ABSTRACT
Strict infection control practices have been implemented for health care visits by cystic fibrosis (CF) patients in an attempt to prevent transmission of important pathogens. This study used whole-genome sequencing (WGS) to determine strain relatedness and assess population dynamics of Staphylococcus aureus isolates from a cohort of CF patients as assessed by strain relatedness. A total of 311 S. aureus isolates were collected from respiratory cultures of 115 CF patients during a 22-month study period. Whole-genome sequencing was performed, and using single nucleotide polymorphism (SNP) analysis, phylogenetic trees were assembled to determine relatedness between isolates. Methicillin-resistant Staphylococcus aureus (MRSA) phenotypes were predicted using PPFS2 and compared to the observed phenotype. The accumulation of SNPs in multiple isolates obtained over time from the same patient was examined to determine if a genomic molecular clock could be calculated. Pairs of isolates with ≤71 SNP differences were considered to be the “same” strain. All of the “same” strain isolates were either from the same patient or siblings pairs. There were 47 examples of patients being superinfected with an unrelated strain. The predicted MRSA phenotype was accurate in all but three isolates. Mutation rates were unable to be determined because the branching order in the phylogenetic tree was inconsistent with the order of isolation. The observation that transmissions were identified between sibling patients shows that WGS is an effective tool for determining transmission between patients. The observation that transmission only occurred between siblings suggests that Staphylococcus aureus acquisition in our CF population occurred outside the hospital environment and indicates that current infection prevention efforts appear effective.
KEYWORDS: MRSA, cystic fibrosis, strain relatedness, whole-genome sequencing
INTRODUCTION
Staphylococcus aureus colonization in populations with chronic illness can often lead to repeated infections and severe disease. Patients with the genetic disease cystic fibrosis (CF) are especially vulnerable due to the very nature of their illness, in which thick mucus in the airways provides for an enriched environment suited to bacterial survival and colonization while allowing for evasion of host immune defenses (1, 2). S. aureus is known to be one of the first pathogens to colonize CF lungs with a 69% prevalence rate, peaking at ages 11 to 15 (3). S. aureus is typically isolated first from the upper airways of CF patients, and in many cases, colonization of the lower airways follows (3, 4). Repeated or chronic antibiotic usage that is common in the CF population may influence colonization by disturbing the normal upper respiratory microbiome, allowing S. aureus to invade and persist. Once colonization is established in the lungs, eradication efforts are typically unsuccessful and single-strain persistence is thought to occur (5, 6), with only a small percentage of colonized CF patients showing evidence of strain replacement (7). Unfortunately, there is an upward trend toward acquisition of methicillin-resistant strains (methicillin-resistant Staphylococcus aureus [MRSA]), with prevalence increasing from 12% in 2003 to 26% in 2013 (8, 29). MRSA colonization is associated with poorer health outcomes in CF patients than colonization with methicillin-sensitive S. aureus (MSSA) (9, 10).
MSSA and MRSA strains are easily spread in daycares and schools and have been implicated in health care-associated outbreaks in intensive care units (ICUs) and nursing homes (11–13). In health care facilities, patients utilize the same clinic space, equipment, and hospital rooms, which increases the risk of transmission of bacteria (14, 15). Because of their vulnerability, there has been a particular effort to decrease the risk of pathogen acquisition from the health care environment among the CF patient population. Infection control guidelines attempt to minimize cross-contamination between patients by enforcing strict rules and protocols, which limit patient interactions, mandate isolation gowns and gloves for staff, and require cleaning and disinfection of high touch surfaces and equipment between patients (16). Breeches in protocol can place patients at risk for acquisition of pathogens, such as MRSA. In order to assess the effectiveness of these infection control guidelines, monitoring the frequency of transmission events between CF patients in health care settings is warranted. A transmission event has occurred when the same strain has been isolated from one or more patients. Recently, outbreak investigators have begun to utilize whole-genome sequencing (WGS) technology to determine relatedness between isolates in a patient population to identify transmission events (17, 18). Since the resolution is at the DNA level, detection of single nucleotide polymorphisms (SNPs) between isolates can be observed and analyzed in order to calculate the degree of genetic relatedness.
We have taken advantage of a 22-month study of 301 S. aureus isolates from 115 patients at the Cystic Fibrosis Center of the Cincinnati Children's Hospital Medical Center (CCHMC) to assess the effectiveness of the infection control program by comparing WGS of those isolates. We compared the relatedness of isolates from the same patient, from sibling patients, and from unrelated patients. The degree of relatedness among a single patient's isolates was used to determine a criterion, measured in number of SNP differences between isolates, for being the “same” strain. We also addressed the issue of genetic variation of a colonizing MRSA strain over time.
RESULTS
From April 2014 to February 2016, 301 S. aureus isolates were collected from 115 unique CF patients, including 13 pairs of siblings. A total of 212 isolates from 70 patients were identified as MRSA by the clinical laboratory, and 60 isolates were identified as MSSA with 45 of these isolates retained from unique patients and the remaining 15 isolates originating from the MRSA patients. A MRSA isolate was retained from all CF patients who had a positive culture during the study period while only a small sample of MSSA isolates were saved. Twenty-four of the isolates were first-time MRSA positives (34%; n = 70). Ten patients had MRSA acquisition a year or less from the saved isolate (14%; n = 70). The remaining 38 patients had established MRSA colonization more than 1 year prior to collection of the study specimen (54%; n = 70). Many of the first time positive and newly positive patients established colonization during the study period by meeting the colonization definition. Demographics of study patients are described in Table 1.
TABLE 1.
Demographics of study patients
| Infection | Patient characteristic |
|||
|---|---|---|---|---|
| Median age (yr [range]) at beginning of study | Gender (male/female) | No. colonized (%) | No. with CFTRa mutation ΔF508 genotype (%) | |
| MSSA (n = 45) | 11.6 (0–20) | 24/21 | 37 (82) | 44 (98) |
| MRSA (n = 70) | 10.3 (0–21) | 30/40 | 48 (69) | 58 (83) |
CFTR, cystic fibrosis transmembrane conductance regulator.
Phylogenetic analysis.
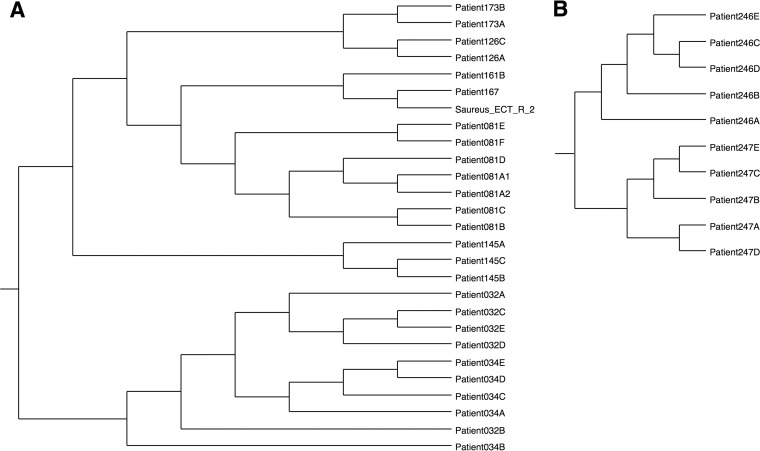
The total data set consisted of 367 S. aureus genome sequences and included 301 sequences from CF patients, 31 sequences from abscesses or blood, and 35 finished (closed) reference sequences from GenBank (see Fig. S1 in the supplemental material). Genomes whose names begin with “Patient” were obtained from CF patients, those whose names begin with “Blood” or “Abscess” were obtained from non-CF isolates, and those whose names begin with “Saureus” are reference genomes. Samples from the same CF patient are identified by the patient number with a letter suffix indicating the order in which they were isolated. Phylogenetic analysis revealed that genomes do not cluster together based on the source of the sample; samples from CF patients are interspersed with blood, abscess, and reference strains. Figure 1A shows several examples of multiple isolates from the same patient. In most cases, those isolates form monophyletic clades. The isolates from patients 032 and 034 are an exception to that rule. These patients are siblings, and the topology of the 032 and 034 clade suggests that these patients have frequently exchanged strains over the course of their colonization. Figure 1B shows a clade that includes two examples of multiple isolates from the same patient. Patients 246 and 247 are siblings, but in this case, the individual patient clades are monophyletic, indicating that they did not exchange strains but that both were initially infected by the same strain.
FIG 1.
Subclades from parsimony tree in Fig. S1 in the supplemental material. (A) Representative clade from the tree shown in Fig. S1 illustrating examples of isolates from the same patient and isolates from sibling patients 032 and 034. All isolates from the same patient are monophyletic, and the isolates from the sibling patients also form a single clade. (B) Another clade from the tree in Fig. S1 showing isolates from a pair of siblings. Each sibling clade is monophyletic, and the two clades are derived from a common ancestor that infected both patients 246 and 247.
What do we mean by “the same strain”?
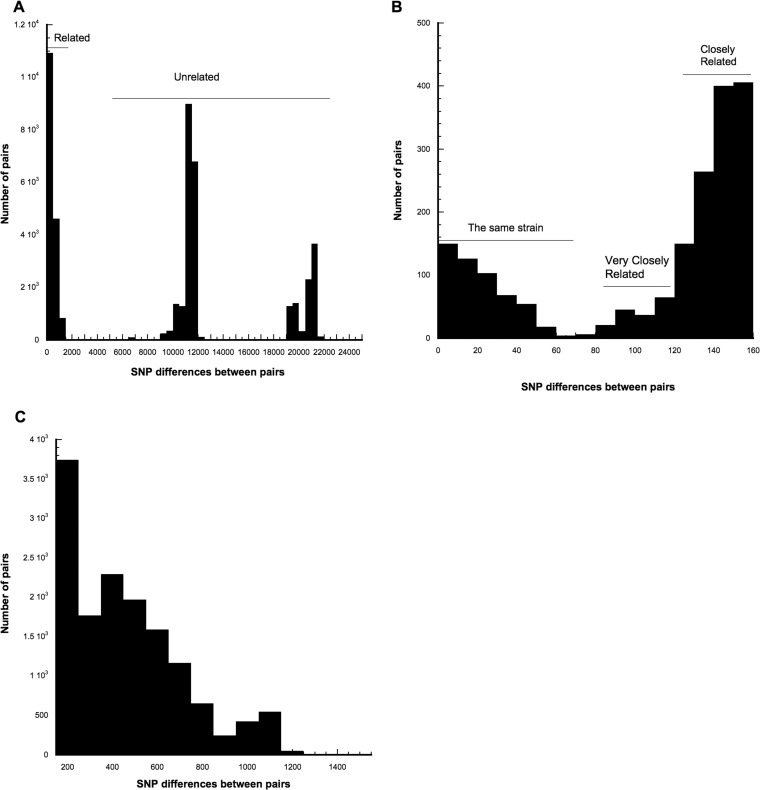
Strain relatedness was investigated by analyzing the number of SNP differences between all pairs of genomes from CF patients, allowing us to directly address the issue of what is meant by the “same” strain. The data set included nine isolates that had been independently sequenced twice. Those are most certainly sequences of the same isolate. Those replicate sequencings are shown in Table S3 in the supplemental material in red type face and have IDs such as Patient053A1 and Patient053A2, indicating that the isolate designated Patient053 was sequenced twice. If genome sequencing was perfect, there would be no differences between replicate sequences of the same isolate. In fact, replicate sequences had a mean of 2.6 ± 0.6 SNP differences (range, 0 to 6 differences). Thus, the “same” cannot be equated to identical sequences. The data set also included serial isolates from 55 patients collected over the 22-month period. These isolates were expected to be the same clone based on previous longitudinal studies. Additionally, there were 13 pairs of genomes from siblings, which in the case of our cohort represented children living together. These data indicate that siblings are likely to exchange S. aureus strains or to have initially been infected by the same S. aureus strain. The WGS results from this data set allowed us to develop definitions for strain relatedness. Table S4 in the supplemental material gives the number of SNP differences for all pairs of CF genomes, with pairs being listed in increasing order of number of SNP differences, and Fig. 2A shows the distribution of those differences. All pairs with ≤71 differences are either from the same patient or from siblings. Thus, it is reasonable to define the “same” strain as isolates with ≤71 SNP differences (Fig. 2B). Of the 525 such pairs, 431 (82%) are from the same patient and 94 (18%) are from siblings. We define pairs with 72 to 123 differences as being “very closely” related (Fig. 2B). Of the 229 very closely related pairs, 7 (3%) are from the same patient, 24 (10.5%) are from siblings, and the remaining 86.5% are from unrelated patients. We define “closely” related pairs as having 124 to 156 differences (Fig. 2B). None of the 1,163 closely related pairs are from the same patient, 2 (0.17%) are from siblings, and the remaining 99.9% are from unrelated patients. We define “distantly” related pairs as having 157 to 1,514 SNP differences (Fig. 2C). Of the 15,618 distantly related pairs, 5 (0.032%) are from the same patient and none are from siblings. The 24,479 pairs with 5,957 to 21,644 differences are considered to be unrelated. The presence of unrelated isolates in a single patient (see Table S5 in the supplemental material) clearly represents superinfection events. There are 47 examples of a patient being superinfected by an unrelated strain. There are also 16 instances in which siblings were infected by unrelated strains.
FIG 2.
Distributions of pairwise SNP differences. (A) All pairs. (B) Pairs with 0 to 156 differences. (C) Distantly related pairs (157 to 1,514 differences).
Predicting the MRSA phenotype.
WGS data were also examined to identify SNPs that are casually associated with the MRSA phenotype. Based on 100 replicate runs, the accuracy with which PPFS2 predicted the MRSA phenotype was 0.986, the positive predictive value (fraction of those predicted to be MRSA that actually were MRSA) was 0.994, and the negative predictive value (fraction of those predicted to be MSSA that actually were MSSA) was 0.967. In Fig. S1, the strains predicted to be MRSA are shown in red typeface while those predicted to be MSSA are shown in black typeface. The three strains in which there is a conflict between the predicted phenotype and the experimentally determined phenotype are enclosed in boxes. The phenotypes of 216 isolates from CF patients were physically determined. The accuracy, PPV, and NPV are based on comparison of predicted phenotypes with those known phenotypes. The Patient013 isolate is predicted to be methicillin sensitive but is in fact methicillin resistant (Table S3). It carries an intact mecA gene, which causes the MRSA phenotype. The Patient013 isolate is located solidly within a methicillin-sensitive clade (Fig. S1), suggesting that mecA may have been recently acquired by horizontal transfer into an otherwise methicillin-sensitive genetic background. Conversely, the Patient092G isolate is predicted to be methicillin sensitive (MSSA), but it is actually methicillin resistant. It also carries the mecA gene and is located within a clade that is entirely MRSA. That clade consists entirely of isolates from the same patient (Patient092). The number of SNP differences between Patient092G and other members of that clade is small enough to be confident that all members of that clade are the same strain. It is unclear why PPFS2 predicted Patient092G to be MSSA. The Patient034B isolate is predicted to be methicillin resistant but is in fact methicillin sensitive. The MSSA phenotype may be accounted for by a deletion of base pairs −43 through −28 upstream of the initiation codon of mecA. We assume that this substantial deletion within the mecA promoter region may account for the MSSA phenotype by a failure to express the intact mecA gene. The phenotypes of the abscess and blood isolates were not physically determined, but the presence or absence of mecA in the genome sequences was determined. In all of those cases, the phenotype predicted by PPFS2 corresponded to the presence or absence of mecA in the genome.
The phylogeny in Fig. S1 was analyzed using MEGA7, which calculated the ancestral state at internal nodes based on the predicted phenotype. In Fig. S1, branches along which the predicted phenotype changed from MSSA to MRSA are shown in red, while those in which the predicted phenotype changed from MRSA to MSSA are shown in blue. There were 13 branches along which the phenotype changed from MSSA to MRSA, and 11 along which the phenotype changed from MRSA to MSSA. The ancestral state reconstruction clearly shows that the root state was MSSA. Methicillin resistance has arisen and has been lost multiple times, and the change in phenotype is primarily accounted for by gain and loss of mecA. All of the branches identified by PPFS2 as having phenotype changes are consistent with the clades of MRSA and MSSA isolates, but PPFS2 failed to identify one such branch (orange in Fig. S1) along which a change from MSSA to MRSA clearly occurred. That failure is attributable to the topological differences between the parsimony tree in Fig. S1 and the maximum likelihood (ML) tree estimated by MEGA7 and used to estimate ancestral states. SNP-based parsimony trees have been shown to be more accurate then SNP-based ML trees (19). PPFS2 identifies as causal SNPs those whose state not only correlates strongly with the MRSA phenotype but those in which the state changes along those branches where the MRSA state changes. PPFS2 identified 125 such causal SNPs, of which 74 had a probability of <10−100 of the allele changing randomly along branches on which the phenotype changed and another 51 had probabilities between 3 × 10−74 and 2 × 10−37. Of those, 49 were nonsynonymous changes. The most frequent genes in which those nonsynonymous changes occurred were the recA and recB recombinases and the mecA peptidoglycan transpeptidase. The identities of the proteins containing the nonsynonymous causal SNPS, and their protein IDs, are provided in Table S6 in the supplemental material.
The existence of so many SNPs that are causally related to the MRSA phenotype suggests that although expression of mecA is the major determinant of methicillin resistance, other genes modify the degree of that resistance and are subject to selection.
How much do strains vary within a patient over the course of colonization?
MRSA strain genetic variability was studied by analyzing serial isolates collected over time. A collection of between 4 and 8 MRSA isolates was retained from 32 study patients over a range of 8 to 22 months, with at least 3 months between most isolates. There were 18 patients with staphylococcal cassette chromosome mec type II (SCCmec II) isolates and 14 with SCCmec IV isolates. The SCCmec II group contained one set of siblings while the SCCmec IV group contained 3 sets of siblings. The average year of colonization for the SCCmec II group was 2010, with only 3 new colonizations during the study period (2014 to 2016) and the longest colonization period being 10 years (2004 acquisition). The SCCmec IV group had an average colonization year of 2013, with 5 new colonizations during the study period and the longest colonization at 4 years (2010). As shown in Fig. S1, all serial isolates clustered within the same clade with only a few exceptions, e.g., Patient192D, Patient045D, Patient201C&D, etc. None of the serial isolates represented acquisition of a new colonizing strain.
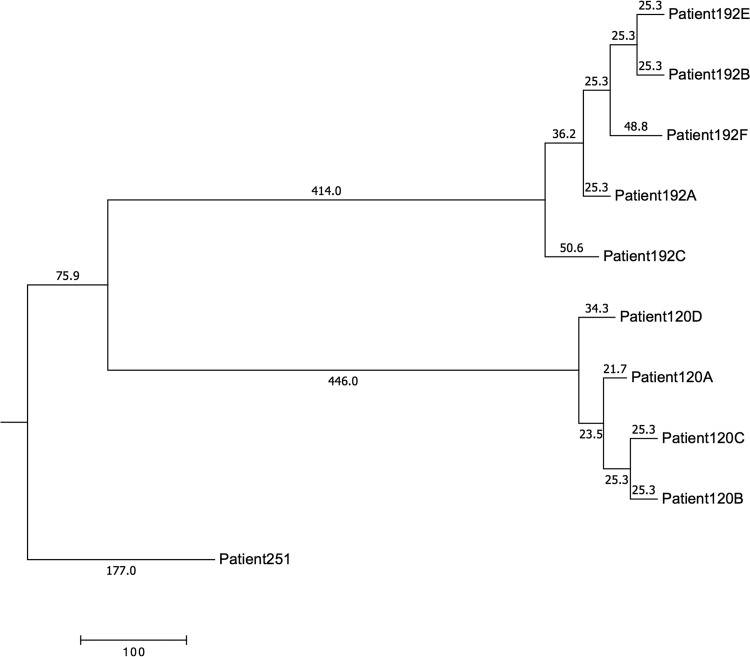
In examining serial isolates, we would assume that SNPs would accumulate chronologically; however, this is not what we observed. Figure 3 shows a portion of the kSNP3 parsimony tree where we attempted to estimate meaningful mutation rates. Branches are labeled with lengths that are the number of SNP changes; i.e., the two branches leading to patients 192E and 192B each represent 25.3 changes (an artifact of kSNP rounding). According to the tree, the total distance from 192B to 192E is 50.6. However, kSNPdist calculates the pairwise distance as 20. Similarly, the phylogeny shows the total distance from 192E to 192C as 162.7, whereas kSNPdist calculates it as 85. The difference lies entirely in the ways that distances and branch lengths are calculated. kSNP counts all changes, including a change from a SNP being present to it being absent. kSNPdist counts only changes from one base to another; i.e., if at a particular position either member of a pair is missing a SNP, that position is ignored. The reason for ignoring missing data is that missing data are not the same as a gap in a traditional alignment. A gap represents an insertion or deletion; thus, a mutation has taken place. A missing SNP is just that, missing data. It may be the result of an indel, or it may be the result of having another SNP within the flanking portion of the kmer (both of which are actual mutational events). If we were dealing only with the finished (closed) genomes, we could safely count missing SNPs as mutational events. However, we are dealing with genome assemblies consisting of many contigs. If a SNP in one genome falls within a kmer length of the end of a contig, it will be missed. More likely, all SNPs that fall within spaces between contigs will be missed. Neither of those is a mutation. Indeed, the replicate sequencing of Patient053A is 0.00029 (52.4) SNPs apart on the tree, but they have zero differences according to kSNPdist. Other replicate sequences have about the same distances on the tree (0.00028 or 51 SNPs).
FIG 3.
kSNP3 parsimony tree example.
If changes were accumulating according to a molecular clock, i.e., at a constant rate, then branch lengths would correspond to time, which would mean that 192C arose before 192A, before 192F, before 192B. But 192A was isolated 3 months before 192B and 4 months before 192C. There are (at least) two explanations for the isolation order not corresponding to the order in which the strains arose according to the phylogeny (1). At the sampling times, there were multiple strains in the population. At the time 192A was isolated, 192C had already arisen, but 192A was picked by chance alone. Since 192B arose last, and at about the same time as 192E, when it was isolated 192C and 192F must also have been present (2). A molecular clock was not operating. Strains arose in isolation order, but there were just more changes along the branch from the common ancestor (CA) to 192A than to 192C.
Table S7 in the supplemental material shows the number of differences accumulated per week for pairs of the same strain from the same patient. The rate at which differences accumulated ranged from a low of 0.043 per week (Patient196A to Patient196B) to a high of 64.4 differences per week (Patient034A to Patient034B).
DISCUSSION
The high resolution of WGS (±2 SNPs) generates an ironic problem in determining whether two isolates are the same strain. Because the S. aureus population is constantly evolving during a persistent infection (colonization) in a CF patient, at any given moment the population will consist of a set of very closely related but distinguishable strains descended from the original infecting organism. Transmission of one of those to another patient is likely to result in isolates from the two patients being nonidentical. Indeed, the population in the recipient patient is likely to have diverged from the original infecting strain by the time the patient isolates are compared. Were identity at the WGS level to be the criterion of transmission, it is likely that no transmission events would ever be detected. Because identity is not a useful criterion for identifying transmission events, it is necessary to decide how different two isolates can be at the WGS level and still be considered the same strain. Analysis of the distribution of SNP differences between pairs of S. aureus strains from CF patients showed that all pairs with ≤71 differences were either from the same patient or from siblings. Based on that, we concluded that pairs that differed by ≤71 SNP differences were the same strain. We found no cases in which the same strain was transmitted between unrelated patients. Therefore, we conclude that the infection control guidelines that were used during the study were very effective in controlling patient-to-patient transmission. Could we have detected transmission events if they had occurred? The fact that we could detect many instances of transmission between siblings, who lived together, indicates that we can indeed detect transmission when it occurs.
It is tempting to try to determine the rate at which strains evolve during long-term infections, but that is not possible from these data. If early isolated strains were the ancestors of late-isolated strains, such estimates would be feasible, but that is not the case. The number of differences between the first isolate from a patient and subsequent isolates does not typically increase with the time between the isolation dates. The phylogeny in Fig. S1 in the supplemental material likewise shows that the order of descent from common ancestors does not typically follow the order of isolation of strains from the same patient. This is most likely because at any time there is a population of different strains that have diverged from the original infecting strain, and the isolate that is sequenced represents only a sample of that population. Were we able to sequence 20 or more isolates from a particular sample, we may be able to describe the diversity at each time and from the population dynamics be able to estimate the rate at which new alleles replace old ones. That level of sampling and genome sequencing is not, however, practical at this time.
There is nothing genetically special about the strains that infect CF patients. The phylogenetic tree (Fig. S1) shows that isolates from CF patients are intermixed with isolates from blood and abscesses. Consistent with that, PPFS2 (20) was unable to identify SNPs that are preferentially associated with being from CF patients (results not shown).
MATERIALS AND METHODS
Study cohort.
The study was conducted at Cincinnati Children's Hospital Medical Center, a 635 bed, free-standing pediatric teaching hospital and medical center. CCHMC's Cystic Fibrosis Center provides medical care to approximately 250 CF patients annually. S. aureus isolates were retained from clinical respiratory cultures (sputum or oropharyngeal) from CF patients that received medical care at CCHMC either as inpatients or outpatients beginning April 2014 until February 2016. Serial isolates were collected from patients colonized with MRSA at least 3 months apart for strain retention and variability studies. Clinical microbiology data from our study participants were reviewed to determine S. aureus history. We applied the accepted definition of at least three positive cultures over a 1-year period to our cohort to determine colonization (9, 21). Demographic data were collected from the Electronic Medical Record system EPIC, and microbiology data were reviewed from surveillance software VigiLanz and Premier SafetySurveillor. Deidentified clinical blood and abscess MRSA isolates were also included for comparison. This study was approved by CCHMC's institutional review board.
Microbiology.
All isolates were identified as S. aureus using the matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and Vitek MS+ system (bioMérieux, St. Louis, MO, USA). Antimicrobial susceptibility testing was performed using the Vitek 2 system for routine isolates. Antimicrobial susceptibility testing of thymidine-dependent isolates was performed using the Etest system (bioMérieux) following manufacturer recommendations. Our primary focus was on methicillin resistance, but resistance or sensitivity to 11 additional antibiotics is provided in Table S1 in the supplemental material. Frozen stocks were made from multiple colonies of pure cultures of isolates on 5% sheep blood agar (Becton Dickinson Diagnostics, Cockeysville, MD, USA). Organisms were frozen in brucella broth with glycerol (Hardy Diagnostics, Santa Maria, CA, USA) and stored at −80°C. Organisms removed from storage were subcultured onto 5% sheep blood agar twice prior to submission for WGS.
Whole-genome sequencing.
Eighteen-hour to 24-h colonies of S. aureus streaked from the frozen cultures were inoculated into Trypticase soy broth (TSB), grown overnight, and submitted to the CCHMC Bioinformatics Core laboratory for whole-genome sequencing (Illumina HiSeq 2000 platform). Raw reads were assembled using Velvet 1.2.10 (22), and gene content was annotated using RAST (23). Phylogenetic relatedness was determined using kSNP3 (24), which identified SNPs and estimated a maximum parsimony phylogenetic tree. SCCmec type was identified by alignment of assembled reads against reference SCCmec sequences using BLAST. S. aureus control isolates from a central sequencing database consisting of known DNA sequences were used to compare against the clinical isolates. For comparison with WGS results, the multilocus sequence types (MLSTs) were determined from the WGS through the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk//services/MLST/) as described in reference 25. The MLSTs are provided in Table S2 in the supplemental material.
Phylogenetic analysis.
kSNP3 (24) was used to identify SNPs and to estimate a maximum parsimony phylogenetic tree. The tree was virtually rooted by using Staphylococcus simiae CCM 7213 as an outgroup. The kmer size was set to 19, the optimum size estimated by Kchooser (26). The accuracy with which kSNP estimates parsimony trees depends on the fractions of core kmers (FCK), in this data set calculated to be 0.454, and on the relative contributions of recombination and mutation (r/m) to diversity (19). Didelot and Falush have estimated r/m in S. aureus as 0.28 (27). The ratio of r/m to FCK was 0.617, giving the parsimony tree an estimated topological accuracy of 97% (19). The SNP alignment file is available from the figshare website at https://doi.org/10.6084/m9.figshare.4779466.v1. PPFS2 (20) was used to identify SNPs that were likely to be causally related to the MRSA phenotype. MEGA 7 was used to estimate the ancestral MRSA states of the internal node of the tree and was thereby used to identify those branches along which the MRSA phenotype changed (28). PPFS2 identifies as causal SNPs those SNPs whose allele state changes are highly nonrandom with respect to phenotype changes along those branches. A brief description of the PPFS2 algorithm is included in the supplemental material. The python script kSNPdist was used to calculate the number of SNP differences between all pairs of CF genomes.
kSNP3 and kSNPdist executables for OS X and Linux are freely available at https://sourceforge.net/projects/ksnp/. PPFS2 executables for OS X and Linux are freely available at https://sourceforge.net/projects/ppfs/.
Accession number(s).
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under accession numbers NDKH00000000 to NDWS00000000. The versions described in the paper are versions NDKH01000000 to NDWS01000000. The BioProject can be accessed at https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=380429.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Barbara Deburger, Cincinnati Children's Hospital, for determining the drug resistance phenotypes of the isolates used in this study. We are grateful to the Cystic Fibrosis group at the Cincinnati Children's Hospital for their support of this project. We are grateful to David Haslam and his lab, Cincinnati Children's Hospital, for performing the whole-genome sequencing.
This project was supported by a grant from the Cystic Fibrosis Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00164-17.
REFERENCES
- 1.Donaldson SH, Boucher RC. 2003. Update on pathogenesis of cystic fibrosis lung disease. Curr Opin Pulm Med 9:486–491. doi: 10.1097/00063198-200311000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Goss CH, Muhlebach MS. 2011. Review: Staphylococcus aureus and MRSA in cystic fibrosis. J Cyst Fibros 10:298–306. doi: 10.1016/j.jcf.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46(Suppl):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Zubeidi D, Hogan PG, Boyle M, Burnham CA, Fritz SA. 2014. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolated in serial cultures from the respiratory tract of children with cystic fibrosis. Pediatr Infect Dis J 33:549–553. doi: 10.1097/INF.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branger C, Gardye C, Lambert-Zechovsky N. 1996. Persistence of Staphylococcus aureus strains among cystic fibrosis patients over extended periods of time. J Med Microbiol 45:294–301. doi: 10.1099/00222615-45-4-294. [DOI] [PubMed] [Google Scholar]
- 7.Renders NH, van Belkum A, Overbeek SE, Mouton JW, Verbrugh HA. 1997. Molecular epidemiology of Staphylococcus aureus strains colonizing the lungs of related and unrelated cystic fibrosis patients. Clin Microbiol Infect 3:216–221. doi: 10.1111/j.1469-0691.1997.tb00980.x. [DOI] [PubMed] [Google Scholar]
- 8.Cystic Fibrosis Foundation. 2011. Patient registry annual data report 2010. Cystic Fibrosis Foundation, Bethesda, MD: http://www.cysticfibrosisdata.org/LiteratureRetrieve.aspx?ID=132651. [Google Scholar]
- 9.Dasenbrook EC, Merlo CA, Diener-West M, Lechtzin N, Boyle MP. 2008. Persistent methicillin-resistant Staphylococcus aureus and rate of FEV1 decline in cystic fibrosis. Am J Respir Crit Care Med 178:814–821. doi: 10.1164/rccm.200802-327OC. [DOI] [PubMed] [Google Scholar]
- 10.Ren CL, Morgan WJ, Konstan MW, Schechter MS, Wagener JS, Fisher KA, Regelmann WE, Investigators Coordinators of the Epidemiologic Study of Cystic Fibrosis. 2007. Presence of methicillin resistant Staphylococcus aureus in respiratory cultures from cystic fibrosis patients is associated with lower lung function. Pediatr Pulmonol 42:513–518. doi: 10.1002/ppul.20604. [DOI] [PubMed] [Google Scholar]
- 11.Lee BY, Bartsch SM, Wong KF, Singh A, Avery TR, Kim DS, Brown ST, Murphy CR, Yilmaz SL, Potter MA, Huang SS. 2013. The importance of nursing homes in the spread of methicillin-resistant Staphylococcus aureus (MRSA) among hospitals. Med Care 51:205–215. doi: 10.1097/MLR.0b013e3182836dc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YC, Lauderdale TL, Lin HM, Chen PC, Cheng MF, Hsieh KS, Liu YC. 2007. An outbreak of methicillin-resistant Staphylococcus aureus infection in patients of a pediatric intensive care unit and high carriage rate among health care workers. J Microbiol Immunol Infect 40:325–334. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2017. Information and advice about MRSA for school and daycare officials. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/mrsa/community/schools/index.html. [Google Scholar]
- 14.Lu PL, Tsai JC, Chiu YW, Chang FY, Chen YW, Hsiao CF, Siu LK. 2008. Methicillin-resistant Staphylococcus aureus carriage, infection and transmission in dialysis patients, healthcare workers and their family members. Nephrol Dial Transplant 23:1659–1665. doi: 10.1093/ndt/gfm806. [DOI] [PubMed] [Google Scholar]
- 15.Zuckerman JB, Zuaro DE, Prato BS, Ruoff KL, Sawicki RW, Quinton HB, Saiman L, Infection Control Study Group. 2009. Bacterial contamination of cystic fibrosis clinics. J Cyst Fibros 8:186–192. doi: 10.1016/j.jcf.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Saiman L, Siegel JD, LiPuma JJ, Brown RF, Bryson EA, Chambers MJ, Downer VS, Fliege J, Hazle LA, Jain M, Marshall BC, O'Malley C, Pattee SR, Potter-Bynoe G, Reid S, Robinson KA, Sabadosa KA, Schmidt HJ, Tullis E, Webber J, Weber DJ, Cystic Fibrous Foundation, Society for Healthcare Epidemiology of America. 2014. Infection prevention and control guideline for cystic fibrosis: 2013 update. Infect Control Hosp Epidemiol 35(Suppl):S1–S67. doi: 10.1086/676882. [DOI] [PubMed] [Google Scholar]
- 17.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price JR, Golubchik T, Cole K, Wilson DJ, Crook DW, Thwaites GE, Bowden R, Walker AS, Peto TE, Paul J, Llewelyn MJ. 2014. Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis 58:609–618. doi: 10.1093/cid/cit807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall BG. 2015. Effects of sequence diversity and recombination on the accuracy of phylogenetic trees estimated by kSNP. Cladistics 32:90–99. doi: 10.1111/cla.12113. [DOI] [PubMed] [Google Scholar]
- 20.Hall BG. 2014. SNP-associations and phenotype predictions from hundreds of microbial genomes without genome alignments. PLoS One 9:e90490. doi: 10.1371/journal.pone.0090490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox DW, Kelly C, Rush R, O'Sullivan N, Canny G, Linnane B. 2011. The impact of MRSA infection in the airways of children with cystic fibrosis; a case-control study. Ir Med J 104:305–308. [PubMed] [Google Scholar]
- 22.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genomes. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 25.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cystic Fibrosis Foundation. 2014. Patient registry annual data report 2013. Cystic Fibrosis Foundation, Bethesda, MD: https://www.cff.org/2013_CFF_Patient_Registry_Annual_Data_Report.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.