ABSTRACT
Dengue is an acute febrile illness caused by dengue virus (DENV) and a major cause of morbidity and mortality in tropical and subtropical regions of the world. The lack of an appropriate small-animal model of dengue infection has greatly hindered the study of dengue pathogenesis and the development of therapeutics. In this study, we conducted mass spectrometry-based serum metabolic profiling from a model using humanized mice (humice) with DENV serotype 2 infection at 0, 3, 7, 14, and 28 days postinfection (dpi). Forty-eight differential metabolites were identified, including fatty acids, purines and pyrimidines, acylcarnitines, acylglycines, phospholipids, sphingolipids, amino acids and derivatives, free fatty acids, and bile acid. These metabolites showed a reversible-change trend—most were significantly perturbed at 3 or 7 dpi and returned to control levels at 14 or 28 dpi, indicating that the metabolites might serve as prognostic markers of the disease in humice. The major perturbed metabolic pathways included purine and pyrimidine metabolism, fatty acid β-oxidation, phospholipid catabolism, arachidonic acid and linoleic acid metabolism, sphingolipid metabolism, tryptophan metabolism, phenylalanine metabolism, lysine biosynthesis and degradation, and bile acid biosynthesis. Most of these disturbed pathways are similar to our previous metabolomics findings in a longitudinal cohort of adult human dengue patients across different infection stages. Our analyses revealed the commonalities of host responses to DENV infection between humice and humans and suggested that humice could be a useful small-animal model for the study of dengue pathogenesis and the development of dengue therapeutics.
IMPORTANCE Dengue virus is the most widespread arbovirus, causing an estimated 390 million dengue infections worldwide every year. There is currently no effective treatment for the disease, and the lack of an appropriate small-animal model of dengue infection has greatly increased the challenges in the study of dengue pathogenesis and the development of therapeutics. Metabolomics provides global views of small-molecule metabolites and is a useful tool for finding metabolic pathways related to disease processes. Here, we conducted a serum metabolomics study on a model using humanized mice with dengue infection that had significant levels of human platelets, monocytes/macrophages, and hepatocytes. Forty-eight differential metabolites were identified, and the underlying perturbed metabolic pathways are quite similar to the pathways found to be altered in dengue patients in previous metabolomics studies, indicating that humanized mice could be a highly relevant small-animal model for the study of dengue pathogenesis and the development of dengue therapeutics.
KEYWORDS: dengue fever, humanized mice, mass spectrometry, metabolomics, systems biology
INTRODUCTION
Dengue is an acute febrile illness caused by dengue virus (DENV) and is a major cause of morbidity and mortality in tropical and subtropical regions of the world. It is arguably the most important arboviral disease globally, with an estimated 390 million infections occurring every year, nearly 100 million of which are clinically apparent (1, 2). In the absence of an effective antiviral drug and with the rapid spread of DENV, which is now circulating in Asia, Africa, and the Americas, dengue has become a major public health threat worldwide (3).
Dengue manifests in a wide spectrum of clinical symptoms ranging from mild dengue fever (DF) to life-threatening dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS). The symptoms of dengue are usually accompanied by hematological changes, such as leukopenia and thrombocytopenia in mild cases and plasma leakage, hemorrhage, or organ impairment, such as liver damage, in severe cases (4). A major challenge in the study of dengue pathogenesis and the development of therapeutics for dengue is the lack of an appropriate small-animal model of dengue infection, despite efforts to develop relevant mouse models (5–7). Among these models, the development of humice, which are immunodeficient mice stably reconstituted with human immune cells, is appealing because it makes it possible to study DENV infection in a human-like context. We have successfully constructed humice by adoptive transfer of human CD34+ fetal liver cells into NOD-scid Il2rg−/− (NSG) mice, which develop significant levels of human platelets, monocytes/macrophages, and hepatocytes (8). Infection of these mice with DENV serotype 2 (DENV2) recapitulates some of the characteristic features of DENV infection in humans, including transient leukopenia, thrombocytopenia, and liver damage (8).
Metabolomics is a rapidly emerging fields of “omics” research that provides an analysis of the global metabolite changes in biological systems in response to biological stimuli or perturbations (9). As the end products of cellular regulatory processes, metabolites not only play critical roles in biology, but also provide a functional readout of cellular biochemistry, which is important for finding metabolic pathways related to disease processes (10, 11). In our previous study, a systematic characterization of serum metabolome changes in a longitudinal cohort of adult dengue patients across three infection stages (febrile, defervescence, and convalescence) was conducted, and many metabolic pathways linked to disease progression were identified (12). In the present study, we extended our metabolomics study to serum samples from humice at multiple time points after DENV infection, with the aim of comparing the similarities and differences of metabolome changes between dengue patients and humice. Our results showed that most perturbed metabolic pathways were common between dengue patients and DENV-infected humice, indicating humice could be a highly relevant small-animal model for the study of dengue pathogenesis and the development of therapeutics.
RESULTS
Systemic DENV infection and characteristic responses in humice.
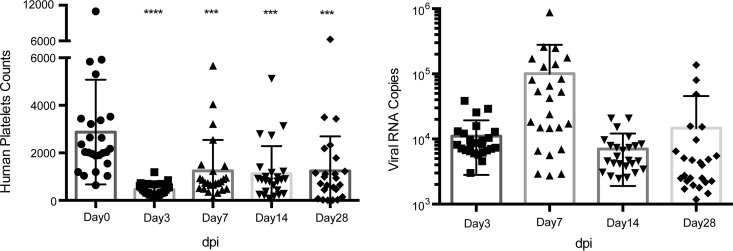
After the construction of humice, mouse peripheral blood cells were analyzed for human leukocyte reconstruction, which showed the presence of human monocytes/macrophages and platelets in the humice. Humice with 10% or more reconstitution levels [using the following formula: human CD45+ cells/(human CD45+ cells + mouse CD45+ cells) × 100] in the blood were used for subsequent experiments. After intravenous infection with 1 × 107 PFU DENV2 07K2861, viremia was detected in the infected humice from 3 days postinfection (dpi) and persisted until 28 dpi by quantitative reverse transcription (qRT)-PCR for the Env gene of viral RNA. The viremia peaked at 7 dpi, which was consistent with our previous studies (Fig. 1A) (8). Human-specific thrombocytopenia was also observed in the humice (Fig. 1B), which specifically identified human and mouse platelets by human CD41 (hCD41) and mouse CD41 (mCD41) cell surface markers. The human platelets dropped significantly from 2,875 (mean value) before infection to 458 (mean value) by day 3 dpi. Overall, based on these model characteristics, serum samples were collected at 0, 3, 7, 14, and 28 dpi, corresponding to the febrile, defervescence, and convalescence phases in humans, for the metabolomics study.
FIG 1.
The thrombocytopenia of human platelets and the viral RNA load were detected in dengue virus-infected humanized mice. Twenty-five humanized mice were infected with 1 × 107 PFU Den2/07K2861 virus intravenously (i.v.). Subsequently, the mice were bled at the indicated times (dpi), and the whole blood was used for platelet counting, while the serum was used for viral load measurement. (A) The human platelets dropped significantly after virus infection from 3 dpi to 28 dpi compared with preinfection levels. Ten microliters of humanized mouse whole blood was stained for human CD41 and mouse CD41, and then human and mouse platelets were counted by flow cytometry using fluorescent beads. (B) Viremia was measured by virus-specific qRT-PCR in serum. The serum samples were collected at the indicated times (dpi) and used to extract viral RNA. The data shown are the numbers of viral RNA copies per microliter of serum. The qRT-PCR detection limit was approximately 1,000 copies.
Metabolomics analysis by LC-MS.
We characterized serum metabolome changes of both humice and NSG mice with DENV infection using liquid chromatography-mass spectrometry (LC-MS). Both reverse-phase ultrahigh performance liquid chromatography (RP-UHPLC) and hydrophilic interaction ultrahigh performance liquid chromatography (HILIC-UHPLC) techniques were adopted here to expand the coverage of both polar and nonpolar metabolites (13). In order to obtain reliable metabolic profiles of the samples, it was important to ensure the robustness of the analytical method. We first evaluated the stability and reproducibility of the LC-MS method by performing principal-components analysis (PCA) on all the samples, including the 7 quality control (QC) samples (14). As shown in Fig. S1 in the supplemental material, the QC samples are clustered in PCA score plots of sera, indicating good stability and reproducibility of the chromatographic separation during the whole sequence.
The transient viremia, leukopenia, and thrombocytopenia of the model with DENV infection suggested the illness was self-limiting in humice. This is similar to DF patients, who recover uneventfully after a period of acute illness. Thus, we adopted the same strategy used in our previous metabolomics study on DF patients by focusing on the identification of the differential metabolites that showed a reversible-change trend along the time course, because these metabolites were more relevant to the disease progression (12).
Metabolome changes between humice and NSG mice without DENV infection.
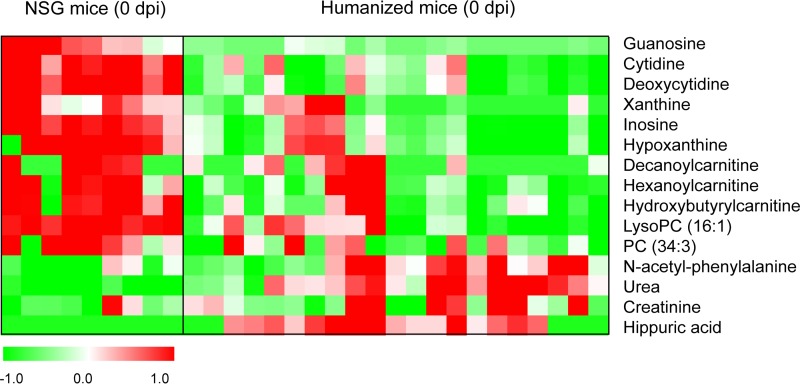
We first studied metabolome differences between humice and NSG mice before DENV infection to determine how the adoptive transfer of human CD34+ fetal liver cells into NSG mice would change their metabolome. About 120 differential features between humice and NSG mice were found, and the structures of 15 metabolites were identified, including purines and pyrimidines, acylcarnitines, and phospholipids (Fig. 2). Compared to NSG mice, most of these differential metabolites showed lower levels in humice, and the levels of purines and pyrimidines decreased most significantly (3 to 4 times). Meanwhile, the levels of reconstitution in humice showed significant negative correlations to the two nucleosides cytidine (r = −0.64; P = 0.002) and deoxycytidine (r = −0.61; P = 0.003) by Pearson correlation analysis (see Fig. S2 in the supplemental material).
FIG 2.
Heat map of identified differential metabolites between humice and NSG mice without dengue infection. Each row shows the ion intensity for a specific metabolite after mean centering and unit variance scaling of the data. Each column shows the serum metabolic profiles of humice and NSG mice at 0 dpi.
Metabolome changes of NSG mice and humice with DENV infection.
Although NSG mice with DENV infection did not show viremia or any of the characteristic features of dengue patients, we still observed about 30 differential features that showed a reversible-change trend between noninfected and DENV-infected NSG mice. Five of these metabolites, hypoxanthine, deoxycytidine, and 3 phospholipids, were identified (see Fig. S3 in the supplemental material). The phospholipids first decreased, reached their lowest levels at around 7 dpi, and then gradually returned to the control levels at 28 dpi. Conversely, hypoxanthine and deoxycytidine were increased, reached their highest levels at 3 or 7 dpi, and then returned to the control levels at 28 dpi.
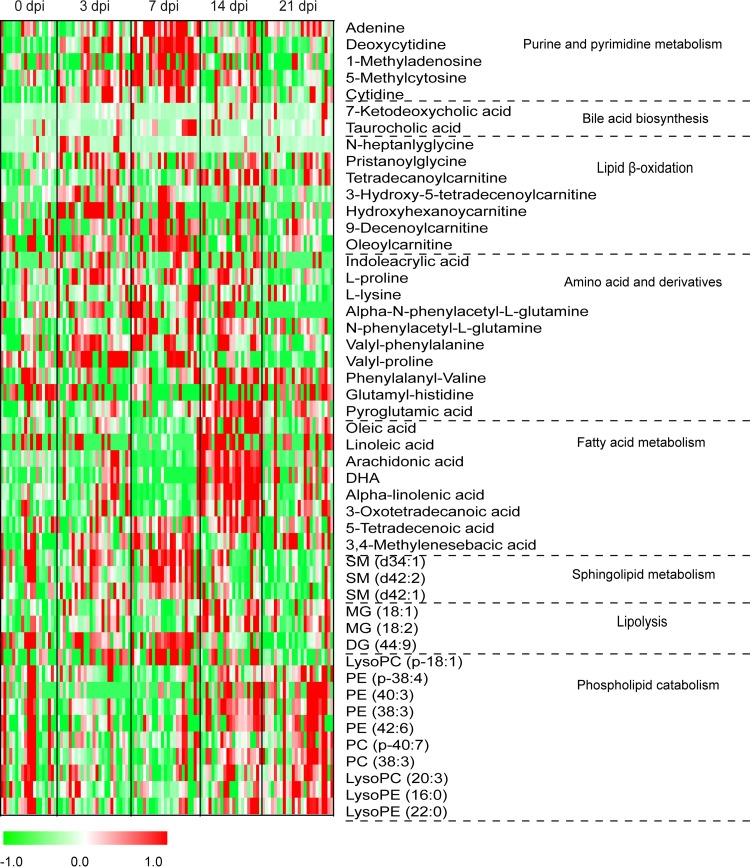
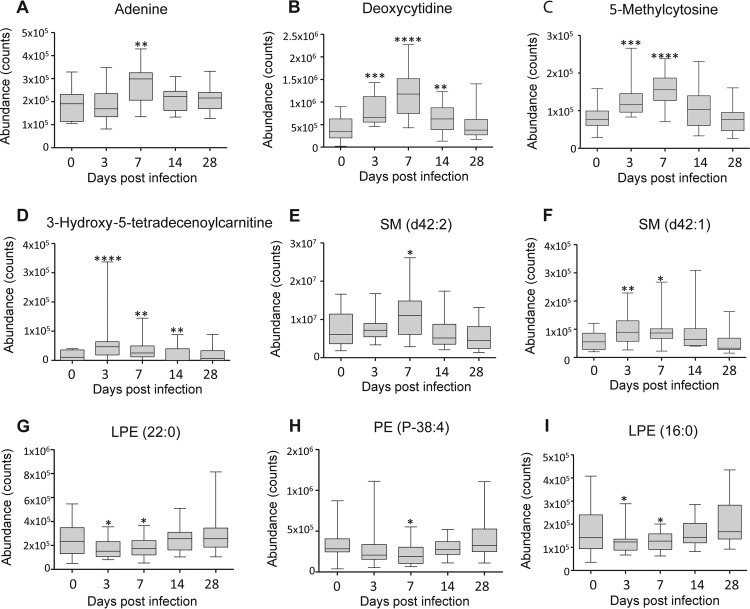
In humice with DENV infection, about 400 differential features showed a reversible-change trend, and 48 metabolites were identified, including purines and pyrimidines, acylcarnitines, acylglycines, phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), lysophosphatidylcholines (lysoPCs), lysophosphatidylethanolamines (lysoPEs), amino acids and derivatives, free fatty acids (FFAs), sphingomyelins (SMs), monoacylglycerides (MGs), diacylglyceride (DG), and bile acids (Fig. 3 and Table 1). Most of these metabolites were significantly perturbed at 3 or 7 dpi and normalized to control levels at 14 or 28 dpi, suggesting that they might serve as prognostic markers of the disease in humice. The metabolites within each metabolite class generally showed similar change trends. For example, the 7 FFAs, 5 purines and pyrimidines, 4 acylcarnitines, and 3 SMs showed an elevated trend at 3 and 7 dpi and returned to the control levels at 14 and 28 dpi (Fig. 4A to F). Conversely, 9 out of the 10 phospholipids showed a decreased trend at 3 and 7 dpi and returned to the control levels at 14 and 28 dpi (Fig. 4G to I). The classes of these different metabolites and their reversible-change trends were similar to our previous findings in a longitudinal cohort of adult dengue patients across three infection stages (12). Specifically, FFA, acylcarnitine, bile acid, purine, SM, MG, and DG showed an increased trend in both humice and dengue patients at the early or acute stage of DENV infection and gradually returned to the control levels at the late stage. Conversely, phospholipids mainly displayed a downward trend in both humice and dengue patients at the early stage and returned to the control levels at the late stage.
FIG 3.
Heat map of identified differential metabolites in humice with dengue infection. Each row shows the ion intensity for a specific metabolite after mean centering and unit variance scaling of the data. The columns show the serum metabolic profiles of humice with DENV infection at 0, 3, 7, 14, and 28 dpi.
TABLE 1.
Identified differential metabolites in humanized mice with dengue infection
| HMDB | Mass (Da) |
Metabolite | Chemical formula | Change trend | Pathway | Dengue patients | |
|---|---|---|---|---|---|---|---|
| Accurate | Theoretical | ||||||
| HMDB00207 | 282.24945 | 282.2558803 | Oleic acid | C18H34O2 | ↑ | Fatty acid biosynthesis | Yes |
| HMDB00673 | 280.2353 | 280.2402302 | Linoleic acid | C18H32O2 | ↑ | Fatty acid biosynthesis | Yes |
| HMDB01043 | 304.2328 | 304.2402302 | Arachidonic acid | C20H32O2 | ↑ | Fatty acid biosynthesis | Yes |
| HMDB02183 | 328.23312 | 328.2402302 | DHA | C22H32O2 | ↑ | Fatty acid biosynthesis | Yes |
| HMDB01388 | 278.219 | 278.2245802 | Alpha-linolenic acid | C18H30O2 | ↑ | Fatty acid biosynthesis | Yes |
| HMDB10730 | 242.18243 | 242.1881946 | 3-Oxotetradecanoic acid | C14H26O3 | ↑ | Fatty acid biosynthesis | No |
| HMDB59729 | 226.1204 | 226.1205091 | 3,4-Methylenesebacic acid | C12H18O4 | ↑ | Fatty acid biosynthesis | No |
| HMDB00521 | 226.19693 | 226.1932801 | 5-Tetradecenoic acid | C14H26O2 | ↑ | Fatty acid oxidation | No |
| HMDB11567 | 356.286 | 356.2926597 | MG(18:1) | C21H40O4 | ↑ | Lipolysis | Yes |
| HMDB11568 | 354.26956 | 354.2770097 | MG(18:2) | C21H38O4 | ↑ | Lipolysis | Yes |
| HMDB07700 | 718.556 | 718.5536255 | DG(22:4/22:5/0:0) | C47H74O5 | ↑ | Lipolysis | Yes |
| HMDB00034 | 135.05618 | 135.0544952 | Adenine | C5H5N5 | ↑ | Purine metabolism | No |
| HMDB00014 | 227.09274 | 227.0906059 | Deoxycytidine | C9H13N3O4 | ↑ | Pyrimidine metabolism | No |
| HMDB03331 | 281.11356 | 281.112404 | 1-Methyladenosine | C11H15N5O4 | ↑ | Pyrimidine metabolism | No |
| HMDB02894 | 125.05924 | 125.0589119 | 5-Methylcytosine | C5H7N3O | ↑ | Pyrimidine metabolism | No |
| HMDB00089 | 243.08305 | 243.0855205 | Cytidine | C9H13N3O5 | ↑ | Pyrimidine metabolism | No |
| HMDB00391 | 406.2621 | 406.2719243 | 7-Ketodeoxycholic acid | C24H38O5 | ↑ | Bile acid biosynthesis | No |
| HMDB00036 | 515.28796 | 515.2916735 | Taurocholic acid | C26H45NO7S | ↑ | Bile acid biosynthesis | No |
| HMDB13010 | 187.1234 | 187.1208434 | N-Heptanoylglycine | C9H17NO3 | ↑ | Acylglycine | No |
| HMDB13303 | 355.30783 | 355.3086442 | Pristanoylglycine | C21H41NO3 | ↑ | Acylglycine | No |
| HMDB05066 | 371.29556 | 371.3035588 | Tetradecanoylcarnitine | C21H41NO4 | ↑ | Lipid β-oxidation | Yes |
| HMDB13330 | 385.27936 | 385.2828234 | 3-Hydroxy-5-tetradecenoylcarnitine | C21H39NO5 | ↑ | Lipid β-oxidation | No |
| HMDB13131 | 275.17087 | 275.1732729 | Hydroxyhexanoycarnitine | C13H25NO5 | ↑ | Lipid β-oxidation | No |
| HMDB13205 | 313.225 | 313.2253085 | 9-Decenoylcarnitine | C17H31NO4 | ↑ | Lipid β-oxidation | Yes |
| HMDB05065 | 425.34814 | 425.350509 | Oleoylcarnitine | C25H47NO4 | ↑ | Lipid β-oxidation | Yes |
| HMDB00267 | 129.04066 | 129.042593 | Pyroglutamic acid | C5H7NO3 | ↑ | Glutathione metabolism | Yes |
| HMDB00734 | 187.0611 | 187.0633285 | Indoleacrylic acid | C11H9NO2 | ↑ | Tryptophan metabolism | Yes |
| HMDB00162 | 115.0609 | 115.0633285 | l-Proline | C5H9NO2 | ↑ | Arginine and proline metabolism | No |
| HMDB00182 | 146.1077 | 146.1055277 | l-Lysine | C6H14N2O2 | ↑ | Lysine degradation | No |
| HMDB06344 | 264.10547 | 264.111007 | Alpha-N-phenylacetyl–l-glutamine | C13H16N2O4 | ↑ | Phenylalanine metabolism | No |
| HMDB00512 | 207.09024 | 207.0895433 | N-Acetyl-l-phenylalanine | C11H13NO3 | ↑ | Phenylalanine metabolism | No |
| HMDB29134 | 264.1464 | 264.1473925 | Valyl-phenylalanine | C14H20N2O3 | ↑ | Dipeptide | No |
| HMDB29135 | 214.1325 | 214.1317425 | Valyl-proline | C10H18N2O3 | ↑ | Dipeptide | No |
| HMDB29134 | 264.15723 | 264.1473925 | Valyl-phenylalanine | C14H20N2O3 | ↓ | Dipeptide | No |
| HMDB28821 | 283.103 | 283.1042446 | Glutamyl-histidine | C11H15N4O5 | ↓ | Dipeptide | No |
| HMDB13464 | 702.5609 | 702.5675748 | SM(d18:0/16:1) | C39H79N2O6P | ↑ | Sphingolipid metabolism | Yes |
| HMDB12107 | 812.6705 | 812.6771252 | SM(d18:1/24:1) | C47H93N2O6P | ↑ | Sphingolipid metabolism | Yes |
| HMDB12095 | 814.6839 | 814.6927753 | SM(d18:0/24:1) | C47H95N2O6P | ↑ | Sphingolipid metabolism | No |
| HMDB13122 | 507.3682 | 507.368875 | LysoPC(P-18:0) | C26H54NO6P | ↑ | Phospholipid catabolism | Yes |
| HMDB05779 | 751.5493 | 751.5515904 | PE(P-38:4) | C43H78NO7P | ↓ | Phospholipid catabolism | Yes |
| HMDB09041 | 797.5837 | 797.5934552 | PE(40:3) | C45H84NO8P | ↓ | Phospholipid catabolism | No |
| HMDB09551 | 769.5633 | 769.5621551 | PE(38:3) | C43H80NO8P | ↓ | Phospholipid catabolism | Yes |
| HMDB09243 | 819.58105 | 819.5778051 | PE(42:6) | C47H82NO8P | ↓ | Phospholipid catabolism | Yes |
| HMDB08754 | 815.5827 | 815.5828905 | PC(P-40:7) | C48H82NO7P | ↓ | Phospholipid catabolism | No |
| HMDB08020 | 811.60394 | 811.6091052 | PC(38:3) | C46H86NO8P | ↓ | Phospholipid catabolism | Yes |
| HMDB10393 | 545.34375 | 545.3481395 | LysoPC(20:3) | C28H52NO7P | ↓ | Phospholipid catabolism | No |
| HMDB11473 | 453.28265 | 453.2855393 | LysoPE(16:0) | C21H44NO7P | ↓ | Phospholipid catabolism | Yes |
| HMDB11490 | 537.3757 | 537.3794397 | LysoPE(22:0) | C27H56NO7P | ↓ | Phospholipid catabolism | No |
FIG 4.
Typical change trends based on differential metabolite classes in humice with dengue infection. (A) Elevated change trend of adenine (purine). (B) Elevated change trend of deoxycytinine (pyrimidine). (C) Elevated change trend of 5-methylcytosine (pyrimidine). (D) Elevated change trend of 3-hydroxy-5-tetradecenoylcarnitine (acylcarnitine). (E) Elevated change trend of SM d42:2 (sphingolipid). (F) Elevated change trend of SM d42:1 (sphingolipid). (G) Decreased change trend of lyoPE (LPE) 22:0 (phospholipid). (H) Decreased change trend of PE P-38:4 (phospholipid). (I) Decreased change trend of LPE 16:0 (phospholipid). The bottom and the top of each box are the 25th and the 75th percentiles, and the black line near the middle of the box is the median peak area of the metabolite. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by the Kruskal-Wallis test. The statistical comparison was with control levels. Whiskers are from minimum to maximum.
We used the pathway analysis tool MetaboAnalyst to determine the underlying biochemical pathways revealed by these different metabolites. Major perturbed metabolic pathways in humice after DENV infection included purine and pyrimidine metabolism, fatty acid β-oxidation, phospholipid catabolism, arachidonic acid and linoleic acid metabolism, sphingolipid metabolism, tryptophan metabolism, phenylalanine metabolism, lysine biosynthesis and degradation, and bile acid biosynthesis (see Fig. S4 in the supplemental material). Most of the altered metabolic pathways were similar to the perturbed pathways in dengue patients in our previous reports (12).
DISCUSSION
Dengue is a major global public health threat, and the lack of an appropriate small-animal model of dengue infection has greatly hindered the study of dengue pathogenesis and the development of therapeutics. We have successfully constructed humice with significant levels of human platelets, monocytes/macrophages, and hepatocytes. Infection of the humice with DENV2 could recapture some of the characteristic features of DENV infection in humans, including transient leukopenia, thrombocytopenia, and liver damage. We have previously studied serum metabolome changes using LC-MS in a longitudinal cohort of adult dengue patients across three prominent infection stages (early febrile, defervescence, and convalescence stages) and identified a variety of metabolic pathways linked to dengue progression. In the present study, a similar metabolomics study was conducted on serum samples from humice at multiple time points after DENV infection with the aim of comparing the similarities and differences of metabolome changes between dengue patients and humice infected with DENV. Our results showed that most disturbed metabolic pathways were common between dengue patients and DENV-infected humice, suggesting humice could be a highly relevant small-animal model for the study of dengue pathogenesis and the development of dengue therapeutics.
DENV-infected NSG mice did not show viremia or any characteristic features of human dengue patients, but we still observed some differential features between infected and noninfected NSG mice, indicating that our analytical approach is sensitive enough to capture the metabolome changes of the NSG mice even without apparent phenotypical changes. Conversely, viremia and certain features of human dengue patients, like transient leukopenia, thrombocytopenia, and liver damage, were observed in infected humice. Correspondingly, a much larger number of differential features were observed in infected humice than in noninfected humice. Thirty-eight differential metabolites were identified, which belonged to different metabolite classes, such as purine and pyrimidine, acylcarnitine, acylglycine, phospholipid, sphingolipid, amino acids and derivatives, free fatty acid, and bile acid. In our previous metabolomics study, we found that metabolites from all these classes were also significantly altered in adult patients with DENV infection compared to healthy controls (12). However, more purines and pyrimidines and amino acids and derivatives but fewer free fatty acids were found in the current study. This could be due to the change of analytical platform from reversed phase to HILIC, which could capture more polar compounds. Nevertheless, these results showed many similarities in host responses to DENV infection between humans and humice.
Dengue in humans is a dynamic disease, and after the incubation period, the illness begins abruptly and is followed by the three phases—febrile (days 0 to 4 after onset of fever), critical (days 5 to 7 after onset of fever), and recovery. The mild form of dengue is generally self-limiting, and patients recover uneventfully after 5 to 7 days of acute illness. Our previous metabolomics study on patients with mild dengue showed that the differential metabolites between dengue patients and healthy controls were significantly perturbed during the early febrile and critical stages and normalized to control levels at the recovery stage, which was reflective of dengue disease progression. Similarly, the transient viremia, leukopenia, and thrombocytopenia in humice with DENV infection suggested the illness was self-limiting in the model, as well. Indeed, we identified 38 differential metabolites between infected and noninfected humice, which showed a reversible-change trend across the disease course—they were significantly perturbed at 3 or 7 dpi and returned to control levels at 14 or 28 dpi. Furthermore, the same metabolite class showed similar change trends between humice and dengue patients. For example, the purines and acylcarnitines showed an elevated trend at early febrile and critical stages in dengue patients, and similarly, they had an elevated trend at 3 and 7 dpi in infected humice. On the other hand, the majority of the phospholipids showed a decreased trend at febrile and critical stages in dengue patients and at 3 and 7 dpi in infected humice. Altogether, the same classes of metabolites were perturbed with similar change trends over the disease course in infected humice and dengue patients, and the differential metabolites found in this study could serve as prognostic markers to monitor dengue progression in humice.
The differential metabolites in the humouse model belonged to a variety of metabolic pathways, most of which were also significantly altered in dengue patients. The commonly altered pathways in both infected humice and dengue patients include fatty acid β-oxidation, phospholipid catabolism, sphingolipid metabolism, purine and pyrimidine metabolism, arachidonic acid and linoleic acid metabolism, tryptophan metabolism, phenylalanine metabolism, and bile acid biosynthesis. Among these pathways, bile acid biosynthesis is closely linked to liver impairment. DENV can infect the hepatocytes and have adverse effects on liver functions in dengue patients, which is monitored by liver enlargement and elevated transaminases among clinical cases (15–17). The levels of liver transaminases were indeed significantly increased in the sera of infected humice. Bile acids are toxic to cells at abnormally high levels (18), and increased bile acid biosynthesis might contribute to liver pathogenesis in infected humice. On the other hand, the steroid hormone biosynthesis and heme degradation pathways were perturbed in dengue patients, but not in humice with DENV infection. DENV infects a multitude of cells and organs in humans (19, 20), and although significant levels of human monocytes/macrophages and hepatocytes are present in humice, many types of cells are still lacking. Thus, although many metabolic pathways were commonly perturbed in infected humice and dengue patients, some altered pathways in dengue patients could not be found in humice.
The main limitation of the study is the relatively large variation of individual humanized mice in response to the infection, and larger numbers of humice, 25 in this study, may be needed. Nevertheless, our analyses provide a detailed description of the metabolome changes in humice with DENV infection, and the data revealed many commonalities of host responses to DENV infection between humans and humice, suggesting that this small-animal model could be used for the study of dengue pathogenesis and the development of dengue therapeutics.
MATERIALS AND METHODS
Ethics statement.
Human fetal livers at 15 to 23 weeks of gestation were collected in accordance with the institutional ethical guidelines of the National University Hospital of Singapore and National University of Singapore (NUS). All the women gave written informed consent for the donation of their fetal tissue for research. All experiments involving mice were carried out in strict accordance with the National Advisory Committee for Laboratory Animal Research (NACLAR) guidelines (Guidelines on the Care and Use of Animals for Scientific Purposes) in facilities licensed by the Agri-Food and Veterinary Authority of Singapore (AVA), the regulatory body of the Singapore Animals and Birds Act. The humouse protocol (R16-0158) was approved by the Institutional Animal Care and Use Committee (IACUC), National University of Singapore. The mice were monitored every day after infection, and any mouse with 30% body weight loss was euthanatized immediately.
Construction of humice and infection with dengue virus.
The humouse model was developed as in our previous report (21). Briefly, human fetal livers at 15 to 23 weeks of gestation were collected, and the human CD34+ cells were isolated and purified using a CD34-positive magnetic selection kit (Stem Cell Technologies). CD34+ cells (2 × 105) were injected into sublethally irradiated NSG pups within 24 to 48 h of birth. The mice were analyzed for the human leukocyte reconstitution rate by staining mouse CD45 and human CD45 at 12 weeks after CD34+ cell injection. Twenty-five humanized mice and 10 NSG mice were infected by injecting 1 × 107 PFU of the concentrated DENV2 (strain 07K2861) in 200 μl of RPMI 1640 medium through the tail vein. Sera were collected from both NSG mice and humice at 0, 3, 7, 14, and 28 dpi for flow cytometry assays, measuring viral RNA levels, platelet counts, and metabolomics analysis. The viral RNA extraction and qRT-PCR assay and platelet counts by fluorescence-activated cell sorting (FACS) were described in our previous paper (8).
Serum sample preparation.
The procedure for serum sample preparation was the same as in our previously published report (12, 22). Briefly, a 50-μl volume of serum was thawed at 4°C, and proteins were precipitated with 200 μl ice-cold methanol, which contained 5 μg/ml 9-fluorenylmethoxycarbonyl-glycine as an internal standard. After vortexing, the mixture was centrifuged at 16,000 rpm for 10 min at 4°C, and the supernatant was collected and evaporated to dryness in a vacuum concentrator. The dry extracts were then resuspended in 100 μl of 98:2 water/methanol for LC-MS analysis. QC samples were prepared by mixing equal amounts of sera from all the samples and processed as real samples. All samples were kept at 4°C and analyzed in a random manner. The QC sample was run after every 15 samples to monitor the stability of the system.
Metabolomics analysis by LC-MS.
The metabolomics analysis followed our previous publications with modifications (12, 22). Briefly, both RP-UHPLC–MS and HILIC-UHPLC–MS analyses were performed using an Agilent 1290 ultrahigh pressure liquid chromatography system (Waldbronn, Germany) equipped with a 6520 quadrupole-time of flight (QTOF) mass detector managed by a MassHunter workstation. For RP-UHPLC, the column used for the separation was an Agilent rapid-resolution HT Zorbax SB-C18 (2.1 by 100 mm by 1.8 mm; Agilent Technologies, Santa Clara, CA, USA). The oven temperature was set at 45°C. The gradient elution involved a mobile phase consisting of 0.1% formic acid in water (A) and 0.1% formic acid in methanol (B). The initial conditions were set at 5% phase B for 2 min with a flow rate of 0.4 ml/min. A 7-min linear gradient to 70% B was applied, followed by a 12-min gradient to 100% B, which was held for 3 min and then returned to the starting conditions over 0.1 min. The sample injection volume of 5 μl was injected, and the oven temperature was set at 40°C. For HILIC-LC, the column used for the separation was an Acquity BEH HILIC (2.1 by 100 mm by 1.7 μm; Waters Corporation, Milford, MA, USA), and the mobile phase was 10 mM ammonium acetate in 95% acetonitrile-5% water containing 0.1% formic acid (A) and 50% acetonitrile-50% water (B). The initial condition of the gradient elution was set at 100% A for 2 min, with a flow rate of 0.4 ml/min. A 13-min linear gradient to 50% B was then applied, followed by a 2-min gradient to 100% B, which was held for 3 min. The sample injection volume was 5 μl, and the oven temperature was set at 40°C.
The electrospray ionization mass spectra were acquired in positive ion mode. Mass data were collected between m/z 100 and 1,000 at a rate of two scans per second. The ion spray voltage was set at 4,000 V, and the heated-capillary temperature was maintained at 350°C. The drying gas and nebulizer nitrogen gas flow rates were 12.0 liters/min and 50 lb/in2, respectively. Two reference masses were continuously infused into the system to allow constant mass correction during the run: m/z 121.0509 (C5H4N4) and m/z 922.0098 (C18H18O6N3P3F24).
Data analysis and compound identification.
Raw spectrometric data were analyzed with MassHunter qualitative analysis software (Agilent Technologies), and the molecular features, characterized by retention time, chromatographic peak intensity, and accurate mass, were obtained by using the Molecular Feature Extractor algorithm. The features were then analyzed with MassHunter Mass Profiler Professional software (Agilent Technologies). Only features with an intensity of ≥20,000 counts (approximately three times the limit of detection of our LC-MS instrument) that were found in at least 80% of the samples at the same sampling time point signal were kept for further processing. Next, a tolerance window of 0.15 min and 2 mDa was used for alignment of retention time and m/z values, and the data were also normalized by an internal standard. For statistical analysis, one-way analysis of variance (ANOVA) (P < 0.05) with Benjamini-Hochberg multiple-testing correction was employed. Fold change (FC) analysis was also performed to further filter the features, and only those features with an FC of >1.5 were selected as potential significantly altered metabolites. Unsupervised multivariate analysis PCA was performed with the Unscrambler-X statistical software package (CAMO Software, Oslo, Norway).
The structure identification of the differential metabolites was based on our published work (23, 24). Briefly, the element compositions of the metabolites were first calculated with MassHunter software from Agilent based on the exact mass, the nitrogen rule, and the isotope pattern. Then, the elemental composition and exact mass were used for open source database searching, including LIPIDMAPS (http://www.lipidmaps.org/), HMDB (http://www.hmdb.ca/), METLIN (http://metlin.scripps.edu/), and MassBank (http://www.massbank.jp/). Next, tandem MS (MS-MS) experiments were performed to obtain structural information via the interpretation of the fragmentation pattern of the metabolite. The MS-MS spectra of possible metabolite candidates in the databases were also searched and compared. Finally, the metabolites were confirmed by comparison with standards when they were commercially available. The metabolites are listed according to the minimum reporting standards for chemical analysis in metabolomics recommended by the Metabolomics Standard Initiative (MSI) (25, 26), briefly, a four-level system ranging from level 1 (identified metabolites) via levels 2 and 3 (putatively annotated compounds and compound classes) to level 4 (unidentified or unclassified metabolites that can still be differentiated based on spectrum data). For metabolic pathway analysis, metaboAnalyst (27) was used to identify relevant pathways.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00386-17.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB, Suaya JA, Shepard DS. 2007. The burden of dengue infection. Lancet 369:1410–1411. doi: 10.1016/S0140-6736(07)60645-X. [DOI] [PubMed] [Google Scholar]
- 3.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2009. Dengue guidelines for diagnosis, treatment, prevention, and control. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 5.Chan KW, Watanabe S, Kavishna R, Alonso S, Vasudevan SG. 2015. Animal models for studying dengue pathogenesis and therapy. Antiviral Res 123:5–14. doi: 10.1016/j.antiviral.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Zompi S, Harris E. 2012. Animal models of dengue virus infection. Viruses 4:62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mota J, Rico-Hesse R. 2009. Humanized mice show clinical signs of dengue fever according to infecting virus genotype. J Virol 83:8638–8645. doi: 10.1128/JVI.00581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sridharan A, Chen Q, Tang KF, Ooi EE, Hibberd ML, Chen J. 2013. Inhibition of megakaryocyte development in the bone marrow underlies dengue virus-induced thrombocytopenia in humanized mice. J Virol 87:11648–11658. doi: 10.1128/JVI.01156-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JK, Lindon JC, Holmes E. 1999. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 10.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. 2009. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11.Vinayavekhin N, Homan EA, Saghatelian A. 2010. Exploring disease through metabolomics. ACS Chem Biol 15:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, Lee YH, Kumar Y, Xu F, Lu K, Ooi EE, Tannenbaum SR, Ong CN. 2013. Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl Trop Dis 7:e2373. doi: 10.1371/journal.pntd.0002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei F, Bowdish DM, McCarry BE. 2014. Comprehensive and simultaneous coverage of lipid and polar metabolites for endogenous cellular metabolomics using HILIC-TOF-MS. Anal Bioanal Chem 406:3723–3733. doi: 10.1007/s00216-014-7797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, Holmes E, Nicholson JK. 2010. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc 5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 15.Marianneau P, Steffan AM, Royer C, Drouet MT, Jaeck D, Kirn A, Deubel V. 1999. Infection of primary cultures of human Kupffer cells by Dengue virus: no viral progeny synthesis, but cytokine production is evident. J Virol 73:5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seneviratne SL, Malavige GN, de Silva HJ. 2006. Pathogenesis of liver involvement during dengue viral infections. Trans R Soc Trop Med Hyg 100:608–614. doi: 10.1016/j.trstmh.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Lee LK, Gan VC, Lee VJ, Tan AS, Leo YS, Lye DC. 2012. Clinical relevance and discriminatory value of elevated liver aminotransferase levels for dengue severity. PLoS Negl Trop Dis 6:e1676. doi: 10.1371/journal.pntd.0001676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann AF. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 159:2647–2658. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 19.Torrentes-Carvalho A, Marinho CF, De Oliveira-Pinto LM, De Oliveira DB, Damasco PV, Cunha RV, de Souza LJ, de Azeredo EL, Kubelka CF. 2014. Regulation of T lymphocyte apoptotic markers is associated to cell activation during the acute phase of dengue. Immunobiology 219:329–340. doi: 10.1016/j.imbio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Green S, Rothman A. 2006. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis 19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- 21.Chen Q, Khoury M, Chen J. 2009. Expression of human cytokines dramatically improves reconstitution of specific human-blood lineage cells in humanized mice. Proc Natl Acad Sci U S A 106:21783–21788. doi: 10.1073/pnas.0912274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui L, Lee YH, Thein TL, Fang J, Pang J, Ooi EE, Leo YS, Ong CN, Tannenbaum SR. 2016. Serum metabolomics reveals serotonin as a predictor of severe dengue in the early phase of dengue fever. PLoS Negl Trop Dis 10:e0004607. doi: 10.1371/journal.pntd.0004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Zhao X, Fritsche J, Yin P, Schmitt-Kopplin P, Wang W, Lu X, Häring HU, Schleicher ED, Lehmann R, Xu G. 2008. Practical approach for the identification and isomer elucidation of biomarkers detected in a metabolomic study for the discovery of individuals at risk for diabetes by integrating the chromatographic and mass spectrometric information. Anal Chem 80:1280–1289. doi: 10.1021/ac702089h. [DOI] [PubMed] [Google Scholar]
- 24.Cui L, Zheng D, Lee YH, Chan TK, Kumar Y, Ho W, Chen J, Tannenbaum SR, Ong CN. 2016. Metabolomics investigation reveals metabolite mediators associated with acute lung injury and repair in a murine model of influenza pneumonia. Sci Rep 6:26076. doi: 10.1038/srep26076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, Nikolau B, Robertson D, Sumner LW, Taylor C, van der Werf M, van Ommen B, Fiehn O. 2007. The metabolomics standards initiative. Nat Biotechnol 25:846–848. [DOI] [PubMed] [Google Scholar]
- 26.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J, Psychogios N, Young N, Wishart DS. 2009. MetaboAnalyst: a Web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.