Abstract
Drosophila phototransduction is mediated by phospholipase C, leading to activation of transient receptor potential (TRP) and TRP-like (TRPL) channels by mechanisms that are unresolved. A role for InsP3 receptors (IP3Rs) had been excluded because IP3R mutants (itpr) appeared to have normal light responses; however, this was recently challenged by Kohn et al. (“Functional cooperation between the IP3 receptor and phospholipase C secures the high sensitivity to light of Drosophila photoreceptors in vivo,” Journal of Neuroscience 35:2530), who reported defects in phototransduction after IP3R-RNAi knockdown. They concluded that InsP3-induced Ca2+ release plays a critical role in facilitating channel activation, and that previous failure to detect IP3R phenotypes resulted from trace Ca2+ in electrodes substituting for InsP3-induced Ca2+ release. In an attempt to confirm this, we performed electroretinograms, whole-cell recordings, and GCaMP6f Ca2+ imaging from both IP3R-RNAi flies and itpr-null mutants. Like Kohn et al., we used GMRGal4 to drive expression of UAS-IP3R-RNAi, but we also used controls expressing GMRGal4 alone. We describe several GMRGal4 phenotypes suggestive of compromised development, including reductions in sensitivity, dark noise, potassium currents, and cell size and capacitance, as well as extreme variations in sensitivity between cells. However, we found no effect of IP3R RNAi or mutation on photoreceptor responses or Ca2+ signals, indicating that the IP3R plays little or no role in Drosophila phototransduction.
Keywords: Gal4, GCaMP6F, GMR, phospholipase C, photoreceptors, TRP channels
Significance Statement
Phototransduction in microvillar photoreceptors such as those of Drosophila is mediated by phospholipase C (PLC), culminating in activation of TRP channels, but how PLC is coupled to channel activation is unresolved. A recent study reported phototransduction defects after InsP3 receptor RNA interference (IP3R-RNAi), supporting a critical role for InsP3-induced Ca2+ release. However, we found that phototransduction was quantitatively unaffected not only after IP3R-RNAi, but also in IP3R-null mutants. Instead, we describe novel phenotypes in photoreceptors from flies expressing the transcription factor (Gal4) used to drive RNAi expression, which potentially account for the reported defects. The results indicate that IP3R plays no significant role in Drosophila phototransduction while emphasizing the need for caution when using Gal4 drivers.
Introduction
Microvillar photoreceptors respond to light using G protein–coupled phospholipase C (PLC) cascades, leading to activation of nonselective cation channels (Yau and Hardie, 2009; Fain et al., 2010). In Drosophila, there are two such “light-sensitive” channels, encoded by the transient receptor potential (trp) and trp-like (trpl) genes. Both are permeable to Ca2+, with TRP being particularly Ca2+ selective (PCa:PNa ∼50:1) and contributing the majority of the light-induced current (Reuss et al., 1997; Liu et al., 2007). First cloned (Montell and Rubin, 1989) and identified as a light-sensitive channel (Hardie and Minke, 1992) more than 20 years ago, TRP is the prototypical member of the TRP ion channel superfamily, with 29 vertebrate isoforms distributed among 7 subfamilies. Of these, TRP and TRPL belong to and define the TRPC subfamily. All TRPCs can be activated via PLC; however, exactly how PLC activity leads to gating of the channels is unclear. Although InsP3-induced Ca2+ release is believed to be important for phototransduction in some microvillar photoreceptors (Brown et al., 1984; Fein et al., 1984; Ziegler and Walz, 1990; Walz and Baumann, 1995), a role in Drosophila had been excluded because light responses appeared to be unaffected in mutants of the InsP3 receptor (IP3R; Acharya et al., 1997; Raghu et al., 2000b). Subsequently, focus centered on other products of PLC activity such as diacylglycerol (Raghu et al., 2000a; Delgado et al., 2014) and its potential polyunsaturated fatty acid metabolites (Chyb et al., 1999; Leung et al., 2008; Lev et al., 2012), or PIP2 depletion and protons (Huang et al., 2010). Our own recent evidence suggested that the channels may be activated by a combination of protons released by the PLC reaction and the physico-mechanical consequences of cleaving PIP2’s bulky headgroup (InsP3) from the microvillar membrane (Hardie and Franze, 2012).
The conclusion that IP3Rs played no role in Drosophila phototransduction was seriously challenged by a recent study using RNA interference (Kohn et al., 2015). Those authors argued that previous failure to detect IP3R mutant phenotypes was due to leakage of trace Ca2+ from patch-clamp recording electrodes, effectively substituting for Ca2+ released from InsP3-sensitive stores. As supporting evidence, although light responses in IP3R-RNAi flies appeared normal in whole-cell recordings made without Ca2+ buffers in the electrode, they reported phenotypes using electrode solutions buffered with EGTA (Kohn et al., 2015). Phenotypes were also reported in electroretinogram (ERG) recordings suggesting a critical role in vivo. The authors proposed that InsP3-sensitive stores at the base of microvilli rapidly released Ca2+ into the microvilli and sensitized channels to activation via alternative products of PLC activity.
Because we found it surprising that Ca2+ leak could so effectively substitute for such a mechanism, we reinvestigated the role of the IP3R using not only RNAi but also IP3R-null mutants, paying particular attention to appropriate controls. In particular, Kohn et al. (2015) used the Gal4-UAS system (Brand and Perrimon, 1993) in which expression of UAS-IP3R-RNAi was driven by the Gal4 transcription factor under the control of the strong eye-specific GMR promoter, but controls from flies expressing GMRGal4 alone were lacking in most cases. We describe a number of novel phenotypes in flies expressing GMRGal4, but could not detect any differences in phototransduction between IP3R-RNAi flies or null IP3R mutants and relevant controls.
Materials and Methods
Flies (Drosophila melanogaster) were reared in the dark at 25°C on standard medium (8.5 g cornmeal, 0.9 g agar, 1.5 g yeast, 7.5 g glucose, and 5 ml nipagin per 100 ml water). The wild-type strain was Oregon; for some experiments white-eyed mutants (w1118) were used, which are indistinguishable in terms of whole-cell electrophysiology, though more sensitive in ERG recordings.
Mutants and transgenic lines used included the following:
w1118; P{w+,GMRGal4} (second chromosome, Bloomington stock 1104); referred to as GMR.
w1118; Sp/Cy; P{w+,GMRGal4},UAS-wRNAi is referred to as GMRw (third chromosome); one copy of GMRGal4, UAS-wRNAi renders eye color almost white (very pale orange) despite presence of the wild-type w + gene or multiple mini w + transgenes (provided by A. Huber).
w1118;;P{w+ UAS-IP3R-RNAi} (third chromosome VDRC stock 6486); homozygous stock appears wild-type in ERG and whole-cell recordings.
w1118;P{w+,UAS-GCaMP6f} (second chromosome, Bloomington stock 47247).
w1118;P{w+,ninaE-GCaMP6f} transgenic flies expressing GCaMP6f in photoreceptors R1-6 under control of the Rh1 opsin (ninaE) promoter: second and third chromosome lines made in-house using the pCaSpeR4 vector and GCaMP6f cDNA from Addgene (Asteriti et al., 2017).
norpAH43;bw;st (white-eyed): expresses near normal PLC protein levels (80%) but has <10% catalytic activity due to a point mutation (Ser347Ala) in the catalytic site, and another (Thr1007Ser) in the C terminus (Yoon et al., 2004; obtained from B. Minke).
l(3)itpr90B.0 larval lethal, null mutation of IP3R due to small deletion; referred to as itpr (Venkatesh and Hasan, 1997); chromosome also has closely linked strong P{w+}.
To generate whole-eye IP3R-null itpr mosaics:
FRT82B, l(3)itpr90B.0/TM6: (i.e., itpr recombined with FRT82B) were crossed to
yw;P{w+, ey-Gal4,UAS-FLP}/CyO;P{ry+,FRT82B}P{w+, GMR-hid},3CLR/TM6 (Bloomington stock 5253, referred to as EGUF;FRT82B). F1 non-Cy and non-TM6 then have itpr-null mosaic eyes (Stowers and Schwarz, 1999; Raghu et al., 2000b).
To obtain similar eye pigmentation for ERGs, red-eyed (w+) FRT82B, itpr/TM6 flies were crossed to yw,EGUF;FRT82B flies and recordings made from red-eyed (w+ males or w+/yw females) itpr mosaics, using sibling itpr/TM6, red-eyed wild-type, and wild-type mosaic eyes (by crossing to w+;;FRT82B) as controls. Further combinations using crosses from these parent lines are described in the text.
Electrophysiology
Whole-cell patch clamp recordings of photoreceptors from dissociated ommatidia from newly eclosed, dark-reared adult flies of either sex were performed as previously described (e.g. Hardie et al., 2002) on inverted Nikon microscopes (Nikon UK). Standard bath contained (in mm): 120 NaCl, 5 KCl, 10 N-Tris-(hydroxymethyl)-methyl-2-amino-ethanesulphonic acid (TES), 4 MgCl2, 1.5 CaCl2, 25 proline, and 5 alanine, pH 7.15. For Ca2+ free solutions, CaCl2 was omitted and 1 mm Na2EGTA was added. Other solutions are described in text. The intracellular pipette solution was (in mm): 140 K gluconate, 10 TES, 4 Mg-ATP, 2 MgCl2, 1 NAD, and 0.4 Na-GTP, with or without 1 or 2 mm K2EGTA, pH 7.15. Chemicals were obtained from Sigma-Aldrich. Recordings were made at room temperature (21 ± 1°C) at –70 mV (including correction for –10-mV junction potential) using electrodes of resistance ∼10-15 MΩ. Series compensation of >80% was applied when required for macroscopic currents, but not for sampling quantum bumps and dark noise. Data were collected and analyzed using Axopatch 200 or HEKA amplifiers and pCLAMP v. 9 or 10 software (Molecular Devices). Quantum bumps and spontaneous dark events were analyzed using Minianalysis (Jaejin Software), analyzing typically at least 50–100 bumps/events per cell. Photoreceptors were stimulated via a green (540-nm) ultrabright light-emitting-diode (LED) controlled by a custom-made LED driver; intensities were calibrated in terms of effectively absorbed photons by counting quantum bumps at low intensities.
ERGs were recorded as described previously (e.g., Satoh et al., 2010) from flies of either sex immobilized with low-melting-point wax in truncated pipette tips using low-resistance (∼10 MΩ) glass microelectrodes filled with standard bath solution, one inserted into the eye and one into the head capsule near the ocelli. Light was delivered by an ultrabright red (640 nm) LED positioned within 5 mm of the eye. Although controls were always performed with flies having the closest possible eye color, the inclusion of variable numbers of P{w +} transgenes (e.g., on GMRGal4 or UAS-RNAi constructs as well as the itpr chromosome) meant it was not always possible to obtain strictly identical pigmentation. However, the use of red light minimizes any effect of variable eye color (note also that flies with two copies of P{w+,UAS-IP3R-RNAi} or two copies of the itpr chromosome would potentially have had darker eye colors than controls). The maximum intensity (100 on figures) corresponded to ∼107 effectively absorbed photons per photoreceptor per second in white-eyed wild-type flies (w1118). Signals were amplified by a DAM60 DC preamplifier (WPI) and sampled and analyzed using pClamp software (Molecular Devices).
GCaMP6f measurements
Fluorescence measurements were made as previously described (Huang et al., 2010; Satoh et al., 2010; Asteriti et al., 2017) on an inverted Nikon microscope (non-confocal) from dissociated ommatidia or in vivo from intact flies via the deep pseudopupil (DPP). Excitation light (470 nm) was delivered from a blue power LED (Cairn Research), and fluorescence of whole ommatidia (via 40× oil objective) or DPP (20× air objective) was measured via a photomultiplier tube (Cairn Research) using 515 nm dichroic and OG515 longpass filters. Background fluorescence was subtracted using estimates from identical measurements from flies lacking fluorescent constructs. For dissociated ommatidia, ΔF/F0 was calculated using the F0 value measured in Ca2+-free solution (see above). To minimize any adverse long-term effects of exposure to Ca2+-free solution (e.g., depletion of Ca2+ stores) and hence to maximize the chance of detecting any Ca2+ release, ommatidia, plated in normal bath, were individually perfused by a nearby (∼10–20 µm) puffer pipette and measurements made within ∼20–40 s of perfusion onset, always confirming that a normal, rapid (latency <10 ms), and large fluorescence signal was recovered on return to normal (1.5 mm Ca2+) bath. The 1 or 2 s blue excitation light used to measure the fluorescence is a supersaturating stimulus and sufficiently bright to photoisomerize 100% of the visual pigment molecules at least once, reaching a photoequilibrium with ∼70% metarhodopsin. After the first measurement (usually made in Ca2+-free solution) the ommatidium was briefly exposed to intense, photoequilibrating red (4 s, 640 nm ultrabright LED) illumination to reconvert metarhodopsin to rhodopsin, returned to the control solution (1.5 mm Ca2+), and allowed to dark-adapt for at least 2 min before the next measurement. The effective intensity of the green illumination for in vivo measurements from the DPP was calibrated in effectively absorbed photons by measuring the rate at which it converted metarhodopsin to rhodopsin, as previously described (Hardie et al., 2015).
Isolation of retinal tissue and PCR
Preparations of nearly pure Drosophila retinal tissue were collected as previously described (Matsumoto et al., 1982; Raghu et al., 2000b). Briefly, whole flies were snap-frozen in liquid nitrogen and dehydrated in acetone at –20° C for 4 d. The acetone was drained off, and retinae were separated as cleanly as possible using a flattened insect pin.
Total RNA was extracted by RNeasy Micro kit (Qiagen). Ten to twenty retinae were collected as described above for each biological group and homogenized by the TissueLyser (Qiagen) with six Zirconia 1 mm beads (Thistle) for 50 s twice, the samples were passed to the Qiashredder column (Qiagen), and standard procedures from RNeasy micro kit were followed. All nucleic acid preparations were quantitated by absorbance measurements at 260 nm using a NanoDrop instrument. The quantitative real time qRT-PCR was performed with a SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) and ABI 7500 fast instrument (Applied Biosystems). The levels of InsP3R transcript were analyzed using the following primers: 5′-GTGTGGCTCTTCACGGATCA-3′ (forward) and 5′-GAACTCCACCTTCGGAATCA-3′ (reverse). The housekeeping gene Drosophila Ef1a48D primers, TCCTCCGAGCCACCATACAG (forward) and GTCTTGCCGTCAGCGTTACC (reverse), were used as internal controls.
Quantitative PCR experiments on genomic DNA (gDNA) were also conducted using the ABI 7500 fast instrument. Ten retinae were collected for each biological group, and gDNA was extracted with 10 mm Tris, 1 mm EDTA, 25 mm NaCl, and 200 µg/ml fresh Proteinase K and homogenized by the TissueLyser with nine Zirconia 1-mm beads for 50 s four times. The samples were then incubated at 37°C for 30 min and 95°C for 3 min (to destroy the proteinase K). The qPCR was set up with Terra qPCR Direct SYBR Premix (Takara) alongside either the IP3R primers, 5′-AAAATGCGTAGCATCGCTCT-3′ (forward) and 5′-CACCACCGGCTTTAGTTGAT-3′ (reverse), or the primers from the Rp132 housekeeping gene, 5′-CAAGAAGCTAGCCCAACCTG-3′ (forward) and 5′-CACTCACCGACAGCTTAGCA-3′ (reverse).
For verifying the presence of the UAS-IP3R-RNAi transgene, single flies were homogenized by the gDNA extraction buffer described above, and the following primers (sequences from VDRC) were used to perform the gDNA PCR: 5′-CGCGAATTCCCTTCGCCAGAGCGTGGAAA-3′ (forward), 5′-CGCTCTAGAACGCCAACATTGCGGAGCAG-3′ (reverse), and 5′-CACAGAAGTAAGGTTCCTTCACAAAGATCC-3′ (reverse).
Statistical analysis
Statistical tests (two-tailed t tests or one-way ANOVAs with posttests as specified in the text and figure legends) were performed in GraphPad Prism5.
Results
GMRGal4 phenotypes
The phenotypes reported by Kohn et al. (2015) came from flies in which IP3R expression was suppressed by UAS-IP3R-RNAi (two copies in most experiments), driven by one copy of GMRGal4. However, in most cases, controls from flies expressing GMRGal4 alone (without UAS-IP3R-RNAi) were lacking. This is an important control, because GMRGal4 homozygotes expressing two copies of GMRGal4 have severe degeneration and developmental phenotypes (Kramer and Staveley, 2003), and although one copy of GMRGal4 is often assumed to cause no phenotype, this has not been thoroughly explored. The GMRGal4 line used by Kohn et al. (2015) had a second chromosome GMRGal4 transgene recombined with UAS-wRNAi, which induces a white-eyed phenotype by suppressing expression of the wild-type w gene or the w+ marker gene in various expression vectors (Kalidas and Smith, 2002). Unfortunately this line has been lost (B. Minke, personal communication); therefore we tested a line with the same GMRGal4,UAS-wRNAi combination inserted on the third chromosome (referred to subsequently as GMRw), as well as the second chromosome GMRGal4 line (Bloomington stock 1104, referred to simply as GMR) but without UAS-wRNAi. We used the same UAS-IP3R-RNAi line as Kohn et al. (2015), VDRC stock 6486, confirming the presence of the IP3R-RNAi transgene in all the backgrounds used by gDNA PCR of diagnostic sequences (from VDRC website). Effective knockdown of IP3R mRNA was validated by qRT-PCR of dissected retinal tissue, with 32.8% ± 1.7% (mean ± SEM, n = 3 independent samples) IP3R mRNA remaining compared with GMR/+ controls. Approximately half of this (∼15%) is likely attributable to contaminating tissue in the dissected retinae (Raghu et al., 2000b).
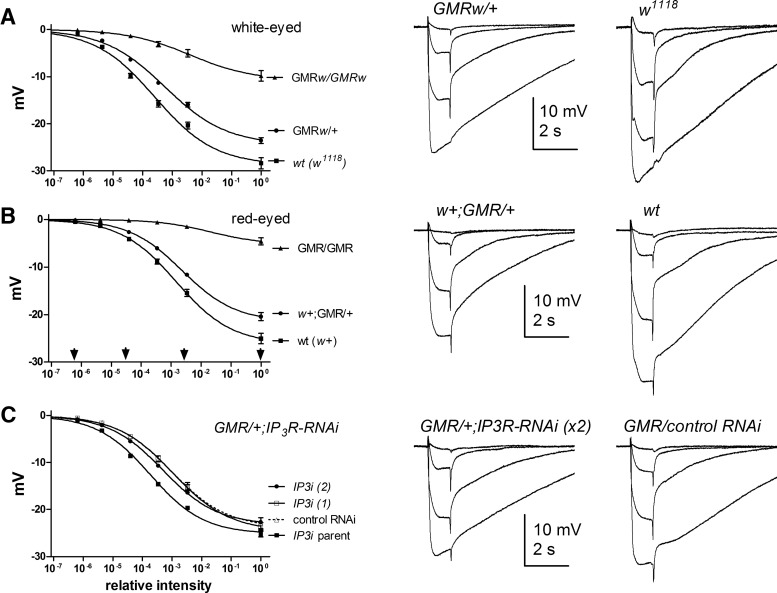
Homozygote flies carrying two copies of GMRGal4 displayed severe defects in retinal morphology (Fig. 1) and physiology (Fig. 2). The surfaces of the eyes were “glassy” with irregular facets, and ERG responses were greatly reduced in amplitude (Fig. 2). Dissociated ommatidia were almost unrecognizable, presenting as roughly spherical clusters, similar to midpupal-stage (∼48-h) ommatidia before the rapid phase of elongation that results in the characteristic adult appearance (Fig. 1A). Eyes of flies expressing just one copy of GMRGal4 appeared superficially normal; however, on preparing dissociated ommatidia from either GMRGal4 line (with or without IP3R-RNAi), it was apparent that GMRGal4/+ photoreceptors did not have the usual wild-type adult appearance. Ommatidia with one copy of GMRGal4, irrespective of IP3R-RNAi, were shorter than in wild-type (∼65 vs. ∼85 µm) and often had a less well-formed appearance (Fig. 1A, B), now somewhat reminiscent of ommatidia prepared from late-stage pupae. In whole-cell recordings, cell capacitances—which largely reflect the area of microvillar membrane—were also significantly reduced in GMRGal4/+ photoreceptors (∼30–50 pF vs. 50–70 pF), again irrespective of IP3R-RNAi (Fig. 1C). There were also marked differences in the voltage-sensitive potassium channel profiles (Fig. 1D, E). In wild-type photoreceptors, the largest component is a fast-inactivating A-current (IA) encoded by the Shaker gene, which typically reaches ∼4 nA (at 20 mV), vs. 2–3 nA for the delayed rectifier encoded by Shab (Hardie, 1991b; Vahasoyrinki et al., 2006). However, in GMR/+ and GMRw/+ flies, the Shaker component was substantially reduced, and in GMR homozygotes, almost undetectable (Fig. 1D, E). During photoreceptor development, the Shaker current appears last, only at late-pupal stages (Hardie, 1991b), so this profile is again suggestive of stunted development.
Figure 1.
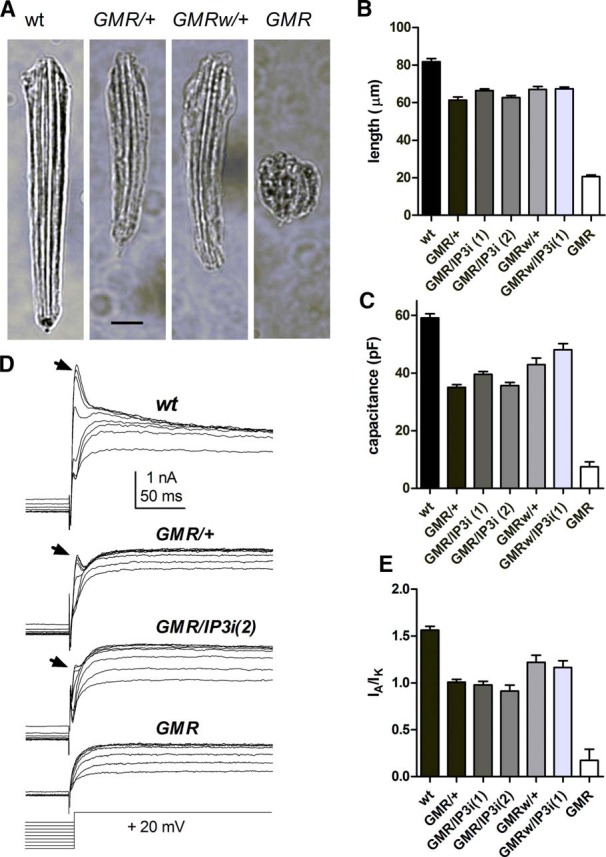
Morphologic and electrophysiological GMRGal4 phenotypes. A, Bright-field micrographs of dissociated ommatidia in wild-type and flies expressing one (GMR/+) or two (GMR) copies of GMRGal4. Scale bar, 10 µm. B, Length of ommatidia in wild type and flies expressing one or two copies of GMRGal4 with or without one or two copies of UAS-IP3R-RNAi (mean ± SEM, n > 10 randomly selected ommatidia from three to four flies per genotype). C, Capacitances in whole-cell recordings from same genotypes (n = 8–34 cells per genotype). Both ommatidia length and capacitance in flies with one copy of GMRGal4 were significantly reduced compared with wild-type (p < 0.001, one-way ANOVA with Dunnett’s multiple comparison test). D, Voltage-sensitive potassium currents at 20 mV after negative 1-s prepulses (–20 to –100 mV) to remove inactivation. Arrows indicate the rapidly inactivating Shaker component (IA), which was greatly reduced in flies expressing one copy of GMRGal4 with or without IP3R-RNAi (two copies) and virtually absent in GMR homozygotes. After 100 ms, the remaining maintained current is largely mediated by delayed rectifier (Shab = IK) channels. E, Ratio of IA (Shaker) peak current to IK (Shab) current measured 100 ms after voltage step: all backgrounds with one copy of GMRGal4 show significantly reduced IA compared with wild-type. Dunnett’s multiple comparison test, p < 0.003, n = 7–17 cells per genotype, except GMRw/IP3R-RNAi (n = 3).
Figure 2.
GMRGal4 ERG phenotypes. Response intensity (V/log I) functions measured from ERG (plateau at end of 1-s stimuli). ERGs from flies with one copy of GMRw (A) or GMR (B) were significantly reduced in amplitude across all intensities compared with respective eye color–matched wild-type controls (mean ± SEM, n = 10–17 flies; p < 0.0001; two-tailed t tests). A, GMRw/+ (F1 of Sp/Cy;GMRw × w1118 compared with w1118). Data from Sp/Cy;GMRw/GMRw parent also plotted. Representative traces on right (at intensities marked by arrows in B). B, F1 of w+;GMR × wild-type (red-eyed) cross compared with red-eyed wild-type (w+) and GMR/GMR homozygote, n = 10–14 flies. C, In contrast, ERGs of GMR/+;IP3R-RNAi (one copy, n = 16, or two copies, n = 19) were similar to a control GMR/+ (control RNAi; n = 10, dotted line). Flies were F1 of GMRGal4;IP3R-RNAi/TM6 × UAS-IP3R-RNAi or UAS-fwd-RNAi (control RNAi line chosen because it has similar eye color, but no effect on photoreceptor physiology). However, all GMR/+ genotypes were less sensitive (p < 0.0001) than the IP3R-RNAi parent stock (n = 7). Maximum intensity (10°) equivalent to ∼107 effectively absorbed photons per photoreceptor in wild-type (w1118).
The ERG is a widely used indicator of in vivo photoreceptor performance, although it is a complex signal reflecting responses of photoreceptors, glia, and second-order neurons as well as extracellular resistance barriers (Heisenberg, 1971; Kohn and Minke, 2011). Kohn et al. (2015) reported that ERGs in GMRGal4/+;IP3R-RNAi flies were reduced compared with wild-type, but did not provide data from GMRGal4/+ controls. We found that ERGs recorded from GMR/+ and GMRw/+ flies were significantly reduced in amplitude and sensitivity compared with eye color–matched wild-type controls. However, GMR/+;IP3R-RNAi with either one or two copies of UAS-IP3R-RNAi showed little or no difference from GMR/+ controls (Fig. 2).
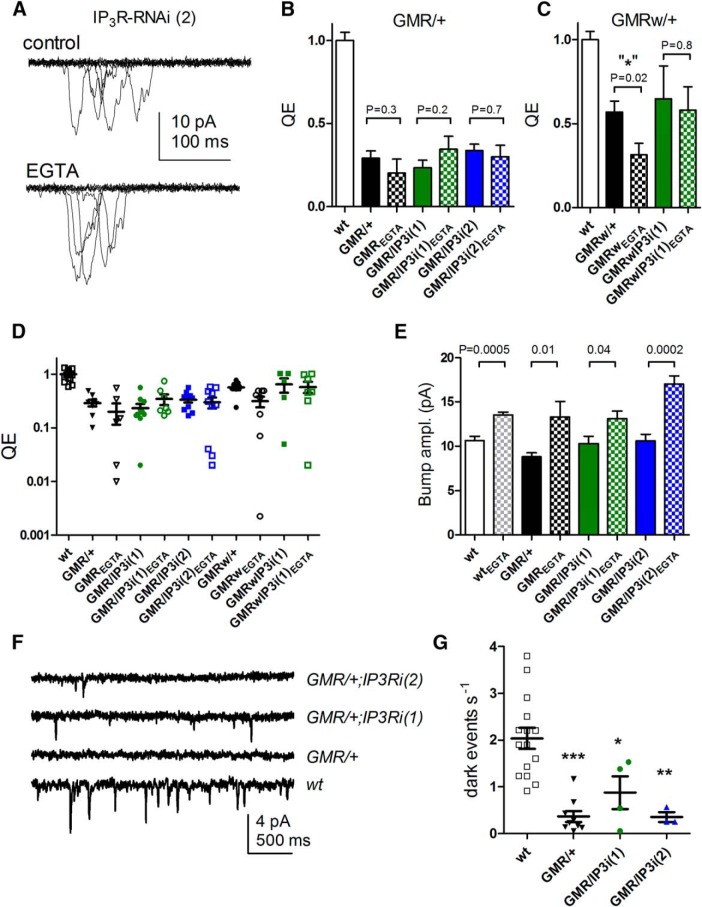
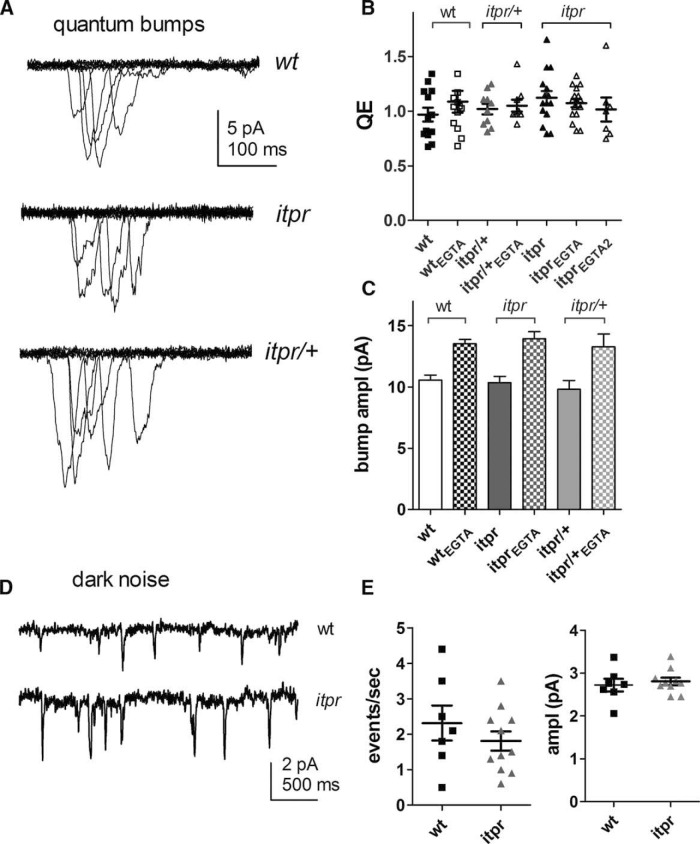
In whole-cell recordings, sensitivity can be defined by the relative quantum efficiency (QE), i.e., the fraction of incident photons evoking a quantum bump, which is determined by the amount of visual pigment (rhodopsin) and the probability that a photoisomerized rhodopsin successfully generates a quantum bump. One of the key findings in Kohn et al. (2015) was that QE was approximately twofold reduced in GMRGal4/+;IP3R-RNAi flies compared with wild-type flies, but only when the electrode solution was buffered with EGTA. When we attempted to confirm this, we found that QE was already reduced by approximately two- to threefold compared with wild-type in all backgrounds with one copy of GMRGal4, irrespective of IP3R-RNAi. However, QE was largely unaffected by the inclusion of EGTA in the electrode (Fig. 3A–D). One apparent exception were GMRw/+ controls (i.e., with one copy of GMRGal4, UAS-wRNAi but without UAS-IP3R-RNAi), which appeared to show an approximately twofold reduction in QE in recordings made with EGTA (n = 8). This was marginally significant (p = 0.02) on a direct t test, but not on an ANOVA including GMRw/IP3R-RNAi data with and without EGTA (p = 0.18). In all lines, inclusion of EGTA in the pipette increased bump amplitude, presumably due to suppression of the negative feedback effects of Ca2+ on the bump wave form (Fig. 3E). This was also reported by Kohn et al. (2015) and confirms the effectiveness of EGTA in our experiments.
Figure 3.
GMRGal4 phenotypes in whole-cell recordings of light-induced currents. A, Quantum bumps (10 superimposed traces, including “failures”) in response to dim flashes containing on average ∼0.5 effective photons recorded from GMR/+;IP3R-RNAi (two copies) photoreceptors with control electrode solution and with 1 mm EGTA (below). Note larger amplitudes with EGTA electrode (see also E). B, C, QE determined from such recordings, normalized to wild-type (n = 20), in flies expressing one copy of GMRGal4 (GMR/+ or GMRw/+) with or without UAS-IP3R-RNAi (one or two copies) and with or without 1 mm EGTA in the electrode (mean ± SEM, n = 5–10 cells per condition; see D). All lines with one copy of GMRGal4 had reduced QE compared with wild-type (p < 0.001, one-way ANOVA, Dunnett’s multiple comparison), but inclusion of EGTA made little or no difference: one exception (in C) was in flies with one copy of GMRw but without IP3R-RNAi. This was marginally significant (p = 0.02) with a direct t test, but not with a one-way ANOVA including data with and without EGTA from GMRw/IP3R-RNAi flies as well. D, Same data showing QE in all cells on log10 plot: note variability in all GMRGal4/+ backgrounds (total n = 78 cells): whereas most cells had QE two- to fourfold lower than in wild type, 11 cells had ≥10-fold lower QE. E, Bump amplitudes were larger in recordings made with 1 mm EGTA in the electrode in all backgrounds (mean ± SEM of average bump amplitudes from n = 4–10 cells, each with 30–100 bumps). F, Dark noise recorded with standard electrode solution (no EGTA). In wild-type cells, spontaneous ∼2-pA events occur at rates of ∼2/s, but backgrounds with one copy of GMRGal4 (with or without IP3R-RNAi) showed far fewer events. G, Summary of data: all lines with one copy of GMRGal4 (GMR/+) had significantly fewer dark events than wild type (*, p < 0.05; **, p < 0.01; ***, p < 0.001), but there was no significant effect of one or two copies of UAS-IP3R-RNAi (one-way ANOVA with Tukey’s posttest).
A conspicuous feature of recordings from flies carrying one copy of GMRGal4 was a large variability in QE (relative SD ∼0.7, vs. ∼0.2 for wild type). Although the majority of cells had QE values clustering within ∼20–60% of wild-type values, cells were regularly encountered (11/78 cells) in which QE was at least 10-fold, and sometimes >100-fold, lower (Fig. 3D). Such large variations are not encountered in recordings from wild-type adult photoreceptors, though they are a feature of recordings from pupal photoreceptors (Hardie et al., 1993). Although numbers were too small for evaluation of statistical significance, we noted that most (8/11) of these conspicuously insensitive cells, but not all of them, were recorded with EGTA-containing electrodes; however, IP3R-RNAi appeared not to make a difference (5/11 cells were from GMR or GMRw controls without IP3R-RNAi). In certain experiments (e.g., Sr2+ substitution; see further below), Kohn et al. (2015) reported large reductions in sensitivity in IP3R-RNAi flies when recorded with EGTA. Given that we were unable to replicate these results even using IP3R-null mutants (see below), one possible explanation for their findings is the fortuitous inclusion of data from such insensitive cells.
Using standard electrode solutions (without EGTA), whole-cell recordings from wild-type photoreceptors exhibit an ongoing spontaneous barrage of miniature (∼2-pA) bump-like events occurring at rates of ∼2/s, and which are believed to arise from spontaneous activation of G proteins (Hardie et al., 2002; Elia et al., 2005; Chu et al., 2013). Kohn et al. (2015) reported that GMRGal4/IP3R-RNAi flies had reduced levels of spontaneous activity even without EGTA in the electrode. However, we found that these spontaneous dark events were greatly reduced in frequency in GMRGal4/+ photoreceptors irrespective of IP3R-RNAi (Fig. 3F, G), suggesting that this result may also be attributable to GMRGal4 expression rather than IP3R knockdown.
In summary, flies expressing one copy of GMRGal4 displayed a number of phenotypes suggestive of compromised development. These are presumably caused by adverse, nonspecific effects of Gal4 in the developing eye and seem potentially able to account for many, if not all, of the results reported by Kohn et al. (2015). We did not, however, detect any additional effects of IP3R-RNAi knockdown using either one or two copies of UAS-IP3R-RNAi. Unfortunately, the GMRGal4,UAS-wRNAi fly stock used by Kohn et al. (2015) has been lost, so we cannot rigorously exclude the possibility that their IP3R-RNAi flies would still have shown some phenotypes when compared with appropriate GMRGal4 controls. Therefore, for further analysis, we turned to IP3R-null mutants (Raghu et al., 2000b), reasoning that these would show significantly more severe phenotypes, should any exist.
Null mutants of the InsP3 receptor (itpr)
Null mutations of the IP3R (l(3)itpr90B.0, referred to as itpr) are larval-lethal; however, whole-eye null mosaics can be generated by inducing mitotic recombination in the developing eye using the flippase-flippase recognition target (FLP-FRT) system under control of ey-Gal4 (Stowers and Schwarz, 1999; Raghu et al., 2000b). As controls, we used wild-type flies, itpr/+ heterozygote siblings from the same cross used to generate the eye mosaics, and “wild-type” mosaic eyes generated using an otherwise wild-type FRT chromosome (see Materials and Methods). Previously, we found that itpr-null eye mosaics expressed no detectable IP3R protein and only trace amounts of genomic DNA or mRNA, from contaminating material in the dissections used to isolate retinal tissue (Raghu et al., 2000b). Because these flies had been left in stock for more than ten years, we first checked the genotype by genomic qPCR and confirmed that the IP3R gene was reduced to trace amounts (15.7 ± 1.8%, n = 3, vs. control wild-type retinae), attributable to contaminating (nonretinal) tissue in dissected retinae (Raghu et al., 2000b). We also confirmed the virtual absence of any mRNA in retinal tissue by qRT-PCR (13.1 ± 3% IP3R mRNA remaining in sample compared with wild-type controls, n = 3).
ERGs of itpr-null mosaic eyes show a variable phenotype
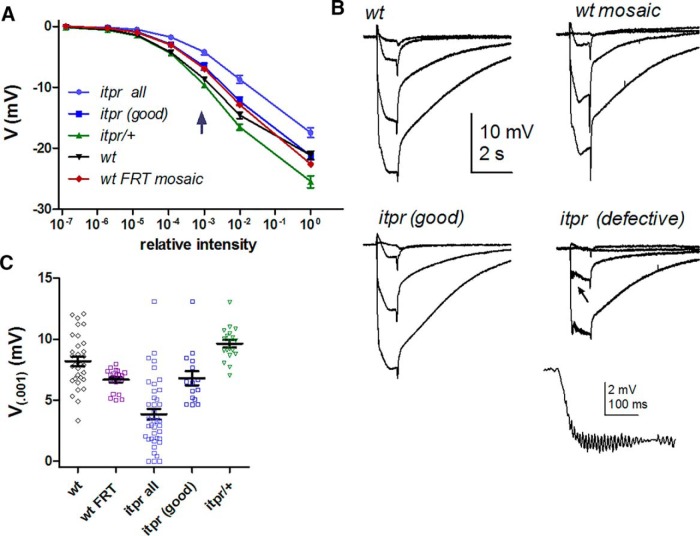
Previously, it was reported that photoreceptor responses from itpr eye mosaics were indistinguishable from controls in intracellular recordings (Acharya et al., 1997) or whole-cell recordings (Raghu et al., 2000b). However, at that time, no measurements were made using ERG recordings. When we measured ERGs from itpr mosaic eyes, amplitudes did in fact appear significantly reduced whether compared with wild-type, itpr/+ sibling controls, or wild-type mosaic eyes (p < 10−5, Fig. 4). However, it was also apparent that there was considerable variability in the ERGs. In ∼40% of flies (15/40), the ERGs resembled those from control flies; however, in others, the ERG was clearly compromised, typically showing reduction of “on” and “off” transients and development of oscillations, which are generally considered to reflect defects at the level of the synapse. If these obviously compromised ERGs were excluded from the analysis, the ERG amplitudes were still reduced compared with wild-type or itpr/+ controls (p = 0.047), but not compared with recordings from wild-type mosaic eyes (p = 0.8, Fig. 4). By implication, ERGs from “wild-type” mosaic eyes were also significantly less sensitive than wild type controls (p = 0.005).
Figure 4.
ERGs in itpr-null mosaic eyes. A, Response intensity (V/log I) functions measured from ERG (plateau at end of 1-s stimuli) in itpr-null mosaics (all flies, n = 40), selected itpr mosaics without obvious ERG defects (itpr “good,” n = 15), wild-type (n = 32), itpr/+ sibling controls (n = 18), and “wild-type” FRT mosaic eyes (n = 18). B, Representative ERG traces, including examples of a “good” and an obviously defective ERG from itpr mosaics: inset shows trace with oscillations (arrow) on expanded scale. C, Scatter plot of ERG amplitudes to relative intensity 10−3 (arrow in A). Overall, w+; itpr mosaic flies showed a significant reduction in amplitude (p ∼10−9 vs. wt, p = 0.0001 vs. wt FRT mosaic; two-tailed unpaired t test). If the obviously compromised recordings (V(0.001) <4 mV, oscillations and reduced synaptic transients) were excluded (itpr “good”), ERG amplitudes were still reduced compared with wild type and itpr/+ controls but now similar (p = 0.8) to recordings from wild-type mosaic eyes generated using an otherwise wild-type FRT82B chromosome (wt FRT).
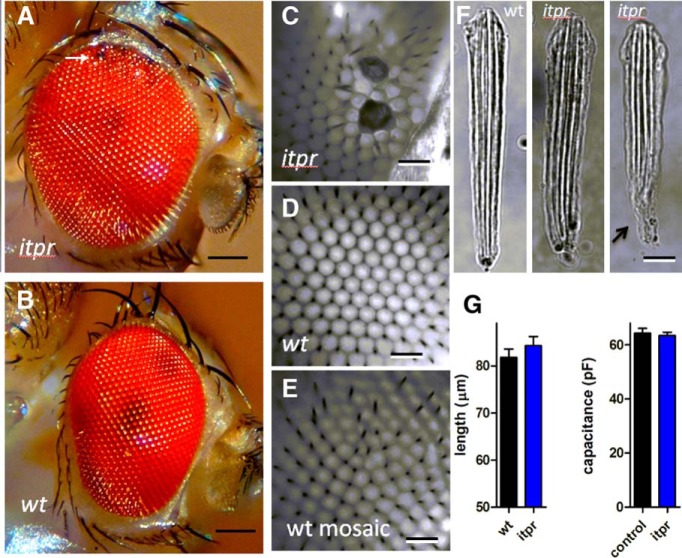
In addition to these variable defects in the ERG, the outward appearance of itpr mosaic eyes revealed clear abnormalities, usually being noticeably larger and more bulbous than wild-type eyes and typically containing a variable number of irregular or darkened facets (Fig. 5). “Wild-type” mosaic eyes occasionally also showed irregular facets, but were not noticeably different in shape or size and never showed the “scorched” facets typical of many itpr mosaic eyes. As described below, we were unable to detect any itpr phenotypes at the level of the photoreceptors in whole-cell recordings (Figs. 6–8) or with completely noninvasive in vivo Ca2+ imaging (see Fig. 10). Hence, we suggest that the variable ERG phenotype reflects defects in the overall structure of the eye, possibly owing to a role of the IP3R during development. For example, ERG amplitude is critically dependent on resistance barriers between the retina, lamina, and hemolymph (Heisenberg, 1971) and if short-circuited, even in a limited region, can be expected to have a potentially severe impact on the ERG. An indication of just such a defect was noted when preparing retinal tissue from freeze-dried heads (see Materials and Methods). In wild-type eyes, the retina separates cleanly and readily from the underlying neuropil (lamina) at a fracture plane near the base of the retina (Matsumoto et al., 1982); however, in itpr mosaics, this separation was noticeably more difficult to achieve, indicating structural differences at or around the base of the retina where the resistance barrier resides (Heisenberg, 1971). Probably related to this, in preparations of dissociated ommatidia from itpr mosaic eyes, ommatidia were frequently seen that had retained part of the axon terminal, whereas in wild-type preparations these are almost invariably broken off at the base of the retina (Fig. 5F).
Figure 5.
Structural abnormalities in itpr mosaic eyes. A, itpr mosaic eyes were noticeably larger and rounder in appearance than wild type (B, D) and frequently had areas of irregular and/or blackened facets (detail in C, D). E, Wild-type mosaic eyes (generated using an otherwise wild-type FRT chromosome) also sometimes showed irregular facets, but not the blackened facets or bulbous appearance of itpr mosaic eyes. F, Most dissociated ommatidia from itpr mosaic eyes appeared wild-type–like in appearance, but characteristically many still retained some of the axon terminal (arrow, right), which was almost invariably broken off in preparations from wild-type eyes. G, Ommatidial lengths and whole-cell capacitances in itpr mosaics were similar to those of controls (control = wild-type and itpr/TM6 pooled; mean ± SEM, n = 11–26 ommatidia/cells). Scale bars; (A, B) 80 µm, (C–E) 30 µm, (F) 10 µm.
Figure 6.
Whole-cell recordings from itpr-null mosaics. A, Quantum bumps: 10 superimposed traces (including failures) from responses to 1-ms flashes containing on average ∼0.5 effective photons, recorded with 1 mm EGTA in electrode in wild-type, itpr mosaic null, and itpr/+ heterozygotes (sibling controls). B, Summary of QE in recordings with and without EGTA, normalized to wild-type values. There was no significant effect of EGTA or itpr-null mutation (mean ± SEM; itpr control electrode solution, n = 15 cells; itpr recorded with 1 mm, n = 15; 2 mm EGTA, n = 7; wt and itpr/+ controls, 9–14 cells; p = 0.77, one-way ANOVA). C, Mean bump amplitude was increased in recordings made with EGTA (n = 4–10 cells). D, Dark noise recorded in wild-type and itpr mutants recorded using normal electrode solution (no EGTA); both showed similar levels of dark noise. E, Dark event rates and amplitudes (mean ± SEM) in wild-type (n = 7 cells) and itpr mosaics (n = 11 cells) were similar (p = 0.34 for rates and 0.64 for amplitude, two-tailed t test).
Figure 7.
Sr2+ substitution affects wt and itpr mosaics similarly. A, Substitution of control bath (1.5 Ca2+, 4 Mg2+ upper traces) with nominally Ca2+- and Mg2+-free solution and 1.5 mm Sr2+ without EGTA in the bath (lower traces from same cells). Left, response to 1-ms flashes containing ∼25 effective photons; right, quantum bumps in response to continuous dim light from itpr-null photoreceptor cells recorded using EGTA (1 mm) in the electrode. B, Summary of QE data from itpr-null mosaics and wild-type controls (+/+). C–F, Bath substitution with EGTA buffered Sr2+ (1 mm EGTA, nominally 0 Ca2+, 0 Mg2+ and 2.5 mm Sr2+: free [Sr2+] = 1.5 mm). Upper traces, before; bottom traces, same cells after perfusing with EGTA-buffered Sr2+ solution. C, Quantum bumps elicited under these conditions showed defects in both controls (itpr/+) and null mutants (itpr), often showing amplification only after a slow ramping phase (arrows). D, However, QE was only slightly affected, with no discernible difference between itpr and itpr/+ controls. E, Macroscopic responses to 1-ms flashes (∼25 effective photons) under the same conditions (slower traces during 0 Ca2+, 0 Mg2+ 1.5 Sr2+ plus EGTA perfusion). F, Peak amplitudes of responses were similarly affected in itpr and itpr/+ controls.
Figure 8.
Effects of Ca2+-free perfusion in itpr-null photoreceptors are similar to those of controls. A, Responses to brief flashes containing ∼25 effective photons recorded with EGTA-buffered electrode solutions before (dotted traces), 10 s after (larger slow responses), and 200 s after perfusing with Ca2+-free solution (1 mm EGTA). B, C, The reduction in peak amplitude (log10 scale) after 10- and 200-s perfusion and slowing of the response (measured by time to peak after 10- to 60-s perfusion) were similarly affected in itpr-null mosaic and itpr/+ sibling controls (p = 0.2–0.7, two-tailed t tests). D, Time course of suppression of the light response in Ca2+-free solution was similar in itpr-null mosaics and controls (mean ± SEM, n = 9–12 cells). E, Same data plotted on log10 scale.
Figure 10.
GCaMP6f signals are unaffected in IP3R-RNAi and itpr mutant flies. A, Average traces of GCaMP6f fluorescence in the presence (1.5 mm) and absence of Ca2+ (perfusion from puffer pipette with 0 Ca2+ 1 mm EGTA) from dissociated ommatidia from flies expressing GMRGal4;UAS-GCaMP6f and two copies of UAS-IP3R-RNAi (mean, n = 15) and control (GMRGal4;UAS-GCaMP6f alone; n = 9 ommatidia); pale traces indicate SEM. ΔF/F0 values for both +Ca2+ and 0 Ca2+ traces were based on F0 values in Ca2+-free solution. B, Ca2+-free responses on expanded scale. C, Summary of ΔF/F0 values measured 1 s after light onset, as well as the dark-adapted level in the presence of Ca2+ estimated from the “pedestal” (arrow in A). D, Ca2+-free ΔF/F0 values replotted, showing all data points: there was no significant difference (p = 0.62, two-tailed unpaired t test) between control and IP3R-RNAi flies. E, Two-pulse paradigm to determine intensity dependence of GCaMP6f signal in vivo from the deep pseudopupil (representative raw traces). Blue excitation was used to measure instantaneous GCaMP6f signal (arrows) in response to green (540 nm) test flashes (2 ms) of variable intensity delivered 300 ms earlier. F, Resulting intensity dependences of GCaMP6f signal in IP3R-RNAi (two copies) and control flies (GMR/+; UAS-GCaMP6f) were essentially identical (mean ± SEM, n = 8 flies). G–L, Similar data from itpr-null mosaics and sibling controls (itpr/+) expressing GCaMP6f under direct control of the Rh1 promoter (ninaE-GCaMP6f): n = 10–15 ommatidia/flies. No significant effects of the itpr–null mutation were detected.
Whole-cell recordings: quantum efficiency and dark noise are unaffected in itpr mutants
Despite abnormalities in the overall eye structure, and in contrast to ommatidia from GMRGal4/+ flies, dissociated ommatidia from itpr mosaic eyes had an essentially wild-type appearance (apart from the occasional retention of some axon terminal) and were of normal length, and in whole-cell recordings, the photoreceptors had capacitances and potassium channel profiles (not shown) similar to wild-type (Fig. 5G; compare Fig. 1).
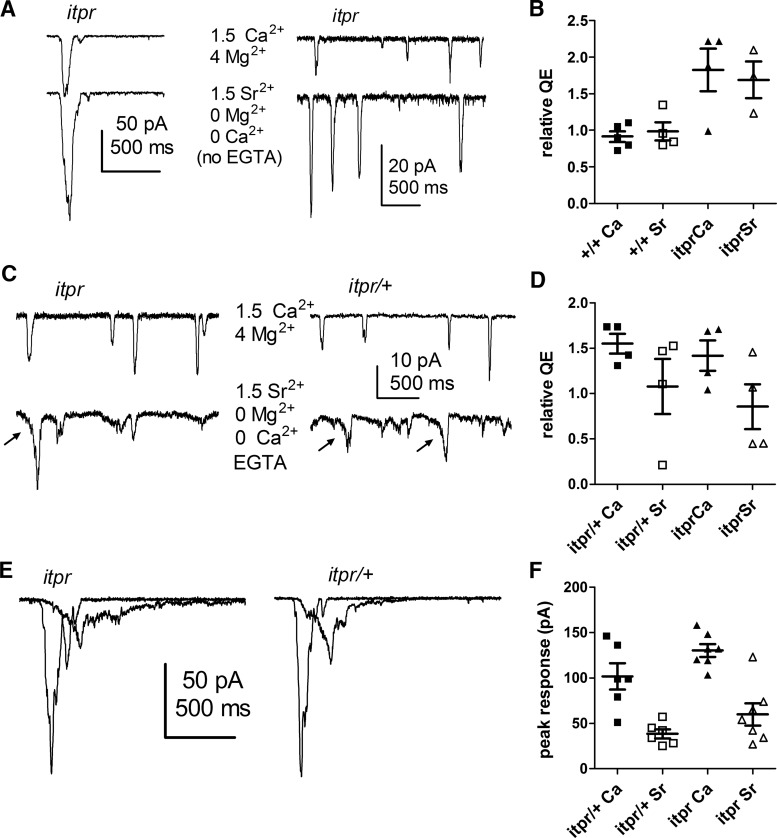
The central argument of Kohn et al. (2015) was that, although recordings from IP3R-RNAi flies made with electrode solutions lacking Ca2+ buffers showed normal light responses, phenotypes, including a twofold reduction in QE, became apparent when using electrode solutions buffered with 1 mm EGTA. However, we found that the QE of itpr-null mosaic photoreceptors recorded with normal electrode solution was indistinguishable from QE in recordings made with electrode solution containing 1 mm or even 2 mm EGTA. Neither were there significant differences in QE between itpr-null and wild-type or itpr/+ heterozygote controls with or without EGTA in the electrode (Fig. 6B). As with GMRGal4/+ flies, we confirmed the larger bump amplitudes recorded using EGTA-buffered electrode solutions (Fig. 6C).
The only phenotype of IP3R-RNAi flies reported by Kohn et al. (2015) in whole-cell recordings made with electrode solutions without EGTA was a reduction in the rate of spontaneous dark events (dark noise). Because we found that such a reduction was a feature of recordings from GMRGal4/+ irrespective of IP3R-RNAi (Fig. 3F, G), we also recorded dark noise in photoreceptors from itpr-null mutant mosaics. However, we found no difference in the dark noise between itpr-null mutants and wild-type controls (Fig. 6D, E), both of which showed spontaneous events of similar amplitudes (∼2 pA) at rates of ∼2 events/s as previously reported (Hardie et al., 2002; Elia et al., 2005; Chu et al., 2013).
itpr mutants are not more profoundly affected by Sr2+ substitution than controls
Arguably, the most dramatic effect of IP3R-RNAi knockdown reported by Kohn et al. (2015) was a severe (∼100-fold) reduction in QE when using EGTA in the electrode and bath Ca2+ substituted with Sr2+. In preliminary experiments, using the same solutions as those authors (1 mm EGTA in electrode; 1.5 mm Sr2+, nominally Ca2+- and Mg2+-free, but no EGTA in bath), QE appeared to be unaffected in either control flies or itpr-null mosaics (Fig. 7A). However, quantum bump amplitudes (and macroscopic responses) in both mutant and control became significantly larger because of the relief of channel block by Mg2+ (Hardie and Mojet, 1995).
Trace levels of Ca2+ in nominally Ca2+-free, unbuffered solutions are typically on the order of a few micromoles, which might still provide sufficient Ca2+ influx to sustain some degree of positive and negative feedback. We therefore proceeded to buffer the external 0 Ca2+, 0 Mg2+ solution with 1 mm EGTA, while increasing total Sr2+ to 2.5 mm. Because EGTA’s affinity for Ca2+ (Kd ∼200 nm) is approximately two orders of magnitude higher than for Sr2+ (∼30 µm), this should reduce trace Ca2+ to low nm levels while leaving ∼1.5 mm free Sr2+ (Xu-Friedman and Regehr, 2000). Under these conditions, bump amplification in both controls and itpr-null cells was significantly impaired after 1- to 2-min perfusion, leaving many bumps reduced in amplitude and with slow, irregular time courses. Bumps that still showed amplification had characteristically altered waveforms in both mutants and controls, with a slow ramping phase often apparent before the onset of rapid amplification (Fig. 7 C, D, arrows). A similar behavior has been observed in solutions containing reduced external Ca2+ or in cells buffered internally with BAPTA, and was attributed to the role of Ca2+ influx in the sequential positive and negative feedback that shapes the bump wave form (Henderson et al., 2000). Again, however, there was no noticeable difference between itpr-null and control cells, with both showing only a minor (<50%) reduction in QE. Because of the difficulty of unequivocally identifying bumps (and hence accurately estimating QE) under these conditions, we also simply recorded macroscopic responses to brief flashes. Peak amplitudes of responses were reduced approximately three- to fivefold after 2- to 3-min perfusion with the EGTA buffered Sr2+ solution, and time to peak slowed from ∼80 ms to >200 ms. Again, however, there was no difference between itpr-null mosaics and controls (Fig. 7E, F).
Previously, Katz and Minke (2012) reported that Sr2+ substitution still supported bump amplification but eliminated dark noise, and argued from this that there must be at least two sites for Ca2+-dependent facilitation (e.g., PLC and the channels). Although we do not exclude the possibility of two sites, we do not believe that this can be concluded from the effects of Sr2+. When trace Ca2+ is buffered with EGTA, our results indicate that Sr2+ influx is also much less effective than Ca2+ influx in supporting light-induced bump amplification (whether in controls or itpr-null mutants), and hence the most parsimonious explanation would be that the same site could also be responsible for the reduction in dark events.
Responses under Ca2+-free conditions are not influenced by IP3R-null mutation
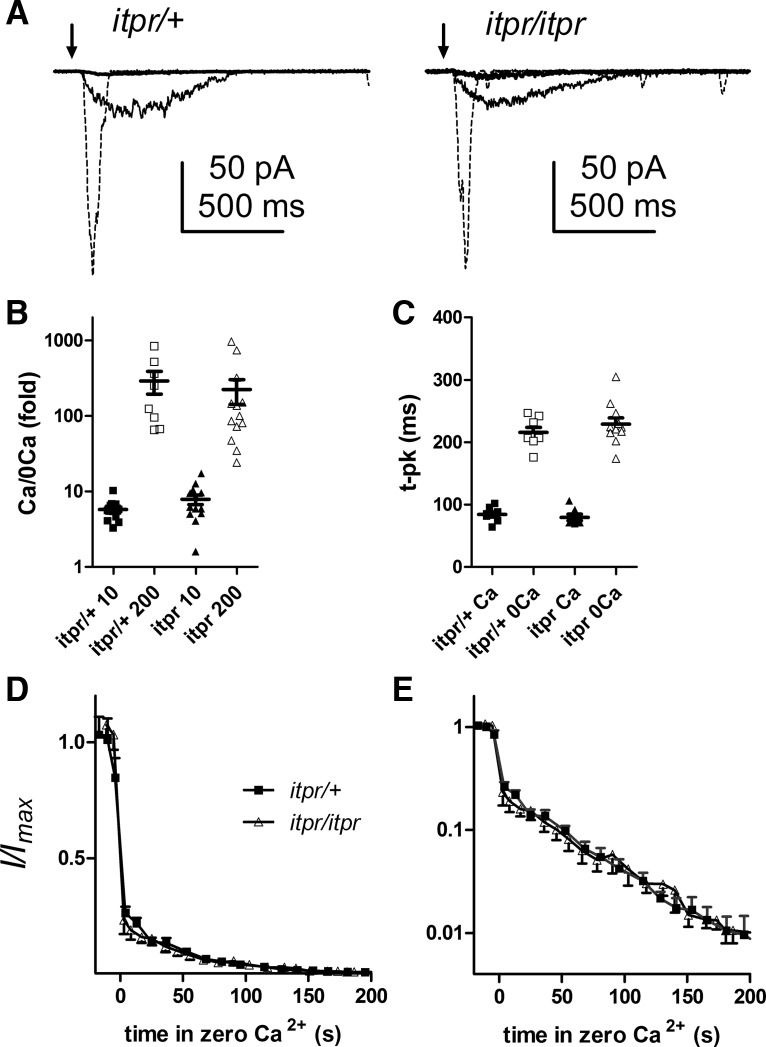
If Ca2+ release from IP3Rs is critical in facilitating phototransduction, then the simplest and most direct test for revealing its role should be testing sensitivity to light in Ca2+-free bath, an experiment not performed by Kohn et al. (2015). Previously, we reported that responses recorded in Ca2+-free bath were unaffected in itpr-null mosaics (Raghu et al., 2000b); however, in those experiments, the pipette solution would have contained trace Ca2+. We therefore repeated these experiments using 1 mm EGTA in the electrode. Within seconds of perfusing with Ca2+-free solution (also buffered with 1 mm EGTA) via a puffer pipette, peak responses were slowed and amplitude was reduced approximately fivefold, which is slightly more severe than the approximately threefold reduction previously reported with normal electrode solutions (e.g., Hardie, 1991a; Ranganathan et al., 1991; Reuss et al., 1997; Raghu et al., 2000b). With time, the sensitivity declined further, presumably as cytosolic Ca2+ levels reequilibrated to lower values, and after 3 min of perfusion, sensitivity (peak amplitude) was reduced by ∼100-fold (Fig. 8). However, the pronounced suppression seen under these conditions was similar, and followed a similar time course, whether recorded from control flies (itpr/+ siblings) or itpr-null mosaics.
In summary, in whole-cell recordings from IP3R-null mutants (itpr), we were unable to detect any of the phenotypes described by Kohn et al. (2015) and attributed to IP3R knockdown in IP3R-RNAi flies. Furthermore, in contrast to the variability we encountered in ERG recordings from itpr mutants or whole-cell recordings from GMRGal4 flies, we found no such variability in responses from single photoreceptors (with or without EGTA), despite making recordings from >60 cells from >30 flies (relative SD for QE ∼0.2–0.25 in both mosaics and controls). This reinforces our view that the variable ERG phenotypes in itpr-null mosaics (Fig. 4) are likely to reflect variable defects in overall eye structure or retinal resistance barriers rather than photoreceptor sensitivity.
Responses in hypomorphic norpA (PLC) mutants are suppressed by GMR-Gal4, but unaffected by IP3R-RNAi
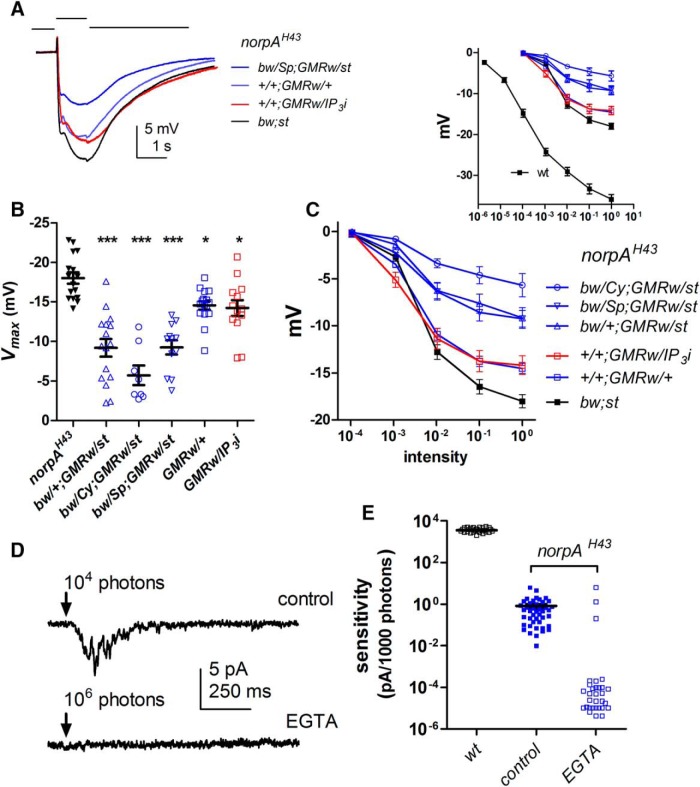
Although null mutants of PLC (norpA) have essentially no light response (Bloomquist et al., 1988), hypomorphic mutants—which still generate finite, albeit compromised, responses—can yield useful information on intermediate steps in the transduction cascade (e.g., Cook et al., 2000; Hardie et al., 2002). Kohn et al. (2015) reported that one copy of IP3R-RNAi driven by GMRGal4 led to a further substantial reduction in the ERG in norpAH43 mutant flies, which have ∼10-fold reduced PLC activity because of a point mutation in the catalytic site. This was attributed to the requirement of residual Ca2+ release from InsP3-sensitive stores to facilitate the weakened response in the PLC hypomorphic background (Kohn et al., 2015). However, controls from norpAH43 bearing one copy of GMRGal4 were again lacking. We therefore repeated these experiments, but now comparing white-eyed norpAH43 flies (norpAH43;bw;st) with norpAH43 flies carrying one copy of GMRw with and without UAS-IP3R-RNAi.
ERGs from norpAH43 flies carrying one copy of the GMRGal4, w-UAS-RNAi chromosome (GMRw/+) appeared rather sensitive to genetic background. In the F1 of three different crosses to introduce one copy of GMRw into a norpAH43 background (Fig. 9), ERG amplitudes were in each case significantly reduced compared with the parent norpAH43;bw;st control. However, norpAH43;+/+;GMRw/UAS-IP3R-RNAi flies actually had the highest sensitivity of any of the crosses and were indistinguishable from their most closely matched control (norpAH43;+/+;GMRw/+). The response amplitudes in norpAH43;bw/+;GMRw/st, norpAH43;bw/Sp;GMRw/st, or norpAH43;bw/Cy;GMRw/st, which represent the genotypes closest to norpAH43;GMRw/bw; IP3R-RNAi/st flies of Kohn et al. (2015), were more severely reduced—in fact, as severely as the data reported in their paper. Kohn et al. (2015) attributed this reduction in sensitivity to IP3R-RNAi knockdown, but according to our results it appears attributable to one copy of GMRGal4.
Figure 9.
GMRGal4, but not IP3R-RNAi, suppresses sensitivity in norpAH43. A, Representative ERG responses to 1-s flashes of submaximal intensity (10−1 on C) in norpAH43 mutant backgrounds. norpAH43 flies carrying one copy of GMRw (generated by three independent crosses) had consistently smaller responses than norpAH43; bw;st control (without GMR). Flies also expressing UAS-IP3R-RNAi (red) had among the largest responses and were indistinguishable from their closest control (norpAH43;+/+;GMRrw/+). B, C, V/log I curve and Vmax values from all genotypes (mean ± SEM, n = 12–16 flies). Vmax in all backgrounds with one copy of GMRGal4 were significantly (*, p < 0.05; ***, p < 0.001) suppressed compared with wild-type (one-way ANOVA, Dunnett’s multiple comparison test). D, Whole-cell recordings from norpAH43 photoreceptors. Top, response to 10-ms flash containing ∼104 wild-type effective photons recorded with control electrode solutions. Bottom, a ∼100× brighter flash (106 photons) elicited no response in a cell recorded with 1 mm EGTA. E, Sensitivity (tested ∼2 min after establishing the whole-cell configuration) expressed in pA/1000 wild-type effective photons in norpAH43 recorded with control electrode solution was approximately four orders of magnitude less than in wild-type (note log10 plot). With EGTA, there was no detectable response in 28 of 31 cells to flashes containing 106 photons (the variation in amplitudes of these data points reflects noise in the baseline).
Kohn et al. (2015) also reported a pronounced effect of IP3R-RNAi in whole-cell patch-clamp recordings from norpAH43. Once again, in recordings made with electrode solution without EGTA, they reported no significant difference between norpAH43 and norpAH43;GMRw/IP3R-RNAi flies, with both having a similar ∼4-log unit reduction in sensitivity compared with wild type. By contrast, when using EGTA-buffered electrode solution, norpAH43 cells were reported to be unaffected, whereas sensitivity in norpAH43;IP3R-RNAi was further drastically reduced, such that cells were essentially completely unresponsive to the brightest lights. However, when we recorded from norpAH43 photoreceptors (no GMRGal4 and no IP3R-RNAi) we found that 1 mm EGTA in the electrode already eliminated or drastically reduced sensitivity to light (Fig 9D, E). Thus, with control electrode solutions (no EGTA), all norpAH43 photoreceptors responded to flashes containing ∼104 effective photons with small but robust responses. With EGTA in the electrode, however, the majority of cells (28/31) gave no response at all to 100× brighter flashes (∼106 photons).
In summary, we found that one copy of GMRGal4 significantly suppressed the ERG in norpAH43 but found no additional effect of IP3R-RNAi, whereas in whole-cell recordings, we already found a profound suppression of sensitivity by EGTA in norpAH43. It is difficult to explain the failure of Kohn et al. (2015) to find an effect of EGTA in norpAH43 control flies. However, we found that norpAH43 photoreceptors can be very sensitive to facilitation, and the few cells (3/31) that failed to show suppressed sensitivity using EGTA in the electrode had a substantial leak currents or low-resistance gigaseals, both likely permeable to Ca2+.
Ca2+ “release” is not affected by InsP3 receptor mutation or RNAi
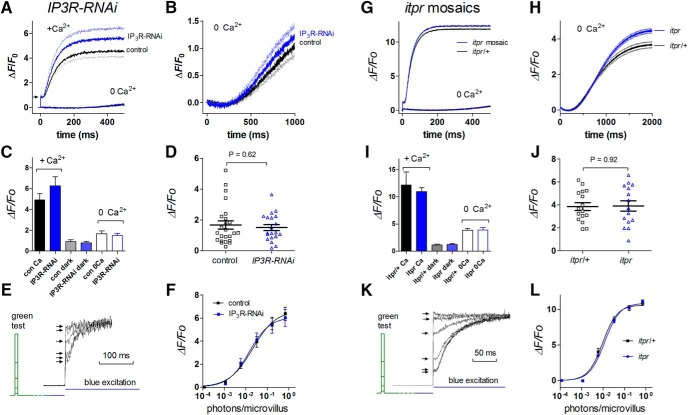
Measurements using fluorescent Ca2+ indicators in dissociated ommatidia show that the Ca2+ signal in response to blue excitation light (a supersaturating stimulus) is dominated by massive Ca2+ influx via light-sensitive channels (Peretz et al., 1994; Ranganathan et al., 1994; Hardie, 1996). In Ca2+-free bath, there is a smaller and slower rise in the fluorescent signal, of uncertain origin. Previously, using a ratiometric Ca2+ indicator dye (INDO-1), this residual “Ca2+-free” signal was reported to be unaffected in itpr-null mosaic mutants, suggesting it was not due to InsP3-induced Ca2+ release from internal stores (Raghu et al., 2000b). However, Kohn et al. (2015) reported that Ca2+ signals in Ca2+-free bath measured in ommatidia expressing GCaMP6f were further substantially reduced and slowed in IP3R-RNAi flies and concluded they were indeed due to InsP3-induced Ca2+ release.
We repeated these measurements using both GMRGal4;UAS-GCaMP6f with and without IP3R-RNAi, as well as ninaE-GCaMP6f under direct control of the Rh1 promoter (Asteriti et al., 2017) expressed in itpr-null mosaics. Our results from itpr mosaics are also reported elsewhere (Asteriti et al., 2017) but are replotted here with different controls (itpr/TM6 siblings) for a comprehensive picture (Fig. 10G–J). Compared with responses in normal bath, responses under Ca2+-free conditions were reduced in amplitude and much slower, with a delay of ∼200 ms before any measurable increase in fluorescence. However, these Ca2+-free responses were at least as large and had a similar time course in flies with IP3R-RNAi (two copies) or itpr-null mutations (Fig. 10). Resting Ca2+ levels in the dark in the presence of extracellular Ca2+, estimated from fluorescence during the brief 10-ms latent period before any Ca2+ rise (Fig 10A, C, arrows), were also not significantly affected by IP3R-RNAi or itpr-null mutation.
Although our measurements in the presence of Ca2+ closely resembled those of Kohn et al. (2015), our signals recorded in Ca2+-free solutions were slower than they reported in control ommatidia, more closely resembling their responses in IP3R-RNAi flies. On the rare occasions that we did see a more rapid Ca2+ signal, it was immediately clear that it was due to failure to adequately perfuse the ommatidium with Ca2+-free solution, and we can only speculate that a similar explanation may account for the signals recorded by Kohn et al. (2015), who used whole-bath perfusion with a lower concentration (0.5 mm) of EGTA.
We also measured the Ca2+ rise in vivo in completely intact flies by monitoring GCaMP6f fluorescence in the deep pseudopupil (Asteriti et al., 2017). By using a two pulse paradigm, this allows accurate determination of the intensity dependence of Ca2+ rises in response to brief flashes of dimmer, physiologically relevant intensities (Fig. 10E, K). The response intensity functions measured in this way should also provide a more direct measure of in vivo photoreceptor sensitivity than the complex signal of the ERG. Data from flies carrying two copies of IP3R-RNAi and itpr-null mosaics were indistinguishable from their relevant controls, although GMRGal4 flies (irrespective of IP3R-RNAi) were somewhat less sensitive than wild type (Fig. 10F, L).
Discussion
Despite extensive experiments, we were unable to detect any effect of RNAi knockdown or genetic elimination (itpr-null mosaic eyes) of the InsP3 receptor on the light response of Drosophila photoreceptors. An apparent exception was the compromised ERG in at least some itpr-null mosaic eyes (Fig. 4). However, as discussed above, we are of the opinion that this results from abnormalities in eye structure (e.g., retinal resistance barriers), possibly indicating a role for IP3R in eye development. In contrast, using a more direct in vivo measure of photoreceptor function (live imaging of GCaMP6f in the DPP), we found no effect of IP3R-RNAi or itpr-null mutation on photoreceptor sensitivity in vivo (Fig. 10F, L). We did, however, find a number of phenotypes attributable to one copy of GMRGal4 (Figs. 1–3), including reductions in sensitivity, dark noise, potassium currents, cell size, and capacitance. In addition, a notable feature of whole-cell recordings from photoreceptors from flies carrying one copy of GMRGal4 was a pronounced variability in sensitivity, with some cells showing massive (up to ∼100-fold) reductions in QE irrespective of IP3R-RNAi (Fig. 3D). These phenotypes, which are suggestive of compromised development, have the potential to explain many, if not all, of the results of Kohn et al. (2015).
Although a clear Ca2+ rise can be detected in the absence of external Ca2+, this was too slow (∼200-ms latency) to influence the onset of the electrical light response, which has a latency of <10 ms and peaks within ∼100 ms even under Ca2+-free conditions at these intensities (e.g., Huang et al., 2010). As previously reported using Ca2+ indicator dyes (Raghu et al., 2000b), we also found that this signal was unaffected by either itpr-null mutation or IP3R-RNAi knockdown (Fig. 10) and is therefore presumably not mediated via InsP3-induced release from internal stores. Previously, we found that this signal all but disappeared in the absence of extracellular Na+ and suggested that the rise might be due to reequilibration of Na+/Ca2+ exchange in response to the massive light-induced Na+ influx that persists under these conditions (Hardie, 1996). This was questioned by Cook and Minke (1999), who proposed that only extracellular Na2+, but not influx, was required for the Ca2+ rise in Ca2+-free solutions. However, in a recent study (Asteriti et al., 2017), we found that not only was this Ca2+-free rise dependent on Na+ influx, but it was also eliminated in mutants of the Na+/Ca2+ exchanger and accelerated by overexpression of the exchanger, strongly supporting our original suggestion.
In conclusion, we were unable to find any phototransduction phenotypes in IP3R-RNAi or itpr-null mutants either in vivo or in whole-cell recordings with or without EGTA in the electrode, and together with a recent study (Asteriti et al., 2017), we found no evidence for significant light and InsP3-induced release of Ca2+ from internal stores. Our results therefore support earlier conclusions that the IP3R plays no significant role in the light response in Drosophila photoreceptors (Acharya et al., 1997; Raghu et al., 2000b). We have however, described a number of significant photoreceptor phenotypes of GMRGal4/+ flies suggestive of compromised development, which we attribute to pleiotropic effects of Gal4 expression in the developing eye, and which should be carefully controlled for in any experiments making use of this widely used driver.
Acknowledgments
Acknowledgments: We are grateful to Drs. Armin Huber and Baruch Minke for providing flies.
Synthesis
Reviewing Editor: Anna Menini, SISSA (International School of Advances Studies)
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Richard Payne
Both reviewers agree that this is a meticulous study that re-evaluates the role (or lack thereof) of the InsP3 receptor in Drosophila phototransduction. The manuscript convincingly disproofs a previous report that suggested a role for the InsP3 receptor in phototransduction. Minor concerns have been fully addressed.
References
- Acharya JK, Jalink K, Hardy RW, Hartenstein V, Zuker CS (1997) InsP3 receptor is essential for growth and differentiation but not for vision in Drosophila . Neuron 18:881–887. [DOI] [PubMed] [Google Scholar]
- Asteriti S, Liu CH, Hardie RC (2017) Calcium signalling in Drosophila photoreceptors measured with GCaMP6f. Cell Calcium 65:40–51. doi: 10.1016/j.ceca.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL (1988) Isolation of putative phospholipase C gene of Drosophila, norpA and its role in phototransduction. Cell 54:723–733. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brown JE, Rubin LJ, Ghalayini AJ, Tarver AP, Irvine RF, Berridge MJ, Anderson RE (1984) Myo-inositol polyphosphate may be a messenger for visual excitation in Limulus photoreceptors. Nature 311:160–163. [DOI] [PubMed] [Google Scholar]
- Chu B, Liu CH, Sengupta S, Gupta A, Raghu P, Hardie RC (2013) Common mechanisms regulating dark noise and quantum bump amplification in Drosophila photoreceptors. J Neurophysiol 109:2044–2055. 10.1152/jn.00001.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyb S, Raghu P, Hardie RC (1999) Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature 397:255–259. 10.1038/16703 [DOI] [PubMed] [Google Scholar]
- Cook B, Minke B (1999) TRP and calcium stores in Drosophila phototransduction. Cell Calcium 25:161–171. 10.1054/ceca.1998.0018 [DOI] [PubMed] [Google Scholar]
- Cook B, Bar-Yaacov M, Ben-Ami HC, Goldstein RE, Paroush Z, Selinger Z, Minke B (2000) Phospholipase C and termination of G-protein-mediated signalling in vivo. 2:296–301. 10.1038/35010571 [DOI] [PubMed] [Google Scholar]
- Delgado R, Muñoz Y, Peña-Cortés H, Giavalisco P, Bacigalupo J (2014) Diacylglycerol activates the light-dependent channel TRP in the photosensitive microvilli of Drosophila melanogaster photoreceptors. J Neurosci 34:6679–6686. 10.1523/JNEUROSCI.0513-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Frechter S, Gedi Y, Minke B, Selinger Z (2005) Excess of G{beta}e over Gq{alpha} in vivo prevents dark, spontaneous activity of Drosophila photoreceptors. J Cell Biol 171:517–526. 10.1083/jcb.200506082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain GL, Hardie R, Laughlin SB (2010) Phototransduction and the evolution of photoreceptors. Curr Biol 20:R114–R124. 10.1016/j.cub.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein A, Payne R, Corson DW, Berridge MJ, Irvine RF (1984) Photoreceptor excitation and adaptation by inositol 1,4,5-trisphosphate. Nature 311:157–160. [DOI] [PubMed] [Google Scholar]
- Hardie RC (1991a) Whole-cell recordings of the light-induced current in Drosophila photoreceptors: evidence for feedback by calcium permeating the light sensitive channels. Proc Roy Soc Lond B 245:203–210. [Google Scholar]
- Hardie RC (1991b) Voltage-sensitive potassium channels in Drosophila photoreceptors. J Neurosci 11:3079–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC (1996) INDO-1 measurements of absolute resting and light-induced Ca2+ concentration in Drosophila photoreceptors. J Neurosci 16:2924–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Minke B (1992) The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron 8:643–651. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Mojet MH (1995) Magnesium-dependent block of the light-activated and trp-dependent conductance in Drosophila photoreceptors. J Neurophysiol 74:2590–2599. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Franze K (2012) Photomechanical responses in Drosophila photoreceptors. Science 338:260–263. 10.1126/science.1222376 [DOI] [PubMed] [Google Scholar]
- Hardie RC, Peretz A, Pollock JA, Minke B (1993) Ca2+ limits the development of the light response in Drosophila photoreceptors. Proc Biol Sci 252:223–229. 10.1098/rspb.1993.0069 [DOI] [PubMed] [Google Scholar]
- Hardie RC, Liu CH, Randall AS, Sengupta S (2015) In vivo tracking of phosphoinositides in Drosophila photoreceptors. J Cell Sci 128:4328–4340. 10.1242/jcs.180364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Martin F, Cochrane GW, Juusola M, Georgiev P, Raghu P (2002) Molecular basis of amplification in Drosophila phototransduction. Roles for G protein, phospholipase C, and diacylglycerol kinase. Neuron 36:689–701. 10.1016/S0896-6273(02)01048-6 [DOI] [PubMed] [Google Scholar]
- Heisenberg M (1971) Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila . J Exp Biol 55:85–100. [DOI] [PubMed] [Google Scholar]
- Henderson SR, Reuss H, Hardie RC (2000) Single photon responses in Drosophila photoreceptors and their regulation by Ca2+ . J Physiol 524:179–194. 10.1111/j.1469-7793.2000.00179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu CH, Hughes SA, Postma M, Schwiening CJ, Hardie RC (2010) Activation of TRP channels by protons and phosphoinositide depletion in Drosophila photoreceptors. Curr Biol 20:189–197. 10.1016/j.cub.2009.12.019 [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP (2002) Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila . Neuron 33:177–184. [DOI] [PubMed] [Google Scholar]
- Katz B, Minke B (2012) Phospholipase C-mediated suppression of dark noise enables single-photon detection in Drosophila photoreceptors. J Neurosci 32:2722–2733. 10.1523/JNEUROSCI.5221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn E, Minke B (2011) Methods for Studying Drosophila TRP Channels. In: TRP Channels. Zhu MX, ed. Boca Raton, FL: CRC Press/Taylor & Francis. [PubMed] [Google Scholar]
- Kohn E, Katz B, Yasin B, Peters M, Rhodes E, Zaguri R, Weiss S, Minke B (2015) Functional cooperation between the IP3 receptor and phospholipase C secures the high sensitivity to light of Drosophila photoreceptors in vivo. J Neurosci 35:2530–2546. 10.1523/JNEUROSCI.3933-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Staveley BE (2003) GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet Mol Res 2:43–47. [PubMed] [Google Scholar]
- Leung HT, Tseng-Crank J, Kim E, Mahapatra C, Shino S, Zhou Y, An L, Doerge RW, Pak WL (2008) DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron 58:884–896. 10.1016/j.neuron.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Katz B, Tzarfaty V, Minke B (2012) Signal-dependent hydrolysis of phosphatidylinositol 4,5-bisphosphate without activation of phospholipase C: implications on gating of Drosophila TRPL (transient receptor potential-like) channel. J Biol Chem 287:1436–1447. 10.1074/jbc.M111.266585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Wang T, Postma M, Obukhov AG, Montell C, Hardie RC (2007) In vivo identification and manipulation of the Ca2+ selectivity filter in the Drosophila transient receptor potential channel. J Neurosci 27:604–615. 10.1523/JNEUROSCI.4099-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, O’Tousa JE, Pak WL (1982) Light-induced modification of Drosophila retinal polypeptides in vivo. Science 217:839–841. [DOI] [PubMed] [Google Scholar]
- Montell C, Rubin GM (1989) Molecular characterization of Drosophila trp locus, a putative integral membrane protein required for phototransduction. Neuron 2:1313–1323. [DOI] [PubMed] [Google Scholar]
- Peretz A, Suss-Toby E, Rom-Glas A, Arnon A, Payne R, Minke B (1994) The light response of Drosophila photoreceptors is accompanied by an increase in cellular calcium: effects of specific mutations. Neuron 12:1257–1267. [DOI] [PubMed] [Google Scholar]
- Raghu P, Usher K, Jonas S, Chyb S, Polyanovsky A, Hardie RC (2000a) Constitutive activity of the light-sensitive channels TRP and TRPL in the Drosophila diacylglycerol kinase mutant, rdgA. Neuron 26:169–179. [DOI] [PubMed] [Google Scholar]
- Raghu P, Colley NJ, Webel R, James T, Hasan G, Danin M, Selinger Z, Hardie RC (2000b) Normal phototransduction in Drosophila photoreceptors lacking an InsP3 receptor gene. Mol Cell Neurosci 15:429–445. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Harris GL, Stevens CF, Zuker CS (1991) A Drosophila mutant defective in extracellular calcium-dependent photoreceptor deactivation and rapid desensitization. Nature 354:230–232. 10.1038/354230a0 [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Bacskai BJ, Tsien RY, Zuker CS (1994) Cytosolic calcium transients: spatial localization and role in Drosophila photoreceptor cell function. Neuron 13:837–848. [DOI] [PubMed] [Google Scholar]
- Reuss H, Mojet MH, Chyb S, Hardie RC (1997) In vivo analysis of the Drosophila light-sensitive channels, TRP and TRPL. Neuron 19:1249–1259. [DOI] [PubMed] [Google Scholar]
- Satoh AK, Xia H, Yan L, Liu CH, Hardie RC, Ready DF (2010) Arrestin translocation is stoichiometric to rhodopsin isomerization and accelerated by phototransduction in Drosophila photoreceptors. Neuron 67:997–1008. 10.1016/j.neuron.2010.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähäsöyrinki M, Niven JE, Hardie RC, Weckström M, Juusola M (2006) Robustness of neural coding in Drosophila photoreceptors in the absence of slow delayed rectifier K+ channels. J Neurosci 26:2652–2660. 10.1523/JNEUROSCI.3316-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh K, Hasan G (1997) Disruption of the IP3 receptor gene of Drosophila affects larval metamorphosis and ecdysone release. Curr Biol 7:500–509. [DOI] [PubMed] [Google Scholar]
- Walz B, Baumann O (1995) Structure and cellular physiology of Ca2+ stores in invertebrate photoreceptors. Cell Calcium 18:342–351. [DOI] [PubMed] [Google Scholar]
- Xu-Friedman MA, Regehr WG (2000) Probing fundamental aspects of synaptic transmission with strontium. J Neurosci 20:4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau KW, Hardie RC (2009) Phototransduction motifs and variations. Cell 139:246–264. 10.1016/j.cell.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Leung HT, Lee S, Geng C, Kim Y, Baek K, Pak WL (2004) Specific molecular alterations in the norpA-encoded phospholipase C of Drosophila and their effects on electrophysiological responses in vivo. J Neurochem 89:998–1008. 10.1111/j.1471-4159.2004.02384.x [DOI] [PubMed] [Google Scholar]
- Ziegler A, Walz B (1990) Evidence for light-induced release of Ca2+ from intracellular stores in bee photoreceptors. Neurosci Lett 111:87–91. [DOI] [PubMed] [Google Scholar]