Abstract
Thus far, identification of functionally important residues in Type II restriction endonucleases (REases) has been difficult using conventional methods. Even though known REase structures share a fold and marginally recognizable active site, the overall sequence similarities are statistically insignificant, unless compared among proteins that recognize identical or very similar sequences. Bsp6I is a Type II REase, which recognizes the palindromic DNA sequence 5′GCNGC and cleaves between the cytosine and the unspecified nucleotide in both strands, generating a double-strand break with 5′-protruding single nucleotides. There are no solved structures of REases that recognize similar DNA targets or generate cleavage products with similar characteristics. In straightforward comparisons, the Bsp6I sequence shows no significant similarity to REases with known structures. However, using a fold-recognition approach, we have identified a remote relationship between Bsp6I and the structure of PvuII. Starting from the sequence–structure alignment between Bsp6I and PvuII, we constructed a homology model of Bsp6I and used it to predict functionally significant regions in Bsp6I. The homology model was supported by site-directed mutagenesis of residues predicted to be important for dimerization, DNA binding and catalysis. Completing the picture of sequence–structure–function relationships in protein superfamilies becomes an essential task in the age of structural genomics and our study may serve as a paradigm for future analyses of superfamilies comprising strongly diverged members with little or no sequence similarity.
INTRODUCTION
Classical Type II restriction endonucleases (REases) are homodimeric enzymes that recognize short DNA sequences (typically 4–8 bp long) and in the presence of Mg2+ cleave the target in both strands at, or in close proximity to the recognition site. Type II enzymes that exhibit structural and functional peculiarities (requirement of more than one target site for cleavage, cleavage at a distance from the asymmetrical target, etc.) have been classified into subtypes [reviewed in (1)]. Because of remarkably high specificity in recognizing and cleaving their target sequences, they are of high interest as model systems for analyzing protein–DNA interactions and one of the most frequently used tools for recombinant DNA technology.
In general, comparison of sequences of Type II REases revealed little or no significant sequence similarities, except for groups of isoschizomers, i.e. enzymes that exhibit identical recognition and cleavage specificities (2,3). This precluded superfamily-wide sequence analysis using standard tools for sequence alignment and raised the question of whether the diversity of amino acid sequences of REases indicates polyphyletic evolution (convergence) or extreme divergence from the common ancestor (2,4). On the one hand, all crystal structures of Type II REases solved to date share a 3D fold, which indicates that they are evolutionarily related [reviewed in (5–7)]. On the other hand, a variety of sequence insertions form a variable ‘shell’ surrounding that conserved core, which indicates extensive divergence and explains problems with sequence alignments (8). The active sites of enzymes, whose structures have been solved, contain a (P)D-Xn-(D/E)-X-K sequence signature, in which two acidic residues and Lys are conserved. Some enzymes, however, have been shown to deviate from this pattern of conservation [reviewed in (9)]. Recently, bioinformatics and biochemical studies provided compelling evidence that some Type II REases belong to unrelated superfamilies of nucleases: Nuc, HNH and GIY-YIG (10–13). The lack of overall sequence conservation, the absence of invariable residues even in the active site and the presence of several alternative folds makes the structure prediction of REases extremely difficult.
Bsp6I is a Type II REase that recognizes the interrupted palindromic sequence GCNGC and cleaves between the cytosine and the unspecified nucleotide in both strands, leaving 5′ overhangs of 1 nt. There are no available crystal structures of REases that recognize similar sequences or produce an identical pattern after cleavage. Furthermore, the Bsp6I sequence shows no similarity to REases with known structures. Therefore, Bsp6I is an example of a protein from the ‘twilight zone’ of the evolutionary landscape of the REase family. In the absence of experimental structural information for Bsp6I, we have built a homology model based on the structure of another enzyme and used the model to guide experiments aimed at identifying residues important for Bsp6I dimerization, DNA binding and catalysis.
MATERIALS AND METHODS
Bioinformatics analysis
Sequence searches of the non-redundant (nr) database and of the putative translations from finished and unfinished microbial genomes were carried out using PSI-BLAST (14). Secondary structure prediction of the protein fold-recognition analysis (matching of the query sequence with known protein structures) was carried out using the GeneSilico MetaServer [with links to the individual methods provided in (15) and at http://genesilico.pl/meta/]. The alignments between the sequence of Bsp6I and the structure of PvuII were used to carry out homology modeling using the ‘FRankenstein's Monster’ approach (16), which comprises cycles of generation of local realignments in uncertain regions, building of alternative models and their evaluation using VERIFY3D (17), realignment in poorly scored regions and merging of the best scoring fragments.
In the model of the Bsp6I dimer, only the area of extensive protein–protein contacts corresponding to the N-terminal region was modeled explicitly, based on the structure of the PvuII dimer (i.e. steric clashes were removed and interactions between the protein side chains were optimized). Protein–DNA interactions have not been modeled at the atomic resolution, because they depend on the mutual orientation of the central and C-terminal parts of the Bsp6I monomers, which could not be predicted with sufficient confidence. Nevertheless, a low-resolution model of Bsp6I monomer–DNA interactions was constructed based on the experimental data.
Bacterial strains and plasmids
Plasmid clones of mutant and wild-type (wt) genes of Bsp6I were propagated in Escherichia coli XL1 Blue MRF′−Δ(mcrA)183, Δ(mcrBC− hsdSMR− mrr)173, endA1, supE44, thi-1, recA1, gyrA96, relA1, lac, [F′ proAB, lacIqZΔM15, Tn10 (tetr)] (Stratagene). λvir was used to test restriction of infecting bacteriophages by the cells harboring the wt Bsp6I and mutant proteins.
The Bsp6I producing plasmid pBsp6IRM2.1B (18) was kindly provided by Dr Arvydas Janulaitis (Institute of Biotechnology, Vilnius) and was used as a vector of wt and mutant genes for in vivo experiments. An additional copy of the bsp6IM gene had been provided on a plasmid pM.Bsp6IAC constructed from a compatible vector pACYC184 (CmR) in order to increase the efficiency of establishment of the complete restriction modification (RM) system (18). Expression plasmids (pBspRET) for protein overproduction and purification were obtained by amplifying pBsp6IRM2.1B insert (wt or mutated) with oligonucleotides: Bsp6IRf, ATCCATGGCACTACATGAT and Bsp6IRr, TTACTCGAGTAACTTGATAATTTTCTTCGTTCG in a PCR reaction and cloning as NcoI–XhoI fragments into the pET28a vector (Novagen), leading to a C-terminal fusion of His6 tag to the recombinant Bsp6IR. Strain E.coli DH10B (Invitrogen) carrying pM.Bsp6IAC was used as a recipient.
Protein expression and purification
Recombinant Bsp6I proteins were overexpressed in E.coli strain ER2566 F− fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 (mcrC−mrr)114::IS10 R(mcr−73::miniTn10–TetS)2 R(zgb-210::Tn10) (TetS) endA1 [dcm] (New England Biolabs) containing both pM.Bsp6IAC and pBspRET plasmids after 1 mM isopropyl-β-d-thiogalactopyranoside induction for 3–5 h at 37°C. Cell pellets were frozen-thawed and lysed in buffer A (20 mM Tris, pH 8.0, 500 mM NaCl, 10 mM β-mercaptoetanol, 10 mM imidazol, 1 mM phenylmethlysulfonyl fluoride and 10% glycerol) by single passage through French press at 20.000 psi. Clarified lysates were batch bound to His-Bind® resin (Novagen) for 1 h at 4°C. After two short washes with 10 vol of binding buffer, resin was applied to empty disposable column and washed sequentially with buffer A with 2 M NaCl and buffer A with 60 mM imidazol. Purified enzyme was eluted with buffer A containing 250 mM imidazol.
For circular dichroism (CD) analysis, the proteins eluted from His-Bind® resin (Novagen) were dialyzed to buffer B (25 mM sodium phosphate buffer, pH 7.6, 0.3 M NaCl, 10% glycerol and 0.25 mM DTT) and further purified on 16/60 Superdex 75 PG gel filtration column (Amersham Pharmacia Biotech) equilibrated with buffer B. Fractions containing the dimeric form of the enzyme were pooled and concentrated with VivaSpin 6 concentrators (VivaScience). Purified proteins were aliquoted and frozen at −70°C. Protein concentration was measured by Bradford assay with NanoQuant kit (Roth) or by densitometry of Coomassie-stained SDS–PAGE gels.
Site-directed mutagenesis of Bsp6I
Site-directed mutagenesis of the Bsp6I gene on the pBsp6IRM2.1B plasmid was performed by a PCR-based technique using the QuikChange Site-Directed Mutagenesis system (Stratagene) following the manufacturer's instruction. The mutant genes were sequenced and found to contain only the desired mutation.
In vivo analysis of the wt Bsp6I REase and the mutant enzymes
Restriction of infecting λvir bacteriophages by wt and mutant Bsp6I REase was assessed by measuring the titer of λvir phage on E.coli strain XL1 Blue MRF′− expressing the Bsp6I methyltransferase (MTase) and comparing it with the wt or mutant REase genes. The extent of phage restriction was determined quantitatively by plating portions of serially diluted phage stock on a lawn of bacteria (19). The strains were grown on Luria–Bertani plates containing 50 μg/ml ampicilin and 20 μg/ml chloramphenicol. The ability to restrict λvir was assessed by measuring the titer (from at least three independent measurements) of λvir phage on E.coli strain XL1 Blue MRF′− expressing wt and mutant Bsp6I genes in the presence of Bsp6I MTase and comparing it with a titer of phage on the strain expressing only the MTase gene and containing the pUC19 vector without the REase gene. Wt Bsp6I restricted the phage growth by nearly four orders of magnitude (1100 ± 80 plaques versus 7.9 × 106 ± 4.2 × 105 plaques for the strain expressing only the MTase gene).
In vitro analysis of the wt Bsp6I REase and selected mutant enzymes
In vitro cleavage assays were set up in 10 μl vol of buffer R (MBI Fermentas) (10 mM Tris–HCl, pH 8.5, 10 mM MgCl2, 100 mM KCl and 0.1 mg/ml BSA) with 0.3 μg of λ DNA (dam−, dcm−) (MBI Fermentas) as a substrate and serial dilutions of purified REase. Digestion was performed for 1 h at 37°C. To compare digestion rates of the wt Bsp6I and partially active E84A mutant, 611 bp PCR-generated fragment of the pBluescript II KS(+) (Stratagene) plasmid with a single Bsp6I site at position 217 was used as a substrate (the fragment was amplified with primers MbspNlaF: TGCTCTTGCCCGGC and MbspNlaR: GCCATCGCCCTGA). Activity tests were carried out in duplicate with the substrate concentration of 20 ng/μl and with several enzyme concentrations, ranging from 16 to 130 nM. For all concentrations, there was a linear relationship between the reaction rate and the enzyme concentration.
DNA-binding assay
Complementary oligonucleotides (5′-GAAATGTGGATGTAAAAGCAGCTAGCAATGATATTCCTGATAGTG-3′) containing Bsp6I recognition sequence (underlined) were end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP 6000 Ci/mmol (PerkinElmer, Life Science). Following incubation for 1 h at 37°C, oligonucleotides were purified by ethanol precipitation. Duplexes were formed by heating equimolar mixture of complementary oligonucleotides in the binding buffer [10 mM Tris–HCl, pH 8.5, 10 mM CaCl2 and 100 mM KCl] at 65°C for 2 min and then cooling down slowly (for 1 h) to the room temperature. Binding reactions contained 20 nM RE, 1 nM labeled duplex with the recognition sequence and 20 nM unlabeled duplex without the Bsp6I recognition sequence (5′-GTAATTACAGGAAATGTCGCGGTAAAAGCAACATCAAATG-3′) in the binding buffer. After 20 min binding at the room temperature, 50 μl aliquots were filtered in triplicate in dot-blot manifold (Scie-Plast) through nitroclellulose filter (Schleicher and Schuell) equilibrated for 1 h in the binding buffer. Filters were washed three times with 150 μl of the binding buffer, dried and exposed to the PhosphorImager screen. The resulting image was scanned on the Storm 820 PhosphorImager and the digitized image was quantified with ImageQuant TL v.2003 software (Amersham Biosciences). Intensities were corrected by subtracting the background intensity measured without any protein present in the binding reaction.
Circular dichroism
CD spectra were collected on the Jasco 710 spectropolarimeter with a temperature controller. The concentration of Bsp6I dimer was 4 μM. Scans were collected at 20°C from 197 to 260, in 1 nm steps, using a 1 mm pathlength cuvette. Secondary structure content was estimated from the CD spectrum using the CDpro server http://lamar.colostate.edu/~sreeram/CDPro/main.html (20).
RESULTS
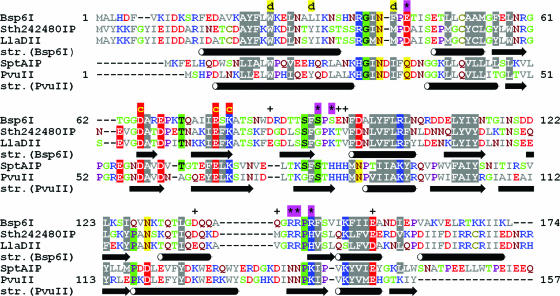
Database searches using PSI-BLAST (14) revealed sequence similarity between Bsp6I, its isoschizomer LlaDII and a putative REase Sth242480IP (Figure 1); however, no significant similarity to other proteins could be detected, which is typical for a highly divergent restriction enzyme. We chose to employ a fold-recognition approach that potentially allows identification of homologs among the known structures even in the absence of sequence similarity [reviewed in (21); for details see Materials and Methods]. Among the fold-recognition programs used to identify a homology modeling template for Bsp6I, only two threading servers reported the PD-(D/E)XK fold: 3DPSSM (22) recognized the PvuII structure (3pvi in PDB) as the best template with a good score of 0.00759. INBGU (23) reported the same structure, albeit with a low score (3.6) at the sixth position of the ranking. Another member of the PD-(D/E)XK fold, the N-terminal domain of Rpb5 from Saccharomyces cerevisiae [1dzf (24)], was also reported by INBGU, with a very low score of 2. None of the potential templates reported by these and other servers gave a score above the documented significance level. Nevertheless, among all protein structures reported, the consensus predictor Pcons2 (25) singled out PvuII as potentially the best template for homology modeling of Bsp6I. It is worth emphasizing that the alignments of Bsp6I and PvuII sequences reported by 3DPSSM and INBGU agreed very well, with only the INBGU alignment being incomplete at the C-terminus. Significantly, the hallmark of PD-(D/E)XK nucleases, i.e. the catalytic triad D-E-K in PvuII appeared to be conserved in Bsp6I (Figure 1). Modeling analysis using other possible templates (all reported with low scores) revealed that the corresponding Bsp6I models would be poorly folded, i.e. that no globular structure could be constructed and/or hydrophobic regions would be exposed to the surface. If these templates were enzymes, their catalytic residues showed no conservation with relation to Bsp6I (data not shown). Therefore, notwithstanding the low sequence similarity between Bsp6I and PvuII and lack of statistical support for the fold-recognition alignment (indicative of the ‘twilight zone’ of homology), the PvuII structure was selected as the most reasonable template for modeling of Bsp6I.
Figure 1.
Refined sequence alignment of Bsp6I and PvuII and their homologs. Amino acid residues are colored according to their physico-chemical features (negative, red; positive, blue; hydrophobic and aromatic, gray; amide, dark red; small hydrophilic, green; and praline, violet). Similar or identical amino acid residues are highlighted. Amino acid residues which were subjected to a mutational analysis in Bsp6I and shown to be essential or at least important are indicated above the alignment by ‘d’, ‘*’ and ‘c’ (presumably involved in dimerization, DNA binding and catalysis, respectively). Residues shown to be non-essential are indicated by ‘+’. Secondary structure (determined experimentally for PvuII and predicted for Bsp6I) is shown below the homologs of the target and the template.
Structures derived from modeling usually contain inaccuracies, though they are often of good enough quality to putatively identify regions important for function, which can then be used to guide hypothesis-driven laboratory experiments. The success of protein tertiary structure prediction by homology modeling depends critically on the quality of the underlying target-template alignment, because the standard algorithms for model refinement are unable to correct errors resulting from misthreading (26). It is known that even if fold-recognition algorithms identify the template with a correct fold, they can produce inaccurate target-template alignments. Thus, we used the ‘FRankenstein's montster’ approach (16) to build a homology model of Bsp6I using the coordinates of PvuII as the template, and at the same time refine the target-template alignment by validation of the sequence-structure fit at the 3D level (for details see Materials and Methods). The final alignment and the corresponding model obtained after several rounds of optimization guided by the structural evaluation and validation with experimental data are shown in Figures 1 and 2.
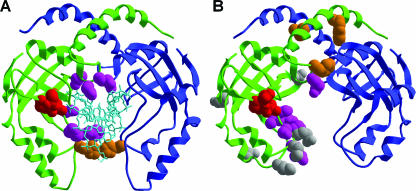
Figure 2.
Comparison of structures of the PvuII template and the homology model of Bsp6I. (A) PvuII dimer complexed with DNA (1pvi). For clarity, only the target hexanucleotide duplex CAGCTG is shown (in cyan). (B) The final model of the Bsp6I dimer based on coordinates of PvuII (orientation of the monomers has been based on the PvuII structure and not modified further). The protein backbone of two monomers is colored in green and blue. The side chains found to be important for the activity of PvuII and Bsp6I are shown for one of the monomers; only the H85 pair (PvuII) is shown for both monomers. Residues involved in dimerization, DNA binding and catalysis are colored in orange, magenta and red, respectively. The side chains non-essential for Bsp6I are indicated in gray.
The catalytic site responsible for Mg2+ binding and phosphodiester bond cleavage
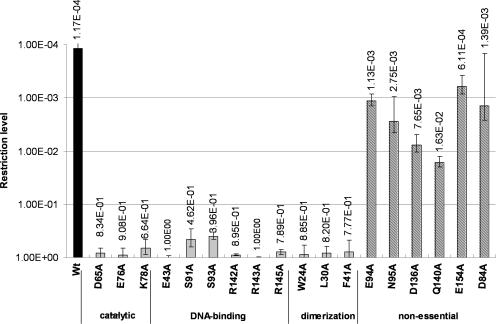
Several enzymatic mechanisms for Type II REases have been proposed [reviewed in (27)]. In the context of PvuII, the consensus mechanism involves two carboxylate groups of D58 and E68 that coordinate the Mg2+ cofactor and an amino group of K70 that stabilizes the transition state and acts as a Lewis acid in the reaction (27). The catalytic triad of PvuII aligns with the corresponding triad (D65, E76 and K78) in the homology model of Bsp6I (Figures 1 and 2), suggesting that the latter may be a part of the active site. To test this prediction, we have replaced these residues with alanine by site-directed mutagenesis and tested the activity of the mutants in vivo. Our operational criterion for an ‘essential’ residue is the reduction of the restriction activity from 1.17 × 10−4 (wt Bsp6I) to at least 5 × 10−1 in the alanine mutant, while mutations of ‘important’ residues reduce the activity by at least three orders of magnitude, i.e. to the value between 1.17 × 10−1 and 5 × 10−1. In agreement with our model, D65, E76 and K78 are essential for Bsp6I activity in vivo (Figure 3). In PD-(D/E)XK enzymes, the two consecutive antiparallel β-strands (amino acids 57–74 in PvuII) that form the structural scaffold for the active site are in a constant position relative to the three catalytic residues. Hence, the similarity of the active sites of PvuII and Bsp6I and the match of observed versus predicted secondary structures suggests that the core of Bsp6I (amino acids 64–82) has been correctly identified by fold-recognition algorithms.
Figure 3.
In vivo λvir phage restriction activity of Bsp6I variants with alanine substitutions of residues of presumptive functional importance (indicated as ‘catalysis’, ‘DNA binding’ and ‘dimerization’) and residues believed to be non-essential for the protein function.
Regions involved in DNA sequence recognition
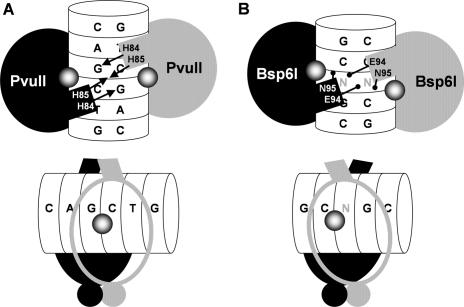
The co-crystal structures of PvuII–DNA, together with mutagenesis studies, have implicated specific residues from three regions in recognition of the C1A2G3↓C4T5G6 sequence: region I: Q33D34N35 in the loop between the first two α-helices; region II: S81 followed by a His triplet H83H84H85; and region III: an Asn doublet N140N141 followed by a K143 residue (28–30).
In PvuII, region I constitutes the floor of the DNA-binding cleft. Q33 and N35 make contacts with backbone phosphates, while D34 probably plays the main role in discrimination of the central GC pair by interacting with the G3 residue via the minor groove (31). In Bsp6I, the sequence-region spanning residues F41-T62 could not be aligned with confidence with the PvuII structure; however, the sequence and predicted structure of the N-terminally located segment of Bsp6I (amino acids D16-N40) unambiguously matched the dimerization helix of PvuII (amino acids D5-N29) (Figure 1, also see below). Thus, we regarded the conserved GIN40 and GGDA66 peptides (GIN29 and GNDA59 in PvuII) as qualified landmarks to guide our efforts to fine-tune the alignment in the uncertain F41-T62 region of Bsp6I. We have built several homology models of Bsp6I using alternative alignments of the segments F41-T62 (Bsp6I) and D30-E55 (PvuII), with the additional constraint on deletions being limited only to the loop/bulge regions in the PvuII structure (D30-N35 and L51-E55). Structure evaluation using VERIFY3D (32) revealed that all models except one exhibited negative scores in the F41-T62 region, suggesting that the corresponding residues make improper contacts (for instance, if too many polar residues are buried and/or hydrophobic residues are exposed to the solvent). In the only model, whose structure has been evaluated as reasonable, region I of PvuII (QDN35) corresponds to the ETI45 tripeptide in Bsp6I, in which the residue E43 is predicted to contact the DNA. In agreement with this prediction, the E43A variant of Bsp6I is inactive in vivo (Figure 3). This result provides strong support for the proposed fold-recognition alignment of the Bsp6I sequence with the PvuII region I structure and for the resulting model. Moreover, none of the alternative models revealed contacts between E43 and the DNA.
Regions II and III correspond to two recognition loops of PvuII that contact DNA via the major groove (28). In region II, substitution of each of the four residues (S81, H83, H84 and H85) interfered with catalysis; however, DNA binding was retained in alanine mutants of each of the His residues (29). H83A and H84A mutants of PvuII exhibited reduction of the cleavage of the unmodified DNA by ∼2 orders of magnitude (29). Interestingly, the H84A mutant cleaved m4C-modified DNA ∼50% faster than the wt enzyme, suggesting that this residue is involved in recognition of native DNA methylation (33). H85 in PvuII forms a hydrogen bond with H85 from the other subunit. This interaction between the PvuII monomers is required to elicit phosphodiester bond cleavage (28,29).
Both 3DPSSM and INBGU fold-recognition methods produced identical, gapless alignment of T87-Y114 from Bsp6I with the T77-Y104 segment of PvuII. Notably, amino acids S91, S93, E94 and N95 were predicted by secondary structure analysis and homology modeling to constitute a region of Bsp6I that corresponds to region II of PvuII (amino acids S81, H83, H84 and H85). In agreement with this prediction, the in vivo cleavage activity of S91A and S93A mutants was significantly reduced. However, the E94A and N95A mutants retained a substantial level of activity (Figure 3). It is noteworthy that in PvuII the N-terminal part of region II (residues S81 and H83) interacts with the DNA and does not participate in monomer–monomer interactions, while the C-terminal part (residues H84 and H85) is involved in the formation of monomer–monomer contacts in the protein–DNA complex (Figure 2). That the effects of mutagenesis of PvuII and Bsp6I are similar in the N-terminal part of region I and different in its C-terminal part suggests that an alteration in the monomer–monomer interactions has occurred in these two enzymes during evolution, and it appears that this distribution of function has created a local change in enzyme–DNA binding.
For the DNA recognition region III and neighboring sequences, each fold-recognition method suggested a different alignment with gaps or insertions in secondary structure elements. Such alignments caused disruption of the protein core in preliminary models and were corrected manually. As for region I, several alternative models were built with variations in the underlying alignment. The target-template alignment was guided primarily by matching hydrophobic amino acids of Bsp6I with those involved in maintaining the hydrophobic core in PvuII (Y124, P142, Y148, V149 and M150). The model with best VERIFY3D score was selected for experimental verification. Alanine substitution of bulky hydrophobic residues that are part of a protein core, regardless of position in the sequence, might interfere with folding and thereby negatively influence the catalytic activity. We therefore decided to test our Bsp6I structural predictions by introducing mutations in the counterparts of DNA-binding residues of region III in PvuII. The correspondence between N140, N141 and K143 in PvuII and R142, R143 and R145 in Bsp6I became apparent only after the model was built, since these residues were not used to guide the refinement of the target-template alignment (Figure 1). Alanine substitution of R142, R143 and R145 in Bsp6I resulted in a greatly diminished enzyme activity in vivo (Figure 3), which is in perfect agreement with our prediction that these residues are the counterpart of DNA-binding region III in Bsp6I. Conversely, other charged or polar residues from the same region of Bsp6I that were predicted to be exposed to the solvent, but remote from the DNA-binding site, and hence not predicted to be important for the REase function (Q140 and E154), could be substituted by alanine with only minor effect on the Bsp6I activity (Figure 3, also see below).
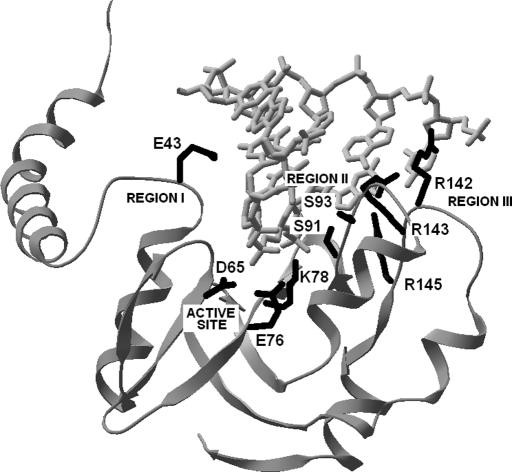
Altogether, our experimental results provide strong support for our theoretical model of Bsp6I structure. We believe that the present model is accurate enough to allocate protein–DNA contacts at the level of low to medium resolution, which is sufficient to guide further, more sophisticated analyses, such as zero-length cross-linking. Figure 4 illustrates our prediction of Bsp6I–DNA interactions, using the DNA molecule copied from the PvuII complex. Since Bsp6I and PvuII recognize different sequence targets and modeling of protein–DNA interactions are at the brink of current techniques, we considered modeling of the quaternary structure of the Bsp6I–DNA complex at the atomic level as unreasonable until more experimental data are available.
Figure 4.
Predicted DNA-binding residues of Bsp6I in stereoview. The DNA molecule (in gray) has been copied from the PvuII complex (1pvi) to illustrate the general orientation of the enzyme and its target. The catalytic site and three DNA-binding regions are indicated.
Amino acid residues involved in Bsp6I dimerization
Characteristic of all PvuII structures and missing from other REase structures is the long N-terminal (amino acids 5–25) kinked α-helix, which is swapped between the two monomers and interacts with its counterpart from the other subunit and with the second α-helix (amino acids 36–46). This structural arrangement provides extensive points of contact between subunits, thus stabilizing the mutual orientation of the N-terminal subdomains (amino acids 1–47) of each monomer near the minor groove of the recognition site, while the catalytic core of each monomer (amino acids 48–157) retains independent conformational freedom (28,34). In Bsp6I, the N-terminal region is also predicted to be α-helical like in PvuII, and the fold-recognition algorithms used to predict the structure of Bsp6I identified a number of hydrophobic amino acid residues conserved between the two REases (Figure 1), suggesting that they may exhibit similar intersubunit contacts on the minor groove side of the target DNA. We analyzed the importance of hydrophobicity in N-terminal residues for dimerization of Bsp6I by replacing them with alanine. Bsp6I mutants W24A, L30A and F41A (Figure 2) exhibited reduced REase activity in vivo (Figure 3), supporting the prediction that tertiary and quaternary structures of PvuII and Bsp6I possess similar N-terminal dimerization regions. This is in contrast to the predicted dissimilarity of the intersubunit contacts on the major groove side of the target DNA, suggested by different effects seen by mutating H84 and H85 in PvuII (29) and compared with mutation of E94 and N95 in Bsp6I (this work, see above).
To provide additional support for the predicted Bsp6I model and eliminate the possibility that introducing alanine mutations anywhere in Bsp6I would lead to reduced REase activity in vivo, we selected several amino acids outside of known critical regions to target for alanine mutagenesis. While the amino acids selected are not predicted to be important for Bsp6I folding or dimerization, they are in close proximity to amino acids predicted to be required for Bsp6I function (Figure 2). In accordance with our model, the substitution of D84, D136, Q140 and E154 with alanine did not substantially affect the REase activity of Bsp6I in vivo (Figure 3). The congruence of fold-recognition and site-directed mutagenesis suggests that the predicted model of Bsp6I is a reasonable approximation of its native structure.
In vitro activity of selected Bsp6I mutants
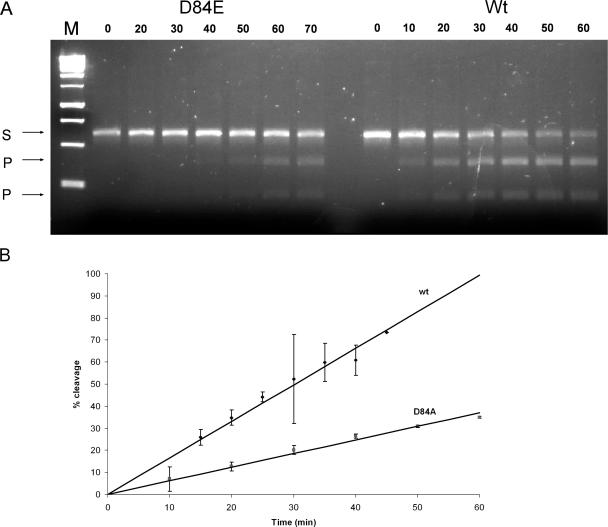
To test whether restriction levels assayed in the phage plating experiments parallel enzyme activity and do not reflect any other possible side-effects of point mutations introduced (such as decreased level of gene expression or protein misfolding leading to insolubility and/or increased rate of degradation), we assayed the in vitro activity of the wt Bsp6I and selected mutant proteins after purification. Mutants E76A (predicted catalytic residue) and R143A (predicted DNA-binding residue) were found to be completely inactive in vitro (<1% of the wt Bsp6I activity) (Supplementary Material, Figure S1) in agreement with the in vivo restriction assays. Mutants of non-essential residues D84A and E94A retained substantial level of enzymatic activity in vitro, in qualitative agreement with the in vivo experiments. Their approximate in vitro activity measured on phage λ DNA was 25 and 50% of the wt level, respectively, whereas their in vivo restriction level was about one order of magnitude lower than for the wt Bsp6I, 1.3 × 10−3 and 1.1 × 10−3, respectively (Figure 3). The steady-state cleavage rate of a PCR-generated DNA substrate measured for the D84A mutant was ∼40% of the cleavage rate determined for the wt Bsp6I (Figure 5).
Figure 5.
DNA cleavage rate of the Bsp6I D84A mutant. (A) Gel assay of DNA cleavage. Numbers above lines indicate time (in minutes). M, molecular weight standard; S, 611 bp PCR-generated DNA substrate; and P, products (219 and 392 bp fragments). (B) Comparison of cleavage rates of the wt Bsp6I and D84A mutant measured in gel assays.
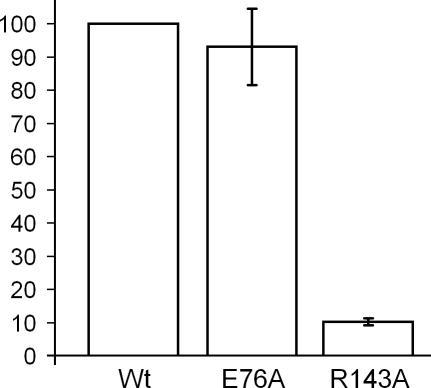
According to our model, observed inactivation of some mutants results from the loss of a catalytic side chain and in others—from the inability of the enzyme to recognize or bind the specific substrate. We used the nitrocellulose filter binding assay to study the ability to bind the specific DNA target by representative mutants predicted to belong to these two classes. The results shown in Figure 6 indicate that mutation of a predicted catalytic residue E76 leads to little, if any, reduction of DNA binding, which is in line with results obtained for catalytic residues of many other restriction enzymes (35–37). On the other hand, mutation of a predicted DNA-binding residue R143 leads to a severe defect in DNA binding, supporting the sequence-function relationships inferred from the structural model of Bsp6I.
Figure 6.
DNA-binding activity of two Bsp6I mutants. DNA-binding activities were measured using the nitrocellulose filter assay (see Materials and Methods) in two independent experiments for alanine substitutions of E76 (predicted to be a catalytic residue) and R143 (predicted to be involved in DNA recognition) of Bsp6I and are expressed as % of wt Bsp6I DNA binding.
Comparison of the wt and mutant Bsp6I structure using CD
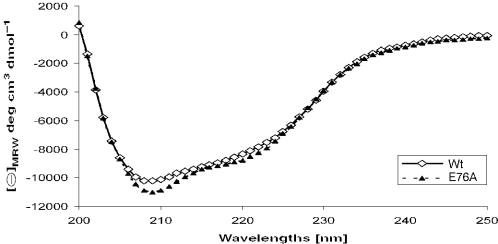
Finally, we used CD spectroscopy to analyze the structure of wt Bsp6I and to demonstrate that a point mutation in the predicted active site (E73A), which completely inactivated the enzyme, results in no gross conformational changes. The CD spectrum of Bsp6I (Figure 7) is characteristic for proteins with an α/β structure and shows two negative bands with extrema in 208 and 220 nm (Figure 7). Using the CDPro server (38), we estimated that the wt Bsp6I contains ∼25.4% of α-helices and 25.1% of β-strands, which is in good agreement with the values predicted based on the theoretical model: 31 and 26%, respectively. Importantly, we observed no significant change between the CD profiles of the wt Bsp6I and the completely inactive mutant E73A (Figure 7). It suggests that the secondary structure composition and presumably the overall structure of the mutant protein is very similar to the wt enzyme, and supports our prediction that E73 is essential for the catalytic activity of Bsp6I, but is not particularly important for its structural stability.
Figure 7.
CD spectra of the wt Bsp6I and the E76A mutant.
DISCUSSION
Using fold-recognition, we detected structural similarities between the monomers of Bsp6I and PvuII, despite that the sequences of these enzymes are dissimilar. Model-based prediction of key residues responsible for dimerization, DNA binding and cleavage in Bsp6I has been confirmed by site-directed mutagenesis and in vivo analysis of the archetypal function of the Type II REase, i.e. the ability to restrict the growth of a lytic phage. These results support our assertion that PvuII and Bsp6I share a PD-(D/E)XK fold, and a similar mechanism of action.
Since PvuII and Bsp6I produce different ends after the cleavage, the question arises whether the architecture of the Bsp6I dimer at the active site is similar to that of PvuII. The structure of PvuII has been solved in the absence [1pvu (34)] and in the presence of DNA [1pvi (28)]. In the free PvuII dimer, the catalytic cores (amino acids 48–157) of each subunit are extended in an open state complex to allow DNA binding. Upon DNA-binding, the catalytic cores clamp onto the DNA by an ∼27° rotation around the DNA axis [superposition of the Cα atoms revealed root-mean-square (RMS) displacement of 7.92 Å between 1pvu and 1pvi] and the H85-H85 interaction is established between the two subunits on the major groove side of the target DNA. The active site residues of the PD-EXK motif are oriented differently in the free enzyme compared with the enzyme complexed with target DNA. In contrast, the extensive dimerization interface formed by the N-terminal subdomain (amino acids 1–47) on the minor groove side remains nearly identical (RMS displacement of 1.33 Å). The movement of the active sites in relation to the N-terminal dimerization region is due to a solvent-exposed loop comprising amino acids 46–49 that acts as a hinge.
In the PvuII–DNA complex, the scissile phosphates are ‘facing each other’ in the two strands of the DNA target, which results in ‘blunt’ cleavage, whereas the phosphodiester bonds targeted by Bsp6I are separated by one non-specific base pair, which is the source of 1 nt extension of the 5′ end generated after the cleavage. Among REases with known structures, those similar to PvuII [i.e. the ‘β’ class of REases (6,39)] typically produce blunt ends after the cleavage. One of the exceptions is BglI, which evolved the ability to produce 3 nt 3′-staggered ends through an alteration of monomer–monomer interactions (40). It is possible that Bsp6I evolved from a typical ‘blunt-end cutting’ β-class ancestor similar to PvuII, by modification of its dimer structure, shifting the active sites away from each other to ‘insert’ one non-specific base pair in the middle of the recognition site. A similar scenario was proposed for SsoII and NgoMIV, which generate 5 and 4 nt staggered ends, respectively (41). However, it should be noted that the target sequences of SsoII and NgoMIV contain common elements (two symmetrical C:G C:G base pairs), while the targets of Bsp6I and PvuII do not. Hence, only a general mechanistic model of the interaction of the Bsp6I dimer with the DNA target can be proposed based on its predicted similarities and differences with respect to PvuII. The proposed evolution of Bsp6I and PvuII from a common ancestor remains hypothetical until more related sequences are identified and/or when the crystal structure of Bsp6I is determined.
Our results provide a strong indication that the Bsp6I monomer shares key structural and functional features with PvuII, including a typical active site (D65, E76 and K78), amino acids involved in DNA binding and the N-terminal dimerization helix. In addition, we identified a region in which apparently homologous residues do not have the same function in PvuII and Bsp6I. The function of PvuII amino acids H84 and H85 of PvuII in target recognition and stabilization of catalytic domains of two monomers in a position suitable for the ‘blunt-end’ cleavage has not been retained in the corresponding residues of Bsp6I, E94 and N95, which are dispensable for the enzyme activity in vivo. We interpret these differences as an indication of a change in the mutual orientation of the Bsp6I monomers in its DNA-bound dimer form that has occurred during evolution (Figure 8).
Figure 8.
Proposed model of Bsp6I quaternary structure in relation to PvuII. (A) A schematic diagram of PvuII dimer (monomers in black and gray) complexed with DNA (white). The target sequence is shown. The scissile phosphate is indicated by a shaded sphere. The side chains of H84 (involved in recognition of G residue in the opposite strand) and H85 (involved in dimerization) are indicated by arrows. (B) Model of the Bsp6I dimer; all labels follow those for PvuII. The structurally mismatched residues E94 and N95 are shown as ‘lollipops’. The non-specific base in the middle of the target sequence is shown in gray.
The structure of PvuII and our hypothetical model of Bsp6I indicate that both REases resemble a kidney, with one end elongated and the other flattened; the concave surface between the ends of the kidney-like structure is involved in DNA binding and cleavage. In the PvuII dimer, the monomers interact tightly with each other through the elongated ends and weakly within the flattened ends. This allows the enzyme to encircle the DNA in such a way that the minor groove is facing the ‘strong’ interface between the monomers and the major groove is facing the ‘weak’ interface. Based on the available data, we propose that the orientation of Bsp6I monomers in complex with the DNA can be deduced from the PvuII–DNA complex by treating the two REase monomers as semi-flexible moieties ‘glued’ to each other by the elongated ends and also separately ‘glued’ to the scissile phosphates (each monomer to one strand). Insertion of 1 bp in the middle of the PvuII DNA recognition sequence forces the scissile phosphates apart. While the elongated ends remain fixed in place, the flattened ends are shifted by a lever mechanism that changes their relative position to each other and the DNA target. This mechanism can be used to explain why the protein–protein interactions mediated by the N-terminal helices and protein–DNA interactions mediated by the active site and the N-terminal part of region II, respectively, are retained in PvuII and Bsp6I (they correspond to the ‘glued’ parts), while at the same time, the protein–protein and protein–DNA interactions mediated by the C-terminal part of region II change (this region forms the flattened end).
Among the DNA-binding residues, S91 and S93 in Bsp6I retain a similar orientation with respect to the recognition of bases in DNA similar to what has been observed for S81 and H83 in PvuII (reference). In contrast, E94 and N95 in Bsp6I cannot make the equivalent contacts made by H84 and H85 in PvuII, because the mutual rotation of the two subunits probably shifts the side chain of E94 too far from the DNA to allow specific contact to be made, and the position of the N95 side chains between subunits is too distant to provide the equivalent contact observed between H85 from the two subunits in PvuII (Figure 2A).
In PvuII, the middle guanine in the CAG↓CTG sequence is recognized by H84 from the subunit that cleaves the complementary strand. Assuming that the malleability of the monomer is limited, residues in the vicinity of the active site would retain the relative orientation and distance to the active site, thus the homolog of H84 should make contact with the base whose phosphate group is liberated after cleavage, i.e. ‘N’ in the GC↓NGC sequence recognized by Bsp6I. Apparently, the side chain of E94 cannot ‘reach’ to the corresponding base; hence, the base remains ‘unrecognized’. This suggests E94 as a tempting target for substitutions that may provide new protein–DNA contacts and thereby generate an enzyme with a novel specificity (with a defined base instead of N in GCNGC). This, however, is beyond the scope of the present study.
CONCLUSIONS
Our work constitutes the first attempt to predict structural features of a Type II restriction enzyme in the so-called ‘twilight zone’ of homology (i.e. no statistically significant sequence similarity to known structures). We have previously studied restriction enzymes by combination of modeling and experimental analyses; however, in all these cases sequence similarities between the target and the template were more pronounced (37,41–44). A similar approach to that described in this work was used to predict the structure of PrfA protein based on the PvuII template; however, the authors did not study the dimerization mode and supported their model with only one mutation of a predicted catalytic residue (45). The fold-recognition analysis of Bsp6I described in this work was used to design 18 alanine substitutions, in regions presumably involved in dimerization, DNA binding, catalysis and in regions presumed to be functionally non-important (as an additional negative control). The mutant variants were tested for the restriction activity in vivo. One caveat of protein genetics is that inactivation by mutations may result from gross structural aberrations in protein structure rather than elimination of the function conferred by the particular side chain. We do not believe this to be a major problem in our study since 9 of 18 Bsp6I mutants analyzed here display a measurable restriction level; hence, they are unlikely to be misfolded. In addition, in vitro cleavage activities of all selected mutants agree with their relative ability to restrict the phage growth in vivo. Moreover, mutant E76A, which is completely inactive in the cleavage assay, maintains the wt level of the DNA-binding activity, as commonly observed for catalytic mutants of REases. Finally, CD spectra of the wt enzyme and the E76A mutant are almost identical, indicating lack of gross conformational differences in the inactive mutant.
Our initial model of Bsp6I based on the PvuII template was in accordance with the experimental results, with the exception of the C-terminal part of region III. Upon consideration of the experimental data and functional differences between Bsp6I and PvuII, our final model of the Bsp6I dimer now agrees with all mutational data. We propose that the 3D fold of Bsp6I is similar to the PvuII crystal structure, but these enzymes differ somehow in the mutual position of the monomers in the dimer. Despite its limitations, the present model of Bsp6I provides insight into the structural and functional organization of this enzyme and will guide future comparative studies of REases. Our analysis can be regarded as a case study of remote homology detection and structure prediction, followed by model validation using a simple, low-resolution experimental methodology. The prediction-validation approach becomes increasingly important in the age of structural genomics, since only a fraction of protein structures will be solved by crystallography, while others have to be modeled using bioinformatics.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online and the Bsp6I model is available from ftp://genesilico.pl/iamb/models/R.Bsp6I/.
Supplementary Material
Acknowledgments
We are grateful to Dr Arvydas Janulaitis (Institute of Biotechnology, Vilnius) for providing us with the plasmids producing the Bsp6I RM system and to Prof. Jacek Otlewski and Dr Daniel Krowarsch for their help with CD analyses. We thank Rich Roberts and Alan Friedman for critical reading of the manuscript and discussions. Special thanks are due to Jim Anderson for his laborious editing of the manuscript to improve the English. This analysis was funded by MNII (grant 3P04A01124 to J.M.B.). M.R. was supported by the Faculty of Biology, Warsaw University Grant BW 1485\16. J.M.B. is an EMBO/HHMI Young Investigator and a fellow of the Foundation for Polish Science. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S., Dryden D.T., Dybvig K., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson G.G., Murray N.E. Restriction and modification systems. Annu. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 3.Jeltsch A., Kroger M., Pingoud A. Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene. 1995;160:7–16. doi: 10.1016/0378-1119(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 4.Bickle T.A., Kruger D.H. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki J.M. Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol. 2000;50:39–44. doi: 10.1007/s002399910005. [DOI] [PubMed] [Google Scholar]
- 6.Bujnicki J.M. Understanding the evolution of restriction-modification systems: clues from sequence and structure comparisons. Acta Biochim. Pol. 2001;48:935–967. [PubMed] [Google Scholar]
- 7.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bujnicki J.M., Rychlewski L. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001;3:69–72. [PubMed] [Google Scholar]
- 9.Pingoud A. Restriction Endonucleases. Berlin, Heidelberg: Springer-Verlag; 2004. [Google Scholar]
- 10.Sapranauskas R., Sasnauskas G., Lagunavicius A., Vilkaitis G., Lubys A., Siksnys V. Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J. Biol. Chem. 2000;275:30878–30885. doi: 10.1074/jbc.M003350200. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L., Makarova K.S., Koonin E.V. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bujnicki J.M., Radlinska M., Rychlewski L. Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci. 2001;26:9–11. doi: 10.1016/s0968-0004(00)01690-x. [DOI] [PubMed] [Google Scholar]
- 13.Saravanan M., Bujnicki J.M., Cymerman I.A., Rao D.N., Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurowski M.A., Bujnicki J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosinski J., Cymerman I.A., Feder M., Kurowski M.A., Sasin J.M., Bujnicki J.M. A “FRankenstein's monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins. 2003;53(Suppl. 6):369–379. doi: 10.1002/prot.10545. [DOI] [PubMed] [Google Scholar]
- 17.Luthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 18.Lubys A., Janulaitis A. Cloning and analysis of the plasmid-borne genes encoding the Bsp6I restriction and modification enzymes. Gene. 1995;157:25–29. doi: 10.1016/0378-1119(94)00795-t. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J., Russell D.W., Sambrook J. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 20.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 21.Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the ‘midnight zone’ of homology. Curr. Protein Pept. Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 22.Kelley L.A., MacCallum R.M., Sternberg M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 23.Fischer D. Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac. Symp. Biocomput. 2000:119–130. [PubMed] [Google Scholar]
- 24.Todone F., Weinzierl R.O., Brick P., Onesti S. Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target. Proc. Natl Acad. Sci. USA. 2000;97:6306–6310. doi: 10.1073/pnas.97.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundstrom J., Rychlewski L., Bujnicki J.M., Elofsson A. Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci. 2001;10:2354–2362. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marti-Renom M.A., Stuart A.C., Fiser A., Sanchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 27.Horton J.R., Blumenthal R.M., Cheng X. In: Restriction Endonucleases. Pingoud A.M., editor. Vol. 14. Berlin: Springer-Verlag; 2004. pp. 361–392. [Google Scholar]
- 28.Cheng X., Balendiran K., Schildkraut I., Anderson J.E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nastri H.G., Evans P.D., Walker I.H., Riggs P.D. Catalytic and DNA binding properties of PvuII restriction endonuclease mutants. J. Biol. Chem. 1997;272:25761–25767. doi: 10.1074/jbc.272.41.25761. [DOI] [PubMed] [Google Scholar]
- 30.Horton J.R., Cheng X. PvuII endonuclease contains two calcium ions in active sites. J. Mol. Biol. 2000;300:1049–1056. doi: 10.1006/jmbi.2000.3938. [DOI] [PubMed] [Google Scholar]
- 31.Horton J.R., Nastri H.G., Riggs P.D., Cheng X. Asp34 of PvuII endonuclease is directly involved in DNA minor groove recognition and indirectly involved in catalysis. J. Mol. Biol. 1998;284:1491–1504. doi: 10.1006/jmbi.1998.2269. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg D., Luthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 33.Rice M.R., Koons M.D., Blumenthal R.M. Substrate recognition by the PvuII endonuclease: binding and cleavage of CAG5mCTG sites. Nucleic Acids Res. 1999;27:1032–1038. doi: 10.1093/nar/27.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Athanasiadis A., Vlassi M., Kotsifaki D., Tucker P.A., Wilson K.S., Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nature Struct. Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 35.Selent U., Ruter T., Kohler E., Liedtke M., Thielking V., Alves J., Oelgeschlager T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992;31:4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- 36.Lagunavicius A., Siksnys V. Site-directed mutagenesis of putative active site residues of MunI restriction endonuclease: replacement of catalytically essential carboxylate residues triggers DNA binding specificity. Biochemistry. 1997;36:11086–11092. doi: 10.1021/bi963125i. [DOI] [PubMed] [Google Scholar]
- 37.Pingoud V., Sudina A., Geyer H., Bujnicki J.M., Lurz R., Luder G., Morgan R., Kubareva E., Pingoud A. Specificity changes in the evolution of Type II restriction endonucleases: a biochemical and bioinformatic analysis of restriction enzymes that recognize unrelated sequences. J. Biol. Chem. 2004. First published on November 24, 2004, 10.1074/jbc.M409020200. [DOI] [PubMed]
- 38.Venyaminov S.Y., Yang J.T. In: Circural Dichroism and the Conformational Analysis of Biomolecules. Fasman G.D., editor. NY: 1996. pp. 69–106. [Google Scholar]
- 39.Huai Q., Colandene J.D., Chen Y., Luo F., Zhao Y., Topal M.D., Ke H. Crystal structure of NaeI—an evolutionary bridge between DNA endonuclease and topoisomerase. EMBO J. 2000;19:3110–3118. doi: 10.1093/emboj/19.12.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman M., Lunnen K., Wilson G., Greci J., Schildkraut I., Phillips S.E. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 1998;17:5466–5476. doi: 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pingoud V., Kubareva E., Stengel G., Friedhoff P., Bujnicki J.M., Urbanke C., Sudina A., Pingoud A. Evolutionary relationship between different subgroups of restriction endonucleases. J. Biol. Chem. 2002;277:14306–14314. doi: 10.1074/jbc.M111625200. [DOI] [PubMed] [Google Scholar]
- 42.Bujnicki J.M. A model of structure and action of Sau3AI restriction endonuclease that comprises two MutH-like endonuclease domains within a single polypeptide. Acta Microbiol. Pol. 2001;50:219–231. [PubMed] [Google Scholar]
- 43.Bujnicki J.M., Rychlewski L. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene. 2001;267:183–191. doi: 10.1016/s0378-1119(01)00405-x. [DOI] [PubMed] [Google Scholar]
- 44.Pingoud V., Conzelmann C., Kinzebach S., Sudina A., Metelev V., Kubareva E., Bujnicki J.M., Lurz R., Luder G., Xu S.Y., et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 45.Rigden D.J., Setlow P., Setlow B., Bagyan I., Stein R.A., Jedrzejas M.J. PrfA protein of Bacillus species: prediction and demonstration of endonuclease activity on DNA. Protein Sci. 2002;11:2370–2381. doi: 10.1110/ps.0216802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.