Abstract
The Liver Kinase B1 (LKB1) tumor suppressor acts as a metabolic energy sensor to regulate AMP-activated protein kinase (AMPK) signaling and is commonly mutated in various cancers, including non-small cell lung cancer (NSCLC). Tumor cells deficient in LKB1 may be uniquely sensitized to metabolic stresses, which may offer a therapeutic window in oncology. To address this question we have explored how functional LKB1 impacts the metabolism of NSCLC cells using 13C metabolic flux analysis. Isogenic NSCLC cells expressing functional LKB1 exhibited higher flux through oxidative mitochondrial pathways compared to those deficient in LKB1. Re-expression of LKB1 also increased the capacity of cells to oxidize major mitochondrial substrates, including pyruvate, fatty acids, and glutamine. Furthermore, LKB1 expression promoted an adaptive response to energy stress induced by anchorage-independent growth. Finally, this diminished adaptability sensitized LKB1-deficient cells to combinatorial inhibition of mitochondrial complex I and glutaminase. Together, our data implicate LKB1 as a major regulator of adaptive metabolic reprogramming and suggest synergistic pharmacological strategies for mitigating LKB1-deficient NSCLC tumor growth.
Keywords: LKB1, cancer metabolism, mitochondria, metabolic flux analysis, phenformin, glutaminase
Introduction
All cells within the human body exhibit distinct metabolic states governed by their environment, tissue of origin, and function (Metallo and Vander Heiden, 2013). Cells must therefore detect and respond to signals indicative of nutrient availability and bioenergetics to reprogram their metabolism and maintain critical tissue functions. Tumor cells must also sense changes in their microenvironment (e.g. nutrient levels, oxygen availability, endocrine signals) in order to support the metabolic demands of unfettered proliferation. An inability to respond to such environmental and intracellular cues can result in energetic stress and ultimately death. A detailed understanding of the molecular pathways involved in cellular stress responses may reveal metabolic dependencies that can be exploited therapeutically.
The tumor suppressor liver kinase B1 (LKB1) is a serine/threonine kinase that lies upstream of AMP-activated protein kinase (AMPK) and is responsible for sensing cellular ATP availability (Shaw et al., 2004). Germline mutations in LKB1 occur in patients with Peutz-Jeghers syndrome (PJS), a disease characterized by the accumulation of noncancerous gastrointestinal polyps and increased risk of cancer (Avizienyte et al., 1999; Bartosova et al., 2007; Hemminki et al., 1998). In addition, somatic mutations in LKB1 occur at high frequencies in human lung, cervical, and breast cancers (Contreras et al., 2008; Sanchez-Cespedes et al., 2002; Shackelford and Shaw, 2009; Wingo et al., 2009). Glucose deprivation, anchorage-independence, EGFR inhibition, and/or biguanide treatment have been shown to activate LKB1-AMPK signaling in cancer cells (Hawley et al., 2002; Jeon et al., 2012; Shaw et al., 2004; Shaw et al., 2005; Whang et al., 2016). In normal cells, AMPK activation results in stimulation of bioenergetic pathways and inhibition of ATP-consuming processes such as biosynthesis and proliferation, in part through regulation of the mammalian target of rapamycin complex 1 (mTORC1) pathway (Bolster et al., 2002; Kimura et al., 2003; Krause et al., 2002). On the other hand, LKB1-deficient tumors exhibit hyper-activated mTORC1 and elevated hypoxia inducible factor (HIF) signaling which, in turn, stimulates aerobic glycolysis and lowers reliance on OXPHOS (Faubert et al., 2014; Shackelford et al., 2009). However, loss of LKB1 also leads to dysfunctional mitochondria and metabolic dysregulation that renders LKB1-deficient tumors hyper-sensitive to pharmacological agents that induce energy stress (Carretero et al., 2007; Jeon et al., 2012; Shackelford et al., 2013; Shaw et al., 2005; Whang et al., 2016).
Anchorage-independence and resistance to anoikis (i.e. apoptosis due to loss of matrix attachment) is a common characteristic of tumor cells, as non-transformed cells rapidly undergo death upon detachment from the ECM. Matrix detachment dramatically reduces glucose uptake, pyruvate dehydrogenase (PDH) flux, and both ATP and NADPH levels in non-transformed cells, while oncogenic factors, such as ERBB2, or antioxidants confer resistance to anoikis by promoting metabolic pathways that sustain ATP and NADPH levels (Grassian et al., 2011; Schafer et al., 2009). Previous studies have shown that AMPK activation, particularly through mTOR inhibition, contributes to anoikis resistance in MEF and NIH3T3 cells transformed with Kras(V12) or oncogenic kinases such as the chimeric tyrosine kinase ETV6-NTRK3 (EN) (Ng et al., 2012). Furthermore, LKB1-deficient non-small cell lung cancer (NSCLC) cells under glucose-deprived or anchorage-independent conditions experience redox stress and undergo apoptosis, presumably due to the inability of such cells to reprogram central metabolism (Jeon et al., 2012). Therefore, a quantitative analysis of how intracellular metabolic fluxes in LKB1-proficient and -deficient NSCLC cells change in response to such stresses may provide insights into potential single agent or combination therapies.
Here we have applied 13C metabolic flux analysis (MFA) to characterize the metabolic impacts of LKB1-deficiency in response to energy stressors, including anchorage-independence and mitochondrial inhibitors. Using isogenic NSCLC cell lines, we observed that LKB1-proficient cells exhibit higher oxidative mitochondrial flux and greater flexibility with respect to mitochondrial substrate utilization as compared to LKB1-deficient cells. Furthermore, LKB1-proficient cells were more flexible with respect to mitochondrial substrate utilization in response to various stresses. This decreased metabolic plasticity resulted in an increased sensitivity of LKB1-deficient cells to combination treatments with phenformin and glutaminase inhibitors. Together, these data provide insights into how loss of LKB1 may be exploited to mitigate the growth and survival of LKB1-deficient tumors.
Results
LKB1 deficiency decreases oxidative mitochondrial metabolism
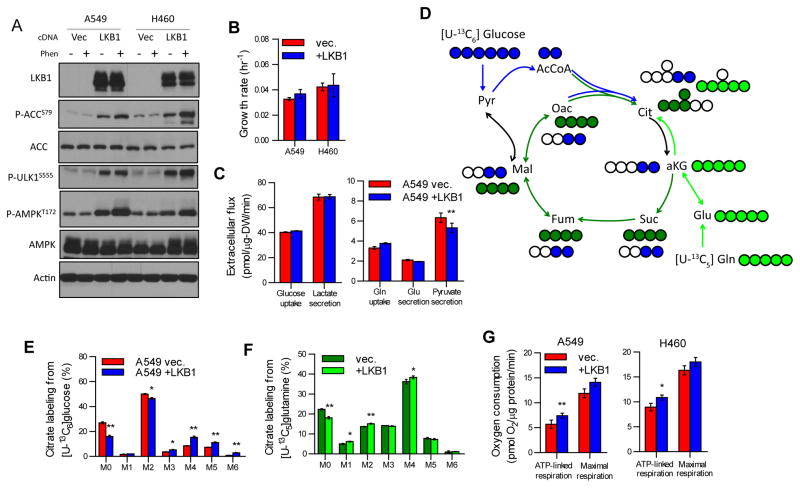
In order to investigate how loss of LKB1 contributes to metabolic reprogramming in NSCLC cells, we stably expressed wild-type LKB1 (+LKB1) or an empty vector (vec.) in the LKB1-deficient A549 and H460 cell lines and characterized AMPK signaling, cell proliferation, and central carbon metabolism in each. In cells expressing wild-type LKB1 we observed functional AMPK signaling, as evidenced by increased phosphorylation of acetyl-coenzyme A carboxylase 1 (ACC1), ULK1, and AMPK (Figure 1A). Notably, ectopic expression of functional LKB1 had minimal impact on cell proliferation in either cell line (Figure 1B). Consistent with the similar growth rates observed in each cell line, LKB1 expression had no significant impact on glucose and glutamine uptake as well as lactate and glutamate secretion (Figure 1C). However, a significant decrease in pyruvate secretion was observed in A549 +LKB1 cells, suggesting mitochondrial pyruvate metabolism was impacted by the re-introduction of functional LKB1 in these cells (Figure 1C).
Fig 1. LKB1 promotes oxidative mitochondrial metabolism.
(A) Immunoblot of LKB1, phosphorylated ACC (S79), total ACC, phosphorylated ULK1 (S555), phosphorylated AMPK (T172), total AMPK, and β-actin on lysates from A549 and H460 cells stably expressing either empty pBabe vector (vec.) or pBabe-LKB1 (LKB1). Lysates from cells treated with vehicle or phenformin (Phen, 2mM) for 4 hours prior to lysis. (B) Maximal exponential growth rate of A549 and H460 vec. and +LKB1 cells. (C) Extracellular flux of glucose, lactate, glutamine, glutamate, and pyruvate in A549 vec. and +LKB1 cells. (D) Atom transition map of [U-13C6]glucose (blue circles) and [U-13C5]glutamine (green circles) used to detect changes in mitochondrial metabolism. Filled circles indicate 13C enrichment. (E) Citrate labeling from [U-13C6]glucose in A549 vec. and +LKB1 cells cultured in ddFBS supplemented with 20 μM palmitate conjugated to BSA for 24 hours. (F) Citrate labeling from [U-13C5]glutamine in A549 vec. and +LKB1 cells cultured in ddFBS supplemented with 20 μM palmitate conjugated to BSA for 24 hours. (G) ATP-linked and maximal FCCP-uncoupled respiration in A549 and H460 vec. and +LKB1 cells.
To characterize the changes in intracellular metabolic pathways we next applied 13C stable-isotope tracing and mass spectrometry to this cell system. Each cell line was cultured in the presence of [U-13C6]glucose (uniformly 13C-labeled glucose), and isotope enrichment (designated as M+0, M+1, M+2, etc. mass isotopologues representing ion fragments containing zero, one, or two 13C atoms, respectively) was quantified using gas chromatography/mass spectrometry (Figure 1D). Significant incorporation of 13C from [U-13C6]glucose was detected in citrate with M+2—derived from flux through pyruvate dehydrogenase and glutamine anaplerosis—being the most highly labeled species (Figure 1E). Citrate isotopologues indicating TCA cycling and glucose anaplerosis (M+4 and M+5, respectively) were significantly higher in LKB1-proficient A549 cells, suggesting increased glucose oxidation in mitochondria (Figure 1E). Furthermore, the mole percent enrichment (MPE) from [U-13C6]glucose of all measured TCA intermediates and associated amino acids was increased (Figure S1A). Given the elevated glucose utilization observed in LKB1-proficient cells, we next investigated whether glutamine metabolism was influenced by LKB1 using [U-13C5]glutamine (Figure 1D). LKB1 deficiency caused only minor changes in the relative extent of glutamine anaplerosis, indicated by similar M+5 labeling of α-ketoglutarate (αKG) and mole percent enrichment of TCA cycle intermediates from [U-13C5]glutamine (Figures S1B–C). Furthermore, LKB1 deficient cells displayed decreased oxidative TCA cycle metabolism as evidenced by significantly decreased M+3 αKG and M+4 citrate isotopologues (Figure 1F and S1B).
Respirometry analysis indicated that ATP-linked oxygen consumption was significantly elevated in LKB1-proficient cells, further highlighting the impact of LKB1 expression on oxidative mitochondrial metabolism (Figure 1G). Previous reports by Faubert et al. carried out in MEF and A427 cells found that cellular respiration was not significantly affected by LKB1 loss (Faubert et al., 2014); however, this measurement represents basal oxygen consumption and includes ATP-linked, proton leak-linked, and non-mitochondrial respiration (Divakaruni et al., 2014a). Here we see that LKB1-proficient A549 and H460 cells exhibit a significant increase in oxygen consumption specifically used for mitochondrial ATP production (Figure 1G).
LKB1 expression reprograms mitochondrial metabolism
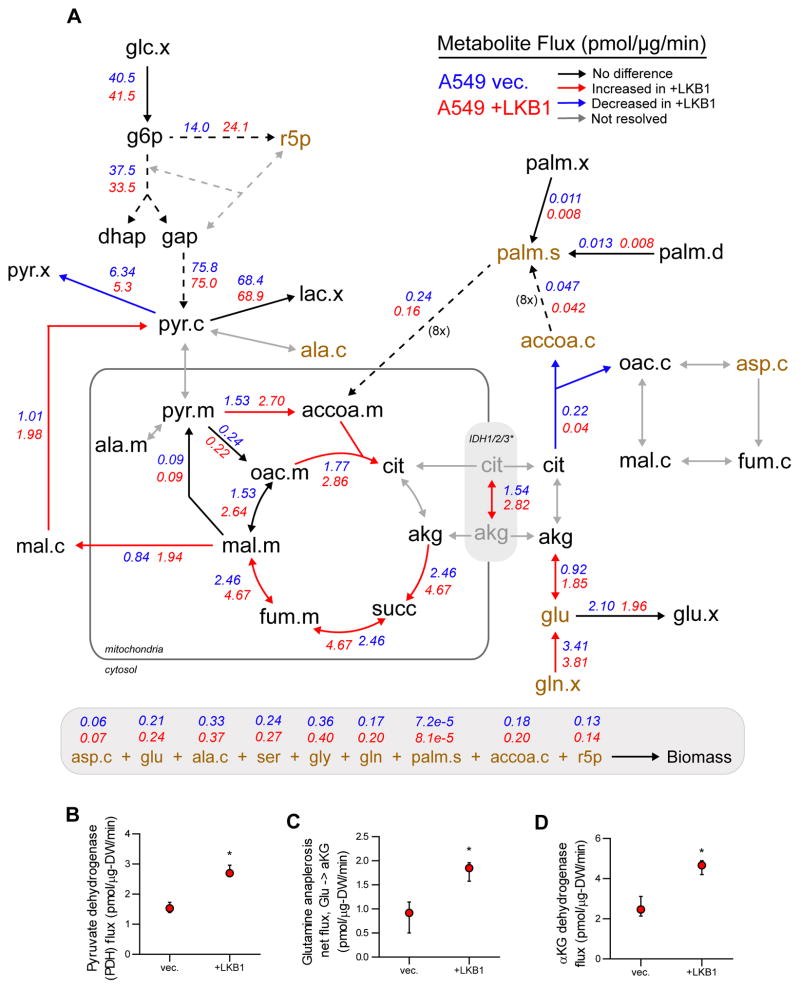
These results highlight the need for comprehensive intracellular flux estimation beyond analysis of substrate utilization, as intracellular (but not extracellular) measurements of metabolism suggest regulation downstream of functional LKB1 expression. Therefore, we employed 13C Metabolic Flux Analysis (MFA) to quantify fluxes throughout central carbon metabolism. Uptake and secretion of major cellular substrates/byproducts such as glucose, lactate, glutamine, glutamate, and pyruvate were incorporated along with isotope enrichment data into a model encompassing glycolysis, the pentose phosphate pathway (PPP), TCA metabolism, fatty acid and biomass biosynthesis, and β-oxidation. Our stable-isotope tracing studies indicated that LKB1 expression had significant impacts on pyruvate and TCA metabolism; therefore, we included isotopologue distributions generated when tracing with either [U-13C6]glucose or BSA-conjugated [U-13C16]palmitate to better resolve flux through pathways fueling tricarboxylic acid (TCA) metabolism. Shunting of glycolytic intermediates through the oxidative branch of the PPP was determined using [1,2-13C2]glucose and incorporated into the model (Figure S2A–B). Finally, an elementary metabolite unit (EMU)-based algorithm was used to estimate fluxes through the metabolic network and associated confidence intervals by assessing flux fit sensitivity to minor flux deviations (Figure 2A) (Antoniewicz et al., 2006; Young, 2014; Young et al., 2008). A detailed description of model parameters and assumptions and tables containing flux estimations are included in Supplementary Materials.
Fig 2. Reprogramming of mitochondrial fluxes by functional LKB1 expression.
(A) Schematic of central carbon metabolism with net flux values estimated by 13C MFA for A549 vec. and +LKB1 cells assigned where resolved. Net flux values for A549 vec. and +LKB1 cells are listed in blue and red, respectively. Arrows are colored to depict significantly different fluxes (based on 95% confidence intervals) and those which could not be resolved by 13C MFA. Dashed arrows represent reactions consisting of multiple steps. The IDH1/2/3* flux represents a single resolved fluxes for the isocitrate dehydrogenase reaction as separate cytosolic IDH1 and mitochondrial IDH2/3 fluxes were not resolved. Metabolites required for biomass synthesis to support proliferation are colored in brown, and sink fluxes of each biomass metabolite are listed below schematic. (B–D) Pyruvate dehydrogenase (B), glutamine anaplerosis (C), and α-ketoglutarate dehydrogenase (D) flux estimates and 95% confidence intervals quantified by the 13C MFA model. Data in B, C, E, F represent mean ± s.d. of at least three replicates. Data in G represent mean ± SEM of four independent experiments. Data in B–D represent mean ± 95% confidence intervals.
Results indicated that molar flux through reactions fueling TCA metabolism, including PDH and glutamine anaplerosis, were significantly higher in LKB1-proficient cells (Figure 2A–C). Furthermore, LKB1-proficient cells exhibited a significantly increased flux through the oxidative TCA cycle, as evidenced by increased αKG-dehydrogenase flux (Figure 2A&D), in agreement with the observed increase in ATP-linked oxygen consumption (Figure 1G). Although LKB1 expression had no effect on cell growth, these results suggest that LKB1-proficient cells have a higher ATP demand, possibly due to increased flux through ATP-dependent processes. Strikingly, LKB1-proficient cells were significantly larger in size and had decreased dry cell weight relative to deficient cells suggesting differences in biomass composition (Figure S2C). These results suggest that LKB1-proficient cells reprogram metabolism to support oxidative mitochondrial metabolism to meet higher ATP demands (or turnover) within the cell.
LKB1 deficiency decreases metabolic flexibility
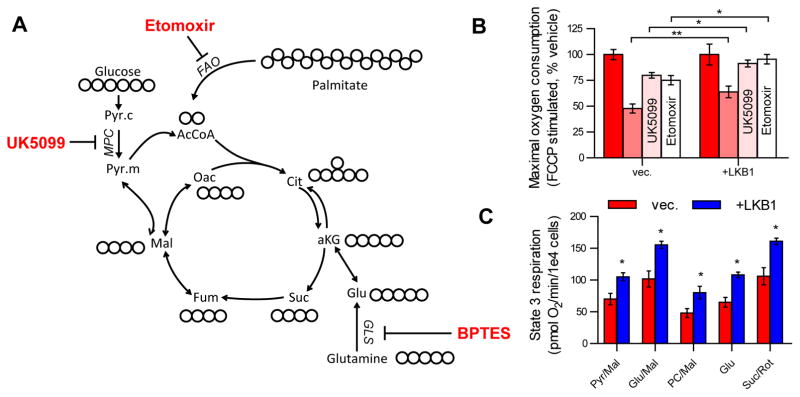
Given the role of LKB1-AMPK signaling in regulating ATP metabolism in response to bioenergetic stress, we next evaluated the metabolic flexibility of LKB1-proficient and -deficient cells in response to pharmacological inhibition of multiple mitochondrial substrate entry points, specifically glutamine, pyruvate, and long chain fatty acids. In this vein, we measured maximal uncoupled respiration after inhibition of glutaminase (BPTES), the mitochondrial pyruvate carrier (MPC; UK5099), or carnitine palmitoyltransferase 1 (CPT1; etomoxir) in A549 vec. and +LKB1 cells (Figure 3A). We observed that maximal respiration in LKB1-proficient A549 cells was less sensitive to any single inhibitor compared to LKB1-deficient cells, suggesting that LKB1 promotes flexibility (or the use of an alternative, untested substrate) in reprogramming metabolism to maintain respiration in response to pharmacological stress (Figure 3B). We also investigated the capacity of permeabilized cells to oxidize major mitochondrial substrates by measuring maximal, uncoupled respiration in the presence of substrates that feed electrons to complex I (pyruvate/malate, glutamate, glutamate/malate, palmitoyl-carnitine/malate) or complex II (succinate/rotenone). Notably, we observed that mitochondria of LKB1-proficient A549 cells were more capable of oxidizing all substrates (Figure 3C). Together, these data suggest that LKB1 promotes metabolic flexibility by maintaining mitochondrial populations capable of responding to stresses that limit substrate availability.
Fig 3. LKB1 increases the metabolic flexibility of NSCLC cells.
(A) Map of central carbon metabolism indicating pharmacological targets of major substrate metabolic pathways. UK5099 inhibits pyruvate metabolism via the mitochondrial pyruvate carrier (MPC), etomoxir inhibits fatty acid oxidation (FAO) via carnitine palmitoyltransferase 1 (CPT1), BPTES inhibits glutamine metabolism via glutaminase (GLS). (B) Maximal FCCP-uncoupled respiration in A549 vec. and +LKB1 cells treated with vehicle, BPTES (3μM), UK5099 (5μM), or etomoxir (5μM). (C) Permeabilized cell respiration in A549 vec. and +LKB1 cells fed with saturating concentrations of specific substrates listed. All data represent mean ± SEM of four independent experiments.
LKB1 expression promotes metabolic flexibility in conditions of energy stress
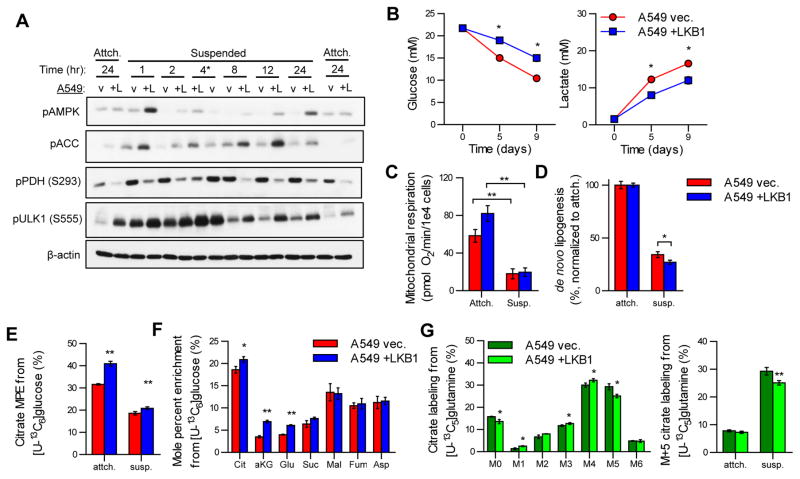
While the above studies were conducted in actively proliferating cells, cancer cells must also sense and respond to exogenous stresses to survive. Given the roles of LKB1-AMPK signaling in responses to changes in energy demand, including that driven by matrix detachment (Jeon et al., 2012), we next investigated how LKB1 expression impacts signaling events and metabolic reprogramming under anchorage-independent conditions by culturing cells in suspension. Significant changes in signaling downstream of LKB1 were induced upon detachment and prolonged culture in suspension. Phosphorylation of AMPK, ACC1, and ULK1 were all induced rapidly under anchorage-independent growth conditions, with significantly more pronounced effects in LKB1-proficient cells, demonstrating that cell detachment promotes LKB1 activation (Figure 4A). On the other hand, phosphorylation of pyruvate dehydrogenase (pPDH), which inactivates the enzymatic activity of this complex, was slightly increased by anchorage-independent conditions regardless of LKB1 expression. However, pPDH was generally decreased in LKB1-proficient compared to LKB1-deficient cells (Figure 4A). Metabolically, LKB1-deficient A549 cells exhibited significantly higher glucose consumption and lactate secretion rates despite no significant difference in cell growth in anchorage-independent conditions (Figure 4B & S4A), consistent with the effects of pPDH on pyruvate metabolism. These data provide evidence that LKB1-deficient cells rely more on aerobic glycolysis for ATP generation compared to LKB1-proficient cells in suspension (Figure 4B). Importantly, the glucose uptake flux in suspension conditions was significantly decreased compared to attached conditions (Figure S4B) suggesting that overall the energy supply and/or demand is decreased in anchorage-independent conditions.
Fig 4. LKB1 coordinates metabolic reprogramming under anchorage-independent growth conditions.
(A) Immunoblot of phosphorylated AMPK (T172), phosphorylated ACC (S79), phosphorylated PDH (S293), phosphorylated ULK1 (S555), and β-actin on lysates from A549 vec. (v) and +LKB1 (+L) cells cultured in monolayer (Attch.) for 24 hours of in anchorage-independent conditions (Suspended) for 1, 2, 4, 8, 12, or 24 hours. A549 vec. and +LKB1 lysates switched at 4 hour time point indicated by *. (B) Glucose and lactate concentrations in spent media of A549 vec. and +LKB1 cells cultured in anchorage-independent conditions over 9 days. (C) Mitochondrial respiration of A549 vec. and +LKB1 cells cultured in attached or suspension conditions. (D) Percentage of fatty acids de novo synthesized in A549 vec. and +LKB1 cells cultured in attached or suspension conditions and normalized to attached values for each respective cell line. Values quantified using Isotopomer Spectral Analysis (ISA) on palmitate labeling from [U-13C6]glucose for 24 hours. (E) Mole percent enrichment (MPE) of citrate from [U-13C6]glucose in A549 vec. and +LKB1 cells cultured in attached or anchorage-independent conditions for 24 hours. (F) MPE of TCA intermediates from [U-13C6]glucose in A549 vec. and +LKB1 cells cultured in anchorage-independent conditions for 24 hours. (G) (left) Citrate labeling from [U-13C5]glutamine in A549 vec. and +LKB1 cells cultured in anchorage-independent conditions for 24 hours. (right) Citrate M+5 labeling from [U-13C5]glutamine in A549 vec. and +LKB1 cells cultured in attached or anchorage-independent conditions for 24 hours. All data (excluding B) are presented as mean ± s.d. of at least three replicates. Data presented in B is mean ± SEM of at least two independent experiments.
We next investigated the capacity of cells to reduce ATP demand in response to energy stress by measuring changes in ATP-linked respiration and de novo lipogenesis (DNL). After 24 hours of anchorage-independence, both cell lines similarly decreased mitochondrial respiration (Figure 4C), in agreement with previous reports of ATP levels and bioenergetic fluxes decreasing in cells cultured in suspension (Grassian et al., 2011; Schafer et al., 2009). Both cells also reduced lipid biosynthesis, as evidenced by decreased DNL measured by isotopomer spectral analysis (ISA) (Figure 4D). Notably, LKB1-proficient A549 cells decreased DNL (and its associated ATP and NADPH demands) to a greater extent than LKB1-deficient cells (Figure 4D), consistent with the observed changes in ACC1 phosphorylation that impact DNL (Figure 4A). Together, these results suggest that LKB1-deficient NSCLCs exhibit increased aerobic glycolysis to meet a greater ATP demand than LKB1-proficient cells in suspension.
To further characterize changes in mitochondrial metabolism we cultured cells with either [U-13C6]glucose or [U-13C5]glutamine for 24 hours in anchorage-independent conditions and quantified enrichment in TCA intermediates. Suspension culture reduced the extent of glucose oxidation in both cells, though LKB1-proficient cells exhibited more labeling from glucose under each condition (Figure 4E), which was expected given the observed differences in pPDH (Figure 4A&E). Indeed, isotope enrichment from [U-13C6]glucose was increased in citrate, αKG, and glutamate in LKB1-proficient cells under these anchorage-independent conditions (Figure 4F). On the other hand, no differences in enrichment were observed in metabolites downstream of ATP-citrate lyase (ACLY; i.e. malate, fumarate, and aspartate-a surrogate for oxaloacetate) (Figure 4D&F). These results suggest that LKB1 facilitates metabolic reprogramming to maintain glucose oxidation by reducing PDH phosphorylation and reducing lipid biosynthesis via ACC phosphorylation under anchorage-independent conditions.
Consistent with results in monolayer culture, we detected no significant difference in glutamine uptake rates under anchorage-independent conditions when comparing LKB1 -proficient and -deficient cells (Figure S4C). However, LKB1 status had significant impacts on intracellular glutamine metabolism in anchorage-independent conditions, as evidenced by labeling of citrate from [U-13C5]glutamine. In this environment both cells increased the percentage of citrate produced from this pathway (Figure 4G), consistent with that observed in other NSCLC cells (Jiang et al., 2016). However, LKB1-proficient cells exhibited a significant decrease in M+5 citrate generated via reductive carboxylation compared to LKB1-deficient cells in anchorage-independent conditions (Figure 4G). These results are consistent with the consequences of increased pPDH and decreased pACC in LKB1-deficient cells, as both would be expected to increase the extent of reductive carboxylation (Figure 4A) (Metallo et al., 2012; Zhang et al., 2016). Together, these data suggest that LKB1-proficient cells more effectively maintain oxidative mitochondrial pathway activity in response to anchorage-independent growth.
LKB1 deficiency sensitizes cells to inhibition of glutamine and mitochondrial metabolism
Our results suggest that expression of functional LKB1 allows cancer cells to be more flexible with respect to mitochondrial metabolic pathway utilization in response to restricted substrate provision by pharmacological agents or changes in energy demand induced by anchorage-independence. Previous studies have shown that LKB1-deficiency sensitizes cells to phenformin, an inhibitor of complex I within the electron transport chain, in part due to an inability to activate ULK1-mediated mitophagy (Shackelford et al., 2013). Studies into the mechanism and specificity of phenformin on mitochondrial function have identified complex I being the primary point of inhibition (Drahota et al., 2014). We observed a cytostatic response in A549 and H460 cells to phenformin at low micromolar concentrations (data not shown) and significant apoptosis at >1 mM concentrations (Figure 5A). Notably, LKB1-deficient cells were significantly more sensitive to phenformin treatment at millimolar doses of phenformin (Figure 5A), consistent with previous studies (Shackelford et al., 2013). These results suggest that phenformin may have multiple impacts on mitochondrial physiology and cell signaling pathways not limited to complex I inhibition, as 50–100 μM treatments inhibited TCA metabolism but not survival while mM concentrations induced cell death. In order to test this, we cultured A549 and H460 cells with cytostatic concentrations of phenformin in the presence of [U-13C5]glutamine to determine if inhibition of the TCA cycle occurred. Indeed, we failed to detect M+3 labeling in αKG, which was almost entirely comprised of M+5 isotopologues, indicating significant inhibition of TCA cycling (Figure 5B–C). Consistent with previous studies using other Complex I inhibitors (Fendt et al., 2013), we observed a marked increase in reductive carboxylation upon phenformin treatment as evidenced by increases in M+3 labeling of fumarate, malate, and aspartate from [U-13C5]glutamine (Figure S5A).
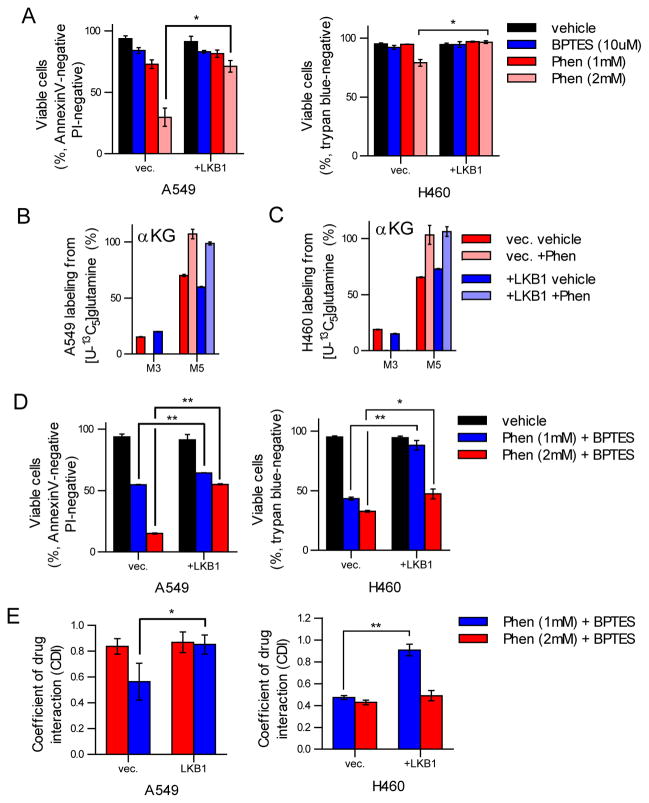
Fig 5. LKB1 deficiency sensitizes cells to inhibition of Complex I and glutaminase.
(A) Cell viability after treatment with vehicle, BPTES (10 μM), or phenformin (1mM or 2mM) after 48 hours treatment. (B–C) Glutamate and α-ketoglutarate (αKG) labeling from [U-13C5]glutamine after treatment with vehicle or phenformin (50 μM) in A549 (B) and H460 (C) vec. and +LKB1 cells. (D) Cell viability after treatment with vehicle, phenformin (1mM) and BPTES (10μM), or phenformin (2mM) and BPTES (10μM) in A549 and H460 vec. and +LKB1 cells. (E) Coefficient of drug interaction (CDI) values for A549 and H460 vec. and +LKB1 cells treated with either phenformin (1mM) and BPTES (10μM) or phenformin (2mM) and BPTES (10μM).
Given that phenformin treatment induced significant reprogramming of TCA metabolism, in particular reductive glutamine metabolism, and LKB1-deficient cells are significantly less metabolically plastic, we hypothesized that LKB1-deficient cells may be more sensitive to combination treatments with glutaminase inhibitors. We treated A549 and H460 cells with single agent BPTES or a combination of BPTES and phenformin and measured cellular viability after 48 hour treatment. Notably, we observed only modest impacts on cellular viability with single agent BPTES treatment in either LKB1-proficient or -deficient A549 or H460 cells (Figure 5A). However, treatment with both phenformin and BPTES yielded significant impacts on cellular viability, with LKB1-deficient cell lines being most sensitive (Figure 5D). Combination index and coefficient of drug interaction (CDI) were calculated to assess potential synergistic effects of phenformin and BPTES on cell viability, where CDI <0.7 indicates a significant synergy (Zhao et al., 2014). Strikingly, we observed that combination treatment with phenformin and BPTES yielded a synergistic response more so in LKB1-deficient cells (Figure 5E). Taken together, these results suggest that phenformin treatment sensitizes LKB1-deficient cells to inhibition of glutamine metabolism. Furthermore, inhibition of multiple mitochondrial substrate entry points may selectively inhibit the growth of less metabolically plastic tumors.
Discussion
Here we demonstrate that LKB1 impacts the metabolic stress response of NSCLC cells, promoting enhanced metabolic flexibility and oxidative mitochondrial flux using numerous substrates. Functional LKB1 expression enabled cells to maintain healthy mitochondrial populations capable of oxidizing pyruvate, glutamine, and fatty acids more effectively than mitochondria in LKB1-deficient cells. Such activity may facilitate the ability of cells to toggle between nutrients in response to stress. Indeed, LKB1-proficient cells more effectively controlled the balance between glycolysis and oxidative phosphorylation through reprogramming of mitochondrial metabolism downstream of intact LKB1-AMPK signaling during anchorage-independent growth. Stress due to nutrient deficiencies or the microenvironment can cause mitochondrial damage, fragmentation, and dysfunction (Mishra and Chan, 2014). AMPK signaling downstream of LKB1 activates ULK1-mediated mitophagy to maintain healthy mitochondrial populations (Egan et al., 2011; Kim et al., 2011) and triggers mitochondrial fission via AMPK-dependent phosphorylation of MFF (Toyama et al., 2016). Mitochondrial fission is believed to be a necessary first step for proper mitophagy, thus AMPK dependent phosphorylation of MFF and ULK1 allows for a coordination of these two facets of proper mitochondrial turnover in response to mitochondrial stressors. Indeed, electron micrograph studies in cells lacking either AMPK or ULK1 revealed an accumulation of mitochondria (Egan et al., 2011), suggesting that cells actively manage mitochondrial populations and those lacking this machinery accumulate dysfunctional mitochondria. Indeed, we observed that LKB1-proficient cells sustained a high level of ULK1 phosphorylation upon growth in anchorage-independent conditions. While LKB1-proficient cells exhibit higher mitochondrial flux under adherent growth conditions, these cells more strongly decreased oxidative glucose metabolism in suspension. These results therefore further link LKB1-mediated signaling and downstream mitophagy to metabolic reprogramming in response to anchorage-independent growth.
The metabolic impact of anchorage-independent growth is well-documented, influencing the redox state and ATP levels of cells (Grassian et al., 2011; Jiang et al., 2016; Schafer et al., 2009). LKB1-deficiency therefore compromises the cellular response to such stresses (Jeon et al., 2012; Momcilovic et al., 2015). We observed that LKB1-proficient cells exhibit increased maximal respiration on numerous mitochondrial substrates and in response to various inhibitors as compared to LKB1-deficient NSCLC cells, suggesting a greater capacity to oxidize multiple mitochondrial substrates in the former population. Ultimately, the metabolic inflexibility displayed by LKB1-deficient cells contributes to their increased susceptibility to the biguanide phenformin and combinatorial treatment with a glutaminase inhibitor. Notably, patients taking another biguanide, metformin, have a significantly reduced risk of cancer (Evans et al., 2005; Landman et al., 2010), sparking interest in metformin and the more cell permeable analog phenformin, as anti-cancer agents (Appleyard et al., 2012). Our results, in agreement with previous reports, indicate that LKB1-deficient cancer cells are significantly more sensitive to phenformin treatment (Shackelford et al., 2013). Biochemically, we demonstrate that phenformin treatment induces significant metabolic reprogramming, increasing reductive carboxylation while inhibiting oxidative TCA metabolism. Indeed, studies into the metabolic effects of biguanides noted a significant depletion of TCA cycle intermediates in breast cancer stem cells, suggesting phenformin treatment significantly impacts mitochondrial metabolism (Janzer et al., 2014). Our results suggest that phenformin treatment at low doses results in complete inhibition of oxidative TCA cycling, further supporting the proposed mechanism of phenformin acting as an ETC inhibitor. Owing to the induction of aerobic glycolysis by phenformin treatment, recent studies have investigated combination strategies using phenformin and oxamate, an inhibitor of lactate dehydrogenase, as an anti-cancer therapeutic strategy (Miskimins et al., 2014). In addition to increased glycolytic flux, we observed that phenformin induces an increased reliance on reductive glutamine metabolism. Given the decreased plasticity of LKB1-deficient cells, we investigated whether combination treatments using phenformin and a glutaminase inhibitor would enhance the cell-killing effects in these cell populations. We observed that BPTES and phenformin synergistically decreased NSCLC cell survival, as evidenced by coefficient of drug interaction (CDI) scores <0.7 in A549 and H460 cell lines (Zhao et al., 2014). Strikingly, LKB1-deficient A549 and H460 cells were significantly more sensitive to combination treatment with phenformin and BPTES compared to LKB1-proficient cells. Notably, a similar strategy employing phenformin and the mTOR catalytic kinase inhibitor MLN0128 have shown efficacy in vitro and in vivo (Momcilovic et al., 2015). Combination treatment with metformin and glutaminase inhibitors has been reported to have antiproliferative effects in prostate cancer cells (Fendt et al., 2013). Furthermore, a more recent study aimed at improving the pharmacokinetics of BPTES by loading into PLGA-PEG nanoparticles (BPTES-NP) reported synergistic effects of BPTES-NP and metformin treatment in orthotopic models of pancreatic cancer (Elgogary et al., 2016). Taken together, these studies suggest that biguanides and glutaminase inhibitors may prove a rational combinatorial antineoplastic therapy for the treatment of many models of cancer.
In contrast to previous observations in distinct cell lines (Faubert et al., 2014), we observed significant increases in the respiration of LKB1-proficient A549 and H460 cells, which resulted in estimations of increased glutamine anaplerosis, PDH, and TCA fluxes in cells with functional LKB1. These discrepancies may be due to distinctions across cell lines or methodological differences and warrant further investigation. Collectively, our metabolic studies demonstrate that LKB1 promotes increased metabolic plasticity in response to bioenergetic stress, and LKB1-deficient cells lack this reprogramming ability, sensitizing them to pharmacological agents that induce metabolic stress. Further investigations into potential synergy between phenformin (or metformin) and glutaminase inhibitors are needed, particularly using in vivo tumor models of NSCLC. Indeed, while this report and others observe minimal impacts of LKB1 expression on cell proliferation in culture (Faubert et al., 2014; Whang et al., 2016), several studies using in vivo cancer models have indicated that LKB1 loss significantly enhances tumor growth (Ji et al., 2007; Kottakis et al., 2016; Shackelford et al., 2013). As significant differences exist in the metabolism of cancer cells growing in vivo versus in vitro (Davidson et al., 2016), care must be taken in translating these results without validating in animal models. However, our data suggest that energy stress induced by the microenvironment may play a significant role in how LKB1 regulates cancer cell metabolism and survival.
Materials and Methods
Cell Culture
For stable cell line generation, A549 and H460 cells were infected with retro-virus (pBabe-Hygro) expressing either empty vector control or full length wild-type human LKB1. Stable integrants were selected with 400 ug/ml of Hygromycin and maintained in Hygromycin for cell culture. Low attachment poly(2-hydroxyethyl methacrylate) coated plates were generated by addition of 7 mL of 1% solution in 95% ethanol to 10cm plates followed by evaporation on rocker for 2–3 days. Plates were irradiated to sterilize following deposition of poly-HEMA. Delipidated and dialyzed serum (ddFBS) was prepared by adding 20 mg/mL fumed silica (Sigma) to dialyzed FBS vortexed overnight at 4°C followed by centrifugation at 2000×g for 15 minute and sterile filtration.
Western Blotting
For biochemical analysis of cells, adherent cells were trypsinized, centrifuged and washed in phosphate buffered saline (PBS) and cells in suspension were centrifuged and washed in PBS. Cell lysates were then prepared in lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Trion X-100, 2.5 mM pyrophosphate, 50 mM NaF, 5mM B-glycero-phosphate, 50 nM Calyculin A, 1 mM Na3VO4 and a complete protease inhibitor cocktail (Roche)), centrifuged and equilibrated for protein levels using a BCA protein assay kit (Pierce). Lysates were resolved on 8% SDS-PAGE gels and immunblotted with the indicated antibodies. pACC (#3661), pULK (#5869), pAMPK (#2535), ACC (#3662) and AMPKα (#2532) antibodies were purchased from Cell Signaling Technologies (CST). pPDHS293 antibody (AP1062) was purchased from Millipore and the Actin antibody (#A5441) was purchased from Sigma-Aldrich.
Reagents
Stable isotopes used in these studies include [1,2-13C2]glucose, [U-13C6]glucose, [U-13C5]glutamine, and [U-13C16]palmitate all acquired from Cambridge Isotope Laboratories. Albumin, phenformin, palmitic acid, and BPTES were acquired from Sigma. Poly(2-hydroxyethyl methacrylate) was acquired from Sigma.
Metabolite Extraction and GC-MS Analysis
Polar and nonpolar metabolites were extracted using methanol/water/chloroform extraction as previously described (Metallo et al., 2012). Derivatization of both polar metabolites and fatty acids were conducted using MOX-tBDMCS derivatization as previously described (Lewis et al., 2014). Derivatized samples were analyzed by GC-MS using a DB-35MS column (30m x 0.25mm i.d. x 0.25 μm) installed in an Agilent 7890A gas chromatograph (GC) interfaced with an Agilent 5975C mass spectrometer (MS) as previously described (Grassian et al., 2014) and corrected for natural abundance using in-house algorithms adapted from (Fernandez et al., 1996)
13C Metabolic Flux Analysis
13C MFA was conducted using INCA, a software package based on elementary metabolite unit (EMU) framework (Young, 2014; Young et al., 2008). Flux through a metabolic network comprising of glycolysis, the pentose phosphate pathway, the TCA cycle, biomass synthesis, and fatty acid synthesis and oxidation were estimated by minimizing the sum of squared residuals between experimental and simulated MIDs and extracellular fluxes using nonlinear least squares regression for A549 LKB1- proficient and deficient cells traced with either [U-13C6]glucose or [U-13C16]palmitate conjugated with BSA (Antoniewicz et al., 2006). The best global fit was found after estimating 100 times using random initial guesses for all fluxes in the metabolic network. A χ2 statistical test was applied to assess the goodness-of-fit using α of 0.01. The 95% confidence intervals for each flux in the metabolic network were estimated by sensitivity analysis due to minor flux variations (Antoniewicz et al., 2006). See Supplementary Methods for further details on assumptions and raw flux values. Mole percent enrichment (MPE) represents the contribution of carbons from a specific source (i.e. glucose, glutamine) to a metabolite and is calculated using the following equation:
Where xi represents the fractional enrichment of an isotopologue (M1 + M2 + … Mn) summed over the number of carbons “n”. For example, the MPE of malate is calculated using the following equation:
Determination of Oxygen Consumption
Oxygen consumption rates were measured using a Seahorse XF96 or XFe96 analyzer. Cells were seeded into Seahorse XF96 plates at either 1.5×104 or 2.0×104 cells/well 24 hours prior to measurements. Respiration in intact cells was measured in DMEM (Sigma #5030) supplemented with 8mM glucose, 2mM pyruvate, 2mM glutamine, and 2mM HEPES. Where indicated, cells were pre-treated with 5μM UK5099 or 3μM BPTES 15 minutes prior to initial measurements. Respiratory rates were measured in response to sequential injections of oligomycin (2μM), FCCP (sequential additions of 400nM) and rotenone (0.5μM)/antimycin A (1μM). When measuring sensitivity to etomoxir (5μM), DMEM was instead supplemented with 0.5mM glucose, 0.25mM glutamine, 17μM albumin-buffered palmitate (Agilent Technologies; free [palmitate] ≈ 100nM), and 2mM HEPES. Data was analyzed and interpreted as in (Divakaruni et al., 2014a). For permeabilized cell respirometry, cells were permeabilized with 3 nM perfringolysin O (Divakaruni et al., 2013) (Agilent Technologies) and phosphorylating (“State 3”) respiration was measured as described in (Divakaruni et al., 2014b). Cells treated overnight on poly-HEMA were taken up, counted, and seeded at 8×104 cells/well onto Seahorse XF96 plates treated with Cell-Tak according to manufacturer’s instructions (Corning).
When correcting wells for protein content, cells were lysed with RIPA immediately after the experiment (20μL/well) and assayed using the BCA method (Pierce). When correcting for cell number, adherent monolayers in matched plates were trypsinized and counted with a Countess automated cell counter (Invitrogen). As confirmation, relative nuclear counts from plates used during assays were measured using CyQuant (Molecular Probes) to ensure any relative differences aligned with quantitative counts from trypsinized, matched plates.
Proliferation and Viability Assays
Cell growth was determined by plating cells in a 96-well dish at approximately 8000 cells/well and cultured for 0, 24, 48, 72 hours. At each time point, media was removed, plates were washed with PBS, cells were fixed in 4% paraformaldehyde solution in PBS for 15 minutes, and cells were dyed with 0.1% crystal violet solution in water for 30 minutes. To quantify relative cell density, dyed plates were washed with water to remove excess dye, plates were dried overnight, dye was solubilized in a solution of ethanol, methanol, and water, and absorbance was measured at approximately 595nm. Maximal cell growth rate was quantified by measuring the change in cell number after 72 hours and estimating growth rate value (hr−1) using an exponential growth model.
Dry weight was measured by isolating cells by trypsinization of monolayer cells followed by resuspension in media and centrifugation. Anchorage-independent cells were pelleted by centrifugation. Pellets were washed with PBS twice and dried at 50°C overnight in pre-weighed tubes.
For viability assays, cells were plated in 6-well plates and incubated in the presence of single or combination treatments for 48 hours following collection by trypsinization and resuspension in DMEM. Viability was determined by staining with AnnexinV and propidium iodide (PI) according to manufacturer instructions (Affymetrix 88-8007-72) and analyzed by FACScan flow cytometer (Becton Dickinson). Flow cytometry data was analyzed using FlowJo software to determine percentage of viable cells (unlabeled by either AnnexinV or PI). Viability was also determine by trypan blue dye exclusion by counting viable and apoptotic cells using a Countess automated cell counter (Thermo Fisher Scientific).
Drug synergism was determined using the combination index (CI) and coefficient of drug interaction (CDI) methods (Hao et al., 2008; Zhao et al., 2014). CI was quantified by calculating the ratio of response compared to vehicle treatment for each single treatment. CDI was calculated using the following equation:
Where CIA and CIB represent the percentage survival compared to vehicle treated cells for phenformin (A) or BPTES (B) treated cells at specified doses. CDI < 1 suggests drugs are synergistic and CDI < 0.7 indicates that combination drug treatment is significantly synergistic (Zhao et al., 2014).
Statistical Analysis
All results shown as averages of multiple independent experiments are presented as mean ± SEM; results shown as averages of technical replicates are presented as mean ± s.d. P-values calculated using a Students two-tailed t test: *, P-value < 0.05; **, P-value < 0.01; ***, P-value < 0.001. All errors associated with 13C MFA are 95% confidence intervals determined via sensitivity analysis.
Supplementary Material
Highlights.
LKB1 deficiency reprograms mitochondrial metabolism in NSCLC
LKB1 promotes metabolic flexibility in the context of energetic stress
LKB1 deficiency sensitizes cells to phenformin and glutaminase inhibitor treatment
Acknowledgments
We thank Nathaniel Vacanti, Courtney R. Green, Christopher Ahn, Kimberly Wang, and Spencer Lee for technical assistance and reagents. This work was supported by DOD Lung Cancer Research Program grant W81XWH-13-1-0105 (to C.M.M.) and NIH grants 1R01CA188652 (to C.M.M.), R01NS087611, and P01DK054441 (to A.N.M.). S.J.P. was supported by NIH grants 1F31CA196066 and 5T32EB009380. A.N.M. and A.S.D. are consultants for Agilent Technologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metabolic engineering. 2006;8:324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Appleyard MV, Murray KE, Coates PJ, Wullschleger S, Bray SE, Kernohan NM, Fleming S, Alessi DR, Thompson AM. Phenformin as prophylaxis and therapy in breast cancer xenografts. British journal of cancer. 2012;106:1117–22. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avizienyte E, Loukola A, Roth S, Hemminki A, Tarkkanen M, Salovaara R, Arola J, Butzow R, Husgafvel-Pursiainen K, Kokkola A, Jarvinen H, Aaltonen LA. LKB1 somatic mutations in sporadic tumors. The American journal of pathology. 1999;154:677–81. doi: 10.1016/S0002-9440(10)65314-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosova Z, Zavodna K, Krivulcik T, Usak J, Mlkva I, Kruzliak T, Hromec J, Usakova V, Kopecka I, Veres P, Bartosova Z, Bujalkova M. STK11/LKB1 germline mutations in the first Peutz-Jeghers syndrome patients identified in Slovakia. Neoplasma. 2007;54:101–7. [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. The Journal of biological chemistry. 2002;277:23977–80. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- Carretero J, Medina PP, Blanco R, Smit L, Tang M, Roncador G, Maestre L, Conde E, Lopez-Rios F, Clevers HC, Sanchez-Cespedes M. Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene. 2007;26:1616–25. doi: 10.1038/sj.onc.1209951. [DOI] [PubMed] [Google Scholar]
- Contreras CM, Gurumurthy S, Haynie JM, Shirley LJ, Akbay EA, Wingo SN, Schorge JO, Broaddus RR, Wong KK, Bardeesy N, Castrillon DH. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer research. 2008;68:759–66. doi: 10.1158/0008-5472.CAN-07-5014. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, Gui DY, Sullivan LB, Wasylenko TM, Subbaraj L, Chin CR, Stephanopolous G, Mott BT, Jacks T, Clish CB, Vander Heiden MG. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism. 2016;23:517–28. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN, Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods in enzymology. 2014a;547:309–54. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Rogers GW, Murphy AN. Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark-Type Oxygen Electrode. Current protocols in toxicology/editorial board, Mahin D. Maines. 2014b;60:25 2 1–16. doi: 10.1002/0471140856.tx2502s60. [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Wiley SE, Rogers GW, Andreyev AY, Petrosyan S, Loviscach M, Wall EA, Yadava N, Heuck AP, Ferrick DA, Henry RR, McDonald WG, Colca JR, Simon MI, Ciaraldi TP, Murphy AN. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5422–7. doi: 10.1073/pnas.1303360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahota Z, Palenickova E, Endlicher R, Milerova M, Brejchova J, Vosahlikova M, Svoboda P, Kazdova L, Kalous M, Cervinkova Z, Cahova M. Biguanides inhibit complex I, II and IV of rat liver mitochondria and modify their functional properties. Physiological research/Academia Scientiarum Bohemoslovaca. 2014;63:1–11. doi: 10.33549/physiolres.932600. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgogary A, Xu Q, Poore B, Alt J, Zimmermann SC, Zhao L, Fu J, Chen B, Xia S, Liu Y, Neisser M, Nguyen C, Lee R, Park JK, Reyes J, Hartung T, Rojas C, Rais R, Tsukamoto T, Semenza GL, Hanes J, Slusher BS, Le A. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E5328–36. doi: 10.1073/pnas.1611406113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. Bmj. 2005;330:1304–5. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, Mamer OA, Avizonis D, Shackelford DB, Shaw RJ, Jones RG. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2554–9. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Davidson SM, Wirth GJ, Fiske B, Mayers JR, Schwab M, Bellinger G, Csibi A, Patnaik A, Blouin MJ, Cantley LC, Guarente L, Blenis J, Pollak MN, Olumi AF, Vander Heiden MG, Stephanopoulos G. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer research. 2013;73:4429–38. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. Journal of mass spectrometry: JMS. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes & development. 2011;25:1716–33. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, Slocum KL, Pu M, Lin F, Vickers C, Joud-Caldwell C, Chung F, Yin H, Handly ED, Straub C, Growney JD, Vander Heiden MG, Murphy AN, Pagliarini R, Metallo CM. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer research. 2014;74:3317–31. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JQ, Li Q, Xu SP, Shen YX, Sun GY. Effect of lumiracoxib on proliferation and apoptosis of human nonsmall cell lung cancer cells in vitro. Chinese medical journal. 2008;121:602–7. [PubMed] [Google Scholar]
- Hawley SA, Gadalla AE, Olsen GS, Hardie DG. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes. 2002;51:2420–5. doi: 10.2337/diabetes.51.8.2420. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–7. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10574–9. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–5. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, Schmidt S, Metallo CM, Dranka BP, Schwartz B, DeBerardinis RJ. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino K, Hara K, Kemp BE, Witters LA, Mimura O, Yonezawa K. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes to cells: devoted to molecular & cellular mechanisms. 2003;8:65–79. doi: 10.1046/j.1365-2443.2003.00615.x. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, Liesa M, Hammerman PS, Wong KK, Hayes DN, Shirihai OS, Dyson NJ, Haas W, Meissner A, Bardeesy N. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539:390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause U, Bertrand L, Hue L. Control of p70 ribosomal protein S6 kinase and acetyl-CoA carboxylase by AMP-activated protein kinase and protein phosphatases in isolated hepatocytes. European journal of biochemistry/FEBS. 2002;269:3751–9. doi: 10.1046/j.1432-1033.2002.03074.x. [DOI] [PubMed] [Google Scholar]
- Landman GW, Kleefstra N, van Hateren KJ, Groenier KH, Gans RO, Bilo HJ. Metformin associated with lower cancer mortality in type 2 diabetes: ZODIAC-16. Diabetes care. 2010;33:322–6. doi: 10.2337/dc09-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55:253–63. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Molecular cell. 2013;49:388–98. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nature reviews. Molecular cell biology. 2014;15:634–46. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins WK, Ahn HJ, Kim JY, Ryu S, Jung YS, Choi JY. Synergistic anti-cancer effect of phenformin and oxamate. PloS one. 2014;9:e85576. doi: 10.1371/journal.pone.0085576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M, McMickle R, Abt E, Seki A, Simko SA, Magyar C, Stout DB, Fishbein MC, Walser TC, Dubinett SM, Shackelford DB. Heightening Energetic Stress Selectively Targets LKB1-Deficient Non-Small Cell Lung Cancers. Cancer research. 2015;75:4910–22. doi: 10.1158/0008-5472.CAN-15-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TL, Leprivier G, Robertson MD, Chow C, Martin MJ, Laderoute KR, Davicioni E, Triche TJ, Sorensen PH. The AMPK stress response pathway mediates anoikis resistance through inhibition of mTOR and suppression of protein synthesis. Cell death and differentiation. 2012;19:501–10. doi: 10.1038/cdd.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer research. 2002;62:3659–62. [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, Shaw RJ. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell. 2013;23:143–58. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews. Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, Vera DR, Shaw RJ. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11137–42. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3329–35. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–6. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–81. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang YM, Park SI, Trenary IA, Egnatchik RA, Fessel JP, Kaufman JM, Carbone DP, Young JD. LKB1 deficiency enhances sensitivity to energetic stress induced by erlotinib treatment in non-small-cell lung cancer (NSCLC) cells. Oncogene. 2016;35:856–66. doi: 10.1038/onc.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo SN, Gallardo TD, Akbay EA, Liang MC, Contreras CM, Boren T, Shimamura T, Miller DS, Sharpless NE, Bardeesy N, Kwiatkowski DJ, Schorge JO, Wong KK, Castrillon DH. Somatic LKB1 mutations promote cervical cancer progression. PloS one. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–5. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnology and bioengineering. 2008;99:686–99. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jager C, Hiller K, Murphy AN, Metallo CM. Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell reports. 2016;16:1536–47. doi: 10.1016/j.celrep.2016.06.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gao JL, Ji JW, Gao M, Yin QS, Qiu QL, Wang C, Chen SZ, Xu J, Liang RS, Cai YZ, Wang XF. Cytotoxicity enhancement in MDA-MB-231 cells by the combination treatment of tetrahydropalmatine and berberine derived from Corydalis yanhusuo W. T. Wang. Journal of intercultural ethnopharmacology. 2014;3:68–72. doi: 10.5455/jice.20140123040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.