Abstract
During processing and digestion, milk proteins are disassembled into peptides with an array of biological functions, including antimicrobial, angiotensin-converting enzyme inhibition, antioxidant, opioid, and immunomodulation. These functions are summarized in numerous reviews, yet information on which peptides have which functions remains scattered across hundreds of research articles. We systematically searched the literature for all instances of bioactive peptides derived from milk proteins from any mammalian source. The data were compiled into a comprehensive database, which can be used to search for specific functions, peptides, or proteins (http://mbpdb.nws.oregonstate.edu). To review this large dataset, the bioactive peptides reported in the literature were visually mapped on the parent protein sequences, providing information on sites with highest abundance of bioactive peptides.
Keywords: ACE, Peptidomics, Digestome, DPP-IV, Casein, Whey
1. Introduction
Milk serves as the primary nutritional source for the mammalian neonate. Beyond basic nutrients, milk provides an array of functional compounds, including bioactive proteins. Degradation of milk proteins releases peptide fragments that have an array of biological effects that can be different from those of the parent protein. These functional peptides are derived from both casein (including αs1-, αs2-, β-, and κ-casein) and whey proteins (including α-lactalbumin, β-lactoglobulin, and lactoferrin). These peptides have an array of activity, including antimicrobial, angiotensin-converting enzyme (ACE) inhibition, dipeptidyl peptidase IV (DPP-IV) inhibition, opioid agonist and antagonist activities, immunomodulation, mineral binding, and antioxidative functions.
Peptides in breast milk or dairy products can be released from milk proteins by native proteases (Dallas, Murray, & Gan, 2015), during production techniques such as fermentation, and during digestion. Whether these peptides exert bioactive effects depends on whether they reach their site of action. In the gut, for example, opioid peptides can bind to gut opioid receptors and alter gastrointestinal motility, and antimicrobial peptides can inhibit pathogen growth. Peptides that are absorbed into the bloodstream can act systemically; for example, the ACE-inhibitory peptides can lower blood pressure. Indeed, some milk peptides are absorbed into the bloodstream as fragments of κ- and αs1-casein were detected in the blood of adult humans for up to eight hours after milk or yogurt consumption (Chabance, Marteau, Rambaud, Migliore-Samour, Boynard, Perrotin, et al, 1998).
Functional milk peptides may be uniquely suited for applications as added food ingredients, supplements, or drugs, as they likely have few negative side effects due to the evolution of milk as food for the mammalian neonate. Peptides in general are increasingly being investigated for their therapeutic potential, as they are often safer and more selective than small-molecule drugs, which can have wide-ranging side effects. Indeed, over 60 peptides are now approved by the Food and Drug Administration and available on the market as therapeutics (Fosgerau & Hoffmann, 2015). Peptides as antimicrobials are particularly attractive as therapeutics as their mechanisms of action resist development of microbial resistance (Jenssen, Hamill, & Hancock, 2006). However, the application of functional milk peptides as therapeutics remains a rarity.
Advances in mass spectrometry (increased sensitivity, dynamic range, and spectral acquisition rate) allow for increasingly comprehensive data collection such that thousands of unique peptides can now be identified from biological sources. These technological advances create an urgent need for a comprehensive bioactive peptide database with which to compare the thousands of identified peptides. Each new peptide discovered must be compared with the entirety of known functional peptides in order to determine which have potential in vivo activity. Bioactive milk peptides have been reviewed numerous times over recent decades, but no comprehensive database of these peptides exists. A partial database (milk AMP) does exist (Théolier, Jérémie, Ismail, Julie, & Riadh, 2013); however, it includes only antimicrobial peptides and was last updated September 2012. Some milk peptides are present in generic bioactive peptide databases such as the Antihypertensive Peptide Database (AHTPDB) (Kumar, Chaudhary, Sharma, Nagpal, Chauhan, Singh, et al., 2015) and BIOPEP (Minkiewicz, Dziuba, Iwaniak, Dziuba, & Darewicz, 2008); however, these databases were far from comprehensive for milk protein-derived peptides and have not been updated within recent years to our knowledge.
The primary aim of the present study was to compile a comprehensive database of functional peptides in milk from mammalian species across the available literature sources. As more and more peptides are discovered, the process of comparing new peptides to previously discovered becomes essential in determining their possible bioactivity. In order to assist with the identification and analysis of novel bioactive peptides, we have constructed a comprehensive functional milk peptide database that allows for examinations of patterns in the data of bioactive peptides: a task that is currently difficult as the data are scattered across hundreds of articles. Our database offers improvement over others in existence in three key areas: it is specific and comprehensive to all milk bioactive peptides across species and proteins, every peptide entry has been thoroughly evaluated in the literature before its inclusion, and it contains several advanced search functions to assist researchers in comparing and analyzing peptidomic data. Using our database, we explored how knowledge of bioactive peptides from the milk of one species can be used to predict yet unidentified functional peptides in the milk of other species. We also formulated visualizations to demonstrate which sites within the milk protein sequences have the most numerous or most potent bioactive peptides to help guide future bioactive peptide research. This database will enable comparison of known functional peptides with biological datasets to explore which bioactive peptides are present in food sources and at various digestive sites.
2. Materials and methods
2.1. Literature search
We constructed our database after searching Web of Science (www.webofknowledge.com) for research articles identifying milk-protein-derived peptides with a biological function. The search was conducted with the terms “biological function” AND peptide AND milk, casein, or whey. The “biological function” terms used were “bioactive” (returned 1099 articles), “antimicrobial” (570 articles), “antihypertensive” (682 articles), “immunomodulatory” (98 articles), “anti-inflammatory” (79 articles), “opioid” (224 articles), “dipeptidyl peptidase IV” (49 articles), “anticancer” (45 articles), and “hypocholesterolemic” (43 articles). We did not specify species in the search terms as we wanted to ensure we identified all bioactive milk peptides from every species that has been investigated.
The search was performed between May and December 2016. We refined this search to only include primary research articles. Each abstract was read, and those describing the biological activity of specific milk peptides were saved in a marked list on Web of Science. Based on the abstracts, we identified original research articles to further review for the identification of peptides with biological function. In total, we identified 258 original research articles that described the biological activity of specific peptides. The milkAMP database (Théolier, Jérémie, Ismail, Julie, & Riadh, 2013), AHTPDB (Kumar, et al., 2015), and BIOPEP database (Minkiewicz, Dziuba, Iwaniak, Dziuba, & Darewicz, 2008) were also searched for any peptides not found using the literature search. For each entry from these three databases, the identified reference was read and the associated data compiled so that our final database cites only original research articles. Those database entries without appropriate references, describing hydrolysates (a mixture of peptides), or with a modified amino acid sequence were excluded. ACE-inhibitory and DPP-IV inhibitory peptides with a half-maximal inhibitory concentration (IC50) value above 1,000 μM were excluded from our database as such peptides represent weak activity and a specific IC50 value was often not provided for these instances in the articles (Hernandez-Ledesma, Quiros, Amigo, & Recio, 2007). Several studies identified the bioactivity of milk protein hydrolysates. Of these hydrolysate studies, only those that identified the exact peptide sequences responsible for the bioactive effect were included in the database.
Peptides were mapped to the parent sequence of the major proteins in human and cow milk proteins using an in-house tool (PepEx) (Guerrero, Dallas, Contreras, Chee, Parker, Sun, et al., 2014). The sequences of specific proteins from different species were aligned using the Protein BLAST alignment tool at blast.ncbi.nlm.nih.gov. Peptides in the database were searched for homology within the same protein of origin but across human, cow, goat, and sheep species with an in-house tool (https://github.com/sdrudn/Peptide_Homology) that returns those peptides with ≥ 80% identical amino acids (identity score). A second function in the tool was also able to count similar amino acids (D = N, I = L = V and Q = E) as identical.
2.2. Online database
The database is SQLite 3.7.17 on a CentOS 7.1.1503 server. The front-end of the site was developed using HTML, Python 2.7.5 and Django 1.9.7 and is served by Apache 2.4.6. The back-end scripts were written in PERL and Python and use Blast+ 2.5.0.
3. Results and discussion
3.1. Online database
The information retrieved from exploring the literature was used to build an online database of human milk and dairy-derived bioactive peptides (Milk Bioactive Peptide Database, MBPDB, http://mbpdb.nws.oregonstate.edu/). MBPDB improves on other databases in three areas. Unlike previous databases, MBPDB is comprehensive for all bioactive milk peptides regardless of species, protein, or function. As milk is likely the most studied source of bioactive peptides with hundreds to thousands of articles, a database specific to it is a necessary tool for identification of these peptides in peptidomics datasets and comparison with newly identified bioactive peptides. Other databases are restricted to a specific peptide function, such as the milkAMP database which only includes antimicrobial peptides. Others still cover bioactive peptides from a wide variety of plant and animal sources and with different functions, and are difficult to search through for peptides of a specific source or function.
MBPDB also has inclusion limits for bioactive peptides that other databases lack. Every new entry is reviewed before inclusion to ensure it meets the proper standards. Adding a new database entry for a bioactive peptide requires the following information: the unique protein ID (e.g P02666) from which the peptide is derived, the peptide sequence, the function, the specific activity of that function (optional), and the original research article reference given as title, author and digital object identifier (DOI). Based on the input information, the database automatically adds the numeric start and end position of the peptide in the protein sequence (including the signal peptide) and the species of origin information. The database identifies errors between protein ID and peptide sequence and only includes the entry if the peptide sequence can be identified in the sequence of the given protein ID. To avoid duplicate entries, the database does not accept input information which is identical to a peptide sequence, function, and DOI combination already existing in the database. Peptides can be added to the peptide library individually or in bulk by uploading a .csv file containing this information. An example of such a file can be obtained through the homepage (http://mbpdb.nws.oregonstate.edu/). Often in our search through other peptide databases, one or more of these features was missing or incorrect for a peptide entry. This incomplete or incorrect data meant that each peptide's information needed to be verified by checking the referenced literature. By limiting peptide inclusion in MBPDB to those for which the complete information set (above) could be provided will ensure that the MBPDB remains reliable.
MBPDB also contains extensive search functions based on NCBI protein searches that are absent from other databases. MBPDB can be used to search for a specific peptide sequence (singly or a batch of peptidomics data), protein ID, species, function or any combination of these four options. If the protein ID is not present in the database it can be added through the add protein function, by uploading the protein information in fasta format. To provide an accurate search system for users, the database accepts multiple ways of searching a species. This means that searching either bos taurus, bovine, or cow gives the same result, and provides a full list of bioactive peptides from the specified species. Furthermore, the search function for peptide sequences includes three options—searching for bioactive peptides matching a sequence query (sequence search), searching for bioactive peptides containing a specific sequence query (truncated search), or searching for bioactive peptides within a specific sequence query (precursor search). The sequence alignments can be searched using two different scoring matrices: either “identical”, which matches only identical amino acids, or using the amino acid substitution matrix BLOSUM62 (Henikoff & Henikoff, 1992). A similarity threshold setting (10% to 100%) filters the result output to only show those subjects matching the searched query sequence at a certain percentage. As peptide functionality may be retained despite minor sequence differences, allowing search for similar sequences and not only the exact match is a major advantage over other available databases.
3.2. Exploring the data
We have created a comprehensive database of peptides from milk proteins for all reported biological functions. The present database, built from a comprehensive literature review, includes 944 entries of bioactive peptides. Every peptide entry in our database is “unique.” “Unique” includes identical peptides from the same protein and species but with different identified activities, shared peptide sequences across different species from the same protein, and short di- and tripeptides that were identified at multiple locations within a protein sequence.
The 944 entries originate from ten different species, 76% (713) cow, 11% (106) human, 7% (63) goat, 5% (47) sheep, 1% (15) from buffalo, donkey, pig, camel, rabbit, and yak. The predominance of cow milk-derived bioactive peptides derives not from a biological reality but rather from cow milk being studied more widely than other milks. However, due to the high sequence homology within milk proteins among species, the extensive studies on cow milk may allow for the translation of bioactive peptide findings to other species. We used the information gathered from the literature to explore how many peptides identified in the milk of one species (either cow, goat, sheep, or human) are also present in another. For this we performed two homology searches using an in-house tool (Table 1). The first homology search looked for exact matches between the peptide and protein sequences. The second homology search included a function that equated select amino acids based on similar physical properties (valine, isoleucine, and leucine; aspartic acid and glutamic acid; arginine and lysine). We excluded peptides from the results that were already identified in the literature to be present in the different species in order for the results to reflect the potential of undiscovered bioactive peptides. From the first search we found 943 peptides identified in one species that had not yet been identified in another. Of the 943 peptides, 588 were exact sequence matches and the remaining 421 peptides were highly similar (≥ 80% identity score). By considering amino acids with similar properties as equal, we identified 118 new peptides, for a total of 1,061 new bioactive peptides with high homology (≥ 80% identity score) to known bioactive peptides (647 exact matches, and 414 highly similar matches). This information can be found in Supplementary Table 1. When sorted by species, bioactive peptides identified in cow milk are potentially identifiable in goat, sheep, and human milks, with 330, 343, and 92 exact homology matches respectively.
Table 1.
Number of peptides predicted from currently identified peptide sequences with a ≥80% identity score across species. The number outside the parentheses represents matches allowing equivalent amino acid substitutions. The number within the parentheses represents matches without allowing amino acid substitutions.
| Known | Predicted | |||
|---|---|---|---|---|
| Cow | Goat | Sheep | Human | |
| Cow | 347 (330) | 356 (343) | 128 (92) | |
| ACE-Inhibitory | 192 (184) | 195 (187) | 80 (62) | |
| Antimicrobial | 71 (68) | 77 (76) | 7 (4) | |
| DPP-IV-Inhibitory | 55 (52) | 63 (53) | 31 (24) | |
| Antioxidant | 24 (23) | 25 (24) | 9 (8) | |
| Anti-Inflammatory | 10 (10) | 8 (8) | 5 (4) | |
| Immunomodulatory | 7 (7) | 7 (7) | 2 (0) | |
| Opioid | 19 (19) | 19 (19) | 4 (4) | |
| Miscellaneous | 69 (68) | 70 (69) | 17 (10) | |
| Goat | 31 (30) | 42 (42) | 4 (2) | |
| ACE-Inhibitory | 11 (11) | 15 (15) | 3 (2) | |
| Antimicrobial | 17 (16) | 25 (25) | 1 (0) | |
| DPP-IV-Inhibitory | 4 (4) | 4 (4) | - | |
| Antioxidant | 2 (2) | - | - | |
| Anti-Inflammatory | - | - | - | |
| Immunomodulatory | - | - | - | |
| Opioid | - | - | - | |
| Miscellaneous | - | - | - | |
| Sheep | 21 (21) | 27 (27) | 7 (4) | |
| ACE-Inhibitory | 16 (16) | 23 (22) | 6 (3) | |
| Antimicrobial | 4 (4) | 4 (4) | - | |
| DPP-IV-Inhibitory | - | - | - | |
| Antioxidant | 1 (1) | 1 (1) | 1 (1) | |
| Anti-Inflammatory | - | - | - | |
| Immunomodulatory | - | - | - | |
| Opioid | - | - | - | |
| Miscellaneous | - | - | - | |
| Human | 34 (19) | 31 (16) | 33 (17) | |
| ACE-Inhibitory | 25 (13) | 24 (13) | 26 (14) | |
| Antimicrobial | - | - | - | |
| DPP-IV-Inhibitory | 0 (0) | 0 (0) | 0 (0) | |
| Antioxidant | 5 (2) | 3 (0) | 3 (0) | |
| Anti-Inflammatory | - | - | - | |
| Immunomodulatory | 1 (1) | 1 (0) | 1 (0) | |
| Opioid | 3 (3) | 3 (3) | 3 (3) | |
| Miscellaneous | - | - | - | |
Most bioactive peptides are derived from the major milk proteins: 36% (338) P-casein, 13% (119) αs1-casein, 11% (105) β-lactoglobulin, 10% (98) κ-casein, 8% (77) αs2-casein, and 5% (43) α-lactalbumin. Of the minor milk proteins, lactoferrin made up 15% (141) of the database, and less than 1% are derived from other minor proteins such as serum albumin. As peptide synthesis is readily available, identified bioactive peptides can easily be modified to improve activity; hence, many studies have investigated analogs of milk protein-derived bioactive peptides (McClean, Beggs, & Welch, 2014). However, this database includes only peptides obtained from milk proteins without modification to the amino acid sequence.
Of the 944 entries, there were 737 unique peptide sequence- and function-combinations, and 606 unique peptide sequences. The generic database BIOPEP contained, to the best of our knowledge, 151 milk-derived bioactive peptides. Of these, 47 peptides were not included in our database due to lacking one or more of the following criteria: the original research article, the bioactive function, the full peptide sequence, the source species and protein, and the start and end positions in the protein. Our database includes 177 unique antimicrobial peptides, of which 108 are also present in the MilkAMP database. The remaining peptides in MilkAMP were not included based on criteria described above. AHTPDB includes 323 unique milk peptides with a known ACE-inhibitory activity (below 1,000 μM). Our database includes 327 unique ACE-inhibitory peptides. However, only 175 of these peptides are present in both databases (Fig. 1). The remaining peptides from AHTPDB not present in our database were deemed ineligible based on criteria described in the methods. Our database also contains 152 published ACE-inhibitory peptides from milk that are not present in the AHTPDB. Six peptides are present in all four databases (MBPDB, BIOPEP, AHTPDB, and MilkAMP), including peptides with both antimicrobial and antihypertensive activity (Fig. 1). The sequences of these six peptides are AVPYPQR, FALPQYLK, LKKISQ, IQY, PYVRYL, and VAGTWY.
Fig. 1.
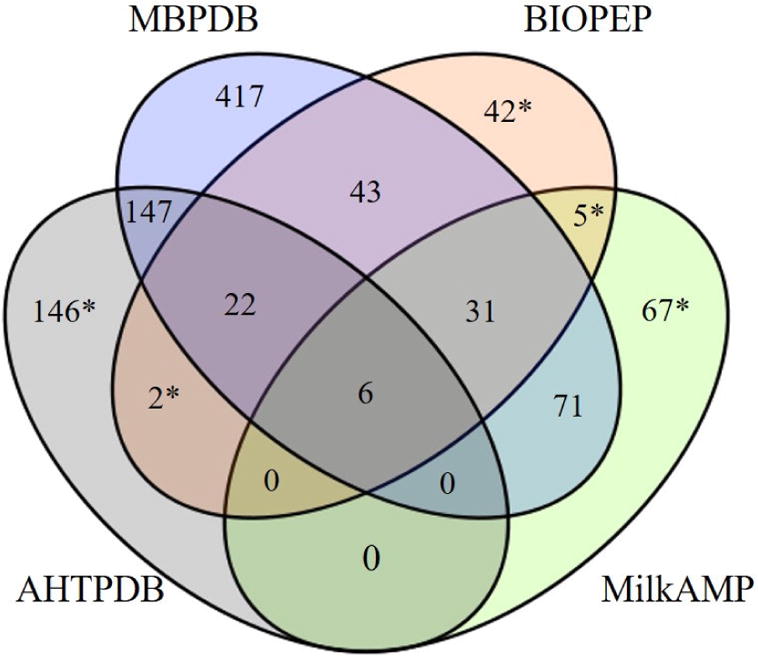
Venn diagram of unique sequence- and function-combination of peptides in the four databases: BIOPEP; AHTPDB, antihypertensive peptide database; MilkAMP, milk antimicrobial peptides; MBPDB, milk bioactive peptide database. *Peptides missing an appropriate reference, false entries, or otherwise did not meet the criteria to be included in MBPDB.
Poor data quality such as incorrect entries or entries from non-reviewed publications in the previous peptide databases may be problematic as these databases have served as the basis for several bioactive peptide prediction models. If large parts of the input data are incorrect, the strength of the prediction models will be greatly reduced.
The bioactive milk peptides identified derive from a variety of milk products. Many of these peptides were identified by in vitro digestion of milk proteins, while others were naturally occurring in different processed milk products, including cheese, yogurt, or kefir, and others were created through peptide synthesis based on milk protein sequences predicted to exert a biological function.
Peptides with ACE-inhibitory and antimicrobial actions are the most abundant in the literature (327 and 177 unique entries, respectively); however, peptides with DPP-IV-inhibitory (64), antioxidant (41), opioid (25), anti-cancer (14), anti-inflammatory (10), immunomodulatory (9), and miscellaneous (70) actions were also identified.
3.3. ACE-inhibitory peptides
ACE-inhibitory peptides from food sources such as milk, cheese, and yogurt have attracted interest for their possible use as a natural alternative to drugs for reducing blood pressure through the binding and inhibition of ACE. Several common factors influence a peptide's capacity to bind ACE, and thus its inhibitor strength. At the C-terminus, aromatic residues (phenylalanine, tyrosine, tryptophan) and proline enhance binding, whereas dicarboxylic residues (glutamic acid, aspartic acid) decrease binding (Cheung, Wang, Ondetti, Sabo, & Cushman, 1980). At the N-terminus, branched-chain amino acids (leucine, isoleucine, valine) enhance binding (Cheung, Wang, Ondetti, Sabo, & Cushman, 1980). Proline at the third-to-last position also enhances ACE binding affinity (Rohrbach, Williams, & Rolstad, 1981). However, quantitative structure-activity relationship studies show that the length of a peptide can change the influence of amino acids at certain positions on ACE-inhibitory activity (Pripp, Isaksson, Stepaniak, & Sørhaug, 2004; Wu, Aluko, & Nakai, 2006) the influence of the C-terminal amino acid is reduced in peptides longer than six amino acid residues due to increased steric effects (Pripp, Isaksson, Stepaniak, & Sørhaug, 2004). Presence of proline can increase peptide resistance to hydrolysis by digestive enzymes (Yang & Russell, 1992). Therefore, peptides with a high number of proline residues likely survive longer in the digestive system and more likely exert in vivo bioactivity.
From the literature, we identified 420 peptides with ACE-inhibitory activity (defined as IC50 <1,000 μM) derived from eight species and nine milk proteins. Of the 420 entries, there are 327 unique peptide sequences. The caseins were the dominant protein source, accounting for 77% of all ACE-inhibitory peptide entries. One explanation for the casein predominance is that research has focused to a large extent on cheese, which is mostly casein. Cheese contains many peptides produced by a wide range of proteases from lactic acid bacteria and other flora introduced during ripening.
ACE-inhibitory peptides from cow's milk proteins represent the largest group in our database with 285 entries, but goat (27 entries), sheep (38), and human (60) milk are also commonly-studied sources of ACE-inhibitory peptides (Table 2). The lower count of peptides identified from these sources likely results from fewer studies investigating peptides from these milks rather than a biological reality. Cow milk ACE-inhibitory peptides were mapped to the sequence to provide a visual overview of their distribution (Figs. 2 and 3). The regions of highest peptide density are indicated by red on a heatmap below the protein sequence.
Table 2.
Count of unique ACE-inhibitory peptide sequences reported in literature by protein and species.
| Species | αs1-casien | αs2-casein | β-casein | κ-casein | α-lactalbumin | β-lactoglobulin | Lactoferrin |
|---|---|---|---|---|---|---|---|
| Cow | 35 | 24 | 86 | 14 | 18 | 32 | 21 |
| Human | - | - | 49 | 10 | - | - | - |
| Sheep | 6 | 3 | 7 | 10 | - | 1 | - |
| Goat | 3 | 4 | 14 | 1 | 1 | 2 | - |
| Buffalo | - | - | 4 | - | - | - | - |
| Camel | - | 2 | 1 | - | - | - | |
| Yak | - | - | 1 | 1 | - | - | - |
| Donkey | - | - | 1 | - | - | - | - |
Fig. 2.
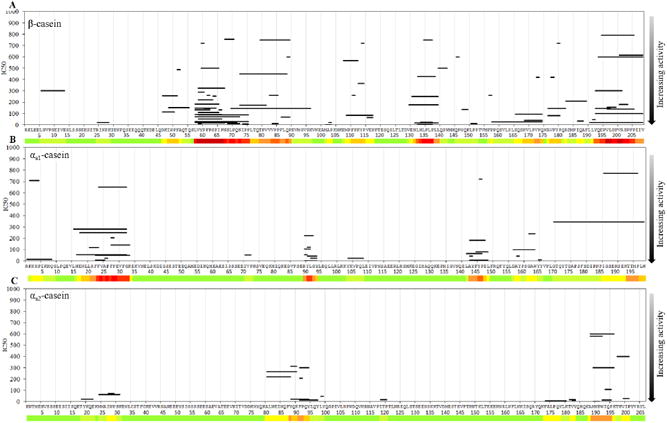
ACE-inhibitory activity of peptides identified in cow β-casein (A), αs1-casein (B), αs2-casein (C). The heatmap below the sequence indicates the density of identified ACE-inhibitory peptides. The y-axis indicates the IC50 values in μM.
Fig. 3.
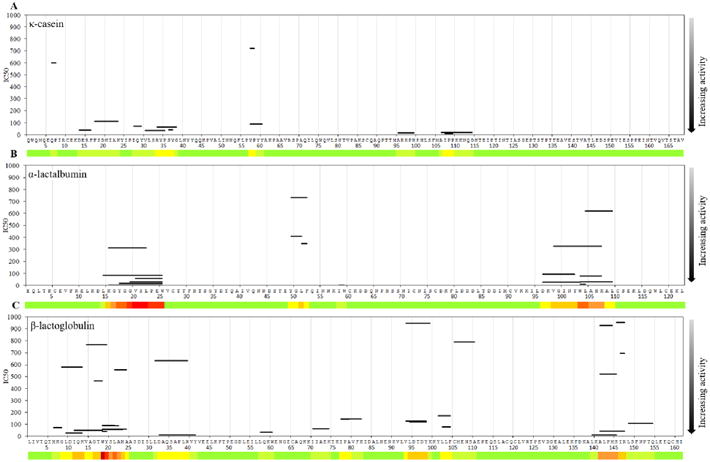
ACE-inhibitory activity of peptides in cow (A) κ-casein, (B) α-lactalbumin, and (C) β-lactoglobulin. The heatmap below the sequence indicates the density of identified ACE-inhibitory peptides. The y-axis indicates the IC50 values in μM.
The amino acid regions f(58–73) (LVYPFPGPIPNSLPQN), f(135–138) (LPLP), and f(197– 204) (VLGPVRGP) are the most studied regions in cow β-casein and are present in the largest numbers in our database (Fig. 2A). β-casein f(58–73) contained 27 unique full or partial sequences, β-casein f(135–138) contained 7, and β-casein f(197–204) contained 10. There are several explanations for the increased abundance of ACE-inhibitory peptides from these regions. These regions have a high frequency of proline residues, which increases the likelihood that peptides may be produced with proline at the C-terminus, a factor which is known to increase ACE-binding. In addition, as the presence of proline in a peptide sequence increases its resistance to digestion (Yang & Russell, 1992), these regions are more likely to survive digestion partially intact, allowing for assessment of their bioactivity. Though these regions have many ACE-inhibitory peptides, the IC50 values of these peptides vary over a wide range.
Peptides from the other proteins have also been studied, though not to the same extent as β-casein. αs1-casein (Fig. 2B) has one region of high peptide density at f(24–29) (FVAPFP) with 10 full or partial peptide sequences. Two other regions, αs1-casein f(91–92) (YL) and αs1-casein f(146–147) (YP), have groupings of peptides with low IC50 values but have been less studied. There are no distinct groupings of peptides from either αs2-casein (Fig. 2C) or κ-casein (Fig. 3A). Only 24 and 14 unique peptides from these proteins, respectively, are present within our database. Though peptide clustering is not apparent in κ-casein, many of the peptides have low IC50 values, with eight of the twelve peptides having an IC50 lower than 100 μM. α-lactalbumin (Fig. 3B) and β-lactoglobulin (Fig. 3C) each have one small region of high peptide density. The sequence α-lactalbumin f(21–23) (VSL) is present in nine peptides, and a single amino acid residue from β-lactoglobulin (tryptophan 19) is present in eight. This tryptophan residue appears predominantly either at the start or the end of a peptide sequence. Contrary to previous findings that tryptophan at the C-terminus increases ACE-inhibition, the peptides with tryptophan at the N-terminus have a lower IC50 than peptides with tryptophan at the C-terminus. Steric effects predicted by QSAR models do not explain this discrepancy (Pripp, Isaksson, Stepaniak, & Sørhaug, 2004), as these peptides are all between two and eight amino acids long and thus too short to be impacted by steric hindrance. Compared with the regions from β-casein, the most studied regions from all other milk proteins have fewer proline residues that might confer positive ACE-inhibitory activity. However, the regions are all hydrophobic and contain many tryptophan, tyrosine, and phenylalanine residues that are also predicted to increase competitive ACE binding.
Although more ACE-inhibitory peptides are found from the above described regions than any other, this disparity does not mean that these peptides have greater activity than peptides from other regions of the milk proteins. The three most potent ACE-inhibitory peptides are all from cow milk, but they all lie outside these regions: αs2-casein f(190–192) (MKP), lactoferrin f(74–76) (LRP), β-casein f(102–104) (MAP). These peptides are all tripeptides that terminate with a proline. The same structural features of these three are present in two well-studied peptides found in fermented milk products, β-casein f(74–76) (IPP) and β-casein f(84–86) (VPP) (Nakamura, Yamamoto, Sakai, & Takano, 1995). Prior studies determined that IPP and VPP can have in vivo antihypertensive effects (Ishida, Shibata, Fukuhara, Yano, Takehara, & Kaneko, 2011). These two peptides are now being included in food products as nutraceuticals for their blood pressure-reducing effects (Cicero, Gerocarni, Laghi, & Borghi, 2011). No in vivo studies have compared the antihypertensive effects of LRP, MAP, and MKP with those of IPP and VPP. A common synthetic antihypertensive drug, captopril, has an IC50 of 0.015 μM (Henda, Labidi, Arnaudin, Bridiau, Delatouche, Maugard, et al., 2013), more than 20 times more active than even the most active milk-derived ACE-inhibitory peptides. Captopril therapy can be accompanied by adverse side-effects, but in human subjects, IPP and VPP consumption showed no adverse effects from continuous use (Ishida, Shibata, Fukuhara, Yano, Takehara, & Kaneko, 2011).
3.4. Antimicrobial peptides
The emergence of bacterial resistance to traditional small molecule antibiotics has raised interest in antimicrobial peptides (AMP) for their unique mechanisms of action that limit resistance development. Milk-derived peptides are particularly interesting as novel antimicrobial therapeutics, as they likely have high tolerability and low toxicity. Milk-derived AMPs could also be applied as an alternative to chemical preservatives in food. Antimicrobial peptides act through various mechanisms, but a common feature is their interaction with the target cell membrane with a resulting increase in membrane permeability (Mohanty, Jena, Choudhury, Pattnaik, Mohapatra, & Saini, 2016). This mechanism explains why AMPs commonly have amphipathic structural properties, a positive net charge, and are mostly longer than 11 amino acids. Peptides with hydrophilic, positively-charged residues initiate attraction with the negatively-charged microbial cell membrane (Mohanty, Jena, Choudhury, Pattnaik, Mohapatra, & Saini, 2016). The hydrophobic regions of the peptide can then interact with the lipid bilayer and penetrate the cell.
We identified 207 AMPs derived from milk proteins in the literature. Of these 207 entries, 177 are unique peptide sequences. The peptides in our database have antimicrobial activity across 49 different species of gram-positive, gram-negative bacteria, fungi, and parasites. However, most peptides identified are antimicrobial against both gram-positive and gram-negative species. Antimicrobial activity was most often reported against E. coli (134 peptides), S. aureus (45 peptides), B. subtilis (36), C. albicans (25), L. monocytogenes (23), L. innocua (19), E. faecalis (14), L. casei (14), B. cereus (13), L. acidophilus (13), and S. epidermidis (10).
The majority of known milk AMPs are derived from lactoferrin, with 37 identified unique peptide sequences from cow lactoferrin and 23 unique peptides from human lactoferrin. In addition to antimicrobial lactoferrin-derived peptides, intact lactoferrin has antimicrobial activity (Bullen, Rogers, & Leigh, 1972). Most AMPs identified in lactoferrin are clustered around two sections, lactoferricin from lactoferrin f(17–41) and lactoferrampin from lactoferrin f(265–284) (Mohanty, Jena, Choudhury, Pattnaik, Mohapatra, & Saini, 2016) (Fig. 4A). Though most studies focused on antimicrobial activity of cow milk peptides, examining which AMPs are released from human milk proteins during infant digestion is essential to understand how these peptides can impact their immune system. Alignment comparison of human and cow lactoferricin shows that these regions have little homology between the two species (41%) (Fig. 4A), despite shared attributes of amphipathic residues and positive charge (Bruni, Capucchio, Biasibetti, Pessione, Cirrincione, Giraudo, et al., 2016). Still, both human and cow lactoferricin exert antimicrobial activity.
Fig. 4.
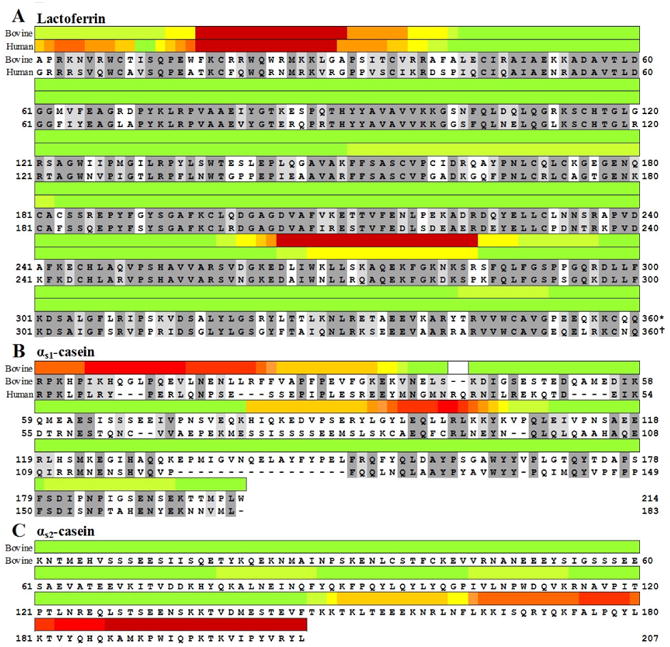
Heatmap of antimicrobial peptides in (A) human and cow lactoferrin sequence 1–360, (B) cow and human αs1-casein sequence, and (C) cow αs2-casein sequence. As no antimicrobial peptides from the last region of lactoferrin are known, they were excluded. The sequence of human and cow lactoferrin are shown underneath the heat map sequence aligned with each other using Protein BLAST at (blast.ncbi.nlm.nih.gov). Dark grey indicates identical amino acids in the sequences. Light grey indicates similar amino acids in the sequence. *Bovine lactoferrin continues from 361 to 708. †Human lactoferrin continues from 361 to 710.
Lactoferrampin, the second cluster of AMPs in lactoferrin, has structural similarity to lactoferricin (amphipathic, positively charged, alpha helix). Lactoferrampin is more homologous between human and cow (74% identity) than lactoferricin, but human lactoferrampin remains far less studied than cow (Fig. 4A).
Both cow αs1-casein and αs2-casein were large sources of AMPs (Fig 4B and 4C, respectively). To the best of our knowledge, no human αs1-casein antimicrobial peptides have been identified. Cow αs1-casein f(1–23), isracidin, is the most studied cluster of AMPs and encompasses caseicin A (αs1-casein f(6–14) (IKHQGLPQE)) and caseicin B (αs1-casein f(15–22) (VLNENLLR)). The alignment reveals little homology between human and cow αs1-casein in this section.
The positively charged region at the C-terminus of cow αs2-casein has high antimicrobial activity (Fig. 4C), and hydrolysis of this section can result in several peptides with antimicrobial activity (Zucht, Raida, Adermann, Mägert, & Forssmann, 1995). The positively charged amino acids in this sequence (e.g., lysine 197, lysine 199, and arginine 205) are important for the antimicrobial function (Alvarez-Ordóñez, Begley, Clifford, Deasy, Considine, & Hill, 2013).
3.5. DPP-IV-inhibitory peptides
Milk proteins also contain peptides with DPP-IV inhibitory activity. DPP-IV is an enzyme that inactivates the incretin hormones glucagon-like peptide 1 and gastric inhibitory polypeptide, and assists in maintaining normal blood glucose levels (Demuth, Hans-Ulrich, McIntosh, & Pederson, 2005).
Our database contains 172 entries for DPP-IV inhibitory milk protein-derived peptides, and 64 unique sequences. There are 43 from lactoferrin (25%), 37 entries from β-casein (22%), 29 from β-lactoglobulin (17%), 23 from κ-casein (13%), 14 from α-lactalbumin (8%), 11 from αs2-casein (6%), and 9 from αs1-casein (5%). These peptides range from 2 to 14 amino acids in length. Half of the unique sequences have a proline at the second position from the N-terminal, indicating a possible structural pattern for DPP-IV-inhibitory peptides. Dipeptides with the general structure Xaa-Pro (Xaa indicates any amino acid), Pro-Xaa, and Xaa-Ala are competitive inhibitors of DPP-IV. Within the major milk proteins, more than 100 different sites exist with a dipeptide structure that allows for DPP-IV-inhibitory activity. Short peptides with a proline in the N2-position position may also have high potential to be DPP-IV inhibitory (Nongonierma & FitzGerald, 2014). As milk proteins are rich in proline residues, a large number of additional, unidentified DPP-IV-inhibitory peptides are likely present within the sequence of milk proteins.
3.6. Antioxidant peptides
Antioxidant peptides from fermented milk products can either prevent the formation of free radicals or scavenge existing free radicals or peroxides implicated in the oxidation of membrane lipids, cellular proteins, DNA, and enzymes (Pihlanto, 2006). Antioxidative peptides from casein can be used in meat processing to help prevent lipid oxidation and the resulting off-flavors that can occur with prolonged shelf life in meat products (Rossini, Noreña, Cladera-Olivera, & Brandelli, 2009).
Our database includes 47 antioxidative peptides of which 41 are unique peptide sequences. There are 32 entries from cow milk protein (68%), 9 from human (19%), 3 from goat (6%), and 3 from sheep (6%). The antioxidant activity of these peptides is through chemical methods, and was determined through radical scavenging assays (Hernandez-Ledesma, Quiros, Amigo, & Recio, 2007). Most antioxidative peptides were from casein (95%), especially β-casein (45%), and αs1-casein (40%). Identified antioxidant peptides range from 2 to 14 amino acids in length. Previous studies found that most antioxidative peptides contained hydrophobic amino acids at the N-terminus and/or C-terminus, and proline, histidine, or tyrosine within the sequence (Li & Li, 2013). Hydrophobic residues may increase the interaction between peptides and fatty acid radicals. Histidine is predicted to chelate and trap radicals, and tyrosine can donate hydrogen to reduce free radicals (Li & Li, 2013). In our database, 30 of the 41 peptides contained hydrophobic amino acids at the N-terminus and/or C-terminus, and all but one contained at least one proline, histidine, or tyrosine in the sequence. A separate study suggests that the C-terminal dipeptide Glu-Leu may also be a feature of some antioxidant peptides (Suetsuna, Ukeda, & Ochi, 2000), which is supported here as 5 unique peptides out of 40 had a C-terminal Glu-Leu. β-lactoglobulin f(19-29) (WYSLAMAASDI) is an example of an antioxidant peptide from milk protein. This peptide has a higher antioxidant activity than butylated hydroxyanisole, a common synthetic antioxidant ingredient used in the food industry to prevent product deterioration (Svenning, Brynhildsvold, Molland, Langsrud, & Elisabeth Vegarud, 2000), and hence is a candidate for use in the food industry.
3.7. Anti-inflammatory peptides
Our database contains ten unique anti-inflammatory peptides. All of the peptides were identified in cow milk, five from β-lactoglobulin, three from β-casein, and two from α-lactalbumin. Except for β-casein f(184–203), all of the peptides are ≤ 5 amino acids long. The activity of many of these peptides was determined based on their inhibition of the nuclear factor-κB (NF-κB) signaling pathway in macrophages (Ma, Liu, Shi, & Yu, 2016). Activation of NF-κB promotes transcription of pro-inflammatory cytokines. Anti-inflammatory peptides can inhibit mRNA expression of the products of NF-κB activation, though their mechanism of action remains unknown. In addition to cytokine pathway inhibition, anti-inflammatory peptides also reduce adhesion of monocytes to endothelial cells in the circulatory system and thus prevent them from contributing to the inflammatory response. Peptides with strong ACE-inhibitory activity are also known to inhibit cellular adhesion (Aihara, Ishii, & Yoshida, 2009). As such, it may be that peptides with characteristics similar to those of potent ACE-inhibitory peptides (hydrophobic tripeptides with proline at the C-terminus) are the most likely candidates for antiinflammatory activity.
Because the number of anti-inflammatory peptides in our database is small, there are no significant regions of anti-inflammatory peptides in the milk protein sequences. More research is needed to determine how these peptides function in modulating the inflammatory response.
3.8. Immunomodulatory peptides
Although all anti-inflammatory peptides can be considered immunomodulatory, some peptides have different means of regulating the immune system. Other functions of immunomodulatory peptides include stimulation of lymphocyte activity and proliferation, and promotion of antibody formation (Migliore-Samour & Jollès, 1988). Nine unique bioactive peptides with immunomodulatory activity are included in our database. Eight of these peptides were identified from cow milk: six from β-casein, one from αs2-casein, one from κ-casein. One was identified from human β-casein. All six cow β-casein peptides were identified either in the regions f(60–68) or f(193–209). Many of the ACE-inhibitory peptides are also derived from these regions.
3.9. Opioid peptides
Opioid peptides are peptides that bind to opiate receptors and exert opiate-like effects. There are 40 entries and 26 unique sequences for bioactive peptides with opioid agonist activity included in our database. Twenty-one peptides were identified from cow casein proteins, eight from β-casein (including β-casomorphins-5 and -7, f(60–64) and f(60–66)), eight from αs1-casein (including αs1-casein-exorphin and αs1-casein f(90–96)), three from κ-casein, one from β-lactoglobulin, and one from lactoferrin. Five peptides were identified from human milk proteins, with three from lactoferrin and one from β-casein.
The database also includes eight unique opioid antagonists. Opioid antagonists suppress the agonist activity of enkephalin. Of these, four are derived from cow κ-casein and are known as the casoxins. One derived from human κ-casein. The common sequence features for both endogenous and exogenous opioid peptides are tyrosine at the N-terminus and phenylalanine or tyrosine in the second, third, or fourth position. These structural elements allow peptides to fit better within the binding pocket of the opioid receptor (Clare & Swaisgood, 2000). Out of the 8 opioid antagonists and 26 opioid agonists, 24 fit one or both of these common sequence features.
3.10. Miscellaneous functional peptides
Besides the biological functions described, there are additional functions identified for milk peptides that are less studied. For example, peptides from κ-casein have antithrombotic activity, which reduces the formation of blood clots by inhibiting fibrinogen binding and platelet aggregation. Antithrombotic peptides can be found in the plasma of infants after ingestion of cow milk-based formula or breastfeeding (Chabance, Jollès, Izquierdo, Mazoyer, Francoual, Drouet, et al., 1995). Other peptides have hypocholesterolemic activity. Four peptides from cow β-lactoglobulin can reduce cholesterol levels by preventing its absorption in the gastrointestinal tract (Nagaoka, Futamura, Miwa, Awano, Yamauchi, Kanamaru, et al., 2001).
There are eighteen anticancer peptides present in our database, all of them from cow milk. The peptide lactoferrin f(17-38) was studied for activity in five separate articles. It was shown to have cytotoxic effects on leukemia and breast cancer cell lines, as well as angiogenesis inhibitory properties. All of the other peptides were identified from casein proteins.
Milk phosphopeptides are primarily derived from the casein proteins and are released upon enzymatic digestion. Phosphorylated serine residues can bind and solubilize calcium ions and improve their bioavailability (FitzGerald, 1998). Casein phosphopeptides have recently been reviewed by Sun et al. (Sun, Wu, Du, Tang, Liu, Fu, et al., 2016).
4. Conclusion
This research meets a critical need by creating a single database for all known bioactive milk peptide sequences. By creating this database, we were able to synthesize information on bioactive peptides from across hundreds of original research articles. Bioactive peptides are present in milk from many species and can have a wide range of functions. This database will enable future comparisons against newly obtained peptidomic data. The resulting database is available for the public to explore and update with data from novel research.
This database will also serve as the basis for the creation of prediction models to determine the likelihood of bioactive milk peptides identified in other types of biological samples. These predictions are based on the relationship between peptide structure and activity. This work will allow for the identification of similar peptides with a high potential for function and guide further therapeutic discovery.
Supplementary Material
Highlights.
A database of bioactive peptides was created from hundreds of published articles.
Peptide mapping displays location and count of peptides within parent protein sequences.
Homology search across species revealed 1,061 potential functional milk peptides.
The database enables bioactive peptide identification from peptidomics data.
The database will serve as the basis for peptide function prediction modelling.
Acknowledgments
The authors thank Nikhil Joshi at the University of California Genome Center Bioinformatics Core, and Shawn O'Neil at Oregon State University, Center for Genome Research and Biocomputing for programming support. All authors read and approved the final manuscript. This project was funded in part by the K99/R00 Pathway to Independence Career Award, Eunice Kennedy Shriver Institute of Child Health & Development of the National Institutes of Health (R00HD079561) (D.C. Dallas).
Footnotes
Conflict of interest statement: The authors have declared no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aihara K, Ishii H, Yoshida M. Casein-derived tripeptide, Val-Pro-Pro (VPP), modulates monocyte adhesion to vascular endothelium. Journal of Artherosclerosis and Thrombosis. 2009;16(5):594–603. doi: 10.5551/jat.729. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ordóñez A, Begley M, Clifford T, Deasy T, Considine K, Hill C. Structure-activity relationship of synthetic variants of the milk-derived antimicrobial peptide αs2-casein f(183-207) Applied and Environmental Microbiology. 2013;79(17):5179–5185. doi: 10.1128/AEM.01394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni N, Capucchio MT, Biasibetti E, Pessione E, Cirrincione S, Giraudo L, Corona A, Dosio F. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules. 2016;21(6):752. doi: 10.3390/molecules21060752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen JJ, Rogers HJ, Leigh L. Iron-binding proteins in milk and resistance to Escherichia coli infection in infants. British Medical Journal. 1972;1(5792):69–75. doi: 10.1136/bmj.1.5792.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabance B, Jollès P, Izquierdo C, Mazoyer E, Francoual C, Drouet L, Fiat AM. Characterization of an antithrombotic peptide from kappa-casein in newborn plasma after milk ingestion. British Journal of Nutrition. 1995;73(4):583–590. doi: 10.1079/bjn19950060. [DOI] [PubMed] [Google Scholar]
- Chabance B, Marteau P, Rambaud JC, Migliore-Samour D, Boynard M, Perrotin P, Guillet R, Jollès P, Fiat AM. Casein peptide release and passage to the blood in humans during digestion of milk or yogurt. Biochimie. 1998;80(2):155–165. doi: 10.1016/s0300-9084(98)80022-9. [DOI] [PubMed] [Google Scholar]
- Cheung HS, Wang FL, Ondetti MA, Sabo EF, Cushman DW. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. Journal of Biological Chemistry. 1980;255(2):401–407. [PubMed] [Google Scholar]
- Cicero AFG, Gerocarni B, Laghi L, Borghi C. Blood pressure lowering effect of lactotripeptides assumed as functional foods: a meta-analysis of current available clinical trials. Journal of Human Hypertension. 2011;25(7):425–436. doi: 10.1038/jhh.2010.85. [DOI] [PubMed] [Google Scholar]
- Clare DA, Swaisgood HE. Bioactive milk peptides: a prospectus. Journal of Dairy Science. 2000;83(6):1187–1195. doi: 10.3168/jds.S0022-0302(00)74983-6. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Murray NM, Gan J. Proteolytic systems in milk: perspectives on the evolutionary function within the mammary gland and the infant. Journal of Mammary Gland Biology and Neoplasia. 2015;20(3-4):133–147. doi: 10.1007/s10911-015-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth HU, Hans-Ulrich D, McIntosh CHS, Pederson RA. Type 2 diabetes—therapy with dipeptidyl peptidase IV inhibitors. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2005;1751(1):33–44. doi: 10.1016/j.bbapap.2005.05.010. [DOI] [PubMed] [Google Scholar]
- FitzGerald RJ. Potential uses of caseinophosphopeptides. International Dairy Journal. 1998;8(5-6):451–457. [Google Scholar]
- Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discovery Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, Dimapasoc LM, Barile D, German JB, Lebrilla CB. Mechanistic peptidomics: factors that dictate the specificity on the formation of endogenous peptides in human milk. Molecular and Cellular Proteomics. 2014;13(12):3343–3351. doi: 10.1074/mcp.M113.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henda YB, Labidi A, Arnaudin I, Bridiau N, Delatouche R, Maugard T, Piot JM, Sannier F, Thiéry V, Bordenave-Juchereau S. Measuring angiotensin-I converting enzyme inhibitory activity by micro plate assays: comparison using marine cryptides and tentative threshold determinations with captopril and losartan. Journal of Agricultural and Food Chemistry. 2013;61(45):10685–10690. doi: 10.1021/jf403004e. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Science of the United States of America. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Ledesma B, Quiros A, Amigo L, Recio I. Identification of bioactive peptides after digestion of human milk and infant formula with pepsin and pancreatin. International Dairy Journal. 2007;17(1):42–49. [Google Scholar]
- Ishida Y, Shibata Y, Fukuhara I, Yano Y, Takehara I, Kaneko K. Effect of an excess intake of casein hydrolysate containing Val-Pro-Pro and Ile-Pro-Pro in subjects with normal blood pressure, high-normal blood pressure, or mild hypertension. Bioscience, Biotechnology, and Biochemistry. 2011;75(3):427–433. doi: 10.1271/bbb.100560. [DOI] [PubMed] [Google Scholar]
- Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clinical Microbiology Reviews. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Chaudhary K, Sharma M, Nagpal G, Chauhan JS, Singh S, Gautam A, Raghava GPS. AHTPDB: a comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Research. 2015;43(Database issue):D956–962. doi: 10.1093/nar/gku1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YW, Li B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: key sequence positions and their amino acid properties. Journal of Theoretical Biology. 2013;318:29–43. doi: 10.1016/j.jtbi.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Ma Y, Liu J, Shi H, Yu LL. Isolation and characterization of anti-inflammatory peptides derived from whey protein. Journal of Dairy Science. 2016;99(9):6902–6912. doi: 10.3168/jds.2016-11186. [DOI] [PubMed] [Google Scholar]
- McClean S, Beggs LB, Welch RW. Antimicrobial activity of antihypertensive food-derived peptides and selected alanine analogues. Food Chemistry. 2014;146:443–447. doi: 10.1016/j.foodchem.2013.09.094. [DOI] [PubMed] [Google Scholar]
- Migliore-Samour D, Jollès P. Casein, a prohormone with an immunomodulating role for the newborn? Experientia. 1988;44(3):188–193. doi: 10.1007/BF01941703. [DOI] [PubMed] [Google Scholar]
- Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M, Darewicz M. BIOPEP database and other programs for processing bioactive peptide sequences. Journal of AOAC International. 2008;91(4):965–980. [PubMed] [Google Scholar]
- Mohanty D, Jena R, Choudhury PK, Pattnaik R, Mohapatra S, Saini MR. Milk derived antimicrobial bioactive peptides: a review. International Journal of Food Properties. 2016;19(4):837–846. [Google Scholar]
- Nagaoka S, Futamura Y, Miwa K, Awano T, Yamauchi K, Kanamaru Y, Tadashi K, Kuwata T. Identification of novel hypocholesterolemic peptides derived from bovine milk β-lactoglobulin. Biochemistry and Biophysics Research Communication. 2001;281(1):11–17. doi: 10.1006/bbrc.2001.4298. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yamamoto N, Sakai K, Takano T. Antihypertensive effect of sour milk and peptides isolated from it that are inhibitors to angiotensin I-converting enzyme. Journal of Dairy Science. 1995;78(6):1253–1257. doi: 10.3168/jds.S0022-0302(95)76745-5. [DOI] [PubMed] [Google Scholar]
- Nongonierma AB, FitzGerald RJ. Susceptibility of milk protein-derived peptides to dipeptidyl peptidase IV (DPP-IV) hydrolysis. Food Chemistry. 2014;145:845–852. doi: 10.1016/j.foodchem.2013.08.097. [DOI] [PubMed] [Google Scholar]
- Pihlanto A. Antioxidative peptides derived from milk proteins. International Dairy Journal. 2006;16(11):1306–1314. [Google Scholar]
- Pripp AH, Isaksson T, Stepaniak L, Sørhaug T. Quantitative structure-activity relationship modelling of ACE-inhibitory peptides derived from milk proteins. European Food Research and Technology. 2004;219(6):579–583. [Google Scholar]
- Rohrbach MS, Williams EB, Rolstad RA. Purification and substrate specificity of bovine angiotensin-converting enzyme. Journal of Biological Chemistry. 1981;256(1):225–230. [PubMed] [Google Scholar]
- Rossini K, Noreña CPZ, Cladera-Olivera F, Brandelli A. Casein peptides with inhibitory activity on lipid oxidation in beef homogenates and mechanically deboned poultry meat. LWT - Food Science and Technology. 2009;42(4):862–867. [Google Scholar]
- Suetsuna K, Ukeda H, Ochi H. Isolation and characterization of free radical scavenging activities peptides derived from casein. Journal of Nutritional Biochemistry. 2000;11(3):128–131. doi: 10.1016/s0955-2863(99)00083-2. [DOI] [PubMed] [Google Scholar]
- Sun N, Wu H, Du M, Tang Y, Liu H, Fu Y, Zhu B. Food protein-derived calcium chelating peptides: a review. Trends in Food Science and Technology. 2016;58:140–148. [Google Scholar]
- Svenning C, Brynhildsvold J, Molland T, Langsrud T, Elisabeth Vegarud G. Antigenic response of whey proteins and genetic variants of β-lactoglobulin — the effect of proteolysis and processing. International Dairy Journal. 2000;10(10):699–711. [Google Scholar]
- Théolier J, Jérémie T, Ismail F, Julie J, Riadh H. MilkAMP: a comprehensive database of antimicrobial peptides of dairy origin. Dairy Science and Technology. 2013;94(2):181–193. [Google Scholar]
- Wu J, Aluko RE, Nakai S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure-activity relationship modeling of peptides containing 4-10 amino acid residues. QSAR and Combinatorial Science. 2006;25(10):873–880. [Google Scholar]
- Yang CM, Russell JB. Resistance of proline-containing peptides to ruminal degradation in vitro. Applied and Environmental Microbiology. 1992;58(12):3954–3958. doi: 10.1128/aem.58.12.3954-3958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucht HD, Raida M, Adermann K, Mägert HJ, Forssmann WG. Casocidin-I: a casein-alpha s2 derived peptide exhibits antibacterial activity. FEBS Letters. 1995;372(2-3):185–188. doi: 10.1016/0014-5793(95)00974-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


