Abstract
Autosomal dominant hyper IgE syndrome (AD-HIES) is a primary immunodeficiency caused by a loss-of-function mutation in the Signal Transducer and Activator of Transcription 3 (STAT3). This immune disorder is clinically characterized by increased susceptibility to cutaneous and sinopulmonary infections, in particular with Candida and Staphylococcus aureus. It has recently been recognized that the skin microbiome of patients with AD-HIES is altered with an overrepresentation of certain Gram-negative bacteria and Gram-positive staphylococci. However, these alterations have not been characterized at the species- and strain-level. Since S. aureus infections are influenced by strain-specific expression of virulence factors, information on colonizing strain characteristics may provide insights into host-pathogen interactions and help guide management strategies for treatment and prophylaxis. The aim of this study was to determine whether the immunodeficiency of AD-HIES selects for unique strains of colonizing S. aureus. Using multi-locus sequence typing (MLST), protein A (spa) typing, and PCR-based detection of toxin genes, we performed a detailed analysis of the S. aureus isolates (n = 13) found on the skin of twenty-one patients with AD-HIES. We found a low diversity of sequence types, and an abundance of strains that expressed methicillin resistance, Panton-Valentine leukocidin (PVL), and staphylococcal enterotoxins K and Q (SEK, SEQ). Our results indicate that patients with AD-HIES may often carry antibiotic-resistant strains that harbor key virulence factors.
Keywords: Staphylococcus aureus, Job’s Syndrome, STAT3, multi-locus sequence typing
1. Introduction
Staphylococcus aureus colonizes human mucosa and skin and is a major human pathogen that causes a strikingly broad range of infections, ranging from relatively minor skin abscesses to potentially life-threatening pneumonia, osteomyelitis, endocarditis, necrotizing fasciitis, septicemia, and toxic shock syndrome [1]. The multitude of virulence factors and immune response modulators expressed by S. aureus contribute to these diverse disease manifestations [2]. In addition, rapid adaptation and development of bacterial resistance to antibiotics impede the effective treatment of S. aureus infections, as demonstrated by the epidemic emergence of community-associated methicillin-resistant S. aureus (CA-MRSA) strains outside the hospital setting [3,4]. The association of certain clonal groups of MRSA, such as USA300 [5], with outbreaks of infection suggests a critical impact of strain-dependent virulence factor expression.
Although S. aureus can infect otherwise healthy individuals, the increased susceptibility of patients with certain primary immunodeficiencies provides insight into the components of a protective immune response. For example, the critical role of neutrophils is highlighted by the increased susceptibility of patients with congenital neutropenia or chronic granulomatous disease [6,7,8]. Autosomal dominant hyper IgE syndrome (AD-HIES; also known as Job’s Syndrome), caused by dominant-negative, loss-of-function mutations in the Signal Transducer and Activator of Transcription 3 (STAT3) [9,10,11], is also characterized by S. aureus susceptibility, which exacerbates eczematoid dermatitis and causes pneumonia and recurrent skin abscesses in these patients. The effects of STAT3 dysfunction on epithelial antimicrobial function, including impaired differentiation of IL-17-secreting CD4+ T (Th17) cells [12,13], likely contribute to this susceptibility.
The skin microbiome of AD-HIES patients is altered compared to unaffected control subjects, with a significant increase in Corynebacteria, Serratia, and Staphylococcus, including S. aureus, S. haemolyticus, and S. epidermidis [14,15]. Molecular characterization of the staphylococcal strains present on the skin of AD-HIES patients has not been previously described. In particular, it remains unclear whether immunodeficiency selects for colonization by unique strains of S. aureus. Here, we identified the species of cultivable, mannitol-fermenting staphylococcal strains present on the skin of patients with AD-HIES. To determine whether patients with AD-HIES are colonized with particular S. aureus strains, all strains identified as S. aureus were genetically characterized by multi-locus sequence typing (MLST) and staphylococcal protein A (spa)-typing; assessed for the presence of lukSF-PV, sek, and seq virulence genes; tested for methicillin resistance; and assessed for their ability to lyse erythrocytes of human, sheep, and rabbit origin. We found a low diversity of sequence types, and an abundance of strains that expressed methicillin resistance, Panton-Valentine leukocidin (PVL), and staphylococcal enterotoxins K and Q (SEK, SEQ). Our results indicate that patients with AD-HIES may often carry antibiotic-resistant strains that harbor key virulence factors.
2. Results
2.1. Patients
We swabbed the antecubital fossa and volar forearm of 36 patients with AD-HIES and isolated mannitol-fermenting staphylococci for further characterization from 21 of these patients. The clinical characteristics of these 21 patients are presented in Table 1. These patients were followed at National Institutes of Health (NIH) and reside in 13 different states from western, mid-western, southern, and northeastern regions of the United States (US). There were 12 females (57%) and nine males (42%). Median age at time of swabbing was 23.8 years (range 1–53 years). All patients had STAT3 mutations confirmed by Sanger sequencing. The mutations were identified to be within the Src homology 2 (SH2) domain (n = 10; 48%), DNA-binding domain (n = 10; 48%), or transactivation domain (n = 1; 5%). Eighteen patients (86%) were taking oral anti-staphylococcal prophylactic antibiotics at the time of swabbing. These antibiotics included but were not limited to trimethoprim/sulfamethoxazole, clindamycin, amoxicillin-clavulanate, and cephalosporins.
Table 1.
Demographics and clinical characteristics of patients with AD-HIES.
| Patient ID | Gender | Age + | Race | STAT3 Mutation (Nucleotide; Amino Acid) | STAT3 Mutation Domain | History of: | On Antibiotics | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Eczema | Skin Abscess | PNA | CMC | |||||||
| P-9 | F | 45 | W | G1909A; V637M | SH2 | Y | Y | Y | Y | N |
| P-10 | M | 27 | B | C1144T; R382W | DNA | Y | Y | Y | Y | Y |
| P-14 | F | 45 | W | T1151C; F384S | DNA | Y | Y | Y | Y | Y |
| P-15 | F | 18 | W | C1144T; R382W | DNA | Y | Y | Y | Y | N |
| P-16 | M | 23 | W | T1997G; L666R | SH2 | Y | Y | Y | Y | Y |
| P-17 | F | 39 | W | C2003T; S668F | SH2 | Y | Y | Y | Y | Y |
| P-18 | F | 43 | W | T1861G; F621V | SH2 | Y | Y | Y | Y | Y |
| P-19 | F | 1 | W | A1939G; N647D | SH2 | Y | N | N | N | Y |
| P-20 | M | 7 | B | C1144T; R382G | DNA | Y | Y | Y | Y | Y |
| P-21 | F | 13 | W | T2117C; L706P | Trans-activation | Y | Y | Y | N | Y |
| P-25 | M | 44 | W | 1387delGTG; V463del | DNA | Y | Y | Y | Y | Y |
| P-26 | M | 9 | B | A1831G; S611G | SH2 | Y | Y | Y | N | Y |
| P-31 | F | 17 | A | G1268A; R423Q | DNA | Y | N | N | N | Y |
| P-32 | M | 17 | W | G1145A; R382Q | DNA | Y | Y | Y | Y | Y |
| P-33 | M | 6 | A | G1909A; V637M | SH2 | Y | Y | Y | N | Y |
| P-34 | M | 20 | W | G1909A; V637M | SH2 | Y | Y | Y | Y | Y |
| P-36 | F | 6 | B | G1909A; V637M | SH2 | Y | Y | Y | Y | Y |
| P-39 | F | 1 | W | G1268A; R423Q | DNA | Y | N | N | N | Y |
| P-40 | F | 36 | W | G1268A; R423Q | DNA | Y | Y | Y | Y | N |
| P-41 | F | 30 | H | G1909A; V637M; G1381C; V461L | SH2 | Y | Y | Y | Y | Y |
| P-53 | M | 53 | W | G1145A; R382Q | DNA | Y | Y | Y | Y | Y |
+ Age in years; M, male; F, female; A, Asian; B, Black; H, Hispanic; W, White; DNA, DNA binding domain; SH2, Src homology 2 domain; PNA, pneumonia; CMC, chronic mucocutaneous candidiasis; Y, yes; N, no.
Staphylococcal isolates from 13 patients with physician-diagnosed atopic dermatitis were included in the study. Patients with atopic dermatitis were chosen as a comparison group for the patients with AD-HIES since they were similarly seen at NIH, were also characterized by S. aureus skin colonization and infection, but did not have the dysfunctional STAT3-dependent immunodeficiency that defines AD-HIES. These patients represented residency in eight states from diverse regions of the US. Their clinical characteristics are presented in Table S1. The majority of patients were male (n = 10; 77%). The median age at time of swabbing was 16 years (range 3–51 years). Disease severity was assessed using the Scoring Atopic Dermatitis (SCORAD) index scale [16], and further classified as mild (SCORAD < 25), moderate (SCORAD 25–50), or severe (SCORAD > 50). Median SCORAD at time of swab was 16 (range 2–56). None of these patients had a history of skin abscesses or were on prophylactic antibiotics at the time of swabbing.
2.2. Strain Isolation and Identification
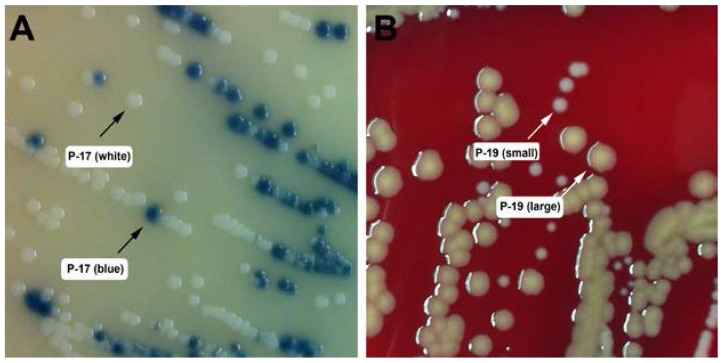
To isolate S. aureus strains from individuals with AD-HIES, skin swabs were plated on mannitol salt agar (MSA). Mannitol-fermenting strains were isolated for further characterization. We obtained 25 mannitol-fermenting staphylococcal strains from the skin of 21 individuals with AD-HIES. An overview of all mannitol-fermenting strains isolated from these patients with AD-HIES is provided in Table 2 and Table 3. Two mannitol-fermenting staphylococcal strains were isolated from each of two individuals with AD-HIES, P-17 and P-19. These were designated P-17 (white) and P-17 (blue) for their appearance on Spectra MRSA plates (Figure 1A), or P-19 (large) and P-19 (small) for their colony size on blood agar (BA) plates (Figure 1B). Furthermore, two patients with AD-HIES (P-10 and P-25) were swabbed on two different occasions, and the resulting non-aureus staphylococcal (NAS) isolates were designated P-10a, P10b, P25a, and P25b, respectively (Table 3).
Table 2.
Coagulase-positive S. aureus strains isolated from patients with AD-HIES.
| S. aureus Isolates from Patients with AD-HIES | |||||
|---|---|---|---|---|---|
| Strain ID | MLST ST | Spa Type | MRSA | PVL | SEK/SEQ |
| P-14 | 88 | t4013 | N | Y | N |
| P-15 | 2148 | t4013 | N | Y | Y |
| P-16 | 88 | t4013 | Y | Y | Y |
| P-17 (blue) | 8 | t064 | Y | N | N |
| P-19 (large) | 88 | t4013 | N | N | N |
| P-20 | 8 | t008 | Y | Y | Y |
| P-21 | 8 | t064 | N | N | N |
| P-31 | N.D. | t189 | N | N | N |
| P-32 | 8 | t008 | Y | Y | Y |
| P-33 | 8 | t118 | N | Y | N |
| P-34 | 8 | t064 | Y | Y | N |
| P-36 | 8 | t451 | Y | Y | Y |
| P-41 | 8 | t008 | Y | Y | Y |
N.D., not determinable; ST, sequence type; N, no; Y, yes.
Table 3.
Mannitol-fermenting, coagulase-negative staphylococcal strains isolated from patients with AD-HIES.
| Non-Aureus Staphylococcal Isolates from Patients with AD-HIES | |
|---|---|
| P-9 | S. warneri |
| P-10 a | S. haemolyticus |
| P-10 b | S. hominis |
| P-17 (white) | S. haemolyticus |
| P-18 | S. haemolyticus |
| P-19 (small) | S. hominis |
| P-25 a | S. haemolyticus |
| P-25 b | S. haemolyticus |
| P-26 | S. simulans |
| P-39 | S. hominis |
| P-40 | S. hominis |
| P-53 | S. haemolyticus |
Figure 1.
Agar plate phenotypes of select staphylococcal isolates from patients with AD-HIES. (A) Isolate P-17 on spectra MRSA plate. The blue colony represents a phosphatase-positive isolate that was confirmed to be S. aureus. The white colony represents a phosphatase-negative strain that was identified as S. haemolyticus. (B) Isolate P-19 on sheep blood agar plate. The large, yellow colony was identified as S. aureus; the small, white colony was identified as S. hominis.
Of all 25 mannitol-fermenting strains isolated from patients with AD-HIES, 13/25 (52%) were S. aureus, as defined by coagulase-positivity (Table 2), and 12/25 (48%) were NAS, such as S. haemolyticus or S. hominis, determined by 16S rRNA sequencing (Table 3). We further determined that 7/13 (54%) S. aureus isolates were MRSA (Table 2). In comparison, 9/13 (59%) manntiol-fermenting isolates obtained similarly from patients with atopic dermatitis were S. aureus, and none of these were MRSA (Table S2).
2.3. Typing of S. aureus Isolates
MLST distinguishes bacterial strains based on DNA sequence variations in seven moderately conserved bacterial housekeeping genes (mlst.saureus.net). We identified only three different MLST types within all 13 S. aureus isolates from individuals with AD-HIES (Table 2). Of these, 61.5% (8/13) of strains were of Sequence Type (ST) 8. The second most frequent type was ST88 (3/13, 23%), accounting for three AD-HIES isolates. One strain (P-31) was non-typeable because of a unique MLST locus profile that did not match any entries in the mlst.net database (Table 2 and Table S3). To further assess the relatedness of strains, we performed spa typing [17] and identified six different spa types (Table 2). The most frequent spa type was t4013 (four isolates); these strains belonged to two MLST types, ST88 and ST2148 (Table 2 and Table S3). Similar analysis of the nine S. aureus isolates from our patients with atopic dermatitis revealed a more diverse distribution of MLST and spa types (Table S2).
2.4. Presence of the Phage-Encoded Toxins Panton-Valentine Leukocidin and Enterotoxins SEK, SEQ
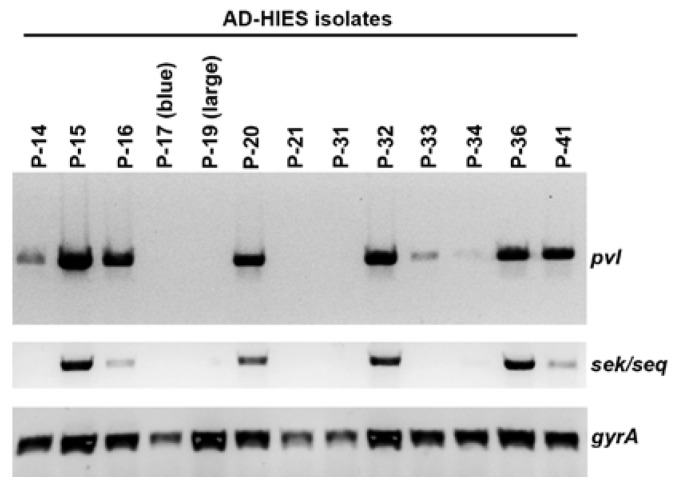
We tested all isolated S. aureus strains for the presence of the genes for lukSF-PV, sek, and seq, encoding for the two-component pore-forming cytotoxin Panton-Valentine leukocidin (PVL) [18] and for the staphylococcal enterotoxins K and Q (SEK, SEQ), respectively. These toxins are often found in USA300 strains that have been associated with CA-MRSA-associated outbreaks of skin and soft tissue infection. We found that a majority of S. aureus isolates from AD-HIES patients (9/13; 69%) contained the lukF/lukS-PV genes (Figure 2 and Table 2). Similarly, some AD-HIES patients harbored strains that were positive for the superantigen genes seq/sek (6/13; 46%). These toxin genes were found in only a striking minority of atopic dermatitis S. aureus isolates (Table S2).
Figure 2.
Detection of select toxin genes in S. aureus strains isolated from patients with AD-HIES. Presence of genes for Panton-Valentine leukocidin (pvl) and two superantigens (sek/seq) were assessed by PCR. As a DNA template control, the housekeeping gene encoding for gyrase A (gyrA) was partially amplified.
2.5. Hemolysis on Sheep, Rabbit, and Human Agar Plates
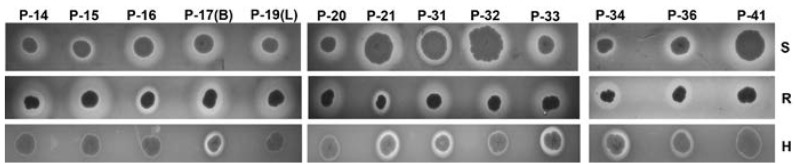
We phenotypically assessed the capacity of S. aureus isolates to lyse red blood cells, an activity ascribed to a number of staphylococcal virulence factors [19]. Because the ability of staphylococcal hemolysins to lyse erythrocytes exhibits species specificity [20], we characterized hemolysis on sheep, rabbit, and human BA plates. We found that all isolates lysed both sheep and rabbit erythrocytes (Figure 3). Robust lysis of human red blood cells was only evident for some strains (5/13; 38%), in particular those belonging to ST8, t064 type (P-17 (blue), P-21, P-34). Weak hemolysis of human BA plates was observed for all four t4013 isolates (P-14, P-15, P-16, P-19 (large)) as well as for the ST8, t008 isolates (P-20 and P-32).
Figure 3.
Hemolysis phenotype of S. aureus strains isolated from patients with AD-HIES. Hemolysis on sheep (S), rabbit (R), and human (H) blood agar is shown.
3. Discussion
The primary aim of this study was to provide a detailed analysis of the S. aureus strains isolated from the skin of patients with AD-HIES. 16S rRNA sequencing has identified altered microbiomes in patients with AD-HIES [14,15], including the enrichment of S. aureus and non-aureus staphylococci. We sought to provide a more detailed, strain-specific analysis of AD-HIES-associated staphylococci, since S. aureus strains are known for their varied expression of colonization and virulence factors that can influence disease frequency and severity [2], and specific strains have been linked to skin infections [4].
We determined three different MLST sequence types in 13 S. aureus isolates from patients with AD-HIES. The most common sequence type was ST8, which accounted for eight of the 13 S. aureus isolates from patients with AD-HIES, and three of the nine S. aureus isolates from STAT3-sufficient patients with atopic dermatitis (3/9). These findings are consistent with ST8 being the most commonly isolated sequence type for MRSA in the US. [5]. The second most frequent sequence type isolated from our AD-HIES cohort was ST88, which has been reported in certain regions of Africa where it accounts for up to 83% of all MRSA isolates [21]. Of note, ST2148 was isolated from one patient with AD-HIES (P-15) and appears to be closely related to ST88, harboring the same spa-type and differing only at one MLST locus.
Among these same 13 AD-HIES-associated S. aureus isolates, we identified six unique spa-types, the most frequent ones being t4013 (4/13, 31%), t064 (3/13, 23%), and t008 (3/13, 23%). Spa-type t4013 was described in 2009 among MRSA strains isolated from wastewater in Sweden [22], and t064 and t008 are common types belonging to MLST ST8, the most frequently isolated ST of MRSA in the US from 2000 to 2013 [5]. Although conclusions are limited by our small sample size, taken together our data suggest that patients with AD-HIES are colonized with prevalent sequence types. We speculate the limited diversity may reflect the patients’ increased exposure to health care facilities and the selective pressure of prophylactic antibiotics.
S. aureus is known for its ability to acquire foreign DNA in the form of extra-chromosomal plasmids, chromosomally-integrated S. aureus pathogenicity islands (SaPI), and other phage-transported DNA regions. These DNA acquisition mechanisms give S. aureus a dynamic genetic variability that may confer advantages under certain circumstances, e.g., antibiotic resistance or acquisition of additional virulence factors [23]. In this study, we tested for the presence of four genes (lukS-PV, lukF-PV, sek and seq) and one resistance marker (methicillin) associated with the accessory gene regions SaPI1, PVL prophages, and staphylococcal cassette chromosome (SCC), respectively. Our findings suggest that patients with AD-HIES frequently carry methicillin-resistant, PVL-positive, and SEK/SEQ-positive strains, indicating significant virulence potential since PVL has been associated with worse outcomes during S. aureus pneumonia [24], and SEK and SEQ have been shown to contribute to T cell activation in a mouse model of pneumonia [25]. However, similar to observations in CA-MRSA infections, bacteremia and disseminated infections are a rare presentation of S. aureus in patients with AD-HIES, indicating that the immune dysfunction is largely limited to epithelial and mucosal sites.
S. aureus expresses a wide variety of pore-forming or membrane-damaging toxins that may influence virulence [20]. We evaluated the broad potential of the isolated strains to lyse blood from animal (sheep, rabbit) and human origin, and found that all strains similarly lysed rabbit and sheep blood. Differences were observed in the lysis of human blood. This might reflect the species-specific activity of alpha-hemolysin [20]. One might also speculate that the expression of PVL by some isolates inhibits the activity of other pore-forming toxins, such as LukED, as recently shown by Yoong and Torres [26], but a more detailed analysis of secreted amounts of PVL and LukED is needed to draw such conclusions and clarify the implications for clinical virulence.
To our knowledge, this is the first detailed characterization of S. aureus strains present on the skin of patients with AD-HIES. Our findings suggest that patients with AD-HIES are colonized with a variety of strains, including those that are known to be prevalent in the US. However, the diversity of AD-HIES strains appears limited compared to what might be expected from the wide number of circulating clinical strains [27,28] and our analysis of atopic dermatitis strains. A high percentage of AD-HIES isolates (7/13, 54%) displayed resistance to methicillin and encoded toxin genes that have been associated with both CA-MRSA strains and pneumonia. Our data suggest that the immunodeficiency of AD-HIES does not predispose to colonization with less virulent strains. In fact, the colonizing strains of patients seem to be enriched for virulence factors and antibiotic resistance, which is likely a reflection of exposure to health care settings and prophylactic antibiotics.
4. Materials and Methods
4.1. Ethics Statement
All subjects were evaluated at the NIH between 1 December 2013 and 30 June 2015. All subjects provided informed consent on clinical research protocols approved by the National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board (clinical trials.gov identifiers: NCT00006150 and NCT02262819).
4.2. Collection and Isolation of Staphylococci
Subjects with AD-HIES (n = 36) or clinically diagnosed atopic dermatitis without a known genetic disorder (n = 13) were swabbed at the antecubital fossa and volar forearm, common sites of eczematous lesions colonized by S. aureus in AD-HIES as well as other forms of atopic dermatitis [14]. The swab was swirled in 2 mL of tryptic soy broth (TSB; Remel Thermo Scientific, Waltham, MA, USA). Aliquots were plated onto MSA (Remel Thermo Scientific) to select for S. aureus, which grows under high salt conditions and ferments mannitol. Unique mannitol-fermenting colonies, identified by their ability to produce yellow zones on MSA, were isolated, resuspended in TSB overnight, and plated onto sheep blood agar plates (Remel Thermo Scientific). S. aureus was identified by a positive slide coagulase test (Sigma-Aldrich, St. Louis, MO, USA). The species for coagulase-negative, mannitol-fermenting bacteria was identified by 16S rRNA sequencing. Subjects were designated by sequential numbering as they were enrolled. Their bacterial isolates were given corresponding numbers; AD-HIES isolates were preceded by ‘P’ (patient) and atopic dermatitis isolates were preceded by ‘AD,’ but overlapping numbers between groups were avoided to prevent confusion (Table 1).
4.3. DNA Isolation from Staphylococci and 16S rRNA Sequencing
DNA was isolated using a protocol described by Krausz and Bose [29]. The 16S rRNA gene was amplified with primers 16S-2F and 16S-1088R (Table 4) using Phusion DNA polymerase (Thermo Scientific) in a reaction volume of 50 μL. The amplification protocol was as follows: 98 °C for 40 s; 30 cycles of 98 °C for 10 s, 56 °C for 15 s, 72 °C for 20 s; and a final extension at 72 °C for 10 min. DNA was purified using the Qiagen PCR purification kit according to manufacturer’s recommendations (Qiagen, Valencia, CA, USA), and a minimum of two sequencing reactions per amplicon were performed (Eurofins MWG Operon, Huntsville, AL, USA) using both forward and reverse primers. Strain identification was performed using NCBI Blast.
Table 4.
Oligonucleotide primers.
| Name | 5′–3′ Sequence | Purpose | Reference |
|---|---|---|---|
| 16S-2F | GCRKGCYTAAYACATGCAAGTCGA | 16S typing | This study |
| 16S-1088R | CACGACACGAGCTGACGACAGCCA | ||
| arcC-up | TTGATTCACCAGCGCGTATTGTC | MLST typing | saureus.mlst.net |
| arcC-dn | AGGTATCTGCTTCAATCAGCG | ||
| aroE-up | ATCGGAAATCCTATTTCACATTC | ||
| aroE-dn | GGTGTTGTATTAATAACGATATC | ||
| glpf-up | CTAGGAACTGCAATCTTAATC | ||
| glpf-dn | TGGTAAAATCGCATGTCCAATTC | ||
| gmk-up | ATCGTTTTATCGGGACCATC | ||
| gmk-dn | TCATTAACTACAACGTAATCGTA | ||
| pta-up | GTTAAAATCGTATTACCTGAAGG | ||
| pta-n | GACCCTTTTCTTCAAAAGCTTAA | ||
| tpi-up | TCGTTCATTCTGAACGTCGTGAA | ||
| tpi-dn | TTTGCACCTTCTAACAATTGTAC | ||
| yqiL-up | CAGATACAGGACACCTATTGGC | ||
| yqiL-dn | CGTTGAGGAATCGATACTGGAAC | ||
| spa-1113f | TAAAGACGATCCTTCGGTGAGC | spa typing | www.ridom.de |
| spa-1514r | CAGCAGTAGTGCCGTTTGCTT | ||
| PVL-1 | ATGTCTGGACATGATCCA | LukSF-PV amplification | [30] |
| PVL-2 | AACTATCTCTGCCATATGGT | ||
| SEQ/SEK1 | GTATGGCGGAATTACGTTGG | sek/seq amplification | This study |
| SEQ/SEK2 | TTGGTAACCCATCATCTCCTG |
4.4. Identification of MRSA Isolates
Mannitol-fermenting staphylococcal isolates were streaked onto Spectra MRSA (Remel Thermo Scientific) plates, which select for MRSA as navy blue colonies.
4.5. MLST and Spa Typing
Primer sequences for MLST typing were obtained from saureus.mlst.net and are listed in Table 4. Internal sequences of seven housekeeping genes were amplified using Phusion polymerase with the same protocol used for 16S rRNA sequencing. Purified PCR products were sequenced and sequences were trimmed according to saureus.mlst.net and compared to the MLST database using the single locus query option. Amplification and typing of the spa gene was performed with the same protocol used for 16S rRNA sequencing and primers spa-1113f and spa-1517r (Table 4). PCR products were sequenced (Eurofins MWG Operon) and spa types were identified using the spatyper (spatyper.fortinbras.us).
4.6. Screening for pvl, sek, and seq
The presence of the lukSF-PV and sek/seq genes was determined by PCR using primers listed in Table 4. The oligonucleotides for sek/seq were derived from the sequence of the USA300 clone FPR3757 [31] and anneal within the coding regions of both sek and seq. Both reactions were performed using Phusion polymerase with the following protocol: 98 °C for 40 s; 35 cycles of 98 °C for 10 s, 56 °C for 15 s, 72 °C for 30 s; and a final extension at 72 °C for 5 min.
4.7. Blood Agar Hemolysis
Bacteria were streaked from a glycerol stock onto sheep blood agar plates and incubated overnight at 37 °C. The next day, single colonies were spotted onto tryptic soy agar (TSA) plates containing 3% of sheep, rabbit (both from Quad Five, Ryegate, MT, USA), or human blood (Department of Transfusion Medicine, NIH, Bethesda, MD, USA). Plates were incubated overnight at 37 °C and photographed the next day.
Acknowledgments
This work was supported by the Intramural Research Program of the NIAID at NIH, Bethesda, MD, USA. We thank Kelly Stone at NIAID for access to patients with atopic dermatitis.
Supplementary Materials
The following are available online at http://www.mdpi.com/2076-0817/6/2/23/s1. Table S1. Demographics and clinical characteristics of patients with physician-diagnosed atopic dermatitis. Table S2. Coagulase-positive S. aureus strains isolated from patients with atopic dermatitis. Table S3. Single locus typing results for all S. aureus isolates.
Author Contributions
I.S., K.W.W., and S.K.D. conceived and designed the experiments; I.S., K.W.W. and E.D.A. performed the experiments; I.S., K.W.W. and E.D.A. analyzed the data; I.A.M., J.D.R. and M.E.-M. contributed reagents/materials/analysis tools; A.F.F. provided patient evaluation and access; and I.S., K.W.W. and S.K.D. wrote the paper. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Ferry T., Perpoint T., Vandenesch F., Etienne J. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 2005;7:420–428. doi: 10.1007/s11908-005-0043-8. [DOI] [PubMed] [Google Scholar]
- 3.Chambers H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno F., Crisp C., Jorgensen J.H., Patterson J.E. Methicillin-resistant Staphylococcus aureus as a community organism. Clin. Infect. Dis. 1995;21:1308–1312. doi: 10.1093/clinids/21.5.1308. [DOI] [PubMed] [Google Scholar]
- 5.Carrel M., Perencevich E.N., David M.Z. USA300 Methicillin-Resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 2015;21:1973–1980. doi: 10.3201/eid2111.150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welte K., Zeidler C. Severe congenital neutropenia. Hematol. Oncol. Clin. N. Am. 2009;23:307–320. doi: 10.1016/j.hoc.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Quie P.G., White J.G., Holmes B., Good R.A. In vitro bactericidal capacity of human polymorphonuclear leukocytes: Diminished activity in chronic granulomatous disease of childhood. J. Clin. Investig. 1967;46:668–679. doi: 10.1172/JCI105568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guide S.V., Stock F., Gill V.J., Anderson V.L., Malech H.L., Gallin J.I., Holland S.M. Reinfection, rather than persistent infection, in patients with chronic granulomatous disease. J. Infect. Dis. 2003;187:845–853. doi: 10.1086/368388. [DOI] [PubMed] [Google Scholar]
- 9.Holland S.M., DeLeo F.R., Elloumi H.Z., Hsu A.P., Uzel G., Brodsky N., Freeman A.F., Demidowich A., Davis J., Turner M.L., et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 10.Minegishi Y., Saito M., Tsuchiya S., Tsuge I., Takada H., Hara T., Kawamura N., Ariga T., Pasic A., Stojkovic O., et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 11.Kane A., Deenick E.K., Ma C.S., Cook M.C., Uzel G., Tangye S.G. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 2014;28:49–57. doi: 10.1016/j.coi.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Milner J.D., Brenchley J.M., Laurence A., Freeman A.F., Hill B.J., Elias K.M., Kanno Y., Spalding C., Elloumi H.Z., Paolson M.L., et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minegishi Y., Saito M., Nagasawa M., Takada H., Hara T., Tsuchiya S., Agematsu K., Yamada M., Kawamura N., Ariga T., et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J. Exp. Med. 2009;206:1291–1301. doi: 10.1084/jem.20082767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh J., Freeman A.F., Program N.C.S., Park M., Sokolic R., Candotti F., Holland S.M., Segre J.A., Kong H.H. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013;23:2103–2114. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smeekens S.P., Huttenhower C., Riza A., van de Veerdonk F.L., Zeeuwen P.L.J.M., Schalkwijk J., van der Meer J.W.M., Xavier R.J., Netea M.G., Gevers D. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J. Innate Immun. 2014;6:253–262. doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Severity scoring of atopic dermatitis: the SCORAD index Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology. 1993;186:23–31. doi: 10.1159/000247298. [DOI] [PubMed] [Google Scholar]
- 17.Hallin M., Friedrich A.W., Struelens M.J. spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol. Biol. 2009;551:189–202. doi: 10.1007/978-1-60327-999-4_15. [DOI] [PubMed] [Google Scholar]
- 18.Panton P.N., Camb M.B., Valentine F.C.O. Staphylococcal Toxin. Lancet. 1932;219:506–508. doi: 10.1016/S0140-6736(01)24468-7. [DOI] [Google Scholar]
- 19.Wiseman G.M. The hemolysins of Staphylococcus aureus. Bacteriol. Rev. 1975;39:317–344. doi: 10.1128/br.39.4.317-344.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandenesch F., Lina G., Henry T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell Infect. Microbiol. 2012;2:12. doi: 10.3389/fcimb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaumburg F., Alabi A.S., Peters G., Becker K. New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 2014;20:589–596. doi: 10.1111/1469-0691.12690. [DOI] [PubMed] [Google Scholar]
- 22.Borjesson S., Matussek A., Melin S., Lofgren S., Lindgren P.E. Methicillin-resistant Staphylococcus aureus (MRSA) in municipal wastewater: an uncharted threat? J. Appl. Microbiol. 2010;108:1244–1251. doi: 10.1111/j.1365-2672.2009.04515.x. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay J.A., Holden M.T. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Gillet Y., Issartel B., Vanhems P., Fournet J.C., Lina G., Bes M., Vandenesch F., Piémont Y., Brousse N., Floret D., et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 25.Parker D., Ryan C.L., Alonzo F., 3rd, Torres V.J., Planet P.J., Prince A.S. CD4+ T cells promote the pathogenesis of Staphylococcus aureus pneumonia. J. Infect. Dis. 2015;211:835–845. doi: 10.1093/infdis/jiu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoong P., Torres V.J. Counter inhibition between leukotoxins attenuates Staphylococcus aureus virulence. Nat. Commun. 2015;6:8125. doi: 10.1038/ncomms9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller L.G., Eells S.J., Taylor A.R., David M.Z., Ortiz N, Zychowski D, Kumar N., Cruz D., Boyle-Vavra S., Daum R.S. Staphylocccus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin. Infect. Dis. 2012;54:1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albrecht V.S., Limbago B.M., Moran G.J., Krishnadasan A., Gorwitz R.J., McDougal L.K., Talan D.A. Staphylococcus aureus Colonization and Strain Type at Various Body Sites among Patients with a Closed Abscess and Uninfected Controls at U.S. Emergency Departments. J. Clin. Microbiol. 2015;53:3478–3484. doi: 10.1128/JCM.01371-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krausz K.L., Bose J.L. Rapid Isolation of DNA from Staphylococcus. Methods Mol. Biol. 2016;1373:59–62. doi: 10.1007/7651_2014_184. [DOI] [PubMed] [Google Scholar]
- 30.Ma X.X., Galiana A., Pedreira W., Mowszowicz M., Christophersen I., Machiavello S., Lope L., Benaderet S., Buela F., Vicentino W., et al. Community-acquired methicillin-resistant Staphylococcus aureus, Uruguay. Emerg. Infect. Dis. 2005;11:973–976. doi: 10.3201/eid1106.041059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diep B.A., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.