Abstract
Background & Aims
Variants in the tumor necrosis factor superfamily member 15 gene (TNFSF15, also called TL1A) have been associated with risk for inflammatory bowel diseases (IBD). TL1A affects expression of multiple cytokines to promote mucosal inflammation. Little is known about the TL1A-response pathways that regulate cytokine expression. We investigated T-cell gene expression patterns to determine the mechanisms by which TL1A regulates cytokine production, and whether these associate with outcomes of patients with Crohn’s disease (CD).
Methods
Peripheral T cells isolated from normal donors were cultured with TL1A. We performed gene expression profile analysis, by RNA sequencing, of subsets of interferon gamma (IFNG)-producing and non-producing cells, purified by flow cytometry. Unsupervised hierarchical clustering analysis was used to identify gene expression differences between these subsets. Ribonuclease T2 gene (RNASET2) expression and methylation were assessed by quantitative trait loci analyses. Clinical characteristics of patients (complications, resistance to therapy, recurrence time) were associated with single nucleotide polymorphisms in RNASET2. We performed motif screening to identify polymorphisms that disrupt transcription factor binding sites. Levels of RNASET2 were knocked down with small interfering RNA in CD4+ T cells and the effect on protein expression was determined by proteomic analysis and cytokine production. Cell aggregation was measured by flow cytometry.
Results
We identified 764 genes with at least a 2-fold difference in TL1A-mediated expression between IFNG-secreting and non-secreting T cells (P<1 × 10−5). Many of these genes were located near IBD susceptibility variants. RNASET2 was the only IBD risk-associated gene with greater than 5-fold downregulation in the IFNG-secreting subset. RNASET2 disease risk variants were associated with decreased expression in peripheral and mucosal tissues and DNA hypermethylation in CD patients requiring surgical intervention. RNASET2 disease risk variants were associated in CD patients with more complicated disease or resistance to therapy, defined in part by failed response to treatment, increased length of intestinal resection, shorter time to repeat surgery, and high Rutgeerts score (>2) in post-operative endoscopy. The RNASET2 variant rs2149092 was predicted to disrupt a consensus binding site for the transcription factor ETS within an enhancer region. Expression of RNASET2 correlated with expression of ETS. RNASET2 knockdown in T cells increased expression of IFNG and ICAM1 and induced T cells aggregation. A blocking antibody against LFA1, disrupting the LFA1-ICAM1 interaction, reduced T-cell production of IFNG.
Conclusions
We identified decreased expression of RNASET2 as a component of TL1A-mediated increase in production of IFNG and as a potential biomarker for patients with severe CD. Further study of the role of RNASET2 in regulating mucosal inflammation may lead to development of novel therapeutic targets.
Keywords: SNP, prognosis, genetics, risk factor
Introduction
IBD is believed to be triggered in genetically susceptible individuals by an inappropriate immune response to the commensal flora. Extensive clinical heterogeneity and complex overlapping genetic associations suggest that the underlying biological pathways differ in subgroups of patients within ulcerative colitis (UC) and CD. Optimal development of targeted therapeutics hinges on subpopulation stratification and prognostic biomarker identification. Although over 200 IBD susceptibility loci have been identified,1,2 little is known regarding their functional significance. Genetic variation in TNFSF15 is associated with CD in multiple populations,3 and the protein it encodes, TL1A, is a key mediator of mucosal inflammation.4, 5 TNFSF15/TL1A are associated with complicated and severe IBD in humans and animal models, 4, 6–11 and is a therapeutic target with compounds currently in development. In vitro, TL1A synergizes with interleukin 12 (IL-12) and interleukin 18 (IL-18), leading to rapid enhancement of IFN-γ production,12 another key mediator of mucosal inflammation. Nevertheless, the pathophysiological mechanism by which TL1A augments inflammatory cytokine secretion by T cells remains unknown.
In this report we identify RNASET2, also an IBD susceptibility gene, as a component of TL1A-mediated enhancement of IFN-γ production. We demonstrate a functional association of RNASET2 disease-risk SNPs with decreased expression and hyper-methylation in T cells isolated from CD patients and an association with clinical parameters suggestive of complicated/resistant disease behavior and rapid recurrence of disease. We show the regulatory potential for ETS TF in modulating RNASET2 expression and the involvement of homotypic T cell aggregation via ICAM1 as a component of RNASET2 mediated upregulation of IFN-γ production. The data distinguish RNASET2 as a potential therapeutic biomarker and identify unique pathways for additional therapeutic modulation within a defined IBD population.
Methods
Study Subjects
Subjects were recruited through the Cedars-Sinai MIRIAD IBD Biobank at the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute. Control subjects had no known personal or family history of autoimmune disease or IBD. Informed consent (approved by the Cedars-Sinai Institutional Review Board) was provided by all participating subjects. Clinical characteristics were collected from 564 CD patients who had undergone surgical resection (index surgery) and who were followed prospectively thereafter. Subjects recruited in the IIBDGC cohort were as described.1, 2, 13
Isolation of Purified Lymphocyte Populations
CD3+ T cells were isolated using CD3-immunomagnetic beads (Miltenyi Biotech, Auburn, CA) and CD4+ T cells using negative selection with magnetic beads (Stemcell Technologies, Vancouver, BC, Canada) and were at least 95% pure.
Infinium 450K Bead Chip Assay
DNA samples from CD3+ T cells were bisulfite converted using the Zymo EZ DNA Methylation kit (Zymo Research). The assay was carried out as per the Illumina Infinium Methylation instructions, using the Infinium HumanMethylation450 BeadChip Kit (Illumina Inc., San Diego, CA). Data were visualized using the GenomeStudio software. The methylation β values were recalculated as the ratio of (methylated probe signal)/(total signal).
IFN-γ Assay
IFN-γ was measured by amplified ELISA as previously described.5
Gene Expression Assay for CD3+ T cells
Expression analysis of CD3+ T cells was performed using the Illumina genome-wide expression BeadChip (HumanHT-12_V4_0_R2) (Illumina) or Nugen human FFPE RNA-seq library system. Illumina gene expression data were processed using BRB array tools (brb.nci.nih.gov/BRB-ArrayTools) and lumi package in R. The data were log2-transformed and normalized using robust spline normalization. Libraries for RNA-Seq were prepared with Nugen human FFPE RNA-seq library system. Reads were mapped to the UCSC transcript set using Bowtie2 version 2.1.0. Gene expression level was estimated using RSEM v1.2.15 and normalized using FPKM.
siRNA Inhibition and Quantitative Proteomic Analysis
CD4+ T cells (15 × 106) were electroporated in the presence of 150 pmole of RNASET2 siRNA or control siRNA using a BTX Electro Square Porator ECM 830 (Genetronics, Inc., San Diego, CA). RNASET2 siRNA-sequence forward 5′-GCAAGAGAAAUUCACAAACUGCAGC-3′ and reverse 5′-GCUGCAGUUUGUGAAUUUCUCUUGCUU-3′. Control siRNA-sequence forward 5′-CUUCCUCUCUUUCUCUCCCUUGUGA-3′ and reverse 5′-UCACAAGGGAGAGAAAGAGAGGAAGGA-3′.
Tandem mass tagging (TMT)-based quantitative proteomics analysis was conducted as described.14 A stringent 1% false discovery rate was set to filter peptide and protein identifications. Peptides with >30% precursor ion interference were excluded from protein quantification.
Flow cytometry and analysis of Cellular Aggregation
IFN-γ-secreting CD4+ T cells were isolated by flow cytometry following activation with recombinant human IL-12 (500 pg/ml, R&D Systems, Minneapolis, MN), IL-18 (50 ng/ml, R&D Systems) and TL1A (100 ng/ml, Fitzgerald Industries International, Acton, MA) for 8h. IFN-γ-secreting cells were detected using an IFN-γ secretion assay cell enrichment and detection kit (Miltenyi Biotec, San Diego, CA) and sorted on a FACS Aria II (BD Biosciences, San Jose, CA).
Intracellular IFN-γ production and analysis of cellular aggregation was conducted essentially as described.15 Cells were either rested or stimulated for 24h with IL12/IL18 and TL1A and Brefeldin A (10ug/ml) was added for the last 4h. Cells were fixed and stained for intracellular IFN-γ (brilliant violet 421-IFN-γ, eBioscience) or isotype control. Samples were washed and stained for cellular aggregation (propidium iodide). Cells were acquired on a LSRII Flowcytometer (BD Biosciences, San Jose) and analyzed with FlowJo software (TreeStar Inc., Ashland, OR). For LFA1 blocking analysis cells were pre-incubated overnight with monoclonal control mouse IgG1k (15ug/ml) or anti-LFA1 (TS1/18) followed by stimulation with IL12/IL18 and TL1A for 24h.
Genotyping
Genotype data were obtained using Illumina HumanImmuno BeadChip array. Markers were excluded based on: test of Hardy—Weinberg Equilibrium with significance threshold of p ≤ 10−3; if genotyping rate was < 100% (for eQTL and mQTL associations) or <98% (for GWAS) and if minor allele frequency was <5%. Identity-by-descent was used to exclude related individuals (Pi-hat scores >0.25) using PLINK16 ADMIXTURE17 was used to perform analysis to obtain ethnicity proportion estimation for individuals. An individual with Caucasian proportion ≥ 0.75 was classified as Caucasian. Independent Caucasian samples were identified based on relatedness check (using cut-off pi-hat scores) and ethnicity analysis from admixture and all subsequent associations were performed using these samples. Principal components in genotype data for independent Caucasian samples were generated using TRACE.18 LDHeatmap R package was used to generate a linkage disequilibrium (LD) plot for the SNPs in RNASET2 locus using genotype data for 139 subjects. Details of the QC and genotyping in IIBDGC cohort can be found in previous reports.1, 2 Of the CD cases from IIBDGC, 13,511 have disease behavior information based on Montreal classification as reported previously13 (described as B1, non-stricturing, non-penetrating, B2, stricturing and B3, penetrating diseases).
Expression data for Small Bowel Surgical Samples
Single channel microarray expression data extracted using Agilent feature extraction software were received from Genome Technology Access Center at Washington University, St. Louis19. Raw expression data available in technical duplicates were normalized using LIMMA package20 implemented in R version 3.2.2. The expression data preprocessing included background correction of the expression data, followed by log2-transformation and quantile-normalization.
EQTL and mQTL mapping
EQTL and mQTL mapping were implemented in Matrix eQTL R package.21 Independent Caucasian samples were used for eQTL and mQTL mapping. Associations between genotype and probe expression level (for eQTL) or methylation β values (for mQTL) were performed using a linear regression model with additive genotype effects. All associations were conducted with gender and first two principal components in genotype data as covariates along with genotype. Around 200 genetic variants within 200 KB of RNASET2 TSS were used to perform associations with RNASET2 gene expression or methylation levels.
Motif Analysis and Identification of Candidate Regulatory SNPs
All variants exhibiting eQTL and mQTL were analyzed for predicted disruption of TF binding motifs using the bioconductor motifbreakR package.22 Only T cell specific TFs identified as being expressed using RNA-seq data from CD patients, were carried forward. Candidate regulatory SNPs were then analyzed for potential functionality based on Roadmap Epigenomics Mapping Consortium (REMC) data.23 Potential active enhancer regions were determined based on overlap of the histone modification H3K4me1 with H3K27ac signals. Potential functionality of TF regulation was determined based on REMC CHIP-seq binding signal and Regulome data.
Pathway Analysis
Pathway analysis was accomplished through the use of Qiagen’s Ingenuity® Pathway Analysis (IPA®, Qiagen, Redwood City, www.qiagen.com/ingenuity) and The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov).
Statistical Analysis
Modeling, data analysis, and data mining were performed using the BRB array tools and R-program (version 2.2.2; www.r-project.org). Class prediction analysis used compound covariate predictor, diagonal linear discriminant analysis, k-nearest neighbor (using k=1 and 3), nearest centroid, and support vector machines, based upon a minimum p value of 0.001. Cluster analysis was performed using Cluster 3.0 and Java Treeview 1.1.6r4. Tests for statistical significance were determined using JMP Statistical Software (Cary, NC). Logistic regression was performed in IIBDGC cohort to evaluate the association with disease and disease severity with PCs included as covariates. Test for clinical association between of rs1819333 and rs9355610 SNPs and therapeutic failure, ANCA sero-positivity, resected bowel length and time to reoperation were calculated by parametric Student’s T test and Pearson correlation; test of association and trend using Fisher’s exact test and Kaplan-Meier Survival Curves. Association with endoscopic recurrence was calculated by Cochran-Armitage trend test. All authors had access to the study data and reviewed and approved the final manuscript.
Results
Decreased RNASET2 is Associated with TL1A-Mediated Enhancement of IFN-γ Production
To identify the underlying molecular pathways involved in TL1A-mediated enhancement of IFN-γ production, CD4+ T cells from normal donors were treated with TL1A, sorted into IFN-γ-secreting and non-secreting subsets and analyzed by RNA-seq (Supplementary Fig. S1, A and B). Unsupervised hierarchical clustering of the set of expressed genes clearly distinguished TL1A-mediated IFN-γ-secreting and non-secreting groups (Supplementary Fig. S1C). Seven hundred and sixty-four “predictor” genes with at least two-fold differential expression between the IFN-γ secreting/non-secreting subsets (p value <1 × 10−5) (Fig. 1A) were identified. Gene ontology analysis indicated that differentially expressed genes were enriched in pathways associated with T cell receptor signaling, apoptosis, and RNA expression, and were downstream targets of infliximab, an anti-TNF biologic drug widely used in IBD (Fig. 1B). Predictor genes were significantly enriched in regions flanking GWAS identified IBD susceptibility variants1–3, 24 (0.25 MB upstream or downstream of the SNP compared to other regions (14% vs. 9%, p value is 3.3 × 10−6, hypergeometric test)). The data suggest that these genes contribute not only to TL1A-mediated modulation of IFN-γ expression, but also overlap with IBD risk-associated loci. Of the IBD-risk associated predictor genes, expression of IFNG was confirmed as the most significantly upregulated and RNASET2 as the most significantly downregulated gene (Fig. 1C). RNASET2 was the only IBD risk associated gene with greater than 5-fold downregulation in the IFN-γ secreting CD4+ subset.
Fig. 1.
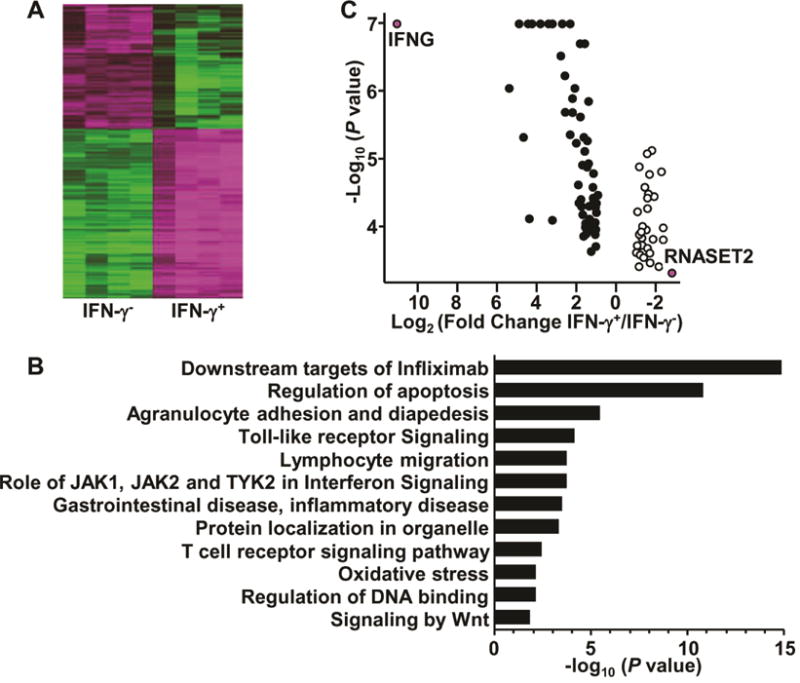
Identification of differentially-expressed genes from IFN-γ-secreting CD4+ T cells. CD4+ T cells were stimulated 8h with IL-12, IL-18 and TL1A, followed by detection and isolation of IFN-γ-secreting cells. RNA was isolated from sorted cells and analyzed through RNA-sequencing (A) Heatmap of 764 predictor genes. (B) Gene ontology analysis of 764 predictor genes. (C) Volcano plot of IBD risk predictor genes.
RNASET2 Variants Displaying eQTL and mQTL Overlap with GWAS Disease-Risk Associated Variants
RNASET2 is the only human member of the Rh/T2/S family of ribonucleases and its expression is decreased in ovarian cancer,25 melanoma26 and non-Hodgkin lymphoma.27 The functional role of RNASET2 in regulation of IFN-γ secretion is unknown. Considering the key role IFN-γ plays in pathogenesis of IBD, 28 RNASET2 expression was examined in freshly-isolated, unstimulated peripheral CD3+ T cells from NL, CD and UC. Because DNA methylation is understood to impact gene expression, particularly in disease-associated genetic variants that map outside transcribed exomes, we examined the DNA methylation status across the RNASET2 locus. RNA-seq analysis demonstrated an inverse correlation between TNFSF15 expression levels and RNASET2 in peripheral T cells from two independent cohorts (a combined total of 138 CD patients, Fig. 2A). Results remained consistent even when each cohort was analyzed separately (Fig. S2). Moreover, there was a significant negative correlation between expression and methylation (Fig. 2B), mainly within 50 kb upstream and downstream from the TSS (Fig. 2C). The strongest correlation of methylation and expression (p = 8.5 × 10−5) was observed at a CpG site (1.4 kb) within the first intron of RNASET2 (Fig. 2C). Additionally, CD genetic risk variants, including the IBD risk SNP tagging the RNASET2 locus in European ancestry populations, rs1819333,1 overlapped with regions correlative for methylation and expression levels (Fig. 2D).
Fig. 2.
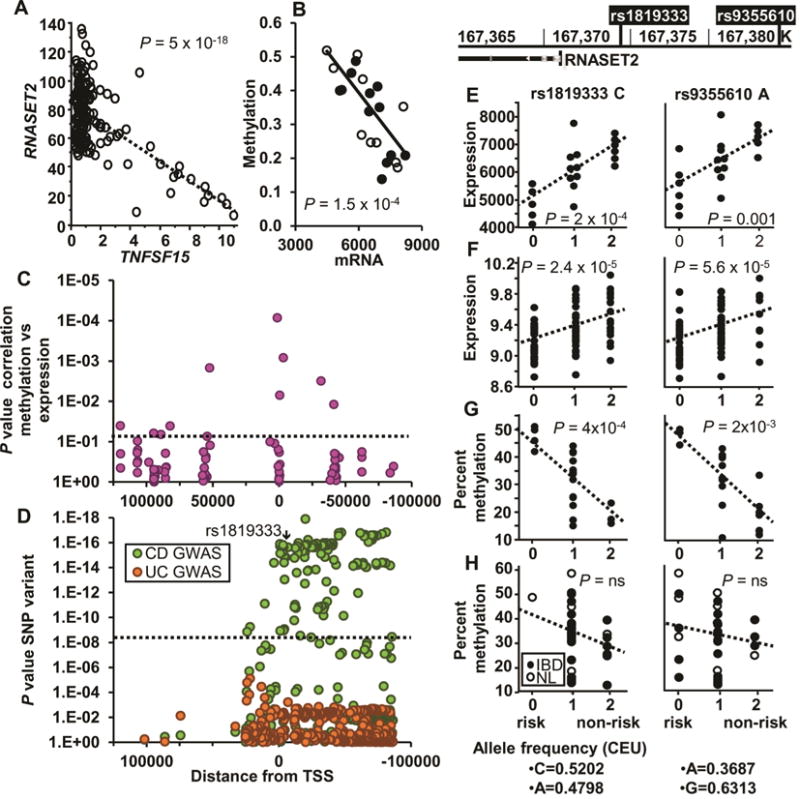
Correlation of RNASET2 and associated SNPs on expression and methylation. (A) Correlation of RNASET2 and TNFSF15 expression in CD3+ peripheral T cells from 138 CD patients requiring surgical intervention for disease management, using RNA-seq. (B) Correlation of RNASET2 expression and methylation (cg25258033, located 1.4 kb within the first intron) in 21 IBD patients. (C) Correlation of methylation and expression located within 100 kb of the RNASET2 TSS for 21 IBD patients. (D) GWAS p values for the same RNASET2 region based upon data from 18729 CD, 14331 UC and 34897 controls. (E–F) eQTL of RNASET2 SNPs (rs1819333 and rs9355610) from (E) CD3+ peripheral T cells from 11 CD and 10 UC patients requiring surgical intervention for disease management, using an Illumina expression array or (F) Ileal surgical resections of 85 CD patients using an Agilent expression array. (G–H) mQTL (cg25258033) of CD3+ peripheral T cells from (G) 20 CD patients requiring surgical intervention for disease management or (H) 16 CD patients who were responsive to IBD therapeutics and 9 normal controls.
RNASET2 Disease-Risk Alleles are Associated with Decreased RNASET2 Expression and Increased DNA Methylation in CD Patients Requiring Surgical Intervention for Disease Management
Gene expression quantitative trait loci (eQTL) analysis was performed to characterize the functional correlation between RNASET2 gene variation and the gene expression level. The disease-associated SNPs for IBD risk in Europeans, rs18193331 and the risk SNP associated with Graves’ disease, rs9355610 29(LD R2 = 0.53), are located −13 kb from the transcriptional start site of RNASET2. The functional correlation between RNASET2 IBD-risk genotypes and the gene transcript expression levels were established in unstimulated peripheral CD3+ T cells isolated from CD and UC patients requiring surgical intervention for disease management. The data demonstrated significantly decreased RNASET2 expression in T cells from subjects carrying the RNASET2 risk alleles rs1819333 and rs9355610 (Fig. 2E). These findings were confirmed with significant eQTL observed for mRNA extracted from uninvolved small bowel tissue obtained from CD subjects at surgical resection (Fig. 2F). The correlation between RNASET2 gene variation and methylation, mQTL was also examined. A significant mQTL was observed with an increase in methylation in CD patients requiring surgical intervention for disease management (Fig. 2G). In contrast, no mQTL was detected in cells isolated from CD patients who were responsive to IBD therapeutics or in normal subjects (Fig. 2H).
Gene expression (eQTL) and DNA methylation (mQTL) were mapped across all informative SNPs spanning the RNASET2 locus (LD plot, Fig. 3A). In T cells isolated from patients requiring surgical intervention for disease management, there is strong overlapping eQTL and mQTL from 10 kb downstream of RNASET2 TSS to −170 kb upstream, spanning fibroblast growth factor receptor 1 oncogene partner (FGFR1OP) to the first intron of chemokine (C-C motif) receptor 6 (CCR6). Likewise, there was a significant overlap in eQTL when comparing RNASET2 expression from unstimulated peripheral T cells to small bowel surgical resection in CD patients requiring surgical intervention for disease management. In contrast, few mQTL associations were detected in CD patients who were responsive to IBD therapeutics or in normal subjects (Fig. 3B). No eQTL association was detected for FGFR1OP or CCR6. These data were further validated in peripheral T cells from a separate cohort of CD patients requiring surgical intervention for disease management. There was significant overlap between RNASET2 risk variants associated with CD and corresponding eQTL (Fig. 3C), suggesting a functional role for RNASET2 in mediating disease.
Fig. 3.
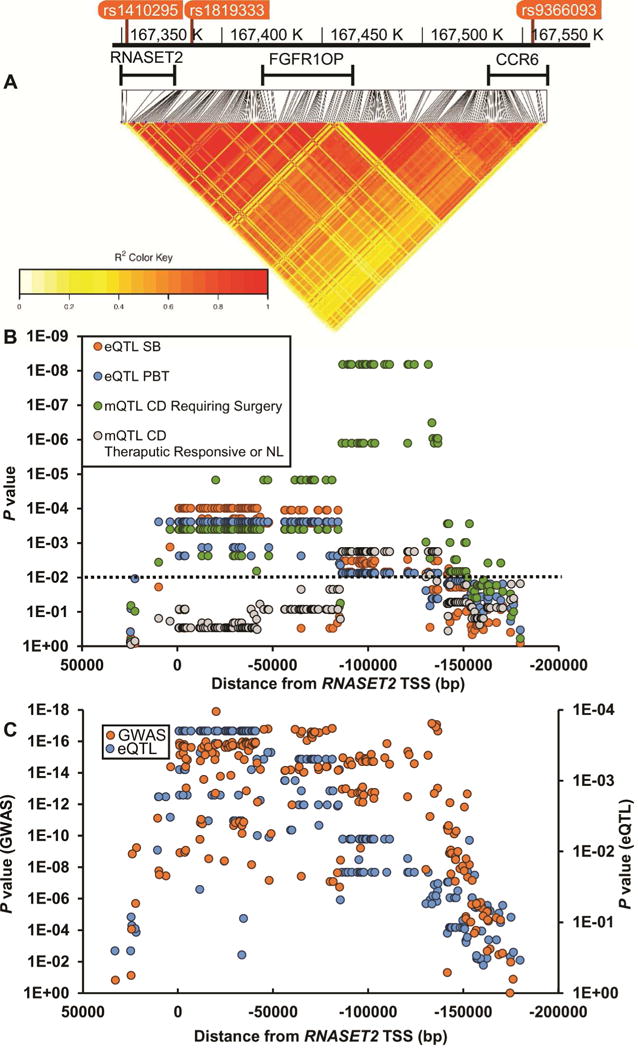
Mapping of eQTL and mQTL across the RNASET2 locus. (A) Heatmap illustrating pairwise LD (measured by the squared allelic correlation coefficient r2) for 210 SNPs around RNASET2 TSS. Boundaries of region examined are defined by rs1410295 and rs9366096. (B) eQTL and mQTL were calculated using CD3+ T cells from both the peripheral and mucosal compartments from patients requiring surgical intervention for disease management compared to those who were responsive to IBD therapeutics or normal controls. (C) Correlation of GWAS p values with eQTL p values over the RNASET2 locus. GWAS values are based upon data from 18729 CD and 34897 controls and eQTL based upon data for 71 CD patients requiring surgical intervention for disease management.
RNASET2 Disease-Risk Alleles are Associated with Complicated and Resistant Disease Behavior
To evaluate the association between RNASET2 and disease activity and severity we utilized a cohort of 564 CD patients who had undergone surgical resection and were followed prospectively. Clinical characteristics including indication for surgery were assessed for association with RNASET2 risk variants (rs1819333 and rs9355610). At the time of index surgery, patients with RNASET2 disease-risk SNPs were associated with therapeutic failure of thiopurine or anti-TNF therapy, ANCA sero-positivity (a marker associated with lack of response to anti-TNF therapy30), and an increased length of intestinal resection characteristic attributed to overall disease severity (Table 1 and Figures S3–S4).31 No association was observed for therapeutic failure on steroids. Moreover, patients with RNASET2 disease-risk SNPs who required more than one resection for disease management exhibited a shorter time between surgeries (Fig. 4A).
Table 1.
Clinical disease parameters associated with RNASET2 risk variants.
| Clinical parameter | rs1819333 | rs9355610 | |||
|---|---|---|---|---|---|
|
| |||||
| p | OR | p | OR | ||
| Disease Behavior | |||||
| B2 vs. B1a | ns | ns | 0.041 | 1.07 | |
| B3 vs. B1a | ns | ns | 0.056 | 1.06 | |
| B2, B3 vs. B1a | 0.051 | 1.05 | 0.016 | 1.07 | |
| Therapeutic failure of thiopurineb | 0.009 | 1.68 | 0.019 | 1.75 | |
| Therapeutic failure of anti-TNFb | 0.039 | 1.46 | 0.042 | 1.56 | |
| ANCA sero-positivityb | 0.009 | 2.24 | 0.047 | 2.07 | |
| Resected segment | >30 cmb | ns | ns | 0.004 | 2.13 |
| >40 cmb | ns | ns | 0.031 | 1.96 | |
| Endoscopic recurrence | p | z score | p | z score | |
|
|
|||||
| Rutgeert’s score 3–4 vs 1–2 | 0.025 | 2.24 | 0.024 | 2.25 | |
IIBDGC cohort CD (B1 = 6278, B2 = 3345, B3 = 3828)
CD patients (n=584) who had undergone surgical resection and followed prospectively.
Fig. 4.
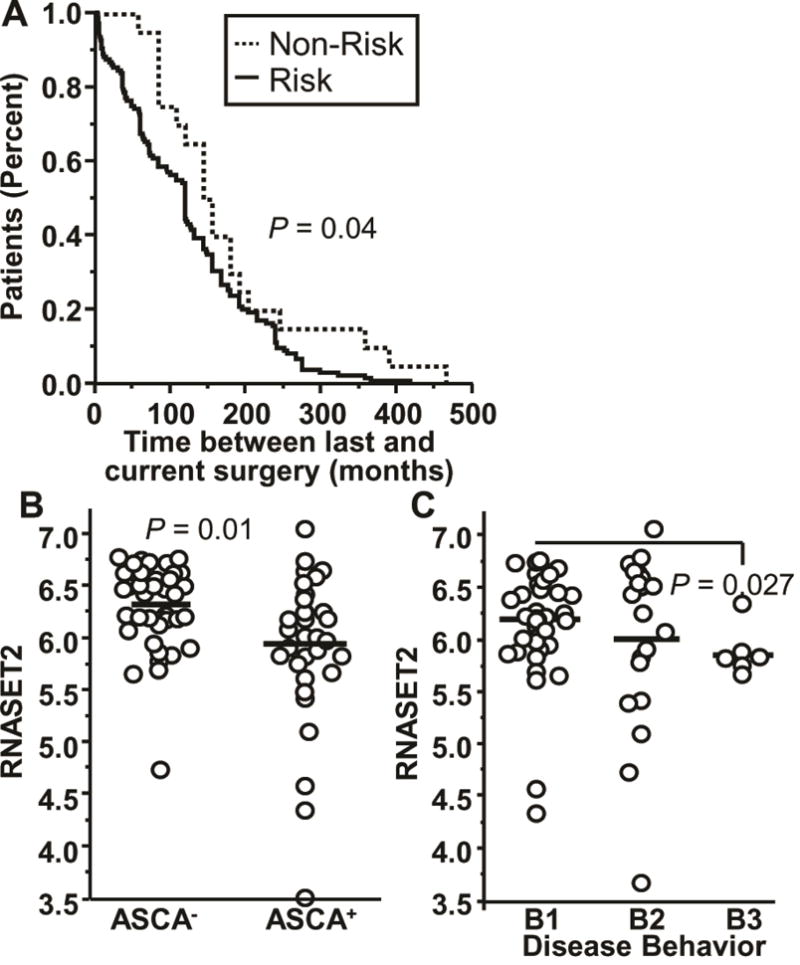
Clinical disease parameters associated with RNASET2 disease-risk variants and expression levels. (A) Survival analysis for time between prior and index surgeries based upon carriage for IBD-risk variant rs9355610 for 154 CD patients who underwent multiple surgeries. (B–C) Expression of RNASET2 by RNA-seq for 71 CD patients based upon (B) IgG ASCA sero-positivity. (C) Montreal disease classification (B1, B2, and B3).
Likewise, RNASET2 risk SNPs were also associated with a more severe disease recurrence. Postoperative endoscopies revealed an association of RNASET2 risk SNPs in patients with a high Rutgeerts score (> 2) who were not receiving postoperative prophylaxis (Table 1). Decreased expression of RNASET2 was also associated with ASCA sero-positivity (Fig. 4B) and a penetrating disease phenotype (Fig. 4C). These data were further confirmed in a separate cohort of CD patients (IIBDGC cohort) in which RNASET2 disease-risk SNPs were associated with a complicated stricturing/penetrating phenotype (Montreal classification B1 vs. B2 and B3), (Table 1). These data support an association of RNASET2 disease-risk SNPs with clinical parameters suggestive of complicated and resistant disease behavior.
RNASET2 Variant in LD with Disease-Tagging SNP Disrupts ETS Transcription Factor Binding Motif
The data presented above demonstrate significant overlap between more than a hundred CD RNASET2 risk variants, many in linkage disequilibrium, associated with eQTL and mQTL creating difficulty in determining functionality/causality. Since the majority of RNASET2 risk variants associated with CD are located in non-coding regions, it is likely that these SNPs alter expression through modulation of regulatory functions. Furthermore, studies suggest 32 that SNPs associated with disease often exist within active enhancer regions of cell types relevant to disease and can disrupt TF binding motifs. REMC data demonstrate that the RNASET2 locus is marked in primary T cells, compared to other tissues, by putative active enhancer histone modifications and active gene expression (Fig. S5). To gain insight into the molecular pathways regulating RNASET2 expression and prioritize the number of candidate functional SNPs, we performed motif analysis to predict TF motif disruptions22 across all SNPs which were associated with eQTL/mQTL. We then selected variants disrupting motifs of TFs expressed in T cells and focused on candidate variants in LD with the RNASET2 disease index SNP rs1819333. The rs2149092 SNP disease-risk variant, located −569 bp from the index SNP (LD R2 = 1), lies within the highly conserved TTCC motif, utilized by most ETS transcription factors, and is predicted to disrupt TF binding. Sequence analysis suggests an overlap of IRF4 and SPI1 binding sites adjacent to a JUN binding site (Fig. 5A). Regulome and REMC data confirm TF occupancy of ETS1, IRF4 and SPI1binding in lymphoblastoid cell lines (Fig. 5B) which overlaps with histone modifications indicative of an active enhancer element. Moreover, there is a strong correlation between expression of RNASET2 with multiple members of ETS and JUNTF (Fig. 5C and S6). No correlation was observed for IRF4 (Fig. S6). These data strengthen the relevance of RNASET2 expression in the immune compartment and support a functional role for ETS and JUN transcription factors in regulating transcription of RNASET2.
Fig. 5.
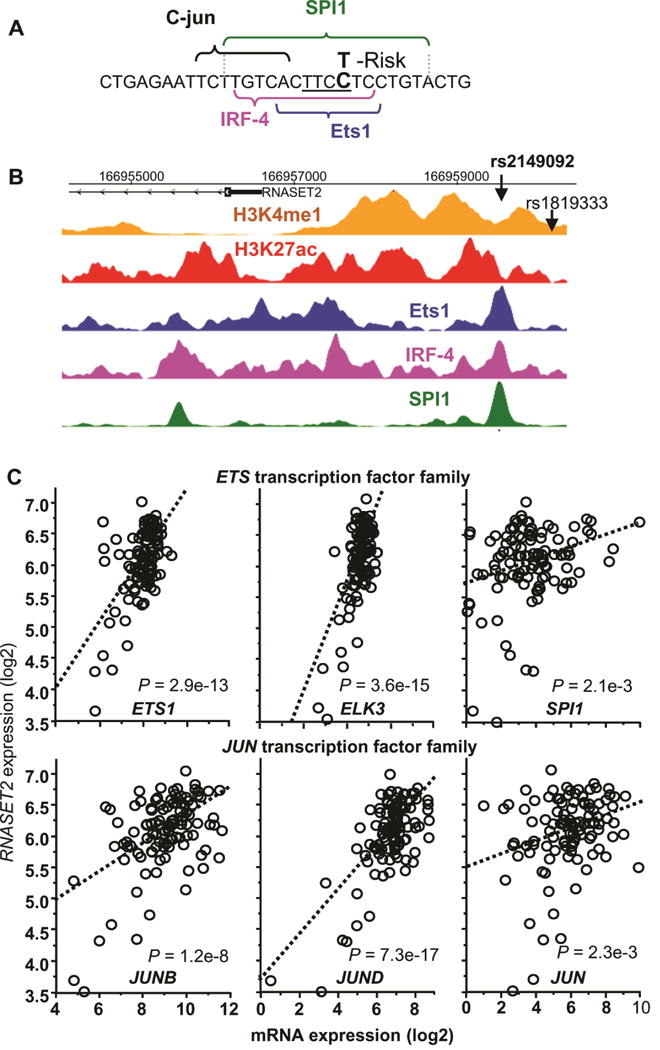
Identification of potential regulatory function of RNASET2 disease-associated variant rs2149092. (A) Predicted disruption of rs2149092 C/T variation in the binding motifs for ETS and IRF4 TF. Central ETS invariant motif is underlined. (B) CHIP-seq and histone modification profiles for ETS1, IRF4 and SPI1 TF binding and histone H3K4me1 and H3K4ac aligned with the genomic sequence surrounding rs2149092 variant. (C) Correlation of expression of RNASET2 and multiple ETS and JUN transcription factors in CD3+ peripheral T cells from 108 CD patients requiring surgical intervention for disease management, using RNA-seq.
Silencing of RNASET2 Enhances IFN-γSecretion via Upregulation of ICAM1 Expression and Homotypic T Cell Aggregation
The functional role of RNASET2 in regulation of IFN-γ secretion was examined using siRNA silencing. CD4+ T cells that were transfected with RNASET2-siRNA or control NC-siRNA, followed by stimulation with TL1A. Cells transfected with siRNA targeting RNASET2 displayed a 60–70% inhibition of RNASET2 expression (Fig. 6A), and a parallel significant enhancement (~1.5 fold) in TL1A mediated IFN-γ secretion, compared to control siRNA (Fig. 6B, Fig. S7). In order to define the signaling pathways involved in this process, proteomic analysis was carried out. Candidate targets were selected on the basis of exhibiting both modulation of expression following siRNA silencing of RNASET2 as well as TL1A-stimulated differential expression when comparing IFN γ secreting and non-secreting CD4+ T cells (data from RNA-seq analysis). ICAM1 was one of the proteins that was upregulated in response to RNASET2 silencing and differentially expressed in the IFN γ secreting compared to non-secreting T cells (Fig. S8). ICAM1 was recently identified as a candidate gene at an IBD susceptibility locus, with upregulated gene expression associated with the disease-risk variant.33 ICAM1 is a transmembrane adhesion protein commonly expressed by vascular endothelium and leukocytes. Binding of ICAM1 to the LFA1 receptor on T cells facilitates and stabilizes cell-cell interactions. Recent studies have demonstrated increased ICAM1 expression on activated T cells34 and proposed a role for ICAM1-LFA1 binding in inducing homotypic T cell aggregation and subsequent T cells differentiation.34–36 To examine the effect of cell-cell contact on TL1A mediated IFN-γ secretion, cells were incubated in flat bottom and conical bottom microwells. A greater than 3 fold increase in IFN-γ production was consistently observed when cells were incubated in close cell-cell conical geometry (data not shown). Flow cytometry was then used to test the hypothesis that TL1A mediated enhancement of IFN-γ production is facilitated by homotypic T cell aggregation. Briefly, T cells were stimulated in the presence or absence of TL1A and then stained with an antibody for intracellular IFN-γ (Fig. 6C and 6D, left panels) and for cellular aggregation using propidium iodide (PI) (Fig 6C and 6D, upper and lower right panels). The PI-labeled peaks correspond to number of cells per event allowing for identifying single cells versus cellular aggregates.15 The first peak in each histogram corresponds to single cell events (black brackets) and the successive peaks, to multicellular aggregates (blue brackets). As expected, only a small percentage of the unstimulated T cells secreted IFN-γ, and these cells were almost equally distributed as single cell events and cellular aggregates (Fig. 6E, left panel). Following TL1A stimulation, there was a significant increase in both the percentage and size of cellular aggregates (upper right panels of Fig. 6C compared to Fig. 6D) as well as, the overall number of IFN-γ producing cells (6-fold) (Fig 6E and 6F) and a 30-fold increase in IFN-γ secretion (data not shown). In contrast, the majority of T cells that do not produce IFN-γ, are comprised of single cell events regardless of whether they were cultured with or without TL1A stimulation (Fig. 6E, right panel). These results suggest that cellular aggregation may contribute to both an increase in the number of cells producing IFN-γ and to overall amount of IFN-γ production, and TL1A stimulation may enhance this process. The functional role of TL1A in mediating cellular aggregation via ICAM1-LFA1 engagement was tested using an LFA-1 blocking antibody. As seen in figure 6G, there was an overall 43% reduction in IFN-γ secretion in response to blocking LFA-1 engagement, compared to IgG control antibody (p value =0.047). Taken together these data indicate that TL1A-mediated downregulation of RNASET2 and concomitant enhancement of ICAM1 expression, promotes homotypic T cell aggregation and augmentation of IFN-γ production. It is intriguing to note that an increase in expression of ICAM1 was associated in CD with ASCA sero-positivity and pre-op therapeutic failure of anti-TNF and thiopurine (Fig. S9), clinical parameters associated with decreased RNASET2 and disease activity.
Fig. 6.
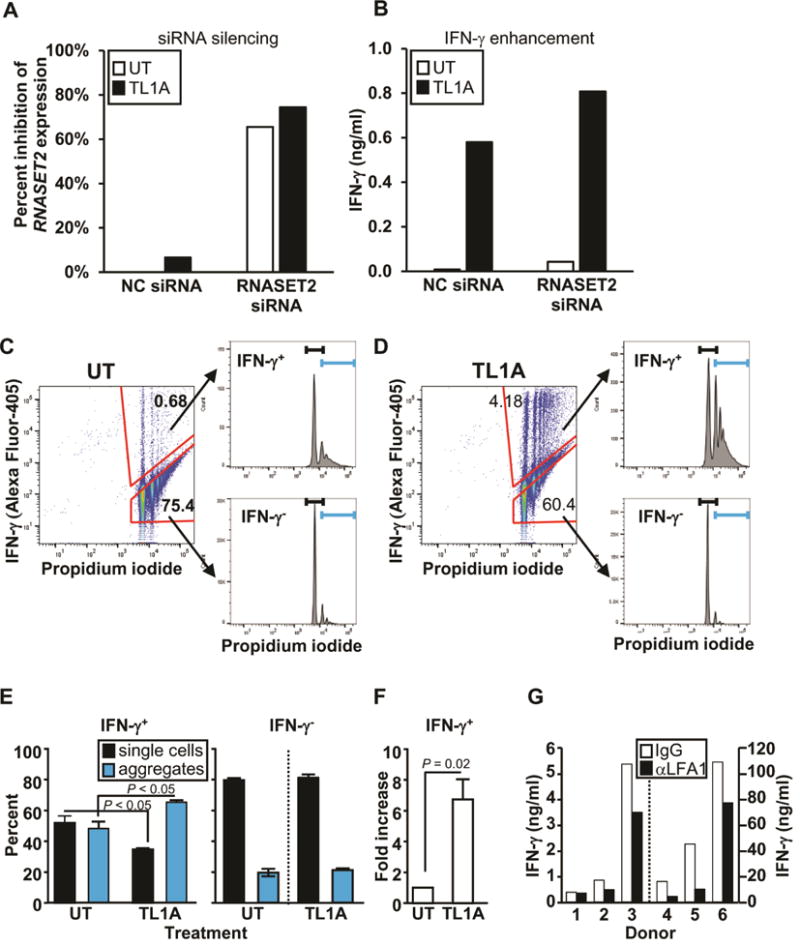
Effect of RNASET2 silencing on IFN-γ secretion and cellular Aggregation. (A) Silencing of RNASET2 expression by RNASET2 or control (NC) siRNA. (B) Effect of RNASET2 silencing on IFN-γ secretion. Panels A and B are representative of 6 out of 7 experiments (Fig. S7) with similar results. (C–D) CD4+ T cells were either (C) not treated (UT) or (D) stimulated with TL1A. Intracellular IFN-γ staining and cellular aggregation were measured by flow cytometry. Cells were gated on IFN-γ secreting and non-secreting populations (left panels) and then using propidium iodide (PI) analyzed for single and aggregate cell fractions (histograms, right panels). The first peak in each histogram corresponds to single cells (black bracket) and the remaining peaks to cellular aggregates (blue bracket). Representative of 4 experiments. (E) Proportion of single cells and cellular aggregates in IFN-γ secreting (IFN-γ+) and non-secreting (IFN-γ−) populations following TL1A stimulation. (F) Fold increase in number of IFN-γ secreting cells (average of 4 experiments). (G) CD4+ T cells were pretreated with control IgG or LFA1 blocking Ab (aLFA1) prior to TL1A stimulation. Overall p value for LFA-mediated blocking of IFN-γ secretion, measured by ELISA, was 0.047.
Discussion
TL1A (TNFSF15) is expressed primarily on activated cells of the immune system following stimulation by immune complexes 37 or through interaction with enteric microorganisms. 38 TNFSF15 disease-associated variants are correlated with increased and sustained expression of TL1A. TNFSF15 has been identified and confirmed in GWAS as an IBD-associated gene1 and is believed to play a role in modulating the location and severity of intestinal inflammation6, 7 as well as development of stricturing disease.9, 39 Transgenic mice with constitutive expression of TL1A developed intestinal inflammation along with ileal and colonic fibrosis, which was reversed by anti-TL1A treatment.9 TNFSF15 is associated with medically refractory UC.7 Despite its importance in IBD, the molecular pathways underlying TL1A enhanced cytokine secretion and inflammation are poorly understood. In this study we investigated TL1A-dependent molecular triggers that induce cytokine expression, particularly IFN-γ, in T cells. This approach identified down-modulation of RNASET2 as a component of TL1A-mediated enhancement of IFN-γ production.
We identified an inverse correlation between the expression of RNASET2 and TNFSF15 among IBD patients. We also demonstrated a functional association between DNA hyper-methylation and decreased expression of RNASET2, and that there was significant eQTL overlap with RNASET2 IBD risk alleles identified through GWAS in samples isolated from the peripheral T cells and small bowel surgical resections. One study reported significant RNASET2 eQTL (rs429083) in whole thymic tissue samples,40 this SNP demonstrated the most significant eQTL in our data as well. Our study provides clinically relevant evidence that decreased expression of RNASET2 correlated with CD clinical parameters suggestive of complicated and resistant disease. Notably, CD patients carrying the RNASET2 disease-risk SNPs exhibited increased penetrating disease behavior. Similarly, RNASET2 disease-risk SNPs display decreased expression in SB mucosal samples, and also in peripheral samples from CD (9 out of 11 were non-responsive to anti-TNF therapy), requiring surgical intervention for disease management. Consistent with our finding, a recent study reported significant RNASET2 eQTL in whole blood from patients resistant to anti-TNF therapy.41 Moreover, RNASET2 disease-associated SNPs correlated with therapeutic failure of anti-TNF therapy, and intestinal resection of >40 cm clinical characteristic of overall disease severity.31 In patients with a history of multiple resections, RNASET2 disease-risk SNP was associated with a shorter time to repeat surgery in CD. Likewise, RNASET2 disease-associated SNPs were associated in patients with endoscopic recurrence characterized by a more severe (>2) Rutgeerts score, predictive for early clinical recurrence and need for reoperation.42
RNASET2 is the only human member of the Rh/T2/S family of acidic hydrolases. These endonucleases are highly conserved among the phyla from viruses to humans suggesting an important evolutionary function. Altered expression of RNASET2, both increased and decreased, is associated with various diseases, including cancers25–27 and autoimmune diseases,29, 40, 43, 44 suggesting a role for RNASET2 in host immune responses. RNASET2 expression is downregulated and likely involved in the pathogenesis of colorectal cancer,33 melanoma,19 anaplastic large cell lymphoma34 and non-Hodgkin lymphoma.27 In human ovarian cancer overexpression of RNASET2 is associated with tumor suppression and is believed to modulate tumor micro-environment cross-talk through recruitment of monocyte/macrophages to the tumor itself.25 Surprisingly, the ribonuclease catalytic activity of RNASET2 is not required for oncosuppressive activity.25 In contrast, vitiligo-enhanced level of RNASET2 is detected in patient specimens and can be induced in vitro in cultured primary human melanocytes and keratinocyte in response to stress.44 Likewise, attenuated level of RNASET2 is associated with multi-organ fibrosis, including lung, heart, liver and kidney.45
The RNASET2 locus has been implicated by GWAS in susceptibility for vitiligo,44 Graves’ disease29 and Crohn’s disease,1, 2 although, the functional roles of RNASET2 in disease pathogenesis remain poorly defined. In cancer cell models, stress-induced apoptosis occurs through the interaction of RNASET2 with tumor necrosis factor receptor-associated factor 2 (TRAF2), supporting a role in caspase-8 activation.44 In ovarian cancer, overexpression of RNASET2 attenuates the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, a key pathway for cell proliferation and differentiation.25 The transcriptional regulatory regions and factors modulating RNASET2 expression are likewise poorly defined. The majority of disease-associated variants identified by GWAS reside within regulatory non-coding regions corresponding to promoters or enhancer sequences. Studies suggest that alteration in transcriptional regulation via disruption of TFBS may play a role in the disease process32. In the present study we utilized TF motif analysis to prioritize and identify, from the large number of variants demonstrating eQTL and mQTL, a prospective regulatory SNP. The rs2149092 disease-associated SNP alters the conserved ETS consensus binding sequence and likely disrupts binding of multiple overlapping TF binding sites including IRF4, SPI1 and ELF1. Moreover, there is a strong positive correlation between the levels of RNASET2 expression and ETS and JUN TF family members. Interestingly, IRF4, SPI1 and ELF1 have been implicated in T cell development and IRF4 and ELF1 have been associated by GWAS with IBD2, 46. These data support a functional role for rs2149092 as a modulator of TF-DNA interactions and set the stage for future studies to determine the mechanistic pathways by which TL1A attenuates expression of RNASET2 in disease.
A role for RNASET2 has been attributed to modulation of cytoskeletal re-organization, cell adhesion and motility45, 47, 48. In the present study we describe a functional relationship between RNASET2 and the cell adhesion molecule, ICAM1. Enhanced IFN–γ secretion in response to TL1A was accompanied by a decrease in RNASET2 expression on the one hand and an increase in ICAM1 levels on the other. TL1A-mediated IFN–γ secretion was inhibited by Ab blockade of the ICAM1-LFA1 interaction. Although ICAM1-LFA1 engagement is classically defined as occurring between endothelial and T cells49, these interactions have more recently been shown to play a critical role in mediating homotypic cellular aggregation of activated T cells. Homotypic T-T aggregates have been shown to promote synaptic-based cytokine delivery of IFN–γ and IL2 from one T cell to another, resulting in IL-2 receptor ligation and subsequent STAT5 phosphorylation.35, 36 In this study, we show that enhanced cellular aggregation is a hallmark of IFN–γ producing cells and TL1A-stimulation increases the number and size of the cellular aggregates. These findings suggest that RNASET2 may act through the integrin signaling pathway to modulate downstream IFN–γ secretion. This hypothesis is in fact supported by a recent study demonstrating that ICAM1-LFA1 signaling in T cells promotes a Th1-dominant response. 50 Further studies are ongoing to define the downstream mechanism by which decreased expression of RNASET2 modulates cytokine secretion in the context of IBD pathogenesis.
In conclusion, this study identifies a novel functional and biological relationship between two IBD susceptibility genes, TNFSF15 and RNASET2. We provide evidence that decreased RNASET2 expression is functionally implicated in both the TL1A driven pro-inflammatory cytokine production by activated T cells and functionally associated with the RNASET2 IBD susceptibility variants. Likewise, the present study demonstrates an association between decreased RNASET2 expression and RNASET2 disease-risk variants and a more complicated and aggressive form of CD inflammation, which might underlie disease pathology triggered by TL1A and its downstream mediated pathways. Thus, RNASET2 expression may serve as a novel disease biomarker of a more severe form of inflammation identifying a patient population not responsive to current treatment strategies, who may benefit from additional alternate RNASET2 mediated therapeutic approaches.
Supplementary Material
Acknowledgments
This work was supported by the National Center for Advancing Translational Sciences, Grants United States Public Health Service Grant DK043211, UL1RR033176 and UL1TR000124 and Cedars-Sinai Medical Center Inflammatory Bowel Disease Research Funds. The MIRIAD IBD Biobank is supported by the Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, United States Public Health Service grant P01DK046763, The European Union Grant 305479, NIDDK Grant DK062413and The Leona M and Harry B Helmsley Charitable Trust. DPBM was supported by DK062413, AI067068, grant 305479 from the European Union, and The Leona M. and Harry B. Helmsley Charitable Trust. The authors are grateful to Cedars Sinai Mass Spectrometry and Biomarker Discovery Core for proteomic analysis.
Abbreviations
- CCR6
chemokine (C-C motif) receptor 6
- CD
Crohn’s disease
- DAVID
the Database for Annotation, Visualization and Integrated Discovery
- ELISA
enzyme-linked immunosorbent assay
- eQTL
expression quantitative trait loci
- FGFR1OP
fibroblast growth factor receptor 1 oncogene partner
- GWAS
genome-wide association study
- IBD
inflammatory bowel diseases
- IFN-γ
interferon gamma
- IL-12
interleukin 12
- IL-18
interleukin 18
- LD
linkage disequilibrium
- mQTL
methylation quantitative trait loci
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NL
normal
- RNA-seq
RNA sequencing
- RNASET2
ribonuclease T2
- siRNA
small interfering RNA
- SNP
single nucleotide polymorphism
- TL1A
TNF-like protein 1A
- TNF
tumor necrosis factor
- TNFSF15
tumor necrosis factor superfamily member 15
- TRAF2
tumor necrosis factor receptor-associated factor 2
- TSS
transcriptional start site
- UC
ulcerative colitis
- SB
small bowel
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Rivkah Gonsky: study concept and design; acquisition of data; analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content; statistical analysis, obtained funding, technical, or material support, study supervision
Phillip Fleshner: acquisition of data; technical, or material support
Richard Deem: study concept and design; acquisition of data; analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content; statistical analysis, technical support
Eva Biener-Ramanujan: study concept and design; acquisition of data; analysis and interpretation of data, critical revision of the manuscript for important intellectual content; statistical analysis
Dalin Li: acquisition of data; analysis and interpretation of data, critical revision of the manuscript for important intellectual content; statistical analysis
Alka A. Potdar: acquisition of data; analysis and interpretation of data, critical revision of the manuscript for important intellectual content; statistical analysis
Janine Bilsborough: critical revision of the manuscript for important intellectual content
Shaohong Yang: acquisition of data; technical and material support
Dermot P.B. McGovern: study concept and design; acquisition of data; analysis and interpretation of data, critical revision of the manuscript for important intellectual content; obtained funding, study supervision
Stephan R. Targan: study concept and design; critical revision of the manuscript for important intellectual content; obtained funding, study supervision
Conflict of Interest
The authors acknowledge having no conflict of interest in connection with the publication of this article.
References
- 1.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 4.Bamias G, Kaltsa G, Siakavellas SI, et al. Differential expression of the TL1A/DcR3 system of TNF/TNFR-like proteins in large vs. small intestinal Crohn’s disease. Dig Liver Dis. 2012;44:30–6. doi: 10.1016/j.dld.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Prehn JL, Mehdizadeh S, Landers CJ, et al. Potential role for TL1A, the new TNF-family member and potent costimulator of IFN-gamma, in mucosal inflammation. Clin Immunol. 2004;112:66–77. doi: 10.1016/j.clim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Michelsen KS, Thomas LS, Taylor KD, et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One. 2009;4:e4719. doi: 10.1371/journal.pone.0004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis. 2010;16:1830–40. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett R, Zhang X, Koon HW, et al. Constitutive TL1A expression under colitogenic conditions modulates the severity and location of gut mucosal inflammation and induces fibrostenosis. Am J Pathol. 2012;180:636–49. doi: 10.1016/j.ajpath.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih DQ, Zheng L, Zhang X, et al. Inhibition of a novel fibrogenic factor Tl1a reverses established colonic fibrosis. Mucosal Immunol. 2014;7:1492–503. doi: 10.1038/mi.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meylan F, Song YJ, Fuss I, et al. The TNF-family cytokine TL1A drives IL-13-dependent small intestinal inflammation. Mucosal Immunol. 2011;4:172–85. doi: 10.1038/mi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takedatsu H, Michelsen KS, Wei B, et al. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–67. doi: 10.1053/j.gastro.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papadakis KA, Prehn JL, Landers C, et al. TL1A synergizes with IL-12 and IL-18 to enhance IFN-gamma production in human T cells and NK cells. J Immunol. 2004;172:7002–7. doi: 10.4049/jimmunol.172.11.7002. [DOI] [PubMed] [Google Scholar]
- 13.Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–67. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu Y, Zhou B, Yang W, et al. Transcriptome and proteome characterization of surface ectoderm cells differentiated from human iPSCs. Sci Rep. 2016;6:32007. doi: 10.1038/srep32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dezorella N, Kay S, Baron S, et al. Measurement of lymphocyte aggregation by flow cytometry-physiological implications in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2016;90:257–66. doi: 10.1002/cyto.b.21263. [DOI] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Zhan X, Liang L, et al. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96:926–37. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–9. doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–9. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleynen I, Gonzalez JR, Figueroa C, et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut. 2013;62:1556–65. doi: 10.1136/gutjnl-2011-300777. [DOI] [PubMed] [Google Scholar]
- 25.Acquati F, Lualdi M, Bertilaccio S, et al. Loss of function of Ribonuclease T2, an ancient and phylogenetically conserved RNase, plays a crucial role in ovarian tumorigenesis. Proc Natl Acad Sci U S A. 2013;110:8140–5. doi: 10.1073/pnas.1222079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monti L, Rodolfo M, Lo Russo G, et al. RNASET2 as a tumor antagonizing gene in a melanoma cancer model. Oncol Res. 2008;17:69–74. doi: 10.3727/096504008784523658. [DOI] [PubMed] [Google Scholar]
- 27.Gaidano G, Hauptschein RS, Parsa NZ, et al. Deletions involving two distinct regions of 6q in B-cell non-Hodgkin lymphoma. Blood. 1992;80:1781–7. [PubMed] [Google Scholar]
- 28.Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–67. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu X, Pan CM, Zhao SX, et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat Genet. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 30.Taylor KD, Plevy SE, Yang H, et al. ANCA pattern and LTA haplotype relationship to clinical responses to anti-TNF antibody treatment in Crohn’s disease. Gastroenterology. 2001;120:1347–55. doi: 10.1053/gast.2001.23966. [DOI] [PubMed] [Google Scholar]
- 31.Siegel CA, Whitman CB, Spiegel BM, et al. Development of an index to define overall disease severity in IBD. Gut. 2016 doi: 10.1136/gutjnl-2016-312648. [DOI] [PubMed] [Google Scholar]
- 32.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zumwalde NA, Domae E, Mescher MF, et al. ICAM-1-dependent homotypic aggregates regulate CD8 T cell effector function and differentiation during T cell activation. J Immunol. 2013;191:3681–93. doi: 10.4049/jimmunol.1201954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabatos CA, Doh J, Chakravarti S, et al. A synaptic basis for paracrine interleukin-2 signaling during homotypic T cell interaction. Immunity. 2008;29:238–48. doi: 10.1016/j.immuni.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerard A, Khan O, Beemiller P, et al. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nat Immunol. 2013;14:356–63. doi: 10.1038/ni.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prehn JL, Thomas LS, Landers CJ, et al. The T cell costimulator TL1A is induced by FcgammaR signaling in human monocytes and dendritic cells. Journal of Immunology. 2007;178:4033–8. doi: 10.4049/jimmunol.178.7.4033. [DOI] [PubMed] [Google Scholar]
- 38.Shih DQ, Kwan LY, Chavez V, et al. Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. European Journal of Immunology. 2009;39:3239–50. doi: 10.1002/eji.200839087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih DQ, Barrett R, Zhang X, et al. Constitutive TL1A (TNFSF15) expression on lymphoid or myeloid cells leads to mild intestinal inflammation and fibrosis. PLoS One. 2011;6:e16090. doi: 10.1371/journal.pone.0016090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabrielsen IS, Amundsen SS, Helgeland H, et al. Genetic risk variants for autoimmune diseases that influence gene expression in thymus. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw152. [DOI] [PubMed] [Google Scholar]
- 41.Di Narzo AF, Peters LA, Argmann C, et al. Blood and Intestine eQTLs from an Anti-TNF-Resistant Crohn’s Disease Cohort Inform IBD Genetic Association Loci. Clin Transl Gastroenterol. 2016;7:e177. doi: 10.1038/ctg.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutgeerts P, Geboes K, Vantrappen G, et al. Predictability of the postoperative course of Crohn’s disease. Gastroenterology. 1990;99:956–63. doi: 10.1016/0016-5085(90)90613-6. [DOI] [PubMed] [Google Scholar]
- 43.Chen XJ, Gong XH, Yan N, et al. RNASET2 tag SNP but not CCR6 polymorphisms is associated with autoimmune thyroid diseases in the Chinese Han population. BMC Med Genet. 2015;16:11. doi: 10.1186/s12881-015-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Wang X, Xiang L. Role and Mechanism of RNASET2 in the Pathogenesis of Vitiligo. J Investig Dermatol Symp Proc. 2015;17:48–50. doi: 10.1038/jidsymp.2015.24. [DOI] [PubMed] [Google Scholar]
- 45.Wenzke KE, Cantemir-Stone C, Zhang J, et al. Identifying common genes and networks in multi-organ fibrosis. AMIA Jt Summits Transl Sci Proc. 2012;2012:106–15. [PMC free article] [PubMed] [Google Scholar]
- 46.Fuyuno Y, Yamazaki K, Takahashi A, et al. Genetic characteristics of inflammatory bowel disease in a Japanese population. J Gastroenterol. 2016;51:672–81. doi: 10.1007/s00535-015-1135-3. [DOI] [PubMed] [Google Scholar]
- 47.Lualdi M, Pedrini E, Rea K, et al. Pleiotropic modes of action in tumor cells of RNASET2, an evolutionary highly conserved extracellular RNase. Oncotarget. 2015;6:7851–65. doi: 10.18632/oncotarget.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesiel-Nuttman L, Doron S, Schwartz B, et al. Human RNASET2 derivatives as potential anti-angiogenic agents: actin binding sequence identification and characterization. Oncoscience. 2015;2:31–43. doi: 10.18632/oncoscience.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grakoui A, Bromley SK, Sumen C, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 50.Verma NK, Fazil MH, Ong ST, et al. LFA-1/ICAM-1 Ligation in Human T Cells Promotes Th1 Polarization through a GSK3beta Signaling-Dependent Notch Pathway. J Immunol. 2016;197:108–18. doi: 10.4049/jimmunol.1501264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


