Abstract
The expression of Rbm3, a glycine-rich RNA-binding protein, is enhanced under conditions of mild hypothermia, and Rbm3 has been postulated to facilitate protein synthesis at colder temperatures. To investigate this possibility, Rbm3 was overexpressed as a c-Myc fusion protein in mouse neuroblastoma N2a cells. Cells expressing this fusion protein showed a 3-fold increase in protein synthesis at both 37°C and 32°C compared with control cells. Although polysome profiles of cells expressing the fusion protein and control cells were similar, several differences were noted, suggesting that Rbm3 might enhance the association of 40S and 60S ribosomal subunits at 32°C. Studies to assess a direct interaction of Rbm3 with ribosomes showed that a fraction of Rbm3 was associated with 60S ribosomal subunits in an RNA-independent manner. It appeared unlikely that this association could explain the global enhancement of protein synthesis, however, because cells expressing the Rbm3 fusion protein showed no substantial increase in the size of their monosome and polysome peaks, suggesting that similar numbers of mRNAs were being translated at approximately the same rates. In contrast, a complex that sedimented between the top of the gradient and 40S subunits was less abundant in cells expressing recombinant Rbm3. Further analysis showed that the RNA component of this fraction was microRNA. We discuss the possibility that Rbm3 expression alters global protein synthesis by affecting microRNA levels and suggest that both Rbm3 and microRNAs are part of a homeostatic mechanism that regulates global levels of protein synthesis under normal and cold-stress conditions.
Keywords: ribosome, polysome, hypothermia, RNA-binding protein
Mammalian cells exposed to mild hypothermia show a general inhibition of protein synthesis (1, 2) and a concomitant increase in the expression of a small number of cold-shock mRNAs and proteins (3). These proteins appear to protect cells by enabling them to more effectively tolerate cold-stress conditions (reviewed in refs. 3 and 4). In addition, it has been suggested that some cold-shock proteins may have important roles in human testis and skin, where cell temperatures are typically 30–34°C and ≈33°C, respectively (3).
One of the mRNAs induced by mild hypothermia encodes the RNA binding motif protein 3 (Rbm3) (5–7). This protein is expressed in a wide variety of human fetal tissues (5) and is subjected to both transcriptional and translational regulation in response to mild hypothermia (6, 8). Rbm3 contains a glycine-rich carboxyl terminus and can bind to both RNA and DNA via an amino-terminal RNA binding domain (ref. 5 and references therein). This protein is a member of a family of glycine-rich RNA binding proteins that include the cold-inducible RNA-binding protein (CIRP) (6, 7, 9, 10). Although the function of this family of proteins is not yet known, it has been suggested that they affect both transcription and translation (11–13) and that they may facilitate translation at colder temperatures by functioning as RNA chaperones (6, 8, 9, 14).
We now provide evidence to show that Rbm3 enhances global protein synthesis at both 37°C and 32°C. The results indicate that Rbm3 may affect translation directly, by binding to 60S ribosomal subunits; however, its small effect on polysome profiles and its association with only a subset of the 60S subunits make it unlikely that this binding can account wholly for its effect on protein synthesis. In contrast, we found that Rbm3 had a major effect on the relative abundance of microRNA-containing complexes in cells, suggesting a potential mechanism of action that may account for its effects on global levels of protein synthesis.
Materials and Methods
DNA Constructs and Infection Studies. A c-Myc epitope tag was added to the amino terminus of a full-length mouse Rbm3 cDNA by PCR (8) and inserted into a retroviral vector replacing the enhanced cyan fluorescent protein (ECFP) gene in murine ES cell virus (MESV)/ECFP/internal ribosome entry site (IRES)/EGFP/Rous sarcoma virus (RSV)pro (15) to generate MESV/c-Myc-Rbm3/IRES/EGFP/RSVpro. A virus designated MESV/IRES/EGFP/RSVpro served as a control. The retroviral plasmids were packaged in Cos1 cells as described in ref. 15, and virus-containing supernatant was added directly to mouse N2a neuroblastoma cells.
Western Blot Analysis and Generation of Antibodies. Protein samples were precipitated with trichloroacetic acid, washed with acetone, and boiled in NuPage sample buffer (pH 8.4) (Invitrogen) for 5 min. Proteins were separated on SDS polyacrylamide minigels (Invitrogen) and transferred to poly(vinylidene difluoride) membranes (Bio-Rad). Membranes were blocked, probed with antibodies, and developed by using the Western Breeze kit (Invitrogen). The antibodies against β-actin (Sigma, mAb), Rbm3, and L4 were used at 1:1,000 dilutions. Rbm3 was recognized by using a polyclonal antiserum raised against a peptide (DRYSGGNYRDNYDN) in its carboxyl terminus. Antibodies against ribosomal protein L4 were produced by immunizing rabbits with isolated L4 protein from purified rat liver 60S subunits.
Coimmunoprecipitation Experiments. Anti-Rbm3 antibodies or preimmune rabbit sera were conjugated to protein A beads by incubation for 4h at 4°C in radioimmunoprecipitation assay buffer (0.15 M NaCl/10 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). Ribosomes were precleared with protein A beads (Amersham Pharmacia) for 2 h at 4°C and separated by centrifugation at 2,500 × g for 5 min. Precleared ribosomal supernatants were incubated with anti-Rbm3 or with preimmune rabbit sera, both conjugated to protein A beads, overnight at 4°C in the presence or absence of RNase (100 μg/ml). Each immunoprecipitation was centrifuged for 5 min at 2,500 × g and washed five times with radioimmunoprecipitation assay buffer. Bound proteins were eluted by using NuPage sample buffer, resolved by SDS/PAGE, transferred to poly(vinylidene difluoride) membranes (Bio-Rad), and Western blotted by using polyclonal antibodies against Rbm3 or L4.
Preparation and Analysis of Polysomes and Ribosomes from N2a Cells. Ribosomal profiles were prepared from a total of 2 × 107 cells that were washed with ice-cold buffer A (20 mM Tris, pH 7.4/100 mM KCl/10 mM MgCl2/2 mg/ml leupeptin/2 mg/ml pepstatin/0.5% aprotinin/0.5 mg/ml heparin/2 mM DTT) containing 100 μg/ml cycloheximide (Sigma) to block ribosomes at the step of elongation. Cells were lysed with buffer A containing 0.3% Igepal-600 (Sigma) and 100 μg/ml cycloheximide. Cytoplasmic extracts were obtained after centrifugation at 30,000 × g for 15 min at 4°C, loaded onto a linear (10–50%) sucrose gradient in buffer A, and centrifuged at 100,000 × g for 3 h at 4°C. Gradients were fractionated by upward displacement with 70% (wt/vol) sucrose, and absorbance at 260 nm was monitored continuously by using an ISCO UA-6 UV monitor.
To produce a polysome-enriched pellet (P100) and a postpolysomal supernatant (S100), cytoplasmic extracts, prepared as described above but without cycloheximide, were centrifuged at 100,000 × g for 3 h at 4°C. Ribosomal subunits were prepared from P100 fractions as described by Blobel and Sabatini (16), and were kindly provided by George Rogers (The Scripps Research Institute).
Where indicated, 60S ribosomal subunit pellets were resuspended in 20 mM Tris, pH 7.4/0.9 M KCl/3 mM MgCl2/2 mM DTT, in the presence or absence of 10 mM EDTA, and the subunits were pelleted by centrifugation at 100,000 × g for 24 h at 4°C.
Preparation of Polysome Profiles and Ribosomes from Rat Tissues. Anesthetized rats were decapitated, and the brains were removed quickly and placed in ice-cold buffer A containing 100 μg/ml cycloheximide. All subsequent operations were performed at 4°C. Tissues were lysed and homogenized in this buffer by using a Dounce homogenizer with a motor-driven Teflon pestle. A postmitochondrial supernatant fraction was prepared by two consecutive centrifugations of the homogenate at 30,000 × g for 15 min. Triton X-100 was added to the supernatant to a concentration of 1%, incubated for 15 min, layered onto a step gradient containing 8 ml of 0.7 M (wt/vol) sucrose on top of 8 ml of 1.6 M (wt/vol) sucrose (both in buffer A), and centrifuged at 100,000 × g for 18 h. The polysome-enriched pellet was resuspended in buffer A containing 100 μg/ml cycloheximide and layered onto 10–50% sucrose gradients prepared in the same buffer. The gradients were centrifuged and fractionated as described above.
For RNase A digestion of rat brain polysomes, polysome-enriched fractions were incubated with RNase A at a final concentration of 100 μg/ml for 15 min on ice. The mixture was loaded onto 10–50% sucrose gradients (in buffer A), which were centrifuged and fractionated as described above.
To completely dissociate 40S and 60S ribosomal subunits from rat tissues, polysome-enriched pellets were resuspended in buffer A containing 3 mM MgCl2, 500 mM KCl, and puromycin was added to a final concentration of 1 mM. Samples were incubated at 37°C for 15 min and centrifuged twice for 15 min at 30,000 × g, and the supernatant was loaded on a linear sucrose gradient (10–30%) in the same buffer. The gradients were centrifuged at 100,000 × g for 6 h at 4°C, and fractions were collected as described above. Fractions corresponding to the indicated subunits were pooled, diluted with buffer A, and pelleted by centrifugation at 100,000 × g for 24 h.
We isolated 18S and 28S rRNAs from purified 40S and 60S subunits, respectively by phenol-chloroform extraction and ethanol precipitation. rRNAs were resuspended in water, separated on 1% denaturing formaldehyde-agarose gel, and visualized with ethidium bromide.
Protein Synthesis Measurements. We plated 2 × 104 control and c-Myc-Rbm3 N2a cells into 24-well microplates, and protein synthesis was measured as described in ref. 17.
RNA Analysis. To isolate RNA from sucrose gradient fractions, the fractions were precipitated with 0.5 M ammonium acetate and 2 vol ethanol, extracted with TRIzol (Sigma), precipitated with ethanol, and resuspended in 10 μl of water. RNA was separated on 15% (wt/vol) acrylamide/7 M urea gels (Invitrogen) and visualized with ethidium bromide. In end-labeling experiments, the 5′ phosphate groups from RNA molecules were removed by using Antarctic phosphatase (New England Biolabs) for 1 h at 37°C. Phosphatase was heat-inactivated at 65°C for 5 min, and RNAs were 5′ end-labeled with [γ-32P]ATP by using polynucleotide kinase (New England Biolabs). The samples were electrophoresed as described above and visualized by using a Molecular Dynamics Storm 860 PhosphorImager. The size of RNA molecules was determined by using molecular weight markers (Decade Marker System, Ambion, Austin, TX).
For Northern blotting, RNA samples were electrophoresed as described above, transferred to nylon membranes, fixed by UV cross-linking, and hybridized with a [32P]ATP end-labeled oligonucleotide probe (mmu-miR-125b; refs. 18 and 19). Hybridizations were performed at room temperature in 7% SDS/0.2M Na3PO4 (pH 7.3) overnight. Membranes were washed twice at room temperature with 2× SSC/0.1% SDS and twice with 0.5× SSE/0.1% SDS.
Results
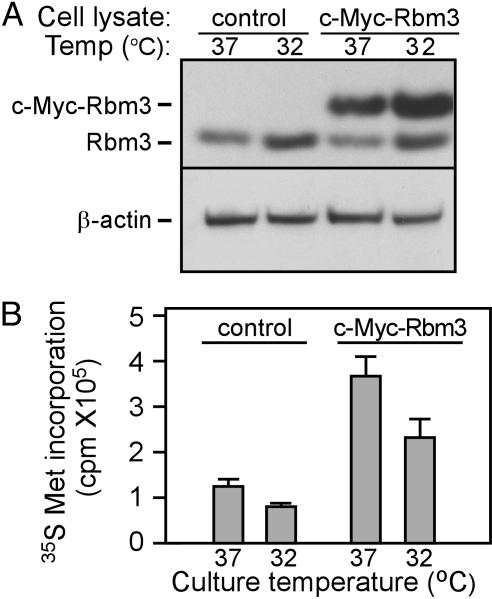
Expression of Rbm3 in Infected Cells. Rbm3 was expressed as a c-Myc fusion protein in N2a cells by using a retrovirus that expressed a dicistronic mRNA encoding c-Myc-Rbm3 as the first cistron and enhanced green fluorescent protein (EGFP) as the second cistron. A stable population of Rbm3-overexpressing cells was obtained after two rounds of cell sorting for EGFP expression. Lysates of cells that were stably infected either with an IRES/EGFP virus (control cells) or with the c-Myc-Rbm3/IRES/EGFP virus (c-Myc-Rbm3 cells) were subjected to immunoblot analysis. Endogenous Rbm3 (Rbm3) was detected in both sets of cells, whereas the Rbm3 fusion protein (c-Myc-Rbm3) was detected only in c-Myc-Rbm3 cells (Fig. 1A). Endogenous Rbm3 protein was expressed more highly at 32°C than at 37°C in agreement with previous findings (5–8). Although the recombinant protein showed a similar expression pattern in this experiment, no consistent differences in the levels of c-Myc-Rbm3 protein expression at 32°C compared with 37°C were observed (e.g., compare Figs. 1 A and 3). This result was consistent with the lack of Rbm3 promoter and noncoding RNA sequences in the expression construct that might enhance its expression under conditions of mild hypothermia (8).
Fig. 1.
Expression of an Rbm3 fusion protein increases protein synthesis in N2a cells. (A) Immunoblot analysis of Rbm3 in control and c-Myc-Rbm3 cells. Cells were grown continuously at 37°C or shifted to 32°C for 24 h. β-actin was used as a loading control. (B) Incorporation of [35S]Met/cysteine into control and c-Myc-Rbm3 cells. Data are the mean of four experiments ± SD. Measurements were normalized with respect to total protein as determined by Bradford assay.
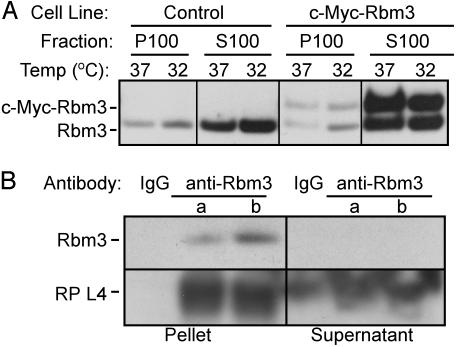
Fig. 3.
Rbm3 and c-Myc-Rbm3 proteins associate with ribosomes. (A) Control or c-Myc-Rbm3 cells were grown at 37°C or shifted to 32°C for 24 h. Postmitochondrial fractions from these cells were centrifuged at 100,000 × g for 3 h. The supernatant (S100) and pellet (P100) fractions were Western blotted by using anti-Rbm3 antibodies. (B) Rbm3 coimmunoprecipitates with rat brain ribosomes. We immunoprecipitated 13 A260 units (a) or 130 A260 units (b) of polysome-enriched fractions of rat brain by using anti-Rbm3 antibodies. The immunoprecipitates and the supernatants of the polysomal materials were analyzed for the presence of Rbm3 and ribosomal protein L4 by Western blotting by using anti-Rbm3 and anti-L4 antibodies, respectively. IgG was used in control experiments.
Expression of c-Myc-Rbm3 Increases Total Protein Synthesis. Control and c-Myc-Rbm3 cells were cultured at 37°C or 32°C and incubated with [35S]Met for 1 h, and the incorporation of radioactivity into proteins was quantified. The results showed that cells expressing c-Myc-Rbm3 incorporated ≈3-fold more [35S]Met than control cells at both temperatures (Fig. 1B). For both control and c-Myc-Rbm3 cells, protein synthesis was reduced by ≈25% at 32°C compared with 37°C. PAGE of the radiolabeled proteins showed a fairly uniform size distribution of these proteins in both control and c-Myc-Rbm3 cells, consistent with a global effect on protein synthesis (data not shown).
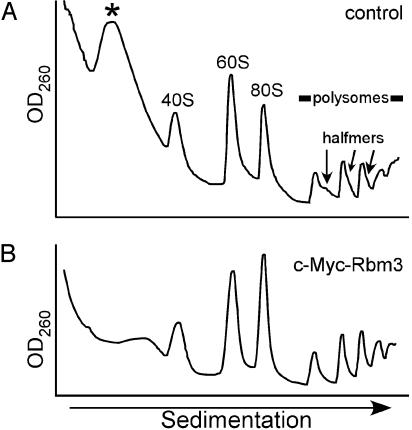
Under Conditions of Mild Hypothermia, the Relative Distribution of Ribosomes in c-Myc-Rbm3 Cells Differs from That of Control Cells. The relative abundance of monosome and polysome peaks from control and c-Myc-Rbm3 cells cultured at 37°C appeared to be similar (data not shown). In contrast, when these cells were cultured at 32°C, a larger fraction of the ribosomal subunits were present in 80S ribosomes and polysomes of the c-Myc-Rbm3 cells as compared with control cells (Fig. 2). In addition, shoulders were evident on the polysome peaks of control cells, which represent stalled translation initiation complexes that have been referred to as halfmers (20). Halfmers are thought to reflect inefficiencies in translation initiation and to comprise monosomes or polysomes with an additional 43S complex (40S ribosomal subunit with attached initiation factors) at the initiation codon, before the addition of a 60S ribosomal subunit (21). Halfmers were not observed in cells expressing the c-Myc-Rbm3 fusion protein, suggesting that this protein might enhance the 60S-joining step at 32°C. This result raised the possibility of a direct interaction between Rbm3 and the translation machinery.
Fig. 2.
Expression of the c-Myc-Rbm3 fusion protein increases monosome and polysome formation during mild hypothermia. Control (A) or c-Myc-Rbm3 (B) cells were cultured at 32°C for 24 h, and ribosomes were analyzed after centrifugation through 10–50% sucrose gradients. The top of the gradient is on the left. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), polysomes, and stalled preinitiation complexes (halfmers) are indicated. An asterisk indicates the position of a complex that sediments between the top of the gradient and 40S subunits.
In addition to the halfmers, there was a difference in the relative amount of an abundant complex that sedimented between the top of the gradient and 40S subunits. This complex was less abundant in the c-Myc-Rbm3 cells. Similar differences in the relative abundance of this complex also were noted at 37°C (data not shown). The position of this peak was reminiscent of a 15S ribonucleoprotein complex observed in HeLa cells (22).
Rbm3 Interacts with Ribosomes. To test whether Rbm3 associates with ribosomes, polysome-enriched fractions (P100) and postribosomal cell lysates (S100) were prepared from control and c-Myc-Rbm3 cells cultured at either 37°C or 32°C. Western blot analyses using anti-Rbm3 antibodies showed that both the native Rbm3 and the c-Myc-Rbm3 fusion protein were predominantly in the ribosome-free fractions of cells cultured at either temperature, but both proteins were also associated with the polysomal fractions (Fig. 3A).
Additional evidence for the association of Rbm3 with the translational apparatus came from coimmunoprecipitation experiments using anti-Rbm3 antibodies and brain polysomes. Ribosomal protein L4, an integral component of 60S ribosomal subunits, coimmunoprecipitated with Rbm3 (Fig. 3B). Although some ribosomal proteins exist in an extraribosomal form as independent polypeptides (23), the source of L4 that coimmunoprecipitated with Rbm3 was unlikely to be extraribosomal because these studies were performed by using a polysome-enriched fraction. After immunoprecipitation, Rbm3 was no longer detected in the supernatant, indicating that all of the Rbm3 in the sample was immunoprecipitated. In contrast, ribosomal protein L4 was still detected in the same supernatant (Fig. 3B), suggesting that Rbm3 was not associated with all of the ribosomes in the sample.
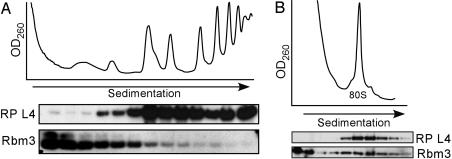
Rbm3 Interacts with Translating Ribosomes in an mRNA-Independent Manner. Western blot analyses of polysome profiles of N2a cells expressing the c-Myc-Rbm3 fusion protein and of rat brain showed the presence of Rbm3 predominantly in the lighter fractions; however, it was distributed throughout the entire sucrose gradient including the polysomal fraction (Fig. 4A). To test the association of Rbm3 with translating ribosomes, a brain polysomal pellet was treated with RNase A, which digests the mRNA between ribosomes and leads to the accumulation of 80S monosomes, which consist of the original monomers and 80S ribosomes derived from the digested polysomes (Fig. 4B). After RNase A treatment, Rbm3 cosedimented with 80S monosomes, suggesting that Rbm3 was associated with translating ribosomes and that mRNA was not a prerequisite for this interaction.
Fig. 4.
Rbm3 associates with translating ribosomes in an RNA-independent manner. (A) A fraction of Rbm3 copurifies with rat brain ribosomes on sucrose gradients. A polysome-enriched fraction of rat brain was centrifuged through a linear 10–50% (wt/vol) sucrose gradient. The top of the gradient is on the left. (B) Ribosomal profile of a rat brain polysome-enriched fraction after treatment with RNase A. For both A and B, the distributions of Rbm3 and ribosomal protein L4 in the fractions of the gradient were analyzed by immunoblotting using anti-Rbm3 and anti-L4 antibodies, respectively.
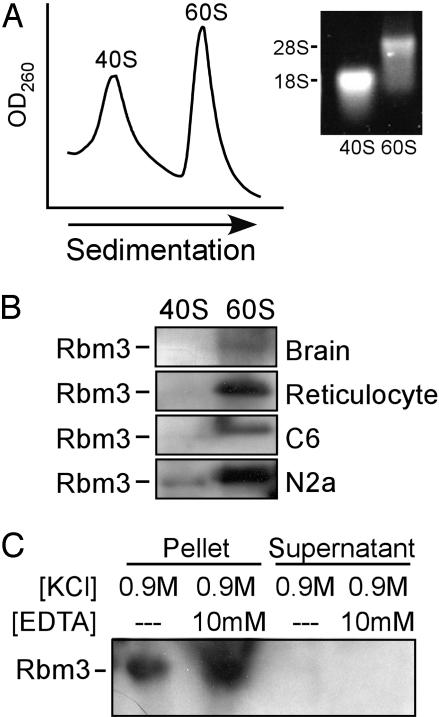
Rbm3 Associates with the Large Ribosomal Subunit. To identify the ribosomal subunit that interacts with Rbm3, polysome-enriched fractions from rat tissue and cell lines were dissociated to 40S and 60S subunits by using puromycin, a protein-synthesis inhibitor that causes premature release of the nascent chains from translating ribosomes (Fig. 5A). Western blots showed that in all cases Rbm3 was associated with the large ribosomal subunits (Fig. 5B).
Fig. 5.
Rbm3 associates tightly with the large ribosomal subunit. (A) Sucrose gradient profiles of rat brain ribosomal subunits treated with 0.5 M KCl. We used 18S and 28S rRNA, isolated from the pooled 60S and 40S fractions, to confirm the purity of the 40S and 60S subunits, respectively. (B) Immunoblot analysis of 60S and 40S subunits from rat brain, reticulocyte, C6 glioma, and N2a neuroblastoma was performed by using anti-Rbm3 antibodies. (C) Purified 60S subunits were treated with 0.9 M KCl in the presence or absence of 10 mM EDTA and subjected to 24-h centrifugation at 100,000 × g to obtain a pellet containing 60S subunits and supernatant fractions. The presence of Rbm3 in these fractions was assessed by immunoblotting using anti-Rbm3 antibodies.
The salt conditions used in our experiments (0.5 M KCl) are known to remove ribosome-associated translation factors, aminoacyl-tRNA synthetases, and some protein kinases, but not intrinsic ribosomal proteins (24). Therefore, the association of Rbm3 with 60S subunits under these conditions suggests a high binding affinity. Moreover, it appears that this interaction is independent of nascent polypeptide chains because it was not disrupted by puromycin.
We further tested the strength of the association of Rbm3 with 60S subunits under higher salt conditions (0.9 M KCl) that are known to remove all of the soluble protein synthesis factors and a few ribosomal proteins (25). The experiments were performed in the presence or absence of EDTA to assess the effect of magnesium cations on the stability of the Rbm3–ribosome complex. Magnesium ions stabilize rRNA higher-order structures, which are recognized as binding sites for some ribosomal proteins, and they directly stabilize intermolecular contacts between ribosomal proteins and rRNA (26). Western blot analyses showed that under all conditions tested, Rbm3 remained bound to 60S subunits and was absent from the supernatant fraction (Fig. 5C), indicating a tight association.
Expression of c-Myc-Rbm3 Was Correlated with Alterations in the Abundance of a microRNA Complex. Polysome profiles revealed the decreased relative abundance of a complex that sedimented between the float and 40S ribosomal subunits in cells expressing the Rbm3 fusion protein compared with control cells (see Fig. 2). This difference was observed reproducibly when the density of the harvested cells was between ≈60% and 90% confluent (data not shown). RNA was extracted from this complex and analyzed by gel electrophoresis (Fig. 6A). The results revealed an RNA component that was estimated to be ≈20 nt in length by using RNA size markers. The size of this RNA was consistent with that of microRNAs, which are ≈22 nt in length (27).
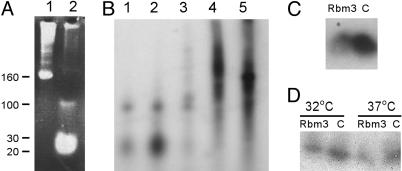
Fig. 6.
Expression of an Rbm3 fusion protein alters the abundance of a miRNA-containing complex. (A) RNA was extracted from fractions of a sucrose gradient profile of control cells at 32°C and stained with ethidium bromide. Lane 1 is RNA from the top of the gradient, and lane 2 is RNA from the complex that sediments between the top of the gradient and 40S subunits (indicated with an asterisk in Fig. 2 A). The positions of RNA size markers are indicated. (B) RNAs from fractions of a sucrose gradient profile of control cells at 32°C were phosphatase treated, [γ-32P]ATP end-labeled, and separated in acrylamide–urea gels. RNAs in lanes 1–5 are from sequential fractions from the top of the gradient to the bottom. Lane 2 corresponds to the fraction represented in A, lane 2. (C) Northern blot analysis of fractions containing small RNAs from control (C) and c-Myc-Rbm3 (Rbm3) cells grown at 32°C by using a microRNA (mmu-miR-125b) probe. (D) Northern blot analysis of total RNA from control (C) and c-Myc-Rbm3 (Rbm3) cells grown at 32 or 37°C by using the mmu-miR-125b probe. The hybridization signals for C and D correspond to RNAs between the 20- and 30-nt size markers.
MicroRNAs have been reported to be present in polysomes, so we asked whether the alterations in the size of the putative microRNA complex observed in our studies (Fig. 2) were due to alterations in the total amount of these RNAs present in the cells or to their redistribution, i.e., being associated with polysomes or not associated with polysomes. To address this question, fractions from polysome profiles of control cells cultured at 32°C were isolated and 5′ end-labeled (Fig. 6B). The results showed that radioactivity corresponding to microRNAs was almost all contained within the complex that sedimented between the float and 40S subunits (Fig. 6B, lane 2). As observed in the polysome profiles (Fig. 2), this complex was less abundant in cells expressing the Rbm3 fusion protein (see Fig. 7, which is published as supporting information on the PNAS web site).
To confirm that the small RNAs observed in these studies were microRNAs, RNAs from the complex that sedimented between the float and 40S subunits were transferred to nylon membrane and probed with a radiolabeled probe to mmu-miR-125b, an abundant mouse microRNA (18, 19). This probe hybridized to the small RNAs, indicating that they were microRNAs (Fig. 6C). The results showed that mmu-miR-125b was less abundant in cells expressing the Rbm3 fusion protein than in control cells. This same probe was hybridized to total RNA from cells expressing the Rbm3 fusion protein and from control cells, and the results also showed a decreased hybridization signal in cells expressing the Rbm3 fusion protein (Fig. 6D), suggesting that the change in the amount of microRNA complex observed in the polysome profiles reflected a corresponding change in the total amount of microRNA rather than a redistribution.
Discussion
Rbm3 protein is a cold-induced member of the glycine-rich RNA-binding protein family. Although a function for this protein has not yet been defined, it has been suggested that Rbm3 may affect gene expression by facilitating translation at colder temperatures (6, 8, 9, 14). Rbm3 also has been implicated in several important cellular processes (28–30). In this study, we showed that expression of a Rbm3 fusion protein in N2a cells increased protein synthesis at both physiological and mild hypothermic temperatures. The expression of this protein led to higher relative levels of 80S monosomes and polysomes consistent with an increased association of ribosomal subunits during the initiation step of protein synthesis. We also found a tight physical association of Rbm3 with 60S ribosomal subunits that did not require RNA or nascent polypeptide chains. In addition, we found that expression of a Rbm3 fusion protein led to a decrease in microRNA levels.
Other RNA binding proteins that associate with mammalian 60S ribosomal subunits include fragile X syndrome protein FMR1 and the fragile X-related proteins FXR1 and FXR2 (31–33). However, these proteins can be removed from ribosomes with 0.5 M KCl (31), indicating that they are less tightly associated than Rbm3, which was still associated with ribosomes after treatment with 0.9 M KCl.
Our studies revealed, nevertheless, that the majority of brain cytoplasmic Rbm3 is not bound to ribosomes. Similar results have been observed for other proteins that associate with ribosomes, including CIRP2 (34), S6 kinase 1 (35), PRMT3 (36), Gemin3 and Gemin4 (37), and the yeast mitochondrial protein OxaI (38, 39). The tight association of a fraction of Rbm3 with ribosomes suggests that the unbound Rbm3 may differ from the ribosome-bound protein in some way or that the ribosome binding sites may be limiting.
It has been reported that mammalian cells exposed to mild hypothermia display a general inhibition of protein synthesis (1, 2). Our findings are consistent with this observation (Fig. 1B); furthermore, the observation that Rbm3 has a positive effect on protein synthesis suggests that this protein may prevent a more dramatic reduction of protein synthesis under this condition. Although the stoichiometry of Rbm3 to ribosomes is not known, the association of Rbm3 with only a subset of ribosomes (Fig. 3B) raises the possibility of a ribosome subpopulation with distinct translational properties (40). The absence of halfmers and the increased level of monosome formation during the initiation step of protein synthesis in cells expressing the fusion protein (Fig. 2) suggest that this subpopulation of ribosomes may initiate translation at a higher rate.
It appears doubtful that a higher rate of translation could account completely for the 3-fold increase in total protein synthesis observed in cells expressing the fusion protein compared with control cells because this increase in protein synthesis was not reflected in the polysome analysis. If the expression of the Rbm3 protein increased protein synthesis solely by affecting translation initiation, elongation, or termination, we would expect a significant change in the size and/or distribution of the polysome peaks. Likewise, any effects of Rbm3 on mRNA levels or on the levels of components of the translation machinery would be expected to alter the size of polysome peaks. One possibility, albeit unlikely, is that extraribosomal Rbm3 enhances protein synthesis by altering the efficiency of multiple processes, e.g., by increasing the efficiency of initiation, elongation, and termination. These types of coincident changes might alter the overall level of protein synthesis without altering the polysome profile.
A more likely scenario is that expression of the Rbm3 affects protein synthesis by a posttranslational mechanism that alters microRNA levels within cells. The relative size of the microRNA complex identified in our profiles (Fig. 2) appears to be comparable to the 15S microRNA complex reported in HeLa cells (22). The abundance of this complex reflects numerous observations in the literature that point to the abundance of this class of RNAs. For example, quantitative Northern blot analyses of several individual microRNAs in Caenorhabditis elegans found that they were present at up to 50,000 copies per cell (41). In addition, the microRNA registry lists >200 different microRNAs in mouse (42).
MicroRNAs were identified in C. elegans (43) and have been shown to block protein production from target mRNAs by means of interactions with 3′ UTR sequences (reviewed in ref. 44). In the case of the lin-4 microRNA from C. elegans, protein expression from the lin-14 mRNA was blocked without altering the polysome distribution of this mRNA (45). It was postulated that lin-4 blocked protein production by either blocking translation elongation or by a mechanism that led to the rapid degradation of the newly synthesized peptides. In the latter case, the rate of translation might be unaltered.
Based on various observations regarding microRNAs, including the number of different microRNAs, their abundance, and the number of potential mRNA target sites, one possibility is that microRNAs may have a general dampening effect on global protein synthesis (44). The results of the present study are consistent with this notion, suggesting that the overall rate of protein synthesis as measured by [35S]Met incorporation is inversely correlated with the number of microRNAs within the cell. Inasmuch as the polysome profiles of cells with radically different overall rates of protein synthesis were very similar, it appears likely that the underlying mechanism involves the stability of the nascent peptide rather than a translational mechanism.
The mechanism by which the Rbm3 fusion protein affected microRNA levels may have involved direct interactions between this protein and microRNAs or ribonucleoprotein complexes containing microRNAs. Alternatively, the Rbm3 protein may have affected microRNA levels indirectly, e.g., by competing for microRNA binding sites on mRNAs or by affecting the expression of a component involved in microRNA synthesis or degradation. Ongoing studies are investigating these and other possibilities and also will assess the effects of the native protein on microRNA levels and protein synthesis under normothermic and hypothermic conditions.
These findings prompt the hypothesis that microRNAs are part of a homeostatic mechanism that controls global protein synthesis. Proteins such as Rbm3 may counteract the dampening effect of microRNAs and increase global protein synthesis under conditions of stress such as mild hypothermia, conditions under which protein synthesis might otherwise be even less efficient.
Supplementary Material
Acknowledgments
We thank Dr. Bruce A. Cunningham for valuable advice regarding these studies. We also thank Dr. Robyn Meech for advice with the RNA analyses and Luke Burman and Kathryn Woodard for excellent technical assistance. This work was supported by National Institutes of Health Grant GM61725, the G. Harold and Leila Y. Mathers Charitable Foundation (to V.P.M.), U.S. Public Health Service Grant NS39837 (to G.M.E.), and the FRAXA Research Foundation (to P.W.V.). J.D. and A.A. were supported by postdoctoral fellowships from The Skaggs Institute for Chemical Biology.
Abbreviations: IRES, internal ribosome entry site; Rbm3, RNA binding motif protein 3.
References
- 1.Burdon, R. H. (1987) Symp. Soc. Exp. Biol. 41, 113–133. [PubMed] [Google Scholar]
- 2.Wassmann, H., Greiner, C., Hulsmann, S., Moskopp, D., Speckmann, E. J., Meyer, J. & Van Aken, H. (1998) Neurol. Res. 20, S61–S65. [DOI] [PubMed] [Google Scholar]
- 3.Fujita, J. (1999) J. Mol. Microbiol. Biotechnol. 1, 243–255. [PubMed] [Google Scholar]
- 4.Phadtare, S., Alsina, J., Inouye, M. (1999) Curr. Opin. Microbiol. 2, 175–180. [DOI] [PubMed] [Google Scholar]
- 5.Derry, J. M., Kerns, J. A. & Francke, U. (1995) Hum. Mol. Genet. 4, 2307–2311. [DOI] [PubMed] [Google Scholar]
- 6.Danno, S., Nishiyama, H., Higashitsuji, H., Yokoi, H., Xue, J.-H., Itoh, K., Matsuda, T. & Fujita, J. (1997) Biochem. Biophys. Res. Commun. 236, 804–807. [DOI] [PubMed] [Google Scholar]
- 7.Danno, S., Itoh, K., Matsuda, T. & Fujita, J. (2000) Am. J. Pathol. 156, 1685–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chappell, S. A., Owens, G. C. & Mauro, V. P. (2001) J. Biol. Chem. 276, 36917–36922. [DOI] [PubMed] [Google Scholar]
- 9.Nishiyama, H., Itoh, K., Kaneko, Y., Kishishita, M., Yoshida, O. & Fujita, J. (1997) J. Cell Biol. 137, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheikh, M. S., Carrier, F., Papathanasiou, M. A., Hollander, M. C., Zhan, Q., Yu, K. & Fornace A. J. Jr., (1997) J. Biol. Chem. 272, 26720–26726. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfuss, G., Matunis, M. J., Pinol-Roma, S. & Burd, C. G. (1993) Annu. Rev. Biochem. 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 12.Ladomery, M. (1997) BioEssays 19, 903–909. [DOI] [PubMed] [Google Scholar]
- 13.Wright, C. F., Oswald, B. W. & Dellis, S. (2001) J. Biol. Chem. 276, 40680–40686. [DOI] [PubMed] [Google Scholar]
- 14.Nishiyama, H., Higashitsuji, H., Yokoi, H., Itoh, K., Danno, S., Matsuda, T. & Fujita, J. (1997) Gene 208, 115–120. [DOI] [PubMed] [Google Scholar]
- 15.Owens, G. C., Chappell, S. A., Mauro, V. P. & Edelman, G. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blobel, G. & Sabatini, D. (1971) Proc. Natl. Acad. Sci. USA 68, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra-Gorur, K., Singer, H. A. & Castellot, J. J., Jr. (2002) J. Biol. Chem. 277, 33537–33540. [DOI] [PubMed] [Google Scholar]
- 18.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 19.Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E. & Ambros, V. (2004) Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helser, T. L., Baan, R. A. & Dahlberg, A. E. (1981) Mol. Cell. Biol. 1, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baronas-Lowell, D. M. & Warner, J. R. (1990) Mol. Cell. Biol. 10, 5235–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wool, I. G. (1996) Trends Biochem. Sci. 21, 164–165. [PubMed] [Google Scholar]
- 24.Spirin, A. S. (1990) in The Ribosome: Structure, Function and Evolution, eds. Hill, W. E., Dahlberg, A., Garrett, R. A., Moore, P. B., Schlessinger, D. & Warner, J. R. (ASM, Washington, DC), pp. 56–70.
- 25.Sherton, C. C. & Wool, I. G. (1972) J. Biol. Chem. 247, 4460–4467. [PubMed] [Google Scholar]
- 26.Klein, D. J., Moore, P. B. & Steitz, T. A. (2004) RNA 10, 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambros, V. (2004) Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- 28.Kita, H., Carmichael, J., Swartz, J., Muro, S., Wyttenbach, A., Matsubara, K., Rubinsztein, D. C. & Kato, K. (2002) Hum. Mol. Genet. 11, 2279–2287. [DOI] [PubMed] [Google Scholar]
- 29.Baghdoyan, S., Dubreuil, P., Eberle, F. & Gomez, S. (2000) Blood 95, 3750–3757. [PubMed] [Google Scholar]
- 30.Wellmann, S., Buhrer, C., Moderegger, E., Zelmer, A., Kirschner, R., Koehne, P., Fujita, J. & Seeger, K. (2004) J. Cell. Sci. 117, 1785–1794. [DOI] [PubMed] [Google Scholar]
- 31.Siomi, M. C., Zhang, Y., Siomi, H. & Dreyfuss, G. (1996) Mol. Cell. Biol. 16, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefani, G., Fraser, C. E., Darnell, J. C. & Darnell, R. B. (2004) J. Neurosci. 24, 7272–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khandjian, E. W., Huot, M. E., Tremblay, S., Davidovic, L., Mazroui, R. & Bardoni, B. (2004) Proc. Natl. Acad. Sci. USA 101, 13357–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto, K., Aoki, K., Dohmae, N., Takio, K. & Tsujimoto, M. (2000) Nucleic Acids Res. 28, 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volarevic, S. & Thomas, G. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 101–127. [DOI] [PubMed] [Google Scholar]
- 36.Bachand, F. & Silver, P. A. (2004) EMBO J. 23, 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, P. T., Hatzigeorgiou, A. G. & Mourelatos, Z. (2004) RNA 10, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia, L., Dienhart, M., Schramp, M., McCauley, M., Hell, K. & Stuart, R. A. (2003) Embo. J. 22, 6438–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szyrach, G., Ott, M., Bonnefoy, N., Neupert, W. & Herrmann, J. M. (2003) EMBO J. 22, 6448–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mauro, V. P. & Edelman, G. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim, L. P., Lau, N. C., Weinstein, E. G., Abdelhakim, A., Yekta, S., Rhoades, M. W., Burge, C. B. & Bartel, D. P. (2003) Genes Dev. 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones, S. (2004) Nucleic Acids Res. 32, D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 44.Bartel, D. P. (2004) Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 45.Olsen, P. H. & Ambros, V. (1999) Dev. Biol. 216, 671–680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.