Abstract
Allelic chromosomal regions totaling more than 2.8 Mb and located on maize (Zea mays) chromosomes 1L, 2S, 7L, and 9S have been sequenced and compared over distances of 100 to 350 kb between the two maize inbred lines Mo17 and B73. The alleles contain extended regions of nonhomology. On average, more than 50% of the compared sequence is noncolinear, mainly because of the insertion of large numbers of long terminal repeat (LTR)-retrotransposons. Only 27 LTR-retroelements are shared between alleles, whereas 62 are allele specific. The insertion of LTR-retrotransposons into the maize genome is statistically more recent for nonshared than shared ones. Most surprisingly, more than one-third of the genes (27/72) are absent in one of the inbreds at the loci examined. Such nonshared genes usually appear to be truncated and form clusters in which they are oriented in the same direction. However, the nonshared genome segments are gene-poor, relative to regions shared by both inbreds, with up to 12-fold difference in gene density. By contrast, miniature inverted terminal repeats (MITEs) occur at a similar frequency in the shared and nonshared fractions. Many times, MITES are present in an identical position in both LTRs of a retroelement, indicating that their insertion occurred before the replication of the retroelement in question. Maize ESTs and/or maize massively parallel signature sequencing tags were identified for the majority of the nonshared genes or homologs of them. In contrast with shared genes, which are usually conserved in gene order and location relative to rice (Oryza sativa), nonshared genes violate the maize colinearity with rice. Based on this, insertion by a yet unknown mechanism, rather than deletion events, seems to be the origin of the nonshared genes. The intergenic space between conserved genes is enlarged up to sixfold in maize compared with rice. Frequently, retroelement insertions create a different sequence environment adjacent to conserved genes.
INTRODUCTION
One of the basic assumptions of genetics is that the genomes of individuals belonging to a single species are colinear at the sequence level and contain the same gene complement. The exceptions include relatively rare single nucleotide polymorphisms (SNPs), insertions/deletions (indels), translocations, and insertions of transposons or other chromosomal abnormalities (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Rafalski, 2002; Yu et al., 2002). Though the amount of intraspecific sequence diversity varies widely between species (Wolfe et al., 1989; Gale and Devos, 1998; Keller and Feuillet, 2000; Bennetzen and Ma, 2003), this fundamental assumption remained unchallenged until recently, as few extensive comparisons of DNA sequences outside of transcribed regions have been performed among individuals of the same species. In rice (Oryza sativa), numerous SNPs and small indels were identified within repetitive DNA and genes in ≈2.3 Mb of orthologous regions from Asian rice subspecies indica and japonica (Feng et al., 2002; Han and Xue, 2003). Genome size increases of >2 and >6%, respectively, were also proposed for the same two rice subspecies since their divergence from a common ancestor (Ma and Bennetzen, 2004b). In wheat, a comparison of two large orthologous loci on chromosome 1AS of Triticum monococcum subsp monococcum and T. turgidum subsp durum showed that the sequence conservation was restricted to small regions containing orthologous genes (Wicker et al., 2003). Fu and Dooner (2002) compared maize (Zea mays) sequences at the bz1 locus in the B73 and McC inbred lines. Surprisingly, much of the intergenic sequence was different, and four of the genes also differed between the two maize inbred lines. Similar local absence of colinearity was observed at the maize z1C-1 locus (Song and Messing, 2003), although, in contrast with the bz1 locus, extensive local duplication of the zein genes appears to have occurred.
The biological implications of a lack of colinearity could be profound. Recombination rates highly increased within genes and reduced in retrotransposon clusters have been noted before in maize (Dooner, 1986; Fu et al., 2002). Obviously, nonshared sequences are excluded from recombination events. Fu and Dooner (2002) proposed that complementation of nonshared genes could be one of the factors contributing to heterosis, whereas Song and Messing (2003) identified unexpected differences in the expression of shared and nonshared genes in reciprocal hybrids. Therefore, analyzing the extent of genomic noncolinearities may help an understanding of recombinational properties and heterosis in maize.
Large contiguous maize sequences have revealed a gene-island structure of 10 to 20 kb (three to four genes), interspersed with long stretches of repetitive DNA that makes up a significant portion of the genome (>80%) (Hake and Walbot, 1980; SanMiguel et al., 1996). The large size of the maize genome has been attributed to long terminal repeat (LTR)-retrotransposons, which occupy most of the nongenic space and are also present in many other plant species at high abundance levels (Flavell, 1992; Voytas et al., 1992; SanMiguel et al., 1996; Kumar and Bennetzen, 1999). In maize, LTR-retrotransposons account for >60% of the nuclear genome length (SanMiguel et al., 1998; Meyers et al., 2001; Messing et al., 2004). The retroelements are classified into numerous distinct families (Kumar and Bennetzen, 1999) and show a tendency to form nested insertions (SanMiguel et al., 1996, 1998; Wicker et al., 2003).
An approximate time for insertion of these retroelements can be estimated from the divergence of the LTR-sequences, which would be identical at the time of insertion of a particular copy of the element (SanMiguel et al., 1998, 2002; Ma and Bennetzen, 2004a). This approach can be used to follow the evolutionary history of specific genomic regions and to date the accumulation of nonshared LTR-retrotransposons in individuals of the species.
To understand better the phenomenon of noncolinearities in the maize genome, including their frequency and biological implications, we analyzed DNA sequences from several allelic genome segments, in the maize inbreds Mo17 and B73.
RESULTS
Comparison of DNA Sequences from the Maize Inbred Lines B73 and Mo17 at Four Genetic Loci Differing in the Extent of Sequence Colinearity
Allelic pairs of BAC contigs (two to three BAC clones each) were identified in the following three genomic regions from the maize inbreds B73 and Mo17 and were sequenced completely (Table 1). Locus9002 on maize chromosome 1L (bin 1.08; markers bz2, An1, and umc1446) is in a high divergence genomic region, as judged by fingerprint overlap (fpc-reported cutoff values between 1E-20 and 1E-40), locus9008 on 2S (bin 2.04; markers umc1326 and umc1448) is in a region of medium divergence (cutoff values between 1E-50 and 1E-70), and locus9009 on 7L (bin 7.04; markers rpot1 and umc1295) shows low intraspecific divergence between B73 and Mo17 (cutoff values between 1E-80 and 1E-100).
Table 1.
Characterization of the Allelic Loci and Their DNA Sequences from the Two Maize Inbred Lines B73 and Mo17
| Maximum Extent of the Shared Sequence (bp)
|
Annotated Genes
|
Repetitive Elements
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BAC Clones and GenBank Accession Numbers
|
Total Analyzed Sequence (bp)
|
Gene Density (kb/Gene)
|
Full-Length LTR-Retroelements
|
Non-LTR-Retroelements
|
||||||||
| Locus | Inbred Line | Total | Nonallelic | Allelic | Total | Allelic | Transposons | |||||
| Locus9002 | B73 | c0189B10 | 317,137 | 295,420 | 21 | 15 | 14 | 12 | – | 3 | ||
| chromosome 1L:bin 1.08 | c178B16 | |||||||||||
| Mo17 | b68b.c4 | 6 | 4 | |||||||||
| b106a.c21 | 366,120 | 338,565 | 6 | – | 56 | 19 | 1 | 3 | ||||
| b101a.c15 | ||||||||||||
| Locus9008 | B73 | c0064G09 | 339,089 | 330,373 | 9 | 2 | 37 | 21 | – | 1 | ||
| chromosome 2S:bin 2.04 | c0088M03 | |||||||||||
| Mo17 | b130b.k18 | 7 | 9 | |||||||||
| b129a.g23 | 282,600 | 255,143 | 7 | – | 36 | 15 | – | 1 | ||||
| b45a.g17 | ||||||||||||
| Locus9009 | B73 | c0187E18 | 323,584 | 323,584 | 17 | – | 19 | 15 | 2 | 3 | ||
| chromosome 7L:bin 7.04 | b0305F08 | |||||||||||
| Mo17 | b4c.h1 | 17 | 8 | |||||||||
| b53c.h1 | 405,672 | 308,206 | 23 | 6 | 13 | 11 | 1 | 3 | ||||
| be82d.f24 | ||||||||||||
| adh1 locus | B73 | AF123535 | 160,480 | 141,724 | 6 | – | 24 | 8 | 2 | – | ||
| chromosome 1L: bin 1.10 | 6 | 5 | ||||||||||
| Mo17 | b161.k19 | 126,039 | 118,523 | 6 | – | 20 | 6 | 2 | – | |||
| Total loci 9002, 9008, 9009, and adh1 | 2,320,721 | 2,111,538 | 95 | 23 | 36 | 22 | 107 | 26 | 8 | 14 | ||
| bz1 locus | McC | AF391808 | 226,001 | 114,076 | 13 | 4 | 9 | 4 | – | – | 1 | |
| chromosome 9S: bin 9.02 | B73 | AF448416 | 106,186 | 78,165 | 9 | – | 9 | 9 | 3 | – | – | |
| Mo17 | b103.c20 | 203,581 | 55,876 | 9 | – | 6 | 2 | 1 | – | – | ||
| Total all loci | 2,856,489 | 2,359,655 | 126 | 27 | 45 | 19 | 116 | 27 | 8 | 15 | ||
The total analyzed sequence segments per locus and the maximum extent of the shared sequence between the inbreds Mo17 and B73 are summarized in columns 4 and 5, respectively. The number of identified genes and their presence in the two inbreds, as well as the gene density, are summarized in columns 6 through 9. The numbers of repetitive elements are indicated in columns 10 through 13.
In addition, we identified and sequenced the Mo17 BAC clones b106.c20 and b161.k19, which are allelic to the published maize sequences of the bz1 locus (Fu and Dooner, 2002) (chromosome 9S, bin 9.02) and the adh1 locus (Tikhonov et al., 1999) (chromosome 1L, bin 1.10), respectively.
A total of 17 maize BAC clones were sequenced from both inbreds, generating >2.3 Mb of sequence (Table 1). Numerous DNA segments, which are not shared by the two inbreds, were identified.
Sequence Comparison of Locus9002
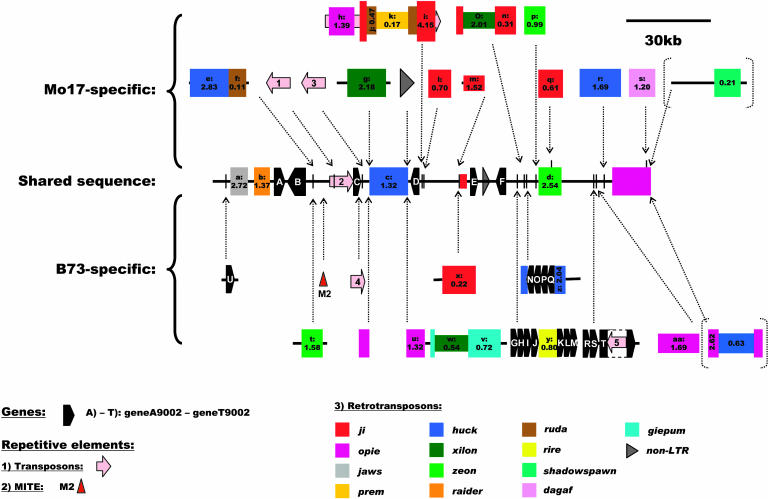
Locus9002 yielded the longest sequence for the intraspecific comparison. The sum of the sequences available for the comparison between the inbreds Mo17 and B73 is 634 kb (Table 1). We identified the second highest number of retroelements among the surveyed regions (Table 1; see Supplemental Table 1 online). Six genes (geneA9002 to geneF9002) are shared between the two inbred lines (Figure 1; see Supplemental Table 2 online). Up to 15 additional genes or genic fragments (geneG9002 to geneU9002) could only be found in inbred line B73 (Figure 1; see Supplemental Table 2 online). GeneG9002 and geneH9002 match a predicted and an expressed protein of rice, respectively, whereas geneI9002 is similar to an Arabidopsis thaliana protein kinase family member, geneJ9002 is homologous to a putative rice PRLI-interacting factor, geneK9002 matches another predicted rice gene, geneL9002 is similar to a putative phosphatidylinositol-phoshpatidylcholine transfer protein of rice, and geneM9002 is homologous to a putative rice AMP deaminase. The first four and the last three of these genes are clustered and all in the same orientation. The two clusters are separated from each other by the insertion of a complete rire LTR-retrotransposon, including a 5-bp target site duplication. This whole arrangement itself is inserted next to indy, a new type of LTR-retrotransposon (see below). Within this indy retroelement, four additional partial genes are present. All are clustered and in the same orientation: geneN9002 partially matches the 40S ribosomal protein S8 of maize, geneO9002 and geneP9002 have some homology to unknown proteins of rice, and geneQ9002 is homologous to a putative rice cytosolic monodehydroascorbate reductase. Another insertion of three nonshared genes has been found in the close vicinity. These three genes are also clustered and in the same orientation and show some homology over a part of the sequence to known genes: geneR9002 is similar to a putative rice hairpin inducing protein, geneS9002 is homologous to the rice origin recognition complex subunit 1, and geneT9002 partially matches a maize Lys-ketoglutarate reductase/saccharopine dehydrogenase bifunctional enzyme. Interestingly, transposon 5, which is present only in inbred B73 and shows homology to a DOPA-like transposon, has inserted into the second intron of geneT9002. Nonshared geneU9002 with homology to a rice hypothetical protein is inserted upstream of LTR-retrotransposon jaws (see below).
Figure 1.
Schematic Representation of the Sequence Relationship between Maize Inbreds B73 and Mo17 at Locus9002 on Chromosome 1L (Bin 1.08; Markers bz2, An1, and umc1446).
The shared sequence between the inbreds is represented by the bar labeled “Shared sequence.” The sequence segments unique to either B73 or Mo17 are represented as “Mo17-specific” and “B73-specific,” with their positions of insertion indicated by dotted arrows, relative to the shared sequence. LTR-retrotransposons are represented by colored boxes, which are also drawn in a different width if nested. Retrotransposon boxes contain labels, which are cross-referenced to the tables, and the insertion times in Myr based on LTR sequence divergence (Ma and Bennetzen, 2004a). Elements in dotted brackets were excluded from the intraspecific comparison. Predicted genes are indicated with black pentagons and transposons with pink arrows. Only nonshared and individually inserted MITE M2 is shown with a red triangle.
Thirteen different families of LTR-retrotransposons were identified in the sequences of locus9002 (Table 2). Retroelements ji, opie, huck, xilon, and zeon are most common. LTR-retrotransposons jaws, raider, and indy could not be assigned to a known retroelement family by BLAST analysis. Jaws shows some homology to a probable Arabidopsis gypsy/Ty3 polyprotein (G86474), and raider is similar to a putative copia-type polyprotein of rice (NP_916659.1), whereas indy matches a putative rice polyprotein (AAK55777).
Table 2.
The Classification of Shared and Nonshared LTR-Retrotransposons at the Loci 9002, 9008, 9009, adh1, and bz1
| LTR-Retrotransposon Family
|
Element Type
|
Locus9002
|
Locus9008
|
Locus9009
|
adh1 Locus
|
bz1 Locus
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B73 | Mo17 | Shared | B73 | Mo17 | Shared | B73 | Mo17 | Shared | B73 | Mo17 | Shared | McC | B73 | Mo17 | Shared | ||
| ji | copia | 1 | 5 | – | 9 | 4 | 3 | 7 | 5 | 3 | 1 | 1 | 1 | – | – | – | – |
| opie | copia | 2 | 1 | – | 3 | 4 | 2 | 5 | 4 | 4 | 3 | 2 | 2 | 2 | – | – | – |
| prem | copia | – | 1 | – | 2 | 1 | – | – | – | – | – | – | – | – | – | – | – |
| fourf | copia | – | – | – | – | – | – | – | – | – | 1 | 1 | 1 | – | – | – | – |
| huck | gypsy | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | – | 1 | – |
| grande | gypsy | – | – | – | – | – | – | 1 | – | – | – | – | – | – | 1 | 1 | 1 |
| zeon | gypsy | 2 | 2 | 1 | 1 | – | – | – | – | – | – | 1 | – | – | – | – | – |
| tekay | gypsy | – | – | – | 1 | 1 | 1 | – | – | – | – | – | – | – | 1 | – | – |
| cinful | gypsy | – | – | – | 1 | 1 | 1 | – | – | – | 1 | – | – | – | – | – | – |
| rire | gypsy | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| milt | gypsy | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – |
| shadowspawn | – | – | – | – | 1 | 2 | 1 | – | – | – | – | – | – | – | – | – | – |
| reina | gypsy | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| ruda | – | – | 2 | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – |
| xilon | – | 1 | 2 | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – |
| dagaf | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| giepum | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| reiver | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – |
| raider | copia | 1 | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – |
| flip | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – |
| jaws | gypsy | 1 | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Total | 12 | 19 | 4 | 21 | 15 | 9 | 15 | 11 | 8 | 8 | 6 | 5 | 4 | 3 | 2 | 1 | |
In total, locus9002 contains 27 different complete LTR-retrotransposons with flanking target site duplications (see Supplemental Table 1 online). Only four of them are shared between B73 and Mo17. Their times of insertion range from 1.32 up to 2.72 million years (Myr) (see Supplemental Table 3 online). By comparison, the times of insertion of the 23 nonshared LTR-retrotransposons varies from 0.11 to 4.15 Myr, but 11 of them are younger than 1 Myr (Figure 1; see Supplemental Table 4 online). In one case, jix3 (labeled with m in Figure 1), the retroelement is complete in Mo17 but remains as a solo-LTR in B73, flanked by target-site duplications. Nested retroelements are common, as previously described (SanMiguel et al., 1998), and their insertion times are consistently more recent than those of the host element. The nested inserted retroelement xilonx1 (labeled with o in Figure 1) violates this rule because the inserted element is older (2.01 Myr) than the host element (0.31 Myr). This can be explained by major rearrangements, such as insertions and deletions, in the xilonx1 LTRs that prevent proper alignment of the two LTR sequences. In several cases, for example, zeonx2 and jix6 (labeled with t and x, respectively, in Figure 1), the retroelement insertions are flanked by additional segments of nonshared DNA. Neither any known gene nor any known repetitive element could be assigned to these sequences, which might represent remnants of retrotransposons that have decayed to a point where they cannot be recognized easily.
Both inbred lines share transposon 2, which is located upstream of geneC9002 and shows nucleotide similarity with transposons 1 and 3 (87.7 and 84.6%, respectively) (Figure 1). These two transposons are orientated in the same direction and have inserted into transposon 2 downstream of the shared geneC9002 in Mo17 (Figure 1). Transposon 1 was classified as a CACTA transposon because of the CACTA motif at the end of the inverted repeat. No such motif was found in the inverted repeat of transposons 2 and 3. The inverted repeats of transposons 1 and 3 are less similar than their internal parts that show ∼94% homology on the nucleotide level. Furthermore, transposon 4, which is located downstream of geneC9002 and only present in B73, was identified because of homology to an En/Spm-like transposon protein from Arabidopsis (Figure 1).
Sequence Comparison of Locus9008
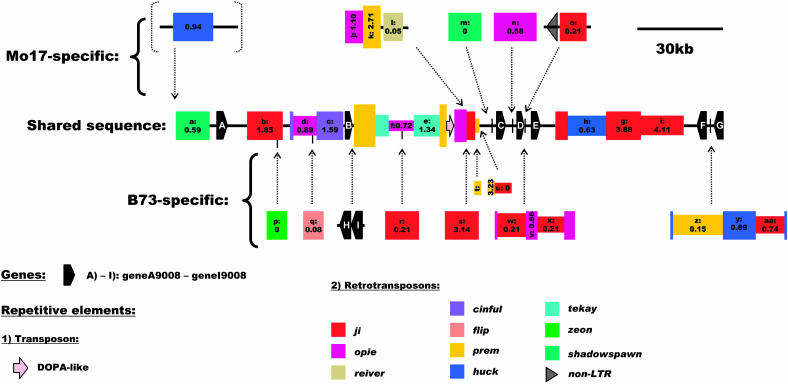
Locus9008 yielded a slightly smaller sequence for the intraspecific comparison than the other two loci. The sum of the sequence available for comparison between the inbreds Mo17 and B73 is ∼586 kb (Table 1). Seven genes (geneA9008 to geneG9008) are shared between Mo17 and B73 in this region (Figure 2; see Supplemental Table 2 online). Unlike in locus 9002, nongenic regions represent most of the differences here. Two genes, geneH9008 and geneI9008, clustered in the same orientation upstream of the prem1y1 retrotransposon (labeled with k in Figure 2) in inbred B73, are not found in Mo17 (Figure 2). GeneH9008 is similar to the rice MADS31 transcription factor, and geneI9008 shows homology to a putative rice phosphoinositide phosphatase (see Supplemental Table 2 online).
Figure 2.
Schematic Representation of the Sequence Relationship between Maize Inbreds B73 and Mo17 at Locus9008 on Chromosome 2S (Bin 2.04; Markers umc1326 and umc1448).
The shared sequence between the inbreds is represented by the bar labeled “Shared sequence.” The sequence segments unique to either B73 or Mo17 are represented as “Mo17-specific” and “B73-specific,” with their positions of insertion indicated by dotted arrows, relative to the shared sequence. LTR-retrotransposons are represented by colored boxes, which are also drawn in a different width if nested. Retrotransposon boxes contain labels, which are cross-referenced to the tables, and the insertion times in Myr based on LTR sequence divergence (Ma and Bennetzen, 2004a). Elements in dotted brackets were excluded from the intraspecific comparison. Predicted genes are indicated with black pentagons, transposons with pink arrows, and non-LTR-retrotransposons with black triangles.
Locus9008 has 27 complete LTR-retroelements with target-site duplications (see Supplemental Table 1 online). These elements are categorized into 10 different families, whereof the copia-like ji and opie families represent more than half of the total (Table 2). Two nonshared retroelements (reiver and flip), with no homology to any described retroelement family, were found. Nine complete LTR-retrotransposons are shared between B73 and Mo17 (Figure 2). Their insertion times range from 0.59 up to 4.11 Myr (see Supplemental Table 3 online). In addition, four other retrotransposons, opiey1, prem1y1, jiy3, and prem1y2 (labeled with j, k, s, and t in Figure 2), are only partially shared and must have been involved in deletion events in one of the inbreds. All other full-length LTR-retroelements are nonshared and have inserted more recently (<0.9 Myr) (Figure 2; see Supplemental Table 4 online). Two inserted retroelements, reiver and jiy1 (labeled with l and o in Figure 2), are flanked by additional segments of nonshared DNA of unknown origin.
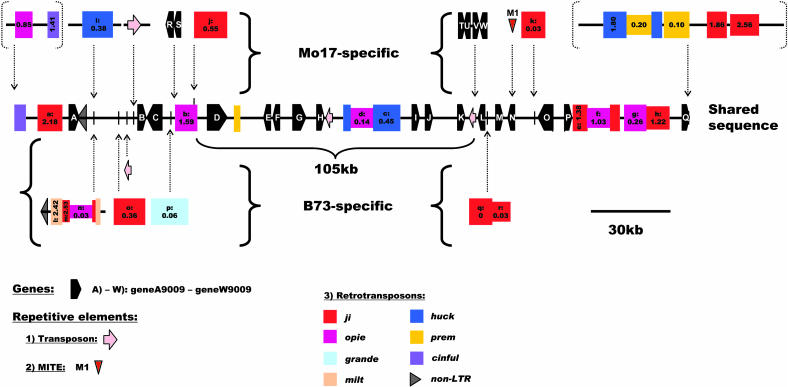
Sequence Comparison of Locus9009
Locus9009 yielded the second largest sequence for the intraspecific comparison. The sum of the sequence available for comparison between the inbreds Mo17 and B73 is ∼631 kb. This region is relatively gene rich (Table 1). Seventeen annotated genes (geneA9009 to geneQ9009) are shared between the two inbreds (Figure 3; see Supplemental Table 2 online). Most of them have a nearly full-length match to a known or predicted gene.
Figure 3.
Schematic Representation of the Sequence Relationship between Maize Inbreds B73 and Mo17 at Locus9009 on Chromosome 7L (Bin 7.04; Markers rpot1 and umc1295).
The sequence segments unique to either B73 or Mo17 are represented as “Mo17-specific” and “B73-specific,” with their positions of insertion indicated by dotted arrows, relative to the shared sequence. LTR-retrotransposons are represented by colored boxes, which are also drawn in a different width if nested. Retrotransposon boxes contain labels, which are cross-referenced to the tables, and the insertion times in Myr based on LTR sequence divergence (Ma and Bennetzen, 2004a). Elements in dotted brackets were excluded from the intraspecific comparison. Predicted genes are indicated with black pentagons, transposons with pink arrows, and non-LTR-retrotransposons with black triangles. Only nonshared and individually inserted MITE M1 is shown with a red triangle.
Six nonshared genes (geneR9009 to geneW9009), with homology over a part of the sequence to known genes, were found in inbred Mo17 (Figure 3; see Supplemental Table 2 online). GeneR9009 and geneS9009 are clustered in the same direction close to retroelement opie9009b (labeled with b in Figure 3). They show homology to the putative rice proteins MAP3K epsilon kinase and splicing factor 3, respectively. GeneT9009, geneU9002, geneV9002, and geneW9009, which are also clustered, are located downstream of geneL9009. They show homology to a rice cell division inhibitor MinD homolog (geneT9009), to a putative rice rRNA processing protein (geneW9009), and to hypothetical and predicted rice proteins (geneU9009 and geneV9009). GeneU9009 and geneW9009 are in opposite direction compared with the other two genes in the cluster.
The 18 complete LTR-retrotransposons with target site duplications at locus9009 were classified in five known families, with ji, opie, and huck being the most common ones (Table 2; see Supplemental Table 1 online). Eight LTR-retrotransposons are shared (Figure 3). The insertion times of five of these shared elements are estimated to be in excess of 1 Myr (see Supplemental Table 3 online).Ten LTR-retrotransposons are found to be nonshared (Figure 3). Eight of them showed insertion time points of 0.55 Myr or less (see Supplemental Table 4 online). The two other retroelements (miltz1 and jiz3, labeled with l and m in Figure 3) are much older and show nested insertion. It is therefore likely that these two elements were not present in the ancestor of Mo17 that contributed that particular region of chromosome 7. The inserted retroelements huckz1 and miltz1 (labeled with i and l in Figure 3) are flanked by nonshared DNA of unknown origin. A block of shared sequence of ∼105 kb, containing eight genes (geneD9009 to geneK9009), uninterrupted by any allelic noncolinearity, is located in the middle of the analyzed genomic segment (Figure 3). Several inbred-specific retroelements as well as the Mo17-specific genes R9009 to W9009 are in the immediate vicinity.
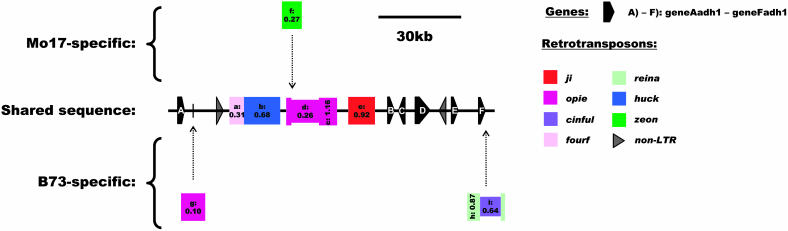
Sequence Comparison of the adh1 Locus
Seven genes are shared at the adh1 locus and therefore make it relatively gene rich (≈18.5 kb/gene) (Figure 4, Table 1). The only differences between inbreds are attributable to four complete nonshared LTR-retrotransposons (one in Mo17 and three in B73), whereof two are nested (Figure 4). These nonshared elements make up 21% of the analyzed sequence (Figure 5; see Supplemental Table 5 online). Copia-type retrotransposons are the major part of the repetitive fraction (Table 2). All LTR-retrotransposons at the adh1 locus are of recent origin (<1.16 Myr) (see Supplemental Tables 3 and 4 online).
Figure 4.
Schematic Representation of the Sequence Relationship between Maize Inbreds B73 and Mo17 at the adh1 Locus on Chromosome 7L (Bin 7.04; Markers rpot1 and umc1295).
The sequence segments unique to either B73 or Mo17 are represented as “Mo17-specific” and “B73-specific,” with their positions of insertion indicated by dotted arrows, relative to the shared sequence. LTR-retrotransposons are represented by colored boxes, which are also drawn in a different width if nested. Retrotransposon boxes contain labels, which are cross-referenced to the tables, and the insertion times in Myr based on LTR sequence divergence (Ma and Bennetzen, 2004a). Elements in dotted brackets were excluded from the intraspecific comparison. Predicted genes are indicated with black pentagons and non-LTR-retrotransposons with black triangles.
Figure 5.
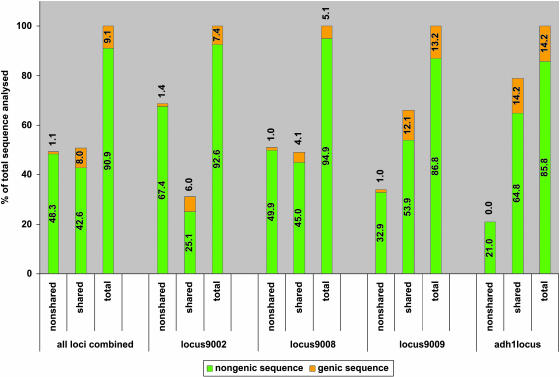
Diagram Showing the Fraction (%) of Genic and Nongenic Sequences in the Shared, Nonshared, and Total Sequence at Loci 9002, 9008, 9009, and adh1 (see Supplemental Table 5 Online for Complete Data Set).
Miniature Inverted Terminal Repeat Analysis at Loci 9002, 9008, and 9009
About 125 miniature inverted terminal repeats (MITEs) have been identified at loci 9002, 9008, and 9009 (see Supplemental Table 6 online). Stowaway elements are more common than Tourist elements (77% versus 23%). The number of shared versus nonshared elements is locus specific, but on average >50% of the MITEs are nonshared. This is comparable to the amount of shared versus nonshared sequences (49% versus 51%). Stowaway elements tend to be proportionally more numerous than Tourist elements in the shared compared with the nonshared sequence fraction (shared, 83% versus 17%; nonshared, 72% versus 28%). Six MITEs (5%) have been found to have integrated into an intron of a gene, which in all cases are shared genes. Interestingly, one such insertion is found only in Mo17 (MITE M1 in Figure 3). Only one additional nonshared MITE (MITE M2 in Figure 1) was identified that has also integrated as a single insertion event. All other inbred-specific MITEs are nested in nonshared sequences. Half of the MITEs have inserted into unknown sequences, whereas the other half integrated into repetitive elements, 13 in transposons and 50 in LTR-retrotransposons. Thirty-four of the later ones are present in nonshared LTR-retrotransposons, whereas the remaining 16 are in shared retroelements (68% versus 32%). This insertion pattern is highly correlated with the number of nonshared versus shared LTR-retrotransposons (71% versus 29%) observed at these loci. MITEs have inserted mostly into the internal part of LTR-retrotransposons, but LTRs were also the target of 13 MITE insertions. In 12 cases, the corresponding MITE is present in both LTRs. Five of the 13 are shared LTR-retrotransposons (times of insertion >3.8 Myr) and seven are nonshared retroelements (five inserted within the last 1.2 Myr). In a single event, the nonshared element jiz19009 (insertion time 0.55 Myr) in Mo17, the insertion is present in only one LTR. Therefore, this MITE insertion must have occurred after the insertion of the LTR-retrotransposon, whereas in all other cases the MITE must have been present in the corresponding retroelement before its insertion at the analyzed loci.
Nonshared Genes Break the Colinearity with Rice
The rice orthologs for all the shared and nonshared genes at loci 9002, 9008, and 9009 were identified in the rice genome by TBLASTN analysis. Rice orthologs of 23 of the 30 (77%) shared maize genes have been assigned to the colinear rice regions (Figure 6; see Supplemental Table 2 online). Exceptions are geneC9008 with no identified rice ortholog and geneD9002 and genes D, F, H, L, and N from locus9009, whose rice orthologs are all located on different rice chromosomes (see Supplemental Table 2 online). The intergenic distance between colinear genes is enlarged in maize compared with rice. Colinear regions in locus9002 and locus9009 are moderately larger in the two maize inbreds than in rice (more than two times the rice sequence), compared with locus9008, which is up to six times enlarged in maize than in rice (Figure 6). Several rice orthologs were identified for most of the nonshared genes, but none of them mapped to the colinear chromosomal region in rice (Figure 6; see Supplemental Table 2 online). Their independent insertion in maize or deletion from rice might explain this interspecific difference.
Figure 6.
Rice–Maize Colinearity of the Analyzed Maize Genes at Locus9002, Locus9008, and Locus9009.
(A) Locus9002.
(B) Locus9008.
(C) Locus9009.
The physical distance between conserved genes is indicated for both species with brackets. Genes that are present in rice but absent in one of the maize inbreds are likely to be deletions in the other inbred. Genes absent in rice but present in one of the maize inbreds are likely to be recent insertions in maize. Genes absent in rice but present in both maize inbreds may have been deleted in the rice lineage or inserted in the common ancestor of B73 and Mo17. cen., centromere; tel., telomere.
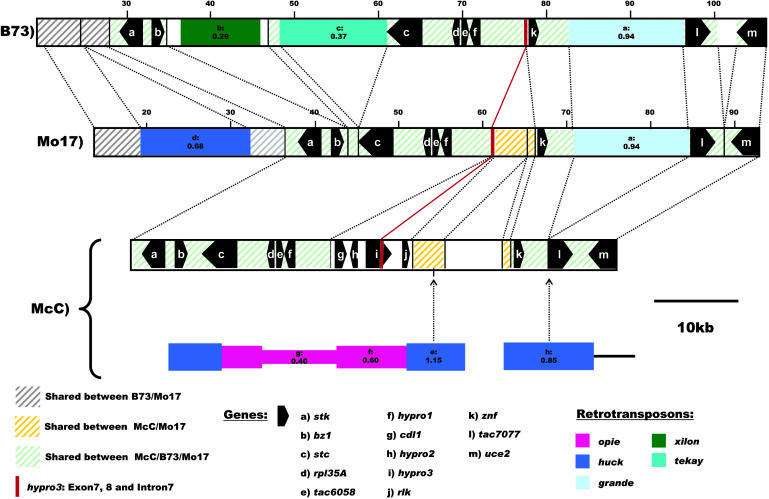
Comparative Sequence Analysis of the bz1 Locus in Maize Inbred Lines B73, McC, and Mo17
The DNA sequence at the bz1 locus on chromosome 9S is available from two maize inbred lines, McC and B73 (Fu and Dooner, 2002). We have sequenced ∼79 kb of the Mo17 allele present on BAC clone b106.c20. The Mo17 sequence is different from both previously sequenced alleles. It shares sequence similarity with B73 in all regions annotated as genes and also lacks entirely three of the four genes, which were reported as missing in B73 compared with McC (Fu and Dooner, 2002): cdl1, hypro2, and rlk (Figure 7). Predicted exons 7 and 8 and intron 7 of the fourth gene hypro3 are however present in all three lines (Figure 7). hypro3, like hypro1 and hypro2, is a predicted gene for which no experimental evidence exists to precisely define its structure. A grande LTR-retrotransposon at position 82 to 95 kb in B73 (Figure 7) is shared between B73 and Mo17. Its insertion was dated to ∼0.94 ± 0.24 Myr in both inbreds (see Supplemental Table 3 online). The time of insertion of all other nonshared retroelements range from 1.15 ± 0.20 to 0.29 ± 0.07 Myr (see Supplemental Table 4 online).
Figure 7.
Schematic Representation of the Sequence Relationship among Maize Inbreds B73, Mo17, and McC at the bz1 Locus on Chromosome 9S (Bin 9.02) (Modified from Fu and Dooner, 2002).
Dashed green lines indicate the shared sequence between all three inbred lines. Dashed gray and orange lines indicate the shared sequence regions between the inbreds B73 and Mo17 and between Mo17 and McC, respectively. Inserted and deleted regions are connected via dashed lines. The LTR-retrotransposons unique to McC or Mo17 and/or B73 are represented by different colored boxes, which are also drawn in a different width if nested, with their positions of insertion indicated by dotted arrows. Retrotransposon boxes contain labels, which are cross-referenced to the tables, and the insertion times in Myr based on LTR sequence divergence (Ma and Bennetzen, 2004a). Genes are represented as black pentagons. The location of exons 7 and 8 and intron 7 of gene hypro3 is labeled with a red line.
The region between the genes stk and uce2 is largely shared between B73 and Mo17, with the only three gross rearrangements: a fragment containing the internal gag/pol domain of a Hopscotch LTR-retroelement that is present in Mo17 but absent from B73 upstream of the znf gene and two retroelements that are found in B73 between the genes bz1 and stc. The Hopscotch-containing fragment is also found in McC. Based on the calculated time of insertion of the LTR-retroelements and assuming all events involving them represent novel insertions rather than deletions, as suggested by the presence of target site duplications for each of them, we estimate that the McC-specific sequences diverged from the common ancestor at least 1.15 ± 0.20 Myr ago and that the bz1 sequences represented by B73 and Mo17 diverged between 0.94 ± 0.24 and 0.37 ± 0.07 Myr ago (see Supplemental Tables 3 and 4 online). If we also consider the fragment preceding gene stk, then the interval for the divergence of the B73- and Mo17-specific sequences is narrowed down to 0.94 ± 0.24 to 0.68 ± 0.13 Myr ago.
Sequence divergence data comparing the exons (synonymous substitutions) and introns (all substitutions but no indels) of each shared gene among the three haplotypes reveal a closer relation between B73 and Mo17 only for genes rpl35A, tac6058, and hypro1 (see Supplemental Figure 1 and Table 7 online). In all other genes, McC is either closer to Mo17 (genes bz1, stc tac7077, and uce2) or to B73 (genes stk and znf). In general, divergence estimates obtained from genes do not agree well with the divergence times we estimated from the retroelement insertions. No rice/maize colinearity was observed for the 13 genes at the maize bz1 locus because the orthologs identified by TBLASTN against the complete genomic sequence of rice are present at various locations on different rice chromosomes (see Supplemental Table 8 online).
Nonshared Sequences Are Gene-Poor and Consist of Clusters of Truncated Genes and Recently Inserted or Incomplete Repetitive Sequences
The sequencing of loci 9002, 9008, 9009, and adh1 resulted in >2.1 Mb of sequence for comparative analysis (Table 1). The sequence shared between the two inbreds at each locus was counted only once in the calculation of the ratios described in Figure 5 and Supplemental Table 5 online. The genic fraction is ∼9.1% when averaged over the four loci (Figure 5; see Supplemental Table 5 online). The gene density within these four regions ranges from 13 to 56 kb/gene (average of 22 kb/gene) (Table 1), which extrapolates to an estimate of ∼113.000 genes for the whole maize genome. This high estimate may be explained by a bias toward gene-rich regions because of the selection of segments with a high density of overgo probes, a selection that is necessary to establish allelic relationships. The majority of the sequence space is made up by retroelements or noncharacterized sequences (Figure 5; see Supplemental Table 5 online). Almost half of the total sequence analyzed is nonshared, but there are large differences in the amount of inbred-specific sequences among the loci (Figure 5; see Supplemental Table 5 online). The nonshared fraction makes only one-fifth of the adh1 locus and one-third of the gene-rich and highly homologous locus9009, but makes up half of locus9008 and more than two-thirds of locus9002. Thus, the sequence composition data confirm the initial high information content fingerprinting (HICF) fingerprinting data.
The nonshared sequences are, on average, more than sevenfold lower in gene content than the shared sequences (loci 9002, 9008, 9009, and adh1: 2.1% versus 15.8%), but this is locus dependent (Figure 5; see Supplemental Table 5 online). In total, 59 genes have been identified at the four loci of which more than one-third (23) are nonshared (Table 1). Nonshared genes are truncated, and the homology of the translated protein products is limited only to N-terminal, C-terminal, or central portions of protein entries from GenBank (see Supplemental Table 2 online). Furthermore, nonshared gene PCR products were amplified from both inbred lines in high-stringency conditions, suggesting that they also may be present elsewhere in the inbred that lacks the sequences in the particular region being studied. However, it is not possible to ascertain if those are, in fact, the closest homologs without further experimentation.
Interestingly, 26 of the 27 nonshared genes (including also the four nonshared genes at the bz1 locus; Fu and Dooner, 2002) are present in seven clusters of 1.8 to 7.6 kb. Statistical tests of the distribution of distances between shared versus nonshared genes identified a denser gene arrangement for nonshared genes (Kolmogorov-Smirnov test, P = 0.001, D = 0.5093; permutation test, P = 8.8 × 10−4) (see Supplemental Figure 2 online). Surprisingly, most of the nonshared genes have the same orientation within clusters. Homologous maize ESTs (cutoff score value of 120) and/or expressed maize massively parallel signature sequencing (MPSS) tags (cutoff value of 2 ppm) were identified for 89% of the nonshared compared with 96% of the shared genes (see Supplemental Table 9 online). Three nonshared genes identified neither any maize ESTs nor any expressed maize MPSS tag. ESTs matching nonshared genes showed homology only over a part of the sequence. Therefore, these ESTs represent, rather, transcripts from expressed homologs than from nonshared genes.
At least 97% of the inbred-specific fraction is composed of repetitive or noncharacterized elements (see Supplemental Table 5 online). We identified 62 nonshared LTR-retrotransposons, including also those from the bz1 locus (Table 2). There are 33 copia-type retroelements, represented by 19 ji, 10 opie, and four prem elements. Twenty-nine elements are of gypsy or unknown type and belong to a variety of families, none of which are present in more than three copies.
No particular repetitive element is unique to the shared fraction, but copia types are more numerous than gypsy ones (17 and 10, respectively), with seven ji and eight opie elements (Table 2). Contingency χ2 analysis did not detect any significant difference in the distribution of copia versus gypsy elements among shared and nonshared elements (P = 0.90) nor of the three most abundant families (ji, opie, and huck) (P = 0.43).
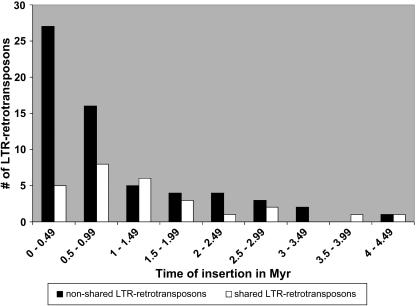
Forty-three of the nonshared LTR-retroelements have inserted within the last 1 Myr, and 27 of these are younger than 0.5 Myr (Figure 8; see Supplemental Table 4 online). The majority (>75%) of the shared LTR-retroelements have inserted within the last 2 Myr (Figure 8; see Supplemental Table 3 online). The distribution of insertion time points of the shared and nonshared LTR-retrotransposons is different (Kolmogorov-Smirnov test, P = 0.027, D = 0.328; nonshared retroelements, mean = 0.91, median = 0.61; shared retroelements, mean = 1.34, median = 1.16). Thus, nonshared retrotransposons are significantly more recent than the shared ones.
Figure 8.
Distribution of the Retrotransposon Insertion Times Calculated from the LTR Sequence Divergence (Ma and Bennetzen, 2004a).
Open bars show the distribution of shared LTR-retrotransposons, and closed bars represent the distribution of nonshared LTR-retrotransposons.
DISCUSSION
Maize Alleles Contain Large Regions of Nonhomology Mainly Consisting of Repetitive Elements
The comparison of sequences from two different inbreds at the bz1 (Fu and Dooner, 2002) and the z1C-1 (Song and Messing, 2003) loci revealed large stretches of nonshared sequences. In both cases, the presence of additional genes in one inbred over the other and of large differences in the compositions of the intergenic regions because of repetitive sequences accounted for the observed noncolinearities. To explore the generality of the violations to sequence microcolinearity among alleles within maize, we characterized several arbitrarily chosen loci in the public B73 and the DuPont Mo17 maize physical contig maps (Plant Genome Initiative at Rutgers, http://pgir.rutgers.edu/, http://www.genome.arizona.edu/fpc/maize; B. Li, personal communication). We included a Mo17 BAC from the bz1 region previously sequenced from B73 and McC (Fu and Dooner, 2002) and a Mo17 BAC from the adh1 locus previously sequenced from B73 (Tikhonov et al., 1999). The existence of two alleles, dramatically different in their DNA sequences, is a common feature of all loci examined. Even at adh1, where the Mo17 and B73 alleles are closely related, four nonshared retroelements and a relatively low level of SNPs and small indels were found, comprising 21% of the sequence (Figure 5; see Supplemental Table 5 online). This is in contrast with the loci 9002, 9008, and 9009, which have, on average, 50% of nonhomologous DNA (68.8, 50.9, and 33.9%, respectively; Figure 5; see Supplemental Table 5 online).
Most of the nonshared sequences consist of LTR-retrotransposons (Table 2) and other mobile elements. The majority of the identified LTR-retrotransposons belong to the ji, opie, and huck types, which have been reported to be the three most abundant retroelements in the maize genome (Meyers et al., 2001).
The differences in LTR-retrotransposon content between lines could have arisen by retrotransposition, leading to insertions, or by recombinational events that would lead to deletions (Devos et al., 2002). Homologous recombination events would mainly produce solo LTRs, whereas nonhomologous events would produce incomplete elements. Homologous unequal and nonhomologous illegitimate recombination events that counteract genome expansion caused by retroelement insertions have been reported in Arabidopsis (Devos et al., 2002), rice (Bennetzen et al., 2005; Ma et al., 2004), and other plant species (SanMiguel et al., 1996; Shirasu et al., 2000; Wicker et al., 2003). The differences that we observed between lines usually encompass entire elements and carry target site duplications and thus appear to be attributable to insertions rather than to deletions. Deletions because of recombinational events should be more likely in older elements than in younger ones, as has been observed in Arabidopsis and rice (Devos et al., 2002; Vitte and Panaud, 2003; Ma and Bennetzen, 2004b; Ma et al., 2004). We observed only one product of a putative homologous recombination event, which affected an older element (jix3 at locus9002).
Most variation in plant genome size is caused by differences in the amounts of repetitive DNA (SanMiguel et al., 1996; Tikhonov et al., 1999; Vicient et al., 2001; Wicker et al., 2001; Ma et al., 2004). Seventy-four percent of sequence differences between sorghum (Sorghum bicolor) and maize are estimated to be a result of the accumulation of retrotransposons since their divergence (Tikhonov et al., 1999). The large majority of the nonshared sequence in maize is also either repetitive or consists of uncharacterized nongenic sequences. Thus, repetitive sequences also accumulate within individuals of the same species, generating large nonshared sequence differences. The amount of nonhomology between some maize alleles is similar to that reported between maize and other grass species (Tikhonov et al., 1999; Bennetzen and Ma, 2003).
Our analysis of MITEs suggests that they preferentially insert into repetitive sequences. Other data indicate that MITEs are common both in the repetitive sequences (Cheng and Lin, 2004) and in the noncoding regions of grass genes (Bureau and Wessler, 1992, 1994a, 1994b; Bureau et al., 1996). In 12 cases, MITEs are present in both LTRs of a retroelement, which means that the MITE was present before the replication of the host element. A single case of a MITE insertion into only one LTR of a nonshared retroelement of recent origin suggests more recent MITE activity in maize, as was observed in rice (Jiang et al., 2003; Kikuchi et al., 2003; Nakazaki et al., 2003). Because only two nonshared MITEs (M1 and M2) are individual insertion events, and not part of a larger nonshared segment, their direct contribution to the evolution of the nonshared fraction is small.
Nonshared Genes in Maize Alleles
A total of 23 putative genes was identified in the nonshared fraction at loci 9002, 9008, 9009, and adh1 (Table 1; see Supplemental Table 2 online). In contrast with the z1C-1 locus (Song and Messing, 2003), where half of the nonshared sequence is genic because of segmental duplications affecting the number of zein gene copies in each haplotype, we did not find any gene duplication associated with the nonhomologous sequences, but a much lower gene density in nonshared segments than in shared ones. The loci we analyzed are therefore closer to the bz1 locus mode of evolution, where four genes make up just 5% of the nonshared sequence (Fu and Dooner, 2002).
In most cases, the nonshared genes are clustered and oriented in the same direction within the clusters. The clustering of nonshared, relative to shared, genes is highly statistically significant. They are all truncated, and their homology to known genes or ESTs is only over a part of the sequence, suggesting that they may be pseudogenes. Such a pattern was observed earlier in maize (Meyers et al., 2001; Ramakrishna et al., 2002) and in an intraspecific comparison in rice (Feng et al., 2002; Han and Xue, 2003) and in other plants (Parniske et al., 1997; Noel et al., 1999; Holub, 2001), where genes were postulated to have arisen from multiple illegitimate and complex break repair events or from retroelements or nonfunctional hypothetical genes. These observations together suggest that clustering may be common among nonshared genes in maize.
It has been postulated that novel genes arise after a gene or genome duplication (Lewis, 1951). As a result of an ancient tetraploidization event (Gaut and Doebley, 1997; Langham et al., 2004), the maize genome contains duplicated chromosomal segments with colinear gene arrangements (Gaut, 2001; Ilic et al., 2003; Lai et al., 2004). Some of the differences between inbred lines may be attributable to the loss of genes in homeologous regions in one inbred lineage that were retained in the other (Ilic et al., 2003; Lai et al., 2004). A detailed analysis of larger homeologous segments could clarify this hypothesis. Fu and Dooner (2002) showed that the genes not shared by McC and B73 are present elsewhere in the maize genome and suggested that they may have arisen by deletion. However, no deletion mechanism, such as intrachromosomal recombination between the 5′ and 3′ LTRs of neighboring LTR-retrotransposons or unequal crossing over between related retrotransposon sequences, was described. The available evidence does not support a deletion hypothesis, even for the genes at the bz1 locus. Preliminary PCR results suggest that all genes, which are not shared between Mo17 and B73 at the investigated loci, are polymorphic and are present elsewhere in the maize genome. Assuming that rice is a representation of the ancestral condition, the consistent lack of colinearity of the nonshared genes and colinearity of the shared ones is best explained by insertion events that occurred in maize after its divergence from rice. This is also supported by the finding that the nonshared genes are incomplete because two successive deletion events would otherwise need to be invoked: the first one partially deleting the gene and the second one erasing the remains of it in one lineage.
Because nonshared genes preserve a normal intron–exon structure, it is unlikely that they are integrated processed pseudogenes. Insertions of multiple nonshared genes can be explained by the activity of retroelements or transposons. This kind of gene trafficking across the genome is well documented in vertebrates and to some extent in plants (Talbert and Chandler, 1988; Bureau et al., 1994; Jin and Bennetzen, 1994; Palmgren, 1994; Martinez-Izquierdo et al., 1997; Le et al., 2000; Pickeral et al., 2000; Elrouby and Bureau, 2001). Interestingly, the four nonshared genes N, O, P, and Q at locus9002 are located within LTR-retrotransposon huckx39002. It is unknown if these four nonshared genes were already part of the retroelement before its insertion or if they have inserted later. The recently uncovered large number of Pack-MULEs, carrying fragments of single or multiple cellular genes, represents a new mechanism for the evolution of genes in higher plants (Jiang et al., 2004). Our data do not indicate the involvement of Pack-MULEs in the insertion of nonshared genes in maize because no Mutator-like sequences were found flanking the nonshared gene clusters.
Nonshared genes are incomplete, and it is unknown if they encompass a promoter, but their expression could still be induced and modulated by promoter elements from neighboring repetitive elements (Kumar and Bennetzen, 1999; Speek, 2001; Vicient et al., 2001; Dunn et al., 2003; Kashkush et al., 2003; Schramke and Allshire, 2003). Although nonshared genes showed homology to many maize ESTs, the similarity was always restricted to a short segment of the EST sequence, implying that the EST was derived from a different, functional, and presumably full-length copy of the gene. The identified maize MPSS tags did not help to distinguish between the expression of nonshared genes and their homologs. The hypothesis that adjacent genic insertions may give rise to novel gene products (Lander et al., 2001; Jiang et al., 2004) could not be confirmed because no EST derived from a transcript across clustered nonshared genes was present in any database.
Analysis of the Three Alleles at the bz1 Locus
Three allelic sequences around the bz1 locus are available: B73, McC (Fu and Dooner, 2002), and Mo17, reported here. As suggested earlier on the basis of DNA gel blot evidence, the three alleles are distinct (Fu and Dooner, 2002). Inferences on the relationship among the three sequences are complicated by the fact that different segments within the same region may evolve differently. The shared genic regions may be differently affected by recombination than nonshared intergenic regions, even assuming mutational rate homogeneity throughout the region. The Mo17 and B73 haplotypes share the grande LTR-retrotransposon and lack the same four genes (cdl1, hypro2, hypro3, and rlk), but they differ by the presence of an internal portion of a Hopscotch element, shared between Mo17 and McC. The divergence of the sequences specific to each of the three haplotypes might be inferred from the LTRs of retrotransposons and from the sequence diversity between alleles of the nine shared genes. Divergence estimates obtained from genes do not agree well with those from the retroelement insertions. This may be explained by differences in recombination rates between retrotransposons and genic sequences. Randomization of genic sequences by recombination could be involved, if recombination within the region is largely restricted to shared genes and intergenic regions. The dating of the retrotransposon insertions relies on a fast molecular clock (Ma and Bennetzen, 2004a). Although the molecular clock used to date the insertion of LTR-retrotransposons is at least twofold faster than synonymous base substitutions within grass genes (Gaut et al., 1996; SanMiguel et al., 1998), the actual molecular drift rate may be even higher (Ma and Bennetzen, 2004a). The nonshared sequences are common in the vicinity of bz1 and are expected to be associated with a reduced recombination rate in inverse proportion to their allelic frequency. By contrast, the recombination rate of shared genes may not be suppressed. For example, bz1 has the highest intragenic recombination rate of any maize gene measured to date (Dooner, 1986). A high recombination rate has also been reported for the genes on the distal side of bz1, whereas it is reduced in retrotransposon clusters, even if they are shared (Fu et al., 2002).
Fu and Dooner (2002) suggested on the basis of hybridization data with rlk-, tac7077-, and bz1-specific probes that the rlk gene is also present at the bz1 locus in Mo17. However, we were unable to identify the rlk gene on the corresponding genomic sequence of Mo17.
Origin of Allelic Nonhomologies
The frequent persistence of major allelic nonhomologies in maize indicates that new allelic variants either are of recent origin or are constantly created, or that balancing selection leads to the maintenance of variants, which would otherwise be fixed or eliminated (Aguilar et al., 2004), or that effective population size (Ne) is sufficiently large. The persistence time of a newly inserted sequence within an allele may be predicted by coalescent theory and is proportional to 2N, where N is the effective population size (Nordborg, 2001). The persistence times of the order of several millions of years are to be expected in maize (Eyre-Walker et al., 1998; Remington et al., 2001; Vigouroux et al., 2002).
By contrast, in genomes lacking major allelic nonhomologies, retroelement insertions have either occurred in the distant past or are occurring at low frequency so that they are very unlikely to be polymorphic in the population at any given time. Alternative hypotheses, such as hybridization between subspecies or populations, which have been subject to a long period of reproductive isolation, would also explain the presence of allelic nonhomologies. Recent phylogenetic data, which postulate a single maize domestication, suggest only modest evidence for increase of diversity by postdomestication gene flow from teosinte into maize (Matsuoka et al., 2002). Therefore, introgressions fail to explain the large amount of observed nonhomologies between the two maize inbreds.
Our data are consistent with the hypothesis of an expanding maize genome, primarily because of the large accumulation of LTR-retrotransposon, which is counteracted by a low frequency of predicted homologous recombination events between LTRs (SanMiguel et al., 1996, 1998; Meyers et al., 2001; Bennetzen, 2002; Messing et al., 2004), as well as with an interpopulation hybridization origin hypothesis, where the present allelic composition could arise from a cross between ancestors that have evolved separately for quite some time (during which time retrotransposon amplification occurred independently in the two lineages) before the cross. The nonshared LTR-retroelements contribute to maize genome expansion, and these elements are of more recent origin than the shared fraction. A few older (>3 Myr) nonshared LTR-retroelements may have originated from a deletion in one of the lineages. Insertion age distribution differences between the shared and nonshared retroelement sets were observed even though only two maize inbreds were sampled. Whereas the nonshared set is unambiguously identified even with two inbreds, the shared set may comprise elements that are absent in other maize inbreds. The observation of a statistically significant difference in the insertion age distribution, despite this uncertainty in assignment to the shared class, indicates that the B73 and Mo17 genomes represent well the elements that are either universally shared (fixed) or close to fixation.
In contrast with barley (Hordeum vulgare) retroelement BARE-1 (Vicient et al., 2001) and rice retroelement Tos17 (Yamazaki et al., 2001; Miyao et al., 2003), no actively transposing maize LTR-retroelements have been described. Retroelement-derived transcripts in maize correspond to low copy types and not to the high copy ones, such as ji, opie, and huck (Meyers et al., 2001). Because the nonshared repetitive fraction consists mainly of high copy number LTR-retroelements, these elements might have transposed more efficiently in a not too distant past.
Biological Implications of the Intraspecific Noncolinearity
Complementation of haplotypes carrying different nonshared genes could contribute to the phenomenon of heterosis (Fu and Dooner, 2002). Although nonshared genic sequences appear to be nonfunctional, they could act through mechanisms similar to transgene cosuppression, siRNA-mediated gene silencing of homologous sequences (Hamilton and Baulcombe, 1999; Hannon, 2002), or interactions with functional proteins forming multimers and causing distinct phenotypic effects (Tsuchisaka and Theologis, 2004).
We advance an alternative hypothesis for the role of nonshared sequences in heterosis, focusing on the differences in the repetitive fraction rather than in the genes. In several instances, conserved and active genes in the two inbreds are flanked by different DNA, for example, by nonconserved retrotransposons inserted nearby (geneB9002, geneC9002, geneD9002, and geneF9002, Figure 1; geneC9008, geneD9008, geneE9008, and geneF9009, Figure 2; geneB9009, geneC9009, geneD9009, geneL9009, and geneO9009, Figure 3). Such retroelements usually are inactive but can be induced by various stresses (Kuff and Lueders, 1988; Pouteau et al., 1991; Hirochika et al., 1996) and may affect the expression of neighboring genes by producing single, chimeric, or antisense transcripts or by acting as enhancers (Medstrand et al., 2001; Speek, 2001; Whitelaw and Martin, 2001; Llave et al., 2002; Nigumann et al., 2002; Dunn et al., 2003; Kashkush et al., 2003; Schramke and Allshire, 2003). It is therefore likely that different repetitive sequence environments affect tissue specificity or temporal regulation of expression. Such differences have been proposed to be the cause of heterotic complementation (Birchler et al., 2003; Song and Messing, 2003) and are comparable to allelic interactions proposed by the overdominance theory explaining hybrid vigor (Crow, 1948; Song and Messing, 2003). Furthermore, shared gene expression might also be altered by a different chromatin state related to the presence of nonshared repetitive sequences nearby (Mette et al., 2002; Plasterk, 2002; Dawe, 2003; Schramke and Allshire, 2003).
Noncolinear regions of the genome cannot engage in homologous recombination, except in a cross with an identical allele. Therefore, the distribution of crossover points will show strong dependence on the specific combination of alleles. Although shared retrotransposon clusters have a reduced recombination rate (Fu et al., 2002), nonshared retrotransposons may contribute significantly to the low recombination rate of the retrotransposon fraction in maize (Yao et al., 2002). Thus, a desired combination of alleles may not be achievable in certain crosses. Genetic-to-physical distance ratios will show extreme local differences between crosses, when examined in sufficient detail. Nonshared sequences will also affect map-based cloning projects, where nonshared genes cannot be cloned from certain BAC libraries.
The effective population size of sequences represented in a fraction of individuals of a population will be different from the value for a genome segment represented in all individuals. Different segments of the genome will therefore behave as if they belonged to different populations, with respect to rate of decay of linkage disequilibrium and other population-dependent parameters. Even if only a fraction of the nonshared genes have measurable biological effects, the implications for maize genetics, breeding, and for maize genome sequencing are enormous.
METHODS
Identification of Allelic BAC Contigs and Comparison of Fingerprints
BAC contigs that represent allelic segments were identified in the public B73 maize (Zea mays) physical map (Plant Genome Initiative at Rutgers, http://pgir.rutgers.edu/, http://www.genome.arizona.edu/fpc/maize) and the DuPont maize physical map of inbred line Mo17 (Fengler et al., 2000) based on shared overgo probes (Gardiner et al., 2004). A selection of B73 BAC clones from each contig were refingerprinted on an ABI377 DNA sequencer (PE-Applied Biosystems, Foster City, CA) using the HICF method (Meyers et al., 2004, for a description of the method) to assure data compatibility with the HICF-fingerprinted Mo17 physical map. The fingerprint patterns of the BACs from B73 and Mo17 contigs were then compared, and allelic pairs were classified based on the number of shared bands as determined by the cutoff value reported by the fpc software (Soderlund et al., 2000). Each divergence class was assigned a distinct range of cutoff values: 1E-20 to 1E-40, high divergence; 1E-50 to 1E-70, medium divergence; 1E-80 to 1E-100, low divergence. Three allelic regions, designated locus9002, locus9008, and locus9009, which represent different levels of divergence, were selected. At each of these loci, at least two BAC clones from B73 and two from Mo17, with an estimated B73–Mo17 overlap of 250 kb at minimum were sequenced (Table 1).
The Mo17 BAC clone b106.c20, which is allelic to the previously sequenced bz1 alleles from maize inbreds B73 and McC (AF391808 and AF448416) (Fu and Dooner, 2002), was identified by hybridizing filters of the Mo17 BAC library with a bz1-specific probe (E. Ananiev, personal communication).
Similarly, the Mo17 BAC clone b161.k19, which is allelic to the published adh1 locus from B73 (AF123535) (Tikhonov et al., 1999), was identified and sequenced as described (Jung et al., 2004).
Sequencing and Assembling of Maize BAC Clones
All BAC clones were sequenced by the shotgun strategy as described (Tarchini et al., 2000). The sequence reads were assembled using Phred and Phrap software (http://www.phrap.org/) (Green, 1996), and the assemblies were viewed and edited in Consed (http://www.phrap.org/consed/consed.html). Vector sequences and bacterial contaminants were masked, and clone-mate information was used to make assessments regarding the validity of the assemblies with the assistance of the program exgap (http://www.genome.ou.edu/informatics.html).
PCR primers were designed to walk across the sequence gaps by extracting the nonrepetitive ends of the relevant contig sequences and importing them together into the Primer 3.0 program (Rozen and Skaletsky, 2000). The following conditions were used in the selection of primers: the smallest allowable product size, primer size of ∼18 bases, annealing temperature of 55°C, ideal GC of 50%, no more than three consecutive identical nucleotides, and a two-base GC clamp. T3 (5′-AATTAACCCTCACTAAAGGG-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′) tags were added to the 5′ ends of the forward and reverse primers, respectively, to facilitate direct sequencing of the PCR products. PCR was performed using a Perkin-Elmer 9700 thermocycler under the following conditions: 95°C for 10 min; 10 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; 35 cycles of 95°C for 30 s and 68°C for 1 min; 92°C for 7 min; and then a constant temperature of 4°C. The 25-μL PCR reaction mix consisted of 2 μL of BAC culture diluted 1:1 with 50% glycerol, 10 mM of each primer, 5% DMSO, 12.5 μL of Hot Star Taq Master Mix (Qiagen, Valencia, CA), and sterile water. PCR products (4 μL) were analyzed via agarose gel electrophoresis. PCR products were prepared for sequencing using exonuclease-I and shrimp alkaline phosphatase (USB, Cleveland, OH) and sequenced directly from both the T3 and T7 primers using an ABI 3700 sequencer (PE-Applied Biosystems) and the BigDye Terminator v3.0 cycle sequencing kit (PE-Applied Biosystems).
Subcontigs robustly connected by clone mates were merged manually where the sequencing failed. Merged sequences were further confirmed by PCR on genomic DNA.
Annotation and Comparative Sequence Analysis
A maize trained version of program FGENESH (Softberry, Mount Kisco, NY) and Repeatmasker (A.F.A. Smit and P. Green http://ftp.genome.washington.edu/RM/RepeatMasker.html) using release 4.0 version of The Institute for Genomic Research maize repeat database (http://www.tigr.org/tdb/tgi/maize/repeat_db.shtml) were used for gene prediction and masking of repetitive elements, respectively. Gene annotation was based on BLAST (BLAST E < 10−7) and BLAT (minimal sequence identity of 80%) analysis against the GenBank and the DuPont maize EST databases, respectively. Predicted genes, which still showed homology to any repetitive element from these databases, were added to the repetitive fraction of the sequences. MITEs were identified using the program FINDMITE (Tu, 2001) with the following parameter settings: target site duplication, TA, TAA, TAC, TGA, TTA, or TCA; length of tandem inverted repeat, 11 bp; number of mismatches, 1; minimum distance, 30 bp; maximum distance, 400 bp; filtering A/T and C/G strings, AT/TA repeats, and terminal inverted repeats composed of >85% of two bases. Programs BESTFIT, GAP, PILEUP, and ASSEMBLE of the GCG Wisconsin package version 10.3 and the program Dotter (Sonnhammer and Durbin, 1995) were used for sequence comparison. Divergence times (DT) for the LTR-retrotransposon were estimated using k = K/2*DT, where k is the proposed mutation rate of 1.3 × 10 to 8 substitutions per site per year (Ma and Bennetzen, 2004a), and K is the estimated number of substitutions per site between sequences using the Kimura two-parameter method (Kimura, 1980). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 2.1 (Kumar et al., 2001). Statistical analysis was performed using Kolmogorov-Smirnov test statistics (http://faculty.vassar.edu/lowry/webtext.html) and permutation methods (Mielke and Berry, 2001).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY664413 (B73_locus9002), AY664417 (Mo17_locus9002), AY664414 (B73_locus9008), AY664418 (Mo17_locus9008), AY664415 (B73_locus9009), AY664419 (Mo17_locus9009), AY664416 (b103.c20), and AY691949 (b161.k19).
Supplementary Material
Acknowledgments
We thank the members of Maureen Dolan's group for the shotgun sequencing efforts, Stéphane Deschamps for help in the assembly process, Mike Hanafey and Nancy Caraher for computer support, Karla Butler and Ada Ching for technical advice, Evgueni Ananiev for the identification of the Mo17 bz1 BAC clone, Sue Wessler for support in MITEs identification, and Enno Krebbers and Barbara Mazur for editorial advice.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Antoni Rafalski (j-antoni.rafalski@cgr.dupont.com).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025627.
References
- Aguilar, A., Roemer, G., Debenham, S., Binns, M., Garcelon, D., and Wayne, R.K. (2004). High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl. Acad. Sci. USA 101, 3490–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2002). Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115, 29–36. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Ma, J. (2003). The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr. Opin. Plant Biol. 6, 128–133. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., Ma, J., and Devos, K.M. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J.A., Auger, D.L., and Riddle, N.C. (2003). In search of the molecular basis of heterosis. Plant Cell 15, 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., Ronald, P.C., and Wessler, S.R. (1996). A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc. Natl. Acad. Sci. USA 93, 8524–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1992). Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4, 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994. a). Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91, 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994. b). Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T.E., White, S.E., and Wessler, S.R. (1994). Transduction of a cellular gene by a plant retroelement. Cell 77, 479–480. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.M., and Lin, B.Y. (2004). Molecular organization of large fragments in the maize B chromosome: Indication of a novel repeat. Genetics 166, 1947–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J.F. (1948). Alternative hypotheses of hybrid vigor. Genetics 33, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K. (2003). RNA interference, transposons, and the centromere. Plant Cell 15, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K.M., and Bennetzen, J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. (1986). Genetic fine structure of the bronze locus in maize. Genetics 113, 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, C.A., Medstrand, P., and Mager, D.L. (2003). An endogenous retroviral long terminal repeat is the dominant promoter for human 1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 100, 12841–12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby, N., and Bureau, T.E. (2001). A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem. 276, 41963–41968. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A., Gaut, R.L., Hilton, H., Feldman, D.L., and Gaut, B.S. (1998). Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 95, 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420, 316–320. [DOI] [PubMed] [Google Scholar]
- Fengler, K.A., Faller, M.L., Meyers, B.C., Dolan, M., Tingey, S.V., and Morgante, M. (2000). Construction of a contig-based physical map of corn using fluorescent fingerprint technology. Plant & Animal Genome VIII Conference, Jan. 9–12, 2000 (San Diego, CA), http://www.intl-pag/8/abstracts/pag8265.html.
- Flavell, A.J. (1992). Ty1-copia group retrotransposons and the evolution of retroelements in the eukaryotes. Genetica 86, 203–214. [DOI] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J., et al. (2004). Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 134, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. (2001). Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 11, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.M., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh1 parallel rate differences at the plastid gene rblL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Green, P. (1996). Towards completely automated sequence assembly. DOE Human Genome Program Contractor-Grantee Workshop V, Jan. 28–Feb. 1, 1996 (Santa Fe, NM), http://www.ornl.gov/sci/techresources/Human_Genome/publicat/96santa/informat/green.html.
- Hake, S., and Walbot, V. (1980). The genome of Zea mays, its organization and homology to related grasses. Chromosoma 79, 251–270. [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Han, B., and Xue, Y. (2003). Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 6, 134–138. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. (2002). RNA interference. Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100, 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Hirochika, H., Eddy, S.R., McCouch, S.R., and Wessler, S.R. (2003). An active DNA transposon family in rice. Nature 421, 163–167. [DOI] [PubMed] [Google Scholar]
- Jin, Y.K., and Bennetzen, J.L. (1994). Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, M., Ching, A., Bhattramakki, D., Dolan, M., Tingey, S., Morgante, M., and Rafalski, A. (2004). Linkage disequilibrium and sequence diversity in a 500-kbp region around the adh1 locus in elite maize germplasm. Theor. Appl. Genet. 109, 681–689. [DOI] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A.A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. [DOI] [PubMed] [Google Scholar]
- Keller, B., and Feuillet, C. (2000). Colinearity and gene density in grass genomes. Trends Plant Sci. 5, 246–251. [DOI] [PubMed] [Google Scholar]
- Kikuchi, K., Terauchi, K., Wada, M., and Hirano, H.Y. (2003). The plant MITE mPing is mobilized in anther culture. Nature 421, 167–170. [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Kuff, E.L., and Lueders, K.K. (1988). The intracisternal A-particle gene family: Structure and functional aspects. Adv. Cancer Res. 51, 183–276. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and Bennetzen, J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33, 479–532. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lai, J., Ma, J., Swigonova, Z., Ramakrishna, W., Linton, E., Llaca, V., Tanyolac, B., Park, Y.J., Jeong, O.Y., Bennetzen, J.L., and Messing, J. (2004). Gene loss and movement in the maize genome. Genome Res. 14, 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Langham, R.J., Walsh, J., Dunn, M., Ko, C., Goff, S.A., and Freeling, M. (2004). Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E.B. (1951). Pseudoallelism and gene evolution. Cold Spring Harb. Symp. Quant. Biol. 16, 159–174. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2004. a). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA. [DOI] [PMC free article] [PubMed]
- Ma, J., and Bennetzen, J.L. (2004. b). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101, 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Izquierdo, J.A., Garcia-Martinez, J., and Vicient, C.M. (1997). What makes Grande1 retrotransposon different? Genetica 100, 15–28. [PubMed] [Google Scholar]
- Matsuoka, Y., Vigouroux, Y., Goodman, M.M., Sanchez, G.J., Buckler, E., and Doebley, J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99, 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand, P., Landry, J.R., and Mager, D.L. (2001). Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C–I genes in humans. J. Biol. Chem. 276, 1896–1903. [DOI] [PubMed] [Google Scholar]
- Messing, J., Bharti, A.K., Karlowski, W.M., Gundlach, H., Kim, H.R., Yu, Y., Wei, F., Fuks, G., Soderlund, C.A., Mayer, K.F., and Wing, R.A. (2004). Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101, 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J.M. (2002). Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 130, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Scalabrin, S., and Morgante, M. (2004). Mapping and sequencing complex genomes: Let's get physical! Nat. Rev. Genet. 5, 578–588. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke, P.W., and Berry, K.J. (2001). Permutation Methods: A Distance Function Approach. (New York: Springer-Verlag).
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozuka, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazaki, T., Okumoto, Y., Horibata, A., Yamahira, S., Teraishi, M., Nishida, H., Inoue, H., and Tanisaka, T. (2003). Mobilization of a transposon in the rice genome. Nature 421, 170–172. [DOI] [PubMed] [Google Scholar]
- Nigumann, P., Redik, K., Matlik, K., and Speek, M. (2002). Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79, 628–634. [DOI] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van Der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M. (2001). Coalescent theory. In Handbook of Statistical Genetics, D.J. Balding, M. Bishop, and C. Cannings, eds (Chichester, UK: John Wiley and Sons), pp. 179–212.
- Palmgren, M.G. (1994). Capturing of host DNA by a plant retroelement: Bs1 encodes plasma membrane H(+)-ATPase domains. Plant Mol. Biol. 25, 137–140. [DOI] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B., and Jones, J.D. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. (2000). Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R.H.A. (2002). RNA silencing: The genome's immune system. Science 296, 1263–1265. [DOI] [PubMed] [Google Scholar]
- Pouteau, S., Huttner, E., Grandbastien, M.A., and Caboche, M. (1991). Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 10, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A. (2002). Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5, 94–100. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P., and Bennetzen, J.L. (2002). Structural analysis of the maize rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D.L., Thornsberry, J.M., Matsuoka, Y., Wilson, L.M., Whitt, S.R., Doebley, J., Kresovich, S., Goodman, M.M., and Buckler IV, E.S. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98, 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and Skaletsky, H.J. (2000). Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology, S. Misener and S.A. Krawetz, eds (Totowa, NJ: Humana Press), pp. 365–386. [DOI] [PubMed]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P.J., Ramakrishna, W., Bennetzen, J.L., Busso, C., and Dubcovsky, J. (2002). Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct. Integr. Genomics 2, 70–80. [DOI] [PubMed] [Google Scholar]
- Schramke, V., and Allshire, R. (2003). Hairpin RNAs and retrotransposon LTRs affect RNAi and chromatin-based gene silencing. Science 301, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Schulman, A.H., Lahaye, T., and Schulze-Lefert, P. (2000). A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. (2000). Contigs built with fingerprints, markers, and FPCV4.7. Genome Res. 10, 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100, 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L., and Durbin, R. (1995). A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167, 1–10. [DOI] [PubMed] [Google Scholar]
- Speek, M. (2001). Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, L.E., and Chandler, V.L. (1988). Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5, 519–529. [DOI] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka, A., and Theologis, A. (2004). Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 101, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z. (2001). Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 98, 1699–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., Jaaskelainen, M.J., Kalendar, R., and Schulman, A.H. (2001). Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, Y., Jaqueth, J.S., Matsuoka, Y., Smith, O.S., Beavis, W.D., Smith, J.S., and Doebley, J. (2002). Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 19, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Vitte, C., and Panaud, O. (2003). Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 20, 528–540. [DOI] [PubMed] [Google Scholar]
- Voytas, D., Cummings, M., Konieczny, A., Ausubel, F., and Rodermel, S. (1992). Copia-like retrotransposons are ubiquitous among plants. Proc. Natl. Acad. Sci. USA 89, 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, E., and Martin, D.I. (2001). Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 27, 361–365. [DOI] [PubMed] [Google Scholar]
- Wicker, T., Stein, N., Albar, L., Feuillet, C., Schlagenhauf, E., and Keller, B. (2001). Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26, 307–316. [DOI] [PubMed] [Google Scholar]
- Wicker, T., Yahiaoui, N., Guyot, R., Schlagenhauf, E., Liu, Z.D., Dubcovsky, J., and Keller, B. (2003). Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 15, 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H.M., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.-H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M., Tsugawa, H., Miyao, A., Yano, M., Wu, J., Yamamoto, S., Matsumoto, T., Sasaki, T., and Hirochika, H. (2001). The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol. Genet. Genomics 265, 336–344. [DOI] [PubMed] [Google Scholar]
- Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B.J., and Schnable, P.S. (2002). Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99, 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.