ABSTRACT
Invariant natural killer T (iNKT) cells are innate T cells with powerful immune regulatory functions that recognize glycolipid antigens presented by the CD1D protein. While iNKT cell-activating glycolipids are currently being explored for their efficacy to improve immunotherapy against infectious diseases and cancer, little is known about the mechanisms that control CD1D antigen presentation and iNKT cell activation in vivo. CD1D molecules survey endocytic pathways to bind lipid antigens in MHC class II-containing compartments (MIICs) before recycling to the plasma membrane. Autophagosomes intersect with MIICs and autophagy-related proteins are known to support antigen loading for increased CD4+ T cell immunity. Here, we report that mice with dendritic cell (DC)-specific deletion of the essential autophagy gene Atg5 showed better CD1D1-restricted glycolipid presentation in vivo. These effects led to enhanced iNKT cell cytokine production upon antigen recognition and lower bacterial loads during Sphingomonas paucimobilis infection. Enhanced iNKT cell activation was independent of receptor-mediated glycolipid uptake or costimulatory signals. Instead, loss of Atg5 in DCs impaired clathrin-dependent internalization of CD1D1 molecules via the adaptor protein complex 2 (AP2) and, thus, increased surface expression of stimulatory CD1D1-glycolipid complexes. These findings indicate that the autophagic machinery assists in the recruitment of AP2 to CD1D1 molecules resulting in attenuated iNKT cell activation, in contrast to the supporting role of macroautophagy in CD4+ T cell stimulation.
KEYWORDS: antigen presentation, autophagy, CD1D1, dendritic cells, glycolipid, innate-like lymphocytes, internalization, NKT cell, T cell
Introduction
Invariant natural killer T (iNKT) cells constitute a subset of T cells that expresses an invariant TCRα chain (Vα24Jα18 in humans and Vα14Jα18 in mice) and recognizes glycolipid antigens in the context of the MHC class I-related glycoprotein CD1D.1 Upon TCR ligation, iNKT cells rapidly secrete large amounts of cytokines, such as IFNG and IL4, and act as an amplification relay at the interface of innate and adaptive immunity.2 Due to their capacity to induce maturation of dendritic cells (DCs) for the initiation and maintenance of adaptive immune responses,3-5 iNKT cells are regarded as a “natural adjuvant.” iNKT cell agonists such as α-galactosylceramide (αGalCer) and its analogs are currently being investigated in clinical trials as part of improved vaccination strategies against infections and cancers.6 The mechanisms controlling lipid antigen loading onto CD1D molecules for iNKT cell activation are incompletely understood. After assembly in the endoplasmic reticulum, CD1 molecules follow the secretory pathway through the Golgi apparatus to the plasma membrane. Some CD1D molecules also associate with MHC class II molecules and the invariant chain, which directs CD1D complexes to endosomal compartments without first reaching the plasma membrane.7,8 After trafficking to the plasma membrane, CD1 molecules including CD1D are internalized to sample antigens in the endosomal-lysosomal system9 and, similar to MHC class II molecules,10 recycle to the cell surface and present bound antigens to iNKT cells. Macroautophagy is an evolutionarily conserved mechanism that delivers cytoplasmic constituents and substrates for MHC class II presentation to lysosomes and late endosomes.11,12 Involvement of autophagy-facilitated transport is confirmed by studies using mice with DC-specific deletion of the essential autophagy gene Atg5 that show impaired CD4+ T cell priming after herpes simplex virus infection.12 Furthermore, human Atg5-deficient macrophages show inefficient MHC class II presentation of extracellular antigens and decreased CD4+ T cell stimulation.13 The relevance of the autophagy machinery in lipid antigen loading within endosomal compartments has not received adequate attention. Here, we investigated its role in dendritic cell-mediated iNKT cell activation.
Results
Loss of Atg5 increases the iNKT cell stimulatory capacity of dendritic cells
To investigate whether macroautophagy regulates CD1D1 trafficking, we initially determined the colocalization of CD1D1 with MAP1LC3A/LC3A (microtubule associated protein 1 light chain 3 α; LC3A), present both on autophagosomes and LC3-associated phagosomes.13,14 In sorted primary splenic DCs, LC3A and CD1D1 colocalized in steady-state as determined by the Pearson correlation coefficient analysis. Activation of DCs with the TLR9 (toll-like receptor 9) agonist CpG, which facilitates DC-mediated iNKT cell activation,15 significantly increased this colocalization (Fig. 1A). To test whether absence of the essential macroautophagy gene Atg5 in DCs inhibits CD1D1 antigen presentation similar to MHC class II presentation,13,16,17 we generated mice with conditional knockout (CKO) of Atg5 in DCs (Atg5f/f × ITGAX-Cre, designated Atg5-DC CKO). FACS-sorted splenic DCs isolated from Atg5-DC CKO mice and Atg5 floxed control (Ctrl) littermates were pulsed with different amounts of the prototypic iNKT agonist αGalCer and used to stimulate the iNKT cell hybridomas A.407 and FF13. Surprisingly, Atg5-DC CKO-derived DCs were more potent in eliciting IL2 production by both iNKT cell lines (Fig. 1B, C). Conversely, induction of the autophagy machinery in wild-type DCs by noncytotoxic concentrations of the MTOR inhibitor rapamycin reduced IL2 production upon co-culture with iNKT cells (Fig. 1D; Fig. S1). These data indicate that autophagy proteins in DCs may regulate iNKT cell responses.
Figure 1.
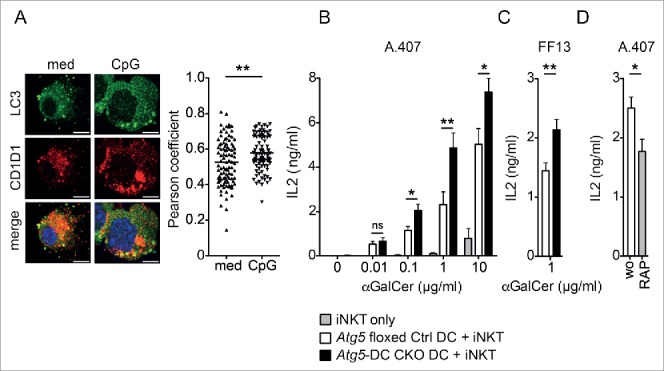
Loss of Atg5 increases the iNKT cell stimulatory capacity of dendritic cells. C57BL/6 derived splenic DCs were either incubated with 2 µg/ml CpG or left untreated. Original magnification with 63×, 1.4 NA oil immersion lens. Scatter dot plot representation and quantification of colocalization between CD1D1 and LC3A (Pearson coefficient analysis). Each symbol represents one cell. Pooled data of 3 independent experiments are shown. Scale bar: 2.5 µm. Statistics: 2-tailed Mann-Whitney U Test (A). FAC-sorted ITGAX and I-Ab double-positive splenic DCs derived from Atg5 floxed Ctrl or Atg5-DC CKO animals were pulsed with increasing concentrations of αGalCer or left untreated and subsequently cocultured with the iNKT hybridoma cell line A.407 (B) or FF13 (C) for 24 h. IL2 concentration in cell culture supernatants was measured via ELISA. Pooled data and SEM of > 3 independent experiments are shown. Each experiment contained at least 3 animals per group. Statistics: 2-tailed unpaired Student t test. C57BL/6 derived purified splenic DCs were treated with rapamycin (RAP; 12.5 μM; 4 h), subsequently pulsed with αGalCer (1 µg/ml; 4 h) and cocultured with iNKT hybridoma cell line A.407 (24 h). IL2 concentration in cell culture supernatants was measured via ELISA. Pooled data and SEM of 3 independent experiments is shown. Each experiment contained at least 3 animals per group. Statistics: 2-tailed unpaired Student t test (D). *** P ≤ 0.001,** P ≤ 0.01,* ≤ 0.05, ns P > 0.05.
To determine the mechanism of Atg5-dependent regulation of iNKT cell activation, we first evaluated whether Atg5-DC CKO DCs differed from Atg5 floxed Ctrl DCs in their acquisition of lipid antigen. Whereas the prototypic iNKT cell agonist αGalCer is taken up via both low density lipoprotein receptor (LDLR)-mediated and scavenger receptor-mediated pathways,18 the nominal antigen digalactosylceramide (GalαGalCer) and the Sphingomonas-derived glucuronylceramide (GSL-1) require selective uptake via the LDLR or MSR1 (macrophage scavenger receptor 1), respectively.18,19 The absence of Atg5 in primary DCs resulted in more efficient stimulation of iNKT cell hybridomas independently of the type of antigen uptake, as shown by stimulation with LDLR-targeted (GalαGalCer), MSR1-targeted (GSL-1), and MSR1 and LDLR-targeted (αGalCer) glycolipid antigens (Fig. 2A). These findings suggest that the increased iNKT cell stimulation by Atg5-deficient DCs did not involve a unique type of internalizing receptor.
Figure 2.
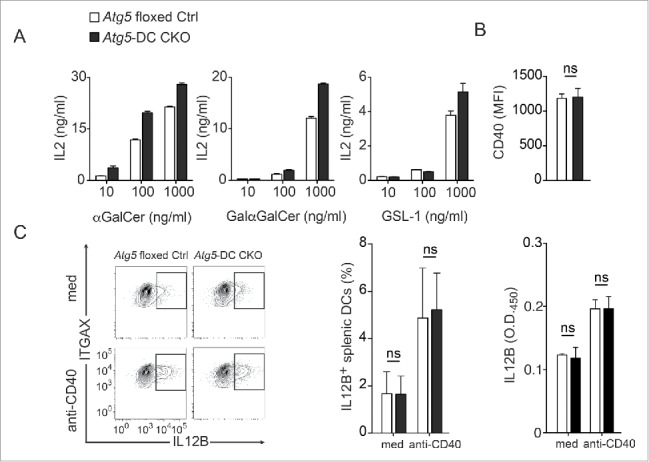
Receptor-mediated uptake of glycolipid antigens and costimulatory properties remain unchanged in Atg5-DC CKO DCs. CD1D1-presentation of indicated iNKT cell agonists by primary splenic DCs was assessed using activation of the iNKT hybridoma A.407 as in Fig. 1B (A). Splenic DCs derived from either Atg5 floxed Ctrl or Atg5-DC CKO animals were stained for CD40 surface expression. Pooled data and SEM of 3 independent experiments are shown. Each experiment contained at least 3 animals per group. Statistics: 2-tailed unpaired Student t test (B). Frequency of IL12B-producing splenic DCs upon CD40 ligation. Purified splenic DCs were either incubated with anti-CD40 overnight or left untreated. IL12B production was measured via intracellular cytokine staining and by ELISA. Results of 3 independent experiments are shown. Each experiment contained at least 3 animals per group. Statistics: 2-tailed unpaired Student t test (C). *** P≤ 0.001,** P≤ 0.01,* ≤ 0.05, ns P> 0.05. MFI, mean fluorescence intensity.
Our findings observed in primary DCs were unexpected and appeared to be in contrast with a recent study in which Atg7-deficient CSF2/GM-CSF-differentiated bone marrow-derived DCs (BMDCs) do not differ from macroautophagy-competent BMDCs in eliciting cytokine production by iNKT cells.20 To investigate this discrepancy we generated BMDCs from Atg5-deficient mice and also found that they elicit IL2 production by iNKT cells following uptake of glycolipid antigens to the same level as BMDCs from wild-type mice (Fig. S2). Because in vitro-derived BMDC cultures contain heterogeneous cell populations including macrophages, DCs, and neutrophils,21,22 we additionally FACS-purified in vitro generated BMDCs for ITGAX and MHC class II double-positive cells which were subsequently loaded with increasing concentrations of αGalCer and coincubated with iNKT cells. Increased IL2 production by iNKT cells were observed only in Atg5-DC CKO-derived ITGAX and MHC class II double-positive BMDC cultures loaded with high αGalCer concentrations (10 µg/ml) (Fig. S3). Even ITGAX and MHC class II double-positive BMDCs contain different myeloid cell subsets including common monocyte precursor-derived macrophages which are inferior to common DC precursor antigen-presenting cells in their T cell priming and stimulatory capacity.22,23 We therefore continued to study DC-mediated iNKT cell activation using FAC-sorted primary splenic DCs. Next, we determined whether Atg5-deficient DCs differed from their Atg5-competent counterparts in providing costimulatory signals required for physiological iNKT cell activation, i.e. CD40 expression and production of heterodimers consisting of IL12A and IL12B.24 Atg5-DC CKO DCs exhibited similar CD40 levels as Atg5 floxed Ctrl DCs (Fig. 2B). Moreover, Atg5 deficiency did not affect the frequency of IL12B-producing DCs or the total amount of IL12B secreted by DCs in response to CD40 ligation (Fig. 2C), in agreement with the reported unaltered expression levels of CD40 and CD86 as well as normal production of IL12B, IL6 and TNF in Atg5-deficient DCs in the steady state.12 Thus, loss of Atg5 in DCs increases their capability for iNKT cell activation independently of receptor-mediated lipid-targeting mechanisms and most costimulatory properties.
Increased CD1D1 surface expression in Atg5-deficient primary DCs
We next investigated whether loss of Atg5 enhances the iNKT cell stimulatory capacity of DCs by affecting CD1D1 expression. We first determined CD1D1 surface expression levels in DCs compared with B cells derived from Atg5-DC CKO mice and their Atg5 floxed Ctrl littermates. Sorted ITGAX and MHC class II double-positive DCs and CD19 and MHC class II double-positive B cells were analyzed for CD1D1 expression (Fig. 3A). Atg5-deficient DCs showed significantly higher expression levels of surface CD1D1 protein (Fig. 3B), whereas no difference was observed for B cells which lacked Cre-expression and, thus, were autophagy-competent (Fig. S4A). To determine whether loss of Atg5 regulates CD1D1 expression in vivo, Atg5-DC CKO and Atg5 floxed Ctrl mice were injected with αGalCer and CD1D1 presentation was investigated using the monoclonal antibody L363 that specifically detects CD1D1-αGalCer-complexes, but does not bind to CD1D1 molecules loaded with other antigens.25 Compared to autophagy-competent DCs, Atg5-deficient DCs showed increased expression levels of CD1D1-glycolipid complexes (Fig. 3C). Levels of stimulatory CD1D1-antigen complexes were similar for B cells in Atg5-DC CKO mice (Fig. 3C). In contrast to CD1D1, the expression levels of ITGAX and MHC class II remained unchanged in Atg5-DC CKO DCs (Fig. 3D).
Figure 3.
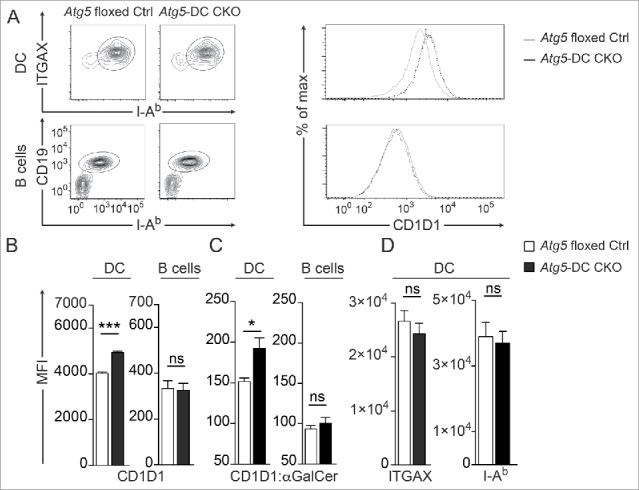
Increased CD1D1 surface expression in Atg5-DC CKO DCs. Flow cytometric analysis of CD1D1 surface levels on splenic DCs. Representative contour plots of splenic DCs or B cells and histograms showing CD1D1 surface expression (A). Quantification of CD1D1 surface expression on splenic DCs and B cells. Pooled data and SEM from >3 independent experiments are shown. Each experiment contained at least 2 animals per group. Statistics: 2-tailed unpaired Student t test (B). CD1D1-αGalCer-complex staining (L363 clone) in splenic DCs or B cells 4 h after i.p. application of either αGalCer or PBS. Pooled data and SEM from 2 independent experiments is shown. Statistics: Unpaired 2-tailed t test (C). DC surface expression of ITGAX or I-Ab. Pooled data and SEM from >3 independent experiments is shown. Each experiment contained at least 2 animals per group. Statistics: Unpaired 2-tailed t test (D). *** P≤ 0.001,** P≤ 0.01,* ≤ 0.05, ns P> 0.05. MFI, mean fluorescence intensity.
Loss of Atg5 impairs CD1D1 internalization but not CD1D1 recycling to the cell surface
CD1D1 expression levels at the cell surface are largely determined by the rates of CD1D1 internalization and recycling through endosomal and lysosomal compartments.26,27 Therefore, we separately analyzed the rates of CD1D1 internalization and CD1D1 recycling to the cell surface in primary Atg5-deficient and Atg5 floxed Ctrl DCs. To evaluate the kinetics of CD1D1 internalization, we determined the remainder of previously biotin-labeled CD1D1 molecules at the cell surface by flow cytometry. To assess CD1D1 recycling rates, we quantified newly resurfaced CD1D1 molecules after the cell surface CD1D1 had been previously blocked with unconjugated anti-CD1D1 antibodies (Fig. 4A). CD1D1 internalization was significantly impaired (Fig. 4B), whereas trafficking of intracellular CD1D1 molecules back to the cell surface remained unaffected in Atg5-DC CKO DCs (Fig. 4C). As expected, B cells derived from Atg5-DC CKO mice and Atg5 floxed Ctrl littermates showed similar kinetics of CD1D1 turnover (Fig. 4B, C). Thus, loss of Atg5 in primary DCs impairs CD1D1 internalization, leading to increased CD1D1 surface expression levels and an enhanced capacity of Atg5-deficient DCs to activate iNKT cells.
Figure 4.
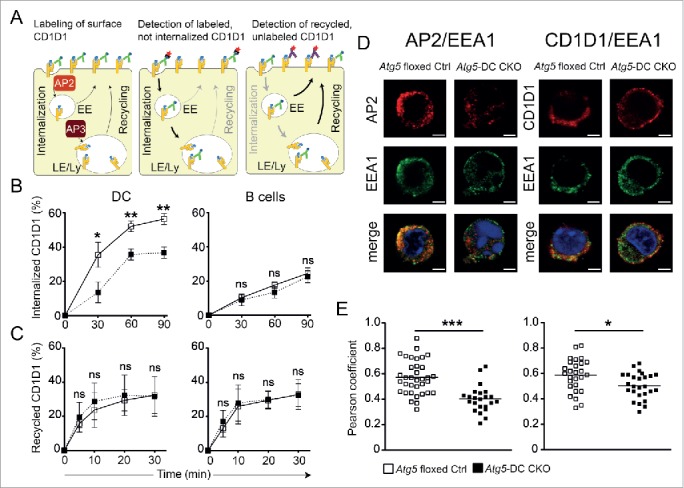
Impaired CD1D1 internalization in Atg5-DC CKO DCs. CD1D1 surface levels are determined by the rates of CD1D1 internalization and recycling through endosomal and lysosomal compartments. EE, early endosomes; LE, late endosomes; Ly, lysosomes (A). Internalization of surface CD1D1 on splenic DCs or splenic B cells was analyzed using a biotin-based flow cytometric endocytosis assay. Pooled data and SEM of at least 3 independent experiments are shown. Each experiment contained at least 2 animals per group. Statistics: 2-tailed unpaired Student t test (B). CD1D1 recycling in splenic DCs or splenic B cells was analyzed using a flow cytometry based recycling assay. Pooled data and SEM of at least 3 independent experiments are shown. Each experiment contained at least 2 animals per group. Statistics: 2-tailed unpaired Student t test (C). Colocalization study of AP2 and EEA1, as well as CD1D1 and EEA1 via confocal microscopy. Original magnification with 63×, 1.4 NA oil immersion lens. Representative photographs from 2 independent experiments per colocalization study are shown. Scale bar: 2.5 µm (D). Scatter dot plot representation and quantification of colocalization between AP2 and EEA1 and between CD1D1 and EEA1 via the Pearson coefficient. Each symbol represents one cell. Pooled data of 2 independent experiments per colocalization study are shown. Statistics: 2-tailed unpaired Student t test (E). *** P≤ 0.001,** P≤ 0.01,* ≤ 0.05, ns P> 0.05.
Reduced endosomal recruitment of CD1D1 and its adaptor protein AP2 in Atg5-deficient primary DCs
Internalization of mouse CD1D1 (Mus musculus CD1D1MmCD1D1), which contains a tyrosine-based endosomal-targeting motif in its cytoplasmic tail,28 follows a clathrin-dependent pathway that allows binding of CD1D1 to adaptor protein complexes (AP). MmCD1D1 molecules bind AP2, which is expressed at the plasma membrane and targets internalized CD1D1 molecules to endosomal compartments.29-31 During macroautophagy, LC3 conjugation to the autophagosomal membrane promotes recruitment of specific substrates into autophagosomes via LC3-binding anchor-proteins which function as selective autophagy receptors in the presence of LC3-interacting regions (LIRs).32,33 AP2 contains a LIR motif within its α subunit, and this LC3 binding by AP2 facilitates delivery of β-amyloid precursors for lysosomal degradation.34 To determine whether loss of autophagy proteins impairs trafficking of AP2 and CD1D1 to endosomal compartments in primary DCs, colocalization of both molecules with EEA1 (early endosomal antigen 1) was evaluated by confocal microscopy (Fig. 4D). Colocalization of AP2 and CD1D1 with EEA1 was significantly reduced in Atg5-deficient DCs as compared with Atg5 floxed Ctrl DCs (Fig. 4E). These data suggest that autophagy proteins assist in the recruitment of CD1D1 molecules into endosomal compartments and provide a mechanistic explanation as to how loss of autophagy proteins stabilizes the cell surface expression of CD1D1-glycolipid complexes on DCs.
Autophagy proteins in DCs regulate iNKT cell responses to lipid immunization and microbial infection in vivo
We investigated whether loss of Atg5 regulates iNKT cell activation and DC maturation upon αGalCer challenge in vivo. Frequencies of iNKT cells, DCs and B cells were comparable in Atg5-DC CKO and Atg5 floxed Ctrl mice at steady state (Fig. 5A, Fig. S4B).
Figure 5.
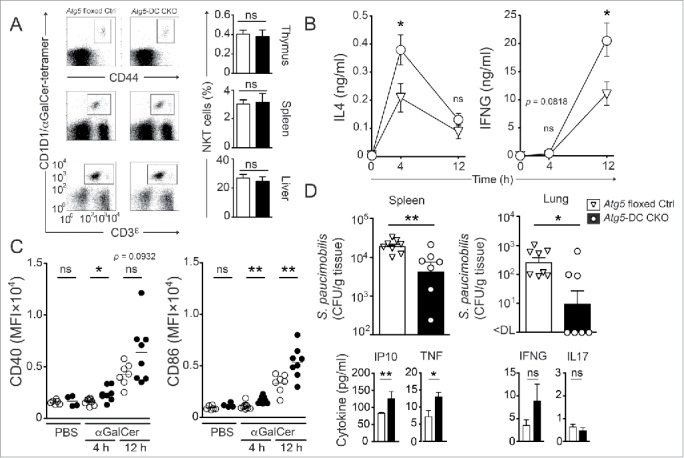
Autophagy proteins in DCs regulate iNKT cell responses to lipid immunization and microbial infection in vivo. Frequencies of iNKT cells (thymus, spleen and liver) from naïve Atg5 floxed Ctrl and Atg5-DC CKO mice were quantified. Representative dot plots from at least 2 independent experiments are shown (A). Serum concentration kinetics of IL4 and IFNG upon glycolipid challenge in vivo measured via ELISA (B). Surface expression kinetics of CD86 and CD40 on splenic DCs. Each symbol represents one animal. Pooled data of at least 2 independent experiments are shown. Statistics: 2-tailed unpaired Student t test (C). Quantification of bacterial titers (CFU/g tissue) in spleen and lung after in vivo infection of mice with S. paucimobilis. Each symbol represents one animal. One representative of 2 experiments with similar results is shown. Quantification of serum cytokines upon in vivo infection with S. paucimobilis. One representative of 2 experiments with similar results is shown. Statistics: 2-tailed Mann-Whitney U Test (D). *** P≤ 0.001,** P≤ 0.01,* ≤ 0.05, ns P> 0.05. MFI, mean fluorescence intensity.
Downregulation of the T cell receptor precluded the evaluation of iNKT cell frequencies and activation markers 12 h after αGalCer challenge.35 Therefore, we determined IFNG and IL4 serum levels after glycolipid challenge in vivo. Following αGalCer injection, Atg5-DC CKO mice showed substantially higher IL4 levels after 4 h and IFNG levels after 12 h, as compared with Atg5 floxed Ctrl mice (Fig. 5B). αGalCer-mediated activation of iNKT cells leads to full DC maturation in vivo within 4 h.3 The increased and prolonged iNKT cell activation observed in Atg5-DC CKO mice was associated with enhanced phenotypic DC maturation as indicated by increased expression levels of CD86 as well as CD40 12 h after αGalCer challenge while costimulatory molecule expression in B cells remained similar in αGalCer-treated Atg5-DC CKO and Atg5 floxed Ctrl mice (Fig. 5C, Fig. S5).
We next investigated whether the enhanced iNKT cell stimulatory capacity of Atg5-deficient DCs as observed upon αGalCer-challenge conferred protection against infection with Sphingomonas paucimobilis (S. paucimobilis). The gram-negative, lipopolysaccharide-free bacterium S. paucimobilis, associated with opportunistic and nosocomial infections in immunocompromised patients,36 contains glycosphingolipids that stimulate iNKT cells through their TCR in a CD1D1-specific manner.37-40 Although the stimulatory potential of S. paucimobilis-derived glycosphingolipids is lower as compared with αGalCer, iNKT cell recognition of these natural microbial glycolipids induces proinflammatory cytokine production in vitro and in vivo.37,39,41 Upon infection with S. paucimobilis, Atg5-DC CKO mice exhibited significantly lower pathogen loads in spleens and lungs as compared with their Atg5 floxed Ctrl counterparts (Fig. 5D). Reduced pathogen loads were associated with increased serum levels of proinflammatory cytokines such as CXCL10/IP10 (chemokine [C-X-C motif] ligand 10) and TNF. Production of IFNG tended to be higher in Atg5-DC CKO mice, whereas IL17 production was unchanged (Fig. 5D). Thus, autophagy proteins in DCs regulate surface expression of stimulatory CD1D1-glycolipid complexes, shape iNKT cell activation and subsequent DC maturation in response to αGalCer and natural microbial glycolipids in vivo.
Discussion
The autophagic machinery enhances adaptive immunity by supporting antigen loading onto MHC class II molecules and is currently being explored for its potential to improve T cell responses upon vaccinations to establish long-lasting T cell memory.42-45 Autophagosomes can fuse with MIICs and deliver intracellular antigens for loading onto MHC class II and MHC class I molecules.44,46-48 In addition, autophagy regulates exogenous antigen processing through the LC3–phosphatidylethanolamine-conjugation machinery during LC3-associated phagocytosis, a process which requires Atg5 and facilitates MHC class II loading.12-14,17 Both pathways support loading of vesicular antigens onto MHC molecules, leading to increased and more efficient CD4+ T cell responses. A fraction of CD1D1 proteins associates with MHC class II and invariant chain complexes which are sorted into late endosomal MIICs before trafficking to the cell surface7,8 However, in contrast to the supportive activity on MHC class II presentation, we found that autophagy proteins negatively regulate the presentation of lipid antigens by CD1D1. Loss of Atg5 in DCs impaired CD1D1 internalization, leading to increased expression of stimulatory CD1D1-glycolipid complexes and enhanced iNKT cell activation.
Internalization of CD1 molecules from the plasma membrane is essential for their ability to sample antigens in the endocytic system.28,49 CD1D1 molecules are internalized in clathrin-coated pits via the interaction of AP2 and AP3 with tyrosine-based sorting motifs present in the cytoplasmic tails of CD1D1.50,51 Tian et al. have identified a LIR motif within the α subunit of AP2 (AP2A1), which is reported to mediate enhanced Alzheimer precursor protein internalization and degradation.34 Once this motif is mutated to alanine repeats, AP2A1 no longer coimmunoprecipitates with LC3.34 In line with this identification of AP2 as an LC3-interacting protein,34 we found that loss of Atg5 impairs trafficking of both AP2 and CD1D1 molecules into endosomal compartments. Altogether, these data indicate that internalization of CD1D1 occurs less efficiently if AP2 cannot be recruited via LC3, leading to increased surface expression of stimulatory CD1D1-glycolipid complexes.
Similar to CD1D1, surface levels of classical MHC class I molecules are elevated due to decreased endocytosis and degradation.52 However, AP2 components were not significantly reduced in immunoprecipitates of classical MHC class I molecules derived from Atg5-deficient cells compared with floxed littermates. Instead, Atg5-deficiency compromised MHC class I association with the AP2-associated kinase 1 which can support AP2-independent, but clathrin-dependent endocytosis pathways.52-54 Furthermore, LIR motifs have also been described in clathrin itself.55 Thus, autophagy proteins appear to affect different clathrin-dependent pathways for the internalization and degradation of classical and nonclassical MHC class I molecules, respectively.
Loss of autophagy proteins has recently been reported to exert a T cell-intrinsic role in regulating iNKT cell development.20,56 Mice with T lymphocyte-specific deletion of Atg5 or Atg7 show reduced progression of thymic iNKT cells through the cell cycle and lower frequencies of both thymic and peripheral iNKT cell populations due to increased apoptosis and impaired survival.20,56 Frequencies of thymic and peripheral iNKT cells were unchanged in Atg5-DC CKO mice, supporting the notion that the reported deficit in iNKT cell differentiation is cell-autonomous. Our data indicate that while autophagy proteins regulate iNKT cell development via T cell-intrinsic effects, they attenuate peripheral iNKT activation through their function in DC-mediated antigen processing and presentation.
In conclusion, our findings indicate that the autophagic machinery regulates iNKT cell responses through CD1D1 internalization. These data broaden the concept of Atg-mediated antigen presentation and indicate that autophagy-related proteins might not only enhance, as described previously with MHC class II-restricted presentation of peptide antigens, but also limit and tune the presentation of lipid antigens and the activation of innate T cells. Its function in tuning lipid antigen presentation could potentially limit the efficacy of vaccination strategies that include iNKT cell ligands as adjuvants and target macroautophagy for improved CD4+ T cell immunity.44,45 A clear and comprehensive concept of how the autophagy machinery couples to antigen-presentation and lymphocyte activation appears to be required to predict the outcome of therapeutic interventions in this pathway to boost adaptive immunity.
Materials and methods
Antibodies, streptavidin, tetramer and iNKT lipid agonists
Unconjugated anti-mouse CD40 (Bio X Cell, BE0016–2; clone: FGK4.5/ FGK45), unconjugated anti-mouse FCGR3A/CD16 and FCGR2A/CD32 (Bio X Cell, CUS-HB-197; clone 2.4G2), PE-conjugated anti-mouse CD1D1 (eBioscience, 12–0011–83; clone: 1B1), PE-conjugated rat IgG2bκ (eBioscience, 12–4031–83; clone: eB149/10H5), PE-conjugated anti-α-GalCer-CD1D1-complex (eBioscience, 12–2019–82; clone: L363), PE-conjugated anti-mouse IL12B (eBioscience, 12–7123–81; clone: C17.8), PerCP-Cy5.5-conjugated anti-mouse CD8A/CD8α (eBioscience, 45–0081–82; clone: 53.6.7), unconjugated anti-mouse CD1D1 (Biolegend, 123515; clone: 1B1), biotinylated anti-mouse CD1D1 (Biolegend, 123506; clone: 1B1), Pacific Blue-conjugated anti-mouse CD19 (Biolegend, 115523; clone: 6D5), Pacific Blue-conjugated anti-mouse CD4 (Biolegend, 100428; clone: GK1.5), PE-conjugated anti-mouse CD40 (Biolegend, 124610; clone: 3/23), PE-Cy7-conjugated anti-mouse ITGAX (Biolegend, 117318; clone: N418), APC-conjugated anti-mouse I-Ab (clone: Biolegend, 107614; M5/114.15.2), APC-conjugated anti-mouse CD3E/CD3ε (Biolegend, 100236; clone: 17A2), PE-conjugated streptavidin (Biolegend, 405204), unconjugated anti-LC3A (MBL International Corporation, PM036), Alexa Fluor 488-conjugated F(ab')2 goat anti-rabbit IgG (H+L) (Thermo Fisher Scientific, A-11070) or -donkey anti-goat IgG (H+L) (Thermo Fisher Scientific, A-11055) and Alexa Fluor 555-conjugated F(ab')2 goat anti-rat (H+L) (Thermo Fisher Scientific, A-21434) or -rabbit anti-mouse IgG (H+L) (Thermo Fisher Scientific, A-21427), unconjugated anti-EEA1 (Antibodies-online, ABIN1439993), unconjugated anti-AP2 (BD Biosciences, 610502; clone 8/Adaptin α), PE-conjugated αGalCer-loaded CD1D1 tetramer (ProImmune, E001–2A), αGalCer (Adipogen, AG-CN2–0013-M001), GSL-1 and GalαGalCer were kindly provided by Paul B. Savage, Brigham Young University, Provo, UT, USA.
Immunocytochemistry and colocalization assays
For assessing colocalization between LC3A and CD1D1, CD1D1 and EEA1 or AP2 and EEA1 in splenic DCs, C57BL/6,-Atg5-DC CKO- or Atg5 floxed Ctrl-derived splenocytes were MAC-sorted via magnetic ITGAX microbeads (Miltenyi Biotech, 130–097–059) according to the manufacturer's recommendation using an autoMACS Pro separator (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany, 130–092–545). MACS-purified ITGAX+ cells were plated out in 0.01% poly-l-lysine (Sigma-Aldrich, P4707–50ML) in ddH2O pretreated 8-well glass chamber slides (Nunc Lab-Tek, NUNC-177402) at 1 × 105/chamber. For assessing colocalization between LC3A and CD1D1, ITGAX+ splenocytes were then either kept in R10 (RPMI-1640 (Life Technologies, 7001612) + 10% fetal calf serum (Sigma-Aldrich, F2442) + 50 U/ml penicillin-streptomycin (Life Technologies, 15070–063) only or R10 supplemented with 2 µg/ml of the B-class mouse TLR9 agonist CpG ODN 1826 (InvivoGen, tlrl-1826–5) for 5 h at 37°C and 5% CO2. Cells were centrifuged (3 min, 300 g), supernatants were carefully aspirated and chambers were washed twice (200 µl/chamber) with cold phosphate-buffered saline (PBS; Thermo Fisher Scientific, 10010023). Cells were fixed for 15 min at room temperature (RT) in 3% paraformaldehyde (PFA; Santa Cruz Biotechnology, sc-281692) in PBS followed by 2 washing steps with PBS (200 µl/chamber). For CD1D1 and EEA1 colocalization studies, cells were incubated with FCGR3A/CD16 and FCGR2A/CD32 blocking antibody (Bio X Cell, CUS-HB-197; clone 2.4G2), labeled for CD1D1 30 min at 4°C, then incubated at 37°C for an additional hour in R10, before fixation. Cells were permeabilized for 5 min at RT in 200 µl of 0.1% Triton X-100 (Sigma-Aldrich, X100–500ML) in PBS followed by 30 min of blocking with 100 µl of 1% bovine serum albumin (Sigma-Aldrich, A9576–50ML), 10% normal goat serum (NGS; Sigma-Aldrich, G9023–5ML) in PBS at RT. Afterwards cells were incubated with primary antibodies (rabbit-anti-LC3A [MBL International Corporation, PM036], rat-anti-CD1D1 [Biolegend, 123515; clone: 1B1], goat anti-EEA1 [Antibodies-online, ABIN1439993], mouse-anti-AP2 [BD Biosciences, 610502; clone 8/Adaptin α]) in 0.1% saponin (Sigma-Aldrich, 47036–50G-F), 10% NGS in PBS for 1 h at RT. Chambers were carefully washed twice with 0.1% saponin in PBS (200 µl/chamber) followed by a 45-min incubation with secondary antibodies in 0.1% saponin, 10% NGS in PBS (Alexa Fluor 488-conjugated or Alexa Fluor 555-conjugated anti-rabbit [Thermo Fisher Scientific, A-11070], anti-rat [Thermo Fisher Scientific, A-21434], anti-goat [Thermo Fisher Scientific, A-11055] and anti-mouse [Thermo Fisher Scientific, A-21427]). Cells were washed twice with PBS (200 µl/chamber), followed by incubation with 4',6-diamidino-2-phenylindole, dihydrochloride (Thermo Fisher Scientific, D1306) for 2 min at RT. Washing steps as above were performed and slides were mounted with ProLong Gold antifade mountant (Thermo Fisher Scientific, P36930), let sit for 24 h at RT in the dark and were then transferred to 4°C in the dark until further analysis. Pictures were acquired using a 63×, 1.4 NA oil immersion lens with an inverted confocal laser scanning microscope (SP5-UV, Leica Microsystems, Heerbrugg, Switzerland). To determine colocalization, the Pearson correlation coefficient values were calculated using JACoP (Just Another Colocalization Plugin) in ImageJ software. A Pearson score >0.6 indicates a statistically relevant colocalization of the signals.
Coculture assays with splenic DCs
CD1D1-mediated presentation of lipid antigens was measured using the mouse Vα14+ Vβ8+ iNKT cell hybridomas A.407 and FF13. NKT cell hybridomas were cultured in hybridoma culture medium (RPMI-1640 supplemented with 10% fetal calf serum, 50 U/ml penicillin-streptomycin, 2 mM L-glutamine [Thermo Fisher Scientific, 25030081] and nonessential amino acids [Thermo Fisher Scientific, 11140050]) at 37°C and 5% CO2. For co-culture assays Atg5 floxed Ctrl- and Atg5-DC CKO-derived splenocytes were purified for ITGAX+ cells using magnetic microbeads according to the manufacturer's recommendation. ITGAX+-enriched fractions were further purified through FAC-sorting using an ARIA III FCF sorter (gated on live ITGAX and I-Ab double-positive cells). Purified splenic DCs were directly sorted in round-bottom 96-well tissue culture plates at 3 × 104 cells/well and kept in R10 at 37°C and 5% CO2 for 45 min to settle. Afterwards, wells were either supplemented with increasing amounts of the glycolipid antigen αGalCer (0.01 - 10 µg/ml) or left untreated for 4 h. Each condition was applied in triplicates. Wells were carefully washed 3× with PBS (200 µl/well) and 5 × 104 NKT hybridoma cells (A.407 or FF13) in hybridoma culture medium were added to a final volume of 200 µl/well. Control wells containing antigen-presenting cells- and iNKT hybridoma cells-only (with or without addition of αGalCer) were included. Wells were incubated at 37°C and 5% CO2. After 24 h cell culture supernatants were collected for analysis of IL2 concentrations via ELISA. In some experiments titrated concentrations of indicated glycolipid antigens were used instead of αGalCer.18
Coculture assays with BMDCs
BMDCs were generated by culturing the bone marrow of Atg5 floxed Ctrl or Atg5-DC CKO mice in presence of 2 ng/ml recombinant CSF2/GM-CSF (Biolegend, 576306) in vitro. After 7 d, differentiated BMDCs were harvested, purified over a 14.5% Histodenz (Sigma-Aldrich, D2158–100G) gradient and pulsed with titrated amounts of lipid antigens for 4 h at 37°C and 5% CO2. Cells were then washed, and 1 × 104 BMDCs were used to stimulate 5 × 104 A.407 iNKT hybridoma cells in triplicate cultures in 96-well tissue culture plates. IL2 concentrations in 24 h cell culture supernatant were determined by ELISA. In some experiments, BMDCs were further purified via FAC-sorting for ITGAX and I-Ab double-positive cells.
Flow cytometry
For surface expression analysis of CD40, CD1D1, ITGAX and I-Ab, Atg5 floxed Ctrl- and Atg5-DC CKO-derived splenocytes were purified for ITGAX+ cells using magnetic microbeads according to the manufacturer's recommendation. After FCGR3A/CD16 and FCGR2A/CD32 block (22.4 µg/ml, 20 min, 4°C) and subsequent washing step, ITGAX-enriched fractions were stained for 20 min with LIVE/DEAD fixable Aqua Dead stain kit (Thermo Fisher Scientific, L34957) in PBS at 4°C in the dark. The ITGAX-depleted fractions were used for CD1D1 surface staining of B cells (gated on CD19 and I-Ab double-positive cells). Samples were washed twice in cold PBS followed by incubation with respective fluorochrome-labeled antibodies in FACS buffer (1% bovine serum albumin, 0.1% NaN3 [Sigma-Aldrich, S8032] in PBS) for 25 min at 4°C in the dark. After 2 washing steps with cold PBS samples were resuspended in 50 µl of FACS buffer before sample acquisition using a BD FACSCanto-II system (BD Biosciences, San Jose, CA, USA). For IL12 intracellular cytokine staining (ICS) using a Fixation/Permeabilization Solution Kit (eBioscience, 88–8824–00), ITGAX-enriched fractions were plated out at 5 × 105 cells/well in 48-well tissue culture plates and incubated for 12 h in R10 either with or without 15 µg/ml anti-CD40 at 37°C and 5% CO2. For flow cytometry analysis of IL12B+ cells, samples were additionally treated with brefeldin A (Sigma-Aldrich, B5936–200UL; used at 10 µg/ml) during the last 5 h of incubation to block secretion. Cells were washed twice with PBS and stained for live cells and the surface molecules ITGAX and I-Ab (see above). Fixation and permeabilization for subsequent ICS were performed according to the manufacturers recommendations (Fixation/Permeabilization Solution Kit). Samples were acquired with a BD FACSCanto-II system and all data were analyzed using FlowJo software v9.3.1 (Tree Star Inc.). The same set up as the ICS experiments was performed omitting the addition of brefeldin A. In these instances, cell culture supernatants were collected after 12 h for analysis of secreted IL12B via ELISA.
iNKT cell detection
iNKT cells were detected using αGalCer-loaded CD1D1 tetramers. Single-cell suspensions from spleens, livers, and thymi were pretreated with FCGR3A/CD16 and FCGR2A/CD32 blocking antibodies and 0.5 mg/ml avidin (Sigma-Aldrich, A9275) in FACS buffer for 10 min. After 2 washes, cells were incubated with α-GalCer-loaded CD1D1 tetramer at RT for 15 min before addition of anti-CD3E/CD3ε and further staining for 30 min. Washed cells were then depleted of erythrocytes, fixed with BD FACS Lysing Solution (BD Biosciences, 349202), and acquired on a BD LSR II system (BD Biosciences, San Jose, CA, USA) using FACSDiva software v6.1.3. Analysis was performed using the FlowJo software v9.3.1 (Tree Star Inc.).
Determination of cytokine concentration
IL2 concentration in coculture supernatants was measured using a Mouse IL2 Ready-SET-GO reagent kit (eBioscience, 88–7024–76). IL12B concentration in cell culture supernatants was measured using the Mabtech Mouse IL12B ELISA development kit (Mabtech, 3451–1H-6). Serum concentrations of IFNG and IL4 were measured using the Mouse IFNG or Mouse IL4 Instant ELISA kit (eBioscience, BMS606INST and BMS613INST). All measurements were performed according to the respective manufacturer's recommendation.
Mice
Eight- to 12-wk-old C57BL/6 (Janvier), Atg5flox/flox,57 Tg(ITGAX-cre,-EGFP)4097Ach designated ITGAX-Cre (Jackson), were bred and housed in the University of Zurich animal facility in individually ventilated cages according to Swiss animal laws and institutional guidelines. Atg5flox/flox mice were crossed to ITGAX-Cre mice to obtain ITGAX-Cre × Atg5flox/flox mice (designated Atg5-DC conditional knockout [CKO]) on a C57BL/6 background. Atg5flox/flox mice (designated Atg5 floxed control [Ctrl]) were used as littermate controls. All animal protocols were approved by the cantonal veterinary office of the canton of Zurich, Switzerland (protocol 25706-ZH209/2014) and the canton of Bern, Switzerland (protocols BE46/14, BE73/14).
Genotyping
Animal genotype was confirmed by PCR analysis on DNA from tail or ear biopsies by the use of the following primer pairs (5′-3′):
| Atg5flox/flox | GAATATGAAGGCACACCCCTGAAATG |
| Atg5flox/flox | GTACTGCATAATGGTTTAACTCTTGC |
| Atg5flox/flox | ACAACGTCGAGCACAGCTGCGCAAGG |
| Atg5flox/flox | CAGGGAATGGTGTCTCCCAC |
| ITGAX-Cre | GCGGTCTGGCAGTAAAAACTATC |
| ITGAX-Cre | GTGAAACAGCATTGCTGTCACTT |
| ITGAX-Cre | CTAGGCCACAGAATTGAAAGATCT |
| ITGAX-Cre | GTAGGTGGAAATTCTAGCATCATCC |
Mouse organ processing
After mice were euthanized with CO2, spleens were removed, collected in cold PBS, dissected into small pieces and digested for 30 min at 37°C and 5% CO2 with collagenase D (Sigma-Aldrich, 11088866001; used at 0.4 mg/ml) and DNAse (Qiagen, 79254; used at µg/ml) in PBS. Digest was stopped with EDTA (Sigma-Aldrich, ED-500G; used at 10 mM) for 5 min at 37°C and 5% CO2. The digested tissue was passed through a 70-µm cell strainer (Fisher Scientific, 08–771–2) to obtain single cell solutions. After washing step with cold PBS a red blood cell lysis (2 min, RT) was performed using 1 ml/spleen ACK lysis buffer (Thermo Fisher Scientific, A1049201) followed by washing with cold PBS. Single cell solutions were kept on ice until further processing.
CD1D1 internalization assay
For assessing internalization of surface CD1D1 in splenic DCs and B cells respectively, Atg5 floxed Ctrl- and Atg5-DC CKO-derived splenocytes were purified for ITGAX+ cells using magnetic microbeads according to the manufacturer's recommendation. After FCGR3A/CD16 and FCGR2A/CD32 blocking (22.4 µg/ml, 20 min, 4°C) and subsequent washing step, the ITGAX+-enriched fraction was incubated for 30 min on ice with a biotinylated-CD1D1 antibody. After extensive washing with ice cold PBS cells were fractionated into the following groups (1 × 105/group): 90 min on ice, 90, 60 and 30 min at 37°C, 5% CO2. To assess CD1D1 internalization in B cells, the negative fraction from the MACS purification was processed accordingly in parallel. After respective incubation time, cells were washed with ice cold PBS and incubated with PE-labeled streptavidin, anti-ITGAX (ITGAX+-enriched fraction for DC analysis) or anti-CD19 (ITGAX+-depleted fraction for B cell analysis), anti-I-Ab and LIVE/DEAD fixable Aqua Dead stain kit for 20 min on ice in the dark. Samples were fixed with 3% PFA in PBS and acquired with a BD FACSCanto-II system. All data were analyzed using FlowJo software (Tree Star Inc.). To obtain % internalized CD1D1 values, the following equation was used (MFI, mean fluorescence intensity):
CD1D1 recycling assay
ITGAX-enriched or -depleted fractions of Atg5 floxed Ctrl- and Atg5-DC CKO-derived splenocytes (see internalization assay for details) were incubated with protein synthesis inhibitor cycloheximide (Sigma-Aldrich, 01810–1G; used at 1 µg/ml) supplemented R10 (60 min at 37°C, 5% CO2). After extensive washing with ice-cold PBS, cells were either incubated with unconjugated anti-CD1D1 antibody for 30 min (preblocked) or left untreated (unblocked) on ice. After a washing step, cells were divided into the following groups (1 × 105/group): 30 min on ice (preblocked), 30 min on ice (unblocked), 30, 20, 10, 5 min at 37°C, 5% CO2. After respective incubation times, cells were washed with ice cold PBS and incubated with PE-labeled anti-CD1D1 antibody (of the same clone used for preblocking), anti-ITGAX (ITGAX+-enriched fraction for DC analysis) or anti-CD19 (ITGAX+-depleted fraction for B cell analysis), anti-I-Ab and LIVE/DEAD fixable Aqua Dead stain kit for 20 min on ice in the dark. An unblocked sample was stained with PE-labeled isotype control instead of the PE-labeled anti-CD1D1 antibody. Samples were fixed with 3% PFA in PBS and acquired with a BD FACSCanto-II system. All data were analyzed using FlowJo software (Tree Star Inc.). To obtain % recycled CD1D1 values, the following equation was used:
Lipid in vivo chase
Atg5 floxed Ctrl and Atg5-DC CKO mice were either injected intraperitoneally (i.p.) with 200 µl PBS or αGalCer (1 µg). Injection time points were chosen as such that animals could be euthanized at the same time point. Blood for serum cytokine analysis was collected via cardiac puncture. Blood was used for serum cytokine analysis (IFNG, IL4). Spleens were processed as described above (see mouse organ processing). MFI of costimulatory molecules was analyzed on either Cre+ (Atg5-DC CKO) or Cre− (Atg5 floxed Ctrl) ITGAX and I-Ab double-positive splenocytes.
Infection with S. paucimobilis
Mice were intravenously (i.v.) infected with 107 colony forming units (CFU) S. paucimobilis (ATCC, 29873) and organs were harvested and weighed at 24 h post infection. After homogenizing the organs in sterile H2O using a tissue lyzer (Qiagen Instruments, Hombrechtikon, Switzerland, 85300), tenfold serial dilutions were plated onto Muller-Hinton agar (Sigma-Aldrich, 70192) containing 200 µg /ml streptomycin (Sigma-Aldrich, S9137) and incubated for 48 h at 30°C. Colonies were then counted and bacterial titers calculated as CFU per g tissue.
Statistics
Statistical significance was determined using the 2-tailed unpaired Student t test and the Mann-Whitney U test in Prism software (GraphPad Software Inc.). P < 0.05 was considered significant.
iNKT cell hybridoma lines
The Vα14+/Vβ8+ iNKT cell hybridoma A.407 was generated by fusion of ex vivo CD1D1 tetramer-sorted iNKT cells with the mouse thymoma BW5147 (S.F. unpublished). The FF13 hybridoma was generated by fusion of Vα14 iNKT cells sorted from αGalCer-treated C57BL/6 mice with mouse BW5147 thymic lymphoma cells.58
Supplementary Material
Abbreviations
- AP2
adaptor protein complex 2
- Atg
autophagy-related
- BMDC
bone marrow-derived DC
- CD
cluster of differentiation
- CKO
conditional knockout
- CSF2/GM-CSF
Colony stimulating factor 2 (granulocyte-macrophage)
- DC
dendritic cell
- EE
early endosome
- EEA1
early endosome antigen 1
- GalαGalCer
digalactosylceramide
- GSL-1
glucuronylceramide
- IFN
interferon
- IL
interleukin
- iNKT
invariant natural killer T cell
- ITGAX
integrin subunit α X
- LDLR
low density lipoprotein receptor
- LE
late endosome
- LIR
LC3-interacting region
- Ly
lysosome
- MAP1LC3A/LC3A
microtubule associated protein 1 light chain 3
- MIIC
MHC class II-containing compartment
- MFI
mean fluorescence intensity
- MHC
major histocompatibility complex
- MSR1
macrophage scavenger receptor 1
- PBS
phosphate-buffered saline
- RAP
rapamycin
- S. paucimobilis
Sphingomonas paucimobilis
- TCR
T cell receptor
- TNF
tumor necrosis factor
- αGalCer
α-galactosylceramide
Disclosure of potential conflicts of interest
The authors have declared that no financial conflict of interest exists.
Acknowledgments
We thank Patrick Weber for expert technical assistance.
Funding
C.W.K. was supported by a scholarship provided by the German Research Foundation (DFG grant KE 1831/1–1 to C.W.K). This work was supported by grants of the Dürmüller-Bol-Stiftung, Uni-Bern-Forschungsstiftung and the Vontobel-Stiftung (to S.F.), EU HORIZON 2020 project “TBVAC 2020” (grant 643381), the Swiss National Foundation (310030–149571), the EU HORIZON 2020 project “TBVAC 2020” (grant 643381), the BMRC-SERC Diagnostic grant call 1121480006, and A*STAR Singapore/Australian NHMRC (1201826277) (to G.D.L.). J.D.L. was supported by the Swiss National Foundation (31003A-169664), the Novartis Foundation for medical-biological research, the Sassella Foundation, the Hartmann Müller Foundation, and the Swiss Multiple Sclerosis Society.
References
- [1].Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol 2007; 25:297-336; PMID:17150027; https://doi.org/ 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- [2].Van Kaer L, Parekh VV, Wu L. Invariant natural killer T cells as sensors and managers of inflammation. Trends Immunol 2013; 34:50-8; PMID:23017731; https://doi.org/ 10.1016/j.it.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fujii S-I, Shimizu K, Smith C, Bonifaz L, Steinman RM. Activation of natural killer T cells by alpha-galactosylceramide rapidly induces the full maturation of dendritic cells in vivo and thereby acts as an adjuvant for combined CD4 and CD8 T cell immunity to a coadministered protein. J Exp Med 2003; 198:267-79; PMID:12874260; https://doi.org/ 10.1084/jem.20030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, Maione D, Volpini G, Finco O, Nuti S, et al.. Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci 2007; 104:3984-9; https://doi.org/ 10.1073/pnas.0700191104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guillonneau C, Mintern JD, Hubert F-X, Hurt AC, Besra GS, Porcelli S, Barr IG, Doherty PC, Godfrey DI, Turner SJ. Combined NKT cell activation and influenza virus vaccination boosts memory CTL generation and protective immunity. Proc Natl Acad Sci USA 2009; 106:3330-5; PMID:19211791; https://doi.org/ 10.1073/pnas.0813309106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol 2014; 32:323-66; PMID:24499274; https://doi.org/ 10.1146/annurev-immunol-032713-120243 [DOI] [PubMed] [Google Scholar]
- [7].Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity 2001; 15:897-908; PMID:11754812; https://doi.org/ 10.1016/S1074-7613(01)00240-0 [DOI] [PubMed] [Google Scholar]
- [8].Kang S-J, Cresswell P. Regulation of intracellular trafficking of human CD1d by association with MHC class II molecules. EMBO J 2002; 21:1650-60; PMID:11927549; https://doi.org/ 10.1093/emboj/21.7.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci 2000; 97:8445-50; https://doi.org/ 10.1073/pnas.150236797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chow A, Toomre D, Garrett W, Mellman I. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature 2002; 418:988-94; PMID:12198549; https://doi.org/ 10.1038/nature01006 [DOI] [PubMed] [Google Scholar]
- [11].Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2007; 26:79-92; PMID:17182262; https://doi.org/ 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010; 32:227-39; PMID:20171125; https://doi.org/ 10.1016/j.immuni.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, Ziegler U, Roesler J, Dengjel J, Reichenbach J, et al.. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol 2013; 203:757-66; PMID:24322427; https://doi.org/ 10.1083/jcb.201308173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan J-L, Tan H, Peng J, et al.. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature Cell Biology 2015; 17:893-906; PMID:26098576; https://doi.org/ 10.1038/ncb3192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [15].Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KCF, Capron M, Ryffel B, Faveeuw C, Leite de Moraes M, et al.. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity 2007; 27:597-609; PMID:17950005; https://doi.org/ 10.1016/j.immuni.2007.08.017 [DOI] [PubMed] [Google Scholar]
- [16].Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010; 32:227-39; PMID:20171125; https://doi.org/ 10.1016/j.immuni.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem 2012; 287:34149-56; PMID:22902620; https://doi.org/ 10.1074/jbc.M112.382812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Freigang S, Landais E, Zadorozhny V, Kain L, Yoshida K, Liu Y, Deng S, Palinski W, Savage PB, Bendelac A, et al.. Scavenger receptors target glycolipids for natural killer T cell activation. J Clin Invest 2012; 122:3943-54; PMID:23064364; https://doi.org/ 10.1172/JCI62267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van den Elzen P, Garg S, León L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng T-Y, Sacks FM, Illarionov PA, et al.. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature 2005; 437:906-10; PMID:16208376; https://doi.org/ 10.1038/nature04001 [DOI] [PubMed] [Google Scholar]
- [20].Salio M, Puleston DJ, Mathan TSM, Shepherd D, Stranks AJ, Adamopoulou E, Veerapen N, Besra GS, Hollander GA, Simon AK, et al.. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci USA 2014; 111:E5678-87; PMID:25512546; https://doi.org/ 10.1073/pnas.1413935112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med 1992; 176:1693-702; PMID:1460426; https://doi.org/ 10.1084/jem.176.6.1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Helft J, Böttcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, Reis E Sousa C. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity 2015; 42:1197-211; PMID:26084029; https://doi.org/ 10.1016/j.immuni.2015.05.018 [DOI] [PubMed] [Google Scholar]
- [23].Guilliams M, Malissen B. A Death Notice for In-Vitro-Generated GM-CSF Dendritic Cells? Immunity 2015; 42:988-90; PMID:26084019; https://doi.org/ 10.1016/j.immuni.2015.05.020 [DOI] [PubMed] [Google Scholar]
- [24].Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, Sato M, Takeda K, Okumura K, Van Kaer L, et al.. The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med 1999; 189:1121-8; PMID:10190903; https://doi.org/ 10.1084/jem.189.7.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu KOA, Im JS, Illarionov PA, Ndonye RM, Howell AR, Besra GS, Porcelli SA. Production and characterization of monoclonal antibodies against complexes of the NKT cell ligand alpha-galactosylceramide bound to mouse CD1d. J Immunol Methods 2007; 323:11-23; PMID:17442335; https://doi.org/ 10.1016/j.jim.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sugita M, Cernadas M, Brenner MB. New insights into pathways for CD1-mediated antigen presentation. Curr Opin Immunol 2004; 16:90-5; PMID:14734115; https://doi.org/ 10.1016/j.coi.2003.11.014 [DOI] [PubMed] [Google Scholar]
- [27].Jayawardena-Wolf J, Bendelac A. CD1 and lipid antigens: intracellular pathways for antigen presentation. Curr Opin Immunol 2001; 13:109-13; PMID:11154926; https://doi.org/ 10.1016/S0952-7915(00)00190-4 [DOI] [PubMed] [Google Scholar]
- [28].Chiu Y-H, Park S-H, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol 2002; 3:55-60; PMID:11731798; https://doi.org/ 10.1038/ni740 [DOI] [PubMed] [Google Scholar]
- [29].Sugita M, Cao X, Watts GFM, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity 2002; 16:697-706; PMID:12049721; https://doi.org/ 10.1016/S1074-7613(02)00311-4 [DOI] [PubMed] [Google Scholar]
- [30].Elewaut D, Lawton AP, Nagarajan NA, Maverakis E, Khurana A, Honing S, Benedict CA, Sercarz E, Bakke O, Kronenberg M, et al.. The adaptor protein AP-3 is required for CD1d-mediated antigen presentation of Glycosphingolipids and development of V 14i NKT Cells. J Exp Med 2003; 198:1133-46; PMID:14557411; https://doi.org/ 10.1084/jem.20030143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lawton AP, Prigozy TI, Brossay L, Pei B, Khurana A, Martin D, Zhu T, Späte K, Ozga M, Höning S, et al.. The mouse CD1d cytoplasmic tail mediates CD1d trafficking and antigen presentation by adaptor protein 3-dependent and -independent mechanisms. J Immunol 2005; 174:3179-86; PMID:15749847; https://doi.org/ 10.4049/jimmunol.174.6.3179 [DOI] [PubMed] [Google Scholar]
- [32].Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 2010; 584:1379-85; PMID:20083108; https://doi.org/ 10.1016/j.febslet.2010.01.018 [DOI] [PubMed] [Google Scholar]
- [33].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; https://doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- [34].Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci USA 2013; 110:17071-6; PMID:24067654; https://doi.org/ 10.1073/pnas.1315110110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wilson MT, Johansson C, Olivares-Villagómez D, Singh AK, Stanic AK, Wang C-R, Joyce S, Wick MJ, Van Kaer L. The response of natural killer T cells to glycolipid antigens is characterized by surface receptor down-modulation and expansion. Proc Natl Acad Sci 2003; 100:10913-8; https://doi.org/ 10.1073/pnas.1833166100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ryan MP, Adley CC. Sphingomonas paucimobilis: a persistent Gram-negative nosocomial infectious organism. J Hosp Infect 2010; 75:153-7; PMID:20434794; https://doi.org/ 10.1016/j.jhin.2010.03.007 [DOI] [PubMed] [Google Scholar]
- [37].Kinjo Y, Wu D, Kim G, Xing G-W, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong C-H, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 2005; 434:520-5; PMID:15791257; https://doi.org/ 10.1038/nature03407 [DOI] [PubMed] [Google Scholar]
- [38].Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol 2005; 35:1692-701; PMID:15915536; https://doi.org/ 10.1002/eji.200526157 [DOI] [PubMed] [Google Scholar]
- [39].Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-dependent versus -independent activation of invariant NKT cells during infection. J Immunol 2014; 192:5490-8; PMID:24813205; https://doi.org/ 10.4049/jimmunol.1400722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al.. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 2005; 434:525-9; PMID:1579125810; https://doi.org/1038/nature03408 [DOI] [PubMed] [Google Scholar]
- [41].Kinjo Y, Pei B, Bufali S, Raju R, Richardson SK, Imamura M, Fujio M, Wu D, Khurana A, Kawahara K, et al.. Natural Sphingomonas glycolipids vary greatly in their ability to activate natural killer T cells. Chem Biol 2008; 15:654-64; PMID:18635002; https://doi.org/ 10.1016/j.chembiol.2008.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Marzo AL, Kinnear BF, Lake RA, Frelinger JJ, Collins EJ, Robinson BW, Scott B. Tumor-specific CD4+ T cells have a major “post-licensing” role in CTL mediated anti-tumor immunity. J Immunol 2000; 165:6047-55; PMID:11086036; https://doi.org/ 10.4049/jimmunol.165.11.6047 [DOI] [PubMed] [Google Scholar]
- [43].Berner V, Liu H, Zhou Q, Alderson KL, Sun K, Weiss JM, Back TC, Longo DL, Blazar BR, Wiltrout RH, et al.. IFN-gamma mediates CD4+ T-cell loss and impairs secondary antitumor responses after successful initial immunotherapy. Nat Med 2007; 13:354-60; PMID:17334371; https://doi.org/ 10.1038/nm1554 [DOI] [PubMed] [Google Scholar]
- [44].Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med 2009; 15:267-76; PMID:19252503; https://doi.org/ 10.1038/nm.1928 [DOI] [PubMed] [Google Scholar]
- [45].Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, Park Y, Jones DP, Chappert P, Davoust J, et al.. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 2014; 343:313-7; PMID:24310610; https://doi.org/ 10.1126/science.1246829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005; 307:593-6; PMID:15591165; https://doi.org/ 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- [47].English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, et al.. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480-7; PMID:19305394; https://doi.org/ 10.1038/ni.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tey S-K, Khanna R. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through M8HC class I pathway. Blood 2012; 120:994-1004; PMID:22723550; https://doi.org/ 10.1182/blood-2012-01-402404 [DOI] [PubMed] [Google Scholar]
- [49].Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity 1999; 11:743-52; PMID:10626896; https://doi.org/ 10.1016/S1074-7613(00)80148-X [DOI] [PubMed] [Google Scholar]
- [50].Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med 1999; 189:103-10; PMID:9874567; https://doi.org/ 10.1084/jem.189.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Roberts TJ, Sriram V, Spence PM, Gui M, Hayakawa K, Bacik I, Bennink JR, Yewdell JW, Brutkiewicz RR. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J Immunol 2002; 168:5409-14; PMID:12023333; https://doi.org/ 10.4049/jimmunol.168.11.5409 [DOI] [PubMed] [Google Scholar]
- [52].Loi M, Müller A, Steinbach K, Niven J, Barreira da Silva R, Paul P, Ligeon L-A, Caruso A, Albrecht RA, Becker AC, et al.. Macroautophagy proteins control MHC Class I levels on dendritic cells and shape anti-viral CD8(+) T cell responses. Cell Reports 2016; 15:1076-87. [DOI] [PubMed] [Google Scholar]
- [53].Gupta-Rossi N, Ortica S, Meas-Yedid V, Heuss S, Moretti J, Olivo-Marin J-C, Israël A. The adaptor-associated kinase 1, AAK1, is a positive regulator of the Notch pathway. J Biol Chem 2011; 286:18720-30; PMID:21464124; https://doi.org/ 10.1074/jbc.M110.190769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Henderson DM, Conner SD. A novel AAK1 splice variant functions at multiple steps of the endocytic pathway. Mol Biol Cell 2007; 18:2698-706; PMID:17494869; https://doi.org/ 10.1091/mbc.E06-09-0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53:167-78; PMID:24462201; https://doi.org/ 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- [56].Pei B, Zhao M, Miller BC, Véla JL, Bruinsma MW, Virgin HW, Kronenberg M. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J Immunol 2015; 194:5872-84; PMID:25926673; https://doi.org/ 10.4049/jimmunol.1402154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al.. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; PMID:16625204; https://doi.org/ 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- [58].Schümann J, Facciotti F, Panza L, Michieletti M, Compostella F, Collmann A, Mori L, De Libero G. Differential alteration of lipid antigen presentation to NKT cells due to imbalances in lipid metabolism. Eur J Immunol 2007; 37:1431-41; PMID:17492806; https://doi.org/ 10.1002/eji.200737160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


