Abstract
The human genome harbors many distinct families of human endogenous retroviruses (HERVs) that stem from exogenous retroviruses that infected the germ line millions of years ago. Many HERV families remain to be investigated. We report in the present study the detailed characterization of the HERV-K14I and HERV-K14CI families as they are represented in the human genome. Most of the 68 HERV-K14I and 23 HERV-K14CI proviruses are severely mutated, frequently displaying uniform deletions of retroviral genes and long terminal repeats (LTRs). Both HERV families entered the germ line ∼39 million years ago, as evidenced by homologous sequences in hominoids and Old World primates and calculation of evolutionary ages based on a molecular clock. Proviruses of both families were formed during a brief period. A majority of HERV-K14CI proviruses on the Y chromosome mimic a higher evolutionary age, showing that LTR-LTR divergence data can indicate false ages. Fully translatable consensus sequences encoding major retroviral proteins were generated. Most HERV-K14I loci lack an env gene and are structurally reminiscent of LTR retrotransposons. A minority of HERV-K14I variants display an env gene. HERV-K14I proviruses are associated with three distinct LTR families, while HERV-K14CI is associated with a single LTR family. Hybrid proviruses consisting of HERV-K14I and HERV-W sequences that appear to have produced provirus progeny in the genome were detected. Several HERV-K14I proviruses harbor TRPC6 mRNA portions, exemplifying mobilization of cellular transcripts by HERVs. Our analysis contributes essential information on two more HERV families and on the biology of HERV sequences in general.
About 8% of the human genome mass consists of sequences of retroviral origin (16). It is thought that in the evolutionary past, exogenous retroviruses formed proviruses in the genomes of germ cells of ancestral primate species. Some of the proviruses were fixed in the population and were inherited as stable genomic components, so-called endogenous retroviruses (ERVs), through generations and the evolution of species. Over millions of years, proviral copy numbers could increase up to several thousand by intragenomic spread of retroviral genomes. In this case, single proviruses produced further proviral copies in the genome by a process probably very similar to retroviral replication but likely excluding an extracellular phase. Multiple clearly distinguishable human ERV (HERV) families indicate repeated germ line infection by a variety of exogenous retroviruses in the evolutionary past. Disagreeing with that view, Belshaw et al. (3) recently suggested that the proliferation of particular HERV families was due to long-term reinfection that involved a persistent infectious pool of endogenous retroviruses throughout primate evolution.
Between 30 and 50 HERV families were recently suggested to reside in the human genome (13, 26, 39), among them various so-called HERV-K families. The letter K indicates that a primer binding site specific for lysine-tRNA was used to prime reverse transcription. In total, 10 HERV-K families have been defined based on sequence similarities, and they have been named HERV-K(HML-1) to HERV-K(HML-10) (for human MMTV [mouse mammary tumor virus]-like) because of some sequence relationship to the mouse mammary tumor virus (1, 31). Repbase, a reference sequence database for repetitive elements (18), employs a slightly different nomenclature for these HERV-K families. Usually, HERVs acquired numerous deleterious mutations over time and became unable to encode retroviral proteins. Although initial germ line fixation occurred ∼30 million years ago, the HERV-K(HML-2) family, also called HERV-K or HTDV/HERV-K, still encodes all major retroviral proteins, Gag, protease (Pro), polymerase (Pol), and envelope (Env), and an HIVREV functional homologue, called Rec or cORF (2, 4, 21, 27, 28, 30, 40). The presence of Gag and Env antibodies in germ cell tumor patients and the physical interaction of Rec with a protein involved in mouse spermatogenesis suggest that HERV-K(HML-2) is involved in the development of germ cell tumors (6, 7, 36, 37). An Env protein encoded by a HERV-W provirus possibly performs an important function in the formation of the human placenta (5, 24, 33).
A large number of HERV families in the human genome remain to be characterized in more detail. While most HERV families may no longer encode retroviral proteins with biological significance, the study of HERV families provides important information, for instance, about the evolution of ERVs after they entered a host genome and about retroviruses that targeted primates millions of years ago (26, 39). The availability of the almost complete human genome sequence is a major advantage for studying HERV families thoroughly. We and others recently characterized the behavior during evolution, evolutionary ages, provirus structures, etc., of various HERV families. The various proviruses of the HERV-W family were shown to consist of three subfamilies that formed in the genome within a short period ∼22 million years ago (mya) (9). The HERV-K(HML-3) family colonized the host genome within a brief period ∼36 mya. Provirus variants carrying uniform deletions within the gag and pol gene regions were amplified to higher numbers during this time (25). In contrast, several studies have revealed that proviruses belonging to the HERV-K(HML-2) family remained active during the evolution of the human lineage, eventually generating a number of human-specific HERV-K(HML-2) loci (2, 8, 32). The HERV-K(HML-5) family was shown to be considerably older than other HERV-K families. The ∼139 proviruses present in the human genome were formed ∼55 million years ago, before the evolutionary split of New World and Old World primates. Also, HERV-K(HML-5) should be designated HERV-M, as the primer binding site is identical to methionine-tRNA (22). As for HERV-K(HML-3), numerous HERV-K(HML-5) proviruses displayed uniform larger deletions within the proviral gag-pro and/or env gene region. Furthermore, monophyletic HERV-K(HML-5) proviral bodies were associated with clearly distinguishable long terminal repeat (LTR) sequences, implying different evolutions of the two proviral regions (22). For both the HERV-K(HML-3) and HERV-K(HML-5) families, we were able to deduce the presumptive former exogenous retroviral sequences from consensus sequences generated from multiple alignments of proviral DNA sequences (22, 25). Thus, studying HERV sequences can significantly increase our knowledge of ancient exogenous retroviruses and the evolution of retroviruses, for instance, the acquisition of protein domains (29).
In the present study, we characterize two further HERV-K families, called HERV-K14I and HERV-K14CI, based on designations in Repbase. The HERV-K14I family has also been named HERV-K(HML-1). We characterized both families in regard to, for example, provirus structures, evolution, and the time periods of provirus formation. We generated fully translatable consensus sequences. In addition, we revealed the mobilization of nonretroviral mRNA portions and hybrid proviruses consisting of two different HERV families for one of the two HERV-K14 families. Our study further reveals characteristics of HERV sequences that appear more and more as variations of a common theme, as well as new characteristics that appear to be specific to the HERV-K14 families investigated.
MATERIALS AND METHODS
The methods in the present work essentially follow strategies established in two previous studies (22, 25). In brief, we collected HERV-K14 sequences from the human genome sequence by doing BLAT searches at the Human Genome Browser (version June 2002) (20) using the relevant HERV-K14 Repbase (version 7.1.1) reference sequences as probes. We retrieved sequences of identified loci, including flanking sequence portions. We identified HERV-K14 sequence portions by Pustell's dot matrix comparisons (35), as implemented in MacVector (Accelrys Inc.), and Repeatmasker analysis. Multiple alignments of proviral sequences were generated by employing DIALIGN2 (34) with standard parameters and were subsequently optimized using Se-Al (provided by Andrew Rambaut [http://evolve.zoo.ox.ac.uk/]). We developed a Perl script to graphically visualize the finished multiple alignments. Basically, the Perl script translates nucleotide-harboring positions in a multiple alignment into dots, thus indirectly translating gaps into blank space. The Perl script is available from the authors on request. The phylogeny of proviral sequences was analyzed using PAUP* version 4.0 with previously described parameters. Proviral ages were estimated based on LTR-LTR divergences and nucleotide divergences from consensus sequences (10). The presence of HERV-K14-homologous sequences in genomic DNA samples from various primate species was investigated by PCR. For amplification of a HERV-K14I gag gene region, we employed PCR primers K14IF (5′-GGACGCATCGTCAAAGGACA-3′) and K14IR (5′-AAATTGCCATGCTTCAAGGTCT-3′). The PCR cycles were as follows: 5 min at 94°C; 34 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C; and 5 min at 72°C. For amplification of a HERV-K14CI gag-pro gene region, we used PCR primers K14CIF (5′-GGCTGAAAGTAAGTTTGCTAAT-3′) and K14CIR (5′-ATTCCCTCACTGTAATTGGCT-3′). The PCR cycles were as follows: 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 59°C, and 45 s at 72°C; and 5 min at 72°C. The PCR products were cloned into the pGEM-T Easy vector (Promega) and were sequenced on a Licor 4000-L automated DNA sequencer.
Nucleotide sequence accession numbers.
The HERV-K14CI consensus sequence (as well as the HERV-K14I consensus sequence) generated in this study was deposited in Repbase.
RESULTS
Mining for HERV-K14 sequences.
To identify HERV-K14 loci in the human genome, we probed the human genome sequence at the Human Genome Browser (20) with the HERV-K14I reference sequence as reported in Repbase. We obtained 68 matches with scores ranging from 5,364 to 297 and match lengths ranging from 5,945 to 336 bp. Matches displayed between 95.6 and 88.1% (mean, 92% ± 1.86%) similarity to the HERV-K14I Repbase sequence. The results of BLAT searches are summarized in Table SA in the supplemental material. Pustell matrix comparisons with HERV-K14I revealed that only six proviruses were intact in structure. Longer deletions in gag, pro, and pol were identified in 47, 40, and 60 proviruses, respectively. Some HERV-K14I loci were represented by only single proviral gene relics: solitary gag, pro, and pol gene remnants were identified 5, 1, and 12 times, respectively. The characteristics of HERV-K14I env gene regions will be addressed below.
HERV-K14CI loci were identified in a similar fashion using the HERV-K14CI reference sequence given in Repbase. Among the 52 matches retrieved by BLAT search, scores ranged from 7,203 to 50 and match lengths were from 7,417 to 277 bp, with similarities ranging from 98.6 to 68.7%. Only three structurally intact proviruses were identified in matrix comparisons. The results of HERV-K14CI BLAT searches are summarized in Table SB in the supplemental material. Further inspection of the loci revealed that a considerable fraction of matches did not represent HERV-K14CI sequences. Matrix comparisons and RepeatMasker analysis confirmed that 29 sequences represented members of other HERV-K families, such as HERV-K(HML-2), HERV-KC4, HERV-K9, HERV-K14I, and HERV-K11D. Very likely, those HERV sequences were detected because of partial sequence similarities of the HERV-K14CI reference sequence to those other HERV families. We conclude that there are 23 HERV-K14CI proviruses, or remains of them, in the human genome.
Chromosomal distribution of HERV-K14 loci.
The various HERV-K14I loci appeared to be randomly distributed along the human chromosomes. The number of matches was proportional to the chromosome size. However, there was an obvious increase in loci on human chromosome 19, as it harbors six loci when a theoretical value of only 1.5 matches was expected.
The chromosomal distribution of HERV-K14CI loci was different. Out of 23 loci, 12 were located on the Y chromosome. Considering the size of the Y chromosome, a theoretical value of only 0.41 proviruses was expected for it. This finding indicates that in the evolutionary past, either HERV-K14CI proviruses were preferentially formed on the Y chromosome or HERV-K14CI proviruses were preferentially deleted from chromosomes other than the Y chromosome. We favor another explanation. The human Y chromosome is known to have undergone a number of intrachromosomal duplication events, forming so-called ampliconic regions (38). When we mapped the various HERV-K14CI loci on the Y chromosome, we found that proviruses were located within previously described intrachromosomally duplicated regions (38) (see Fig. SD in the supplemental material). Although, only two provirus pairs (17CI-23CI and 21CIa-22CI [see Table SB in the supplemental material]) displayed similar flanking sequences. The sequences flanking the remaining proviruses were dissimilar, suggesting that in those cases, only the proviruses themselves were amplified in copy numbers. Nevertheless, intrachromosomal duplication events on the Y chromosome may also have acted on HERV-K14CI proviruses, amplifying their copy numbers as well. More detailed analysis will be required to reveal the dynamics of HERV-K14CI sequences on the Y chromosome.
HERV-K14 provirus structures and consensus sequences.
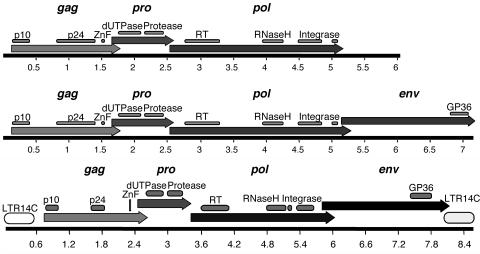
We included 66 out of 68 HERV-K14I sequences in a multiple alignment totaling 189,323 bp. We subsequently generated a consensus sequence from the multiple alignment. The complete alignment has been graphically visualized in Fig. SA in the supplemental material. An annotated HERV-K14I consensus sequence can be found in Fig. SE in the supplemental material. The HERV-K14I consensus sequence generated in the present study was 6,079 bp in length, 133 bp longer than the consensus sequence in Repbase and 3% different from that sequence. The new consensus sequence displayed open reading frames (ORFs) for the major retroviral proteins Gag, Pro, and Pol, including expected protein motifs (Fig. 1, top, and 2) There was no evidence of an env gene in the ∼896-bp sequence downstream from the pol gene. No Env similarities were found for that region when we subjected the sequence to DNA searches and translated BLAST searches. We conclude that the HERV-K14I variant, comprising the majority of HERV-K14I loci in the human genome, is entirely lacking an env gene and harbors ∼900 bp of a sequence unrelated to env downstream from the pol gene.
FIG. 1.
Maps of HERV-K14 ORFs as generated from fully translatable consensus sequences. (Top) HERV-K14I ORF maps. The variant without env is depicted in the upper part, while the variant with a putative env gene is shown in the lower part. (Bottom) HERV-K14CI ORF map. The locations of retroviral gag, pro, pol, and env reading frames are shown, as well as the locations of typical protein domains. LTR14C, associated with HERV-K14CI, is included in the bottom panel. LTRs were omitted in the top panel, as HERV-K14I is associated with three distinct LTR families (see the text). The scales are in kilobases.
FIG. 2.
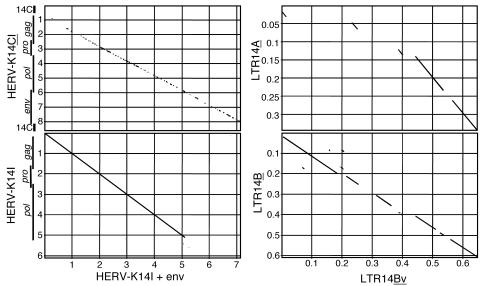
Dot matrix comparison of HERV-K14I and associated LTR variants with different env gene regions. (Left) Comparison of an HERV-K14I provirus with a putative env gene with the HERV-K14I reference sequence (lacking LTRs) in Repbase (bottom) and with the HERV-K14CI reference sequence (flanked by LTR14C) (top). The locations of proviral LTRs and genes are indicated. (Right) Comparison of variant LTR14Bv sequences that flank HERV-K14I env+ proviruses with the LTR14B (bottom) and LTR14A (top) reference sequences. Note that the LTR14C sequence is much less similar to the three LTR sequences shown. The dot matrix comparisons were generated with MacVector, employing a window size of 30 nt and a minimum similarity of 60%. The scales are in kilobases.
However, we detected seven HERV-K14I variants, including relics (proviruses 10, 11, 14, 17, 21, 22, and 29 [see Table SA in the supplemental material]), in the human genome sequence that were considerably longer. The consensus sequence of these proviral elements was 7,121 bp. Notably, while the gag, pro, and pol sequences were very similar to the sequence of the short HERV-K14I variant, only the sequence region downstream from the pol gene contributed to the length difference. DNA sequence comparisons revealed that the region downstream from pol displayed no sequence similarities with the non-env region in the short HERV-K14I variant. Rather, the sequence was more similar to the 3′ sequence portion—the putative env gene (see below)—in the HERV-K14CI consensus sequence (Fig. 2). More importantly, the “long” HERV-K14I 3′ region was significantly similar to those of other retroviral Env proteins in translated BLAST searches, including a GP36 motif typical of Env. Hence, a minority of HERV-K14I proviruses in the human genome harbor a putative env gene and therefore appear more intact than the majority of short HERV-K14I proviruses lacking env.
HERV-K14I proviruses with putative env genes also display LTR sequences that characteristically differed from both the LTR14A and the LTR14B sequences usually associated with HERV-K14I proviruses. The overall similarity of that LTR14 variant to LTR14B was higher than to LTR14A. We therefore named these variants LTR14Bv (for variant) (Fig. 2). The LTR14Bv sequence contributes an additional 14 amino acids to the env reading frame.
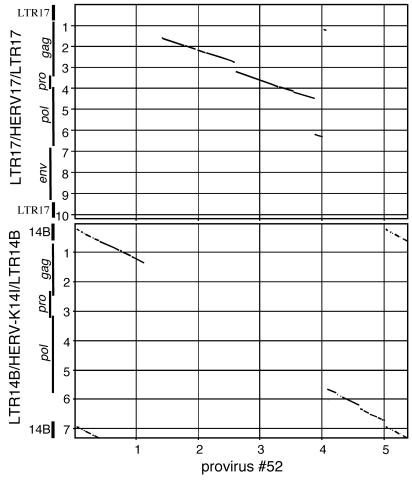
We also detected, in total, seven HERV-K14I proviruses in the human genome, belonging to the variant lacking env, that uniformly harbored HERV17 (also called HERV-W) sequences. In the HERV-K14I loci in particular, ∼2.8 kb of the central HERV-K14I proviral portions was replaced by ∼2.7 kb of HERV-W sequences. As for the missing HERV-K14I portions, the HERV-W sequence portions corresponded to presumptive gag, pro, and pol gene regions in the HERV-W proviral sequence. The HERV-K14I-HERV-W fusion proviruses were flanked by LTR14B. Thus, the proviral fusion structures were ∼5 kb in total length and consisted of ∼2.3 kb of HERV-K14I sequences and ∼2.7 kb of HERV-W sequences (Fig. 3).
FIG. 3.
Dot matrix comparison exemplifying a hybrid provirus between HERV-K14I and HERV17 (HERV-W) that was identified multiple times in the human genome. Provirus number 52 (see Table SA in the supplemental material) is compared to a sequence consisting of HERV-K14I (lacking env; see the text) flanked by LTR14B (bottom) and to a sequence consisting of the Repbase HERV17 sequence flanked by LTR17 (top). The locations of proviral LTRs and genes are indicated. The locations of proviral genes in HERV-17 were estimated from TBLASTN database search results. Parameters of comparisons are given in Fig. 2. The scales are in kilobases.
For HERV-K14CI, we aligned 23 proviral loci totaling 135,388 bp. The multiple alignment is depicted in Fig. SB in the supplemental material. The consensus sequence generated from the alignment was 7,436 bp in length, while the previously reported sequence in Repbase was 7,417 bp long. An annotated HERV-K14CI consensus sequence is depicted in Fig. SF in the supplemental material. The new HERV-K14CI consensus sequence displayed overlapping ORFs for the Gag, Pro, Pol, and Env proteins, including characteristic protein domains. The env ORF reached 44 amino acids into the 3′ LTR (Fig. 1, bottom). In total, 9 HERV-K14CI proviral loci uniformly displayed a larger deletion of ∼2.75 kb within the presumptive pol gene region. The multiple sequence alignments for HERV-K14I and HERV-K14CI are available from the authors on request.
HERV-K14-associated LTRs and evolutionary ages.
HERV-K14I proviruses were flanked by either LTR14A or LTR14B sequences. Out of 66 loci examined, 25 were flanked by LTR14A. Among those, eight displayed both LTRs. Another nine and eight loci displayed only 5′ or 3′ LTRs, respectively. Thirty-nine proviruses were flanked by LTR14B, with 18 loci displaying both LTRs and 12 and 9 loci displaying only the 5′ or the 3′ LTR, respectively. One provirus was flanked by LTR14B. There, the 3′ LTR14B displayed an inserted LTR14A element. This particular provirus structure may be due to secondary provirus insertion and mutation events. Probably, a HERV-K14I provirus flanked by LTR14A inserted into the preexisting HERV-K14I provirus flanked by LTR14B. Homologous recombination then reduced the latter provirus to a solitary LTR.
We calculated the nucleotide divergence between 5′ and 3′ LTR sequences for proviruses with both LTRs (10), excluding gaps and CpG dinucleotides from the analysis. LTR14A sequences, on average, diverged from each other by 10.12% (±2.17%). LTR14B sequences yielded a mean divergence of 10.28% (±3.25%). Taking previously established considerations into account (10, 23), those divergences correspond to evolutionary ages of 38.92 (±8.3) and 39.54 (±12.5) million years for the examined HERV-K14I proviruses that were flanked by LTR14A or LTR14B sequences, respectively. There was no significant difference between the values. Thus, the HERV-K14I proviruses were formed in the germ line ∼39 mya. Similar data were obtained when proviral ages were estimated according to nucleotide divergences from the consensus sequence (19, 22). We obtained a mean value of 6.66% (±2.743%) divergence from the consensus, corresponding to proviral ages of ∼41.6 (±17.2) million years. The calculated age is also supported by the presence of Alu-Sp elements in some HERV-K14I proviruses. The Alu-Sp family is known to have expanded in the genome ∼37 million years ago (19).
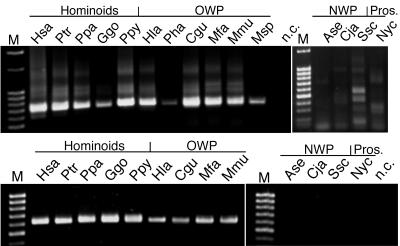
Furthermore, we performed PCR experiments to examine the presence of HERV-K14I-homologous sequences in various primate species. Specific PCR products were obtained only from hominoids and Old World primates, not from New World primates and prosimians (Fig. 4, top). Old World and New World primates separated from each other evolutionarily ∼40 mya (14). PCR products amplified from prosimians were determined to be nonretroviral and therefore nonspecific after being sequenced. Thus, the PCR results coincide very well with the evolutionary ages calculated from LTR divergence data. HERV-K14I appears to have entered the primate genome ∼39 mya, after the evolutionary separation of Old World from New World primates.
FIG. 4.
Presence of HERV-K14-homologous sequences in various primate species, as revealed by PCR. (Top) Presence of HERV-K14I homologues in hominoids and Old World primates but not in New World primates and prosimians. Specific PCR products of ∼740 bp were amplified from positive species. PCR products in New World primates and prosimians proved to be nonretroviral after being sequenced. The lower PCR yields in lanes Pha and Msp are probably due to suboptimal genomic DNA quality for those species. (Bottom) Similar species distributions of HERV-K14CI homologues, as revealed by a specific PCR product of ∼620 bp. Species abbreviations are as follows. Hominoidea: Hsa (Homo sapiens), Ptr (Pan troglodytes), Ppa (Pan paniscus), Ggo (Gorilla gorilla), and Ppy (Pongo pygmaeus). Old World primates (OWP): Hla (Hylobates lar), Pha (Papio hamadryas), Cgu (Colobus guereza), Mfa (Macaca fascicularis), Mmu (Macaca mulatta), Msp (Mandrillus sphinx). New World primates (NWP): Ase (Alouatta seniculus), Cja (Callithrix jacchus), Ssc (Saimiri sciureus). Prosimian (Pros.): Nyc (Nycticebus coucang).
HERV-K14CI proviruses were found to be exclusively associated with LTR14C sequences. Out of 23 proviruses examined, 14 displayed both LTRs. The average sequence divergence between 5′ and 3′ LTR14C sequences was determined to be 14.07% (±3.95%), corresponding to a proviral age of ∼54 (±15.2) million years. While the age calculated from LTR divergences indicated the presence of HERV-K14CI in both Old World and New World primates, PCR analysis of various primate species failed to detect HERV-K14CI-homologous sequences in New World primates (Fig. 4, bottom). When we investigated the LTR divergence data in more detail, we found a significant discrepancy between LTR divergences of proviral loci on the Y chromosome and other chromosomes. Proviruses located on the Y chromosome displayed a mean LTR divergence of 16.75% (±2.31%), corresponding to an evolutionary age of ∼64 (±8.9) million years. Proviruses on other chromosomes diverged, on average, 10.5% (±2.51%), equaling 40.4 (±9.7) million years.
Considering HERV-K14CI species distribution as determined by PCR, LTR divergences for proviruses located on the Y chromosome provide inconsistent ages, as opposed to LTR divergences for non-Y proviruses. Moreover, nucleotide divergences of the various proviral sequences from the HERV-K14CI consensus sequence yielded evolutionary ages of ∼45.1 (±15.8) million years. There was no significant difference for proviral sequences on the Y chromosome. We therefore conclude that LTR divergence data obtained from proviruses on the Y chromosome are inaccurate and thus misleading in regard to the evolutionary ages of HERV-K14CI proviruses. We suggest that HERV-K14CI proviruses first entered the primate lineage ∼40 million years ago, after the evolutionary separation of Old World from New World primates.
HERV-K14 phylogenies.
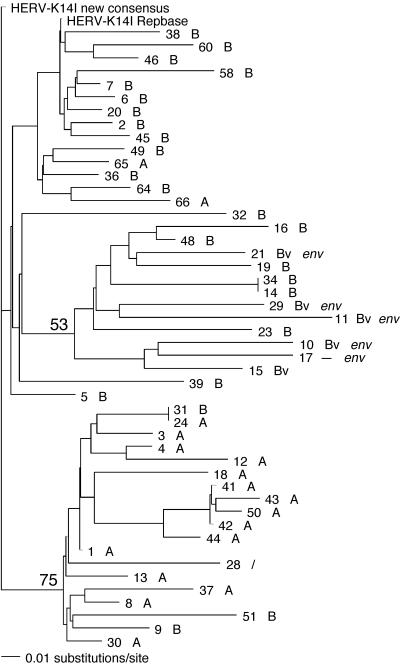
We analyzed the phylogenetic relationships of HERV-K14I proviruses for a total of seven different proviral regions. The HERV-K14I proviruses tended to form two subgroups. When a proviral region ranging from nucleotides (nt) 1 to 693 relative to the HERV-K14I consensus sequence (generated in this study) was analyzed, the first subgroup displayed 75% bootstrap support while the second subgroup displayed only weak bootstrap support of 53%. Furthermore, the first subgroup displayed 94% bootstrap support for a proviral region ranging from nt 2962 to 3973 in the HERV-K14I consensus sequence. The majority of sequences (15 of 19) in the first subgroup consisted of proviruses that were flanked by LTR14A sequences. A minority (3 of 19) were flanked by LTR14B. One provirus lacked LTRs. The proviruses in the second subgroup, as well as other proviruses not belonging to either subgroup, were flanked by LTR14B or LTR14Bv. Thus, proviruses with putative env gene regions also grouped in the second subgroup (because those proviruses are flanked by LTR14Bv; see above) (Fig. 5). However, it must be considered that the second subgroup is only weakly supported. Therefore, only the first subgroup should be considered a valid subgroup. In the various phylogenetic trees, branch lengths in the different proviral sequences were very similar. Thus, the HERV-K14I proviral sequences accumulated similar numbers of nucleotide differences over time, eventually indicating similar evolutionary ages for the proviruses. This finding is also supported by very similar nucleotide distances of proviral sequences from the consensus sequence, which likewise did not reveal significant subgroups.
FIG. 5.
Neighbor-joining analysis of HERV-K14I sequences for the proviral intergenic and gag gene regions. The provirus numbers correspond to the numbers in Table SA in the supplemental material. When possible, each provirus is followed by information regarding the LTR14 family flanking the provirus (LTR14A [A], LTR14B [B], or LTR14Bv [Bv]) and the presence of a true env gene. Note that a subgroup of HERV-K14I sequences, supported by a bootstrap value of 75% in 1,000 replicates, is almost exclusively flanked by LTR14A. Consensus sequences, as generated in this study and as given in Repbase, were also included in the analysis.
Phylogenetic analysis of HERV-K14CI proviral sequences was performed for two different proviral regions, one located in the HERV-K14CI gag gene region and the other located in the env gene region. Each region included 20 proviral sequences. For both regions, the various proviral sequences did not form subgroups that were supported by bootstrap values (data not shown). Therefore, the HERV-K14CI sequences investigated in this study comprise a monophyletic group. Branch lengths were very similar, again indicating similar evolutionary ages of HERV-K14CI proviruses. According to the HERV-K14CI reference sequence information in Repbase, that reference is derived from one provirus. Our phylogenetic analysis revealed that the HERV-K14CI consensus sequence in Repbase is identical to the sequence of provirus number 1CI identified in the present study. Thus, that provirus sequence served as the reference sequence for the HERV-K14CI family. The HERV-K14CI consensus sequence generated in the present study is more similar to the various HERV-K14CI proviral sequences than is the less representative HERV-K14CI reference sequence in Repbase. The HERV-K14CI consensus sequence (as well as the HERV-K14I consensus sequence) generated in this study was deposited in Repbase.
TRPC6 mRNA sequences in some HERV-K14I proviruses.
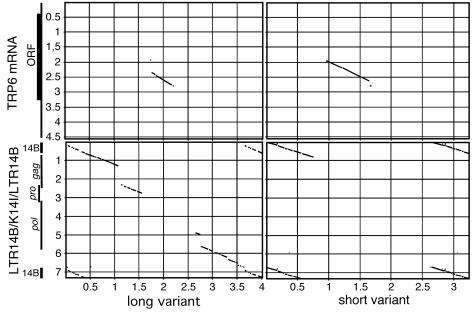
Non-HERV-K14I-related insertions ∼1.8 or 1.1 kb in length were detected in a total of 13 HERV-K14I loci. The proviruses were flanked by LTR14B and displayed larger mutations within the gag, pro, and pol gene regions. The insertion sequences were uniformly located at nt 2958 relative to the HERV-K14I consensus sequence. We found that the 5′ portions of the insertion sequences were similar to sequence portions of the transient receptor potential channel 6 (TRPC6) mRNA (GenBank accession number AJ006276) (11). About 900 bp of insertion sequence was similar to a central protein-coding portion of the TRPC6 mRNA. Two proviral variants with sequences similar to TRPC6 mRNA were detected that differed both in the TRPC6 mRNA sequence and the HERV-K14I proviral sequence portions. One longer variant lacked TRPC6 mRNA sequence portions and displayed additional HERV-K14I sequence portions compared to the other, shorter variant. Seven proviral loci were identified for the first, long variant, and five proviral loci were identified for the second, short variant (Fig. 6). An alignment of all HERV-K14I proviruses harboring TRPC6 mRNA portions can be found in Fig. SC in the supplemental material. The remaining non-HERV-K14I portions upstream from the region similar to TRPC6 were annotated by Repeatmasker as ArturI and as stretches of simple repeats. The sequence portions downstream from the TRPC6-homologous region were annotated as HERV-K14CI, HERV-KC4, or HERV-K3.
FIG. 6.
Dot matrix comparisons of HERV-K14I proviruses associated with TRPC6 mRNA portions. Two proviruses in which the sizes of HERV-K14I and TRPC6 sequence portions differed are depicted. The long variant (left) contains additional HERV-K14I and fewer TRPC6 portions than the short variant (right). Each variant is compared to a sequence consisting of HERV-K14I flanked by LTR14B (bottom) and to a recently reported full-length TRPC6 mRNA (11) (top). The locations of proviral LTRs and genes, as well as the open reading frame within the TRPC6 mRNA, are indicated. The parameters of comparisons are identical to the ones given for Fig. 2. The scales are in kilobases.
Thus, a number of HERV-K14I proviruses in the human genome harbor portions from the TRPC6 mRNA within their proviral bodies. Very likely, a single provirus once acquired TRPC6 mRNA sequence portions and then formed new proviral copies in other genomic locations. It is not clear whether the two observed variants are due to subsequent mutational events or to two independent recombination events between HERV-K14I proviruses and the TRPC6 mRNA. Also, the possible involvement of non-HERV-K14I sequences flanking the TRPC6 sequence portion in the creation of the particular structures is unknown. In any case, HERV-K14I proviruses once acquired nonretroviral cellular mRNA sequence portions and subsequently shuffled them to new genomic locations.
DISCUSSION
We characterize in the present study two so far little investigated HERV families. It should be stressed that although the family designations HERV-K14I and HERV-K14CI, as introduced by Repbase, imply relatively high similarity between the two HERV families, pairwise sequence comparison shows that the two families are not much more closely related to each other than to other HERV-K families (Fig. 2).
While BLAT search results for HERV-K14I identified “true” HERV-K14I proviruses or relics, closer examination of HERV-K14CI BLAT search matches showed that the majority of matches (29 out of 52) were detected only because of partial sequence similarities between the probe sequence and other non-HERV-K14CI proviruses in the human genome. Thus, simply taking the quantitative results of similarity searches as the actual ERV copy number in the human genome, or another genome, might produce inaccurate results. Closer examination of the HERV family assignments of detected proviruses should be performed.
As previously seen for other HERV-K families (22, 25), the majority of HERV-K14I and HERV-K14CI proviruses appear to be severely mutated; 5′ or 3′ portions were missing, and numerous HERV-K14I family loci displayed large and uniformly deleted regions within the gag, pro, and pol genes. About half of the HERV-K14CI loci displayed deleted pro-pol or pol gene regions. Again, coding-deficient proviruses formed at one point in the genome and were amplified in copy number. Such structural features of HERV-K14 proviruses appear as variations of an obviously common theme among HERVs, and potentially ERVs in other species. We previously discussed possible mechanisms by which these proviral deletion variants were generated, e.g., provirus formation from spliced transcripts or deletion events on the genomic level (22, 25). Despite the relatively high level of mutations, we were able to generate completely translatable consensus sequences for both HERV-K14 families, displaying all major retroviral proteins and protein domains. Those ancestral retroviral protein sequences, which can be expected to be closely related to the former exogenous retroviral sequence, may serve to study the evolution of retroviruses in more detail.
The proviruses of the two HERV-K14 families both appear to have been fixed in the germ line genome ∼39 mya. We did not find evidence, either from LTR divergence data, divergence from the consensus, or phylogenetic analysis, that HERV-K14I proviruses were formed in the genome in much more recent time periods. Both families appear to have been amplified in copy numbers in the human genome in a relatively brief period. The HERV-K(HML-2) family remains the only HERV family that has formed proviruses up to recent human evolution (see the introduction).
Our study also shows that age calculations for HERV proviruses based on LTR divergence data (10) can produce biased and thus inaccurate results. The majority of HERV-K14CI proviruses (12 out of 23) were present on the human Y chromosome. As discussed above, we believe that the higher number is due to passive provirus amplification in the course of intrachromosomal duplications (38) rather than selective provirus formation on the Y chromosome or selective provirus deletions on other chromosomes. While proviruses on non-Y chromosomes yielded mean evolutionary ages of ∼40 million years, proviruses on the Y chromosome yielded mean ages of ∼64 million years. The latter number was not supported by divergence values of both Y and non-Y proviruses from the consensus sequence or by the primate species distribution of HERV-K14CI homologues. Thus, HERV-K14CI proviruses on the Y chromosome appear older than they actually are. A possible explanation for the discrepancy could be that (at least) one provirus on the Y chromosome accidentally accumulated more mutations than expected from a molecular clock between the 5′ and the 3′ LTRs. Such an erroneously old-appearing provirus could then have been amplified in copy number on the Y chromosome in the course of intrachromosomal duplications, thus mimicking a much higher evolutionary age for the entire family.
The various HERV-K14CI proviruses were uniformly associated with LTR14C sequences. Similar to previous findings for HERV-K(HML-5) (22), the HERV-K14I proviruses were associated with two clearly distinguishable LTR families, namely, LTR14A and LTR14B. In addition, proviral variants with putative env genes were associated with LTR14Bv sequences that are similar to, yet clearly different from, LTR14B. While monophyletic HERV-K(HML-5) proviral bodies were associated with distinct LTR sequences (22), there was some indication from phylogenetic analysis for a subgroup of HERV-K14I proviruses that were predominantly associated with LTR14A. Thus, association with different LTR sequences is also reflected to some extent in the proviral body sequences. However, the proviral body sequences differ less dramatically from each other than the LTR sequences. The LTRs appear to have undergone many more sequence changes than the proviral bodies. To the best of our knowledge, there is presently no explanation for the significantly different sequence alterations in the LTRs versus the proviral bodies.
We previously revealed frequent deletion of proviral env gene regions for the HERV-K(HML-3) and HML-5 families (22, 25) that potentially contributed to the fading of exogenous retroviral counterparts in the evolutionary past. Again, HERV-K14I proviruses present mutations within the env gene region. However, these mutations are more remarkable than env gene deletions in HML-3 and HML-5 sequences. The majority of HERV-K14I proviruses in the human genome display ∼800 bp of non-env-related sequence downstream from the pol gene. No env similarities were found for that region when different analysis approaches were employed. In contrast, a minority of only six HERV-K14I proviruses displayed a significantly longer sequence of ∼1.8 kb downstream from the pol gene that was somewhat similar in sequence to the HERV-K14CI env gene region and that displayed unambiguous similarities to other retroviral Env proteins. The remaining proviral gag, pro, and pol regions were identical in sequence to the HERV-K14I proviruses lacking env. Thus, a minority of HERV-K14I proviruses in the human genome display a provirus structure that resembles proviruses with “true” retroviral origins. The majority of HERV-K14I proviruses in the human genome, however, structurally rather resemble LTR retrotransposons (lacking env). Based on our findings we suggest that proviruses with putative env genes are ancestral to the proviruses lacking env and that the latter group of proviruses arose by replacement of the putative env gene region by a sequence unrelated to env. The mutational events also included sequence alterations in the LTRs. HERV-K14I proviruses lacking env were found to be associated with the LTR14B, while proviruses with putative env were associated with the variant LTR14Bv defined in this study. Again, the provirus variants lacking env amplified to much higher copy numbers in the host genome.
Our study also identified seven proviruses in the human genome that apparently represent hybrids between HERV-K14I and HERV-W (called HERV17 in Repbase) sequences. An ∼2.7-kb-long HERV-W portion derived from the presumptive HERV-W gag, pro, and pol gene region uniformly replaced ∼2.3 kb of HERV-K14I sequence, also representing gag, pro, and pol gene regions. Potentially, a recombination event—either on the RNA level during reverse transcription of retroviral RNAs or on the genomic-DNA level—resulted in the identified hybrid structure. It is not clear whether the fusion proviruses once encoded, at least to some extent, functional proteins. In any case, for the flanking cellular sequences of the seven hybrid proviruses that were dissimilar in sequence, the initial hybrid provirus was obviously able to form provirus progeny in the genome, eventually requiring transcriptional activity. That is, the fusion proviruses were functional to such an extent that a proviral transcript could be transcribed that subsequently was able to form a new provirus in a different genomic location. The identification of such a fusion provirus provides further evidence that potentially functional new retroviral variants could have been created in the human evolutionary lineage via recombination events.
Finally, our study revealed that HERV-K14I proviruses moved sequence portions from a cellular mRNA derived from the TRPC6 gene to a total of 14 positions in the human genome. TRP proteins provide localized Ca2+ increases for spatially defined signal transduction processes (12). TRPC6 has been suggested to form a store-independent calcium entry channel (15). Two different hybrid structures between HERV-K14I and TRPC6 mRNA were detected that differed in the HERV-K14I and TRPC6 sequence portions. In addition, stretches of non-HERV-K14I sequence portions were present downstream from the TRPC6 portion. It is not clear whether the shorter hybrid proviruses represent a deletion and thus a secondary variant of the longer hybrid proviruses. In any case, it appears that one (or two independent) recombination event resulted in the recombination detected between TRPC6 mRNA and HERV-K14I sequences. Such recombined proviruses were subsequently amplified in copy number in the host genome. It can be concluded that, in order to produce provirus progeny, one or several HERV-K14I-TRPC6 hybrid proviruses were transcriptionally active at that time in evolution. Whether the expression of the TRPC6 mRNA portion on the RNA or potentially even the protein level had a biological effect is unknown. A similar finding was recently reported, and discussed in detail, for HERV-K14 sequences that shuffled FAM8A1 mRNA portions to several positions in the human genome (17). In the present study, we report another domain from a cellular mRNA that was shuffled in the genome by HERV-K14I proviruses. It is possible that other hitherto little investigated HERV families—and there are plenty—harbor similar discoveries.
Our study provides new insight into the biology of human endogenous retroviruses. Besides the basic characterization of HERV families, such as evolutionary ages, improved consensus sequences, and subfamilies, as established in previous work by us and others, unexpected features of HERV-K14 proviruses, such as mobilization of cellular mRNA portions and obviously mobile fusion proviruses of different HERV families, were revealed. We believe that further examination of other HERV families will add more hitherto unknown aspects of HERVs, thus providing more information on the overall biological role of endogenous retroviruses in general.
Supplementary Material
Acknowledgments
W.S., E.M., and J.M. are supported by DFG grants SCH214/7-3, Me917/16-1, and Ma2298/2-1, respectively. S.B. is supported by the Future and Emerging Technologies Program of the European Union (IST-1999-14186 ALCOM-FT).
We thank Laurence Lavie for comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andersson, M. L., M. Lindeskog, P. Medstrand, B. Westley, F. May, and J. Blomberg. 1999. Diversity of human endogenous retrovirus class II-like sequences. J. Gen. Virol. 80:255-260. [DOI] [PubMed] [Google Scholar]
- 2.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 3.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., M. Jebbink, and J. Zsiros. 1999. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J. Virol. 73:2365-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328-4336. [DOI] [PubMed] [Google Scholar]
- 7.Boller, K., O. Janssen, H. Schuldes, R. R. Tonjes, and R. Kurth. 1997. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J. Virol. 71:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzdin, A., S. Ustyugova, K. Khodosevich, I. Mamedov, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics 81:149-156. [DOI] [PubMed] [Google Scholar]
- 9.Costas, J. 2002. Characterization of the intragenomic spread of the human endogenous retrovirus family HERV-W. Mol. Biol. Evol. 19:526-533. [DOI] [PubMed]
- 10.Dangel, A. W., B. J. Baker, A. R. Mendoza, and C. Y. Yu. 1995. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics 42:41-52. [DOI] [PubMed] [Google Scholar]
- 11.D'Esposito, M., M. Strazzullo, M. Cuccurese, C. Spalluto, M. Rocchi, M. D'Urso, and A. Ciccodicola. 1998. Identification and assignment of the human transient receptor potential channel 6 gene TRPC6 to chromosome 11q21→q22. Cytogenet. Cell Genet. 83:46-47. [DOI] [PubMed] [Google Scholar]
- 12.Freichel, M., S. Philipp, A. Cavalie, and V. Flockerzi. 2004. TRPC4 and TRPC4-deficient mice. Novartis Found. Symp. 258:189-199. [PubMed] [Google Scholar]
- 13.Gifford, R., and M. Tristem. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291-315. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, M. 1999. The genomic record of humankind's evolutionary roots. Am. J. Hum. Genet. 64:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassock, S. R., M. X. Zhu, C. Trost, V. Flockerzi, and K. S. Authi. 2002. Expression and role of TRPC proteins in human platelets: evidence that TRPC6 forms the store-independent calcium entry channel. Blood 100:2801-2811. [DOI] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 17.Jamain, S., M. Girondot, P. Leroy, M. Clergue, H. Quach, M. Fellous, and T. Bourgeron. 2001. Transduction of the human gene FAM8A1 by endogenous retrovirus during primate evolution. Genomics 78:38-45. [DOI] [PubMed] [Google Scholar]
- 18.Jurka, J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- 19.Kapitonov, V., and J. Jurka. 1996. The age of Alu subfamilies. J. Mol. Evol. 42:59-65. [DOI] [PubMed] [Google Scholar]
- 20.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura, Y., T. Ayukawa, T. Ishikawa, T. Kanda, and K. Yoshiike. 1996. Human endogenous retrovirus K10 encodes a functional integrase. J. Virol. 70:3302-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavie, L., P. Medstrand, W. Schempp, E. Meese, and J. Mayer. 2004. Human endogenous retrovirus family HERV-K(HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J. Virol. 78:8788-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebedev, Y. B., O. S. Belonovitch, N. V. Zybrova, P. P. Khil, S. G. Kurdyukov, T. V. Vinogradova, G. Hunsmann, and E. D. Sverdlov. 2000. Differences in HERV-K LTR insertions in orthologous loci of humans and great apes. Gene 247:265-277. [DOI] [PubMed] [Google Scholar]
- 24.Mallet, F., O. Bouton, S. Prudhomme, V. Cheynet, G. Oriol, B. Bonnaud, G. Lucotte, L. Duret, and B. Mandrand. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. USA 101:1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer, J., and E. Meese. 2002. The human endogenous retrovirus family HERV-K(HML-3). Genomics 80:331-343. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, J., and E. Meese. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 27.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Chromosomal assignment of human endogenous retrovirus K (HERV-K) env open reading frames. Cytogenet. Cell Genet. 79:157-161. [DOI] [PubMed] [Google Scholar]
- 28.Mayer, J., E. Meese, and N. Mueller-Lantzsch. 1997. Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet. Cell Genet. 78:1-5. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, J., and E. U. Meese. 2003. Presence of dUTPase in the various human endogenous retrovirus K (HERV-K) families. J. Mol. Evol. 57:642-649. [DOI] [PubMed] [Google Scholar]
- 30.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257-258. [DOI] [PubMed] [Google Scholar]
- 31.Medstrand, P., and J. Blomberg. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J. Virol. 67:6778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 34.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 35.Pustell, J., and F. C. Kafatos. 1982. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 10:4765-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter, M., K. Roemer, B. Best, M. Afting, S. Schommer, G. Seitz, M. Hartmann, and N. Mueller-Lantzsch. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362-4365. [PubMed] [Google Scholar]
- 37.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dolken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier, L. G. Brown, S. Repping, T. Pyntikova, J. Ali, T. Bieri, A. Chinwalla, A. Delehaunty, K. Delehaunty, H. Du, G. Fewell, L. Fulton, R. Fulton, T. Graves, S. F. Hou, P. Latrielle, S. Leonard, E. Mardis, R. Maupin, J. McPherson, T. Miner, W. Nash, C. Nguyen, P. Ozersky, K. Pepin, S. Rock, T. Rohlfing, K. Scott, B. Schultz, C. Strong, A. Tin-Wollam, S. P. Yang, R. H. Waterston, R. K. Wilson, S. Rozen, and D. C. Page. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825-837. [DOI] [PubMed] [Google Scholar]
- 39.Sverdlov, E. D. 2000. Retroviruses and primate evolution. Bioessays 22:161-171. [DOI] [PubMed] [Google Scholar]
- 40.Tonjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73:9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.