Abstract
This review focuses on the construction and application of structural chemokine receptor models for the elucidation of molecular determinants of chemokine receptor modulation and the structure-based discovery and design of chemokine receptor ligands. A comparative analysis of ligand binding pockets in chemokine receptors is presented, including a detailed description of the CXCR4, CCR2, CCR5, CCR9, and US28 X-ray structures, and their implication for modeling molecular interactions of chemokine receptors with small-molecule ligands, peptide ligands, and large antibodies and chemokines. These studies demonstrate how the integration of new structural information on chemokine receptors with extensive structure–activity relationship and site-directed mutagenesis data facilitates the prediction of the structure of chemokine receptor–ligand complexes that have not been crystallized. Finally, a review of structure-based ligand discovery and design studies based on chemokine receptor crystal structures and homology models illustrates the possibilities and challenges to find novel ligands for chemokine receptors.
1. Introduction
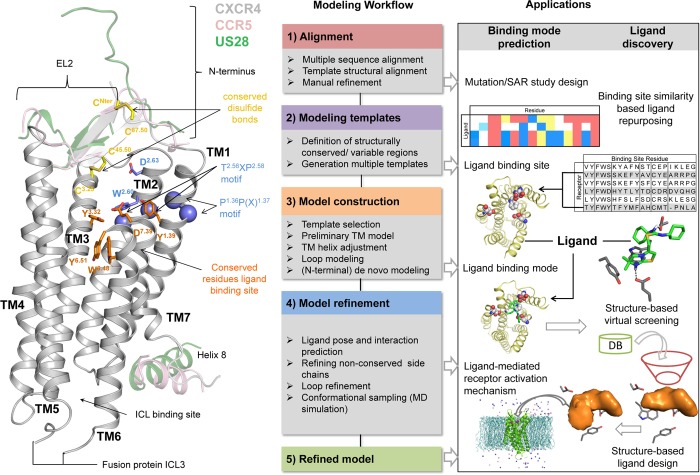
Chemokines and chemokine receptors play an important role in the immune defense system by controlling the migration, activation, differentiation, and survival of leukocytes.1,2 The 50 human chemokines are divided into C, CC, CXC, and CX3C classes based on the number and spacing of conserved cysteine residues in their N-terminus region. Chemokine receptors belong to the family A of G-protein coupled receptors (GPCRs), characterized by a seven transmembrane (7TM) helical domain (Figure 1). There are 18 human chemokine receptors that are primarily activated by different subfamilies of chemokines: C (XCR1), CC (CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, CCR10), CXC (CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, CXCR6), or CX3C (CX3CR1), and four atypical decoy chemokine receptors (ACKRs: ACKR1, ACKR2, ACKR3/CXCR7, and ACKR4).3 Chemokine receptors are considered to interact with their chemokine ligands via a two-step binding mechanism in which: (i) the structured C-terminal region of the chemokine first binds the N-terminus region and extracellular loops (ECLs) of the receptor (chemokine recognition site 1, CRS1), allowing (ii) the unstructured N-terminus of the chemokine to target the 7TM helical bundle (chemokine recognition site 2, CRS2) and stabilize the receptor in an active conformation that facilitates intracellular signal transduction by, e.g., G-proteins or arrestins.1,4 Because of their crucial role in cell migration chemokine receptors are important therapeutic targets for inflammatory diseases and cancer.5,6 Herpesviruses contain DNA that encodes for receptors that are similar to human chemokine receptors, including ORF74, BILF1, and US28, to hijack chemokine receptor-mediated cellular signaling networks of the host.7 Hence, these viral chemokine receptors can therefore be considered as promising antiviral drug targets as well.8 A variety of proteins, peptides, and small-molecule ligands have been identified that can modulate the activity of chemokine receptors1 by targeting the minor or major pockets in the 7TM helical bundle or intracellular binding pocket (Figures 1–2). Examples of small nonpeptide ligands are the clinically approved drugs 16 (Maraviroc, CCR5 antagonist, Figures 3 and 11)9 and 1 (plerixafor/AMD3100, CXCR4 antagonist, Figure 11),10 used for the treatment of HIV and stem cell mobilization, respectively. Molecular pharmacological, medicinal chemistry, and molecular modeling studies have provided insights into molecular determinants of chemokine receptor modulation1,2,4 and in the past few years the first high-resolution crystal structures of chemokine receptors have been solved that give more detailed structural information on the interaction of chemokine receptors and their ligands.11−16 The current review describes how the combination of these three-dimensional structural templates with extensive pharmacological data provide new possibilities to investigate the determinants of chemokine receptors modulation and ligand binding in more detail and to exploit this knowledge for computer-aided discovery of new chemokine receptor ligands.
Figure 1.
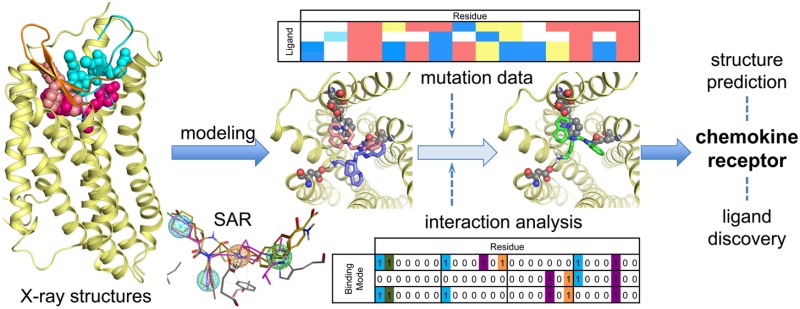
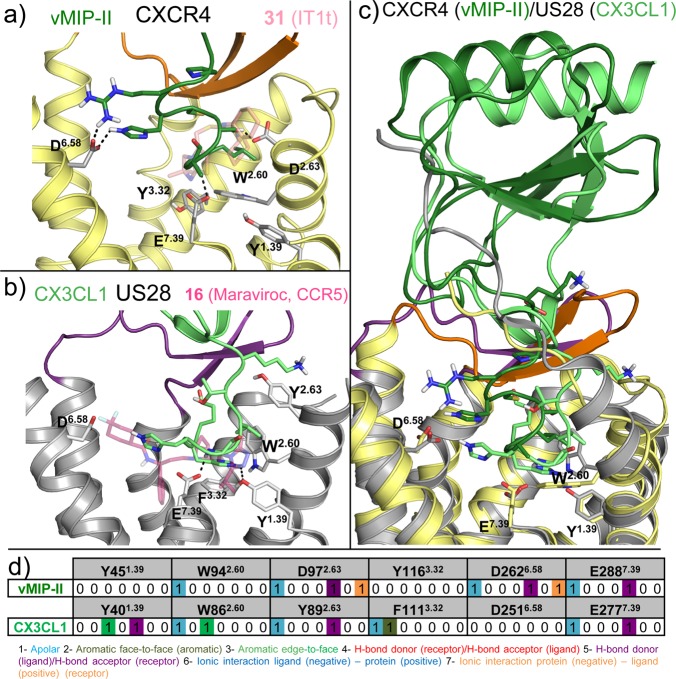
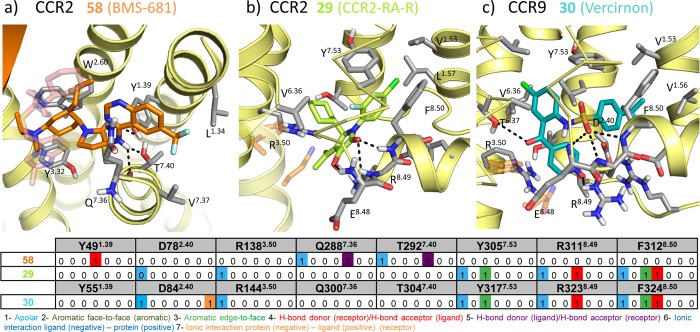
Chemokine receptor X-ray structures. (a) Alignment of 31 (PDB 3ODU;11 pink spheres), CVX15 (PDB 3OE0;11 cyan spheres), and (b) vMIP-II (PDB 4RWS;13 dark-green cartoon and spheres) bound CXCR4 crystal structures. The receptor is colored for a better interpretation: 3ODU in light yellow, 3OE0 in gray. TM helices align well in the three different reported structures with subtle differences: TM1 is one turn longer (R30N-ter–N33N-ter) and laterally shifted outward in the vMIP-II bound CXCR4 structure, TM6 is half turn shorter in the 31 bound CXCR4 structure (H2326.28–Q2336.29), helix 8 is missing in all the structures, and the C-terminus has only been solved for the 31 bound CXCR4 structure (A307C-ter–S319C-ter). vMIP-II targets both the chemokine recognition site 1 (CRS1, comprising the N-terminus and extracellular loops of the receptor) and the chemokine recognition site 2 (CRS2, including the TM domain binding site) of CXCR4, consistent with the two-step binding model. (c) An active conformation of US28, a viral chemokine-like receptor, binding the human CX3CL1 chemokine in the extracellular binding site, and a nanobody (Nb7, purple cartoon) in the intracellular binding site (PDB 4XT1;14 green cartoon and spheres). Both chemokines vMIP-II (a) and CX3CL1 (c) are shown as spheres on their N-terminus coils, and their globular cores are shown as a cartoon for a better visualization of their secondary structure. (d) CCR5 crystal structure bound to the small ligand 16 (PDB 4MBS;12 magenta spheres), occupying both the transmembrane site 1 (TMS1), also known as small pocket, and transmembrane site 2 (TMS2), or major pocket. (e) CCR9 crystal structure bound to the small allosteric antagonist 30 (PDB 5LWE,16 dark-cyan spheres) targeting an intracellular allosteric intracellular pocket and thereby blocking G-protein coupling. (f) CCR2 crystal structure bound to the orthosteric antagonist 58 (orange spheres) and the allosteric antagonist 29 (lime spheres) targeting an intracellular binding pocket (PDB 5T1A(15)). (g) Summary of interactions observed in the CXCR4, CCR5, US28, CCR2, and CCR9 crystal structures. The background of the amino acid residue positions is colored according to the different binding site regions (defined in panel a), amino acid residues involved in receptor–ligand interactions are depicted in bold and colored according to the cocrystallized ligand with which they interact. More detailed analyses of the structural receptor–ligand interactions are provided in Figures 3–5. Two-dimensional representations of the chemical structures of the small-molecule ligands 16, 29, 30, 31, and 58 are provided in Figures 11–13
Figure 2.
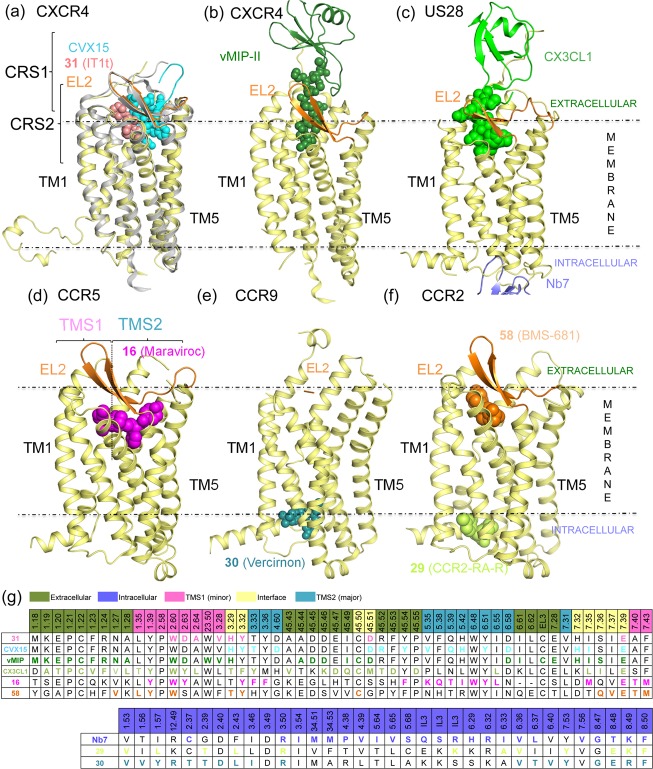
Structure-based sequence alignment (in line with GPCRdb49) of chemokine receptors for which crystal structure and/or site-directed mutagenesis information on small-molecule ligand binding is available (described in sections 2–5), including CCR1,50,51 CCR2,15,52,53 CCR5,54−58 CCR8,59 CCR9,16 CXCR2,60−62 CXCR3,63−67 CXCR4,21,68−74 and US28.75 Amino acid residues in CXCR4, CCR2, CCR5, CCR9, and US28 that are involved in receptor–ligand interactions are highlighted in bold and colored corresponding to the cocrystallized ligands 29 and 58 (in CCR2), 16 (CCR5), 30 (CCR9), 31, CVX15 and vMIP-II (CXCR4), and CX3CL1 and Nb7 (US28) according to the color coding in Figure 1. More detailed analyses of the structural receptor–ligand interactions are provided in Figures 3–5. Two-dimensional representations of the chemical structures of the small-molecule ligands 16, 29, 30, 31, and 58 are provided in Figures 11–13. The background of residues for which site-directed mutagenesis data have been reported is marked gray.
Figure 3.
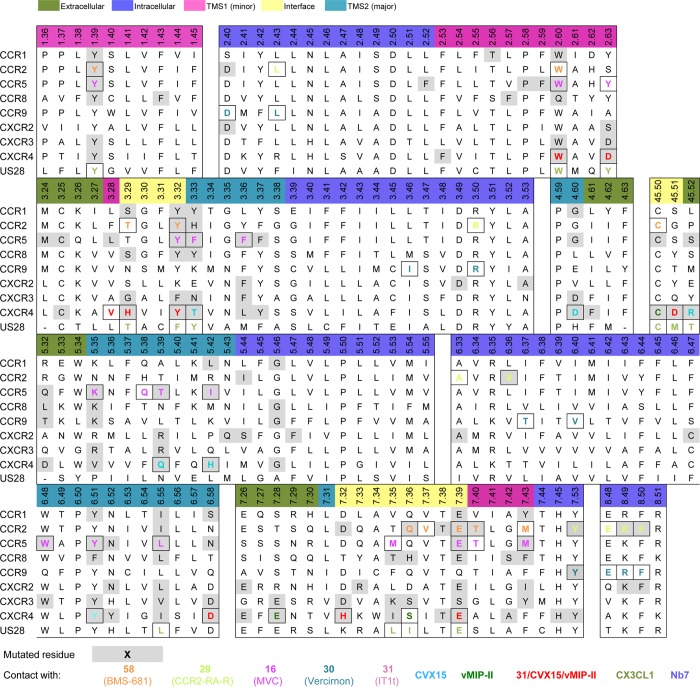
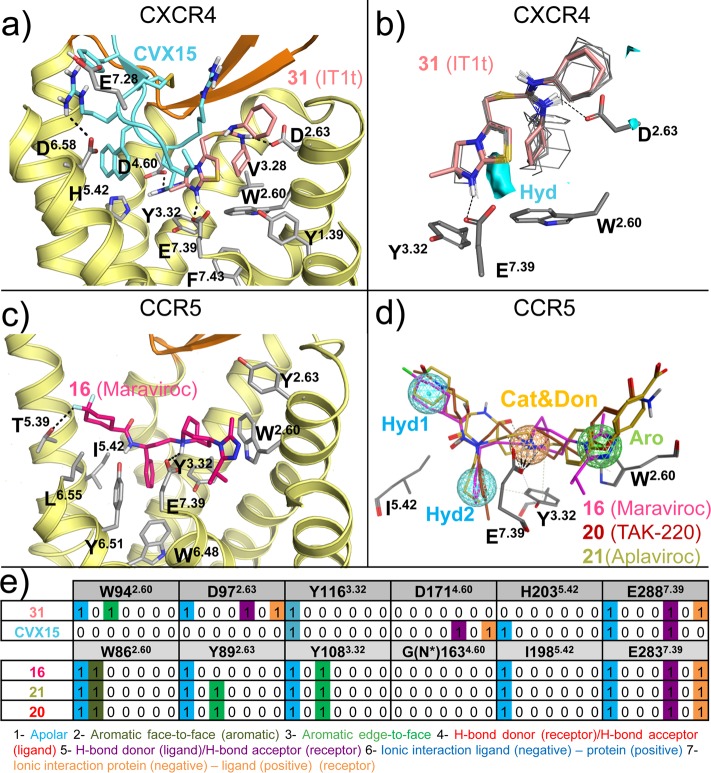
(a) Detailed comparison of the binding modes of 31 (PDB 3ODU) and CVX15 (PDB 3OE0) in CXCR4 crystal structures.11 The small-molecule antagonist 31 (pink carbon atoms) binds the minor binding pocket (TMS1) of CXCR4, whereas the peptide antagonist CVX15 (cyan) mainly targets in the major binding pocket (TMS2). (b) Three-dimensional quantitative structure–activity relationship (3D-QSAR) model of 13 analogues of CXCR4 antagonist 31(78) constructed using FLAP79,80 based on an alignment to the cocrystallized pose of 31 in CXCR4,11 indicating that a hydrophobic interaction field between the methyl groups of the imidazothiazole ring system (cyan surface) and the six-membered ring is an important determinant for binding the minor binding pocket of CXCR4. (c) Detailed analysis of the binding mode of 16 (magenta carbon atoms) targeting both the minor and the major binding pockets in the CCR5 crystal structure (PDB 4MBS).12 (d) Ligand-based pharmacophore model of some of the most representative CCR5 small ligands 16,8120 (TAK-220),82 and 21 (Aplaviroc),83 including four pharmacophore features: two apolar/hydrophobic moieties (Hyd1, Hyd2), a hydrogen bond acceptor/cationic feature (Cat&Don), and an aromatic (Aro) feature. The residues corresponding to the 16 bound CCR5 crystal structure potentially interacting with the model are shown as gray sticks. (e) Comparative structural interaction fingerprint (IFP) analysis of the binding modes of 31 and CVX15 in CXCR4 and 16, 20, and 21 in CCR5, presented in panels a, c, and d. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between the ligand and the different CXCR4/CCR5 amino acid residues. Two-dimensional representations of the chemical structures of the small-molecule ligands 16, 20, 21, and 31 are provided in Figures 11,12.
Figure 11.
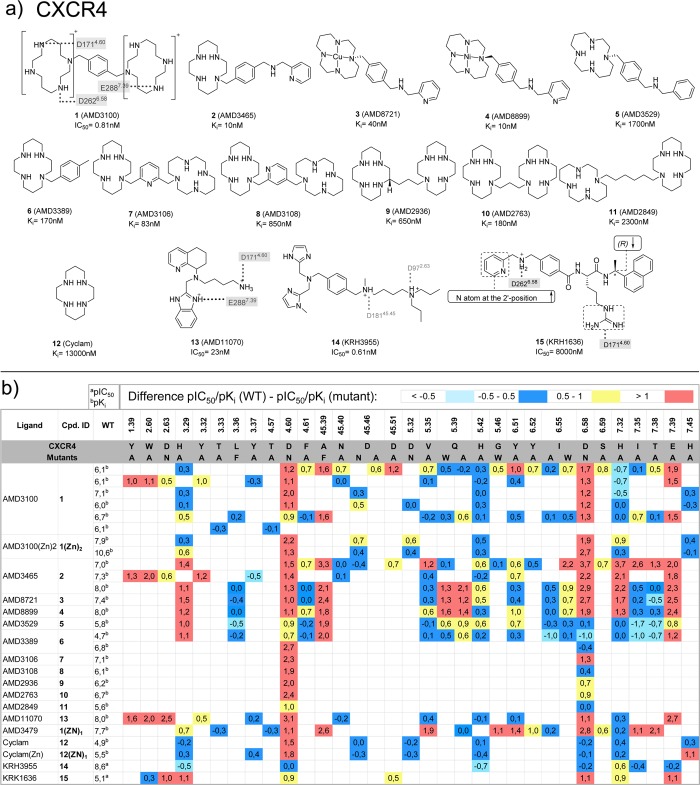
(a) Chemical structures of CXCR4 ligands investigated in CXCR4 mutation studies.21,68−73 Interactions between the ligands and specific residues derived from CXCR4 X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (b) Differences between the pIC50a or pKib values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 276 CXCR4 mutant–ligand combinations covering ligands 1–15 and 33 CXCR4 mutants (annotated mutation data set included in Supporting Information).
NMR and X-ray crystallography structures of CX3CL1, CCL11, CXCL8, CXCL12, and vMIP-II chemokines have provided insights into the secondary structure of the different chemokine families and the structural interactions of chemokines with the N-terminal regions of US28, CCR3, CXCR1, and CXCR4 chemokine receptors.17−19 The first crystal structures of the 7TM domains of chemokine receptors (CXCR4, CCR2, CCR5, CCR9, and US28) have been solved only in the past six years.11−16 These small-molecule,12,15,16 peptide,11 and chemokine13,14 bound structures have provided a structural basis to validate and improve chemokine receptor homology modeling studies,20 to rationalize SAR data21,22 and to perform structure-based virtual screening and ligand design studies.23−30 Chemokine homology models and de novo receptor models have already been successfully used to identify new ligands for CCR3,31 CCR4,32,33 CCR5,34,35 CXCR3,26 CXCR4,24,26−30,36,37 and CXCR7,38 and the recently released crystal structures have increased the possibilities to study and predict structural chemokine receptor–ligand interactions. This review will present a comparative analysis of ligand binding pockets in chemokine receptors, including a review of the CXCR4, CCR2, CCR5, CCR9, and US28 X-ray structures (section 2), and their implication for modeling interactions between chemokine receptors and small-molecules and larger peptide-like and chemokine ligands (sections 3–6). We will demonstrate how the combination of these chemokine receptor structures with extensive structure–activity relationship and site-directed mutagenesis data can be used to rationalize and predict structural determinants of chemokine receptor modulation (sections 3 and 4) and can facilitate the construction of structural models of chemokine receptor–ligand complexes that have not been crystallized yet (section 5). Finally, a review of virtual screening studies based on chemokine receptor crystal structures and homology models will be provided as a basis to discuss the possibilities and challenges of structure-based chemokine receptor ligand discovery (section 6).
2. Analysis of Chemokine Receptor Crystal Structures
To allow systematic comparison of the residues at different positions in the TM helices of different GPCRs, receptor residue numbers are annotated throughout this review by their Uniprot numbers (for specific receptors) as well as their Ballesteros–Weinstein residue number and secondary structure motif (as superscript).39,40 According to the Ballesteros–Weinstein rhodopsin family (class A) GPCR39 residue numbering schemes, the single most conserved residue in each TM helix is designated X.50.40 For the ECL2 and ECL3, similar residue numbering schemes have been applied. ECL2 residues are labeled 45.X, and the reference residue C45.50 is a conserved cysteine forming a disulfide bridge with C3.25 in TM3.41 ECL3 residues are labeled 67.X, and the reference residue C67.50 is a conserved cysteine forming a disulfide bridge with a conserved cysteine residue in the N-terminus of most chemokine receptors (with the exception of CXCR6). To distinguish receptor residues from chemokine/peptide ligand residues, receptor residues are annotated as single-letter amino acid codes, while peptide and chemokine ligand residues are annotated as three-letter amino acid codes with residue number as superscripts (e.g., Arg1, arginine at position 1 in the 16-residue cyclic peptide ligand CVX15, His6 in the chemokine ligand vMIP-II).
2.1. CXCR4, CCR2, CCR5, CCR9, and US28 Chemokine Receptor Crystal Structures
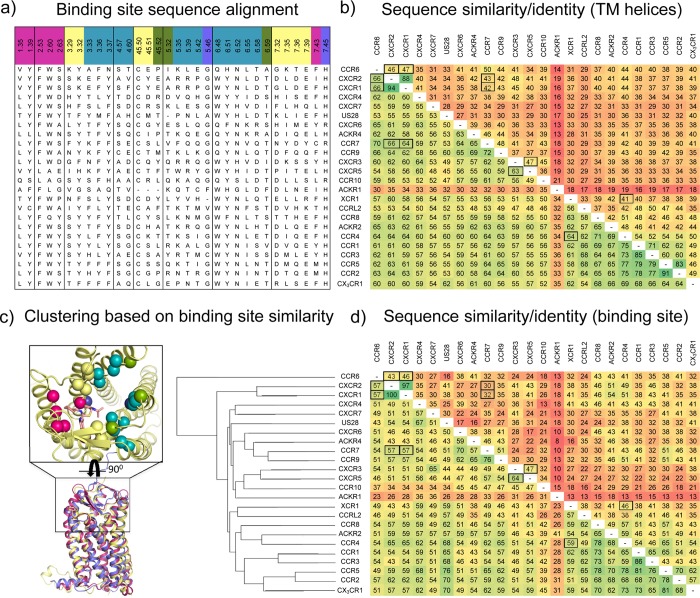
The endogenous chemokine ligand of the CXCR4 chemokine receptor is CXC chemokine ligand 12 (CXCL12), also known as stromal derived factor-1 (SDF-1). CXCR4:CXCL12 signaling axis plays a role in several inflammatory diseases and cancers.42 CXCR4 was the first chemokine receptor reported to be a coreceptor for HIV-1.43 The marketed drug 1(10) targets CXCR4 to mobilize hematopoietic stem cells in the treatment of patients with non-Hodgkin lymphoma (NHL) and multiple myeloma (MM). Three crystal structures have been reported for CXCR4 fused to the T4 lysozyme (T4L) with different cocrystallized ligands (Figure 1a):11,13 the small ligand 31 (IT1t) (PDBs 3ODU, 3OE6, 3OE8, and 3OE9, Figures 1a, 3a), the 16-residue cyclic peptide ligand CVX15 (PDB 3OE0, Figures 1a, 3a), and the viral CC chemokine vMIP-II (PDB 4RWS, Figure 1b, 4a,c)). CCR2 binds CCL2, CCL7, CCL9, CCL11, CCL12, CCL13, CCL24, and CCL26 CC chemokines1 and is implicated in inflammatory and neurodegenerative diseases.44 Recently a crystal structure of T4 lysozyme fused CCR2 has been reported in a ternary complex with an orthosteric antagonist 58 (BMS-681)45 and an allosteric antagonist 29 (CCR2-RA-[R])46 bound to an intracellular pocket (PDB 5T1A, Figures 1f, 5).15 CCR5 binds CCL2, CCL3, CCL4, CCL5, CCL8, CCL11, CCL13, and CCL14 CC chemokines and is the major HIV-1 coreceptor.1 The FDA approved CCR5 antagonist 16(9) is used for the treatment of patients with HIV (R5-tropic HIV-1), and a crystal structure of rubredoxin fused CCR5 bound to 16 (PDB 4MBS, Figures 1d, 3b) has been reported.12 CCR9 activation by CCL25 plays a key role in leukocyte recruitment to the gut and is a therapeutic target in inflammatory bowel disease.47 A CCR9 crystal structure has recently been solved, including seven thermostabilizing mutations, in complex with the selective allosteric CCR9 antagonist 30 (Vercirnon) bound to the same intracellular pocket as 29 in the CCR2 crystal structure (PDB 5LWE, Figures 1e, 5).16 US28 is a chemokine-like receptor encoded by the human cytomegalovirus that binds different chemokines such as CX3CL1, CCL5, CCL2, and CCL3, among others, in part as a strategy to evade the host immune system.48 Two crystal structures have been reported for US28 bound to the human CX3CL1 chemokine,14 one with the nanobody 7 (Nb7) (PDB 4XT1, Figure 1c) bound to the intracellular surface of the receptor, and the other one without the nanobody (PDB 4XT3). US28 shares lower sequence similarity with CXCR4 (30%) and CCR5 (27%) than with the human CX3CR1 chemokine receptor (38%). The US28 crystal structure nevertheless shares a similar fold with CXCR4, CCR2, CCR5, and CCR9 structures, which allows comparison of the ligand binding modes and receptor conformations (Figure 1–4). Figure 2 shows a structure-based sequence alignment of chemokine receptors.
Figure 4.
Details of chemokine binding to CXCR4 (PDB 4RWS(13)) and US28 (PDB 4XT1(14)). (a) vMIP-II N-terminus binding to CXCR4 (pale yellow).13 The N-terminus of vMIP-II (dark-green sticks) binds primarily in the minor pocket that is also targeted by 31 (transparent pink sticks), interacting with W942.60, D972.63, and E2887.39 but also partially binds the major binding site, interacting with D2626.58. (b) CX3CL1 N-terminus (light green sticks) binding to US28 (gray).14 CX3CL1 N-terminus, as well as vMIP-II, binds mainly in the small binding site, interacting with Y401.39, T17545.52, and E2777.39 but also partially occupies the major binding site. CCR5 antagonist 16 is shown as transparent magenta sticks as reference. (c) vMIP-II (dark green) and CX3CL1 (green) superimposition. The overall architecture is conserved: the N-terminus inside the TM domain and the core to the extracellular surface (CRS1). (d) Comparative structural interaction fingerprint (IFP) analysis of the binding modes of vMIP in CXCR4, and CX3CL in US28, presented in panels a–c. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between the ligand and the different CXCR4/US28 amino acid residues. Two-dimensional representations of the chemical structures of the small-molecule ligands 16 and 31 are presented in Figures 11, 12
Figure 5.
Details of small ligand binding to CCR2 (PDB 5T1A(15)) and CCR9 (PDB 5LWE(16)). (a) Structural interactions between the orthosteric antagonist 58 (orange sticks) and the minor binding pocket of CCR2. CXCR4 antagonist 31 (pink sticks) is shown transparent as reference. (b) Structural interactions between the allosteric antagonist 29 (lime sticks) and the allosteric intracellular pocket of CCR2. Residues from the G-protein in the G-protein bound ADRB2 structure87 are shown in transparent orange sticks as reference. (c) Structural interactions between the allosteric antagonist 30 (dark-cyan sticks) and the intracellular allosteric pocket of CCR9. Residues from the G-protein in the G-protein bound ADRB2 structure87 are shown in transparent orange sticks as reference. (d) Comparative structural interaction fingerprint (IFP) analysis of the binding modes of 58 and 29 in CCR2, and 30 in CCR9, presented in panels a–c. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between the ligand and the different CCR2/CCR9 amino acid residues. Two-dimensional representations of the chemical structures of the small-molecule ligands 29, 30, 31, and 58 are provided in Figure 13.
2.2. Comparison of Ligand Binding Modes in Chemokine Receptor Crystal Structures
The 7TM ligand binding pockets of CXCR4, CCR5, US28, CCR2, and CCR9 chemokine receptor crystal structures are less buried and more solvent accessible than in the crystal structures of other crystallized class A GPCRs.76,77 The more open pocket of chemokine receptors is covered to a lesser extent by the extracellular loop 2 (ECL2) than in most other GPCR crystal structures (not solved in CCR9 crystal structure16), which can be related to the fact that chemokine receptors bind chemokines that are larger than the natural ligands of other receptors. In addition, the binding pockets of chemokine receptors contain several negatively charged residues that are involved in ligand binding, as observed in crystal structures (Figures 1–5) and indicated by mutation studies (Figures 10–13, 18–20).
Figure 10.
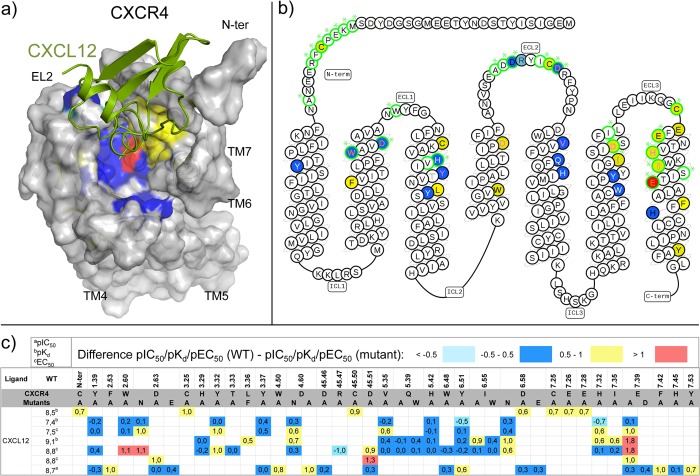
CXCL12 binding to CXCR4 based on site-directed mutagenesis assays. Differences between the pIC50a, pKdb, or pEC50c values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 90 CXCR4 mutant-CXCL12 combinations covering 42 residues (annotated mutation data set included in Supporting Information).68,69,105,106,108,111,112 Maximum mutation effects are mapped on: (a) CXCL12 (green) bound CXCR4 structure (modeled based on the vMIP-II bound CXCR4 crystal structure (PDB 4RWS),13 and (b) CXCR4 snakeplot adapted from GPCRdb.143 Residues involved in ligand interactions for the vMIP bound CXCR4 crystal structure are encircled in green. Effects on CXCL12 affinity and potency are annotated by background and amino acid color, respectively. Mutation data derived from antibodies inhibition binding is not shown. (c) Summary of CXCR4 site-directed mutagenesis effects on CXCL12 binding/potency in individual studies. A recent study has been published reporting single-point binding and functional data of all CXCR4 residues mutants, which has indicated that in particular W942.60 and D972.63 are critical for CXCL12-mediated signaling.74
Figure 13.
(a) Chemical structures of CCR252,53,93 and CCR916 ligands with related mutation data. Interactions between the ligands and specific residues derived from CCR5 X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (b) Affinities from site-directed mutagenesis studies on chemokine receptors. Differences between the pIC50a, pKib, or pKdc values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 57 CCR2 mutant–ligand combinations covering ligands 19, 25–29, and 18 mutants, and five CCR9 mutants on ligand 30 (annotated mutation data set included in Supporting Information).
Figure 18.
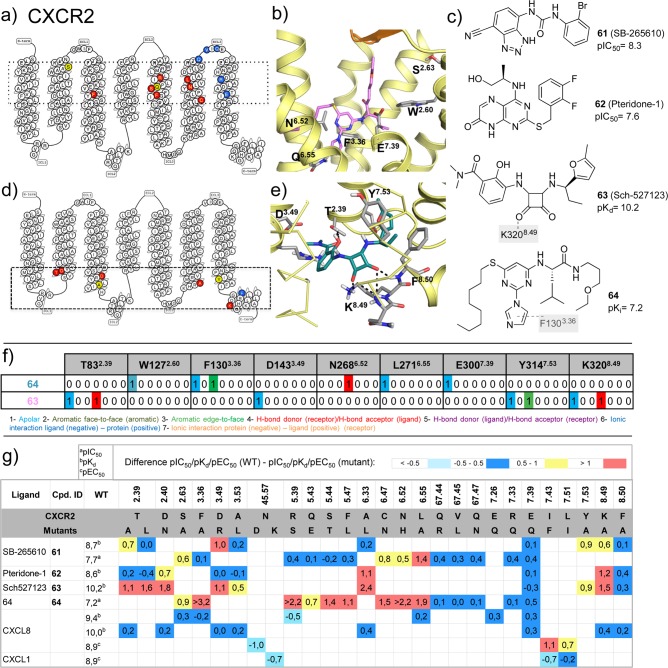
CXCR2 ligand binding model predictions based on molecular docking and site-directed mutagenesis assays.60−62 (a,d) CXCR2 snakeplots (adapted from GPCRdb49) summarizing the effects of mutation effects on affinity/potency of (a) 64 (imidazolylpirimidine) and (d) 63, suggesting that 64 and 63 target extracellular and intracellular binding sites of CXCR2, respectively (indicated by dotted boxes). (b,e) Structural details of the predicted binding modes of (b) 64 (pink) and (e) 63 (turquoise). (c) Chemical structures of CXCR2 ligands that have been investigated in mutation studies. Interactions between the ligands and specific residues derived from X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (f) Comparative structural interaction fingerprint (IFP) analysis of the predicted binding modes of 64 and 63 in CXCR2 presented in panels b and e. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between the ligand and the different CXCR2 amino acid residues. (g) Differences between the pKia, pKdb, or pEC50c values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 76 CXCR2 mutant–ligand combinations covering ligands 61–64, CXCL8, CXCL1, and 28 CXCR2 mutants (annotated data set included in Supporting Information).60−62
Figure 20.
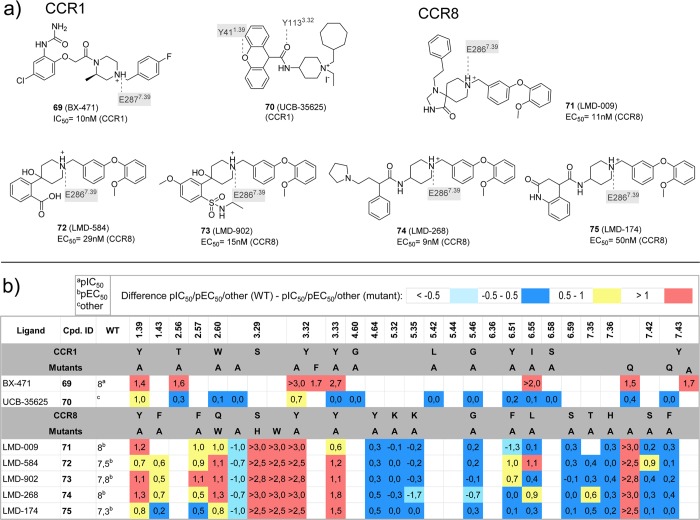
(a) Chemical structures of ligands investigated in CCR150,51 and CCR859 mutation studies. Interactions between the ligands and specific residues derived from mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (b) Differences between the pIC50a, pEC50b, −log[fold change % inhibition CCL3-induced chemotactic response] (indicated as other)c values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 22 CCR1 mutant–ligand combinations covering ligands 69–70 and 16 mutants,, and 103 CCR8 mutant–ligand combinations covering ligands 71–75 and 21 mutants (annotated mutation data set included in Supporting Information).
Small-molecule antagonists 31 and 58 exclusively bind the minor pockets (TMS1) of CXCR4 (Figures 1a, 2, 3a) and CCR2 (Figures 1f, 2, 5a), respectively, located in the core of the TM domain, comprised by TM helices 1–3 and 7, whereas the small-molecule antagonist 16 binds both the minor and the major pocket (TMS2, including residues of TM 3–7) of CCR5 (Figures 1d, 2, 3b). The N-termini of vMIP-II and CX3CL1 chemokines also bind both minor and major pockets of CXCR4 and US28, respectively (Figures 1, 2, 4). The peptidomimetic ligand CVX15 primarily targets the major pocket, (Figures 1a, 2, 3a). The globular cores of the chemokines interact with the extracellular surface of the receptors, including the top of the TM helices, the EC loops, and the receptor N-terminus (Figures 1, 3c, 8). Small-molecule allosteric antagonists 29(46) and 30 and the G-protein mimic Nb7 bind intracellular binding pockets (Figure 5) of CCR2, CCR9, and US28, respectively.
Figure 8.
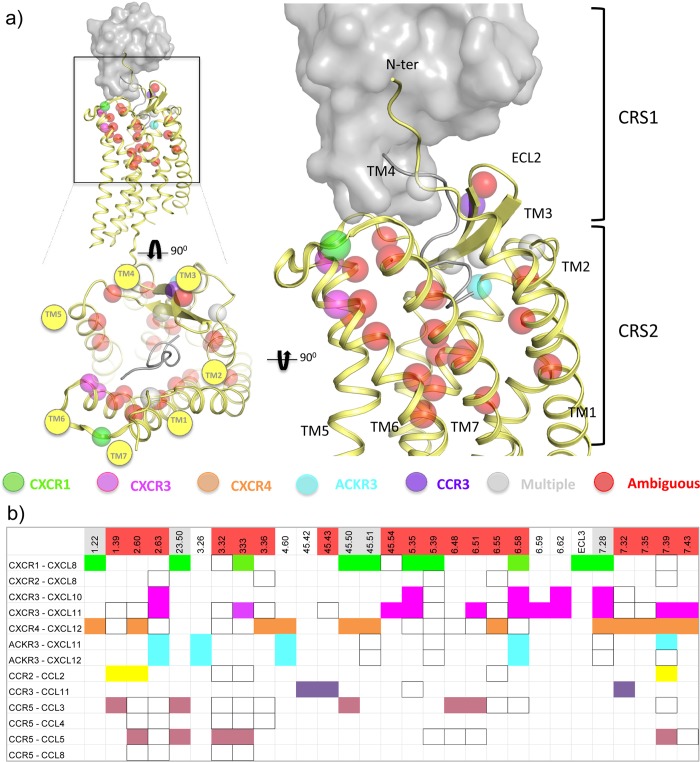
Molecular determinants of chemokine binding supported by site-directed mutagenesis data.52,53,55,58−61,63−66,68,69,90,94,95,103,105,108,112,115,125,126 (a) 3D representation of the reported receptor residues involved in chemokine binding. (b) Summary of the determinants of receptor:chemokine binding. Mutated positions that significantly decrease the binding of the chemokine are colored as follows: green for CXCR1, magenta for CXCR3, orange for CXCR4, yellow for CCR2, purple for CCR3, and blue for CCR5. Mutations of positions that affect the binding of more than one receptor:chemokine pair (multiple) are colored in gray, and those positions that when mutated affect the binding of some receptor:chemokine pairs but not other pairs (ambiguous), are colored in red. Those positions that have been mutated but do not decrease the binding of the chemokine are not colored, but squared.
2.2.1. Minor Pocket (TMS1)
In the minor pocket, one of the positively charged nitrogens of the isothiourea group of 31 and the N-terminus amino group of vMIP-II form a salt bridge with CXCR4 D972.63 (Figures 3–4). The other crystallized chemokine receptor ligands that bind in the minor pocket, 16, 58, and CX3CL1, do not interact with the residue at position 2.63 (a serine in CCR2, and a tyrosine in CCR5 and in US28). The conserved W2.60 residue is involved in interactions with all the ligands binding the minor pocket, including an edge-to-face aromatic interaction with one of the phenyl rings in the isothiourea group of 31 and an hydrophobic interaction with Leu1 of vMIP-II for CXCR4, a face-to-face aromatic interaction with the triazole ring of antagonist 16 in CCR5, an hydrophobic packing with the trisubstituted cyclohexane of compound 58 (Figure 5), and an edge-to-face aromatic interaction with His1 of CX3CL1 in US28 (Figures 2–4). Another conserved residue in the minor pocket, E7.39 (Figure 2), is involved in ionic and/or hydrogen-bond (H-bond) interactions with four of the five cocrystallized ligands that target TMS1, forming: (i) a salt bridge with the protonated nitrogen of the imidazothiazole feature of 31, (ii) an H-bond with Ala3 backbone of vMIP-II in CXCR4, (iii) a salt bridge with the protonated nitrogen of the tropane group of 16 in CCR5, and (iv) a hydrogen bond with His2 of CX3CL1 (Figures 2–4) for US28. In contrast, compound 58 does not form a direct interaction with E2917.39 in CCR2. Residues W2.60, V/L3.28 in the minor pocket, and the Y/F3.32 of the interface between TMS1 and TMS2 form a hydrophobic pocket that accommodates 31 in the CXCR4 crystal structure, 16 in the CCR5 crystal structure, and CX3CL1 in US28 (Figures 3,4) but is not involved in binding of antagonist 58 to CCR2 (Figure 5). The 6-trifluoromethyl quinazoline moiety of 58 protrudes from the minor pocket of CCR2 toward the membrane bilayer, interacting with the membrane-oriented residues L441.34 and V2897.37 (Figure 5).
2.2.2. Major Pocket (TMS2)
In the major pocket (TMS2), the negatively charged D4.60 forms a salt bridge with Arg2 of CVX15 in the CVX15 bound CXCR4 crystal structure (Figure 3a).11 The residue at position 4.60 is however not conserved among chemokine receptors (it corresponds to a glycine in CCR5 and to a histidine in US28, Figure 2) and it does not interact with other cocrystallized ligands. Figure 1–4 show that several other residues in TMS2 interact with cocrystallized ligands in CXCR4 (H1133.29, Y1163.32, H2035.42, Y2556.51, D2626.58, and E2887.39 with CVX15, and H1133.29, D2626.58, and E2887.39 with chemokine ligand vMIP-II), CCR5 (T1053.29, Y1083.32, F1123.36, T1955.39, I1985.42, 2486.48, Y2516.51, T2596.59, and E2837.39 with small-molecule antagonist 16), and US28 (Y1113.32, Y1123.33, I2747.36, and E2777.39 with chemokine ligand CX3CL1). T1955.39 in CCR5 forms hydrogen bonds with one of the fluorine atoms of 16 (Figure 3c), while Q2005.39 in CXCR4 and N1895.39 in US28 are not involved in interactions with CVX15, vMIP-II, or CX3CL1. Residues H2035.42 in CXCR4 and I1985.42 in CCR5 establish hydrophobic contacts with the naphthalene ring of Nal3 in CVX15 (Figure 3a) and the benzene moiety of 16 (Figure 3c), respectively, but are not interacting with the cocrystallized chemokines in CXCR4 or US28. The negatively charged D6.58 that is conserved within CXC chemokine receptors (Figure 2) forms a salt bridge with CVX15 (Arg14, Figure 3a) and forms two hydrogen bonds with vMIP-II (His6 and Arg7, Figure 4a) in CXCR4 but is not involved in CX3CL1 chemokine binding to US28. An additional feature in the major pocket includes a hydrophobic subpocket located deep in the TM domain (defined by Y1083.32, F1093.33, F1123.36, W2486.48, and Y2516.51) that is targeted by the phenyl group of 16 in CCR5 but not by other cocrystallized chemokine receptor ligands.
2.2.3. Extracellular Region
Crystal structure and site-directed mutagenesis studies suggest that the extracellular surface of chemokine receptors is an important interaction site for chemokine and peptide ligands, including the cocrystallized chemokines vMIP-II (CXCR4)13 and CX3CL1 (US28)14 and the peptidomimetic CXCR4 ligand CVX1511 (Figures 1–4). Small ligands like the cocrystallized 31 (in CXCR4), 58 (in CCR2), and 16 (in CCR5) primarily interact with the minor and/or major pocket in the TM domain and make few interactions with, in particular, the extracellular loop 2 (Figures 1–3, 5).1,2,4 The positively charged Arg1 residue of the peptidomimetic ligand CVX15 forms a salt bridge with the negatively charged D18745.51 residue in ECL2 of CXCR4, while Arg2 forms H-bond interactions with the backbone of R18845.52 and Y19045.54 (Figure 1g). The solved residues of the N-terminus of CXCR4 (S23N-ter–E31N-ter) interact with the N-loop (Leu13–Leu20) and third beta strand (Gln49–Cys51) of the chemokine via H-bonds (including Gln16:K25N-ter and Asp9:R30N-ter) and hydrophobic contacts (including Gln16:M24N-ter, Val50:K25N-ter, and F29N-ter/R30N-ter:Arg7). D18145.45 and D18245.46 of the ECL2 of the receptor are involved in nonpolar contacts with Asp9, Lys10, and Ser32 of the chemokine. D18745.51 has been mutated to a cysteine residue in the vMIP-II bound CXCR4 crystal structure and forms a disulfide bridge with Cys5 of the chemokine.84 The N-terminus of US28 (D15N-ter–F25N-ter) interacts with the N-loop (Thr11–Lys18) and the third beta strand (Leu48–Ala51) of CX3CL1, making H-bond interactions between D15N-ter and Lys18 and between E18N-ter and Met15. There are also interactions between a mini-helix (Gln31-Gly35) and a loop fragment (Lys36-Ile39) of CX3CL1 and the ECL2 (K16945.46–D17845.55) of US28, including hydrogen bonds between Q17245.49 of US28 and Gln31 and Ser33 of CX3CL1 and between D17845.55 (US28) and Arg37 (CX3CL1).
2.2.4. Intracellular Region
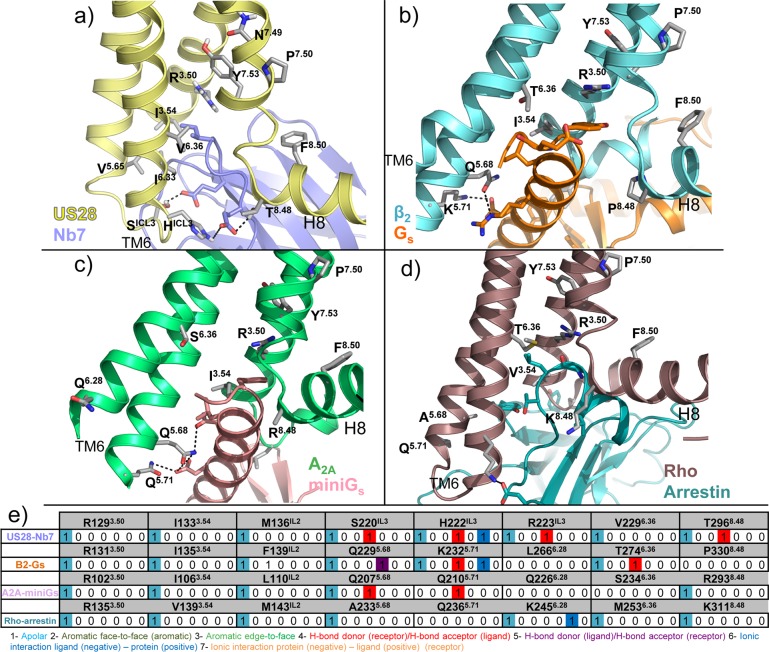
The intracellular binding sites of 29(15,46) and 30(16) are located between TM helices 1–3 and 6–7 and H8 of CCR2 and CCR9, respectively (Figures 1, 5). The small allosteric antagonist 29 targets a hydrophobic pocket in CCR2 including V631.53, L671.57, L812.43, L1343.46, A2416.33, V2446.36, I2456.37, Y3057.53, and F3128.50 and forms hydrogen bonds with the backbone amides of E3108.48, K3118.49, and F3128.50 via its hydroxyl and pyrrolone carbonyl groups. The allosteric antagonist 30 targets hydrophobic binding sites in CCR9 with its butylphenyl group (V691.53, V721.56, Y731.57, L872.43, Y3177.53, and F3248.50) and its chlorophenyl moiety (L872.43, I1403.46, A2556.36, and V2596.40) and forms an extensive hydrogen bond network with CCR9 via its sulfone group (backbone amino groups of E3228.48, R3238.49, and F3248.50), pyridine-N-oxide (T812.37 and R3238.49), and ketone moieties (T2566.37). The intracellular pocket of US28 is occupied by the nanobody Nb7 (Figure 6b), which is stabilizing the active-like conformation of the receptor (section 2.3). The 116 residues of Nb7 occupy a large volume in the intracellular part of the receptor, interacting with the intracellular half of all TMs, intracellular loops (ICLs), and with the C-terminus. The binding mode of Nb7 is similar to other reported nanobody-bound GPCR structures.85,86 The conserved R1293.50 of the DRY motif in US28 (Figure 2) interacts with Nb7 via van der Waals interactions and water-mediated H-bond interactions with the side chain and backbone of Ile,101 respectively (Figure 6b). Similarly, R1313.50 of the β2-adrenoceptor (ADRB2) interacts with Tyr391 of the alpha subunit of the Gs-protein (PDB 3SN6)87 as well as R1023.50 of the A2A adenosine receptor does with Tyr391 of the engineered mini G-protein in a recently published structure (PDB 5G53);88 R1353.50 of rhodopsin also interacts with Gly2076 and Leu2077 of a visual arrestin (PDB 4ZWJ).89 Additionally, S220ICL3 and H222ICL3/T2968.48 of US28 form H-bond interactions with Nb7 Glu99 and Glu104, respectively. These polar interactions are not conserved in the binding of Gs-protein and arrestin.
Figure 6.
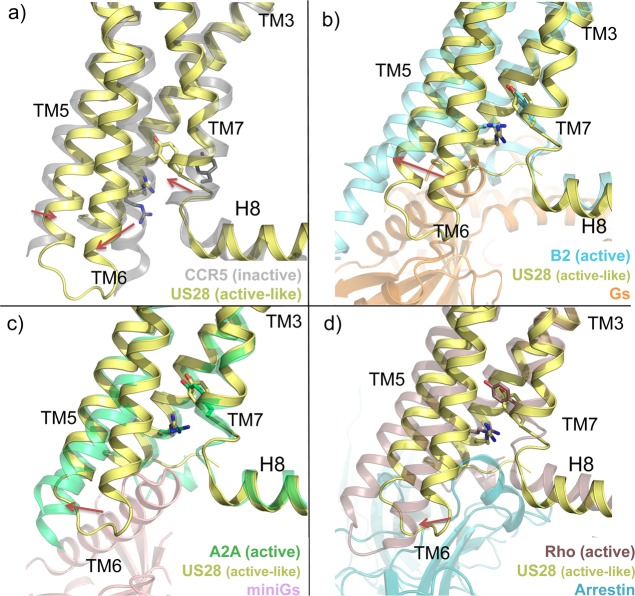
Structural changes associated with the active state of class A GPCRs. (a) Conformational changes from an inactive conformation (CCR5, transparent gray cartoon, PDB 4MBS(12)) to an active-like conformation (US28, yellow cartoon, PDB 4XT1(14)). (b–d) Conformational changes from an active-like conformation (US28, yellow cartoon) to a fully active conformation (beta 2 adrenergic receptor, transparent cyan cartoon, PDB 3SN6;87 A2A adenosine receptor, transparent limegreen cartoon, PDB 5G53;88 rhodopsin, transparent violet cartoon, PDB 4ZWJ(89)). Gs protein (orange), mini Gs protein (pink), and arrestin (blue) are shown in transparent cartoon for a better visualization. The structural alignments reveal an outward position of TM6 (red arrows), a lateral shift of TM5, and an inward movement of TM7 in the active-like conformation of US28 in comparison to the inactive conformation of CCR5. The outward shift of TM6 is significantly bigger for beta 2AR, A2a, and rhodopsin receptors. R3.50 of the DRY motif and Y7.53 of the NPxxY motif are shown as sticks as reference: in an active-like conformation, as well as in the full active conformations, the side chains of both residues are pointing toward the center of the TM bundle, while in the inactive conformation they are not.
2.3. Comparison of Chemokine Receptor Conformations
2.3.1. Extracellular Region
The N-terminus of the receptor adopts slightly different conformations in every crystal structure, but in general these conformations are similar for the crystals binding nonchemokine ligands: the N-terminus is oriented toward the center of the TM domain, partially covering the access to the binding site. The N-terminal of CCR9 adopts an α helix conformation (F33N-ter:C38N-ter) that orients toward ECL2. The N-terminal of CCR2 has not been solved. For the crystal structures binding chemokines, the N-terminus is almost perpendicular to the membrane due to the size of the chemokine, which is occupying the extracellular vestibule (ECL1, 2, 3, and N-terminus). ECL1 has the same conformation in CCR2, CCR5 and CXCR4, stabilized by a tryptophan residue (W23.50) that is highly conserved among the chemokine receptor subfamily90 and which is present in most class A GPCRs. The ECL1 of US28 does not contain this stabilizing W23.50 residue and has not been solved in the US28 crystal structure, possibly reflecting a relatively higher flexibility of ECL1 in this receptor. ECL1 and ECL2 have not been solved in the CCR9 crystal structure. ECL2 contains a double beta strand, and it adopts a more open conformation in all the crystal structures than in nonpeptide binding GPCRs (e.g., rhodopsin89). In ECL2, a disulfide bridge is highly conserved between the most conserved residue, C45.50, and C3.25 in the top of TM3 (Figure 2). A structural alignment between CCR2, CCR5, and CXCR4 crystal structures reveals that the loop connecting the beta hairpin of ECL2 of CXCR4 and CCR2 (A18045.44–R18345.47, and E18445.44 −V18745.47, respectively) is bent inward compared to the more extended beta hairpin loop of CCR5 (E17245.44–H17545.47), reflected by a 3–6 Å shift of the position of the Cα atoms of D18145.45 (CXCR4) and D18545.45 (CCR2) compared to G17345.45 (CCR5). The bent conformation of this ECL2 loop region is stabilized by a salt bridge between R18345.47 and D972.63 in CXCR4. Structural alignment of all the CXCR4 structures shows that in the vMIP-II bound CXCR4 crystal structure, the Cα atoms of D18145.45 and D18245.46 are shifted 3–5 Å compared to the 31 and CVX15 bound CXCR4 structures, allowing these negatively charged residues to form a salt bridge with Lys10 in vMIP-II. A structural alignment of all chemokine receptors crystal structures reveals that the ECL2 of US28 adopts the most different conformation between the crystallized structures: the beta strands (V16645.43–K16945.46 and Q17245.49–T17545.52) and the loop connecting them (D17045.47–N17145.48) are smaller and bent more outward compared to the other structures. This conformation permits US28 to accommodate the mini-helix (Gln31–Gly35) of CX3CL1 (reflected by a 12 Å shift of the position of the Cα atoms of K16645.45 in US28 and D18145.45 in the 31 bound CXCR4 structure). Superimposed with the vMIP-II bound CXCR4 structure, CX3CL1 would clash with the ECL2 of CXCR4. The N-terminus region of TM7 of the chemokine receptor crystal structures is 1–2 helical turns longer than in most other class A GPCR crystal structures and is stabilized by a conserved disulfide bridge between C67.50 at the top of TM7 and a cysteine in the N-terminus of the receptor. Comparison of the released chemokine receptor crystal structures (Figure 1) shows that ECL3 has a similar conformation in all structures. Within the TM domain of chemokine receptors, TM1 top is one turn longer in the vMIP-II bound CXCR4 structure and CX3CL1 bound US28 structure and it is laterally shifted outward (e.g., ∼6 Å between the Cα atoms of E32N-ter in the 31 and vMIP-II bound CXCR4 structures). There is a unique helical kink in TM2, induced by the motif S/T2.56XP2.58, that places the residues 2.60 and 2.63 toward the ligand-binding site instead of to the membrane interface as in other GPCRs.91 The top of TM3 in the CCR9 crystal structure is tilted inward the TM domain compared with the other structures (e.g., ∼7 Å between the Cα atoms of C1193.25 in CCR9 and C1093.25 in the 31 bound CXCR4 structure). In CCR5 and US28, the top of TM4 is more bent inward than in CXCR4 (reflected by a 3–4 Å shift of the Cα atoms of N1634.60 and H1624.60 compared to D1714.60, respectively), resulting in a more closed binding pocket between TM3, TM4, and TM5 in CCR5 and US28 compared to CXCR4. In CCR9, the extracellular part of TM5 is bended outward the TM domain compared to the other structures (e.g., ∼8.5 Å between the Cα atoms of K2095.33 in CCR9 and L2105.33 in the 31 bound CXCR4 structure). The extracellular half of TM6 and TM7 adopt a slightly different conformation for all the crystal structures, but there are not significant differences due to the conserved disulfide bridge between TM7 and the N-terminus, with the exception of the top of TM6 in CCR9, laterally tilted and subtly bended inward the TM bundle (e.g., ∼5 Å between the Cα atoms of A2816.62 in CCR9 and L2666.62 in the 31 bound CXCR4 structure).
2.3.2. Intracellular Region
The conformation of the intracellular half of the receptors varies between the CXCR4, CCR2, CCR5, and CCR9 structures representing inactive receptor states and the nanobody bound US28 structure representing an active-like receptor state. The nanobody Nb7 stabilizes US28 in a conformation in which:
-
(i)
TM5 is laterally tilted toward TM6 in the active-like conformation (Figure 6a), as reflected by a 4 Å shift between the Cα atoms of residues S2125.62 of US28 and I2235.62 in the 31 bound CXCR4 structure. This shift is stabilized by interactions between P2005.50 and F2376.44 in the signal transmission switch region91 of US28 and a water-mediated H-bond interaction network of Y2085.58 with R1293.50 and I1223.43. In the inactive CXCR4, CCR2, CCR5, and CCR9 structures, V/I3.40 obstructs the interaction between the homologous P5.50 and F/Y6.44 residues.
-
(ii)
TM6 is shifted outward from the TM bundle (Figure 6a), resulting in a shift of 5 Å between the Cα atoms of G2246.31 in US28 and R2356.31 in the 31 bound CXCR4 structure, facilitating interactions with Nb7 or helix 8 (H8) in US28 structures. This shift is considerably bigger in crystallized full-active GPCRs, including beta-2 adrenergic receptor,87 A2A adenosine receptor,88 and rhodopsin89 (resulting in a distance of 8, 6.3, and 3.2 Å in between the respective Cα of residue 6.31) as illustrated in Figure 6. R3.50 of the conserved DRY motif (Figure 2) stabilizes the inactive conformation of most class A GPCRs via an “ionic lock”92 with an acidic residue at position 6.30 that is present in most class A GPCRs but not in chemokine receptors or US28 (K2236.30). Nevertheless R1293.50 is oriented inward toward the center on the TM domain in US28 (Figure 6a and 7a) in a similar way as in the active beta-2 adrenergic receptor structure (Figures 6b and 7b),86 A2A adenosine receptor (Figures 6c and 7c),88 and rhodopsin (Figures 6d and 7d),89 completing the hydrogen-bond network between TM helices 3 (R3.50, X3.43), 5 (Y5.58), and 7 (Y7.53), observed in other active-state GPCR structures.87
-
(iii)
TM7 is bent inward toward the axis of the TM bundle (Figure 6a), as revealed by a 7 Å shift between the Cα atoms of the NPXXY residue Y7.53 (Y2917.53 in US28 and Y3027.53 in the 31 bound CXCR4 structure) (Figure 6a). This shift of TM7 enables Y2917.53 to form a water-mediated H-bond network with Y2085.58 and I1223.43 that is not possible in the inactive conformations of CXCR4 and CCR5.
Figure 7.
Binding details of cocrystallized Nb714 (a, purple, PDB 4XT1(14)) and intracellular effectors (b, Gs, orange, PDB 3SN6;87 c, mini-Gs, pink, PDB 5G53;88 d, beta-arrestin, blue, PDB 4ZWJ(89)). The side chains of residues interacting with the intracellular binders and the residues of the two important motifs stabilizing the active conformation (DRY and NPxxY) are shown as sticks. (b) Structural interaction fingerprint (IFP) analysis of the binding mode of the intracellular binders presented in panels a–d. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between Nb7 and the different US28 amino acid residues.
In addition to these conformational differences associated with the receptor activation state, there are several structural differences in the intracellular loops and H8 of CCR2, CCR5, CCR9, CXCR4, and US28 crystal structures that can be related to differences in protein constructs and/or crystallization. ICL1 has not been solved in CVX15 and vMIP-II bound CXCR4 structures, but a structural alignment of the other structures shows that the end of TM2 in CXCR4 is bent inward, reflected by a relative shift of 5 Å of the Cα atoms of CXCR4 S71ICL1 and US28 C66ICL1. ICL2 adopts an alpha helical conformation in CCR2, CCR5, and CCR9 that runs parallel to the membrane (A145ICL2–R152ICL2 in CCR2, A133ICL2–R140ICL2 in CCR5, and A151ICL2–E159ICL2 in CCR9) and is not observed in CXCR4 (A141ICL2–S144ICL2) and US28 (V134ICL2–P140ICL2) structures. ICL3 has been replaced by lysozyme and rubredoxin fusion proteins in CXCR4 and CCR2 and CCR5, respectively, and has been solved for CCR9 and US28. In CCR2, CCR5, CCR9, and US28 structures, H8 has an α helix conformation that runs in parallel to the membrane as observed in most class A GPCRs crystal structures. In the nanobody-free US28 structure, H8 is oriented toward the intracellular binding site, stabilized by crystal packing. In CVX15 and vMIP-II bound CXCR4 structures, H8 has not been solved, but in the 31 bound CXCR4 structure, the unstructured C-terminus of H8 interacts with the TM pocket of the neighboring CXCR4 protein in the crystal lattice.
3. Crystal Structure-Based Analysis of the Effects of Site-Directed Mutation on CXCR4, CCR5, and US28 Ligand Binding
A large amount of site-directed mutagenesis studies, covering almost all chemokine receptors (CCR1,50,51 CCR2,15,52,53,93,94 CCR3,95 CCR5,54−58,90,96−100 CCR8,59 CCR9,16 CXCR1,101−103 CXCR2,60,61 CXCR3,63−67,104 CXCR4,13,21,68−72,74,105−114 CXCR7,115 and US2875) have resulted in 2709 mutation data points, covering 343 different mutants (Figures 10–13, 18–20). About half of the mutation data (1389 data points, covering 238 different mutants) have resulted from studies with 63 unique small-molecule ligands (molecular weight ≤650), of which 46 are shown in Figures 11–13, 18–20.16,21,50−61,63,64,67−72,93,100,116 A total number of 236 mutants have been investigated to study a total of 24 different chemokine ligands resulting in 645 data points,13,21,52−55,58−61,63−69,72,74,75,90,94,97−99,105,106,108−113,115 while 606 mutation data points have been used to study the epitopes of nine different antibodies.21,68,71,72,96,106,107Sections 3.1 and 3.2 will provide crystal structure-based analyses of CXCR4, CCR2, CCR5, CCR9, and US28 mutation data, while section 5 will discuss how receptor mutation data can be used to model interactions between small-molecule ligands and chemokine receptors for which no crystal structure has been reported.
3.1. Structural Determinants of Chemokine Binding
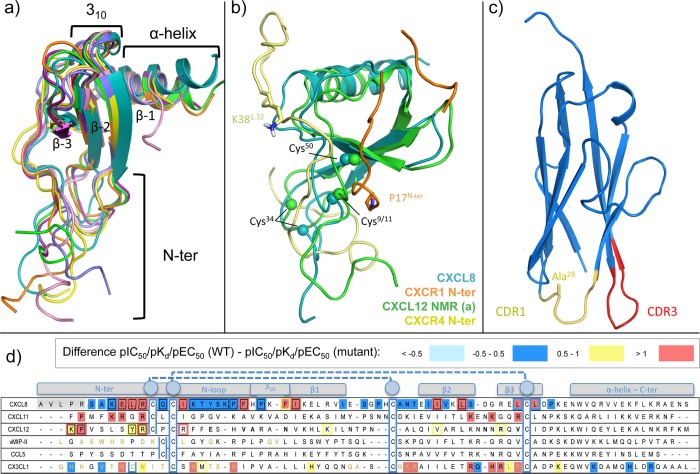
Chemokines are soluble proteins of low molecular mass (7–12 kDa) and about 70–90 residues77,117 that share a conserved structural fold observed in the different chemokine crystal structures118−120 (Figure 9a). The conformation of chemokines is stabilized by two disulfide bonds: a N-terminus coil of variable length, followed by the cysteine motif (C, CC, CXC, or CX3C), linked through an N-loop to the globular core of the chemokine, consisting on a 310 helix turn, three antiparallel beta strands, and followed by an α helix on the C-terminus. The disulfide bonds connect the N-terminus of the chemokine to (1) the loop between the first and the second beta strand and (2) to the third beta strand (e.g., Cys11–Cys35 and Cys12–Cys51 in vMIP-II, Cys8–Cys34 and Cys12–Cys50 in CX3CL1). The vMIP-II bound CXCR4 and CX3CL1 bound US28 crystal structures show that the N-terminus region of the chemokine (e.g., Leu1–Cys12 in vMIP, pGlu1–Cys12 in CX3CL1) interacts with the TM pocket, while the core of the chemokine interacts with the extracellular part of the receptor. This topology is also supported by a large amount of experimental data, including disulfide trapping experiments, dimer dilution experiments, bioluminescence resonance energy transfer (BRET), and coimmunoprecipitation, mutation studies, among others,84,111,121 and is consistent with NMR structure of CXCL8 bound to the N-terminus (residues 9–29) of CXCR1122 (Figure 9b). The experimentally supported topology does not seem to be compatible with the geometry of NMR models of CXCL12 bound to the N-terminus of CXCR4.18 In these NMR structures the N-terminus peptide of CXCR4 (residues 1–38) is interacting with the chemokine in a way that is not compatible with the crystal structure topology. As shown in Figure 9b, the N-terminus peptide, including the C-terminal residue K381.32, is oriented in parallel to the membrane, instead of pointing into the membrane (where TM1 is) as P17N-ter of the CXCR1 model. The geometry of these models assumes that CXCR4 adopts a bent conformation, facilitating extensive interactions with the TM domain.123 The accumulated pharmacological, biophysical, and structural biology data indicate that chemokines bind their receptors via a two-step process involving two different sites,2,124 chemokine recognition sites 1 and 2 (CRS1, CRS2). This two-step binding model is consistent with the recently solved chemokine bound CXCR4 and US28 crystal structures, as described in sections 3.1.1 and 3.1.2.
Figure 9.
Insights into chemokine structure–activity relationships based on X-ray crystal structures, NMR structures, and site-directed mutagenesis studies. (a) Structural alignment of X-ray structures and NMR structures of: CXCL8 (turquoise, PDB 1ILQ(17)), CXCL12 (purple, PDB 3GV3;118 light green and orange, PDB 2J7Z(141)), vMIP-II (pink, PDB 4RWS(13)), CCL5 (blue, PDB 1U4M(142)), and CX3CL1 (yellow, PDB 4XT1(14)). The main structural motifs are conserved (including the C-terminal α helix, the three antiparallel beta strands, and the 310 turn), but the N-terminus of the different chemokines adopts many different conformations, usually pointing, however, in the TM domain direction. (b) NMR models structure superimposition of the CXCR4 N-terminus (pale yellow) binding CXCL12 (light green), and CXCR1 N-terminus (orange) binding CXCL8 (turquoise). The structural alignment shows that the N-terminal regions of the chemokines adopt have different orientations with respect to the conserved chemokine core region. The C-terminal residue side chain of the N-terminus is shown as sticks for a better interpretation (P17N-ter for CXCR1, K381.32 for CXCR4). The Cα atoms of three of the conserved cysteine residues in both chemokines are shown as spheres. (c) Engineered I-body scaffold based in human neural cell adhesion molecule (NCAM) immunoglobulin domain 1. Complementarity determining-like binding regions 1 and 3 (CDR1 and CDR3) are colored in yellow and red, respectively. In CDR1, Ala28 is shown as sticks, which corresponds with the position of a conserved arginine in the derivatives. CDR3 is variable in length for each i-body and also contains different highly conserved arginines. (d) Chemokines sequence alignment. Differences between the pIC50, pKd, or pEC50 values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) log units for chemokines are reported and color coded (annotated data set included in Supporting Information).18,111,121,134,135 The secondary structure motifs are indicated in boxes. Residues of vMIP-II and CX3CL1 interacting with the receptors CXCR4 and US28 respectively in the corresponding crystal structures are highlighted in bold gold. The aligned cysteines involved in the disulfide bridges that stabilize the chemokine tertiary structure are surrounded by a blue box. The first CXCL8 residues in a gray background correspond to an alternative but minority isoform also active in physiological conditions.
3.1.1. Chemokine Recognition Site 1 (CRS1)
First the globular core of the chemokine (including the N-loop) binds to the N-terminus, the extracellular loops, and some residues on the top of the TM domain of the receptor (step 1), which is called chemokine recognition site 1 (CRS1). Mutations in CRS1 are shown to reduce the affinity of the chemokine (Figure 8, 10, 18–20): D11N-ter, T18N-ter, C30N-ter, W103ECL1, G106ECL1, C18745.50, and E275ECL3 for CXCR1;125,126 I3047.43 and L3147.51 for CXCR2;62 V16N-ter, Y27N-ter, Y29N-ter, R19745.44, F20745.54, and E2937.28 for CXCR3;65,66 Y21N-ter, C28N-ter, D18745.51, E2757.26, and E2777.28 for CXCR4;68,105,108 K1183.26, D1794.60, K206ECL2, and D2756.58 for ACKR3;115 E17545.42, E17645.43, and D2807.32 for CCR3.95 Mutations of the chemokine core also reduce the affinity of the chemokines for their receptors (Figure 9d): for CXCL8, those positions that, when mutated, significantly reduce the affinity for the receptor, are also the positions that interact with the receptor N-terminus according to the NMR model, including Phe17, Phe21, Leu43, and Leu49,127,128 The same occurs with the CX3CL1 bound CX3CR1,129,130 where mutations of Lys14, Lys18, and Phe49, for example, reduce the affinity of the chemokine for the receptor more than 10-fold. This first interaction site has been reported to be the mainly responsible for receptor affinity, while it is less involved in receptor activation and function.111
Sulfation of Y21N-ter in the N-terminal region of CXCR4 increases the binding affinity for CXCL12,18 and homologous sulfated tyrosine residues are suggested to play a similar important role in the recognition of chemokine ligands by other chemokine receptors.131 Sulfation of tyrosine residues in the CXCR4 N-terminal has been shown to be relevant in CXCR4-mediated metastasis.132 A sulfate ion observed in CXCL12 crystal structures (PDBs 1A15, 1QG7, 4UAI) has been proposed to mimick the sulfated sY21N-ter residue.133,134 This sulfate ion forms an H-bond network with Arg20, Ala21, and Asn22, therefore targeting some common residues of the binding site of heparin disaccharide (PDB 2NWG)135 and a small-molecule ligand (PDB 4UAI)136 in other CXCL12 crystal structures. It should be noted that the binding pocket of the small ligand (PDB 4UAI), a derivative of ZINC 310454, is comprised between three monomers of different crystal lattices: Arg8 belongs to one crystal neighbor, Phe14 and Ser16 belong to another crystal neighbor, and Glu15, Val18, Ala19, Asn22, Asn45, Arg47, and Val49 to a third crystal neighbor. The small ligand is not observed in (unpublished) crystal structures of CXCL12 in which binding site residues Arg8 and/or His17 have been mutated to alanine, further validating the ligand binding mode in WT CXCL12.136 A predicted interaction interface between the CXCR4 N-terminus and CXCL12 has been used to identify low affinity binders of CXCL12 in virtual screening studies.137
3.1.2. Chemokine Recognition Site 2 (CRS2)
According to the two step binding model, chemokine receptor activation is linked to the second binding step in which the N-terminal coil of the chemokine interacts with the core of the TM domain (minor pocket TMS1 and partially the major pocket TMS2) and ECL2.2 In the CXCR4:vMIP-II crystal structure, the main interactions within the 7TM binding pocket and the chemokine include hydrogen bonds with residues D972.63, D2626.58, and E2887.39, and apolar interactions with several residues, including W942.60, H2817.32, and I2847.35 (Figures 1–3). Mutation studies confirm the importance of D972.63,74 D2626.58, and E2887.39 for CXCL12 binding affinity and/or potency for CXCR4 (Figure 10).21,68,69,105,106,111−113 Mutation studies furthermore suggest that also the anionic D1714.60 (which does not interact with vMIP-II in the CXCR4 crystal structure) and D18745.51 (mutated to a cysteine residue for a covalent attachment with Cys5 in vMIP-II) residues are also important anionic residues for CXCL12 affinity and/or potency for CXCR473,74,78,84,105,111 (Figure 10). Consistent with the vMIP-III bound CXCR4 crystal structure, mutation studies show that W942.60 plays an important role in binding CXCL12 in the minor pocket of CXCR4,69,74 whereas mutation of Y1163.32 at the interface between the minor and major pockets does not affect CXCL12 affinity (Figure 10c).69 The main interactions in the TM domain of the CX3CL1 bound US28 structure are hydrogen bonds with Y401.39, Y1123.33, T17545.52, and E2777.39 and apolar contacts with W892.60, L932.64, F1113.32, L2737.35, and I2747.36.
Many of the residue positions involved in chemokine binding in CXCR4:vMIP-II and/or US28:CX3CL1 complexes also play a role in chemokine binding to other receptors. Mutation of W2.60 also affects binding affinity of CCL2 for CCR253,94 and CCL5 affinity for CCR5,55,99 whereas mutation of D18745.51 affects the affinity of CXCL8 for CXCR1,126 and residue D6.58 plays an important role in binding CXCL8 to CXCR1,103 CXCL10 to CXCR3,65 CXCL11 to CXCR3,67 CXCL11 to ACKR3,115 and CXCL12 to ACKR3.115 Comparative analysis of chemokine receptor mutation data show however that the role of residue positions 1.39, 2.63, 3.32, 6.48, 6.51, 6.55, 7.39, and 7.43 are highly chemokine–receptor complex dependent (Figure 8). Mutation of Y1.39 in the minor pocket does not affect the affinity of CXCR4 for CXCL1269 (Figure 10c) or the affinity of CXCR3 for CXCL1163 (Figure 19f) but does affect CCR2 binding affinity for CCL253 nor CCR5 affinity for CCL3.54 Mutation of Y892.63 in the minor pocket of CCR5 decreases the potency of CCL4 and CCL8 but does not affect the potency of CCL3 and CCL5,55,90,99 whereas mutation of D1122.63 affects binding of CXCL11 to CXCR363,64 and mutation of S1032.63 decreases the binding affinity of CXCL11 and CXCL12 for ACKR3.115 Mutation of the residue at position 3.32 at the interface between the minor and major pockets affects binding affinity of CCL2 for CCR2,52,53,94 CCL5 for CCR5,90 and potency of CCL1 for CCR8.59 Substitution of residue 3.32 however does not affect the binding affinity of CXCL8 for CXCR1125 or CXCL11 affinity for CXCR3.63,67 Mutation of the residue at position 7.39 negatively affects binding affinity and/or potency of CXCL11 for CXCR3 (S3047.39),65 CXCL11 for ACKR3 (Q3017.39),115 CCL2 for CCR2,53,94 CCL3 for CCR5,58 and CCL5 for CCR5,55 whereas mutation of E7.39 does not affect binding affinity/potency of CXCL8 for CXCR1,125,126 CXCL8 for CXCR2,60,61 CXCL12 for ACKR3,115 or CCL1 for CCR8.59 The effects of single point mutations in the major binding pocket on chemokine ligand binding are receptor dependent (Figure 8), including W6.48, F/Y6.51, and residues at positions 6.55 and 7.43. For example, W2486.48A and Y2516.51A/I mutations affect CCL3 binding to CCR5 by 10-fold and 7-fold respectively54 and Y2716.51A decreases the binding of CXCL11 to CXCR3 more than 10-fold.64 However, W2526.48A and Y2556.51A do not significantly affect CXCL12 binding to CXCR4.68,69,105,106,112 The I6.55A mutant affects CXCL12 binding to CXCR4,70 but mutation of the homologous hydrophobic residue in CCR5,54 CCR2,53 CXCR2,61 and CXCR364 does not affect chemokine binding. It should be noted however that mutations of W6.48, Y6.51, and I/L6.55, which only have small effects on chemokine ligand binding affinity, can have a large effect on chemokine ligand potency and/or efficacy. The I2596.55A mutant of CXCR4 for example does not affect CXCL12 binding, but affects CXCL12 potency by 8-fold,68 whereas W2686.48A of CXCR3 decreases the potency of CXCL11 more than 10-fold.67 Finally mutation studies indicate that I3047.43 is important for the binding of CXCL8 but not for the binding of CXCL1 to CXCR2.62
Figure 19.
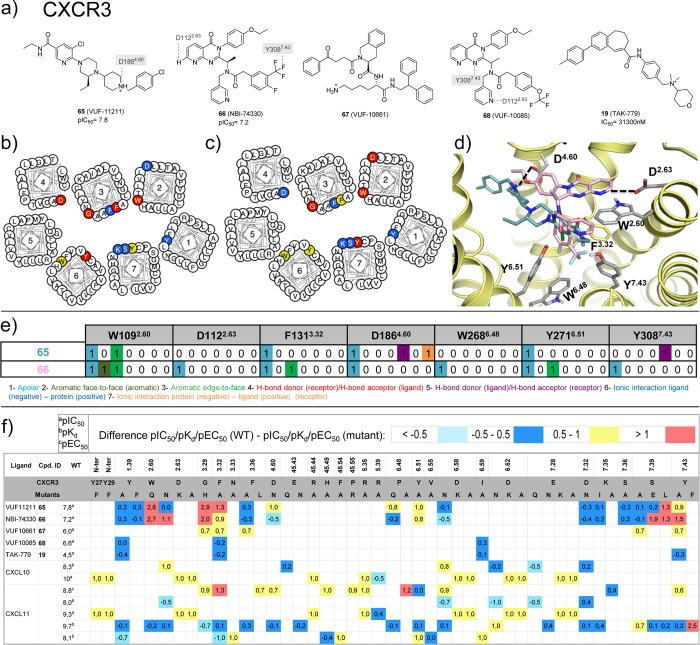
CXCR3 ligand binding models based on molecular docking and site-directed mutagenesis studies.63−67 (a) Chemical structures of CXCR3 ligands with mutation data. Interactions between the ligands and specific residues derived from X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (b,c) CXCR3 mutation effects on 65 and 66 binding mapped on helical box diagram (adapted from GPCRdb49). The helical box of TM2 does not reflect the T2.56XP2.58 kink of chemokine receptors, depicting the residues of 2.60 and 2.63 toward the membrane surface while they are in fact pointing toward the TM binding site. (d) Predicted binding modes of 65 and 66, targeting both the minor and the major pockets of CXCR3. (e) Comparative structural interaction fingerprint (IFP) analysis of the predicted binding modes of 65 and 66 in CXCR3 presented in panel d. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between the ligand and the different CXCR3 amino acid residues. (f) Differences between the pIC50a, pKdb, or pEC50c values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 125 CXCR3 mutant–ligand combinations covering ligands 19, 65–68, CXCL10, CXCL11, and 46 CXCR3 mutants (annotated mutation data set included in Supporting Information).
Mutation studies indicate that residues in the N-terminus of chemokines are important for both binding affinity and receptor activation (Figure 9d) and that most residues in the CRS2 of chemokine receptors play a more important role in chemokine mediated receptor activation than in chemokine binding.111 The binding of small ligands occurs also in CRS2 (minor and major pocket of the 7TM helical domain) and will be described in more detail in the next sections. Recently, a human single-domain antibody-like scaffold (i-body) with antagonistic activity for CXCR4 (to nanomolar) has been reported138 that is proposed to target a similar binding site as chemokines (PDB 5AEA, Figure 9c). Epitope mapping revealed that i-bodies bind the transmembrane binding pocket of CXCR4 (including residues V1123.28, F18945.53, D2626.58), whereas CXCR4 antibodies139 and nanobodies140 have been reported to target the extracellular loops (E17945.43, D18145.45) and the N-terminus (C28N-ter) of the receptor. The best hits of engineered i-bodies all possess conserved positively charged Arg residues, complementary to the negatively charged binding pocket of CXCR4.
3.2. Structural Determinants of Small Ligand Binding to CCR2, CCR5, CCR9, CXCR4, and US28
CXCR4, CCR5, US28, CCR2, and CCR9 crystal structures (Figure 1) and chemokine receptor mutagenesis studies (Figures 10–13 and 17–20) indicate that small-molecule ligands can bind chemokine receptors in the extracellular part of the TM domain, the minor pocket between TM1, TM2, TM3, and TM7, the major pocket between TM3, TM4, TM5, TM6, and TM7, and in the intracellular part of the receptor.
Figure 17.
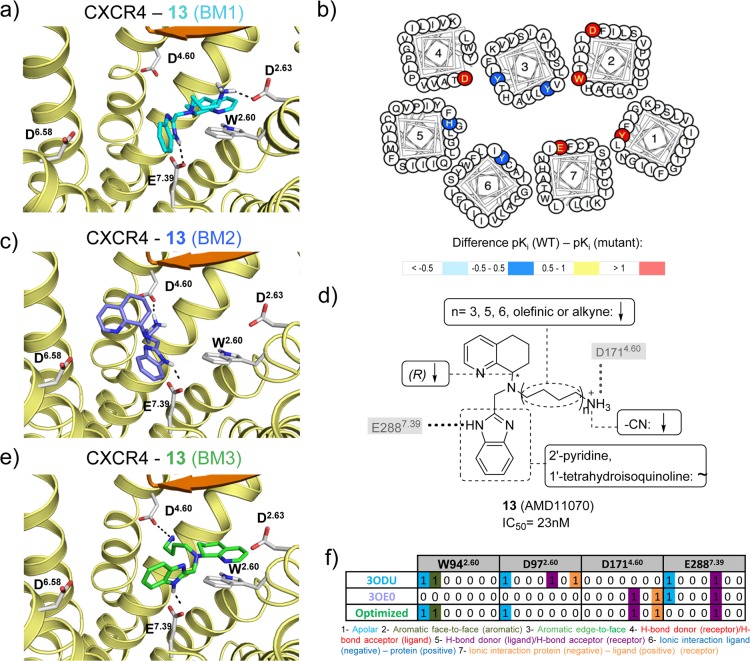
Optimization and evaluation of molecular docking based binding mode prediction studies of compound 13 in CXCR4 using on site-directed mutagenesis69 and SAR153,203,204 data. (a) Docking pose of 13 in the binding pocket of the 31 bound CXCR4 crystal structure, (c) docking pose in the binding pocket of the CVX15 bound CXCR4 crystal structure, (e) optimized docking pose based on mutation and SAR data. (d) Effects of CXCR4 mutations on the binding affinity of 13, mapped on a helical box diagram (adapted from GPCRdb49). Note that (i) the mutational effect of D2626.58 (see Figure 11b) is not shown, and (ii) that the helical box of TM2 does not reflect the T2.56XP2.58 kink of chemokine receptors, depicting the residues of 2.60 and 2.63 toward the membrane surface while they are in fact pointing toward the TM binding site. Differences between the pKi values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) are reported in logarithmic units (annotated data set included in Supporting Information). (d) Summary of structure–activity relationships of analogues of 13. The interactions with key residues derived from mutation studies (gray background) are depicted by a gray dotted line. (f) Comparative structural interaction fingerprint (IFP) analysis of the different binding modes of 13 in CXCR4 presented in panels a, c, and e. The structural receptor–ligand interaction patterns are described by IFP bit strings encoding different interaction types between 13 and the amino acid residues of CXCR4.
3.2.1. Minor Pocket in TM Domain (TMS1)
Within the minor pocket (TMS1), the most important features are
-
(i)
Ionic interactions between D2.63 and the cationic isothiourea group of the small ligand 31 at the top of the minor binding pocket are observed in the CXCR4 crystal structures (Figure 3a). Mutation studies indicate that D2.63 is important for the binding of ligands 15 (KRH-1636)21 and 13(69) to CXCR4 (Figure 11)21,69 and also play a role in small-molecule ligand binding to CXCR3 (section 5.5).63 It should be noted however that D2.63 is not conserved within the chemokine receptor subfamily.
-
(ii)
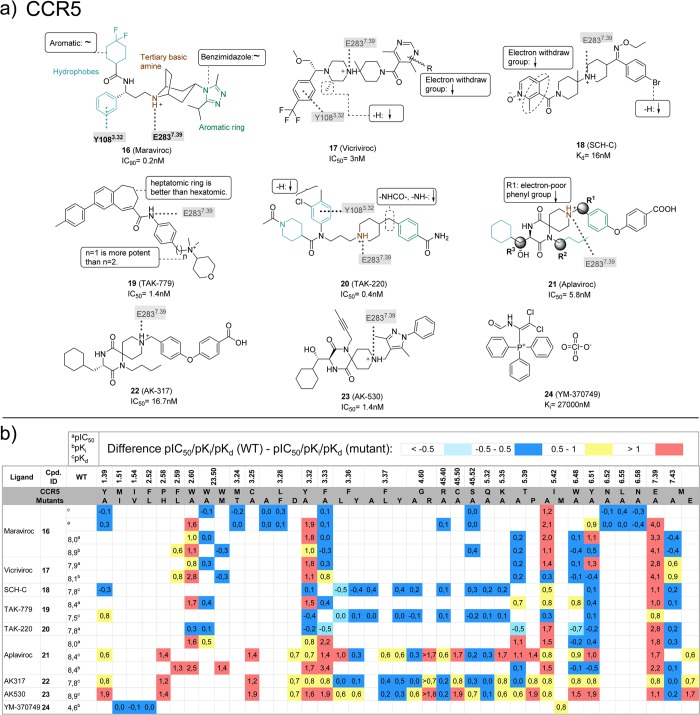
A polar interaction between T2927.40 and the γ-lactam secondary exocyclic amine of the small ligand 58 at the interface of the minor pocket and the membrane bilayer. The role of T2927.40 in CCR2 ligand binding is supported by T2927.40A/V mutation studies, resulting in a significant decrease in the affinity of 19 (TAK-779), 19-1 (TAK-779 base), 25, 26 (RS-504393), and 27 (Teijin) for CCR2 (Figure 13).52,53,93
-
(iii)
A hydrophobic subpocket in the lower region of the minor pockets of CXCR4, CCR5, and CCR2 contains conserved hydrophobic and aromatic residues, including Y1.39, W2.60, and F/M7.43 (Figure 3a, 5a). Mutation of Y1.39, W2.60, and F7.43 affects ligand binding to CXCR4 (Figure 11),69 CCR5 (Figure 12),54−57 CCR1 (section 5.6),50,51 and CCR8 (section 5.8).59 The 31 bound CXCR4, 16 bound CCR5, and 58 bound CCR2 crystal structures suggest that the highly conserved W2.60 (Figure 2) plays an important role in ligand binding by forming hydrophobic/aromatic interactions (Figures 3, 5a), and this role is supported by mutation studies on CXCR4 binding to 1,692,69 and 13(69) (Figure 11) and CCR5 binding to 16,54,55,10017 (Vicriviroc),55,10019,55 and 21(55,100) (Figure 12), whereas no mutation studies of this residue position have been reported for CCR2. The conserved Y1.39 residue (Figure 2) does not interact with any of the cocrystallized CXCR4 ligands, but according to mutation data it is relevant for the binding of some CXCR4 small ligands, such as 1,692,69 and 13(69) (Figure 11). Y1.39 is relevant for the binding of other CCR5 small ligands (19,5621,5722 (AK317),57 and 23 (AK530),57Figure 12) according to mutant studies, validating the observed interaction with antagonist 16 in the CCR5 crystal structure. Y491.39 in CCR2 is involved in a hydrogen bond with the carbonyl oxygen of the γ-lactam of 58, and it has been confirmed to decrease the affinity of 19 in site-directed mutagenesis studies.52 F/M2877.43 is located deep in the hydrophobic pocket and does not interact with any of the cocrystallized CXCR4 ligands. However, the mutant M2877.43E has been reported to decrease the binding of some CCR5 small ligands, including 21,55,5722,57 and 23(57) (Figure 12). M2957.43 makes a hydrophobic contact with 58 in the CCR2 crystal structure, but no mutant data have been reported providing additional information on the role of this residue in CCR2 binding.
Figure 12.
(a) Chemical structures of CCR5 ligands with related mutation data.54−58 Interactions between the ligands and specific residues derived from CCR5 X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. (b) Affinities from site-directed mutagenesis studies on chemokine receptors. Differences between the pIC50a, pKib, or pKdc values of wild-type and mutant <−0.5 (cyan), −0.5 to 0.5 (blue), 0.5 to 1.0 (yellow), and >1.0 (red) logarithmic units are reported for 217 CCR5 mutant–ligand combinations covering ligands 16–24 and 38 CCR5 mutants (annotated mutation data set included in Supporting Information).
3.2.2. Interface of Minor and Major Pockets in the TM Domain
The definition of a minor and a major binding pocket implies that some residues overlap between both pockets on what we called the “interface”. The interface area comprises residues in TM3, TM7, and ECL2 and includes the following features:
-
(i)
Hydrophobic interactions with residue 3.32, which lines both the minor and major binding site and interacts with 31 and CVX15 in CXCR4, with 16 in CCR5, and with 58 in the CCR2 crystal structure (Figures 3, 5a). Mutation studies demonstrated that Y1163.32 indeed is an important determinant of CXCR4 binding for ligands 1,692,69 and 13(69) (Figure 11), whereas F1113.32 plays an important role in binding small-molecule ligands 16,54,55,10017,55,10021,55,57,10022,57 and 23(57) to CCR5 (Figure 12). Similarly, mutation Y1203.32A in CCR2 significantly decreases the affinity of 25,5219-1,5219,52 and 27(52) (Figure 13).
-
(ii)
Polar interactions with residue E7.39, which is present in most chemokine receptors (Figure 2). Mutation studies show that this residue is important for small-molecule ligand binding to CXCR4 (Figure 11),68,144 CCR5 (Figure 12),54−57 CCR2 (Figure 13),52 and US28 (Figure 14),145 consistent with the 31 bound CXCR4 and 16 bound CCR5 structures in which respectively E2887.39 and E2837.39 are involved in a H-bond and ionic interactions with the cocrystallized ligands.11,12 However, no direct interaction is observed between E2917.39 and the cocrystallized CCR2 ligand 58.15 D45.51 is also located at the interface between the minor and major binding sites (one position downstream from the conserved C45.50 in ECL2) and interacts with the peptide ligand CVX15 in the CXCR4 crystal structure (Figure 3a). Mutation of D45.51 affects ligand binding to CXCR4 ligands, including ligands 1,682,68 and 15(21) (Figure 11), suggesting that this residue plays a role in binding small-molecule ligands as well. The residue at position 45.51 is however not conserved among chemokine receptors (Figure 2) and mutation of this residue seems to be receptor and ligand dependent.21,54,56,57,68
Figure 14.
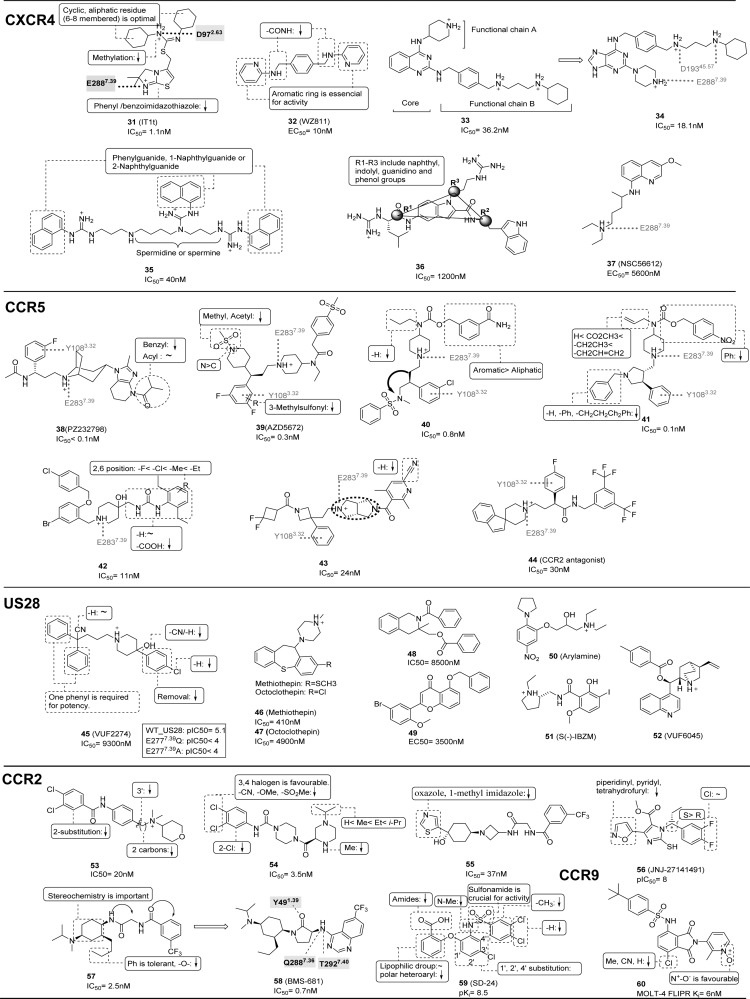
Summary of structure–activity relationship (SAR) of CXCR4, CCR2, CCR5, CCR9, and US28 ligands. The representative CXCR4 ligands include compounds 31,8032,15433–34,155,15635,157,18736,158 and 37;36 CCR5 ligands include compounds, 38,17239,18840,15941,18942,16143,162 and 44.163 US28 ligands include compounds 45,18046, 47,18148,18249,18150,18451,185 and 52.186 CCR2 ligands include compounds 53,17354,17455,17556,17757, 58,4559,178 and CCR9 ligands 60.179 Interactions between the ligands and specific residues derived from X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by a dotted line, interacting groups are surrounded by a dotted line, and key features are summarized by a solid box. Mutation data for the US28 ligand 45(145) is also included in a squared box. For each ligand, the binding affinity (IC50, Kd, Ki) or potency (EC50, IC90) is reported (except for compound 35, for which functional IC50 is reported, and for compound 60Ki is based on MOLT-4 cells).
3.2.3. Major Pocket in TM Domain (TMS2)
Important chemokine receptor–ligand interactions features within the major binding pocket are polar interactions with the top of the major binding site and apolar interactions in a hydrophobic supocket at the bottom of major pocket.
-
(i)
Polar interactions with D4.60and D6.58. Mutation of D1714.60 in CXCR4 leads to a decrease in affinity for almost all the small ligands with which it has been tested (Figure 11): 1, 1(Zn)2 (AMD3100(Zn)2), 1(ZN)1 (AMD3100(Zn)1), 2, 3 (AMD8721), 4 (AMD8899), 5 (AMD3529), 6 (AMD3389), 7 (AMD3106), 8 (AMD3108), 9 (AMD2936), 10 (AMD2763), 11 (AMD2849), 12 (cyclam), 12(ZN)1 (cyclam(Zn)1), 13, and 15.21,68−73 In the CVX15 bound CXCR4 crystal structure, D1714.60 forms a salt bridge with CVX15 Arg2, confirming the importance of this residue. However, this position is not highly conserved (Figure 2), neither in physicochemical properties, meaning that this position cannot be involved in polar interactions in all the receptors. Thus, for CCR5 (glycine), most of the tested small ligands are not affected by 4.60 mutation to alanine (G1634.60A) (Figure 12); only in the case in which this residue has been mutated to arginine (G1634.60R) it decreases the affinity of 21–2357 (Figure 12). Additionally, D/G4.60 is not interacting with any of the two chemokines in vMIP-II bound CXCR4 and CX3CL1 bound US28 crystal structures. CXCL12 affinity and potency are decreased by the D1714.60N mutant in CXCR4, while the homologous D1864.60N mutation in CXCR3 does not affect affinity for CXCL11.63,67 D6.58 is conserved among CXCRs, while position 6.58 corresponds to a less conserved residue in CCRs (Figure 2). Mutation studies show that this residue plays an important role in binding small-molecule ligands to CXCR4 (Figure 11).21,68−72
-
(ii)
A hydrophobic subpocket located in the major pocket of CXCR4 (Y1163.32, W2526.48, Y2556.51, I2596.55), CCR5 (Y1083.32, W2486.48, Y2516.51, L2556.55, Figure 3c), and US28 (F1113.32, W2416.48, Y2446.51, L2486.55) plays a role in the binding of the naphthalene ring of CVX15 in CXCR4, the phenyl ring of 16 in CCR5 (Figure 3b). Mutation studies indicate that W2486.48 is an important residue for CCR5 binding of small-molecule ligands 19,5521,55−5722,57 and 23(57) (Figure 12), while Y2556.51 and I2596.55 are involved in binding of small-molecule ligands 1–5 to CXCR4 (Figure 11).68,69,71 Mutation of I1985.42, which interacts with the cyclohexyl group of 16 in the CCR5 in the same hydrophobic binding site (Figure 3b), results in a 3–125-fold decrease in affinity and/or potency of all small ligands evaluated in CCR5 studies (16,57,5517,5518,5519,5521,55−5722,5723,57 and 24 (YM-370749),58 whereas mutation of the homologous H2035.42 only has a moderate 3–5-fold effect on affinity/potency of 4 of the 11 small ligands evaluated in CXCR4 studies (2, 3, 5, 13).68,69
3.2.4. Intracellular Region
The recent CCR2 and CCR9 crystal structures, as well as chimera studies, cysteine accessibility scanning, site-directed mutagenesis, and radioligand displacement studies have indicated that several small-molecule ligands bind an intracellular pocket of chemokine receptors, including CXCR2 (ligands 61–64, Figure 18),60,61 CCR2 (ligand 29, Figures 5b, 13),15,134,146 CCR4,147,148 CCR5,147 CCR9 (ligand 30, Figures 5c, 13),16 and CX3CR1149 (sections 5.3 and 5.4). This allosteric small-molecule ligand binding pocket overlaps with the nanobody Nb7 binding site in the US28 crystal structure14 (Figure 3d), as well as the G-protein interaction site in ADRB287 and A2A150 crystal structures:
-
(i)
A hydrophobic subpocket comprised of TM3, TM5, TM6, and H8. This subpocket includes in US28 the residues I1333.54, V2155.65, I2266.33, V2296.36, L2306.37, and F2988.50 that mainly interact with the beta hairpin Ala24–Ile31 and the beta hairpin Glu99–Thr112 of Nb7 (Figure 7a,e). Several of these residue positions comprise the Gs binding site in beta-2 adrenergic receptor (I1353.54, T2746.36), the miniGs binding site in adenosine A2A receptor (I1063.54), and the beta-arrestin binding site of bovine rhodopsin (V1393.54, M2536.36) crystal structures (Figure 7). Few residues in this hydrophobic subpocket form hydrophobic contacts with 29 in CCR2 (A2526.33, V2556.36) or 30 in CCR9 (I952.51, T2566.37, V2596.40) (Figure 5).15,16 The mutation I2266.33A significantly decreases the binding of 62 (pteridone-1) and 63 (Sch527123), while it does not affect the binding of 61 (SB-265610)60 in CXCR2, implying that different compounds may be able to adopt different binding modes in the intracellular pocket.
-
(ii)
An additional hydrophobic subpocket comprised between TM1, TM2, TM3, TM7, and H8. Both crystallized intracellular ligands mostly bind in this subpocket, which includes residues I/V1.53, I/V1.56, L2.43, L/I3.46, Y7.53, and F8.50. As described in section 2.2.4, 29 and 30 bind the intracellular pocket. This was predicted before for 29 by mutagenesis data that confirmed the relevance of residues 7.53 and 8.50 for binding affinity of this compound to CCR2,93 and it has been validated for 30 also through site-directed mutagenesis assays.16
-
(iii)
Polar interaction with ICLs and H8. S220ICL3 of US28 forms an H-bond interaction with Glu99 in Nb7. The polar Glu104 side chain of Nb7 is located between the end of TM6-ICL3 and the end of TM7-H8 and makes a polar interaction with H222ICL3 and T2968.48 (Figure 7a,e). These residues also make polar interactions with the Gs and miniGs in beta 2AR and A2A, respectively (Figure 7e). Regarding the small molecules 29 and 30, important polar interactions involve the backbones of K3118.49 and F3128.50 for CCR2,15,93 the side chains of T812.37, and T2566.37, and backbones of E3228.48, R3238.49, and F3248.50 for CCR9. Mutant studies have confirmed the relevance of T812.37 for compound 30 binding to CCR9.16
4. Crystal Structure-Based Analysis CXCR4, CCR2, CCR5, CCR9, and US28 Ligand Structure–Activity Relationships
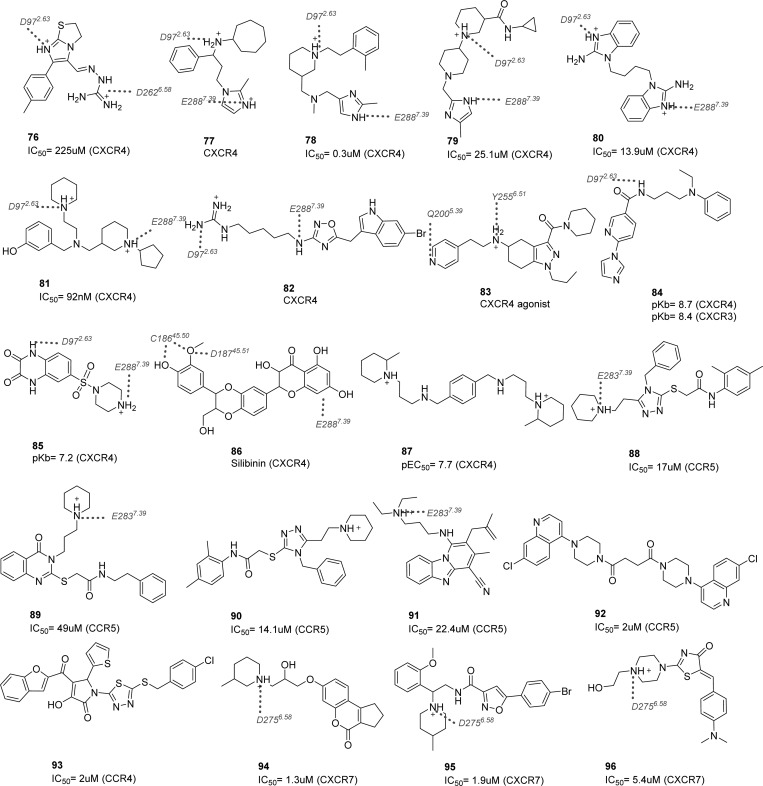
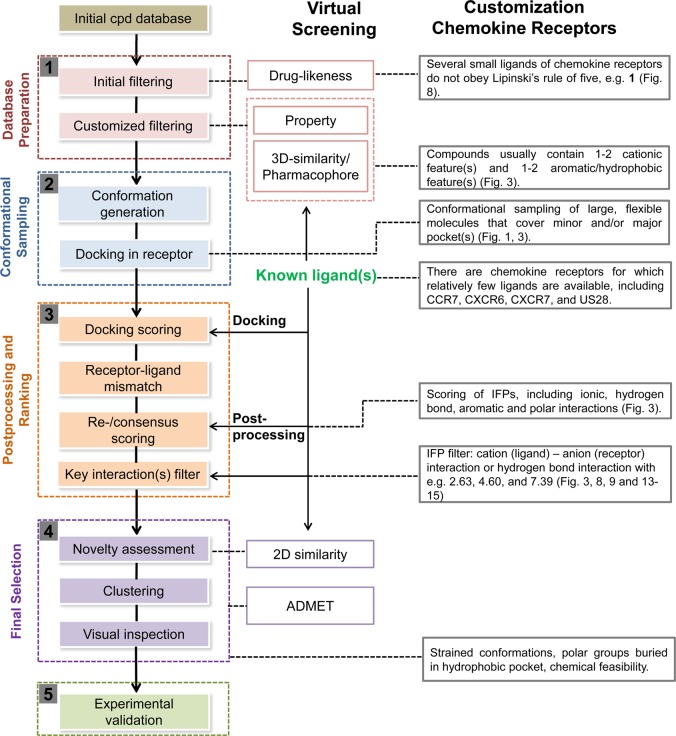
The chemokine receptor crystal structures with different cocrystallized ligands constitute a valuable source for retrospective ligand search through structure–activity relationship studies, and they provide an important clue to the binding mode prediction for derivatives and other ligand chemotypes.77 The current section is dedicated to the compilation and analysis of the different strategies and results regarding binding mode prediction and its mutual contribution to drug design.
4.1. CXCR4–Ligand Structure–Activity Relationships
For CXCR4, 949 mutation data points have been determined covering 96 different residues positions, 20 chemokines/antibodies, and 18 different small ligands (including 1–15, Figure 11). D1714.60N, D2626.58N, and E2887.39Q mutations resulted in a significant decrease of binding of 1–1368−72 and 15,21 supporting a binding mode in which these negatively ionizable residues are involved in ionic interactions with the positively charged basic nitrogen atoms of ligands 1–13 and 15 (Figure 11). SAR studies have indeed indicated that the cationic basic moieties of 15,151,15213,15331,7832 (WZ811),15433, and 34(155,156) are essential for CXCR4 binding (Figure 11). In addition, Y451.39A, W902.60A, and Y1163.32A mutations affected the affinity of 1, 2, and 13,69 while Q2005.39A and H2817.32A mutation affected potency of 2, 3, and 4.68 All together, the mutation data suggest that ligands 3–11 and 14 primarily target the major pocket, while ligands 1, 2, 13, and 15 target the minor and the major pocket simultaneously (Figure 11). CXCR4 binding affinity of 14 (KRH-3955) is not affected by mutation of D1714.60, D2626.58, and E2887.39 but may be dependent on interactions with other negatively charged residues in CXCR4, such as D972.63 and D18145.45, as proposed in CXCR4 modeling studies.73 SAR studies provide insights into the determinants of binding affinity of 31 analogues to CXCR478 that are consistent with the 31 bound CXCR4 crystal structure (Figure 3e),11 showing that (i) the two protonated nitrogen moieties interacting with D972.63 and E2887.39 are essential for ligand binding affinity, (ii) two aliphatic six-, seven-, or eight-membered rings are essential for optimal binding of the hydrophobic subpocket between W902.60, H1133.29, and Y1163.32 (Figure 14). The 3D-QSAR model presented in Figure 3b, derived from FLAP based on an alignment of analogues of 31 to the crystallized conformation of 31,78 shows that a tight fit of apolar moieties in the hydrophobic subpocket between W942.60 and Y1163.32 is required for high affinity binding, consistent with other druggability assessments of the minor CXCR4 binding site.76 Combined SAR,151,152 mutagenesis, and modeling studies21 suggest a binding mode in which the guanidine and the amine group of antagonist 15 interact with D1714.60 and D2626.58 residues, respectively (Figure 11), consistent with SAR studies showing that the guanidine moiety, the position of the nitrogen atom in the pyridine, and the S-stereo configuration of 15 are important for CXCR4 binding. The cyclam and pyridine moieties of 1 and its derivatives 2, 5, and 6 (Figure 11a) play important roles in CXCR4 binding affinity and are proposed to interact with Y451.39, W942.60, H1133.29, Y1163.32, and D1714.60 based on SAR and mutation data.21 SAR studies indicated that the basicity of the benzoimidazole, the length of the aliphatic amine group, and the S stereoconfiguration are important requirements for 13 binding affinity. Different putative binding modes of 13 in relation to SAR and mutagenesis data will be discussed in more detail in section 5.3 (Figure 17).69,153 The replacement of any of the two basic amines with amides and the modification of the pyridine rings into phenyl, methyl, or guanidine groups significantly reduce the potency of antagonist 32, indicating that these cationic and/or aromatic groups are important for 32 binding and/or CXCR4 activation.154 The central phenyl ring and linker length between the aromatic ring and the terminal hexenamine moiety are important determinants of the binding affinity of compounds 33 and 34 for CXCR4.155,156 On the basis of this SAR data, binding modes have been proposed for 33 and 34 in which the cationic piperazine/piperidine interact E2887.39 and D19345.57, respectively. The combination of CXCR4 mutation data and SAR studies suggest that the essential aromatic quinazoline of 33, purine of 34,155,156 naphthalene of 35,157 and indolyl of 36(158) enable these ligands to interact with aromatic residues in the CXCR4 binding site, including Y451.39, W902.60, Y1163.32, and/or H2817.32.
4.2. CCR5–Ligand Structure–Activity Relationships
For CCR5, 1021 mutation data points have been determined, including 101 different residue positions, 13 chemokines/antibodies ligands, and nine different small ligands (including 16–24, Figure 12). E2837.39A mutation resulted in a significant decrease of binding of ligands 16–23, suggesting a binding mode in which the negatively ionizable E2837.39 residue is involved in an ionic interaction with the positively charged basic nitrogen atoms of these ligands (Figure 12a). Y371.39A, P842.58H, I1985.42A, W2486.48A, and Y2516.51A mutations affected the potency of 21, 22, and 23,57 while W862.60A, Y1083.32A, and I1985.42A mutations affected the affinity of 16, 17, and 19,55 indicating that these ligands are in a similar way targeting the minor and major pocket simultaneously (Figure 3d). On the basis of this structure-based alignment a pharmacophore model for CCR5 ligands can be defined consisting of: (i) one basic feature (the basic amine of 16 that interacts with E2837.39), (ii) one aromatic moiety (the triazole of 16 that interacts with W862.60), (iii) hydrophobic feature 1 (the phenyl ring of 16 interacts with Y1083.32), and (iv) hydrophobic feature 2 (the difluorocyclohexane of 16 interacts with T1955.39 and I1985.42), presented in Figure 3. 16 shares a similar spatial distribution of cationic, aromatic, and hydrophobic features as several other CCR5 ligands, including 17–23, 38 (PZ232798), 39 (AZD5672), 40,15941,16042,16143,162 and 44,163 although this does not necessarily mean that all these ligands adopt a similar binding mode. Moreover, the symmetric distribution of hydrophobic/aromatic features compared to the central cationic feature offer a challenge to predicting CCR5–ligand binding modes in a similar way as CXCR4, as discussed in section 5.3. Despite these caveats, the structure-based CCR5 pharmacophore model facilitates a structure-based analysis of CCR5 mutagenesis (Figure 12) and SAR (Figure 14) data. Modification of the aromatic pyridine ring of 17,16418,164,16521,83,166 and 42(161) by electron withdrawing substituents decrease the affinity of these ligands, consistent with mutation and modeling studies, suggesting that this ring may form π–π interactions with W862.6055,57 (Figure 3d). The seven-membered ring of 19(167,168) is preferred over the six-membered ring, while the optimal linker between the phenyl and quaternary ammonium groups is one carbon atom, suggesting that the relative distance and directionality of the cationic and aromatic/hydrophobic groups are essential for CCR5 binding. The hydrophobic substituents of the benzene ring of 20(82) are favorable for CCR5 affinity, emphasizing the importance of this hydrophobic feature in the CCR5 ligand pharmacophore (Figure 14). The modification of the carbon linker between piperidine and benzene of 20 into a nitrogen atom results in a decreased affinity,82 which may suggest that the conformational flexibility of the aromatic benzene ring to target the minor pocket is an important determinant of CCR5 affinity. The six-membered aliphatic ring and triazole groups of 16 can be replaced by aromatic169 and benzimidazole170,171 moieties, respectively, without affecting CCR5 binding affinity. Modification of the tetrahydro pyridinoimidazole of 38(172) into a phenyl moiety results in a 290-fold decrease in CCR5 affinity, which may be caused by the fact that the bulky moiety affects the aromatic interactions between the tetrahydro pyridinoimidazole and W862.60.
4.3. CCR2–Ligand Structure–Activity Relationships
More than 40 mutation data points have been determined for CCR2, covering 15 different residues positions (Y1203.32, H1213.33, E2917.39, and T2927.40, among others) and seven different small ligands (19, 19–1, 25, 26, 27, 28 (INCB3344), and 29 (Figure 13).52,53 The E2917.39Q mutation diminishes binding of 19 and its derivative 19-1,52 supporting a binding mode in which the negatively ionizable E2917.39 is involved in ionic interactions with the positively charged quaternary amine of these ligands. SAR analysis of 19 and derivatives 53(173) and 54(174) demonstrate that CCR2 binding affinity is sensitive to small variations in the distance between the cationic and hydrophobic features and small structural changes close to the essential cationic feature (Figure 14). Y1203.32A, H1213.33, and T2927.40A mutations resulted in significant decrease of binding of ligands 19, 19–1, 25, 26, and 27,52 indicating that (at least parts of) these ligands occupy the minor pocket. SAR studies show the importance of hydrophobic interactions (increased affinity for more apolar substituents in chemical series around 54,17455,175 and 57/5845) and steric and conformational compatibility (decreased affinity for 2-chlorine substituted analogues of 53(173) and 54(174)) with the minor and/or major hydrophobic binding pocket of CCR2. SAR studies indicate that the stereochemistry of trisubstituted cyclohexane of 57 is important for CCR2 affinity,45 in line with the tight packing of the corresponding cyclohexane ring of 58 against W982.60 in the CCR2 crystal structure (Figure 5). The binding mode of 29 in the intracellular binding pocket observed in the CCR2 crystal structure15 is consistent with earlier mutation studies showing the negative effects of V2446.36A, Y3057.53A, K3118.49A, and F3128.50A on CCR2 binding affinity.93 SAR studies furthermore show the favorable role of the hydrophobic halogen substituents of 29(176) and several other small-molecule ligands (56177 and 59(178)) that are proposed to target the same hydrophobic intracellular binding pocket of CCR2 formed by V631.53, L671.57, T772.39, L812.43, and L1343.46.
4.4. CCR9–Ligand Structure–Activity Relationships
SAR studies demonstrate that the chlorine and nitroxy substituents of the CCR9 antagonist 60 are important for potency,179 consistent with the recently solved CCR9 crystal structure showing that the chlorine of the chemically similar allosteric antagonist 30 targets the hydrophobic subpocket composed by L872.43, I1403.46, and V2596.40, whereas the nitroxy group of 30 forms H-bond interactions with T812.37 and R3238.49.16
4.5. US28–Ligand Structure–Activity Relationships
For US28, 25 mutation data have been determined, covering seven different residues positions (T11N-terA, T12N-terA, E13N-terA, F14N-terA, D15N-terA, and Y16N-terA in the N-terminus region and E2777.39A in TM7) for four chemokines (CCL3, CCL4, CCL5, and CX3CL1) and one small ligand (45 (VUF2274)) (Figure 14). The F14N-terA mutant significantly decreased binding affinity of CCL3, CCL4, and CCL5 but not CX3CL1.75 Y16N-terF mutation also has a moderate effect on CCL4 binding potency. E2777.39A mutation regarding the small ligand 45 indicates that E2777.39 is a crucial interaction partner for 45, and it was proposed that the carboxylate group of E2777.39 forms an ionic interaction with the piperidine amine of the ligand.2045 derivatives are the first reported nonpeptidergic inverse agonists of US28, and SAR studies indicate that the hydroxyl, chloro, and chlorobenzene features are essential for affinity, while the cyano group and the two phenyls are not necessary to maintain the affinity (Figure 14).180 Other small US28 ligands, 46 (methiothepin), 47 (octoclothepin) (Figure 14),181 and series of dihydroisoquinolinone, 48 (tetrahydroisoquinoline)182 and 49 (flavonoid)181 containing US28 agonists have similar low potency,183 hampering SAR analysis. There are furthermore several US28 small ligand reported in patents, including 50 (arylamine),18451 (S(−)-IBZM),185 and 52 (VUF6045),186 but clear, quantitative SAR data is missing for these series.
5. Experimentally Informed Modeling of Chemokine Receptor–Ligand Structure
The community-wide GPCR DOCK 2010 assessment20 to predict the three-dimensional coordinates of the 31 and CVX15 bound CXCR4 structures before release of the crystal structures, highlighted some of the challenges of predicting ligand binding modes in chemokine receptors. None of the GPCR DOCK 2010 participants were able to predict the binding pose of the large peptide CVX15 in the major pocket of CXCR4, demonstrating that the prediction of GPCR–peptide interactions is particularly difficult, especially in the absence of experimental modeling constraints. The key interactions of the small-molecule ligand 31 with D972.63 and E2887.39 in the minor pocket of CXCR4190 could only be correctly predicted by the explicit incorporation of CXCR4 mutation studies69,90 and ligand structure–activity relationship (SAR)78 to guide the modeling of CXCR4 structure and CXCR4–ligand interactions. The GPCR DOCK 2010 challenge and other modeling studies have demonstrated that compared to other GPCRs families, chemokine receptor structures have a couple of peculiarities that require careful considerations. These include the positioning of the helix bundle, the extracellular domain, and the ligand binding sites, which pose different challenges for model construction. Furthermore, it should be noted that small-molecules ligand of different chemokine receptors are chemically diverse (Figures 11, 12, 13, and 18–20) and that the effects of mutation studies are highly ligand dependent, limiting the possibilities to translate binding mode hypotheses between chemokine receptors and/or ligand chemotypes. While sections 3 and 4 provide a retrospective analysis of protein mutation and ligand SAR data of crystallized CXCR4, CCR5, and US28 receptors, the following sections 5 and 6 provide an overview of the implications of the new crystal structures on the construction of chemokine receptor models for the elucidation of the structural determinants of chemokine–ligand interactions (described in section 5) and the structure-based discovery and design of new chemokine receptor modulators (section 6). In the current section, the use of experimental anchors to steer the modeling procedure and address challenges associated with different steps along the modeling workflow (Figure 15) will be discussed, including (step 1) amino acid sequence alignment, (step 2) template selection, (step 3) TM bundle and loop modeling, (step 4) ligand binding pocket prediction, and (step 5) ligand binding mode prediction. Although the workflow is sequential, the sequence alignment, template selection, and model building steps are interconnected and will be therefore described together in section 5.2. Different sequence alignment approaches will be described, based on GPCRdb structure-based sequence alignments,49 manually curated based on the analyses of structural alignments. After building the initial model, more detailed information can be used to refine the binding pocket and the ligand binding mode, which will be discussed in section 5.3. We will provide an overview how protein site-directed mutagenesis studies (section 3) and ligand structure–activity relationships (section 4) have been used to map ligand binding sites, resulting in proposed binding mode models for numerous ligands in different chemokine receptors (CXCR2, section 5.4; CXCR3, section 5.5; CCR1, section 5.6; CCR2, section 5.7; and CCR8, section 5.8). It should be noted that the modeling requirements and challenges dependent on the purpose of the chemokine receptor model. Some models have been constructed with the purpose to closely capture receptor–ligand interactions or are optimized for structure-based virtual screening but nevertheless contain inaccuracies in specific regions in the TM domain. Other models provide an accurate structural architecture of the receptor–chemokine complex but do not offer insights into interactions on residue or atomic level. A schematic overview of the applicability domain of different chemokine receptor models is provided in Figure 15.
Figure 15.
General GPCR molecular modeling workflow with specific details in chemokine receptor modeling customization and applications. For each step, specific details from experimental and in silico data concerning the target to model may be used to improve the approach. The left panel shows specific details on chemokine receptors modeling, including considerations in length, conserved residues (represented in orange), or conserved motifs (represented in pink) that can influence the orientation of specific residues in or out of the binding site (colored in green). The right panel summarizes the applicability domains of the structural models generated along the modeling workflow, ranging from the design of SAR and mutation studies and the generation of ligand repurposing hypotheses based on crystal structure-based sequence alignments, the identification of ligand binding sites, elucidation of ligand binding modes, and the application of structural models for structure-based virtual screening, structure-based ligand design, and the elucidation of ligand-mediated receptor activation mechanisms.
5.1. Sequence Alignment and Template Selection for Chemokine Receptor Modeling
The prediction and modeling of the 31 and CVX15 bound CXCR4 crystal structures in the GPCR Dock 201020 was especially challenging because its similarity in both sequence and function compared to the GPCRs structures available at that time was distant (ADRβ1, 26% identity). A multiple sequence alignment of the target sequence with potential homologues is an important step toward the prediction of the structure of the target (step 1, Figure 15). Sequence motifs containing glycines, prolines, serines, and threonines are of particular interest because they are known to be involved in helix kinks/bulges.191 An important challenge in the GPCR DOCK 2010 modeling assessment was the prediction of alternative conformation of the top of TM2, stabilized by the T2.56xP2.58 motif that is conserved in chemokine receptors (section 2.3.1, Figure 2 and Figure 15)1,20 and introduces a one-residue gap resulting in a 100° rotation of the top of TM2 compared to the closest crystal structure template at that time ADRβ1.23 The T2.56xP2.58 induced helical bulge was correctly predicted by a molecular dynamics (MD) simulation based conformational analysis of TM2 and the consideration of mutation studies indicating that residues 2.60 and 2.63 (W942.60 and D972.63 in CXCR4) play a role in ligand binding to CXCR4 and other chemokine receptors and should therefore be oriented inside of the TM bundle (Figure 15).20,23,69,191 A structure-based alignment of the CXCR4, CCR2, CCR5, CCR9, and US28 crystal structures now facilitates the sequence alignment of the TM helices, as well as ECL1, ECL3, and large parts of ECL2. Alignment of the N-terminus, ECL2 (in particular upstream of the conserved C45.50), ICL3, and C-terminal still remains challenging because of the variation of loop length and limited availability of structural templates for these regions (steps 1 and 2, Figure 15).
The overall conserved structural fold of the TM helical bundle of chemokine receptor crystal structures implies that the CCR5, CXCR4, CCR2, CCR9, and US28 structures represent useful templates for modeling the 7TM domains of other chemokine receptors. CCR5 shares sufficiently high sequence identity (≥40%) and similarity (≥60%)20 to model the TM helices of CCR1, CCR3, CCR4, CCR8, ACKR2, and CCRL2, whereas CXCR4 shares sufficiently high sequence identity and similarity with CXCR1, CXCR2, and CXCR3, CCR2 shares sufficiently high sequence identity and similarity with CCR1, CCR3, CCR4, CCR7, CCR8, CCRL2, and ACKR2, CCR9 shares sufficiently high sequence identity and similarity with CCR1, CCR4, CCR6, CCR7, CXCR1, CXCR2, CXCR6, and ACKR4, while US28 provides a template for modeling the homologous CX3CR1 (Figure 16c). Homology models or new crystal structures of CCR4, CXCR1, or CXCR2, and CXCR3 would provide homologous templates for chemokine receptors with lower sequence identity (<40%) or similarity (<60%) to the currently crystallized chemokine receptors. A crystal structure of CCR4 would be a homologous template to model XCR1, while CXCR1 or CXCR2 structures would provide a homologous template to construct models of CCR6 and CCR7, and an experimentally determined structure of CXCR3 would facilitate CXCR5 modeling. CCR10, CXCR6, ACKR1, and ACKR3/CXCR7 do not have high TM sequence identity/similarity with the three crystallized receptors or the 13 receptors for which high resolution homology models can be directly derived from chemokine receptor crystal structures. Binding site similarities show more distinct chemokine receptor clusters (Figure 16), and several receptors share significantly higher binding site similarity than TM helix similarity. For example, CCR5 provides a good template to accurately model the bindings site of CX3CR1 (62% identity), while it shares relatively high sequence similarity with ACKR3/CXCR7 (65% similarity) and US28 provides a good template to model the binding site of CCR8 (62% similarity). Variability of the length and sequence identity of the extracellular and intracellular regions, which conformations have been discussed in sections 2.3.1 and 2.3.2, may also influence template selection The implications of this variability for extracellular regions on prediction of interactions with chemokines will be described in section 5.2, and for interactions with intracellular effectors such as nanobodies, G-proteins, or beta-arrestin will be described in section 5.3.
Figure 16.
(a) Chemokine receptors binding site alignment, (b,d) amino acid sequence similarity and identitity matrices based on alignments of (b) TM helices (TM1- TM7) and ECL2 and (d) binding sites, and (c) binding site sequence similarity based clustering of 23 human chemokine receptors (CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR8, CCR9, CCR10, CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, CXCR6, CX3CR1, XCR1, ACKR1, ACKR2, ACKR3/CXCR7, ACKR4, and CCRL2) and viral chemokine receptor US28 (calculated using GPCRdb49). The residues selected for the binding site alignment, sequence similarity/identity calculations, and clustering include TM1 (1.35, 1.39), TM2 (2.53, 2.60, 2.63), TM3 (3.29, 3.32, 3.33, 3.36, 3.37), TM4 (4.57, 4.60), TM5 (5.32, 5.35, 5.39, 5.42, 5.46), TM6 (6.48, 6.51, 6.52, 6.55, 6.58, 6.59), TM7 (7.32, 7.35, 7.36, 7.39, 7.43, 7.45), and ECL2 (45.50, 45.51, 45.52). Binding site residue Cα atoms are depicted as spheres in panel c and colored according to binding site region (see panel a). Pairwise sequence similarity (lower-left) and identity (upper right) percentages are reported based on sequence alignment of (b) TM helices and (d) binding sites and are gradually color-coded from red to green. Cells surrounded by black squares correspond to data described in the text. Chemokine receptor binding site based clustering (c) has been performed based on the UPGMA algorithm with 100 replicas of bootstrapping (calculated using GPCRdb49).
In addition to sequence similarity in the TM helices and putative binding pockets, several other criteria can be considered to select the appropriate (or combination of) homology modeling templates (step 2–3), including: (i) sequence motifs that determine the helical conformation, (ii) local sequence similarity in specific ligand binding sites, (iii) chemical similarity between the crystallized and modeled ligand, (iv) conformational state (active/inactive), and (v) crystallographic artifacts. In addition to the conserved T2.56XP2.58 motif that determines the fold of TM2, there are other sequence motifs that determine the conformation of helices in chemokine receptors, including the P1.36P1.37 motif present in CCR5 and conserved also in CCR1, CCR2, CCR3, CCR4, and CCR9. This motif induces a different kink in TM1 than the alternative P1.36X1.37 motif that CXCR4 shares with CCR6, CCR7, and CCR10. CXCR4 may therefore be a better template to model the conformation of TM1 of the latter three CC chemokine receptors. CXCR2 has a lower overall sequence identity with CCR5 (37%) than with CXCR4 (40%) but share residues F3.36, N6.52, and L6.55 that are reported to be important for binding compounds 61 and 64 based on mutagenesis studies.61 CCR5 may therefore be a better template to model 61 and 64 bound CXCR2. A chemical similarity analysis of known chemokine receptor ligands and cocrystallized class A GPCR ligands allows the identification of the most relevant template for ligand-bound chemokine receptor models. For example, the predicted binding orientation of 19 in the CCR5 crystal structure can be used to model the binding mode of 19 in the homologous CCR2 (70% binding site similarity, Figure 16), whereas a refined binding pose of 19 in the CXCR4 crystal structure would be a more suitable template to predict the ligand binding mode in the more homologous CXCR3. Derivatives of 19 have also been proven to bind CCR5 and CCR2. CCR5 would provide an accurate template to construct a 19 bound CCR2 homology model.22 As pointed out before by others,23 the 31 bound CXCR4 crystal structure (PDB 3ODU) contains two protruding lipids in the major pocket between TM5 and TM6, which constitute a crystallization artifact that conditions the orientation of these two helices. For ligand-bound models in which important interactions are characterized in the major pocket, the CVX15-bound structure may constitute a better template. The location of the receptor C-terminal into the TM bundle of the symmetry neighbor receptor in the crystal lattice may explain the presence of the lipids and the opened conformation of this structure (section 5.3).
5.2. Modeling Structural Interactions between Chemokine Receptor and Chemokine Ligands
Comparison of the vMIP-II bound CXCR4 and CX3CL1 bound US28 crystal structures (section 3.1), the CXCL8, CXCL12, CX3CL1, and vMIP-II chemokine crystal structures and NMR models (section 3.1), chemokine/chemokine receptor mutation data (section 3), and chemokine/chemokine receptor sequences (Figures 1d and 4d), indicate that the chemokine bound CXCR4 and US28 structures provide useful templates but also highlight potential challenges in the modeling of structural chemokine–receptor interactions. These challenges include first of all modeling of interactions between the chemokine C-terminal core region with the first 23 and 15 residues of each receptor N-terminus (missing in the CXCR4 and US28 structures, respectively) and the extracellular loops (in particular ECL2 that is differently positioned in the CXCR4 and US28 structures). To address this fact, the overall geometry of the chemokine bound chemokine receptor structure has to be defined a priori. The chemokine bound CXCR4 and US28 crystal structures provide evidence that supports the two-sites binding model in which the N-terminus of the chemokine interacts with the minor and major pockets in the 7TM helical bundle, while the chemokine core interacts with the receptor N-terminus and ECLs.111 NMR models supporting alternative binding modes have been reported (Figure 9b), suggesting that chemokines may adopt different binding orientations in complex with chemokine receptors.18 The different binding modes of the N-terminus regions of the vMIP-II and CX3CL1 with the TM domain of CXCR4 and US28 indicate that both the structure and receptor interactions of the chemokine N-terminus will differ between chemokine bound receptor complexes. The N-terminus of chemokine receptors is highly variable in length and composition, and it has not been fully solved (step 3, Figure 15). For instance, CCR2 N-terminus is 12 residues longer than CCR5 N-terminus, while the CXCR2 N-terminus is 10 residues longer than the N-terminus of CXCR4. Modeling of a large part of the receptor N-terminus has to be done de novo, which is complicated because of the lack of knowledge of interactions between the chemokine and the N-terminus of the receptor and requires structural refinement by, e.g., MD simulations (step 4, Figure 15). For the modeling de novo of the N-terminus, there is a potential restraint to be used: the sulfotyrosine in position 21 in CXCR4 which has been predicted to bind in the disaccharide binding site, as explained in section 3.1. Mutation studies indicate that in particular ECL2 is important for chemokine binding (Figure 10).2 The length and structure of the ECL2 of chemokine receptors is relatively more conserved than for other GPCR subfamilies (e.g., aminergic GPCRs receptors91), which allows the combination of modeling the TM helices and loops as an integrated part of the model construction step 3. There are nevertheless differences in the sequences of the ECL2 of chemokine receptors in length, sequence, and structure. The length of the downstream region from the conserved C45.50 ECL2 to the start of TM5 is one residue longer in CCR5 than in CXCR4, and it is predicted to be even eight residues longer in ACKR3/CXCR7 compared to CXCR4. Significant differences in the sequence and therefore in the overall structural properties and molecular interaction features of ELC2 can have important effects on ligand binding as has been demonstrated for CCR3,192 which has an acidic motif (173-ETEELFEET-181) that may explain why ligand binding to this receptor is highly pH dependent.192 Finally, structural differences in ECL2 between CXCR4 and US28 crystal structures indicate ligand-induced conformational flexibility of this region in chemokine receptors, which has to be properly considered in homology modeling of other chemokine receptor–ligand complexes. As an example of the ligand-induced conformational flexibility of ECL2, a structural superimposition of vMIP-II bound CXCR4 and CX3CL1 bound US28 reveals a clash between the mini-helix of CX3CL1 and CXCR4 ECL2.
5.3. Modeling Structural Interactions between Chemokine Receptors and Intracellular Effectors
The potential bias of the T4L (CXCR4) and rubredoxin (CCR5) fusion proteins and the influence of Nb7 nanobody (US28) should be considered for modeling of the intracellular pocket of chemokine receptors. ICL3 has only been solved in the US28 structure, but the conformation of ICL3 in the US28 structure may be only relevant for the modeling of an active conformation. Of special interest is the modeling of the intracellular pocket for intracellular binding ligands (which will be further described in upcoming sections), as well as for intracellular effectors, including nanobodies, G-proteins, and beta-arrestin, whose main interactions have been described in section 2.2.4. Therefore, for the selection of the most relevant structural template for the modeling of the intracelular pocket, those structures in which this pocket is biased for the presence of stabilizing elements or crystal artifacts should be avoided. There are a few templates to model intracellular effector-bound structures, including complexes between the adrenergic beta-2 receptor and Gs protein (PDB 3SN6),87 the A2A adenosine receptor and an engineered mini Gs protein comprising a truncated GTPase domain of the Gαs subunit (PDB 5G53),88 bovine rhodopsin and visual arrestin (PDB 4ZWJ),89 and the C-tail of the vasopressin V2 receptor and beta-arrestin1 (PDB 4JQI).193 However, the templates are still limited to specific GPCRs and incomplete (the beta-arrestin1 bound V2 structure only contains the C-tail of the receptor) or do not have a high resolution (the beta-arrestin bound opsin structure has a 7.7 Å resolution). Therefore, modeling of intracellular effectors binding is still challenging and needs more and better templates. However, some models have been already described of chemokine receptors interacting with intracellular effectors.194 Moreover, important intracellular effectors with no reported GPCR-bound structure are G protein-coupled receptor kinases (GRKs), which are difficult to crystallize due to their flexibility and fast kinetics, being also difficult to model due to their sequence variability.195 Finally, the available chemokine receptor templates have some characteristics to consider to model intracelular binders: for example, modeling of active conformations may be addressed using US28 structure as template, but it is important to consider also other templates in a fully activated state, especially for modeling a G-protein bound conformation, in which case the ADRB2 structure may represent a better template. Also the best resolution crystal structure of CXCR4 (PDB 3ODU) lacks H8 and has a nonstructured C-terminal that may affect the modeling of intracellular effector interactions.
5.4. Modeling Structural Interactions between Chemokine Receptors and Small-Molecule Ligands
Predicting the residues that conform the binding pocket is one of the most important steps of the chemokine receptor modeling workflow (step 4, Figure 15), especially for those receptors for which no crystal structure has been reported. The availability of experimental data, including site-directed mutagenesis studies, NMRs, or fluorescent measurements, is therefore, important to support the prediction of structural chemokine receptor–ligand interactions. In the GPCR DOCK 2010 challenge, site-directed mutagenesis data67,69 were valuable, indicating that residues such as D2.63, D4.60, D6.58, and E7.39 play a key role in CXCR4 antagonist binding. Although the CXCR4, CCR2, CCR5, CCR9, and US28 X-ray structures provide useful templates for chemokine receptor modeling, binding pocket prediction is still challenging due to the prevalence of multiple potential binding sites in these receptors. For small ligands, it is known that some bind exclusively the minor (CXCR4 antagonist 31) or the major pocket (CXCR4 antagonist CVX15), while others may target both simultaneously (CCR5 antagonist 16). A wide amount of chemokine receptor ligands share a cationic feature (or even two like the CXCR4 antagonist 31), and both the minor and the major pocket have anionic groups with which ligands can interact (residue positions 2.63 and/or 7.39 in the minor pocket, 4.60 and/or 6.58 in the major pocket) as discussed in section 4. The recently solved 29 bound CCR2 and 30 bound CCR9 crystal structures confirm that small-molecule ligands can also target the intracellular binding sites of chemokine receptors, consistent with earlier radioligand binding and mutation studies.60,61,93,134,146−149 The diversity of different possible binding sites pose a challenges to the prediction of structural chemokine receptor–ligand interactions. Intracellular ligands, including allosteric antagonists of CXCR2 (61–64, Figure 18),60,61 CCR2 (29, Figure 5, 13),15,134,146 CCR4,147,148 CCR5,147 CCR9 (30, Figure 5, 13),16 and CX3CR1149 combine electronegative H-bond acceptor moieties (to interact with the amide backbone of residue positions 8.48, 8.49, and 8.50) positioned between at least two medium sized aromatic/hydrophobic ring systems (that target different hydrophobic subpockets), and can therefore be distinguished from cationic ligands that target the orthosteric binding site of chemokine receptors. Putative binding regions of novel small-molecule ligands can furthermore be predicted based on integrated SAR and site directed mutagenesis data that is available for similar ligands and/or similar receptors. It should be noted however that the additional, previously unexplored ligand binding sites in chemokine receptors may be accessible, including for example the extracellular vestibule (like the allosteric modulator binding site in the muscarinic M2 receptor crystal196) or the interface with the membrane outlier (e.g., as observed in P2Y1197 receptor and GCGR251 crystal structures).
The prediction of the putative ligand binding site(s) is followed by the prediction of ligand binding poses (step 4, Figure 15). Molecular docking and pharmacophore tools can generate binding poses for ligands, especially small ligands.26,198 The docking of large peptide and protein ligands, including chemokines, increases challenges in sampling of protein–ligand conformations. Poses obtained from docking programs are ranked based on scoring functions. The main challenge in scoring and ranking binding poses is related to the conformation of the side chains in the binding pockets due to rotamer variants. Priority filters can be used as postprocessing method to prioritize interactions, and MD simulations can be useful to determine the most stable conformations of the residues, as well as of the ligands, to improve the confidence on the protein–ligand interactions.199
Despite the availability of multiple chemokine receptor crystal structures, symmetry in both the ligand (prevalence of two basic moieties) and the binding site (prevalence of multiple acidic residues) constitute a challenge for the chemokine receptor–ligand binding mode prediction (section 5.3).77 The evaluation of different putative binding modes of compound 13 in CXCR4 presented in Figure 17 illustrates the caveats of the extrapolation of binding mode information on cocrystallized ligands to other small-molecule ligands and emphasize that structural modeling should be guided and evaluated using experimental data, in line with the GPCR Dock 2010 challenge.20Figure 17a shows a docking pose of compound 13 in the minor binding site of CXCR4 with a similar structural interaction fingerprint (IFP) pattern as the cocrystallized ligand 31 (Figure 3a), whereas Figure 17b presents a docking pose of 13 in the major binding site of CXCR4 that is targeted by CVX15 (Figure 3a). IFP representation of compound 13 (Figure 17f) illustrates common interactions (W942.60, E2887.39) as well as specific interactions derived from the pocket selection of the reference ligand (D972.63 for 31, D1714.60 for CVX15). SAR studies (Figure 17d) reveal a significantly decrease in affinity if one of the basic amino groups (predicted to interact with D1714.60) is changed to a cyano group, confirming an ionic interaction between the amino group and a negatively charged acidic residue in the binding site. Changing the benzimidazole to a 2-pyridine or 1-tetrahydroisoquinoline is tolerated, consistent with a binding mode in which the basic amine in the benzimidazole group is interacting with an acidic residue (predicted to be E2887.39). The relevance of these acidic residues (D972.63, D1714.60, D2626.58, and E2887.39) in binding 13 is reflected by mutation studies (Figures 10b and 15b).69 Moreover, the length and flexibility of the aliphatic side chain and the stereochemistry of the chiral carbon also affect the potency of compound 13. Both predicted docking poses partially fit the described interactions, but none of them fit all the experimental data, suggesting that these poses may represent different populations of CXCR4–13 conformations or that the docking results should be optimized (Figure 17e). Optimization includes postprocessing filters, applying restraints on those interactions known to be important for ligand binding and molecular dynamics simulations.200 Molecular dynamics simulations can be used for the extended sampling of receptor–ligand conformations, the assessment of the relative distribution of different binding mode populations, and the possible role of residues in molecular dissociation and/or association. Despite the challenges that chemokine receptors constitute for molecular modellers, three-dimensional chemokine receptor models have been successfully applied for the identification and design of new chemokine receptor ligands, binding pocket, and ligand binding prediction for CCR5 and CXCR4,36,201 and other chemokine receptors, including CXCR2,60 CXCR3,63 CCR1,50 CCR3,95 CCR4,33 and CCR8.202
Challenges in modeling the binding mode of large chemokine ligands in chemokine receptors subfamily include the consideration of protein flexibility in protein–protein docking and the consideration of alternative binding mode hypotheses to guide the docking process and/or evaluate the docking poses. The vMIP-II bound CXCR4 and CX3CL1 bound US28 crystal structures and reported mutation data indicate that chemokine receptors can accommodate a large variety of chemokine ligand binding modes in different TM subpockets,13,14,111 and the integrated analysis of NMR structures, in silico models, and mutation data indicate multiple binding modes may be possible for the same receptor:chemokine complex.111,123,205 Model evaluation therefore may require complementary in silico approaches and experimental data, such as extensive molecular dynamics simulations and the consideration of combined mutation data of both the receptor and the chemokine ligand.206
5.5. Structural CXCR2–Ligand Interaction Models
For CXCR2, more than 90 mutant data points have been determined, covering about 26 different residue positions (including, among others, D1433.49 of the DRY motif and Y3147.53 from the NPXXY motif), and four different small ligands (61, 63, and 64) (Figure 18).60−62 Mutations in the intracellular region of CXCR2 significantly decreased binding affinity of 61, 62, and 63, whereas the E3007.39Q mutant in the extracellular side did not affect binding affinity, supporting an intracellular binding of all three ligands (Figure 18c,d). T832.39A, D1433.49R, Y3147.53A, and K3208.49A mutations resulted in a significant decrease of binding of 61,60 whereas D842.40N, A2496.33L, and K3208.49A mutations significantly reduced 62 binding, and T832.39A, D842.40N, D1433.49R, A2496.33L, Y3147.53A, and K3208.49A significantly decreased the binding of 63 (Figure 18). The CXCR2 mutation data indicate that 61, 62, and 63 target a similar binding site as 29 in the CCR2 crystal structure15 and 30 in the CCR9 crystal structure.16 The negative effect of the K3208.49A mutant, for example, supports a binding mode in which the acidic groups of 61, 62, and 63 (Figure 18b)60 are involved in polar interactions with the positively ionizable K3208.49 residue and simultaneously form H-bond interactions with the backbone amide nitrogen of residues R/K8.49 and F8.50, like 29(15) and 30 in the CCR2 and CCR916 crystal structures (Figure 5b–d). Compound 64 does not displace the radiolabeled intracellular binder 61,207 and F1303.36A, F2205.47L, and N2686.52H mutants decrease CXCR2 affinity of compound 64 (Figure 18a,g),61 indicating that this ligand targets the extracellular binding site in CXCR2 and supporting a binding mode in which the imidazole nitrogen of 64 accepts an H-bond from N2686.52, while the pyrimidine and imidazole moieties of the ligand may be involved π–π stacking interactions with F1303.36 and F2205.47 (Figure 18b,c,f). CXCR2 therefore represents an experimentally supported example of a chemokine receptor that is targeted by small-molecule modulators that target both extracellular and intracellular binding sites.
5.6. Structural CXCR3–Ligand Interaction Models
For CXCR3, there are about 50 mutant points to assess the small ligand binding mode predictions of 65 (VUF11211), 66 (NBI-74330), 67 (VUF10661), 68 (VUF10085), and 19 (Figure 19a), including residues W1092.60, D1122.63, and S3047.39. The predicted binding modes of 65 and 66 (Figure 19d) share common interactions with W1092.60, F1313.32, Y2716.51, S3047.39, and Y3087.43 (Figures 19d, and 17f) but also include distinct interaction features supported by ligand-dependent mutation effects.63 The D1122.63N mutation data suggest that the anionic carboxylic acid moiety of D1122.63 plays a role in binding of 66 but is less important for binding 65 (Figure 19b). The decreased affinity of 65 for the D1864.60N mutant supports a binding mode in which the positively ionizable amine group of 65 forms a salt bridge with the negatively charged D1864.60 residue. In contrast, the affinity of 66 is not significantly affected by the same D1864.60N mutation, suggesting that this residue does not play an important role in binding compound 66 (Figure 19c).63 The differential effect of the F1313.32A mutant on CXCR3 binding affinity of the chemically similar 68(64) and 66(63) (Figure 19f) may be explained by subtle differences in substitution pattern of the 4-(trifluoromethoxy)phenyl group of 68 versus the 4-fluoro-3-(trifluoromethyl)phenyl group of 66 that is indeed proposed to target the deep hydrophobic binding site formed by F1313.32 (Figure 19d). CXCR3 mutation studies suggest that D1122.63 and D1864.60 are also involved in ionic/H-bond interactions with compound 67 and that also the apolar F1353.36 residue plays an important role in binding 67.104 None of the three mutants tested in combination with 19 (Y601.39A, F1313.32A, I2796.59A) affected CXCR3 binding.64 The comparative analysis of effects of CCR5 (section 4.2)56 and CCR2 (section 5.7)53 mutations on 19 binding affinity may be useful to guide the design of complementary future CXCR3 mutation studies to elucidate the structural determinants of CXCR3 binding by this promiscuous chemokine receptor ligand.
5.7. Structural CCR1–Ligand Interaction Models
For CCR1 about 20 mutation data points have been determined, covering 13 different residue positions (including, among others, Y411.39 and Y1133.32) and two different ligands: 69 (BX-471) and 70 (UCB-35625) (Figure 20).50,51 Mutation studies indicated that Y411.39, Y1133.32, Y1143.33, I2596.55, E2877.39, and Y2917.43 play an important role in 69 binding and suggest that the ligand occupies both the minor and major binding pockets.50 The significant negative effect of the E2877.39Q mutant furthermore suggests that this residue is involved in an ionic interaction with the basic piperazine nitrogen atom in the ligand. The binding affinity of 70 is only significantly affected by Y411.39A and Y1133.32A mutations in the minor pocket but not by mutations in the major pocket (including Y1143.33A, I2596.55A, and E2877.39Q),51 suggesting that this ligand primarily targets the minor pocket of CCR1.
5.8. Structural CCR8–Ligand Interaction Models
The effect of a set of 100 mutations on CCR8 potency has been tested for the high-affinity nonpeptide ligand 71 (LMD-009) and its analogues 72 (LMD-584), 73 (LMD-902), 74 (LMD-268), and 75 (LMD-174), including residues Y421.39, Y1133.32, and E2867.39, among others.59 These analogues share a common scaffold including a positively charged amino within a piperidine-ring linked to a biphenyl ether on one side and to a variable group to the other site. The Y421.39A, Y1133.32A, or E2867.39A mutation data suggest that all four ligands share interactions with the minor pocket of CCR8 (Figure 20b).59 The negative effect of the E2867.39A mutation supports a ligand binding mode in which the charged amine of the piperidine-ring of 71–75 forms an H-bond and ionic interaction with the carboxylate side chain of E2867.39. The differential effects of F2546.51A and L2576.55A mutants on the potency of 71, 72, 73, 74, and 75 suggest that the variable left-hand side of these ligands (Figure 20b) is targeting the major pocket.
6. In Silico Chemokine Receptor Ligand Discovery and Design
As described in section 5, many challenges have to be overcome in the chemokine receptor modeling process. Despite the inevitable structural inaccuracies of refined chemokine receptor models, they have successfully been used to guide site-directed mutagenesis studies (Figure 11, 12, 13, 18–20) and design novel chemokine receptor ligands through structure-based in silico methods24,26−29 (Figure 22 and Table 1). Retrospective virtual screening experiments24,38,208 have been used to validate and select optimal conformations of chemokine receptor homology models by assessing the ability of the model (and in silico screening method) to discriminate known receptor ligands from inactive molecules and/or decoy molecules (randomly selected molecules that have not been experimentally tested on a specific protein target) with similar physicochemical properties. Ligand-based virtual screening (LBVS) has also significantly contributed in identifying new chemokine ligands, as well as other ligand-based approaches including three dimension quantitative structure–activity relationships,37 ligand-based pharmacophore modeling,37 similarity search34 (including shape matching or topological fingerprints), and Bayesian models.209 However, the current section will focus on prospective structure-based virtual screening (SBVS) studies to find novel chemokine receptor ligands, as well as discuss the challenges and possibilities of SBVS along the different steps of the SBVS workflow (Figure 21).
Figure 22.
Representative ligands obtained in structure-based virtual screening (design) studies against chemokine receptors homology models and X-ray structures, including CXCR4 ligands 76,2477,2478,2479,2480,2481,2782,2883,3085,2686,2987,37 and CXCR4/CXCR3 dual ligands 84;26 CCR5 ligands 88,3489,3490,3491,3492,35 and CCR4 ligand 93,32,33 and ACKR3/CXCR7 ligands 94,3895,38 and 96.38 Interactions between the ligands and specific residues derived from X-ray structures (bold), mutation studies (gray), or models without support from experimental data (gray italics) are depicted by dotted lines. The affinity or potency values of ligands are reported.
Table 1. Overview of Prospective Structure-Based Virtual Screening (SBVS) against Chemokine Models and Crystal Structuresa.
| receptor | templateb | focused database creationc | conf samplingd | (re)scoringe | interaction filterf | prospective initial dbg | validated hitsh (tested) | refi |
|---|---|---|---|---|---|---|---|---|
| CCR3 | de novo | dl | ad | score+3D | 120000 | 5 (43) | (31) | |
| CCR4 | bRho | 1D+2D | ad | score | 450000 | 16 (116) | (32,33) | |
| CCR5 | bRho | dl+2D | ad | score+clust+3D | 44524 | 10 (59) | (34) | |
| CCR5 | bRho | ad | score | 80000 | 1 (95) | (35) | ||
| CXCR4 | de novo | none | ad | score | D972.63/H1133.29/D1714.60/D2726.58/E2887.39 | 350000 | 1 (32) | (36) |
| CXCR4 | bRho | 1D+2D | ad | score | D1714.60 | 5 (5) | (37) | |
| D2626.58 | ||||||||
| E2887.39 | ||||||||
| CXCR4 | bRho ADRB1 ADRB2 A2A | ll | ad | score | E2887.39 | 3300000 | 1 (24) | (24) |
| CXCR4 | X-ray | ll | ad | score | E2887.39 | 420000 | 4 (23) | (24) |
| CXCR4 | X-ray | 2D+3D | ad | score | D972.63/Y1163.32 | 750 | 3 (16) | (27) |
| F1744.63/A1754.64 | ||||||||
| D18245.46/D18745.51 | ||||||||
| R18845.52/Y19045.54 | ||||||||
| D2626.58/E2887.39 | ||||||||
| CXCR4 | X-ray | MNP | 3D+ad | D972.63/D1714.60 | 250 | 1 (1) | (28) | |
| D18745.51/D2626.58 | ||||||||
| E2887.39 | ||||||||
| CXCR4 | X-ray | ll | ad | score+2D | D972.63/R18845.52 | 2400000 | 5 (11) | (26) |
| E2887.39 | ||||||||
| CXCR4 | X-ray | NP | ad | score | D972.63/D18745.51 | 8000 | 1 (1) | (29) |
| CXCR4 | X-ray | GF | ad | score | 13000 | 9 (3) | (30) | |
| CXCR3 | CXCR4 | ll | ad | score+2D | W1092.60/D1122.63 | 2400000 | 8 (11) | (26) |
| Q20445.51 | ||||||||
| ACKR3/CXCR7 | CXCR4 | Dl+clust. | ad+3D | score | D2756.58 | 800000 | 21 (626) | (38) |
| bRho | E2135.39 | |||||||
| ADRB1 ADRB2 A2A | D1794.60 | |||||||
| W1002.60 |
Only structure-based virtual screening studies targeting the TM domain are included.
Crystal structure template(s) used to construct the chemokine receptor homology model or cases in which the crystal structure are used are indicated (X-ray).
Consecutive filters (dl (drug-like physicochemical properties), ll (lead-like physicochemical properties), fl (fragment-like physicochemical properties), 1D (physicochemical properties known ligands), 2D (two-dimensional topological/chemical similarity/pharmacophoric features/subgroups), and 3D (three-dimensional pharmacophore) used to compile database for docking/3D conformer search, MNP marine natural product, NP natural product, GF GPCR-focused library.
Conformer search method: ((H-bond) constr(ained)) automated docking (ad), protein-based, or docked ligand-based 3D pharmacophore search (3D).
Method to score, rank and/or filter conformers: clust (scaffold clustering), (c)-score ((consensus) docking scoring function), 2D (two-dimensional topological/chemical similarity/pharmaphoric features/subgroups), and 3D (three-dimensional pharmacophore).
Key interactions with the listed residues were used to filter the docking poses.
Prospective validation: initial database (db).
Number of experimentally confirmed hits with detectable affinity/activity (of the total number of tested compounds).
References to homology modeling, virtual screening, and structure-based ligand optimization studies are provided.
Figure 21.
Customization of chemokine receptor structure-based virtual screening workflow. Different steps of the general structure-based virtual screening workflow (left, adapted from ref (212)) and their customization for chemokine receptors (right, described in section 6). The main steps in the structure-based virtual screening workflow are depicted in colored boxes and are complemented with chemokine receptor customized information in gray boxes on the right.
6.1. Challenges in Structure-Based Virtual Screening to Identify Chemokine Receptor Ligands
Before the release of CXCR4, CCR5, and US28 crystal structures, refined and customized chemokine homology models32−35,37 based on rhodopsin,92 ADRB2,210 and A2A211 crystal structures, as well as de novo receptor models,31,36 have been successfully used to identify new ligands for CCR3,31 CCR4,32,33 CCR5,34,35 and CXCR4.24,27−29,37 For the past five years, the first structure-based virtual screening studies against CXCR4 crystal structures,24,26−29 CXCR3,26 and ACKR3/CXCR738 homology models (based on CXCR4 crystal structures) have been reported. Virtual screening hit rates (the percentage of experimentally confirmed ligands among all tested in silico hits) based on chemokine receptor homology models (4%)24 and the CXCR4 crystal structures (18%)24,27 are somewhat lower than the hit rates reported for other GPCRs,212,213 including ADRB2 (24–60%),214,215 DRD3 (20–40%),216 A2A (10–41%),217,218 and H1R (73%).219 Moreover, one SBVS study31 performed against de novo 3D models of five different GPCRs (5-HT1A, 5-HT4, DRD2, NK1R, and CCR3) yielded hit rates of 12–21% (at a 5 μM cutoff) and several novel nanomolar affinity ligands for 5-HT1A, 5-HT4, DRD2, and NK1R, but only one low affinity hit (IC50 = 12 μM) for CCR3.
Despite the noticeable challenges of in silico discovery of chemokine receptor ligands, therapeutically interesting molecules that modulate chemokine receptors have recently been discovered in structure-based virtual screening campaigns. The binding mode prediction case study of 13 in CXCR4 discussed in section 5.4 (Figure 17) reflects the challenges of binding pocket residues selection, as well as pose selection in docking approaches. Because of ligand fit, residues often show different conformations between structures, even within the same receptor. For example, Y1163.32, H2035.42, W942.60, D972.63, D1714.60, and E2887.39 in the 31 bound CXCR4 structure adopt different conformations in the CVX15 bound CXCR4 structure. These conformational variations of binding site residues poses constitute both a challenge and an opportunity for structure-based virtual screening to identify CXCR4 ligands.
6.2. Hierarchical Workflow for Chemokine Receptor Structure-Based Ligand Discovery
A five-step structure-based virtual screening workflow can be defined customized to identify new ligands for chemokine receptors (Figure 21). The database preparation step 1 includes prefiltering of the initial compound library (e.g., commercially available compounds in ZINC (http://zinc.docking.org/)220) based on physical–chemical properties (including molecular weight, number of rotatable bonds, rings, hydrogen bond donor/acceptors, positively/negatively ionizable atoms) in order to select molecules that are, e.g., drug-like,221 fragment-like,222,223 and/or reflect physical–chemical properties of known ligands of a the chemokine receptor target(s). Chemokine receptor ligands are relatively large compared to, e.g,. aminergic receptor ligands224 and some known chemokine receptor ligands do not pass drug-likeness filters such as Lipinski’s rule of five (molecular weight <500 Da, <5 H-bond donors, <10 H-bond acceptors, log P < 5). An example is the CXCR4 antagonist 1 (Figure 11) with a molecular weight of 502 Da and 6 H-bond donors. The definition of physical chemical property filters based on known chemokine receptor ligands with high ligand efficiency225 can be a strategy to focus the virtual screening campaign on the identification of useful chemical starting points for further ligand optimization. An additional chemokine receptor specific filter would be to only consider molecules that contain a positively ionizable group, as this feature is present in most chemokine receptor ligands (Figures 10, 12, 13, and 17–20) and proposed to target negatively ionizable residues in the binding pocket of chemokine receptors (Figure 1–3). An alternative could be to consider only neutral or negatively ionizable molecules that are proposed to interact with the intracellular binding site of chemokine receptors (Figures 5 and 18).14,60 Challenges in the preparation of three-dimensional molecular structures of chemokine receptor focused databases using the physical chemical property and/or pharmacophore filters described above include (i) the definition of correct chemical topology and generation of relevant tautomers and protonation states of molecules containing multiple positively ionizable groups, and (ii) the generation of relevant three-dimensional ring conformations of basic nitrogen containing aliphatic ring systems. Both issues are important for the accurate sampling of ligand conformational space, scoring of receptor–ligand interactions, and definition of protein–ligand interaction fingerprints for docking pose postprocessing.226 In several virtual screening studies targeting chemokine receptors, additional ligand-based similarity filters have been used for database preprocessing based on substructures, 2D chemical similarity descriptors, 3D-shape similarity, or pharmacophore model features (Table 1). Shape-based and/or pharmacophore filters can for example be derived from crystal structure bound conformations of compounds 31 (in CXCR4) or 16 (in CCR5) (Figure 3), from receptor–ligand docking studies (Figure 17), and/or from ligand-based alignments (Figure 3).34,37 Pharmacophore models compatible with the interaction features of residues that play an important role in ligand binding based on chemokine receptor crystal structures and mutation studies (Figure 11, 12, 13, 18–20) are predicted to contain 1–2 cationic feature(s) and 1–2 aromatic/hydrophobic feature(s) (Figure 3d).
In step 2, the database of three-dimensional structures of small molecules is automatically docked into receptor structures/models, generating and ranking multiple conformations227 in a predefined binding site. Preparation of the protein for docking simulations includes careful definition of: (i) the binding site that will be targeted in the docking simulation, (ii) the assessment of tautomers and ionization states of receptor residues (i.e., H2817.32 in CXCR4) considering the interplay with ligand ionization states (e.g., the charged/neutral isothiourea moiety of CXCR ligand 31 interacting with E2887.39 in CXCR4), (iii) the position of polar hydrogen atoms (e.g., Y371.39 in CCR5), (iv) atom typing of cofactors and/or unnatural amino acids (e.g., sY12N-ter in CXCR4), and (v) the consideration of relevant water molecules that can be targeted and/or displaced (e.g., water molecules 1629, 1646, and 1720 in the 31 bound CXCR4 crystal structure). A possible strategy could be to target only the minor pocket (e.g., defined based on the binding pocket of 31 in the CXCR4 crystal structure), the major pocket (e.g., based on the CVX15 bound CXCR4 structure), the minor and major pocket simultaneously (e.g., based on the 16 bound CCR5 structure), or other putative binding sites (e.g., extracellular vestibule, intracellular region, membrane interface) depending on ligand similarity, mutation data, SAR information, and/or radioligand displacement data.
In step 3, the docking poses are postprocessed and ranked. Most structure-based virtual screening studies targeting chemokine receptors have primarily employed docking scoring functions to rank docking poses (Table 1), but more and more structure-based in silico screening protocols now include additional filters to postprocess docking results that combine consensus scoring strategies,228−230 topological filters,231 receptor–ligand IFP scoring methods,41,219,231−233 and receptor–ligand interaction post-processing filters.24,26−28,36,38,234 Site-directed mutagenesis studies (Figure 10–12, 18–20) and ligand structure–activity relationships (Figure 14) have provided information to define knowledge-based chemokine receptor–ligand interaction filters to postprocess docking poses (Table 1). For example, ionic and/or H-bond interactions involving D972.63 and E2887.39 (Figure 11) have been used as a filter for postprocessing docking poses in structure-based virtual screening studies against CXCR4.24,26−28,36,234 Crystal structures and mutation studies have identified several key residues that are essential for binding affinity and interactions with these residues, which can be used to define postprocess/filter docking poses: Y451.39, W942.60, D972.63, Y1163.32, H1133.29, D1714.60, D2626.58, and E2887.39 in CXCR4 (Figures 3a, 4a, 10–11); Y371.39, P842.58, W862.60, C1013.25, Y1083.32, I1985.42, Y2516.51, and E2837.39 in CCR5 (Figures 3c, 12); S1072.63, D1433.49, R2125.39, Q2165.43, S2175.44, C2636.47, and N2686.52 for CXCR2 (Figure 18); W1092.60, D1122.63, G12883.29, F1313.32, R2125.35, W2686.48, S3047.39, and Y3087.43 for CXCR3 (Figure 19); Y411.39, Y1133.32, Y1143.33, I2596.55, E2877.39, and Y2927.43 for CCR1 (Figure 20); Y1203.32, H1213.33, E2917.39, and Y2927.40 for CCR2 (Figure 13); and Y421.39, F461.43, F882.57, Q912.60, S1103.29, Y1133.32, Y1143.33, and E2867.39 for CCR8 (Figure 20). It should be noticed however that the binding mode and role of specific residues can be ligand dependent. It is therefore necessary to perform retrospective virtual screening studies to evaluate the effect of docking pocket definition and postprocessing filter on the possibility to discriminate known ligands from inactive or decoy molecules.
In step 4, the final hits can be selected based on clustering, chemical novelty, visual inspection of the protein–ligand binding mode, and other properties associated with the feasibility to optimize the ligand in subsequent medicinal chemistry projects (including drug-likeness (see also step 1 and synthetic tractability). The novelty of the discovered hits can be assessed by a 2D or 3D similarity search against known chemokine receptor ligands. For example, the highest Tanimoto coefficients (Tc) of hits identified in a virtual screening study against CXCR424 was 0.36 to the most chemically similar known CXCR4 ligand (at the time of the screen) based on the ECFP-4 chemical fingerprint.235 Clustering virtual screening hits by chemical diversity before visual selection is an efficient way of analysis when a lot of ligands are retrieved. Furthermore, pan-assay interference compounds (PAINS) or those that have poor oral bioavailability221 can be excluded. The CXCR7 virtual screening hit 96(38) for example contains a rhodanine PAINS scaffold (137:ene_five_het_B) which is enriched in high-throughput screening hit sets236 and has been shown to undergo light-induced reactions that irreversibly modify proteins.237 Another criteria for the final selection of hits is the visual inspection of docking poses for the detailed analysis of protein–ligand interactions that are challenging to automate and/or score, including for example the analysis of strained ligand conformations,238 unfavorable protein–ligand interactions such as buried polar groups in hydrophobic pockets,212,213 or scoring of specific interactions that are not yet incorporated in most scoring functions, such as halogen bonds.239 For this purpose, 3D-QSAR models may be used to assist the prioritization of docking poses in a more systematic way. For example, the 3D-QSAR model presented in Figure 3b shows favorable hydrophobic interaction field that can facilitate the prioritization of docking poses of other compounds in the minor pocket of CXCR4. In addition to optimization of protein–ligand interactions, other criteria like protein selectivity (e.g., emphasizing receptor specific interactions and/or incompatibility with off-targets) or synthetic tractability (e.g., consideration of accessible reaction routes, avoiding potential complexity of multiple stereoisomer) can be considered in the final hit prioritization. In most published chemokine receptor structure-based virtual screening studies, a small subset of the compounds is purchased and experimentally validated (step 5, see 76–96 in Figure 22).240 Typically, on the order of 10–100 hits24,26,27,31−36 are evaluated (though smaller28,29,37 or larger38 sets have been reported), yielding 1–20 experimentally validated ligands. Structure-based virtual screening hits have been optimized by structure-based design for several GPCRs (e.g., A2A receptor antagonist,241,242 5-HT receptor antagonist,175 and 5-HT1A agonist243), but so far only few studies27 have described the structure-based optimization of virtual screening hits for chemokine receptors.
6.3. Prospective VS Targeting Chemokine Receptors
Table 1 summarizes 12 successfully SBVS studies against CCR3,31 CCR4,32,33 CCR5,34,35 CXCR3,26 CXCR4,24,26−29,36,37 and ACKR3/CXCR7.38 Becker et al.31 identified 43 compounds through virtual screening of 120000 compounds against a de novo constructed structural model of CCR3, yielding five experimentally validated hits (Ki< 20 μM). Bayry et al. docked 13000 molecules (selected from a pharmacophore filtered database of 45000 compounds) in a CCR4 homology model based on the bovine rhodopsin (bRho) crystal structure. This resulted in 116 hits, of which 16 were experimentally confirmed as potent CCR4 antagonist, including 93.32,33 Kellenberger et al. identified new CCR5 agonists,34 including 90 and 91, by virtual screening against an antagonist-customized receptor homology model combining bRho-based homology modeling and refinement of the T2.56XP2.58 induced kink of TM2 and validated by retrospective virtual screening using two different docking programs. A focused library of 44524 drug-like compounds selected based on a 2D pharmacophore filter was docked into the receptor model by two different docking programs. A total of 59 molecules representing scaffold classes enriched in the top 5% of the docking scoring lists were selected by visual inspection of the docking poses and tested; 10 compounds exhibited a detectable binding affinity for CCR5, of which four molecules had an IC50 in the high micromolar range. Surprisingly, three molecules were characterized as agonists, illustrating that agonists can also be found by structure-based virtual screening in inactive GPCR models.233 Another CCR5 virtual screening study conducted by Liu et al.35 through docking screening (92) and followed by fragment assembly (from the known CCR5 antagonist 16), design, and synthesis resulted in a series of novel small CCR5 antagonists (IC50: 0.2–10 μM). Kim et al.36 performed docking-based virtual screening of 350000 compounds against a de novo CXCR4 model and selected molecules that were docked in close proximity to the acidic residues D1714.60, D2626.58, and E2887.39. One of the 32 experimentally validated virtual screening hits (compound 37, which is chemically similar to the antimalarial drugs chloroquine and hydroxychloroquine) was confirmed as an antagonist of CXCR4-mediated signaling and cell proliferation in pancreatic cancer cells. Perez-Nueno et al.37,234 used an optimized and retrospectively validated combination of ligand-based and protein structure-based approaches to design five compounds that were synthesized and experimentally confirmed as CXCR4 ligands (including 87) with anti-HIV activities of 0.022–4.4 μg/mL. Mysinger et al.24 used the CXCR4 crystal structure and CXCR4 homology models (constructed for the GPCR Dock 2010 Assessment20) in a comparative virtual screening study. In a retrospective validation of their procedure to discriminate 60 actives from 2000 decoys, the crystal structure (28) shows higher enrichment factor (1% of decoys) than the models (the best is 22). Then libraries of >3 million of lead-like compounds were docked against the models and crystal structures. Experimental validation yielded one and four hit compounds (a hit rate of 4% and 17%) from homology models and the crystal structure, respectively (with affinities ranging from 0.31 μM (78) to 225 μM (76)). Das et al.27 identified a high affinity CXCR4 ligand (81) by combining Tanimoto shape similarity search, docking, and filtering docking poses that made interactions with at least two critical residues (D972.63/Y1163.32/F1744.63/A1754.64/D18245.46/D18745.51/R18845.52/Y19045.54/D2626.58/E2887.39). Finally, three of the 16 hits were confirmed experimentally (IC50: 92–161 nM). Recently, Mishra et al. identified one CXCR4 agonist (83) and two antagonists with micromolar potency by CXCR4 crystal structure-based virtual screening of a GPCR focused library of 13000 compounds.30 Other structure-based virtual screening targeting CXCR4 which focus on natural product databases also resulted in discovering novel CXCR4 ligands such as 82 (PHIA)28 and 86 (silibinin).29 Yoshikawa et al. combined a 3D pharmacophore filter and docking simulations to identify 21 novel ACKR3/CXCR7 ligands (94–96) (IC50: 1–11 μM).38 Docking poses that formed polar interaction with D1794.60, D2756.58, and a hydrophobic interaction with W1002.60 were prioritized in the virtual screening protocol. Schmidt et al. demonstrated that structure-based virtual screening can be successfully used in the discovery of multitarget chemokine ligands.26 Docking simulations against a CXCR3 homology model and CXCR4 crystal structure resulted in the identification of six CXCR3 selective ligands, three CXCR4 selective ligands, and two CXCR3/CXCR4 dual binders (including 84). Overlay of the CXCR3 homology model and CXCR4 crystal structure showed similarities and differences in binding site residues, including: (i) CXCR4 residue E2887.39 is a conserved (glutamate) in most chemokine receptors but a serine (S3047.39) in CXCR3, (ii) aromatic residues F1313.32 and Y3087.43 in CXCR3 align with aromatic residues Y1163.32 and F2927.43 in CXCR4, (iii) H1133.29 of CXCR4 interacts with CVX15, whereas the homologous G1283.29 CXCR3 offers more space for ligands and has been proposed as a potential selectivity site, and (iv) the cationic R2165.39 in CXCR3 versus the neutral Q2005.39 in CXCR4 represent an additional potential determinant of CXCR3/CXCR4 selectivity.
7. Conclusions and Outlook
The CXCR4, CCR2, CCR5, CCR9, and US28 X-ray crystal structures offer useful structural templates for the comparative analysis and prediction of the interactions between chemokine receptors and small molecules and large peptide and protein ligands. The integrated analysis of the new structural information on chemokine receptors with extensive structure–activity relationship and site-directed mutagenesis data has facilitated the construction of structural models of chemokine receptor–ligand complexes that have not been crystallized. This experimentally enhanced chemokine receptor modeling strategy helps to overcome the challenges in predicting structural chemokine receptor–ligand interactions. Chemical tools such as small-molecule radioligands are required to complement integrated structural modeling and mutation studies to accurately characterize the many distinct extracellular and intracellular small-molecule allosteric binding sites in chemokine receptors and identify other alternative binding sites such as for example the interface of the TM domain and the membrane bilayer, observed in recent GPCR crystal structures.197,244
Chemokine bound crystal structures of CXCR4 and US28 share similar binding modes of the chemokine core but lack structural information regarding chemokine interactions with the N-terminal region of the receptor and show different binding modes of the N-terminus of the chemokine in the TM bundle. These differential binding modes are in line with reported chemokine selectivity profiles, ligand dependent mutation effects, and the different conformations of the flexible chemokine N-terminus in X-ray and NMR structures. Specific challenges in modeling chemokine bound structures therefore include: (i) the prediction of interactions with the receptor N-terminus of chemokines, (ii) modeling chemokine binding induced conformational changes of in particular the extracellular loop region, and (iii) describing the dynamics of the flexible binding mode of the N-terminal region of the chemokine in the minor and/or major 7TM binding site(s).
The currently available crystal structures of CXCR4, CCR2, CCR5, CCR9, and US28 provide useful templates to accurately model the TM helical fold, including the chemokine receptor specific T2.56XP2.58 stabilized bulge in TM2, and offer structural model templates with sufficiently high sequence identity to model the TM helices and/or binding sites of other chemokine receptors. More crystal structures of chemokine bound receptor structures will provide new insights into receptor-bound chemokine structural alignment, ELs conformation in a chemokine-bound state, and the bound conformation of N-terminus of the chemokine in the 7TM helical bundle of the receptor. Moreover, it is known that various chemokines can bind more than one chemokine receptor, as well as a receptor can bind more than one different chemokine.1 More chemokine-bound crystal structures will provide insights into chemokine selectivity/redundancy because the key interactions that promote chemokine selectivity remain still unknown. However, specific regions have lower similarity to their subfamily reference crystallized chemokine receptors and therefore using as a template the structure of the alternative subfamily should be considered. Although the length of extracellular loops are similar among chemokine receptors, the CXCR4, CCR2, CCR5, CCR9, and US28 crystal structures show that their conformation is influenced by ligand binding, especially by binding of large molecules such as chemokines.
Structure-based ligand discovery and design studies based on chemokine receptor crystal structures and homology models illustrate not only the possibilities but also the challenges to find novel ligands for chemokine receptors. Important pitfalls that have to be addressed in structure-based virtual screening against chemokine receptors are (i) defining an effective scoring method for hit selection and prioritization, (ii) availability of structural data of the receptor, including X-ray, NMR, SAR, and site-directed mutagenesis data, (iii) existence of multiple and/or allosteric binding pockets in the same receptor, (iv) making enough conformational sampling considering ligands flexibility, and (v) properly preparation of ligands and receptor, especially regarding their protonation state. Structure-based virtual approaches have been successfully applied for chemokine receptors but still require optimization to efficiently address the challenges associated with the symmetric distribution of pharmacophore features in chemokine receptors ligands and the large, open binding sites of chemokine receptors, compared to for example the more druggable, occluded binding sites in aminergic GPCRs. The structural alignment comparison of active US28 vs inactive CXCR4 and CCR5 structures provide clues into the structural features that are significantly representative of active and inactive states. More detailed insights into the structural determinants of functional efficacy of chemokine and small-molecule ligands would, for example, facilitate the rational discovery and design of biased agonists104 or antagonists149 by stimulating or inhibiting specific signaling pathways mediated by chemokine receptors. Crystal structures bound to ligands with different functional activities therefore will give new insights that may be used to rationalize interesting structure–function relationships of chemokines121 and small-molecule ligands,245 and may be used to develop customized structure-based virtual screening methods to rationally identify ligands with a specific functional effect, as demonstrated for, e.g., aminergic GPCRs.246 Whereas the currently available (mini) G-protein and beta-arrestin bound GPCR crystal structures87−89 provide templates for low resolution modeling of chemokine receptor–transducer interactions,247 crystal structures of intracellular effector bound chemokine receptors will be required for high resolution investigation of the structural determinants of chemokine receptor mediated signal transduction. Although the CXCR4 crystal structure may represent a possible conformation of a chemokine receptor homodimer,248 several other GPCR oligomerization models have been proposed,249 and more information is needed to identify and validate chemokine receptors oligomerization models that are pharmacologically relevant.250 The recently solved allosteric antagonist bound CCR9 crystal structure and CCR2 chemokine receptor crystal structure simultaneously targeted by orthosteric and allosteric antagonists provide high resolution structural templates to investigate structural interactions with distinct ligand binding pockets in chemokine receptors and will increase our understanding of the interplay between orthosteric and allosteric ligand binding. The accumulated information on the extracellular and intracellular binding sites of chemokine receptors, as well as so far unexploited extrahelical binding sites identified in other GPCRs197,251 can open up new possibilities for the computer-aided discovery and design of novel chemokine receptor ligands with complementary modes of action. These future developments can include parallel or sequential252 structure-based virtual screening against multiple binding sites, the design of bitopic ligands that simultaneously target different binding pockets by, e.g., fragment linking approaches,253 or the structure-based design of bivalent ligands that target different chemokine receptor monomers254 simultaneously. Moreover, complementing the available structural information with MD simulations may help to reveal ligand association and dissociation pathways and provide insights into the structural determinants of chemokine receptor–ligand binding kinetics.100,255 It is expected that experimentally enhanced structural modeling of structural chemokine receptor–ligand interactions will provide new insights into complex GPCR structure–function relationships by the computer-aided integration of structural, pharmacological, and chemical information.
Acknowledgments
This work was supported by a European Union’s Horizon2020 MSCA Programme under grant agreement 641833 (ONCORNET to M.J.S. and R.L.), The Netherlands eScience Center (NLeSC)/NWO (Enabling Technologies project: 3D-e-Chem, grant 027.014.201 to C.d.G.), NWO CW TOP-PUNT grant 718.014.002, 7 ways to 7TMR modulation (7-to-7), to R.L. and M.J.S.) and China Scholarship Council (CSC grant to S.S.). M.A., S.S., M.J.S., R.L., I.J.P.d.E., and C.d.G. participate in the European Cooperation in Science and Technology Action CM1207 [GPCR–Ligand Interactions, Structures, and Transmembrane Signaling: A European Research Network (GLISTEN)] and the GPCR Consortium (gpcrconsortium.org).
Glossary
Abbreviations Used
- A2A
adenosine receptor A2A
- ACKR
atypical decoy chemokine receptor
- ADRB
adrenoceptor B
- BRET
bioluminescence resonance energy transfer
- Cα
α carbon
- CDR
complementarity determining-like binding regions
- CRS
chemokine recognition site
- C-ter
c terminal
- ECL
extracellular loop
- GPCR
G-protein coupled receptor
- GRK
G-protein coupled receptor kinase
- H8
helix 8
- H-bond
hydrogen bond
- ICL
intracellular loop
- IFP
interactions fingerprint
- LBVS
ligand-based virtual screening
- MD
molecular dynamics
- MM
multiple myeloma
- Nb7
nanobody 7
- NCAM
neural cell adhesion molecule
- NHL
non-Hodgkins lymphoma
- N-ter
N-terminal
- PAINS
pan assay interface
- QSAR
quantitative structure–activity relationships
- SBVS
structure-based virtual screening
- SDF-1
stromal derived factor-1
- T4L
T4 lysozyme
- Tc
Tanimoto coefficient
- TM
transmembrane
- TMS
transmembrane site
Biographies
Marta Arimont obtained her B.Sc. degree in Biomedical Sciences and her M.Sc. degree in Bioinformatics at Universitat Autonoma de Barcelona. Since 2015, she has been carrying out her Ph.D. under the supervision of Dr. Chris de Graaf, Prof. Dr. Rob Leurs, and Prof. Dr. Iwan de Esch in the Division of Medicinal Chemistry at Vrije Universiteit Amsterdam. Her research focuses on in silico guided identification of molecular determinants for modulation of CXCR4 and CXCR7, developing three-dimensional molecular models to predict interaction of ligands of several natures, identifying molecular determinants and unraveling mechanisms of orthosteric and allosteric modulation of CXCR4 and CXCR7 function, and working on computer-aided discovery and design of novel modulators, including docking, virtual screening, SAR design, and homology modeling.
Shan-Liang Sun obtained his B.Sc. degree in Information Management and Information System and his M.Sc. degree in Medicinal Chemistry at China Pharmaceutical University. During his Master’s, he worked on the modeling, design, and synthesis of Polo-like kinase 1 inhibitors in the supervision of Prof. Dr. Tao Lu. Since 2014, he has been carrying out his Ph.D. under the supervision of Dr. Chris de Graaf, Prof. Dr. Iwan de Esch, and Prof. Dr. Rob Leurs in the Division of Medicinal Chemistry at Vrije Universiteit Amsterdam. In his Ph.D. project, computational techniques are combined with medicinal chemistry approaches to develop new small-molecule ligands for the chemokine receptors CXCR4 and CXCR7.
Rob Leurs performed his Ph.D. research on G-protein coupled receptors at Vrije Universiteit Amsterdam. After a postdoctoral fellowship at INSERM, he was awarded with a Royal Netherlands Academy of Arts and Sciences fellowship. He was awarded the Galenus Research Prize (1997), the Organon Award for Pharmacology (2000), a Pfizer Academic Award (2001), a STW/NOW Pionieer grant (2001), and was appointed academy member of the Royal Netherlands Academy of Arts and Sciences (KNAW) (2016). Rob Leurs is full professor and head of the Division of Medicinal Chemistry at Vrije Universiteit Amsterdam and cofounder of Griffin Discoveries, a company that valorizes the GPCR expertise. Prof. Dr. Leurs is project leader of the EU FP7-funded project PDE4NPD, phosphodiesterase inhibitors for the treatment of neglected parasitic diseases (www.PDE4NPD.eu).
Martine Smit obtained her Ph.D. in Molecular Pharmacology at the VU University Amsterdam, After a postdoctoral fellowship at Mount Sinai School of Medicine, she established a new research line on (viral) chemokine receptors (fellowship Royal Netherlands Academy of Arts and Sciences), funded by the Dutch Science Foundation (NWO-VIDI-VICI) at Vrij Universiteit Amsterdam. She was awarded the Galenus Research Prize (2002) and the Organon Award for Pharmacology (2007). She was appointed professor in “Target and Systems Biochemistry” in 2005. Her research focuses at novel concepts of GPCRs (e.g., constitutive/ligand-biased signaling) and understanding/targeting of (oncogenic) signaling networks activated by (virally encoded) chemokine receptors. Prof. Dr. Smit is project leader of the European Union’s Horizon2020 MSCA Programme ITN project Oncogenic GPCR Network of Excellence and Training (www.oncornet.eu).
Iwan J. P. de Esch performed his Ph.D. research at Vrije Universiteit Amsterdam. He became a research associate in the drug design group at the University of Cambridge in 1998 and cofounded De Novo Pharmaceuticals in 2000. In 2003, Iwan de Esch returned to academia and is now full professor Biocomputational Chemistry for Drug Innovation at Vrije Universiteit Amsterdam. Prof. Dr. De Esch is cofounder of IOTA Pharmaceuticals and cofounder of Griffin Pharmaceuticals, contributing to the valorization of the fragment-based drug discovery and the GPCR research lines of the academic group. In 2011, De Esch was awarded the Galenus Research Prize for his work on fragment-based drug discovery, and since 2016 he is project leader of the European Union’s Horizon2020 MSCA Programme ITN project FragNet (www.fragnet.eu).
Chris de Graaf performed his Ph.D. research at Vrije Universiteit Amsterdam on computational ligand binding mode and affinity predictions in cytochrome P450 enzymes (2002–2006). As postdoctoral fellow in the Structural Chemogenomics group (Dr. Rognan) at Universitéde Strasbourg, he worked in collaboration with AstraZeneca Pharmaceuticals on the development and application of novel G protein-coupled receptor modeling techniques (2006–2008). In 2009, Chris de Graaf obtained an NWO Veni grant to develop a research line in the computational prediction of structural protein–ligand interactions and was appointed assistant professor in the Division Medicinal Chemistry at Vrije Universiteit Amsterdam. In this interdisciplinary research environment, Dr. De Graaf is developing chemo/bioinformatics methods to complement synthetic medicinal chemistry and molecular pharmacology programs, including e-science technologies for structure-based polypharmacology prediction (http://3d-e-chem.github.io).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.6b01309. The chemokine receptor mutation data has furthermore been uploaded to Zenodo (https://zenodo.org/record/268416) and the GPCRdb (http://gpcrdb.org) to allow searching and comparing the data with other GPCR mutation data (including links to the original publications and review).
The rendered structural alignments (PDB and Pymol files), interaction fingerprint (IFP) analyses, and chemokine receptor mutation data used for the analyses (ZIP)
Author Contributions
† Marta Arimont and Shan-Liang Sun contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Scholten D. J.; Canals M.; Maussang D.; Roumen L.; Smit M. J.; Wijtmans M.; de Graaf C.; Vischer H. F.; Leurs R. Pharmacological modulation of chemokine receptor function. Br. J. Pharmacol. 2012, 165, 1617–43. 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I.; Salanga C. L.; Handel T. M. Chemokine and chemokine receptor structure and interactions: implications for therapeutic strategies. Immunol. Cell Biol. 2015, 93, 372–83. 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelerie F.; Graham G. J.; Locati M.; Mantovani A.; Murphy P. M.; Nibbs R.; Rot A.; Sozzani S.; Thelen M. An atypical addition to the chemokine receptor nomenclature: IUPHAR Review 15. Br. J. Pharmacol. 2015, 172, 3945–9. 10.1111/bph.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S. J.; Crown S. E.; Handel T. M. Chemokine: receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 2007, 25, 787–820. 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- Bongers G.; Maussang D.; Muniz L. R.; Noriega V. M.; Fraile-Ramos A.; Barker N.; Marchesi F.; Thirunarayanan N.; Vischer H. F.; Qin L.; Mayer L.; Harpaz N.; Leurs R.; Furtado G. C.; Clevers H.; Tortorella D.; Smit M. J.; Lira S. A. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Invest. 2010, 120, 3969–78. 10.1172/JCI42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger E.; Maussang D.; Schreiber A.; Siderius M.; Rahbar A.; Fraile-Ramos A.; Lira S. A.; Soderberg-Naucler C.; Smit M. J. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signaling 2010, 3, ra58. 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- Vischer H. F.; Siderius M.; Leurs R.; Smit M. J. Herpesvirus-encoded GPCRs: neglected players in inflammatory and proliferative diseases?. Nat. Rev. Drug Discovery 2014, 13, 123–39. 10.1038/nrd4189. [DOI] [PubMed] [Google Scholar]
- Smit M. J.; Vink C.; Verzijl D.; Casarosa P.; Bruggeman C. A.; Leurs R. Virally encoded G protein-coupled receptors: targets for potentially innovative anti-viral drug development. Curr. Drug Targets 2003, 4, 431–41. 10.2174/1389450033491000. [DOI] [PubMed] [Google Scholar]
- Woollard S. M.; Kanmogne G. D. Maraviroc: a review of its use in HIV infection and beyond. Drug Des., Dev. Ther. 2015, 9, 5447–5468. 10.2147/DDDT.S90580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatse S.; Princen K.; Bridger G.; De Clercq E.; Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002, 527, 255–62. 10.1016/S0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- Wu B.; Chien E. Y.; Mol C. D.; Fenalti G.; Liu W.; Katritch V.; Abagyan R.; Brooun A.; Wells P.; Bi F. C.; Hamel D. J.; Kuhn P.; Handel T. M.; Cherezov V.; Stevens R. C. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010, 330, 1066–71. 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q.; Zhu Y.; Li J.; Chen Z.; Han G. W.; Kufareva I.; Li T.; Ma L.; Fenalti G.; Li J.; Zhang W.; Xie X.; Yang H.; Jiang H.; Cherezov V.; Liu H.; Stevens R. C.; Zhao Q.; Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–90. 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L.; Kufareva I.; Holden L. G.; Wang C.; Zheng Y.; Zhao C.; Fenalti G.; Wu H.; Han G. W.; Cherezov V.; Abagyan R.; Stevens R. C.; Handel T. M. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 2015, 347, 1117–22. 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg J. S.; Ingram J. R.; Venkatakrishnan A. J.; Jude K. M.; Dukkipati A.; Feinberg E. N.; Angelini A.; Waghray D.; Dror R. O.; Ploegh H. L.; Garcia K. C. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 2015, 347, 1113–7. 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.; Qin L.; Zacarias N. V.; de Vries H.; Han G. W.; Gustavsson M.; Dabros M.; Zhao C.; Cherney R. J.; Carter P.; Stamos D.; Abagyan R.; Cherezov V.; Stevens R. C.; Ijzerman A. P.; Heitman L. H.; Tebben A.; Kufareva I.; Handel T. M. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 2016, 540, 458–461. 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald C.; Rappas M.; Kean J.; Dore A. S.; Errey J. C.; Bennett K.; Deflorian F.; Christopher J. A.; Jazayeri A.; Mason J. S.; Congreve M.; Cooke R. M.; Marshall F. H. Intracellular allosteric antagonism of the CCR9 receptor. Nature 2016, 540, 462–465. 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- Skelton N. J.; Quan C.; Reilly D.; Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure 1999, 7, 157–68. 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Veldkamp C. T.; Seibert C.; Peterson F. C.; De la Cruz N. B.; Haugner J. C. 3rd; Basnet H.; Sakmar T. P.; Volkman B. F. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci. Signaling 2008, 1, ra4. 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard C. J.; Ludeman J. P.; Canals M.; Bridgford J. L.; Hinds M. G.; Clayton D. J.; Christopoulos A.; Payne R. J.; Stone M. J. Structural basis of receptor sulfotyrosine recognition by a CC chemokine: the N-terminal region of CCR3 bound to CCL11/eotaxin-1. Structure 2014, 22, 1571–81. 10.1016/j.str.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Kufareva I.; Rueda M.; Katritch V.; Stevens R. C.; Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure 2011, 19, 1108–1126. 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariassen Z. G.; Karlshoj S.; Haug B. E.; Rosenkilde M. M.; Vabeno J. Probing the Molecular Interactions between CXC Chemokine Receptor 4 (CXCR4) and an Arginine-Based Tripeptidomimetic Antagonist (KRH-1636). J. Med. Chem. 2015, 58, 8141–53. 10.1021/acs.jmedchem.5b00987. [DOI] [PubMed] [Google Scholar]
- Junker A.; Kokornaczyk A. K.; Zweemer A. J.; Frehland B.; Schepmann D.; Yamaguchi J.; Itami K.; Faust A.; Hermann S.; Wagner S.; Schafers M.; Koch M.; Weiss C.; Heitman L. H.; Kopka K.; Wunsch B. Synthesis, binding affinity and structure-activity relationships of novel, selective and dual targeting CCR2 and CCR5 receptor antagonists. Org. Biomol. Chem. 2015, 13, 2407–22. 10.1039/C4OB02397H. [DOI] [PubMed] [Google Scholar]
- Roumen L.; Scholten D. J.; de Kruijf P.; de Esch I. J.; Leurs R.; de Graaf C. C(X)CR in silico: Computer-aided prediction of chemokine receptor-ligand interactions. Drug Discovery Today: Technol. 2012, 9, e281–91. 10.1016/j.ddtec.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Mysinger M. M.; Weiss D. R.; Ziarek J. J.; Gravel S.; Doak A. K.; Karpiak J.; Heveker N.; Shoichet B. K.; Volkman B. F. Structure-based ligand discovery for the protein-protein interface of chemokine receptor CXCR4. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 5517–22. 10.1073/pnas.1120431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B. D.; Prosser A. R.; Sun Y.; Li Z.; Lee S.; Huang M. B.; Bond V. C.; Snyder J. P.; Krystal M.; Wilson L. J.; Liotta D. C. Pyrazolo-Piperidines Exhibit Dual Inhibition of CCR5/CXCR4 HIV Entry and Reverse Transcriptase. ACS Med. Chem. Lett. 2015, 6, 753–7. 10.1021/acsmedchemlett.5b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.; Bernat V.; Brox R.; Tschammer N.; Kolb P. Identifying modulators of CXC receptors 3 and 4 with tailored selectivity using multi-target docking. ACS Chem. Biol. 2015, 10, 715–24. 10.1021/cb500577j. [DOI] [PubMed] [Google Scholar]
- Das D.; Maeda K.; Hayashi Y.; Gavande N.; Desai D. V.; Chang S. B.; Ghosh A. K.; Mitsuya H. Insights into the mechanism of inhibition of CXCR4: identification of Piperidinylethanamine analogs as anti-HIV-1 inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1895–904. 10.1128/AAC.04654-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale R. M.; Gatti M.; Carbone M.; Barbieri F.; Felicita V.; Gavagnin M.; Florio T.; Amodeo P. Minimalist hybrid ligand/receptor-based pharmacophore model for CXCR4 applied to a small-library of marine natural products led to the identification of phidianidine a as a new CXCR4 ligand exhibiting antagonist activity. ACS Chem. Biol. 2013, 8, 2762–70. 10.1021/cb400521b. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Liang W. C.; Pan W. L.; Law W. K.; Hu J. S.; Ip D. T.; Waye M. M.; Ng T. B.; Wan D. C. Silibinin, a novel chemokine receptor type 4 antagonist, inhibits chemokine ligand 12-induced migration in breast cancer cells. Phytomedicine 2014, 21, 1310–7. 10.1016/j.phymed.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Mishra R. K.; Shum A. K.; Platanias L. C.; Miller R. J.; Schiltz G. E. Discovery and characterization of novel small-molecule CXCR4 receptor agonists and antagonists. Sci. Rep. 2016, 6, 30155. 10.1038/srep30155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker O. M.; Marantz Y.; Shacham S.; Inbal B.; Heifetz A.; Kalid O.; Bar-Haim S.; Warshaviak D.; Fichman M.; Noiman S. G protein-coupled receptors: in silico drug discovery in 3D. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 11304–9. 10.1073/pnas.0401862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. N.; Bayry J.; Tchilian E. Z.; Vani J.; Shaila M. S.; Forbes E. K.; Draper S. J.; Beverley P. C.; Tough D. F.; Flower D. R. Toward the discovery of vaccine adjuvants: coupling in silico screening and in vitro analysis of antagonist binding to human and mouse CCR4 receptors. PLoS One 2009, 4, e8084. 10.1371/journal.pone.0008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayry J.; Tchilian E. Z.; Davies M. N.; Forbes E. K.; Draper S. J.; Kaveri S. V.; Hill A. V.; Kazatchkine M. D.; Beverley P. C.; Flower D. R.; Tough D. F. In silico identified CCR4 antagonists target regulatory T cells and exert adjuvant activity in vaccination. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10221–6. 10.1073/pnas.0803453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E.; Springael J. Y.; Parmentier M.; Hachet-Haas M.; Galzi J. L.; Rognan D. Identification of nonpeptide CCR5 receptor agonists by structure-based virtual screening. J. Med. Chem. 2007, 50, 1294–303. 10.1021/jm061389p. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Zhou E.; Yu K.; Zhu J.; Zhang Y.; Xie X.; Li J.; Jiang H. Discovery of a novel CCR5 antagonist lead compound through fragment assembly. Molecules 2008, 13, 2426–41. 10.3390/molecules13102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Yip M. L.; Shen X.; Li H.; Hsin L. Y.; Labarge S.; Heinrich E. L.; Lee W.; Lu J.; Vaidehi N. Identification of anti-malarial compounds as novel antagonists to chemokine receptor CXCR4 in pancreatic cancer cells. PLoS One 2012, 7, e31004. 10.1371/journal.pone.0031004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Nueno V. I.; Pettersson S.; Ritchie D. W.; Borrell J. I.; Teixido J. Discovery of novel HIV entry inhibitors for the CXCR4 receptor by prospective virtual screening. J. Chem. Inf. Model. 2009, 49, 810–23. 10.1021/ci800468q. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y.; Oishi S.; Kubo T.; Tanahara N.; Fujii N.; Furuya T. Optimized method of G-protein-coupled receptor homology modeling: its application to the discovery of novel CXCR7 ligands. J. Med. Chem. 2013, 56, 4236–51. 10.1021/jm400307y. [DOI] [PubMed] [Google Scholar]
- Ballesteros J. A.; Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- Isberg V.; de Graaf C.; Bortolato A.; Cherezov V.; Katritch V.; Marshall F. H.; Mordalski S.; Pin J. P.; Stevens R. C.; Vriend G.; Gloriam D. E. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015, 36, 22–31. 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C.; Foata N.; Engkvist O.; Rognan D. Molecular modeling of the second extracellular loop of G-protein coupled receptors and its implication on structure-based virtual screening. Proteins: Struct., Funct., Genet. 2008, 71, 599–620. 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 2004, 14, 171–9. 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Feng Y.; Broder C. C.; Kennedy P. E.; Berger E. A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–7. 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- O’Connor T.; Borsig L.; Heikenwalder M. CCL2-CCR2 Signaling in Disease Pathogenesis. Endocr., Metab. Immune Disord.: Drug Targets 2015, 15, 105–18. 10.2174/1871530315666150316120920. [DOI] [PubMed] [Google Scholar]
- Carter P. H.; Brown G. D.; Cherney R. J.; Batt D. G.; Chen J.; Clark C. M.; Cvijic M. E.; Duncia J. V.; Ko S. S.; Mandlekar S.; Mo R.; Nelson D. J.; Pang J.; Rose A. V.; Santella J. B. 3rd; Tebben A. J.; Traeger S. C.; Xu S.; Zhao Q.; Barrish J. C. Discovery of a Potent and Orally Bioavailable Dual Antagonist of CC Chemokine Receptors 2 and 5. ACS Med. Chem. Lett. 2015, 6, 439–44. 10.1021/ml500505q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasse O. A.; Evans J. L.; Zhai H. X.; Zou D.; Kintigh J. T.; Chan F.; Hamilton K.; Hill E.; Eckman J. B.; Higgins P. J.; Volosov A.; Collart P.; Nicolas J. M.; Kondru R. K.; Schwartz C. E. Novel, Acidic CCR2 Receptor Antagonists: Lead Optimization. Letters in Drug Design & Discovery 2007, 4, 263–271. 10.2174/157018007784619989. [DOI] [Google Scholar]
- Wendt E.; Keshav S. CCR9 antagonism: potential in the treatment of Inflammatory Bowel Disease. Clin. Exp. Gastroenterol. 2015, 8, 119–130. 10.2147/CEG.S48305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. L.; Murphy P. M. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 1994, 269, 28539–28542. [PubMed] [Google Scholar]
- Isberg V.; Mordalski S.; Munk C.; Rataj K.; Harpsøe K.; Hauser A. S.; Vroling B.; Bojarski A. J.; Vriend G.; Gloriam D. E. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016, 44, D356–D364. 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidehi N.; Schlyer S.; Trabanino R. J.; Floriano W. B.; Abrol R.; Sharma S.; Kochanny M.; Koovakat S.; Dunning L.; Liang M.; Fox J. M.; de Mendonca F. L.; Pease J. E.; Goddard W. A. 3rd; Horuk R. Predictions of CCR1 chemokine receptor structure and BX 471 antagonist binding followed by experimental validation. J. Biol. Chem. 2006, 281, 27613–20. 10.1074/jbc.M601389200. [DOI] [PubMed] [Google Scholar]
- de Mendonca F. L.; da Fonseca P. C.; Phillips R. M.; Saldanha J. W.; Williams T. J.; Pease J. E. Site-directed mutagenesis of CC chemokine receptor 1 reveals the mechanism of action of UCB 35625, a small molecule chemokine receptor antagonist. J. Biol. Chem. 2005, 280, 4808–16. 10.1074/jbc.M412267200. [DOI] [PubMed] [Google Scholar]
- Berkhout T. A.; Blaney F. E.; Bridges A. M.; Cooper D. G.; Forbes I. T.; Gribble A. D.; Groot P. H.; Hardy A.; Ife R. J.; Kaur R.; Moores K. E.; Shillito H.; Willetts J.; Witherington J. CCR2: characterization of the antagonist binding site from a combined receptor modeling/mutagenesis approach. J. Med. Chem. 2003, 46, 4070–86. 10.1021/jm030862l. [DOI] [PubMed] [Google Scholar]
- Hall S. E.; Mao A.; Nicolaidou V.; Finelli M.; Wise E. L.; Nedjai B.; Kanjanapangka J.; Harirchian P.; Chen D.; Selchau V.; Ribeiro S.; Schyler S.; Pease J. E.; Horuk R.; Vaidehi N. Elucidation of binding sites of dual antagonists in the human chemokine receptors CCR2 and CCR5. Mol. Pharmacol. 2009, 75, 1325–36. 10.1124/mol.108.053470. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez J.; Rueda P.; Alcami J.; Rognan D.; Arenzana-Seisdedos F.; Lagane B.; Kellenberger E. Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5). J. Biol. Chem. 2011, 286, 33409–21. 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondru R.; Zhang J.; Ji C.; Mirzadegan T.; Rotstein D.; Sankuratri S.; Dioszegi M. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 2008, 73, 789–800. 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- Maeda K.; Das D.; Ogata-Aoki H.; Nakata H.; Miyakawa T.; Tojo Y.; Norman R.; Takaoka Y.; Ding J.; Arnold G. F.; Arnold E.; Mitsuya H. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 2006, 281, 12688–98. 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- Maeda K.; Das D.; Yin P. D.; Tsuchiya K.; Ogata-Aoki H.; Nakata H.; Norman R. B.; Hackney L. A.; Takaoka Y.; Mitsuya H. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J. Mol. Biol. 2008, 381, 956–74. 10.1016/j.jmb.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saita Y.; Kodama E.; Orita M.; Kondo M.; Miyazaki T.; Sudo K.; Kajiwara K.; Matsuoka M.; Shimizu Y. Structural basis for the interaction of CCR5 with a small molecule, functionally selective CCR5 agonist. J. Immunol. 2006, 177, 3116–22. 10.4049/jimmunol.177.5.3116. [DOI] [PubMed] [Google Scholar]
- Jensen P. C.; Nygaard R.; Thiele S.; Elder A.; Zhu G.; Kolbeck R.; Ghosh S.; Schwartz T. W.; Rosenkilde M. M. Molecular interaction of a potent nonpeptide agonist with the chemokine receptor CCR8. Mol. Pharmacol. 2007, 72, 327–40. 10.1124/mol.107.035543. [DOI] [PubMed] [Google Scholar]
- Salchow K.; Bond M. E.; Evans S. C.; Press N. J.; Charlton S. J.; Hunt P. A.; Bradley M. E. A common intracellular allosteric binding site for antagonists of the CXCR2 receptor. Br. J. Pharmacol. 2010, 159, 1429–39. 10.1111/j.1476-5381.2009.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kruijf P.; Lim H. D.; Roumen L.; Renjaan V. A.; Zhao J.; Webb M. L.; Auld D. S.; Wijkmans J. C.; Zaman G. J.; Smit M. J.; de Graaf C.; Leurs R. Identification of a novel allosteric binding site in the CXCR2 chemokine receptor. Mol. Pharmacol. 2011, 80, 1108–18. 10.1124/mol.111.073825. [DOI] [PubMed] [Google Scholar]
- Catusse J.; Liotard A.; Loillier B.; Pruneau D.; Paquet J. L. Characterization of the molecular interactions of interleukin-8 (CXCL8), growth related oncogen alpha (CXCL1) and a non-peptide antagonist (SB 225002) with the human CXCR2. Biochem. Pharmacol. 2003, 65, 813–21. 10.1016/S0006-2952(02)01619-2. [DOI] [PubMed] [Google Scholar]
- Scholten D. J.; Roumen L.; Wijtmans M.; Verkade-Vreeker M. C.; Custers H.; Lai M.; de Hooge D.; Canals M.; de Esch I. J.; Smit M. J.; de Graaf C.; Leurs R. Identification of overlapping but differential binding sites for the high-affinity CXCR3 antagonists NBI-74330 and VUF11211. Mol. Pharmacol. 2014, 85, 116–26. 10.1124/mol.113.088633. [DOI] [PubMed] [Google Scholar]
- Nedjai B.; Viney J. M.; Li H.; Hull C.; Anderson C. A.; Horie T.; Horuk R.; Vaidehi N.; Pease J. E. CXCR3 antagonist VUF10085 binds to an intrahelical site distinct from that of the broad spectrum antagonist TAK-779. Br. J. Pharmacol. 2015, 172, 1822–33. 10.1111/bph.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedjai B.; Li H.; Stroke I. L.; Wise E. L.; Webb M. L.; Merritt J. R.; Henderson I.; Klon A. E.; Cole A. G.; Horuk R.; Vaidehi N.; Pease J. E. Small molecule chemokine mimetics suggest a molecular basis for the observation that CXCL10 and CXCL11 are allosteric ligands of CXCR3. Br. J. Pharmacol. 2012, 166, 912–23. 10.1111/j.1476-5381.2011.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin R. A.; Campanella G. S.; Manice L. A.; Luster A. D. CXCR3 requires tyrosine sulfation for ligand binding and a second extracellular loop arginine residue for ligand-induced chemotaxis. Mol. Cell. Biol. 2006, 26, 5838–5849. 10.1128/MCB.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkilde M. M.; Andersen M. B.; Nygaard R.; Frimurer T. M.; Schwartz T. W. Activation of the CXCR3 chemokine receptor through anchoring of a small molecule chelator ligand between TM-III, -IV, and -VI. Mol. Pharmacol. 2007, 71, 930–941. 10.1124/mol.106.030031. [DOI] [PubMed] [Google Scholar]
- Rosenkilde M. M.; Gerlach L. O.; Hatse S.; Skerlj R. T.; Schols D.; Bridger G. J.; Schwartz T. W. Molecular mechanism of action of monocyclam versus bicyclam non-peptide antagonists in the CXCR4 chemokine receptor. J. Biol. Chem. 2007, 282, 27354–65. 10.1074/jbc.M704739200. [DOI] [PubMed] [Google Scholar]
- Wong R. S.; Bodart V.; Metz M.; Labrecque J.; Bridger G.; Fricker S. P. Comparison of the potential multiple binding modes of bicyclam, monocylam, and noncyclam small-molecule CXC chemokine receptor 4 inhibitors. Mol. Pharmacol. 2008, 74, 1485–95. 10.1124/mol.108.049775. [DOI] [PubMed] [Google Scholar]
- Gerlach L. O.; Jakobsen J. S.; Jensen K. P.; Rosenkilde M. R.; Skerlj R. T.; Ryde U.; Bridger G. J.; Schwartz T. W. Metal ion enhanced binding of AMD3100 to Asp262 in the CXCR4 receptor. Biochemistry 2003, 42, 710–7. 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]
- Rosenkilde M. M.; Gerlach L. O.; Jakobsen J. S.; Skerlj R. T.; Bridger G. J.; Schwartz T. W. Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: transfer of binding site to the CXCR3 receptor. J. Biol. Chem. 2004, 279, 3033–41. 10.1074/jbc.M309546200. [DOI] [PubMed] [Google Scholar]
- Gerlach L. O.; Skerlj R. T.; Bridger G. J.; Schwartz T. W. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J. Biol. Chem. 2001, 276, 14153–14160. [DOI] [PubMed] [Google Scholar]
- Murakami T.; Kumakura S.; Yamazaki T.; Tanaka R.; Hamatake M.; Okuma K.; Huang W.; Toma J.; Komano J.; Yanaka M.; Tanaka Y.; Yamamoto N. The novel CXCR4 antagonist KRH-3955 is an orally bioavailable and extremely potent inhibitor of human immunodeficiency virus type 1 infection: comparative studies with AMD3100. Antimicrob. Agents Chemother. 2009, 53, 2940–8. 10.1128/AAC.01727-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wescott M. P.; Kufareva I.; Paes C.; Goodman J. R.; Thaker Y.; Puffer B. A.; Berdougo E.; Rucker J. B.; Handel T. M.; Doranz B. J. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 9928–9933. 10.1073/pnas.1601278113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa P.; Waldhoer M.; LiWang P. J.; Vischer H. F.; Kledal T.; Timmerman H.; Schwartz T. W.; Smit M. J.; Leurs R. CC and CX3C chemokines differentially interact with the N terminus of the human cytomegalovirus-encoded US28 receptor. J. Biol. Chem. 2005, 280, 3275–85. 10.1074/jbc.M407536200. [DOI] [PubMed] [Google Scholar]
- Mason J. S.; Bortolato A.; Congreve M.; Marshall F. H. New insights from structural biology into the druggability of G protein-coupled receptors. Trends Pharmacol. Sci. 2012, 33, 249–60. 10.1016/j.tips.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Kufareva I.; Abagyan R.; Handel T. M.. Role of 3D Structures in Understanding, Predicting, and Designing Molecular Interactions in the Chemokine Receptor Family. In Chemokines: Chemokines and Their Receptors in Drug Discovery; Tschammer N., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp 41–85. [Google Scholar]
- Thoma G.; Streiff M. B.; Kovarik J.; Glickman F.; Wagner T.; Beerli C.; Zerwes H. G. Orally bioavailable isothioureas block function of the chemokine receptor CXCR4 in vitro and in vivo. J. Med. Chem. 2008, 51, 7915–20. 10.1021/jm801065q. [DOI] [PubMed] [Google Scholar]
- Baroni M.; Cruciani G.; Sciabola S.; Perruccio F.; Mason J. S. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J. Chem. Inf. Model. 2007, 47, 279–94. 10.1021/ci600253e. [DOI] [PubMed] [Google Scholar]
- Nijmeijer S.; Vischer H. F.; Sirci F.; Schultes S.; Engelhardt H.; de Graaf C.; Rosethorne E. M.; Charlton S. J.; Leurs R. Detailed analysis of biased histamine H(4) receptor signalling by JNJ 7777120 analogues. Br. J. Pharmacol. 2013, 170, 78–88. 10.1111/bph.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani A.; Tagat J. R. Discovery and development of small-molecule chemokine coreceptor CCR5 antagonists. J. Med. Chem. 2006, 49, 2851–7. 10.1021/jm060009x. [DOI] [PubMed] [Google Scholar]
- Imamura S.; Ichikawa T.; Nishikawa Y.; Kanzaki N.; Takashima K.; Niwa S.; Iizawa Y.; Baba M.; Sugihara Y. Discovery of a piperidine-4-carboxamide CCR5 antagonist (TAK-220) with highly potent Anti-HIV-1 activity. J. Med. Chem. 2006, 49, 2784–93. 10.1021/jm051034q. [DOI] [PubMed] [Google Scholar]
- Habashita H.; Kokubo M.; Hamano S.; Hamanaka N.; Toda M.; Shibayama S.; Tada H.; Sagawa K.; Fukushima D.; Maeda K.; Mitsuya H. Design, synthesis, and biological evaluation of the combinatorial library with a new spirodiketopiperazine scaffold. Discovery of novel potent and selective low-molecular-weight CCR5 antagonists. J. Med. Chem. 2006, 49, 4140–52. 10.1021/jm060051s. [DOI] [PubMed] [Google Scholar]
- Kufareva I.; Gustavsson M.; Holden L. G.; Qin L.; Zheng Y.; Handel T. M. Disulfide Trapping for Modeling and Structure Determination of Receptor: Chemokine Complexes. Methods Enzymol. 2016, 570, 389–420. 10.1016/bs.mie.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert D.; Kruse A. C.; Manglik A.; Hiller C.; Zhang C.; Hubner H.; Kobilka B. K.; Gmeiner P. Covalent agonists for studying G protein-coupled receptor activation. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 10744–8. 10.1073/pnas.1410415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. G.; Choi H. J.; Fung J. J.; Pardon E.; Casarosa P.; Chae P. S.; Devree B. T.; Rosenbaum D. M.; Thian F. S.; Kobilka T. S.; Schnapp A.; Konetzki I.; Sunahara R. K.; Gellman S. H.; Pautsch A.; Steyaert J.; Weis W. I.; Kobilka B. K. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 2011, 469, 175–80. 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. G.; DeVree B. T.; Zou Y.; Kruse A. C.; Chung K. Y.; Kobilka T. S.; Thian F. S.; Chae P. S.; Pardon E.; Calinski D.; Mathiesen J. M.; Shah S. T.; Lyons J. A.; Caffrey M.; Gellman S. H.; Steyaert J.; Skiniotis G.; Weis W. I.; Sunahara R. K.; Kobilka B. K. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 2011, 477, 549–55. 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B.; Nehme R.; Warne T.; Leslie A. G.; Tate C. G. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 2016, 536, 104–7. 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.; Zhou X. E.; Gao X.; He Y.; Liu W.; Ishchenko A.; Barty A.; White T. A.; Yefanov O.; Han G. W.; Xu Q.; de Waal P. W.; Ke J.; Tan M. H.; Zhang C.; Moeller A.; West G. M.; Pascal B. D.; Van Eps N.; Caro L. N.; Vishnivetskiy S. A.; Lee R. J.; Suino-Powell K. M.; Gu X.; Pal K.; Ma J.; Zhi X.; Boutet S.; Williams G. J.; Messerschmidt M.; Gati C.; Zatsepin N. A.; Wang D.; James D.; Basu S.; Roy-Chowdhury S.; Conrad C. E.; Coe J.; Liu H.; Lisova S.; Kupitz C.; Grotjohann I.; Fromme R.; Jiang Y.; Tan M.; Yang H.; Li J.; Wang M.; Zheng Z.; Li D.; Howe N.; Zhao Y.; Standfuss J.; Diederichs K.; Dong Y.; Potter C. S.; Carragher B.; Caffrey M.; Jiang H.; Chapman H. N.; Spence J. C.; Fromme P.; Weierstall U.; Ernst O. P.; Katritch V.; Gurevich V. V.; Griffin P. R.; Hubbell W. L.; Stevens R. C.; Cherezov V.; Melcher K.; Xu H. E. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–7. 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts C.; Bondue A.; Springael J. Y.; Olivella M.; Deupi X.; Le Poul E.; Wodak S. J.; Parmentier M.; Pardo L.; Blanpain C. Activation of CCR5 by chemokines involves an aromatic cluster between transmembrane helices 2 and 3. J. Biol. Chem. 2003, 278, 1892–903. 10.1074/jbc.M205685200. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan A. J.; Deupi X.; Lebon G.; Tate C. G.; Schertler G. F.; Babu M. M. Molecular signatures of G-protein-coupled receptors. Nature 2013, 494, 185–194. 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Palczewski K.; Kumasaka T.; Hori T.; Behnke C. A.; Motoshima H.; Fox B. A.; Trong I. Le; Teller D. C.; Okada T.; Stenkamp R. E.; Yamamoto M.; Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 2000, 289, 739–745. 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Zweemer A. J.; Bunnik J.; Veenhuizen M.; Miraglia F.; Lenselink E. B.; Vilums M.; de Vries H.; Gibert A.; Thiele S.; Rosenkilde M. M.; Ijzerman A. P.; Heitman L. H. Discovery and mapping of an intracellular antagonist binding site at the chemokine receptor CCR2. Mol. Pharmacol. 2014, 86, 358–368. 10.1124/mol.114.093328. [DOI] [PubMed] [Google Scholar]
- Gavrilin M. A.; Gulina I. V.; Kawano T.; Dragan S.; Chakravarti L.; Kolattukudy P. E. Site-directed mutagenesis of CCR2 identified amino acid residues in transmembrane helices 1, 2, and 7 important for MCP-1 binding and biological functions. Biochem. Biophys. Res. Commun. 2005, 327, 533–40. 10.1016/j.bbrc.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Duchesnes C. E.; Murphy P. M.; Williams T. J.; Pease J. E. Alanine scanning mutagenesis of the chemokine receptor CCR3 reveals distinct extracellular residues involved in recognition of the eotaxin family of chemokines. Mol. Immunol. 2006, 43, 1221–31. 10.1016/j.molimm.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Olson W. C.; Rabut G. E.; Nagashima K. A.; Tran D. N.; Anselma D. J.; Monard S. P.; Segal J. P.; Thompson D. A.; Kajumo F.; Guo Y.; Moore J. P.; Maddon P. J.; Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 1999, 73, 4145–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C.; Doranz B. J.; Vakili J.; Rucker J.; Govaerts C.; Baik S. S.; Lorthioir O.; Migeotte I.; Libert F.; Baleux F.; Vassart G.; Doms R. W.; Parmentier M. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J. Biol. Chem. 1999, 274, 34719–27. 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- Zhou N.; Luo Z.; Hall J. W.; Luo J.; Han X.; Huang Z. Molecular modeling and site-directed mutagenesis of CCR5 reveal residues critical for chemokine binding and signal transduction. Eur. J. Immunol. 2000, 30, 164–73. . [DOI] [PubMed] [Google Scholar]
- Dragic T.; Trkola A.; Lin S. W.; Nagashima K. A.; Kajumo F.; Zhao L.; Olson W. C.; Wu L.; Mackay C. R.; Allaway G. P.; Sakmar T. P.; Moore J. P.; Maddon P. J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 1998, 72, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D. C.; Beavis P.; Chuang K. T.; Zheng Y.; Lee I.; Gee P.; Deval J.; Rotstein D. M.; Dioszegi M.; Ravendran P.; Zhang J.; Sankuratri S.; Kondru R.; Vauquelin G. A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands. Br. J. Pharmacol. 2014, 171, 3364–75. 10.1111/bph.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Feng Y.; Chen X.; Gerard C.; Boisvert W. A. Characterization of G protein coupling mediated by the conserved D134(3.49) of DRY motif, M241(6.34), and F251(6.44) residues on human CXCR1. FEBS Open Bio 2015, 5, 182–90. 10.1016/j.fob.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Tachado S. D.; Koziel H.; Boisvert W. A. Leu128(3.43) (l128) and Val247(6.40) (V247) of CXCR1 are critical amino acid residues for g protein coupling and receptor activation. PLoS One 2012, 7, e42765. 10.1371/journal.pone.0042765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetomi K.; Rojo D.; Navarro J. Identification of a signal transduction switch in the chemokine receptor CXCR1. J. Biol. Chem. 2002, 277, 31563–31566. 10.1074/jbc.M204713200. [DOI] [PubMed] [Google Scholar]
- Scholten D. J.; Canals M.; Wijtmans M.; de Munnik S.; Nguyen P.; Verzijl D.; de Esch I. J.; Vischer H. F.; Smit M. J.; Leurs R. Pharmacological characterization of a small-molecule agonist for the chemokine receptor CXCR3. Br. J. Pharmacol. 2012, 166, 898–911. 10.1111/j.1476-5381.2011.01648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.; Tai H. H. Expression and functional characterization of mutant human CXCR4 in insect cells: role of cysteinyl and negatively charged residues in ligand binding. Arch. Biochem. Biophys. 2000, 373, 211–7. 10.1006/abbi.1999.1555. [DOI] [PubMed] [Google Scholar]
- Thiele S.; Mungalpara J.; Steen A.; Rosenkilde M. M.; Vabeno J. Determination of the binding mode for the cyclopentapeptide CXCR4 antagonist FC131 using a dual approach of ligand modifications and receptor mutagenesis. Br. J. Pharmacol. 2014, 171, 5313–29. 10.1111/bph.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelot A.; Heveker N.; Montes M.; Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 2000, 275, 23736–44. 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- Zhou N.; Luo Z.; Luo J.; Liu D.; Hall J. W.; Pomerantz R. J.; Huang Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 2001, 276, 42826–42833. 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- Berchiche Y. A.; Chow K. Y.; Lagane B.; Leduc M.; Percherancier Y.; Fujii N.; Tamamura H.; Bachelerie F.; Heveker N. Direct assessment of CXCR4 mutant conformations reveals complex link between receptor structure and G(alpha)(i) activation. J. Biol. Chem. 2007, 282, 5111–5. 10.1074/jbc.C600270200. [DOI] [PubMed] [Google Scholar]
- Saini V.; Marchese A.; Tang W. J.; Majetschak M. Structural determinants of ubiquitin-CXC chemokine receptor 4 interaction. J. Biol. Chem. 2011, 286, 44145–52. 10.1074/jbc.M111.298505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I.; Stephens B. S.; Holden L. G.; Qin L.; Zhao C.; Kawamura T.; Abagyan R.; Handel T. M. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: molecular modeling and experimental validation. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E5363–72. 10.1073/pnas.1417037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S.; Choi W. T.; Liu D.; Pesavento J.; Wang Y.; An J.; Sodroski J. G.; Huang Z. Distinct functional sites for human immunodeficiency virus type 1 and stromal cell-derived factor 1alpha on CXCR4 transmembrane helical domains. J. Virol 2005, 79, 12667–73. 10.1128/JVI.79.20.12667-12673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. T.; Tian S.; Dong C. Z.; Kumar S.; Liu D.; Madani N.; An J.; Sodroski J. G.; Huang Z. Unique ligand binding sites on CXCR4 probed by a chemical biology approach: implications for the design of selective human immunodeficiency virus type 1 inhibitors. J. Virol 2005, 79, 15398–404. 10.1128/JVI.79.24.15398-15404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulais P. E.; Escher E.; Leduc R. Analysis by substituted cysteine scanning mutagenesis of the fourth transmembrane domain of the CXCR4 receptor in its inactive and active state. Biochem. Pharmacol. 2013, 85, 541–50. 10.1016/j.bcp.2012.11.022. [DOI] [PubMed] [Google Scholar]
- Benredjem B.; Girard M.; Rhainds D.; St-Onge G.; Heveker N. Mutational Analysis of Atypical Chemokine Receptor 3 (ACKR3/CXCR7) Interaction with its Chemokine Ligands CXCL11 and CXCL12. J. Biol. Chem. 2017, 292, 31–42. 10.1074/jbc.M116.762252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. B.; Navenot J. M.; Haribabu B.; Tamamura H.; Hiramatu K.; Omagari A.; Pei G.; Manfredi J. P.; Fujii N.; Broach J. R.; Peiper S. C. A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40–4C are weak partial agonists. J. Biol. Chem. 2002, 277, 24515–21. 10.1074/jbc.M200889200. [DOI] [PubMed] [Google Scholar]
- Murphy P. M.; Baggiolini M.; Charo I. F.; Hebert C. A.; Horuk R.; Matsushima K.; Miller L. H.; Oppenheim J. J.; Power C. A. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000, 52, 145–176. [PubMed] [Google Scholar]
- Murphy J. W.; Yuan H.; Kong Y.; Xiong Y.; Lolis E. J. Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family. Proteins: Struct., Funct., Genet. 2010, 78, 1331–7. 10.1002/prot.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover D. M.; Boulegue C.; Yang D.; Oppenheim J. J.; Tucker K.; Lu W.; Lubkowski J. The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J. Biol. Chem. 2002, 277, 37647–37654. 10.1074/jbc.M203907200. [DOI] [PubMed] [Google Scholar]
- Xue X.; Lu Q.; Wei H.; Wang D.; Chen D.; He G.; Huang L.; Wang H.; Wang X. Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor. PLoS Pathog. 2011, 7, e1002162. 10.1371/journal.ppat.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump M. P.; Gong J. H.; Loetscher P.; Rajarathnam K.; Amara A.; Arenzana-Seisdedos F.; Virelizier J. L.; Baggiolini M.; Sykes B. D.; Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997, 16, 6996–7007. 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton N. J.; Aspiras F.; Ogez J.; Schall T. J. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry 1995, 34, 5329–42. 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- Kleist A. B.; Getschman A. E.; Ziarek J. J.; Nevins A. M.; Gauthier P. A.; Chevigne A.; Szpakowska M.; Volkman B. F. New paradigms in chemokine receptor signal transduction: moving beyond the two-site model. Biochem. Pharmacol. 2016, 114, 53–68. 10.1016/j.bcp.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. R.; Bolt A.; Chapman G. A.; Knight R. L.; Meissner J. W. G.; Owen D. A.; Watson R. J. Identification and structure-activity relationships of 1-aryl-3-piperidin-4-yl-urea derivatives as CXCR3 receptor antagonists. Bioorg. Med. Chem. Lett. 2007, 17, 697–701. 10.1016/j.bmcl.2006.10.088. [DOI] [PubMed] [Google Scholar]
- Hebert C. A.; Chuntharapai A.; Smith M.; Colby T.; Kim J.; Horuk R. Partial functional mapping of the human interleukin-8 type A receptor. Identification of a major ligand binding domain. J. Biol. Chem. 1993, 268, 18549–18553. [PubMed] [Google Scholar]
- Leong S. R.; Kabakoff R. C.; Hebert C. A. Complete mutagenesis of the extracellular domain of interleukin-8 (IL-8) type A receptor identifies charged residues mediating IL-8 binding and signal transduction. J. Biol. Chem. 1994, 269, 19343–19348. [PubMed] [Google Scholar]
- Williams G.; Borkakoti N.; Bottomley G. A.; Cowan I.; Fallowfield A. G.; Jones P. S.; Kirtland S. J.; Price G. J.; Price L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J. Biol. Chem. 1996, 271, 9579–9586. 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- Falsone A.; Wabitsch V.; Geretti E.; Potzinger H.; Gerlza T.; Robinson J.; Adage T.; Teixeira M. M.; Kungl A. J. Designing CXCL8-based decoy proteins with strong anti-inflammatory activity in vivo. Biosci. Rep. 2013, 33, 743–754. 10.1042/BSR20130069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoue L. S.; Sullivan S. K.; King D. S.; Kledal T. N.; Schwartz T. W.; Bacon K. B.; Handel T. M. Molecular determinants of receptor binding and signaling by the CX3C chemokine fractalkine. J. Biol. Chem. 2001, 276, 33906–14. 10.1074/jbc.M101348200. [DOI] [PubMed] [Google Scholar]
- Harrison J. K.; Fong A. M.; Swain P. A.; Chen S.; Yu Y. R.; Salafranca M. N.; Greenleaf W. B.; Imai T.; Patel D. D. Mutational analysis of the fractalkine chemokine domain. Basic amino acid residues differentially contribute to CX3CR1 binding, signaling, and cell adhesion. J. Biol. Chem. 2001, 276, 21632–41. 10.1074/jbc.M010261200. [DOI] [PubMed] [Google Scholar]
- Liu J.; Louie S.; Hsu W.; Yu K. M.; Nicholas H. B. Jr.; Rosenquist G. L. Tyrosine sulfation is prevalent in human chemokine receptors important in lung disease. Am. J. Respir. Cell Mol. Biol. 2008, 38, 738–43. 10.1165/rcmb.2007-0118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Deng X.; Tang M.; Li L.; Xiao L.; Yang L.; Zhong J.; Bode A. M.; Dong Z.; Tao Y.; Cao Y. Tyrosylprotein sulfotransferase-1 and tyrosine sulfation of chemokine receptor 4 are induced by Epstein-Barr virus encoded latent membrane protein 1 and associated with the metastatic potential of human nasopharyngeal carcinoma. PLoS One 2013, 8, e56114. 10.1371/journal.pone.0056114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dealwis C.; Fernandez E. J.; Thompson D. A.; Simon R. J.; Siani M. A.; Lolis E. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 ″fusin″ coreceptor. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 6941–6. 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y.; Senda T.; Nandhagopal N.; Sugimoto K.; Shioda T.; Nagai Y.; Mitsui Y. Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: an attempt at a more comprehensive interpretation of accumulated structure-activity relationship data. J. Interferon Cytokine Res. 2000, 20, 691–700. 10.1089/10799900050116390. [DOI] [PubMed] [Google Scholar]
- Murphy J. W.; Cho Y.; Sachpatzidis A.; Fan C.; Hodsdon M. E.; Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1 alpha) binding to heparin. J. Biol. Chem. 2007, 282, 10018–27. 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. W.; Liu Y.; Getschman A. E.; Peterson F. C.; Ziarek J. J.; Li R.; Volkman B. F.; Chen Y. Structural analysis of a novel small molecule ligand bound to the CXCL12 chemokine. J. Med. Chem. 2014, 57, 9693–9. 10.1021/jm501194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. W.; Nevins A. M.; Qiao Z.; Liu Y.; Getschman A. E.; Vankayala S. L.; Kemp M. T.; Peterson F. C.; Li R.; Volkman B. F.; Chen Y. Structure-Based Identification of Novel Ligands Targeting Multiple Sites within a Chemokine-G-Protein-Coupled-Receptor Interface. J. Med. Chem. 2016, 59, 4342–4351. 10.1021/acs.jmedchem.5b02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths K.; Dolezal O.; Cao B.; Nilsson S. K.; See H. B.; Pfleger K. D.; Roche M.; Gorry P. R.; Pow A.; Viduka K.; Lim K.; Lu B. G.; Chang D. H.; Murray-Rust T. A.; Kvansakul M.; Perugini M. A.; Dogovski C.; Doerflinger M.; Zhang Y.; Parisi K.; Casey J. L.; Nuttall S. D.; Foley M. I-bodies: human single domain antibodies that antagonize chemokine receptor CXCR4. J. Biol. Chem. 2016, 291, 12641–12657. 10.1074/jbc.M116.721050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnec X.; Quan L.; Olson W. C.; Hazan U.; Dragic T. Anti-CXCR4 monoclonal antibodies recognizing overlapping epitopes differ significantly in their ability to inhibit entry of human immunodeficiency virus type 1. J. Virol 2005, 79, 1930–3. 10.1128/JVI.79.3.1930-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnichen S.; Blanchetot C.; Maussang D.; Gonzalez-Pajuelo M.; Chow K. Y.; Bosch L.; De Vrieze S.; Serruys B.; Ulrichts H.; Vandevelde W.; Saunders M.; De Haard H. J.; Schols D.; Leurs R.; Vanlandschoot P.; Verrips T.; Smit M. J. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 20565–70. 10.1073/pnas.1012865107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu E. K.; Kim T. G.; Kwon T. H.; Jung I. D.; Ryu D.; Park Y. M.; Kim J.; Ahn K. H.; Ban C. Crystal structure of recombinant human stromal cell-derived factor-1alpha. Proteins: Struct., Funct., Genet. 2007, 67, 1193–7. 10.1002/prot.21350. [DOI] [PubMed] [Google Scholar]
- Shaw J. P.; Johnson Z.; Borlat F.; Zwahlen C.; Kungl A.; Roulin K.; Harrenga A.; Wells T. N.; Proudfoot A. E. The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure 2004, 12, 2081–93. 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Horn F.; Bettler E.; Oliveira L.; Campagne F.; Cohen F. E.; Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003, 31, 294–297. 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.; Beevers A. J.; Kukol A.; Dupree R.; Smith M. E. Solid-state 17O NMR spectroscopy of a phospholemman transmembrane domain protein: implications for the limits of detecting dilute 17O sites in biomaterials. Solid State Nucl. Magn. Reson. 2008, 33, 72–5. 10.1016/j.ssnmr.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Casarosa P.; Menge W. M.; Minisini R.; Otto C.; van Heteren J.; Jongejan A.; Timmerman H.; Moepps B.; Kirchhoff F.; Mertens T.; Smit M. J.; Leurs R. Identification of the first nonpeptidergic inverse agonist for a constitutively active viral-encoded G protein-coupled receptor. J. Biol. Chem. 2003, 278, 5172–8. 10.1074/jbc.M210033200. [DOI] [PubMed] [Google Scholar]
- Zweemer A. J.; Nederpelt I.; Vrieling H.; Hafith S.; Doornbos M. L.; de Vries H.; Abt J.; Gross R.; Stamos D.; Saunders J.; Smit M. J.; Ijzerman A. P.; Heitman L. H. Multiple binding sites for small-molecule antagonists at the CC chemokine receptor 2. Mol. Pharmacol. 2013, 84, 551–61. 10.1124/mol.113.086850. [DOI] [PubMed] [Google Scholar]
- Andrews G.; Jones C.; Wreggett K. A. An intracellular allosteric site for a specific class of antagonists of the CC chemokine G protein-coupled receptors CCR4 and CCR5. Mol. Pharmacol. 2008, 73, 855–867. 10.1124/mol.107.039321. [DOI] [PubMed] [Google Scholar]
- Slack R. J.; Russell L. J.; Barton N. P.; Weston C.; Nalesso G.; Thompson S. A.; Allen M.; Chen Y. H.; Barnes A.; Hodgson S. T.; Hall D. A. Antagonism of human CC-chemokine receptor 4 can be achieved through three distinct binding sites on the receptor. Pharmacol. Res. Perspect. 2013, 1, e00019. 10.1002/prp2.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederblad L.; Rosengren B.; Ryberg E.; Hermansson N. O. AZD8797 is an allosteric non-competitive modulator of the human CX3CR1 receptor. Biochem. J. 2016, 473, 641–9. 10.1042/BJ20150520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B.; Nehme R.; Warne T.; Leslie A. G.; Tate C. G. Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 2016, 536, 104–107. 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K.; Yokoyama-Kumakura S.; Tanaka Y.; Tanaka R.; Hirose K.; Bannai K.; Edamatsu T.; Yanaka M.; Niitani Y.; Miyano-Kurosaki N.; Takaku H.; Koyanagi Y.; Yamamoto N. A duodenally absorbable CXC chemokine receptor 4 antagonist, KRH-1636, exhibits a potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 4185–90. 10.1073/pnas.0630420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi T.; Tanaka T.; Hashimoto C.; Nomura W.; Aikawa H.; Sohma A.; Itotani K.; Kawamata M.; Murakami T.; Yamamoto N.; Tamamura H. Pharmacophore-based small molecule CXCR4 ligands. Bioorg. Med. Chem. Lett. 2012, 22, 4169–72. 10.1016/j.bmcl.2012.04.032. [DOI] [PubMed] [Google Scholar]
- Miller J. F.; Gudmundsson K. S.; D’Aurora Richardson L.; Jenkinson S.; Spaltenstein A.; Thomson M.; Wheelan P. Synthesis and SAR of novel isoquinoline CXCR4 antagonists with potent anti-HIV activity. Bioorg. Med. Chem. Lett. 2010, 20, 3026–3030. 10.1016/j.bmcl.2010.03.118. [DOI] [PubMed] [Google Scholar]
- Zhan W.; Liang Z.; Zhu A.; Kurtkaya S.; Shim H.; Snyder J. P.; Liotta D. C. Discovery of small molecule CXCR4 antagonists. J. Med. Chem. 2007, 50, 5655–64. 10.1021/jm070679i. [DOI] [PubMed] [Google Scholar]
- Wu C. H.; Song J. S.; Chang K. H.; Jan J. J.; Chen C. T.; Chou M. C.; Yeh K. C.; Wong Y. C.; Tseng C. T.; Wu S. H.; Yeh C. F.; Huang C. Y.; Wang M. H.; Sadani A. A.; Chang C. P.; Cheng C. Y.; Tsou L. K.; Shia K. S. Stem cell mobilizers targeting chemokine receptor CXCR4: renoprotective application in acute kidney injury. J. Med. Chem. 2015, 58, 2315–25. 10.1021/jm501769r. [DOI] [PubMed] [Google Scholar]
- Wu C. H.; Wang C. J.; Chang C. P.; Cheng Y. C.; Song J. S.; Jan J. J.; Chou M. C.; Ke Y. Y.; Ma J.; Wong Y. C.; Hsieh T. C.; Tien Y. C.; Gullen E. A.; Lo C. F.; Cheng C. Y.; Liu Y. W.; Sadani A. A.; Tsai C. H.; Hsieh H. P.; Tsou L. K.; Shia K. S. Function-oriented development of CXCR4 antagonists as selective human immunodeficiency virus (HIV)-1 entry inhibitors. J. Med. Chem. 2015, 58, 1452–65. 10.1021/jm501772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson R. A.; Pincus S. H.; Song K.; Shepard J. B.; Weaver A. J. Jr.; Labib M. E.; Teintze M. Improved guanide compounds which bind the CXCR4 co-receptor and inhibit HIV-1 infection. Bioorg. Med. Chem. Lett. 2013, 23, 2197–201. 10.1016/j.bmcl.2013.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S.; Kato M.; Inuki S.; Ohno H.; Evans B.; Wang Z. X.; Peiper S. C.; Izumi K.; Kodama E.; Matsuoka M.; Nagasawa H.; Oishi S.; Fujii N. Identification of novel non-peptide CXCR4 antagonists by ligand-based design approach. Bioorg. Med. Chem. Lett. 2008, 18, 4124–9. 10.1016/j.bmcl.2008.05.092. [DOI] [PubMed] [Google Scholar]
- Finke P. E.; Oates B.; Mills S. G.; MacCoss M.; Malkowitz L.; Springer M. S.; Gould S. L.; DeMartino J. A.; Carella A.; Carver G.; Holmes K.; Danzeisen R.; Hazuda D.; Kessler J.; Lineberger J.; Miller M.; Schleif W. A.; Emini E. A. Antagonists of the human CCR5 receptor as anti-HIV-1 agents. Part 4: synthesis and structure-activity relationships for 1-[N-(methyl)-N-(phenylsulfonyl)amino]-2-(phenyl)-4-(4-(N-(alkyl)-N-(benzyloxycar Bonyl)amino)piperidin-1-yl)butanes. Bioorg. Med. Chem. Lett. 2001, 11, 2475–9. 10.1016/S0960-894X(01)00492-9. [DOI] [PubMed] [Google Scholar]
- Hale J. J.; Budhu R. J.; Holson E. B.; Finke P. E.; Oates B.; Mills S. G.; MacCoss M.; Gould S. L.; DeMartino J. A.; Springer M. S.; Siciliano S.; Malkowitz L.; Schleif W. A.; Hazuda D.; Miller M.; Kessler J.; Danzeisen R.; Holmes K.; Lineberger J.; Carella A.; Carver G.; Emini E. 1,3,4-Trisubstituted pyrrolidine CCR5 receptor antagonists. Part 2: lead optimization affording selective, orally bioavailable compounds with potent anti-HIV activity. Bioorg. Med. Chem. Lett. 2001, 11, 2741–5. 10.1016/S0960-894X(01)00545-5. [DOI] [PubMed] [Google Scholar]
- Lu S. F.; Chen B.; Davey D.; Dunning L.; Jaroch S.; May K.; Onuffer J.; Phillips G.; Subramanyam B.; Tseng J. L.; Wei R. G.; Wei M.; Ye B. CCR5 receptor antagonists: discovery and SAR of novel 4-hydroxypiperidine derivatives. Bioorg. Med. Chem. Lett. 2007, 17, 1883–7. 10.1016/j.bmcl.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Lemoine R. C.; Petersen A. C.; Setti L.; Wanner J.; Jekle A.; Heilek G.; deRosier A.; Ji C.; Berry P.; Rotstein D. Evaluation of secondary amide replacements in a series of CCR5 antagonists as a means to increase intrinsic membrane permeability. Part 1: Optimization of gem-disubstituted azacycles. Bioorg. Med. Chem. Lett. 2010, 20, 704–8. 10.1016/j.bmcl.2009.11.072. [DOI] [PubMed] [Google Scholar]
- Pasternak A.; Marino D.; Vicario P. P.; Ayala J. M.; Cascierri M. A.; Parsons W.; Mills S. G.; Maccoss M.; Yang L. Novel, orally bioavailable gamma-aminoamide CC chemokine receptor 2 (CCR2) antagonists. J. Med. Chem. 2006, 49, 4801–4. 10.1021/jm060439n. [DOI] [PubMed] [Google Scholar]
- Tagat J. R.; McCombie S. W.; Nazareno D.; Labroli M. A.; Xiao Y.; Steensma R. W.; Strizki J. M.; Baroudy B. M.; Cox K.; Lachowicz J.; Varty G.; Watkins R. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. IV. Discovery of 1-[(4,6-dimethyl-5-pyrimidinyl)carbonyl]- 4-[4-[2-methoxy-1(R)-4-(trifluoromethyl)phenyl]ethyl-3(S)-methyl-1-piperazinyl]- 4-methylpiperidine (Sch-417690/Sch-D), a potent, highly selective, and orally bioavailable CCR5 antagonist. J. Med. Chem. 2004, 47, 2405–8. 10.1021/jm0304515. [DOI] [PubMed] [Google Scholar]
- McCombie S. W.; Tagat J. R.; Vice S. F.; Lin S. I.; Steensma R.; Palani A.; Neustadt B. R.; Baroudy B. M.; Strizki J. M.; Endres M.; Cox K.; Dan N.; Chou C. C. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. III: synthesis, antiviral and pharmacokinetic profiles of symmetrical heteroaryl carboxamides. Bioorg. Med. Chem. Lett. 2003, 13, 567–71. 10.1016/S0960-894X(02)00918-6. [DOI] [PubMed] [Google Scholar]
- Nishizawa R.; Nishiyama T.; Hisaichi K.; Matsunaga N.; Minamoto C.; Habashita H.; Takaoka Y.; Toda M.; Shibayama S.; Tada H.; Sagawa K.; Fukushima D.; Maeda K.; Mitsuya H. Spirodiketopiperazine-based CCR5 antagonists: Lead optimization from biologically active metabolite. Bioorg. Med. Chem. Lett. 2007, 17, 727–31. 10.1016/j.bmcl.2006.10.084. [DOI] [PubMed] [Google Scholar]
- Baba M.; Nishimura O.; Kanzaki N.; Okamoto M.; Sawada H.; Iizawa Y.; Shiraishi M.; Aramaki Y.; Okonogi K.; Ogawa Y.; Meguro K.; Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 5698–703. 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M.; Aramaki Y.; Seto M.; Imoto H.; Nishikawa Y.; Kanzaki N.; Okamoto M.; Sawada H.; Nishimura O.; Baba M.; Fujino M. Discovery of novel, potent, and selective small-molecule CCR5 antagonists as anti-HIV-1 agents: synthesis and biological evaluation of anilide derivatives with a quaternary ammonium moiety. J. Med. Chem. 2000, 43, 2049–63. 10.1021/jm9906264. [DOI] [PubMed] [Google Scholar]
- Ernst J.; Dahl R.; Lum C.; Sebo L.; Urban J.; Miller S. G.; Lundstrom J. Anti-HIV-1 entry optimization of novel imidazopiperidine-tropane CCR5 antagonists. Bioorg. Med. Chem. Lett. 2008, 18, 1498–501. 10.1016/j.bmcl.2007.12.058. [DOI] [PubMed] [Google Scholar]
- Kazmierski W. M.; Aquino C.; Chauder B. A.; Deanda F.; Ferris R.; Jones-Hertzog D. K.; Kenakin T.; Koble C. S.; Watson C.; Wheelan P.; Yang H.; Youngman M. Discovery of bioavailable 4,4-disubstituted piperidines as potent ligands of the chemokine receptor 5 and inhibitors of the human immunodeficiency virus-1. J. Med. Chem. 2008, 51, 6538–46. 10.1021/jm800598a. [DOI] [PubMed] [Google Scholar]
- Duan M.; Aquino C.; Ferris R.; Kazmierski W. M.; Kenakin T.; Koble C.; Wheelan P.; Watson C.; Youngman M. [2-(4-Phenyl-4-piperidinyl)ethyl]amine based CCR5 antagonists: derivatizations at the N-terminal of the piperidine ring. Bioorg. Med. Chem. Lett. 2009, 19, 1610–3. 10.1016/j.bmcl.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Stupple P. A.; Batchelor D. V.; Corless M.; Dorr P. K.; Ellis D.; Fenwick D. R.; Galan S. R.; Jones R. M.; Mason H. J.; Middleton D. S.; Perros M.; Perruccio F.; Platts M. Y.; Pryde D. C.; Rodrigues D.; Smith N. N.; Stephenson P. T.; Webster R.; Westby M.; Wood A. An imidazopiperidine series of CCR5 antagonists for the treatment of HIV: the discovery of N-{(1S)-1-(3-fluorophenyl)-3-[(3-endo)-3-(5-isobutyryl-2-methyl-4,5,6,7-tetrahydr o-1H-imidazo[4,5-c]pyridin-1-yl)-8-azabicyclo[3.2.1]oct-8-yl]propyl}acetamide (PF-232798). J. Med. Chem. 2011, 54, 67–77. 10.1021/jm100978n. [DOI] [PubMed] [Google Scholar]
- Lagu B.; Gerchak C.; Pan M.; Hou C.; Singer M.; Malaviya R.; Matheis M.; Olini G.; Cavender D.; Wachter M. Potent and selective CC-chemokine receptor-2 (CCR2) antagonists as a potential treatment for asthma. Bioorg. Med. Chem. Lett. 2007, 17, 4382–4386. 10.1016/j.bmcl.2007.01.115. [DOI] [PubMed] [Google Scholar]
- Cumming J. G.; Bower J. F.; Waterson D.; Faull A.; Poyser P. J.; Turner P.; McDermott B.; Campbell A. D.; Hudson J.; James M.; Winter J.; Wood C. The design and synthesis of novel, potent and orally bioavailable N-aryl piperazine-1-carboxamide CCR2 antagonists with very high hERG selectivity. Bioorg. Med. Chem. Lett. 2012, 22, 3895–3899. 10.1016/j.bmcl.2012.04.118. [DOI] [PubMed] [Google Scholar]
- Lin X.; Huang X. P.; Chen G.; Whaley R.; Peng S.; Wang Y.; Zhang G.; Wang S. X.; Wang S.; Roth B. L.; Huang N. Life beyond kinases: structure-based discovery of sorafenib as nanomolar antagonist of 5-HT receptors. J. Med. Chem. 2012, 55, 5749–59. 10.1021/jm300338m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasse O. A.; Evans J. L.; Zhai H.-X.; Zou D.; Kintigh J. T.; Chan F.; Hamilton K.; Hill E.; Eckman J. B.; Higgins P. J.; Volosov A.; Collart P.; Nicolas J.-M.; Kondru R. K.; Schwartz C. E. Novel, Acidic CCR2 Receptor Antagonists: Lead Optimization. Lett. Drug Des. Discovery 2007, 4, 263–271. 10.2174/157018007784619989. [DOI] [Google Scholar]
- Doyon J.; Coesemans E.; Boeckx S.; Buntinx M.; Hermans B.; Van Wauwe J. P.; Gilissen R. A.; De Groot A. H.; Corens D.; Van Lommen G. Discovery of potent, orally bioavailable small-molecule inhibitors of the human CCR2 receptor. ChemMedChem 2008, 3, 660–9. 10.1002/cmdc.200700276. [DOI] [PubMed] [Google Scholar]
- Peace S.; Philp J.; Brooks C.; Piercy V.; Moores K.; Smethurst C.; Watson S.; Gaines S.; Zippoli M.; Mookherjee C.; Ife R. Identification of a sulfonamide series of CCR2 antagonists. Bioorg. Med. Chem. Lett. 2010, 20, 3961–4. 10.1016/j.bmcl.2010.04.142. [DOI] [PubMed] [Google Scholar]
- Kalindjian S. B.; Kadnur S. V.; Hewson C. A.; Venkateshappa C.; Juluri S.; Kristam R.; Kulkarni B.; Mohammed Z.; Saxena R.; Viswanadhan V. N.; Aiyar J.; McVey D. A New Series of Orally Bioavailable Chemokine Receptor 9 (CCR9) Antagonists; Possible Agents for the Treatment of Inflammatory Bowel Disease. J. Med. Chem. 2016, 59, 3098–111. 10.1021/acs.jmedchem.5b01840. [DOI] [PubMed] [Google Scholar]
- Hulshof J. W.; Casarosa P.; Menge W. M.; Kuusisto L. M.; van der Goot H.; Smit M. J.; de Esch I. J.; Leurs R. Synthesis and structure-activity relationship of the first nonpeptidergic inverse agonists for the human cytomegalovirus encoded chemokine receptor US28. J. Med. Chem. 2005, 48, 6461–71. 10.1021/jm050418d. [DOI] [PubMed] [Google Scholar]
- Vischer H. F.; Hulshof J. W.; Hulscher S.; Fratantoni S. A.; Verheij M. H.; Victorina J.; Smit M. J.; de Esch I. J.; Leurs R. Identification of novel allosteric nonpeptidergic inhibitors of the human cytomegalovirus-encoded chemokine receptor US28. Bioorg. Med. Chem. 2010, 18, 675–88. 10.1016/j.bmc.2009.11.060. [DOI] [PubMed] [Google Scholar]
- Kralj A.; Wetzel A.; Mahmoudian S.; Stamminger T.; Tschammer N.; Heinrich M. R. Identification of novel allosteric modulators for the G-protein coupled US28 receptor of human cytomegalovirus. Bioorg. Med. Chem. Lett. 2011, 21, 5446–50. 10.1016/j.bmcl.2011.06.120. [DOI] [PubMed] [Google Scholar]
- Kralj A.; Nguyen M. T.; Tschammer N.; Ocampo N.; Gesiotto Q.; Heinrich M. R.; Phanstiel O. t. Development of flavonoid-based inverse agonists of the key signaling receptor US28 of human cytomegalovirus. J. Med. Chem. 2013, 56, 5019–32. 10.1021/jm4003457. [DOI] [PubMed] [Google Scholar]
- McMaster B. E.; Schall T. J.; Penfold M. E.; Wright J. J.; Dairaghi D. J.. Arylamines as inhibitors of chemokine binding to US28. Patent WO03020029, 2003.
- Schall T. J.; McMaster B. E.; Dairaghi D. J.. Reagents and methods for the diagnosis of CMV dissemination. Patent WO0217969, 2002.
- McMaster B. E.; Schall T. J.; Penfold M. E.; Wright J. J.; Dairaghi D. J.. Bicyclic compounds as inhibitors of chemokine binding to US28. Patent WO03018549, 2003.
- Wilkinson R. A.; Pincus S. H.; Shepard J. B.; Walton S. K.; Bergin E. P.; Labib M.; Teintze M. Novel compounds containing multiple guanide groups that bind the HIV coreceptor CXCR4. Antimicrob. Agents Chemother. 2011, 55, 255–63. 10.1128/AAC.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming J. G.; Tucker H.; Oldfield J.; Fielding C.; Highton A.; Faull A.; Wild M.; Brown D.; Wells S.; Shaw J. Balancing hERG affinity and absorption in the discovery of AZD5672, an orally active CCR5 antagonist for the treatment of rheumatoid arthritis. Bioorg. Med. Chem. Lett. 2012, 22, 1655–9. 10.1016/j.bmcl.2011.12.117. [DOI] [PubMed] [Google Scholar]
- Hale J. J.; Budhu R. J.; Mills S. G.; MacCoss M.; Malkowitz L.; Siciliano S.; Gould S. L.; DeMartino J. A.; Springer M. S. 1,3,4-Trisubstituted pyrrolidine CCR5 receptor antagonists. Part 1: discovery of the pyrrolidine scaffold and determination of its stereochemical requirements. Bioorg. Med. Chem. Lett. 2001, 11, 1437–40. 10.1016/S0960-894X(01)00232-3. [DOI] [PubMed] [Google Scholar]
- Roumen L.; Sanders M. P. A.; Vroling B.; De Esch I. J. P.; De Vlieg J.; Leurs R.; Klomp J. P. G.; Nabuurs S. B.; De Graaf C. In Silico Veritas: The Pitfalls and Challenges of Predicting GPCR-Ligand Interactions. Pharmaceuticals 2011, 4, 1196–1215. 10.3390/ph4091196. [DOI] [Google Scholar]
- Govaerts C.; Blanpain C.; Deupi X.; Ballet S.; Ballesteros J. A.; Wodak S. J.; Vassart G.; Pardo L.; Parmentier M. The TXP motif in the second transmembrane helix of CCR5. A structural determinant of chemokine-induced activation. J. Biol. Chem. 2001, 276, 13217–13225. 10.1074/jbc.M102244200. [DOI] [PubMed] [Google Scholar]
- Dairaghi D. J.; Oldham E. R.; Bacon K. B.; Schall T. J. Chemokine receptor CCR3 function is highly dependent on local pH and ionic strength. J. Biol. Chem. 1997, 272, 28206–9. 10.1074/jbc.272.45.28206. [DOI] [PubMed] [Google Scholar]
- Shukla A. K.; Manglik A.; Kruse A. C.; Xiao K.; Reis R. I.; Tseng W. C.; Staus D. P.; Hilger D.; Uysal S.; Huang L. Y.; Paduch M.; Tripathi-Shukla P.; Koide A.; Koide S.; Weis W. I.; Kossiakoff A. A.; Kobilka B. K.; Lefkowitz R. J. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 2013, 497, 137–41. 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik S. M.; Kooistra A. J.; van Offenbeek J.; Nijmeijer S.; de Graaf C.; Smit M. J.; Leurs R.; Vischer H. F. The Viral G Protein-Coupled Receptor ORF74 Hijacks beta-Arrestins for Endocytic Trafficking in Response to Human Chemokines. PLoS One 2015, 10, e0124486. 10.1371/journal.pone.0124486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan K. T.; Tesmer J. J. Molecular basis for small molecule inhibition of G protein-coupled receptor kinases. ACS Chem. Biol. 2015, 10, 246–56. 10.1021/cb5003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A. C.; Ring A. M.; Manglik A.; Hu J.; Hu K.; Eitel K.; Hubner H.; Pardon E.; Valant C.; Sexton P. M.; Christopoulos A.; Felder C. C.; Gmeiner P.; Steyaert J.; Weis W. I.; Garcia K. C.; Wess J.; Kobilka B. K. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 2013, 504, 101–6. 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Gao Z. G.; Zhang K.; Kiselev E.; Crane S.; Wang J.; Paoletta S.; Yi C.; Ma L.; Zhang W.; Han G. W.; Liu H.; Cherezov V.; Katritch V.; Jiang H.; Stevens R. C.; Jacobson K. A.; Zhao Q.; Wu B. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 2015, 520, 317–21. 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadhe C. G.; Kim M. H. Insights into the binding modes of CC chemokine receptor 4 (CCR4) inhibitors: a combined approach involving homology modelling, docking, and molecular dynamics simulation studies. Mol. BioSyst. 2015, 11, 618–34. 10.1039/C4MB00568F. [DOI] [PubMed] [Google Scholar]
- Fano A.; Ritchie D. W.; Carrieri A. Modeling the structural basis of human CCR5 chemokine receptor function: from homology model building and molecular dynamics validation to agonist and antagonist docking. J. Chem. Inf. Model. 2006, 46, 1223–35. 10.1021/ci050490k. [DOI] [PubMed] [Google Scholar]
- Slynko I.; Scharfe M.; Rumpf T.; Eib J.; Metzger E.; Schule R.; Jung M.; Sippl W. Virtual screening of PRK1 inhibitors: ensemble docking, rescoring using binding free energy calculation and QSAR model development. J. Chem. Inf. Model. 2014, 54, 138–50. 10.1021/ci400628q. [DOI] [PubMed] [Google Scholar]
- Metz M.; Bourque E.; Labrecque J.; Danthi S. J.; Langille J.; Harwig C.; Yang W.; Darkes M. C.; Lau G.; Santucci Z.; Bridger G. J.; Schols D.; Fricker S. P.; Skerlj R. T. Prospective CCR5 small molecule antagonist compound design using a combined mutagenesis/modeling approach. J. Am. Chem. Soc. 2011, 133, 16477–85. 10.1021/ja2043722. [DOI] [PubMed] [Google Scholar]
- Gadhe C. G.; Balupuri A.; Cho S. J. In silico characterization of binding mode of CCR8 inhibitor: homology modeling, docking and membrane based MD simulation study. J. Biomol. Struct. Dyn. 2015, 33, 2491–510. 10.1080/07391102.2014.1002006. [DOI] [PubMed] [Google Scholar]
- Gudmundsson K. S.; Sebahar P. R.; Richardson L. D.; Miller J. F.; Turner E. M.; Catalano J. G.; Spaltenstein A.; Lawrence W.; Thomson M.; Jenkinson S. Amine substituted N-(1H-benzimidazol-2ylmethyl)-5,6,7,8-tetrahydro-8-quinolinamines as CXCR4 antagonists with potent activity against HIV-1. Bioorg. Med. Chem. Lett. 2009, 19, 5048–52. 10.1016/j.bmcl.2009.07.037. [DOI] [PubMed] [Google Scholar]
- Gudmundsson K. S.; Boggs S. D.; Catalano J. G.; Svolto A.; Spaltenstein A.; Thomson M.; Wheelan P.; Jenkinson S. Imidazopyridine-5,6,7,8-tetrahydro-8-quinolinamine derivatives with potent activity against HIV-1. Bioorg. Med. Chem. Lett. 2009, 19, 6399–403. 10.1016/j.bmcl.2009.09.056. [DOI] [PubMed] [Google Scholar]
- Kufareva I. Chemokines and their receptors: insights from molecular modeling and crystallography. Curr. Opin. Pharmacol. 2016, 30, 27–37. 10.1016/j.coph.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutolo P.; Basdevant N.; Bernadat G.; Bachelerie F.; Ha-Duong T. Interaction of chemokine receptor CXCR4 in monomeric and dimeric state with its endogenous ligand CXCL12: Coarse-grained simulations identify differences. J. Biomol. Struct. Dyn. 2017, 35, 399–412. 10.1080/07391102.2016.1145142. [DOI] [PubMed] [Google Scholar]
- de Kruijf P.; van Heteren J.; Lim H. D.; Conti P. G.; van der Lee M. M.; Bosch L.; Ho K. K.; Auld D.; Ohlmeyer M.; Smit M. J.; Wijkmans J. C.; Zaman G. J.; Smit M. J.; Leurs R. Nonpeptidergic allosteric antagonists differentially bind to the CXCR2 chemokine receptor. J. Pharmacol. Exp. Ther. 2009, 329, 783–90. 10.1124/jpet.108.148387. [DOI] [PubMed] [Google Scholar]
- Huang D.; Gu Q.; Ge H.; Ye J.; Salam N. K.; Hagler A.; Chen H.; Xu J. On the value of homology models for virtual screening: discovering hCXCR3 antagonists by pharmacophore-based and structure-based approaches. J. Chem. Inf. Model. 2012, 52, 1356–66. 10.1021/ci300067q. [DOI] [PubMed] [Google Scholar]
- Cox B. D.; Prosser A. R.; Sun Y.; Li Z.; Lee S.; Huang M. B.; Bond V. C.; Snyder J. P.; Krystal M.; Wilson L. J.; Liotta D. C. Pyrazolo-Piperidines Exhibit Dual Inhibition of CCR5/CXCR4 HIV Entry and Reverse Transcriptase. ACS Med. Chem. Lett. 2015, 6, 753–757. 10.1021/acsmedchemlett.5b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V.; Rosenbaum D. M.; Hanson M. A.; Rasmussen S. G.; Thian F. S.; Kobilka T. S.; Choi H. J.; Kuhn P.; Weis W. I.; Kobilka B. K.; Stevens R. C. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–65. 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Chun E.; Thompson A. A.; Chubukov P.; Xu F.; Katritch V.; Han G. W.; Roth C. B.; Heitman L. H.; Ijzerman A. P.; Cherezov V.; Stevens R. C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 2012, 337, 232–236. 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra A. J.; Leurs R.; de Esch I. J.; de Graaf C. From three-dimensional GPCR structure to rational ligand discovery. Adv. Exp. Med. Biol. 2014, 796, 129–57. 10.1007/978-94-007-7423-0_7. [DOI] [PubMed] [Google Scholar]
- Kooistra A. J.; Roumen L.; Leurs R.; de Esch I. J.; de Graaf C. From heptahelical bundle to hits from the Haystack: structure-based virtual screening for GPCR ligands. Methods Enzymol. 2013, 522, 279–336. 10.1016/B978-0-12-407865-9.00015-7. [DOI] [PubMed] [Google Scholar]
- Kolb P.; Rosenbaum D. M.; Irwin J. J.; Fung J. J.; Kobilka B. K.; Shoichet B. K. Structure-based discovery of beta2-adrenergic receptor ligands. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6843–8. 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra A. J.; Vischer H. F.; McNaught-Flores D.; Leurs R.; de Esch I. J.; de Graaf C. Function-specific virtual screening for GPCR ligands using a combined scoring method. Sci. Rep. 2016, 6, 28288. 10.1038/srep28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J.; Coleman R. G.; Setola V.; Irwin J. J.; Fan H.; Schlessinger A.; Sali A.; Roth B. L.; Shoichet B. K. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 2011, 7, 769–78. 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J.; Yoo L.; Gao Z. G.; Irwin J. J.; Shoichet B. K.; Jacobson K. A. Structure-based discovery of A2A adenosine receptor ligands. J. Med. Chem. 2010, 53, 3748–55. 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V.; Jaakola V. P.; Lane J. R.; Lin J.; Ijzerman A. P.; Yeager M.; Kufareva I.; Stevens R. C.; Abagyan R. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J. Med. Chem. 2010, 53, 1799–809. 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C.; Kooistra A. J.; Vischer H. F.; Katritch V.; Kuijer M.; Shiroishi M.; Iwata S.; Shimamura T.; Stevens R. C.; de Esch I. J.; Leurs R. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H(1) receptor. J. Med. Chem. 2011, 54, 8195–206. 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J. J.; Shoichet B. K. ZINC--a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–82. 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- de Graaf C.; Vischer H. F.; de Kloe G. E.; Kooistra A. J.; Nijmeijer S.; Kuijer M.; Verheij M. H.; England P. J.; van Muijlwijk-Koezen J. E.; Leurs R.; de Esch I. J. Small and colorful stones make beautiful mosaics: fragment-based chemogenomics. Drug Discovery Today 2013, 18, 323–30. 10.1016/j.drudis.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Congreve M.; Carr R.; Murray C.; Jhoti H. A ’rule of three’ for fragment-based lead discovery?. Drug Discovery Today 2003, 8, 876–7. 10.1016/S1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- Paolini G. V.; Shapland R. H.; van Hoorn W. P.; Mason J. S.; Hopkins A. L. Global mapping of pharmacological space. Nat. Biotechnol. 2006, 24, 805–15. 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- Schultes S.; de Graaf C.; Haaksma E. E. J.; de Esch I. J. P.; Leurs R.; Krämer O. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discovery Today: Technol. 2010, 7, e157–e162. 10.1016/j.ddtec.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Kooistra A. J.; Kanev G. K.; van Linden O. P.; Leurs R.; de Esch I. J.; de Graaf C. KLIFS: a structural kinase-ligand interaction database. Nucleic Acids Res. 2016, 44, D365–71. 10.1093/nar/gkv1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitessier N.; Englebienne P.; Lee D.; Lawandi J.; Corbeil C. R. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br. J. Pharmacol. 2008, 153 (S1), S7–S26. 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissantz C.; Bernard P.; Hibert M.; Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-Protein Coupled Receptors suitable targets?. Proteins: Struct., Funct., Genet. 2003, 50, 5–25. 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- Chen J. Z.; Wang J.; Xie X. Q. GPCR structure-based virtual screening approach for CB2 antagonist search. J. Chem. Inf. Model. 2007, 47, 1626–37. 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- Evers A.; Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J. Med. Chem. 2005, 48, 1088–1097. 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- de Graaf C.; Rein C.; Piwnica D.; Giordanetto F.; Rognan D. Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors. ChemMedChem 2011, 6, 2159–69. 10.1002/cmdc.201100317. [DOI] [PubMed] [Google Scholar]
- de Graaf C.; Rognan D. Selective structure-based virtual screening for full and partial agonists of the beta2 adrenergic receptor. J. Med. Chem. 2008, 51, 4978–85. 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- de Graaf C.; Rognan D. Customizing G Protein-coupled receptor models for structure-based virtual screening. Curr. Pharm. Des. 2009, 15, 4026–48. 10.2174/138161209789824786. [DOI] [PubMed] [Google Scholar]
- Perez-Nueno V. I.; Ritchie D. W.; Rabal O.; Pascual R.; Borrell J. I.; Teixido J. Comparison of ligand-based and receptor-based virtual screening of HIV entry inhibitors for the CXCR4 and CCR5 receptors using 3D ligand shape matching and ligand-receptor docking. J. Chem. Inf. Model. 2008, 48, 509–33. 10.1021/ci700415g. [DOI] [PubMed] [Google Scholar]
- Rogers D.; Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–54. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–40. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Voss M. E.; Carter P. H.; Tebben A. J.; Scherle P. A.; Brown G. D.; Thompson L. A.; Xu M.; Lo Y. C.; Yang G.; Liu R. Q.; Strzemienski P.; Everlof J. G.; Trzaskos J. M.; Decicco C. P. Both 5-arylidene-2-thioxodihydropyrimidine-4,6(1H,5H)-diones and 3-thioxo-2,3-dihydro-1H-imidazo[1,5-a]indol-1-ones are light-dependent tumor necrosis factor-alpha antagonists. Bioorg. Med. Chem. Lett. 2003, 13, 533–8. 10.1016/S0960-894X(02)00941-1. [DOI] [PubMed] [Google Scholar]
- Tirado-Rives J.; Jorgensen W. L. Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. J. Med. Chem. 2006, 49, 5880–4. 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- Wilcken R.; Zimmermann M. O.; Lange A.; Joerger A. C.; Boeckler F. M. Principles and applications of halogen bonding in medicinal chemistry and chemical biology. J. Med. Chem. 2013, 56, 1363–88. 10.1021/jm3012068. [DOI] [PubMed] [Google Scholar]
- de Wit R. H.; de Munnik S. M.; Leurs R.; Vischer H. F.; Smit M. J. Molecular Pharmacology of Chemokine Receptors. Methods Enzymol. 2016, 570, 457–515. 10.1016/bs.mie.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Langmead C. J.; Andrews S. P.; Congreve M.; Errey J. C.; Hurrell E.; Marshall F. H.; Mason J. S.; Richardson C. M.; Robertson N.; Zhukov A.; Weir M. Identification of novel adenosine A(2A) receptor antagonists by virtual screening. J. Med. Chem. 2012, 55, 1904–9. 10.1021/jm201455y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M.; Andrews S. P.; Dore A. S.; Hollenstein K.; Hurrell E.; Langmead C. J.; Mason J. S.; Ng I. W.; Tehan B.; Zhukov A.; Weir M.; Marshall F. H. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J. Med. Chem. 2012, 55, 1898–903. 10.1021/jm201376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker O. M.; Dhanoa D. S.; Marantz Y.; Chen D.; Shacham S.; Cheruku S.; Heifetz A.; Mohanty P.; Fichman M.; Sharadendu A.; Nudelman R.; Kauffman M.; Noiman S. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J. Med. Chem. 2006, 49, 3116–35. 10.1021/jm0508641. [DOI] [PubMed] [Google Scholar]
- Srivastava A.; Yano J.; Hirozane Y.; Kefala G.; Gruswitz F.; Snell G.; Lane W.; Ivetac A.; Aertgeerts K.; Nguyen J.; Jennings A.; Okada K. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 2014, 513, 124–7. 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- Wijtmans M.; Scholten D. J.; Roumen L.; Canals M.; Custers H.; Glas M.; Vreeker M. C.; de Kanter F. J.; de Graaf C.; Smit M. J.; de Esch I. J.; Leurs R. Chemical subtleties in small-molecule modulation of peptide receptor function: the case of CXCR3 biaryl-type ligands. J. Med. Chem. 2012, 55, 10572–83. 10.1021/jm301240t. [DOI] [PubMed] [Google Scholar]
- Kooistra A. J.; Leurs R.; de Esch I. J.; de Graaf C. Structure-Based Prediction of G-Protein-Coupled Receptor Ligand Function: A beta-Adrenoceptor Case Study. J. Chem. Inf. Model. 2015, 55, 1045–61. 10.1021/acs.jcim.5b00066. [DOI] [PubMed] [Google Scholar]
- Simpson L. M.; Taddese B.; Wall I. D.; Reynolds C. A. Bioinformatics and molecular modelling approaches to GPCR oligomerization. Curr. Opin. Pharmacol. 2010, 10, 30–7. 10.1016/j.coph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez D.; Gutierrez-de-Teran H. Characterization of the homodimerization interface and functional hotspots of the CXCR4 chemokine receptor. Proteins: Struct., Funct., Genet. 2012, 80, 1919–1928. 10.1002/prot.24099. [DOI] [PubMed] [Google Scholar]
- Ferre S.; Casado V.; Devi L. A.; Filizola M.; Jockers R.; Lohse M. J.; Milligan G.; Pin J. P.; Guitart X. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 2014, 66, 413–34. 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vischer H. F.; Castro M.; Pin J. P. G Protein-Coupled Receptor Multimers: A Question Still Open Despite the Use of Novel Approaches. Mol. Pharmacol. 2015, 88, 561–71. 10.1124/mol.115.099440. [DOI] [PubMed] [Google Scholar]
- Jazayeri A.; Dore A. S.; Lamb D.; Krishnamurthy H.; Southall S. M.; Baig A. H.; Bortolato A.; Koglin M.; Robertson N. J.; Errey J. C.; Andrews S. P.; Teobald I.; Brown A. J.; Cooke R. M.; Weir M.; Marshall F. H. Extra-helical binding site of a glucagon receptor antagonist. Nature 2016, 533, 274–7. 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- Lane J. R.; Chubukov P.; Liu W.; Canals M.; Cherezov V.; Abagyan R.; Stevens R. C.; Katritch V. Structure-based ligand discovery targeting orthosteric and allosteric pockets of dopamine receptors. Mol. Pharmacol. 2013, 84, 794–807. 10.1124/mol.113.088054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass M.; Agai-Csongor E.; Horti F.; Keseru G. M. Multiple fragment docking and linking in primary and secondary pockets of dopamine receptors. ACS Med. Chem. Lett. 2014, 5, 1010–4. 10.1021/ml500201u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.; Nomura W.; Narumi T.; Masuda A.; Tamamura H. Bivalent ligands of CXCR4 with rigid linkers for elucidation of the dimerization state in cells. J. Am. Chem. Soc. 2010, 132, 15899–901. 10.1021/ja107447w. [DOI] [PubMed] [Google Scholar]
- Springael J. Y.; de Poorter C.; Deupi X.; Van Durme J.; Pardo L.; Parmentier M. The activation mechanism of chemokine receptor CCR5 involves common structural changes but a different network of interhelical interactions relative to rhodopsin. Cell. Signalling 2007, 19, 1446–56. 10.1016/j.cellsig.2007.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.