Abstract
A simple statistical method for predicting the functional differentiation of duplicate genes was developed. This method is based on the premise that the extent of functional differentiation between duplicate genes is reflected in the difference in evolutionary rate because the functional change of genes is often caused by relaxation or intensification of functional constraints. With this idea in mind, we developed a window analysis of protein sequences to identify the protein regions in which the significant rate difference exists. We applied this method to MIKC-type MADS-box proteins that control flower development in plants. We examined 23 pairs of sequences of floral MADS-box proteins from petunia and found that the rate differences for 14 pairs are significant. The significant rate differences were observed mostly in the K domain, which is important for dimerization between MADS-box proteins. These results indicate that our statistical method may be useful for predicting protein regions that are likely to be functionally differentiated. These regions may be chosen for further experimental studies.
INTRODUCTION
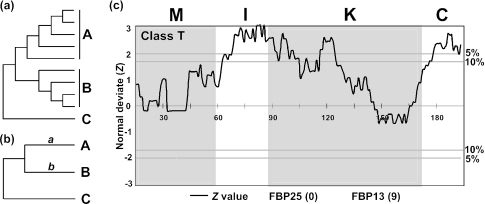
The functional differentiation of duplicate genes is thought to be an important mechanism of evolution of organisms (1–4). This differentiation is often associated with the relaxation or intensification of purifying selection in certain regions of protein sequences. Therefore, comparison of the evolutionary rates of paralogous protein sequences may give some insights into the functional differentiation. With this idea, a number of authors have developed statistical methods for predicting functional differentiation by examining the evolutionary rates. Dermitzakis and Clark (5) suggested that this functional differentiation may be revealed by examining the heterogeneity of substitution rate between two pairs of duplicated genes. Considering two groups of paralogous duplicate proteins (A and B in Figure 1a), Gu (6) and Knudsen and Miyamoto (7) respectively proposed a Bayesian and a maximum likelihood method of detecting amino acid sites that show a significant rate difference between the two groups. In these methods, the number of sequences in each group (A or B) must be relatively large to have reliable results. When the groups A and B include only one sequence, their methods are not applicable. This is also true with Dermitzakis and Clark's method.
Figure 1.
(a) Gu (6) and Knudsen and Miyamoto's tests (7). Groups A and B include many sequences to be compared, and C is the outgroup. (b) A and B represent two sequences to be compared, and C is the outgroup. (c) Comparison of the protein sequences of 2 class T MADS-box genes from petunia. For the outgroup two rice sequences were used. If FBP25 (the former) evolved faster than FBP13 (the latter) in a window, the Z-value is positive, and if the former evolved slower than the latter, the Z-value becomes negative. The number in a parenthesis for each gene is the number of interacting protein partners given by Immink et al. (29). Horizontal lines with ‘5%’ or ‘10%’ correspond to the cutoff Z-value of 1.96 or 1.65, respectively. The amino acid positions are given on the Z = 0 line. M, I, K and C represent the M, I, K and C domains. Window size (w) and skipping size (s) are 30 amino acids and one amino acid, respectively.
In real experimental studies, it is customary to identify the functional difference by comparing a sequence with known functional domains with a new sequence by using domain swapping or site-directed mutagenesis. However, it is time-consuming and expensive to use this method for a large number of pairs of sequences. It is therefore useful to develop a statistical method for identifying protein domains that are likely to be functionally differentiated. For this reason, we propose a new method in which only two sequences are compared at a time after construction of a rooted tree. This method will then be illustrated by an analysis of a number of MIKC-type MADS-box genes that control the development of flowers in plants.
METHODS AND RESULTS
Statistical methods
In our method, two protein sequences to be compared (A and B in Figure 1b) and an outgroup sequence (C) will be used after sequence alignment (Figure 1b). A phylogenetic tree for the sequences is constructed to determine the root of the sequences A and B. Here, we suggest that the p-distance (proportion of different amino acids) (8) be used, because the sequences to be compared are usually closely related and the p-distance has a smaller variance than any other distance measure. However, if a pair of divergent sequences is to be tested (e.g. p-distance > 0.3), the Poisson-correction or some other distance may be used (8). To identify the protein regions that show a significant rate difference, we use a sliding window analysis. Let n be the total number of amino acid (codon) sites used and w be the window size (the number of amino acids considered for one window). This window analysis may be done by sliding the window by one amino acid position consecutively or by skipping s amino acid positions each time. The total number of windows to be considered (T) is then (n − w)/s + 1. Here, T should be an integer. For example, if T happens to be 55.2, it should be reset to 55.
For each window, we now estimate the number of amino acid substitutions a and b for branch (sequence) A and B in Figure 1b, respectively. The branch lengths a and b may be estimated by the least squares method and are given by
where dAB, dAC, etc., are the observed distances between sequences A and B, A and C, etc, respectively. We are now interested in testing the significance level of the difference that is,
The variance of this D can be obtained by the formula given by Takezaki et al. (9). We can then consider
where V(D) is the variance of D. This Z is approximately normally distributed as long as w is about 30 or greater. Therefore, the significance level can easily be determined. When w < 30, the above Z is distributed as the t distribution with (w − 1) degrees of freedom (10). In reality, unless w ≥ 30, the statistical power of the window test is not very high. We therefore recommend that the window size is equal to or greater than 30.
It should be noted that in this sliding window analysis, the Z-values obtained for consecutive windows are highly correlated. Therefore, the significance levels of Z-values for consecutive window analyses may not be accurate. However, if the Z-value for one of the windows is significant, one can take it seriously. Furthermore, our purpose is to identify protein regions that should be subjected to experimental tests. Therefore, any consecutive windows showing significant Z-values should be considered biologically important. Actually, for this purpose, even a region showing Z-values with a significant level of 10% may be considered for experimentation.
In the above method, we considered the case where each of A, B and C contains only one sequence. However, the above approach can easily be extended to the case where protein sequences are classified into two groups, A and B, and the average rate difference between the two groups of proteins is studied. In this case, because the above test is a special case of two-cluster method by Takezaki et al. (9), we can directly apply the two-cluster method to test the rate difference between the two groups for each window. The outgroup may also contain many sequences. This is true even when A and B contain one sequence each.
Another extension of the above method is to consider the number of nonsynonymous nucleotide substitutions per nonsynonymous site (dN) (11) or the number of radical nucleotide substitutions (substitutions causing the changes of amino acid charge, hydrophobicity and size) per site (12,13). At present, however, it is unclear how useful these quantities are, because they generate rather large variances.
Application to MADS-box genes controlling flower development in plants
Floral MIKC-type MADS-box genes encode transcription factors that control flower development in plants. Major floral MADS-box genes can be classified into at least eight classes (14) in terms of their function and evolutionary relationships, i.e. A, B, C, D, E, F, G and T classes according to the simplified notation in (15). Each of these classes of genes encodes a protein consisting of the MADS (M) domain (DNA-binding site with about 60 amino acids), intervening (I) domain (∼30 amino acids), keratin-like (K) domain (∼70 amino acids), and C-terminal (C) domain (variable number of amino acids) (16) (Figure 1c). The M domain is composed of DNA-binding α helices, carries a nuclear localization signal and is involved in dimerization of proteins together with the I domain (17,18). The K domain mediates protein–protein interaction, whereas the C domain possesses transcriptional activation function in some MADS-box proteins (17,19–22) and might also be involved in protein dimerization (23) or formation of multimeric complexes (24). Among these domains, the I and K domains are most well known for determining the pattern of homodimerization or heterodimerization of MADS-box proteins. The K domain is involved in protein–protein interaction and is characterized by three strings of heptad repeats (abcdefg)n which are potentially forming coiled coils, with hydrophobic amino acids predominantly in positions a and d (22,25). The proteins encoded by different classes of floral MADS-box genes interact with one another or with some other proteins to form a particular organ. According to the floral Quartet model, the formation of petals is controlled by a combination of tetramers of class A, B and E proteins, and that of stamens is by tetramers of class B, C and E proteins (19,26–28). However, to explain the development of various forms of flowers in different species, we have to know detailed aspects of protein–protein interaction within each class of proteins. For this reason, many experimentalists are now studying protein–protein interaction by using techniques such as yeast two-hybrid analysis, domain swapping and site-specific mutagenesis.
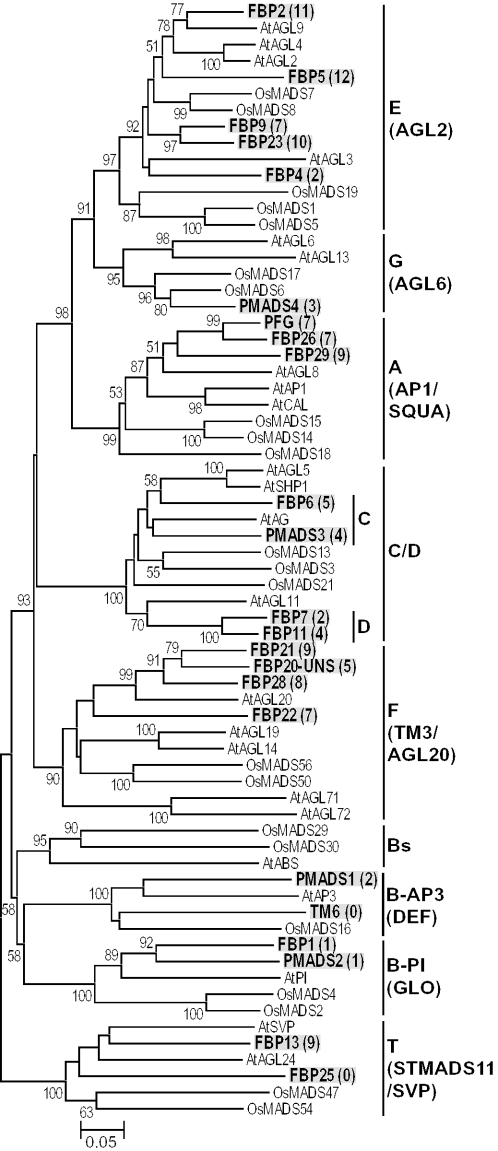
Immink et al. (29) studied the gene expression and protein–protein interaction patterns of 23 floral MADS-box genes in petunia using northern hybridization and yeast Cytotrap experiments. They identified a number of MADS-box proteins interacting with each other (see Figure 2). Their results showed that even closely related MADS-box proteins often have different numbers of protein interaction partners. This suggests that there was some kind of functional differentiation between these MADS-box proteins. We therefore decided to apply our new statistical method for predicting protein regions responsible for the functional differentiation using our Perl script (see http://www.bio.psu.edu/People/Faculty/Nei/Lab/software.htm). We first constructed a phylogenetic tree for the 23 petunia MADS-box protein sequences together with 22 rice and 23 Arabidopsis sequences. The rice and Arabidopsis sequences were used to classify the petunia sequences into the eight classes of genes mentioned above and to find proper outgroup genes.
Figure 2.
Phylogenetic tree of 68 MIKC-type MADS-box genes from petunia, Arabidopsis and rice. This tree was constructed by the neighbor-joining method with p-distance. One hundred fifty one amino acids were used after removing all alignment gaps. The number for each interior branch is the percent bootstrap value (500 bootstraps). The bootstrap values <50% are not shown. The genes in bold characters with gray shadows are from petunia, and ‘At’ and ‘Os’ indicate Arabidopsis and rice genes, respectively. The numbers in parentheses refer to the numbers of interacting protein partners in the yeast Cytotrap system described in (29).
Figure 2 shows the phylogenetic tree obtained by the neighbor-joining method (30) for all 68 genes (see the Supplementary Material for the sequence alignment). Parsimony analysis (31) produced essentially the same tree (see the Supplementary Material for the maximum-parsimony tree). The topology of this tree for major gene classes is essentially the same as that of our previous trees for floral MADS-box genes (15,32), and the eight gene classes (A, B, C, D,…,T) form separate monophyletic classes, though class C and D genes often form a mixed group and class B genes are decomposed into three classes (Bs, B-AP3 and B-PI) in Figure 2. This indicates that petunia also has all classes of genes (Figure 2). The number of sequences for classes A, B-AP3, B-AP1, C, D, E, F, G and T were 3, 2, 2, 2, 2, 5, 4, 1 and 2, respectively. We applied our statistical method for all gene pairs within each gene class, testing 23 pairs of genes (see the Supplementary Material for the 23 data sets). In this analysis, we considered consecutive windows with s = 1 and w = 30. We used the p-distance for this analysis. According to this analysis, 14 out of the 23 pairs of genes studied contained protein regions that showed at least one window with a Z-value exceeding 1.96 (5% level).
The results of our test for a pair of class T genes (FBP25 and FBP13) are given in Figure 1c. In this case, the rice gene OsMADS47 and OsMADS54 were used as outgroups. The Z-value line in this figure shows three peaks in which Z exceeds the 5 and 10% (Z = 1.65) significance levels (one each in the I, K and C domains). As mentioned earlier, the I and K domains are important for homodimerization and heterodimerization of MADS-box proteins, whereas the C domain is involved in transcriptional activation in some proteins. It is possible that all the three domains are involved in the functional differentiation between FBP25 and FBP13. It is also interesting to note that protein FBP13 is known to have nine protein interaction partners, whereas protein FBP25 has no known interacting partners (29).
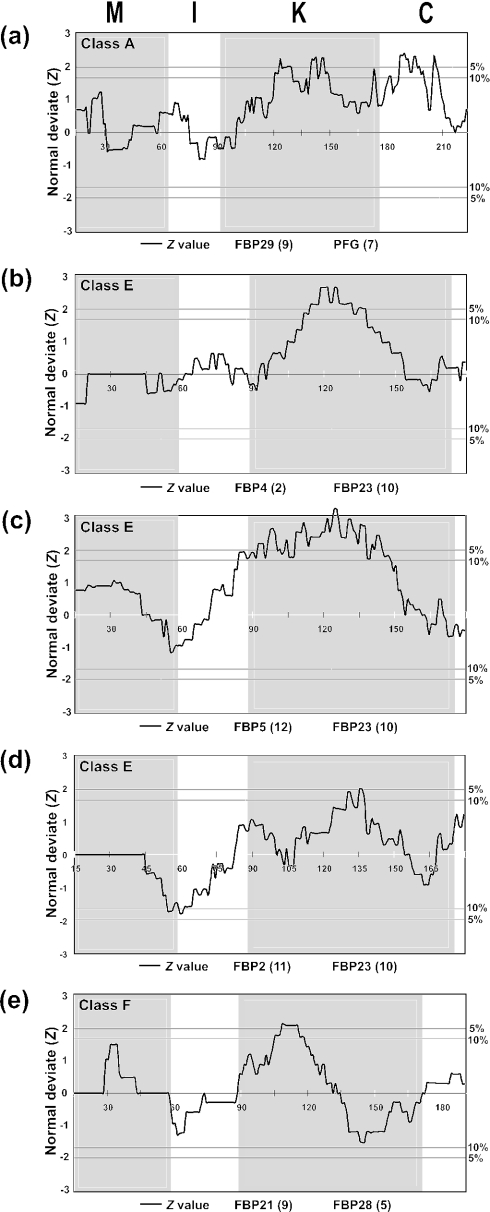
Figure 3 shows five more examples of our test in which Z became significant at the 5% level. The results of this analysis for a pair of class A genes (FBP29 and PFG) are presented in Figure 3a. In this case, the K domain has two peaks in which Z exceeds the 10% level. These peaks are located in a 30 amino acids region of the K domain. Therefore, experimentalists may focus on this region if they are interested in finding functional differentiation. The C domain also has two peaks of Z values which are significant at the 10% level. Therefore, the C domain may also be tested for the possible functional differentiation. In the other four examples given in Figure 3, only the K domain appears to have diverged significantly.
Figure 3.
Five more cases in which significant rate differences were observed at the 5% level. The outgroup sequences used for each analysis are as follows: (a) OsMADS14/15/18, (b–d) OsMADS1/5/19 and (e) AGL20.
As mentioned above, there were eight more cases in which our test gave positive results (see the Supplementary Material). In most of these cases, the K domain again showed a Z-value significant at the 5 or 10% level, though the M or I domain occasionally showed a significant region.
DISCUSSION
In our statistical analysis, we implicitly assumed that different amino acid sites have evolved independently. If there are highly conserved regions or hyper-variable regions in the proteins studied, our test would not give accurate significance levels, and the test will be too liberal or too conservative depending on the data set. For example, if conserved protein regions are studied, the test results may be too liberal because some amino acid sites may not have changed at all and therefore the actual number of degrees of freedom may be smaller than w − 1. In contrast, if a differentiated protein region is longer than the window size, the test will be conservative because a test based on the entire protein region would give a higher Z-value than the regular window test due to the smaller sampling variance of D. However, since our test is intended to identify approximate protein regions to be tested biochemically, it does not need to be very accurate in terms of the statistical significance.
It should be noted that the positive results of our test do not necessarily mean that the identified regions are functionally differentiated. Therefore, if the biochemical test to be used is available, it is always recommended that both statistical and biochemical tests should be conducted. It should also be noted that our functional differentiation test is not necessarily related to the positive Darwinian selection examined by the ratio of the number of nonsynonymous nucleotide substitutions per nonsynonymous site (dN) to the number of synonymous nucleotide substitutions per synonymous site (dS). We are only interested in the functional differentiation of duplicate genes whether the dN/dS is higher than 1 or not. Actually, the functional change of a gene may have been caused by a few amino acid changes in the functionally important region or by many substitutions in other regions. Here, dS is irrelevant under the assumption that synonymous nucleotide substitutions are neutral. Strictly speaking, this assumption is incorrect [e.g. (33,34)], but for our purpose the violation of this assumption is not important.
When we applied our method to floral MADS-box genes, we found that the extent of the difference of evolutionary rate is not necessarily correlated with the number of interacting protein partners. This is different from some of the previous observations that the extent of evolutionary rate differences is sometimes negatively correlated with the number of protein partners (35). This difference could be due to the fact that we studied a specific protein group or may mean that our test does not necessarily detect the region where functional differentiation has occurred. These problems should be studied experimentally in the future.
It is interesting to note that functional specificity of class A, B and C genes in Arabidopsis is not determined by their DNA binding domain (36). Therefore, I, K and C domains may be critical for determining functional specificity of floral MADS-box genes. In our study, the difference of evolutionary rate was often observed in the K domain, while it was not observed as often in the I and C domains. It has been proposed that internal repeats of proteins give favorable conditions for evolutionary change, because their functional constraint may change with time (37). Therefore, the frequent observation of significant rate differences in the K domain may be related to the presence of heptad repeats, which can be subdivided into the K1, K2 and K3 regions (25). Because the M, I and C domains also showed significant rate differences in some pairs, it is possible that these domains have also been subject to the functional differentiation. If functional differentiation occurs in different domains of a protein, the effect of such combinatorial differentiation on the regulatory network may be more significant than the case where only one domain is functionally differentiated. Our method may be useful for studying this problem as well.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Hong Ma, Jianzhi (George) Zhang, Yoshihito Niimura, Xun Gu and two anonymous reviewers for valuable comments on an earlier version of this paper. This study was supported by National Institutes of Health Grant GM20293 (to M.N.). J.N. had a scholarship from the Rotary Foundation, and K.K. a short-term fellowship from the German Academic Exchange Service (DAAD). Funding to pay the Open Access publication charges for this article was provided by NIH grant GM20293.
REFERENCES
- 1.Lewis E.B. Pseudoallelism and gene evolution. Cold Spring Harbor Symp. Quant. Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Nei M. Gene duplication and nucleotide substitution in evolution. Nature. 1969;221:40–42. doi: 10.1038/221040a0. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 4.Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- 5.Dermitzakis E.T., Clark A.G. Differential selection after duplication in mammalian developmental genes. Mol. Biol. Evol. 2001;18:557–562. doi: 10.1093/oxfordjournals.molbev.a003835. [DOI] [PubMed] [Google Scholar]
- 6.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 7.Knudsen B., Miyamoto M.M. A likelihood ratio test for evolutionary rate shifts and functional divergence among proteins. Proc. Natl Acad. Sci. USA. 2001;98:14512–14517. doi: 10.1073/pnas.251526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nei M., Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Press; 2000. [Google Scholar]
- 9.Takezaki N., Rzhetsky A., Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol. Biol. Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 10.Rao P.V. Statistical Research Methods in the Life Sciences. Pacific Grove CA: The Brooks/Cole Publishing Company; 1998. [Google Scholar]
- 11.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 12.Hughes A.L., Ota T., Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol. Biol. Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
- 14.Becker A., Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 15.Nam J., dePamphilis C.W., Ma H., Nei M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003;20:1435–1447. doi: 10.1093/molbev/msg152. [DOI] [PubMed] [Google Scholar]
- 16.Ma H., Yanofsky M.F., Meyerowitz E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- 17.Shore P., Sharrocks A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 18.Riechmann J.L., Wang M., Meyerowitz E.M. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honma T., Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 20.Moon Y.H., Kang H.G., Jung J.Y., Jeon J.S., Sung S.K., An G. Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol. 1999;120:1193–1204. doi: 10.1104/pp.120.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S., Jang S., Chae S., Chung K.M., Moon Y.H., An G., Jang S.K. Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol. 1999;40:419–429. doi: 10.1023/a:1006273127067. [DOI] [PubMed] [Google Scholar]
- 22.Fan H.Y., Hu Y., Tudor M., Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997;12:999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x. [DOI] [PubMed] [Google Scholar]
- 23.Tzeng T.Y., Liu H.C., Yang C.H. The C-terminal sequence of LMADS1 is essential for the formation of homodimers for B function proteins. J. Biol. Chem. 2004;279:10747–10755. doi: 10.1074/jbc.M311646200. [DOI] [PubMed] [Google Scholar]
- 24.Egea-Cortines M., Saedler H., Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus . EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Fanning L., Jack T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003;33:47–59. doi: 10.1046/j.0960-7412.2003.01473.x. [DOI] [PubMed] [Google Scholar]
- 26.Weigel D., Meyerowitz E.M. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 27.Ma H., dePamphilis C. The ABCs of floral evolution. Cell. 2000;101:5–8. doi: 10.1016/S0092-8674(00)80618-2. [DOI] [PubMed] [Google Scholar]
- 28.Theissen G. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 2001;4:75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 29.Immink R.G., Ferrario S., Busscher-Lange J., Kooiker M., Busscher M., Angenent G.C. Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics. 2003;268:598–606. doi: 10.1007/s00438-002-0781-3. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Swofford D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 32.Nam J., Kim J., Lee S., An G., Ma H., Nei M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl Acad. Sci. USA. 2004;101:1910–1915. doi: 10.1073/pnas.0308430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 34.Akashi H. Inferring weak selection from patterns of polymorphism and divergence at ‘silent’ sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hahn M.W., Conant G.C., Wagner A. Molecular evolution in large genetic networks: does connectivity equal constraint? J. Mol. Evol. 2004;58:203–211. doi: 10.1007/s00239-003-2544-0. [DOI] [PubMed] [Google Scholar]
- 36.Riechmann J.L., Meyerowitz E.M. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell. 1997;8:1243–1259. doi: 10.1091/mbc.8.7.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrade M.A., Perez-Iratxeta C., Ponting C.P. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.