Abstract
Inositol pyrophosphates physiologically regulate vesicular endocytosis, ribosomal disposition, and directly phosphorylate proteins. Here we demonstrate roles in cell death and regulation of telomere length. Lethal actions of wortmannin and caffeine are selectively abolished in yeast mutants that cannot synthesize inositol pyrophosphates. Wortmannin and caffeine appear to act through the phosphoinositide 3-kinase-related protein kinases Tel1 and Mec1, known regulators of telomere length. Inositol pyrophosphates physiologically antagonize the actions of these kinases, which is demonstrated by the fact that yeast mutants with reduced or elevated levels of inositol pyrophosphates, respectively, display longer and shorter telomeres.
Keywords: wortmannin, yeast, caffeine
Inositol polyphosphates mediate diverse forms of cell signaling, with the best characterized, inositol 1,4,5-trisphosphate (IP3), releasing calcium from intracellular stores (1). Inositol hexakisphosphate (IP6) (also known as phytic acid), the most abundant inositol polyphosphate present in eukaryotes (2), is the precursor of diphosphoinositol polyphosphates, a class of phosphorylated inositols containing one or more pyrophosphate moieties on the inositol ring (3, 4). The pyrophosphate moieties of diphosphoinositol-pentakisphosphate (PP-IP5 or IP7) and bis-diphosphoinositol-tetrakisphosphate ([PP]2-IP4 or IP8) contain energetic bonds that turn over rapidly (3, 4), possibly indicating a molecular switching role. Recently, we showed that IP7 physiologically transfers the β-phosphate of the pyrophosphate moiety to several target proteins (5), implying a major role in intracellular signaling. Yeast lacking the inositol pyrophosphate-forming enzyme, IP6 kinase (yIP6K; also designated KCS1; ORF DRO17c) (6, 7), display defective vesicular endocytosis (8). yIP6K-deficient (ip6kΔ) yeast also manifest slow cell growth (9), sensitivity to environmental stresses (10), and abnormal ribosomal functions (5). In the present study, we demonstrate the involvement of inositol pyrophophates in signaling cascades that mediate cell death and telomere length. ip6kΔ yeast are resistant to the lethal effects of wortmannin and caffeine, drugs that inhibit the activity of phosphoinositide 3-kinase (PI3K)-related protein kinases. Mutant yeast with elevated or reduced levels of inositol pyrophosphates respectively possess shorter and longer telomeres.
Materials and Methods
Yeast Growth Conditions and Drug Treatments. Yeast were grown in synthetic medium supplemented with a complete or appropriate amino acid mixture of the Synthetic Complete supplement purchased from Qbiogene (Carlsbad, Ca). Wortmannin and caffeine (Sigma) were added to the synthetic media from concentrated stocks prepared in DMSO and water, respectively.
Yeast Strains, Plasmid Preparation, and HPLC Analyses of Inositol Phosphates. Yeast strains used in this study were described in refs. 7–9 or generated by standard mating and tetrad dissection techniques. The plasmids p415yIP6K and p415IPK1 were generated by using standard recombinant DNA techniques. The yeast IP6K gene was amplified by using the oligos 5′-GCTGCGGCCGCTTTAACCTTAAACCAAACAT-3′ and 5′-GCTGCGGCCGCATGTACATATATCCTCACA-3′ and cloned into the NotI site of pRS415. Yeast IPK1 gene was amplified by using the oligos 5′-GCAGCGGCCGCTCTTGGCCATTACTTTTCTTCTC-3′ and 5′-CGTGCGGCCGCCCTCACTTACTTATGGTTTTCTTGTTC-3′ and cloned into the NotI site of pRS415. HPLC analyses were performed as described in ref. 8.
Analysis of Telomere Length. Yeast genomic DNA was extracted from 5 ml of an overnight yeast culture. The cells were washed with water and resuspended in 0.5 ml of lysis buffer (50 mM Tris·HCl, pH7.4/20 mM EDTA/1% SDS). Samples were vortexed twice for 1 min with acid-washed glass beads and incubated at 70°C for 10 min. After incubation, 200 μl of 5 M KOAc and 150 μl of 5 M NaCl was added to the lysates. Lysates were incubated on ice for 20 min and spun at 16,000 × g for 10 min. DNA was precipitated by addition of two volumes of ethanol. Precipitated DNA was resuspended in water, digested by PstI, and subjected to agarose gel electrophoresis followed by transfer onto Hybond N+ membranes (Amersham Pharmacia). The blot was probed with the yeast Y′ subtelomeric DNA (11) obtained by PCR amplification from yeast DNA by using the oligos 5′-ACACACTCTCTCACATCTACC-3′ and 5′-TTGCGTTCCATGACGAGCGC-3′. Probe sequence was confirmed by direct sequencing.
Results and Discussion
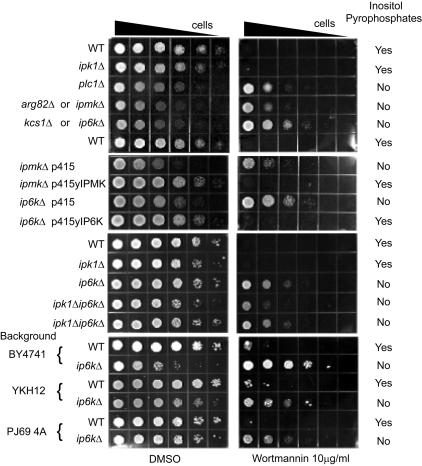
We compared the effects of a variety of drugs on the growth rates of WT and several yeast strains possessing impairments in inositol polyphosphate metabolism (Fig. 1). ip6kΔ yeast were markedly resistant to the lethal actions of wortmannin. The resistance is associated with the loss of inositol pyrophosphates, because yeast lacking inositol polyphosphate multikinase (yIMPK; also designated ARG82; ORF YDR173c) (6, 12, 13) or lacking phospholipase C (PLC1; ORF YPL268w) (14–16) are also resistant. In contrast, yeast lacking inositol polyphosphate kinase 1 (IPK1; ORF YDR315c) (16) are not resistant to wortmannin. Although IPK1-deficent yeast could not synthesize IP6, IP7, or IP8, they did accumulate alternative IP5-derived inositol pyrophosphates, PP-IP4 and [PP]2-IP3 (Fig. 2) (8). The resistance to wortmannin of ip6kΔ and ipmkΔ yeast was reversed in cells transformed with yIP6K and yIPMK, respectively (Fig. 1).
Fig. 1.
Absence of inositol pyrophosphates confers wortmannin resistance. Serial dilutions (7-fold) of cells were planted in synthetic media containing 10 μg/ml wortmannin (Right) or vehicle alone (Left) and incubated at 30°C for 72 h. Indicated to the left are the different genotypes and strain backgrounds analyzed. Strains ipmkΔ and ip6kΔ were transformed with empty vector (p415) or a vector carrying their respective WT gene (p415yIPMK and p415yIP6K). Indicated to the right are the presence (yes) or absence (no) of inositol pyrophosphates in the different strains as described in Fig. 2 or as published in ref. 8.
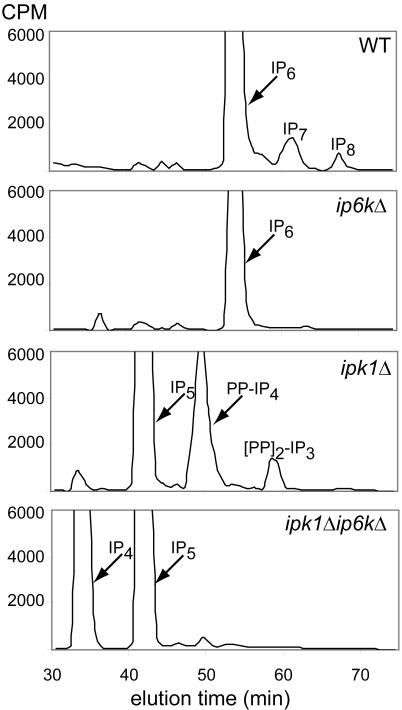
Fig. 2.
Intracellular levels of inositol polyphosphates. WT, yIP6K-null (ip6kΔ), IPK1-null (ipk1Δ), and double-mutant yIP6K- and IPK1-null (ipk1Δip6kΔ) yeast were radiolabeled with [3H]inositol. Inositol polyphosphates were extracted and analyzed by HPLC as described in ref. 9; standards used to identify the different inositol polyphosphate species were described in ref. 8.
To further confirm the role of inositol pyrophosphates in wortmannin-mediated cell death, we evaluated double-mutant ipk1Δip6kΔ yeast. These cells possessed high levels of IP4 and IP5 but lacked IP6, IP7, IP8, as well as PP-IP4 and [PP]2-IP3 (Fig. 2). The yeast manifested a similar resistance to wortmannin as ip6kΔ yeast (Fig. 1), which confirmed the requirement of pyrophosphate-containing inositols for wortmannin sensitivity.
Recent genome-wide screening of wortmannin sensitivity in yeast identified ip6kΔ yeast as wortmannin hypersensitive (17). This discrepancy may reflect sample misidentification, because the hyposensitive phenotype we observed in ip6kΔ yeast was conserved across three different genetic backgrounds (Fig. 1).
Our experiments implicate inositol pyrophosphates in the lethal actions of wortmannin. We ruled out direct effects of wortmannin on yIP6K, because wortmannin (10–20 μM) failed to inhibit either yIP6K enzymatic activity in yeast lysates or recombinant mouse IP6K1 (data not shown). Moreover, treatment for 30–60 min with wortmannin (10 μg/ml) did not alter cellular levels of [3H]IP7 or [3H]IP8 in intact WT yeast labeled with [3H]inositol in vivo (data not shown).
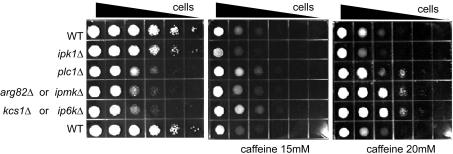
Caffeine, which, like wortmannin, inhibits PI3K-related protein kinases (18–20), retarded yeast growth (Fig. 3). ip6kΔ yeast are resistant to this effect, as are plc1Δ and ipmkΔ yeast, whereas ipk1Δ remain sensitive. The similar response of yeast mutants to caffeine and wortmannin ensures that the findings with wortmannin are not unique to a single drug but reflect actions common to caffeine and wortmannin.
Fig. 3.
Absence of inositol pyrophosphates confers caffeine resistance. Serial dilutions (7-fold) of cells were planted in synthetic media containing 15 mM (Center) or 20 mM(Right) of caffeine or vehicle alone (Left) and incubated at 30°C for 72 h (vehicle and 15 mM caffeine) or 96 h (20 mM caffeine). Indicated to the left are the genotypes analyzed.
What molecular actions of wortmannin and caffeine are responsible for the effects on cell growth that are prevented by yIP6K deletion? Wortmannin, which potently inhibits mammalian PI3K (Ki, low nM) (21) is a very weak inhibitor of the yeast PI3-kinase, Vps34 (Ki, μM) (22). Yeast contain two phosphatidylinositol 4-kinases, enzymes necessary for the formation of phosphatidylinositol 4, 5 bisphosphate, the precursor of IP3. One of these enzymes, Pik1, is wortmannin-insensitive (23), whereas the other, Stt4, is inhibited by wortmannin, with a Ki of 10–20 nM (23). We think it unlikely that inhibition of this enzyme explains wortmannin lethality, because phosphatidylinositol 4, 5 bisphosphate levels are elevated in ipmkΔ yeast but markedly reduced in ip6kΔ yeast (8), but both mutants are resistant to the lethality of wortmannin.
Wortmannin also inhibits PI3K-related protein kinases (24–26), whose yeast forms are Tel1 (homologous to mammalian ataxia-telangiectasia mutated, ATM), Mec1 (homologous to mammalian ATM-related, ATR), and targets of rapamycin Tor1/2 (homologous to mammalian TOR) with 20–200 nM potencies (24–26). Like wortmannin, caffeine inhibits the kinase activities of ATM and ATR (18–20). In our experiments, caffeine inhibits Tel1 kinase activity, with PHAS-I as substrate (Ki of 0.4 mM) (data not shown). Moreover, the mutagenic actions of caffeine in fission yeast reflect its inhibitory action on Mec1/Rad3 (27).
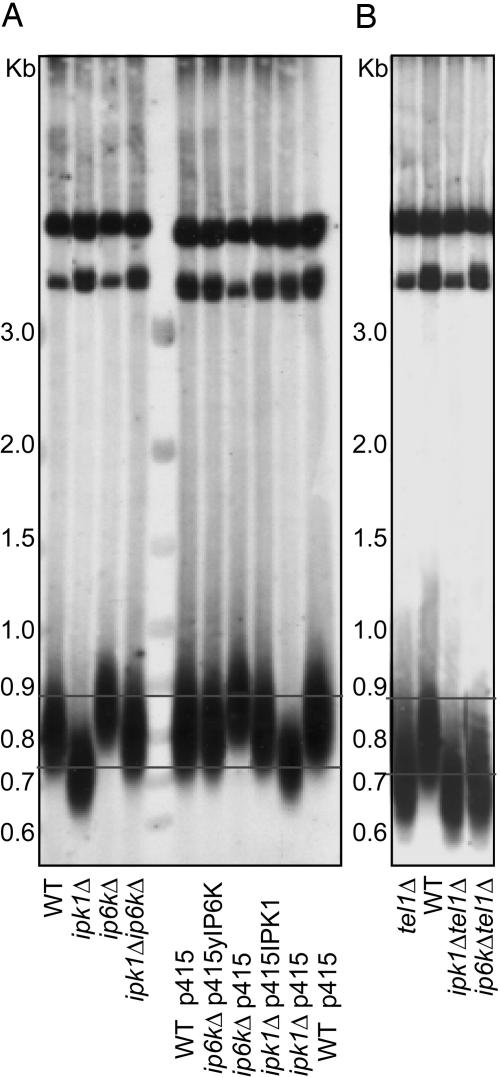
We wondered whether inositol pyrophosphates influence signaling by members of the PI3K-related protein kinase family. If yIP6K physiologically inhibits Tel1 and Mec1 signaling and this inhibition accounts for the lethal actions of wortmannin, then ip6kΔ yeast would resist wortmannin effects as we observe. Tel1 regulates telomere length, which is decreased in the tel1Δ strain (28). yIP6K deletion should have consequences opposite to TEL1 deletion, and as predicted, we find increased telomere length in ip6kΔ yeast (Fig. 4A). If yIP6K acts by generating inositol pyrophosphates to antagonize Tel1 activity, then yeast with elevated levels of inositol pyrophosphates should have shorter telomeres. Although inositol phosphate metabolism is altered considerably in ipk1Δ yeast, formation of inositol pyrophosphates still occurs and, in fact, is considerably greater than in WT yeast (Fig. 2). As predicted, telomere length is reduced in ipk1Δ yeast (Fig. 4A). The increased telomere length of ip6kΔ yeast and the shortened telomere length in ipk1Δ yeast are reversed by transformation with plasmids encoding yIP6K and IPK1, respectively (Fig. 4A). Yeast with deletions of IPK1 and yIP6K (ipk1Δip6kΔ) display an intermediate phenotype with almost normal telomere length.
Fig. 4.
Inositol pyrophosphates control telomere length. Genomic DNA was extracted from yeast grown in yeast extract/peptone/dextrose, digested by PstI, and analyzed by using a telomere probe as described in Materials and Methods. (A) Yeast with defects in inositol pyrophosphate synthesis (described in Fig. 2) display associated telomere defects that were rescued by the introduction of specified plasmids. Strains ipk1Δ and ip6kΔ were transformed with empty vector (p415) or a vector carrying their respective gene (p415IPK1 and p415yIP6K). (B) The analysis of the double-mutants ipk1Δtel1Δ and ip6k1Δtel1Δ reveals that the shortened telomeres present in tel1Δ yeast are not affected by changes in inositol pyrophosphate levels. The lines indicate the range of the WT telomere lengths observed.
Our findings imply a direct interaction between Tel1 and inositol pyrophosphate signaling in regulating telomere length. Telomere length is markedly reduced in tel1Δ yeast (Fig. 4B) (28). To clarify how inositol pyrophosphates regulate Tel1 influences on telomere length, we generated the double-mutant yeast strains tel1Δipk1Δ and tel1Δip6kΔ. The two double mutants display short telomeres identical to those found in tel1Δ yeast (Fig. 4B). These findings imply that inositol pyrophosphates and Tel1 share the same signaling pathway in controlling telomere length.
Very recently, York et al. (29) independently obtained evidence for inositol pyrophosphate modulation of Tel1-dependent telomere length. In agreement with our analyses, their results directly link yIP6K-dependent production of PP-IP4 with shortened telomeres in ipk1Δ yeast.
Because MEC1 deletion is lethal (30), we could not directly examine the influences of inositol pyrophosphates on Mec1 function in yeast. This lethality can be suppressed by the combined deletion of SML1 (31), but the triple deletion of MEC1, SML1, and yIP6K posed technical challenges, and we were unable to generate the mec1Δsml1Δip6kΔ strain. To evaluate links between Tor1/2 and inositol pyrophosphate signaling, we investigated the lethal effects of rapamycin, which kills yeast by inhibiting Tor1/2 signaling. Concentration-response relationships for rapamycin lethality in ip6kΔ yeast are the same as in WT yeast (data not shown).
In summary, we have shown that inositol pyrophosphates physiologically inhibit signaling by Tel1, and possibly Mec1. Loss of this inhibition underlies the resistance of inositol pyrophosphate-deficient yeast to wortmannin and caffeine toxicity. Mammals possess three distinct IP6K enzymes. One of these, IP6K2, regulates cell death, given that its overproduction enhances apoptosis and its deletion is antiapoptotic (32).
Tor1/2 do not appear to be involved in the actions of inositol pyrophosphates on cell death because rapamycin toxicity does not correlate with inositol pyrophosphate levels. However, the precise effects of rapamycin on Tor1/2 kinase activities have yet to be fully elucidated and many Tor1/2 functions are rapamycin-insensitive. The Tor1/2 proteins may participate in the regulation by inositol pyrophosphates of vesicular endocytosis (8). Consistent with this hypothesis are reports of Tor2 localization to membranes of yeast vacuoles (33) and of Tor1 to the plasma membrane of yeast (34).
The PI3K-related protein kinases Tel1 and Mec1 control DNA damage checkpoints during cell cycle so that their deletion or inhibition elicits hypersensitivity to DNA-damaging agents (35–37). Earlier, we showed that inositol pyrophosphates are required for DNA hyper recombination in certain yeast (38). Conceivably, inositol pyrophosphates influence hyper recombination through actions on Tel1 and Mec1. IP6 has been reported to influence end joining by DNA-dependent protein kinase (DNA-PK) (39). DNA-PK is an exclusively metazoan protein whose close yeast counterparts are Tel1 and Mec1. Conceivably, the reported effects of IP6 on DNA-PK reflect the actions of inositol pyrophosphates on Tel1 signaling that we have described.
Acknowledgments
We thank A. Riccio and R. Bhandari for comments, suggestions, and discussions and Dr. L. S. Symington (Columbia University, New York) for providing yeast strains. This work was supported by U.S. Public Health Service Grant MH18501, Conte Center Grant MH068830-02, and Research Scientist Award DA00074 (to S.H.S.).
Author contributions: A.S., A.C.R., A.M.S., B.W., and S.H.S. designed research; A.S., A.C.R., and A.M.S. performed research; B.W. contributed new reagents/analytic tools; A.S., A.C.R., A.M.S., and S.H.S. analyzed data; and A.S., A.C.R., B.W., and S.H.S. wrote the paper.
Abbreviations: IP3, inositol 1,4,5-trisphosphate; IP4, inositol tetrakisphosphate; IP5, inositol pentakisphosphate; IP6, inositol hexakisphosphate; PP, diphosphoinositol; IP7, PP-IP5; IP8, [PP]2-IP4; Tor, target of rapamycin; yIPMK, yeast inositol polyphosphate multikinase; yIP6K, yeast IP6 kinase; PI3K, phosphoinositide 3-kinase; IPK1, inositol polyphosphate kinase 1.
See Commentary on page 1811.
References
- 1.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Science 287, 1604–1605. [DOI] [PubMed] [Google Scholar]
- 2.Shears, S. B. (2001) Cell. Signaling 13, 151–158. [Google Scholar]
- 3.Stephens, L., Radenberg, T., Thiel, U., Vogel, G., Khoo, K. H., Dell, A., Jackson, T. R., Hawkins, P. T. & Mayr, G. W. (1993) J. Biol. Chem. 268, 4009–4015. [PubMed] [Google Scholar]
- 4.Menniti, F. S., Miller, R. N., Putney, J. W., Jr., & Shears, S. B. (1993) J. Biol. Chem. 268, 3850–3856. [PubMed] [Google Scholar]
- 5.Saiardi, A., Bhandari, R., Resnick, A. C., Snowman, A. M. & Snyder, S. H. (2004) Science 306, 2101–2105. [DOI] [PubMed] [Google Scholar]
- 6.Saiardi, A., Erdjument-Bromage, H., Snowman, A. M., Tempst, P. & Snyder, S. H. (1999) Curr. Biol. 9, 1323–1326. [DOI] [PubMed] [Google Scholar]
- 7.Huang, K. N. & Symington, L. S. (1995) Genetics 141, 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiardi, A., Sciambi, C., McCaffery, J. M., Wendland, B. & Snyder, S. H. (2002) Proc. Natl. Acad. Sci. USA 99, 14206–14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saiardi, A., Caffrey, J. J., Snyder, S. H. & Shears, S. B. (2000) J. Biol. Chem. 275, 24686–24692. [DOI] [PubMed] [Google Scholar]
- 10.Dubois, E., Scherens, B., Vierendeels, F., Ho, M. M., Messenguy, F. & Shears, S. B. (2002) J. Biol. Chem. 277, 23755–23763. [DOI] [PubMed] [Google Scholar]
- 11.Shampay, J., Szostak, J. W. & Blackburn, E. H. (1984) Nature 310, 154–157. [DOI] [PubMed] [Google Scholar]
- 12.Saiardi, A., Caffrey, J. J., Snyder, S. H. & Shears, S. B. (2000) FEBS Lett. 468, 28–32. [DOI] [PubMed] [Google Scholar]
- 13.Odom, A. R., Stahlberg, A., Wente, S. R. & York, J. D. (2000) Science 287, 2026–2029. [DOI] [PubMed] [Google Scholar]
- 14.Flick, J. S. & Thorner, J. (1993) Mol. Cell. Biol. 13, 5861–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoko-o, T., Matsui, Y., Yagisawa, H., Nojima, H., Uno, I. & Toh-e, A. (1993) Proc. Natl. Acad. Sci. USA 90, 1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York, J. D., Odom, A. R., Murphy, R., Ives, E. B. & Wente, S. R. (1999) Science 285, 96–100. [DOI] [PubMed] [Google Scholar]
- 17.Zewail, A., Xie, M. W., Xing, Y., Lin, L., Zhang, P. F., Zou, W., Saxe, J. P. & Huang, J. (2003) Proc. Natl. Acad. Sci. USA 100, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasina, A., Price, B. D., Turenne, G. A. & McGowan, C. H. (1999) Curr. Biol. 9, 1135–1138. [DOI] [PubMed] [Google Scholar]
- 19.Hall-Jackson, C. A., Cross, D. A., Morrice, N. & Smythe, C. (1999) Oncogene 18, 6707–6713. [DOI] [PubMed] [Google Scholar]
- 20.Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M. & Abraham, R. T. (1999) Cancer Res. 59, 4375–4382. [PubMed] [Google Scholar]
- 21.Powis, G., Bonjouklian, R., Berggren, M. M., Gallegos, A., Abraham, R., Ashendel, C., Zalkow, L., Matter, W. F., Dodge, J. & Grindey, G. (1994) Cancer Res. 54, 2419–2423. [PubMed] [Google Scholar]
- 22.Stack, J. H. & Emr, S. D. (1994) J. Biol. Chem. 269, 31552–31562. [PubMed] [Google Scholar]
- 23.Cutler, N. S., Heitman, J. & Cardenas, M. E. (1997) J. Biol. Chem. 272, 27671–27677. [DOI] [PubMed] [Google Scholar]
- 24.Alarcon, C. M., Heitman, J. & Cardenas, M. E. (1999) Mol. Biol. Cell 10, 2531–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallory, J. C. & Petes, T. D. (2000) Proc. Natl. Acad. Sci. USA 97, 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkaria, J. N., Tibbetts, R. S., Busby, E. C., Kennedy, A. P., Hill, D. E. & Abraham, R. T. (1998) Cancer Res. 58, 4375–4382. [PubMed] [Google Scholar]
- 27.Moser, B. A., Brondello, J. M., Baber-Furnari, B. & Russell, P. (2000) Mol. Cell. Biol. 20, 4288–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwell, P. W., Kronmal, S. L., Porter, S. E., Gassenhuber, J., Obermaier, B. & Petes, T. D. (1995) Cell 82, 823–829. [DOI] [PubMed] [Google Scholar]
- 29.York, S. J., Armbruster, B. N., Greenwell, P., Petes, T. D. & York, J. D. (November 23, 2004) J. Biol. Chem., 10.1074/jbc.M412070200. [DOI] [PubMed]
- 30.Weinert, T. A., Kiser, G. L. & Hartwell, L. H. (1994) Genes Dev. 8, 652–665. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, X., Muller, E. G. & Rothstein, R. (1998) Mol. Cell 2, 329–340. [DOI] [PubMed] [Google Scholar]
- 32.Nagata, E., Luo, H. R., Saiardi, A., Il Bae, B., Suzuki, N. & Snyder, S. H. (2005) J. Biol. Chem., 280, 1634–1640. [DOI] [PubMed] [Google Scholar]
- 33.Kunz, J., Schneider, U., Howald, I., Schmidt, A. & Hall, M. N. (2000) J. Biol. Chem. 275, 37011–37020. [DOI] [PubMed] [Google Scholar]
- 34.Cardenas, M. E. & Heitman, J. (1995) EMBO J. 14, 5892–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennaneach, V. & Kolodner, R. D. (2004) Nat. Genet. 36, 612–617. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, Y., Desany, B. A., Jones, W. J., Liu, Q., Wang, B. & Elledge, S. J. (1996) Science 271, 357–360. [DOI] [PubMed] [Google Scholar]
- 37.Vialard, J. E., Gilbert, C. S., Green, C. M. & Lowndes, N. F. (1998) EMBO J. 17, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, H. R., Saiardi, A., Yu, H., Nagata, E., Ye, K. & Snyder, S. H. (2002) Biochemistry 41, 2509–2515. [DOI] [PubMed] [Google Scholar]
- 39.Hanakahi, L. A., Bartlet-Jones, M., Chappell, C., Pappin, D. & West, S. C. (2000) Cell 102, 721–729. [DOI] [PubMed] [Google Scholar]