Abstract
In mammals, the unicellular zygote starts the process of embryogenesis and differentiates into all types of somatic cells, including both fetal and extraembryonic lineages—in a highly organized manner to eventually give rise to an entire multicellular organism comprising more than 200 different tissue types. This feature is referred to as totipotency. Upon fertilization, oocyte maternal factors epigenetically reprogram the genomes of the terminally differentiated oocyte and spermatozoon and turn the zygote into a totipotent cell. Today, we still do not fully understand the molecular properties of totipotency. In this review, we discuss recent findings on the molecular signature and mechanism of transcriptional regulation networks in the totipotent mouse embryo.
Keywords: Totipotency, Zygote, Transcription factors
Introduction
Multicellular organisms typically originate from a single totipotent cell, the zygote. In plants, structurally and functionally specialized cells of leaves, roots, stem, floral parts, and endosperm retain the potential to revert back to the undifferentiated state and form entire new plants, irrespective of their ploidy level (haploid, diploid, or triploid). The potential of terminally differentiated cells to regenerate whole plants was referred to as “cellular totipotency” by the remarkable German plant physiologist Göttlieb Haberlandt in his famous address to the German Academy in 1902 [1]. Now, regeneration of totipotency from isolated single plant cells is well demonstrated [2]. However, in the mouse, totipotency seems to be restricted up to two-cell embryos. The term “Totipotency” is defined by two related but different criteria: (1) the ability of a single cell to contribute to all cell lineages, including the TE, of an organism; and (2) more stringently, the ability of a single cell to develop into a complete organism [3, 4]. The zygote is the ultimate totipotent cell (Fig. 1). Blastomeres from two-cell–stage embryos also fulfill the more stringent definition for totipotency [5–7]. Prior to the first lineage segregation, totipotency is lost gradually [8]. Some blastomeres from eight-cell–stage embryos contribute to the development of all lineages in chimeric mice [9–11], and thus provide evidence for totipotency based on the less stringent definition. In this review, we discuss present-day understanding of the transcription factor networks and epigenetic reprogramming involved in the emergence of totipotency in the mammalian embryo.
Fig. 1.
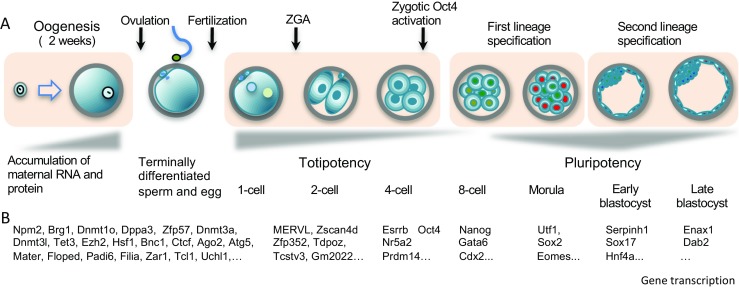
Mouse preimplantation development. a Mature oocytes are ovulated from the ovary into the oviduct and fertilized by sperm to establish totipotent zygotes that divide and become blastocysts, and finally implant in the uterus at embryonic day 4.5. b After fertilization, stored maternal factors trigger zygotic genome activation (ZGA) that results in formation of a totipotent zygote with a unique two-cell-specific gene-expression profile, followed by waves of transcription activations of lineage specific genes during preimplantation development
Establishment of totipotency
Zygotic genome activation
Following fertilization, maternal factors play a leading role in epigenetically resetting the parental DNA and histones across the genome of the zygote, thereby preparing for whole-genome activation and establishment of totipotency. A burst of transcription—known as zygotic genome activation (ZGA)—begins at the late one-cell stage and peaks at the two-cell stage in the mouse [12]. ZGA is characterized by more efficient use of TATA-less promoters [13]; activation of repetitive elements [14], particularly endogenous retrotransposons, e.g., murine endogenous retrovirus with a leucine tRNA primer binding site (MERVL) at the two-cell stage as a marker for totipotent cells [15]; uncoupling of transcription and translation in zygotes [16]; and activation of enhancers for transcription in two-cell embryos [17]. ZGA provides the first step in the establishment of totipotency.
Maternal factor storage
During oogenesis, the volume of oocytes dramatically increases to accommodate the storage of maternal factors (RNA, proteins) required for establishing totipotency and ZGA, such as nucleoplasmin (NPM) 2 [18], and the subcortical maternal complex (SCMC, including Mater, Tle6, Floped, Padi6, Filia) [19]. In the growing oocytes, subcortical ribonucleoprotein (RNP) particle domains (SCRDs) are formed to serve as the storage compartment of maternal messenger RNA (mRNA) [20]. Maternally accumulated yes-associated protein (YAP) has recently been identified to play a critical role in ZGA [21]. However, the paucity of biological materials from mouse oocytes and zygotes has hampered our effort to understand how maternal factors reprogram cells to totipotency [22]. Further identification of key maternal regulators and their functions could greatly facilitate studies for improving chromatin reprogramming [23, 24].
Histone modifications
Hyperaccessibility of chromatin by transcriptional machinery is a prerequisite for ZGA. Chromatin accessibility is largely determined by histone modifications of its N-terminal tails (“marks”), which acts as a fundamental epigenetic regulator to control the gene expression during embryo development in mammals.
There are two major types of histone modifications involved in regulation of gene expression during the ZGA: lysine acetylation and lysine (tri)methylation. H4 acetylation makes pronucleus permissive for active transcription [25]. Loss of the maternal Brg1, a component of the ATP-dependent chromatin remodeling SWI/SNF complex, results in reduced levels of 30% of zygotic genes and arrest at two-cell, demonstrating that chromatin remodelers that induce to acetylation are required for mouse embryogenesis [26].
The opposing marks histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 27 trimethylation (H3K27me3) at gene promoter regions are associations with active and repressed genes, respectively. Following fertilization, H3K4me3 and H4 acetylation in the paternal genome are responsible for a minor ZGA. They are depleted in late zygotes stage but reestablished on promoter regions during the major ZGA at the late two-cell stage [27, 28]. On the maternal genome, a noncanonical (nc) form of H3K4me3 (ncH3K4me3) is present broadly in oocytes and zygote and overlaps almost exclusively with partially methylated DNA domains. The ncH3K4me3 is erased in the late two-cell embryos [27]. Active removal of broad H3K4me3 domains by the lysine demethylases KDM5A and KDM5B is required for ZGA and is essential for early embryo development [29].
Protamine-to-histone replacement
At the time of fertilization, the chromatin molecules of the paternal and maternal genomes exhibit different epigenetic marks and organization. The paternal genome is haploid, and most of it is packaged densely, with protamines rather than histones, while the maternal genome is diploid, as it arrests at metaphase II, and is packaged with histones. After a sperm penetrates the cytoplasm of the oocyte, the paternal genome decondenses, enabling protamine removal and repackaging with the stored maternal histones in the absence of DNA replication, while the maternal genome completes meiosis. These newly integrated histones possess a transcriptionally permissive pattern of modifications, including H4 hyperacetylation [25] and H3K9 and H3K27 monomethylation [30]. Of note, when round spermatids, which contain DNA that is still associated with histones, are injected into oocytes by round spermatid injection (ROSI), paternal genome failed to undergo active DNA demethylation, but when mature sperm, which contain DNA associated mainly with protamines, are injected into oocytes by intracytoplasmic sperm injection (ICSI), active paternal genome demethylation is observed [31]. These results indicate that the protamine-histone exchange may cause the pronounced demethylation of the paternal DNA in the zygote. However, as both ROSI- and ICSI-derived embryos have the same likelihood of developing to term, paternal genome demethylation mediated by protamine-histone exchange is not an essential step in ZGA and establishment of totipotency [32].
Histone variant H3.3
During preimplantation development, striking changes in epigenetic modifications in the form of deposition of histone variants, reestablishment of histone marks, and DNA demethylation occur throughout the genome.
Both canonical histones (H2A, H2B, H3, and H4) and variant histones (which have sequence homology and structural similarity with canonical histones, but harbor specialized functions and play essential roles in chromatin reprogramming) are incorporated into chromatin throughout the first cell cycle of the zygote. Variant histones preferentially are deposited into specific genomic regions to form nucleosomes with unique biophysical characteristics. As one of the three variants of histone H3 in mammals, H3.3 differs from canonical H3 in only four amino acids and incorporates into chromatin in both a replication-independent and a replication-coupled manner. H3.3 interacts with the chaperones HIRA and Daxx/ATRX and is enriched in transcriptionally active regions [24]. H3.3 also localizes to telomeres, where its presence depends upon ATRX [33]. Following fertilization in the mouse, maternal H3.3 is deposited by HIRA onto paternal chromatin during the protamine-to-histone exchange [34], an essential step for oocyte-mediated reprogramming [35]. H3.3 is also required for maintaining chromatin in the decondensed state in early mouse embryos by antagonizing linker H1, an activity dependent on H3.3 lysine 36 [36].
Active DNA demethylation
The genome-wide cytosine methylation profile differs among cell types, and it functions as a form of memory of the cell’s identity [37]. 5-methylcytosine (5mC) is present mostly in CpG sequences [37–39]. Methylation occurs globally in mammalian genomes at various loci including genes, transposons, repeat sequences, and intergenic DNA [40]. The enzymes that methylate cytosine to form 5mC have been well characterized. DNA methyltransferase (DNMT) 1 preferentially methylates hemi-methylated cytosines in CpG sequences and thus acts as a methyltransferase that maintains genome-wide methylation patterns during replication [41–43]. DNMT3A and DNMT3B can methylate unmethylated CpG sequences and hence function as de novo methyltransferases [44]. DNMT3L has no catalytic activity but recruits DNMT3A and DNMT3B to their target sequences by recognizing nucleosomes that carry unmethylated histone H3 lysine 4 (H3K4) [45–49].
In concordance with histone acquisition, the paternal genome undergoes genome-wide loss of DNA methylation via an active mechanism prior to the start of DNA replication [50, 51].
Recent studies have found a new mechanism of active demethylation involving prior modification of methylated cytosine and nucleotide excision and repair. 5-hydroxy-methylcytosine (5hmC), a stable hydroxylated metabolite of 5mC, was first identified in the genome of T-even bacteriophages [52], and it is produced as an oxidation damage product of 5mC [53, 54]. Subsequent studies have found this to actually be a physiologically relevant DNA modification in mammals, e.g., in mouse neurons and embryonic stem cells (ESCs) [55, 56]. The hydroxylation of 5mC into 5hmC is catalyzed by a family of dioxygenases—the ten-eleven translocation (TET) 1/2/3 proteins. TET proteins convert 5mC into 5hmC [56], and further into 5-formylcytosine (5fC) and 5-carboxymethylcytosine (5caC) for excision [57, 58]. As 5hmC has a significantly lower affinity for methyl-CpG binding proteins [59], it may be directly involved in epigenetic regulation. Indeed, genome-wide DNA demethylation in the zygote is accompanied by Tet3-driven genome-wide oxidation of 5mC into 5hmC [60–62]. Such 5hmC formation does not account for the initial loss of paternal 5mC in the early pronuclear stage, but it is dependent on the activity of zygotic Dnmt3a and Dnmt1, suggesting that Tet3 is targeting de novo methylated sites for the accumulation of 5hmC [63].
Although recent sequence data has shown active demethylation in maternal DNA as well [64, 65], high levels of 5hmC are detected only in the paternal genome of the zygote [60, 66]. A maternal knockout of Tet3 has been shown to prevent both elevation of 5hmC and reduction of 5mC levels in the paternal genome, impair promoter demethylation of Oct4 (Pou5f1) and Nanog, delay the activation of a paternally derived Oct4 transgene, and cause frequent death of the resulting embryos [62]. These findings suggest that during normal development, TET3 converts 5mC into 5hmC in the paternal genome, and that TET3-mediated hydroxylation of 5mC accounts for at least some of the active DNA demethylation of the paternal genome. The ubiquitin ligase Cullin-ring finger ligase-4 (CRL4) has recently been reported to induce TET3 activity and plays an essential role in female fecundity [67], further strengthening the importance of active DNA demethylation during embryonic development.
Activation of embryonic Oct4 expression
The maternal octamer-binding transcription factor 4 (Oct4), encoded by the gene Pou5f1 hereafter referred to as Oct4, is at the top of the pluripotency regulatory hierarchy in pluripotent cells [62, 63]. However, several recent studies using conditional genetic depletion of maternal Oct4 have found that the oocytes of Oct4 flox/flox /ZP3 Cre/+ female mice are capable of completing full-term development after fertilization, indicating that Oct4 is not required for initiating totipotency or pluripotency in embryos [68–70]. The two cell-like ESCs are found to lose Oct4 expression at the protein level [15], suggesting that Oct4 activation in early embryos demarcates pluripotency and totipotency. Still, Oct4 is at the top of the pluripotency regulatory hierarchy in pluripotent cells [71, 72]. It forms a positive feedback loop [73] and is essential for maintaining pluripotency [74]. Therefore, identifying upstream factors of Oct4 activation in early embryos is critical for understanding the molecular regulation network of totipotency and transition from totipotency to pluripotency. There are a few transcriptional factors found to be involved in the regulation of Oct4 expression. In proliferating stem cells, Promyelocytic leukemia (Pml) protein, along with the transcription factors TR2, SF1, and Sp1, and the Brg1-dependent chromatin remodeling complex (BRGC), associates with the Oct4 promoter to maintain a nucleosome-free region for Oct4 gene expression [75]. Cancer-associated factor Tpt1 has been reported to activate the transcription of Oct4 and Nanog in transplanted somatic nuclei in Xenopus oocytes [76], but knockdown of Tpt1 by small interfering RNA (siRNA) does not reduce Oct4 expression in mouse embryos [68]. The maternal transcription factor spalt-like transcription factor 4 (Sall4) binds to the Oct4 distal enhancer (DE), and evidence shows that injection of Sall4 siRNA into zygotes knocking down Sall4 mRNA levels by 50% leads to a 70% reduction of Oct4 expression levels, suggesting that Sall4 is a transcriptional activator of Oct4 expression [77]. Contradictorily, knockdown of Sall4 by injection of more efficient Sall4 siRNA into maternal Oct4-deficient zygotes—to avoid any possible effect of maternal Oct4 as a positive autoregulator—does not lead to any Oct4 expression changes at the blastocyst stage [68]. The nuclear receptor subfamily 5, group A, member 2 (Nr5a2), also known as liver receptor homolog-1 (LRH-1), was found to maintain Oct4 expression at the epiblast stage of embryonic development, by binding to the proximal enhancer (PE) and proximal promoter (PP) regions of Oct4, but to play no evident role in the self-renewal of ESCs [78]. However, Nr5a2 can induce epiblast stem cells into ground-state pluripotency—a basal proliferative state that is free of epigenetic restriction [79], and to replace Oct4 in the reprogramming of somatic cells into pluripotent cells [80]. As a component of an active DNA demethylase, activation-induced cytidine deaminase (AID) has also been shown to be required for Oct4 activation during reprogramming [81]. A genome-scale RNA interference (RNAi) screen in ESCs has identified components of the Pol II-associated factor 1 (Paf1) complex that have strong effects on Oct4 expression, and shown that Paf1C overexpression blocks the differentiation of ESCs while Paf1C knockdown causes expression changes similar to those caused by Oct4 or Nanog depletion [82]. Studies in search for oocyte master genes have revealed a novel oocyte-specific eukaryotic translation initiation factor 4E (Eif4eloo) [83] and a large number of oocyte-specific genes with yet unknown functions, such as those belonging to the homeodomain transcription factor Obox family [84]. To this day, it is unclear how Oct4 expression is activated in the embryo.
Molecular signature of totipotency
Unlike the case for pluripotency, the mechanism underlying the molecular regulation of totipotency remains largely unknown. In mice, only the zygote and two-cell-stage blastomeres can generate an entire organism on their own, and are therefore regarded as totipotent cells [6]. The morphology of two-cell embryos is characterized by lack of 4,6-diamidino-2-phenylindole (DAPI)–stained chromocenters in the nucleus [85], and the high chromatin mobility at the two-cell stage progressively decreases with development [86]. The transcriptional profile of two-cell embryos is characterized by activation of major satellites, MERVL, and two-cell-specific genes, such as Eif1a-like genes (which include Gm5662, Gm2022, Gm4027, BB287469, Gm2016, Gm21319, Gm8300, and Gm10264), Zscan4 genes (Zscan4b–Zscan4f), Zfp352, and Tdpoz genes (Tdpoz1–Tdpoz5) [87]. A recent study has demonstrated that depletion of either the p150 or p60 subunit of chromatin assembly factor-1 (CAF-1) in ESCs leads to the formation of 2-cell-like cells with a morphology and transcriptional profile similar to those of two-cell-stage embryos [88]. As CAF-1 performs the first step of the chromatin assembly process by bringing H3 and H4 in close proximity to the daughter DNA strands [89] and as the absence of functional CAF-1 delays nucleosome assembly [90], the inefficiency of chromatin assembly in two-cell embryos has been proposed to be a key mechanism in establishing totipotency [88].
Retrotransposon transcripts contribute a significant portion to the transcriptome during ZGA. Retrotransposons can also act as alternative promoters in the activation of protein-coding genes by generating chimeric transcripts with retrotransposon gene junctions [14]. The most active LINE-1 retrotransposons form a stimulatory auto-enhancing loop, indicating that maternal retrotransposon transcripts could activate endogenous retrotransposons after fertilization [91].
Zscan4 is activated during ZGA [92] and can act as an activator of spontaneous telomere sister chromatid exchange (T-SCE) and telomere elongation in mouse ESCs [93]. Knockdown of Zscan4 by siRNAs delays progression from the two-cell to four-cell stage, and thus leads to the formation of blastocysts that fail to implant or proliferate in blastocyst outgrowth culture [92]. Zscan4 is essential for generation of induced pluripotent stem cells (iPSCs), and its ectopic expression can activate early embryonic genes and improve the efficiency of iPSC generation [94]. Expression of the Zscan4 gene family plays important roles in genome stability and maintenance of telomeres [93]. Of note, the absence of nuclear receptor subfamily 0, group B, member 1 (Nr0b1), also known as Dax1, which is an important component of the transcription factor network that governs pluripotency in mouse ESCs, also leads to the overexpression of two-cell embryo-specific transcripts, including Zscan4c, preventing normal self-renewal by inducing arrest at the G2 phase followed by cell death [95].
Furthermore, another recent study has described a small transient ESC/iPSC population with fluctuating expression of a particular retrotransposon, MERVL and a transcriptome that closely resembles that observed in the blastomeres of the totipotent, two-cell embryos [15], indicating that some features of totipotent cells can be regained occasionally in pluripotent cells. This phenomenon provides us with a novel way of studying certain aspects of totipotency. However, the study did not prove these two-cell like cells to be totipotent according to the stringent criteria, in which a single totipotent cell can develop into a complete organism. Moreover, near-complete (95%–99%) knockdown of muERV-L transcripts by three different siRNA duplexes did not interfere with full-term embryonic development (unpublished data), suggesting that the transcripts of retrotransposon elements are not involved in the regulation of totipotency, but rather occur as a consequence of global DNA demethylation prior to ZGA.
Recent progress in identifying two-cell marker genes and new players in the genome-wide demethylation process has shed light on the molecular mechanism governing totipotency. Nevertheless, many questions still remain unanswered. To date, no totipotent cell lines have been established in vitro. The upstream “master” signals that trigger the establishment of totipotency have not yet been identified. Given the distinct phases of ZGA in the mouse, it is likely that multiple regulators/cofactors are associated with regulators of ZGA to ensure temporal gene activation for establishing totipotency. It would be interesting to see how the upstream signals relate to histone replacement or modification, genome-wide DNA demethylation, ZGA, and the expression of two-cell-specific genes, particularly those of maternal origin. The identification of two-cell marker genes has moved us closer to solving the fundamental question in developmental biology of how totipotency is established.
Acknowledgements
Open Access funding provided by Max Planck Society. This work was supported by the Max Planck Society, DFG grants DFG SI 1695/1-2 (SPP1356) and SCHO 340/7-1, and grant NIH R01HD059946-01 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank Areti Malapetsas for the final editing.
References
- 1.Haberland G. Kultureversuche mit isolierten Pflanzenzellen. Sitzungsber Kaiserl Akad Wiss Wien, Math-Naturwiss Cl, Abt J. 1902;111:69–92. [Google Scholar]
- 2.Vasil IK, Vasil V. Totipotency and embryogenesis in plant cell and tissue cultures. In Vitro. 1972;8:117–127. doi: 10.1007/BF02619487. [DOI] [PubMed] [Google Scholar]
- 3.Ishiuchi T, Torres-Padilla ME. Towards an understanding of the regulatory mechanisms of totipotency. Curr Opin Genet Dev. 2013;23:512–518. doi: 10.1016/j.gde.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Edwards RG, Beard HK. Oocyte polarity and cell determination in early mammalian embryos. Mol Hum Reprod. 1997;3:863–905. doi: 10.1093/molehr/3.10.863. [DOI] [PubMed] [Google Scholar]
- 5.Papaioannou VE, Mkandawire J, Biggers JD. Development and phenotypic variability of genetically identical half mouse embryos. Development. 1989;106:817–827. doi: 10.1242/dev.106.4.817. [DOI] [PubMed] [Google Scholar]
- 6.Tarkowski AK. Experiments on the development of isolated blastomeres of mouse eggs. Nature. 1959;184:1286–1287. doi: 10.1038/1841286a0. [DOI] [PubMed] [Google Scholar]
- 7.Tsunoda Y, McLaren A. Effect of various procedures on the viability of mouse embryos containing half the normal number of blastomeres. J Reprod Fertil. 1983;69:315–322. doi: 10.1530/jrf.0.0690315. [DOI] [PubMed] [Google Scholar]
- 8.Tarkowski AK, Suwinska A, Czolowska R, Ozdzenski W. Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev Biol. 2010;348:190–198. doi: 10.1016/j.ydbio.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Balakier H, Pedersen RA. Allocation of cells to inner cell mass and trophectoderm lineages in preimplantation mouse embryos. Dev Biol. 1982;90:352–362. doi: 10.1016/0012-1606(82)90384-0. [DOI] [PubMed] [Google Scholar]
- 10.Kelly SJ. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool. 1977;200:365–376. doi: 10.1002/jez.1402000307. [DOI] [PubMed] [Google Scholar]
- 11.Rossant J. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J Embryol Exp Morphol. 1976;36:283–290. [PubMed] [Google Scholar]
- 12.Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- 13.Davis W, Jr, Schultz RM. Developmental change in TATA-box utilization during preimplantation mouse development. Dev Biol. 2000;218:275–283. doi: 10.1006/dbio.1999.9486. [DOI] [PubMed] [Google Scholar]
- 14.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- 17.Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol. 1991;147:403–414. doi: 10.1016/0012-1606(91)90298-H. [DOI] [PubMed] [Google Scholar]
- 18.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemr M, Ma J, Schultz RM, Svoboda P. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod. 2010;82:1008–1017. doi: 10.1095/biolreprod.109.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, Yan LY, Qiao J, Tang F, Fan HY. Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 2016;26:275–287. doi: 10.1038/cr.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci U S A. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Munoz E, Arboleda-Estudillo Y, Otu HH, Cibelli JB. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 24.Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y, et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14:217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 26.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 29.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 31.Kurotaki YK, Hatanaka Y, Kamimura S, Oikawa M, Inoue H, Ogonuki N, Inoue K, Ogura A. Impaired active DNA demethylation in zygotes generated by round spermatid injection. Hum Reprod. 2015;30:1178–1187. doi: 10.1093/humrep/dev039. [DOI] [PubMed] [Google Scholar]
- 32.Polanski Z, Motosugi N, Tsurumi C, Hiiragi T, Hoffmann S. Hypomethylation of paternal DNA in the late mouse zygote is not essential for development. Int J Dev Biol. 2008;52:295–298. doi: 10.1387/ijdb.072347zp. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen D, Banaszynski LA, Liu Y, Geng F, Noh KM, Xiang J, Elemento O, Rosenwaks Z, Allis CD, Rafii S. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc Natl Acad Sci U S A. 2014;111:7325–7330. doi: 10.1073/pnas.1406389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 41.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 42.Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 44.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 45.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Kawasaki K, Minoshima S, Krohn K, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 46.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 47.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 48.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 49.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 51.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 52.Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burdzy A, Noyes KT, Valinluck V, Sowers LC. Synthesis of stable-isotope enriched 5-methylpyrimidines and their use as probes of base reactivity in DNA. Nucleic Acids Res. 2002;30:4068–4074. doi: 10.1093/nar/gkf520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuo S, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;23:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 61.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 63.Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15:447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Iqbal K, Jin S-G, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C, Zhang YL, Pan WW, Li XM, Wang ZW, Ge ZJ, Zhou JJ, Cang Y, Tong C, Sun QY, et al. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science. 2013;342:1518–1521. doi: 10.1126/science.1244587. [DOI] [PubMed] [Google Scholar]
- 68.Wu G, Han D, Gong Y, Sebastiano V, Gentile L, Singhal N, Adachi K, Fischedick G, Ortmeier C, Sinn M, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frum T, Halbisen MA, Wang C, Amiri H, Robson P, Ralston A. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Bin GC, Muñoz-Descalzo S, Kurowski A, Leitch H, Lou X, Mansfield W, Etienne-Dumeau C, Grabole N, Mulas C, Niwa H, et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 2014;141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 72.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 73.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 74.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 75.Chuang YS, Huang WH, Park SW, Persaud SD, Hung CH, Ho PC, Wei LN. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem Cells. 2011;29:660–669. doi: 10.1002/stem.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koziol MJ, Garrett N, Gurdon JB. Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr Biol. 2007;17:801–807. doi: 10.1016/j.cub.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 78.Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 81.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 83.Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rajkovic A, Yan C, Yan W, Klysik M, Matzuk MM. Obox, a family of homeobox genes preferentially expressed in germ cells. Genomics. 2002;79:711–717. doi: 10.1006/geno.2002.6759. [DOI] [PubMed] [Google Scholar]
- 85.Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma. 2007;116:403–415. doi: 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- 86.Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, et al. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res. 2004;105:240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- 88.Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME (2015) Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22:662–671 [DOI] [PubMed]
- 89.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell. 2007;18:129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 92.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujii S, Nishikawa-Torikai S, Futatsugi Y, Toyooka Y, Yamane M, Ohtsuka S, Niwa H. Nr0b1 is a negative regulator of Zscan4c in mouse embryonic stem cells. Sci Rep. 2015;5:9146. doi: 10.1038/srep09146. [DOI] [PMC free article] [PubMed] [Google Scholar]


