Abstract
Activity of the Met4 transcription factor is antagonized by the SCFMet30 ubiquitin ligase by degradation-dependent and degradation-independent mechanisms, in minimal and rich nutrient conditions, respectively. In this study, we show that the heavy metal Cd2+ over-rides both mechanisms to enable rapid Met4-dependent induction of metabolic networks needed for production of the antioxidant and Cd2+-chelating agent glutathione. Cd2+ inhibits SCFMet30 activity through rapid dissociation of the F-box protein Met30 from the holocomplex. In minimal medium, dissociation of SCFMet30 complex is sufficient to impair the methionine-induced degradation of Met4. In rich medium, dissociation of the SCFMet30 complex is accompanied by a deubiquitylation mechanism that rapidly removes inhibitory ubiquitin moieties from Met4. Post-translational control of SCFMet30 assembly by a physiological stress to allow rapid induction of a protective gene expression program represents a novel mode of regulation in the ubiquitin system.
Keywords: cadmium, ligase, methionine, sulfur, ubiquitin
Introduction
The covalent modification of proteins by the small protein ubiquitin regulates myriad cellular processes (Hershko and Ciechanover, 1998). The best understood function of ubiquitylation is to target proteins for destruction by the 26S proteasome, a large compartmentalized protease particle that recognizes the polyubiquitin tag (Hochstrasser, 1996). More recently, nonproteolytic functions have been ascribed to ubiquitin conjugation, including targeting to different subcellular compartments and allosteric control of enzymatic events (Pickart, 2001). How ubiquitylation leads to different outcomes in different contexts is not fully understood.
Conjugation of ubiquitin to substrate proteins is achieved through a cascade of E1, E2 and E3 enzymes, which activate and then serially transfer ubiquitin to substrates (Hershko and Ciechanover, 1998). E3 enzymes, also called ubiquitin ligases, select specific substrates for ubiquitylation. The SCF (Skp1–Cdc53/cullin–F-box protein) and SCF-like complexes form a diverse family of ubiquitin ligases that regulate the cell cycle, the immune response, signaling cascades and developmental programs (Willems et al, 2004). SCF complexes recruit substrates through a repertoire of specific adapter subunits, termed F-box proteins, which are linked to a core ubiquitylation apparatus composed of the scaffold protein Cdc53/cullin, the RING finger protein Rbx1/Hrt1/Roc1, the F-box-binding protein Skp1 and an E2 enzyme (Skowyra et al, 1999; Willems et al, 2004). In budding yeast, over 20 different F-box proteins can be identified, but only three of these have been studied in detail, namely Cdc4, Grr1 and Met30 (Patton et al, 1998). SCFCdc4 and SCFGrr1 each target multiple substrates in a manner that depends on substrate phosphorylation (Willems et al, 2004). To date, the only known substrate of the SCFMet30 complex is the Met4 transcriptional activator, which governs the MET gene network responsible for the biosynthesis of the sulfur-containing amino acids methionine and cysteine (Patton et al, 1998; Rouillon et al, 2000; McMillan et al, 2002). While SCFMet30 may target substrates other than Met4, for instance in the regulation of interorganelle phosphatidylserine trafficking (Schumacher et al, 2002), restraint of the Met4-dependent transcriptional activity is the only essential function of Met30 (Patton et al, 2000).
Ubiquitylation controls the MET gene network by both degradation-dependent and degradation-independent mechanisms. When yeast cells are grown in minimal medium and exposed to a high concentration of methionine, SCFMet30-dependent ubiquitylation targets Met4 for degradation by the 26S proteasome (Rouillon et al, 2000). In contrast, when the cells are grown in rich medium, SCFMet30 oligoubiquitylates Met4 but without subsequent degradation (Kaiser et al, 2000; Kuras et al, 2002; Flick et al, 2004). In this circumstance, Met4 remains localized within the nucleus but is selectively excluded from most but not all MET gene promoters (Kuras et al, 2002). In particular, oligoubiquitylated Met4 retains activity at the SAM genes needed for the biosynthesis of S-adenosylmethionine (Kuras et al, 2002), a sulfur-containing compound widely used in intermediary metabolism as a methyl donor (Cantoni, 1977). The Met4–Met30 system thus enables selective activation of different promoters under different growth conditions. How nutrient conditions dictate the fate of ubiquitylated Met4 is not understood.
In addition to its regulation by methionine and by rich medium, control of Met4 activity was recently hypothesized to be modulated by other environmental changes (Fauchon et al, 2002; Aranda and Del Olmo, 2004). More especially, Met4 appears to mediate the adaptive response to cadmium (Cd2+), a nonessential heavy metal that is highly toxic to eukaryotic cells (Fauchon et al, 2002). The stress response to Cd2+, like that of many agents that induce oxidative stress, involves activation of sulfur metabolism pathways that generate compensating reducing equivalents (Jamieson, 2002; Pocsi et al, 2004). In yeast, Cd2+ activates the MET gene network in a Met4-dependent fashion, presumably to help build glutathione reserves necessary to complex and detoxify Cd2+ (Fauchon et al, 2002). Given this link, we thus examined if and how Cd2+ exposure might modify SCFMet30-dependent regulation of Met4. We find that Cd2+ impairs both degradation-dependent and degradation-independent inhibition of Met4 by dissociation of Met30 from the core SCF complex. Further, in rich medium, Cd2+ elicits deubiquitylation of pre-existing pools of inactive oligoubiquitylated Met4. The Cd2+ adaptive response thus entails novel modes of transcriptional regulation by the ubiquitin system.
Results
Cd2+ inhibits degradation-dependent regulation of Met4 activity
To address how Cd2+ might affect the MET gene regulatory network, we first analyzed MET gene transcription in cells grown in minimal B medium and simultaneously exposed to methionine (1 mM) and Cd2+ (100 μM). The presence of Cd2+ compromised MET gene repression normally triggered by high methionine, as shown by the failure to repress MET3, MET16 and MET25 expression (Figure 1A). Cd2+ also blocked repression of GSH1, a Met4-dependent gene that encodes γ-glutamylcysteine synthetase, the rate-limiting enzyme in glutathione biosynthesis from cysteine (Dormer et al, 2000; Wheeler et al, 2003). In all cases, the Cd2+ chelating agent EGTA prevented derepression of Met4-dependent genes by Cd2+.
Figure 1.
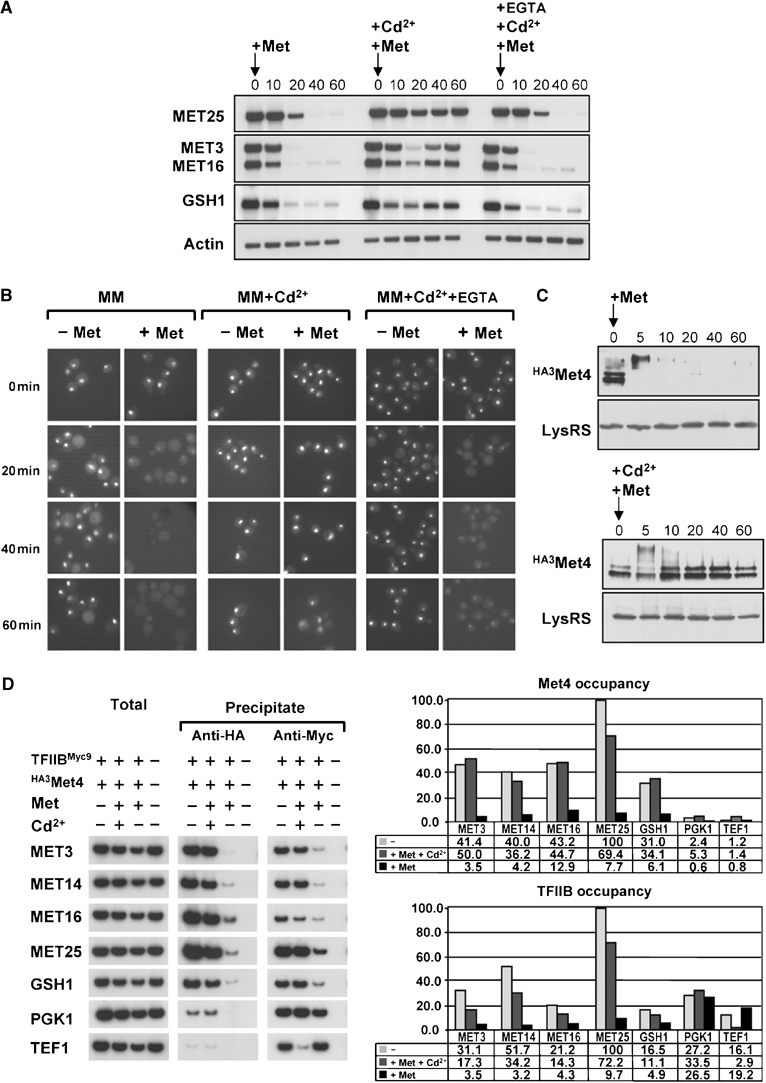
Cd2+inhibits the degradation-dependent regulation of Met4. (A) Cd2+ prevents MET gene repression by methionine in minimal medium. Total RNA was extracted at the indicated times after the addition of 1 mM L-methionine, in the presence (+Cd2+) or absence of 100 μM Cd2+, or in the presence of 100 μM of both Cd2+ and EGTA (+Cd2+, +EGTA), and MET gene expression was assessed by Northern analysis. (B) Cd2+ blocks elimination of endogenous GFP-Met4 by methionine. met4∷GFP-MET4 cells (strain CD240) were imaged at the indicated times after the addition (+Met) or not (−Met) of 1 mM L-methionine, in the presence (+Cd2+) or absence of 100 μM Cd2+, or in the presence of 100 μM of both Cd2+ and EGTA (+Cd2+, +EGTA). (C) Cd2+ stabilizes endogenous Met4 protein. met4∷HA3MET4 cells (strain CD233) were treated with (+Met) 1 mM L-methionine, in the presence (+Cd2+) or absence of 100 μM Cd2+, and total protein from TCA extracts was immunoblotted for the HA epitope or, as a control, lysyl-tRNA synthetase. (D) Met4p and TFIIB occupancy at MET promoters is elevated in response to Cd2+ exposure. A strain expressing both HA3Met4 and TFIIB9MYC (CD269) or an untagged isogenic control strain (W303-1A) were grown in B minimal medium and exposed to 1 mM L-methionine, in the presence or absence of 100 μM Cd2+. After crosslinking and immunoprecipitation with anti-HA or anti-Myc antibody, total DNA was analyzed by quantitative PCR with primer-pairs specific to the indicated Pol II promoters. ChIPs were performed at least twice and yielded similar results.
In minimal medium, Met4 is actively degraded upon exposure to high methionine (Kuras et al, 2002). To test whether the Cd2+-mediated abrogation of MET gene repression might be due to Met4 stabilization, the stability of Met4 was analyzed in the presence of both methionine and Cd2+. The rapid SCFMet30-dependent elimination of Met4 can be measured in live cells by using a GFP-Met4 fusion protein expressed from the endogenous Met4 promoter (Kuras et al, 2002). Concurrent addition of Cd2+ and methionine to minimal medium prevented the disappearance of the GFP-Met4 fluorescent signal observed when methionine alone was added (Figure 1B). To corroborate this result, we determined the stability of an HA epitope-tagged version of Met4 expressed from the chromosomal locus. Consistently, the HA3Met4 protein abundance was maintained when Cd2+ was added together with methionine to cells grown in minimal medium (Figure 1C), whereas in the absence of Cd2+, the HA3Met4 protein was rapidly degraded upon methionine exposure.
Chromatin immunoprecipitation (ChIP) was used to assess the abundance of Met4 at individual promoters in cells exposed to Cd2+. Association of the general transcription factor TFIIB was analyzed in parallel to determine the correlation between the presence of Met4 and transcriptional activity. For this purpose, we used a strain in which the MET4 and SUA7 genes (encoding TFIIB) were replaced at the chromosomal locus with epitope-tagged derivatives under control of the endogenous promoters (met4∷HA3MET4 and sua7∷SUA7MYC9, respectively). Crosslinked chromatin was prepared from cells grown in minimal B medium before and 40 min after the addition of 1 mM methionine, in the presence and absence of 100 μM Cd2+. Addition of Cd2+ prevented the four- to 25-fold decrease in both Met4 and TFIIB occupancy at MET gene promoters caused by addition of repressive amounts of methionine in the absence of Cd2+ (Figure 1D). Taken together, these results demonstrate that Cd2+ impairs SCFMet30-mediated degradation of Met4 that is normally induced by methionine in minimal medium.
Cd2+-induced Met4 DNA recruitment in complete medium
In rich medium, Met4 is oligoubiquitylated by SCFMet30 but not degraded (Kaiser et al, 2000). Under these conditions, MET gene expression is repressed as ubiquitylated forms of Met4 are selectively excluded from most MET promoters (Kuras et al, 2002). To determine if Cd2+ affected this alternate mode of Met4 regulation, cells were grown in rich medium, exposed to Cd2+ and assessed for MET gene transcription. We note that in rich medium, a higher concentration of Cd2+ (0.5 mM) was needed to elicit a biological effect, perhaps because of sequestration by unknown components in this medium. We found that transcription of the MET3, 10, 14, 16 and 25 genes was rapidly activated upon addition of 0.5 mM Cd2+ (Figure 2A). As expected, this activation was dependent upon a functional Met4 protein, as it was not observed in met4Δ cells (Figure 2B). Consistent with the observed transcriptional re-activation, Met4 occupancy at MET gene promoters was increased at least five-fold in cells grown in rich medium in the presence of Cd2+, as compared to cells grown in the absence of Cd2+ (Figure 2C). The degree of TFIIB occupancy at these promoters was similarly increased by the presence of Cd2+, in agreement with the high-level expression of MET genes under these conditions. Finally, the specificity of the Cd2+ effect on Met4 activity was assessed by exposing cells grown in rich medium to other heavy metals including cobalt, copper, manganese, mercury, silver and zinc. Northern and quantitative real-time (RT)-PCR assays showed that in contrast to Cd2+, these other heavy metals did not significantly activate MET gene expression in rich medium (Figure 2D). Cd2+ thus specifically reprieves Met4 from inactivation.
Figure 2.
Cd2+ activates MET gene expression in rich medium. (A) Rapid induction of MET genes by Cd2+. MET gene expression in wild-type cells was assessed at the indicated times after the addition of 0.5 mM Cd2+ in the absence or presence of 0.5 mM EGTA. (B) Cd2+-mediated activation of MET genes depends on Met4. MET gene expression was assessed in wild-type and met4Δ (CC849-8A) cells at the indicated times after the addition of 0.5 mM Cd2+. (C) Met4 and TFIIB recruitment to MET gene promoters in response to Cd2+. ChIP experiments were carried out as in Figure 1 on wild-type cultures before and 40 min after the addition of 0.5 mM Cd2+. (D) Cd2+ specifically induces MET gene expression. MET gene expression was assessed in wild-type cells before and 40 min after addition of the indicated heavy metals by Northern analysis (left) and quantitative real-time RT–PCR (right). Heavy metals were added at the following concentrations: Cu2+ 1 mM, Zn2+ 1 mM, Co2+ 2 mM, Hg2+ 0.3 μM, Mn2+ 0.5 mM and Ag2+ 30 μM.
Cd2+ inhibits ubiquitylation of Met4
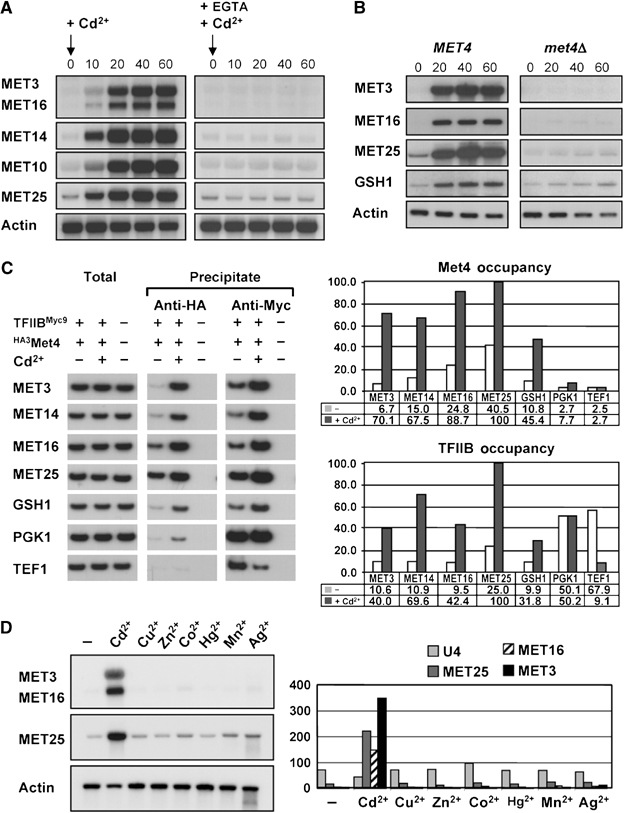
In rich medium, Met4 is stable but rendered inactive because of its oligoubiquitylation by SCFMet30 (Kaiser et al, 2000; Kuras et al, 2002). The above results suggested that Cd2+ might abrogate this effect. Indeed, the high-molecular-weight ubiquitylated forms of an HA epitope-tagged version of Met4 observed in rich medium were rapidly replaced by unmodified Met4 upon exposure to Cd2+ (Figure 3A). As expected, this collapse was prevented by concomitant addition of EGTA. After the initial collapse of the Met4 isoforms caused by Cd2+, new higher molecular weight forms gradually appeared. To investigate the nature of these subsequent isoforms, endogenous Met4 was immunoprecipitated with an anti-Met4 polyclonal antibody, followed by immunoblot with either anti-Met4 or anti-ubiquitin antibodies. This experiment confirmed that while Met4 was oligoubiquitylated in rich medium, the secondary modified forms induced after Cd2+ addition were not due to ubiquitylation (Figure 3B). Rather, the Cd2+-dependent modifications were due to phosphorylation, as shown by collapse to the lowest mobility form upon lambda phosphatase treatment (Figure 3C). Although the function of these phosphorylation events remains to be deciphered, similar phosphorylated Met4 species were observed in cells that lack Met30 when grown in rich medium in the absence of Cd2+ (Figure 3C), as reported recently (Flick et al, 2004). All told, these observations suggest that Cd2+ acts by inhibiting SCFMet30 activity.
Figure 3.
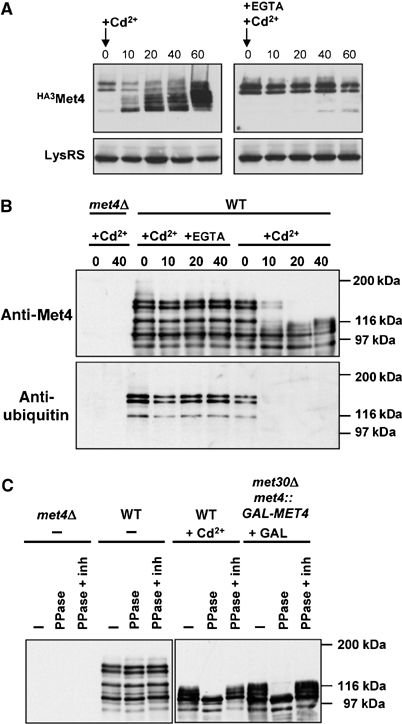
Cd2+ impairs SCFMet30-dependent ubiquitylation of Met4 in rich medium. (A) Collapse of Met4 isoforms in response to Cd2+. met4∷HA3MET4 cells (strain CD233) were exposed to 0.5 mM Cd2+ in the absence or presence of 0.5 mM EGTA and extracted proteins analyzed for Met4 and lysyl-tRNA synthetase abundance by immunoblot. (B) Loss of ubiquitinated Met4 species upon Cd2+ treatment. Endogenous Met4 was immunoprecipitated with anti-Met4 antibody and then immunoblotted with anti-Met4 and anti-ubiquitin antibodies. (C) Accumulation of phosphorylated Met4 species in response to Cd2+ and in the absence of Met30. Wild-type, met4Δ and met4∷GAL1-MET4, met30Δ strains were grown in rich medium containing 2% raffinose and harvested before and after 40 min of the indicated treatment with either 0.5 mM Cd2+ or galactose. Anti-Met4 immunoprecipitates were treated with lambda phosphatase in the presence or absence of phosphatase inhibitors and then immunoblotted with anti-Met4 antibody.
Cd2+ induces dissociation of the SCFMet30 ubiquitin ligase
The above results suggest that SCFMet30 activity is a target of the Cd2+ signal, regardless of whether cells are grown in minimal medium and exposed to high methionine concentrations, or are grown in rich medium. One possible mechanism for Cd2+-mediated inhibition of SCFMet30 might be the complete elimination of Met30 itself. However, a promoter shutoff experiment showed that while Met30 was a moderately unstable protein, as shown previously (Galan and Peter, 1999), the in vivo half-life of Met30 was unaffected by Cd2+ (Figure 4A). We next considered whether the interactions between Met4 and Met30 and/or the assembly of the SCFMet30 complex might be affected by Cd2+. To test the former possibility, we overexpressed Met30 to facilitate detection of the Met4–Met30 interaction, which is constitutive but is normally limited by the low-level expression of MET30 in minimal medium (Rouillon et al, 2000). In this context, Cd2+ treatment did not affect formation of the Met4–Met30 complex, in either minimal medium (Figure 4B) or in rich medium (data not shown). We therefore determined if the critical interaction between Met30 and the F-box-binding protein Skp1 was altered by Cd2+ treatment. As judged by co-immunoprecipitation from yeast extracts, the interaction between Skp1 and Met30 was abolished in cells exposed to Cd2+ (Figure 4C and D). This result was obtained with cells grown in either minimal or rich medium and by immunoprecipitating first either Met30 or Skp1 immune complexes. The loss of the interaction between Skp1 and Met30 was specific since another interaction within the SCFMet30 complex, that between Skp1 and Cdc53, was not altered by Cd2+ (Figure 4C and D). Thus, exposure of cells to Cd2+ cripples the ability of the SCFMet30 complex to ubiquitylate Met4 by disrupting the interaction between the substrate-binding subunit Met30 and the core catalytic apparatus.
Figure 4.
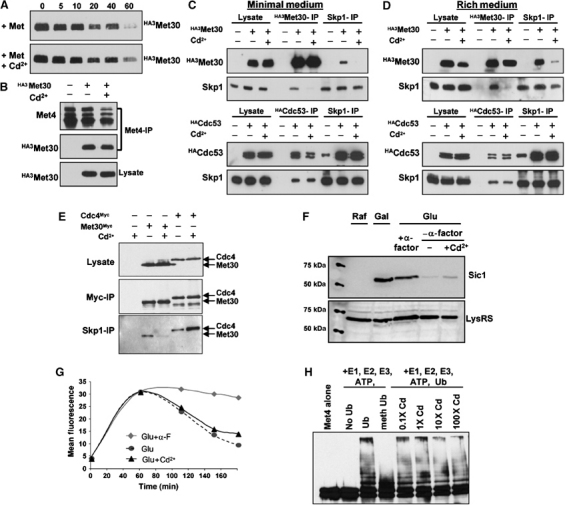
Cd2+ induces dissociation of the SCFMet30 ubiquitin ligase. (A) Met30 half-life is not altered by Cd2+ exposure. A strain bearing a GAL1-HA3MET30 construct was induced in galactose medium for 2 h, then transferred from galactose to 2% glucose- and 1 mM methionine-containing medium, in the presence or absence of 100 μM Cd2+, and HA3Met30 abundance assessed by anti-HA immunoblot at the indicated times after glucose repression. (B) The Met4–Met30 interaction is unaffected by Cd2+. An HA3Met30 fusion protein was expressed from a GAL1 promoter in B medium containing 2% galactose in the presence or absence of 100 μM Cd2+, Met4 immunoprecipitated from extracts with anti-Met4 antibody and immunoblotted with anti-Met4 and anti-HA antibodies. (C) The Skp1–Met30 interaction is specifically abolished by Cd2+ in minimal medium. HA3Met30 and HA3Cdc53 fusion proteins were expressed as in (B), extracts subjected to immunoprecipitation with the indicated antibodies and immunoblotted with anti-Skp1 and anti-HA antibodies. (D) The Skp1–Met30 interaction is specifically abolished upon Cd2+ exposure in rich medium. The experiments were performed as in (C) but cells were grown in rich medium and in the presence of 0.5 mM Cd2+. (E) The Skp1–Cdc4 interaction is not abolished upon Cd2+ exposure in rich medium. Met30MYC3 and Cdc4MYC3 fusion proteins were expressed from the constitutive CDC53 promoter (Ho et al, 2002) in rich medium in the presence or absence of 0.5 mM Cd2+ and protein interactions tested as in (C) with anti-MYC and anti-Skp1 antibodies. (F) Sic1 instability is not affected by Cd2+. A strain (CYS37) expressing a GFP-Sic1 fusion protein from the GAL1 promoter was grown in minimal medium, arrested in G1 phase by α-factor mating pheromone for 1.5 h, released into glucose medium in the presence and absence of 100 μM Cd2+, with or without α-factor, and GFP-Sic1 abundance assessed 120 min after the promoter shutoff by immunoblot with anti-GFP or, as a control, anti-lysyl-tRNA synthetase (LysRS) antibodies. (G) Quantification by flow cytometry of GFP-Sic stability in either an α-factor block or after release from the block in the presence and absence of 100 μM Cd2+. Cells were grown as described in (F) and the GFP-Sic1 fluorescent signal recorded and quantified at the indicated times using a Beckton Dickinson FacScalibur™ flow cytometer. (H) Met4 ubiquitylation by recombinant SCFMet30 is not affected by Cd2+. Purified recombinant Met4 and SCFMet30 produced in insect cells were incubated with E1 enzyme, Cdc34, ubiquitin and ATP, either in the absence or presence of the indicated amounts of Cd2+. Methyl-ubiquitin served as a control to demonstrate polyubiquitin chain formation on Met4.
As Skp1 recruits many other F-box-containing proteins to the SCF core apparatus, we tested the specificity of the Cd2+ effect by examining the assembly and activity of another yeast SCF complex, SCFCdc4, which mediates degradation of the CDK inhibitor Sic1 at the G1/S transition (Willems et al, 2004). Unlike the Skp1–Met30 complex, addition of Cd2+ to cells in rich medium did not disrupt the Skp1–Cdc4 complex (Figure 4E). If anything, the amount of Skp1 bound to Cdc4 was increased by Cd2+, perhaps because more Skp1 was available upon its liberation from Met30. As a further control for specificity of the response, Cd2+ also did not affect abundance of the Cdc4 substrate Sic1 (Figure 4F). Moreover, quantitative flow cytometry of a GFP-Sic1 reporter construct showed that the kinetics of Sic1 degradation upon release from a G1 arrest imposed by mating pheromone was not impaired by Cd2+ (Figure 4G).
To begin to address the mechanism of Cd2+ action, we examined whether Cd2+ affected the activity of a recombinant SCFMet30 complex. For this purpose, an active SCFMet30 complex was affinity purified from insect cells that had been coinfected with Skp1, Cdc53, Rbx1 and Met30 baculovirus constructs. This SCFMet30 complex efficiently ubiquitylated recombinant Met4, which was also produced in insect cells (Figure 4H). Despite its potent effects in vivo, Cd2+ did not discernibly inhibit the ability of the recombinant complex to ubiquitylate Met4 in vitro (Figure 4H). Addition of crude cell extracts, from either untreated or Cd2+-treated cultures, also did not impair the extent of Met4 ubiquitylation in the in vitro system (data not shown). SCFMet30 is thus specifically dissociated in response to Cd2+ in a manner that appears to require active cellular metabolism.
Cd2+ triggers Met4 deubiquitylation
In rich medium, highly ubiquitylated forms of Met4 are localized within the nucleus of the cells and are stable, being apparently immune to the proteasome (Kaiser et al, 2000; Kuras et al, 2002). However, when cells grown in rich medium are exposed to Cd2+, ubiquitylated Met4 species disappear very quickly (Figure 3B). As Met4 forms are stable in rich medium, these rapid changes in Met4 modification pattern could not be explained simply by the inhibition of SCFMet30 activity. We therefore examined whether Cd2+ might also induce an active Met4 deubiquitylation response. That is, pre-existing transcriptionally inactive ubiquitylated isoforms of Met4 might be converted into transcriptionally active nonubiquitylated forms. To test this hypothesis, Met4 was conditionally expressed from the GAL1 promoter, which was then repressed by glucose for 60 min prior to addition of Cd2+. Northern analysis demonstrated that MET gene expression was strongly activated by Cd2+ in the complete absence of any GAL1-MET4 transcript, consistent with the conversion of pre-existing inactive Met4 into transcriptionally competent Met4 (Figure 5B). Parallel immunoprecipitation analysis of Met4 revealed that pre-existing ubiquitylated Met4 species were indeed converted into nonubiquitylated forms upon Cd2+ exposure (Figure 5C).
Figure 5.
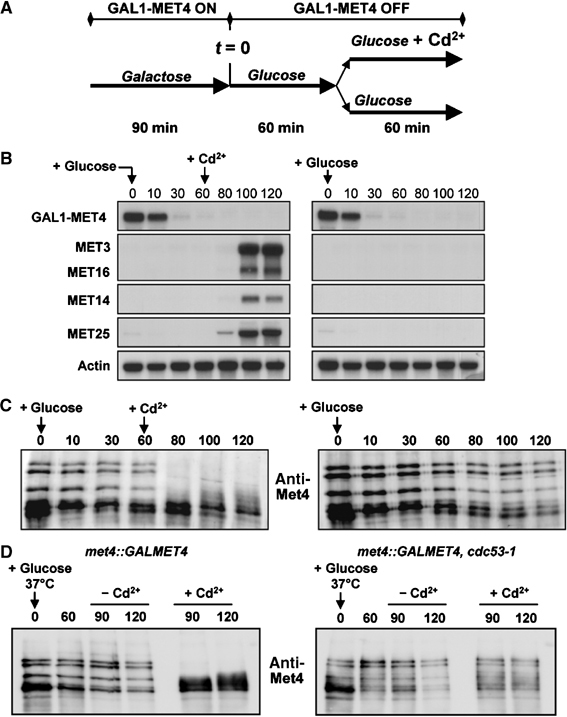
Cd2+ triggers deubiquitylation of Met4 in rich medium in the absence of de novo Met4 synthesis. (A) Schematic representation of the experiment. (B) Cd2+-activated MET gene expression does not require de novo Met4 synthesis. A met4∷GAL1-MET4 strain (CC932-6D) was subjected to the regimen in (A) and assessed for MET gene expression at the indicated time points. (C) Cd2+ induces Met4 deubiquitylation. The same experiment as in (B) was performed except that Met4 was immunoprecipitated and immunoblotted with anti-Met4 antibody. (D) Inactivation of SCFMet30 does not induce Met4 deubiquitylation. Cultures of wild-type and cdc53-1 strains were shifted to the nonpermissive temperature of 37°C at the same time as repression of GAL1-MET4 by glucose. Cd2+ was either added or not at 60 min postshift.
In principle, the deubiquitylation of Met4 in rich medium might result from a basal level of deubiquitylation that dominates only when SCFMet30 is fully inhibited, or from an induction of ubiquitin hydrolase activity by Cd2+. To discriminate between these possibilities, we inactivated SCFMet30 by shifting cdc53-1 strain to the restrictive temperature and examined the status of preformed Met4–ubiquitin conjugates. Inactivation of Cdc53 did not trigger Met4 deubiquitylation (Figure 5D). From this result, we conclude that Cd2+ induces a Met4-specific ubiquitin hydrolase activity. Intriguingly, Cd2+ did not induce Met4 deubiquitylation when the cdc53-1 strain was inactivated, perhaps suggesting that an intact SCFMet30 complex is needed for recognition of Met4 by the ubiquitin hydrolase. In summary, in rich medium, MET gene activation by Cd2+ appears to result from both the disassembly of the SCFMet30 complex and the rapid deubiquitylation of Met4.
Met4 is required for growth in the presence of Cd2+
To corroborate the inferred role of Met4 in the detoxification of Cd2+ (Fauchon et al, 2002), we assessed the Cd2+ sensitivity of cells that do not express a functional Met4 protein or one of its associated cofactors, Cbf1, Met28, Met31 and Met32, which variously tether Met4 to promoter DNA in a gene-specific manner (Kuras and Thomas, 1995; Blaiseau and Thomas, 1998). Serial dilutions of met4Δ, met28Δ, met31Δ, met32Δ and the double met31Δ met32Δ mutant strains on minimal medium plates containing 20 μM Cd2+ demonstrated that the single met4Δ and the double met31Δ met32Δ deleted cells are exquisitely sensitive to Cd2+ (Figure 6). The Cd2+ sensitivity of the met31Δ met32Δ strain further suggests that Met31/Met32 factors furnish the predominant DNA-binding platform for Met4 (Blaiseau and Thomas, 1998), consistent with genome-wide expression profiles of met31Δ met32Δ and cbf1Δ strains (TA Lee and M Tyers, unpublished data). We note that Cd2+ sensitivity of these strains is not particularly manifest in rich medium, perhaps because ample reducing equivalents are supplied by the complex bacterial and yeast extracts used in this formulation (data not shown). Met4 thus plays an essential role in Cd2+ detoxification under limiting nutrient conditions.
Figure 6.
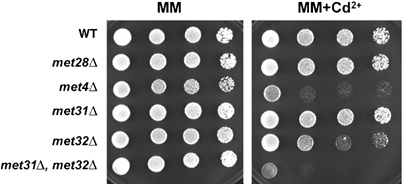
Cd2+ sensitivity of met4Δ and met31Δ met32Δ strains. Serial dilutions of wild-type (WT, W303-1A), met28Δ (CC769-7D), met4Δ (CC849-1B), met31Δ (CC867-1C), met32Δ (CC845-1C) and met31Δ met32Δ (CC845-1A) strains were plated onto YNB minimal medium in the presence and absence of 20 μM Cd2+ and grown for 2 days.
Discussion
The diverse roles of the ubiquitin system derive from its ability to selectively modify many different substrates in a highly regulated manner, a feat achieved by the large number of E3 ubiquitin ligases in the cell (Hershko and Ciechanover, 1998; Pickart, 2001). Regulation of E3–substrate interactions may occur at the level of substrate modification, E3 expression or E3 modification and assembly. One of the most highly regulated E3 activities is that of the anaphase-promoting complex/cyclosome (APC/C), which targets many substrates in mitosis and G1 phase. The APC/C is controlled primarily by the availability of substrate recruitment factors and their phosphorylation-dependent association with the core APC/C particle (Willems et al, 2004). In contrast, substrate ubiquitylation by SCF complexes is usually dictated by substrate level modification, predominantly phosphorylation, and/or by F-box protein expression.
Met30 differs from most characterized F-box proteins in that it appears to recognize its substrate Met4 in a constitutive manner. Regulation is instead conferred at the level of MET30 expression, which itself is controlled by Met4 in an autoregulatory loop (Rouillon et al, 2000), and by an as yet undiscovered mechanism that links the fate of Met4–ubiquitin conjugates to nutrient conditions (Kaiser et al, 2000; Rouillon et al, 2000; Kuras et al, 2002). Here we report that the activity of Met4 is also modulated by the heavy metal Cd2+ so as to allow constitutive activation of the sulfate assimilation pathway and build the glutathione reserves required to chelate and detoxify Cd2+. The ubiquitylation status of Met4 and its attendant transcriptional activity are dictated by at least two Cd2+-dependent pathways. First, in both minimal and rich medium, Cd2+ triggers the specific disengagement of Met30 from its binding partner Skp1, thereby preventing the ubiquitylation of Met4. Second, in rich medium, pre-existing inactive Met4–ubiquitin conjugates are rapidly deubiquitylated in the presence of Cd2+. This deubiquitylation activity appears to be induced in response to Cd2+. The redundancy of the 17 predicted deubiquitinylating enzymes in yeast has so far precluded genetic identification of the relevant activity. Regardless of the precise mechanism, the rapid deubiquitylation of Met4 poises cells to respond rapidly to Cd2+ in all nutrient conditions. These supernumerary regulatory mechanisms serve to link tightly the regulation of sulfur metabolism to the heavy metal stress response.
Cd2+-induced dissociation of the SCFMet30 ubiquitin ligase
The fact that Cd2+ compromised both the degradation-dependent and degradation-independent regulation of Met4, under minimal and rich nutrient conditions respectively, suggested that the divalent metal might target the common mediator of the two mechanisms, the SCFMet30 ubiquitin ligase. Of several possible targets in the SCFMet30 pathway, we found that Cd2+ impairs the assembly of the Skp1–Met30 subcomplex. The dissociation of the Skp1–Met30 interaction in response to Cd2+ is specific, as the Skp1–Cdc53 interaction is not compromised by Cd2+ nor is the assembly and in vivo activity of SCFCdc4 affected. The interaction of Skp1 with F-box proteins occurs via a bipartite interface comprised of a conserved core F-box domain and an N-terminal variable region (Schulman et al, 2000). The variable region of Met30 is thus the logical potential target of the Cd2+ pathway. Based on the structures of the SCFSkp2, SCFCdc4 and SCFβ-TrCP complexes, all of which indicate fixed juxtaposition of the substrate to the E2 by the F-box protein (Zheng et al, 2002; Orlicky et al, 2003; Wu et al, 2003), perturbation of the F-box protein–Skp1 interaction would afford a specific means to inactivate any given SCF complex. Indeed, under some circumstances, carbon source appears to control the interaction between Skp1 and the F-box protein Grr1, which helps dictate the response to glucose (Li and Johnston, 1997). The mechanism whereby the Skp1–Met30 interface is disrupted is unknown. Because Cd2+ appears not to affect purified recombinant SCFMet30 activity in vitro, direct binding of the metal ion to Met30 seems unlikely. Whether Cd2+ triggers post-translational modification of Met30 or the association of an inhibitory factor remains to be seen.
The regulation of SCFMet30 by Cd2+ also bears on the somewhat controversial issue of degradation-dependent versus degradation-independent regulation of Met4. While our data strongly suggest that SCFMet30-dependent ubiquitylation of Met4 leads to Met4 degradation specifically in minimal media (Rouillon et al, 2000; Kuras et al, 2002), whereas Met4 is stably ubiquitylated in rich medium (Kaiser et al, 2000; Kuras et al, 2002), the degradation of Met4 under minimal conditions has been disputed (Flick et al, 2004). Experiments in this study demonstrate that Cd2+ causes the accumulation of unmodified forms of Met4, association of Met4 with cognate promoters and transactivation of MET genes in the presence of methionine in minimal medium (see Figure 3). Because a single agent can switch Met4 from a highly unstable to stable state in otherwise identical experimental conditions, it is unlikely that the instability of Met4 derives from artifacts of epitope tags, media conditions or protein extraction (Kaiser et al, 2000). Rather, the central role of sulfur metabolism appears to have selected for multiple modes of Met4-dependent transcriptional regulation, each attuned to respond to particular environmental conditions.
Role of Met4 in Cd2+ detoxification
In yeast cells, the primary Cd2+ detoxification mechanism involves the formation of a Cd2+-glutathione chelate, which is subsequently sequestered in the vacuole (Li et al, 1997). Efficient elimination of Cd2+ therefore requires high levels of glutathione, a sulfur-containing tripeptide whose biosynthesis is rate limited by the cysteine consuming step. Cd2+ detoxification is dependent upon a fully active sulfate assimilation pathway, which provides the reduced sulfur atom required for cysteine and hence glutathione biosynthesis. The transcriptional regulation of the sulfate assimilation pathway requires Met4 and its cofactors Cbf1, Met28, Met31 and Met32 (Thomas and Surdin-Kerjan, 1997). Of these factors, only Met4 is endowed with intrinsic transcription activation function, while Cbf1, Met28, Met31 and Met32 act by tethering Met4 to promoter DNA (Kuras and Thomas, 1995; Blaiseau and Thomas, 1998). Thus, the direct regulation of Met4 activity is an efficient means to meet the requirements for reduced sulfur in glutathione biosynthesis during Cd2+ detoxification. As the Met4–Met30 interaction is not perturbed by Cd2+, the presence of Met30 apparently does not interfere with the activity of Met4 transcriptional complexes, a feature that may also enable dynamic regulation of Met4 activity without the need for de novo synthesis. The fact that the Cd2+ signaling pathway is epistatic to both degradation-dependent and degradation-independent regulation of Met4 allows the cell to cope with Cd2+ toxicity regardless of nutrient conditions (Figure 7). In addition to direct regulation of sulfur metabolism under Cd2+ stress, Met4 also helps control a proteome-wide response to Cd2+, whereby the production of abundant sulfur-rich proteins is repressed, thus redirecting reduced sulfur equivalents toward glutathione biosynthesis (Fauchon et al, 2002; Jamieson, 2002). The major role of Met4 in Cd2+ detoxification is underscored by the sensitivity of met4Δ and met31Δ met32Δ strains to this heavy metal. The cellular response to Cd2+ in yeast thus occurs at a system-wide level that relies on metabolic, transcriptional and post-transcriptional effectors.
Figure 7.
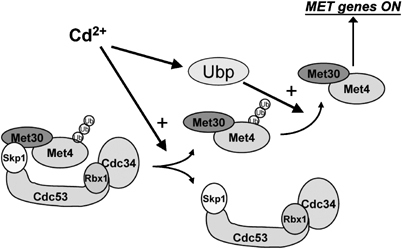
Model for Cd2+-mediated activation of the MET gene network through the inhibition of the SCFMet30 ubiquitin ligase and activation of an uncharacterized deubiquitinylating enzyme (Ubp).
Conservation of the oxidative stress response
Cd2+ causes oxidative stress by depletion of intracellular glutathione pools (Ercal et al, 2001). The oxidative stress response in mammals and that in yeast bear striking similarities. In mammalian cells, upon oxidative stress, the bZIP transcription factor Nrf2, in association with a family of coactivators, directs the expression of phase II detoxification enzymes, which in part mediate glutathione biosynthesis (Nguyen et al, 2004). Nrf2 activity is controlled in part through its regulated localization (Itoh et al, 1999), and in part through its degradation by an SCF-related ubiquitin ligase complex composed of Cul3, Rbx1 and a BTB-Kelch domain adaptor protein called Keap1 (Cullinan et al, 2004; Kobayashi et al, 2004). Similar to the rescue of met30 lethality by a met4 deletion, the lethality of Keap1−/− mice is rescued by codeletion of Nrf2 (Wakabayashi et al, 2003). In a manner also analogous to the Cd2+-induced dissociation of Met30 from the core SCF complex, recent evidence suggests that oxidative stress specifically impairs the interaction between Cul3 and Keap1 (Zhang et al, 2004). As a consequence, Nrf2 is stabilized by Cd2+ and other oxidative stresses, and thereby able to drive transcription of stress response genes (Stewart et al, 2003). While the mechanism whereby oxidative stress inhibits the Keap1–Cul3 interaction is not precisely known, several cysteine residues on Keap1 are critical for the degradation of Nrf2 (Zhang et al, 2004), perhaps serving as sites of modification for oxidative metabolites (Levonen et al, 2004) and/or disulfide bridges (Wakabayashi et al, 2004). In addition, phosphorylation of Nrf2 appears to impair its interaction with Keap1 (Huang et al, 2002). Whether or not Cd2+ or other oxidative stresses lead to similar modifications on Met30 and/or Met4 remains to be determined. The striking parallels between the Nrf2–Keap1 and Met4–Met30 systems should afford further cross-species insights into the oxidative stress response.
Materials and methods
Yeast culture
Saccharomyces cerevisiae strains used in this study are listed in Table I. Minimal B medium is a synthetic medium that lacks organic and inorganic sulfur sources (Cherest and Surdin-Kerjan, 1992). Unless indicated, cells were grown in the presence of 0.2 mM DL-homocysteine as a sulfur source, and amino acids needed to complement the auxotrophic requirements for each strain were added (i.e., drop-in medium). YPD rich medium contains 0.5% yeast extract (Difco), 0.5% bacto-peptone (Difco) and 3% glucose. Transformation was by the lithium acetate method (Gietz et al, 1992). To test the specificity of the Cd2+ response, yeast cells were exposed to other heavy metals including cobalt, copper, manganese, mercury, silver and zinc. The heavy metals were used at concentrations that were shown to be toxic to yeast cells (Thorvaldsen et al, 1993; Ciriolo et al, 1994; Blackwell et al, 1998; Li and Kaplan, 1998).
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| CC769-7D | MATα, ade2, his3, leu2, trp1, ura3, met28∷LEU2 | Kuras et al (1997) |
| CC845-1A | MATa, ade2, his3, leu2, trp1, ura3, met31∷LEU2, | |
| met32∷URA3 | This study | |
| CC845-1C | MATa, ade2, his3, leu2, trp1, ura3, met32∷URA3 | This study |
| CC849-1B | MATa, ade2, his3, leu2, trp1, ura3, met4∷TRP1 | Rouillon et al (2000) |
| CC867-1C | MATα, ade2, his3, leu2, trp1, ura3, met31∷LEU2 | Blaiseau et al (1997) |
| CC932-6D | MATa, ade2, his3, leu2, ura3, met4∷GAL1-MET4 | Patton et al (2000) |
| CC932-8B | MATa, ade2, his3, leu2, ura3, met4∷GAL1-MET4, met30∷LEU2 | Patton et al (2000) |
| CD233 | MATα, his3, leu2, trp1, ura3, met4∷HA3MET4 | Kuras et al (2002) |
| CD240 | MATα, his3, leu2, trp1, ura3, met4∷GFP-MET4 | Kuras et al (2002) |
| CD269 | MATα, his3, leu2, trp1, ura3, met4∷HA3MET4, | |
| sua7∷SUA79MYC∷TRP1 | Kuras et al (2002) | |
| CYS37 | MATa, his3, leu2, trp1, ura3∷pGAL1-GFPmut3-Sic1∷URA3 | This study |
| MT1885 | MATa, met4∷GAL1-MET4 | This study |
| MT3341 | MATα, met4∷GAL1-MET4, cdc53-1 | This study |
| W303-1A | MATa, ade2, his3, leu2, trp1, ura3, | R Rothstein |
Fluorescence microscopy and cytometry
GFP-Met4 fusion protein signals were monitored in living cells on a Nikon Eclipse fluorescence microscope using an Omega XF116 filter. All images were collected with a Princeton CCD camera using identical settings and analyzed with the Meta-Imaging V4.5 software (Universal Imaging Corporation, Downingtown, PA). Nuclei were stained using the dye HOECHST no. 33342 (Sigma), which was added at 1 μg/ml to the culture 20 min prior to imaging. Levels of the GFP-Sic1 fusion protein were assessed by quantitative fluorescence of live cells using a Beckton Dickinson FacsScalibur flow cytometer.
Chromatin immunoprecipitation and RNA analysis
Crosslinked chromatin preparation and immunoprecipitation (ChIP) was performed as described previously (Kuras et al, 2002) using the following antibodies: mouse monoclonal anti-HA antibody F-7 from Santa Cruz, mouse monoclonal anti-Myc antibody PL14 from StressGen, and a rabbit polyclonal Met4 antiserum produced against full-length Met4 produced in insect cells. Immunoprecipitated and total DNA samples were analyzed by quantitative PCR in the presence of [α-32P]dATP. Linearity of the PCR reaction was established in multiple independent dilutions of each sample. PCR products were separated on an 8% TBE polyacrylamide gel and quantified on a PhosphorImager (Molecular Dynamics). The occupancy level at a given promoter was defined as the ratio of immunoprecipitated DNA over total DNA for each PCR product. The occupancy level at MET25 was arbitrarily set to 100 and all other values were represented relative to this standard. For Northern analysis, total RNA was extracted by the hot phenol method, separated and probed as described (Rouillon et al, 2000).
Protein analysis
Total proteins were extracted either by a TCA procedure (Rouillon et al, 2000) or by glass bead lysis (Kuras et al, 2002). For co-immunoprecipitation, total protein was extracted by glass bead lysis in buffer containing 1% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 250 mM NaCl, 50 mM Tris–HCl (pH 7.5), 10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 1 mM DTT, 5 mM NEM, 1 mM PMSF, 1 μg/ml leupeptin and pepstatin. A 1 μl portion of crude anti-Met4 antiserum was added to 5 mg of extract, incubated on ice for 1 h, bound to 30 μl of 50% protein A bead slurry for 1 h at 4°C and washed. Samples were separated on a 7.5% acrylamide gel, transferred to a PVDF membrane and probed with anti-Met4 polyclonal antibody (1:200 dilution) followed by peroxidase-conjugated anti-rabbit secondary antibody (1:10 000 dilution) and detected by SuperSignal West Pico chemiluminescent substrate (Pierce). A GFP-Sic1 protein fusion was detected with anti-GFP antibody. Equal loading was established by detection with a lysyl-tRNA synthetase antibody.
In vitro ubiquitylation assays
Recombinant proteins were produced in insect cells by baculovirus coinfection and used for in vitro ubiquitination assays as described (Skowyra et al, 1997). A MycCdc53–Rbx1Myc–Skp1FLAG–GSTMet30 complex was affinity purified from infected insect cell lysate on anti-FLAG-M2 agarose resin (Sigma). Recombinant His6-HAMet4 produced in insect cells was affinity purified on nickel resin and treated with lambda phosphatase. Immobilized SCFMet30 was washed three times with 1 ml of ubiquitination buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM ATP, 50 μM DTT), incubated with His6Uba1 (E1), His6Cdc34 (E2), ubiquitin (Sigma) or methylated ubiquitin (Affiniti) and Met4 in 10 μl ubiquitylation buffer at 30°C for 1 h. Products were separated by SDS–PAGE and immunoblotted with anti-HA (12CA5) antibody.
Acknowledgments
We thank Laurent Kuras for advice on ChIP. This work was supported by funds from the Centre National de la Recherche Scientifique and the Association de la Recherche sur le Cancer to DT and by grants from the National Cancer Institute of Canada, the Canadian Institutes of Health Research (CIHR) and the Human Frontiers Science Program to MT. PBC is supported by the Leukemia and Lymphoma Society of America, TAL is supported by a training fellowship from the CIHR and MT is supported by a Canada Research Chair in Functional Genomics and Bioinformatics.
References
- Aranda A, Del Olmo ML (2004) Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol 70: 1913–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell KJ, Tobin JM, Avery SV (1998) Manganese toxicity towards Saccharomyces cerevisiae: dependence on intracellular and extracellular magnesium concentrations. Appl Microbiol Biotechnol 49: 751–757 [DOI] [PubMed] [Google Scholar]
- Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D (1997) Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol 17: 3640–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau PL, Thomas D (1998) Multiple transcription activation complexes tether the yeast activator Met4 to DNA. EMBO J 17: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni GL (1977) S-adenosylmethionine: present status and future perspectives. In The Biochemistry of S-adenosylmethionine, Salvatore F, Borek E, Zappia V, Williams-Ashman HG, Schlenk F (eds) pp 557–577. New York: Columbia University Press [Google Scholar]
- Cherest H, Surdin-Kerjan Y (1992) Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriolo MR, Civitareale P, Carri MT, De Martino A, Galiazzo F, Rotilio G (1994) Purification and characterization of Ag,Zn-superoxide dismutase from Saccharomyces cerevisiae exposed to silver. J Biol Chem 269: 25783–25787 [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormer UH, Westwater J, McLaren NF, Kent NA, Mellor J, Jamieson DJ (2000) Cadmium-inducible expression of the yeast GSH1 gene requires a functional sulfur-amino acid regulatory network. J Biol Chem 275: 32611–32616 [DOI] [PubMed] [Google Scholar]
- Ercal N, Gurer-Orhan H, Aykin-Burns N (2001) Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem 1: 529–539 [DOI] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J (2002) Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell 9: 713–723 [DOI] [PubMed] [Google Scholar]
- Flick K, Ouni I, Wohlschlegel JA, Capati C, McDonald WH, Yates JR, Kaiser P (2004) Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol 6: 634–641 [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M (1999) Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, StJean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB (2002) Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 277: 42769–42774 [DOI] [PubMed] [Google Scholar]
- Itoh K, Ishii T, Wakabayashi N, Yamamoto M (1999) Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res 31: 319–324 [DOI] [PubMed] [Google Scholar]
- Jamieson D (2002) Saving sulfur. Nat Genet 31: 228–230 [DOI] [PubMed] [Google Scholar]
- Kaiser P, Flick K, Wittenberg C, Reed SI (2000) Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102: 303–314 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Barbey R, Thomas D (1997) Assembly of a bZIP/bHLH transcription activation complex: formation of the yeast Cbf1–Met4–Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J 16: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Rouillon A, Lee T, Barbey R, Tyers M, Thomas D (2002) Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol Cell 10: 69–80 [DOI] [PubMed] [Google Scholar]
- Kuras L, Thomas D (1995) Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol Cell Biol 15: 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM (2004) Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J 378: 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Johnston M (1997) Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J 16: 5629–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kaplan J (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 273: 22181–22187 [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Theesfeld CL, Harrison JC, Bardes ES, Lew DJ (2002) Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol Biol Cell 13: 3560–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Yang CS, Pickett CB (2004) The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic Biol Med 37: 433–441 [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems AR, Tyers M, Sicheri F (2003) Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- Patton EE, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D (2000) SCFMet30-mediated control of the transcriptional activator Met4 is required for the G1–S transition. EMBO J 19: 1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M (1998) Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev 12: 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pocsi I, Prade RA, Penninckx MJ (2004) Glutathione, altruistic metabolite in fungi. Adv Microb Physiol 49: 1–76 [DOI] [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D (2000) Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J 19: 292–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Schumacher MM, Choi JY, Voelker DR (2002) Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J Biol Chem 277: 51033–51042 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284: 662–665 [DOI] [PubMed] [Google Scholar]
- Stewart D, Killeen E, Naquin R, Alam S, Alam J (2003) Degradation of transcription factor Nrf2 via the ubiquitin–proteasome pathway and stabilization by cadmium. J Biol Chem 278: 2396–2402 [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61: 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Sewell AK, McCowen CL, Winge DR (1993) Regulation of metallothionein genes by the ACE1 and AMT1 transcription factors. J Biol Chem 268: 12512–12518 [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA 101: 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 35: 238–245 [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Trotter EW, Dawes IW, Grant CM (2003) Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J Biol Chem 278: 49920–49928 [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochem Biophys Acta 1695: 133–170 [DOI] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP (2003) Structure of a beta-TrCP1–Skp1–beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 11: 1445–1456 [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]


