Abstract
Intestinal tumorigenesis in the ApcMin/+ model is initiated by aberrant activation of Wnt pathway. Increased IL-4 expression in human colorectal cancer tissue and growth of colon cancer cell lines implied that IL-4–induced Stat6-mediated tumorigenic signaling likely contributes to intestinal tumor progression in ApcMin/+ mice. Stat6 also appears to promote expansion of myeloid-derived suppressor cells (MDSCs) cells. MDSCs promote polyp formation in the ApcMin/+ model. Hence, Stat6 could have a broad role in coordinating both polyp cell proliferation and MDSC expansion. We found that IL-4–induced Stat6-mediated proliferation of intestinal epithelial cells is augmented by platelet-derived growth factor–BB, a tumor-promoting growth factor. To determine whether polyp progression in ApcMin/+ mice is dependent on Stat6 signaling, we disrupted Stat6 in this model. Total polyps in the small intestine were fewer in ApcMin/+ mice lacking Stat6. Furthermore, proliferation of polyp epithelial cells was reduced, indicating that Stat6 in part controlled polyp formation. Stat6 also promoted expansion of MDSCs in the spleen and lamina propria of ApcMin/+ mice, implying regulation of antitumor T-cell response. More CD8 cells and reduced PD-1 expression on CD4 cells correlated with reduced polyps. In addition, a strong CD8-mediated cytotoxic response led to killing of tumor cells in Stat6-deficient ApcMin/+ mice. Therefore, these findings show that Stat6 has an oncogenic role in intestinal tumorigenesis by promoting polyp cell proliferation and immunosuppressive mediators, and preventing an active cytotoxic process.
Introduction
Colorectal cancer (CRC) is one of the leading causes of death among cancer-related deaths worldwide [1]. Familial adenomatous polyposis is genetically acquired and always results in colon cancer. As familial adenomatous polyposis develops due to mutations in the adenomatous polyposis coli (APC) gene, a relevant genetic model is provided by ApcMin/+ mice which have a mutation resulting in truncated APC protein [2]. This mutation in ApcMin/+ mice causes aberrant Wnt signaling leading to polyp development in the small intestine and colon. Hence, this model allows study of earlier stages of CRC which will provide important clues about the mechanism leading to CRC.
Expression of constitutively activated Stat6 in leukemia, lymphomas, and prostate cancer suggests that it contributes to tumor cell proliferation [3]. In T cells, IL-4–induced Stat6 directly upregulates growth factor independence–1 expression, which correlates with decreased expression of cell cycle inhibitor p27Kip1, and results in increased cell proliferation [3], [4]. IL-4–dependent growth of murine and human colon tumor cell lines and primary colorectal cancer cells suggested that it could promote proliferation of intestinal epithelial cells (IECs) similar to that of lymphocytes [5]. As IL-4 was highly expressed in human CRC tissue, lack of correlation between IL-4 expression and overall survival suggested a probable causal role for IL-4 in CRC [6]. Phosphorylated Stat6 was observed in IECs of patients with inflammatory bowel disease; however, as colitis progressed to colitis-associated cancer [7], phosphorylated Stat3 was detected. Hence, further investigation is required to evaluate the role of IL-4–induced Stat6 signaling in intestinal tumorigenesis.
Stat6 is activated by several ligands in addition to its canonical stimuli IL-4 and IL-13. Other Stat6-activating ligands include PDGF, IL-15, IFNα, IL-3, kit ligand, angiotensin II, and obese receptor [8]. Among these ligands, PDGF is noteworthy due to its established tumor-promoting role in glioma, Kaposi's sarcoma, prostate cancer, and pancreatic cancer [9] and because it activates Stat6 in fibroblasts and promotes their proliferation [10]. It is secreted by endothelial cells, epithelial cells, and platelets [9]. Due to vascularization of tumors, endothelial cells constituting blood vessels and platelets are abundant in the tumor microenvironment. PDGF most likely activates Stat6 in stromal cells and IECs in a paracrine manner due to the close proximity of PDGF sources such as endothelial cells and platelets with polyp epithelial cells. Hence, Stat6 could be activated in IECs due to the combined effect of IL-4 and PDGF.
In addition to having a tumor-proliferating function, Stat6 appears to promote the expansion of myeloid suppressor cells similar to myeloid-derived suppressor cells (MDSCs) in a murine model of induced injury and in a murine breast cancer metastasis model [11], [12]. However, there was no difference in MDSC accumulation in IL-4Rα−/− or wild-type (WT) mice with implanted murine colon cancer [13]. MDSCs are immunosuppressive cells that expand in tumor-bearing mice and are tumorigenic in ApcMin/+ mice [14], [15]. They suppress T-cell responses by several mechanisms including the production of IL-4–dependent arginase [15], [16]. Therefore, MDSC expansion in the ApcMin/+ model could be Stat6 dependent.
Stat6 could emerge as a potent tumorigenic factor regulating both polyp cell proliferation and MDSC expansion. To explore this rationale, we genetically disrupted Stat6 in ApcMin/+mice. Disruption of Stat6 function significantly reduced polyps in ApcMin/+ mice due to reduced proliferation of polyp epithelial cells and MDSC accumulation, and increased cytotoxic effectors. Overall, we show that a tumor microenvironment with elevated IL-4 is ideal for the tumorigenic effect of Stat6 in intestinal tumorigenesis.
Materials and Methods
Mice
C57BL/6J and ApcMin/+ mice were purchased from The Jackson Laboratory. Stat6−/− mice on C57BL/6J background were kindly provided by Dr. Joe Craft (Yale University). Both male and female mice at 15 weeks were used in all experiments. WT littermates were used as controls. All mouse protocols were approved by the Yale University Institutional Animal Care and Use Committee.
Histology and Immunohistochemistry
Five-micrometer sections of paraffin-embedded polyps were processed for hematoxylin and eosin staining or immunohistochemistry. Antigen retrieval was done with sodium citrate solution. Antibodies to Ki67 (Cell Signaling), phospho-Stat6 (Thermofisher Scientific), or isotype control were used for immunohistochemistry. Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) assay was done using the Apoptag kit (Millipore). Signal was visualized using the ABC reagent and DAB peroxidase substrate (Vector laboratories).
Flow Cytometry
Splenocytes obtained by mechanical disruption were stained with CD45, CD4, CD8, TCRβ, PD-1, CD11b, Gr-1, Ly6C, Ly6G, CD11c, F4/80, and CD19. For intracellular cytokine staining, splenocytes were activated with 0.5 μg/ml of anti-CD3 and anti-CD28 (eBioscience) for 48 hours; brefeldin A was added during the last 6 hours of culture before fixation. Cells were surface stained, permeabilized with permeabilization buffer (eBioscience), and stained with anti-IFNγ. Lamina propria lymphocytes were prepared by digestion of small intestine fragments with collagenase (Sigma) and DNase (Roche) at 37°C for 30 minutes; live lymphocytes were separated on a Percoll density gradient and stained for flow cytometry as described. Viable CD45+ population was used for all analysis.
Immunoblotting
A total of 3 × 105 HT29 cells obtained from ATCC were used between 3 and 10 passages after thawing. Cells were serum starved for 3 hours followed by treatment with recombinant human IL-4 at 1 ng/ml and human PDGF–BB at 1, 10, and 30 ng/ml for 30 minutes. After lysis with RIPA buffer, equal amounts of protein were loaded on a 4% to 15% polyacrylamide gel and detected for phospho-Stat6, total Stat6 (Cell Signaling), or β-actin (Sigma-Aldrich). ImageJ was used for densitometry.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (PCR)
RNA was extracted with Trizol from size-matched polyps and nontumor small intestine tissue. cDNA was reverse-transcribed from DNase-treated RNA using Superscript III reverse transcriptase (Invitrogen). Target gene amplification was done by SYBR green PCR master mix (Biorad). Expression of target genes was normalized to rpl32. Relative quantification of gene expression was done using the standard curve method, where the same control RNA sample was used to generate standard curve for all assays. Primer sequences are listed in Supplementary Table 1.
Statistical Analysis
GraphPad prism software was used to determine statistical significance by analysis of variance unless otherwise specified. Data are presented as means ± SEM. P value less than .05 was considered significant.
Results
Stat6 Promotes Polyp Progression in the Small Intestine of ApcMin/+ Mice
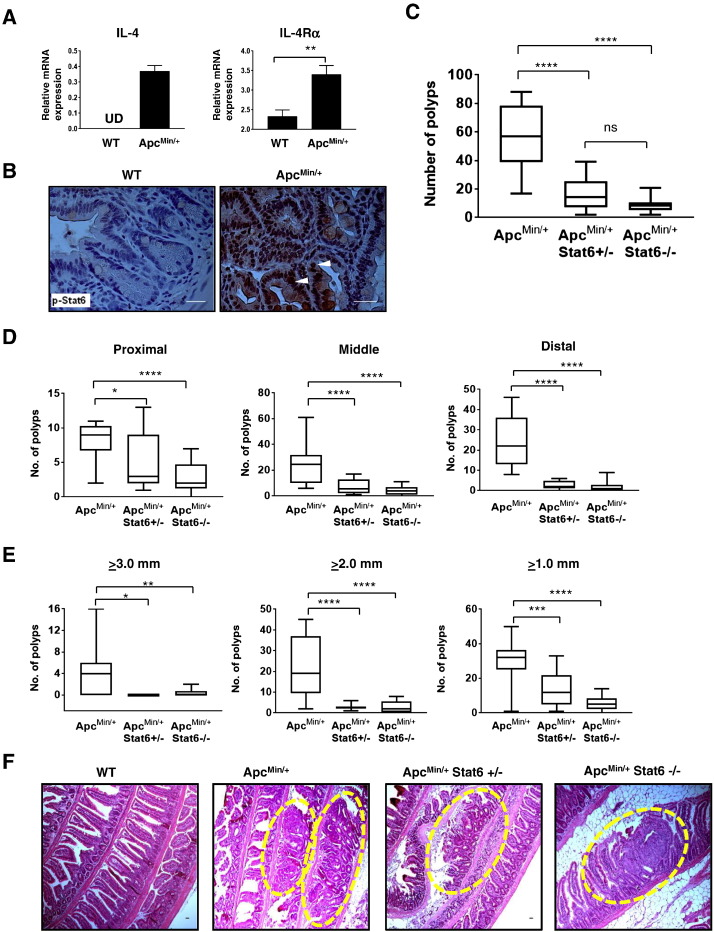
Increased IL-4 and IL-4Rα expression and Stat6 phosphorylation were observed in ApcMin/+ polyps compared to nontumor small intestine tissue from WT mice (Figure 1, A and B). To assess whether Stat6 signaling in ApcMin/+ mice promotes polyp development, we generated ApcMin/+ mice lacking Stat6. Homozygous and heterozygous deletion of Stat6 in ApcMin/+ mice resulted in 86% and 74% reduction in polyps, respectively, compared to ApcMin/+ mice (Figure 1, C and F). There was stronger reduction of polyps in the proximal small intestine in ApcMin/+ Stat6−/− mice compared to ApcMin/+ and ApcMin/+ Stat6+/− mice (Figure 1D). However, ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice had fewer polyps in the middle and distal small intestine than ApcMin/+ mice. Few large polyps (≥3.0 mm) were seen in ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice compared to ApcMin/+mice (Figure 1E). All small polyps (≥2.0 and ≥1.0 mm) were reduced in both ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice; however, there was greater reduction of polyps ≥1.0 mm in the total absence of Stat6. Splenomegaly, loss of Peyer’s patches, and lymphodepletion [17] are characteristic features of tumor progression in ApcMin/+ mice. Total splenocytes were reduced in Stat6-deficient ApcMin/+ mice, indicating reduced splenomegaly compared to ApcMin/+ mice that had more total splenocytes. Further, there were five to eight Peyer’s patches and an overall increase in T and B cells in Stat6-deficient ApcMin/+ mice compared to ApcMin/+ mice with two or less Peyer’s patches and fewer T and B cells (Supplementary Figure 1). Together, these results show that absence or reduction of Stat6 inhibits polyp progression and related features of tumorigenesis in ApcMin/+ mice.
Figure 1.
Deficiency of Stat6 reduces intestinal polyps in ApcMin/+ mice. (A) IL-4 and IL-4Rα mRNA expression in ApcMin/+ polyps (n = 5). UD, undetected. (B) Representative images (630× magnification) of immunohistochemical analysis of phospho-Stat6 in polyps from ApcMin/+ mice and nontumor small intestine tissue from WT. (C) Total polyp counts from each small intestine of ApcMin/+ (n = 14), ApcMin/+ Stat6+/− (n = 12), and ApcMin/+ Stat6−/− mice (n = 16). (D and E) Polyp counts in proximal, middle, and distal segments of the small intestine and number of polyps ≥3.0 mm, 2.0 mm, and 1.0 mm in the groups described above. Data in C to E are presented as box and whiskers plot, where median is shown by the center black line inside the box and min to max values are represented by the closed error bars. (F) Representative images of hematoxylin and eosin–stained sections of small intestine “Swiss rolls” at 100× magnification show intestinal polyps in mice from each group described above. Dashed areas indicate polyps. Scale bars = 1 mm. * P < .05, ** P < .005, *** P < .0005, ****P < .0001.
Stat6 Activation in Tumor-Derived IECs Is Driven by IL-4 and PDGF-BB
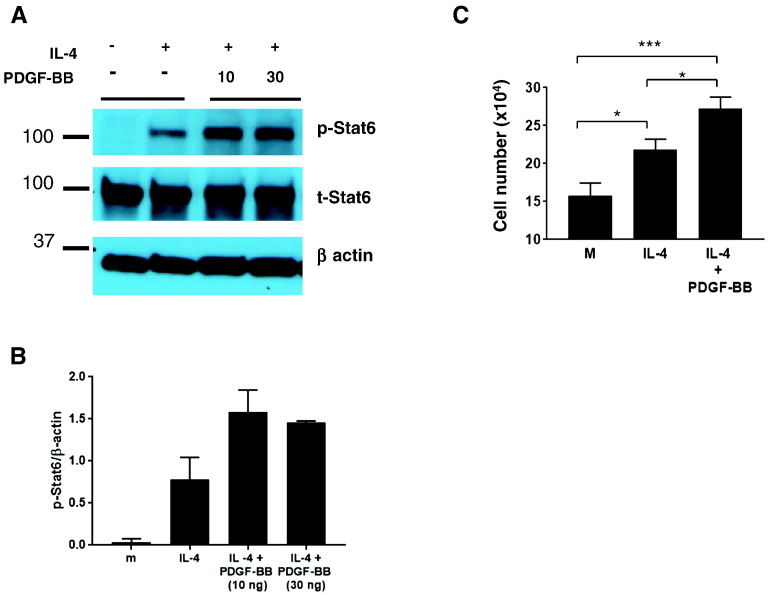
As both IL-4 and PDGF-BB are Stat6-activating agonists, we tested whether they induced Stat6 phosphorylation in IECs. HT29 colorectal cancer–derived IECs were treated with IL-4 alone or IL-4 plus PDGF-BB. IL-4 induced Stat6 phosphorylation, whereas PDGF-BB treatment alone induced no response (Figure 2, A and B; and data not shown). However, combined treatment with IL-4 and PDGF-BB induced stronger Stat6 phosphorylation relative to treatment with IL-4 alone. Further, IL-4 enhanced growth of HT29 cells, and PDGF-BB augmented IL-4–induced proliferation (Figure 2C). Collectively, these results show Stat6 activation in IECs and IEC growth is enhanced by combined action of IL-4 and PDGF-BB.
Figure 2.
Stat6 is activated by IL-4 and PDGF-BB in intestinal epithelial cells. (A) Representative immunoblot of phospho-Stat6 in HT29 cells activated by hIL-4 (1 ng/ml) and hPDGF-BB (10 and 30 ng/ml). Total Stat6 and β-actin were used as loading controls. (B) Densitometry analysis of p-Stat6 normalized to β-actin. (C) HT29 cells treated with hIL-4 (5 ng/ml) and hPDGF-BB (10 ng/ml) for 48 hours were counted by trypan blue assay. Statistical significance was determined by two-tailed Student’s t test. * P < .05, ** P < .005, *** P < .0005.
Stat6 Increased Epithelial Cell Proliferation in ApcMin/+ Polyps
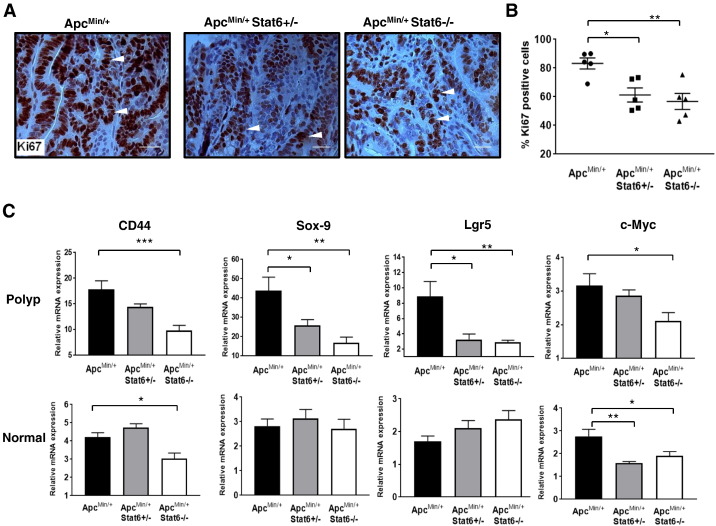
Stat6 phosphorylation in ApcMin/+ polyps and in IL-4 treated HT29 cells, and IL-4 and PDGF-BB–induced proliferation of HT29 cells implied Stat6-mediated proliferation of polyp epithelial cells. Hence, to assess if absence of Stat6 affects proliferation of polyp epithelial cells, we analyzed Ki67+ cells in polyps. Fewer Ki67+ cells were seen in ApcMin/+Stat6+/− and ApcMin/+Stat6−/− polyps than in ApcMin/+ polyps (Figure 3, A and B). Stronger reduction of proliferation in ApcMin/+Stat6−/− mice showed that tumor cell proliferation is partly regulated by Stat6. Further, Wnt target genes associated with rapid proliferation of IECs including CD44, Sox-9, Lgr5, and c-Myc were reduced in polyps of ApcMin/+Stat6−/− mice, but only Sox-9 and Lgr5 were significantly reduced in ApcMin/+Stat6+/− mice polyps (Figure 3C). Further, Vegf-A was unaltered in polyps from ApcMin/+ mice and those lacking Stat6 (Figure 4), suggesting that Stat6 provides a key proliferative signal for polyp epithelial cells other than growth factors supplied by increased vascularization induced by Vegf-A. Notably, Vegf-A was highly expressed in the nontumor small intestinal tissue of ApcMin/+ mice, indicating that aberrant Wnt signaling potentiates a tumorigenic gut environment, which is consistent with higher IL-6, CD44, and c-Myc expression in the nontumor small intestinal tissue of ApcMin/+ mice. Overall, Stat6 promotes polyp development by inducing proliferation of polyp epithelial cells and regulating expression of tumor promoting factors.
Figure 3.
Proliferation of epithelial cells in ApcMin/+ polyps is mediated by Stat6. (A) Representative images (630× magnification) of Ki67+ cells in individual polyps from ApcMin/+, ApcMin/+Stat6+/−, and ApcMin/+ Stat6−/− mice (n = 5/group). (B) Percent Ki67+ cells per field. Four fields were counted per polyp per mouse. (C) mRNA expression of Wnt target genes CD44, Lgr5, Sox-9, and c-Myc in size-matched polyps from ApcMin/+, ApcMin/+ Stat6+/−, and ApcMin/+ Stat6−/− mice (n = 4/group). * P < .05, ** P < .005, *** P < .0005.
Figure 4.
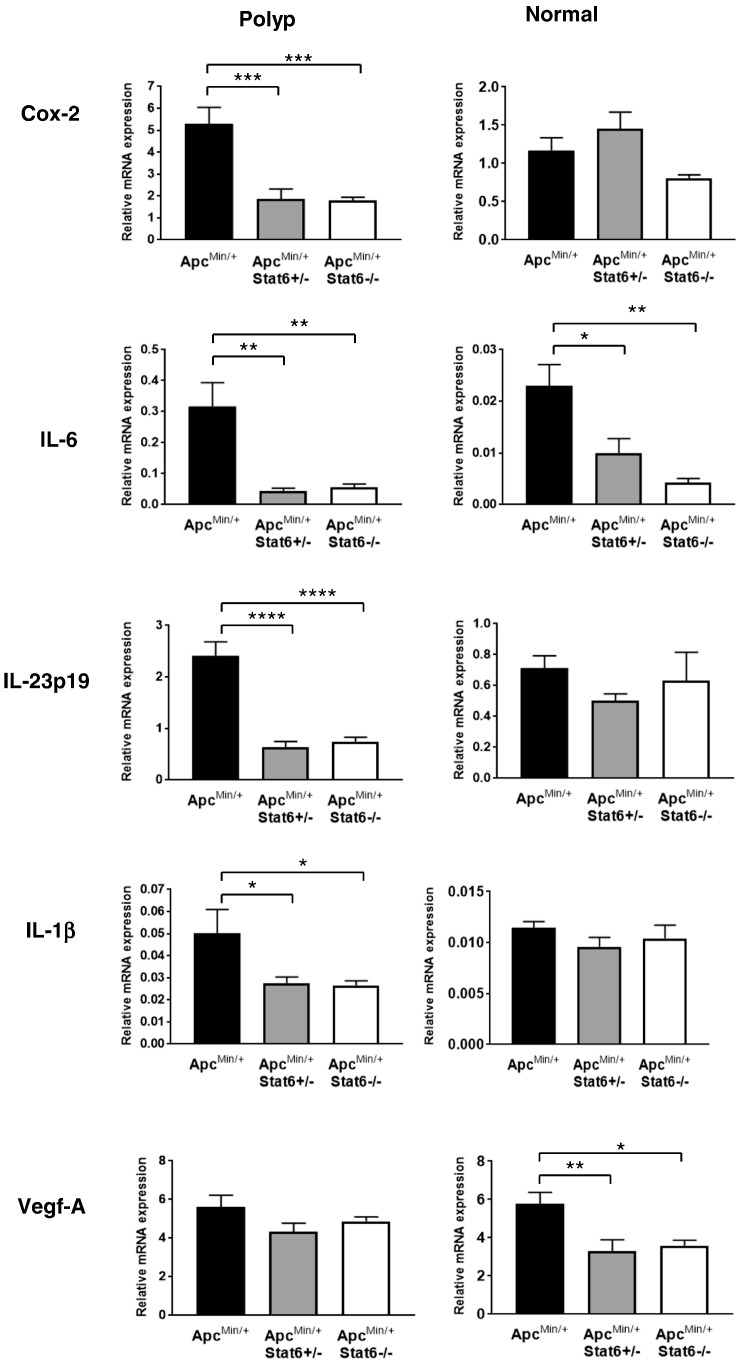
Inflammation is decreased in the small intestine of ApcMin/+ mice deficient in Stat6. Size-matched polyps and nontumor small intestine tissue from ApcMin/+, ApcMin/+ Stat6+/−, and ApcMin/+ Stat6−/− mice (n = 4/group) were analyzed for mRNA expression of Cox-2, IL-1β, IL-6, IL-23p19, and Vegf-A. P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Stat6 Deletion in ApcMin/+ Mice Limits Inflammation in the Small Intestine
To determine whether Stat6 deficiency alters proinflammatory mediators including cytokines in the small intestine of ApcMin/+ mice, mRNA expression of cyclooxygenase-2 (Cox-2), IL-6, IL-23p19, and IL-1β in polyps and nontumor small intestine tissue was assessed. In ApcMin/+ polyps, Cox-2, IL-6, IL-23p19, and IL-1β were upregulated (Figure 4), which is consistent with our previous findings [18]. These proinflammatory mediators were downregulated in polyps from ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice, which correlates with reduced polyps in these mice. IL-6 was lower in the nontumor small intestine tissue of Stat6-deficient ApcMin/+ compared to ApcMin/+ mice, but there was no difference in Cox-2, IL- 23p19, and IL-1β. However, expression of Cox-2, IL-23p19, and IL-1β in the nontumor small intestine tissue of ApcMin/+ mice with and without Stat6 was comparatively lower than that in ApcMin/+ polyps, indicating reduced inflammation. Collectively, absence of Stat6 lowers overall inflammation in polyps and surrounding small intestine tissue, resulting in an unfavorable environment for polyp progression.
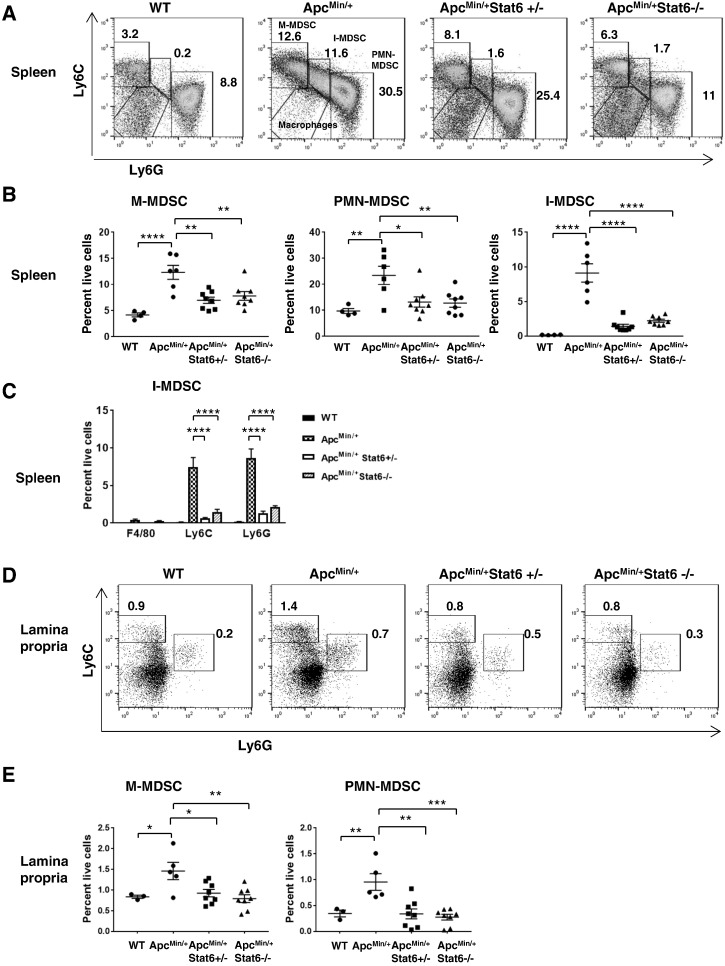
Accumulation of MDSCs in ApcMin/+ Mice Is Stat6 Dependent
MDSCs secrete many tumor-promoting factors including IL-6 and Vegf-A [19]. They suppress T-cell responses via IL-4–dependent arginase and by engaging PD-1 with its ligand PD-L1 [20]. Monocytic MDSCs (M-MDSCs: CD11b+Ly6G--Ly6Chi) and polymorphonuclear-MDSCs (PMN-MDSCs: CD11b+Ly6G+Ly6Clo) accumulated more in both the spleen and lamina propria of ApcMin/+ mice compared to WT monocytes and neutrophils (Figure 5). An additional population that is CD11b+Ly6GloLy6Cint (intermediate-MDSCs or I-MDSCs) which is also CD11b+Gr-1int was observed in ApcMin/+ spleen (Figure 5, A and B, and Supplementary Figure 2C). Similar populations of CD11b+Gr-1int cells were observed in other implanted tumor models [16], [21], [22]. As these cells have medium Ly6C, low Ly6G, but negligible F4/80 expression, they closely resemble monocytes (Figure 5C). Consistent with the strong increase in M-MDSCs and PMN-MDSCs, there was a significant increase in total CD11b+Gr-1+ cells in ApcMin/+ mice compared to WT (Supplementary Figure 2, A and B). Together, these results show that tumor progression in ApcMin/+ mice is associated with expansion of MDSCs. Splenic M-MDSCs and PMN-MDSCs (Figure 5, A and B) were reduced in Stat6-deficient ApcMin/+ mice, which is consistent with an overall decrease of CD11b+Gr1+ cells (Supplementary Figure 2, A and B). I-MDSCs were also significantly reduced in ApcMin/+ mice lacking Stat6 (Figure 5B). A fourth population that is CD11b+Ly6CloLy6G−− appears in ApcMin/+Stat6+/− mice and is increased in ApcMin/+Stat6−/− (Figure 5A and Supplementary Figure 2D), which is the CD11b+F4/80+ macrophages. In the lamina propria, both M-MDSCs and PMN-MDSCs were reduced in Stat6-deficient ApcMin/+ mice (Figure 5, D and E). These findings show that MDSCs do not accumulate in the spleen or lamina propria of ApcMin/+ mice in the absence of Stat6, which correlates with polyp inhibition.
Figure 5.
Reduction of MDSCs in ApcMin/+ mice deficient in Stat6. (A and B) Representative plots of M-MDSCs (CD11b+Ly6G−Ly6Chi), I-MDSCs (CD11b+Ly6GloLy6Cint), and PMN-MDSCs (CD11b+Ly6G+Ly6Clo) gated on CD45+CD11bhiCD11clo live splenocytes from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), ApcMin/+ Stat6−/− mice (n = 8), and WT (n = 4). (C) Expression of Ly6C, Ly6G, and F4/80 on splenic I-MDSCs in the groups described above. (D and E) CD45+CD11b+CD11clo lamina propria cells from ApcMin/+ (n = 5), ApcMin/+ Stat6+/− mice (n = 8), ApcMin/+ Stat6−/− mice (n = 8), and WT (n = 3) were analyzed for M-MDSCs and PMN-MDSCs. Percentage of total CD45 cells is shown in plots. In WT mice, CD11b+Ly6G−Ly6C+ cells are monocytes and CD11b+Ly6G+Ly6Clo cells are neutrophils. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
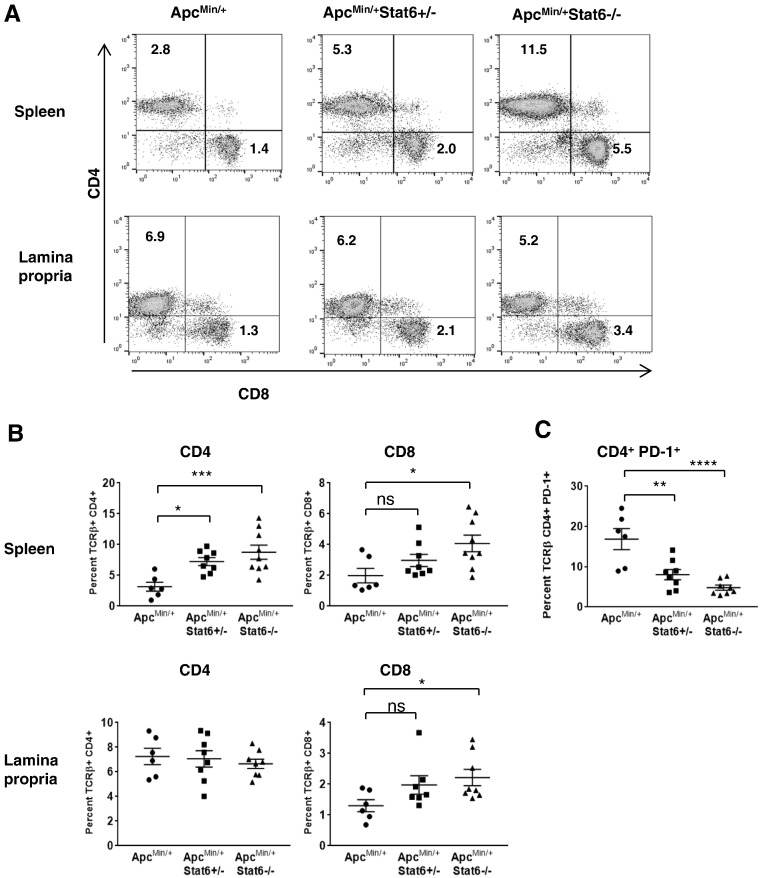
PD-1 Expression on CD4 Cells Is Reduced in ApcMin/+ Mice Deficient in Stat6
Fewer splenic lymphocytes including CD4 and CD8 cells were observed in ApcMin/+ mice than in Stat6-deficient ApcMin/+ mice (Figure 6, A and B, and Supplementary Figure 1, C and D). However, the proportion of lamina propria CD4 cells was similar between ApcMin/+ and Stat6-deficient ApcMin/+ mice. To determine if lamina propria CD4 cells were qualitatively altered by the absence of Stat6 in ApcMin/+mice, we analyzed PD-1 expression on T cells as it is associated with functional exhaustion and inhibition of immune response. Lamina propria CD4 T cells from ApcMin/+mice had high PD-1 expression, whereas it was downregulated on CD4 cells from ApcMin/+ mice lacking Stat6 (Figure 6C). Hence, CD4 cells in ApcMin/+Stat6−/− have the potential to function normally by producing sufficient IL-2 to favor an effective CD8-mediated cytotoxic effect.
Figure 6.
Elevated CD8 T cells in Stat6-deficient ApcMin/+ correlates with downregulation of PD-1 on CD4 T cells. (A and B) Splenocytes and lamina propria cells from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), and ApcMin/+ Stat6−/− mice (n = 8) were analyzed for CD4 and CD8 T cells. Student’s t test was used to determine statistical significance. (C) Lamina propria cells from the above groups were analyzed for CD4 T cells expressing PD-1. * P < .05, ** P < .005, *** P < .0005, **** P < .0005.
Cytotoxic CD8 Cells Inhibit Polyp Progression in the Absence of Stat6
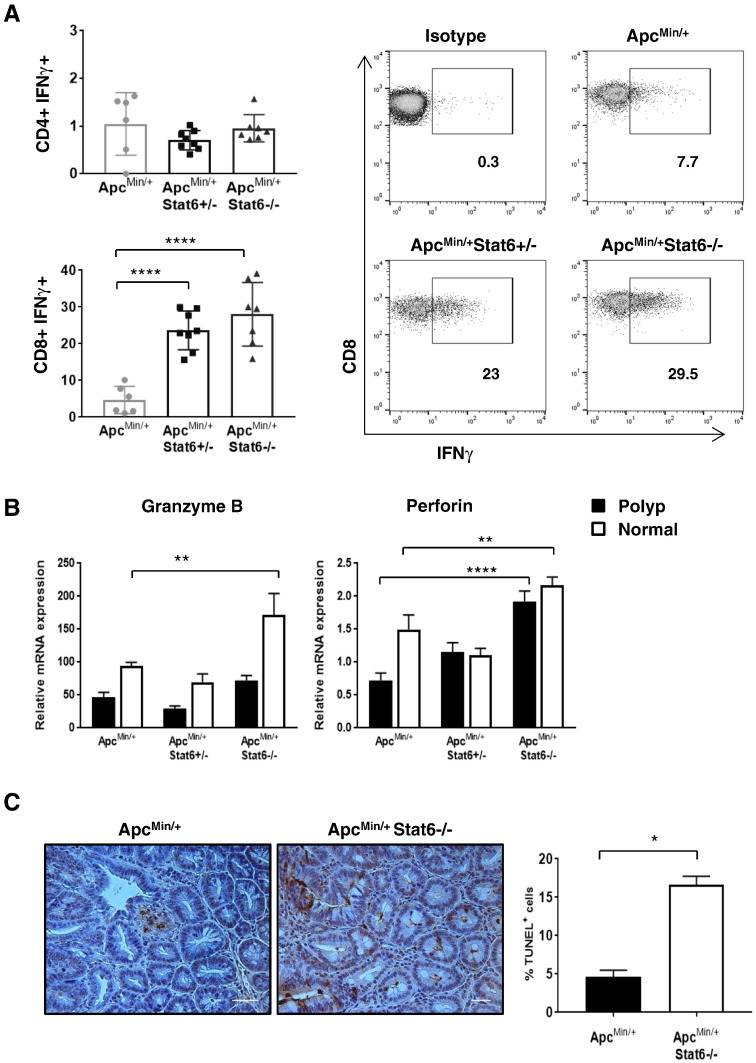
To assess if reduction of MDSCs in the lamina propria and spleen improved T-cell function, T cells and their effector functions were analyzed. More CD8 T cells accumulated in the spleen and lamina propria of ApcMin/+Stat6−/− mice (Figure 6, A and B) than in ApcMin/+Stat6+/− and ApcMin/+ groups. Splenic CD4 T cells were significantly higher in ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice compared to ApcMin/+. Next, we found that CD8 cells from ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice produced more IFNγ compared to those from ApcMin/+ mice (Figure 7A). CD4 cells from all groups were poor producers of IFNγ. Perforin was increased in the polyps and nontumor small intestine tissue of ApcMin/+Stat6−/− mice compared to ApcMin/+ polyps, whereas granzyme B was elevated in the nontumor small intestine tissue from ApcMin/+Stat6−/− mice (Figure 7B). Further, more apoptosis in Stat6-deficient ApcMin/+ polyps than in ApcMin/+ polyps showed killing of polyp cells (Figure 7C). Together, our results show that, in the absence of Stat6, CD8 T cells in ApcMin/+ mice acquire cytotoxic capacity and inhibit tumor cells which effectively limit polyp progression.
Figure 7.
CD8-mediated cytotoxic mediators inhibit polyp growth in Stat6-deficient ApcMin/+ mice. (A), IFNγ expression by splenocytes from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), and ApcMin/+ Stat6−/− mice (n = 7) that were stimulated in vitro with 0.5 μg/ml of CD3 and CD28 antibodies. (B) mRNA expression of granzyme B and perforin in polyps and nontumor small intestine tissue of ApcMin/+ (n = 5), ApcMin/+ Stat6+/− mice (n = 4), and ApcMin/+ Stat6−/− mice (n = 5). Statistical significance was determined by two-way analysis of variance. (C) TUNEL assay showing apoptosis of polyp epithelial cells in ApcMin/+ (n = 3) and ApcMin/+ Stat6−/− mice (n = 3). TUNEL-positive cells were counted in three to four fields per polyp/mouse. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Discussion
In the ApcMin/+ model, initiation of intestinal tumorigenesis by aberrant Wnt signaling is promoted by IL-17 [18], EGF-activated ERK signaling [23], or TLR activation [24]. The abundance of IL-4 in ApcMin/+ polyps [25] and the tumorigenic function of PDGF suggested that Stat6 might be important for promoting intestinal tumors. We demonstrate that Stat6 provides a proliferative signal promoting polyp epithelial cell growth and regulates expansion of M-MDSCs, I-MDSCs, and PMN-MDSCs, thereby contributing to intestinal tumorigenesis.
In chemically induced murine colon cancer, although IL-4Rα deletion reduced colonic tumors [26], there was no difference in tumors arising from MC38 cells with knockdown of IL-4Rα and unaltered MC38 cells. Formation of more aberrant crypt foci in the absence of IL-4Rα indicated that IL-4 could be inhibiting polyps [27]. However, IL-4 was ineffective as a therapeutic [28]. Therefore, by deleting Stat6 which is the main downstream signaling mediator of IL-4, we targeted transcriptional control of polyp formation. Furthermore, on finding that Stat6 could be costimulated by IL-4 and PDGF-BB, we reasoned that Stat6 activation by more than one ligand contributes to its central role in regulating polyp epithelial cell proliferation. This is consistent with the potent tumorigenic effect of PDGF in glioma, Kaposi's sarcoma, prostate cancer, and pancreatic cancer, and elevated PDGF receptor expression on tumor and stromal cells [9]. To our knowledge, we have shown for the first time that PDGF-BB augments IL-4–induced proliferation of a tumor-derived intestinal cell line.
Although ApcMin/+ mice with heterozygous or homozygous deletion of Stat6 resulted in similar reduction of total polyps, ApcMin/+Stat6+/− mice had more tumors in the distal segment of the small intestine (P < .05 vs. P < .0001) and more small polyps (>1.0 mm; P < .005 vs. P < .0001) compared to ApcMin/+Stat6−/−. This could be due to dampened cytotoxic effect in ApcMin/+ mice with heterozygous Stat6 expression. However, there was similar reduction of total polyps in ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice. This is consistent with reduced proliferation of polyp epithelial cells and reduced expression of stem cell markers Lrgr5 and Sox-9 in both ApcMin/+Stat6+/− and ApcMin/+Stat6−/− mice compared to ApcMin/+ mice, suggesting that Stat6-mediated proliferation is critical for polyp formation. Similarly, in an implanted pancreatic cancer model, both heterozygous and homozygous deletion of host Stat6 resulted in tumor reduction compared to WT tumor-bearing mice [29]. Notably, Vegf-A does not fully compensate for the absence of Stat6-induced proliferation in ApcMin/+ polyps. Therefore, Stat6 signaling is critical for polyp epithelial cell proliferation.
MDSCs and immune checkpoint inhibitor PD-1 curtail antitumor immunity by immunosuppression [30]. MDSCs suppress T cells by depleting essential arginine with IL-4–dependent arginase or by engaging PD-1 [15], [20]. Elevation of M-MDSCs in the lamina propria suggests that T cells could be suppressed by IL-4–dependent arginase as the immunosuppressive capacity of M-MDSCs is dependent on IL-4Rα [16]. Previously, it was shown that blocking PD-1 resulted in CD8-mediated rejection of MC38 tumors [31]. Increased MDSCs and CD4+PD-1+ cells in ApcMin/+ lamina propria imply inhibition of CD4 function by MDSCs via PD-1 interaction. Absence of CD4-mediated help may diminish the antitumor effect of CD8 T cells. Reduced M-MDSCs and PMN-MDSCs, and strong reduction of PD-1 expression on CD4 cells in the local tumor environment of Stat6-deficient ApcMin/+ mice likely allowed development of an efficient cytotoxic response resulting in increased apoptotic killing of polyp epithelial cells. We are currently investigating whether MDSCs directly suppress CD8 cells or inhibit CD8 cells via CD4 T-cell suppression.
Accumulation of splenic MDSCs is linked to increased tumor burden as shown previously in lung, colon, and breast cancer models [32]. In addition to suppressing immune response, MDSCs secrete tumorigenic factors including IL-6 and Vegf-A [19]. In ApcMin/+ mice, more splenic MDSCs are reflected by increased MDSCs in the lamina propria and increased expression of IL-6 and Vegf-A in polyps, providing evidence that MDSCs contribute to tumorigenesis. Our finding is supported by a study where inhibition of CCL2-mediated recruitment of MDSCs to the intestine of ApcMin/+ mice reduced polyps [14]. In our study, expansion of all MDSC subsets is Stat6 dependent in the ApcMin/+ model, implying an association between Stat6-dependent intestinal tumorigenesis and MDSC expansion. As IL-4 signaling on host stromal cells was ineffective in modifying MDSC expansion in an implanted murine colon cancer model [13], Stat6 signaling in ApcMin/+ polyp cells might be critical for expanding MDSCs. As MDSCs eventually differentiate to macrophages or neutrophils [15], reduction of I-MDSCs, which have monocytic characteristics, in the absence of Stat6 could be due to differentiation into a specific macrophage subset or failure to accumulate. In our system, increased accumulation of CD11bhi F4/80+ macrophages in the absence of Stat6 could be due to I-MDSCs or M-MDSCs differentiating into M1-like macrophages. These cells are unlikely to be M2-like TAMs as classical M2 markers arginase, Fizz1, CD206, and Ym1 are Stat6 dependent [33], [34]. Instead, they could be aiding antitumor immunity as shown in an implanted breast cancer induced metastasis model where breast cancer metastasis was likely prevented by tumoricidal M1 macrophages in the absence of Stat6 [12].
Stat6 mediates two processes contributing to intestinal polyp progression. First, it directly promotes proliferation of polyp epithelial cells, and second, it mediates expansion of MDSCs, which possibly limits effective antitumor CD8 cytotoxicity. Furthermore, positive correlation between polyp numbers and MDSCs suggests that these immunosuppressive cells promote intestinal polyps. In this model, we found I-MDSCs that are equivalent to the previously identified CD11b+Gr-1int cells. Further analysis of Stat6-dependent genes in I-MDSCs could reveal novel therapeutic targets. Collectively, our findings show that disruption or reduction of Stat6 could significantly reduce the progression of adenomatous polyposis by inhibiting polyp growth and downregulating immunosuppressive mechanisms. Therefore, we show that Stat6 signaling is critical for the preliminary stages of intestinal tumorigenesis.
Conclusions
-
•
Stat6 promotes proliferation of polyp epithelial cells determining the progression of early tumorigenesis.
-
•
M-MDSC, I-MDSC, and PMN-MDSC expansion is promoted by Stat6 in the ApcMin/+ model.
-
•
Cytotoxic CD8 response inhibits polyp epithelial cells and is associated with reduction of MDSCs.
The following are the supplementary data related to this article.
List of Primers Used in Quantitative Reverse Transcriptase PCR Analysis
Distribution of lymphocyte populations and secondary lymphoid structures in ApcMin/+ and Stat6-deficient ApcMin/+ mice. (A) Absolute cell number from spleens of ApcMin/+ (n = 12), ApcMin/+ Stat6+/− (n = 7), and ApcMin/+ Stat6−/− (n = 12) mice were counted. (B) Total Peyer’s patches per mouse from ApcMin/+ (n = 7), ApcMin/+ Stat6+/− (n = 5), and ApcMin/+ Stat6−/− (n = 10) mice were counted. (C and D) Splenic T and B cells from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− (n = 8), and ApcMin/+ Stat6−/− (n = 8) mice were analyzed by flow cytometry. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Analysis of splenic MDSCs based on Gr-1 expression and splenic macrophages in ApcMin/+ and Stat6-deficient ApcMin/+ mice. (A and B) CD11b+Gr-1+ MDSCs in ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), and ApcMin/+ Stat6−/− mice (n = 8), and CD11b+Gr-1+ cells in WT (n = 4). Representative plots are shown. (C) CD11b+Gr-1hi, CD11b+Gr-1int, and CD11b+Gr-1lo cells with varying expression of Ly6C and Ly6G. (D) F4/80, Ly6C, and Ly6G expression on CD11b+ cells that are negative for Ly6G or Ly6C from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), ApcMin/+ Stat6−/− mice (n = 8), and WT (n = 4) was analyzed. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Gating strategy for lamina propria MDSCs. (A) MDSCs were gated on live CD45+CD11b+CD11clo viable lamina propria cells. Subgates separated them into M-MDSCs (CD11b+ Ly6G− Ly6Chi) and PMN-MDSCs (CD11b+ Ly6G+ Ly6Clo). Similar gating was used for analyzing spleen MDSCs.
Acknowledgements
We would like to thank Dr. Marcus Bosenberg and Dr. Jemin Choi for their helpful suggestions and critical reading of the manuscript, and Dr. Tiago Castilho for his advice on ApcMin/+ genotyping. Many thanks to all our lab members for helpful discussions and the Yale Histology core facility for tissue processing.
References
- 1.Half E, Bercovich D, Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis. 2009;4:22. doi: 10.1186/1750-1172-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 3.Bruns HA, Kaplan MH. The role of constitutively active Stat6 in leukemia and lymphoma. Crit Rev Oncol Hematol. 2006;57:245–253. doi: 10.1016/j.critrevonc.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 5.Todaro M, Lombardo Y, Francipane MG, Alea MP, Cammareri P, Iovino F, Di Stefano AB, Di Bernardo C, Agrusa A, Condorelli G. Apoptosis resistance in epithelial tumors is mediated by tumor-cell–derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 6.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pages F. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 7.Wick EC, LeBlanc RE, Ortega G, Robinson C, Platz E, Pardoll DM, Iacobuzio-Donahue C, Sears CL. Shift from pStat6 to pStat3 predominance is associated with inflammatory bowel disease–associated dysplasia. Inflamm Bowel Dis. 2012;18:1267–1274. doi: 10.1002/ibd.21908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y. Multifarious functions of PDGFs and PDGFRs in tumor growth and metastasis. Trends Mol Med. 2013;19:460–473. doi: 10.1016/j.molmed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Patel BK, Wang LM, Lee CC, Taylor WG, Pierce JH, LaRochelle WJ. Stat6 and Jak1 are common elements in platelet-derived growth factor and interleukin-4 signal transduction pathways in NIH 3T3 fibroblasts. J Biol Chem. 1996;271:22175–22182. doi: 10.1074/jbc.271.36.22175. [DOI] [PubMed] [Google Scholar]
- 11.Munera V, Popovic PJ, Bryk J, Pribis J, Caba D, Matta BM, Zenati M, Ochoa JB. Stat 6-dependent induction of myeloid derived suppressor cells after physical injury regulates nitric oxide response to endotoxin. Ann Surg. 2010;251:120–126. doi: 10.1097/SLA.0b013e3181bfda1c. [DOI] [PubMed] [Google Scholar]
- 12.Sinha P, Clements VK, Ostrand-Rosenberg S. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol. 2005;174:636–645. doi: 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- 13.Sinha P, Parker KH, Horn L, Ostrand-Rosenberg S. Tumor-induced myeloid-derived suppressor cell function is independent of IFN-gamma and IL-4Ralpha. Eur J Immunol. 2012;42:2052–2059. doi: 10.1002/eji.201142230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, Soucy G, Odze R, Glickman JN, Garrett WS. CCL2 promotes colorectal carcinogenesis by enhancing polymorphonuclear myeloid-derived suppressor cell population and function. Cell Rep. 2015;12:244–257. doi: 10.1016/j.celrep.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmgaard RB, Zamarin D, Li Y, Gasmi B, Munn DH, Allison JP, Merghoub T, Wolchok JD. Tumor-expressed IDO recruits and activates MDSCs in a Treg-dependent manner. Cell Rep. 2015;13:412–424. doi: 10.1016/j.celrep.2015.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coletta PL, Muller AM, Jones EA, Muhl B, Holwell S, Clarke D, Meade JL, Cook GP, Hawcroft G, Ponchel F. Lymphodepletion in the ApcMin/+ mouse model of intestinal tumorigenesis. Blood. 2004;103:1050–1058. doi: 10.1182/blood-2003-03-0707. [DOI] [PubMed] [Google Scholar]
- 18.Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:5540–5544. doi: 10.1073/pnas.0912675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, Geilich M, Winkels G, Traggiai E, Casati A. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol. 2010;40:22–35. doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- 22.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Lee SH, Hu LL, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, Herdman S, Varki N, Corr M, Lee J. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi Y, Nakatsuji M, Seno H, Ishizu S, Akitake-Kawano R, Kanda K, Ueo T, Komekado H, Kawada M, Minami M. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis. 2011;32:1333–1339. doi: 10.1093/carcin/bgr128. [DOI] [PubMed] [Google Scholar]
- 26.Koller FL, Hwang DG, Dozier EA, Fingleton B. Epithelial interleukin-4 receptor expression promotes colon tumor growth. Carcinogenesis. 2010;31:1010–1017. doi: 10.1093/carcin/bgq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko CW, Cuthbert RJ, Orsi NM, Brooke DA, Perry SL, Markham AF, Coletta PL, Hull MA. Lack of interleukin-4 receptor alpha chain-dependent signalling promotes azoxymethane-induced colorectal aberrant crypt focus formation in Balb/c mice. J Pathol. 2008;214:603–609. doi: 10.1002/path.2316. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki A, Leland P, Joshi BH, Puri RK. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine. 2015;75:79–88. doi: 10.1016/j.cyto.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 29.Yan D, Wang HW, Bowman RL, Joyce JA. STAT3 and STAT6 signaling pathways synergize to promote cathepsin secretion from macrophages via IRE1alpha activation. Cell Rep. 2016;16:2914–2927. doi: 10.1016/j.celrep.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusmartsev S, Gabrilovich DI. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer Immunol Immunother. 2006;55:237–245. doi: 10.1007/s00262-005-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72:917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WFt, Cavassani KA, Li X, Lukacs NW. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Primers Used in Quantitative Reverse Transcriptase PCR Analysis
Distribution of lymphocyte populations and secondary lymphoid structures in ApcMin/+ and Stat6-deficient ApcMin/+ mice. (A) Absolute cell number from spleens of ApcMin/+ (n = 12), ApcMin/+ Stat6+/− (n = 7), and ApcMin/+ Stat6−/− (n = 12) mice were counted. (B) Total Peyer’s patches per mouse from ApcMin/+ (n = 7), ApcMin/+ Stat6+/− (n = 5), and ApcMin/+ Stat6−/− (n = 10) mice were counted. (C and D) Splenic T and B cells from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− (n = 8), and ApcMin/+ Stat6−/− (n = 8) mice were analyzed by flow cytometry. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Analysis of splenic MDSCs based on Gr-1 expression and splenic macrophages in ApcMin/+ and Stat6-deficient ApcMin/+ mice. (A and B) CD11b+Gr-1+ MDSCs in ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), and ApcMin/+ Stat6−/− mice (n = 8), and CD11b+Gr-1+ cells in WT (n = 4). Representative plots are shown. (C) CD11b+Gr-1hi, CD11b+Gr-1int, and CD11b+Gr-1lo cells with varying expression of Ly6C and Ly6G. (D) F4/80, Ly6C, and Ly6G expression on CD11b+ cells that are negative for Ly6G or Ly6C from ApcMin/+ (n = 6), ApcMin/+ Stat6+/− mice (n = 8), ApcMin/+ Stat6−/− mice (n = 8), and WT (n = 4) was analyzed. * P < .05, ** P < .005, *** P < .0005, **** P < .0001.
Gating strategy for lamina propria MDSCs. (A) MDSCs were gated on live CD45+CD11b+CD11clo viable lamina propria cells. Subgates separated them into M-MDSCs (CD11b+ Ly6G− Ly6Chi) and PMN-MDSCs (CD11b+ Ly6G+ Ly6Clo). Similar gating was used for analyzing spleen MDSCs.