LETTER
Even in the genomic era, multilocus sequence typing (MLST) remains a method of choice for bacterial typing, quickly revealing relationships within and between bacterial lineages. There are two Acinetobacter MLST schemes, one devised by Bartual et al. (1), now referred to as the Oxford scheme, and the other is the Institut Pasteur scheme (2). Sequence type (ST) numbering is inevitably independent, and to avoid confusion, the scheme used should be stated; here we use STOx and STIP to distinguish them. The databases for both of these schemes can be found at http://pubmlst.org/abaumannii. Each scheme uses seven genes, three of which are shared (Fig. 1). However, much of the chromosome is not sampled (Fig. 1), particularly in the Oxford scheme, where the seven genes are all in one half of the chromosome. In addition, some genes in each scheme (e.g., rpoD, gltA, and cpn60 in the Oxford scheme or gtlA and cpn60 in the Institut Pasteur scheme) are close together (Fig. 1) and can potentially be replaced via a single recombination event.
FIG 1.
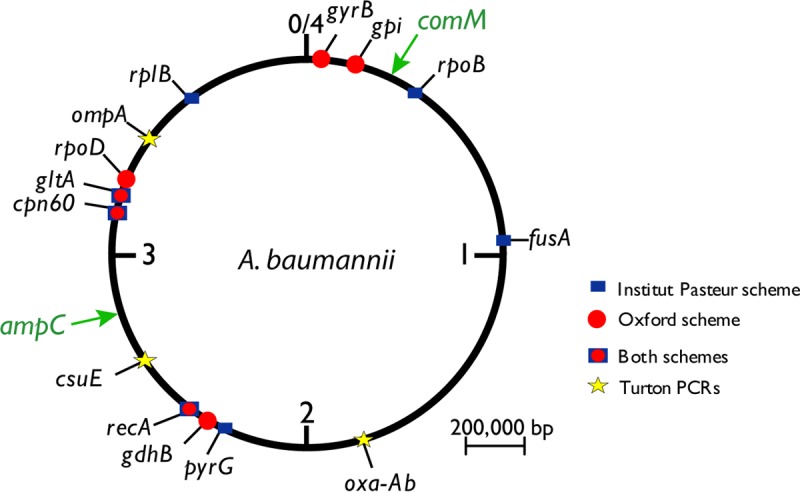
Circular map of the A. baumannii chromosome showing the locations of genes used in the Oxford and Institut Pasteur MLST schemes. Genes detected by trilocus typing by PCR (7) are also indicated by stars (Turton PCRs). The locations of the comM and ampC genes are also indicated.
Overall, the Institut Pasteur scheme more readily identifies members of clonal complexes (CCs), particularly CC1 and CC2, which correspond to global clone 1 (GC1) and GC2 (international clone [IC] I and IC II), the most important clones made up mainly of multiple-antibiotic-resistant isolates. In contrast, the Oxford scheme reveals the diversity in members of the same clone in the region of the capsule locus. The gpi gene is near one end of the capsule gene cluster, and variants that differ only in the gpi allele or double-locus variants that have different alleles for both gpi and the nearby gyrB gene are common. This variation, first noted by Hamouda et al. (3), was later traced to the existence of many recombinational replacements of this region that cause the structure of the capsular polysaccharide to differ (4).
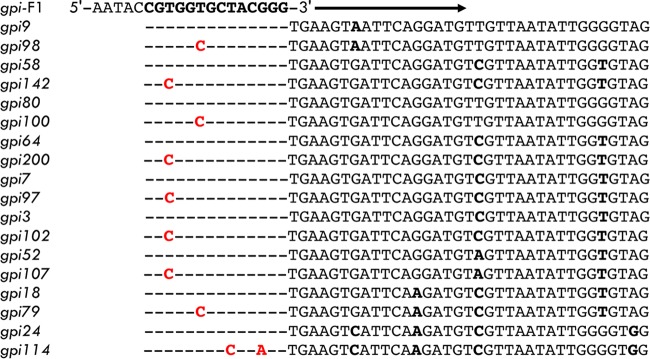
A serious problem was revealed when STs in the Oxford scheme generated from whole-genome sequence data were compared to values determined with the primers specified by Bartual et al. (1). We first encountered a discrepancy between STs determined both ways in 2011 while analyzing GC2 isolate WM99c (5). We had determined the MLST profile of a closely related isolate, A91, as ST92 (1-3-3-2-2-7-3) (6). However, the genome sequence data indicated that WM99c was ST208 (1-3-3-2-2-97-3), which differs from ST92 by a single base at one end of the gpi gene. We sequenced the gpi amplicon generated by using the specified primers, and the sequence was for allele 7, and we published the ST of WM99c as ST92, rather than ST208 (5). Subsequently, we encountered similar problems with other isolates (Table 1). This led us to reexamine this issue, and we found that part of the forward primer sequence has been included in the region used for allele determination for the gpi gene (Fig. 2). The STs of all of the strains for which we had previously published STOxs (Table 1) were reexamined by using whole-genome sequence data. In addition to ST92, which has been frequently reported for GC2 (CC2IP) isolates and is, in fact, ST208, several were affected (Table 1). ST109, a commonly reported ST for GC1 (CC1IP) isolates, was ST231. In all cases, the forward primer had a single base difference from the actual sequence (Fig. 2).
TABLE 1.
Discrepancies in Oxford MLST (gltA-gyrB-gdhB-recA-cpn60-gpi-rpoD)
| Strain(s) | GCa | ST by original PCR methodb | Reference(s) | ST determined in silico from genomesb | Reference |
|---|---|---|---|---|---|
| A1, G7, A297, WM98 | 1 | 109 (10-12-4-11-4-9-5) | 8–11 | 231 (10-12-4-11-4-98-5) | 12 |
| D36 | 1 | 247 (10-12-4-11-4-58-5) | 13–15 | 498 (10-12-4-11-4-142-5) | 12 |
| D78, D81 | 1 | 347 (10-12-4-11-4-80-5) | 12 | 441 (10-12-4-11-4-100-5) | 12 |
| A85 | 1 | 126 (10-53-4-11-4-64-5) | 13–15 | 781 (10-53-4-11-4-200-5) | 12 |
| A91, WM99c | 2 | 92 (1-3-3-2-2-7-3) | 5, 6, 16 | 208 (1-3-3-2-2-97-3) | 17 |
| RBH44 | 2 | 69 (1-46-3-2-2-58-3) | 18 | 423 (1-46-3-2-2-142-3) | This study |
| A320 | 2 | 98 (1-12-3-2-2-3-3) | 19 | 350 (1-12-3-2-2-102-3) | This study |
| D46 | 110 (1-15-2-28-1-52-32) | 9 | 229 (1-15-2-28-1-107-32) | This study | |
| RBH2 (F2) | 125 (1-52-59-12-1-18-44) | 9 | 1134 (1-52-59-12-1-79-44) | This study | |
| RCH51 | 253 (1-52-29-28-18-24-7) | 9 | 514 (1-52-29-28-18-114-7) | This study |
Global clones.
Discrepancies are in bold type.
FIG 2.
Comparison of actual sequences at the 5′ end of the gpi allele to that of the gpi forward primer, gpi-F1. The gpi-F1 primer sequence is shown above with the bases in the segment used as the gpi allele in bold. Sequences obtained with gpi-F1 are interspersed with the actual sequences with the correct base shown in red.
A broader investigation revealed that the amplification primers overlapped the region analyzed for another gene, namely, the reverse primer for the cpn60 gene. We reported these problems to the curator of the MLST database, and subsequently, the primers amplifying these two genes were changed so that they lie outside the region used for allele determination. Unfortunately, these changes left the original problem in place. Because the regions analyzed were not altered to remove the problem primer sequences, many of the alleles and many STs in the Oxford database are not real. Prior reports or current ones where labs unaware of the change have continued using the original primers yield one ST while the genome sequence and the replacement primers yield another.
In our experience, the problem arises most often in the gpi gene, where the magnitude of the problem is amplified by the fact that the gpi allele lies within the capsule biosynthesis gene cluster. When the capsule locus is replaced, which is known to be a common occurrence (4), a different gpi sequence is introduced.
Hence, though the use of CC92 and CC109 continues, ST92 and ST109 may not actually exist. It would be useful if this problem were recorded on the MLST website.
ACHNOWLEDGEMENTS
At the time this work was undertaken, S.J.N. and M.H. were supported by Australian Postgraduate Awards.
REFERENCES
- 1.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamouda A, Evans BA, Towner KJ, Amyes SG. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla (OXA-51-like) genes. J Clin Microbiol 48:2476–2483. doi: 10.1128/JCM.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nigro SJ, Farrugia DN, Paulsen IT, Hall RM. 2013. A novel family of genomic resistance islands, AbGRI2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J Antimicrob Chemother 68:554–557. doi: 10.1093/jac/dks459. [DOI] [PubMed] [Google Scholar]
- 6.Nigro SJ, Hall RM. 2012. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother 67:1342–1346. doi: 10.1093/jac/dks037. [DOI] [PubMed] [Google Scholar]
- 7.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 13:807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 8.Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamidian M, Nigro SJ, Hall RM. 2012. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother 67:2833–2836. doi: 10.1093/jac/dks318. [DOI] [PubMed] [Google Scholar]
- 10.Hamidian M, Wynn M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. Identification of a marker for two lineages within the GC1 clone of Acinetobacter baumannii. J Antimicrob Chemother 69:557–558. doi: 10.1093/jac/dkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt KE, Hamidian M, Kenyon JJ, Wynn MT, Hawkey J, Pickard D, Hall RM. 2015. Genome sequence of Acinetobacter baumannii strain A1, an early example of antibiotic-resistant global clone 1. Genome Announc 3:e00032-15. doi: 10.1128/genomeA.00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt KE, Kenyon JJ, Hamidian M, Schultz MB, Pickard D, Dougan G, Hall RM. 2016. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2:e000052. doi: 10.1099/mgen.0.000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 14.Hamidian M, Hawkey J, Holt KE, Hall RM. 2015. Genome sequence of Acinetobacter baumannii strain D36, an antibiotic-resistant isolate from lineage 2 of global clone 1. Genome Announc 3:e01478-15. doi: 10.1128/genomeA.01478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. 2014. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother 69:2625–2628. doi: 10.1093/jac/dku188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigro SJ, Post V, Hall RM. 2011. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother 66:1504–1509. doi: 10.1093/jac/dkr163. [DOI] [PubMed] [Google Scholar]
- 17.Nigro SJ, Hall RM. 2016. Loss and gain of aminoglycoside resistance in global clone 2 Acinetobacter baumannii in Australia via modification of genomic resistance islands and acquisition of plasmids. J Antimicrob Chemother 71:2432–2440. doi: 10.1093/jac/dkw176. [DOI] [PubMed] [Google Scholar]
- 18.Hamidian M, Hall RM. 2013. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 68:2682–2683. doi: 10.1093/jac/dkt233. [DOI] [PubMed] [Google Scholar]
- 19.Nigro SJ, Hall RM. 2012. Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J Antimicrob Chemother 67:335–338. doi: 10.1093/jac/dkr447. [DOI] [PubMed] [Google Scholar]