Abstract
Genetic and genomic approaches have been successfully used to assign genes to distinct regulatory networks. However, the present challenge of distinguishing differentially regulated genes within a network is particularly hard because members of a given network tend to have similar regulatory features. We have addressed this challenge by developing a method, termed Gene Promoter Scan, that discriminates coregulated promoters by simultaneously considering both multiple cis promoter features and gene expression. Here, we apply this method to probe the regulatory networks governed by the PhoP/PhoQ two-component system in the enteric bacteria Escherichia coli and Salmonella enterica. Our analysis uncovered members of the PhoP regulon and interactions with other regulatory systems that were not discovered in previous approaches. The predictions made by Gene Promoter Scan were experimentally validated to establish that the PhoP protein uses multiple mechanisms to control gene transcription, regulates acid resistance determinants, and is a central element in a highly connected network.
Keywords: promoter, machine learning, gene transcription, acid pH
Acritical challenge of the postgenomic era is to understand how genes are differentially regulated. Because the prevalent mechanisms controlling gene expression operate at the level of transcription initiation, computational techniques have been developed that identify cis regulatory features (1, 2) and map such features into expression patterns (3, 4) to classify genes into distinct networks. However, these methods are not focused on the more challenging problem of distinguishing between differentially regulated genes within a network. Here, we use a unsupervised machine learning method (5–7), Gene Promoter Scan (GPS), to investigate the regulatory interactions governed by the PhoP/PhoQ two-component system, a central element in one of the most interconnected bacterial genetic networks thus far described.
The PhoQ protein is a sensor for extracytoplasmic Mg2+ that modifies the phosphorylated state of the DNA-binding protein PhoP (8–10). The PhoP protein controls the expression of a large number of genes that mediate adaptation to low Mg2+ environments and/or virulence in several bacterial species including Salmonella enterica, Shigella flexneri, Yersinia pestis, Erwinia carotovora, Neisseria meningitidis, Photorhabdus luminescens and Escherichia coli (see ref. 11 for a review). It has been proposed that the PhoP protein recognizes the direct hexanucleotide repeat (T/G)GTTTA, separated by five nucleotides, which has been termed the PhoP box (12). Indeed, experiments carried out with the PhoP-activated mgtA promoter of Escherichia coli demonstrated the critical role that certain PhoP box nucleotides play in mgtA expression and that the purified PhoP protein and RNA polymerase were sufficient to promote mgtA transcription in vitro (13). However, there is uncertainty about what constitutes a PhoP binding site because many PhoP-regulated promoters do not conform to the mgtA promoter model (14, 15). Moreover, the identification of PhoP-regulated targets is confounded by the fact that many genes are indirectly regulated by PhoP, which controls other two-component regulatory systems at the transcriptional (e.g., RstA/RstB) (14), posttranscriptional (e.g., SsrB/SpiR) (J. Bijlsma and E.A.G., unpublished results), and posttranslational (e.g., PmrA/PmrB) (16) levels. In this article, we use GPS to uncover previously uncharacterized members of the PhoP regulatory network and demonstrate that PhoP controls transcription from different classes of promoters and has an unsuspected role in the regulation of acid resistance genes.
Materials and Methods
Bacterial Strains, Growth Conditions, and Enzymatic Assays. Strains used in this study are listed in Table 1. The Escherichia coli K-12 phoP strain EG12976 was constructed from the wild-type MG1655 strain by P1 transduction with a lysate prepared by using the phoP strain FS1002 (17) as a donor and selected on a LB plate containing 50 μg/ml kanamycin. Construction of other strains is described in Supporting Text, which is published as supporting information on the PNAS web site. For the GeneChip (Affymetrix, Santa Clara, CA) experiment, strains MG1655 and EG12976 were grown overnight in N-minimal medium (pH 7.7) and 10 mM MgCl2 and washed three times with N-minimal medium without Mg2+. The OD600 was adjusted to 0.5 and 1/100 volume of cells was added to 30 ml of N-minimal medium (pH 7.7) and 10 μM MgCl2 and was shaken at 250 rpm (Innova Incubator Shaker Model 4230, New Brunswick Scientific) until OD600 equals 0.25. Cells were chilled on ice and immediately harvested to prepare total RNA. β-gal activity was carried out after growth of bacteria in N-minimal medium (pH 7.7) and 10 mM Mg2+, 10 μM Mg2+, or 10 μM Mg2+ plus 100 μM Fe3+ as described in ref. 18.
Table 1. Bacterial strains used in this study.
| Strain | Description | Source |
|---|---|---|
| MG1655 | Wild-type | 44 |
| EG12976 | phoP::KanR | This article |
| EG13922 | phoP-HA pmarA-FLAG | 22 |
| EG14307 | hdeA+-lacZY+ | This article |
| EG14316 | hdeA+-lacZY+ phoP::Tn10dCm | This article |
| EG14312 | yhiW+-lacZY+ | This article |
| EG14321 | yhiW+-lacZY+ phoP::CmR | This article |
| EG14356 | hdeA+-lacZY+ ΔyhiW::CmR | This article |
| EG14574 | yrbL+-lacZY+ | This article |
| EG15594 | yrbL+-lacZY+ phoP::KmR | This article |
| EG15595 | yrbL+-lacZY+ pmrA::TetR | This article |
| FS1002 | phoP::KanR | 17 |
RNA Isolation, Processing, and Analysis. Total Escherichia coli RNA was isolated by using 600 μl of the Tissue and Cell Lysis Solution of the Masterpure RNA Purification Kit (Epicentre Technologies, Madison, WI). Enrichment of mRNA was done according to the manufacturer's instructions (Affymetrix). Alternatively, total RNA was isolated as described above with the following modification: after DNaseI treatment, total RNA was purified by using the Megaclear Purification Kit (Ambion, Austin, TX). mRNA enrichment was performed for this total RNA (50 μg) by using the MICROBExpress Bacterial mRNA Purification Kit (Ambion). Biotin labeling, hybridization, and scanning were done as described in the manufacturer's instructions (Affymetrix). We used the Affymetrix GeneChip to examine four experiments by using strains MG1655 and EG12976 grown in low Mg2+, which were carried out with different cultures and conducted on four different occasions. Two of the four experiments were conducted in duplicate to determine the error due to technical aspects of the process. Systematic error (19) was treated by a consensus approach among dchip (20), sam (21) and microarray suite 5.0 (Affymetrix) software. Array data were normalized and filtered, and replicas were treated by the default values of dchip software and by the use of the PM-only model (20). Then, we compared samples within the four experiments and combined these comparisons to increase sensitivity and to avoid side effects due to potential variations between experiments. The candidate genes were further filtered in agreement between dchip selection, a false discovery rate of 5% in sam software and default significant P values of the Wilcoxon test performed by microarray suite 5.0 for comparison calls. Genes that exhibited differential expression all through the four experiments (i.e., satisfying a statistical power of 80%) were selected.
Chromatin Immunoprecipitation Assays. Chromatin immunoprecipitation assays were performed as described in ref. 22 by using strain EG13922 (Table 1) grown in N-minimal medium (pH 7.7) and supplemented with 10 mM or 10 μM MgCl2 to mid-exponential phase (OD600 = 0.3). Analysis of the immunoprecipitated DNA was performed by using quantitative PCR as described in ref. 22. All promoter-specific primers used in this study are listed in Table 2.
Table 2. List of promoter-specific primers used in chromatin immunoprecipitation assay.
| Primer | Target promoter | Sequence |
|---|---|---|
| 2779 | mgtA forward | 5′-GGCGATGTCTTTGATAGTGAGCCG-3′ |
| 2780 | mgtA reverse | 5′-CCTCCGGTAAGTAAATAATTTGCGC-3′ |
| 2791 | ugd forward | 5′-TTCAGGCGCAGCGTGACTACTTTG-3′ |
| 2792 | ugd reverse | 5′-AATCATGATGTATTTCGCATCCTG-3′ |
| 2891 | mig-14 forward | 5′-TCGCTACTGGATGGTTAAATACAG-3′ |
| 2892 | mig-14 reverse | 5′-GGTTAATATACGCTTAACTTCTTG-3′ |
| 2895 | pagC forward | 5′-ACCGGTTACCTAAATGAGCGATAG-3′ |
| 2896 | pagC reverse | 5′-AAAGGCGTTAGTATCGGCCTGTGC-3′ |
| 3461 | rpoD forward | 5′-TGCTGCGACCCTTGAAAAATTATCG-3′ |
| 3462 | rpoD reverse | 5′-TTTCTGGCCAGCTCCTGGTTTAAC-3′ |
| 3552 | mgtC forward | 5′-TACGTGCAGGCATCATAACAGAGC-3′ |
| 3553 | mgtC reverse | 5′-TAGCGTGCGTTAACTTGCCAGATG-3′ |
DNaseI Footprinting Assays. Footprinting was conducted as described in ref. 18 by using purified PhoP-His-6 protein from Salmonella enterica and DNA fragments generated by the PCR with primers 3849 (5′-GAACCACCCTATAAAATTAAGAAG-3′) and 3851 (5′-CGTAATATCCTCAACTATAAAG-3′) for hdeA and 3856 (5′-TAGAGTTTTACTCAGACA-3′) 3858 (5′-GATTATCCCTTATATTTCATACTGCG-3′) for yhiW, and MG1655 chromosomal DNA was used as the template.
Acid Resistance Assays. To determine the acid resistance of exponential-phase cells, a single colony of Escherichia coli was inoculated in 1 ml of LB broth and was grown overnight with aeration at 37°C. The following day, 20 ml of LB broth was inoculated with 0.1 ml of the overnight culture and was grown at 37°C with aeration. When the cultures reached a cell number of 2 × 108 colony-forming units (cfu)/ml, 50 μl of the culture was transferred to 2 ml of PBS (pH 7.2) and 2 ml of warmed LB broth (pH 2.5, adjusted with HCl) and incubated for 1 hour at 37°C. The number of cfu per milliliter in PBS was determined by plating serial dilutions in PBS buffer (pH 7.2) on LB agar and was used as initial cell populations. The LB broth (pH 2.5) inoculated with Escherichia coli was incubated further at 37°C, and the number of cfu per milliliter in LB broth (pH 2.5) was determined as described above and used as final cell populations. The percent of cells surviving acid pH was then calculated as the number of cfu per milliliter remaining after the acid treatment divided by the initial number of cfu per milliliter at time 0. Two or three repetitions were performed for each experiment. Percent of survival values were converted to logarithmic values (log10 x, where x equals the percent survival) for calculation of geometric means and standard errors. The survival levels of the wild-type strain were normalized to 100% and those of the mutant strains are displayed relative to those of the wild-type strain.
Genome-Wide Searches. Searches of the intergenic regions of the Escherichia coli and Salmonella genomes and groupings of promoters into profiles consisting of promoters with common sets of features was carried with the GPS method (unpublished data)
Results
Identification of PhoP-Regulated Genes. We examined the genome-wide transcription profile of wild-type and phoP Escherichia coli strains experiencing low Mg2+, which is the physiological signal that activates the PhoP/PhoQ system (11). We identified 33 genes whose expression differed statistically between the two strains (20, 21). These genes were clustered by their expression similarity: E1 and E2, consisting of up-regulated genes, and E3, harboring down-regulated genes (Fig. 3A, which is published as supporting information on the PNAS web site). Then, we classified all of the genes in the Escherichia coli genome based on the similarity of their expression to that of models built for each of the three expression groups, permitting individual genes to belong to more than one group (i.e., E1 and/or E2) (23, 24). This guideline allowed the recovery of weakly expressed genes that would have otherwise gone undetected by using strict statistical filters (20, 21).
To identify promoters that are directly regulated by the PhoP protein, we built a position weight matrix model (25) for the PhoP binding site by using Escherichia coli promoters exhibiting similar expression patterns (Fig. 3A) as well as a set of 10 Salmonella enterica promoters that are the orthologs of the Escherichia coli genes uncovered in the gene expression experiment. We used the model to analyze the intergenic regions of the Escherichia coli (40) and S. enterica (41) genomes by using relaxed thresholds (25), which allowed the recovery of PhoP-regulated promoters with weak matching to the PhoP box consensus, such as the Salmonella pmrD promoter, that could not be detected by using consensus cutoffs (2, 25) despite being regulated and footprinted by the PhoP protein (18, 26).
Classification of PhoP-Regulated Promoters. The use of relaxed thresholds resulted in the expected increased recovery of false positive candidates. Thus, we incorporated additional promoter features into the analysis in an effort to retrieve and classify bona fide PhoP-regulated promoters. Some of these features are as follows: the discrimination of PhoP box submotifs (M1–M4) (Fig. 3B), the orientation (O1–O2) (Fig. 3C) and distance of the PhoP box relative to the RNA polymerase site (P1–P3) (Fig. 3D), the class of σ70 promoter (because σ70 is responsible for transcription of PhoP-regulated genes; ref. 13) (P1–P3) (Fig. 3D), the presence of potential binding sites for 60-plus transcription factors (27) (I0–I4) (Fig. 3E), and whether the position of the PhoP box suggests a promoter is activated or repressed (A1–A3) (Fig. 3F).
We built models for each of these features by using the 43 promoters described above and 12 promoters identified in the initial analysis and known to be regulated by PhoP (Table 3, which is published as supporting information on the PNAS web site). Then, we generated profiles consisting of overlapping groups of promoters with common sets of features (Fig. 3 G and H). For example, one of the profiles encompassed promoters that belong to the same expression (E1), PhoP box submotif (M2), and promoter class (P1) (Fig. 1H) and harbored not only the prototypical PhoP-regulated phoP and mgtA promoters (13, 14) but also the yhiW promoter, which was not known to be under PhoP control. Yet, another profile comprised promoters that share the promoter class (P2), PhoP box orientation (O1), and regulatory interactions (I3) and included the ompT, pipD, ugtL, and ybjX promoters. We used the generated promoter profiles to search the intergenic regions of the Escherichia coli and S. enterica genomes and recovered additional putative promoters belonging to the PhoP regulon. Finally, we revised the original profiles by incorporating the latter promoters, which increased the robustness of the profiles (for examples of identified profiles, see Fig. 4, which is published as supporting information on the PNAS web site).
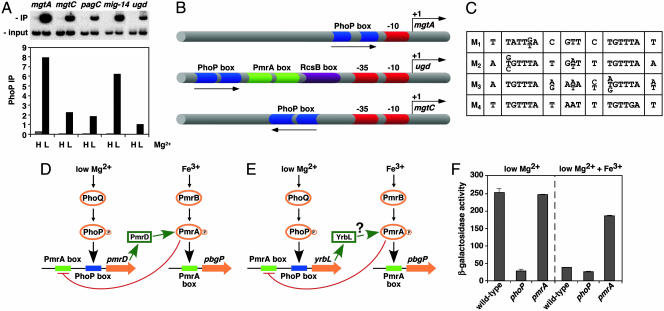
Fig. 1.
Experimental validation of GPS predictions and schematic representation of PhoP-regulated promoters. (A) Chromatin immunoprecipitation (ChIP) of PhoP-regulated promoters predicted by GPS demonstrates binding of the PhoP-HA protein to the PhoP-activated mgtA, mgtC, pagC, mig-14, and ugd promoters, which are found in different profiles. Binding is detected in organisms experiencing low (L) (i.e., 10 μM) but not high (H) (i.e., 10 mM) Mg2+, which correspond to conditions that activate and repress, respectively, the PhoP/PhoQ system. PhoP immunoprecipitation (IP) values correspond to the DNA bound by the PhoP protein (IP) divided by the total DNA before precipitation (input). (B) Representation of PhoP-regulated promoters identified by GPS and experimentally validated. Arrows, transcription start sites; red symbols, RNA polymerase sites; blue symbols, PhoP binding sites; green symbols, PmrA binding sites; purple symbols, RcsB binding sites. Drawing is not to scale. (C) The PhoP binding site submotifs. We built a model for the PhoP binding site by applying an extension of the consensus/patser program (25) by using promoters exhibiting coherent expression patterns (Fig. 3A). This initial model was used for identifying promoter motifs, which were clustered into four subsets based on their degree of sequence similarity to one another (M1–M4). All four submotifs have an hexameric direct repeat separated by 5 bp and preserved those conserved positions critical for PhoP-promoted transcription of the mgtA promoter (13). (D) Model for a multicomponent feedback loop in Salmonella involving the regulatory protein PmrA and its posttranslational activator PmrD. Transcription of the pmrD gene is induced in low Mg2+ in a PhoP-dependent fashion and repressed in the presence of Fe3+ in a PmrA-dependent manner. The PhoP and PmrA proteins exert their regulatory effects directly on the pmrD promoter, which has binding sites for these two proteins. (E) Model for regulatory interactions at the Escherichia coli yrbL promoter, which harbors PhoP- and PmrA-binding sites arranged in a fashion similar to those present in the Salmonella pmrD promoter. (F) Transcription of the yrbL gene is induced in low Mg2+ in a PhoP-dependent fashion and repressed by Fe3+ in a PmrA-dependent manner. β-gal activity (Miller units) of Escherichia coli strains harboring a lac-transcriptional fusion immediately downstream of the yrbL gene was determined in wild type, phoP, and pmrA backgrounds by using organisms grown in N-minimal medium (pH 7.7) and 10 μMMg2+ with or without 100 μMFe3+.
Our search successfully retrieved 92% of a group of 26 PhoP-regulated promoters that had not been used in the development of the models (P < 10–5). Analysis of 487 additional promoters known to be regulated by transcription factors other than PhoP (27) indicated that our method recovered PhoP-regulated promoters with a specificity of 94.6% and a sensitivity of 92.3% (correlation coefficient, 93.9%).
Experimental Validation of Regulatory Interactions Mediated by PhoP. Transcriptional control of atypical promoters. The PhoP-activated mig-14, mgtC, virK, and pagC promoters of Salmonella do not harbor a typical PhoP box in place of the –35 region, as found in archetypal PhoP-regulated promoters such as the mgtA promoter (13). This finding has led to the proposal that PhoP regulates these promoters indirectly through another regulatory protein(s) (15). However, we determined that these promoters do harbor a PhoP box but in the opposite orientation and located further upstream of the RNA polymerase site than the archetypal PhoP-regulated promoters (15, 28). Moreover, the statistical inclusion of these promoters within a profile supported the notion that they are regulated directly by PhoP despite their unusually oriented PhoP box.
We verified the predictions described above by using chromatin immunoprecipitation to probe promoter occupancy by the PhoP protein in vivo. We established that when Salmonella experiences low Mg2+, the PhoP protein binds to both the archetypal mgtA promoter as well as the mig-14, mgtC, and pagC promoters (Fig. 1 A). In contrast, there was no binding to these promoters when Salmonella experienced high Mg2+, a condition that represses transcription of PhoP-activated genes. Furthermore, we conducted site-directed mutagenesis of the hexameric repeat sequences that makes up the PhoP box in the mgtC promoter (see Supporting Text) and demonstrated that, despite its orientation, the PhoP box is essential for the low-Mg2+ PhoP-dependent expression of the mgtC gene (data not shown). These results demonstrate that the PhoP protein can regulate transcription directly from promoters harboring PhoP binding sites in either orientation and at various distances from the RNA polymerase binding site (Fig. 1B).
A possible multicomponent loop involving the PhoP-activated yrbL gene. We have previously identified a multicomponent loop in Salmonella where the PhoP-dependent PmrD protein activates the regulatory protein PmrA, and activated PmrA represses transcription from the pmrD promoter, which harbors binding sites for both the PhoP and PmrA proteins (18) (Fig. 1D). We hypothesized that the PhoP-activated yrbL gene of Escherichia coli might participate in an analogous loop with the PmrA protein because both the Salmonella pmrD and Escherichia coli yrbL promoters can be found in the same promoter profile and harbor similarly arranged PhoP and PmrA boxes (Fig. 1 D and E). In agreement with this notion, transcription of the Escherichia coli yrbL gene was induced in low Mg2+ in a PhoP-dependent fashion and repressed by PmrA in response to Fe3+ (Fig. 1F), which is the specific signal that activates the PmrA protein (11).
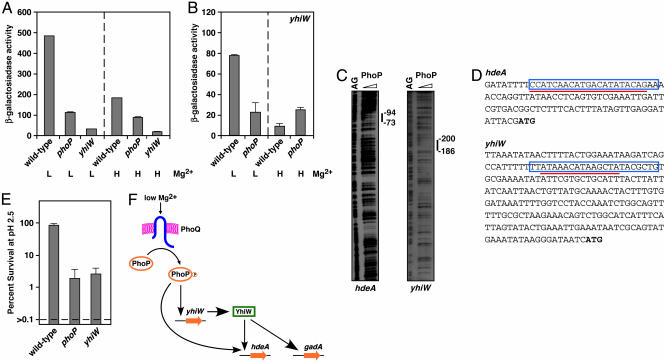
The PhoP/PhoQ system controls acid resistance genes in Escherichia coli. We classified the PhoP-activated genes identified in the genome-wide transcription experiment into two groups: E1 and E2, corresponding to genes that were up-regulated strongly and mildly, respectively. Interestingly, most of the genes in expression group E2 (Fig. 3A) encode proteins implicated in acid resistance in Escherichia coli (29–31), suggesting a role of PhoP in the control of acid resistance. Although these acid resistance genes displayed similar expression, they can be grouped into three profiles based on the presence of a PhoP box, the PhoP box submotif (M1–M4), the presence of binding sites for other regulatory proteins (I0–I4), and the distance from the PhoP box to the RNA polymerase site (P1–P3) (Fig. 5, which is published as supporting information on the PNAS web site). One profile contained acid resistance structural genes lacking a recognizable PhoP box, such as dps and gadA. A second profile comprised a different set of acid resistance structural genes, including hdeD and hdeAB, which are predicted to have a class II promoter with a PhoP box close to the RNA polymerase site (P1). The third profile, which harbors the PhoP box-containing promoters of the acid resistance regulatory genes yhiE and yhiW (also termed gadE and gadW, respectively), shared the promoter class and distance of the PhoP box to the RNA polymerase site (P3).
We verified that PhoP plays a critical role in the control of acid resistance in a series of experiments demonstrating that: (i) expression of the hdeA (Fig. 2A) and yhiW (Fig. 2B) genes is regulated by Mg2+ in a PhoP-dependent fashion; (ii) the low Mg2+ activation of hdeA requires the regulatory gene yhiW (Fig. 2A), which encodes a protein that can bind to the gadA promoter (32); (iii) the PhoP protein footprints the hdeA and yhiW promoters (Fig. 2C), protecting regions that correspond to the PhoP-binding sites predicted by GPS (Fig. 2D), indicating that PhoP regulates these targets directly; and (iv) the phoP mutant was hypersensitive to acid pH after logarithmic growth, like the yhiW mutant (Fig. 2E).
Fig. 2.
The PhoP protein controls expression of acid resistance genes in Escherichia coli. (A) Transcription of the acid resistance hdeA gene is induced in low Mg2+ in a PhoP- and YhiW-dependent fashion. β-gal activity (Miller units) of strains harboring a lac-gene transcriptional fusion to the hdeA gene was determined in wild-type, phoP, and yhiW backgrounds by using organisms grown in N-minimal media (pH 7.4) and 10 μM low (L) or 10 mM high (H) Mg2+.(B) Transcription of the acid resistance regulatory gene yhiW is modulated by Mg2+ in a PhoP-dependent manner. β-gal activity (Miller units) of strains harboring lac-transcriptional fusions to the yhiW gene was determined in wild-type and phoP backgrounds by using organisms grown in N-minimal media (pH 7.4) and 10 μM (L) or 10 mM (H) Mg2+.(C) The PhoP protein footprints the hdeA and yhiW promoters. We used 0, 25, 50, and 100 pmol of Salmonella PhoP-His-6 (from left to right). AG corresponds to a sequencing ladder showing the A and G nucleotides. (D) Nucleotide sequence of the promoter regions of the hdeA and yhiW genes. Boxed sequences correspond to PhoP binding sites predicted by GPS. The red underline corresponds to DNA sequences protected by the purified PhoP-H6 protein as shown in C. Bold ATG sequences correspond to the predicted start codons. (E) The phoP and yhiW mutants are hypersensitive to acid pH. Logarithmically growing wild-type, phoP, and yhiW strains were exposed to pH 2.5 in LB broth adjusted with HCl for 1 hour at 37°C and then diluted and plated to determine the number of cfu. The percent survival was calculated as described in Materials and Methods. Data correspond to three repeats of the experiment. Percent survival values were converted to logarithmic values (log10 x, where x equals the percent survival) for calculation of geometric means and standard errors. The survival levels of the wild-type strain were normalized to 100% and those of the mutant strains are displayed relative to those of the wild-type strain. The actual survival of the wild-type strain was 5.6%. (F) Model for the control of acid resistance genes by the PhoP protein in Escherichia coli. The hdeA gene is regulated in a feed-forward loop by the PhoP and YhiW proteins, whereas the gadA gene is regulated in a classical transcriptional cascade where the PhoP protein promotes transcription of the regulatory gene yhiW and the YhiW protein binds to the gadA promoter.
Discussion
The use of the GPS methodology to scan, analyze and group regulatory regions allowed us to unveil previously uncharacterized PhoP-regulated promoters, thereby uncovering an unsuspected complexity in the regulatory targets that are under direct and indirect transcriptional control of the PhoP protein. Our analysis revealed several classes of PhoP-regulated promoters that differ in the location and orientation of the PhoP box relative to the RNA polymerase binding site (Fig. 1B) and the presence of a –35 region and/or binding sites for other transcription factors (Fig. 3E). In addition, by distinguishing classes of PhoP box submotifs (Fig. 1C) supported by many promoters (Fig. 3B) and clarifying the exceptions to the original consensus (12), we could increase the specificity of the PhoP binding-site model without affecting its sensitivity to weak sites. This approach allowed us to discover promoters with a weak degree of matching to the consensus sequence. The experimental verification that the PhoP protein binds to different classes of promoters in vivo (Fig. 1 A) and in vitro (Fig. 2C), together with the demonstration that the PhoP box is necessary for mgtC transcription despite its unusual orientation (data not shown), argues against proposals that atypical PhoP-dependent promoters are regulated by the PhoP protein only indirectly (15). Moreover, as transcription from certain PhoP-dependent promoters requires additional regulatory proteins, such as RcsB (33), SlyA (34), and YhiW (Fig. 2 A), our results indicate that the PhoP protein uses multiple mechanisms to promote transcription. The control of different classes of promoters may allow the PhoP protein to coordinate the expression of a variety of products that may be required in different amounts and/or for different extents of time.
We established that the PhoP/PhoQ system controls expression of acid resistance genes in Escherichia coli. These results were unexpected because acid resistance genes were not identified as PhoP-regulated genes in two previous microarray experiments that compared expression of wild-type and phoP Escherichia coli strains (14, 35) nor had mutations in phoP been uncovered in screenings for acid regulatory genes in Escherichia coli. The difference with these reports might be due to the fact that we used defined media instead of complex media to grow the bacteria. Alternatively or in addition, it may reflect the ability of the GPS method to recover promoters, even if they are not strongly up-regulated by a transcription factor. The PhoP-dependent induction of acid resistance genes occurred at the mildly alkaline pH 7.4, suggesting that these genes may also help alleviate low Mg2+ stress.
The GPS method considers gene expression as one feature among many. This attribute of GPS allowed us to distinguish between acid resistance genes (Fig. 5), which otherwise would have stayed undifferentiated within the same expression group if analyzed by methods that back correlate gene expression with cis regulatory features (3, 4). Moreover, it predicted that PhoP uses at least two modes of regulation to control transcription of acid resistance genes: a feed-forward loop and classical transcriptional cascade (Fig. 2F). This prediction was based on: first, the identification of a PhoP box in the promoter of only a subset of acid resistance genes; second, considering different PhoP submotifs (as opposed to determining regulon membership by using a strict statistical cut off to examine the degree of matching to a consensus sequence); and third, by incorporating multiple promoter features into the analysis to recover profiles comprising small numbers of promoters that would otherwise be subsumed by “typical” profiles encompassing large numbers of promoters. And it was also based on a model for the sequences recognized by the YhiW protein that we generated from analyzing promoters that are coordinately regulated by the YhiW protein (36). We provided experimental evidence for our model by demonstrating that a functional phoP gene is necessary for transcription of acid resistance genes after growth in low Mg2+ (Fig. 2 A and B), the PhoP protein footprints the promoters of both structural and regulatory acid resistance genes (Fig. 2C), and inactivation of the phoP gene rendered Escherichia coli hypersusceptible to acid pH, as susceptible as mutants in the well characterized acid resistance regulatory gene yhiW (Fig. 2E).
A distinguishing characteristic of our approach is that promoters for orthologous genes are considered individually. This tenet is in contrast to some phylogenetic footprinting methods (1) that often ignore regulatory differences among closely related organisms due to their strict reliance on the conservation of regulatory motifs across bacterial species. Thus, we could uncover cases of phenotypic differences between closely related species resulting from the differential regulation of homologous genes. For example, the ugd promoter of Salmonella (Fig. 1B), but not of Escherichia coli, harbors a PhoP box that is located much further upstream from an RNA polymerase binding site than in other PhoP-regulated promoters (Table 3). Despite the lack of conservation in this atypically located regulatory site, we demonstrated that the PhoP protein binds to the Salmonella ugd promoter in vivo (Fig. 1 A) and in vitro (33), inactivation of the PhoP box in the ugd promoter abolishes PhoP-dependent ugd transcription (33), and Escherichia coli is unable to promote ugd transcription under the PhoP-dependent conditions that activate ugd expression in Salmonella (C. Mouslim and E.A.G., unpublished results).
In addition to controlling transcription directly at a variety of different promoters, the PhoP protein regulates several dozen genes indirectly through transcriptional, posttranscriptional, and posttranslational cascades promoting expression and/or activity of other two-component systems and regulatory proteins. This conclusion suggests that the PhoP/PhoQ system is a central element in a highly connected network and places PhoP in the same group of transcriptional regulators as Crp, Fnr, and ArcA, which also govern expression of large number of targets in Escherichia coli, often by working with additional regulatory proteins (37). The high connectivity of the PhoP/PhoQ system may be due to its role in maintaining Mg2+ homeostasis, and the fact that the essential role that Mg2+ plays in all cells demands the adjustment of numerous cellular activities when an organism experiences Mg2+-limiting environments. Alternatively or in addition, the high connectivity could reflect the apparent use of Mg2+ as a signal to denote extracellular and intracellular environments during infection of a mammalian host (38).
In summation, we have described previously uncharacterized regulatory targets of and interactions by the bacterial PhoP protein. The analysis of individual networks in eukaryotes is likely to be even more complex because a single nucleotide difference in the binding site for a transcription factor can dictate the requirement for coactivator proteins (39). Thus, such analysis will require approaches like the one presented here that can uncover subtle differences between regulatory targets.
Supplementary Material
Acknowledgments
We thank G. Stormo and K. Tan for initial discussions, E. Ruspini and C. Gu for methodological suggestions, H. Salgado for the Salmonella operon information, and H. Ochman and J. Bijlsma for comments on the manuscript. This work was supported in part by National Institutes of Health Grant AI49561 (to E.A.G.). E.A.G. is an investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cfu, colony-forming units; GPS, Gene Promoter Scan.
References
- 1.McCue, L., Thompson, W., Carmack, C., Ryan, M. P., Liu, J. S., Derbyshire, V. & Lawrence, C. E. (2001) Nucleic Acids Res. 29, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stormo, G. D. (2000) Bioinformatics 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 3.Beer, M. A. & Tavazoie, S. (2004) Cell 117, 185–198. [DOI] [PubMed] [Google Scholar]
- 4.Conlon, E. M., Liu, X. S., Lieb, J. D. & Liu, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman, P. & Oldford, R. W. (1994) Selecting Models from Data: Artificial Intelligence and Statistics IV (Springer-Verlag, New York).
- 6.Cook, D. J., Holder, L. B., Su, S., Maglothin, R. & Jonyer, I. (2001) IEEE Eng. Med. Biol. Mag. 20, 67–74. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell, T. M. (1997) Machine Learning (McGraw-Hill, New York).
- 8.Castelli, M. E., Garcia Vescovi, E. & Soncini, F. C. (2000) J. Biol. Chem. 275, 22948–22954. [DOI] [PubMed] [Google Scholar]
- 9.Montagne, M., Martel, A. & Le Moual, H. (2001) J. Bacteriol. 183, 1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamnongpol, S., Cromie, M. J. & Groisman, E. A. (2003) J. Mol. Biol. 325, 795–807. [DOI] [PubMed] [Google Scholar]
- 11.Groisman, E. A. (2001) J. Bacteriol. 183, 1835–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, A., Tanabe, H. & Utsumi, R. (1999) J. Bacteriol. 181, 5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto, K., Ogasawara, H., Fujita, N., Utsumi, R. & Ishihama, A. (2002) Mol. Microbiol. 45, 423–438. [DOI] [PubMed] [Google Scholar]
- 14.Minagawa, S., Ogasawara, H., Kato, A., Yamamoto, K., Eguchi, Y., Oshima, T., Mori, H., Ishihama, A. & Utsumi, R. (2003) J. Bacteriol. 185, 3696–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lejona, S., Aguirre, A., Cabeza, M. L., Garcia Vescovi, E. & Soncini, F. C. (2003) J. Bacteriol. 185, 6287–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, A. & Groisman, E. A. (2004) Genes Dev. 18, 2302–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groisman, E. A., Heffron, F. & Solomon, F. (1992) J. Bacteriol. 174, 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, A., Latifi, T. & Groisman, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadon, R. & Shoemaker, J. (2002) Trends Genet. 18, 265–271. [DOI] [PubMed] [Google Scholar]
- 20.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin, D. & Groisman, E. A. (2005) J. Biol. Chem. 280, 4089–4094. [DOI] [PubMed] [Google Scholar]
- 23.Gasch, A. P. & Eisen, M. B. (2002) Genome Biol. 3, RESEARCH0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedrycz, W., Bonissone, P. P. & Ruspini, E. H. (1998) Handbook of Fuzzy Computation (Institute of Physics, Bristol, U.K.).
- 25.Hertz, G. Z. & Stormo, G. D. (1999) Bioinformatics 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 26.Kox, L. F., Wosten, M. M. & Groisman, E. A. (2000) EMBO J. 19, 1861–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salgado, H., Santos-Zavaleta, A., Gama-Castro, S., Millan-Zarate, D., Diaz-Peredo, E., Sanchez-Solano, F., Perez-Rueda, E., Bonavides-Martinez, C. & Collado-Vides, J. (2001) Nucleic Acids Res. 29, 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eguchi, Y., Okada, T., Minagawa, S., Oshima, T., Mori, H., Yamamoto, K., Ishihama, A. & Utsumi, R. (2004) J. Bacteriol. 186, 3006–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda, N. & Church, G. M. (2002) J. Bacteriol. 184, 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda, N. & Church, G. M. (2003) Mol. Microbiol. 48, 699–712. [DOI] [PubMed] [Google Scholar]
- 31.Tucker, D. L., Tucker, N. & Conway, T. (2002) J. Bacteriol. 184, 6551–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, Z., Richard, H., Tucker, D. L., Conway, T. & Foster, J. W. (2002) J. Bacteriol. 184, 7001–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouslim, C., Latifi, T. & Groisman, E. A. (2003) J. Biol. Chem. 278, 50588–50895. [DOI] [PubMed] [Google Scholar]
- 34.Shi, Y., Latifi, T., Cromie, M. J. & Groisman, E. A. (2004) J. Biol. Chem. 279, 39146–39154. [DOI] [PubMed] [Google Scholar]
- 35.Oshima, T., Aiba, H., Masuda, Y., Kanaya, S., Sugiura, M., Wanner, B. L., Mori, H. & Mizuno, T. (2002) Mol. Microbiol. 46, 281–291. [DOI] [PubMed] [Google Scholar]
- 36.Tucker, D. L., Tucker, N., Ma, Z., Foster, J. W., Miranda, R. L., Cohen, P. S. & Conway, T. (2003) J. Bacteriol. 185, 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Antonio, A. & Collado-Vides, J. (2003) Curr. Opin. Microbiol. 6, 482–489. [DOI] [PubMed] [Google Scholar]
- 38.Groisman, E. A. (1998) Bioessays 20, 96–101. [DOI] [PubMed] [Google Scholar]
- 39.Leung, T. H., Hoffmann, A. & Baltimore, D. (2004) Cell 118, 453–464. [DOI] [PubMed] [Google Scholar]
- 40.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C, K., Mayhew, G. F., et al. (1997) Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 41.McClelland, M., Sanderson, K. E., Spieth, J. Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. (2001) Nature 413, 852–856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.