Abstract
Biological nanopores are a class of membrane proteins that open nanoscale water conduits in biological membranes. When they are reconstituted in artificial membranes and a bias voltage is applied across the membrane, the ionic current passing through individual nanopores can be used to monitor chemical reactions, to recognize individual molecules and, of most interest, to sequence DNA. In addition, a more recent nanopore application is the analysis of single proteins and enzymes. Monitoring enzymatic reactions with nanopores, i.e. nanopore enzymology, has the unique advantage that it allows long-timescale observations of native proteins at the single-molecule level. Here, we describe the approaches and challenges in nanopore enzymology.
This article is part of the themed issue ‘Membrane pores: from structure and assembly, to medicine and technology’.
Keywords: nanopore enzymology, single-molecule, protein trapping, review
1. Introduction
In recent years, nanopores have emerged as powerful tools to observe single molecules. In nanopore analysis, the signal is given by the flux of ions passing through individual nanopores embedded in an insulating membrane under an externally applied bias voltage. Molecules diffusing inside the nanopore can be recognized or studied by monitoring the changes in the nanopore current. Initial work with nanopores focused on the detection of single molecules [1,2], investigated mechanisms of chemical reactions at the single-molecule level [3] and, most notably, developed sensors for nucleic acids [4]. Only recently researchers have started using nanopores for the detection and analysis of more complex analyte molecules such as proteins. The aim of this review is to highlight the work that has been done towards the analysis of enzymatic reactions with nanopores. We will start by describing the nanopores used to monitor proteins and then we will introduce three different approaches used to monitor enzymatic reactions with nanopores: (i) the engineering of a nanopore with an enzymatic function, (ii) the monitoring of enzymes moving a polymer across a nanopore, and (iii) the observation of the binding of ligands to enzymes confined inside a nanopore.
2. Nanopores
Nanopores can now be fabricated using a variety of materials, including solid-state membranes [5–8], glass [9,10], carbon nanotubes [11–13] or DNA origami [14–19]. Solid-state nanopores have the advantage that their size and the thickness of the insulating material can be readily controlled, and that their surface properties can be chemically modified to acquire new properties [20]. Notably, mono- and bi-atomic membranes can now be used to form nanopores [21–27]. However, the reproducible fabrication of artificial nanopores with sub-nanometre precision, which is important to obtain consistent molecular recognition, is still a major challenge. While DNA nanopores have just begun to reveal their potential [28], the ease with which protein nanopores can be reproducibly fabricated and engineered with atomic-level precision means that, at least for now, they remain superior to their artificial counterparts.
Owing to an early available crystal structure [29], the alpha-hemolysin (αHL) nanopore has been widely engineered over the last 20 years. The rigidity of its β-barrel made αHL particularly amenable to mutagenesis [30,31], chemical engineering [32] and rational design of binding elements [33]. Later, other β-barrel pores, such as MspA [34], OmpG [35], aerolysin [36] and FhuA [37] were also characterized by single-molecule electrophysiology. The Guo laboratory showed that a Φ29 portal protein can be engineered to reversibly insert itself into lipid bilayers, demonstrating that biological nanopores may be obtained from soluble proteins [38]. Recently, we showed that the properties of α-helical Cytolysin A (ClyA) [39] and Fragaceatoxin C (FraC) [40] nanopores could be improved or tuned by directed evolution. Most notably, ClyA nanopores with higher oligomeric states could be isolated [39]. The difficulty modulating the size and shape of protein nanopores is one of their biggest limitations, as the ability to obtain nanopores with a larger diameter is particularly sought after.
3. Approach 1: engineering nanopores with catalytic functions
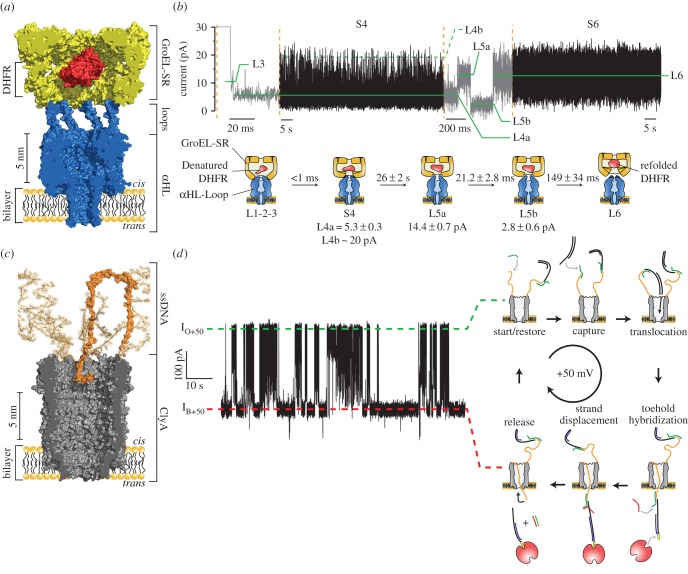
One possibility for sampling enzymatic reactions with a nanopore is to engineer an enzymatic function into the nanopore itself. In a first approach to build a catalytic nanopore, we designed an αHL nanopore with the co-catalytic function of GroES (figure 2a,b) [49]. In bacteria, the GroEL/GroES chaperonin pair forms a folding chamber of approximately 65 Å diameter, inside which many cellular proteins reach their native fold. The co-chaperonin GroES associates as a lid to GroEL through the interaction of seven unstructured loops that slide into seven hydrophobic pockets in GroEL [50]. We created a chimeric nanopore with the co-catalytic activity of GroES by fusing the flexible loops of GroES to the seven subunits of αHL (figure 2a). The αHL-GroES nanopores were then used to assist and simultaneously monitor protein folding by GroEL. Crucially, carefully designed αHL-GroES chimeras were as efficient in assisting GroEL protein folding as GroES alone. Our designed system allowed the single-molecular study of protein folding and gave insights not achievable with ensemble studies such as the observation of intermediate states (figure 2b) [49]. A similar approach might be used to pair other ring-shaped proteins to nanopores. Proteins with toroidal structures are also found in processivity factors, DNA replication initiators, helicases, transcription terminators, DNA-binding protease, proteasome proteins and the AAA+ (ATPases associated with various cellular activities) protein family.
Figure 2.
Nanopores with enzymatic functions. (a,b) A co-chaperonin nanopore. (a) A nanopore-GroES was engineered by inserting the flexible loop of GroES into the αHL nanopore. (b) Catalytic cycle of dihydrofolate reductase (DHFR) refolding by a single-ring GroEL molecule atop of a nanopore-GroES nanopore in the presence of ATP. The L1 to L6 current level indicate the intermediates of the enzymatic reaction. Adapted from [49]. Reprinted with permission from AAAS. (c,d) A catalytic translocase nanopore. (c) A ClyA nanopore (grey) with a ring of ssDNA molecules (orange) attached to its cis entry. (d) Ionic current trace (left) showing the sequential transport of selected DNA molecule under a constant applied potential of +50 mV, as indicated by the cartoon on the right of the trace. Adapted by permission from Macmillan Publishers Ltd: Nature Communications [51], copyright © 2013.
In another example, a ClyA nanopore was engineered to catalyse the active transport of selected DNA molecules across a membrane (figure 2c,d) [51]. It was found that while dsDNA molecules can translocate through the nanopore under a positive applied potential, ssDNA is too flexible to be readily confined inside the nanopore. This physical property of ssDNA was exploited to catalyze the transport across the nanopore. ssDNA molecules were covalently attached to the mouth of ClyA, which then created a physical barrier outside the nanopore preventing unspecific DNA translocation (figure 2c). At positive applied potentials, the ssDNA loops recognized a target ‘cargo’ DNA molecule in solution that was presented on one side of the pore. The newly formed double-stranded DNA complex on top of ClyA could then translocate to the other side of the membrane through the nanopore. DNA strand displacement on the opposite side of the nanopore by another single-stranded DNA molecule released the cargo and restored the transporter to its initial state, ready for the next cycle (figure 2d). As observed in cells, the transport occurred under a constant applied transmembrane potential. Although this system did not work against the applied potential and thus was categorized as a catalytic translocase [52], analogous to biological active transporters, it also carried the ability to accumulate the DNA cargo against a chemical gradient [51].
4. Approach 2: observing enzymes that ratchet a polymer across a nanopore
Ever since its conception, the potential of sequencing DNA with nanopores has been an important driving force in the nanopore field. After the first experiment showing the electrophoretic transport of single-stranded nucleic acids across an αHL nanopore [4] and the recognition of individual nucleobases in immobilized strands [53,54], researchers focused their attention on using enzymes to control the transport of DNA across the nanopore. Initial efforts mainly focused on establishing the binding or immobilization of a DNA-processing enzyme on top of the pore [55–57]. Then, in 2008, using ssDNA molecules immobilized inside a nanopore, Ghadiri's group showed the first example of an enzyme-mediated transport of a DNA molecule across an αHL nanopore [58]. However, because the selected TopoTaq enzyme could not polymerize DNA against the applied potential, the system had to be regularly cycled between a negative applied potential where the enzyme could bind to the DNA and incorporate one nucleotide, and a positive applied potential where the incorporation could be monitored. During each cycle the enzyme was released from the DNA, thus this approach did not allow monitoring the action of the same, single enzyme.
5. Enzymes ratcheting DNA in real time
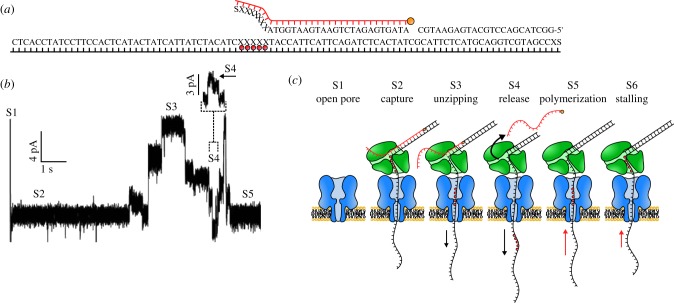
Several studies sampled various DNA polymerases under an applied potential [59–61]; however, it was not until the bacteriophage Φ29 DNA polymerase was introduced by the Akeson group that an enzymatic reaction could be observed in real time with an αHL pore (figure 3) [62]. The DNA template consisted of a double-stranded DNA extended with a long 5′ overhang, which allowed capture by the nanopore, and a 3′ phosphorylated end that permitted the initiation of the DNA polymerization reaction. A stretch of abasic nucleobases was also incorporated in the 5′ overhang to help monitoring the transport of DNA across an αHL nanopore. An additional blocking primer, hybridizing just before the 3′ phosphorylated end of the hairpin, was used to prevent the bulk phase replication of the template DNA (figure 3a) [63]. Upon capture of the DNA construct complexed with Φ29 polymerase, the blocking DNA was unzipped by the applied voltage, making the 3′ phosphorylated end accessible for the DNA polymerase (figure 3b,c). DNA polymerization above the nanopore revealed different states in the polymerization reaction as the position of the abasic insert moved inside the αHL pore. Further work showed that this system can be used to reveal the details of enzymatic reactions, such as the dynamics of the pre- and post-translocation steps of binary and ternary complex formation [64,65] and the primer strand transfer from the polymerase to the exonuclease site within the enzyme [66]. Using an identical approach but with MspA (figure 1b), a nanopore with a narrower constriction than αHL, the Gundlach group studied DNA polymerization at a higher resolution [67]. This approach revealed the occasional back-stepping of Φ29 polymerase, which is most probably due to the 3′-5′ proofreading activity or a failure to incorporate a nucleotide.
Figure 3.
Enzymatic ratcheting of DNA across a nanopore. (a) The DNA substrate protected by a blocking oligomer used in the study, with the blocking oligomer highlighted in red. The five consecutive X letters (red dots) indicate abasic residues in the 94-mer DNA template that cause an ionic current increase as they traverse the nanopore. (b,c) Forward and reverse ratcheting of DNA through the nanopore. (b) Ionic current trace of the experiment. (c) Detailed schematic overview of the ratcheting process. S1–S5 correspond to each other in (b) and (c). An αHL nanopore is in blue and a phi29 polymerase in green. Adapted by permission from Macmillan Publishers Ltd: Nature Biotechnology [63], copyright © 2012.
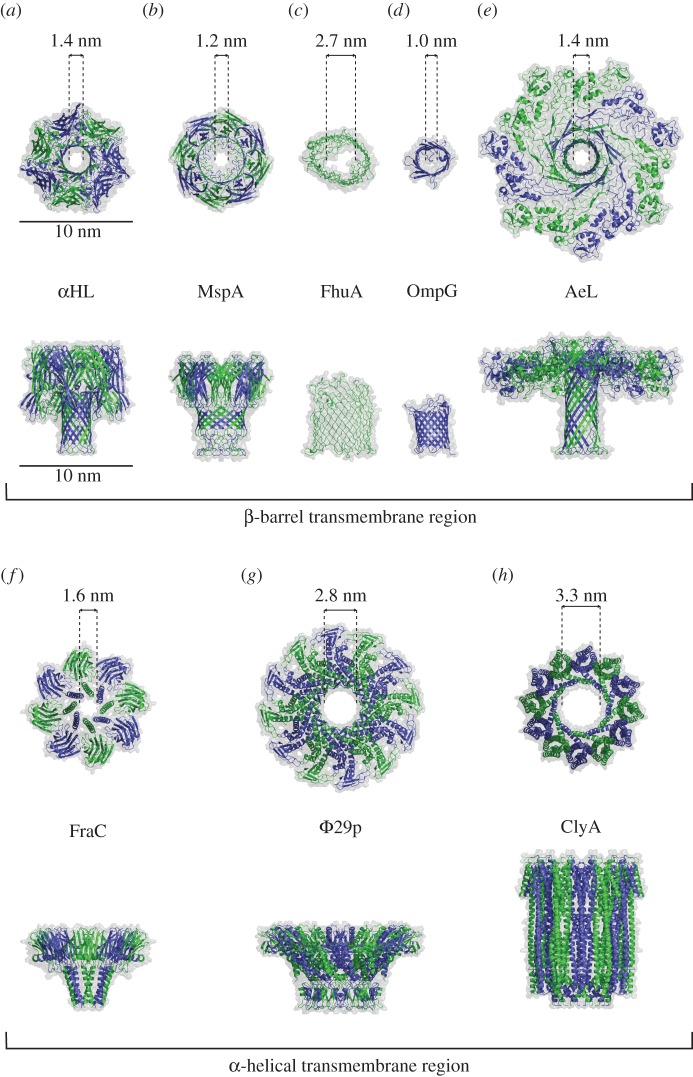
Figure 1.
Biological nanopores. Cartoon representation and estimated nanopore diameters of: (a) α-hemolysin (αHL, PDB ID = 7AHL) [29], (b) Mycobacterium smegmatis porin A (MspA, PDB ID = 1UUN) [41], (c) Ferric hydroxamate uptake A (FhuA, PDB ID = 1BY3) [42], (d) outer membrane porin G (OmpG, PDB ID = 2IWV) [43], (e) aerolysin (AeL, PDB ID = 5JZT) [44], (f) Fragaceatoxin C (FraC, PDB ID = 4TSY) [45], (g) Φ29 portal protein (Φ29p, PDB ID = 1FOU) [46], and (h) Cytolysin A (ClyA, PDB ID = 2WCD) [47] nanopores. The figures were prepared with PyMOL [48].
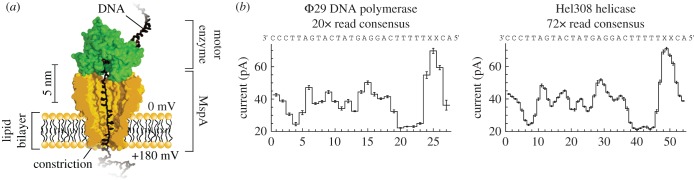
The Gundlach group also investigated the enzymatic activity of an ATP-dependent DNA helicase Hel308 (figure 4a) [68]. By comparing the current steps produced by the DNA movement with a reference curve obtained by moving the same strand with Φ29 DNA polymerase (figure 4b, left), the current signal was translated into the position of the base in the pore. The authors observed an additional step in the motion of Hel308 along a DNA strand (figure 4b, right). This confirmed a previously made hypothesis based on the crystal structure [69], where the additional step corresponds to domain 2 in the helicase that is pushed upwards upon ATP binding, thus making DNA move up in the pore. This is then followed by ATP hydrolysis, which will move the DNA inside the nanopore by one nucleotide. The motion of DNA in the pore therefore was monitored with a resolution of half a nucleotide, thereby resolving smaller motions than FRET.
Figure 4.
Single-molecule picometer-resolution nanopore tweezers (SPRNT). (a) A ssDNA:helicase complex is captured in the MspA pore. (b) Consensus of current level patterns for the shown DNA sequence using Φ29 DNA polymerase (left) or Hel308 helicase (right). Φ29 polymerase moves DNA base-by-base through the pore, while Hel308 shows intermediate current steps. Adapted by permission from Macmillan Publishers Ltd: Nature Biotechnology [68], copyright © 2015.
In summary, DNA-processing enzymes operating above the nanopore appear not to perturb the ionic current through the nanopore, enabling the measurement of their enzymatic activity as they move a DNA/RNA strand across the nanopore. However, only enzymes that are able to sustain a relatively strong force (approx. 10 pN for a ssDNA inside an αHL nanopore under + 160 mV [70]) can be used with this method.
6. Enzymatic unfolding and ratcheting of proteins
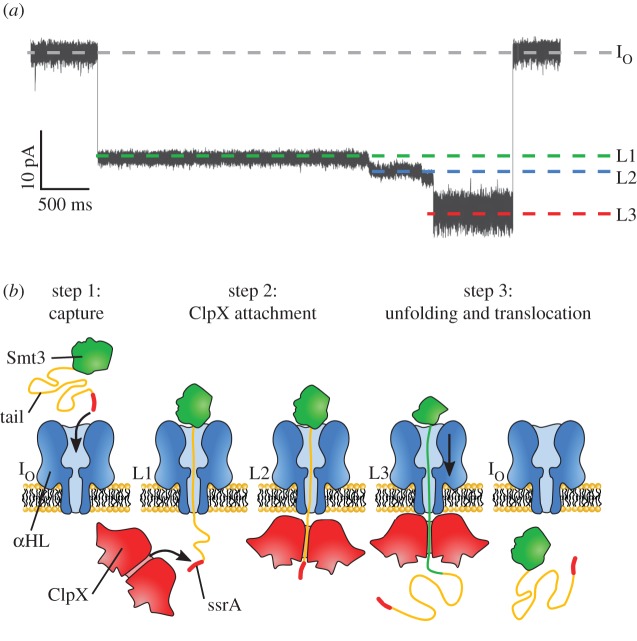
An approach to actively transport proteins through the pore has been attempted with the bacterial AAA+ unfoldase ClpX (figure 5) [71]. ClpX forms a hexameric ring that uses the chemical energy of ATP hydrolysis to unfold a protein through the pore. Although such a system was developed to sequence proteins, it can also be employed to study the activity of the unfoldase enzyme. The ubiquitin-like protein Smt3 was chosen as a target protein and modified with a negatively charged tail in order to be readily captured by the αHL pore. A ClpX targeting motif (ssrA) was also added, enabling the specific binding to ClpX on the other side of the membrane. In the presence of ATP, the ionic current signal revealed at least three clear steps (figure 5a,b): step 1 (IO to L1) reflected the capture of the Smt3 complex, step 2 (L1 to L2) represented the binding of ClpX to the protein, and step 3 (L2 to L3) showed the sequential unfolding and translocation of the Smt3 protein. In the absence of ClpX and ATP, steps 2 and 3 were not observed, confirming the protein is actively unfolded and then transported by the ClpX motor protein. The authors also showed that the ionic current fluctuations reflected the amino acid composition and that ClpX-dependent translocation was relatively unaffected by changes in the applied voltage. In subsequent work, the authors showed that different protein domains and variations within a protein domain could be discriminated in titin and GFP proteins [72].
Figure 5.
Enzymatic ratcheting of proteins across nanopores. (a) Current trace showing the enzymatic unfolding of ClpX monitored by a αHL nanopore. IO corresponds to the open pore current, L1 to the capture of the protein complex, L2 to the binding of ClpX and L3 to the protein unfolding. (b) Working model of ClpX-mediated unfolded translocation of the protein substrate. The substrate protein is a ubiquitin-like protein Smt3 (green) extended by a negatively charged polyanion tail (yellow) and capped by a ssrA targeting motif (red). Adapted by permission from Macmillan Publishers Ltd: Nature Biotechnology [71], copyright © 2013.
7. Approach 3: enzyme reactions inside a nanopore
A more generalized approach to study enzymes with nanopores is to observe enzymatic reactions while the enzyme is lodged inside the nanopore lumen. It has been extensively shown that once proteins diffuse inside the capture radius of a nanopore, the combined action of the electrophoretic and the electro-osmotic forces promote their entry inside the nanopore [7,73–82]. In order to be studied by ionic currents, however, the proteins should remain inside the nanopore for at least a few seconds. Although the confinement of a protein inside a nanopore of comparable size will probably result in a greatly reduced diffusive and electrophoretic mobility (up to two or three orders of magnitude) [83–85], a significant number of studies with artificial nanopores showed that proteins still translocate too quickly to be properly sampled [73–81]. Furthermore, the events that are observed in such experiments are consistent with high protein–pore interactions, which might interfere with enzyme activity [80,81,86].
8. Protein immobilization: solid-state nanopores
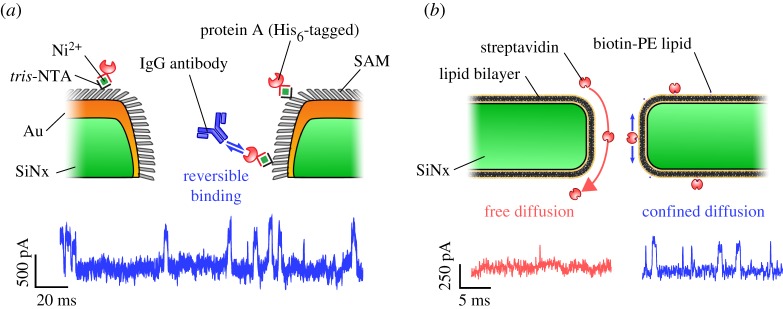
In one attempt to increase the residence time of proteins inside a nanopore, the Rant group trapped His6-tagged proteins by chemically modifying the walls of a gold-coated SiN nanopore with sparse nitrilotriacetic acid (NTA) moieties (figure 6a) [7]. Depending on the multivalency of the NTA groups, observation times were increased from 1.2 ± 0.3 ms (no NTA) to 49.8 ± 5.8 ms (mono-NTA), 3.8 ± 0.3 s (bis-NTA) and over 1000s (tris-NTA). Using the tris-NTA nanopore, the kinetics of two different IgG antibodies binding to single immobilized Protein A molecules could be observed. The Dekker group constructed an artificial nuclear pore complex (NPC) by functionalizing the SiN nanopore walls with a monolayer of FG domains derived from either the Nup98 or the Nup153 proteins [87]. The authors observed the selective transport of importin β over bovine serum albumin, together with a 10-fold increase in the average translocation time from approximately 200 µs to approximately 3 ms. In another biomimetic approach, the Mayer group coated the walls of their SiN nanopores with a fluid lipid bilayer (figure 6b) [78]. Besides virtually eliminating non-specific binding, the tethering of proteins to a biotinylated lipid anchor reduced diffusive translocation times of streptavidin, anti-biotin Fab fragments and anti-biotin antibodies by at least 2 orders of magnitude to approximately 50–200 µs. Notably, the lipid-coated nanopore permitted the accurate determination of both the excluded volume occupied by the protein (via the blockade amplitude) and the protein charge (via the translocation time). Tuning of the bilayer lipid composition and temperature also enabled a flexible way of modulating the nanopore size and translocation times through bilayer thickness and lipid viscosity, respectively. In a follow-up paper, a detailed analysis of the current distribution of each individual protein translocation event was performed. Using the theory of rotational dynamics, the authors succeeded in determining the approximate shape, volume, charge, rotational diffusion coefficient and dipole moment of 10 individual proteins [88].
Figure 6.
Protein immobilization inside solid-state nanopores. (a) NTA-moieties attached to a gold-coated silicon nitride (SiN) nanopore enabled the immobilization of protein A molecules baring a poly-Histidine tag inside the pore. The ionic current shows the reversible binding kinetics of IgG to single protein. Adapted by permission from Macmillan Publishers Ltd: Nature Nanotechnology [7], copyright ©2012. (b) The coating of SiN nanopores with a lipid bilayer enabled the detection of single avidin molecules bound to biotin lipid anchors. The ionic current shows the greatly reduced diffusion rate of the bound compared to the unbound protein molecules. Adapted by permission from Macmillan Publishers Ltd: Nature Nanotechnology [78], copyright © 2011.
9. Protein immobilization: ClyA biological nanopore
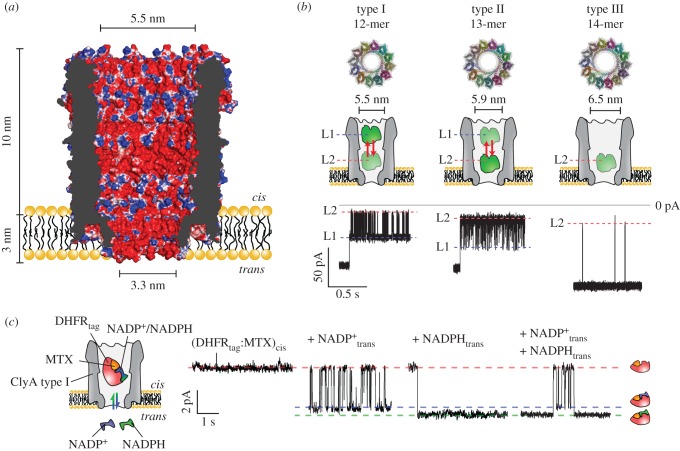
Biological nanopores with an internal diameter wide enough to accommodate proteins in their natively folded configuration, such as the ClyA [89] or Φ29 [38] nanopores (figure 1), might also be used to sample proteins. Our research group recently showed that proteins can be trapped inside the ClyA nanopore (figure 7a) depending on their size [89]. Conveniently, owing to its modular structure, higher order ClyA multimers can be isolated, thus producing variants with pore lumen diameters of 5.5, 5.9 and 6.5 nm (figure 7b) [39]. Proteins such as lysozyme (14 kDa) quickly translocated across the dodecameric nanopore. However, proteins smaller than the cis entry of the nanopore (approx. 5.5 nm), but larger than the trans constriction (approx. 3.3 nm), such as thrombin (37 kDa), were trapped inside the nanopore by the electro-osmotic flow for tens of minutes at selected applied potentials (figure 7b). While the electrical potential usually has a limited role in dragging proteins inside the nanopore, it can be exploited to increase the residence time of proteins. For example, the interaction of a DNA aptamer with human thrombin at high applied potential (−100 mV) increased the dwell time by three orders of magnitude [93]. This effect was explained by an increase of the electrophoretic force, introduced with the binding to the DNA aptamer, that opposed the electro-osmotic force [93]. In another experiment, the dwell time of dihydrofolate reductase (DHFR) 19 kDa was increased by introducing a positively charged polypeptide tag at the C-terminus [92,94]. This tag was designed to neutralize the relatively high negative charge of the protein (pI = 4.8, −11 net charge at pH 8). It was found that the binding of the inhibitor methotrexate, which carries two additional negative charges and binds at the opposite end of the added polypeptidic tag, prolonged the dwell time by three orders of magnitude, most probably by immobilizing the protein at an electrostatic minimum inside the nanopore [92]. Similar results were obtained with unfolded peptides during translocation through αHL nanopores [95,99].
Figure 7.
Single-molecule enzymology with ClyA nanopores. (a) Cross-section of the dodecameric ClyA-AS nanopore with its molecular surface coloured according to electrostatic potential in 150 mM NaCl (APBS [95], −2 to +2 kBT/e). The highly negatively charged interior generates a strong electro-osmotic flow [96]. (b) Top: Top down view of homology models of the three different sizes of ClyA. Middle: cartoon representation of the three ClyA nanopores. Bottom: characteristic time traces from a thrombin trapping experiment for each ClyA variant. The L1 and L2 current levels likely reflect the residence of human thrombin in two residence sites within the ClyA lumen (L1 and L2). Adapted with permission from [39]. Copyright ©2013 American Chemical Society. (c) Binding and unbinding of NADP+ and NADPH to a single DHFR:MTX complex trapped inside a dodecameric ClyA nanopore. Adapted with permission from [91]. Copyright ©2015 American Chemical Society.
10. Observing ligands binding to proteins inside ClyA
To observe enzymatic reactions with a nanopore, the ionic current should report information about enzyme kinetics and dynamics. Many enzymes undergo a conformational change before or after association with their substrate. It has been shown that the accurate detection and analysis of translocation events using a high bandwidth detector results in a wealth of information about proteins, including their concentration, charge, size and shape, protein–protein interactions and even interaction energy with the pore [85], suggesting that small differences in the protein structure or overall charge should be reflected by changes in the ionic current. Most probably, however, in order to detect the minute structural changes during the enzymatic cycle in the internalized protein, one should employ nanopores with diameters close to that of the protein of interest. Recently, we have shown that ClyA nanopores can report the binding of substrates to internalized proteins [92]. Using DHFR, which adopts different conformations during the catalytic progression, and AlkB, which folds upon binding to its substrate, we showed that in the presence of ligands, the internalized proteins show step-wise current signals (figure 7c) whose frequency increases with the ligand concentration. Importantly, protein variants with a strongly decreased affinity for the ligands did not elicit step-wise current blockades, proving that the ligand-induced signal is specific to the binding of proteins to their ligands. Although it is not clear if the signal arises from different protein conformations or from different positions/orientations/interactions of the protein within the nanopore upon binding their charged ligands, these results indicate that enzymatic reactions can be reported by a nanopore. Remarkably, the binding of NADPH and NADP+, which differ by only one hydride ion, elicited a different current block signal when binding to a DHFR:methotrexate complex inside ClyA (figure 7c), suggesting that minute differences in protein:ligand complexes might be observed when sampling enzymatic reactions with nanopores.
11. The environment of the ClyA nanopore
One important consideration when measuring enzymatic reactions with nanopores is the role of the nanopore walls and the relatively strong electrostatic and electrophoretic forces inside the nanopore. The crystal structure of the oligomeric ClyA protein revealed that the nanopore has a plethora of negatively charged residues lining its interior walls (figure 7a). Such a negatively charged nanocavity is reminiscent of the well-studied E. coli GroEL/GroES chaperone [51]. Although the precise mechanism by which GroEL aids protein refolding remains controversial [97–100], it is likely that the confinement prevents misfolded proteins from aggregating. Another explanation could be that the rich electrostatic environment inside the GroEL cavity increases the water density and hence enhances folding due to an enhanced hydrophobic effect compared with bulk [97,98]. The diffusivity and the structuring of the water molecules confined inside GroEL in the absence of substrate protein are likely to be similar to that in bulk solution [101]. When there is a protein confined inside the chaperonin cavity, however, these properties might change depending on the interaction strength between the protein and the internal walls of the chaperonin [100]. Interestingly, the folding pathways inside the chaperonin chamber have been proposed to be similar [102] or different [103] compared with bulk [104]. Regardless of the precise folding mechanism and the behaviour of the confined solvent, a charged, hydrophilic cage appears to promote the native state of proteins. Therefore, no detrimental effects on the conformation of proteins trapped inside ClyA are to be expected.
Although the nanoscale confinement of proteins inside ClyA is likely not to affect the folding of the protein, the role of the applied potential across the pore, and the resulting electrophoretic and electro-osmotic forces, is not known. To estimate their magnitude, the lumen of ClyA may be described as two cylindrical electrolyte chambers in series. The strength of the electrical field along the central nanopore axis inside the wide cis chamber of dodecameric ClyA can then be estimated to be approximately 3 mV nm−1, as calculated from the resistance of the cis chamber for a bias voltage of −50 mV (Rcis ≈ 2.6 × 108 Ω, Rtrans ≈ 2.2 × 108 Ω, ΔVcis ≈ 27 mV, ΔVtrans ≈ 23 mV, lcis = 10 nm, ltrans = 3 nm [39]). Thus, if an immobilized protein bares a significant net charge, e.g. 10 ec, it would be subject to a Coulomb force of approximately 4 pN. Forces of this magnitude have been shown before to partially unfold or deform proteins by AFM [105]. However, the Pelta group showed that within the 50–250 mV voltage range, the fractional residual current (defined as the blocked pore current divided by the open pore current) of maltose binding protein translocating through a solid-state nanopore remains constant, as expected for a protein that remains folded [106]. Using ClyA and sampling several proteins (human thrombin, AlkB and DHFR), we did not observe a change in fractional residual current up to −200 mV, confirming that the protein normally should not unfold under moderate applied potentials.
It is worth noticing that the net force experienced by a protein immobilized inside a nanopore will also depend on the magnitude and direction of the electro-osmotic flow. The negative charges lining the interior walls of ClyA make the pore highly cation-selective and induce a strong electro-osmotic flow [91], enabling the capture of proteins even against the electrical field [89]. While the precise magnitude of the water velocity inside ClyA is currently unknown, experimental data using αHL nanopores [107] and simulations performed with solid-state [108,109] and αHL [110,111] nanopores, revealed velocities in the order of 100 mm s−1. Assuming a similar water velocity inside ClyA and applying Stokes' law, a static and uncharged sphere of 5 nm in diameter would endure a force of approximately 4 pN. Thus, under negatively applied potentials (trans) the electrophoretic and electro-osmotic forces acting on negatively charged proteins would oppose and cancel each other out, resulting in less stress on the protein and a more diffusion-dominated environment [73]. For positively charged molecules, these two forces will add and increase the net force, resulting in additional stress on the captured protein.
12. Outlook
Using the ideas and approaches outlined above, several potential applications can be envisioned. Engineering nanopores with enzymatic functions could yield additional building blocks for the bottom-up creation of artificial cells, while computational design of nanopores holds great promise for enhancing engineering efforts and to expand the functional nanopore repertoire [112,113]. Currently the biggest challenge in nanopore enzymology is to control the transit time of proteins across the nanopore. Native enzymes might be trapped by steric and electrostatic forces, locked by rotaxanes [94], or immobilized covalently inside a nanopore to allow their single-molecular study for extended periods of time. Alternatively, substrate molecules could be covalently attached inside the nanopore and the binding catalysis performed by enzymes monitored by current recordings. The nanopore approach could yield potentially unprecedented long observation times of individual enzymes. This will most probably provide new insights into the role of dynamics and structure heterogeneity in enzymatic reactions and will allow studying rare intermediate molecule species not accessible with ensemble studies. Notably, such systems could also be used for biomolecule sensing applications by exploiting enzymes as molecular adapters with intrinsic affinities to target substrates [92]. As nanopore currents can be easily interfaced with electronic devices, such approaches could allow manufacturing of low-cost and portable sensors to monitor the concentration of a large panel of biologically relevant molecules or to detect disease-linked biomarkers.
Acknowledgements
The authors appreciate discussions on the electro-osmotic flow with Mr Ujjal Barman and the help of Dr John M. O'Callaghan with proofreading the manuscript.
Authors' contributions
K.W., V.V.M., C.W. and G.M. co-wrote the paper. K.W. and V.V.M. prepared the figures. K.W. and V.V.M. contributed equally to this work.
Competing interests
We have no competing interests.
Funding
This work was supported by ERC 260884 to G.M. and C.W., IWT 3E130054 to K.W. and FWO 3E130053 to V.V.M.
References
- 1.Bezrukov SM, Vodyanoy I, Parsegian VA. 1994. Counting polymers moving through a single ion channel. Nature 370, 279–281. ( 10.1038/370279a0) [DOI] [PubMed] [Google Scholar]
- 2.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. 1997. Designed protein pores as components for biosensors. Chem. Biol. 4, 497–505. ( 10.1016/S1074-5521(97)90321-5) [DOI] [PubMed] [Google Scholar]
- 3.Luchian T, Shin SH, Bayley H. 2003. Single-molecule covalent chemistry with spatially separated reactants. Angew. Chem. Int. Ed. 42, 3766–3771. ( 10.1002/anie.200351313) [DOI] [PubMed] [Google Scholar]
- 4.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. 1996. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13 770–13 773. ( 10.1073/pnas.93.24.13770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker C. 2007. Solid-state nanopores. Nat. Nanotechnol. 2, 209–215. ( 10.1038/nnano.2007.27) [DOI] [PubMed] [Google Scholar]
- 6.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. 2001. Ion-beam sculpting at nanometre length scales. Nature 412, 166–169. ( 10.1038/35084037) [DOI] [PubMed] [Google Scholar]
- 7.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. 2012. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 7, 257–263. ( 10.1038/nnano.2012.24) [DOI] [PubMed] [Google Scholar]
- 8.Venkatesan BM, Bashir R. 2011. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 6, 615–624. ( 10.1038/nnano.2011.129) [DOI] [PubMed] [Google Scholar]
- 9.Li W, Bell NA, Hernández-Ainsa S, Thacker VV, Thackray AM, Bujdoso R, Keyser UF. 2013. Single protein molecule detection by glass nanopores. ACS Nano 7, 4129–4134. ( 10.1021/nn4004567) [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Zhang B, Wayment JR, Harris JM, White HS. 2006. Electrostatic-gated transport in chemically modified glass nanopore electrodes. J. Am. Chem. Soc. 128, 7679–7686. ( 10.1021/ja061357r) [DOI] [PubMed] [Google Scholar]
- 11.Choi W, Ulissi ZW, Shimizu SF, Bellisario DO, Ellison MD, Strano MS. 2013. Diameter-dependent ion transport through the interior of isolated single-walled carbon nanotubes. Nat. Commun. 4, 2397 ( 10.1038/ncomms3397) [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Yang C, Zhao K, Li J, Wu HC. 2013. Ultrashort single-walled carbon nanotubes in a lipid bilayer as a new nanopore sensor. Nat. Commun. 4, 2989 ( 10.1038/ncomms3989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, et al. 2010. Translocation of single-stranded DNA through single-walled carbon nanotubes. Science 327, 64–67. ( 10.1126/science.1181799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei R, Martin TG, Rant U, Dietz H. 2012. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 51, 4864–4867. ( 10.1002/anie.201200688) [DOI] [PubMed] [Google Scholar]
- 15.Bell NA, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. 2012. DNA origami nanopores. Nano Lett. 12, 512–517. ( 10.1021/nl204098n) [DOI] [PubMed] [Google Scholar]
- 16.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. 2012. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936. ( 10.1126/science.1225624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns JR, Stulz E, Howorka S. 2013. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 13, 2351–2356. ( 10.1021/nl304147f) [DOI] [PubMed] [Google Scholar]
- 18.Hernández-Ainsa S, Bell NA, Thacker VV, Gopfrich K, Misiunas K, Fuentes-Perez ME, Moreno-Herrero F, Keyser UF. 2013. DNA origami nanopores for controlling DNA translocation. ACS Nano 7, 6024–6030. ( 10.1021/nn401759r) [DOI] [PubMed] [Google Scholar]
- 19.Messager L, Burns JR, Kim J, Cecchin D, Hindley J, Pyne ALB, Gaitzsch J, Battaglia G, Howorka S. 2016. Biomimetic hybrid nanocontainers with selective permeability. Angew. Chem. Int. Ed. 55, 11 106–11 109. ( 10.1002/anie.201604677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wanunu M, Meller A. 2007. Chemically modified solid-state nanopores. Nano Lett. 7, 1580–1585. ( 10.1021/nl070462b) [DOI] [PubMed] [Google Scholar]
- 21.Fischbein MD, Drndic M. 2008. Electron beam nanosculpting of suspended graphene sheets. Appl. Phys. Lett. 93, 113107 ( 10.1063/1.2980518) [DOI] [Google Scholar]
- 22.Merchant CA, et al. 2010. DNA translocation through graphene nanopores. Nano Lett. 10, 2915–2921. ( 10.1021/nl101046t) [DOI] [PubMed] [Google Scholar]
- 23.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae MH, Aluru NR, Pop E, Bashir R. 2012. Stacked graphene-Al2O3 nanopore sensors for sensitive detection of DNA and DNA-protein complexes. ACS Nano 6, 441–450. ( 10.1021/nn203769e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Liu K, Bulushev RD, Khlybov S, Dumcenco D, Kis A, Radenovic A. 2015. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotechnol. 10, 1070–1076. ( 10.1038/nnano.2015.219) [DOI] [PubMed] [Google Scholar]
- 25.Feng J, et al. 2015. Electrochemical reaction in single layer MoS2: Nanopores opened atom by atom. Nano Lett. 15, 3431–3438. ( 10.1021/acs.nanolett.5b00768) [DOI] [PubMed] [Google Scholar]
- 26.Liu K, Feng J, Kis A, Radenovic A. 2014. Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano 8, 2504–2511. ( 10.1021/nn406102h) [DOI] [PubMed] [Google Scholar]
- 27.Waduge P, et al. 2015. Direct and scalable deposition of atomically thin low-noise MoS2 membranes on apertures. ACS Nano 9, 7352–7359. ( 10.1021/acsnano.5b02369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernández-Ainsa S, Keyser UF. 2014. DNA origami nanopores: developments, challenges and perspectives. Nanoscale 6, 14 121–14 132. ( 10.1039/c4nr04094e) [DOI] [PubMed] [Google Scholar]
- 29.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. 1996. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866. ( 10.1126/science.274.5294.1859) [DOI] [PubMed] [Google Scholar]
- 30.Kasianowicz JJ, Burden DL, Han LC, Cheley S, Bayley H. 1999. Genetically engineered metal ion binding sites on the outside of a channel's transmembrane β-barrel. Biophys. J. 76, 837–845. ( 10.1016/S0006-3495(99)77247-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayub M, Bayley H. 2016. Engineered transmembrane pores. Curr. Opin. Chem. Biol. 34, 117–126. ( 10.1016/j.cbpa.2016.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Boersma AJ, Boudreau MA, Cheley S, Daltrop O, Li J, Tamagaki H, Bayley H. 2016. Semisynthetic nanoreactor for reversible single-molecule covalent chemistry. ACS Nano 10, 8843–8850. ( 10.1021/acsnano.6b04663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington L, Alexander LT, Knapp S, Bayley H. 2015. Pim kinase inhibitors evaluated with a single-molecule engineered nanopore sensor. Angew. Chem. Int. Ed. 54, 8154–8159. ( 10.1002/anie.201503141) [DOI] [PubMed] [Google Scholar]
- 34.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. 2008. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl Acad. Sci. USA 105, 20 647–20 652. ( 10.1073/pnas.0807514106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, Khalid S, Sansom MSP, Bayley H. 2008. Outer membrane protein G: engineering a quiet pore for biosensing. Proc. Natl Acad. Sci. USA 105, 6272–6277. ( 10.1073/pnas.0711561105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacovache I, Paumard P, Scheib H, Lesieur C, Sakai N, Matile S, Parker MW, van der Goot FG. 2006. A rivet model for channel formation by aerolysin-like pore-forming toxins. EMBO J. 25, 457–466. ( 10.1038/sj.emboj.7600959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. 2012. Engineering a rigid protein tunnel for biomolecular detection. J. Am. Chem. Soc. 134, 9521–9531. ( 10.1021/ja3043646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo PX. 2009. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat. Nanotechnol. 4, 765–772. ( 10.1038/Nnano.2009.259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soskine M, Biesemans A, De Maeyer M, Maglia G. 2013. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 135, 13 456–13 463. ( 10.1021/ja4053398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wloka C, Mutter NL, Soskine M, Maglia G. 2016. Alpha-helical fragaceatoxin C nanopore engineered for double-stranded and single-stranded nucleic acid analysis. Angew. Chem. Int. Ed. 55, 12 494–12 498. ( 10.1002/anie.201606742) [DOI] [PubMed] [Google Scholar]
- 41.Faller M, Niederweis M, Schulz GE. 2004. The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192. ( 10.1126/science.1094114) [DOI] [PubMed] [Google Scholar]
- 42.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor. Cell 95, 771–778. ( 10.1016/s0092-8674(00)81700-6) [DOI] [PubMed] [Google Scholar]
- 43.Yildiz O, Vinothkumar KR, Goswami P, Kühlbrandt W. 2006. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 25, 3702–3713. ( 10.1038/sj.emboj.7601237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iacovache I, De Carlo S, Cirauqui N, Dal Peraro M, van der Goot FG, Zuber B. 2016. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat. Commun. 7, 12062 ( 10.1038/ncomms12062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K, Caaveiro JM, Morante K, González-Mañas JM, Tsumoto K. 2015. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 6, 6337 ( 10.1038/ncomms7337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson AA, et al. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408, 745–750. ( 10.1038/35047129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. 2009. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature 459, 726–730. ( 10.1038/nature08026) [DOI] [PubMed] [Google Scholar]
- 48.Schrodinger L. 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 49.Ho CW, Van Meervelt V, Tsai KC, De Temmerman PJ, Mast J, Maglia G. 2015. Engineering a nanopore with co-chaperonin function. Sci. Adv. 1, e1500905 ( 10.1126/sciadv.1500905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franceschini L, Soskine M, Biesemans A, Maglia G. 2013. A nanopore machine promotes the vectorial transport of DNA across membranes. Nat. Commun. 4, 2415 ( 10.1038/ncomms3415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z, Horwich AL, Sigler PB. 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741–750. ( 10.1038/41944) [DOI] [PubMed] [Google Scholar]
- 52.Watson MA, Cockroft SL. 2016. Man-made molecular machines: membrane bound. Chem. Soc. Rev. 45, 6118–6129. ( 10.1039/c5cs00874c) [DOI] [PubMed] [Google Scholar]
- 53.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. 2005. Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angew. Chem. Int. Ed. 44, 1401–1404. ( 10.1002/anie.200462114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. 2009. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl Acad. Sci. USA 106, 7702–7707. ( 10.1073/pnas.0901054106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benner S, Chen RJ, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. 2007. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat. Nanotechnol. 2, 718–724. ( 10.1038/nnano.2007.344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Astier Y, Kainov DE, Bayley H, Tuma R, Howorka S. 2007. Stochastic detection of motor protein-RNA complexes by single-channel current recording. ChemPhysChem 8, 2189–2194. ( 10.1002/cphc.200700179) [DOI] [PubMed] [Google Scholar]
- 57.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. 2007. Single-molecule analysis of DNA-protein complexes using nanopores. Nat. Methods 4, 315–317. ( 10.1038/nmeth1021) [DOI] [PubMed] [Google Scholar]
- 58.Cockroft SL, Chu J, Amorin M, Ghadiri MR. 2008. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J. Am. Chem. Soc. 130, 818–820. ( 10.1021/ja077082c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gyarfas B, Olasagasti F, Benner S, Garalde D, Lieberman KR, Akeson M. 2009. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 Å resolution. ACS Nano 3, 1457–1466. ( 10.1021/nn900303g) [DOI] [PubMed] [Google Scholar]
- 60.Hurt N, Wang H, Akeson M, Lieberman KR. 2009. Specific nucleotide binding and rebinding to individual DNA polymerase complexes captured on a nanopore. J. Am. Chem. Soc. 131, 3772–3778. ( 10.1021/ja809663f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. 2010. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat. Nanotechnol. 5, 798–806. ( 10.1038/nnano.2010.177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. 2012. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat. Biotechnol. 30, 344–348. ( 10.1038/nbt.2147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. 2010. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 132, 17 961–17 972. ( 10.1021/ja1087612) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lieberman KR, Dahl JM, Mai AH, Akeson M, Wang H. 2012. Dynamics of the translocation step measured in individual DNA polymerase complexes. J. Am. Chem. Soc. 134, 18 816–18 823. ( 10.1021/ja3090302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lieberman KR, Dahl JM, Mai AH, Cox A, Akeson M, Wang H. 2013. Kinetic mechanism of translocation and dNTP binding in individual DNA polymerase complexes. J. Am. Chem. Soc. 135, 9149–9155. ( 10.1021/ja403640b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahl JM, Lieberman KR, Wang H. 2016. Modulation of DNA polymerase noncovalent kinetic transitions by divalent cations. J. Biol. Chem. 291, 6456–6470. ( 10.1074/jbc.M115.701797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. 2012. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353. ( 10.1038/nbt.2171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derrington IM, et al. 2015. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 33, 1073–1075. ( 10.1038/nbt.3357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buttner K, Nehring S, Hopfner KP. 2007. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 14, 647–652. ( 10.1038/nsmb1246) [DOI] [PubMed] [Google Scholar]
- 70.Stoddart D, Franceschini L, Heron A, Bayley H, Maglia G. 2015. DNA stretching and optimization of nucleobase recognition in enzymatic nanopore sequencing. Nanotechnology 26, 084002 ( 10.1088/0957-4484/26/8/084002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nivala J, Marks DB, Akeson M. 2013. Unfoldase-mediated protein translocation through an alpha-hemolysin nanopore. Nat. Biotechnol. 31, 247–250. ( 10.1038/nbt.2503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nivala J, Mulroney L, Li G, Schreiber J, Akeson M. 2014. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano 8, 12 365–12 375. ( 10.1021/nn5049987) [DOI] [PubMed] [Google Scholar]
- 73.Firnkes M, Pedone D, Knezevic J, Doblinger M, Rant U. 2010. Electrically facilitated translocations of proteins through silicon nitride nanopores: conjoint and competitive action of diffusion, electrophoresis, and electroosmosis. Nano Lett. 10, 2162–2167. ( 10.1021/nl100861c) [DOI] [PubMed] [Google Scholar]
- 74.Fologea D, Ledden B, McNabb DS, Li J. 2007. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 91, 539 011–539 013. ( 10.1063/1.2767206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han A, Creus M, Schurmann G, Linder V, Ward TR, de Rooij NF, Staufer U. 2008. Label-free detection of single protein molecules and protein–protein interactions using synthetic nanopores. Anal. Chem. 80, 4651–4658. ( 10.1021/ac7025207) [DOI] [PubMed] [Google Scholar]
- 76.Stefureac RI, Trivedi D, Marziali A, Lee JS. 2010. Evidence that small proteins translocate through silicon nitride pores in a folded conformation. J. Phys. Condens. Matter 22, 454133 ( 10.1088/0953-8984/22/45/454133) [DOI] [PubMed] [Google Scholar]
- 77.Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton JM, Pelta J. 2012. Sensing proteins through nanopores: fundamental to applications. ACS Chem. Biol. 7, 1935–1949. ( 10.1021/cb300449t) [DOI] [PubMed] [Google Scholar]
- 78.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. 2011. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 6, 253–260. ( 10.1038/nnano.2011.12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Japrung D, Dogan J, Freedman KJ, Nadzeyka A, Bauerdick S, Albrecht T, Kim MJ, Jemth P, Edel JB. 2013. Single-molecule studies of intrinsically disordered proteins using solid-state nanopores. Anal. Chem. 85, 2449–2456. ( 10.1021/ac3035025) [DOI] [PubMed] [Google Scholar]
- 80.Niedzwiecki DJ, Grazul J, Movileanu L. 2010. Single-molecule observation of protein adsorption onto an inorganic surface. J. Am. Chem. Soc. 132, 10 816–10 822. ( 10.1021/ja1026858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C. 2013. Fast translocation of proteins through solid state nanopores. Nano Lett. 13, 658–663. ( 10.1021/nl3042678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong CT, Muthukumar M. 2007. Polymer capture by electro-osmotic flow of oppositely charged nanopores. J. Chem. Phys. 126, 164903 ( 10.1063/1.2723088) [DOI] [PubMed] [Google Scholar]
- 83.Dechadilok P, Deen WM. 2006. Hindrance factors for diffusion and convection in pores. Ind. Eng. Chem. Res. 45, 6953–6959. ( 10.1021/ie051387n) [DOI] [Google Scholar]
- 84.Muthukumar M. 2014. Communication: Charge, diffusion, and mobility of proteins through nanopores. J. Chem. Phys. 141, 081104 ( 10.1063/1.4894401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larkin J, Henley RY, Muthukumar M, Rosenstein JK, Wanunu M. 2014. High-bandwidth protein analysis using solid-state nanopores. Biophys. J. 106, 696–704. ( 10.1016/j.bpj.2013.12.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oukhaled A, et al. 2011. Dynamics of completely unfolded and native proteins through solid-state nanopores as a function of electric driving force. ACS Nano 5, 3628–3638. ( 10.1021/nn1034795) [DOI] [PubMed] [Google Scholar]
- 87.Kowalczyk SW, Kapinos L, Blosser TR, Magalhães T, van Nies P, Lim RY, Dekker C. 2011. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 6, 433–438. ( 10.1038/nnano.2011.88) [DOI] [PubMed] [Google Scholar]
- 88.Yusko EC, et al. 2016. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. ( 10.1038/nnano.2016.267) [DOI] [PubMed] [Google Scholar]
- 89.Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. 2012. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 12, 4895–4900. ( 10.1021/nl3024438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10 037–10 041. ( 10.1073/pnas.181342398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franceschini L, Brouns T, Willems K, Carlon E, Maglia G. 2016. DNA translocation through nanopores at physiological ionic strengths requires precise nanoscale engineering. ACS Nano 10, 8394–8402.( 10.1021/acsnano.6b03159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soskine M, Biesemans A, Maglia G. 2015. Single-molecule analyte recognition with ClyA nanopores equipped with internal protein adaptors. J. Am. Chem. Soc. 137, 5793–5797. ( 10.1021/jacs.5b01520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van Meervelt V, Soskine M, Maglia G. 2014. Detection of two isomeric binding configurations in a protein-aptamer complex with a biological nanopore. ACS Nano 8, 12 826–12 835. ( 10.1021/nn506077e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Biesemans A, Soskine M, Maglia G. 2015. A protein rotaxane controls the translocation of proteins across a ClyA nanopore. Nano Lett. 15, 6076–6081. ( 10.1021/acs.nanolett.5b02309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asandei A, Chinappi M, Kang HK, Seo CH, Mereuta L, Park Y, Luchian T. 2015. Acidity-mediated, electrostatic tuning of asymmetrically charged peptides interactions with protein nanopores. ACS Appl. Mater. Interfaces 7, 16 706–16 714. ( 10.1021/acsami.5b04406) [DOI] [PubMed] [Google Scholar]
- 96.Asandei A, Chinappi M, Lee J, Ho Seo C, Mereuta L, Park Y, Luchian T. 2015. Placement of oppositely charged aminoacids at a polypeptide termini determines the voltage-controlled braking of polymer transport through nanometer-scale pores. Sci. Rep. 5, 10419 ( 10.1038/srep10419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.England JL, Lucent D, Pande VS. 2008. A role for confined water in chaperonin function. J. Am. Chem. Soc. 130, 11 838–11 839. ( 10.1021/ja802248m) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.England JL, Pande VS. 2008. Potential for modulation of the hydrophobic effect inside chaperonins. Biophys. J. 95, 3391–3399. ( 10.1529/biophysj.108.131037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Motojima F, Motojima-Miyazaki Y, Yoshida M. 2012. Revisiting the contribution of negative charges on the chaperonin cage wall to the acceleration of protein folding. Proc. Natl Acad. Sci. USA 109, 15 740–15 745. ( 10.1073/pnas.1204547109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weber JK, Pande VS. 2013. Functional understanding of solvent structure in GroEL cavity through dipole field analysis. J. Chem. Phys. 138, 165101 ( 10.1063/1.4801942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franck JM, Sokolovski M, Kessler N, Matalon E, Gordon-Grossman M, Han SI, Goldfarb D, Horovitz A. 2014. Probing water density and dynamics in the chaperonin GroEL cavity. J. Am. Chem. Soc. 136, 9396–9403. ( 10.1021/ja503501x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Horst R, Fenton WA, Englander SW, Wuthrich K, Horwich AL. 2007. Folding trajectories of human dihydrofolate reductase inside the GroEL–GroES chaperonin cavity and free in solution. Proc. Natl Acad. Sci. USA 104, 20 788–20 792. ( 10.1073/pnas.0710042105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jewett AI, Baumketner A, Shea JE. 2004. Accelerated folding in the weak hydrophobic environment of a chaperonin cavity: creation of an alternate fast folding pathway. Proc. Natl Acad. Sci. USA 101, 13 192–13 197. ( 10.1073/pnas.0400720101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Apetri AC, Horwich AL. 2008. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl Acad. Sci. USA 105, 17 351–17 355. ( 10.1073/pnas.0809794105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Best RB, Li B, Steward A, Daggett V, Clarke J. 2001. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 81, 2344–2356. ( 10.1016/s0006-3495(01)75881-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Talaga DS, Li J. 2009. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 131, 9287–9297. ( 10.1021/ja901088b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paula S, Akeson M, Deamer D. 1999. Water transport by the bacterial channel α-hemolysin. Biochim. Biophys. Acta Biomembr. 1418, 117–126. ( 10.1016/S0005-2736(99)00031-0) [DOI] [PubMed] [Google Scholar]
- 108.van Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. 2009. Origin of the electrophoretic force on DNA in solid-state nanopores. Nat. Phys. 5, 347–351. ( 10.1038/nphys1230) [DOI] [Google Scholar]
- 109.Luan B, Aksimentiev A. 2008. Electro-osmotic screening of the DNA charge in a nanopore. Phys. Rev. E 78, 021912 ( 10.1103/PhysRevE.78.021912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aksimentiev A, Schulten K. 2005. Imaging α-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 88, 3745–3761. ( 10.1529/biophysj.104.058727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pederson ED, Barbalas J, Drown BS, Culbertson MJ, Keranen Burden LM, Kasianowicz JJ, Burden DL. 2015. Proximal capture dynamics for a single biological nanopore sensor. J. Phys. Chem. B 119, 10 448–10 455. ( 10.1021/acs.jpcb.5b04955) [DOI] [PubMed] [Google Scholar]
- 112.Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN. 2014. Computational design of water-soluble α-helical barrels. Science 346, 485–488. ( 10.1126/science.1257452) [DOI] [PubMed] [Google Scholar]
- 113.Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF. 2014. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524. ( 10.1126/science.1261172) [DOI] [PMC free article] [PubMed] [Google Scholar]