Abstract
Substantial evidence indicates that amino acid conjugates of indole-3-acetic acid (IAA) function in auxin homeostasis, yet the plant enzymes involved in their biosynthesis have not been identified. We tested whether several Arabidopsis thaliana enzymes that are related to the auxin-induced soybean (Glycine max) GH3 gene product synthesize IAA–amino acid conjugates. In vitro reactions with six recombinant GH3 enzymes produced IAA conjugates with several amino acids, based on thin layer chromatography. The identity of the Ala, Asp, Phe, and Trp conjugates was verified by gas chromatography–mass spectrometry. Insertional mutations in GH3.1, GH3.2, GH3.5, and GH3.17 resulted in modestly increased sensitivity to IAA in seedling root. Overexpression of GH3.6 in the activation-tagged mutant dfl1-D did not significantly alter IAA level but resulted in 3.2- and 4.5-fold more IAA-Asp than in wild-type seedlings and mature leaves, respectively. In addition to IAA, dfl1-D was less sensitive to indole-3-butyric acid and naphthaleneacetic acid, consistent with the fact that GH3.6 was active on each of these auxins. By contrast, GH3.6 and the other five enzymes tested were inactive on halogenated auxins, and dfl1-D was not resistant to these. This evidence establishes that several GH3 genes encode IAA-amido synthetases, which help to maintain auxin homeostasis by conjugating excess IAA to amino acids.
INTRODUCTION
Throughout their life cycle, plants adjust their growth and development in response to a variety of internal and external cues. A critical mechanism to coordinate these responses is regulation of the quantity and activity of the phytohormone indole-3-acetic acid (IAA). The amount of active IAA in specific tissues is determined by an array of metabolic processes, including regulation of its synthesis, transport to or from specific cells or tissues, IAA inactivation and reactivation, and degradation via multiple oxidative pathways (for reviews, see Bartel, 1997; Normanly, 1997; Bartel et al., 2001; Ljung et al., 2002). In addition to the free acid, IAA occurs in a variety of modified forms, such as glycosyl esters and amide-linked conjugates with various amino acids and peptides. Only free IAA is established to be the direct biologically active compound, but its conjugates help to maintain IAA homeostasis, both by inactivating IAA and by serving as a reservoir of IAA that can be released upon hydrolysis.
IAA conjugates undoubtedly occur in all higher plants and in at least some lower plant species. Besides the ester conjugates, amide conjugates with Asp, Glu, Ala, Gly, Val, and Leu have been found in a variety of plants, and conjugates with other amino acids may also occur (Normanly, 1997; Ljung et al., 2002; Rampey et al., 2004). The amount of conjugated IAA is generally greater than, and sometimes far exceeds, the level of the free acid in plant tissues (see Bandurski et al., 1995). Conjugate formation and hydrolysis is developmentally regulated and varies significantly among plant tissues (for review, see Klezkowski and Schell, 1995). Perturbing IAA homeostasis can also influence IAA conjugate level. For example, the exogenous application of IAA or other auxins generally increases IAA conjugate level, and the expression of transgenes involved in IAA synthesis or metabolism can also alter the amount of conjugates that accumulate (Sitbon et al., 1992; Normanly et al., 1993).
A substantial quantity of IAA in plants can exist as amide conjugates with peptides that range in size from 3.6 to 35 kD. In fact, IAA peptides are the major amide conjugate in bean (Phaseolus vulgaris) (Bialek and Cohen, 1986) and Arabidopsis thaliana seeds (Ljung et al., 2002), the proportion relative to free IAA varying among tissues and during development. By contrast, IAA-Asp and IAA-Glu account for only 1% of the total amide conjugates of IAA in Arabidopsis (Tam et al., 2000), and even lower quantities of IAA-Ala and IAA-Leu occur in this species (Kowalczyk and Sandberg, 2001; Rampey et al., 2004). Nevertheless, evidence indicates that amino acid conjugates play important roles in IAA metabolism, particularly as temporary storage reserves and by initiating the catabolism of IAA (Cohen and Bandurski, 1982; Bandurski et al., 1995).
Although some IAA-amino acid conjugates have auxin-like activity when applied exogenously, this activity generally correlates with the ability of plant tissues to hydrolyze these conjugates (Hangarter et al., 1980; Bialek et al., 1983). This suggests it is the released IAA that functions in signaling, rather than the conjugates themselves. Recent mutational analysis of an IAA-amido hydrolase family in Arabidopsis confirms this. Eliminating the activity of specific hydrolases reduced the level of free IAA in seeds and seedlings and produced an auxin-deficient phenotype (LeClere et al., 2002; Rampey et al., 2004). By contrast, the levels of IAA-Ala and IAA-Leu were elevated, providing convincing evidence that these conjugates contribute to the pool of free IAA through the action of IAA-amido hydrolases.
By contrast, IAA-Asp and probably IAA-Glu function in IAA turnover. IAA-Asp appears soon after the addition of IAA to plant tissues, and metabolic labeling has shown that formation of this conjugate leads irreversibly to oxidation, followed by catabolism of IAA (see Normanly, 1997; Ljung et al., 2002). Unlike the Ala and Leu conjugates, IAA-Asp was not elevated in the Arabidopsis hydrolase mutants, supporting the idea that this conjugate is not hydrolyzed appreciably in plants (Rampey et al., 2004). Further evidence that amino acid conjugates function in auxin homeostasis comes from the expression of a bacterial IAA-Lys synthetase gene (iaaL) in transgenic tobacco (Nicotiana tabacum). The resulting epinastic plants with decreased apical dominance and inhibited vascular differentiation are indicative of an auxin imbalance (Romano et al., 1991; Spena et al., 1991).
Besides storage and degradation, it has been suggested that the varied chemical properties of the constituents conjugated to IAA might determine the fate of IAA in other ways, such as auxin transport (Cohen and Bandurski, 1978). Biological activity for IAA conjugates also cannot be ruled out entirely. IAA-Ala may be a weak auxin, independent of its contribution to free IAA by hydrolysis (Hangarter et al., 1980). Consistent with this, IAA hydrolase mutants retained greater sensitivity to IAA-Ala compared with their sensitivity to IAA-Leu and IAA-Phe (Rampey et al., 2004).
Besides IAA, several closely related plant compounds also have auxin-like activity. These include 4-chloroindole-3-acetic acid (4-Cl-IAA) and indole-3-butyric acid (IBA) (for reviews, see Epstein and Ludwig-Müller, 1993; Bandurski et al., 1995). Like IAA, IBA occurs as the free acid as well as in a variety of conjugated forms, although both are generally less abundant than IAA. IBA also functions in IAA homeostasis through interconversion between IBA and IAA (Ljung et al., 2002), which complicates our understanding of the direct role of IBA as an auxin signal. Indole-3-pyruvic acid (IPyA) is also found in plants (Cooney and Nonhebel, 1989; Tam and Normanly, 1998), as is the weak auxin phenylacetic acid (PAA), but their physiological roles are unclear. A variety of indolic compounds and their conjugates occur in plant pathogenic and symbiotic microorganisms, but their roles are also largely unknown (Costacurta and Vanderleyden, 1995). The function of iaaL in Pseudomonas savastanoi may be related to pathogenesis, but the mechanism is presently not clear (Romano et al., 1991; Spena et al., 1991).
Despite the fact that amino acid conjugates of IAA were discovered in plants nearly half a century ago (Andrea and Good, 1955), the biosynthetic pathway leading to their formation has eluded discovery (Ljung et al., 2002). Synthases that produce IAA-glucosyl esters have been characterized (Szerszen et al., 1994), as have hydrolases that release free IAA from glucosyl esters (Jakubowska et al., 1993). As mentioned earlier, hydrolases that cleave amide conjugate bonds have also been well documented (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002; Rampey et al., 2004). However, although bacterial IAA-Lys synthetase was identified some time ago (Roberto et al., 1990), this has not revealed the nature of the plant IAA-amido conjugating enzymes.
We have studied an Arabidopsis gene family that is closely related to the auxin-responsive soybean (Glycine max) GH3 gene (Hagen and Guilfoyle, 1985; Wright et al., 1987; Hagen et al., 1991). Structural modeling (Staswick et al., 2002) revealed that plant GH3 proteins belong to the firefly luciferase family of adenylate-forming enzymes, found in several diverse organisms (Kleinkauf and Von Dohren, 1996). We recently determined that Arabidopsis GH3.11 (JAR1) is a jasmonic acid–amido synthetase that conjugates several amino acids to jasmonic acid (JA) (Staswick and Tiryaki, 2004). Importantly, JA-Ile complemented the defect in the JA response mutant jar1, establishing that amino acid conjugation to JA is necessary for some jasmonate responses in Arabidopsis. Based on these findings, we have now examined whether six other Arabidopsis GH3 enzymes previously shown to adenylate IAA (Staswick et al., 2002) are in fact IAA-amido synthetases. We report here that these enzymes synthesize a variety of IAA-amino acid conjugates in vitro and that overexpression and insertional mutation of various GH3 genes leads to altered auxin sensitivity in Arabidopsis. These results also suggest a functional rationale for the rapid transcriptional activation of some GH3 genes by auxins.
RESULTS
Several Arabidopsis GH3 Enzymes Conjugate Amino Acids to IAA
We examined whether six Arabidopsis enzymes previously shown to adenylate IAA (Staswick et al., 2002) catalyze the synthesis of IAA-amino acid conjugates. The nomenclature of Hagen and Guilfoyle (2002) is adopted here, except that the hyphen in gene names is replaced by a period to avoid confusion with the standard designation for alleles of Arabidopsis genes. The relationship of the GH3 enzymes to the previously reported BAC open reading frames (Staswick et al., 2002) and gene identifiers is as follows: GH3.2 (BAC F6G17.40, At4g37390), GH3.3 (BAC T20D16.20, At2g23170), GH3.4 (T30E16.2, At1g59500), GH3.5 (M4I22.70, At4g27260), GH3.6 (F24B18.13, At5g54510), and GH3.17 (F3H9.21, At1g28130). GH3.9 (BAC F17A22.14, At2g47750) consistently gave low protein yields as a glutathione S-transferase (GST) fusion protein in Escherichia coli and was not analyzed in this study. Recombinant GH3.6 was tested in a reaction with IAA and Asp to determine whether the commonly occurring IAA-Asp conjugate was formed. Figure 1A shows that the reaction yielded a product with the same RF as IAA-Asp on thin layer chromatography (TLC). Consistent with the previously described IAA adenylating activity of this enzyme (Staswick et al., 2002), synthesis of the product was dependent on ATP. In contrast with GH3.6, the JAR1 enzyme that conjugates amino acids to JA did not yield a product with these substrates.
Figure 1.
TLC Analysis of Amino Acid Conjugates of IAA Synthesized by Recombinant GST:GH3s.
(A) Assays for enzymatic formation of IAA-Asp with indicated reagents included in the reaction mixtures. The JAR1 enzyme conjugates amino acids to JA. Standards are shown in the two left lanes.
(B) Evaluation of GH3.6 enzyme reactions with 20 amino acids (single letter abbreviations).
(C) Assays for GH3.2, GH3.3, GH3.4, GH3.5, and GH3.17 reactions with the indicated amino acids. The spot near the origin at the bottom for the reactions with Trp is the unreacted amino acid. All plates were stained for indoles with van Urk-Salkowski reagent as described in the text.
The pH optimum for GH3.6 was determined using protein that was enzymatically cleaved from GST. Results shown in Table 1 demonstrate that no activity was found at or below pH 7.5. Highest activity was seen at pH 9, where the formation of IAA-Asp was 244 nmol/min/mg. We did not test for activity above pH 9.0 because this would seem biologically irrelevant. Activity was also greater at pH 8.8 than at 7.5 for reactions with Glu, Gln, and Trp when evaluated by TLC, and similar results were found for the other enzymes used in this study (data not shown). All further reactions were performed at pH 8.8.
Table 1.
Effect of pH on GH3.6 Activity
| pH | Synthetase Activity (nmol IAA-Asp Formed/min/mg Protein) |
|---|---|
| 7.0 | ND |
| 7.5 | ND |
| 8.0 | 97 (0) |
| 8.5 | 237 (2) |
| 9.0 | 244 (8) |
Values are means (±se) for IAA-Asp formed in two independent experiments. ND, not detected (detection limit, 2 nmol IAA-Asp/min/mg).
Next, GH3.6 activity on IAA and each of 20 amino acids was monitored by TLC. Many reactions yielded products that migrated with a lower mobility than IAA (Figure 1B), albeit with variable staining intensity. For several reactions, a clear correlation between loss of free IAA and the appearance of a product was evident. Putative conjugates for the reactions with Ala, Ile, Phe, and Val had the same RF as the standards for these IAA conjugates (data not shown). Products from the reactions with Ala, Asp, Phe, and Trp were methylated and analyzed by gas chromatography–mass spectrometry (GC-MS). Each sample contained a major chromatographic peak with a mass spectrum that included the major quinolinium ion at a mass-to-charge ratio (m/z) of 130. The expected molecular ions at 260, 318, 336, and 375 m/z for the derivatized IAA-Ala, -Asp, -Phe, and -Trp, respectively, were also evident. The complete mass spectral data for these conjugates are available in the supplemental data online.
Reactions with each of the other five enzymes were also analyzed by TLC, and the results were similar in terms of relative amount of each product formed (Figure 1C). Based on staining intensity, Asp and/or Glu were strongly reactive for each of the enzymes tested. GH3.17 appeared to favor Glu over Asp under these conditions. Met and Trp also yielded a substantial amount of product for each enzyme, although the latter contains two indole rings, which may overestimate the amount of product compared with other reactions. Phe, Gly, Gln, Tyr, Ile, and Val also yielded appreciable amounts of product with most enzymes. Weakly stained products were evident for several other amino acids, although some of these were only evident for one or two enzymes. As for GH3.6, the other five enzymes gave little or no evidence for product formation with His, Ser, Thr, Arg, Lys, or Cys (data not shown). Together, these results demonstrate that the family of Arabidopsis enzymes previously shown to adenlyate IAA is in fact IAA-amido synthetases that are capable of forming a variety of amino acid conjugates with IAA in vitro.
IAA-Amido Synthetases Are Active on Several Auxins
To further examine the substrate specificities of these enzymes, we tested their activity on several other auxins and other compounds having structures related to auxins. Substrates were evaluated with in vitro assays for adenylation by monitoring exchange of [32P]-PPi into ATP. Results summarized in Figure 2 show that each enzyme was active on several substrates in addition to IAA. In general, the relative activity among the substrates tested was similar for each enzyme. In addition to IAA, enzymes were active on the pyruvic and butyric acid analogs of IAA. PAA and the synthetic auxin naphthaleneacetic acid (NAA) were also strong substrates for adenylation. By contrast, the 4-chloro derivative of IAA was a poor substrate, as was the indole-containing amino acid Trp. Two chlorinated synthetic auxin herbicides, 2,4-D and 3,6-dichloro-o-anisic acid (dicamba), were also ineffective in stimulating isotope exchange with these enzymes. The major difference in substrate specificity among enzymes was in their activity on IPyA and IBA relative to IAA. These were comparable to IAA for GH3.6 and greater than IAA for GH3.5. The other enzymes had lower activity on IPyA and IBA; activity on the latter being only about one-fourth that of IAA for GH3.4 and GH3.17.
Figure 2.
Adenylation Activity of IAA-Amido Synthetases on Several Auxins.
Percentage of total radioactivity exchanged into ATP for each substrate is expressed as the mean of three independent experiments. Bars indicate standard errors. Cl-IAA, 4-chloro-3-indole acetic acid.
GH3.2, GH3.3, GH3.4, GH3.5, and GH3.6 were also tested for their ability to form amino acid conjugates with IBA. Analysis by TLC showed that products were formed with the different amino acids at approximately the same relative levels as for IAA. An example of the TLC results for GH3.6 is shown in the supplemental data online. Products of the GH3.6 reactions for IBA with Ala, Phe, or Trp were analyzed by GC-MS, and these gave mass spectra that included the major quinolinium ion and the expected molecular ion in each case (see supplemental data online). These results suggest that in addition to IAA, the IAA-amido synthetases might also metabolize other natural or synthetic auxins in vivo.
Overexpression of GH3.6 Increases IAA-Asp Accumulation
To determine whether a member of this enzyme family forms IAA conjugates in Arabidopsis, we examined the level of IAA-Asp in the activation-tagged mutant dfl1-D, which overexpresses GH3.6 (Nakazawa et al., 2001). Two-week-old seedlings were grown in culture boxes, and four-week-old leaves were harvested from plants grown in soil. Figure 3 shows that the amount of IAA in the wild type and dfl1-D was 5 to 10 pg/g tissue fresh weight from both tissue sources. Although the mean IAA level for dfl1-D was lower than the wild type in both cases, the differences were not statistically significant (one-tailed t test; P < 0.05). By contrast, the content of IAA-Asp was 3.2- and 4.5-fold greater in dfl1-D than in the wild type for seedlings and leaves, respectively. This provides evidence that GH3.6 synthesizes IAA-Asp in vivo and that expression of this gene quantitatively affects the accumulation of IAA-Asp.
Figure 3.
Quantitation of IAA and IAA-Asp in the Wild Type and dfl1-D.
Hormone level in 2-week-old Arabidopsis seedlings and 4-week-old leaves is shown as pg/g tissue fresh weight (FW) obtained from extraction of three independent tissue samples. Error bars indicate se of the means, and asterisks denote significant difference from the wild type (P < 0.05).
Elevated Expression of GH3.6 Confers Resistance to Other Auxins
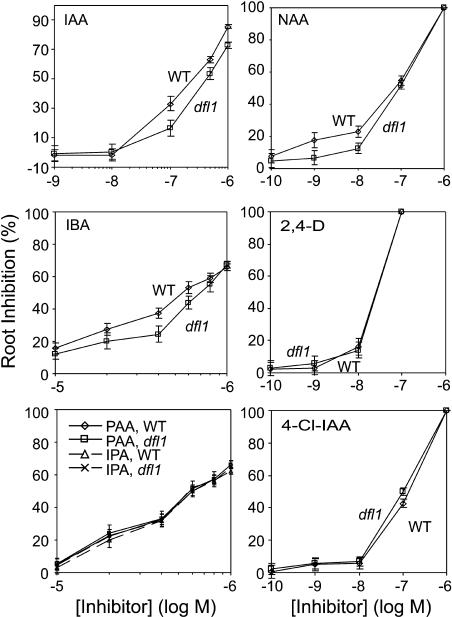
Our results suggest that the basis for the decreased sensitivity to exogenous IAA found earlier for dfl1-D (Nakazawa et al., 2001) is because of an enhanced ability to conjugate IAA in this mutant, thereby removing it from the pool of active auxin. To assess whether GH3.6 is active on other auxins in vivo, we compared the resistance of dfl1-D with the wild type grown in the presence of several different auxins. Figure 4 confirms that dfl1-D is resistant to IAA. In addition, seedling root growth in this mutant is less inhibited by IBA and NAA, two auxins on which GH3.6 shows adenylating activity (Figure 2). IBA was a much weaker inhibitor of root growth than IAA, but roots of dfl1-D were less sensitive over the range of 10−6 to 10−5 M. By contrast, resistance to NAA was seen at 10−9 and 10−8 M but not at higher concentrations. We detected no resistance to IPyA or PAA in dfl1-D, despite the fact that GH3.6 showed adenylating activity on these two substrates in vitro. dfl1-D was also not resistant to 4-Cl-IAA or to the synthetic auxin 2,4-D. This latter result is consistent with the inactivity of these compounds as substrates in the adenylation assay.
Figure 4.
Root Inhibition by Several Auxins in dfl1-D and Wild-Type Arabidopsis.
Seedling roots were grown in agar media for 9 d with the indicated concentration of inhibitor. Inhibition is expressed as a percentage of each genotype grown in the absence of inhibitor. Error bars indicate 95% confidence intervals for each ratio of the means (n = 18 to 27 seedlings/data point).
Mutations in IAA-Amido Synthetase Genes Increase Auxin Sensitivity
We next examined whether insertional mutations in IAA-amido synthetase genes affected auxin response in Arabidopsis. Several putative T-DNA or transposon insertions in GH3s were analyzed, and as illustrated in Figure 5A, inserts were found in the coding sequences of GH3.1 (SGT4174), GH3.2 (SGT5949), and GH3.17 (SALK 050597). A fourth, SALK 014376, contained a T-DNA insert 260 bp upstream of the predicted ATG start codon of GH3.5. These mutants are called gh3.1-1, gh3.2-1, gh3.17-1, and gh3.5-1, respectively. Although we have not previously documented that GH3.1 (At2g14960) is active on IAA, we would predict that this is the case based on its sequence similarity with the other IAA-amido synthetases (Staswick et al., 2002).
Figure 5.
Analysis of DNA Insertion Mutants for IAA-Amido Synthetase Genes.
(A) Insertion sites (open triangles) for T-DNAs (gh3.5-1 and gh3.17-1) or transposons (gh3.1-1 and gh3.2-1) were verified by amplification of genomic DNA using primers described in the text. Arrows indicate primer orientation. T-DNA mutants contain inverted tandem insertions yielding Lba1 primer sites at each end. Closed bars, coding regions; open bars, introns.
(B) Auxin inhibition of root growth in insertion mutants. Inhibition and errors are expressed as in Figure 4 (n = 23 seedlings/data point). T-DNA mutants and transposon mutants are in the Col and Ler backgrounds, respectively.
Homozygous lines of each mutant were tested for seedling root sensitivity to IAA. Figure 5B shows results for gh3.5-1 and gh3.17-1, which are in the Columbia (Col) background. Compared with the wild type, both mutants were more sensitive to IAA. Although the concentration for 50% inhibition was only approximately twofold less for gh3.17-1 and ∼1.5-fold less for gh3.5-1, these results were consistent in three independent experiments. Mutants gh3.1-1 and gh3.2-1 are both in the Landsberg erecta (Ler) background, and compared with the wild type, they too were more sensitive to IAA (Figure 5B). The differences were again small, especially for gh3.2-1, but they were consistent in independent experiments. The inactivity of IAA-amido synthetases on 2,4-D suggested that in contrast with IAA, mutations in these genes might not alter sensitivity to the herbicide. Indeed, root growth in the presence of 2,4-D was not different from the wild type in these mutants (Figure 5B). This provides further evidence that members of the IAA-amido synthetase family metabolize excess IAA in plant tissues by conjugating it to amino acids.
We next examined whether auxin-mediated developmental processes such as root growth and lateral root formation (Wightman and Thimann, 1980; Klee and Estelle, 1991) were affected in the insertion mutants in the absence of added auxin. Table 2 shows that the rate of seedling root growth between days 4 and 7 did not vary for any of the mutants compared with their respective wild genotypes. Similarly, lateral root formation was not significantly different from the wild type in 11-d-old seedlings (Table 2), based on an analysis of variance (α = 0.05). Thus, although sensitivity to exogenous IAA is marginally affected in each of these mutants, the defect in single gene family members is apparently not sufficient to cause a noticeable phenotype during normal seedling root development.
Table 2.
Comparison of Root Growth and Lateral Root Formation in Wild-Type and GH3 Mutants
| Genotype | Root Growth (mm/d) | Lateral Roots (no./plant) |
|---|---|---|
| Col | 4.3 (0.1) | 4.9 (0.7) |
| gh3.5-1 | 4.4 (0.1) | 3.5 (0.5) |
| gh3.17-1 | 4.1 (0.1) | 4.5 (0.6) |
| Ler | 4.8 (0.1) | 4.0 (0.8) |
| gh3.1-1 | 4.7 (0.1) | 5.3 (0.7) |
| gh3.2-1 | 4.7 (0.2) | 4.8 (0.7) |
Values are the means (±se) for seedling root growth rate over 3 d (n = 20) and number of lateral roots/seedling (n = 22). Col and Ler are the wild types for mutants with which they are grouped.
DISCUSSION
GH3 Transcriptional Regulation and Auxin Homeostasis
The first GH3 gene was discovered approximately two decades ago as a soybean transcript that was induced by low concentrations of natural or synthetic auxins (Hagen and Guilfoyle, 1985; Wright et al., 1987). In fact, GH3 is among the most rapidly responding genes when exogenous auxins are applied (Hagen and Guilfoyle, 2002). Many additional GH3-like proteins have since been identified in other plant species, but until recently, their biochemical function remained a mystery. We have now determined that several Arabidopsis GH3s encode IAA-amido synthetases that conjugate amino acids to IAA. We also recently determined that the soybean GH3 enzyme has similar biochemical activity (P.E. Staswick, unpublished data). Based on these observations, the simplest explanation for why several GH3 genes are induced by auxins is that this provides a mechanism for plants to cope with the presence of excess auxin. In this model, transcriptional activation would lead to more IAA-amido synthetase, which then converts excess auxin to amino acid conjugates that are either inactive or degraded.
Evidence from Arabidopsis supports the idea that regulated expression of IAA-amido synthetases helps to maintain IAA homeostasis. Conjugation of IAA to Asp and Glu is markedly induced by exogenously applied IAA in Arabidopsis (Östin et al., 1998). Overexpression of GH3.6 in dfl1-D yields dwarf plants with characteristics of auxin imbalance, including epinastic growth, leaves that are misshaped, and reduced apical dominance (Nakazawa et al., 2001). dfl1-D plants are also resistant to added auxins (Figures 3 and 4) and have an elevated level of IAA-Asp. Conversely, insertional mutations in four other Arabidopsis IAA-amido synthetase genes increased sensitivity to IAA (Figure 5). Sensitivity to 2,4-D was not affected in either mutant class, consistent with the fact that the synthetases are not active on 2,4-D.
Published studies (Nakazawa et al., 2001; Sawa et al., 2002; Tian et al., 2002; Zhao et al., 2003; Goda et al., 2004) and publicly available gene expression data (GEO accession GSM9969, http://www.ncbi.nlm.nih.gov/geo/) indicate that several Arabidopsis IAA-amido synthetase genes are induced by auxin. GH3.1, GH3.2, GH3.3, and GH3.4 were elevated >10-fold in seedlings within an hour or two of IAA treatment. Although induced somewhat less (2.5-fold to eightfold), GH3.5 and GH3.6 were also responsive, whereas GH3.9 and GH3.17 showed little or no induction under these conditions. Some of these studies revealed similar patterns of induction with various synthetic auxins as well. By contrast, Arabidopsis GH3 genes that apparently do not encode IAA-amido synthetases (Staswick et al., 2002, Staswick and Tiryaki, 2004) showed little or no induction by auxin. It should be noted that although GH3.9 and GH3.17 were not auxin inducible in seedlings, this does not preclude their response to auxin at other developmental stages.
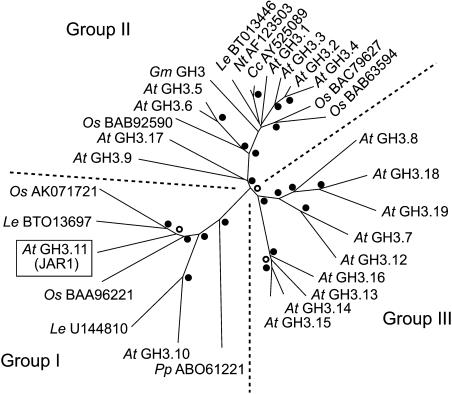
The differential seedling response to auxin among the Arabidopsis GH3 genes matches the phylogenetic grouping of the corresponding enzymes shown in Figure 6. The four showing strongest transcriptional induction (GH3.1 to GH3.4) are more closely related, whereas the two showing weaker response (GH3.5 and GH3.6) are more similar to each other. The two that did not show significant induction in seedlings are also more alike. This may indicate distinct functions based on gene regulation. A tobacco homolog of soybean GH3 also responds to auxin (Roux and Perrot-Rechenmann, 1997), and IAA-Asp formation is auxin-inducible in other species as well (Venis, 1972; Slovin et al., 1999). Together, this evidence suggests that regulated expression of IAA-amido synthetase genes may be a common mechanism that plants use to feedback-regulate the level of active auxin.
Figure 6.
Phylogenetic Analysis of Plant GH3 Proteins.
Proteins are identified by a two-letter genus/species designations followed by the gene name or database accession number for the nucleotide sequence from which the protein was translated. The Arabidopsis GH3 nomenclature is as described in the text. Le U144810 was constructed from unigenes SGN-U144810 build 2 (5′ end) and SGN-U54065 build 3 (3′ end) with ∼700 nucleotides of identical overlapping sequence (www.sgn.cornell.edu). The boxed enzyme JAR1 conjugates amino acids to JA. Dotted lines separate enzymes categorized as Group I, II, or III, based on sequence similarity and enzymatic activity (Staswick et al., 2002; Staswick and Tiryaki, 2004). Bootstrap values (1000 replicates) for individual nodes are represented by dots (≥95%), open circles (50 to 94%), or unlabeled (<50%). At, Arabidopsis thaliana; Cc, Capsicum chinense; Gm, Glycine max; Le, Lycopersicon esculentum; Nt, Nicotiana tabacum; Os, Oryza sativa; Pp, Physcomitrella patens.
Gene Redundancy and Function
We previously divided the Arabidopsis GH3s into three groups based on their sequence similarities and substrate specificities (Staswick et al., 2002). An extension of this grouping to include the phylogenetic relationship of all plant GH3s for which complete predicted amino acid sequences were available is shown in Figure 6. With the exception of GH3.1, which we have not analyzed, all Arabidopsis Group II enzymes (Figure 6) are demonstrated to be active on IAA (Staswick et al., 2002; this study). Soybean GH3 also falls within this group, which agrees with its biochemical activity (P.E. Staswick, unpublished data). We would predict that the other Group II proteins from tobacco, tomato (Lycopersicon esculentum), rice (Oryza sativa), and Capsicum chinense are also IAA-amido synthetases. Surprisingly, Group III enzymes have only been identified in Arabidopsis to date. Their function has not been determined, but several do not appear to be active on IAA or other plant hormones (Staswick et al., 2002). Arabidopsis JAR1 (GH3.11) is the only Group I enzyme for which a function has been documented (Staswick and Tiryaki, 2004), although we have evidence that some others are also JA-amido synthetases (P.E. Staswick, unpublished data). Importantly, neither of the Arabidopsis enzymes JAR1 or GH3.10 is active on IAA. Thus, it appears that the Arabidopsis IAA-amido synthetases are restricted to the eight enzymes of Group II.
Although it is presently not clear why Arabidopsis has multiple IAA-amido synthetases, this situation parallels the Arabidopsis IAA-amido hydrolase family. Biochemically, the hydrolases provide a counterpart to the IAA inactivating function of the conjugate synthetases. A summary of the complementary role of these enzymes in the maintenance of IAA homeostasis is presented in Figure 7. The conjugate hydrolases overlap in their amino acid specificities, but distinct differences also occur (LeClere et al., 2002; Rampey et al., 2004). The apparent lack of major differences in substrate specificity among the Arabidopsis synthetases, on the other hand, suggests that the reason for multiple genes may be more related to tissue-specific or developmental roles than to different biochemical properties of the enzymes themselves. The IAA-conjugate hydrolase genes vary in their spatial and temporal expression (Rampey et al., 2004), and limited analysis suggests this is also the case for some of the synthetase genes. For example, GH3.2 promoter activity was evident throughout seedling roots, except for root tips, whereas GH3.6 activity was present in all root tissues (Takase et al., 2004). GH3.2 activity was also detected in pollen, whereas GH3.6 was not active in flowers. The variation in auxin inducibility discussed earlier also supports the idea that regulatory differences exist among these genes.
Figure 7.
The Role of Amido Synthetase and Hydrolase Activity in IAA Homeostasis.
Dotted arrow indicates transcriptional activation for several IAA-amido synthetase genes. The specific role of other IAA-amino acid conjugates in plants is unclear.
Nevertheless, the fact that Arabidopsis IAA-amido synthetase genes have not been previously identified in knockout mutant screens for auxin activity suggests that there is significant redundancy among them. Our finding of only minor increases in sensitivity to IAA and no morphological differences in the GH3 insertion mutants supports the idea of functional redundancy for these genes. This is also similar to the hydrolase genes, for which single gene mutations cause only modest decreases in sensitivity to IAA-amino acid conjugates, whereas multiple gene mutants have more severe phenotypes (Rampey et al., 2004). We are currently pyramiding gene mutations for the synthetases to further explore their roles in Arabidopsis. On the other hand, it should not be surprising that overexpression of a single IAA-amido synthetase gene has a dramatic effect on plant phenotype, as evidenced in the activation tagged mutants dfl1-D (Nakazawa et al., 2001) and ydk1-D (Takase et al., 2004).
IAA-Synthetase Activity and IAA Level
Because IAA-Asp is viewed as a committed step in IAA degradation, one might have expected that this conjugate would not have accumulated in dfl1-D and that free IAA level would be depressed. However, this was not the case. That IAA-Asp was elevated approximately fourfold suggests that the mechanisms for catabolizing this conjugate could not keep pace with its increased synthesis or that some of the IAA-Asp is sequestered or otherwise protected from oxidative degradation. Nevertheless, IAA-Asp would still be a minor component in dfl1-D, comprising only ∼4% of total amide IAA if other amides were not also changed appreciably.
The increased IAA-Asp in dfl1-D would require about as much IAA for its synthesis as is present in the free IAA pool of the wild type. Synthesis of other conjugates by GH3.6 may require substantially more IAA, and yet the level of free IAA was not significantly reduced in this mutant. Spena et al. (1991) also reported that expression of iaaL in tobacco did not significantly affect free IAA level, despite the marked accumulation of IAA-Lys in these plants. And although overexpression of a bacterial IAA-Asp hydrolase gene in Arabidopsis substantially reduced the level of IAA-Asp, there was relatively little effect on free IAA, apparently because the excess was rapidly oxidized through other pathways (Chou et al., 1996; Tam and Normanly, 2002). These results indicate that to some degree plants can compensate for perturbations in the normal rates of amide conjugate synthesis or metabolism. On the other hand, Klee and Lanahan (1995) found that iaaL expression in tobacco resulted in up to a 20-fold reduction in free IAA in some transgenic plants, which was regarded as consistent with the observed epinastic phenotypes (Romano et al., 1991). The IAA level was also reduced approximately twofold in an Arabidopsis IAA-amido hydrolase triple mutant (Rampey et al., 2004). The reason for the apparent discrepancy in free IAA level among these experiments may center on the degree to which conjugate metabolism is altered in each case, although more complex changes to metabolism may also occur.
The fact that IAA level was not reduced significantly in dfl1-D raises an important question. What is the physiological basis for the strong phenotype in this mutant? Possibly, a small undetected decline in free IAA is responsible. More substantial IAA deficiencies may also occur in a limited number of cells or tissues that are critical to development, but these may not be seen in assays of whole seedlings or leaves. Finally, although generally regarded as biologically inactive, we cannot rule out the possibility that specific auxin–amino acid conjugates synthesized by GH3.6 play a direct role in developmental signaling. Evidence for the possible biological activity of IAA-Ala was discussed earlier (Hangarter et al., 1980; Rampey et al., 2004). The apparent broad specificity of the Arabidopsis GH3 enzymes for both indole and phenyl carboxyl donors, as well as for amino acids, suggests these enzymes could synthesize a diversity of amide conjugates in plants. Some of these might directly or indirectly influence IAA signaling pathways in ways that alter normal growth and development when they overaccumulate. It is relevant to repeat here that synthesis of the JA-Ile conjugate by a JA-amido synthetase is required for some jasmonate responses, whereas several other JA conjugates appear to be inactive (Staswick and Tiryaki, 2004). Thus JA-amido synthetases may play a dual role, both activating JA and removing it from the free JA pool. The same could be true of IAA-amido synthetases.
Unlike Arabidopsis dfl1-D, tobacco plants expressing iaaL were not dwarf, but actually had increased internode lengths (Romano et al., 1991; Spena et al., 1991). Because GH3.6 does not appear to synthesize IAA-Lys appreciably, this may reflect differences in the way plants respond to overproduction of specific conjugates, or it could indicate species differences in their response to perturbed auxin balance.
IAA-Amido Synthetase Substrate Specificities
Convincing evidence discussed earlier indicates that IAA-Ala and IAA-Leu can serve as IAA storage reserves, whereas IAA-Asp and IAA-Glu irreversibly remove IAA from the active pool. Therefore, regulation of which conjugates are formed would appear to be an important mechanism determining the fate of IAA in plants. The endpoint reactions analyzed by TLC suggested that IAA-Asp was a major conjugate formed by GH3.6 and several other enzymes we tested. By contrast, of the four Arabidopsis hydrolases examined by LeClere et al. (2002), none showed particularly strong activity on IAA-Asp. This is consistent with the evidence that this conjugate does not serve as a significant reservoir of IAA in plants, but provides one important entry into the oxidative degradation pathways for IAA (Östin et al., 1998). Maximal activity for GH3.6 was approximately pH 8.5 to 9 for the formation of IAA-Asp, and no activity was detected at pH 7.5 or below. It is presently unclear which pH is biologically relevant for these enzymes. IAA-Asp was synthesized by GH3.6 at 244 nmol IAA-Asp produced/min/mg enzyme, which is comparable to the rate of hydrolysis found for the GST fusion of ILL2 on certain IAA-amino acid conjugates (LeClere et al., 2002).
The IAA-amido synthetases formed several conjugates in vitro that have not been reported to occur in plants. In addition to those previously identified (IAA-Asp, -Glu, -Ala, -Gly, -Val, and -Leu), amino acids Phe, Trp, Tyr, Met, Ile, and Gln were effective substrates for several enzymes. However, these conjugates may not accumulate significantly in Arabidopsis (Kowalczyk and Sandberg, 2001) because their accumulation would also depend on the availability of individual amino acids, the activity of hydrolases, and the degradation rate for specific conjugates. It will be important to further examine whether any of the novel conjugates do occur, and if so, what is their abundance and role in IAA metabolism. Unlike bacterial iaaL, which is active on basic amino acids (Roberto et al., 1990), the Arabidopsis enzymes showed little activity on Lys and Arg, nor on Asn, Pro, His, Ser, or Thr, consistent with the fact that these conjugates have not been found in plants.
In contrast with JAR1 (GH3.11), which is specific for JA (Staswick and Tiryaki, 2004), the enzymes tested here were not specific for the carboxyl donor IAA. Several other indoles, as well as nonindoles like NAA and PAA, were effective in the adenylation assay (Figure 2). On the other hand, we found no evidence that these synthetases are active on JA or other plant hormones. By contrast, the IAR3 hydrolase apparently cleaves some JA as well IAA conjugates (LeClere et al., 2002), possibly providing a link between the regulation of JA and IAA activity.
In general, the synthetase activity on specific substrates agreed with the resistance to these compounds seen in dfl1-D. In addition to the IAA resistance that was previously reported (Nakazawa et al., 2001), dfl1-D was less sensitive to IBA and NAA. Although we demonstrated that IBA-amino acid conjugates are formed by GH3.6 in vitro, we cannot rule out the possibility that IBA is converted to IAA in vivo (Ljung et al., 2002), and it is the conjugation of IAA that confers resistance to IBA in dfl1-D. The simplest explanation for resistance to NAA is the ability of GH3.6 to act directly on this substrate. It is not clear why dfl1-D showed wild-type sensitivity to PAA and IPyA, despite the fact that these were effective substrates for adenylation by GH3.6. The expression level of GH3.6 in dfl1-D may be insufficient to metabolize the relatively high concentrations of PAA and IPyA needed for root inhibition. Alternatively, conjugates with these substrates may be efficiently metabolized after their synthesis.
Inactivity on Halogenated Auxins
Although 4-Cl-IAA-Asp occurs in plants (Bandurski et al., 1995), the enzymes we tested showed only weak activity on 4-Cl-IAA. This agreed with the fact that dfl1-D exhibited no increased resistance to this halogenated IAA. If conjugation to amino acids is an important way for plants to remove excess auxin from the active pool, then the low activity of the IAA-amido synthetases on the 4-Cl analog may help to explain why this auxin is 10- to 100-fold more active than IAA in some bioassays (Bandurski et al., 1995; Reinecke et al., 1995). The chlorinated synthetic auxin herbicides 2,4-D and dicamba also showed little activity in our enzyme assays. Consistent with this, dfl1-D was not resistant to 2,4-D, and the insertion mutants were not more sensitive to this auxin. Possibly the inactivity of GH3 enzymes on halogenated auxin-like herbicides contributes to their efficacy as plant herbicides.
In summary, the identification of a previously unknown family of IAA-amido synthetases is an important advance in our understanding of the biochemical mechanisms of IAA metabolism and its effect on auxin activity. Consistent with earlier suggestions, we have provided evidence that the synthesis of IAA-amino acid conjugates plays an important role in IAA homeostasis. Several GH3s are regulated by light or are involved in light-regulated growth (Nakazawa et al., 2001; Tepperman et al., 2001; Hagen and Guilfoyle, 2002; Tanaka et al., 2002; Takase et al., 2004), indicating that conjugating IAA to amino acids may be an important mechanism to control growth in response to light. Although the apparent functional redundancy and broad substrate specificities of the GH3 enzymes complicates our ability to understand their roles, targeted disruption of multiple genes by RNA interference strategies and by pyramiding multiple insertion mutants should be useful future approaches. A more detailed characterization of GH3 activation tagged mutants should also help to reveal their overall role in auxin metabolism and function.
METHODS
Plant Material, Growth Conditions, and Sequence Analysis
Seedlings were grown on agar media containing the inhibitors indicated using surface-sterilized seeds as previously described (Staswick et al., 2002). Plates containing seeds were incubated at 4°C for 4 d, then placed vertically in an incubator at 23°C, and grown under 12-h-fluorescent-light/12-h-dark cycles for the times indicated. Root lengths for inhibitor treatments were expressed as percentage of inhibition relative to the untreated control for each genotype. Confidence intervals (95%) were calculated using the delta method. Each assay was done three times with similar results. Number of lateral roots was determined on 11-d-old seedlings. Root growth rate in the absence of added hormone was determined by measuring growth over 72 h, beginning after 3 d of growth. Tissue for auxin isolation was grown in soil in a growth chamber for 4 weeks, and excised leaves were frozen immediately and stored at −80°C. Alternatively, surface-sterilized seeds were grown aseptically on 3MM gel blot paper (Whatman, Maidstone, UK) supported nearly vertically by a glass plate placed in a Magenta box that contained 50 mL of liquid MS basal-salt medium. Seedlings were grown in an incubator under the conditions described above.
Insertion mutants were obtained from the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre. The presence of insertions was verified by amplifying genomic border DNA flanking each end of the inserts, using methods previously described (Staswick et al., 2002). Primer pairs for transposon inserts (Parinov et al., 1999) were Ds3′-1A plus 5′-GCGGTAGACTCCAACCTCTCTTC-3′ and Ds5′-1A plus 5′-TTATCTCCTTCTCTCCGGCG-3′ for SGT4174 (GH3.2); Ds3′-1A plus 5′-CTAACGACGTCGTTCTGG-3′ and Ds5′-1A plus 5′-ATGGCCGTTGATTCACCTC-3′ for SGT5949 (GH3.1). SALK 14376 and SALK 050597 (Alonso et al., 2003) apparently contained tandem inverted T-DNA inserts because the left border primer Lba1 (http://signal.salk.edu/T-DNArecovery.pdf) yielded products of the expected size from both ends, whereas right border primers did not yield products from either end. Gene primers were 5′-CGGAAAGAGAGAAAA-3′ for the upstream and 5′-CGATCCTGTTGATCTCAGGC-3′ for the downstream side of the insert in GH3.5 and 5′-TTCAACATCCTTCAAGCCTC-3′ for the upstream and 5′-CGAAAAAGAGAGGGAGACAAAG-3′ for the downstream side of the insert in GH3.17. Sequencing of PCR products confirmed the presence of the expected genomic borders at each end. Homozygous lines for each insertion mutant were identified by confirming the presence of the insert in all of at least 12 progeny in the subsequent generation.
For phylogenetic analysis, GH3-related plant sequences were identified by BlastP searches (P value ≤ 10−20) of the nonredundant translated database using Arabidopsis thaliana JAR1 (GH3.11) as the query sequence. Nonredundant sequences that were judged to be full length based on comparison with Arabidopsis and soybean (Glycine max) GH3 sequences were used for the analysis. Sequence alignment was done with ClustalW using the Gonnet series matrix (Thompson et al., 1994). Minor adjustments to the alignment were done manually, and phylogenetic relationships were determined using the neighbor-joining method with bootstrap analysis (1000 replicates). The tree was displayed with TreeView 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html).
Isolation and Quantitation of Auxins
IAA-amino acid conjugates were from Sigma-Aldrich (St. Louis, MO). [13C6] IAA was purchased from Cambridge Isotope Laboratories (Andover, MA), and [13C6] IAA-Asp was a gift from J. Cohen. Solvents were HPLC grade, and chloroform contained 1% ethanol as stabilizer. The general purification methods adapted from Kramell et al. (2000) for JA conjugate quantitation were followed (Staswick and Tiryaki, 2004), with the modifications described here. Weighed frozen tissue (∼200 mg) was added to 7 mL of 80% MeOH with known amounts of 13C standards as internal controls. Elution from DEAE ion exchange was with 21 mL of MeOH containing 12% (v/v) of 88% formic acid. Residue was dissolved in 0.2% acetic acid and loaded onto a SPE RP C18 column (Burdick and Jackson, Muskegon, MI; 500 mg, 8 mL) equilibrated in the same. The column was washed with 6 mL of 0.2% acetic acid and then eluted with 6 mL of 80% MeOH, 0.2% acetic acid. Derivatization was with 2,3,4,5,6-pentafluorobenzyl bromide as described (Epstein and Cohen, 1981), followed by silica gel SPE chromatography. IAA and IAA-Asp were fractionated by reverse-phase HPLC on a 250 × 4.6-mm Luna 5-μm C18 column (Phenomenex, Torrance, CA) with isocratic 80% MeOH, 0.2% acetic acid (1 mL/min). Dried fractions were resuspended in chloroform for analysis on an HP6890 gas chromatograph with a DB-5 ht column (15 m × 0.25 mm, 0.1 μm) from J.W. Scientific (Folsom, CA). The injector port was at 280°C and the column gradient was 60 to 300°C in 25 min. Detection was with an HP5973 EI mass selective detector operated in SIM mode. Quantitative data was obtained from integrated peak areas for the 130 and 136 ions as described by Cohen et al. (1986). For identification of conjugates produced by in vitro reactions using recombinant GH3.6 enzyme, reactions were acidified with acetic acid and extracted with chloroform, followed by drying and derivatization with diazomethane. GC-MS analysis was then done in scan mode under conditions specified above.
Enzyme Assays
The GH3 GST-fusion proteins were described previously (Staswick et al., 2002). Isotope exchange assays were performed as previously described (Staswick et al., 2002) using glutathione agarose-bound enzyme at ∼0.17 ug/uL. Radioactivity in ATP and PPi was determined by cutting the appropriate area from the poly(ethyleneimine) TLC sheet and counting by liquid scintillation. To assure that enzymes did not synthesize appreciable levels of adenosine dinucleoside polyphosphates (Plateau and Blanquet, 1992) that could interfere with the chromatographic analysis, reactions were tested on silica gel 60 F260 TLC plates (EM Sciences, Darmstadt, Germany) developed in water:dioxane:ammonia (4:6:1, v/v) (Guranowski et al., 1990). No product other than ATP was found, at a sensitivity of 1% of the total radioactivity loaded for each sample. Assays for conjugate formation were done at room temperature in 50 mM Tris-HCl, pH 8.6, 3 mM MgCl2, 3 mM ATP, 1 mM DTT, 1 mM IAA, and 1 mM amino acid. Reactions were analyzed on silica gel 60 F260 plates developed in chloroform:ethyl acetate:formic acid (35:55:10, v/v), except that 2-propanol:ammonium hydroxide:water (8:1:1, v/v) was used for reactions with amino acids His, Ser, Thr, Arg, Lys, and Cys. Staining for indoles was done with Van Urk-Salkowski reagent (Ehmann, 1977). Quantitative analysis of GH3.6 activity was done with enzyme cleaved from GST with thrombin according to the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). Assays contained 0.33 mg/mL of enzyme in the same reaction mixtures as above, except that pH was varied as indicated. Reactions were done at room temperature and aliquots assayed for the IAA-Asp peak area by HPLC. The linear correlation coefficient over the 5-, 10-, and 15-min time points was ≥0.985 at each of the three pH levels yielding data and the reported values were calculated from the activity between 5 and 15 min.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge J. Cohen for providing the [13C6] IAA-Asp and for his invaluable advice concerning auxin quantitation. M. Nakazawa generously provided the Arabidopsis dfl1-D mutant. This is Journal Series Paper 14710, a contribution of the University of Nebraska Agricultural Research Division, Lincoln, NE 68583. Funding was provided by the Nebraska Research Initiative and the National Science Foundation (Award MCB-0130868).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Paul E. Staswick (pstaswick1@unl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026690.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Andrea, W.A., and Good, N.E. (1955). The formation of indoleacetyl aspartic acid in pea seedlings. Plant Physiol. 30, 380–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandurski, R.S., Cohen, J.D., Slovin, J.P., and Reinecke, D.M. (1995). Auxin biosynthesis and metabolism. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 118–139.
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 51–66. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Fink, G. (1995). ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748. [DOI] [PubMed] [Google Scholar]
- Bartel, B., LeClere, S., Magidin, M., and Zolman, B.K. (2001). Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 20, 198–216. [Google Scholar]
- Bialek, K., and Cohen, J.D. (1986). Isolation and partial characterization of the major amide-linked conjugate of indole-3-acetic acid from Phaseolus vulgaris L. Plant Physiol. 80, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek, K., Meudt, W.J., and Cohen, J.D. (1983). Indole-3-acetic acid (IAA) and IAA conjugates applied to bean stem sections. IAA content and growth response. Plant Physiol. 73, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, J.C., Kuleck, G.A., Cohen, J.D., and Mulbry, W.W. (1996). Partial purification and characterization of an inducible indole-3-acetyl-L-aspartic acid hydrolase from Enterobacter agglomerans. Plant Physiol. 112, 1281–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.D., Baldi, B.G., and Slovin, J.P. (1986). 13C6-[benzene ring]-indole-3-acetic acid. A new internal standard for quantitative mass spectral analysis of indole-3-acetic acid. Plant Physiol. 80, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.D., and Bandurski, R.S. (1978). The bound auxins: Protection of indole-3-acetic acid from peroxidase-catalyzed oxidation. Planta 139, 203–208. [DOI] [PubMed] [Google Scholar]
- Cohen, J.D., and Bandurski, R.S. (1982). Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 33, 403–430. [Google Scholar]
- Cooney, T.P., and Nonhebel, H.M. (1989). The measurement and mass spectral identification of indole-3-pyruvate from tomato shoots. Biochem. Biophys. Res. Commun. 162, 761–766. [DOI] [PubMed] [Google Scholar]
- Costacurta, A., and Vanderleyden, J. (1995). Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 21, 1–18. [DOI] [PubMed] [Google Scholar]
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann, A. (1977). The van Urk-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. 132, 267–276. [DOI] [PubMed] [Google Scholar]
- Epstein, E., and Cohen, J.D. (1981). Microscale preparation of pentafluorbenzyl esters. Electron-capture gas chromatographic detection of indole-3-acetic acid from plants. J. Chromatogr. 209, 413–420. [Google Scholar]
- Epstein, E., and Ludwig-Müller, J. (1993). Indole-3-butyric acid in plants: Occurrence, synthesis, metabolism and transport. Physiol. Plant. 88, 382–389. [Google Scholar]
- Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guranowski, A., Sillero, M.A., and Sillero, A. (1990). Firefly luciferase synthesizes P1,P4-bis(5′-adenosyl)tetraphosphate (Ap4A) and other dinucleoside polyphosphates. FEBS Lett. 271, 215–218. [DOI] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T.J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T.J. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Hangarter, R.P., Peterson, M.D., and Good, N.E. (1980). Biological activities of indoleacetylamino acids and their use as auxins in tissue culture. Plant Physiol. 65, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska, A., Kowalczyk, S., and Leznicki, A.J. (1993). Enzymatic hydrolysis of 4-O and 6-O-indole-3-ylacetyl-β-D-glucose in plant tissues. J. Plant Physiol. 142, 61–66. [Google Scholar]
- Klee, H., and Estelle, M. (1991). Molecular genetic approaches to plant hormone biology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 529–551. [Google Scholar]
- Klee, H.J., and Lanahan, M.B. (1995). Transgenic plants in hormone biology. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 340–353.
- Kleinkauf, H., and Von Dohren, H. (1996). A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236, 335–351. [DOI] [PubMed] [Google Scholar]
- Klezkowski, K., and Schell, J. (1995). Phytohormone conjugates: Nature and function. Crit. Rev. Plant Sci. 14, 283–298. [Google Scholar]
- Kowalczyk, M., and Sandberg, G. (2001). Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis thaliana. Plant Physiol. 127, 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Kramell, R., Miersch, O., Atzorn, R., Parthier, B., and Wasternack, C. (2000). Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol. 123, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere, S., Tellez, R., Rampey, R.A., Matsuda, S.P.T., and Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Kowalczyk, M., Marchant, A., Celenza, J., Cohen, J.D., and Sandberg, G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y.Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Normanly, J. (1997). Auxin metabolism. Physiol. Plant. 100, 431–442. [Google Scholar]
- Normanly, J., Cohen, J.D., and Fink, G.R. (1993). Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 90, 10355–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östin, A., Kowalyczk, M., Bhalerao, R., and Sandberg, G. (1998). Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 118, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plateau, P., and Blanquet, S. (1992). Synthesis of NpnN′ (n = 3 or 4) in vitro and in vivo. In Ap4A and Other Dinucleoside Polyphosphates, A.G. McLennan, ed (Boca Raton, FL: CRC Press), pp. 63–79.
- Rampey, R.A., LeClere, S., Kowalczyk, M., Ljung, K., Sandberg, G., and Bartel, B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke, D., Ozga, J.A., and Magnus, V. (1995). Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry 40, 1361–1366. [Google Scholar]
- Roberto, F.F., Klee, H., White, F., Nordeen, R., and Kosuge, T. (1990). Expression and fine structure of the gene encoding N epsilon-(indole-3-acetyl)-L-lysine synthetase from Pseudomonas savastanoi. Proc. Natl. Acad. Sci. USA 87, 5797–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, C.P., Hein, M.B., and Klee, H.J. (1991). Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 5, 438–446. [DOI] [PubMed] [Google Scholar]
- Roux, C., and Perrot-Rechenmann, C. (1997). Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3 related to GH3. FEBS Lett. 419, 131–136. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S., and Koshiba, T. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32, 1011–1022. [DOI] [PubMed] [Google Scholar]
- Sitbon, F., Hennion, S., Sundberg, B., Little, C.H.A., Olsson, O., and Sandberg, G. (1992). Transgenic tobacco plants coexpressing the Agrobacterium tumefaciens iaaM and iaaH genes display altered growth and indoleacetic acid metabolism. Plant Physiol. 99, 1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovin, J., Bandurski, R., and Cohen, J. (1999). Auxin. In Biochemistry and Molecular Biology of Plant Hormones, Vol. 33, P. Hooykaas, M. Hall, and K. Libbenga, eds (Amsterdam: Elsevier Science), pp. 115–140.
- Spena, A., Prinsen, E., Fladung, M., Schulze, S.C., and Van Onckelen, H. (1991). The indoleacetic acid-lysine synthetase gene of Pseudomonas syringae subsp. savastanoi induces developmental alterations in transgenic tobacco and potato plants. Mol. Gen. Genet. 227, 205–212. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen, J.B., Szczyglowski, K., and Bandurski, R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701. [DOI] [PubMed] [Google Scholar]
- Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N., Shimada, H., Manabe, K., and Matsui, M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483. [DOI] [PubMed] [Google Scholar]
- Tam, Y.Y., Epstein, E., and Normanly, J. (2000). Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol. 123, 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, Y.Y., and Normanly, J. (1998). Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J. Chromatogr. A. 800, 101–108. [DOI] [PubMed] [Google Scholar]
- Tam, Y.Y., and Normanly, J. (2002). Overexpression of a bacterial indole-3-acetyl-L-aspartic acid hydrolase in Arabidopsis thaliana. Physiol. Plant. 115, 513–522. [DOI] [PubMed] [Google Scholar]
- Tanaka, S., Mochizuki, N., and Nagatani, A. (2002). Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 43, 281–289. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin regulated gene expression. Plant Cell 14, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venis, M. (1972). Auxin-induced conjugation systems in peas. Plant Physiol. 49, 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, F., and Thimann, K.V. (1980). Hormonal factors controlling the initiation and development of lateral roots. I. Physiol. Plant. 49, 13–20. [Google Scholar]
- Wright, R.M., Hagen, G., and Guilfoyle, T. (1987). An auxin-induced polypeptide in dicotyledonous plants. Plant Mol. Biol. 9, 625–634. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Dai, X., Blackwell, H.E., Schreiber, S.L., and Chory, J. (2003). SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301, 1107–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.