SUMMARY
Invasive fungal infections cause significant morbidity and mortality in part due to a limited antifungal drug arsenal. One therapeutic challenge faced by clinicians is the significant host toxicity associated with antifungal drugs. Another challenge is the fungistatic mechanism of action of some drugs. Consequently, the identification of fungus-specific drug targets essential for fitness in vivo remains a significant goal of medical mycology research. The trehalose biosynthetic pathway is found in a wide variety of organisms, including human-pathogenic fungi, but not in humans. Genes encoding proteins involved in trehalose biosynthesis are mechanistically linked to the metabolism, cell wall homeostasis, stress responses, and virulence of Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus. While there are a number of pathways for trehalose production across the tree of life, the TPS/TPP (trehalose-6-phosphate synthase/trehalose-6-phosphate phosphatase) pathway is the canonical pathway found in human-pathogenic fungi. Importantly, data suggest that proteins involved in trehalose biosynthesis play other critical roles in fungal metabolism and in vivo fitness that remain to be fully elucidated. By further defining the biology and functions of trehalose and its biosynthetic pathway components in pathogenic fungi, an opportunity exists to leverage this pathway as a potent antifungal drug target. The goal of this review is to cover the known roles of this important molecule and its associated biosynthesis-encoding genes in the human-pathogenic fungi studied to date and to employ these data to critically assess the opportunities and challenges facing development of this pathway as a therapeutic target.
KEYWORDS: fungal pathogenesis, fungal virulence, trehalose, antifungal agents, antifungal therapy, carbon metabolism, cell wall
INTRODUCTION
The incidence of invasive fungal infections (IFIs) has increased significantly over the last 4 decades, largely due to increased use of aggressive chemotherapies for malignancies, potent immunosuppressive regimens for organ transplantation, and the HIV/AIDS pandemic (1–4). Cryptococcus, Candida, Aspergillus, and Pneumocystis are the most common genera that cause so-called opportunistic invasive fungal infections, but the incidence of many other fungal diseases is also increasing across the globe (1–9).
There are many clinical challenges in promoting positive patient outcomes for these human fungal infections that are too often lethal. For instance, a major challenge is that fungi are eukaryotes that share cellular structures and metabolic pathways with humans. Therefore, current antifungal drug options are limited and are fraught with serious side effects in humans. Currently, there are four main treatment classes of antifungal drugs for invasive disease. These classes target the fungal membrane (azoles and polyenes), cell wall (echinocandins), and RNA/DNA synthesis (flucytosine) (5). Although newer antifungal drugs (voriconazole, posaconazole, and isavuconazole) have been developed from previous antifungal drug structures, these are still limited by the route of administration, the spectrum of activity, reduced fungicidal properties, drug-drug interactions, toxicity, and bioavailability (6). For instance, drug-drug interactions with agents metabolized by the P450 cytochrome system may limit the use of triazoles, such as voriconazole (5, 7). Recently, multiple case reports of antifungal drug resistance, especially against the azoles and now the echinocandins, have been reported (5, 6, 8–11). Further, a multidrug-resistant species of Candida, Candida auris, is now emerging in clinics across the world (12). Thus, there is a critical need for novel antifungal drugs that are fungicidal but have reduced off-target side effects for the patients who desperately need them.
One strategy for discovering new and potent antifungal drugs is to target unique fungal metabolic pathways important for fungal fitness and virulence in vivo. Trehalose biosynthesis is one of the pathways that broadly exists in fungi but not in humans (13, 14). Recent studies on the molecular genetics of this pathway have revealed that components of the trehalose biosynthesis pathway are essential for Candida, Cryptococcus, and Aspergillus species to cause invasive diseases in vertebrates (13–21). However, much remains to be learned about the mechanisms through which trehalose biosynthesis affects fungus-host interactions. This knowledge is essential not only for determining the therapeutic efficacy of targeting this pathway for each respective disease but also for defining the best target for each fungal pathogen. Consequently, the goal of this review is to analyze and present the data on the role of the trehalose pathway in human fungal pathogenesis in order to assess the opportunities and challenges facing therapeutic development of this pathway.
TREHALOSE PATHWAY
What Is Trehalose?
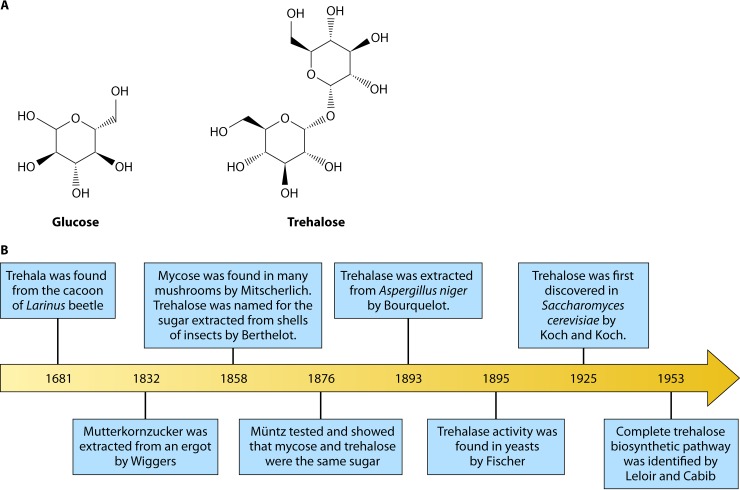
Trehalose is a nonreducing sugar containing two glucose subunits with an α,α-1,1-glycosidic linkage (Fig. 1A). It is found in plants, fungi, lichens, algae, a wide variety of bacteria, insects, and invertebrates but not in mammals (outside their microbiomes) (13, 14). Interestingly, trehalases (trehalose-degrading enzymes) are found in the kidney and the brush border of the small intestine in mammals, including humans (14). In fungi, trehalose is present in spores, fruiting bodies, and vegetative cells such as hyphae. Trehalose is rapidly depleted after germination and used in central fungal metabolism and in response to specific environmental stresses (13).
FIG 1.
(A) Chemical structures of glucose and trehalose. Trehalose consists of two glucose molecules with an α,α-1,1-glycosidic linkage. (B) Timeline of trehalose and trehalose-related enzyme discovery.
History of Trehalose
The Pharmacopoea Persica, written in 1681 by Friar Ange of Toulouse, contains the first record of trehalose, recorded as trehala in the cocoons of Larinus beetles (22–24). Around a century later, in 1832, H. A. Wiggers discovered that undisturbed solutions of ergot formed crystals of an odorless nonreducing sugar that he named “mutterkornzucker” (an ergot sugar) (24, 25). This new sugar was resistant to hydrolysis and polarized light to a greater extent than sucrose (24). In 1858, Mitscherlich found the same sugar in mushrooms and called it mycose, while Berthelot also found this sugar in cocoon-like shells of various insects from the Middle East (24). Berthelot extracted this sugar from the shell and named it trehalique glucose or trehalose (24). He also found similarity between trehalose and mycose (24). In 1876, Müntz tried to find trehalose in brewer's yeast, i.e., Saccharomyces cerevisiae. However, he could not extract trehalose by using a water and alcohol separation approach and erroneously concluded that this sugar did not exist in this important yeast. Yet he also tested and concluded that mycose and trehalose were the same sugar (26). Thereafter, Bourquelot extracted trehalase from the filamentous fungus Aspergillus niger and Fischer discovered trehalase activity in yeasts, in 1893 and 1895, respectively (26, 27). By 1925, Koch and Koch had observed an unknown sugar on the sides of a flask from an alcohol extract of Fleischmann's yeast, S. cerevisiae, that had been undisturbed for several months (26). The complete trehalose biosynthetic pathway in Saccharomyces was identified and carefully described in 1953 and 1958 by Leloir and Cabib (28, 29). Lemieux and Bauer successfully synthesized trehalose in 1954 (24, 30). The amount of trehalose in a given sample can be detected by monitoring glucose levels after treatment with trehalase, a highly specific enzyme, which cleaves trehalose into two d-glucose units (14, 31). Given the seminal role that S. cerevisiae has played in the understanding of trehalose biosynthesis and function in fungi, it is used for comparative purposes throughout this review. The timeline for trehalose discovery is summarized in Fig. 1B.
Biochemical Properties of Trehalose
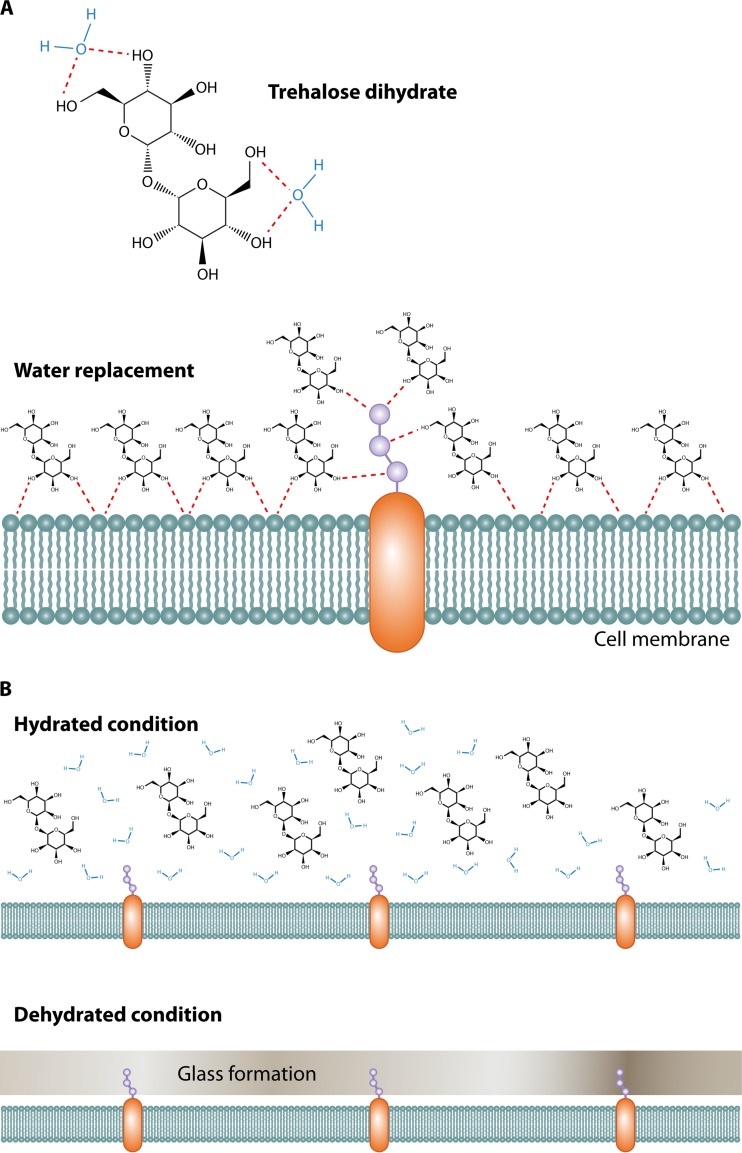
Trehalose's formal name is α-d-glucopyranosyl α-d-glucopyranoside (C12H22O11·2H2O [trehalose dihydrate] or C12H22O11 [anhydrous trehalose]) (Fig. 1A). The functions of trehalose proposed by Colaço and Roser in 1995 are myriad and include water replacement, glass transformation, and chemical stability (32). Water replacement by molecules forms hydrogen bonds and protects the surface structure. Trehalose does not form direct internal hydrogen bonds but instead forms four hydrogen bonds with two water molecules. This structure allows special molecular flexibility around the disaccharide bond and may allow trehalose to interact with phospholipid or polar groups of certain macromolecules (Fig. 2A) (32). However, surprisingly, there is still no direct evidence of specific interactions between trehalose and proteins (32). For pathogenic fungi, interactions between trehalose and other molecules are unreported and an area for further investigation.
FIG 2.
(A) Trehalose forms hydrogen bonds with two water molecules and functions as a replacement for water by interacting with the phospholipids or other macromolecules on the cell membrane to protect their structures under stress conditions. (B) Trehalose forms a glass state without any water retention or crystallization, thereby protecting the cell membrane under dehydrated conditions.
With regard to glass transformation, sugars may solidify in a glass state that helps biomolecules to stabilize and protect small hydrophobic volatile esters from evaporation in cold and desiccating environments. The glass state of trehalose is different from other sugars because it does not retain water molecules and thus does not form crystals as other sugars do (Fig. 2B). Due to this unique property, the trehalose glass state is stable at high temperatures and under desiccation conditions (33, 34). What role this property of trehalose has, if any, on fungal biology is unclear.
As a nonreducing sugar with a low free energy of activation of the glycosidic linkage, trehalose is more resistant to hydrolysis than other disaccharides. Under mildly acidic conditions, other disaccharides go through a Maillard (browning) reaction that forms many compounds, e.g., furans, imidazoles, and N-nitroso derivatives, which may negatively affect dried food nutrition. However, trehalose is relatively stable in this context and does not hydrolyze (32). In fact, O'Brien observed in 1996 that under suboptimal conditions, trehalose undergoes ∼2,000-fold fewer Maillard reactions than those observed for sucrose (35). Consequently, although there is still some controversy about the mechanisms of trehalose's protective properties, it is well known and widely used in food preservative processes and in mammalian cell and plant preservations (35). For example, it is used as a cryogenic preservative for spermatozoa and ovarian tissue and also in the formulation of commercial products such as Herceptin and Avastin. Its unique properties that preserve protein structures allow its use in everyday products (36). Further experiments to unravel the biological properties of trehalose are necessary to fully reveal its biological mechanisms and its potential for preservation of fragile cells and molecules. This is particularly true for the role of trehalose itself in the pathogenesis mechanisms of human-pathogenic fungi.
Trehalose Biosynthesis in Fungi
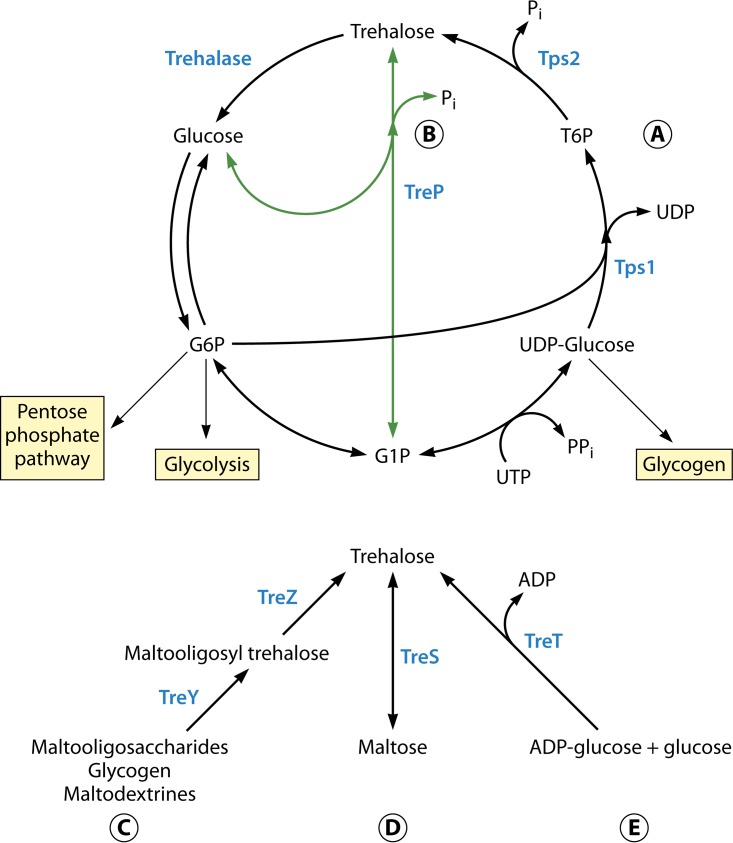
Several trehalose biosynthesis pathways have been identified throughout the tree of life, and five of these pathways are currently well described (37, 38). The first pathway consists of two main enzymes: trehalose-6-phosphate synthase (TPS or Tps1p) and trehalose-6-phosphate phosphatase (TPP or Tps2p) (Fig. 3A) (28, 29). TPS converts UDP-glucose and glucose-6-phosphate (G6P) into UDP and trehalose-6-phosphate (T6P). T6P is converted by the TPP into trehalose and free inorganic phosphate (Pi). This canonical pathway is found in a wide variety of organisms, such as eubacteria, archaea, fungi, plants, and insects (13). It is the most well-studied trehalose biosynthesis pathway in human-pathogenic fungi. The second pathway found in some fungi was reported in 1971 (39) and was characterized for the mushrooms Agaricus bisporus (40), Grifola frondosa (41), and Schizophyllum commune (42). It is also found in some bacteria (43–46) and protists (47). This pathway is found in the model filamentous fungus Neurospora crassa, as evidenced by the identification and characterization of the ccg-9 gene (48), which encodes a trehalose phosphorylase enzyme (TreP). TreP converts glucose-1-phosphate (G1P) and glucose into trehalose (Fig. 3B). This pathway may be reversible, but to date, that property has been shown only in vitro (40, 49). This noncanonical pathway of trehalose biosynthesis in fungi is likely present in the human fungal pathogen Aspergillus fumigatus, as suggested by the presence of 2 TreP-encoding genes, but so far has not been characterized fully for A. fumigatus or other pathogenic fungi (21). Thus, it remains unclear whether trehalose can be produced through TreP in human-pathogenic fungi, and the role of the TreP genes in the pathogenesis of fungi is unknown. In the context of assessing the value of targeting trehalose biosynthesis for therapeutic development, it seems warranted to define all possible trehalose biosynthesis pathways in human-pathogenic fungi. This is a significant challenge, as a noncanonical pathway may be functional only under specific environmental or cellular conditions. Regardless, if an alternative pathway can complement inhibition of the primary pathway, this must be taken into consideration in targeting these pathways for therapeutic development.
FIG 3.
Trehalose biosynthesis pathways in fungi. (A) The Tps1/Tps2 (TPS/TPP) pathway consists of two main enzymes: trehalose-6-phosphate synthase (Tps1) and trehalose-6-phosphate phosphatase (Tps2). (B) The alternative trehalose phosphorylase pathway (TreP) consists of a trehalose phosphorylase enzyme that reversibly converts glucose and glucose-1-phosphate (G1P) into trehalose and inorganic phosphate (Pi). (C) The TreY/TreZ pathway consists of a maltooligosyl trehalose synthase enzyme (TreY) and a maltooligosyl trehalose trehalohydrolase (TreZ). TreY converts maltooligosaccharides, glycogen, and starch (maltodextrins) into maltooligosyl trehalose, and TreZ releases trehalose from maltooligosyl trehalose. (D) Trehalose synthase (TreS) changes maltose directly into trehalose. (E) ADP-glucose and glucose are reversibly converted into trehalose by trehalose glycosyl-transferring synthase (TreT).
A third pathway is found in Arthrobacter spp. and Sulfolobus spp. (50–52) and is related to the maltooligosyl trehalose synthase enzyme TreYp. TreYp changes the maltooligosaccharides glycogen and starch (maltodextrins) into maltooligosyl trehalose. Trehalose is released from maltooligosyl trehalose by the enzyme maltooligosyl trehalose trehalohydrolase (TreZp) (Fig. 3C). There are some bacteria, e.g., Pimelobacter sp. (53, 54), that can change maltose directly into trehalose by using a trehalose synthase (TreSp). The TreSp pathway is the fourth trehalose biosynthesis pathway (Fig. 3D). The fifth pathway was discovered recently in Thermococcus litoralis and other extremophilic archaea, e.g., Pyrococcus spp. (55, 56). This pathway utilizes a trehalose glycosyl-transferring synthase (TreTp) to reversibly generate trehalose by using ADP-glucose and glucose (Fig. 3E) (55, 56). However, the last three pathways have not been described in fungi. Importantly, the existence of multiple pathways of trehalose biosynthesis highlights the central role of trehalose production in a variety of organisms capable of causing human disease. This broad importance of trehalose in the physiology of many microbes may have therapeutic implications that need to be considered at a deeper level (discussed later in this review).
Trehalose is degraded into two glucose molecules by trehalase enzymes (57). Fungal trehalases are divided into two groups, namely, nonregulatory and regulatory trehalases (57). Nonregulatory trehalases, found in the ascomycetes and basidiomycetes, function under acidic conditions (pH 3.5 to 5.5) and have high heat stability (57). They do not possess rapid activity changes during periods of fast trehalose mobilization and are not activated by phosphorylation (57). Regulatory trehalases, found in Phycomyces blakesleeanus, Mucor rouxii, Pichia pastoris, Piptocephalis spp., and S. cerevisiae, function under neutral conditions (pH 6.0 to 7.5) and have low heat stability (57). Unlike nonregulatory trehalases, they are rapidly activated during trehalose mobilization (57). A summary of the known genes encoding proteins important for trehalose metabolism in fungi is presented in Table 1.
TABLE 1.
Genes involved in the trehalose pathway in fungia
| Fungal species | Gene name | Gene product | Function/properties of gene product | Phenotype of mutant | Reference(s) |
|---|---|---|---|---|---|
| Yeasts | |||||
| Saccharomyces cerevisiae | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | Inability to grow on glucose and other fermentable sugars | 64 |
| TPS2 | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Inability to grow at temp above 34°C and accumulation of T6P | 115, 116 | |
| TSL1 | Regulatory subunit | Controlling TPS complex activity | Decreased trehalose synthase activity | 96, 123, 124, 126 | |
| TPS3 | Regulatory subunit | Controlling TPS2 activity | No effect on trehalose synthase activity | 124–126 | |
| NTH1 | Neutral trehalase | Degrading trehalose into glucose at pH 7.0, controlling intracellular trehalose degradation | Susceptible to heat shock | 137, 138 | |
| NTH2 | Neutral trehalase | No significant trehalase activity | Susceptible to heat shock | 137, 138 | |
| ATH1 | Acid trehalase | Degrading trehalose into glucose at pH 5.0, controlling trehalose utilization | Inability to grow on trehalose as a carbon source | 142 | |
| Schizosaccharomyces pombe | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | No growth defect on glucose-containing medium | 208 |
| TPP1 | Main trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Inability to grow at temp above 37°C and accumulation of T6P | 209 | |
| Candida albicans | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | Inability to grow on glucose at 42°C, hyphal transformation defect at 37°C, increased sensitivity to oxidative stress, and attenuation of virulence in systemic mouse model | 15, 16 |
| TPS2 | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Decreased growth at 42°C and attenuation of virulence and dissemination in systemic mouse model | 17, 18 | |
| TPS3 | Putative regulatory subunit | Binding site for Cap1p at the promoter related to oxidative stress tolerance; involved with Efg1p, Tsa1p, and biofilm formation | ND | 127–130 | |
| NTC1 | Cytosolic trehalase | Degrading trehalose into glucose, inhibited by ATP, increased activity in stationary phase | No virulence defects in systemic mouse model | 57, 145, 146 | |
| ATC1 | Cell wall-linked trehalase | Degrading trehalose into glucose, increased activity in resting cells or in trehalose- or glycerol-containing media | Significant virulence defect in systemic mouse model | 57, 145, 146 | |
| Cryptococcus neoformans | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | Prominent growth defect on a glucose-containing medium at 37°C, increased susceptibility to oxidative stress, osmotic, stress, and antifungal drugs, decreased virulence in animal models | 19 |
| TPS2 | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Accumulation of T6P and increased sensitivity to high temp | 19 | |
| NTH1 | Trehalase | Degrading trehalose into glucose | No apparent phenotypes related to virulence | 19 | |
| Cryptococcus gattii | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | Prominent growth defect on glucose-containing medium at 37°C, defects in cell wall integrity and melanin and capsule synthesis, decreased virulence in animal models | 20 |
| TPS2 | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Accumulation of T6P and increased sensitivity to high temp, decreased virulence in animal models | 20 | |
| NTH1 | Trehalase | Degrading trehalose into glucose | No apparent phenotypes related to virulence | 20 | |
| Molds | |||||
| Aspergillus nidulans | tpsA | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | Lack of trehalose, reduced viability under constant exposure to sublethal heat or oxidative stress | 65–67 |
| orlA | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Defects in chitin production and lysis at 42°C, reduced activity of GFAT at 28°C | 75–77 | |
| treA | Acid trehalase | Degrading trehalose into glucose | Essential for growing on trehalose but not related to intracellular mobilization of the trehalose pool | 67 | |
| Aspergillus niger | tpsA | Trehalose-6-phosphate synthase | Most important for trehalose production during conidiation, induced by different derepressing carbon sources | Reduced trehalose content | 109, 110 |
| tpsB | Trehalose-6-phosphate synthase | Induced by heat shock | No apparent phenotypes | 109, 110 | |
| tpsC | Trehalose-6-phosphate synthase | Expressed at low level at 72 h | No apparent phenotypes | 110 | |
| tppA | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Reduction of T6P and trehalose and abnormal growth, conidiation, and accumulation of T6P | 110 | |
| tppB | Putative regulatory subunit | Possible alternative T6P phosphatase | Increased sensitivity to oxidative, salt, and acid stresses | 110 | |
| tppC | Putative regulatory subunit | Possible alternative T6P phosphatase | No apparent phenotypes | 110 | |
| Aspergillus fumigatus | tpsA | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | No apparent phenotypes | 68 |
| tpsB | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P | No apparent phenotypes, lack of trehalose with loss of both tpsA and tpsB | 68 | |
| tpsC | Putative trehalose-6-phosphate synthase | Low expression at 37°C and under other stress conditions | No apparent phenotypes | 68 | |
| tpsD | Putative trehalose-6-phosphate synthase | Low expression at 37°C and under other stress conditions | No apparent phenotypes | 68 | |
| orlA | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | Accumulation of T6P, defects in cell wall integrity, no lysis of germlings under temp stress, and significant virulence defect in invasive aspergillosis mouse model | 21 | |
| Magnaporthe oryzae | TPS1 | Trehalose-6-phosphate synthase | Converting UDP-glucose and G6P into T6P, regulating G6P metabolism, pentose phosphate pathway, and carbon and nitrogen source utilization | Inability to grow in glucose-containing medium, inability to cause invasive disease in rice | 204, 207, 210–215 |
| TPS2 | Trehalose-6-phosphate phosphatase | Dephosphorylating T6P into trehalose and inorganic phosphate | ND | 213 | |
| TPS3 | Regulatory subunit | Regulating trehalose synthesis | Decreased T6P and trehalose levels, defect in glycogen metabolism, inability to cause rice blast disease | 213 | |
| NTH1 | Neutral trehalase | Degrading trehalose into glucose | Ability to infect plants normally but decreased ability to colonize plant tissue | 204 | |
| TRE1 | Cell wall-localized trehalase | Degrading trehalose into glucose | Inability to grow on trehalose, no effect on virulence | 204 |
Abbreviations: G6P, glucose-6-phosphate; T6P, trehalose-6-phosphate; ND, not described; GFAT, glutamine:fructose-6-phosphate amidotransferase.
Specific Functions of Trehalose in Fungi
Trehalose has critical functions in fungal biology. There is a hypothesis that trehalose serves as an alternative carbon source for some fungi, though how this relates to pathogenesis is unclear (57). One possibility that has not been tested experimentally is that trehalose produced by the microbiota of the human body provides a carbon source for fungi under certain pathological or homeostatic conditions. Trehalose is critically important for filamentous fungal conidium survival and germination, likely due to its function as a carbon source (57). An energy reserve, i.e., glycogen, usually accumulates when nutrients are rich (58). In contrast, trehalose accumulates in yeast after glucose depletion, at the beginning of the stationary phase. In the stationary phase, yeast cells use glycogen initially, but trehalose is utilized under extreme starvation conditions (58, 59). From these data, it can be concluded that trehalose is less likely to be a main reserve carbohydrate in S. cerevisiae but rather serves as an alternative carbon source under severe stress conditions. In human-pathogenic fungi, the role of trehalose as a reserve carbohydrate or an alternative carbon source is still unclear, and this area still remains to be explored in depth.
Trehalose has a critical role as a general stress protectant in fungi, especially in response to dehydration and thermal stress. For example, trehalose is proposed to form hydrogen bonds with proteins and to interact with the polar head groups of phospholipids to maintain membrane structure under dehydration conditions. Trehalose also inhibits both fusion and lipid-phase transitions under anhydrobiotic conditions (60, 61). Under these conditions, trehalose may form hydrogen bonds with proteins, but as discussed above, specific molecular interactions remain elusive and it is unclear whether these conditions and mechanisms influence fungus-host interactions in vivo. However, growth at human body temperature may be considered an extreme stress for many fungi. In S. cerevisiae at suboptimal environmental temperatures, the total amount of trehalose (e.g., after 1 h of exposure to 40°C) increases dramatically (62, 63). Under the same conditions, the trehalose phosphate synthase (TPS) and neutral trehalase enzymes have increased activity to both produce and degrade trehalose during heat stress and the recovery phase, respectively (62, 63). The trehalose phosphate synthase complex accumulates under heat stress conditions, and a tps1 null mutant of S. cerevisiae has a growth defect in glucose-containing medium at high temperatures (64).
Consequently, it is perhaps not surprising that trehalose is also important for thermal stress survival of human-pathogenic fungi, including Candida albicans (15), Cryptococcus neoformans (19), and the rarely pathogenic fungus Aspergillus nidulans (65–67). A trehalose-deficient mutant (ΔtpsA/B) of A. fumigatus is sensitive to heat shock, as suggested by a significant decrease in viability at 50°C (68). However, at normal human body temperature, loss of TpsA and TpsB in A. fumigatus does not affect viability. Thus, mechanisms other than trehalose biosynthesis per se appear to support the ability of A. fumigatus to thrive at human body temperature. Conversely, trehalose biosynthesis-encoding genes are important for cold shock in Escherichia coli (69). Trehalose also accumulates in S. cerevisiae after 12 h of exposure to 10°C (late cold response), but a tps1/tps2 double null mutant still survives at 10°C (70). This result suggests the existence of an unknown pathway for yeast cold adaptation. Nonetheless, it has been hypothesized that trehalose may still prevent the aggregation of denatured proteins under cold conditions and thus keep the cell membrane intact (13).
Trehalose also plays an important role as a free radical scavenger under oxidative stress conditions (71). This likely has direct relevance to human fungal pathogenesis, as reactive oxygen species (ROS) play a critical role in the immune system defense against fungi and other pathogenic microbes. A trehalose-deficient mutant of S. cerevisiae is more susceptible to hydrogen peroxide (H2O2) and accumulates more oxidized proteins than the wild type (71). A lack of trehalose in human-pathogenic fungi, e.g., C. albicans (16, 72), C. neoformans (19), and A. nidulans (65), also affects survival under oxidative stress conditions. For example, the A. fumigatus tpsA/tpsB null mutant has reduced viability after exposure to 100 mM H2O2 (68). Therefore, prevention of trehalose biosynthesis in vivo may enhance the efficacy of ROS-dependent host defense mechanisms that are often compromised in the setting of immune suppression therapies, such as treatment with corticosteroids. Consequently, a more thorough investigation of the role of fungal trehalose biosynthesis in immune cell interactions is warranted.
As an important component of the cell wall, trehalose-containing glycolipids, such as trehalose-dimycolate, are involved in virulence and cell wall homeostasis in Mycobacterium species (73, 74). However, to date, there are no robust reports of trehalose contributing directly to structural components of the fungal cell wall. Nonetheless, loss of tps2 (called orlA in aspergilli) in A. nidulans and A. fumigatus affects cell wall homeostasis, likely through a reduction in chitin content leading to cell lysis at high temperatures (21, 75–77). This observation raises a critical point for consideration in identifying promising drug targets in the trehalose biosynthesis pathway. The canonical trehalose biosynthesis pathway utilizes substrates, i.e., glucose-6-phosphate and UDP-glucose, that intersect with glycolysis, the pentose phosphate pathway, and, consequently, cell wall biosynthesis. Therefore, disruption of this pathway leads to dysregulation of other key metabolic pathways in the cell, and as such, the overall consequences and impact of trehalose biosynthesis on each fungal pathogen remain to be fully defined. Given the importance of the cell wall in host-fungus interactions and fungal survival, the mechanistic connection(s) between the trehalose biosynthesis pathway, trehalose itself, and cell wall biosynthesis is an important area of ongoing research.
As suggested above, trehalose and associated molecules needed for its synthesis have impacts on central carbon metabolism. For example, disruption of tps1 in S. cerevisiae results in a significant growth defect on glucose-containing media (64, 78). Ultimately, loss of TPS in S. cerevisiae results in altered energy homeostasis at the level of glycolysis, though the mechanisms remain to be fully elucidated. In C. albicans, loss of tps1 does not affect growth on glucose at 30°C but does at 42°C (15). In Cryptococcus neoformans and Cryptococcus gattii, loss of Tps1p function impairs growth on glucose at 37°C (19, 20). In contrast, no defect in growth on glucose-containing medium was observed for the A. fumigatus tpsA/tpsB null mutant (68). For a pathogenic filamentous fungus such as A. fumigatus, accumulation of the signaling molecule T6P inhibits hexokinases and regulates the influx of glucose (21).
As discussed in part above, preliminary molecular genetic analyses have revealed important and complex roles for the trehalose biosynthesis pathway in fungal biology that extend beyond the biosynthesis of trehalose itself. However, the specific mechanisms remain to be elucidated and fully appreciated. Surprisingly, the amino acid sequences of the proteins involved in trehalose biosynthesis are highly conserved among TPS1, TPS2, and the regulatory subunits (TPS3/TSL1). In some pathogenic fungi, there is an expansion of the number of these genes, but their functions remain to be elucidated fully. A similar situation exists in Arabidopsis thaliana, which contains 11 putative AtTPSs and 10 putative AtTPPs (79–82). These observations highlight the importance of these gene products in metabolic homeostasis and fitness across a diverse array of organisms and environments. Consequently, they also provide support for the hypothesis that this pathway is an attractive antifungal drug target whose inhibition will severely diminish the fitness and virulence of pathogenic fungi.
TPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE)
Tps1p, the trehalose-6-phosphate synthase, is found in both bacteria and eukaryotes. Tps1p converts UDP-glucose and glucose-6-phosphate (G6P) into trehalose-6-phosphate (T6P). For S. cerevisiae, tps1 mutants were initially characterized in the late 1970s and were called by many names, e.g., FDP1, CIF1, BYP1, and GLC6 (83–87). Later it was shown that these mutations are all allelic with the same gene, tps1 (64, 88). fdp1 and cif1 mutants cannot grow on glucose and other fermentable sugars, such as fructose, mannose, and sucrose. Furthermore, these mutants show a defect in glucose-induced inactivation of fructose-1,6-bisphosphatase and have very low trehalose levels with a change in the regulation of glycogen synthesis (84, 85, 89, 90). The byp1 mutant was previously believed to bypass glycolysis, and it shares similar phenotypes with fdp1 and cif1 mutants (86, 91, 92). The byp1 mutant displays slow growth and is completely blocked for growth on both glucose and fructose (86, 91). There is also no growth on glucose of byp1 mutants deficient in pfk1 or pfk2, encoding phosphofructokinase enzymes catalyzing the phosphorylation of fructose-6-phosphate into fructose-1,6-bisphosphate or fructose-2,6-bisphosphate, respectively (86, 91). Because of the common involvement of these alleles in glucose-induced signaling, FDP1 was renamed GGS1 (general glucose sensor 1) (93).
GLC6 is related to glycogen accumulation and is allelic to CIF1, FDP1, BYP1, and GGS1 (87). While GGS1 was believed to function as a glucose sensor responsible for glucose influx, the same gene, called tps1, has been characterized as the smallest subunit of the trehalose synthase complex (64). Tps1p is an important component of trehalose-6-phosphate synthase, and deletion of tps1 in S. cerevisiae causes a growth defect on glucose-containing medium similar to those of the fdp1 and cif1 mutants (64). Moreover, expression of tps1 restores trehalose production in an Escherichia coli otsA mutant lacking a T6P synthase enzyme (the gene product of otsA) (88).
The growth defects of tps1 mutants and mutants in associated alleles provide a rich opportunity to explore the critical role of the trehalose biosynthesis pathway in the regulation of carbon metabolism. A detailed understanding of the effects of Tps1p loss on fungal metabolism in each respective organism is critical for evaluating the impact of targeting Tps1p on therapeutic development. To this end, it seems clear that a loss of TPS1 has multiple effects on carbon metabolism that are organism specific. Extensive research with S. cerevisiae has been undertaken to understand the impact of tps1 loss on regulation of glycolysis. To explain the growth defect on glucose-containing medium, three non-mutually exclusive hypotheses were initially proposed (89). The first hypothesis is that Tps1p regulates glucose influx to prevent an accumulation of sugar-phosphate intermediates upstream of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (93). In support of this hypothesis, deletion of hxk2 decreased sugar-phosphate accumulation and restored tps1 mutant growth on glucose (94). The second hypothesis proposes that trehalose synthesis provides a mechanism for free phosphate recovery (89). Reductions in intracellular phosphate after addition of glucose to the tps1 mutant and inhibition of the trehalose synthase complex by free phosphate support this hypothesis (92, 95, 96). In further support of this hypothesis, stimulation of glycerol production, which increases phosphate recovery by reducing the accumulation of sugar-phosphate intermediates, restores the tps1 mutant growth defect on glucose after a longer lag phase (89, 94). However, a byp1 tps2 double mutant shows increased growth on glucose compared to that of the single byp1 mutant, thereby contradicting the phosphate recovery hypothesis (97). Intriguingly, the byp1 tps2 double mutant still accumulates sugar-phosphate intermediates. This observation is similar to data from a tps1 mutant strain that expresses the E. coli otsA gene (98). These data argue for a model whereby T6P, trehalose, or Tps1p has additional roles in regulating the second half of glycolysis (83).
The third hypothesis proposes that T6P controls glucose influx through its inhibition of hexokinase activity (89). T6P competitively inhibits hexokinases in S. cerevisiae (99). However, overexpression of hexokinase activity does not result in a glucose growth defect in wild-type S. cerevisiae cells (100). Recently, Walther et al. observed significant acidification of the cytosol in the absence of Tps1p due to loss of a plasma membrane H+ ATPase (101). In summary, the loss of Tps1p has significant effects on the regulation of glucose influx and glycolysis in yeast cells that affect cell fitness, which bodes well for targeting this protein for antifungal drug development. However, the complexity of Tps1p function and loss in yeast highlights a potential challenge in targeting this pathway for antifungal drug development. This challenge must be met with additional research to fully define Tps1p function in respective pathogenic organisms.
Importantly, while the function of Tps1p in human-pathogenic fungi is heterogeneous across species, studies to date suggest clear roles in fungal pathogenesis that bode well for therapeutic development. In C. albicans, tps1 encodes a trehalose-6-phosphate synthase, and the promoter of this gene contains four copies of the stress response element (STRE). As stated above, the tps1/tps1 null mutant grows normally at 30°C (15). However, at 42°C, the mutant cannot grow on glucose, but it is able to grow on glycerol or galactose at higher temperatures. The mutant has a defect in hyphal formation in serum-containing medium at 37°C. It also has decreased intracellular ATP levels and no trehalose accumulation at stationary phase or after heat shock. Importantly, the mutant is attenuated in virulence in a mouse model of systemic infection (15). The tps1/tps1 null mutant displays a severe reduction in cell viability after exposure to high concentrations of hydrogen peroxide, while the wild type retains high cell viability associated with an increase in intracellular trehalose (16, 72). However, pretreatment of the C. albicans wild type or the tps1/tps1 null mutant with a nonlethal concentration of hydrogen peroxide (0.5 mM H2O2) or mild heat stress (37°C) protects both strains against oxidative stress, to the same extent (16). From this result, it can be inferred that trehalose accumulation plays a major role in protecting yeast cells from an immediate direct exposure to severe oxidative stress. Direct exposure to oxidative stress occurs during interactions with immune cells. Other pathways related to the oxidative stress response, e.g., the HOG–mitogen-activated protein kinase (MAPK) pathway and enzymatic (catalase and superoxide dismutase) and nonenzymatic (glutathione and thioredoxin) components, may protect cells after mild stress exposure (16, 102). In contrast to oxidative stress, osmotic or heat stress has no effect on cell viability of the C. albicans tps1/tps1 null mutant (72).
Perhaps consistent with the in vitro oxidative stress phenotype, the tps1/tps1 null mutant is more susceptible to macrophage killing than the wild type (103). Intriguingly, this phenotype is dependent on the culture conditions and cell wall composition. Electron microscopy reveals a difference in the outer cell wall layer in the mutant compared to the wild type (103). In liquid culture medium, the mutant increases its β-mannosylation (overglycosylation) on the outer cell wall, leading to reduced hydrophobicity and increased resistance to macrophage killing, whereas on solid medium the mutant has no difference in overglycosylation on the cell wall and is more susceptible to macrophage killing (104). These observations have direct relevance to C. albicans Tps1p as a drug target. While the reduction in virulence of the tps1/tps1 mutant in a systemic murine model is promising, the environment-specific impacts on the cell wall and phagocyte interactions warrant further investigation in in vivo models. The latter observation is important given the phagocyte resistance phenotype and altered cell wall observed under specific in vitro conditions. Importantly, this theme of trehalose biosynthesis affecting cell wall composition and integrity is found across the human-pathogenic fungi, although the mechanism(s) remains ill defined.
In C. neoformans, tps1 encodes a 671-amino-acid trehalose-6-phosphate synthase with eight putative STREs in the promoter region. A tps1 null mutant displays a prominent growth defect on glucose-containing medium at 37°C, but the growth defect is restored in a galactose-containing medium, similar to the case with C. albicans (19). The tps1 null mutant grows in glucose-containing medium supplemented with 1 M sorbitol at 37°C, which suggests an impact on the fungal cell wall (19). Trehalose and T6P are not detected in the mutant (19). The C. neoformans tps1 mutant is more susceptible to oxidative stress by hydrophobic peroxides, e.g., t-BOOH, to osmotic stress (1 M sorbitol), and, importantly, to antifungal drugs (amphotericin B and caspofungin) (19). The increased susceptibility of the mutant to current antifungal drugs raises the possibility of a combination therapy approach targeting trehalose biosynthesis in addition to current antifungal drugs.
A critical relationship between trehalose and its synthesis through Tps1p and the virulence of C. neoformans is establishing Tps1p as a potential antifungal drug target in this important human pathogen (105–107). tps1 was one of the most highly expressed transcripts in a global transcriptional analysis of yeast cells from cerebrospinal fluid (CSF) from rabbits with cryptococcal meningitis (107). Nuclear magnetic resonance (NMR) studies show that trehalose is one of the most abundant metabolites in cryptococcomas (105, 106). To further support the importance of trehalose in the virulence of C. neoformans, the tps1 null mutant is avirulent in both rabbit and murine models of infection (19). Moreover, significant attenuation of the virulence of the tps1 null mutant is also observed in Caenorhabditis elegans and zebrafish models (19, 108). These results in model organisms with body temperatures approximating room temperature reveal that the effect of Tps1p loss on C. neoformans virulence goes beyond growth inhibition at mammalian body temperature (19, 108). Additional research on the C. neoformans tps1 null mutant in the context of interactions with the immune system is warranted.
Tps1p is also critical in the related pathogenic yeast, Cryptococcus gattii, which causes disease in immunocompetent hosts. However, there are important aspects of the tps1 mutants of C. neoformans and C. gattii that differ (20). For example, growth of the tps1 mutant in C. gattii is not restored at 37°C in galactose-containing media, and it exhibits significant defects in cell wall integrity and melanin and capsule synthesis that are not observed in C. neoformans (20). Importantly, the C. gattii tps1 null mutant possesses profound virulence defects in both invertebrate and mammalian hosts. These species-specific differences in the trehalose pathway may affect the differences in disease presentation and/or ecological diversity of the Cryptococcus species complex. However, more studies are needed to determine the precise mechanisms and causality. A major future research direction alluded to above is interrogation of these mutants in vivo in the setting of active host responses under dynamic environmental conditions that occur in an active infection. Importantly, these data highlight and emphasize the importance of studying the trehalose pathway and its role in the biology of each fungal species. This conclusion is emphasized when one turns to studies of this pathway in the genus Aspergillus.
Unlike the yeast species studied to date, in the genus Aspergillus multiple copies of the TPS1 gene exist in several species, with the relatively nonpathogenic species A. nidulans being an interesting exception. In A. nidulans, which has only one trehalose-6-phosphate synthase (tpsA), trehalose is found in conidia at high concentrations and is rapidly used during germination (65). A. nidulans also accumulates trehalose during conidial differentiation, nutrient starvation, heat shock, and oxidative shock (65–67). A tpsA null mutant lacks production of trehalose and shows reduced conidial viability during constant exposure to sublethal stresses, i.e., prolonged high temperature and oxidative stress (65). In Aspergillus niger, two copies of tps1 exist and are named tpsA and tpsB (109). TpsA is the most important for trehalose production during conidiation and has increased expression on derepressing carbon sources (109). tpsA is expressed constitutively, while tpsB is induced by heat shock and has a role during differentiation (110). A. fumigatus also contains tpsA and tpsB, and loss of both genes eliminates trehalose production (68). Intriguingly, in this common human fungal pathogen of immunocompromised patients, two additional genes encoding proteins with amino acid sequence similarity to TpsA and TpsB are found in the genome and have been named tpsC and tpsD (68). However, mRNA levels of tpsC and tpsD are low under the in vitro environmental conditions examined to date. Consequently, their role in A. fumigatus pathobiology is not currently clear. Importantly, unlike C. albicans, C. neoformans, and S. cerevisiae tps1 null mutants, the A. fumigatus double tpsA/tpsB mutant grows normally in the presence of glucose at 37°C (68). The ability to grow in the presence of glucose in the absence of genes encoding catalytic TPS may indicate a regulatory or structural role for TpsC and TpsD in regulating glycolytic and/or carbon flux (68). Alternatively, or in addition, A. fumigatus contains a highly active glucokinase (glkA) that activates glucose and is likely resistant to inhibition by T6P (111). Despite the normal growth on glucose of the tpsA/tpsB null mutant, carbon metabolism is significantly altered, as clear changes in the cell wall of the mutant are observed (68). From transmission electron microscopy (TEM) images, the electron-dense outer layer on the cell walls of both hyphae and conidia of the tpsA/tpsB null mutant is absent, and conidia of the mutant have an enhanced electrolucent zone compared to that of the wild type (68). In addition to the abnormal cell wall structure from TEM images, the expression of ags3, encoding an α-glucan synthase, is significantly reduced in the double null mutant, and its reduction might contribute to the abnormal cell wall structure (68). Surprisingly, despite increased sensitivity to oxidative stress, the A. fumigatus tpsA/tpsB null mutant displays a modest increase in virulence in a cortisone acetate murine model of invasive pulmonary aspergillosis. While the mechanism behind this hypervirulence is not fully understood, cell wall changes in the null mutant may promote increased immunopathogenesis through activation of detrimental host immune responses. Alternatively, enhanced in vivo fitness of the null mutant due to alterations in carbon metabolism and stress responses cannot be ruled out. Regardless, the virulence persistence of the tpsA/tpsB null mutant of A. fumigatus is striking in comparison to the results of TPS studies of other pathogenic fungi and is worth further investigation in the context of therapeutic development.
On the surface, these results for A. fumigatus appear to argue against targeting Tps1p for antifungal drug development from the perspective of developing a broad-spectrum target. However, the latter conclusion must be tempered by the observation that two additional Tps1p-like proteins exist in A. fumigatus that remain to be characterized. While the loss of TpsA and TpsB is clearly not sufficient to negatively affect A. fumigatus virulence, elimination of all 4 TPS1-like proteins in a strain has yet to be investigated. Moreover, one cannot rule out that in vivo, at the infection site microenvironment, the tpsA/tpsB null mutant is somehow complemented by host or fungal factors to produce trehalose and/or other virulence factors. These results in conjunction with the yeast data suggest that TPS proteins play multiple roles beyond trehalose biosynthesis that warrant further investigation. In vivo analyses of fungal genetic null mutants remain a significant technical challenge that is beginning to be met with novel techniques, such as Nanostring nCounter gene expression analyses and conditional promoter systems, among other approaches (112–114). Thus, despite the above challenges, unraveling the differences in the functions of TPSs between A. fumigatus and pathogenic yeasts has the potential to yield new insights into the Achilles' heels of these important pathogens.
TPS2 (TREHALOSE-6-PHOSPHATE PHOSPHATASE)
Tps2p is the trehalose-6-phosphate phosphatase (TPP) that dephosphorylates T6P into trehalose and inorganic phosphate. The first described TPP was Tps2p of S. cerevisiae (115). Investigators observed that a tps2 null mutant could not grow at temperatures above 34°C (116). There are now multiple reports that suggest T6P accumulation as the cause of the temperature-sensitive phenotype of the tps2 mutant of S. cerevisiae (115, 117). Furthermore, T6P accumulates in this mutant during heat shock (115, 116). Thus, under stress conditions, it seems likely that accumulation of T6P in tps2 mutants is toxic to cells (116). For human-pathogenic fungi, tps2 null mutants consistently display severe virulence attenuation and other fitness defects that portend well for Tps2-targeted antifungal drug development.
In C. albicans, Tps2p contains 878 amino acid residues, with two phosphohydrolase domains, and shares 67% sequence similarity with S. cerevisiae Tps2p and 73% similarity with A. nidulans OrlA (17, 18). Van Dijck et al. observed that a C. albicans tps2/tps2 null mutant accumulates T6P with a temperature-sensitive phenotype at 44°C (18). In contrast to the C. albicans tps1/tps1 null mutant, the tps2/tps2 null mutant does not have hyphal formation defects on glucose-containing medium at 30°C (18). The tps2/tps2 null mutant has a significant decrease in growth rate at 42°C and aggregates in stationary phase, with a defect in cell wall integrity, when the pH is over 7.0 (17). Importantly, the mutant has a significant decrease in virulence, highlighted by a decrease in fungal dissemination to the kidneys and livers, in a systemic murine model (17, 18). The mechanism(s) for the decreased dissemination and virulence in the absence of Tps2p is currently unclear but is an important area of research for further validation of Tps2p as an antifungal drug target.
Along these lines, as mentioned earlier, the importance of trehalose biosynthesis in fungal stress responses and virulence suggests that therapeutic development of this pathway should also be considered in the context of combination approaches. For example, loss of C. albicans Gpr1p, a nutrient receptor activating cyclic AMP-protein kinase A (cAMP-PKA)-mediated signaling, results in a significant increase in trehalose levels and morphological defects on hypha-inducing media (118, 119). However, a C. albicans gpr1/gpr1 null mutant displays only a slight decrease in virulence in a systemic murine model (120). In contrast, loss of tps2 in the gpr1/gpr1 null mutant completely attenuates virulence (120). The gpr1 tps2 double null mutant accumulates very high levels of T6P under stress conditions, with a concomitant growth defect at high temperatures (120). These results support the observation that an increase in intracellular T6P levels has an enormous effect on the virulence and fitness of C. albicans under specific conditions, particularly those that promote TPS activity. Consequently, treatments that promote increased TPS activity may be synergistic in vivo with a drug targeting Tps2p to drive accumulation of toxic T6P levels. Given that TPS activity is induced by many host-associated stresses in vivo, Tps2p-targeting drugs may be expected to have increased potency in vivo, beyond in vitro MIC testing results.
In C. neoformans, tps2 encodes a predicted 988-amino-acid protein with the presence of four STREs in the promoter region (19). C. gattii tps2 encodes a 990-amino-acid protein with 89% sequence similarity to C. neoformans Tps2p (20). The tps2 null mutants of both C. neoformans and C. gattii accumulate T6P and have severe growth defects at 37°C on glucose-containing medium, and in fact, they die at this temperature (19, 20). The growth and survival defects are rescued by growth on galactose- or sorbitol-containing medium at 37°C (19, 20). As in S. cerevisiae, C. gattii has a strong connection between the trehalose pathway and control of glycolytic fluxes via hexokinases. For instance, the loss of hxk2 in a tps2 null mutant background suppresses the high-temperature growth defect, possibly by reducing the pool of glucose-6-phosphate that is a building block for toxic T6P accumulation (20). From a therapeutic perspective, the toxic effect of Tps2p loss in Cryptococcus is tremendously exciting and further highlights the possibility of therapeutically targeting this important virulence-associated fungal enzyme. However, the C. gattii tps2 null mutant has only a slight decrease in virulence in the C. elegans model, though a profound virulence defect in the murine inhalation model is observed (20). Consequently, the lack of full virulence attenuation in the C. elegans model warrants further investigation, as specific host conditions may feasibly inhibit or overcome the loss of Tps2p activity. However unlikely, this hypothesis can be tested experimentally with the development of Tps2-inhibiting small molecules in relevant animal models.
For the filamentous aspergilli, the story is somewhat different but no less impactful. For A. nidulans, the Tps2 gene ortholog has been characterized and was named orlA (osmotic-remediable lysis strain) (75–77). orlA encodes a 908-amino-acid protein that shares the same predicted protein domains as S. cerevisiae Tps2p (77). The A. nidulans orlA null mutant has a defect in chitin production and lyses when grown at 42°C, whereas growth is partially recovered on osmotic stabilizers or an N-acetylglucosamine-containing medium (75). Furthermore, glutamine:fructose-6-phosphate amidotransferase (GFAT), the first step in amino sugar synthesis, has reduced activity in the A. nidulans orlA null mutant at 28°C (75, 77). It is hypothesized that the lysis phenotype of the A. nidulans orlA null mutant occurs from a defect in chitin synthesis (76). The orlA null mutant accumulates significant levels of T6P but, surprisingly, still produces trehalose at both 32 and 42°C (77). For A. niger, a tppA (tps2) null mutant shows abnormal growth, conidiation, and accumulation of T6P (110). The A. fumigatus orlA mutant exhibits defects in colony morphology and conidiation on glucose minimal medium (GMM) at 37°C (21). However, unlike that of the yeast tps2 null mutants, A. fumigatus growth at 37°C is affected only minimally on GMM. A substantial accumulation of T6P and significant changes in cell wall integrity are observed in the A. fumigatus orlA mutant, but similar to A. nidulans and C. albicans, trehalose production is not lost. The mechanism behind the persistence of trehalose production in some fungal tps2 null mutants remains enigmatic. Unlike A. nidulans, A. fumigatus orlA null mutant germlings do not lyse under temperature stress, possibly highlighting important cell wall differences between these two species. Importantly for therapeutic considerations, the orlA null mutant has a striking attenuation in virulence in a leukopenic invasive pulmonary aspergillosis murine model (21). Initial attempts to characterize metabolic defects in the orlA null mutant revealed a reduction in hexokinase activity along with a reduction in pyruvate decarboxylase activity required for ethanol fermentation (21). Intriguingly, loss of ethanol production through loss of the alcohol dehydrogenase alcC gene significantly reduces lung fungal burdens in murine models of invasive pulmonary aspergillosis (121).
In summary, loss of Tps2p consistently results in severe temperature-sensitive growth defects and/or attenuated virulence in human-pathogenic fungi. Thus, Tps2p orthologs in pathogenic fungi are potential broad-spectrum antifungal drug targets that are absent in humans. However, the mechanisms through which Tps2p mediates growth and virulence in pathogenic fungi still remain to be fully defined. While loss of Tps2p in yeast is conditionally lethal, the consequences of Tps2p loss are more nuanced in molds. For example, it is unclear if the reduction in chitin content in the A. fumigatus orlA null mutant is compensated by overproduction of the proinflammatory pathogen-associated molecular pattern (PAMP) beta glucan. If this is so, use of a Tps2p-inhibitory drug in certain patient populations may unexpectedly promote immunopathogenesis. These hypotheses remain to be tested experimentally. A major recent advance directly relevant to targeting Tps2p is the solving of high-resolution protein structures from three human-pathogenic fungi (discussed more below). The careful assessment of the germane protein structures, recently completed, is expected to catalyze the discovery of specific inhibitors of this phosphatase (122).
REGULATORY SUBUNITS
TPS3/TSL1
In S. cerevisiae, two additional proteins with high amino acid sequence similarity to Tps1p and Tps2p are found (96, 123–125). Tsl1p (trehalose synthase long chain) is found in the same protein complex as Tps1p and Tps2p (TPS complex) (96, 123), while Tps3p is also found to be a part of the complex (124). During heat shock, Tsl1p and Tps3p have similar regulatory roles in the TPS complex of S. cerevisiae (125). However, Tsl1p and Tps3p have different impacts on trehalose synthase activity under other conditions (124). While a tsl1 null mutant has reduced trehalose synthase activity, deletion of tps3 does not affect trehalose synthase activity (124). Tsl1p is important for TPS complex activity, while Tps3p is a target for phosphorylation that regulates Tps2 activity (126). It has been proposed that Tsl1 might also play a structural role in formation of the TPS complex, as no TPS activity is detected in the absence of Tsl1p (126). Consequently, these data strongly support a model in which trehalose-producing enzymes function as a complex in S. cerevisiae. Whether a similar or identical model occurs in pathogenic fungi is unclear, as biochemical work on the respective trehalose proteins in these fungi is thus far sparse. Given some of the divergent phenotypes associated with TPS/TPP mutants in pathogenic fungi and the presence/absence of specific genes, alternative models are possible, if not likely. It will be important to define these mechanisms and structures in the respective pathogenic fungi to maximize antifungal drug development opportunities. To this end, genetic analysis of regulatory subunit homologs has been conducted in some human-pathogenic fungi.
For C. albicans, a putative regulatory subunit called Tps3p is found encoded in the genome by amino acid sequence similarity searches, with 41% identity to S. cerevisiae Tps3p. However, it is still unclear how or whether C. albicans Tps3p regulates trehalose production in this important human-pathogenic yeast. The tps3 promoter may contain a binding site for Cap1p, which is a transcription factor related to oxidative stress tolerance in C. albicans. tps3 mRNA levels are induced significantly when C. albicans is exposed to hydrogen peroxide (127). C. albicans tps3 is positively regulated by Efg1p, which is an APSES (Asm1, Phd1, Sok2, Efg1, and StuA family) transcriptional regulator involved in the yeast-to-hypha transition and cell shape during white-opaque switching, among other important phenotypes (128). A cell wall protein in hyphal cells, Tsa1p, is related to oxidative stress resistance and hyphal cell wall integrity, and a tsa1 null mutant contains reduced tps3 mRNA levels under oxidative stress conditions (129). Furthermore, tps3 mRNA levels are reduced in C. albicans during biofilm formation (130). To date, a potential C. albicans Tsl1p homolog has not been identified by sequence comparison with BLAST algorithms. For Cryptococcus gattii, a putative Tps3p (CGB_I4320W) homolog has been identified by BLAST searches, with 33% identity to S. cerevisiae Tps3p, but it has not been characterized. Interestingly, a Tps3p homolog has not been identified in C. neoformans by sequence analyses.
In A. niger, TppB and TppC are the homologs of Tps3p and Tsl1p, respectively. The tppB null mutant is more susceptible to thermal stress and has reduced internal trehalose levels, but it does not show increased sensitivity to oxidative stress, osmotic stress, or acidic stress (110). Because TppB and TppC share significant amino acid sequence similarity with the TPP TppA (Tps2/OrlA), it has been hypothesized that they may function as alternative phosphatases in the absence of TppA, which would explain the persistence of trehalose production in tps2 null mutants (110). Importantly, however, the potential TPP activity of both TppB and TppC remains to be investigated to support this hypothesis. In A. fumigatus, as in A. niger, two proteins encoded in the genome share protein domains similar to those in OrlA and are the likely A. fumigatus Tps3p and Tsl1p homologs. The functions of these two proteins in trehalose biosynthesis and fungal metabolism are currently unknown but are under investigation in our laboratory (our unpublished data).
The catalytic residues required for TPP activity have been defined for the archaeon Thermoplasma acidophilum (TaT6PP) (aspartates 7, 9, 179, 180, and 183, threonines 11, 45, and 182, arginine 47, and lysine 161) (131) and the filariasis-causing parasite Brugia malayi (motif I active sites, aspartates 213 and 215; motif II active site, threonine 253; motif III active site, lysine 398; and motif IV active sites, aspartates 424 and 428) (132). These amino acid residues are found in the Mg2+-dependent haloacid dehalogenase (HAD) phosphatase domain superfamily (131). S. cerevisiae Tps3p and Tsl1p have a HAD-like domain but no known catalytic activity (133). In the plant A. thaliana, Tps3p also contains the HAD domain but also has no TPP activity (133, 134). Consequently, additional biochemical experiments are needed to clarify the roles of HAD domains and specific amino acid residues in regulatory-like proteins in the aspergilli and other human-pathogenic fungi. In summary, much remains to be learned about the functions of trehalose biosynthesis and associated proteins in human-pathogenic fungi. Additional genetic and biochemical investigations into potential complex formation and the function(s) of the putative regulatory proteins previously characterized for S. cerevisiae are needed to further our understanding of these proteins in human fungal pathogenesis. A particularly important area of future investigation is exploration of the so-called moonlighting roles of these proteins outside canonical trehalose biosynthesis that may directly affect virulence-associated functions, such as cell wall biosynthesis and stress responses to host defense mechanisms.
NONCANONICAL TREHALOSE BIOSYNTHESIS IN FUNGI
The persistence of trehalose production in the absence of TPP function in pathogenic fungi remains an enigma and potentially suggests the existence of a noncanonical trehalose biosynthesis pathway in some species. As discussed above, one hypothesis is that other proteins with shared domains in the trehalose pathway contribute to dephosphorylation of T6P to produce trehalose. Van Dijck and colleagues suggested that nonspecific phosphatase activity may account for the persistence of trehalose in tps2 null mutants (18). Yet there are hints that additional biosynthetic routes may exist in fungi that do not depend on the canonical TPS-TPP-mediated pathway. For example, in the model filamentous fungus Neurospora crassa, clock-controlled gene 9 (ccg-9) functions as a gene important for trehalose production and stress protection (48). The ccg-9 gene product has high sequence similarity to a novel trehalose synthase (TSase) in the basidiomycete mushroom Grifola frondosa. The G. frondosa TSase functions as a trehalose phosphorylase that reversibly catalyzes glucose-1-phosphate and glucose into trehalose and inorganic phosphate (135). Sequence similarity is observed with trehalose phosphorylases in other basidiomycetes, including the mushrooms Agaricus bisporus (40) and Schizophyllum commune (42). Consequently, trehalose phosphorylase activity is found in many fungi within the Basidiomycota, Zygomycota, and Ascomycota (135). However, in the ascomycete yeasts, e.g., S. cerevisiae and C. albicans, TSase homologs have not been found by sequence-based comparisons to date. Based on the identification of two putative TSase proteins encoded in the A. fumigatus genome sequence, it has been proposed that this noncanonical pathway may be induced in this pathogen upon phosphate depletion caused by T6P accumulation in the absence of TPP activity to help to mitigate the accumulation of potentially toxic sugar phosphates and also to resupply the cell with inorganic phosphate (21). This model remains to be tested experimentally in A. fumigatus. Another potential explanation for trehalose production in the absence of detectable TPP activity is the presence of an unknown pathway for trehalose biosynthesis in these fungi. The pathway would be activated by loss of Tps2 and/or T6P accumulation but not in the absence of trehalose per se because tps1 mutants remain deficient in trehalose. These data illustrate a main theme of this review in that much remains to be learned about the complexity of the trehalose biosynthetic pathway and its impact on pathogenic fungal biology. Yet these studies are critical to realize the full therapeutic potential and challenges facing any attempts to target this pathway.
TREHALASES
Fungi utilize trehalose through trehalase-mediated degradation. In S. cerevisiae, there are three different trehalases: Nth1p, Nth2p, and Ath1p (136). Nth1p, the neutral trehalase, is located in the cytoplasm and functions at pH 7.0 (13, 136–138). Nth2p is also found in the cytoplasm but has no significant trehalase activity under conditions examined to date (137, 138). Nth1p has maximal activity during early growth stages. NTH1 expression increases under stress conditions due to the presence of three STREs in its promoter (139). Nth1p is also regulated by PKA-mediated posttranslational modification (phosphorylation) (140, 141). Furthermore, Nth1p also needs Bmh2p (a 14-3-3 protein family member) and a Ca2+/calmodulin-dependent kinase II (DscIp) to control its activity (141). In contrast, the acid trehalase, Ath1p, functions at pH 5.0 and is important for utilizing trehalose as the sole carbon source (142). Both trehalose levels and its transport activity increase during glucose starvation, which suggests that trehalose transport and utilization are important under stress conditions that perturb carbon utilization. Crowe and Crowe (60) and Eleutherio et al. (143, 144) observed that trehalose is necessary at both intracellular and extracellular sites. The complexity of trehalase activity regulation in fungal cells is striking and further illustrates the central role of trehalose in fungal biology.
Similar to the situation for regulatory subunits, there are unfortunately, to date, relatively few studies on trehalases in pathogenic fungi. In C. albicans, there are two trehalase enzymes: Ntc1p (cytosolic trehalase) and Atc1p (cell wall-linked trehalase) (57, 145, 146). Ntc1p activity is not dependent on pH and is strongly inhibited by high ATP levels associated with glucose-replete conditions (145). Ntc1p activity is weakly activated by divalent cations (Ca2+ or Mn2+) but does not change in the presence of cAMP (145). While Ntc1p activity decreases during stationary phase, with growth on glycerol media being an exception, Atc1p activity increases in resting cells and/or in trehalose- or glycerol-containing media (145). Atc1p activity is not increased in the absence of glucose and is not dependent on ATP, cAMP, divalent cations, or pH (145, 146). Importantly, the C. albicans ntc1 null mutant has no virulence defect in a systemic murine model (147). However, in contrast to the C. albicans Ntc1p mutant, an atc1 null mutant has a significant virulence defect in a systemic murine model that is associated with Atc1p roles in dimorphism and stress resistance (148). This promising result in the context of Atc1p loss warrants further investigation in C. albicans and perhaps other pathogenic yeast species.
For C. neoformans, only nth1, encoding an 826-amino-acid protein with identities to other fungal species proteins of between 45 and 62%, has been characterized (19). Nth1p in C. gattii has also been characterized and has approximately 89% sequence similarity to C. neoformans Nth1p (20). As with the C. albicans ntc1 null mutant, the nth1 null mutants of both C. neoformans and C. gattii have no apparent phenotypes related to virulence or in vivo fitness (19, 20).
In A. nidulans, an acid trehalase (treA) is essential for growth on trehalose but is not related to intracellular mobilization of the trehalose pool (67). However, the S. cerevisiae homolog of neutral trehalase in A. nidulans is involved in the mobilization of the intracellular trehalose pool (67). No studies to date have characterized the putative trehalases present in the A. fumigatus genome sequence and their potential role in host-pathogen interactions. Thus, with the exception of C. albicans Atc1p, fungal trehalases remain unclear as potential drug targets. Importantly, however, a trehalase inhibitor, validamycin, does exist and has been used to uncover a role for trehalose in the regulation of HSP90 function in C. albicans (149). As a critical chaperone involved in antifungal drug resistance, HSP90 is under active investigation as a therapeutic target (150, 151). Additional studies on the role of Atc1p in C. albicans-host interactions and in the context of HSP90-mediated drug resistance seem warranted. Along these lines, the role of fungal trehalases in shaping interactions with the microbiota and the subsequent impact on host immunity is still completely unexplored.
TREHALOSE TRANSPORTERS
In S. cerevisiae, Agt1p (also called Mal11p) is a high-affinity H+-trehalose symporter (152). The deletion of this gene causes decreased tolerance to peroxide and heat shock (153). Agt1p exports cytosolic trehalose to the extracellular space, and Ath1p degrades this trehalose pool during stress recovery (136). For pathogenic fungi, the trehalose transporter(s) has yet to be identified and characterized. However, from sequence-based analyses with BLASTp searches, many maltose permease transporters and other major facilitator superfamily (MFS) transporters that may potentially function as trehalose transporters are found in C. albicans, C. neoformans, C. gattii, and A. fumigatus. The relevance of trehalose transport in human invasive fungal infections is unclear but may conceivably affect potential interactions with the microbiota, where trehalose is produced by other resident microbes.
REGULATION OF THE TREHALOSE PATHWAY
Another viable strategy for targeting trehalose biosynthesis in pathogenic fungi is to identify conserved critical regulatory mechanisms. Studies examining the regulation of trehalose biosynthesis and its associated genes and encoded proteins remain in their infancy for pathogenic fungi. While early studies on trehalose biosynthesis in pathogenic fungi clearly revealed differences in model organisms, such as S. cerevisiae, we briefly review here what is known for this model system as a basis for future investigations of these pathogens.
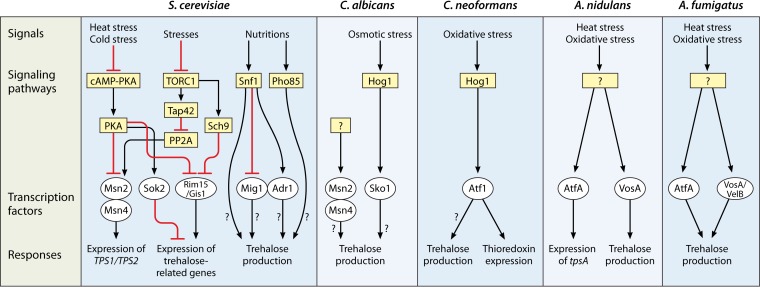
The HOG-MAPK pathway in fungi consists of two important components: a two-component phosphorelay system and a MAPK module. The phosphorelay system contains hybrid sensor kinases, a histidine-containing phosphotransfer (HPt) protein, and response regulators that can sense and send an environmental signal to activate the MAPK pathway (154–156). For S. cerevisiae, there are many studies on the HOG-MAPK cascade response to osmotic, oxidative, or heat stress (154, 155). Under these stress conditions, especially osmotic stress, the HOG pathway is important for yeast cells to grow and survive. The Hog1 MAPK is conserved in pathogenic yeasts and molds, i.e., C. albicans (Hog1p), C. neoformans (Hog1p), and A. fumigatus (SakA). While hog1 mutants of S. cerevisiae and sakA mutants of A. fumigatus do not show decreased trehalose levels (157, 158), a Cryptococcus hog1 mutant contains less trehalose than that in wild-type yeast cells (J. R. Perfect and R. G. Brennan, unpublished data). However, a clear link between trehalose metabolism and the SakA/MpkC pathway has been suggested for A. fumigatus and remains to be explored further (159).
S. cerevisiae requires the Msn2/4 transcription factors to express tps1 and tps2 at low temperatures (below 10°C) (160). These transcription factors are regulated via phosphorylation by the cAMP-PKA pathway, which plays an important role in the regulation of a wide range of stress conditions in addition to glucose signaling (161–166). Under stress conditions, e.g., heat and cold stresses, the Msn2/4p proteins localize to the nucleus and activate stress response genes that contain the canonical STRE sequence in their promoters, e.g., tps1, tps2, and hsp12/26 (167). In the recovery stage or under nutrient-replete conditions, PKA-dependent phosphorylation of Msn2/4p restricts these proteins to the cytosol and prevents them from activating stress response genes (168). The PKA pathway also regulates the transcriptional repressor Sok2p, and intriguingly, trehalose-related genes also contain a binding site for Sok2p in their promoter regions (169). In addition, PKA can repress trehalose production through phosphorylation of the Rim15-Gis1p pathway, which binds to an upstream activating sequence (UASpds) in most trehalose-related genes (Fig. 4) (169).
FIG 4.
Comparison of mechanisms of regulation of the trehalose pathway in S. cerevisiae, C. albicans, C. neoformans, A. nidulans, and A. fumigatus.
In C. albicans, Msn2/4-like transcription factors do not appear to be directly related to stress responses (170). However, C. albicans Sko1p is involved in the osmotic stress response regulated by Hog1p and is also critical for the cell wall damage response (Fig. 4) (171, 172). A connection between the trehalose pathway and Sko1p or Msn2/4p is uncharacterized for C. albicans. Similarly, both C. neoformans and A. fumigatus lack apparent Msn2/4 and Sko1 homologs encoded in their genomes, though functional analogs may remain to be identified (173). C. neoformans does have the Atf1p transcription factor, which is not found in C. albicans (173, 174). C. neoformans Atf1p is transcriptionally regulated by Hog1p and is important for the oxidative stress response, in part through its regulation of thioredoxin expression (Fig. 4) (173, 174).
In A. nidulans and Aspergillus oryzae, the ATF/CREB-type transcription factor AtfA is a critical regulator of the conidial stress response (175–178). In A. nidulans, AtfA protects the fungus from oxidative and heat stresses (175). The expression of tpsA and a conidium-specific catalase gene, catA, in conidia is regulated by A. nidulans AtfA (Fig. 4) (175). A. oryzae contains three ATF-like proteins, two of which (AtfA and AtfB) have been studied (177, 178). Conidia of atfA and atfB null mutants are susceptible to oxidative, UV, and heat stresses. Trehalose levels in conidia and the expression of tpsA and tpsC, which are homologs of Sctps1 and Sctps3, respectively, are lower in both atfA and atfB null mutants than in the wild type (177, 178). Recently, Hagiwara et al. observed that AtfA in A. fumigatus is important for the fungus to survive under heat and oxidative stress conditions (158). As in other aspergilli, AtfA is involved in trehalose accumulation in the conidia of A. fumigatus (Fig. 4) (158). However, connections between AtfA and the virulence of A. fumigatus remain to be studied in an in vivo model.
TOR and TORC1 complexes in S. cerevisiae play a critical role in carbohydrate storage and metabolism in part by increasing the expression of glycogen- and trehalose-related genes (179, 180). TORC1 inhibits the Ser/Thr protein phosphatase 2A (PP2A) via phosphorylation of Tap42p. Therefore, inhibition of PP2A prevents dephosphorylation of Msn2p, which consequently remains in the cytosol in the phosphorylated form. While TORC1 can inhibit PP2A, it can also inhibit the Rim15-Gis1p pathway via activation of Sch9p to phosphorylate and sequester Rim15 in the cytosol (181). There is a connection between the PKA and TOR pathways through Msn2p, but the mechanisms that regulate and control the balance of these two pathways are unclear (168). Moreover, Pho85p and Snf1p also regulate trehalose and glycogen levels through transcriptional and posttranslational mechanisms (182, 183). It has been proposed that the Snf1 kinase regulates the transcription of Adr1p or Mig1p (a carbon catabolite repressor). However, the regulation of trehalose gene expression through Pho85p is unclear (Fig. 4). Determining the connections between these critical metabolism and fungal fitness regulatory pathways and trehalose biosynthesis in human-pathogenic fungi is a promising future research direction.
One family of regulatory proteins tied mechanistically to trehalose biosynthesis that is conserved in the pathogenic fungi is the velvet domain family. The velvet family proteins are conserved throughout the fungal kingdom, especially in the ascomycetes and basidiomycetes (184). This family is well characterized in A. nidulans and consists of four members, i.e., VelA, VelB, VosA, and VelC. VosA (viability of spores A) is a regulator of asexual development and is necessary for trehalose accumulation in A. nidulans conidia (184). Conidia of a vosA null mutant of A. nidulans display increased sensitivity to heat and oxidative stresses and lack trehalose (Fig. 4) (184). A. fumigatus velvet family vosA or velB null mutants also have reductions in conidial trehalose levels and a subsequent loss of conidial tolerance of oxidative and UV stresses (185). However, in contrast to those of A. nidulans, conidia of the A. fumigatus vosA mutant are not completely depleted of trehalose (Fig. 4) (184, 185). Further research is needed to investigate the impacts of velvet family-mediated trehalose biosynthesis and stress responses in the context of fungal pathogenesis.
In summary, the regulation of trehalose production in fungi is complex and involves multiple pathways, many of which remain to be investigated fully for human-pathogenic fungi. This complexity further reflects the importance of trehalose in the survival and adaptation of fungi in response to a wide variety of stresses. It is unclear if specific regulatory mechanisms that affect trehalose biosynthesis are promising antifungal drug targets, but it is clear that additional research into these mechanisms is warranted, with the potential to yield new insights into drug development.
FUTURE RESEARCH DIRECTIONS AND CLINICAL POTENTIAL
A brief review of the development of antifungal drugs highlights the challenges and paucity of current antifungal drug development. In the 1950s, the first systemic antifungal drug, amphotericin B deoxycholate, was licensed for clinical use (186, 187). After that important event, it took more than 2 decades to discover and use additional antifungal drug classes, including the azoles and echinocandins. In the 1980s, the discovery and development of the triazoles dramatically improved the treatment of invasive fungal infections, especially for polyene-refractory infections (188). In the 1990s, triazoles and amphotericin B formulations were further developed (188). Even though the newest antifungal drug class, the echinocandins, was discovered in the 1970s, it took almost 30 years for these drugs to be used in the treatment of invasive fungal infections (approved by the FDA in 2002) (187, 188). The latest FDA-approved antifungal drugs are posaconazole (2006), anidulafungin (2006), and isavuconazole (isavuconazonium sulfate) (2015) (http://www.accessdata.fda.gov/scripts/cder/daf).
From this brief history of antifungal drug development, it is clear that there are many gaps in time between drug class discovery and development of a usable drug (189–192). Given the increasing number of fungal infections and the emergence of drug resistance and innate resistance of select fungal pathogens to some drugs, a potential shortage of effective antifungal drugs in the near future is a very real cause for concern. It remains a great challenge to identify and develop new fungicidal drugs that target these eukaryotic pathogens effectively, with minimal adverse host side effects.
A current approach for antifungal drug development is to identify and target essential processes in fungi. However, it is very difficult to discover targets that, when inhibited, have a killing effect only in the fungi, not against human cells. In addition to these challenges, it is argued that targeting essential genes may lead to rapid emergence of drug resistance, though this remains a topic for robust conversation (193). Another perspective is that organisms will be pathogens when they express virulence attributes that function to induce disease (194). Therefore, instead of targeting essential genes, targeting attributes involved in virulence is an alternative approach to the development of novel antifungal drugs specific to fungi but safe for humans (195). Additional approaches to drug development must be encouraged, as we need a continuous discovery of new agents to ensure that current antifungal drugs reach their full therapeutic potential against human fungal pathogens. Therefore, to keep up with drug resistance and new emerging causes of disease, we need to fully consider and appreciate a pathogenesis-based approach as an additional option for antifungal drug development (195).
Several reports suggest that targeting virulence factors can serve as a viable approach to the development of novel antifungal agents (195–197). For instance, an inhibitor of glycosylphosphatidylinositol (GPI) biosynthesis, which is important for GPI-modified proteins that provide cell wall integrity and membrane homeostasis (198), was discovered and developed into the oral broad-spectrum antifungal inhibitor E1210 (199, 200). E1210 is effective in murine models of oropharyngeal and disseminated candidiasis, pulmonary aspergillosis, and disseminated fusariosis (200). It has activities against fluconazole-resistant Candida strains, Pseudallescheria boydii, Scedosporium prolificans, and Paecilomyces lilacinus (201–203). While the GPI biosynthesis target is not a canonical virulence factor in the traditional definition of the term, and its inhibition directly affects fungal growth, its use highlights that studies on virulence-associated processes can identify targets that ultimately affect fungal fitness in vivo.
Along these lines, unique metabolic pathways that exist only in fungi and function when saprophytes cause disease should be a point of focus for therapeutic development. The trehalose pathway is one of these unique metabolic pathways. As discussed throughout this review and elsewhere, proteins involved in trehalose biosynthesis are critically involved in many aspects of the biology and virulence of pathogenic fungi (15, 18–21, 68, 204). Importantly, we are now beginning to develop the preliminary infrastructure in these major fungal pathogens to make insightful discoveries with therapeutic potential, with the trehalose pathway serving as one potential model (122). The search for inhibitors of this pathway as potential antifungal drugs has begun and represents both the synthesis and degradative portions of trehalose biology (205). There are already inhibitors of the trehalase enzymes, such as validamycin A, that have been identified (206). Many studies of plants and parasites have characterized the structure of trehalose-related enzymes to screen for small-molecule inhibitors (132, 207). For example, Xue et al. (207) characterized the structure of TPS1 from the plant pathogen Magnaporthe oryzae and screened for potential small-molecule inhibitors by using a chemical database in silico. These studies are now possible for human-pathogenic fungi.
Recently, Miao et al. provided a new level of understanding of the major enzymes within the fungal trehalose pathway (122). In their studies, structures of the trehalose-6-phosphate phosphatase (Tps2) at different stages of the catalytic process were solved. These structures unveil the specific conformational changes and enzymatic components needed for substrate recognition and subsequent phosphate removal. Moreover, they reveal the essentially identical catalytic pockets, and therefore identical substrate recognition and enzyme mechanisms, of the Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus Tps2 proteins. Biochemical, in vivo, and structural investigations show how these enzymes are substrate specific (Fig. 5), thereby protecting the fungal cell from the potential cytotoxicity of T6P. The finding that fungal Tps2 proteins possess extreme substrate specificity as phosphatases for T6P bodes well for the creation of a specific inhibitor with limited “off target” phosphatase-inhibitory activity and thus toxicity. As an added “bonus” to this substrate specificity, structure-guided mutagenesis of residues involved in T6P binding and the very poor T6P phosphatase activity of the single-site mutants suggest that simple single-site Tps2 mutations within the catalytic pocket will not allow the generation of resistant strains. Although Tps2 mutants with changes within the catalytic pocket might avoid drug binding, they will not be able to produce trehalose from T6P, resulting in high levels of cytotoxic T6P. Finally, the structures of Tps1 proteins of several pathogenic fungi have been determined in complex with substrates and substrate analogs (Y. Miao, J. R. Perfect, and R. G. Brennan, unpublished data). Therefore, the high-resolution structures of the core targets of canonical fungal trehalose biosynthesis are now available such that aggressive drug discovery programs to identify novel inhibitors, and ultimately drugs, are able to commence, with the goal of disrupting this key pathway and thereby leading to novel, broad-spectrum fungicidal drugs. At minimum, the expected discovery of small-molecular inhibitors of these proteins will allow rigorous hypothesis testing in relevant animal models of infection to address many of the challenges facing therapeutic targeting of this pathway that are discussed throughout this review.
FIG 5.
Structure of Tps2 in complex with its substrate, trehalose-6-phosphate. (A) Structure of Cryptococcus neoformans Tps2 (CnTps2), depicting the cap domain in magenta and the core domain in cyan. Substrate-bound Tps2 takes a “closed” conformation, bringing the cap and core domains closer, whereas substrate-free Tps2 assumes an “open” conformation whereby the cap and core domains are more splayed. Trehalose-6-phosphate (T6P) and residue N24 are labeled and depicted as atom-colored sticks, and the catalytically important magnesium ion (Mg2+) is shown as a yellow sphere and labeled. In wild-type CnTps2, residue 24 is an aspartate and is responsible for the nucleophilic attack on the phosphate group of T6P. (B) View of the active site of Tps2 bound to T6P. All residues that interact with either T6P or the Mg2+ ion or play a key part in forming the substrate-binding pocket are shown as atom-colored sticks and labeled. The carbon atoms of residues from the cap domain are colored magenta, while those from the core domain are colored cyan. T6P is shown as atom-colored sticks, with its carbon atoms colored white, and the catalytically important Mg2+ ion is shown as a yellow sphere. All solvent molecules and dashes indicating protein-substrate or protein-Mg2+ interactions have been omitted for the sake of clarity. Note the tripartite stacking of the side chains of residues F70 and R66 and the glucose-6-phosphate ring of the T6P disaccharide.
Consequently, the future of the trehalose biosynthesis pathway as a highly desirable antifungal drug target seems bright. Further mechanistic investigations into the functions of key proteins in the pathway are needed for the respective human fungal pathogens to help in determining additional therapeutic leverage points and to address concerns about the potential limitations of inhibitors of this pathway. With regard to the latter, in addition to some of the issues addressed throughout this review, because this pathway is found in a wide variety of organisms, inhibitors of this pathway may affect other human commensals, which should be investigated further. In fact, the role of trehalose biosynthesis in microbe-microbe interactions is an intriguing area of investigation. Depending on the specificity of the inhibitors' structural design and routes of administration, it seems, however, that this potential limitation is addressable if needed. In addition, in filamentous fungi, such as A. fumigatus, redundant protein functions, including other alternative pathways or unknown phosphatases (21), may affect the efficacy of inhibitors, and this simply requires more investigation as highlighted by the example of the A. fumigatus tpsA/B null mutant (68).
One additional concern that is receiving more attention in the field is the efficacy of a given agent in the context of an established infection microenvironment. While the data discussed in this review suggest that trehalose biosynthesis proteins likely have roles in established infections, this remains to be fully validated experimentally. The use of conditional promoter systems in vivo may be a viable approach to confirm the role of these proteins in established infections, as will the identification of small-molecule inhibitors. All of these concerns emphasize one of the main themes of this review, i.e., that further studies are essential to move drug discovery forward to meet the challenges that await. A particularly attractive area worth further emphasis is the link between trehalose biosynthesis and fungal cell wall homeostasis given the critical importance of this fungus-specific structure to virulence and host immune responses. In this arena, inhibitors of trehalose metabolism may prove to be highly effective adjunctive therapeutics in combination with existing cell wall-targeting drugs. These hypotheses and associated key in vivo experiments are on the cusp of being realized, as emphasized by the determination of key biosynthesis protein structures and the associated search for small-molecule inhibitors.
ACKNOWLEDGMENTS
Research in the Cramer laboratory is supported by NIH grants (NIAID grant R01AI081838 and NIH General Medical Science grant 1P30GM106394-01 [to Bruce Stanton]). R.A.C. holds an Investigators in the Pathogenesis of Infectious Diseases award from the Burroughs Wellcome Fund. A.T. is supported by a scholarship from the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand. Support from U.S. Public Service grants was given for J.R.P. (grants AI73896, AI93257, and AI04533) and R.G.B. (grant AI04533).
Biographies
Arsa Thammahong received his M.D. from the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, in 2010. He continued his internship as a lecturer and research assistant in the Mycology Unit, Department of Microbiology, Faculty of Medicine, Chulalongkorn University, from 2010 to 2012. He has received a Ph.D. scholarship from the Faculty of Medicine, Chulalongkorn University, since 2012 for study in the fungal pathogenesis field, and he is currently a 5th-year Ph.D. candidate in Robert Cramer's laboratory at the Geisel School of Medicine at Dartmouth. His project focuses on the trehalose biosynthetic pathway and its impact on metabolism and virulence in Aspergillus fumigatus.
Srisombat Puttikamonkul graduated with a B.Sc. in microbiology from Chulalongkorn University, Bangkok, Thailand, in 1997 and obtained her M.Sc. in biotechnology from Mahidol University, Bangkok, Thailand, in 2002. She worked as a biomedical research technician at the Department of Immunology, AFRIMS, Thailand, from 2002 to 2007. She was awarded a Thai government scholarship for her Ph.D. work and received her Ph.D. in 2012 from Robert Cramer's lab at Montana State University, Bozeman, MT, with the thesis “Trehalose-6-phosphate is required for metabolism and virulence in the human fungal pathogen, Aspergillus fumigatus.” After finishing her Ph.D., she continued her career as a lecturer and a researcher in the Department of Microbiology, Faculty of Medicine, Srinakharinwirot University, Bangkok, Thailand. Her research focuses on the role of the trehalose-6-phosphate phosphatase enzyme OrlA and the trehalose biosynthesis pathway in the regulation of cell wall homeostasis and pathogenesis in Talaromyces (formerly Penicillium) marneffei.
John R. Perfect is the James B. Duke Professor of Medicine and Chief of Infectious Diseases at Duke University Medical Center. He is Director of the Duke University Mycology Research Unit. His research is sponsored by both the NIH and the pharmaceutical industry and has focused on cryptococcal pathogenesis and both translational and clinical aspects of invasive mycoses for over 35 years. He is a fellow of the Infectious Diseases Society of America (IDSA), the American Academy of Microbiology, and the American Association for the Advancement of Science and is a member of the American Academy of Physicians. In 2010, he led a group who revised the IDSA guidelines for the management of cryptococcosis. He is or has been on the advisory groups of a variety of organizations and study sections, but specifically, he is a member of the advisory panel for the International Mycoses Study Group for clinical trials and education on fungal infections.
Richard G. Brennan received his Ph.D. in biochemistry from the University of Wisconsin-Madison in 1984. After postdoctoral studies with Brian W. Matthews at the University of Oregon, he joined the Department of Biochemistry and Molecular Biology in 1989 as an Assistant Professor at the Oregon Health & Science University. He was promoted to Associate Professor in 1995 and to Professor in 2000. In 2001, he was named the Richard T. Jones Professor of Structural Biology. In 2005, he moved to the Department of Biochemistry and Molecular Biology at the University of Texas MD Anderson Cancer, where he was named the Robert A. Welch Distinguished University Professor in Chemistry. In 2011, he assumed the position of the Chair of the Department of Biochemistry at Duke University School of Medicine, where he is also a James B. Duke Professor of Biochemistry. Dr. Brennan is a fellow of the American Academy of Microbiology and the American Association for the Advancement of Science. His research focuses on the biochemical and structural mechanisms of microbial pathogenesis, transcriptional and posttranscriptional gene regulation, multidrug resistance, and bacterial multidrug tolerance and persistence.
Robert A. Cramer was awarded a Ph.D. from Colorado State University in 2004 for thesis work on pathogenesis mechanisms of fungal plant pathogens in the genus Alternaria under the guidance of Dr. Christopher Lawrence. In 2004, he moved to Duke University Medical Center and, as an NIH Molecular Mycology postdoctoral fellow, conducted research on pathogenesis mechanisms of the human fungal pathogen Aspergillus fumigatus with Dr. John Perfect and Dr. William Steinbach. In 2007, he joined the faculty of the Department of Veterinary Molecular Biology at Montana State University, where his studies focused on the role of oxygen and metabolism in the pathogenesis of A. fumigatus. In 2012, he was recruited to the Geisel School of Medicine at Dartmouth, where he currently resides as an Associate Professor in the Department of Microbiology and Immunology. In 2014, he was awarded a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases fellowship for studies on Aspergillus sp. bioenergetics and virulence.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 2.Nucci M, Queiroz-Telles F, Tobon AM, Restrepo A, Colombo AL. 2010. Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis 51:561–570. doi: 10.1086/655683. [DOI] [PubMed] [Google Scholar]
- 3.Lehrnbecher T, Frank C, Engels K, Kriener S, Groll AH, Schwabe D. 2010. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J Infect 61:259–265. doi: 10.1016/j.jinf.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Slavin MA, Chakrabarti A. 2012. Opportunistic fungal infections in the Asia-Pacific region. Med Mycol 50:18–25. doi: 10.3109/13693786.2011.602989. [DOI] [PubMed] [Google Scholar]
- 5.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 6.Denning DW, Hope WW. 2010. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol 18:195–204. doi: 10.1016/j.tim.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Segal BH, Steinbach WJ, Stevens DA, van Burik JA, Wingard JR, Patterson TF, Infectious Diseases Society of America. 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis 46:327–360. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 8.Verweij PE, Howard SJ, Melchers WJ, Denning DW. 2009. Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat 12:141–147. doi: 10.1016/j.drup.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Howard SJ, Pasqualotto AC, Denning DW. 2010. Azole resistance in allergic bronchopulmonary aspergillosis and Aspergillus bronchitis. Clin Microbiol Infect 16:683–688. doi: 10.1111/j.1469-0691.2009.02911.x. [DOI] [PubMed] [Google Scholar]
- 10.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 13.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 14.Elbein AD. 1974. The metabolism of alpha,alpha-trehalose. Adv Carbohydr Chem Biochem 30:227–256. doi: 10.1016/S0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- 15.Zaragoza O, Blazquez MA, Gancedo C. 1998. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J Bacteriol 180:3809–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Peral FJ, Zaragoza O, Pedreno Y, Arguelles JC. 2002. Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology 148:2599–2606. doi: 10.1099/00221287-148-8-2599. [DOI] [PubMed] [Google Scholar]
- 17.Zaragoza O, de Virgilio C, Ponton J, Gancedo C. 2002. Disruption in Candida albicans of the TPS2 gene encoding trehalose-6-phosphate phosphatase affects cell integrity and decreases infectivity. Microbiology 148:1281–1290. doi: 10.1099/00221287-148-5-1281. [DOI] [PubMed] [Google Scholar]
- 18.Van Dijck P, De Rop L, Szlufcik K, Van Ael E, Thevelein JM. 2002. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect Immun 70:1772–1782. doi: 10.1128/IAI.70.4.1772-1782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. 2006. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun 74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, Malik R, Daniel HM, Toffaletti D, Djordjevic JT, Mylonakis E, Meyer W, Perfect JR. 2009. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect Immun 77:4584–4596. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puttikamonkul S, Willger SD, Grahl N, Perfect JR, Movahed N, Bothner B, Park S, Paderu P, Perlin DS, Cramer RA Jr. 2010. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol Microbiol 77:891–911. doi: 10.1111/j.1365-2958.2010.07254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanbury D. 1859. Note on two insect-products from Persia. J Proc Lin Soc London 3:178–183. [Google Scholar]
- 23.Richards AB, Krakowka S, Dexter LB, Schmid H, Wolterbeek AP, Waalkens-Berendsen DH, Shigoyuki A, Kurimoto M. 2002. Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem Toxicol 40:871–898. doi: 10.1016/S0278-6915(02)00011-X. [DOI] [PubMed] [Google Scholar]
- 24.Singer MA, Lindquist S. 1998. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol 16:460–468. doi: 10.1016/S0167-7799(98)01251-7. [DOI] [PubMed] [Google Scholar]
- 25.Harding T. 1923. History of trehalose. Its discovery and methods of preparation. Sugar 25:476–478. [Google Scholar]
- 26.Koch EM, Koch FC. 1925. The presence of trehalose in yeast. Science 61:570–572. [DOI] [PubMed] [Google Scholar]
- 27.Fischer E. 1895. Ueber den Einfluss der Konfiguration auf die Wirkung der Enzyme III. Ber Dtsch Chem Ges 28:1429–1438. [Google Scholar]
- 28.Leloir LF, Cabib E. 1953. The enzymic synthesis of trehalose phosphate. J Am Chem Soc 75:5445–5446. [Google Scholar]
- 29.Cabib E, Leloir LF. 1958. The biosynthesis of trehalose phosphate. J Biol Chem 231:259–275. [PubMed] [Google Scholar]
- 30.Lemieux RU, Bauer HF. 1954. A chemical synthesis of d-trehalose. Can J Chem 32:340–344. doi: 10.1139/v54-044. [DOI] [Google Scholar]
- 31.Dellamora-Ortiz GM, Ortiz CH, Maia JC, Panek AD. 1986. Partial purification and characterization of the interconvertible forms of trehalase from Saccharomyces cerevisiae. Arch Biochem Biophys 251:205–214. doi: 10.1016/0003-9861(86)90067-6. [DOI] [PubMed] [Google Scholar]
- 32.Colaço CALS, Roser B. 1995. Trehalose—a multifunctional additive for food preservation, p 123–140. In Mathlouthi M. (ed), Food packaging and preservation. Blackie Professional, London, United Kingdom. [Google Scholar]
- 33.Crowe JH, Crowe LM. 2000. Preservation of mammalian cells—learning nature's tricks. Nat Biotechnol 18:145–146. doi: 10.1038/72580. [DOI] [PubMed] [Google Scholar]
- 34.Crowe JH, Carpenter JF, Crowe LM. 1998. The role of vitrification in anhydrobiosis. Annu Rev Physiol 60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien J. 1996. Stability of trehalose, sucrose and glucose to nonenzymatic browning in model systems. J Food Sci 61:679–682. doi: 10.1111/j.1365-2621.1996.tb12180.x. [DOI] [Google Scholar]
- 36.Ohtake S, Wang YJ. 2011. Trehalose: current use and future applications. J Pharm Sci 100:2020–2053. doi: 10.1002/jps.22458. [DOI] [PubMed] [Google Scholar]
- 37.Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. 2006. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6:109. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walmagh M, Zhao R, Desmet T. 2015. Trehalose analogues: latest insights in properties and biocatalytic production. Int J Mol Sci 16:13729–13745. doi: 10.3390/ijms160613729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belocopitow E, Marechal LR, Gros EG. 1971. Enzymic synthesis of 6-deoxy-d-glucopyranosyl-d-glucopyranoside and -d-xylopyranosyl-d-glucopyranoside. Carbohydr Res 19:268–271. doi: 10.1016/S0008-6215(00)81631-6. [DOI] [PubMed] [Google Scholar]
- 40.Wannet WJ, Op den Camp HJ, Wisselink HW, van der Drift C, Van Griensven LJ, Vogels GD. 1998. Purification and characterization of trehalose phosphorylase from the commercial mushroom Agaricus bisporus. Biochim Biophys Acta 1425:177–188. doi: 10.1016/S0304-4165(98)00066-X. [DOI] [PubMed] [Google Scholar]
- 41.Saito K, Yamazaki H, Ohnishi Y, Fujimoto S, Takahashi E, Horinouchi S. 1998. Production of trehalose synthase from a basidiomycete, Grifola frondosa, in Escherichia coli. Appl Microbiol Biotechnol 50:193–198. doi: 10.1007/s002530051276. [DOI] [PubMed] [Google Scholar]
- 42.Eis C, Nidetzky B. 1999. Characterization of trehalose phosphorylase from Schizophyllum commune. Biochem J 341:385–393. [PMC free article] [PubMed] [Google Scholar]
- 43.Aisaka K, Masuda T, Chikamune T, Kamitori K. 1998. Purification and characterization of trehalose phosphorylase from Catellatospora ferruginea. Biosci Biotechnol Biochem 62:782–787. doi: 10.1271/bbb.62.782. [DOI] [PubMed] [Google Scholar]
- 44.Ren Y, Dai X, Zhou J, Liu J, Pei H, Xiang H. 2005. Gene expression and molecular characterization of a thermostable trehalose phosphorylase from Thermoanaerobacter tengcongensis. Sci China C Life Sci 48:221–227. [DOI] [PubMed] [Google Scholar]
- 45.Han SE, Kwon HB, Lee SB, Yi BY, Murayama I, Kitamoto Y, Byun MO. 2003. Cloning and characterization of a gene encoding trehalose phosphorylase (TP) from Pleurotus sajor-caju. Protein Expr Purif 30:194–202. doi: 10.1016/S1046-5928(03)00104-9. [DOI] [PubMed] [Google Scholar]
- 46.Inoue Y, Ishii K, Tomita T, Yatake T, Fukui F. 2002. Characterization of trehalose phosphorylase from Bacillus stearothermophilus SK-1 and nucleotide sequence of the corresponding gene. Biosci Biotechnol Biochem 66:1835–1843. doi: 10.1271/bbb.66.1835. [DOI] [PubMed] [Google Scholar]
- 47.Marechal LR, Belocopitow E. 1972. Metabolism of trehalose in Euglena gracilis. I. Partial purification and some properties of trehalose phosphorylase. J Biol Chem 247:3223–3228. [PubMed] [Google Scholar]
- 48.Shinohara ML, Correa A, Bell-Pedersen D, Dunlap JC, Loros JJ. 2002. Neurospora clock-controlled gene 9 (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot Cell 1:33–43. doi: 10.1128/EC.1.1.33-43.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiraldi C, Di Lernia I, De Rosa M. 2002. Trehalose production: exploiting novel approaches. Trends Biotechnol 20:420–425. doi: 10.1016/S0167-7799(02)02041-3. [DOI] [PubMed] [Google Scholar]
- 50.Streeter JG, Bhagwat A. 1999. Biosynthesis of trehalose from maltooligosaccharides in Rhizobia. Can J Microbiol 45:716–721. doi: 10.1139/w99-050. [DOI] [PubMed] [Google Scholar]
- 51.Nakada T, Maruta K, Tsusaki K, Kubota M, Chaen H, Sugimoto T, Kurimoto M, Tsujisaka Y. 1995. Purification and properties of a novel enzyme, maltooligosyl trehalose synthase, from Arthrobacter sp. Q36. Biosci Biotechnol Biochem 59:2210–2214. doi: 10.1271/bbb.59.2210. [DOI] [PubMed] [Google Scholar]
- 52.Nakada T, Ikegami S, Chaen H, Kubota M, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y. 1996. Purification and characterization of thermostable maltooligosyl trehalose synthase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biosci Biotechnol Biochem 60:263–266. doi: 10.1271/bbb.60.263. [DOI] [PubMed] [Google Scholar]
- 53.Tsusaki K, Nishimoto T, Nakada T, Kubota M, Chaen H, Sugimoto T, Kurimoto M. 1996. Cloning and sequencing of trehalose synthase gene from Pimelobacter sp. R48. Biochim Biophys Acta 1290:1–3. doi: 10.1016/0304-4165(96)00023-2. [DOI] [PubMed] [Google Scholar]
- 54.Nishimoto T, Nakano M, Nakada T, Chaen H, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y. 1996. Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp. R48. Biosci Biotechnol Biochem 60:640–644. doi: 10.1271/bbb.60.640. [DOI] [PubMed] [Google Scholar]
- 55.Qu Q, Lee SJ, Boos W. 2004. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 279:47890–47897. doi: 10.1074/jbc.M404955200. [DOI] [PubMed] [Google Scholar]
- 56.Ryu SI, Park CS, Cha J, Woo EJ, Lee SB. 2005. A novel trehalose-synthesizing glycosyltransferase from Pyrococcus horikoshii: molecular cloning and characterization. Biochem Biophys Res Commun 329:429–436. doi: 10.1016/j.bbrc.2005.01.149. [DOI] [PubMed] [Google Scholar]
- 57.Thevelein JM. 1984. Regulation of trehalose mobilization in fungi. Microbiol Rev 48:42–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiemken A. 1990. Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Van Leeuwenhoek 58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]
- 59.Lillie SH, Pringle JR. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crowe LM, Crowe JH. 1988. Trehalose and dry dipalmitoylphosphatidylcholine revisited. Biochim Biophys Acta 946:193–201. doi: 10.1016/0005-2736(88)90392-6. [DOI] [PubMed] [Google Scholar]
- 61.Rudolph AS, Chandrasekhar E, Gaber BP, Nagumo M. 1990. Molecular modeling of saccharide-lipid interactions. Chem Phys Lipids 53:243–261. doi: 10.1016/0009-3084(90)90050-2. [DOI] [Google Scholar]
- 62.Hottiger T, Boller T, Wiemken A. 1987. Rapid changes of heat and desiccation tolerance correlated with changes of trehalose content in Saccharomyces cerevisiae cells subjected to temperature shifts. FEBS Lett 220:113–115. doi: 10.1016/0014-5793(87)80886-4. [DOI] [PubMed] [Google Scholar]
- 63.Hottiger T, Schmutz P, Wiemken A. 1987. Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J Bacteriol 169:5518–5522. doi: 10.1128/jb.169.12.5518-5522.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell W, Klaassen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, Van der Zee P, Wiemken A. 1992. Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem 209:951–959. doi: 10.1111/j.1432-1033.1992.tb17368.x. [DOI] [PubMed] [Google Scholar]
- 65.Fillinger S, Chaveroche MK, van Dijck P, de Vries R, Ruijter G, Thevelein J, d'Enfert C. 2001. Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology 147:1851–1862. doi: 10.1099/00221287-147-7-1851. [DOI] [PubMed] [Google Scholar]
- 66.Noventa-Jordao MA, Couto RM, Goldman MH, Aguirre J, Iyer S, Caplan A, Terenzi HF, Goldman GH. 1999. Catalase activity is necessary for heat-shock recovery in Aspergillus nidulans germlings. Microbiology 145:3229–3234. doi: 10.1099/00221287-145-11-3229. [DOI] [PubMed] [Google Scholar]
- 67.d'Enfert C, Fontaine T. 1997. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol Microbiol 24:203–216. doi: 10.1046/j.1365-2958.1997.3131693.x. [DOI] [PubMed] [Google Scholar]
- 68.Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CM, Campoli P, Chabot J, Filler SG, Sheppard DC. 2010. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun 78:3007–3018. doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kandror O, DeLeon A, Goldberg AL. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci U S A 99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY. 2004. Cold adaptation in budding yeast. Mol Biol Cell 15:5492–5502. doi: 10.1091/mbc.E04-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benaroudj N, Lee DH, Goldberg AL. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem 276:24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- 72.Zaragoza O, Gonzalez-Parraga P, Pedreno Y, Alvarez-Peral FJ, Arguelles JC. 2003. Trehalose accumulation induced during the oxidative stress response is independent of TPS1 mRNA levels in Candida albicans. Int Microbiol 6:121–125. doi: 10.1007/s10123-003-0119-y. [DOI] [PubMed] [Google Scholar]
- 73.Lederer E. 1976. Cord factor and related trehalose esters. Chem Phys Lipids 16:91–106. doi: 10.1016/0009-3084(76)90001-3. [DOI] [PubMed] [Google Scholar]
- 74.Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64:29–63. [DOI] [PubMed] [Google Scholar]
- 75.Borgia PT. 1992. Roles of the orlA, tsE, and bimG genes of Aspergillus nidulans in chitin synthesis. J Bacteriol 174:384–389. doi: 10.1128/jb.174.2.384-389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borgia PT, Dodge CL. 1992. Characterization of Aspergillus nidulans mutants deficient in cell wall chitin or glucan. J Bacteriol 174:377–383. doi: 10.1128/jb.174.2.377-383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borgia PT, Miao Y, Dodge CL. 1996. The orlA gene from Aspergillus nidulans encodes a trehalose-6-phosphate phosphatase necessary for normal growth and chitin synthesis at elevated temperatures. Mol Microbiol 20:1287–1296. doi: 10.1111/j.1365-2958.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 78.Bell W, Sun W, Hohmann S, Wera S, Reinders A, De Virgilio C, Wiemken A, Thevelein JM. 1998. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem 273:33311–33319. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- 79.Blazquez MA, Santos E, Flores CL, Martinez-Zapater JM, Salinas J, Gancedo C. 1998. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13:685–689. doi: 10.1046/j.1365-313X.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- 80.Vogel G, Aeschbacher RA, Muller J, Boller T, Wiemken A. 1998. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J 13:673–683. doi: 10.1046/j.1365-313X.1998.00064.x. [DOI] [PubMed] [Google Scholar]
- 81.Vogel G, Fiehn O, Jean-Richard-dit Bressel L, Boller T, Wiemken A, Aeschbacher RA, Wingler A. 2001. Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J Exp Bot 52:1817–1826. doi: 10.1093/jexbot/52.362.1817. [DOI] [PubMed] [Google Scholar]
- 82.Eastmond PJ, Li Y, Graham IA. 2003. Is trehalose-6-phosphate a regulator of sugar metabolism in plants? J Exp Bot 54:533–537. doi: 10.1093/jxb/erg039. [DOI] [PubMed] [Google Scholar]
- 83.Gancedo C, Flores CL. 2004. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res 4:351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- 84.van de Poll KW, Kerkenaar A, Schamhart DH. 1974. Isolation of a regulatory mutant of fructose-1,6-diphosphatase in Saccharomyces carlsbergensis. J Bacteriol 117:965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Navon G, Shulman RG, Yamane T, Eccleshall TR, Lam KB, Baronofsky JJ, Marmur J. 1979. Phosphorus-31 nuclear magnetic resonance studies of wild-type and glycolytic pathway mutants of Saccharomyces cerevisiae. Biochemistry 18:4487–4499. doi: 10.1021/bi00588a006. [DOI] [PubMed] [Google Scholar]
- 86.Breitenbach-Schmitt I, Schmitt HD, Heinisch J, Zimmermann FK. 1984. Genetic and physiological evidence for the existence of a second glycolytic pathway in yeast parallel to the phosphofructokinase-aldolase reaction sequence. Mol Gen Genet 195:536–540. doi: 10.1007/BF00341459. [DOI] [Google Scholar]
- 87.Cannon JF, Pringle JR, Fiechter A, Khalil M. 1994. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics 136:485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McDougall J, Kaasen I, Strom AR. 1993. A yeast gene for trehalose-6-phosphate synthase and its complementation of an Escherichia coli otsA mutant. FEMS Microbiol Lett 107:25–30. doi: 10.1111/j.1574-6968.1993.tb05998.x. [DOI] [PubMed] [Google Scholar]
- 89.Thevelein JM, Hohmann S. 1995. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci 20:3–10. doi: 10.1016/S0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- 90.Charlab R, Oliveira DE, Panek AD. 1985. Investigation of the relationship between sst1 and fdp mutations in yeast and their effect on trehalose synthesis. Braz J Med Biol Res 18:447–454. [PubMed] [Google Scholar]
- 91.Hohmann S, Huse K, Valentin E, Mbonyi K, Thevelein JM, Zimmermann FK. 1992. Glucose-induced regulatory defects in the Saccharomyces cerevisiae byp1 growth initiation mutant and identification of MIG1 as a partial suppressor. J Bacteriol 174:4183–4188. doi: 10.1128/jb.174.12.4183-4188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Aelst L, Hohmann S, Bulaya B, de Koning W, Sierkstra L, Neves MJ, Luyten K, Alijo R, Ramos J, Coccetti P. 1993. Molecular cloning of a gene involved in glucose sensing in the yeast Saccharomyces cerevisiae. Mol Microbiol 8:927–943. doi: 10.1111/j.1365-2958.1993.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 93.Thevelein JM. 1992. The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 62:109–130. doi: 10.1007/BF00584466. [DOI] [PubMed] [Google Scholar]
- 94.Hohmann S, Neves MJ, de Koning W, Alijo R, Ramos J, Thevelein JM. 1993. The growth and signalling defects of the ggs1 (fdp1/byp1) deletion mutant on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet 23:281–289. doi: 10.1007/BF00310888. [DOI] [PubMed] [Google Scholar]
- 95.Vandercammen A, Francois J, Hers HG. 1989. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase of Saccharomyces cerevisiae. Eur J Biochem 182:613–620. doi: 10.1111/j.1432-1033.1989.tb14870.x. [DOI] [PubMed] [Google Scholar]
- 96.Londesborough J, Vuorio OE. 1993. Purification of trehalose synthase from baker's yeast. Its temperature-dependent activation by fructose 6-phosphate and inhibition by phosphate. Eur J Biochem 216:841–848. [DOI] [PubMed] [Google Scholar]
- 97.Hohmann S, Bell W, Neves MJ, Valckx D, Thevelein JM. 1996. Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol Microbiol 20:981–991. doi: 10.1111/j.1365-2958.1996.tb02539.x. [DOI] [PubMed] [Google Scholar]
- 98.Bonini BM, Van Vaeck C, Larsson C, Gustafsson L, Ma P, Winderickx J, Van Dijck P, Thevelein JM. 2000. Expression of Escherichia coli otsA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem J 350:261–268. [PMC free article] [PubMed] [Google Scholar]
- 99.Blazquez MA, Lagunas R, Gancedo C, Gancedo JM. 1993. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett 329:51–54. doi: 10.1016/0014-5793(93)80191-V. [DOI] [PubMed] [Google Scholar]
- 100.Ernandes JR, De Meirsman C, Rolland F, Winderickx J, de Winde J, Brandao RL, Thevelein JM. 1998. During the initiation of fermentation overexpression of hexokinase PII in yeast transiently causes a similar deregulation of glycolysis as deletion of tps1. Yeast 14:255–269. [DOI] [PubMed] [Google Scholar]
- 101.Walther T, Mtimet N, Alkim C, Vax A, Loret MO, Ullah A, Gancedo C, Smits GJ, Francois JM. 2013. Metabolic phenotypes of Saccharomyces cerevisiae mutants with altered trehalose 6-phosphate dynamics. Biochem J 454:227–237. doi: 10.1042/BJ20130587. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez-Parraga P, Hernandez JA, Arguelles JC. 2003. Role of antioxidant enzymatic defences against oxidative stress H2O2 and the acquisition of oxidative tolerance in Candida albicans. Yeast 20:1161–1169. doi: 10.1002/yea.1029. [DOI] [PubMed] [Google Scholar]
- 103.Martinez-Esparza M, Aguinaga A, Gonzalez-Parraga P, Garcia-Penarrubia P, Jouault T, Arguelles JC. 2007. Role of trehalose in resistance to macrophage killing: study with a tps1/tps1 trehalose-deficient mutant of Candida albicans. Clin Microbiol Infect 13:384–394. doi: 10.1111/j.1469-0691.2007.01663.x. [DOI] [PubMed] [Google Scholar]
- 104.Martinez-Esparza M, Tapia-Abellan A, Vitse-Standaert A, Garcia-Penarrubia P, Arguelles JC, Poulain D, Jouault T. 2011. Glycoconjugate expression on the cell wall of tps1/tps1 trehalose-deficient Candida albicans strain and implications for its interaction with macrophages. Glycobiology 21:796–805. doi: 10.1093/glycob/cwr007. [DOI] [PubMed] [Google Scholar]
- 105.Himmelreich U, Dzendrowskyj TE, Allen C, Dowd S, Malik R, Shehan BP, Russell P, Mountford CE, Sorrell TC. 2001. Cryptococcomas distinguished from gliomas with MR spectroscopy: an experimental rat and cell culture study. Radiology 220:122–128. doi: 10.1148/radiology.220.1.r01jl25122. [DOI] [PubMed] [Google Scholar]
- 106.Himmelreich U, Allen C, Dowd S, Malik R, Shehan BP, Mountford C, Sorrell TC. 2003. Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes Infect 5:285–290. doi: 10.1016/S1286-4579(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 107.Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, Perfect JR, Kronstad J. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell 2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tenor JL, Oehlers SH, Yang JL, Tobin DM, Perfect JR. 2015. Live imaging of host-parasite interactions in a zebrafish infection model reveals cryptococcal determinants of virulence and central nervous system invasion. mBio 6:e01425-15. doi: 10.1128/mBio.01425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolschek MF, Kubicek CP. 1997. The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase-encoding genes, tpsA and tpsB. J Biol Chem 272:2729–2735. doi: 10.1074/jbc.272.5.2729. [DOI] [PubMed] [Google Scholar]
- 110.Svanstrom A, van Leeuwen MR, Dijksterhuis J, Melin P. 2014. Trehalose synthesis in Aspergillus niger: characterization of six homologous genes, all with conserved orthologs in related species. BMC Microbiol 14:90. doi: 10.1186/1471-2180-14-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fleck CB, Brock M. 2010. Aspergillus fumigatus catalytic glucokinase and hexokinase: expression analysis and importance for germination, growth, and conidiation. Eukaryot Cell 9:1120–1135. doi: 10.1128/EC.00362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu W, Solis NV, Ehrlich RL, Woolford CA, Filler SG, Mitchell AP. 2015. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol 13:e1002076. doi: 10.1371/journal.pbio.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pongpom M, Liu H, Xu W, Snarr BD, Sheppard DC, Mitchell AP, Filler SG. 2015. Divergent targets of Aspergillus fumigatus AcuK and AcuM transcription factors during growth in vitro versus invasive disease. Infect Immun 83:923–933. doi: 10.1128/IAI.02685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Allert S, Brunke S, Hube B. 2016. In vivo transcriptional profiling of human pathogenic fungi during infection: reflecting the real life? PLoS Pathog 12:e1005471. doi: 10.1371/journal.ppat.1005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.De Virgilio C, Burckert N, Bell W, Jeno P, Boller T, Wiemken A. 1993. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem 212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- 116.Piper PW, Lockheart A. 1988. A temperature-sensitive mutant of Saccharomyces cerevisiae defective in the specific phosphatase of trehalose biosynthesis. FEMS Microbiol Lett 49:245–250. doi: 10.1111/j.1574-6968.1988.tb02724.x. [DOI] [Google Scholar]
- 117.Elliott B, Haltiwanger RS, Futcher B. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, Thevelein JM, Van Dijck P. 2005. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol Biol Cell 16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maidan MM, Thevelein JM, Van Dijck P. 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem Soc Trans 33:291–293. doi: 10.1042/BST0330291. [DOI] [PubMed] [Google Scholar]
- 120.Maidan MM, De Rop L, Relloso M, Diez-Orejas R, Thevelein JM, Van Dijck P. 2008. Combined inactivation of the Candida albicans GPR1 and TPS2 genes results in avirulence in a mouse model for systemic infection. Infect Immun 76:1686–1694. doi: 10.1128/IAI.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, Cramer RA. 2011. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog 7:e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miao Y, Tenor JL, Toffaletti DL, Washington EJ, Liu J, Shadrick WR, Schumacher MA, Lee RE, Perfect JR, Brennan RG. 2016. Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis. Proc Natl Acad Sci U S A 113:7148–7153. doi: 10.1073/pnas.1601774113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vuorio OE, Kalkkinen N, Londesborough J. 1993. Cloning of two related genes encoding the 56-kDa and 123-kDa subunits of trehalose synthase from the yeast Saccharomyces cerevisiae. Eur J Biochem 216:849–861. doi: 10.1111/j.1432-1033.1993.tb18207.x. [DOI] [PubMed] [Google Scholar]
- 124.Ferreira JC, Silva JT, Panek AD. 1996. A regulatory role for TSL1 on trehalose synthase activity. Biochem Mol Biol Int 38:259–265. [PubMed] [Google Scholar]
- 125.Reinders A, Burckert N, Hohmann S, Thevelein JM, Boller T, Wiemken A, De Virgilio C. 1997. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol Microbiol 24:687–695. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- 126.Trevisol ET, Panek AD, De Mesquita JF, Eleutherio EC. 2014. Regulation of the yeast trehalose-synthase complex by cyclic AMP-dependent phosphorylation. Biochim Biophys Acta 1840:1646–1650. doi: 10.1016/j.bbagen.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 127.Cao Y, Wang Y, Dai B, Wang B, Zhang H, Zhu Z, Xu Y, Cao Y, Jiang Y, Zhang G. 2008. Trehalose is an important mediator of Cap1p oxidative stress response in Candida albicans. Biol Pharm Bull 31:421–425. doi: 10.1248/bpb.31.421. [DOI] [PubMed] [Google Scholar]
- 128.Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell 15:3167–3180. doi: 10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Urban C, Xiong X, Sohn K, Schroppel K, Brunner H, Rupp S. 2005. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol Microbiol 57:1318–1341. doi: 10.1111/j.1365-2958.2005.04771.x. [DOI] [PubMed] [Google Scholar]
- 130.Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d'Enfert C. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rao KN, Kumaran D, Seetharaman J, Bonanno JB, Burley SK, Swaminathan S. 2006. Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications. Protein Sci 15:1735–1744. doi: 10.1110/ps.062096606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Farelli JD, Galvin BD, Li Z, Liu C, Aono M, Garland M, Hallett OE, Causey TB, Ali-Reynolds A, Saltzberg DJ, Carlow CK, Dunaway-Mariano D, Allen KN. 2014. Structure of the trehalose-6-phosphate phosphatase from Brugia malayi reveals key design principles for anthelmintic drugs. PLoS Pathog 10:e1004245. doi: 10.1371/journal.ppat.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schluepmann H, Berke L, Sanchez-Perez GF. 2012. Metabolism control over growth: a case for trehalose-6-phosphate in plants. J Exp Bot 63:3379–3390. doi: 10.1093/jxb/err311. [DOI] [PubMed] [Google Scholar]
- 134.Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F. 2010. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 3:406–419. doi: 10.1093/mp/ssp114. [DOI] [PubMed] [Google Scholar]
- 135.Saito K, Kase T, Takahashi E, Horinouchi S. 1998. Purification and characterization of a trehalose synthase from the basidiomycete Grifola frondosa. Appl Environ Microbiol 64:4340–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eleutherio E, Panek A, De Mesquita JF, Trevisol E, Magalhaes R. 2015. Revisiting yeast trehalose metabolism. Curr Genet 61:263–274. doi: 10.1007/s00294-014-0450-1. [DOI] [PubMed] [Google Scholar]
- 137.Nwaka S, Kopp M, Holzer H. 1995. Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J Biol Chem 270:10193–10198. doi: 10.1074/jbc.270.17.10193. [DOI] [PubMed] [Google Scholar]
- 138.Nwaka S, Mechler B, Destruelle M, Holzer H. 1995. Phenotypic features of trehalase mutants in Saccharomyces cerevisiae. FEBS Lett 360:286–290. doi: 10.1016/0014-5793(95)00105-I. [DOI] [PubMed] [Google Scholar]
- 139.Zahringer H, Burgert M, Holzer H, Nwaka S. 1997. Neutral trehalase Nth1p of Saccharomyces cerevisiae encoded by the NTH1 gene is a multiple stress responsive protein. FEBS Lett 412:615–620. doi: 10.1016/S0014-5793(97)00868-5. [DOI] [PubMed] [Google Scholar]
- 140.Schepers W, Van Zeebroeck G, Pinkse M, Verhaert P, Thevelein JM. 2012. In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J Biol Chem 287:44130–44142. doi: 10.1074/jbc.M112.421503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Veisova D, Macakova E, Rezabkova L, Sulc M, Vacha P, Sychrova H, Obsil T, Obsilova V. 2012. Role of individual phosphorylation sites for the 14-3-3-protein-dependent activation of yeast neutral trehalase Nth1. Biochem J 443:663–670. doi: 10.1042/BJ20111615. [DOI] [PubMed] [Google Scholar]
- 142.Nwaka S, Mechler B, Holzer H. 1996. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett 386:235–238. doi: 10.1016/0014-5793(96)00450-4. [DOI] [PubMed] [Google Scholar]
- 143.Eleutherio EC, Araujo PS, Panek AD. 1993. Protective role of trehalose during heat stress in Saccharomyces cerevisiae. Cryobiology 30:591–596. doi: 10.1006/cryo.1993.1061. [DOI] [PubMed] [Google Scholar]
- 144.Eleutherio EC, Araujo PS, Panek AD. 1993. Role of the trehalose carrier in dehydration resistance of Saccharomyces cerevisiae. Biochim Biophys Acta 1156:263–266. doi: 10.1016/0304-4165(93)90040-F. [DOI] [PubMed] [Google Scholar]
- 145.Sanchez-Fresneda R, Gonzalez-Parraga P, Esteban O, Laforet L, Valentin E, Arguelles JC. 2009. On the biochemical classification of yeast trehalases: Candida albicans contains two enzymes with mixed features of neutral and acid trehalase activities. Biochem Biophys Res Commun 383:98–102. doi: 10.1016/j.bbrc.2009.03.134. [DOI] [PubMed] [Google Scholar]
- 146.Pedreno Y, Maicas S, Arguelles JC, Sentandreu R, Valentin E. 2004. The ATC1 gene encodes a cell wall-linked acid trehalase required for growth on trehalose in Candida albicans. J Biol Chem 279:40852–40860. doi: 10.1074/jbc.M400216200. [DOI] [PubMed] [Google Scholar]
- 147.Eck R, Bergmann C, Ziegelbauer K, Schonfeld W, Kunkel W. 1997. A neutral trehalase gene from Candida albicans: molecular cloning, characterization and disruption. Microbiology 143:3747–3756. doi: 10.1099/00221287-143-12-3747. [DOI] [PubMed] [Google Scholar]
- 148.Pedreno Y, Gonzalez-Parraga P, Martinez-Esparza M, Sentandreu R, Valentin E, Arguelles JC. 2007. Disruption of the Candida albicans ATC1 gene encoding a cell-linked acid trehalase decreases hypha formation and infectivity without affecting resistance to oxidative stress. Microbiology 153:1372–1381. doi: 10.1099/mic.0.2006/003921-0. [DOI] [PubMed] [Google Scholar]
- 149.Serneels J, Tournu H, Van Dijck P. 2012. Tight control of trehalose content is required for efficient heat-induced cell elongation in Candida albicans. J Biol Chem 287:36873–36882. doi: 10.1074/jbc.M112.402651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cowen LE. 2009. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog 5:e1000471. doi: 10.1371/journal.ppat.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cowen LE. 2013. The fungal Achilles' heel: targeting Hsp90 to cripple fungal pathogens. Curr Opin Microbiol 16:377–384. doi: 10.1016/j.mib.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 152.Plourde-Owobi L, Durner S, Parrou JL, Wieczorke R, Goma G, Francois J. 1999. AGT1, encoding an alpha-glucoside transporter involved in uptake and intracellular accumulation of trehalose in Saccharomyces cerevisiae. J Bacteriol 181:3830–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.da Costa Morato Nery D, da Silva CG, Mariani D, Fernandes PN, Pereira MD, Panek AD, Eleutherio EC. 2008. The role of trehalose and its transporter in protection against reactive oxygen species. Biochim Biophys Acta 1780:1408–1411. doi: 10.1016/j.bbagen.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 154.Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, Cardenas ME. 2007. Sensing the environment: lessons from fungi. Nat Rev Microbiol 5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 155.Xu JR. 2000. Map kinases in fungal pathogens. Fungal Genet Biol 31:137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- 156.Kruppa M, Calderone R. 2006. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res 6:149–159. doi: 10.1111/j.1567-1364.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 157.Schmidt M, Akasaka K, Messerly JT, Boyer MP. 2012. Role of Hog1, Tps1 and Sod1 in boric acid tolerance of Saccharomyces cerevisiae. Microbiology 158:2667–2678. doi: 10.1099/mic.0.060590-0. [DOI] [PubMed] [Google Scholar]
- 158.Hagiwara D, Suzuki S, Kamei K, Gonoi T, Kawamoto S. 2014. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet Biol 73:138–149. doi: 10.1016/j.fgb.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 159.Pereira Silva L, Alves de Castro P, Reis TF, Paziani MH, Von Zeska Kress MR, Riano-Pachon DM, Hagiwara D, Ries LN, Brown NA, Goldman GH. 27 October 2016. Genome-wide transcriptome analysis of Aspergillus fumigatus exposed to osmotic stress reveals regulators of osmotic and cell wall stresses that are SakAHog1 and MpkC dependent. Cell Microbiol doi: 10.1111/cmi.12681. [DOI] [PubMed] [Google Scholar]
- 160.Kandror O, Bretschneider N, Kreydin E, Cavalieri D, Goldberg AL. 2004. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell 13:771–781. doi: 10.1016/S1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- 161.Thevelein JM, de Winde JH. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 162.Garreau H, Hasan RN, Renault G, Estruch F, Boy-Marcotte E, Jacquet M. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146:2113–2120. doi: 10.1099/00221287-146-9-2113. [DOI] [PubMed] [Google Scholar]
- 163.Estruch F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 164.Wilson WA, Roach PJ. 2002. Nutrient-regulated protein kinases in budding yeast. Cell 111:155–158. doi: 10.1016/S0092-8674(02)01043-7. [DOI] [PubMed] [Google Scholar]
- 165.Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aguilera J, Randez-Gil F, Prieto JA. 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol Rev 31:327–341. doi: 10.1111/j.1574-6976.2007.00066.x. [DOI] [PubMed] [Google Scholar]
- 167.Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 168.François J, Walther T, Parrou JL. 2012. Genetics and regulation of glycogen and trehalose metabolism in Saccharomyces cerevisiae, p 29–55. In Liu Z. (ed), Microbial stress tolerance for biofuels: systems biology. Springer, New York, NY. [Google Scholar]
- 169.Ward MP, Gimeno CJ, Fink GR, Garrett S. 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol 15:6854–6863. doi: 10.1128/MCB.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJ. 2004. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell 3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 17:1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, Smith FJ, Nantel A, Mitchell AP. 2008. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell 19:2741–2751. doi: 10.1091/mbc.E08-02-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bahn YS. 2008. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell 7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Missall TA, Lodge JK. 2005. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol Microbiol 57:847–858. doi: 10.1111/j.1365-2958.2005.04735.x. [DOI] [PubMed] [Google Scholar]
- 175.Hagiwara D, Asano Y, Yamashino T, Mizuno T. 2008. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci Biotechnol Biochem 72:2756–2760. doi: 10.1271/bbb.80001. [DOI] [PubMed] [Google Scholar]
- 176.Emri T, Szarvas V, Orosz E, Antal K, Park H, Han KH, Yu JH, Pocsi I. 2015. Core oxidative stress response in Aspergillus nidulans. BMC Genomics 16:478. doi: 10.1186/s12864-015-1705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sakamoto K, Iwashita K, Yamada O, Kobayashi K, Mizuno A, Akita O, Mikami S, Shimoi H, Gomi K. 2009. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet Biol 46:887–897. doi: 10.1016/j.fgb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 178.Sakamoto K, Arima TH, Iwashita K, Yamada O, Gomi K, Akita O. 2008. Aspergillus oryzae atfB encodes a transcription factor required for stress tolerance in conidia. Fungal Genet Biol 45:922–932. doi: 10.1016/j.fgb.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 179.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zurita-Martinez SA, Cardenas ME. 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell 4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.De Virgilio C, Loewith R. 2006. The TOR signalling network from yeast to man. Int J Biochem Cell Biol 38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 182.Timblin BK, Bergman LW. 1997. Elevated expression of stress response genes resulting from deletion of the PHO85 gene. Mol Microbiol 26:981–990. doi: 10.1046/j.1365-2958.1997.6352004.x. [DOI] [PubMed] [Google Scholar]
- 183.Parrou JL, Enjalbert B, Francois J. 1999. STRE- and cAMP-independent transcriptional induction of Saccharomyces cerevisiae GSY2 encoding glycogen synthase during diauxic growth on glucose. Yeast 15:1471–1484. [DOI] [PubMed] [Google Scholar]
- 184.Ni M, Yu JH. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Park HS, Bayram O, Braus GH, Kim SC, Yu JH. 2012. Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol 86:937–953. doi: 10.1111/mmi.12032. [DOI] [PubMed] [Google Scholar]
- 186.Ostrosky-Zeichner L, Marr KA, Rex JH, Cohen SH. 2003. Amphotericin B: time for a new “gold standard.” Clin Infect Dis 37:415–425. [DOI] [PubMed] [Google Scholar]
- 187.Butts A, Krysan DJ. 2012. Antifungal drug discovery: something old and something new. PLoS Pathog 8:e1002870. doi: 10.1371/journal.ppat.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- 189.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bailly S, Maubon D, Fournier P, Pelloux H, Schwebel C, Chapuis C, Foroni L, Cornet M, Timsit JF. 2016. Impact of antifungal prescription on relative distribution and susceptibility of Candida spp.—trends over 10 years. J Infect 72:103–111. doi: 10.1016/j.jinf.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 191.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis 62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wiederhold NP, Patterson TF. 2015. Emergence of azole resistance in Aspergillus. Semin Respir Crit Care Med 36:673–680. doi: 10.1055/s-0035-1562894. [DOI] [PubMed] [Google Scholar]
- 193.Mdluli KE, Witte PR, Kline T, Barb AW, Erwin AL, Mansfield BE, McClerren AL, Pirrung MC, Tumey LN, Warrener P, Raetz CR, Stover CK. 2006. Molecular validation of LpxC as an antibacterial drug target in Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:2178–2184. doi: 10.1128/AAC.00140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Alksne LE, Projan SJ. 2000. Bacterial virulence as a target for antimicrobial chemotherapy. Curr Opin Biotechnol 11:625–636. doi: 10.1016/S0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- 195.Gauwerky K, Borelli C, Korting HC. 2009. Targeting virulence: a new paradigm for antifungals. Drug Discov Today 14:214–222. doi: 10.1016/j.drudis.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 196.Perfect JR. 1996. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agents Chemother 40:1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roemer T, Krysan DJ. 2014. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med 4:a019703. doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, Popolo L, Hartland RP, Latge JP. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem 275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- 199.Tsukahara K, Hata K, Nakamoto K, Sagane K, Watanabe NA, Kuromitsu J, Kai J, Tsuchiya M, Ohba F, Jigami Y, Yoshimatsu K, Nagasu T. 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol Microbiol 48:1029–1042. doi: 10.1046/j.1365-2958.2003.03481.x. [DOI] [PubMed] [Google Scholar]
- 200.Hata K, Horii T, Miyazaki M, Watanabe NA, Okubo M, Sonoda J, Nakamoto K, Tanaka K, Shirotori S, Murai N, Inoue S, Matsukura M, Abe S, Yoshimatsu K, Asada M. 2011. Efficacy of oral E1210, a new broad-spectrum antifungal with a novel mechanism of action, in murine models of candidiasis, aspergillosis, and fusariosis. Antimicrob Agents Chemother 55:4543–4551. doi: 10.1128/AAC.00366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Watanabe NA, Miyazaki M, Horii T, Sagane K, Tsukahara K, Hata K. 2012. E1210, a new broad-spectrum antifungal, suppresses Candida albicans hyphal growth through inhibition of glycosylphosphatidylinositol biosynthesis. Antimicrob Agents Chemother 56:960–971. doi: 10.1128/AAC.00731-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pfaller MA, Hata K, Jones RN, Messer SA, Moet GJ, Castanheira M. 2011. In vitro activity of a novel broad-spectrum antifungal, E1210, tested against Candida spp as determined by CLSI broth microdilution method. Diagn Microbiol Infect Dis 71:167–170. doi: 10.1016/j.diagmicrobio.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 203.Miyazaki M, Horii T, Hata K, Watanabe NA, Nakamoto K, Tanaka K, Shirotori S, Murai N, Inoue S, Matsukura M, Abe S, Yoshimatsu K, Asada M. 2011. In vitro activity of E1210, a novel antifungal, against clinically important yeasts and molds. Antimicrob Agents Chemother 55:4652–4658. doi: 10.1128/AAC.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Foster AJ, Jenkinson JM, Talbot NJ. 2003. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J 22:225–235. doi: 10.1093/emboj/cdg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Perfect JR, Tenor JL, Miao Y, Brennan RG. 2016. Trehalose pathway as an antifungal target. Virulence doi: 10.1080/21505594.2016.1195529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guirao-Abad JP, Sanchez-Fresneda R, Valentin E, Martinez-Esparza M, Arguelles JC. 2013. Analysis of validamycin as a potential antifungal compound against Candida albicans. Int Microbiol 16:217–225. [DOI] [PubMed] [Google Scholar]
- 207.Xue Y, Shui G, Wenk MR. 2014. TPS1 drug design for rice blast disease in Magnaporthe oryzae. Springerplus 3:18. doi: 10.1186/2193-1801-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Blazquez MA, Stucka R, Feldmann H, Gancedo C. 1994. Trehalose-6-P synthase is dispensable for growth on glucose but not for spore germination in Schizosaccharomyces pombe. J Bacteriol 176:3895–3902. doi: 10.1128/jb.176.13.3895-3902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Franco A, Soto T, Vicente-Soler J, Guillen PV, Cansado J, Gacto M. 2000. Characterization of tpp1 (+) as encoding a main trehalose-6P phosphatase in the fission yeast Schizosaccharomyces pombe. J Bacteriol 182:5880–5884. doi: 10.1128/JB.182.20.5880-5884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Badaruddin M, Holcombe LJ, Wilson RA, Wang ZY, Kershaw MJ, Talbot NJ. 2013. Glycogen metabolic genes are involved in trehalose-6-phosphate synthase-mediated regulation of pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS Pathog 9:e1003604. doi: 10.1371/journal.ppat.1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fernandez J, Wilson RA. 2012. Why no feeding frenzy? Mechanisms of nutrient acquisition and utilization during infection by the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 25:1286–1293. doi: 10.1094/MPMI-12-11-0326. [DOI] [PubMed] [Google Scholar]
- 212.Wilson RA, Gibson RP, Quispe CF, Littlechild JA, Talbot NJ. 2010. An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc Natl Acad Sci U S A 107:21902–21907. doi: 10.1073/pnas.1006839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wilson RA, Jenkinson JM, Gibson RP, Littlechild JA, Wang ZY, Talbot NJ. 2007. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J 26:3673–3685. doi: 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang ZY, Jenkinson JM, Holcombe LJ, Soanes DM, Veneault-Fourrey C, Bhambra GK, Talbot NJ. 2005. The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem Soc Trans 33:384–388. doi: 10.1042/BST0330384. [DOI] [PubMed] [Google Scholar]
- 215.Fernandez J, Wright JD, Hartline D, Quispe CF, Madayiputhiya N, Wilson RA. 2012. Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1-3, and a MATE-family pump regulate glucose metabolism during infection. PLoS Genet 8:e1002673. doi: 10.1371/journal.pgen.1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]