASTELLANI F, REHBEIN K, DIEHL A, OSCHKINAT H. Characterization of 1H-1H distances in a uniformly 2H,15N-labeled SH3 domain by MAS solid-state NMR spectroscopy. Journal of the American Chemical Society. 2003;125(6):1488–1489. doi: 10.1021/ja0283697. [DOI] [PubMed] [Google Scholar]
- RIENSTRA CM, TUCKER-KELLOGG L, JARONIEC CP, HOHWY M, REIF B, MCMAHON MT, TIDOR B, LOZANO-PEREZ T, GRIFFIN RG. De novo determination of peptide structure with solid-state magic-angle spinning NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10260–10265. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROGERS SL, ROGERS GC, SHARP DJ, VALE RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. Journal of Cell Biology. 2002;158(5):873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSEN MK, GARDNER KH, WILLIS RC, PARRIS WE, PAWSON T, KAY LE. Selective methyl group protonation of perdeuterated proteins. Journal of Molecular Biology. 1996;263(5):627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- ROZOVSKY S, MCDERMOTT AE. The time scale of the catalytic loop motion in triosephosphate isomerase. Journal of Molecular Biology. 2001;310(1):259–270. doi: 10.1006/jmbi.2001.4672. [DOI] [PubMed] [Google Scholar]
- SALMOND GP, FINERAN PC. A century of the phage: past, present and future. Nature Reviews Microbiology. 2015;13(12):777–786. doi: 10.1038/nrmicro3564. [DOI] [PubMed] [Google Scholar]
- SALZWEDEL K, MARTIN DE, SAKALIAN M. Maturation inhibitors: a new therapeutic class targets the virus structure. Aids Reviews. 2007;9(3):162–172. [PubMed] [Google Scholar]
- SAMOSON A, TUHERM T, GAN Z. High-field high-speed MAS resolution enhancement in 1H NMR spectroscopy of solids. Solid State Nuclear Magnetic Resonance. 2001;20(3–4):130–136. doi: 10.1006/snmr.2001.0037. [DOI] [PubMed] [Google Scholar]
- SAMOSON A, TUHERM T, PAST J, REINHOLD A, ANUPOLD T, HEINMAA I. New horizons for magic-angle spinning NMR. New Techniques in Solid-State NMR. 2005;246:15–31. doi: 10.1007/b98647. [DOI] [PubMed] [Google Scholar]
- SBORGI L, RAVOTTI F, DANDEY VP, DICK MS, MAZUR A, RECKEL S, CHAMI M, SCHERER S, HUBER M, BOCKMANN A, EGELMAN EH, STAHLBERG H, BROZ P, MEIER BH, HILLER S. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(43):13237–13242. doi: 10.1073/pnas.1507579112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHAEFER J, MCKAY RA, STEJSKAL EO. Double-cross-polarization NMR of solids. Journal of Magnetic Resonance. 1979;34(2):443–447. [Google Scholar]
- SCHANDA P, MEIER BH, ERNST M. Quantitative analysis of protein backbone dynamics in microcrystalline ubiquitin by solid-state NMR spectroscopy. Journal of the American Chemical Society. 2010;132(45):15957–15967. doi: 10.1021/ja100726a. [DOI] [PubMed] [Google Scholar]
- SCHANDA P, TRIBOULET S, LAGURI C, BOUGAULT CM, AYALA I, CALLON M, ARTHUR M, SIMORRE JP. Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan. Journal of the American Chemical Society. 2014;136(51):17852–17860. doi: 10.1021/ja5105987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUTZ AK, SORAGNI A, HORNEMANN S, AGUZZI A, ERNST M, BOCKMANN A, MEIER BH. The amyloid-Congo red interface at atomic resolution. Angewandte Chemie-International Edition. 2011;50(26):5956–5960. doi: 10.1002/anie.201008276. [DOI] [PubMed] [Google Scholar]
- SCHWIETERS CD, KUSZEWSKI JJ, CLORE GM. Using Xplor-NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48(1):47–62. [Google Scholar]
- SCHWIETERS CD, KUSZEWSKI JJ, TJANDRA N, CLORE GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- SERGEYEV IV, BAHRI S, DAY LA, MCDERMOTT AE. Pf1 bacteriophage hydration by magic angle spinning solid-state NMR. Journal of Chemical Physics. 2014;141(22) doi: 10.1063/1.4903230. [DOI] [PubMed] [Google Scholar]
- SERGEYEV IV, DAY LA, GOLDBOURT A, MCDERMOTT AE. Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. Journal of the American Chemical Society. 2011;133(50):20208–20217. doi: 10.1021/ja2043062. [DOI] [PubMed] [Google Scholar]
- SHEN Y, BAX A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Protein NMR Techniques. 2015;1260:17–32. doi: 10.1007/978-1-4939-2239-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN Y, DELAGLIO F, CORNILESCU G, BAX A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN Y, LANGE O, DELAGLIO F, ROSSI P, ARAMINI JM, LIU G, ELETSKY A, WU Y, SINGARAPU KK, LEMAK A, IGNATCHENKO A, ARROWSMITH CH, SZYPERSKI T, MONTELIONE GT, BAKER D, BAX A. Consistent blind protein structure generation from NMR chemical shift data. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(12):4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHI C, FRICKE P, LIN L, CHEVELKOV V, WEGSTROTH M, GILLER K, BECKER S, THANBICHLER M, LANGE A. Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR. Science Advances. 2015;1(11) doi: 10.1126/sciadv.1501087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIN R, TZOU YM, KRISHNA NR. Structure of a monomeric mutant of the HIV-1 capsid protein. Biochemistry. 2011;50(44):9457–9467. doi: 10.1021/bi2011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHON KJ, KIM YG, COLNAGO LA, OPELLA SJ. NMR studies of the structure and dynamics of membrane-bound bacteriophage Pfl coat protein. Science. 1991;252(5010):1303–1304. doi: 10.1126/science.1925542. [DOI] [PubMed] [Google Scholar]
- SMITH AE, HELENIUS A. How viruses enter animal cells. Science. 2004;304(5668):237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- SMITH GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- SPEIR JA, JOHNSON JE. Nucleic acid packaging in viruses. Current Opinion in Structural Biology. 2012;22(1):65–71. doi: 10.1016/j.sbi.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPERLING LJ, BERTHOLD DA, SASSER TL, JEISY-SCOTT V, RIENSTRA CM. Assignment strategies for large proteins by magic-angle spinning NMR: The 21-kDa disulfide-bond-forming enzyme DsbA. Journal of Molecular Biology. 2010;399(2):268–282. doi: 10.1016/j.jmb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN S, SIGLIN A, WILLIAMS JC, POLENOVA T. Solid-state and solution NMR studies of the CAP-Gly domain of mammalian dynactin and its interaction with microtubules. Journal of the American Chemical Society. 2009;131(29):10113–10126. doi: 10.1021/ja902003u. [DOI] [PubMed] [Google Scholar]
- SZEVERENYI NM, SULLIVAN MJ, MACIEL GE. Observation of spin exchange by two-dimensional fourier-transform 13C cross polarization-magic-angle spinning. Journal of Magnetic Resonance. 1982;47(3):462–475. [Google Scholar]
- TAKEGOSHI K, NAKAMURA S, TERAO T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344(5–6):631–637. [Google Scholar]
- TAN WM, JELINEK R, OPELLA SJ, MALIK P, TERRY TD, PERHAM RN. Effects of temperature and Y21M mutation on conformational heterogeneity of the major coat protein (pVIII) of filamentous bacteriophage fd. Journal of Molecular Biology. 1999;286(3):787–796. doi: 10.1006/jmbi.1998.2517. [DOI] [PubMed] [Google Scholar]
- TANG M, NESBITT AE, SPERLING LJ, BERTHOLD DA, SCHWIETERS CD, GENNIS RB, RIENSTRA CM. Structure of the disulfide bond generating membrane protein DsbB in the lipid bilayer. Journal of Molecular Biology. 2013;425(10):1670–1682. doi: 10.1016/j.jmb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIRIOT DS, NEVZOROV AA, OPELLA SJ. Structural basis of the temperature transition of Pf1 bacteriophage. Protein Science. 2005;14(4):1064–1070. doi: 10.1110/ps.041220305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIRIOT DS, NEVZOROV AA, ZAGYANSKIY L, WU CH, OPELLA SJ. Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. Journal of Molecular Biology. 2004;341(3):869–879. doi: 10.1016/j.jmb.2004.06.038. [DOI] [PubMed] [Google Scholar]
- TOLLINGER M, SIVERTSEN AC, MEIER BH, ERNST M, SCHANDA P. Site-Resolved measurement of microsecond-to-millisecond conformational-exchange processes in proteins by solid-state NMR spectroscopy. Journal of the American Chemical Society. 2012;134(36):14800–14807. doi: 10.1021/ja303591y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TORCHIA DA, SZABO A. Information content of powder lineshapes in the fast motion limit. Journal of Magnetic Resonance. 1985;64:135–141. [Google Scholar]
- TYCKO R. Solid-state NMR studies of amyloid fibril structure. Annual Review of Physical Chemistry. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TZENG SR, KALODIMOS CG. Protein activity regulation by conformational entropy. Nature. 2012;488(7410):236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- ULLRICH SJ, GLAUBITZ C. Perspectives in enzymology of membrane proteins by solid-state NMR. Accounts of Chemical Research. 2013;46(9):2164–2171. doi: 10.1021/ar4000289. [DOI] [PubMed] [Google Scholar]
- ULRICH EL, AKUTSU H, DORELEIJERS JF, HARANO Y, IOANNIDIS YE, LIN J, LIVNY M, MADING S, MAZIUK D, MILLER Z, NAKATANI E, SCHULTE CF, TOLMIE DE, KENT WENGER R, YAO H, MARKLEY JL. BioMagResBank. Nucleic Acids Research. 2008;36:D402–408. doi: 10.1093/nar/gkm957. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- VALE RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112(4):467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- VAN DER WEL PCA, LEWANDOWSKI JR, GRIFFIN RG. Structural characterization of GNNQQNY amyloid fibrils by magic angle spinning NMR. Biochemistry. 2010;49(44):9457–9469. doi: 10.1021/bi100077x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VASA S, LIN L, SHI C, HABENSTEIN B, RIEDEL D, KUHN J, THANBICHLER M, LANGE A. β-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(2):E127–E136. doi: 10.1073/pnas.1418450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAUGHAN PS, MIURA P, HENDERSON M, BYRNE B, VAUGHAN KT. A role for regulated binding of p150Glued to microtubule plus ends in organelle transport. Journal of Cell Biology. 2002;158(2):305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEREL R, ERNST M, MEIER BH. Adiabatic dipolar recoupling in solid-state NMR: The DREAM scheme. Journal of Magnetic Resonance. 2001;150(1):81–99. doi: 10.1006/jmre.2001.2310. [DOI] [PubMed] [Google Scholar]
- VINOGRADOV E, MADHU PK, VEGA S. High-resolution proton solid-state NMR spectroscopy by phase-modulated Lee-Goldburg experiment. Chemical Physics Letters. 1999;314(5–6):443–450. [Google Scholar]
- VOLKMAN BF, LIPSON D, WEMMER DE, KERN D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291(5512):2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- WANG JF, KIM S, KOVACS F, CROSS TA. Structure of the transmembrane region of the M2 protein H+ channel. Protein Science. 2001;10(11):2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG S, MUNRO RA, KIM SY, JUNG KH, BROWN LS, LADIZHANSKY V. Paramagnetic relaxation enhancement reveals oligomerization interface of a membrane protein. Journal of the American Chemical Society. 2012;134(41):16995–16998. doi: 10.1021/ja308310z. [DOI] [PubMed] [Google Scholar]
- WANG S, MUNRO RA, SHI L, KAWAMURA I, OKITSU T, WADA A, KIM SY, JUNG KH, BROWN LS, LADIZHANSKY V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nature Methods. 2013;10(10):1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- WANG SL, LADIZHANSKY V. Recent advances in magic angle spinning solid state NMR of membrane proteins. Progress in Nuclear Magnetic Resonance Spectroscopy. 2014;82:1–26. doi: 10.1016/j.pnmrs.2014.07.001. [DOI] [PubMed] [Google Scholar]
- WANG YA, YU X, OVERMAN S, TSUBOI M, THOMAS GJ, JR, EGELMAN EH. The structure of a filamentous bacteriophage. Journal of Molecular Biology. 2006;361(2):209–215. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- WASMER C, LANGE A, VAN MELCKEBEKE H, SIEMER AB, RIEK R, MEIER BH. Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- WASMER C, SCHUTZ A, LOQUET A, BUHTZ C, GREENWALD J, RIEK R, BOCKMANN A, MEIER BH. The molecular organization of the fungal prion HET-s in its amyloid form. Journal of Molecular Biology. 2009;394(1):119–127. doi: 10.1016/j.jmb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- WATERMAN-STORER CM, KARKI S, HOLZBAUR EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATT ED, RIENSTRA CM. Recent advances in solid-state nuclear magnetic resonance techniques to quantify biomolecular dynamics. Analytical Chemistry. 2014;86(1):58–64. doi: 10.1021/ac403956k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEINGARTH M, BALDUS M. Solid-state NMR-based approaches for supramolecular structure elucidation. Accounts of Chemical Research. 2013;46(9):2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- WEINGARTH M, VAN DER CRUIJSEN EAW, OSTMEYER J, LIEVESTRO S, ROUX B, BALDUS M. Quantitative analysis of the water occupancy around the selectivity filter of a K+ channel in different gating modes. Journal of the American Chemical Society. 2014;136(5):2000–2007. doi: 10.1021/ja411450y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WICKRAMASINGHE NP, SHAIBAT MA, JONES CR, CASABIANCA LB, DE DIOS AC, HARWOOD JS, ISHII Y. Progress in 13C and 1H solid-state nuclear magnetic resonance for paramagnetic systems under very fast magic angle spinning. Journal of Chemical Physics. 2008;128(5) doi: 10.1063/1.2833574. [DOI] [PubMed] [Google Scholar]
- WILLIAMSON MP. Secondary-structure dependent chemical-shifts in proteins. Biopolymers. 1990;29(10–11):1428–1431. doi: 10.1002/bip.360291009. [DOI] [PubMed] [Google Scholar]
- WISHART DS, SYKES BD. The 13C chemical-shift index – a simple method for the identification of protein secondary structure using 13C chemical-shift data. Journal of Biomolecular NMR. 1994;4(2):171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- WOOD KW, BERGNES G. Mitotic kinesin inhibitors as novel anti-cancer agents. Annual Reports in Medicinal Chemistry. 2004;39:173–183. [Google Scholar]
- WORTHYLAKE DK, WANG H, YOO S, SUNDQUIST WI, HILL CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallographica Section D, Biological Crystallography. 1999;55(Pt 1):85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- WRIGHT AK, BATSOMBOON P, DAI J, HUNG I, ZHOU HX, DUDLEY GB, CROSS TA. Differential binding of rimantadine enantiomers to influenza A M2 proton channel. Journal of the American Chemical Society. 2016;138(5):1506–1509. doi: 10.1021/jacs.5b13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYLIE BJ, BHATE MP, MCDERMOTT AE. Transmembrane allosteric coupling of the gates in a potassium channel. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):185–190. doi: 10.1073/pnas.1319577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYLIE BJ, SCHWIETERS CD, OLDFIELD E, RIENSTRA CM. Protein structure refinement using 13Cα chemical shift tensors. Journal of the American Chemical Society. 2009;131(3):985–992. doi: 10.1021/ja804041p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYLIE BJ, SPERLING LJ, NIEUWKOOP AJ, FRANKS WT, OLDFIELD E, RIENSTRA CM. Ultrahigh resolution protein structures using NMR chemical shift tensors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):16974–16979. doi: 10.1073/pnas.1103728108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, GUO C, HOU G, ZHANG H, LU X, WILLIAMS JC, POLENOVA T. Atomic-resolution structure of the CAP-Gly domain of dynactin on polymeric microtubules determined by magic angle spinning NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2015a;112(47):14611–14616. doi: 10.1073/pnas.1509852112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, HOU GJ, SEHWIETERS CD, AHMED S, WILLIAMS JC, POLENOVA T. Three-dimensional structure of CAP-Gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy: Conformational plasticity and interactions with end-binding protein EB1. Journal of Molecular Biology. 2013a;425(22):4249–4266. doi: 10.1016/j.jmb.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, SUITER CL, HOU GJ, ZHANG HL, POLENOVA T. Probing structure and dynamics of protein assemblies by magic angle spinning NMR spectroscopy. Accounts of Chemical Research. 2013b;46(9):2047–2058. doi: 10.1021/ar300309s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, ZHANG HL, HOU GJ, AHMED S, WILLIAMS JC, POLENOVA T. Internal dynamics of dynactin CAP-Gly is regulated by microtubules and plus end tracking protein EB1. Journal of Biological Chemistry. 2015b;290(3):1607–1622. doi: 10.1074/jbc.M114.603118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, TASAYCO ML, POLENOVA T. Magic angle spinning NMR experiments for structural studies of differentially enriched protein interfaces and protein assemblies. Journal of the American Chemical Society. 2008;130(17):5798–5807. doi: 10.1021/ja711304e. [DOI] [PubMed] [Google Scholar]
- YANG J, TASAYCO ML, POLENOVA T. Dynamics of reassembled thioredoxin studied by magic angle spinning NMR: Snapshots from different time scales. Journal of the American Chemical Society. 2009;131(38):13690–13702. doi: 10.1021/ja9037802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG RF, AIKEN C. A mutation in α-helix 3 of CA renders human immunodeficiency virus type 1 cyclosporin A resistant and dependent: Rescue by a second-site substitution in a distal region of CA. Journal of Virology. 2007;81(8):3749–3756. doi: 10.1128/JVI.02634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG RF, SHI J, BYEON IJL, AHN J, SHEEHAN JH, MEILER J, GRONENBORN AM, AIKEN C. Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects. Retrovirology. 2012;9 doi: 10.1186/1742-4690-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO LS, VOGELI B, YING JF, BAX A. NMR determination of amide N-H equilibrium bond length from concerted dipolar coupling measurements. Journal of the American Chemical Society. 2008;130(49):16518–16520. doi: 10.1021/ja805654f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO XL, HONG M. Dipolar filtered 1H-13C heteronuclear correlation spectroscopy for resonance assignment of proteins. Journal of Biomolecular NMR. 2001;20(3):263–274. doi: 10.1023/a:1011251924874. [DOI] [PubMed] [Google Scholar]
- YAO XL, SCHMIDT-ROHR K, HONG M. Medium- and long-distance 1H-13C heteronuclear correlation NMR in solids. Journal of Magnetic Resonance. 2001;149(1):139–143. [Google Scholar]
- YLINEN LM, SCHALLER T, PRICE A, FLETCHER AJ, NOURSADEGHI M, JAMES LC, TOWERS GJ. Cyclophilin A levels dictate infection efficiency of human immunodeficiency virus type 1 capsid escape mutants A92E and G94D. Journal of Virology. 2009;83(4):2044–2047. doi: 10.1128/JVI.01876-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU TY, SCHAEFER J. REDOR NMR characterization of DNA packaging in bacteriophage T4. Journal of Molecular Biology. 2008;382(4):1031–1042. doi: 10.1016/j.jmb.2008.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZECH SG, OLEJNICZAK E, HAJDUK P, MACK J, MCDERMOTT AE. Characterization of protein-ligand interactions by high-resolution solid-state NMR spectroscopy. Journal of the American Chemical Society. 2004;126(43):13948–13953. doi: 10.1021/ja040086m. [DOI] [PubMed] [Google Scholar]
- ZERI AC, MESLEH MF, NEVZOROV AA, OPELLA SJ. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6458–6463. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Q, SUN XY, WATT ED, AL-HASHIMI HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311(5761):653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- ZHANG R, DAMRON J, VOSEGAARD T, RAMAMOORTHY A. A cross-polarization based rotating-frame separated-local-field NMR experiment under ultrafast MAS conditions. Journal of Magnetic Resonance. 2015;250:37–44. doi: 10.1016/j.jmr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO G, PERILLA JR, YUFENYUY EL, MENG X, CHEN B, NING J, AHN J, GRONENBORN AM, SCHULTEN K, AIKEN C, ZHANG P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497(7451):643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU DH, RIENSTRA CM. Rapid analysis of organic compounds by proton-detected heteronuclear correlation NMR spectroscopy with 40 kHz magic-angle spinning. Angewandte Chemie-International Edition. 2008;47(38):7328–7331. doi: 10.1002/anie.200802108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU DH, SHAH G, CORMOS M, MULLEN C, SANDOZ D, RIENSTRA CM. Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. Journal of the American Chemical Society. 2007a;129(38):11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- ZHOU DH, SHEA JJ, NIEUWKOOP AJ, FRANKS WT, WYLIE BJ, MULLEN C, SANDOZ D, RIENSTRA CM. Solid-state protein-structure determination with proton-detected triple-resonance 3D magic-angle-spinning NMR spectroscopy. Angewandte Chemie-International Edition. 2007b;46(44):8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZINKEVICH T, CHEVELKOV V, REIF B, SAALWACHTER K, KRUSHELNITSKY A. Internal protein dynamics on ps to mu s timescales as studied by multi-frequency N-15 solid-state NMR relaxation. Journal of Biomolecular NMR. 2013;57(3):219–235. doi: 10.1007/s10858-013-9782-2. [DOI] [PubMed] [Google Scholar]


