Abstract
Myxoid liposarcoma (MLS) is a soft tissue sarcoma characterized by a recurrent t(12;16) translocation. Although tumors are initially radio- and chemosensitive, the management of inoperable or metastatic MLS can be challenging. Therefore, our aim was to identify novel targets for systemic therapy. We performed an in vitro high-throughput drug screen using three MLS cell lines (402091, 1765092, DL-221), which were treated with 273 different drugs at four different concentrations. Cell lines and tissue microarrays were used for validation. As expected, all cell lines revealed a strong growth inhibition to conventional chemotherapeutic agents, such as anthracyclines and taxanes. A good response was observed to compounds interfering with Src and the mTOR pathway, which are known to be affected in these tumors. Moreover, BIRC5 was important for MLS survival because a strong inhibitory effect was seen at low concentration using the survivin inhibitor YM155, and siRNA for BIRC5 decreased cell viability. Immunohistochemistry revealed abundant expression of survivin restricted to the nucleus in all 32 tested primary tumor specimens. Inhibition of survivin in 402-91 and 1765-92 by YM155 increased the percentage S-phase but did not induce apoptosis, which warrants further investigation before application in the treatment of metastatic MLS. Thus, using a 273-compound drug screen, we confirmed previously identified targets (mTOR, Src) in MLS and demonstrate survivin as essential for MLS survival.
Introduction
Myxoid liposarcoma (MLS) is a malignant soft tissue tumor accounting for 20% to 30% of the liposarcomas and roughly 5% of all soft tissue sarcomas [1]. These tumors are histopathologically characterized by a proliferation of stellate spindle cells with monomorphic ovoid nuclei, embedded in a myxoid matrix with a plexiform vasculature [1]. High-grade tumors are defined by having more than 5% of closely packed small blue round cells with high nuclear/cytoplasm ratio and scant stroma. MLS is genetically characterized by a reciprocal translocation t(12;16)(q13;p11), generating a fusion product of FUS and DDIT3. The chimeric fusion oncoprotein acts as an aberrant transcription factor and is known to influence the expression of several genes, including inhibition of adipogenic transcription factors C/EBPα and PPARγ [2], [3].
MLS tumors are initially sensitive to conventional chemo- and radiation therapy, but despite adequate local treatment, up to 40% can progress to local or distant relapse [4], [5], [6], [7]. MLS exhibits a unique metastatic pattern, as tumor cells tend to spread to other soft tissue sites before metastasizing to the lungs. The disease can become quite extensive, and management of metastatic or otherwise inoperable tumors often is challenging. This is reflected by the variable 5-year survival rates reported in several studies, which range from 8% for advanced disease to around 83% to 93% for cases with purely myxoid and localized tumors [5], [6], [7], [8], [9].
In addition to doxorubicin and ifosfamide, recently, eribulin, a microtubule-dynamics inhibitor, was shown to offer a survival benefit when compared with dacarbazine in the third-line setting in liposarcomas and is now FDA approved [10]. Moreover, MLS was shown to be sensitive to trabectedin (ET-743, Ecteinascidin), a natural alkylating agent derived from a marine tunicate [11]. The drug has a complex mechanism of action that is not entirely elucidated but involves binding to the DNA-minor groove, interaction with DNA repair complexes, and additional effects on the tumor microenvironment [12]. Unfortunately, similar to other systemic therapies, resistance develops, and the antitumor effect of trabectedin has been shown to diminish after some time on treatment [13]. Therefore, new therapeutic approaches are warranted to improve the outcome of advanced or metastatic MLS.
Over the past decades, therapeutic progress has been hampered by the sparse availability of representative preclinical models. For many years, only two published cell lines (403-91 and 1765-92) were widely available, both of which were SV40 immortalized [14], [15]. Recently, we reported on the generation of a novel MLS cell line (DL-221) and ancillary mouse xenograft model [16]. This newly established cell line is so far the only known MLS cell line that underwent spontaneous immortalization.
Here we used all three available MLS cell lines in an in vitro high-throughput drug screen to search for novel therapeutic agents that have the potential to enter future clinical trials. Drug screens are regularly used and contribute to the discovery of new candidate targets in cancer therapies [17], [18]; furthermore, the pathways targeted by effective drugs can yield insights into tumor biology. In addition to the conventional chemotherapeutic agents used in daily practice, such as anthracyclines and taxanes, we found that YM155, a survivin inhibitor, also strongly decreased tumor growth. Strong nuclear accumulation of survivin was observed in 100% of MLSs and confirmed to be essential for tumor growth.
Materials and Methods
Cell Culture
The MLS cell lines 402-91 and 1765-92 (generated using SV40 transformation and kindly provided by Pierre Åman, Sahlgrenska Cancer Centre, Department of Pathology, Institute of Biomedicine, University of Gothenburg, Sweden) were cultured in RPMI supplemented with 10% fetal bovine serum (Fisher Scientific, Landsmeer, the Netherlands) and 1% penicillin-streptomycin (100 U/ml). DL-221 was cultured with Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. All included cell lines have been well characterized for possible alterations in MLSs. All three cell lines are PIK3CA wild type, 402-91 and 1765-92 are TP53 wild type, and only DL-221 has two TP53 mutations (T125R and N239D) [16]. Cells were maintained in a humidified 5% CO2 incubator at 37°C. Cell lines were tested on a regular basis for mycoplasma infections. Short tandem repeat typing was performed before and after the experiments to confirm cell line identity using the Cell ID GenePrint 10 system (Promega, Leiden, the Netherlands).
Drug Screen
A high-throughput drug screen was performed in which a selection of 273 drugs (Supplementary Table 1) out of 2100 of the Bioactive Compound Library L1700 (Selleckchem, Houston, TX) was tested on the three MLS cell lines. Selection was based on potential clinical relevance of drugs. Drug stocks were dissolved in DMSO and stored in aliquots at −80°C. Cells were seeded in 96-well plates 24 hours before treatment. Addition of the drugs was performed with the Freedom EVO 200 liquid handling platform (Tecan, Männedorf, Switzerland), and final treatment concentrations were 1, 10, 100, and 1000 nM, each in triplicate. Cells were treated for 72 hours, and thereafter, CellTiter-Blue Cell Viability Assay (#G8081, Promega, Madison, WI) was added and incubated for 3 or 4 hours. Plates were read at room temperature using Fluoroskan Ascent FL fluorometer (Thermo Fisher Scientific Inc., Pittsburgh, PA), measuring the fluorescence at 530 Ex/604 Em. Data were analyzed by Tableau (Tableau, Seattle, WA). A compound was considered effective if able to reduce the cell viability >50% at a drug concentration of 100 nM in at least two cell lines.
Compounds and Cell Viability Assays
For single-agent validation studies, the survivin suppressant YM155, sepantronium bromide (S1130, Selleckchem), everolimus (S1120, Selleckchem), panobinostat (LBH-589, 13280, Cayman Chemical, Ann Arbor, MI), and trabectedin (ET-743, PharmaMar, Madrid, Spain) were dissolved in DMSO according to the manufacturers' instructions. Z-vad-FMK (550377, BD bioscience) was used as a general caspase inhibitor and also dissolved in DMSO. MLS cell lines were plated in 96-well plates 24 hours before treatment. Cells were treated with YM155 using concentrations from 0.01 until 5000 nM, in 11 steps, for 72 hours. Cell viability experiments were performed three times, in triplicate. As MLSs are known to respond well to trabectedin, combination treatments with YM155 were performed to investigate possible synergism or antagonism of both drugs. Concentrations used for combination treatment were for both drugs selected around the IC25, IC50, and IC75 values of single drug treatment for each cell line. This resulted in nine different dose combinations per cell line, and drugs were added simultaneously. After 72 hours of incubation, a Presto blue assay (Life-Technologies, Scotland, UK) was performed. Fluorescence was measured at 590 nm on a fluorometer (Victor3V 1420 multilabel reader, Perkin Elmer, the Netherlands). Combination experiments were performed twice, in triplicate. To evaluate whether the combination treatments were synergistic, a simplified version of the Bliss independence model was applied in which the Bliss expectation was calculated with the equation (A + B) − A × B, in which A and B are the fractional growth inhibitions caused by compounds A and B at a given dose, respectively [19].
siRNA Knockdown
For siRNA knockdown experiments, validated SMARTpool siRNAs targeting BIRC5 and control siRNAs (siGAPDH and siPLK1) were purchased from Dharmacon (GE Life Sciences, Landsmeer, the Netherlands). For the BIRC5 SMARTpool siRNA, we previously showed that results of the four individual siRNAs were identical to the SMARTpool [20]. Reversed siRNA (50 nM) transfection was carried out using DharmaFECT3 transfection reagent (Thermo Fisher Scientific Inc.) in triplo using 5000 to 10,000 cells/well; the experiments were performed twice. After 72 hours, Presto blue assay and protein analysis by Western blot analysis were performed.
Immunohistochemistry
Survivin expression was investigated in MLS tissue samples using a previously constructed tissue microarray with tumor samples from 32 patients diagnosed in the Leiden University Medical Center [21]. The tumors of 15 patients consisted of a purely myxoid (M), intermediate cell density (I), or round cell (RC) morphology. The other 17 patients had tumors containing a combination of two histological areas (M + I, M + RC, or I + RC); in these cases, both areas were included, resulting in a total of 49 tumor samples. All samples were handled in a coded fashion, and all procedures were performed according to the ethical guidelines, “Code for Proper Secondary Use of Human Tissue in the Netherlands” (Dutch Federation of Medical Scientific Societies). Cell pellets of the three untreated MLS cell lines were fixed in formalin and embedded in paraffin by using Shandon Cytoblock (Thermo Fisher Scientific Inc.). Sections were incubated with rabbit anti-survivin monoclonal antibody (71G4B7, #2808; Cell Signaling Technology, Leiden, the Netherlands) in a 1:100 dilution. Staining was visualized with DAB+ substrate Chromogen System (DAKO, Heverlee, Belgium). Placenta tissue was included as a control. Both nuclear and cytoplasmic expression was scored separately by two independent observers (J.V.M.G.B., M.A.G.). A semiquantitative scoring system was used, combining the staining intensity (0 = negative, 1 = weak, 2 = moderate, 3 = strong) and the percentage of positive tumor cells (0 = 0%, 1 = 1%-25%, 2 = 25%-50%, 3 = 51%-75%, and 4 = 76%-100%) as described previously [22].
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (PCR)
RNA was isolated from MLS cell lines using TRIzol (Invitrogen, Carlsbad, CA) followed by a purification procedure using the RNeasy mini kit (Qiagen, Venlo, the Netherlands) according to manufacturer's instructions. Quantitative PCR was performed for the isoforms wild-type (WT) survivin, survivin 2b, and survivin Δex3 with primers described previously [23]. Expression levels were normalized towards housekeeping genes HPRT, GAPDH, and TBP. Results are depicted relative to survivin WT expression for each cell line.
Apoptosis
Induction of apoptosis was assessed using the Caspase-Glo 3/7 assay (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, MLS cells were plated into white-walled 96-well plates (Corning, Fisher Scientific, Landsmeer, the Netherlands) and incubated with YM155 at the IC75 concentrations, as determined by cell viability assays. After 24-hour treatment, the substrate was added in a 1:1 dilution and incubated for 30 minutes at room temperature. Luminescence was measured with a luminometer (Victor3V 1420 multilabel reader). Experiments were performed twice.
Western Blot
After 24 hours YM155 treated and untreated MLS cells or 72 hours after siRNA transfection MLS cells were collected in hot-SDS buffer (1% SDS, 10 mM Tris/EDTA with complete inhibitor and phosSTOP) for protein analysis as described previously [24]. Briefly, 5 μg or 10 μg of protein of each sample was size-fractioned on 12% (TGX Stain-Fast Cast Acrylamide kit, BIO-RAD Laboratories) separating gels. Jurkat cell lysate treated with 25 μM etoposide [Cell Signaling (#2043)] was used as positive control for PARP and cleaved PARP detection. Proteins were transferred to a PVDF membrane and incubated overnight at 4°C with primary antibodies (all from Cell Signaling) PARP antibody (clone 46D11), survivin antibody (clone 71G4B7), or GAPDH antibody (clone D16H11). As a loading control, α-tubulin staining (clone DM1A; Sigma-Aldrich, Zwijndrecht, the Netherlands) was included. After incubation with the secondary HRP-conjugated antibody for 30 minutes at room temperature, membranes were treated with ECL2 substrate (Pierce, Thermo Scientific) as per manufacturer's instructions, and chemiluminescence was captured using ECL hyperfilm (Amersham, GE Healthcare Life Sciences, Eindhoven, the Netherlands).
Cell Cycle Analysis
Cell lines were cultured in T25 flasks in amounts ensuring 70% to 90% confluency when harvested and treated with the YM155 IC50 concentrations. After 48-hour treatment, cells were prepared for flow cytometric analysis. In short, cells were harvested, counted, and fixed in ice-cold methanol. Next, cells were washed, treated with RNase, and stained for DNA using propidium iodide and stored overnight at 4°C. Next day, cells were analyzed using an LSRII flowcytomer (BD biosciences). A blue 488-nm, 20-mW laser was used for excitation. Propidium iodide fluorescence was collected using a 610/20 band pass filter. Data analysis was performed by using WinList 8 remotely connected to ModFit LT 4 (Verity Software House, Topsham, ME) [25]. Each data file contained at least 10,000 single cell events. A one-compartment polynomial model was used for calculating the percentage G1, S, and G2M phase of the cell cycle. This statistical model showed the best fit.
Statistical Analysis
Dose-response curves and IC50 values were determined using GraphPad Prism (version 6.05; GraphPad Software, La Jolla, CA). Mann-Whitney testing was performed to investigate differences in survivin expression (GraphPad Prism). Statistically significant differences in cell cycle phases before and after treatment with YM155 were determined using two-way analysis of variance testing using Tukey's multiple-comparisons testing.
Results
Identification of 27 Compounds Effective Against MLS Tumor Cells
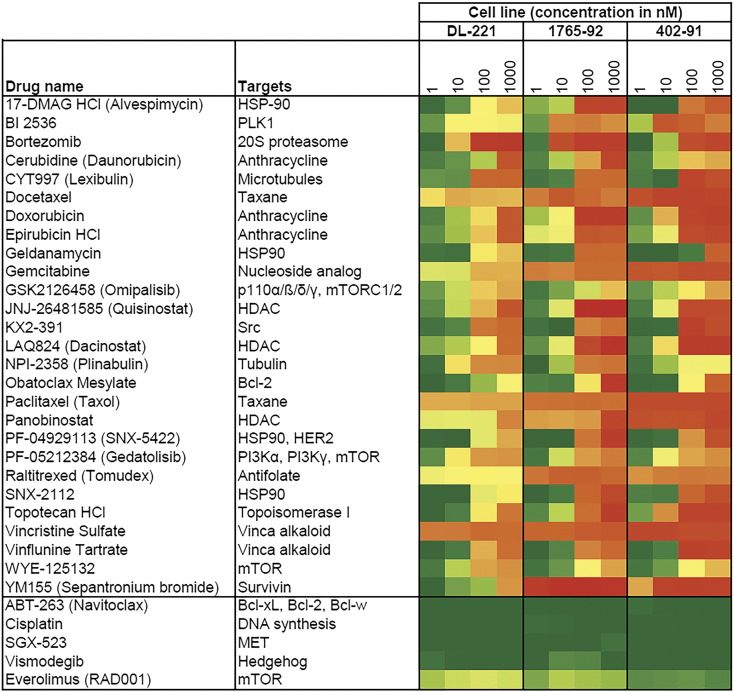
Two hundred seventy-three compounds were tested in three MLS tumor cell lines. Twenty-seven drugs were shown to be effective, which was defined as a loss of >50% cell viability in two or all three cell lines at a drug concentration of 100 nM (Figure 1, Supplementary Table 1). All three MLS cell lines showed a strong response (>80% loss of cell viability) to treatment with the anthracyclines doxorubicin and epirubicin at dose concentrations of both 100 nM and 1000 nM (402-91 and 1765-92) and 1000 nM (DL-221) (Figure 1). The tested taxanes, docetaxel and paclitaxel, as well as a member of another class of mitotic inhibitors, vincristine, revealed a strong decrease in cell viability at the lowest concentration of 1 nM (average cell viabilities of the three cell lines, respectively, ~22%, ~24%, and ~19% after 72 hours of treatment). In the drug screen, YM155, a survivin suppressant, showed in the 402-91 and 1765-92 cell line a very strong decrease in cell viability (average ~13% cell viability) at all four drug concentrations. In the DL-221 cell line, there was a moderate reduction of the cell viability after treatment with YM155 at concentrations of 10, 100, and 1000 nM (ranging from 81% to 30% cell viability). Other interesting agents in the list are 17-DMAG HCl, geldanamycin, SNX-5422, and SNX-2112, all targeting HSP90. The list also contains around 15 drugs affecting the PI3K/AKT/mTOR pathway, and although only omipalisib, gedatolisib, and WYE-132 did meet the selection criteria of a reduction in cell viability of more than 50% at 100 nM, also several other drugs affecting the mTOR pathway demonstrated a response to treatment. The response patterns of everolimus, temsirolimus, and sirolimus were comparable, and they all showed a reduction in cell viability at the lowest dose and almost no further reduction in cell viability at the higher treatment concentrations (data not shown). The HDAC inhibitors quisinostat, dacinostat, and panobinostat showed a strong inhibitory effect in all three cell lines. KX2-391, an agent interfering with the proto-oncogene Src, also was effective in all three cell lines.
Figure 1.
Hits of high throughput in vitro drug screen of three MLS cell lines. List of 27 drugs with a reduction in cell viability of >50% in two or all three cell lines at a drug concentration of 100 nM. Strong inhibitory effect of the survivin inhibitor YM155 is observed in two out of three MLS cell lines. Also, a good response is observed to several conventional chemotherapeutics, like doxorubicin, gemcitabine, and paclitaxel. Per drug, four concentrations (1, 10, 100, and 1000 nM) are tested; green boxes correspond to a high cell viability (~100%) and red boxes to a loss of cell viability (~0%). For comparison, at the bottom, five compounds are randomly shown that did not meet the criteria.
Validation of Selected Hits Using Cell Viability Assays
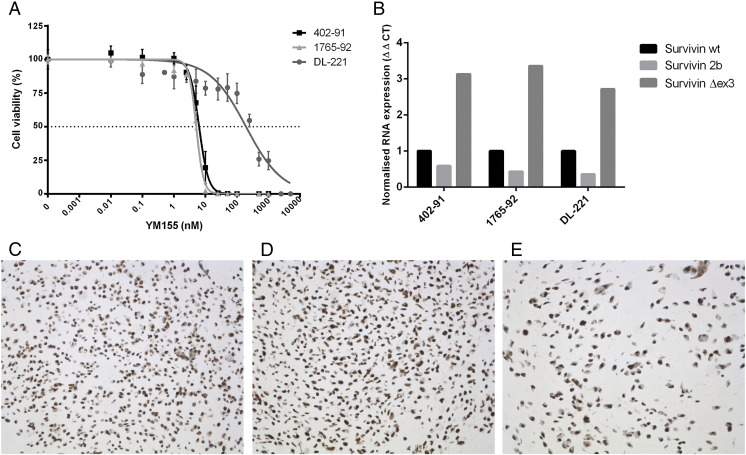
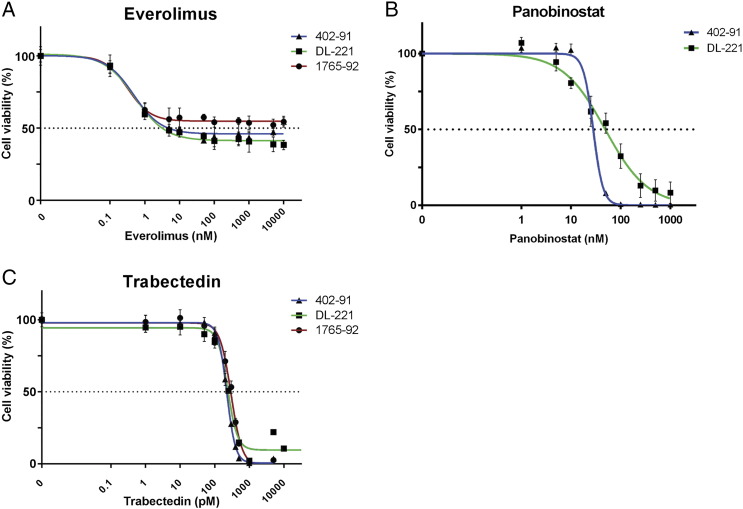
We selected the the survivin inhibitor YM155, the mTOR inhibitor everolimus, and the broad-spectrum HDAC inhibitor panobinostat (LBH-589) for further validation using cell viability assays. YM155 was of particular interest because this was the drug with the best antitumor effect at the lowest treatment dose in two of the three cell lines. The 402-91 and 1765-92 cell lines were confirmed to be highly sensitive to YM155 and showed IC50 values of 6.3 nM and 5.1 nM, respectively (Figure 2A). The IC50 value of DL-221 was 194 nM, which is consistent with the results observed in the drug screen. In all three cell lines, an inhibition of cell viability (~50%) was observed at a low dose concentration (~5 nM) of everolimus (Supplementary Figure 1A). However, with increasing dose, no further decrease of cell viability was observed. The tested cell lines 402-91 and DL-221 demonstrated an IC50 value of 28 nM and 49 nM, respectively, after treatment with panobinostat (Supplementary Figure 1B).
Figure 2.
Role of survivin in MLS cell lines. Dose-response curves for YM155 (72 hours) in MLS cell lines. Error bars represents three experiments performed in triplicates (A). Normalized RNA expression of three survivin isoforms in cell lines showing relative abundance of the Δex3 isoform (B). Survivin immunohistochemistry of FFPE cell pellets revealed a strong nuclear survivin expression in the cell lines 402-91 (C), 1765-92 (D), and DL-221 (E) (20× magnification).
Supplementary Figure 1.
Dose-response curves of MLS cell lines after monotherapy with everolimus (A) and panobinostat (B) after 72 hours of treatment.
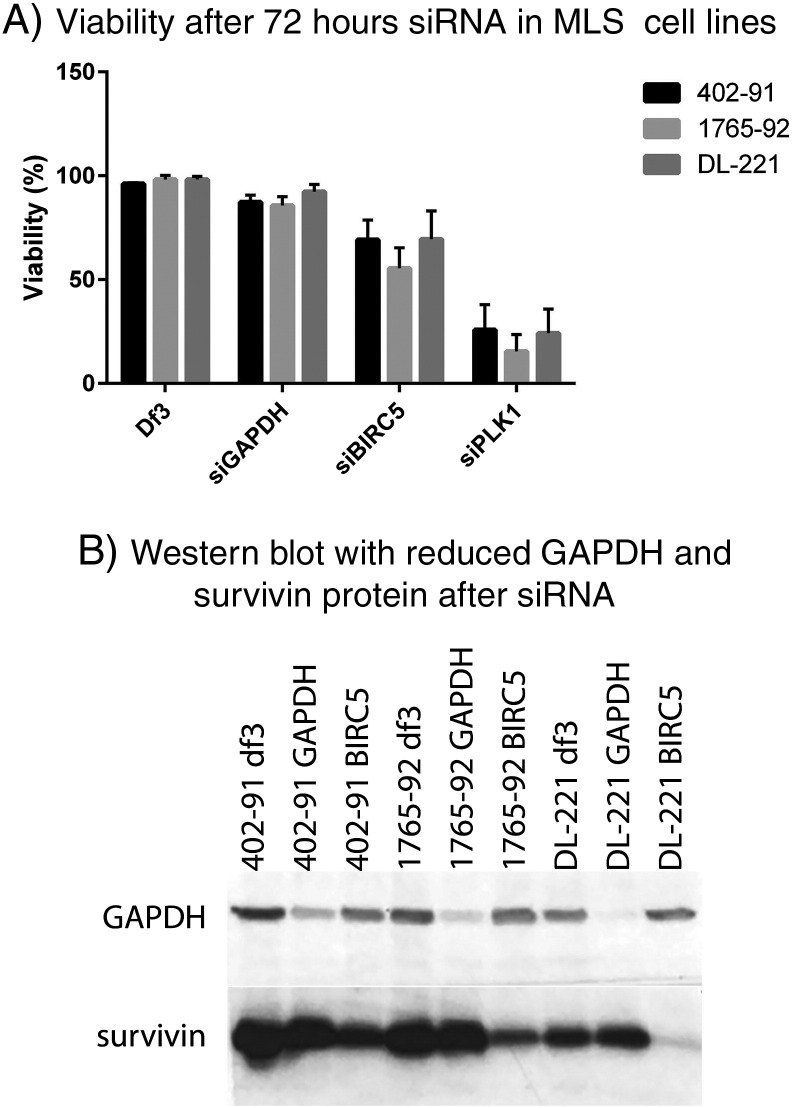
To confirm the importance of BIRC5 in MLS cell survival, siRNA experiments were performed. Knockdown of the survivin gene by SMARTpool siBIRC5 resulted in a partial decrease of viability in all three cell lines. Western blot analysis after transfection with siBIRC5 confirmed a decrease in survivin protein in all three cell lines with variable knockdown efficiency (Supplementary Figure 2).
Supplementary Figure 2.
Knockdown BIRC5. BIRC5 siRNA resulted in a partial decrease of cell viability (402-91, 69%; 1765-92, 55%; and DL-221, 69%). The transfection was successful as all three MLS cell lines and the control siRNA GAPDH showed a minor reduction of cell viability, whereas transfection with the positive control siPLK1 resulted in all three cell lines with a pronounced decrease in cell viability. The transfection reagent alone hardly affected cell viability (A). A reduction in GAPDH and survivin protein expression was found after transfection with, respectively, siGAPDH and siBIRC5 in all three cell lines, although knockdown efficiency was variable (B).
No Synergistic Effect of Everolimus or YM155 and Trabectedin
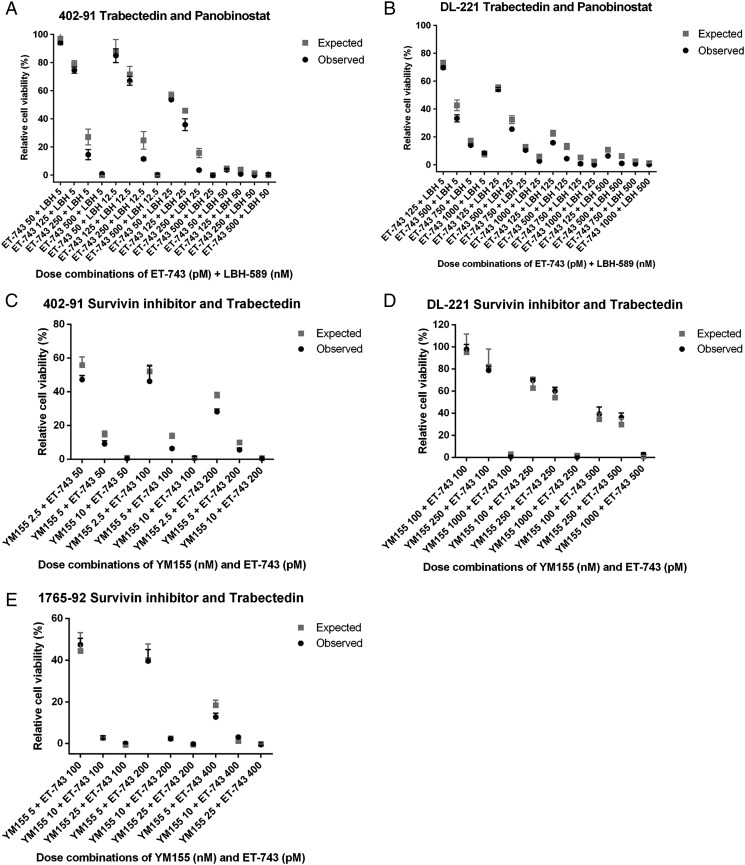
As trabectedin (ET-743) is effective and therefore often used in the treatment of MLS, we investigated possible synergy between trabectedin and two of the validated screen hits, YM155 or panobinostat. The MLS cell lines were confirmed to be sensitive to trabectedin (Supplementary Figure 1C). Combination treatment of trabectedin with YM155 showed no statistically significant difference between the expected and the observed cell viability in all three tested cell lines, indicating absence of synergism or antagonism for this drug combination (Supplementary Figure 3, A, C, and E). The combination of trabectedin and panobinostat was evaluated in the 402-91 and DL-221 cell lines, but again, no statistical significant synergism or antagonism was observed (Supplementary Figure 3, B and D).
Supplementary Figure 3.
Combination treatment of cell line 402-91 and DL-221 with panobinostat and trabectedin (A and B) showed no synergism or antagonism. Combination therapy of three MLS cell lines with trabectedin and YM155 also demonstrated no synergism or antagonism (C-E).
Strong Nuclear Survivin Expression in All MLS Cell Lines and Primary Tumors
Because YM155 was most efficient in decreasing MLS cell viability, we evaluated the protein expression of survivin in the MLS cell lines. The FFPE cell pellets of the three cell lines showed strong nuclear survivin expression and a weak cytoplasmic expression in all three cell lines (Figure 2, C-E). No difference in the expression levels was observed.
Moreover, we examined the relative distribution of the three most common survivin isoforms: survivin WT, survivin 2b, and survivin Δex3, by quantitative reverse transcriptase PCR. All cell lines revealed a similar mRNA expression pattern, with survivin Δex3 being most highly expressed, followed by survivin WT, whereas the lowest expression was found for survivin 2b (Figure 2B).
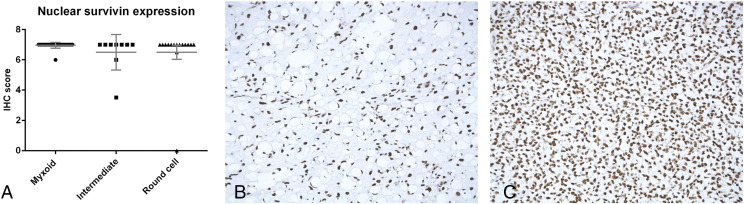
Next, we studied a cohort of MLS primary tumor samples. The tumor samples of all 32 patients (100%) on the tissue microarrays demonstrated strong nuclear expression of survivin protein (Figure 3A). No statistically significant differences were observed between the myxoid, intermediate, and round cell components (P > .05) (Figure 3, B and C). Cytoplasmic staining was absent, with the exception of a single round cell liposarcoma with moderate cytoplasmic staining.
Figure 3.
High nuclear expression of survivin in MLS. Immunohistochemical analysis of nuclear survivin expression in 32 MLS patients showing high expression in tumor components with myxoid, intermediate, and round cell morphology (A). High nuclear survivin expression in tumor with myxoid morphology (B). High nuclear survivin expression in tumor with round cell morphology (C) (20× magnification).
No Apoptosis But Increased S-Phase by YM155 in Two of Three MLS Cell Lines
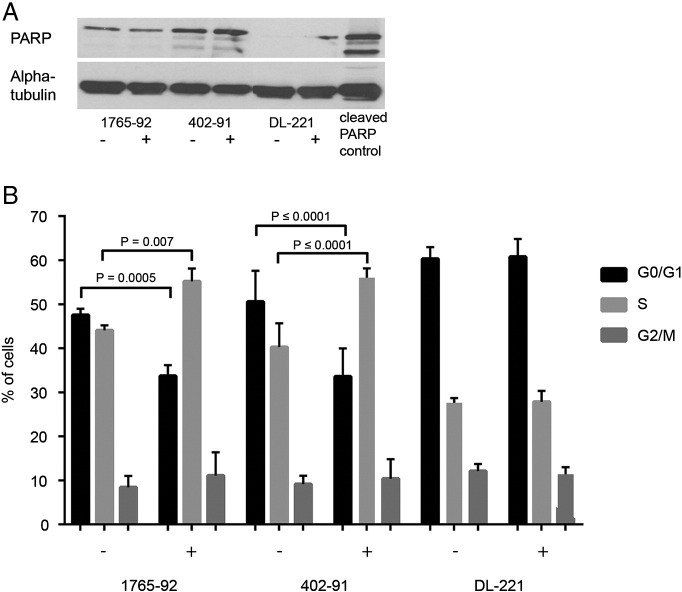
To further evaluate the mechanism of action of YM155 in MLS cell lines, we evaluated PARP expression in all three MLS cell lines using Western blot analysis, but no cleaved PARP was found in the cell lines (Figure 4A) after 24 hours of YM155 treatment. Caspase 3/7 activity after treatment with the survivin inhibitor YM155 at IC75 concentration also revealed no significant increase in caspase 3/7 activity after 24 hours (data not shown). These results indicate that YM155 does not induce apoptosis in MLS.
Figure 4.
Effect of YM155 on apoptosis and cell cycle. YM155 does not cause PARP-dependent apoptosis but increases S-phase in two of the MLS cell lines. Western blot analysis for PARP and cleaved PARP expression in MLS cell lines (A). FACS cell cycle analysis of MLS cell lines treated with YM155 for 48 hours, measured in two independent experiments. Two cell lines, 402-91 and 1765-92, show a decrease in G1 and an increase in S-phase after treatment. DL-221 does not show a difference in cell cycle distribution (B).
We subsequently performed cell cycle analysis to evaluate a possible effect of YM155 on cell cycle regulation. After 48 hours, YM155-treated cells revealed an increase in the S-phase fraction and a decrease in the G1 fraction as compared with untreated 402-91 and 1765-92 cells. In DL-221, no clear changes were observed in cell cycle distribution after YM155 treatment (Figure 4B).
Discussion
MLS initially often responds well to treatment with radiotherapy, conventional chemotherapeutics, or trabectedin; however, after several treatment cycles, the effect of treatment is observed to decrease. Therefore, for patients with locally advanced or metastatic disease, new therapeutic options are highly warranted to improve survival. In this study, our aim was to identify novel candidate targets for treatment by performing a high-throughput drug screen with multiple targeted (tyrosine kinase) inhibitors as well as substances of other classes.
The 27 most effective compounds (Figure 1) include the conventional chemotherapeutic agents used in daily practice, such as anthracyclines and taxanes, as anticipated. In addition, the results included three potent inhibitors of the PI3K/AKT/mTOR pathway (omipalisib, gedatolisib, and WYE-125132). A subset of MLSs demonstrates dysregulation of this pathway by PIK3CA mutation, loss of PTEN expression, or Akt activation [26], [27], [28]. No large series on mTOR inhibition in MLS patients is available, although in two cases, a minor response has been observed [29], and a clinical trial is ongoing (COSYMO www.clinicaltrials.gov NCT02821507). In the current study, we found a decrease of cell viability down to 50% at low-dose treatment with everolimus. Unlike three other inhibitors of the mTOR pathway, everolimus did not end up in the list of most effective drugs of the drug screen, which is caused by the fact that, at the higher drug concentrations, no reduction of cell viability above 50% was achieved. This suggests that, in clinical studies, the drug may need to be combined with another (chemotherapeutic) drug. Interestingly, BI2536, a potent PLK1 inhibitor, was also one of the hits of the drug screen, and we confirm that inhibition of PLK1 results in a decrease in cell viability (Supplementary Figure 2A). As PLK1 was very recently shown to interact with the mTOR pathway by inhibiting MTORC1, it might be interesting to further explore combination treatment in MLS [30]. Moreover, dual treatment of temsirolimus with YM155 demonstrated an improved antitumor effect in a renal cancer model [53].
The Src inhibitor KX2-391 is another interesting hit confirming previous preclinical results, as two previous studies reported that the Src pathway is highly active in MLS [31], [32]. KX2-391 is the first clinical Src inhibitor targeting the peptide substrate-binding site, with higher in vitro potency than dasatinib [33]. In a small phase I study with several solid malignancies, favorable pharmacokinetics and antitumor activity were observed [34].
We demonstrate that inhibition of survivin by YM155 results in a significant decrease of cell viability in two out of three cell lines (402-91 and 1765-92), whereas DL-221 seems less dependent on survivin. A similar trend was seen using siRNA for BIRC5 in DL221; despite almost complete knockdown, the effect on viability is comparable to 402-91, in which knockdown was ~50% (Supplementary Figure 2). YM155 blocks BIRC5 at the promoter region. In addition to this inhibition at the transcriptional level, also at the posttranscriptional level, survivin can be downregulated by the addition of CDK or HSP90 inhibitors which are known to interact with the expression of BIRC5 in the cells [35]. Interestingly, 4 of the 27 hits emerging from the compound screen target HSP90 (Figure 1) and demonstrate a strong inhibitory effect on cell viability. A recent study also demonstrates an important role for HSP90 inhibitor 17-DMAG resulting in decreased phosphorylation of several receptor tyrosine kinases and demonstrating massive tumor cell death in a xenograft model [36].
Survivin (BIRC5) is a member of the inhibitor of apoptosis family and is a multifunctional protein that is involved in several important cellular processes in both the nucleus and the cytoplasm. Survivin interacts with aurora B kinase, and both are part of the chromosomal passenger complex, which forms at the kinetochore in the nucleus and regulates microtubule-kinetochore attachment, ensuring proper segregation of the sister chromatids during mitosis [37], [38]. Cytoplasmic survivin is involved in two processes. On one hand, it is involved in the binding of XIAP, which inhibits caspase-9 and prevents activation of the apoptotic pathway. In addition, survivin is able to activate AKT and to upregulate α5 integrin, resulting in stimulation of cell motility. Normally, survivin is expressed during fetal development and also in certain differentiated tissues. High expression of survivin has been found in several malignancies, including sarcomas such as malignant peripheral nerve sheath tumor, pleomorphic liposarcoma, uterine leiomyosarcoma, chondrosarcoma, and Ewing sarcoma [20], [39], [40], [41], [42], [43].
All primary MLS tumor samples, as well as the three cell lines, showed high nuclear expression of survivin. There are multiple survivin isoforms described. Several studies show a particular overexpression of survivin Δex3 in several malignancies, including, for example, breast and colon carcinoma and soft tissue sarcomas [44], [45]. The expression of this isoform is also associated with a worse prognosis. We here show that, also in the MLS cell lines, the survivin Δex3 isoform is relatively abundant.
Previously, we and others showed that inhibition of survivin by YM155 is effective in vitro in several other malignancies, including squamous cell carcinoma, gastrointestinal stromal tumor, and chondrosarcoma [20], [46], [47]. Evaluation of the effect of YM155 in in vivo models for gastric carcinoma and osteosarcoma demonstrated a suppression of tumor growth in mice [48], [49]. The mechanism of action of YM155 as anticancer drug is still a matter of debate. Although initially proposed as a BIRC5 transcriptional repressor, YM155 has been subsequently shown to induce DNA damage [50] and to inhibit double-strand break repair [51]. In most of the carcinoma models, survivin inhibition induces apoptosis, whereas for MLS as well as for chondrosarcoma [20], survivin expression was predominantly nuclear. Based on its role in maintaining proper sister chromatid segregation during mitosis, depletion of BIRC5 has indeed been described to induce a prominent defect in mitosis without direct induction of apoptosis [52]. This has led to the hypothesis that the antiapoptotic function of BIRC5 is secondary to its role in mitosis. Strikingly, after treatment with YM155, we observed an increase in percentage of cells in S-phase in 402-91 and 1765-92. This phenomenon has also been observed in other cancer models [20], [53] and may be interpreted in the context of “mitotic catastrophe,” a type of cell death that occurs during mitosis and is being controlled by numerous molecular players including survivin [52]. However, a high S-phase is significantly associated with a poor prognosis [54], [55]. Furthermore, survivin knockdowns by shRNA increased chromosomal instability, irrespective of p53 [56]. As, together with a high S-phase, this might lead to clonal selection and outgrowth of resistant clones, this warrants further investigation.
In conclusion, using a large compound screen, we identified 27 compounds that are effective in decreasing cell viability of MLS cells. We identified survivin as being essential for MLS tumor growth. Survivin is expressed in 100% of tumor samples. Although the S-phase increased, a strong decrease in cell viability was observed in two of three tumor cell lines, which warrants further investigation for possible treatment of advanced MLS patients with YM155 targeting survivin.
The following are the supplementary data related to this article.
Complete List of 273 Compounds Tested on the Three MLS Cell Lines.
Footnotes
Financial support: This work was financially supported by the Liddy Shriver Sarcoma Initiative.
Disclosure/conflict of interest: The authors have no disclosure or conflict of interest to declare.
References
- 1.Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. Pathology and Genetics of Tumours of Soft Tissue and Bone. 4th ed. IARC Press; Lyon: 2013. World Health Organisation Classification of Tumours. [Google Scholar]
- 2.Perez-Mancera PA, Bermejo-Rodriguez C, Sanchez-Martin M, Abollo-Jimenez F, Pintado B, Sanchez-Garcia I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS One. 2008;3(7):e2569. doi: 10.1371/journal.pone.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aman P, Dolatabadi S, Svec D, Jonasson E, Safavi S, Andersson D. Regulatory mechanisms, expression levels and proliferation effects of the FUS-DDIT3 fusion oncogene in liposarcoma. J Pathol. 2016;238(5):689–699. doi: 10.1002/path.4700. [DOI] [PubMed] [Google Scholar]
- 4.ten Heuvel SE, Hoekstra HJ, van Ginkel RJ, Bastiaannet E, Suurmeijer AJ. Clinicopathologic prognostic factors in myxoid liposarcoma: a retrospective study of 49 patients with long-term follow-up. Ann Surg Oncol. 2007;14(1):222–229. doi: 10.1245/s10434-006-9043-7. [DOI] [PubMed] [Google Scholar]
- 5.Fuglo HM, Maretty-Nielsen K, Hovgaard D, Keller JO, Safwat AA, Petersen MM. Metastatic pattern, local relapse, and survival of patients with myxoid liposarcoma: a retrospective study of 45 patients. Sarcoma. 2013;2013:548628. doi: 10.1155/2013/548628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreau LC, Turcotte R, Ferguson P, Wunder J, Clarkson P, Masri B. Myxoid\round cell liposarcoma (MRCLS) revisited: an analysis of 418 primarily managed cases. Ann Surg Oncol. 2012;19(4):1081–1088. doi: 10.1245/s10434-011-2127-z. [DOI] [PubMed] [Google Scholar]
- 7.Nishida Y, Tsukushi S, Nakashima H, Ishiguro N. Clinicopathologic prognostic factors of pure myxoid liposarcoma of the extremities and trunk wall. Clin Orthop Relat Res. 2010;468(11):3041–3046. doi: 10.1007/s11999-010-1396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman A, Ghadimi MP, Demicco EG, Creighton CJ, Torres K, Colombo C. Localized and metastatic myxoid/round cell liposarcoma: clinical and molecular observations. Cancer. 2013;119(10):1868–1877. doi: 10.1002/cncr.27847. [DOI] [PubMed] [Google Scholar]
- 9.Baxter KJ, Govsyeyev N, Namm JP, Gonzalez RJ, Roggin KK, Cardona K. Is multimodality therapy necessary for the management of pure myxoid liposarcomas? A multi-institutional series of pure myxoid liposarcomas of the extremities and torso. J Surg Oncol. 2015;111(2):146–151. doi: 10.1002/jso.23786. [DOI] [PubMed] [Google Scholar]
- 10.Schoffski P, Chawla S, Maki RG, Italiano A, Gelderblom H, Choy E. Eribulin versus dacarbazine in previously treated patients with advanced liposarcoma or leiomyosarcoma: a randomised, open-label, multicentre, phase 3 trial. Lancet. 2016;387(10028):1629–1637. doi: 10.1016/S0140-6736(15)01283-0. [DOI] [PubMed] [Google Scholar]
- 11.Gordon EM, Sankhala KK, Chawla N, Chawla SP. Trabectedin for soft tissue sarcoma: current status and future perspectives. Adv Ther. 2016;33(7):1055–1071. doi: 10.1007/s12325-016-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol Cancer Ther. 2010;9(8):2157–2163. doi: 10.1158/1535-7163.MCT-10-0263. [DOI] [PubMed] [Google Scholar]
- 13.Grosso F, Sanfilippo R, Virdis E, Piovesan C, Collini P, Dileo P. Trabectedin in myxoid liposarcomas (MLS): a long-term analysis of a single-institution series. Ann Oncol. 2009;20(8):1439–1444. doi: 10.1093/annonc/mdp004. [DOI] [PubMed] [Google Scholar]
- 14.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosomes Cancer. 1992;5(4):278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 15.Thelin-Jarnum S, Lassen C, Panagopoulos I, Mandahl N, Aman P. Identification of genes differentially expressed in TLS-CHOP carrying myxoid liposarcomas. Int J Cancer. 1999;83(1):30–33. doi: 10.1002/(sici)1097-0215(19990924)83:1<30::aid-ijc6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.de Graaff MA, Yu JS, Beird HC, Ingram DR, Nguyen T, Juehui Liu J. Establishment and characterization of a new human myxoid liposarcoma cell line (DL-221) with the FUS-DDIT3 translocation. Lab Invest. 2016;96(8):885–894. doi: 10.1038/labinvest.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halvorson KG, Barton KL, Schroeder K, Misuraca KL, Hoeman C, Chung A. A high-throughput in vitro drug screen in a genetically engineered mouse model of diffuse intrinsic pontine glioma identifies BMS-754807 as a promising therapeutic agent. PLoS One. 2015;10(3):e0118926. doi: 10.1371/journal.pone.0118926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriksson A, Osterroos A, Hassan S, Gullbo J, Rickardson L, Jarvius M. Drug screen in patient cells suggests quinacrine to be repositioned for treatment of acute myeloid leukemia. Blood Cancer J. 2015;5:e307. doi: 10.1038/bcj.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong M, Tan N, Zha J, Peale FV, Yue P, Fairbrother WJ. Navitoclax (ABT-263) reduces Bcl-x(L)–mediated chemoresistance in ovarian cancer models. Mol Cancer Ther. 2012;11(4):1026–1035. doi: 10.1158/1535-7163.MCT-11-0693. [DOI] [PubMed] [Google Scholar]
- 20.de Jong Y, van Oosterwijk JG, Kruisselbrink AB, Briaire-de Bruijn IH, Agrogiannis G, Baranski Z. Targeting survivin as a potential new treatment for chondrosarcoma of bone. Oncogene. 2016;5:e222. doi: 10.1038/oncsis.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endo M, de Graaff MA, Ingram DR, Lim S, Lev DC, Briaire-de Bruijn IH. NY-ESO-1 (CTAG1B) expression in mesenchymal tumors. Mod Pathol. 2015;28(4):587–595. doi: 10.1038/modpathol.2014.155. [DOI] [PubMed] [Google Scholar]
- 22.Bovee JV, Cleton-Jansen AM, Kuipers-Dijkshoorn NJ, van den Broek LJ, Taminiau AH, Cornelisse CJ. Loss of heterozygosity and DNA ploidy point to a diverging genetic mechanism in the origin of peripheral and central chondrosarcoma. Genes Chromosomes Cancer. 1999;26(3):237–246. [PubMed] [Google Scholar]
- 23.Pavlidou A, Dalamaga M, Kroupis C, Konstantoudakis G, Belimezi M, Athanasas G. Survivin isoforms and clinicopathological characteristics in colorectal adenocarcinomas using real-time qPCR. World J Gastroenterol. 2011;17(12):1614–1621. doi: 10.3748/wjg.v17.i12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willems SM, Schrage YM, Baelde JJ, Briaire-de BI, Mohseny A, Sciot R. Myxoid tumours of soft tissue: the so-called myxoid extracellular matrix is heterogeneous in composition. Histopathology. 2008;52(4):465–474. doi: 10.1111/j.1365-2559.2008.02967.x. [DOI] [PubMed] [Google Scholar]
- 25.van Haaften C, Boot A, Corver WE, van Eendenburg JD, Trimbos BJ, van WT. Synergistic effects of the sesquiterpene lactone, EPD, with cisplatin and paclitaxel in ovarian cancer cells. J Exp Clin Cancer Res. 2015;34:38. doi: 10.1186/s13046-015-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demicco EG, Torres KE, Ghadimi MP, Colombo C, Bolshakov S, Hoffman A. Involvement of the PI3K/Akt pathway in myxoid/round cell liposarcoma. Mod Pathol. 2012;25(2):212–221. doi: 10.1038/modpathol.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negri T, Virdis E, Brich S, Bozzi F, Tamborini E, Tarantino E. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clin Cancer Res. 2010;16(14):3581–3593. doi: 10.1158/1078-0432.CCR-09-2912. [DOI] [PubMed] [Google Scholar]
- 28.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42(8):715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanfilippo R, Dei Tos AP, Casali PG. Myxoid liposarcoma and the mammalian target of rapamycin pathway. Curr Opin Oncol. 2013;25(4):379–383. doi: 10.1097/CCO.0b013e32836227ac. [DOI] [PubMed] [Google Scholar]
- 30.Ruf S, Heberle AM, Langelaar-Makkinje M, Gelino S, Wilkinson D, Gerbeth C. PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy. 2017;13(3):486–505. doi: 10.1080/15548627.2016.1263781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willems SM, Schrage YM, Bruijn IH, Szuhai K, Hogendoorn PC, Bovee JV. Kinome profiling of myxoid liposarcoma reveals NF-kappaB-pathway kinase activity and casein kinase II inhibition as a potential treatment option. Mol Cancer. 2010;9:257. doi: 10.1186/1476-4598-9-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievers E, Trautmann M, Kindler D, Huss S, Gruenewald I, Dirksen U. SRC inhibition represents a potential therapeutic strategy in liposarcoma. Int J Cancer. 2015;137(11):2578–2588. doi: 10.1002/ijc.29645. [DOI] [PubMed] [Google Scholar]
- 33.Lau GM, Lau GM, Yu GL, Gelman IH, Gutowski A, Hangauer D. Expression of Src and FAK in hepatocellular carcinoma and the effect of Src inhibitors on hepatocellular carcinoma in vitro. Dig Dis Sci. 2009;54(7):1465–1474. doi: 10.1007/s10620-008-0519-0. [DOI] [PubMed] [Google Scholar]
- 34.Naing A, Cohen R, Dy GK, Hong DS, Dyster L, Hangauer DG. A phase I trial of KX2-391, a novel non-ATP competitive substrate-pocket–directed SRC inhibitor, in patients with advanced malignancies. Invest New Drugs. 2013;31(4):967–973. doi: 10.1007/s10637-013-9929-8. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Duan N, Zhang C, Zhang W. Survivin and tumorigenesis: molecular mechanisms and therapeutic strategies. J Cancer. 2016;7(3):314–323. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safavi S, Jarnum S, Vannas C, Udhane S, Jonasson E, Tomic TT. HSP90 inhibition blocks ERBB3 and RET phosphorylation in myxoid/round cell liposarcoma and causes massive cell death in vitro and in vivo. Oncotarget. 2016;7(1):433–445. doi: 10.18632/oncotarget.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenzie JA, Grossman D. Role of the apoptotic and mitotic regulator survivin in melanoma. Anticancer Res. 2012;32(2):397–404. [PubMed] [Google Scholar]
- 38.van der Waal MS, Hengeveld RC, van der Horst A, Lens SM. Cell division control by the chromosomal passenger complex. Exp Cell Res. 2012;318(12):1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Wurl P, Kappler M, Meye A, Bartel F, Kohler T, Lautenschlager C. Co-expression of survivin and TERT and risk of tumour-related death in patients with soft-tissue sarcoma. Lancet. 2002;359(9310):943–945. doi: 10.1016/S0140-6736(02)07990-4. [DOI] [PubMed] [Google Scholar]
- 40.Hingorani P, Dickman P, Garcia-Filion P, White-Collins A, Kolb EA, Azorsa DO. BIRC5 expression is a poor prognostic marker in Ewing sarcoma. Pediatr Blood Cancer. 2013;60(1):35–40. doi: 10.1002/pbc.24290. [DOI] [PubMed] [Google Scholar]
- 41.Ghadimi MP, Liu P, Peng T, Bolshakov S, Young ED, Torres KE. Pleomorphic liposarcoma: clinical observations and molecular variables. Cancer. 2011;117(23):5359–5369. doi: 10.1002/cncr.26195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghadimi MP, Young ED, Belousov R, Zhang Y, Lopez G, Lusby K. Survivin is a viable target for the treatment of malignant peripheral nerve sheath tumors. Clin Cancer Res. 2012;18(9):2545–2557. doi: 10.1158/1078-0432.CCR-11-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lusby K, Savannah KB, Demicco EG, Zhang Y, Ghadimi MP, Young ED. Uterine leiomyosarcoma management, outcome, and associated molecular biomarkers: a single institution's experience. Ann Surg Oncol. 2013;20(7):2364–2372. doi: 10.1245/s10434-012-2834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taubert H, Kappler M, Bache M, Bartel F, Kohler T, Lautenschlager C. Elevated expression of survivin-splice variants predicts a poor outcome for soft-tissue sarcomas patients. Oncogene. 2005;24(33):5258–5261. doi: 10.1038/sj.onc.1208702. [DOI] [PubMed] [Google Scholar]
- 45.Necochea-Campion R, Chen CS, Mirshahidi S, Howard FD, Wall NR. Clinico-pathologic relevance of survivin splice variant expression in cancer. Cancer Lett. 2013;339(2):167–174. doi: 10.1016/j.canlet.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Liu Y, Li YF, Yue Y, Yang X, Peng L. Targeting of survivin pathways by YM155 inhibits cell death and invasion in oral squamous cell carcinoma cells. cell Physiol Biochem. 2016;38(6):2426–2437. doi: 10.1159/000445594. [DOI] [PubMed] [Google Scholar]
- 47.Falkenhorst J, Grunewald S, Muhlenberg T, Marino-Enriquez A, Reis AC, Corless C. Inhibitor of apoptosis proteins (IAPs) are commonly dysregulated in GIST and can be pharmacologically targeted to enhance the pro-apoptotic activity of imatinib. Oncotarget. 2016 doi: 10.18632/oncotarget.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng XJ, Lin JC, Ding YF, Zhu L, Ye J, Tu SP. Survivin inhibitor YM155 suppresses gastric cancer xenograft growth in mice without affecting normal tissues. Oncotarget. 2016;7(6):7096–7109. doi: 10.18632/oncotarget.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Ma L, Wang J. YM155 exerts a growth inhibitory effect on human osteosarcoma in vitro and in vivo. Oncol Rep. 2015;34(2):1074–1080. doi: 10.3892/or.2015.4067. [DOI] [PubMed] [Google Scholar]
- 50.Glaros TG, Stockwin LH, Mullendore ME, Smith B, Morrison BL, Newton DL. The "survivin suppressants" NSC 80467 and YM155 induce a DNA damage response. Cancer Chemother Pharmacol. 2012;70(1):207–212. doi: 10.1007/s00280-012-1868-0. [DOI] [PubMed] [Google Scholar]
- 51.Hong M, Ren MQ, Silva J, Paul A, Wilson WD, Schroeder C. YM155 inhibits topoisomerase function. Anticancer Drugs. 2017;28(2):142–152. doi: 10.1097/CAD.0000000000000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23(16):2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 53.Mehta A, Zhang L, Boufraqech M, Liu-Chittenden Y, Zhang Y, Patel D. Inhibition of survivin with YM155 induces durable tumor response in anaplastic thyroid cancer. Clin Cancer Res. 2015;21(18):4123–4132. doi: 10.1158/1078-0432.CCR-14-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stal O. DNA and cell cycle analysis as prognostic indicators in breast tumors revisited. Clin Lab Med. 2001;21(4):875–895. [PubMed] [Google Scholar]
- 55.Dayal JH, Sales MJ, Corver WE, Purdie CA, Jordan LB, Quinlan PR. Multiparameter DNA content analysis identifies distinct groups in primary breast cancer. Br J Cancer. 2013;108(4):873–880. doi: 10.1038/bjc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiedemuth R, Klink B, Topfer K, Schrock E, Schackert G, Tatsuka M. Survivin safeguards chromosome numbers and protects from aneuploidy independently from p53. Mol Cancer. 2014;13:107. doi: 10.1186/1476-4598-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete List of 273 Compounds Tested on the Three MLS Cell Lines.