Abstract
Approximately 5% of the Arabidopsis (Arabidopsis thaliana) proteome is predicted to be involved in the ubiquitination/26S proteasome pathway. The majority of these predicted proteins have identity to conserved domains found in E3 ligases, of which there are multiple types. The RING-type E3 is characterized by the presence of a cysteine-rich domain that coordinates two zinc atoms. Database searches followed by extensive manual curation identified 469 predicted Arabidopsis RING domain-containing proteins. In addition to the two canonical RING types (C3H2C3 or C3HC4), additional types of modified RING domains, named RING-v, RING-D, RING-S/T, RING-G, and RING-C2, were identified. The modified RINGs differ in either the spacing between metal ligands or have substitutions at one or more of the metal ligand positions. The majority of the canonical and modified RING domain-containing proteins analyzed were active in in vitro ubiquitination assays, catalyzing polyubiquitination with the E2 AtUBC8. To help identity regions of the proteins that may interact with substrates, domain analyses of the amino acids outside the RING domain classified RING proteins into 30 different groups. Several characterized protein-protein interaction domains were identified, as well as additional conserved domains not described previously. The two largest classes of RING proteins contain either no identifiable domain or a transmembrane domain. The presence of such a large and diverse number of RING domain-containing proteins that function as ubiquitin E3 ligases suggests that target-specific proteolysis by these E3 ligases is a complex and important part of cellular regulation in Arabidopsis.
The covalent attachment of the 76-amino acid protein ubiquitin to target proteins is a central and essential part of numerous cellular processes in eukaryotes. Ubiquitin attachment is a multistep reaction involving three enzymes referred to as E1, E2, and E3 (Glickman and Ciechanover, 2002). The conjugation cascade begins with the formation of thioester-linked ubiquitin by the ubiquitin-activating enzyme or E1 in an ATP-dependent reaction. Thioester-linked ubiquitin is then transferred to a cysteinyl residue of an ubiquitin-conjugating (UBC) enzyme or E2. Ubiquitin is then transferred from the E2-ubiquitin intermediate to a lysyl group on the substrate. This final step is mediated by the ubiquitin ligase or E3 enzyme. Uniquely, E3 ligases of the HECT domain class form a ubiquitin thioester prior to transfer to the substrate, whereas all the other ligases interact noncovalently with the E2 carrying a thioester-linked ubiquitin. The E3 is also responsible for recruiting the target protein for ubiquitination, and hence is considered the major substrate recognition component of the pathway.
Ubiquitin-dependent protein degradation selectively targets a diverse range of substrates, including receptors, nuclear transcription activators and repressors, abnormal proteins, and other short-lived regulatory proteins for degradation by the 26S proteasome (Glickman and Ciechanover, 2002). Ubiquitin modifications also regulate proteins in a proteasome-independent manner and have been implicated in the activation of signaling proteins, endocytosis, and histone modification (Schnell and Hicke, 2003). The fate of the modified substrate is determined by the nature of the ubiquitin linkage (Weissman, 2001). Ubiquitination may result in the attachment of a single (monoubiquitination), a few, or many ubiquitins (polyubiquitination). The choice of the Lys residue used to form the ubiquitin chain also influences the fate of the ubiquitinated protein (Weissman, 2001). For example, the major fate of Lys-48 polyubiquitinated proteins is degradation by the 26S proteasome, whereas protein modification with a polyubiquitin Lys-63 chain has been implicated in protein activation (Deng et al., 2000).
More than 5% of the predicted Arabidopsis (Arabidopsis thaliana) proteome is postulated to be involved in the ubiquitination/26S proteasome pathway (Smalle and Vierstra, 2004). The E1 enzyme (of which there are 2 isoforms), the 37 predicted E2s, the components of the 26S proteasome, and other factors such as deubiquitinating enzymes account for only a small portion of the proteins that are thought to be involved in this process. With more than 1,300 genes predicted to encode for E3 components, ubiquitin ligases account for the majority of the proteins involved in the ubiquitnation/26S pathway (Smalle and Vierstra, 2004). The large number and diversity of E3 ligases confer specificity upon the ubiquitination pathway. The level of specificity is further increased by the different possible E2-E3 combinations, thus allowing not only for the attachment of different types of ubiquitin conjugates but also for the specific regulation of a large number of target proteins.
The Arabidopsis E3 ligases can be grouped into three defined classes based on the presence of the HECT, U-box, or RING domain (Smalle and Vierstra, 2004). The RING-type E3s can be further subdivided into simple and complex E3s, with the latter class including the anaphase-promoting complex and the well-characterized, multisubunit Skp1-Cullin-F-box (SCF)-type ligase (Smalle and Vierstra, 2004). In SCF complexes, the F-box protein is the substrate recognition subunit and the Arabidopsis genome is predicted to encode for approximately 700 F-box proteins (Gagne et al., 2002; Kuroda et al., 2002; Risseeuw et al., 2003). The RING-containing protein of SCF complexes, Rbx/Roc/Hrt, recruits the E2-ubiquitin intermediate to the SCF complex (Kuroda et al., 2002; Lechner et al., 2002; Risseeuw et al., 2003). By contrast, the simple RING E3 ligases contain both the substrate-binding domain and the E2-binding RING domain in a single protein, or as a homodimer or heterocomplex with another RING protein.
The Cys-rich RING domain was first identified in a protein encoded by the Really Interesting New Gene, hence the domain name (Freemont et al., 1991). The RING domain is similar to the zinc finger in that Cys and/or His residues coordinate two zinc ions (referred to here as metal ligand residues). However, in contrast with the known DNA-binding zinc finger domains, the RING domain appears to function as a protein-protein interaction domain (Lovering et al., 1993; Borden, 2000). The RING domain is distinct from other zinc fingers in that the eight metal ligand residues coordinate the zinc ions in a cross-brace structure (Barlow et al., 1994; Borden et al., 1995; Borden, 2000). While both of the two previously characterized RING domains contain a His at metal ligand position 4, the C3HC4 and C3H2C3 types (referred to here RING-HC and RING-H2, respectively) differ in the presence of either a Cys or His residue, respectively, at metal ligand position 5 (Freemont, 1993; Lovering et al., 1993). These two RING domain types were found to be essential for catalyzing E3 ligase activity of RING-containing proteins (Lorick et al., 1999).
The Arabidopsis genome is predicted to encode for a considerable number of RING proteins of which very little is known (Kosarev et al., 2002). RING proteins with predicted or known biological function include SINAT5 (auxin signaling), COP1 and COP1-interacting protein 8 (CIP8; photomorphogenesis), ATL2 (plant defense), BRH1 (brassinosteroid responsive/pathogen response), TED3/AtPex2p (light signaling), RMA1 (secretory pathway), and RIE1 (seed development; Matsuda et al., 2001; Hardtke et al., 2002; Holm et al., 2002; Molnar et al., 2002; Xie et al., 2002; Xu and Li, 2003; Serrano and Guzman, 2004). E2-dependent ubiquitin ligase activity has been demonstrated and targets identified for only a few RING proteins, including RMA1, PRT1, COP1, CIP8, and SINAT5 (Matsuda et al., 2001; Hardtke et al., 2002; Holm et al., 2002; Xie et al., 2002; Stary et al., 2003).
The large number of potential RING proteins and other types of E3 ligases suggests that target specific ubiquitination plays an important role in protein regulation in Arabidopsis. However, there is little evidence demonstrating that the Arabidopsis family of RING domain-containing proteins functions as E3 ligases. Therefore, in this study, we carried out an extensive search of the Arabidopsis genome to identify potential RING-containing proteins and tested whether representative family members possess E3 ligase activity. In addition to the canonical RING domains, the search also led to the identification of a number of modified RING domains. Here, we also describe the domain organization of each RING and modified RING domain-containing protein. Finally, we provide biochemical evidence that a significant number of the 469 RING and modified RING proteins identified have the capacity for E2-dependent protein ubiquitination and that the RING domain is required for this activity.
RESULTS
Predicted RING Domain-Containing Proteins in Arabidopsis Can Be Divided into Three RING Types and Five Modified RING Types
Our analysis of the translated Arabidopsis genome resulted in a final collection of 477 RING domains in 469 predicted proteins containing one or more of the various types of RING domains (Fig. 1A). A complete list of all RING domain-containing proteins identified can be found in Supplemental Table I. RING domain-containing proteins were identified by database searches via InterPro (release v7.2), which compiles the results from multiple domain-searching programs. A number of additional loci were identified using reiterative BLAST searches of the nonredundant Arabidopsis genome (The Arabidopsis Information Resource [TAIR] database, release date April 16, 2003; Rhee et al., 2003) using the RING domains of known proteins. However, the domains retrieved by these searches did not necessarily meet all the criteria that define a RING-HC or RING-H2 domain (Borden, 2000). Therefore, each potential RING domain was analyzed manually for the presence of, and the distance between, each of the eight zinc-coordinating Cys and/or His residues. RING domains that lacked one or more of the defining characteristics were further analyzed so as to detect possible modified RING domains (Fig. 1A). One such modified RING domain, the RING-v domain, was identified by the Simple Modular Architecture Research Tool (SMART) and named for RING-variant domain (Fig. 1A).
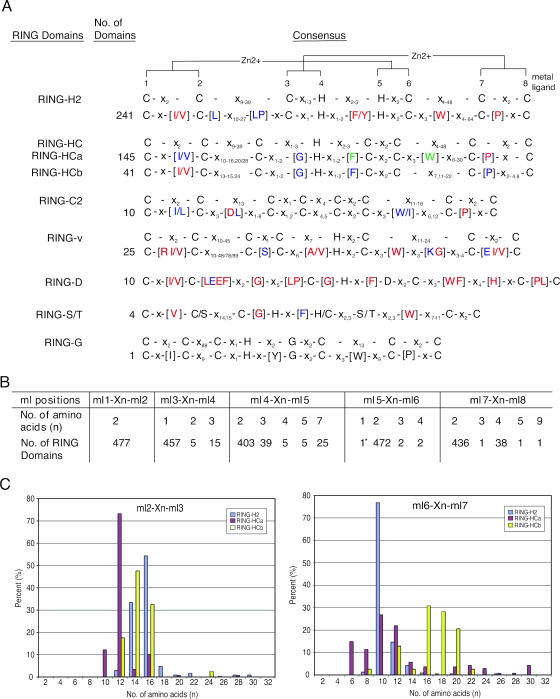
Figure 1.
Arabidopsis RING and modified RING domains. A, Consensus and number of each type of RING domain identified in Arabidopsis. Consensus previously characterized canonical RING domains (RING-H2 and RING-HC) and modified RING domains (RING-C2, RING-v, and RING-G) are shown above that of Arabidopsis. The conserved metal ligand positions and zinc (Zn2+) coordinating amino acid pairs are illustrated using the RING-H2 consensus. The eight conserved Cys (C) and His (H) residues of each RING type and metal ligand substitutions in the RING-D, RING-S/T, and RING-G domains are shown. Other conserved amino acids are shown in brackets. For each RING type, amino acids in red are found in ≥80% of domains, and blue and green in ≥50% and ≤50% of domains, respectively. X(n) indicates number of spacing amino acids between conserved amino acids. B, Spacing variation observed between zinc-coordinating metal ligand (ml) pairs in Arabidopsis RING domains. Number of amino acids between each metal ligand and the number of domains in which the spacing variation is found are shown. *, Spacing unique to RING-HCa protein At2g42160, metal ligand position 5 assignment uncertain. C, Comparison of the number of amino acids in the loops between metal ligands 2 and 3 and metal ligands 6 and 7 of RING-H2, RING-HCa, and RING-HCb domains.
The 477 RING domains were classified into one of eight RING types based on the nature of the metal ligand residues present and/or the number of amino acids between them (Fig. 1A). The largest was the RING-H2 type with 241 domains, followed by the RING-HC type with 186 domains (Fig. 1A). The RING-H2 class includes AtRBX1a and AtRBX1b, which, as observed in other Rbx/Roc/Hrt proteins, has an Asp residue at metal ligand position 8 instead of a Cys residue and a number of additional Cys residues between metal ligands 2 and 3 that have been shown to form a third zinc ion-binding site (Supplemental Fig. 1; Kamura et al., 1999; Chen et al., 2000; Zheng et al., 2002). This variation is unique to the Rbx family of proteins and is not observed in any other RING-H2 protein (data not shown). The RING-HC class was divided into two subgroups: the RING-HCa and RING-HCb, with 145 and 41 domains, respectively (Figs. 1A and 2). All RING-HCb domains contain additional amino acids between metal ligand residues C7 and C8 (Fig. 1A; Supplemental Table I). The division of the RING-HC class is further supported by the difference in the range of amino acids found between metal ligands 2 and 3 and metal ligands 6 and 7 (Fig. 1C). More than 70% of the RING-HCa domains contain 12 amino acids between metal ligands 2 and 3, whereas only approximately 18% of RING-HCb domains have 12 amino acids between these metal ligands. The majority of RING-HCb domains have 14 to 16 amino acids between metal ligands 2 and 3. In addition, the number of amino acids between metal ligands 6 and 7 of the RING-HCb domain is greater than that of the RING-HCa (Fig. 1C). Finally, when the entire amino acid sequences of RING-HCb proteins are compared, these proteins share additional predicted protein-protein interaction domains (Supplemental Table I), again suggesting a closer relationship among them than to RING-HCa proteins. The RING-HCb group includes members of the ARIADNE (ARI) protein family described previously (Mladek et al., 2003).
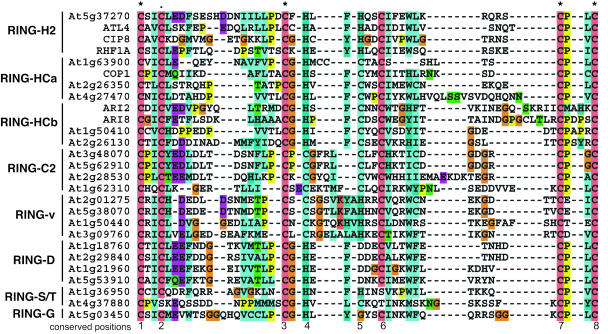
Figure 2.
Multiple sequence alignment of representative Arabidopsis RING domains. Sequences are grouped according to RING domain type. Conserved metal ligand positions are numbered. Arabidopsis Genome Initiative (AGI) loci/code of putative proteins and names of known proteins are shown. Multiple sequence alignment of all RING domains identified can be found in Supplemental Figures 1 and 3. Dashes (-) denote gaps introduced to maximize the alignment, asterisks (*) indicate conserved residues, and the black dot indicates residue conserved in all RING types except for RING-S/T amino acids.
The modified RING types represent only 11% of the total predicted RING domains identified (Fig. 1A). Previously described modified RING domains found in Arabidopsis include the RING-v, RING-C2, and RING-G types. The RING-v domain (Hewitt et al., 2002) is characterized by a cysteinyl residue at metal ligand position 4 and a histinyl residue at metal ligand position 5, rather than the reverse found in RING-HC domains. In addition, there are seven amino acids between metal ligands 4 and 5 as opposed to two to three in RING-HC and RING-H2 domains. Two of the RING-v domains have a larger number of amino acids between metal ligands 2 and 3 than found previously (Supplemental Fig. 3). Twenty-five predicted RING-v domains were identified in Arabidopsis. The two other modified RING domains previously described in other organisms, RING-G (Dasgupta et al., 2004) and RING-C2 (Albert et al., 2002), have metal ligand substitutions, a glycyl residue for C5 and a cysteinyl residue for H4, respectively (Fig. 1A). Both of these modified RING domain types can be found in the predicted Arabidopsis proteome, with 1 and 10 members, respectively (Table I; Supplemental Table I). The Arabidopsis RING-C2 domains differ from the previous RING-C2 consensus (Albert et al., 2002), with a more variable region between metal ligands 2 and 3, metal ligands 3 and 4, and metal ligands 4 and 5 (Fig. 1A). Two previously unrecognized modified RING domains, RING-D and RING-S/T types, were identified in the Arabidopsis predicted proteome. The RING-D domain present in 10 proteins contains an Asp residue at metal ligand 5, whereas the RING-S/T type in 4 proteins contains either a Ser or Thr at one or both metal ligand positions 2 and 6.
Table I.
Representative Arabidopsis RING and modified RING domain-containing proteins
E2 AtUBC8 was used to determine E3 ligase activity. Representative Arabidopsis proteins that contain RING and modified RING domains are listed. Please see Supplemental Table I for a complete list of all Arabidopsis RING and modified RING proteins. AGI Loci/Code, Unique gene identification number. RING Type, Indicates the type of RING domain found in each protein. The different types of RING domains are illustrated in Figure 1A. Group No., Specifies the group to which each protein belongs, as determined by domain organization. See Table II for a description of each group. E3 Activity, In vitro ubiquitin ligase activity of selected proteins was tested using Arabidopsis AtUBC8; yes, polyubiquitination; no, no polyubiquitination observed (see Fig. 4). a.a., Amino acids.
| AGI Loci/Code | RING Type | Group No. | E3 Activity | Comments |
|---|---|---|---|---|
| At1g02860a | HCa | 23 | Nob | |
| At1g10650 | HCa | 1 | Noc | 3 a.a. between metal ligands 4 and 5 |
| At1g12760 | H2 | 24 | Yesc | |
| At1g14260 | v | 24 | Yesc | |
| At1g15100d | H2 | 25 | Yese | RHA2a |
| At1g18760f | D | 1 | Yesc | |
| At1g18780f | D | 25 | Noc | |
| At1g22500 | H2 | 24 | Yese | ATL5-like |
| At1g49210 | H2 | 24 | Yesi | |
| At1g50440 | v | 1 | Yesb | |
| At1g60610 | HCa | 6 | Noe | 3 a.a. between metal ligands 4 and 5 |
| At1g61620 | HCa | 1 | Noe | |
| At1g63900 | HCa | 1 | Yese | 3 a.a. between metal ligands 4 and 5 |
| At1g65430g | HCb | 11.1 | Yesh | ARI8 |
| At1g68180 | H2 | 24 | Yesc | |
| At1g74370 | HCa | 24 | Yese | |
| At1g74410 | H2 | 24 | Noe | |
| At1g74760d | H2 | 1 | Nob | |
| At2g15580 | H2 | 1 | Noe | |
| At2g18670 | H2 | 24 | Noe | |
| At2g22680 | H2 | 27.1 | Yesc | |
| At2g22690 | S/T | 15 | Nob | |
| At2g28530a | C2 | 20 | Yesh | 4 a.a. between metal ligands 4 and 5 |
| At2g28840 | HCa | 4.1 | Noi | |
| At2g34000 | D2 | 25 | Yesc | |
| At2g35000 | H2 | 24 | Yese | |
| At2g42360 | H2 | 24 | Yesc | |
| At2g44330a | H2 | 1 | Yesc | |
| At2g47700 | H2 | 1 | Yesi | |
| At3g05200 | H2 | 24 | Yesi | ATL6-like |
| At3g05250 | HCa | 1 | Yesh | |
| At3g05545 | H2 | 1 | Yesh | |
| At3g06330a | v | 24 | Yesb | |
| At3g09760 | v | 24 | Yesb | |
| At3g09770 | HCa | 1 | Yese | 3 a.a. between metal ligands 4 and 5 |
| At3g16720 | H2 | 24 | Nob | ATL2 |
| At3g19140 | S/T | 24 | Noc | |
| At3g23280d | HCa | 4.1 | Yesh | 3 a.a. between metal ligands 4 and 5 |
| At3g29270 | HCa | 24 | Yesi | |
| At3g45555 | HCb | 11.1 | Noc | |
| At3g47160 | HCa | 1 | Yesh | |
| At3g48070a | C2 | 1 | Yesh | |
| At3g60220 | H2 | 24 | Yese | ATL4 |
| At3g60300 | H2 | 1 | Noh | |
| At4g02075 | v | 24 | Yesb | |
| At4g10160 | H2 | 24 | Yesh | |
| At4g11360 | H2 | 1 | Yese | RHA1b |
| At4g11680 | H2 | 24 | Yesb | |
| At4g14220 | H2 | 1 | Yese | RHF1a |
| At4g14365 | HCa | 4.1 | Noh | 3 a.a. between metal ligands 4 and 5 |
| At4g21070 | HCa | 5 | Yesh | |
| At4g23450 | H2 | 1 | Yese | |
| At4g27470 | HCa | 24 | Yese | |
| At4g28890 | H2 | 24 | Yese | |
| At4g39140 | H2 | 1 | Noe | |
| At5g01520 | HCa | 1 | Noe | |
| At5g07270 | HCa | 4.1 | Yesh | |
| At5g14420 | HCa | 27.2 | Yesi | |
| At5g20910 | H2 | 6 | Yese | ABI3-interacting protein 2 |
| At5g22920 | H2 | 1 | Noe | |
| At5g37270 | H2 | 1 | Yesc | |
| At5g38070a | v | 24 | Yesb | |
| At5g42200 | H2 | 25 | Yesc | |
| At5g53910df | D | 1 | Yesc |
RING domains previously determined to be false positive due to a difference in ligand spacing.
ORF was isolated by RT-PCR from a mixture of RNA from leaves, seedlings, and flowers (from 6- to 7-week-old plants).
ORF isolated from leaf (from 2- to 4-week-old plants) DNA.
Previously thought not to be a RING domain due to the presence of an aromatic residue two amino acids upstream of metal ligand 7.
ORF was isolated by RT-PCR from leaves of 2- to 4-week-old plants.
Previously characterized as incomplete RING/metal ligand at position 4 or 5 missing or substituted.
Proteins with RING domain previously characterized as incomplete/metal ligand at position 6 or 7 missing or substituted.
ORF was isolated by RT-PCR from 10-d-old seedlings.
EST clones obtained from ABRC.
Characteristic of RING domains is the spacing between metal ligand residues, with short distances between all except between metal ligands 2 and 3 and 6 and 7. For all RING types, the most frequent spacing between the amino acids of each zinc-coordinating pair is two between position metal ligands 1 and 2 (100%), one between metal ligands 3 and 4 (96%), two between metal ligands 5 and 6 (99%), and two between metal ligands 7 and 8 (91%; Fig. 1B). The spacing variation observed between the last zinc-coordinating pair, metal ligands 7 and 8, is due to the expanded spacing in the 41 RING-HCb types (with 3–9 amino acids). Only 84% of the RING domains have 2 amino acids between metal ligands 4 and 5 (Fig. 1B). This lowest percentage of conserved number of amino acids is because all 25 RING-v domains have 7 amino acids, the RING-C2 domains differ with 4 or 5 amino acids, and 39 of the RING-HCa domains have 3 amino acids in this region (Fig. 1B; Supplemental Table I).
For each Arabidopsis RING type, the length of the variable loops between metal ligands 2 and 3 and metal ligands 6 and 7 showed some differences compared to the known RING domains (Fig. 1A). For example, the previously published consensus for the RING-H2 domain has 9 to 39 and 4 to 48 amino acids between metal ligands 2 and 3 and metal ligands 6 and 7, respectively (Borden, 2000). However, the majority of the Arabidopsis RING-H2 have only 14 or 15 amino acids between metal ligands 2 and 3 and only 10 amino acids between metal ligands 6 and 7 (Figs. 1, A and C, and 2). The Arabidopsis RING-G domain does not have the large spacing present between metal ligands 2 and 3 of the yeast (Saccharomyces cerevisiae) San1 protein, the defining member of this type (Dasgupta et al., 2004; Fig. 1A).
Eight proteins were found to contain multiple RING domains (Supplemental Table I). The Arabidopsis ARI proteins, though previously characterized as having two RING domains (Mladek et al., 2003), were not included in this category because the second RING domains were missing one or more metal ligand amino acids, lacked consensus spacing, and failed to fall into one of the modified RING types. Our decision to exclude the second RING domain in the ARI proteins is supported by data demonstrating that only the first RING domain of mammalian Ariadne proteins has been shown to be required for E2 interaction and ubiquitin ligase activity (Moynihan et al., 1999; Ardley et al., 2001). In addition, the structure of the second RING domain was recently shown to be a distinct zinc-binding domain, which only chelates one zinc ion (Capili et al., 2004). Furthermore, a number of Arabidopsis ARI-related proteins do not contain a second RING domain (Supplemental Tables I and II; RING group 11.1).
Several loci with incomplete RING domains in the publicly released annotation were included in this study because analysis of the genomic sequence indicated that the lack of a complete RING domain might be due to an error in the splicing or premature translation stop predictions. No complementary DNA (cDNA) sequences are available for these loci (GenBank, July 2004), and reverse transcription (RT)-PCR experiments failed to amplify products that could be sequenced to determine the endogenous splicing pattern (data not shown). However, gene prediction programs produce a spliced version that is predicted to encode for proteins with complete RING domains with significant similarity to other predicted RING proteins (Supplemental Table III). Conversely, using this same approach, we were unable to detect a complete RING domain at a number of other loci, even though the predicted proteins were related to a particular family of RING proteins (Supplemental Table IV).
BLAST searches of the translated Arabidopsis genome also identified RING domain-encoding sequences in unannotated regions of the genome (Supplemental Table V). RING domains that were previously classified as false positive or as having an incomplete RING were also analyzed (Table I; Supplemental Table I; Kosarev et al., 2002). A significant number of these domains were included in our collection because they belonged to a particular RING type or they were found to possess E2-dependent ubiquitin ligase activity (Table I). For example, the RING-HCb and RING-D domains were previously characterized as incomplete and the RING-v domains were characterized as false positives. Previously characterized proteins such as HOS1 (Lee et al., 2001) and CER3 (Hannoufa et al., 1996) were not included in our collection because the RING domain failed to meet a number of defining criteria (Supplemental Table VI).
The RING-D Domain May Be Specific to Arabidopsis
Except for the RING-D, all other seven types of RING domains identified were also found in the genomes of other organisms. BLAST searches using representative RING-D domains did not retrieve a similar type of domain from any other genome, including other plants such as Oryza sativa. This suggests that this type of modified RING domain is taxonomically more restricted. By contrast, proteins similar to the Arabidopsis RING-S/T-containing proteins (Table II, RING group 15) were found in other species such as yeast, human, and Plasmodium falciparum. However, the RING-S/T domains of many of these proteins were missing metal ligand position 5 or 6 (data not shown). The RING-G domain identified in the yeast San1 (Schnell et al., 1989; Dasgupta et al., 2004) protein has only one representative in Arabidopsis and other species such as Drosophila melanogaster.
Table II.
Arabidopsis RING domain-containing protein groups based on domain presence and organization
| Group No. | No. of Proteina | Additional Domainsb | Domain Description and/or Predicted Function | Examplec | Speciesd |
|---|---|---|---|---|---|
| 1 | 140 | None | Zinc-binding domain; protein-protein interactions; ubiquitin ligase domain (Lorick et al., 1999) | At2g47700 | At, Bn, Ce, Dm, Hs, Mm, Os, Sc, Sp, Xl, and Zm |
| 2.1 | 3 | PHD | Plant homeodomain C4HC3 type; found in proteins involved in chromatin-mediated transcriptional regulation; probable ubiquitin ligase domain (Aasland et al., 1995; Capili et al., 2001; Lu et al., 2002) | At3g05670 | At, Ce, Dm, Hs, Mm, Os, Sc, Sp, and Xl |
| 2.2 | 1 | ZNF_ZZ | Zinc-finger domain C-x2-C-x5-C-x2-C type; found in Dystophin and CREB-binding protein (Ponting et al., 1996) | At3g24800 | At |
| 3 | 4 | AAA ATPase | ATPase associated with diverse cellular activities; contains an ATP-binding site (Neuwald et al., 1999) | At2g44950 | At |
| 4.1 | 6 | ANK | Ankyrin repeats; protein-protein interaction domain (Bork, 1993) | At5g07270 | At, Ce, Dm, Hs, Mm, Os, and Xl |
| 4.2 | 1 | Protein kinase | Ser/Thr and tyrosine protein kinase; protein phosphorylation | At5g13530 | At |
| 5 | 2 | BRCT | Breast cancer carboxy terminus domain; phospho-protein-binding domain; found in proteins involved in cell cycle checkpoint or DNA damage repair (Callebaut and Mornon, 1997; Yu et al., 2003) | At4g21070 | At, Ce, Hs, Mm, Os, and Xl |
| 6 | 20 | Coiled coil | Protein-protein interaction/oligomerization motif made up of two to five α-helices (Burkhard et al., 2001) | At5g47050 | At, Bn, Ce, Dm, Hs, Mm, Os, Sc, Sp. and Xl |
| 7 | 2 | CUE | Ubiquitin-binding motif; found in proteins involved in ubiquitination (Kang et al., 2003; Shih et al., 2003) | At5g51450 | At, Ce, Hs, Mm, and Xl |
| TM | Transmembrane domain | ||||
| 8.1 | 10 | DEXDc | DEAD-like helicases; ATP-dependent helicase activity; found in proteins involved in DNA damage repair (Rocak and Linder, 2004) | At3g16600 | At, Hs, Mm, Os, Sc, and Sp |
| 8.2 | 2 | HELICc | Helicase superfamily C-terminal domain; found in a number of helicases (Rocak and Linder, 2004) | At5g10370 | At |
| 8.3 | 1 | KH | K homology RNA-binding domain; first identified in hnRNP K; proteins usually contain at least two copies (Ostareck-Lederer et al., 1998) | At3g54460 | At and Os |
| HA2 | Helicase-associated domain; found in a variety of RNA helicases and may be involved in nucleic acid binding | ||||
| F box | F-box domain found in proteins that are the substrate-binding component of the SCF E3 ligase; interacts with SKP1 (Kipreos and Pagano, 2000) | ||||
| IBR | See group 11.1 | ||||
| ZNF_C2H2 | See group 29.1 | ||||
| 9 | 1 | HHE | Function unknown; found as tandem repeats in plant, bacteria, and yeast proteins; implicated in cell wall physiology (Brunskill et al., 1997) | At3g18290 | At and Os |
| 10 | 1 | HIG_1_N | Hypoxia induced protein conserved region; found at the N terminus of TM proteins predicted to be involved in the response to hypoxia (Gracey et al., 2001) | At3g48030 | At |
| TM | Transmembrane domain | ||||
| 11.1 | 29 | IBR | In between ring fingers; Cys-rich region (C6HC) usually found between two RING domains; function unknown. (van der Reijden et al., 1999) | At3g14250 | At, Dm, Hs, Mm, Os, and Sp |
| 11.2 | 1 | RWD | RING finger- and WD repeat-containing proteins and DEXDc-like helicases; shows similarity to the UBCc domain; protein interaction domain. (Doerks et al., 2002) | At1g32340 | At, Ce, Hs, Mm, Os, and Sp |
| 12 | 5 | JmjC | Jumonji C domain; predicted to be cupin metalloenzyme that may regulate chromatin remodeling (Clissold and Ponting, 2001) | At1g62310 | At, Mt, and Ps |
| 13 | 2 | KISc | Kinesin motor catalytic domain; involved in cell division and organelle transport (Block, 1998) | At2g21380 | At and Os |
| Coiled coil | See group 6 | ||||
| 14 | 1 | LON | ATP-dependent protease La (LON) domain; ATP-dependent Ser peptidases (Van Dyck et al., 1994) | At1g18660 | At, Dm, Hs, Mm, and Sp |
| 15 | 2 | LisH | Lissencephaly type-1 (Lis1)-like homology motif; may be involved in regulating microtubule dynamics (Reiner, 2000) | At2g22690 | At and Dd |
| CTLH | C-terminal to LisH motif; function unknown | ||||
| 16 | 6 | PA | Protease-associated domain; found in plant vacuolar sorting proteins and E3 ligases (Borchers et al., 2002) | At4g09560 | At, Dm, Hs, Mm, Os, Sp, and Xl |
| TM | Transmembrane domain | ||||
| 17.1 | 1 | Pep3_Vps18 | Region found in Pep3/Vps18 proteins; found in protein involved in Golgi function and vacuolar sorting; usually precedes a RING domain (Warner et al., 1998) | At1g12470 | At, Ce, Dm, Os, Sc, and Sp |
| 17.2 | 1 | Clathrin/CLH | Region in Clathrin and VPS; found as one or two repeats in vacuolar membrane proteins (Warner et al., 1998) | At1g08190 | At, Dm, Le, and Os |
| WD40 | See group 28 | ||||
| Coiled Coil | See group 6 | ||||
| 18 | 2 | PEX | Similar to the N-terminal region of Pex2/12 peroxisomal biogenesis proteins (Sparkes and Baker, 2002) | At1g79810 | At, Ce, Dd, Dm, Hs, Mm, Os, Sc, and Sp |
| 19 | 1 | PPR | Pentatricopeptide repeat; found as four or more tandem repeats in plant proteins (Small and Peeters, 2000) | At4g17910 | At |
| TM | Transmembrane domain | ||||
| 20 | 2 | RRM | RNA recognition motif; putative RNA-binding motif; found in proteins implicated in regulation of alternative splicing and transcriptional regulation (Birney et al., 1993) | At3g45630 | At, Dm, Hs, Mm, Os, and Xl |
| 21 | 13 | SINA | Seven in absentia protein family; similar to the Drosophila Sina E3 ligase (Hu et al., 1997) | At2g41980 | At, Bn, Ce, Dm, Hs, Mm, Os, and Xl |
| 22 | 1 | SPRY | Domain in SPla and the ryanodine receptor; function unknown (Ponting et al., 1997) | Atg2g22010 | At, Ce, Dm, and Hs |
| 23 | 2 | SPX | Domain named after SYG1/Pho81/XPR1 proteins; usually found at the amino terminus of a variety of proteins; function unknown. | At1g02860 | At, Os, and Sp |
| 24 | 107 | One/more TM | Transmembrane domains | At5g57750 | |
| 25 | 29 | Signal peptide | At1g76410 | ||
| 26 | 6 | SRA | SET- and RING finger-associated domain; found in SET domain-containing proteins; histone- and chromatin-binding domain (Citterio et al., 2004) | At5g39550 | At, Hs, Mm, and Os |
| PHD | See group 2.1 | ||||
| 27.1 | 9 | VWA N-terminal | von Willebrand factor type A domain; protein-protein interactions; found in cell adhesion and extracellular matrix/intracellular proteins (Whittaker and Hynes, 2002) | At2g22680 | At and Os |
| 27.2 | 5 | VWA C-terminal | At1g79380 | At and Os | |
| 28 | 2 | WD40 | Found as tandem repeats each containing a central Trp/Asp motif; Phospho-Ser/Thr-binding domain (Yaffe and Elia, 2001) | At1g21655 | At, Ce, Dm, Hs, Le, Mm, Os, Sc, and Xl |
| 29.1 | 4 | ZNF_C2H2 | Zinc-finger domain C-x2-C-x12-H-x3-H type; implicated in nucleic acid binding (Yaffe and Elia, 2001) | At1g77770 | At, Ce, Dm, Hs, Mm, Os, Sc, Sp, and Xl |
| 29.2 | 3 | ZNF_C3H1 | Zinc-finger domain C-x8-C-x5-C-x3-H type; nucleic acid binding (Worthington et al., 1996) | At5g06420 | At, Ce, Dm, Hs, Mm, Os, Ps, Sc, and Sp |
| 29.3 | 2 | ZNF_UBP | Ubiquitin C-terminal hydrolase-like zinc finger; found in ubiquitin-specific proteases and HDAC6; ubiquitin-binding domain (Seigneurin-Berny et al., 2001; Hook et al., 2002) | At2g42160 | At, Ce, Dm, Hs, Mm, Os, Sc, Sp, and Xl |
| Coiled coil | See group 6 | ||||
| 30 | 3 | DUF1117 | Domain of unknown function | At5g59550 | At and Os |
Numbers do not include the RING-v domain-containing and RING-D domain-containing proteins. These types of RING proteins either did not contain any other known/detectable domains or were found to contain predicted transmembrane domains.
Domains found in RING proteins as determined by SMART, Pfam, or Prosite databases.
See Supplemental Table II for schematic of representative Arabidopsis proteins from each group.
Species that have RING proteins with similar domain architecture. BLAST searches and SMART and Pfam databases were used to determine the species distribution of each RING group. At, Arabidopsis; Bn, Brassica napus; Ce, C. elegans; Dd, Dictyostelium discoideum; Dm, D. melanogaster; Hs, human; Le, Lycopersicon esculentum; Mm, M. musculus; Mt, M. truncatula; Os, O. sativa; Ps, P. sativum; Sc, yeast; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis; Zm, Zea mays.
Other Residues in Addition to Metal Ligand Amino Acids Are Conserved in RING Domains
Amino acid sequences of representative RING domains from each type are shown in Figure 2. An alignment of all RING domains, except for the RING-v type, can be found in Supplemental Figure 1. The RING-v domains were analyzed separately due to the high sequence similarity within the group and spacing variations (Supplemental Fig. 3). Close examination of the multiple sequence alignment identified a number of amino acids, outside of the metal ligand residues, that are fairly conserved (Figs. 1A and 2). In the majority of RING-H2, RING-HCa/b, RING-v, and RING-D domains, an Ile or Val precedes metal ligand 2. More than 80% of all RING-H2 and RING-D domains have a Phe or Tyr residue in front of metal ligand 5. The Phe is also often present in front of metal ligand 5 of the RING-HCa/b domains. The RING-v domain has an Ala or Val instead of a Phe in front of metal ligand 5. These amino acids are less conserved or rarely found in the RING-C2 domain. In all RING types, except for the RING-HCb and RING-C2, a Trp is usually the fourth amino acid C-terminal to metal ligand 6. Interestingly, the Pro residue present right after metal ligand 7 in all other RING domain types is absent in RING-v proteins. Within the RING-v domain, the amino acid residues observed between metal ligands 7 and 8 are usually Glu followed by Ile or Val. Within the RING-v and RING-D groups, other amino acids also are well conserved (Figs. 1A and 2). Notably, the amino acid after the first metal ligand of the RING-v domain is almost always an Arg. Also, between metal ligand positions 6 and 7, the following amino acids are well conserved: C6-x3-[W]-x3-[KG]-x3-4-C7. A Glu and Ile/Val are observed between metal ligands 7 and 8 in more than 50% and 80% of the RING-v domains, respectively. We also observed that the amino acid immediately following the C8 position of all RING domains is usually a Lys or Arg (data not shown).
Additional Protein-Protein Interaction Domains in RING Proteins
To define related groups of RING proteins, we analyzed a phylogenetic tree generated using the various types of RING domains. Domains of similar RING type clustered together, although within each RING type no large clade of related RING domains was observed (Supplemental Fig. 2). The RING-H2, RING-HCa, RING-HCb, RING-C2, and RING-S/T domains were grouped into small clades containing two to eight domains. The RING-D domains form a single distinct clade (Supplemental Fig. 2). The RING domains consistently fell into the same clades after several independent trials of tree generation. The results of the phylogenetic analysis suggest that, though the RING domains share several key features, outside of the conserved metal ligand residues the RING proteins are very distinct.
To further define the RING proteins, we examined each protein for the presence of other known domains. Domain predictions resulted in the identification of 30 groups of RING proteins based upon domain presence and organization (Table II; Supplemental Table II). With the exception of a few groups, such as groups 1 (contains no known domain) and 24 (contains predicted transmembrane domain[s]), the number of proteins within each group is small, with some groups containing only one representative. RING groups with larger numbers usually contain proteins encoded by genes that are arranged in tandem repeats. For example, RING group 11.1 contains two sets of tandem repeated genes on chromosomes 2 and 3, containing three and six genes, respectively (Table II; Supplemental Table I). Overall, there are more than 20 groups of tandemly repeated genes, containing two to six loci that encode for RING-containing proteins.
Proteins within each distinctive group or subgroup contain the same type of RING domain. For example, all the RING domains of subgroup 27.1 are of the RING-H2 type, and those of subgroup 27.2 are of the RING-HCa type (Table I). The only obvious difference between these two subgroups is the position of the RING domain within the protein (Table II; Supplemental Table II). Even more striking is the fact that the majority of domains of the multiple RING-containing proteins are of the RING-HCa type (Supplemental Table I). For example, PRT1 (At3g24800) contains two RING-HCa domains. The findings from our domain analysis are reflected in the phylogenetic tree, where RING domains of proteins within a particular group tend to clade together (Supplemental Fig. 2). This gives further support to the relationships defined by the RING domain phylogenetic tree.
The RING proteins are predicted to contain a surprisingly diverse array of previously described domains. The majority of these are protein-protein interaction domains, which may function as the substrate-binding domain of the E3 ligase (Table II). Predicted protein interaction domains include Ankyrin repeats, BRCT, vWA, coiled-coil domains, and WD40 repeats. Thirteen of the RING groups or subgroups were found only in plant genomes. For example, proteins containing a RING domain in combination with PPR repeats, vWA, or JmjC domains were found only in the Arabidopsis proteome or also in other plant species, such as O. sativa, Medicago truncatula, or Pisum sativum (for domain definitions, see Table II). The most prevalent type of RING proteins are those predicted to contain a transmembrane domain (group 24). Another 29 RING proteins are predicted to contain only a signal peptide (group 25). Three types of zinc fingers were found associated with the RING domain: ZNF_C2H2 (group 29.1), ZNF_C3H1 (group 29.2), and ZNF_ZZ (group 2.2). Both ZNF_C2H2 and ZNF_C3H1 are characterized as nucleic acid-binding zinc fingers. ZNF_ZZ is thought to mediate protein-protein interactions. RING proteins were also found to contain the ubiquitin-binding domains CUE (group 7) or ZNF_UBP (group 29.3). Other domains associated with ubiquitination were also identified. For example, a single DEXDc/RING/HELICc protein (group 8.3) contains an F-box domain, and another was found to contain a PHD domain (group 2.1). The RWD domain, which shows homology to the catalytic domain of UBCs but lacks the conserved Cys required for activity, was also found associated with the RING domain (group 11.2).
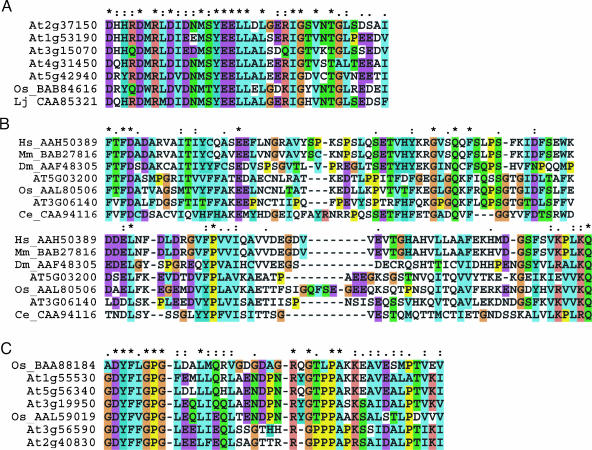
A significant number of RING proteins did not contain any other detectable, previously described domains (Table II; group 1). Sequence analysis of these RING proteins revealed regions of similarity between a few proteins outside of the RING domain. These putative domains are referred to as DAR1, DAR2, and DAR3 (for domain associated with RING). Representative domains are shown in Figure 3, and a complete list of all putative DAR domains identified is presented in Supplemental Figure 4. DAR1 and DAR3 are approximately 40 amino acids in length, and DAR2 consists of more than 120 amino acids. BLAST searches with the putative DAR domains retrieved predicted proteins from other species with similar regions (Fig. 3; Supplemental Fig. 4). DAR1 and DAR3 were found only in O. sativa and/or Lotus japonicus predicted proteins, whereas DAR2 was found in predicted proteins from a number of genomes, including human, Caenorhabditis elegans, D. melanogaster, and O. sativa. A number of the DAR3-containing Arabidopsis proteins show a low level of similarity to CIP8 (data not shown). In almost all proteins, the putative DAR domains were found associated with a RING domain. In these cases, the DAR domain was found N-terminal to the RING domain.
Figure 3.
Sequence alignment of potential novel domains detected in RING proteins. The amino acid sequence of representative DAR1 (A), DAR2 (B), and DAR3 (C) domains from Arabidopsis are compared to those found in other species. A complete list of DAR-containing RING proteins can found in Supplemental Figure 4. Species abbreviations: Os, O. sativa; Lj, L. japonicus; Hs, human; Dm, D. melanogaster; Mm, M. musculus; and Ce, C. elegans. AGI loci/code of Arabidopsis proteins and accession numbers for all other putative proteins are given. Dashes (-) denote gaps introduced to maximize the alignment, asterisks indicate conserved residues, and “:” and single black dots indicate semiconserved amino acids.
Arabidopsis RING Domains Possess E3 Ubiquitin Ligase Activity
RING proteins are involved in numerous cellular processes including transcription, signal transduction, and recombination. Functions attributed to the RING domain itself include protein-protein interaction and more recently protein ubiquitination (Lorick et al., 1999; Borden, 2000). Although sequence analysis can be used to identify RING domains, it does not provide proof that a RING domain-containing protein will function as an E3 ubiquitin ligase. To determine if representatives from the Arabidopsis family of RING proteins had the capacity for E2-dependent protein ubiquitination, we used an in vitro ubiquitination assay (Hardtke et al., 2002) to test for activity in 64 RING and modified RING-containing proteins. RING domain-containing proteins representative of the different subgroups that were used in biochemical analyses are shown in Table I. Each glutathione S-transferase (GST)-tagged full-length RING protein was tested in an in vitro ubiquitination assay consisting of recombinant yeast E1, 6×His-tagged recombinant Arabidopsis E2 AtUBC8, and ubiquitin. More than 70% of the RING proteins tested were capable of mediating polyubiquitination, as evident by the high molecular smear of ubiquitinated proteins detected by western-blot analysis using anti-ubiquitin antibodies (Table I; Fig. 4). Omission of the GST-RING protein, AtUBC8, or ubiquitin from the activity assay resulted in a loss of protein ubiquitination (Fig. 4). AtUBC8 catalyzes a low and variable level of E3-independent ubiquitination, presumably to itself (Fig. 4, −E3 lanes). Thus, a significant number of the GST-tagged RING proteins are capable of mediating E2-dependent protein ubiquitination in vitro.
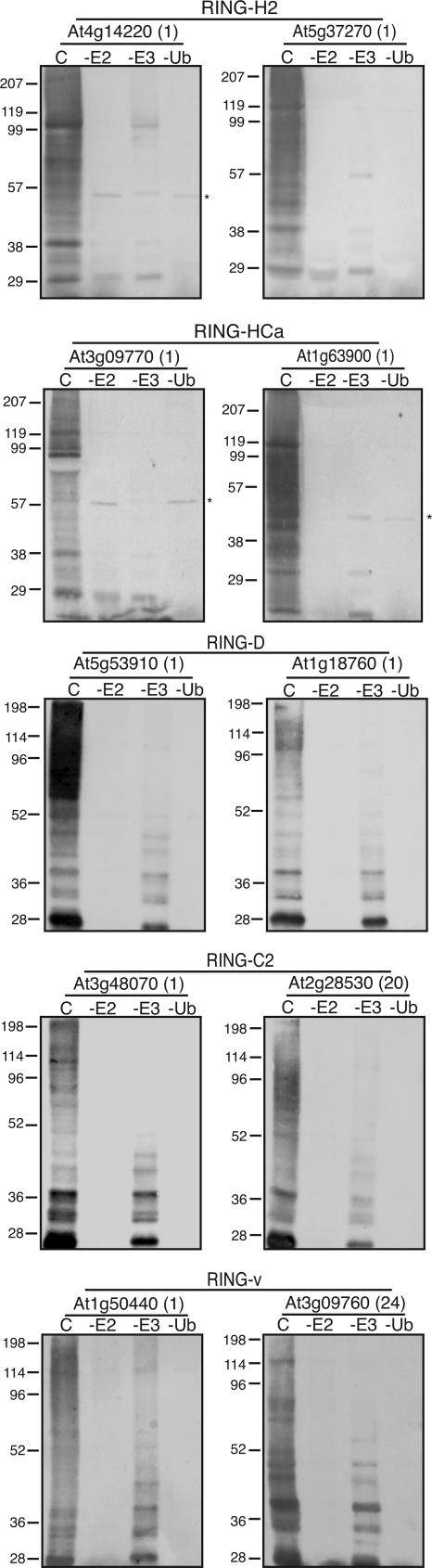
Figure 4.
Analysis of E3 ligase activity of representative RING proteins. GST-tagged full-length RING-H2-, RING-HCa-, RING-HCb-, RING-C2-, RING-D-, and RING-v-containing proteins are capable of mediating protein ubiquitination in an E2-dependent manner. Complete (C) in vitro ubiquitination assays contained recombinant yeast E1 enzyme, recombinant 6×His-tagged Arabidopsis E2 enzyme AtUBC8, and ubiquitin. Omission of AtUBC8 (−E2 lanes), GST-RING protein (−E3 lanes), or ubiquitin (−Ub lanes) from the assay resulted in a loss of protein polyubiquitination. Ubiquitinated proteins were visualized via western-blot analysis using ubiquitin antibodies. AGI loci/code and RING type for each protein analyzed are indicated above each blot. Numbers in parenthesis indicate the group (see Table II) to which the RING protein belongs. Position and size of molecular mass markers are shown to the left of each blot. *, Indicates nonspecific proteins that cross-react with the ubiquitin antibody. Proteins detected in the −E3 lanes represent AtUBC8-Ub(n).
We tested the biochemical activity of representative full-length proteins from each RING and modified RING type. GST-tagged RING-H2, RING-HCa, RING-HCb, RING-v, RING-C2, and RING-D domain-containing proteins were all found to possess E2-dependent E3 ligase activity in vitro, whereas we were unable to detect ligase activity for the RING-S/T domain-containing proteins (Fig. 4; Table I). Twenty-two of the 29 GST-RING-H2 proteins analyzed were active in the in vitro assay. Of the 19 RING-HCa types tested, polyubiquitination was not observed for 8 (Fig. 4; Table I). RING-HCb protein At1g65430/ARI8 was active in the in vitro assay. However, no polyubiquitination was observed for the other RING-HCb protein, At3g45555, tested. Two of the four RING-S/T proteins identified were tested, and no E3 ubiquitin ligase activity was observed for either GST-RING-S/T protein (Table I). Of the six GST-RING-v proteins tested, all were capable of promoting polyubiquitination (Fig. 4; Table I). Four RING-D proteins were chosen for biochemical analysis. Ubiquitin ligase activity was observed for three of the four GST-RING-D proteins (Fig. 4; Table I). Both GST-RING-C2 proteins analyzed were active in the in vitro ubiquitination assay (Fig. 4). The RING-G-containing protein was not analyzed. However, the RING-G-containing San1 protein from yeast has been shown to mediate protein ubiquitination in vitro (Dasgupta et al., 2004).
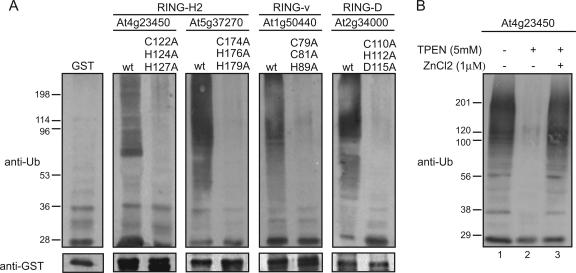
To test the requirement of the RING and modified RING domains for ubiquitin ligase activity, well-conserved amino acids in the RING domains of selected proteins were mutated and the mutated proteins assayed for ubiquitination activity. Mutation of one or more of the metal ligands has been shown previously to disrupt the ability of the RING domain to promote protein ubiquitination (Lorick et al., 1999). The amino acids at metal ligand positions C3, H4, and H/C5 of the RING-H2 proteins At4g23450 and At5g37270 and RING-v protein At1g50440 were mutated to Ala, as were the amino acids at positions C3, H4, and D5 of the RING-D protein At2g34000. GST-tagged full-length RING domain-containing proteins bearing these amino acid changes had a greatly diminished ability to promote protein ubiquitination (Fig. 5A). Thus, the RING and modified RING domains of each protein are necessary for the E2-dependent protein ubiquitination observed. Treatment of RING proteins with zinc chelators such as N,N,N′,N′-tetrakis(2-pyridylmethyl) ethylenediamine (TPEN) has also been shown to disrupt the ubiquitin ligase activity of the RING domain (Fang et al., 2000). To confirm the results obtained by the mutagenesis study, a zinc-chelating experiment was carried out using At4g23450. The TPEN-treated GST-RING-H2 domain-containing protein lost the ability to promote protein ubiquitination (Fig. 5B, lane 2). However, ubiquitin ligase activity was regained once the TPEN-treated RING protein was incubated with zinc chloride (Fig. 5B, lane 3). Thus, these Arabidopsis RING proteins require an intact zinc-coordinating RING domain to mediate E2-dependent protein ubiquitination.
Figure 5.
An intact RING domain is required for the protein ubiquitin ligase activity of the Arabidopsis RING proteins. A, Mutation of conserved metal ligand positions C3, H/C4, and H/C/D5 to an Ala abrogates the E3 ligase activity of the full-length RING-H2-, RING-D-, and RING-v-containing proteins. AGI loci/code and RING type for representative proteins used are indicated above each blot. Except for At2g34000 that belongs to RING group 25, all RING proteins used are from group 1 (see Table II). wt, Wild-type protein. B, Effects of cation chelation on full-length RING-H2 protein (At4g23450) E3 ligase activity. Treatment of the GST-RING protein with 5 mm TPEN abolished E3 activity. Activity was regained after the TPEN-treated protein was incubated with zinc chloride.
DISCUSSION
Previous analysis of the RING-containing protein family reported that the Arabidopsis genome encodes for 387 RING and modified RING domains (Kosarev et al., 2002). Our analysis identified 90 additional RING domains. Therefore, the Arabidopsis genome contains at least 469 genes encoding for RING domain-containing proteins. The RING genes account for approximately 2% of the predicted protein-encoding genes of Arabidopsis. Unlike the F-box family, which contains twice as many proteins, the size of the Arabidopsis RING family is comparable to the numbers found in other eukaryotes such as human and Mus musculus, which contain 385 and 305 RING domains, respectively (Semple, 2003). The D. melanogaster and C. elegans genomes are predicted to encode for 121 and 170 RING proteins, respectively, and the yeast genome encodes for 14. Although many RING proteins are unique, the number of RING domains encoded by tandemly repeated genes suggests that the size of the Arabidopsis RING family may be due in part to gene duplication. Therefore, the RING family contains both unique and tandemly duplicated members. However, functional studies could uncover unique roles for closely linked genes.
A variety of protein-protein interaction domains, ubiquitin and nucleic acid-binding domains, and a diverse array of domains with unknown function were found associated with the RING domain. A number of these domains are also found in functional RING E3 ligases from other eukaryotes. For example, the domain organization of the Arabidopsis CUE-containing RING proteins is similar to the mammalian transmembrane receptor gp78, an endoplasmic reticulum E3 ligase (Fang et al., 2001). The RWD domain is found in the mammalian E3 ligase AO7 (Lorick et al., 1999; Doerks et al., 2002). The Arabidopsis PA-containing RING protein is similar to the E3 ligase GREUL1 involved in Xenopus development, the D. melanogaster GOLIATH protein family, and the human GRAIL E3 ligase involved in regulation cytokine gene transcription (Borchers et al., 2002; Anandasabapathy et al., 2003). The RING-RRM combination is present in the yeast CNOT4 E3, which is a component of the CCR4-NOT transcription repressor complex (Albert et al., 2002). Domains found associated with the RING are also found in other E3 ligases; for example, the JmjC, WD40, and DEAD-like helicases are present in Arabidopsis F-box proteins (Gagne et al., 2002). The diverse array of domains found associated with RINGs may link ubiquitination to a variety of cellular processes.
The domain organization of a number of RING-containing proteins is plant-specific. The Znf_ZZ-containing multiple RING domain-containing E3 ligase protein PRT1, which is involved in the N-end rule ubiquitination pathway, is only detected in the Arabidopsis genome (Potuschak et al., 1998; Stary et al., 2003). Other RING-containing proteins with domain combinations specific to plant species were also found in this study. For example, vWA, KISc, and PPR containing RING proteins were only detected in plant genomes. The PPR domain is present mainly in plant proteins (Small and Peeters, 2000), while the vWA, KISc, and Znf_ZZ domains are widely dispersed among different species (Ponting et al., 1996; Block, 1998; Whittaker and Hynes, 2002). However, the association of these domains with the RING domain seems to be limited to plants. This suggests that the domain combination of these RING proteins has biological roles that are specific to plants.
Almost 500 proteins representing 8 types of RING domains were identified. Five of the eight types identified are modified RING domains that display variation in spacing between, or have amino acids substitutions at, conserved zinc-coordinating residues. Biochemical analysis of representative RING domain-containing proteins from each RING type demonstrates that certain modifications do not affect E3 ligase activity in vitro, whereas others seem to abrogate E3 activity. As shown here and in previous studies, RING domains utilizing a Cys at all metal ligand positions (RING-C2) and domains with spacing variations (RING-v) are functional E3 ligase domains (Albert et al., 2002; Hewitt et al., 2002). All the RING-D-containing proteins tested here were capable of mediating protein ubiquitination. Therefore, an Asp substitution at metal ligand position 5 does not disrupt ubiquitin ligase activity. RING-D domains were not found in any other species, suggesting that the in vivo ubiquitination reaction in which this RING variation functions may be specific to Arabidopsis. A glycyl residue at metal ligand position 5 is also an acceptable substitution, as observed with the RING-G domain-containing San1 protein (Dasgupta et al., 2004). It should also be noted that the U-box ubiquitin ligase domain only has a few of the conserved Cys/His residues found in the RING domain and does not coordinate zinc ions (Andersen et al., 2004; Mudgil et al., 2004). However, hydrogen bonds and salt bridges formed by residues at metal ligand positions allow the U-box domain to have a similar structure to that of the RING domain. Other functional variations on the RING include the RING-H2 domain of the Rbx/Roc/Hrt protein, which is the E2-recruiting subunit of the SCF and other related E3 ligase complexes. In addition to having an Asp at metal ligand position 8, the Rbx/Roc/Hrt RING domain also coordinates a third zinc ion using amino acid residues in the loop between metal ligands 2 and 3 (Chen et al., 2000; Zheng et al., 2002). Human Rbx1 RING domain has a cross-brace structure similar to that of the canonical RING domains, and mutations within the third zinc-coordinating site do not affect protein function compared with mutations within the first or second zinc-binding sites (Kamura et al., 1999; Ohta et al., 1999). Thus, the significance of the third zinc ion-binding site is unknown.
The RING-S/T-containing proteins were not functional in our in vitro assays, indicating that Ser or Thr substitutions at metal ligand position 2 or 6 are not acceptable, at least in the context of this protein in combination with AtUBC8. We also failed to detect activity for the RING-S/T proteins in in vitro assays with other AtUBCs, such as AtUBC10 and AtUBC11, which are similar to AtUBC8, and AtUBC35 and AtUBC36 (S. Stone, E. Kraft, and J. Callis, unpublished data). The RING-S/T domains of the LisH and CTLH protein family may not represent a bona fide RING domain, or the RING may have lost the ability to function as an ubiquitin ligase domain. However, not all UBCs have been tested, and these RING domains could function with one specific E2. Alternatively, RING-S/T domains could require an additional protein/cofactor/modification that is not present in our in vitro assays. Similarly, a number of RING-H2- and RING-HCa/b-containing proteins analyzed also showed no activity in the biochemical assay. To function as E3 ligases in vitro, these proteins may also require cofactors or a specific type of E2 to facilitate protein ubiquitination. Therefore, we cannot dismiss these RING or modified RING proteins as nonfunctional or as not containing a bona fide RING domain.
In addition to the eight zinc-coordinating amino acids, other residues are important for the RING domain to function as an E3 ligase. For many RING domains, the amino acid preceding metal ligand position 2 is usually a hydrophobic amino acid, such as Leu, Ile, or Val. This hydrophobic amino acid at this position, located within the binding site of the first zinc ion, is utilized by E3 ligases CNOT4 (RING-C2), c-CBL (RING-HCa), and BRCA1 (RING-HCa) during E2 binding and is required for E3 activity, as mutation of this amino acid abrogates activity (Zheng et al., 2000; Albert et al., 2002; Brzovic et al., 2003). A hydrophobic amino acid at this position is conserved in the eight types of RING domains of Arabidopsis including the RING-S/T, for which no in vitro E3 ligase activity was observed. Trp-408 of c-CBL is also involved in E2 binding and is required for E3 ligase activity (Joazeiro et al., 1999; Zheng et al., 2000). The corresponding amino acids, Leu-51 of BRCA1 (RING-HCa), Ile-45 of CNOT4 (RING-C2), and Trp-269 of San1 (RING-G), have been shown to be required for E3 ligase activity and/or E2 interactions (Albert et al., 2002; Brzovic et al., 2003; Dasgupta et al., 2004). Except for the RING-HCb, this position is well conserved in all other Arabidopsis RING types identified. RING proteins harboring the RING-HCb domain are functional E3 ligases. For example, the Arabidopsis RING-HCb-containing protein, ARI8, and similar proteins such as HHAR1, a Parkin/Ariadne-like ubiquitin ligase, are functional ubiquitin ligases (Moynihan et al., 1999; Ardley et al., 2001). This suggests that this type of RING domain may utilize other amino acids during E2 binding and/or protein ubiquitination.
The information provided in this study demonstrates that the Arabidopsis RING-type family of E3 ligases is quite diverse, and, in addition to the previously defined types of RING domain, Arabidopsis utilizes a number of variations on the canonical RING domain. Therefore, searches for RING domain-containing proteins should not be limited to the known types. However, more characterization of the Arabidopsis RING types is needed so as to further define the requirements for a functional RING domain. The presence of the various types of RING domains found may represent another level of specificity within the ubiquitination pathway, in which each type of RING domain may function in vivo with a particular subset of the more than 37 Arabidopsis E2 enzymes (Bachmair et al., 2001). Further biochemical analysis utilizing the different families of Arabidopsis E2s to define functional E2-E3 combination would address this issue as well as give insight onto the requirements of specificity of E2-E3 interactions. The number of RING domain-containing proteins, the different types of RING domains, and the presence of a diverse array of protein-protein interaction domains in the RING proteins allude to a role for the RING-type E3 ligase in different cellular processes via the targeted regulation of numerous substrates. The family of RING-type E3 ligases described here adds to the emerging complexity of protein ubiquitination and degradation observed in Arabidopsis.
MATERIAL AND METHODS
Identification of Arabidopsis RING Domain-Containing Proteins
InterPro (Integrated Documentation Resource of Protein Families, release v7.2, March 29, 2004; http://www.ebi.ac.uk/interpro/) database was used to retrieve Arabidopsis (Arabidopsis thaliana) proteins containing a RING domain [InterPro domain IPR001841 (Znf_RING); Prosite domain PS00518 (ZF_RING_1); Pfam domain PF00097 (zf-C3HC4); SMART domain SM00184 (RING)]. RING domains of known Arabidopsis and mammalian RING proteins were used in BLAST searches against the complete nonredundant Arabidopsis genome (TAIR, April 16, 2003; http://www.arabidopsis.org). Retrieved sequences were analyzed by the SMART database (version 4.0, May 28, 2004; http://smart.embl-heidelberg.de/), followed by manual inspection. Each potential RING domain was analyzed manually for the presence of, and distance between, each of the eight zinc-coordinating Cys and/or His residues.
Database search for RING domains also retrieved proteins with related Cys-rich domains, such as the PHD (C4HC3) and LIM (C2HC4C/H; Capili et al., 2001). These Cys-rich domains typically lacked one or more of the defining characteristics of the canonical RING domains and were not considered further. Other identified Cys-rich domains that lacked one or more of the defining characteristics were further analyzed so as to detect possible modified RING domains. If a significant number of proteins containing a particular type of modified RING domain were found, the domain was used to define a new subgroup.
Sequence Analysis and Domain Definition
The ClustalX program using the PAM350 protein matrix, with gap opening and gap extension penalty parameters of 35.0 and 0.75, respectively, in pairwise alignment and 15.0 and 0.3, respectively, in the multiple alignments was used to generate the initial alignment of RING domains. Se-Al sequence editor (Evolutionary Biology Group, University of Oxford, UK) was used to manually edit the alignment. Unrooted phylogenetic trees were created by PAUP* (Phylogenetic Analysis Using Parsimony) version 4.0 using the neighbor-joining method with 1,000 bootstrap replicates.
Gene annotation database GENSCAN (http://genes.mit.edu/GENSCAN.html) and/or manual editing were used to analyzed genomic sequences to predict alternate open reading frames (ORFs) for potentially misannotated genes. For domain identification, SMART and Pfam (Protein families database of alignments and HMMs, version 14.0, June 2004; http://www.sanger.ac.uk/Software/Pfam/) databases were used to analyze each protein sequence. To identify potential novel domains, sequence alignments and BLAST searches were performed using the entire amino acid sequence of each RING protein. Phylogenetic analysis was used to confirm the amino acid sequence similarity observed between proteins.
Plant Material
Seeds from Arabidopsis ecotype Col-0 were either sown on soil and grown under photoperiodic cycles of 16 h light and 8 h dark at 16°C with 50% relative humidity, or seeds surface sterilized with 30% (v/v) bleach and 0.1% (v/v) Triton X-100 were grown on 1% (w/v) agar with 1× Murashige and Skoog and 1% (w/v) Suc under continuous light.
Cloning and Mutagenesis
Total RNA isolated from Arabidopsis ecotype Col-0, 10-d-old seedlings, leaves (2- to 4-week-old plants), or a mixture of RNA from leaves, seedlings, and flowers (6- to 7-week-old plants) was used in reverse-transcription reactions followed by PCRs to amplify the complete predicted ORF for each RING gene with one or more introns. The Qiagen RNeasy plant RNA extraction kit (Qiagen, Valencia, CA) was used to isolate total RNA as per the manufacturer's instructions. For intronless genes, the complete ORF was isolated from leaf (2- to 4-week-old plants) DNA. Otherwise, the ORF was isolated from expressed sequence tag (EST) clones obtained from the Arabidopsis Biological Resource Center (ABRC). The source of RNA/DNA used to isolate the ORF for each particular gene is outlined in Table I.
The Gateway cloning system (Invitrogen, Carlsbad, CA) was used to clone each RING gene. Full-length RING cDNAs were first introduced into the Gateway entry vector pDONR and the DNA sequence determined. Sequences of each RING cDNA were compared with the predicted ORF available on the TAIR (http://www.arabidopsis.org) and The Institute for Genomic Research (http://www.tigr.org) Arabidopsis genome annotation databases. cDNAs matching the predicted ORFs were introduced into the pDEST15 (Invitrogen) protein expression vector to produce in-frame fusions with the GST tag. AtUBC8 was cloned in a similar manner and introduced into the pDEST17 (Invitrogen) vector to produce an in-frame fusion with the 6×His tag. For RING mutational analysis, site-directed mutagenesis (Stratagene, La Jolla, CA) was used to make a series of point mutations within the region of the gene predicted to encode for the RING domain. pDONR vector containing the RING cDNA of interest was used as a PCR template. Sequence analysis was used to confirm nucleotide changes. The mutated RING cDNA was introduced, via the Gateway system, into the pDEST15 vector to produce an in-frame fusion with the GST tag.
Protein Expression, Purification, and Western-Blot Analysis
GST-RING or mutated RING fusions were expressed in Escherichia coli strain BL21 (DE3) pLysS in 50-mL cultures. Transformed cells were grown at 37°C for 2 to 3 h or to an OD600 of 0.4 to 0.6 before induction with 0.5 mm isopropylthio-β-galactoside for 3 to 4 h at 37°C. Cells were harvested by centrifugation and lysed in 2 mL of lysis buffer containing 25 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 0.01% Triton X-100. For purification, 100 μL of glutathione agarose beads (Sigma, St. Louis) was added to cleared lysates and incubated for 2 h at 4°C. Beads were then washed four times with 1 mL of wash buffer containing 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 0.01% (v/v) Triton X-100. GST fusion proteins were eluted with 100 μL of elution buffer containing 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.01% (v/v) Triton X-100 supplemented with 15 mm reduced glutathione, or the GST fusion proteins were left bound to the beads. Forty microliters of glycerol was added to the eluted protein. Bead-bound GST proteins were stored in 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.01% (v/v) Triton X-100, and 40% (v/v) glycerol. Proteins were stored at −80°C until needed. The 6×His-tagged AtUBC8 was expressed and purified in a similar manner. The 6×His fusion was expressed in E. coli strain BL21 AI (Invitrogen) and induced with 0.2% (w/v) Ara. Lysis and wash buffer were supplemented with 5 mm imidazole and elution buffer supplemented with 300 mm imidazole.
SDS-PAGE electrophoresis followed by Coomassie blue staining and Bradford assays (Bio-Rad, Hercules, CA) were used to quantify purified proteins. Western-blot analysis using GST antibodies or His antibodies (Amersham, Buckinghamshire, UK) was also used to confirm the presence and integrity of each fusion protein.
In Vitro Ubiquitination Assay
Ubiquitination assays were carried out as described previously (Hardtke et al., 2002). Reactions (30 μL) containing 50 mm Tris-HCl, pH 7.5; 10 mm MgCl2; 0.05 mm ZnCl2; 1 mm ATP; 0.2 mm dithiothreitol; 10 mm phosphocreatine; 0.1 unit of creatine kinase (Sigma); 50 ng of yeast E1 (Boston Biochem, Cambridge, MA); 250 ng of purified E2 AtUBC8; 250 ng of eluted/bead-bound GST-RING, GST-mutated RING protein, or GST-CIP8 (positive control); and 2 μg ubiquitin (Sigma) were incubated at 30°C for 2 h. Reactions were stopped by adding 6 μL of 5× SDS-PAGE sample buffer (125 mm Tris-HCl, pH 6.8, 20% [v/v] glycerin, 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) and analyzed by SDS-PAGE electrophoresis followed by western blotting using ubiquitin antibodies. For zinc-chelating experiments, bead-bound GST-RING protein was either incubated in 50 mm Tris-HCl, pH 7.4, containing 5 mm TPEN or 0.5% (v/v) ethanol (vehicle; mock treatment) for 16 h at 4°C with at least three solution changes. Beads were then washed three times in 50 mm Tris-HCl, pH 7.4. An aliquot of TPEN-treated bead-bound GST-RING protein was incubated with 1 μm ZnCl2 for 4 h at 4°C with at least three solution changes, followed by three washes in 50 mm Tris-HCl, pH 7.4. Mock-, TPEN-, and TPEN plus ZnCl2-treated bead-bound GST-RING protein were then used in ubiquitination assays.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank M. Wogulis (University of California, Davis) for the gift of E1 protein and helpful discussions, and members of the Callis lab for helpful discussions and editing. We thank Dr. Richard Gardner (Fred Hutchinson Cancer Research Center, Seattle) for helpful information regarding the Arabidopsis RING-G protein. We would also like to thank Charles Forsyth and Colin Wu (University of California, Davis) for assistance in the assembly of the RING database.
This work was supported by the National Science Foundation (2010 grant no. MCB–00115870). E.K. also was partially supported by the National Institute of Health Training (grant no. GM0007377–27), and S.L.S. was supported by the Natural Sciences and Engineering Research Council of Canada and the International Human Frontier Science Program fellowships.
The online version of this article contains Web-only data.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56–59 [DOI] [PubMed] [Google Scholar]
- Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT (2002) Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J 21: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L (2003) GRAIL: An E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 18: 535–547 [DOI] [PubMed] [Google Scholar]
- Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K (2004) Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem 279: 40053–40061 [DOI] [PubMed] [Google Scholar]
- Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA (2001) Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. J Biol Chem 276: 19640–19647 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Barlow PN, Luisi B, Milner A, Elliott M, Everett R (1994) Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol 237: 201–211 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM (1998) Kinesin: What gives? Cell 93: 5–8 [DOI] [PubMed] [Google Scholar]
- Borchers AG, Hufton AL, Eldridge AG, Jackson PK, Harland RM, Baker JC (2002) The E3 ubiquitin ligase GREUL1 anteriorizes ectoderm during Xenopus development. Dev Biol 251: 395–408 [DOI] [PubMed] [Google Scholar]
- Borden KL (2000) RING domains: master builders of molecular scaffolds? J Mol Biol 295: 1103–1112 [DOI] [PubMed] [Google Scholar]
- Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS (1995) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J 14: 1532–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P (1993) Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17: 363–374 [DOI] [PubMed] [Google Scholar]
- Brunskill EW, de Jonge BL, Bayles KW (1997) The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiol 143: 2877–2882 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D III, Fukuda M, Ohta T, Klevit R (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82–88 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett 400: 25–30 [DOI] [PubMed] [Google Scholar]
- Capili AD, Edghill EL, Wu K, Borden KL (2004) Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J Mol Biol 340: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher IF, Borden KL (2001) Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J 20: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Wu K, Fuchs SY, Tan P, Gomez C, Pan ZQ (2000) The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J Biol Chem 275: 15432–15439 [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM (2004) Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol 24: 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP (2001) JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci 26: 7–9 [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Ramsey KL, Smith JS, Auble DT (2004) Sir Antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem 279: 26830–26838 [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang LY, Braun A, You JX, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P (2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA 98: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Freemont PS (1993) The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci 684: 174–192 [DOI] [PubMed] [Google Scholar]
- Freemont PS, Hanson IM, Trowsdale J (1991) A novel cysteine-rich sequence motif. Cell 64: 483–484 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Gracey AY, Troll JV, Somero GN (2001) Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc Natl Acad Sci USA 98: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoufa A, Negruk V, Eisner G, Lemieux B (1996) The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J 10: 459–467 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW (2002) Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J 30: 385–394 [DOI] [PubMed] [Google Scholar]
- Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ (2002) Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J 21: 2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Orian A, Cowley SM, Eisenman RN (2002) Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc Natl Acad Sci USA 99: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER (1997) Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46: 103–111 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, et al (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284: 657–665 [DOI] [PubMed] [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I (2003) Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113: 621–630 [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1: REVIEWS3002 [DOI] [PMC free article] [PubMed]
- Kosarev P, Mayer KF, Hardtke CS (2002) Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol 3: RESEARCH0016 [DOI] [PMC free article] [PubMed]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Lechner E, Xie D, Grava S, Pigaglio E, Planchais S, Murray J, Parmentier Y, Mutterer J, Bureucq B, Shen W-H, et al (2002) The AtRbx1 protein is part of plant SCF complexes and its down-regulation causes severe growth and developmental defects. J Biol Chem 277: 50069–50080 [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleocytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering R, Hanson IM, Borden KL, Martin S, O'Reilly NJ, Evan GI, Rahman D, Pappin DJ, Trowsdale J, Freemont PS (1993) Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA 90: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell 9: 945–956 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Suzuki T, Tanaka K, Nakano A (2001) Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci 114: 1949–1957 [DOI] [PubMed] [Google Scholar]
- Mladek C, Guger K, Hauser MT (2003) Identification and characterization of the ARIADNE gene family in Arabidopsis. A group of putative E3 ligases. Plant Physiol 131: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar G, Bancos S, Nagy F, Szekeres M (2002) Characterisation of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta 215: 127–133 [DOI] [PubMed] [Google Scholar]
- Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA (1999) The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J Biol Chem 274: 30963–30968 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43 [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell 3: 535–541 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Hentze MW (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 23: 409–411 [DOI] [PubMed] [Google Scholar]
- Ponting C, Schultz J, Bork P (1997) SPRY domains in ryanodine receptors (Ca2+-release channels). Trends Biochem Sci 22: 193–194 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ (1996) ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci 21: 11–13 [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlogelhofer P, Becker F, Nejinskaia V, Bachmair A (1998) PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA 95: 7904–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O (2000) LIS1. let's interact sometimes… (part 1). Neuron 28: 633–636 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Schnell R, D'Ari L, Foss M, Goodman D, Rine J (1989) Genetic and molecular characterization of suppressors of SIR4 mutations in Saccharomyces cerevisiae. Genetics 122: 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol 21: 8035–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple CA (2003) The comparative proteomics of ubiquitination in mouse. Genome Res 13: 1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Guzman P (2004) Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Prag G, Francis SA, Sutanto MA, Hurley JH, Hicke L (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J 22: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26s proteasome proteolytic pathway. Annu Rev Plant Physiol Plant Mol Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Baker A (2002) Peroxisome biogenesis and protein import in plants, animals and yeasts: enigma and variations? (Review). Mol Membr Biol 19: 171–185 [DOI] [PubMed] [Google Scholar]
- Stary S, Yin XJ, Potuschak T, Schlogelhofer P, Nizhynska V, Bachmair A (2003) PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol 133: 1360–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH (1999) TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci 8: 1557–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck L, Pearce DA, Sherman F (1994) PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem 269: 2206–2214 [PubMed] [Google Scholar]
- Warner TS, Sinclair DA, Fitzpatrick KA, Singh M, Devlin RH, Honda BM (1998) The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome 41: 236–243 [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO (2002) Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13: 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington MT, Amann BT, Nathans D, Berg JM (1996) Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc Natl Acad Sci USA 93: 13754–13759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang SY, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu R, Li QQ (2003) A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Mol Biol 53: 37–50 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Elia AE (2001) Phosphoserine/threonine-binding domains. Curr Opin Cell Biol 13: 131–138 [DOI] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song LZ, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al (2002) Structure of the Cul1-Rbx1-Skp1-F box(Skp2) SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.