ABSTRACT
Although genotype 4 (GT4)-infected patients represent a minor overall percentage of the global hepatitis C virus (HCV)-infected population, the high prevalence of the genotype in specific geographic regions coupled with substantial sequence diversity makes it an important genotype to study for antiviral drug discovery and development. We evaluated two direct-acting antiviral agents—grazoprevir, an HCV NS3/4A protease inhibitor, and elbasvir, an HCV NS5A inhibitor—in GT4 replicons prior to clinical studies in this genotype. Following a bioinformatics analysis of available GT4 sequences, a set of replicons bearing representative GT4 clinical isolates was generated. For grazoprevir, the 50% effective concentration (EC50) against the replicon bearing the reference GT4a (ED43) NS3 protease and NS4A was 0.7 nM. The median EC50 for grazoprevir against chimeric replicons encoding NS3/4A sequences from GT4 clinical isolates was 0.2 nM (range, 0.11 to 0.33 nM; n = 5). The difficulty in establishing replicons bearing NS3/4A resistance-associated substitutions was substantially overcome with the identification of a G162R adaptive substitution in NS3. Single NS3 substitutions D168A/V identified from de novo resistance selection studies reduced grazoprevir antiviral activity by 137- and 47-fold, respectively, in the background of the G162R replicon. For elbasvir, the EC50 against the replicon bearing the reference full-length GT4a (ED43) NS5A gene was 0.0002 nM. The median EC50 for elbasvir against chimeric replicons bearing clinical isolates from GT4 was 0.0007 nM (range, 0.0002 to 34 nM; n = 14). De novo resistance selection studies in GT4 demonstrated a high propensity to suppress the emergence of amino acid substitutions that confer high-potency reductions to elbasvir. Phenotypic characterization of the NS5A amino acid substitutions identified (L30F, L30S, M31V, and Y93H) indicated that they conferred 15-, 4-, 2.5-, and 7.5-fold potency losses, respectively, to elbasvir. The activity profiles of grazoprevir and elbasvir supported the testing of the direct-acting antivirals in clinical studies.
KEYWORDS: HCV, genotype 4, resistance, antiviral, elbasvir, grazoprevir, NS5A, NS3/4A, antiviral agents, drug resistance mechanisms, hepatitis C virus
INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, with an estimated 170 million people infected globally (1, 2). The World Health Organization estimates that more than 350,000 people die every year from hepatitis C-related liver diseases (3). Approximately 20% of individuals chronically infected with HCV can be expected to develop liver cirrhosis and, of these, 6% will decompensate to end-stage liver disease, with an additional 4% developing hepatocellular carcinoma. A number of treatment options are now available with combinations of interferon-free direct-acting antiviral agents (DAAs) that inhibit distinct viral targets, including NS3/4A protease, NS5A protein, and NS5B polymerase, have now become the standard of care (4). However, there are patient populations that are not adequately treated by the available regimens, such as those with decompensated cirrhosis or those for whom access to approved drugs remains an issue.
HCV displays a high degree of genetic heterogeneity and can be classified into seven major genotypes that have distinct geographic distributions. The major genotypes—genotype 1 (GT1), GT2, and GT3—account for 70 to 85% of all HCV infections (1, 5) and receive significant attention for drug discovery and therefore treatment options given their overrepresentation in clinical trials. The other genotypes, while not prevalent globally, can occur at high rates in certain geographic regions; genotype 4 (GT4) is of particular interest among these minor genotypes. The distribution of GT4 is highly localized, with a noted high prevalence. It is estimated that ∼14% of the Egyptian population is infected with HCV GT4. Overall, about 70% of the global GT4-infected population is localized in North Africa and the Middle East (1). The diversity of GT4 is also substantial, with at least 17 subtypes identified (6). Since the HCV genotype or subtype of an infected HCV patient, among other factors, impacts treatment response and may also influence disease progression, the significant diversity among GT4 subtypes is a concern. Although combinations of DAAs such as elbasvir/grazoprevir (7), sofosbuvir/ledispavir (8), simeprevir/sofosbuvir (9), daclatasvir/sofosbuvir (10), and ombitasvir/paritaprevir/ritonavir (11) have now been approved for treating GT4 infections, there is limited information about how GT4 subtypes may impact disease progression. In a report published in 2007 in which 131 subjects were investigated, it was noted that among Egyptian GT4-infected patients, there was a strong correlation between subtype 4o infections and hepatocellular carcinoma (12). In addition, the prevalence of resistance-associated substitutions (RASs) among these subtypes and potential impact on treatment response is poorly understood. It was therefore of interest to profile the direct-acting antiviral agents grazoprevir and elbasvir in GT4 to determine their potential clinical activity.
Grazoprevir is a potent, pangenotype HCV NS3/4A protease macrocyclic inhibitor (13, 14). HCV NS3/4A is a serine protease responsible for processing the polyprotein precursor following translation of the viral RNA (15). The viral NS3/4A protease performs four specific cleavages to release functional viral nonstructural proteins (NS3, NS4A, NS4B, NS5A, and NS5B) that are required for replication. Therefore, inhibition of this critical activity blocks viral replication. Elbasvir is a potent inhibitor of the HCV NS5A protein with broad activity against HCV genotypes (16). Although the precise function of HCV NS5A remains to be established, it is critical for replication and virus assembly; both activities are inhibited by elbasvir in replicon cells (17). Thus, grazoprevir and elbasvir are potent DAAs targeting two independent pathways of HCV replication. In combination, grazoprevir and elbasvir have demonstrated at least additive effects in blocking HCV RNA replication and potently suppressed the emergence of resistance by creating a higher genetic barrier to resistance in GT1a replicons (18). Early clinical virology data indicated that the cell-based replicon activity of elbasvir and grazoprevir in GT1 and GT3 translated well into the clinic (19). Hence, the activities of elbasvir and grazoprevir were investigated in GT4 replicons as a prelude to clinical studies.
RESULTS
The diversity of HCV sequences in GT4 is enormous, with several subtypes compared to GT1, the most prevalent genotype, for example. There are at least 17 subtypes described for GT4. Prior to generating replicons bearing patient isolates for phenotypic characterization, a phylogenetic analysis of annotated full-length GT4 sequences available in the Los Alamos National Laboratories (LANL) and GenBank HCV databases were conducted using both NS3 and NS5A sequences. Using NS3 and NS5A sequences for construction of phylogenetic trees resulted in the identification of 17 subtypes of GT4 from the data set (see Fig. S1 and S2 in the supplemental material). On the basis of sequence availability and positioning of subtypes on the phylogenetic tree, a number of sequences were selected as representative for the broad genotype diversity to establish stable replicon cell lines. Generally, it was more difficult establishing chimeric cell lines for replicons with NS3 (which included the NS4A sequence of the subtype) gene sequences than for those with NS5A sequences. Including the reference GT4a (ED43) sequence, a total of 5 unique stable cell lines representing three subtypes were successfully established for NS3 and NS4A sequences, whereas a total of 14 unique stable cell lines representing eight different subtypes were generated for NS5A sequences. All the replicons were generated as chimeras in the GT2a (JFH1) background.
Activity of grazoprevir and elbasvir in GT4 subtypes. (i) Grazoprevir.
All the NS3/4A replicons (including three generated with the G162R adaptive mutation for transient expression) from the GT4 clinical isolates were potently inhibited by grazoprevir. The potency of the NS3/4A protease inhibitor was within a 3-fold 50% effective concentration (EC50) range across the clinical isolates suggesting grazoprevir will be broadly active against GT4 isolates. A median EC50 of 0.2 nM was computed (range, 0.11 to 0.33 nM; n = 5) for the clinical isolates for which stable replicons were generated, which was comparable to that obtained for the reference GT4a (ED43) reference sequence. The data are summarized in Table 1. Two JFH-1-based replicon cell lines bearing NS3 sequences from clinical isolates (DQ418786 [GT4d] and JX227977 [GT4o]) did not successfully grow. These sequences were introduced into the GT4a (ED43) subgenomic replicon bearing firefly luciferase and a cell culture adaptive mutation (G162R) in NS3 for transient expression in an alternative assay to evaluate compound potency. Using this model, it was possible to characterize grazoprevir potency for the two isolates; the EC50 values were comparable and within 3-fold of the parental construct also bearing the G162R adaptive mutation (Table 1).
TABLE 1.
Activity of grazoprevir in HCV GT4 subtypes
| Replicon (GenBank accession no.)a | Mean EC (nM) ± SDb |
|
|---|---|---|
| EC50 | EC90 | |
| Group 1 | ||
| GT4a ED43 (GU814265) | 0.7 ± 0.4 | 1.4 ± 0.7 |
| GT4b (FJ025854) | 0.17 ± 0.04 | 0.27 ± 0.05 |
| GT4b (FJ025855) | 0.11 ± 0.04 | 0.32 ± 0.13 |
| GT4g (JX227963) | 0.15 ± 0.09 | 0.71 ± 0.78 |
| GT4g (JX227971) | 0.33 ± 0.1 | 0.6 ± 0.16 |
| Group 2 | ||
| GT4a ED43 (GU814265) | 3.7 ± 1.3 | 9 ± 3.33 |
| GT4d (DQ418786) | 1.1 ± 0.8 | 4.8 ± 1.4 |
| GT4o (JX227977) | 1.2 ± 0.6 | 2.8 ± 0.1 |
For group 1, chimeric replicons bearing patient isolates were generated in a GT2a_JFH1 background, and potencies were determined using a TaqMan-based assay. For group 2, the GT4 NS3 subtype sequences are in a full-length ED43 background with the G162R adaptive mutation and were assayed by monitoring the luciferase activity.
Potencies were determined by using a TaqMan-based assay (group 1) or luciferase assay (group 2).
(ii) Elbasvir.
When tested against the established replicons bearing the GT4 clinical isolates, elbasvir was potent against the majority of them, with EC50s in the subpicomolar to low nanomolar range. The data demonstrate that elbasvir will be broadly active in HCV patients infected with GT4. The median EC50 of elbasvir against these chimeric replicons encoding NS5A sequences from clinical isolates was 0.7 pM for genotype 4 (range, 0.0002 to 34 nM; n = 14). The potency data for all the subtypes tested are summarized in Table 2. There was a difference in potency for replicons with NS5A genes from subtype 4b. One GT4b NS5A sequence (FJ025854) was relatively sensitive to elbasvir (EC50 = 0.017 nM). The NS5A sequence for the most resistant isolate GT4b (FJ025855; EC50 = 34 nM) retained amino acid substitutions at three key positions known to confer resistance to NS5A inhibitors in other genotypes: M28, S30, and S93 (see Fig. S3 in the supplemental material). A total of three nucleotide changes are required to generate these substitutions relative to the sensitive sequence from GT4b (FJ025854). Another fairly resistant isolate GT4b (FJ462435; EC50 = 3.6 nM) retained amino acid substitutions at S30 and H93 (see Fig. S3 in the supplemental material), thereby requiring three nucleotide changes to generate the mutations compared to the sensitive FJ025854 sequence. A fourth GT4b NS5A sequence was identified in publicly available databases (FJ025856); with the same M28/S30/S93 substitutions as GT4b (FJ025855), this isolate was not established as a replicon but, given the available data, it would be predicted to be less susceptible to elbasvir. Thus, the data suggest that amino acid substitutions at the key positions (28, 30, and 93) previously associated with resistance to elbasvir in GT1 (18) are also needed to elicit resistance in GT4.
TABLE 2.
Activity of elbasvir in GT4 subtypes
| Replicon (GenBank accession no.)a | Mean EC (nM) ± SDb |
|
|---|---|---|
| EC50 | EC90 | |
| GT4a (GU814265) | 0.0002 ± 0.0001 | 0.0008 ± 0.0001 |
| GT4a (DQ418784) | 0.0002 ± 0.0001 | 0.0005 ± 0.0002 |
| GT4b (FJ025854) | 0.017 ± 0.017 | 0.030 ± 0.016 |
| GT4b (FJ025855) | 34 ± 23 | >100 |
| GT4b (FJ462435) | 3.6 ± 2.3 | 6.2 ± 3.8 |
| GT4d (DQ418786) | 0.0005 ± 0.0001 | 0.003 ± 0.001 |
| GT4d (EU392172) | 0.0004 ± 0.0001 | 0.0011 ± 0.0002 |
| GT4f (EF589161) | 0.0019 ± 0.0014 | 0.018 ± 0.028 |
| GT4g (JX227963) | 0.0006 ± 0.0002 | 0.0017 ± 0.001 |
| GT4g (JX227971) | 0.072 ± 0.035 | 0.25 ± 0.13 |
| GT4m (FJ462433) | 0.0004 ± 0.0001 | 0.0024 ± 0.0016 |
| GT4m (JX227972) | 0.0007 ± 0.0005 | 0.0043 ± 0.0024 |
| GT4o (JX227977) | 0.0022 ± 0.001 | 0.016 ± 0.011 |
| GT4q (FJ462434) | 0.0005 ± 0.0001 | 0.0014 ± 0.0003 |
All replicons have the GT4a NS5A sequences in a JFH1 background.
Averages of ≥3 analyses are shown.
De novo resistance selections in GT4. (i) Grazoprevir.
Studies were conducted with the GT4a (ED43) reference sequence to identify potential pathways of resistance to grazoprevir. Concentrations of grazoprevir up to 30-fold the EC50 in the reference replicon were used. Population sequencing of RNA isolated from resistant colonies showed mutations coding for amino acid substitutions at position 168 (D168) were primarily observed with increasing grazoprevir inhibitor concentrations (Table 3). D168G and D168V emerged at a concentration of 10× EC50 for grazoprevir, while D168V and D168A emerged at a higher concentration of 30 × EC50 D168 mutations were not observed at lower drug concentrations (1× to 3× EC50). Two substitutions, A61T and G90R, were observed in a number of the selections; however, these amino acid substitutions were also present in the dimethyl sulfoxide (DMSO) control (without inhibitor) and were considered genetic drifts and not compound related; they were not studied further. G162R was observed as a minor population in one of the selections with a low concentration (3× EC50) of grazoprevir.
TABLE 3.
De novo resistance selections with grazoprevir in the GT4a replicon
| Grazoprevir concn (nM) | Treatment (fold EC50) | Mutations (% population)a |
|---|---|---|
| No drug | 0 | A61T (100), G90R (5) |
| 1 | 1 | A61T (100), G90R (5) |
| 3 | 3 | A61T (100), G90R (5), G162R (5) |
| 10 | 10 | A61T (70), G90R (5), D168V (30), D168G (30) |
| 30 | 30 | A61T (95), G90R (5), D168V (50), D168A (50) |
Key amino acid substitutions deduced from population sequencing of resistant colonies and quantified based on peaks from the electropherogram are indicated.
(ii) Elbasvir.
In de novo resistance selection studies in GT4a replicon cells, elbasvir showed a concentration-dependent reduction in the number of resistant colonies up to the highest concentration tested, 30× EC90 (Table 4). Population sequencing revealed that amino acid substitutions at positions 30, 31, and 93 were most prevalent. In addition to these changes, a novel N69K substitution was also observed. At the highest dose tested, all colonies retained a L30S substitution. The mutation coding for the L30S substitution requires two nucleotide changes and underscores a higher genetic barrier to resistance for the compound in GT4, although a single substitution engendered resistance. The extent of resistance was also evaluated by testing the activity of elbasvir against pooled resistant colonies. The pooled resistant colonies selected at the highest dose were 40-fold more resistant than selections conducted with DMSO alone (Table 4).
TABLE 4.
De novo resistance selections with elbasvir in the GT4a (ED43) replicon
| Treatment (fold multiple of EC90) | No. of resistant coloniesa | Activity (EC90 [nM]) in pooled colonies | Fold shift (relative to DMSO) | Substitution(s) observedb |
|---|---|---|---|---|
| 3 | TMTCa | 0.003 | 6 | L30F, M31V, Y93H |
| 10 | 144 | 0.017 | 34 | L30F, L30P, M31V, N69K, Y93H |
| 30 | 35 | 0.02 | 40 | L30S |
TMTC, too many to count.
Key amino acid substitutions deduced from population sequencing of resistant colonies are listed.
Impact of amino acid substitutions. (i) Grazoprevir.
Efforts to introduce the amino acid substitutions identified in NS3 from the resistance selection studies for phenotypic characterization failed as replicon cells bearing the substitutions at position 168 failed to grow. However, the other change investigated, G162R, was successfully generated and did not confer a reduction in potency to grazoprevir. The EC50 for grazoprevir in the G162R replicon was comparable to the wild-type replicon (Table 5). Upon further characterization, it was discovered that the G162R substitution in NS3 increases the replicative capacity of replicons signifying that it could serve as a cell line adaptive substitution. The introduction of the desired substitutions at position 168, along with the G162R adaptive substitution, enabled the successful construction of stable D168A/V replicons for phenotypic characterization. The D168A and D168V substitutions (in the context of G162R) conferred potency reductions of 137- and 47-fold, respectively, for grazoprevir relative to the EC50 for the wild-type replicon (Table 5). Despite the incorporation of the G162R substitution, the D168G variant remained unfit; hence, a stable replicon could not be generated. Gratifyingly, the introduction of the G162R substitution in combination with D168G enabled the determination of a fitness value of ca. 0.2% relative to the wild-type replicon in the transient assay. However, the transient assay for the G162R and D168G double substitution was not robust enough to generate potency values.
TABLE 5.
Activity of grazoprevir in GT4 NS3 RASsa
| Replicon | EC (nM) ± SD and fold shift |
|||
|---|---|---|---|---|
| EC50 | EC50 fold shift | EC90 | EC90 fold shift | |
| Wild type | 0.7 ± 0.4 | 1 | 1.4 ± 0.7 | 1 |
| G162R | 0.7 ± 0.5 | 1 | 1.6 ± 1 | 1.1 |
| G162R_D168A | 96 ± 12 | 137.1 | 216 ± 40 | 154.3 |
| D168G | Unfit | NA | Unfit | NA |
| G162R_D168V | 33 ± 20 | 47.1 | 97 ± 31 | 69.3 |
The chimeric replicons bearing the resistance-associated substitutions were generated in a GT2a (JFH1) background, and potencies were determined using a TaqMan-based assay. The fold shift is based on the wild-type value. NA, not applicable.
(ii) Elbasvir.
All the substitutions selected in NS5A by elbasvir were successfully introduced into replicon cells for phenotypic characterization. Based on the impact of the pooled resistant colonies on the potency of elbasvir, it was not surprising that the impact of the independent substitutions in NS5A on elbasvir potency was minimal. The reduction in elbasvir potency by the substitutions ranged from 1.5- to 15-fold as summarized in Table 6. The L30S change caused a 4-fold reduction in elbasvir EC50 potency when introduced into replicon cells, while the novel N69K substitution did not impact inhibitor potency (Table 6). Interestingly, L30F, which gave the greatest fold reduction in potency (15-fold) against elbasvir, was not selected at the highest concentration tested.
TABLE 6.
Activity of elbasvir in GT4 NS5A RASsa
| Replicon | EC (nM) ± SD and fold shift |
|||
|---|---|---|---|---|
| EC50 | EC50 fold shift | EC90 | EC90 fold shift | |
| Wild type (ED43) | 0.0002 ± 0.0002 | 1 | 0.0008 ± 0.0001 | 1 |
| L30F | 0.003 ± 0.002 | 15 | 0.015 ± 0.011 | 18.8 |
| L30P | 0.0002 ± 0.0001 | 1 | 0.0004 ± 0.0002 | 0.5 |
| L30S | 0.0008 ± 0.0005 | 4 | 0.0043 ± 0.0030 | 5.4 |
| M31V | 0.0005 ± 0.0004 | 2.5 | 0.0009 ± 0.0007 | 1.1 |
| N69K | 0.0003 ± 0.0002 | 1.5 | 0.0003 ± 0.0001 | 0.4 |
| Y93H | 0.0015 ± 0.0002 | 7.5 | 0.0032 ± 0.0030 | 4 |
The chimeric replicons bearing the resistance-associated substitutions were generated in a GT2a (JFH1) background, and potencies were determined using a TaqMan-based assay. The fold shift is based on the wild-type value.
NS3 and NS5A polymorphisms in GT4.
Given the potency shifts observed for a few of the clinical isolates particularly for elbasvir, an analysis of GT4 sequences was conducted to determine the prevalence of substitutions at positions normally associated with resistance for both the NS3/4A protease and the NS5A inhibitor classes. Fourteen amino acid positions in NS3 were evaluated: 36, 54, 55, 56, 80, 107, 122, 132, 155, 156, 158, 168, 170, and 175. For NS5A, the four amino acid positions (28, 30, 31, and 93) most commonly seen in elbasvir resistance studies (18, 20) were evaluated. As reported in Table 7, there was a high global conservation among ≥187 NS3 sequences obtained from the public databases. At each of the positions evaluated, a conservation of >93% relative to the reference GT4a (ED43) strain was computed. The conservation of NS5A sequences at the four positions was not uniform. The highest conservation among >142 sequences was at positions 31 and 93, with 94.4 and 98.6%, respectively; position 30 showed the least conservation at 43% (Table 8). The data are consistent with position 30 being most prone to mutational changes in the resistance studies; however, reductions in elbasvir potency from single amino acid substitutions were minimal.
TABLE 7.
Global prevalence of polymorphisms within NS3 in HCV GT4 population
| Parameter | Amino acid position |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 36 | 54 | 55 | 56 | 80 | 107 | 122 | 132 | 155 | 156 | 158 | 168 | 170 | 175 | |
| GT4 ED43 | L | T | V | Y | Q | V | T | I | R | A | V | D | V | L |
| GT4 conservation (%)a | 100 | 97.3 | 99.5 | 100 | 99.5 | 95.9 | 93.6 | 96.8 | 100 | 100 | 99.5 | 99.5 | 93.6 | 100 |
| GT4 substitutionb | L | T | V | Y | Q | V | T/S | I | R | A | V | D | V/I | L |
| No. of database sequences evaluated | 219 | 218 | 219 | 219 | 219 | 219 | 218 | 219 | 196 | 196 | 195 | 187 | 188 | 188 |
That is, the percent conservation of the reference sequence.
That is, the substitution within the specified positions that has ≥5% of the sequences in the database. Substitutions are presented in order of decreasing frequency.
TABLE 8.
Global prevalence of polymorphisms within NS5A in the HCV GT4 population
| Parameter | Amino acid position |
|||
|---|---|---|---|---|
| 28 | 30 | 31 | 93 | |
| GT4 ED43 | L | L | M | Y |
| GT4 conservationa | 86.6 | 43 | 94.4 | 98.6 |
| GT4 substitution(s)b | L/M | R/L | M/L | Y |
| No. of database sequences evaluated | 142 | 142 | 143 | 144 |
That is, the percent conservation of the reference sequence.
That is, the substitution(s) within the specified positions that has ≥5% of the sequences in the database. Substitutions are presented in order of decreasing frequency.
DISCUSSION
Given the high global prevalence of HCV GT1, -2, and -3, a significant proportion of clinical and nonclinical studies have understandably focused on these genotypes. From these studies, particularly in the highly prevalent GT1, certain amino acid positions in NS3 and NS5A have been identified to be associated with the emergence of resistance. For example, the presence of resistance-associated substitutions in NS5A can in part account for lower response rates, particularly in GT1a (21, 22). Since there is limited information on the prevalence and impact of NS3 and NS5A amino acid substitutions coupled with a dearth of knowledge on the contribution of GT4 subtypes to disease progression and clinical response, the activities of elbasvir, an NS5A inhibitor, and grazoprevir, an NS3/4A inhibitor, were investigated in GT4 clinical isolates prior to commencing clinical studies.
Chimeric GT4 replicons bearing NS3 and NS4A sequences from clinical isolates were constructed for phenotypic characterization. Grazoprevir potently inhibited replicons bearing sequences representing seven clinical isolates from subtypes 4a, 4b, 4d, 4g, and 4o. Although it was more difficult to obtain additional NS3 replicons from other subtypes compared to NS5A (see below), the highly prevalent subtypes 4a and 4d that account for the majority (∼90%) of GT4 infections were generated along with 4o, which has been associated with high levels of hepatocellular carcinoma. Grazoprevir was equipotent across the subtypes with potency differences within a narrow 3- to 5-fold range, suggesting the NS3/4A inhibitor may have broad activity across GT4 subtypes. Twice as many replicons were successfully generated with NS5A sequences, which also included all the subtypes generated for the NS3 sequences. Elbasvir was potent across seven of the eight GT4 subtypes (4a, 4b, 4d, 4f, 4g, 4m, 4o, and 4q) represented, which included the most prevalent subtypes 4a and 4d, but less so with two subtype 4b sequences (one GT4b sequence, FJ025854, was sensitive to elbasvir). The least susceptible subtype 4b sequence (FJ025855) harbored amino acid substitutions at positions 28 (M), 30 (S), and 93 (S) and would require three nucleotide changes to establish the mutations in the sensitive FJ025854 GT4b sequence. Similarly, the resistant 4b (FJ462435) sequence also harbored changes at positions 31 (S) and 93 (H) that would require three nucleotide change to generate the mutations in the sensitive isolate. Given the limited distribution of GT4b (23), GT4b sequences with these RASs are not anticipated to be prevalent within the population. Certainly, the combination of an NS3/4A protease inhibitor and an NS5A replication inhibitor is expected to increase the overall barrier to resistance to variant viruses, as seen, for example, using elbasvir and grazoprevir in GT1a replicons (18).
Since sequences that were associated with resistance were identified among the clinical isolates, particularly within NS5A, we investigated the common resistance pathways by de novo selections in susceptible GT4a replicon cells. Two compound-dependent pathways were selected with grazoprevir. The changes at position 168 (D168A/G/V) in NS3 were observed previously in GT1a resistance selection studies (18) and therefore were not surprising. Indeed, previous studies of GT4a viruses bearing substitutions at position 168 were poorly fit (24). However, the amino acid change at position 162 was novel; this unexpected substitution was investigated in detail. Phenotypic characterization indicated that the G162R did not confer resistance to grazoprevir. We therefore hypothesized that it potentially influenced the replicative capacity of the replicon. By introducing the substitution into full-length replicons that also expressed firefly luciferase that can be monitored as an index of replication, we determined the fitness of transiently expressed replicons relative to the parental replicon. The introduction of the G162R substitution increased the replicative capacity of the replicon by ∼4-fold. This discovery provided a path forward to phenotypically characterize NS3 substitutions that confer a low replicative fitness capacity as the replication-boosting G162R substitution did not impact grazoprevir potency (although we cannot exclude the possibility that G162R could affect the extent of resistance associated with a RAS). This approach enabled the phenotypic characterization of the D168A and D168V NS3 substitutions which conferred 137- and 47-fold potency losses to grazoprevir (relative to the EC50 of the parental replicon). Despite the introduction of the G162R substitution in NS3, the combination with the D168G amino acid change could not be established as a stable replicon or provide a robust enough transient replication for characterization; hence, its impact on grazoprevir antiviral activity remains unknown.
Low replicative fitness was not an issue for the identified NS5A substitutions. All the amino acid changes selected with elbasvir were successfully established as replicons. Most of the changes occurred at position 30 (L30F/P/S) with the L30F conferring the highest potency loss (15-fold) to elbasvir. An unexpected N69K substitution (on the basis of previous studies in GT1) was detected in resistant colonies. However, this change did not confer a reduction in inhibitor potency to elbasvir. It also did not appear to influence replicon fitness. The Y93H change that usually engenders >1,000-fold resistance to elbasvir in GT1a stable replicons did not impact the inhibitor substantially in GT4 since it conferred only a 7.5-fold loss in potency. Although the pool of resistant colonies selected at the highest dose of elbasvir demonstrated a potency reduction of 40-fold, none of the individual RASs tested conferred this level of resistance. Since the mutations were identified using population sequencing (with a sensitivity threshold for the detection of minority variants of ∼20%), it is possible that HCV genomes in the population with changes below the level of detection may have contributed to the overall reduction in potency for the pool of resistant colonies. It is also possible that a combination of such undetected RASs may influence this reduction in potency as observed for the less-susceptible GT4b isolates. This would suggest that more than a single nucleotide change would be required for substantial resistance. Future studies will investigate the impact of combinations of RASs in NS5A from GT4 on inhibitor potency.
While the de novo resistance selection studies with both grazoprevir and elbasvir suggested a potential broad activity among GT4 subtypes, we investigated this further given the significant diversity among GT4 sequences. An analysis of the sequences available in the public databases showed a high conservation among NS3 sequences at 14 key positions associated with resistance within the inhibitor class. The least-conserved amino acid position, i.e., position 170, still showed a 93.6% conservation relative to the reference GT4a (ED43) sequence; this was consistent with the broad activity of grazoprevir in the five subtypes evaluated. For NS5A sequences, the least conservation occurred at position 30 of the four critical positions associated with resistance to elbasvir and other NS5A inhibitors in the well-studied GT1a. The conservation of 43% at position 30 was in line with the amino acid substitutions observed in the de novo resistance selection studies where the most changes were found at this position.
In summary, our studies in GT4 demonstrated a broad activity for grazoprevir and elbasvir among GT4 subtypes and provided nonclinical support for the evaluation of the compounds in GT4 patients in the clinic. In the phase 3 C-EDGE clinical trials (25–27), a fixed-dose combination of elbasvir and grazoprevir administered for 12 weeks resulted in a sustained virologic response 12 weeks after treatment (SVR12) of 95% among HCV treatment-naive GT4-infected patients. The elbasvir-grazoprevir combination is a valuable option, among others (4, 28), for patients chronically infected with GT4 HCV.
MATERIALS AND METHODS
Compounds.
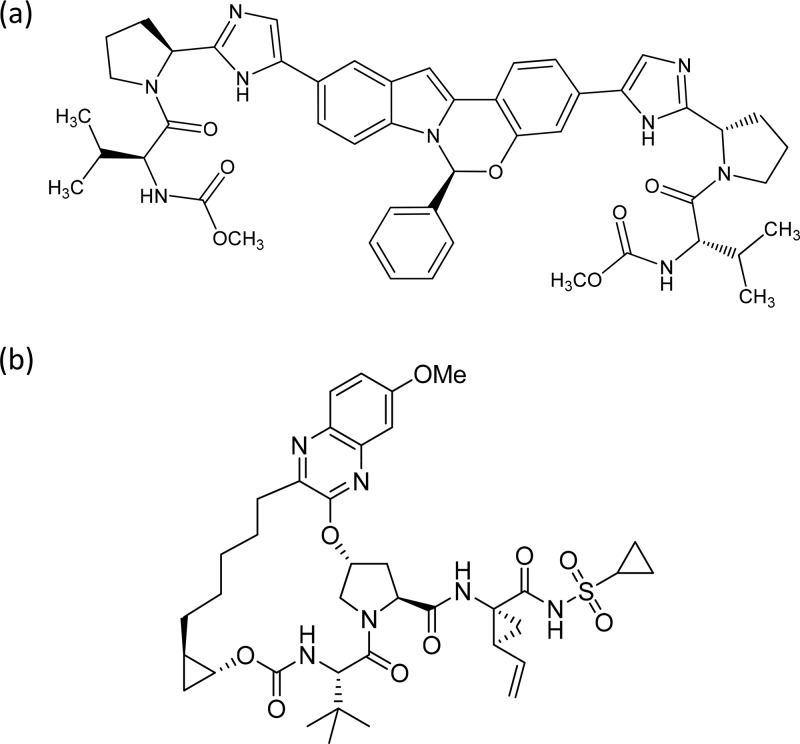
Elbasvir, N,N′-[[(6S)-6-phenyl-6H-indolo[1,2-c][1,3]benzoxazine-3,10-diyl]bis[1H-imidazole-5,2-diyl-(2S)-2,1-pyrrolidinediyl[(1S)-1-(1-methylethyl)-2-oxo-2,1-ethanediyl]]]bis[carbamic acid] C,C′-dimethyl ester (Fig. 1a) was prepared as reported previously (16). Grazoprevir, N-[[[(1R,2R)-2-[5-(3-hydroxy-6-methoxy-2-quinoxalinyl)pentyl]cyclopropyl]oxy]carbonyl]-3-methyl-l-valyl-(4R)-4-hydroxy-l-prolyl-(1R,2S)-1-amino-N-(cyclopropylsulfonyl)-2-ethenylcyclopropanecarboxamide cyclic (1→2)-ether (Fig. 1b), was also prepared as reported previously (13, 14). All cell culture reagents (unless otherwise indicated in the text) were obtained from BioWhittaker (Radnor, PA).
FIG 1.
(a) Chemical structure of elbasvir 1; (b) chemical structure of grazoprevir.
Cell culture.
Human hepatoma cell line Huh-7 or Huh-7.5 (19) was cultured in Dulbecco minimal essential medium supplemented with 2 mM glutamine, nonessential amino acids, 0.075% sodium bicarbonate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS) in 10 mM HEPES at a pH of 7.5. Stable replicons generated in Huh7 or Huh7.5 cells were cultured in G418 (Cellgro, Manassas, VA) at 0.5 mg/ml. The generation and establishment of HCV replicons in Huh cell lines was previously described (29).
Phylogenetic analyses.
A phylogenetic analysis was conducted to generate a representative and varied set of GT4 replicons from available sequences. For the GT4 NS5A gene, there were 130 GT4 patient (PT) sequences from the elbasvir-grazoprevir phase 2/3 development program, 35 North American (NA), and 45 other (OT) sequences from diverse geographic regions were available in the LANL and GenBank HCV databases. For the NS3 gene, there were 134 PT, 29 NA, and 45 OT sequences available. All sequences from both NS5A and NS3 sets were aligned using the software package MUSCLE (30). The alignment was then used to estimate a maximum-likelihood phylogeny in the software package PhyML (31) under a GTR+G+I model of nucleotide evolution. The tree was visualized using the software package Figtree (http://tree.bio.ed.ac.uk/software/figtree/). Genetic distances were parsed from the tree to generate the beanplots via the R package “Beanplot” (32). This information (see Fig. S1 and S2 in the supplemental material) was used as described in “Generation of replicons” (below) to create replicons (referred to as lab strains in Fig. S1 and S2) that included 14 NS5A and 7 NS3 sequences, based on selected NA and OT sequences. Sequences from patients with GT4 infections in the phase 2/3 program were compared to lab strains and the publicly available GT4 sequences. The comparison indicated that the genetic variation within the captured clinical samples were similar to the genetic variation existing in the NA and OT sequences. In addition, the genetic variation from the patients (PT) also captures the genetic variation within the lab strains (not shown).
Alignment of Lab NS5A protein sequences (see Fig. S3 in the supplemental material) was generated using the AlignX program from the VectorNTI suite (Invitrogen).
Generation of replicons.
Chimeric cDNAs were designed and made by gene synthesis (Genewiz, South Plainfield, NJ) as cassettes for cloning into the GT2a (JFH-1) subgenomic replicon background vector (19). Chimeric HCV replicons bearing sequences of NS3 protease (with the associated NS4A sequence) and NS5A patient isolates from GT4 infections were created using the evolutionary analysis (see “Phylogenetic analyses”) to select a broad representation of the NS3 and NS5A genes (see Fig. S1 and S2 in the supplemental material). Representative sequences were selected, and maps were designed to replace the cognate NS3 protease/NS4A or the NS5A gene in the GT2a (JFH-1) replicon. Modified NS3 genes included NS3 residues 1 to 180 and the complete NS4A sequences from the genotype and/or subtype of interest, whereas replicons bearing new NS5A genes carried the complete NS5A sequence. In JFH-1, the NS5A sequence segment for each chimeric genome spanning the restriction sites NsiI and BsrGI (at nucleotides 4115 and 6599, respectively, in the wild-type JFH-1 replicon) was synthesized and sequence confirmed (GeneWiz, South Plainfield, NJ). In addition, to ensure proper cleavage site recognition by the NS3/4A protease in the NS3 chimeric constructs, the first 9 bp of each NS5A sequence were changed to match the GT4 sequence. Replicon cDNAs with RASs were generated by the same strategy. RNA was transcribed from XbaI-linearized plasmids using T7 MEGAscript (Ambion/Life Technologies) according to the manufacturer's protocol, as described previously (33). Ten micrograms of purified transcript RNA was electroporated into Huh7 cells as reported previously (29). Cells were placed under G418 selection at 0.5 mg/ml, and media were refreshed until colonies containing the replicon genomes were obtained. Established cell lines were validated for drug susceptibility against a panel of control inhibitors in subsequent studies.
Inhibition studies in replicon cells using a TaqMan assay.
To measure the cell-based inhibitory activity of elbasvir or grazoprevir, Huh-7 (or Huh-7.5) replicon-containing cells were seeded at 1,000 (or 2,000) cells/well in 384-well collagen I-coated Biocoat plates (Becton Dickinson). At 24 h postseeding, the inhibitor was added to the cells in a final concentration of 0.5% (vol/vol) DMSO in 5% FBS with no addition of G418. The cells were treated with inhibitor for 3 days, at which point they were washed with phosphate-buffered saline and lysed in cell lysis buffer (Ambion). The amount of replicon RNA level was measured by real-time quantitative PCR (TaqMan) assay (29, 34). The PCR primers for GT1a replicons were located in the HCV internal ribosome entry site (IRES): IRES_F (5′-TGCGGAACCGGTGAGTACA-3′) and IRES_R (5′-GCGGGTTTATCCAAGAAAGGA-3′). The probe sequence was FAM-labeled 5′-CGGAATTGCCAGGACGACCGG-3′. The PCR primers for GT1b were located in NS5B: 5B_F (5′-ATGGACAGGCGCCCTGA-3′) and 5B.2_R (5′-TTGATGGGCAGCTTGGTTTC-3′). The probe sequence was FAM-labeled 5′-CACGCCATGCGCTGCGG-3′. Real-time reverse transcription-PCR (RT-PCR) analyses were run on an ABI Prism 7900HT sequence detection system according to the following program: 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The CT values were plotted against log inhibitor concentration and fitted to the sigmoid dose response model using an assay data analyzer (Merck & Co., Inc., Kenilworth, NJ) or Activity Base v8.0.1.3 software (ID Business Solutions, Ltd.). The EC50 was the drug dose necessary to achieve ΔCT = 1 over the projected baseline. The EC90 was the drug dose necessary to achieve ΔCT = 3.2 over the baseline. All TaqMan reagents were obtained from Life Technologies.
Inhibition studies in replicon cells using a luciferase assay.
Alternatively, the cell-based inhibitory activity of elbasvir or grazoprevir was determined using a GT4 HCV replicon bearing the firefly luciferase gene (F-luc) integrated in frame with the viral nonstructural genes (Apath, New York, NY) (35). This construct was used to analyze the phenotypes of diverse subtypes of patient isolated genomes and various amino acid substitutions. The substitutions of interest were produced by cloning a cassette of the modified target gene made by gene synthesis into the GT4a ED43 background. Transfection of this replicon RNA, transcribed from cDNA using T7 MEGAscript, into Huh 7.5 cells results in replication of the HCV RNA and expression of the F-luc protein. The levels of expressed F-luc protein directly correlate with HCV RNA copy number and viral protein translation. The transiently expressed replicon system allows for growth and characterization of viruses bearing mutations that may have detrimental effects on viral fitness and an inability to establish stable replicons. In this protocol, 5 × 106 Huh 7.5 cells were transfected by electroporation with 5 μg of replicon RNA on a Bio-Rad gene pulser Xcell using the exponential protocol at 270 V, a 950-μF capacitance, and a resistance of 100 Ω. Cells were transferred to 75-ml flasks with 1/100 transferred to six wells of a 24-well plate. Cell lysate prepared using luciferase cell culture lysis reagent (Promega, catalog no. E1531) was collected from one well after 6 h and each day thereafter. All collected cell lysates were stored at −20°C until luciferase measurement. After 7 days, all lysate samples were assayed for firefly luciferase activity using a luciferase assay system (Promega, catalog no. E1501) according to the manufacturer's protocol.
To measure the effect of changes in genomes on the compound potency, the cells were transferred into 96-well plates 3 days after transfection and allowed to adhere overnight. The following day, serial 2-fold dilution of compounds in 0.5% DMSO was added. After 72 h, the cells were lysed as described above, and the plates were stored at −20°C until luciferase measurement. The luciferase activity was measured on an Envision plate reader (model 2104) from Perkin-Elmer. The concentrations of compounds reducing the luciferase measurements by 50 or 90% compared to the DMSO only control were taken as the EC50 and the EC90, respectively, using Prism analysis of a sigmoidal dose-response curve.
De novo resistance selection studies.
To select replicon cells bearing genomes resistant to elbasvir or grazoprevir, subconfluent monolayers of replicon cells were cultured with concentrations of the compounds at multiples of the EC90 values (18). All cells were passaged at a 1:10 ratio when they were ∼95% confluent. The colonies that survived selection were pooled and expanded for further analysis. Total cellular RNA was isolated from pooled colonies and amplified by RT-PCR. The RT-PCR products were purified by using a QIAquick PCR purification kit (Qiagen), and the NS5A region was sequenced. Alternatively, the RT-PCR products were cloned into the TOPO TA vector (Invitrogen), and the plasmid DNA from 12 bacterial colonies was sequenced.
Relative replicative fitness of viral genomes.
The replicative fitness of transiently expressed GT4 F-luc viral genomes was determined by measuring the firefly luciferase activity in cell lysates collected 4 to 7 days after transfection, according to a previously described strategy (36). This value was then normalized to the luciferase activity measured in the cell culture lysate collected 6 h after transfection to exclude the contribution of input RNA. The increase in luciferase signal for an RAS-containing genome or a subtype chimera is subsequently compared to the equivalent increase in signal observed for the parental GT4a ED43 sequence bearing a cell culture adaptive G162R substitution in NS3. The ratio of the normalized signal observed for a RAS or chimeric replicon relative to that of the parental GT4 ED43 NS3 G162R defines the relative fitness of that construct. For the purpose of comparison, the parental GT4a ED43 replicon with the NS3 G162R substitution was assigned a fitness value of 1.
Supplementary Material
ACKNOWLEDGMENTS
Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, has developed grazoprevir (MK-5172) and elbasvir (MK-8742) as components of a combination therapy for chronic HCV infections. The design, execution, and interpretation of this study were performed by the authors, who are or were employees of Merck & Co., Inc., Kenilworth, NJ. As present or former employees of Merck & Co., Inc., Kenilworth, NJ, authors may own stock and/or stock options in the company. All authors had full access to any pertinent data upon request. Each coauthor approved a final version of the manuscript.
The opinions expressed here represent the consensus of the authors and do not necessarily reflect the formal position of Merck & Co., Inc., Kenilworth, NJ.
We thank Stuart Black for helpful discussions. We also thank Michele McColgan for assistance with the figure files.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00363-17.
REFERENCES
- 1.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. 2014. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2014. Guidelines for the screening, care and treatment of persons with hepatitis C infection. World Health Organization, Geneva, Switzerland: http://www.who.int/hiv/pub/hepatitis/hepatitis-c-guidelines/en/. [PubMed] [Google Scholar]
- 4.Bastos J, Padilla M, Caserta L, Miotto N, Vigani A, Arns C. 2016. Hepatitis C virus: promising discoveries and new treatments. World J Gastroenterology 22:6393–6401. doi: 10.3748/wjg.v22.i28.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messina J, Humphreys I, Flaxman A, Brown A, Cooke G, Pybus O, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merck & Co, Inc. 2016. ZepatierTM (elbasvir and grazoprevir) tablets, for oral use. Initial US approval. Merck & Co, Inc, Kenilworth, NJ. [Google Scholar]
- 8.Gilead Sciences, Inc. 2015. HarvoniTM (ledipasvir and sofosbuvir) tablets, for oral use. Prescribing information. Gilead Sciences, Inc, Foster City, CA. [Google Scholar]
- 9.Willemse SB, Baak L, Kuiken SD, van der Sluys Veer A, Lettinga KD, van der Meer JT, Depla AC, Tuynman H, van Nieuwkerk CM, Schinkel CJ, Kwa D, Reesink HW, van der Valk M. 2016. Sofosbuvir plus simeprevir for the treatment of HCV genotype 4 patients with advanced fibrosis or compensated cirrhosis is highly efficacious in real life. J Viral Hepat 23:950–954. doi: 10.1111/jvh.12567. [DOI] [PubMed] [Google Scholar]
- 10.Hezode C, Abergel A, Chas J, Conti F, Cotte L, Tateo M, Alric L, Vergniol J, Tomei C, Bernard P-H, Loustaud-Ratti V, Arpurt J-P, Blaison D, Larrey D, Fedchuk L, Bennai Y, Filipovics A, Fontaine H, Pageaux G-P. 2019. Sustained virologic response to daclatasvir and sofosbuvir, with or without ribavirin, among patients in the French daclatasvir ATU programme infected with HCV genotypes 4, 5, and 6. J Hepatol 64:S755. doi: 10.1016/S0168-8278(16)01471-9. [DOI] [Google Scholar]
- 11.AbbVie, Inc. 2015. Technivie (ombitasvir/paritaprevir/ritonavir), copackaged for oral use. Initial US approval. AbbVie, Inc, North Chicago, IL. [Google Scholar]
- 12.Abdel-Hamid M, El-Daly M, Molnegren V, El-Kafrawy S, Abdel-Latif S, Esmat G, Strickland GT, Loffredo C, Albert J, Widell A. 2007. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol 88:1526–1531. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 13.Harper S, McCauley J, Rudd MT, Ferrara M, DiFilippo M, Crescenzi B, Koch U, Petrocchi A, Holloway MK, Butcher JW, Romano JJ, Bush KJ, Gilbert KF, McIntyre CJ, Nguyen KT, Nizi E, Carroll SS, Ludmerer SW, Burlein C, DiMuzio JM, Graham DJ, McHale CM, Stahlhut MW, Olsen DB, Monteagudo E, Cianetti S, Giuliano C, Pucci V, Trainor N, Fandozzi CM, Rowley M, Coleman PJ, Vacca JP, Summa V, Liverton NJ. 2012. Discovery of MK-5172, a macrocyclic hepatitis C virus NS3/4a protease inhibitor. ACS Med Chem Lett 3:332–336. doi: 10.1021/ml300017p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summa V, Ludmerer S, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, Dimuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, Huang Q, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2014. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolykhalov A, Mihalik K, Feinstone SM, Rice CM. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol 74:2046–2051. doi: 10.1128/JVI.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn CA, Meinke P, Chang W, Fandozzi CM, Graham DJ, Hu B, Huang Q, Kargman S, Kozlowski J, Liu R, McCauley JA, Nomeir AA, Soll RM, Vacca JP, Wang D, Wu H, Zhong B, Olsen DB, Ludmerer SW. 2013. Discovery of MK-8742: an HCV NS5A inhibitor with broad genotype activity. Chem Med Chem 8:1930–1940. doi: 10.1002/cmdc.201300343. [DOI] [PubMed] [Google Scholar]
- 17.McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, Lemon SM. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147:453–462. doi: 10.1053/j.gastro.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahser FC, Bystol K, Curry S, McMonagle P, Xia E, Ingravallo P, Chase R, Liu R, Black T, Hazuda D, Howe AY, Asante-Appiah E. 2016. The combination of grazoprevir, a hepatitis C virus (HCV) NS3/4A protease inhibitor, and elbasvir, an HCV NS5A inhibitor, demonstrates a high genetic barrier to resistance in HCV genotype 1a replicons. Antimicrob Agents Chemother 60:2954–2964. doi: 10.1128/AAC.00051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Curry S, McMonagle P, Yeh WW, Ludmerer SW, Jumes PA, Marshall WL, Kong S, Ingravallo P, Black S, Pak I, DeNubile MJ, Howe AY. 2015. Susceptibilities of genotype 1a, 1b, and 3 hepatitis C virus variants to the NS5A inhibitor elbasvir. Antimicrob Agents Chemother 59:6922–6929. doi: 10.1128/AAC.01390-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu TE, Boyd S, Sherwat A, Tracy L, Naeger LK, O'Rear JJ, Harrington PR. 2017. Regulatory analysis of effects of hepatitis C virus NS5A polymorphisms on efficacy of elbasvir and grazoprevir. Gastroenterology 152:586–597. doi: 10.1053/j.gastro.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson IM, Asante-Appiah E, Wong P, Black T, Howe A, Wahl J, Robertson MN, Nguyen B-Y, Shaughnessy M, Hwang P, Barr E, Hazuda D. 2015. Prevalence and impact of baseline NS5A resistance-associated variants (RAVs) on the efficacy of elbasvir/grazoprevir (EBR/GZR) against GT1a infection. Hepatology 62:LB-22. [Google Scholar]
- 22.Zeuzem S, Mizokami M, Pianko S, Mangia A, Han KH, Martin R, Svarovskaia E, Dvory-Sobol H, Doehle B, Hedskog C, Yun C, Brainard DM, Knox S, McHutchison JG, Miller MD, Mo H, Chuang WL, Jacobson I, Dore GJ, Sulkowski M. 2017. NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: prevalence and effect on treatment outcome. J Hepatol doi: 10.1016/j.jhep.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Kamal SM, Nasser I. 2007. Hepatitis C genotype 4: what we know and what we don't yet know. Hepatology 47:1371–1383. doi: 10.1002/hep.22127. [DOI] [PubMed] [Google Scholar]
- 24.Jensen SB, Serre S, Humes DG, Ramirez S, Li YP, Bukh J, Gottwein JM. 2015. Substitutions at NS3 residue 155, 156, or 168 of hepatitis C virus genotypes 2 to 6 induce complex patterns of protease inhibitor resistance. Antimicrob Agents Chemother 59:7426–7436. doi: 10.1128/AAC.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dore GJ, Altice F, Litwin AH, Dalgard O, Gane EJ, Shibolet O, Luetkemeyer A, Nahass R, Peng CY, Conway B, Grebely J, Howe AY, Gendrano IN, Chen E, Huang HC, Dutko FJ, Nickle DC, Nguyen BY, Wahl J, Barr E, Robertson MN, Platt HL; C-EDGE CO-STAR Study Group. 2016. Elbasvir-grazoprevir to treat hepatitis C virus infection in persons receiving opioid agonist therapy: a randomized trial. Ann Intern Med 165:625–634. doi: 10.7326/M16-0816. [DOI] [PubMed] [Google Scholar]
- 26.Rockstroh JK, Nelson M, Katlama C, Lalezari J, Mallolas J, Bloch M, Matthews GV, Saag MS, Zamor PJ, Orkin C, Gress J, Klopfer S, Shaughnessy M, Wahl J, Nguyen BY, Barr E, Platt HL, Robertson MN, Sulkowski M. 2015. Efficacy and safety of grazoprevir (MK-5172) and elbasvir (MK-8742) in patients with hepatitis C virus and HIV coinfection (C-EDGE COINFECTION): a non-randomised, open-label trial. Lancet HIV 2:e319–e327. doi: 10.1016/S2352-3018(15)00114-9. [DOI] [PubMed] [Google Scholar]
- 27.Zeuzem S, Ghalib R, Reddy KR, Pockros PJ, Ben Ari Z, Zhao Y, Brown DD, Wan S, DiNubile MJ, Nguyen BY, Robertson MN, Wahl J, Barr E, Butterton JR. 2015. Grazoprevir-elbasvir combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: a randomized trial. Ann Intern Med 163:1–13. doi: 10.7326/M15-0785. [DOI] [PubMed] [Google Scholar]
- 28.Hathorn E, Elsharkawy A. 2016. Management of hepatitis C genotype 4 in the directly acting antivirals era. BMJ Open Gastro 3:e000112. doi: 10.1136/bmjgast-2016-000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong X, Bogen S, Chase R, Girijavallabhan V, Guo Z, Njoroge FG, Prongay A, Saksena A, Skelton A, Xia E, Ralston R. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res 77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 32.Kampstra P. 2008. Beanplot: a boxplot alternative for visual comparison of distributions. J Statistical Software 28:1. doi: 10.18637/jss.v028.c01. [DOI] [Google Scholar]
- 33.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res 70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Gu Z, Graci J, Lahser F, Breslin JJ, Jung SP, Crona JH, McMonagle P, Xia E, Liu S, Karp G, Zhu J, Huang S, Nomeir A, Weetall M, Almstead NG, Peltz SW, Tong X, Ralston R, Colacino JM. 2013. Identification of PTC725, an orally bioavailable small molecule that selectively targets the hepatitis C virus NS4B protein. Antimicrob Agents Chemother 57:3250–3261. doi: 10.1128/AAC.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed M, Scheel T, Gottwein J, Marukian S, Dustin L, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.