Abstract
Alternative pre-mRNA splicing affects a majority of human genes and plays important roles in development and disease. Alternative splicing (AS) events conserved since the divergence of human and mouse are likely of primary biological importance, but relatively few of such events are known. Here we describe sequence features that distinguish exons subject to evolutionarily conserved AS, which we call alternative conserved exons (ACEs), from other orthologous human/mouse exons and integrate these features into an exon classification algorithm, acescan. Genome-wide analysis of annotated orthologous human–mouse exon pairs identified ≈2,000 predicted ACEs. Alternative splicing was verified in both human and mouse tissues by using an RT-PCR-sequencing protocol for 21 of 30 (70%) predicted ACEs tested, supporting the validity of a majority of acescan predictions. By contrast, AS was observed in mouse tissues for only 2 of 15 (13%) tested exons that had EST or cDNA evidence of AS in human but were not predicted ACEs, and AS was never observed for 11 negative control exons in human or mouse tissues. Predicted ACEs were much more likely to preserve the reading frame and less likely to disrupt protein domains than other AS events and were enriched in genes expressed in the brain and in genes involved in transcriptional regulation, RNA processing, and development. Our results also imply that the vast majority of AS events represented in the human EST database are not conserved in mouse.
Keywords: exon skipping, regulatory element, cassette exon, transcriptome, comparative genomics
The processing of human primary transcripts to produce the mRNAs that will direct protein synthesis is often variable, producing multiple alternatively spliced (AS) mRNA products, most commonly by alternative inclusion or exclusion (“skipping”) of individual exons (1–3). Alternative pre-mRNA splicing plays a major role in expanding protein diversity and regulating gene expression in higher eukaryotes (4, 5). Regulated AS is crucial in fruit fly development (3) and in the physiology of the heart, skeletal muscle, brain, and other tissues, and misregulation of AS is associated with human disease (6–8).
EST and cDNA sequence databases provide a rich source of information about splicing events occurring in the human and mouse transcriptomes. Considering the set of human ESTs and cDNAs that can be reliably aligned to a human gene locus overlapping a particular exon, this set can be subdivided into transcripts that include the exon and those that exclude, or skip, the exon in question. Here, the skipping of an exon refers to the situation in which a transcript aligns consecutively to an upstream exon and a downstream exon of a gene, omitting the given exon. This consideration can be applied to all of the exons in a human gene, and an analogous subdivision can be made of the mouse transcripts that align to exons of the orthologous mouse gene. Each orthologous human/mouse exon pair can then be assigned to one of four categories, SH,m, Sh,M, SH,M, or Sh,m, depending on whether exon skipping has been observed only in human transcripts (SH,m), only in mouse (Sh,M), in both human and mouse (SH,M), or not observed in either species (Sh,m).
By using publicly available EST databases totaling over 5 million human and over 3 million mouse ESTs and databases of ≈94,000 and ≈91,600 human and mouse cDNAs, respectively, thousands of alternative exons can be inferred in each species. However, the overlap between these sets is relatively small; i.e., for only ≈240 (≈1 in 18) of the ≈4,500 conserved human–mouse exons observed to be skipped in human was transcript evidence found supporting alternative usage (skipping) of the orthologous mouse exon, as discussed below (9–11). This observation raises the question of how many of the AS events observable in the human transcriptome are evolutionarily conserved and, therefore, presumably contribute to organismal fitness and how many are aberrant, disease- or allele-specific, or highly lineage-restricted events, which may or may not affect fitness. Although study of the latter types of events may lead to important insights and applications, a significant fraction of these events may constitute biochemical “noise” or transient evolutionary fluctuations. On the other hand, conservation of a specific pattern of AS over the ≈90 million years since divergence of the mouse and human lineages provides strong evidence of biological function. Therefore, defining the set of AS events conserved between human and mouse is of primary interest in efforts to understand the biological importance of splicing regulation.
Alternative inclusion/exclusion of exons is known to be influenced by a number of factors, such as intron length, exon length, splice site strength and pre-mRNA secondary structure (1, 3, 12). Certain cis-regulatory elements, including exonic splicing enhancers (ESEs), intronic splicing enhancers, exonic splicing silencers (ESSs), and intronic splicing silencers can also control exon skipping by recruiting trans-acting splicing factors (4, 13). Computational studies have identified other sequence features that differ between skipped exons (also known as cassette exons) and constitutive exons in human and mouse genes, including increased conservation in the introns flanking exons skipped in human and mouse (9, 10, 14–16). These observations motivated us to systematically identify, characterize, and integrate sequence features into a classifier that could be used to identify exons subject to evolutionarily conserved exon skipping, here termed alternative conserved exons (ACEs).
Materials and Methods
Regularized Least-Squares Classification. The regularized least-squares classifier was used to learn the features from SH,M and Sh,m exons and to derive a real-valued output for unlabeled conserved exon pairs. The regularized least-squares classifier has a quadratic loss function and requires the solution of a single system of linear equations, (K + λLW–1) c = y, in matrix notation. The goal is to obtain an optimal vector c, defined as c = [c1... cL]T, where L is the size of the training set, K is the L × L kernel matrix, λ is the tradeoff between generalization and over-fitting, W is the diagonal matrix of penalties wi (equal to β for positive examples and equal to 1 for negative examples), and y is the column vector of labels (+1,–1). The algorithm, cross-validation, sampling, and performance measures are described in further detail in Supporting Text, which is published as supporting information on the PNAS web site.
Experimental Validation. The SuperScript III First-Strand synthesis system for RT-PCR (Invitrogen) was used to generate cDNAs from normal human (fetal brain, fetal liver, cerebellum, heart, whole brain, prostate, liver, lung, kidney, bone marrow, skeletal muscle, and testis) and normal mouse (embryonic mix, whole brain, kidney, skeletal muscle, liver, lung, heart, and testis) tissues by using oligo(dT) primers. The TaqDNA polymerase kit (Invitrogen) was used with primers targeted to exons flanking candidate ACEs (further details are given in Supporting Text). PCR products of the expected size were gel-purified with the QIAquick Gel Extraction kit (Qiagen, Valencia, CA) and sequenced.
Results and Discussion
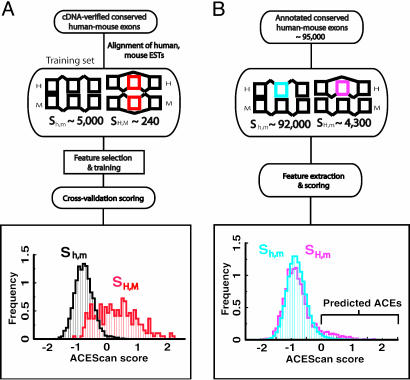
Outline of Strategy for Identification of ACEs. Our scheme for identifying ACEs consisted of three phases: learning, prediction, and validation (Fig. 1). In the learning phase, a set of sequence features was identified, including exon and intron length, splice site strength, sequence conservation, and region-specific oligonucleotide composition, which differed between training sets of 241 exons of the class SH,M and ≈5,000 exons of the class Sh,m defined above (Fig. 2). For training purposes, exons of the Sh,m class were chosen from genes containing at least one other exon with evidence for AS, because genes lacking AS may experience differing selective pressures than AS genes (17). Next, these features were incorporated into a discriminant classifier, acescan, which was used in the prediction phase to predict which of ≈96,000 annotated orthologous human/mouse exon pairs not previously known to exhibit conserved AS are, in fact, ACEs. Finally, in the validation phase, a subset of candidate exons with positive acescan scores (designated acescan[+] exons) was chosen for experimental testing, together with two sets of negative control exons with negative acescan scores (acescan[–] exons): one set with previous transcript evidence for exon skipping in human (SH category) and one set lacking such evidence (Sh category).
Fig. 1.
Schematic overview of the learning and prediction stages of the acescan procedure. (A) Learning. Sequence features that differed between sets SH,M and Sh,m were identified as described (Supporting Text). Random subsets of SH,M and Sh,m were used to train the acescan algorithm, and cross-validation scores were calculated for the unseen subsets of SH,M and Sh,m. The cross-validated acescan score distributions for SH,M (red) and Sh,m (black) are shown. (B) Prediction. Spliced alignments of transcript sequences were used to assign ensembl-annotated exons from ≈10,000 human–mouse orthologous gene pairs (not necessarily alternatively spliced) to one of two sets: SH,m and Sh,m. acescan score distributions for SH,m (pink) and Sh,m (blue) are shown.
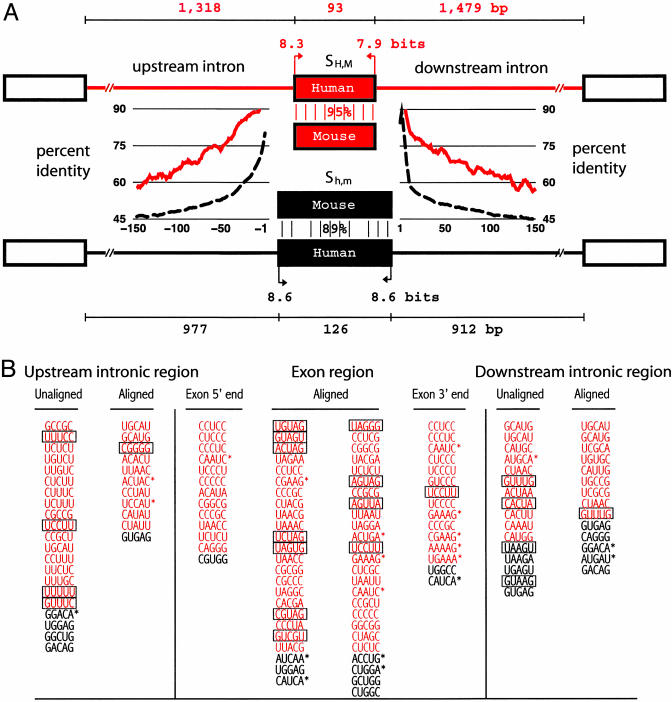
Fig. 2.
Sequence features that differ between conserved alternative and constitutive human–mouse exons. (A) Features typical of exons of the SH,M (alternatively spliced) and Sh,m (constitutive) training sets are depicted. SH,M exons had shorter median exon length (93 versus 126 bases, P < 10–22), longer upstream intron length (P < 0.005), longer downstream intron length (P < 10–5), weaker 5′ and 3′ splice site scores [P < 10–5 and P < 0.02, respectively; maxentscan (http://genes.mit.edu)], higher exon sequence conservation (percent identity; P < 10–46), and higher conservation (clustal w alignment score) in the 150-base intron regions immediately upstream and downstream of the exon (P < 10–63 and P < 10–66, respectively). For each feature, the Kolmogorov–Smirnov test was used to test the null hypothesis of independent samples drawn from the same underlying population. Length and splice site score values are shown for human exons/introns; mouse values were similar. Average percent identity for alignments of flanking intron regions are shown in a 9-base sliding window for SH,M (red trace) and Sh,m (black dashed trace) exons. (B) Pentanucleotides used by acescan. Overrepresented (red) and underrepresented (black) pentamers in exon or 150-base flanking intron regions of SH,M versus Sh,m exons. Pentamer frequencies were analyzed separately for clustal w-aligned regions only (aligned) or the entire region (unaligned). Exon 5′ and 3′ ends refer to the first and last 100 bases of exon, respectively. Boxed oligonucleotides indicate overlap with ESS hexamers (20), and oligonucleotides with asterisks indicate overlap with RESCUE-ESE hexamers (19).
The following features were initially incorporated into acescan: (i) exon length, (ii) upstream intron length, (iii) downstream intron length, (iv)5′ splice site score, (v)3′ splice site score, (vi) nucleotide percent identity between orthologous human and mouse exons, (vii) human–mouse intronic sequence conservation within the last 150 bases upstream, and (viii) human–mouse intronic sequence conservation within the first 150 bases downstream of the exon. In general, exon pairs skipped in both human and mouse (set SH,M) were observed to be shorter than unskipped exon pairs (Sh,m), were flanked by longer upstream and downstream introns, and possessed significantly weaker splice sites (Fig. 2). Strikingly, exon pairs in SH,M have significantly higher sequence identity and higher flanking intronic conservation as compared with exon pairs in Sh,m (Fig. 2). High levels of sequence conservation in the exons and flanking introns is suggestive of conservation of regulatory motifs or RNA structure. These observations are similar to and consistent with previous studies (10, 14–16).
Oligonucleotides Useful in Discrimination of ACEs. Oligonucleotide features designed to score potential cis-regulatory elements consisted of the highest-ranking (most biased) overrepresented and underrepresented oligonucleotides of length k (k-mers) in different exon and intron regions. The regions considered were the first and last 100 bases of exons and the proximal 150 bases in the upstream and downstream introns flanking the exon, because of the high levels of sequence conservation in these regions and their proximity to the regulated splice junctions. Counts of conserved oligonucleotides in human–mouse nucleotide alignments of the 150 bases of upstream and downstream intronic sequence and in the entire exon were scored for enrichment in the SH,M set versus the Sh,m set. Inclusion of oligonucleotide counts from aligned and unaligned sequences permits scoring of cis-regulatory elements that do and do not require strict spatial constraints for function.
Oligonucleotides were ranked by their enrichment as measured by a χ2 value. Several of the overrepresented intron elements were similar to known intronic regulatory elements (e.g., UGCAUG and UC-rich repeats; Table 1, which is published as supporting information on the PNAS web site, and Supporting Text). We propose that a significant fraction of the remaining elements may represent previously uncharacterized intronic regulatory sequences. A number of the overrepresented and underrepresented exon elements (Fig. 2) were similar to ESE hexamers (Table 2, which is published as supporting information on the PNAS web site) identified by using the RESCUE-ESE method or systematic evolution of ligands by exponential enrichment (SELEX) (18, 19) or to ESS elements identified through a recent cell fluorescence-based in vivo screen for splicing silencers (20). The relative distribution of these elements suggests that ACEs may have a higher density of ESS sequences than constitutive exons, which would tend to facilitate exclusion by the splicing machinery. The increased frequency of ESS sequences in ACEs relative to constitutive exons might reflect differing selective pressures, with constitutive exons presumably being under selection for efficient exon inclusion, whereas alternative exons are presumably selected for inefficient inclusion under at least some conditions (e.g., in specific cell types or developmental stages).
Integration and Selection of Features for Accurate Exon Classification. The task of integrating the general features and oligonucleotide features described above into an algorithm that distinguishes exon pairs in SH,M (positively labeled) from those in Sh,m (negatively labeled) was posed as a supervised binary classification problem. We adapted a regularized least-squares classifier, which finds the optimal separating hyperplane in a high-dimensional space that distinguishes two classes of samples (21). Because it was not known apriori which of the 8,245 general and oligonucleotide features were most important in the classification scheme, models using different combinations of the eight general features and the region-specific oligonucleotide features were compared, and a feature selection protocol was used to reduce the number of parameters and to retain only the most relevant oligonucleotide features.
To determine the optimal features and parameters for the classifier, the training data were used to generate several models by varying (i) the choice of general features, (ii) the exon or intron regions from which oligonucleotide features were generated, and (iii) the number of most discriminative oligonucleotide features included. The model with the best performance used all of the general sequence features and 240 oligonucleotides with lengths of 4 and 5 bases (shown in Fig. 5, which is published as supporting information on the PNAS web site). This model assigned correct labels to ≈90 exon pairs for every 100 exon pairs drawn equally likely from SH,M and Sh,m. For an individual exon, the acescan score was defined as the mean of the classifier outputs over 50 random samplings of the training data. The distribution of acescan scores for the exon pairs in SH,M ranged from approximately –0.8 to 2.0 (arbitrary units), compared to a range of approximately –1.8 to 0 for most of the exons in Sh,m (Fig. 1). At a cutoff score of zero, only ≈2% of Sh,m exons had positive acescan scores, compared with ≈61% of the exons in SH,M, suggesting that acescan[+] exon pairs are highly enriched for ACEs.
Experimental Validation of Conserved AS for 21 of 30 acescan[+] Exon Pairs. A combination of experimental tests and bioinformatic approaches was used to explore the features of acescan[+] and acescan[–] exon pairs. First, the splicing patterns of a set of 30 arbitrarily chosen acescan[+] exons were tested in a battery of human and mouse tissues by RT-PCR with primers targeted to flanking exons. acescan[+] exons were selected from four intervals: I1 (acescan score range, 0.0–0.5); I2 (acescan score range, 0.5–1.0); I3 (acescan score range, 1.0–1.5); and I4 (acescan score, >1.5), spanning the range of scores of most SH,M exons. Panels of 12 normal human tissues and 8 normal mouse tissues were assayed. To avoid the undesired detection of aberrant or disease-specific splicing, tumor or other diseased tissues were not used. The products of these 600 RT-PCRs (30 exons × 20 tissues) were analyzed by gel electrophoresis, and the identities of PCR products with expected sizes for mRNAs including or excluding the test exon were confirmed by sequencing (Fig. 3A). In all, four of nine, seven of eight, six of eight, and four of five candidate ACEs in intervals I1, I2, I3 and I4, respectively, were observed to undergo skipping in both human and mouse, whereas, for another two exons (both from interval I1), exon skipping was observed only in human tissues (Fig. 3; complete results are shown in Table 3, which is published as supporting information on the PNAS web site). Thus, of 30 predicted ACEs interrogated by RT-PCR, 21 were observed to be skipped in human and mouse tissues, and high rates of validation of AS were seen in all four score intervals. These data support the presence of conserved AS in a majority of acescan[+] exons. Although the 30 acescan[+] candidates had no previous transcript evidence for skipping, searches of the literature and low-stringency searches of the cDNA and EST databases (August, 2004) identified possible evidence for a fraction of the AS events observed by RT-PCR, most often consisting of a single EST in only one species. In the examples studied, exon skipping was observed in many different combinations of human and mouse tissues, suggesting that many of the features used by acescan are characteristic of skipped exons generally, regardless of tissue specificity. Variations in tissue specificity of AS were observed between human and mouse for several tested exons. However, a general tendency to conserve exon skipping in corresponding tissues was apparent, e.g., 9 of 10 predicted ACEs observed to be skipped in human whole brain or cerebellum were also skipped in mouse brain tissue (Table 3).
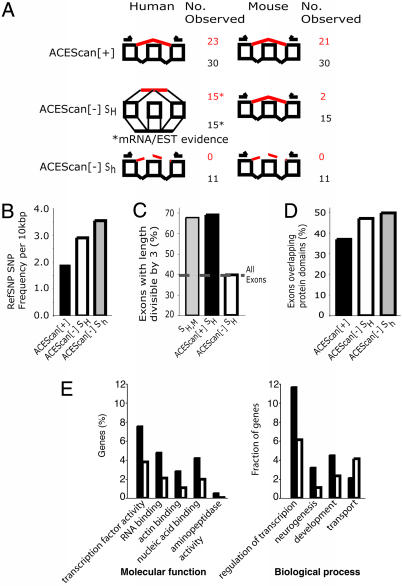
Fig. 3.
Validation and analysis of acescan[+] predictions. (A) Experimental validation by means of RT-PCR and sequencing of subsets of candidate acescan[+] exons and negative control acescan[–] exons in panels of normal human and mouse tissues with primers in flanking exons. Graphical representations of splicing patterns (inclusion/exclusion) and the number of exon pairs observed to be excluded and included are designated in red and black, respectively. The three randomly selected subsets tested were (i)30 acescan[+] exon pairs; (ii) as negative controls, 15 acescan[–]SH exon pairs (with EST/cDNA evidence for inclusion and exclusion of the human exon indicated by horizontal lines representing spliced transcripts); and (iii)11 acescan[–]Sh exon pairs (with no transcript evidence for skipping in either human or mouse). (B) SNP density in acescan[+], acescan[–]SH, and acescan[–]Sh exons. The number of stringently filtered SNPs per 10,000 bases was computed for each exon set. (C) Fraction of SH,M exons, acescan[+]SH exons, and acescan[–]SH exons that had lengths that were multiples of three and the background fraction of frame-preserving constitutive exons. (D) Analysis of protein domain preservation of acescan[+], acescan[–]SH, and acescan[–]Sh exons that maintain reading frame (i.e., length divisible by three). Maximum exon size cutoffs (150, 110, and 108 bases for acescan[+], acescan[–]SH, and acescan[–]Sh exons, respectively) were used to avoid exon length biases. The median length of exons in each subset was 84 bases, with no significant difference in the distribution of sizes among the sets (by a Kruskal–Wallis nonparametric test). The minimum number of exonic bases overlapping the protein domain was set to 30 bases. (E) GO “molecular function” and “biological process” categories, which differed significantly (P < 0.05), in the representation between genes containing predicted ACEs (black bars) and genes not containing predicted ACEs (white bars) are shown. Statistical significance was assessed by using χ2 statistics with Bonferroni correction for multiple hypothesis testing. GO categories are ordered from right to left in order of increasingly significant bias toward genes containing predicted ACEs. Only one category (transport) was significantly biased toward genes without predicted ACEs.
Low Detection of Conserved AS for acescan[–] Exon Pairs. As a negative control, 11 acescan[–] exon pairs from the set Sh were chosen from the five score intervals, C1 (–0.5 to 0), C2 (–1.0 to –0.5), C3 (–1.5 to –1.0), C4 (–2.0 to –1.5), and C5 (less than –2.0), with at least one pair per interval. By using the same RT-PCR sequencing assay and the same sets of human and mouse tissues, we did not observe exon skipping for any of the 11 negative control exons in any of the 12 human or 8 mouse tissues studied (Table 3). Thus, considering the human and mouse exons tested, exon skipping was detected for 44 of 60 acescan[+] exons (including 21 orthologous pairs), compared with 0 of 22 acescan[–] exons, a highly significant difference (P < 0.0001, Fisher exact test). Of course, for either group of exons, failure to detect exon skipping by our RT-PCR assay is not proof that exon skipping does not occur, and some exons not skipped in the tissues studied might be skipped in other untested tissues. However, low-stringency searches of the August 2004 human and mouse EST databases failed to detect any evidence of skipping of the 11 acescan[–] exons tested.
As a second type of negative control, an arbitrary set of 15 acescan[–] exon pairs was chosen from the score intervals C2–C4, with the added requirement that transcript evidence of exon skipping was present for the human member of each exon pair. By using the same RT-PCR sequencing assay in the same set of eight mouse tissues as above, we detected exon skipping for only 2 of the 15 mouse exons tested, suggesting that a substantial majority of these exon pairs are not ACEs. To explore the potential biological roles of the 13 remaining exons that undergo possible human-specific AS, we examined the tissue sources of the transcripts that showed exon skipping. In 9 of the 13 cases, these transcripts derived exclusively from cancer cell lines or diseased tissues, suggesting that many of these exons may be skipped primarily in disease states rather than in normal human tissues. The difference in the rate of RT-PCR validation of exon skipping in mouse tissues for the acescan[+] exons tested (21 of 30, 70%), relative to the acescan[–] exons tested (2 of 26, ≈8%), was also highly significant (P < 0.002, Fisher exact test), demonstrating the power of acescan to discriminate evolutionarily conserved AS exons from those that are either constitutively spliced or skipped in a species-specific (or disease-specific) manner.
Many Literature-Derived AS Events Correspond to acescan[+] Exons. The principle that important regulatory elements are usually evolutionarily conserved is well established and forms the basis of a number of successful comparative genomics approaches for identifying such elements (22). To explore the extent to which this principle applies to AS events, we extracted known exon skipping events from the Manually Annotated Alternatively Spliced Events (MAASE) database (23), representing AS events that are curated from published works. A total of 29 exon skipping events in mouse were identified from this database, for which both the human and mouse orthologous exons were available. Strikingly, almost all of the extracted exons had acescan scores greater than –0.5 (28 of 29), and 62% (18 of 29) were acescan[+]. Thus, although small in scale, this analysis of published AS events suggests that a majority of interesting (i.e., interesting enough to be described in the scientific literature) exon skipping events are acescan[+] and, therefore, that most such events are conserved between human and mouse (Table 4, which is published as supporting information on the PNAS web site).
Approximately 11% of EST-Derived AS Events Are Likely to Be Evolutionarily Conserved. Of the ≈4,300 exon pairs with transcript evidence of skipping in human but not mouse (class SH,m), only ≈7% had positive acescan scores (Fig. 1). Together with the observation that ≈61% of SH,M exons were acescan[+], this low fraction suggests that for only ≈11% (0.07/0.61) of the SH,m exons is AS likely to be conserved in mouse. Thus, a surprising implication of these data is that the vast majority of the AS events inferable from human EST/cDNA-genomic alignments are not evolutionarily conserved in mouse. Instead, most of these events may represent aberrant, disease-specific, or allele-specific splicing (24) or events for which phylogenetic distribution is highly restricted.
Functional Differences Between acescan[+] and acescan[–] Exons. To assess potential functional differences between acescan[+] and acescan[–] exons that either have or do not have EST or cDNA evidence of exon skipping in human, we analyzed the density of SNPs and the frequency of reading frame preservation and protein domain disruption for each of these three classes of exon. Selective pressure on nucleotide sequence was assayed by mapping stringently filtered reference SNPs onto exons that had been scored by acescan (Fig. 3B). This analysis found a ≈50% higher density of SNPs in acescan[–] SH exons than in acescan[+] exons (this difference is significant at P < 10–5, χ2 test), suggesting that ACEs have been under much more stringent selection to conserve nucleotide sequence in recent human evolution than other exons. By contrast, acescan[–] SH exons appear to have experienced a degree of selection that was more similar to constitutive exons than to ACEs.
Further evidence for the functional roles of many acescan[+] SH exons came from the observation that a far higher fraction of these exons had lengths that were multiples of three (68%, comparable with that seen in the training set of SH,M exons) than was seen for acescan[–] SH exons, for which only ≈43% had lengths divisible by three, near background levels for constitutive internal exons (Fig. 3C). This difference is highly significant (P < 10–15, χ2 test) and implies the existence of strong selection on the alternative protein products derived from alternative splicing of acescan[+] exons. Notably, divisibility of the exon length by three was not used in the predictions (only the general size of the exon, with shorter lengths favored over longer lengths).
The frequency of disruption or removal of a protein domain by AS has been studied by several groups (e.g., refs. 25–27). We found that only ≈37% of acescan[+] exons overlapped ORF regions encoding interpro-annotated protein domains by 30 bases (10 codons) or more, a significantly lower fraction than for acescan[–] exons studied of either the SH or Sh classes (Fig. 3D), both of which had similar frequencies of domain disruption (≈50%). Reducing the minimum overlap to 15 bases gave similar results (data not shown). This finding is generally consistent with the results of Kriventseva et al. (26), who observed that protein isoforms arising from AS are more likely to preserve protein domain structure than is expected by chance. Taken together, the data shown in Fig. 3 consistently demonstrate that acescan[+] exons are under strong selection to conserve function, both at the nucleotide level (Fig. 3B) and at the level of the encoded alternative protein isoform (Figs. 3 C and D). In contrast, acescan[–] exons show less evidence of selective constraints at the nucleotide level (Fig. 3B), and there is little if any evidence of additional constraints on the protein products derived from exon skipping of acescan[–] exons, even when there is transcript evidence that such skipping occurs (Figs. 3 C and D).
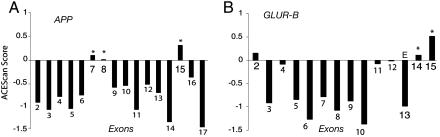
Applications of acescan at the Gene Level. Application of acescan to well studied genes illustrates some of the strengths and limitations of our approach (APP and GLUR-B shown in Fig. 4; PTB and CACNA1G shown in Fig. 6, which is published as supporting information on the PNAS web site). Of the identifiable orthologous human/mouse exon pairs in these genes, known exon skipping events (asterisks in Fig. 4) all received positive acescan scores, implying that their skipping is likely to be conserved in mouse. Skipping of exons 7 and 8 of the β-amyloid precursor protein gene (APP) implicated in Alzheimer's disease was detected successfully in a recent large-scale microarray analysis of AS in human tissues (28). These exons, as well as exon 15 of the APP gene, received positive acescan scores (Fig. 4A); all three of these exons are known to undergo exon skipping (29, 30). The GLUR-B gene, one of the four GluR subunits that assemble to form the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid glutamate receptor, contains two well known skipped exons (flip and flop, exons 14 and 15, respectively), both of which received positive acescan scores, as well as an exon (exon 13, which is marked with an E in Fig. 4) that undergoes RNA editing (31). This edited exon and the downstream intron form an RNA hairpin and are highly conserved in sequence (31). Despite this high level of exonic and intronic sequence conservation, this exon received a negative acescan score (Fig. 4B), providing an example of the specificity of our method for AS exons. A web server has been set up (http://genes.mit.edu/acescan) that provides access to the training sets of all acescan plots for ensembl-annotated orthologous human/mouse gene pairs.
Fig. 4.
acescan scores for internal exons of well known alternatively spliced genes. Known alternative exons are indicated by asterisks; the known RNA edited exon of GLUR-B is indicated by the letter E. The following known AS exons are illustrated: exons 7 (168 bases), 8 (57 bases) and 15 (54 bases) of the human β-amyloid precursor protein precursor gene (APP, ensembl Gene ID ENSG00000142192) (A) and exons 14 (115 bases) and 15 (249 bases) of the human glutamate receptor, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid 2 gene (GLUR-B, ensembl Gene ID ENSG00000120251) (B).
Recently, Bejerano et al. (32) reported 111 exonic ultraconserved regions of ≥200 bases with 100% sequence identity between the human and mouse genomes, most of unknown function. Comparing these with our predicted ACEs, 33 of the 37 ultraconserved regions (≈89%) that mapped to internal exons that could be scored by acescan received positive acescan scores, suggesting that a number of these elements correspond to ACEs.
Functional Characteristics of acescan[+] Genes. In total, 1,550 genes were identified, containing 2,092 acescan[+] exons, ≈85% of which lacked prior transcript (EST/cDNA) evidence for exon skipping. Initial comparisons to the partially annotated rat genome showed a high correlation between human–mouse and human–rat acescan scores, as expected (data not shown). To determine whether genes that contain acescan[+] exons, which we refer to as acescan[+] genes, are biased toward particular biological activities, we compared these genes to the set of genes not found to contain any acescan[+] exons (acescan[–] genes) by using Gene Ontology (GO) classifications (www.geneontology.org), as in refs. 32 and 33. The results showed that acescan[+] genes are enriched for transcription factors and aminopeptidase activity and for the “actin-binding,” “RNA-binding,” and “nucleic acid-binding” GO molecular function categories (Fig. 3E). In terms of GO biological process categories, acescan[+] genes were more likely to be involved in transcriptional regulation, neurogenesis, and development and less likely to be involved in transport than acescan[–] genes. Only slight biases in GO category representation were present in the training set of SH,M genes (Fig. 7, which is published as supporting information on the PNAS web site). Closer examination of the acescan[+] genes that encode RNA-binding factors identified acescan[+] exons in genes encoding many of the heterogeneous nuclear ribonucleoproteins, a majority of which (including PTB) are candidates for nonsense-mediated mRNA decay, suggesting frequent regulation of expression level through regulated AS in this gene family (Fig. 6 and Table 5, which is published as supporting information on the PNAS web site).
To explore the expression patterns of genes containing predicted ACEs, we used microarray data from the Gene Atlas survey of 47 diverse human tissues and cell lines (34). Overwhelmingly, acescan[+] genes were more likely to be differentially expressed in a spectrum of nervous system tissues, including spinal cord and fetal and adult whole brain, and in several brain regions, compared with acescan[–] genes (Fig. 8, which is published as supporting information on the PNAS web site). Only two cell lines (both ovarian) of the 47 tissue/cell lines studied exhibited similar biases. These results imply an unusually high frequency of conserved AS events in the brain.
While this work was in progress, two other groups have demonstrated that conserved sequence features can be used to identify alternative exons in fruit fly (35) and human genes (14, 36). Our computational approach differs in a number of important ways: (i) acescan associates a real-valued score to orthologous human–mouse exon pairs, rather than associating a binary label to an exon pair, which grants much greater flexibility in adjusting the algorithm's sensitivity/specificity compared to the methods used in refs. 14 and 35. (ii) acescan does not use the length of the exon modulo three in its predictions (14, 36). This generality allows us to assess the degree of selection on ACEs to preserve protein reading frame (Fig. 3C) rather than assuming that reading frame must always be preserved, and it enables acescan to identify the subset of ACEs that create mRNAs that encode truncated proteins or that are subject to nonsense-mediated mRNA decay, an emerging class of regulated AS events (37). Supporting the validity of this subset of predictions, approximately half of the ACEs validated by our RT-PCR sequencing protocol had lengths that were not divisible by three (Fig. 3A and Table 3). (iii) A much larger set of discriminatory features was used in acescan, including oligonucleotide features (compared with refs. 14 and 35), many of which are likely to represent splicing regulatory elements, and inclusion of these features enhanced the performance of our algorithm (36). Experimental validation of predicted AS exons and negative control exons is important in providing estimates for the reliability and accuracy of any computational approach. A comparison of sensitivity and specificity based on experimental validation demonstrates that acescan has higher accuracy than previously published approaches (Table 6, which is published as supporting information on the PNAS web site, compares computational approaches and the extent of experimental validation). Finally, the accuracy and relatively large numbers of ACEs predicted by acescan allow us to identify functional and expression biases in the set of genes containing high-confidence ACEs.
Comparative genomics, machine-learning techniques, and rigorous experimental validation have facilitated the accurate prediction of ≈2,000 ACEs (Table 7, which is published as supporting information on the PNAS web site). The predictive power of acescan can likely be improved in the future through the use of larger training sets of known ACEs, improved genome assemblies and annotations, and by incorporating tiling array and/or splice junction array data. The set of predicted ACEs holds the potential for further elucidating the roles of AS in modulating the expression of mammalian genomes.
Supplementary Material
Acknowledgments
We thank P. Sharp and Z. Wang for helpful discussions. This work was supported by National Science Foundation Grant 0218506 (to C.B.B.), a grant from the National Institutes of Health (to C.B.B.), and a Lee Kuan-Yew Graduate Fellowship from Singapore (to G.W.Y.).
Abbreviations: AS, alternative splicing; ACE, alternative conserved exon; ESE, exonic splicing enhancer; ESS, exonic splicing silencer; GO, Gene Ontology.
References
- 1.Black, D. L. & Grabowski, P. J. (2003) Prog. Mol. Subcell. Biol. 31, 187–216. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis, T. & Tasic, B. (2002) Nature 418, 236–243. [DOI] [PubMed] [Google Scholar]
- 3.Lopez, A. J. (1998) Annu. Rev. Genet. 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 4.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 5.Black, D. L. (2000) Cell 103, 367–370. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F. & Kornblihtt, A. R. (2002) Trends Genet. 18, 186–193. [DOI] [PubMed] [Google Scholar]
- 7.Musunuru, K. (2003) Trends Cardiovasc. Med. 13, 188–195. [DOI] [PubMed] [Google Scholar]
- 8.Faustino, N. A. & Cooper, T. A. (2003) Genes Dev. 17, 419–437. [DOI] [PubMed] [Google Scholar]
- 9.Thanaraj, T. A., Clark, F. & Muilu, J. (2003) Nucleic Acids Res. 31, 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorek, R. & Ast, G. (2003) Genome Res. 13, 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurtdinov, R. N., Artamonova, I. I., Mironov, A. A. & Gelfand, M. S. (2003) Hum. Mol. Genet. 12, 1313–1320. [DOI] [PubMed] [Google Scholar]
- 12.Bell, M. V., Cowper, A. E., Lefranc, M. P., Bell, J. I. & Screaton, G. R. (1998) Mol. Cell. Biol. 18, 5930–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladd, A. N. & Cooper, T. A. (2002) Genome Biol. 3, reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorek, R., Shemesh, R., Cohen, Y., Basechess, O., Ast, G. & Shamir, R. (2004) Genome Res. 14, 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufmann, D., Kenner, O., Nurnberg, P., Vogel, W. & Bartelt, B. (2004) Eur. J. Hum. Genet. 12, 139–149. [DOI] [PubMed] [Google Scholar]
- 16.Sugnet, C. W., Kent, W. J., Ares, M., Jr., & Haussler, D. (2004) Pac. Symp. Biocomp. 9, 66-77. [DOI] [PubMed] [Google Scholar]
- 17.Iida, K. & Akashi, H. (2000) Gene 261, 93–105. [DOI] [PubMed] [Google Scholar]
- 18.Liu, H. X., Zhang, M. & Krainer, A. R. (1998) Genes Dev. 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. (2002) Science 297, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 20.Wang, Z., Rolish, M. E., Yeo, G., Tung, V., Mawson, M. & Burge, C. B. (2004) Cell 119, 831–845. [DOI] [PubMed] [Google Scholar]
- 21.Rifkin, R., Yeo, G. & Poggio, T. (2003) in Advances in Learning Theory: Methods, Model, and Applications, ed. Suykens, J. A. K., Horvath, G., Basu, S., Micchelli, C. & Vandewalle, J. (IOS, Amsterdam), Vol. 190, pp. 131–154. [Google Scholar]
- 22.Loots, G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- 23.Zheng, C. L., Nair, T. M., Gribskov, M., Kwon, Y. S., Li, H. R. & Fu, X. D. (2004) Pac. Symp. Biocomput. 9, 78–88. [DOI] [PubMed] [Google Scholar]
- 24.Nembaware, V., Wolfe, K. H., Bettoni, F., Kelso, J. & Seoighe, C. (2004) FEBS Lett. 577, 233–238. [DOI] [PubMed] [Google Scholar]
- 25.Xing, Y., Xu, Q. & Lee, C. (2003) FEBS Lett. 555, 572–578. [DOI] [PubMed] [Google Scholar]
- 26.Kriventseva, E. V., Koch, I., Apweiler, R., Vingron, M., Bork, P., Gelfand, M. S. & Sunyaev, S. (2003) Trends Genet. 19, 124–128. [DOI] [PubMed] [Google Scholar]
- 27.Cline, M. S., Shigeta, R., Wheeler, R. L., Siani-Rose, M. A., Kulp, D. & Loraine, A. E. (2004) Pac. Symp. Biocomput. 9, 17–28. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- 29.Ponte, P., Gonzalez-DeWhitt, P., Schilling, J., Miller, J., Hsu, D., Greenberg, B., Davis, K., Wallace, W., Lieberburg, I. & Fuller, F. (1988) Nature 331, 525–527. [DOI] [PubMed] [Google Scholar]
- 30.Konig, G., Monning, U., Czech, C., Prior, R., Banati, R., Schreiter-Gasser, U., Bauer, J., Masters, C. L. & Beyreuther, K. (1992) J. Biol. Chem. 267, 10804–10809. [PubMed] [Google Scholar]
- 31.Cha, J. H., Kinsman, S. L. & Johnston, M. V. (1994) Brain Res. Mol. Brain Res. 22, 323–328. [DOI] [PubMed] [Google Scholar]
- 32.Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W. J., Mattick, J. S. & Haussler, D. (2004) Science 304, 1321–1325. [DOI] [PubMed] [Google Scholar]
- 33.Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. (2003) Cell 115, 787–798. [DOI] [PubMed] [Google Scholar]
- 34.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipps, D. L., Park, J. W. & Graveley, B. R. (2004) RNA 10, 1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dror, G., Sorek, R. & Shamir, R. (2005) Bioinformatics, in press. [DOI] [PubMed]
- 37.Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A. & Smith, C. W. (2004) Mol. Cell 13, 91–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.