Abstract
Oncogene-specific changes in cellular signaling have been widely observed in lung cancer. Here, we investigated how these alterations could affect signaling heterogeneity and suggest novel therapeutic strategies. We compared signaling changes across six human bronchial epithelial cell (HBEC) strains that were systematically transformed with various combinations of TP53, K-RAS, and MYC—oncogenic alterations commonly found in non-small cell lung cancer (NSCLC). We interrogated at single-cell resolution how these alterations could affect classic readouts (β-CATENIN, SMAD2/3, phospho-STAT3, P65, FOXO1 and phospho-ERK1/2) of key pathways commonly affected in NSCLC. All three oncogenic alterations were required concurrently to observe significant signaling changes, and significant heterogeneity arose in this condition. Unexpectedly, we found two mutually exclusive altered subpopulations: one with STAT3 up-regulation and another with SMAD2/3 down-regulation. Treatment with a STAT3 inhibitor eliminated the up-regulated STAT3 subpopulation, but left a large surviving subpopulation with down-regulated SMAD2/3. A bioinformatics search identified BCL6, a gene downstream of SMAD2/3, as a novel pharmacologically accessible target of our transformed HBECs. Combination treatment with STAT3 and BCL6 inhibitors across a panel of NSCLC cell lines and in xenografted tumors significantly reduced tumor cell growth. We conclude that BCL6 is a new therapeutic target in NSCLC and combination therapy that targets multiple vulnerabilities (STAT3 and BCL6) downstream of common oncogenes and tumor suppressors may provide a potent way to defeat intra-tumor heterogeneity.
Keywords: Lung cancer, oncogene, signaling, heterogeneity, therapeutics
Introduction
Lung cancer is a disease of immense complexity with ~140 oncogenes and tumor suppressor gene abnormalities identified (1). These genetic alterations can appear in different combinations even within classically defined cancer subtypes, such as the lung adenocarcinoma subtype of non-small cell lung cancer (NSCLC) (2). It is apparent that no one therapy can fit all oncogenotypes.
The success of targeted therapy directed at EGFR mutant or EML4-ALK fusion lung cancers (despite the non-curative nature of such targeting) has led to a commonly applied approach, namely to search for therapy that is effective for specific oncogenotypes (3). When direct targeting of the oncogenes is not feasible, the goal is to identify vulnerabilities (i.e. molecular alterations crucial for cancer cell survival) within each genetically defined cancer subtype (4). The search for these vulnerabilities arises from the hypothesis that even if the oncogenic alterations themselves are not pharmacologically targetable, downstream, targetable signaling pathways on which cancer cells critically depend may also be altered and represent vulnerabilities. This approach, however, has two major challenges. First, it is difficult to identify specific oncogene-induced signaling alterations without a baseline reference of signaling for a specific cell type. Second, cancer cell populations are notoriously phenotypically heterogeneous (5). In fact, it is possible that even within a well-defined oncogenotype and identified signaling alteration, in an individual tumor the cancer cell population may be comprised of multiple subpopulations in different signaling states.
Here, we pursue a strategy of investigating changes in signaling for a common oncogenotype found in NSCLC, involving alterations in TP53, KRAS and MYC using a simplified preclinical model of such changes. Our preclinical model used immortalized human bronchial epithelial cells (HBECs) manipulated to contain various combinations of these three important oncogenic drivers to yield fully tumorigenic derivatives (6). We use this model HBEC system with defined oncogenic changes to search for pathways whose signaling has been differentially altered due to these defined genetic manipulations. Specifically, we first analyzed the pathways at single-cell level to identify intra-tumor subpopulations in altered signaling states. Next, we searched for targetable vulnerabilities in identified subpopulations. Finally, we tested the ability of small molecule inhibitors targeting these vulnerabilities—alone or in combination—to eliminate cancer cells both in vitro and in vivo. Thus, in contrast to previous studies that investigated cellular heterogeneity after drug treatment (5), we studied pre-treatment signaling heterogeneity to identify subpopulation-specific therapeutic targets and utilize this information to design a novel combination therapy targeting intra-tumor heterogeneity.
Materials and Methods
Cell lines and basal culture conditions
Normal and oncogenically manipulated immortalized Human Bronchial Epithelial Cells (HBECs) were cultured with Keratinocyte Serum Free Medium (KSFM; Life Technologies Inc., Carlsbad, CA) media containing 50 μg/mL of Bovine Pituitary Extract (BPE; Life Technologies Inc.) and 5 ng/mL (or, 50 pg/mL if otherwise stated) of Epidermal Growth Factor (EGF; Life Technologies Inc.). Parental and oncogenically manipulated HBECs were established between 2003 and 2009. Lung cancer cell lines, established in the laboratories of John Minna and Adi Gazdar between 1987 and 1994, were maintained in RPMI-1640 (Life Technologies Inc.) with 5% fetal bovine serum. The approximate number of passages for all cell lines between collection and thawing is 10. All cell lines were DNA fingerprinted (PowerPlex 1.2 Kit, Promega, Madison, WI) and mycoplasma-free (e-Myco Kit, Boca Scientific, Boca Raton, FL).
Viral transfection and transduction of Omomyc construct
MYC target gene knockdown was achieved using the Omomyc construct with pTripZ vector backbone. This construct was originally made by Laura Soucek (Vall d’Hebron Institute of Oncology (VHIO), Edifici Mediterrània, Hospital Vall d’Hebron, 08035 Barcelona, Spain Universitat Autònoma de Barcelona, Bellaterra (Cerdanyola del Vallès), 08193 Barcelona, Spain). Cell lines were transduced as described previously (6) and single cell clones with high level of RFP (inducible by doxycycline) intensity were selected.
qRT-PCR
The mRNA was isolated using Qiagen kit (Qiagen Inc., Valencia, CA), the cDNA was made by iScript (Life Sciences Research). Quantitative Reverse Transcription PCR (qRT-PCR) was performed using validated Taqman primers and probes (Applied Biosystems, Foster City, CA) using Applied Biosystems 7500 qRT-PCR machine and relative expression was calculated using the 2−ΔΔCT method. Number of technical replicates is as described in figure legends. Wells flagged as problematic by the qRT-PCR machine were dropped from the final calculation.
Treatment with small molecule inhibitors
STAT3 inhibitors BBI-608 (Tocris Bioscience, UK), Stattic (Calbiochem, EMD Millipore) and BCL6 inhibitor FX-1 (provided by Dr. Leandro Cerchietti, Weill Cornel Medical College, NY) were used in varying concentration in MTS drug sensitivity and colony formation assays as previously described (6). For anchorage-dependent liquid colony formation assay, 500 cells were seeded in each well of 6-well-plates (in triplicates for each cell line) for varying concentration of EGF at 0, 0.5, 5, 50, 500, 5000 pg/mL in Keratinocyte Serum Free Media (KSFM). The cells were cultured for 2 weeks and then the colonies were stained with crystal violet. Empty wells, defined as having intensity less than one standard deviation below that of blank wells, were omitted.
siRNA mediated knockdown
siRNA transfections were performed as described in (7). Cells were harvested 48 h post-transfection to seed in in vitro tumorigenicity assays. 6-well siRNA invasion assays were performed with siRNAs at 20nM, RNAi MAX lipid (Life Technologies, Carlsbad, CA), and 50000 cells per well. At 48 hours cells were washed with PBS, trypsinized and mRNA was isolated using RNeasy Mini Kit (Qiagen).After 96 hours, cells were assessed for viability. siRNA oligos were utilized along with positive siPLK1 (positive cell death phenotype), negative (non-silencing) controls (Qiagen, Hilden, Germany) and C911 controls ((7), Sigma).
Signaling readouts, immunofluorescence and immunoblot assay
Six signaling readouts were selected (Suppl. Table 1). Hoechst 33342 was used to identify nuclear regions. Cells were fixed with 4% paraformaldehyde for 5 minutes, permeabilized with ice cold 100% methanol at −20°C for 10 minutes, washed with 0.1% TBST, blocked with 5% BSA solution in 0.1% TBST at room temperature for 30 minutes. 5% BSA in 0.1% TBST was used for primary and secondary antibody dilutions. Plates stained with primary antibodies were incubated at 4C overnight. They were washed with 0.1% TBST three times. Next, plates were incubated with secondary antibodies in the dark at room temperature for two hours and then washed again with TBST three times. After the final washing step, 100μl of TBST containing 0.1% sodium azide was added to each well. Immunoblots were performed as described in (6) with primary antibodies shown in supplementary table 2.
Image acquisition, processing and quality control
All fluorescence images were acquired using a TE-2000 epifluorescence microscope (Nikon) equipped with integrated Perfect-Focus System (PFS), Nikon Plan Apochromat 20× objective lens and Photometrics Cool SNAP HQ camera. Image acquisition was controlled by NIS-Elements software (Nikon). Image background correction was done using the National Institute of Health ImageJ rolling-ball background subtraction plug-in (8). Cellular regions were determined using a watershed-based segmentation algorithm (9) which first retrieves nuclear regions using DNA staining then combines multiple cytosolic region markers to identify cellular boundaries. Images were visually inspected, and images with severe focus, staining, or cell-segmentation artifacts were discarded. We identified ~1,000 cellular regions per marker/well after applying automated cell segmentation to our image data.
Determination of altered cellular signaling state
To understand the activity in each signaling pathway, we focused on a specific intensity feature for each marker, namely the ratio of the average nuclear to average cytoplasmic intensities. The exception was β-CATENIN, for which we measured total intensity in the cytoplasm to capture its loss in the cell membrane and cytoplasm. For each cell we extracted this intensity feature. Approximately, 104 cells were analyzed per marker per cell line. Hence, each cell line gave us a distribution for a specific marker. In short, for any marker in parental HBEC, cells that fall within 5th and 95th percentile were defined to constitute the baseline signaling in parental HBEC. The top and bottom 5% (Total 10% of cells) were defined as outliers. The distribution of oncogenically manipulated HBECs, when superimposed on this control parental distribution, gave us the altered fraction of cells from total population. The altered fractions for a condition were calculated by combining cells across multiple replicate wells.
List of TGFβ downstream target genes used in microarray data analysis
P15, P21, P57, 4EBP1, BIK, BIM, DAPK, FAS, GADD45b, ATG5, ATG7, BECLIN 1/ATG6, THROMBOSPONDIN, FOXP3, FOXC2, FOXK1, CD25A, E2F-1, ID1-3, MYC, BCL-XL, BCL2, BCL6, HGF, MSP, TGFα, GATA-3, T-BET, SOX2, SOX4, SOX15, MMP7, MMP19, MMP2, MMP9, PDGF-B, VEGF, LIF, SNAIL1/2, ZEB1/2, HMGA2, HDM2, TIMP, IFNγ, MICA, NKG2D, NKP30, PERFORIN, T-BET
Drug treatment in xenografted tumors
106 H1993 cells were injected subcutaneously to each of 40 female NON-SCID mice (4–6 weeks old) following the previously described protocol (6). The tumor volume and body weight were routinely measured. After the tumors reached a volume between 50 – 100 cu.mm, they were randomized in 4 groups of 10. 20 mg/kg of BBI-608 was administered intraperitoneally with 1x PBS vehicle as described previously (10,11). 25 mg/kg of FX-1 was administered intraperitoneally with vehicle (5% Tween-80, 30% PEG-400, 65% Dextrose solution 5%) (12). The treatment dose and schedule for single agent and combination are described in details in supplementary table 3. After the tumors reached approximately 2000 cu.mm. in the vehicle treated group, all the mice were sacrificed following appropriate humane protocols described before (6) and the tumors were surgically resected, measured (1/2*length*width*width) and weighed. P-values were calculated by two-way Anova with Tukey’s multiple comparison test using GraphPad Prism (version 7.01, GraphPad Software, La Jolla California USA). Error bars represent standard deviation (n = 10 technical replicates). All studies were conducted on approval by the University of Texas Southwestern Medical Center Institutional Animal Care and Research Advisory Committee.
Data availability
The microarray data discussed in this publication have been made available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) public repository <http://www.ncbi.nlm.nih.gov/geo/> and are accessible through GEO Series accession number GSE40828. Raw images of a subset or complete dataset are available upon request.
Results
Oncogenically manipulated HBECs provide a unique model system to search for intra-tumor heterogeneity
To decipher signaling alterations in the context of commonly observed NSCLC oncogenotypes, we used our collection of immortalized HBECs with defined combinations of TP53 knockdown, KRASV12 and MYC overexpression. We previously showed that while each change and various combinations progressed the cells toward malignancy, only after introduction of all three oncogenic changes did HBEC3KT cells become fully tumorigenic (6). We refer to this triply manipulated, transformed derivative as HBECPKM while the other derivatives include TP53 knockdown alone (HBECP), TP53 knockdown and KRASV12 (HBECPK), MYC overexpression alone (HBECM), TP53 knockdown and MYC overexpression (HBECPM). Consistent with variability in gene expression across tumor cells observed in vivo (13–15), HBECPKM cells show a high degree of cell-to-cell differences in the expression of TP53, KRAS, and MYC (Suppl. Fig. 1). Taken together, the cohort of HBECs provided us with a unique in vitro model system to study single-cell variability in multiple signaling pathway alterations during oncogenic progression of lung cancer.
Oncogenically transformed HBECPKM cells reveal significant alterations in SMAD2/3 and STAT3 signaling
To identify signaling markers altered in different HBEC oncogenotypes, we focused on somatic mutation-related signaling readouts in NSCLC. In particular, to facilitate microscopy-based single-cell studies, we selected six signaling readouts whose levels and/or intra-cellular localization (cytoplasm vs. nucleus) change upon pathway alteration, namely: β-CATENIN, SMAD2/3, phospho-STAT3, P65, FOXO1 and phospho-ERK1/2 (16)(Fig. 1A; Suppl. Table 1). For β-CATENIN, we measured total intensity in the cytoplasm to capture its loss in the cell membrane and cytoplasm (as in other studies that make use of β-CATENIN as readout of Epithelial to Mesenchymal Transition (EMT)(17–19)). For the other five markers, we measured the ratio of average nuclear to average cytoplasmic intensities from individually identified cells in the immunofluorescence images (20).
Figure 1. Oncogenically transformed HBECPKM cells reveal significant alterations in SMAD2/3 and STAT3 signaling.
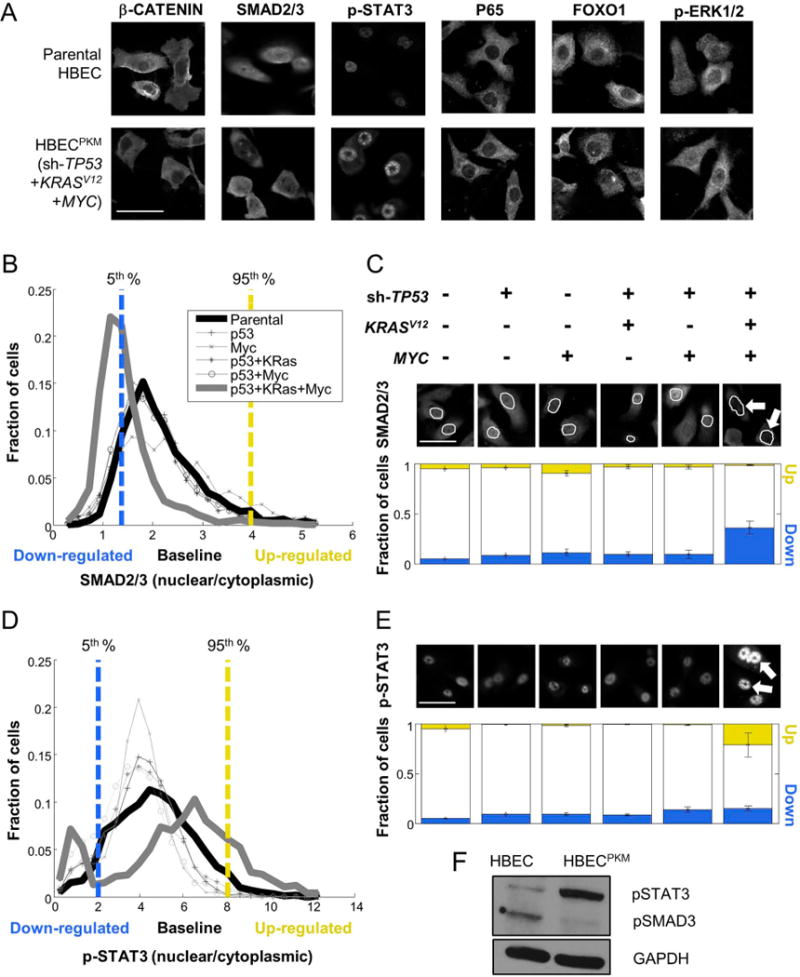
A. Immunofluorescence (IF) images of parental and HBECPKM with antibodies against β-CATENIN, SMAD2/3, p-STAT3, P65, FOXO1, p-ERK1/2 (scale bar = 50 μm).
B. Definition of downregulated (blue), baseline and upregulated (yellow) fraction of cells in SMAD2/3 signaling in oncogenically manipulated HBECs compared to the parental HBEC. For each cell line, shown are the single cell distributions of nuclear to cytoplasmic SMAD2/3 intensity. The vertical lines denote 5th (blue) and 95th (yellow) percentiles of the parental HBEC distribution. Cells below and above these lines are considered down-regulated and up-regulated respectively.
C. Quantification of signaling alteration for total SMAD2/3 in parental and oncogenically manipulated HBECs. Top: Shown are the oncogenic manipulations performed on each cell line. Middle: Sample IF images with antibodies against SMAD2/3, with cell nuclei outlined in white. The white arrows point to lower SMAD2/3 in the nuclei of HBECPKM cells. Bottom: For each cell line, blue and yellow bars indicate the fraction of up-regulated (“up”) and down-regulated (“down”) cells. Error bars represent standard deviations (n = 8 technical replicates) for fractions of altered subpopulation measured across technical replicate wells.
D–E. As in (B-C) for p-STAT3 (n = 6 technical replicates). Growth conditions are as described in text.
F. Western blot for parental HBEC and HBECPKM cells with antibodies against p-SMAD3, p-STAT3 and GAPDH (loading control).
We adopted a high-throughput, single-cell approach that can detect oncogene-induced signaling alterations, even if they are present only in a sub-population of cells. For each HBEC derivative, we used our quantitative microscopy approach to profile ~104 cells per condition, across multiple experimental conditions involving growth factors and pharmacological inhibitors. Each HBEC derivative provided a distribution of marker expression within a population. For each marker, we defined cells to be in a “baseline signaling” state if their feature values were between the 5th to 95th percentile range of all feature values observed within the parental HBEC population (Methods). Our analysis of heterogeneity focused on the enrichment or de-enrichment of outlier subpopulations. Cells with feature values below or above the basal signaling range were defined to be in a down- or up-regulated state (respectively). We measured the degree of signaling alteration based on the deviation of down- or up-regulated cellular subpopulations from 5% in the parental HBECs (Fig. 1B, 1C, 1D, 1E).
Three of our signaling readouts (P65, FOXO1, phospho-ERK1/2) showed no appreciable alteration in any oncogenically manipulated HBECs. P65 showed strong cytoplasmic localization in all cell lines, suggesting possible inactivation of TNFα pathway in HBECs. FOXO1 was distributed mostly in cytoplasm throughout all the cell lines, suggesting no obvious alteration during oncogenic progression. In addition, compared to parental HBECs, we also did not observe upregulation of phospho-ERK1/2, despite the presence of the constitutively activated (and upstream) oncogenic KRASV12 (Suppl. Fig. 2) (21). The unaltered level of ERK signaling is consistent with the similar sensitivity of parental HBEC and HBECPKM cells to the MEK (upstream of ERK) inhibitor (Suppl. Fig. 3).
The three remaining pathway readouts (β-CATENIN, SMAD2/3, phospho-STAT3) did show substantial signaling alterations, with the most dramatic change occurring for HBECPKM (Fig. 1, Suppl. Fig. 2). β-CATENIN showed dramatic loss of intensity from the cell membrane and cytoplasm, as expected from the (previously observed) mesenchymal nature of HBECPKM (Fig. 1A and Suppl. Fig. 2)(6). SMAD2/3 showed a considerable increase of down-regulated cells (from 5% to ~40% of the total population) in HBECPKM cells (Fig. 1B, 1C). This down-regulation in SMAD2/3 signaling in HBECPKM cells were also observed with phospho-SMAD2/3 antibody (Suppl. Fig. 4,). Our initial studies of phospho-STAT3 showed no obvious change in the number of up-regulated cells (Suppl. Fig. 5). This was not expected, as MYC-dependent up-regulation of phospho-STAT3 has been reported in breast cancer (22–24). We wondered whether this was due to a high concentration of added EGF (5,000 pg/mL) in our defined media. We found by lowering the concentration of EGF (50 pg/mL vs. 5000 pg/mL), which reduced EGF-related STAT3 upregulation in HBECs, that the fraction of up-regulated cells increased from 5% to ~30% (Fig. 1D, 1E, Suppl. Fig. 6). The lowered EGF concentration maintained the altered SMAD2/3 signaling differences between HBEC to HBECPKM cells. Thus, throughout the remainder of our studies, we used the low (50 pg/mL) concentration of EGF. Immunoblots using antibodies against phospho-SMAD2/3 confirmed down-regulation in SMAD2/3 signaling in HBECPKM vs. parental HBEC (of course, immunoblots cannot detect both the down- and up-regulation of signaling observed in single-cell measurements of STAT3; Fig. 1F, Suppl. Table 2). Taken together, we identified two specific signaling pathways that are altered during TP53, KRASV12 and MYC oncogenic progression (SMAD2/3 down-regulation and pSTAT3 upregulation). Importantly we found that these alterations in signaling state are not graded with the addition of oncogenic changes, but are rather “switch-like” occurring only after three oncogenic changes are in place.
Upregulation of STAT3 signaling can be utilized as a targetable vulnerability in HBECPKM
STAT3 is a known druggable target (25). We found that the HBECPKM showed increased sensitivity to STAT3 inhibitor Stattic compared to the non-manipulated HBECs (Fig. 2A, Suppl. Fig. 7). However, no cells with up-regulated STAT3 signaling were present after Stattic treatment, although the fraction of cells down-regulated simultaneously for both SMAD2/3 as well as STAT3 signaling increased (Fig. 2B). Our data suggested we need to further characterize the alterations in SMAD2/3 signaling.
Figure 2. STAT3 inhibitor diminishes the subpopulation of cells up-regulated in STAT3 signaling but leaves the subpopulation of cells down-regulated in SMAD2/3 signaling.
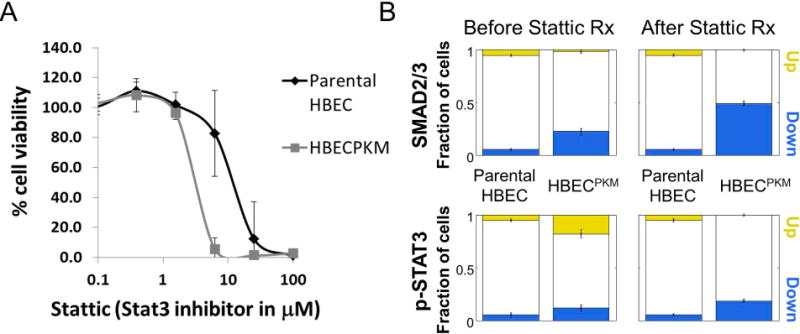
A. MTS assay of parental HBEC and HBECPKM cells for their response to STAT3 inhibitor Stattic. x-axis: concentration of Stattic in log scale; y-axis: percent viability, with 100% corresponding to DMSO control conditions for each respective cell line. Error bars represent standard deviation (n = 8 technical replicates). Solid curves were constructed using a sigmoidal curve fit.
B. Quantification of downregulated (blue) and upregulated (yellow) fraction of cells in SMAD2/3 and p-STAT3 signaling in HBECPKM cells (relative to parental HBEC) before and after Stattic treatment. Error bars as in Fig 1C (n = 6 technical replicates).
SMAD2/3 signaling alteration is MYC- dependent but not related to cell cycle or cell size changes
Because the HBECPKM cell line shows mesenchymal-like behavior (6), the SMAD2/3 down-regulated subpopulation was not expected (26,27). Thus, we next chose to investigate if alterations in SMAD2/3 signaling are genuinely oncogene-related.
First, we used a previously developed approach to identify cells in the G1 stage of the cell cycle based on the distribution of total DNA intensity (5). We found that oncogene-induced down-regulation in SMAD2/3 signaling is apparent even within the subpopulation of G1 cells (Fig. 3A). Thus, the observed changes in signaling are unlikely due to changes in the fractions of cells in different cell cycle stages. We additionally found that signaling alterations were not simply due to changes in cell size between the parental and oncogenically manipulated cells (Suppl. Fig. 8).
Figure 3. Down-regulation in SMAD2/3 signaling is oncogene-dependent.
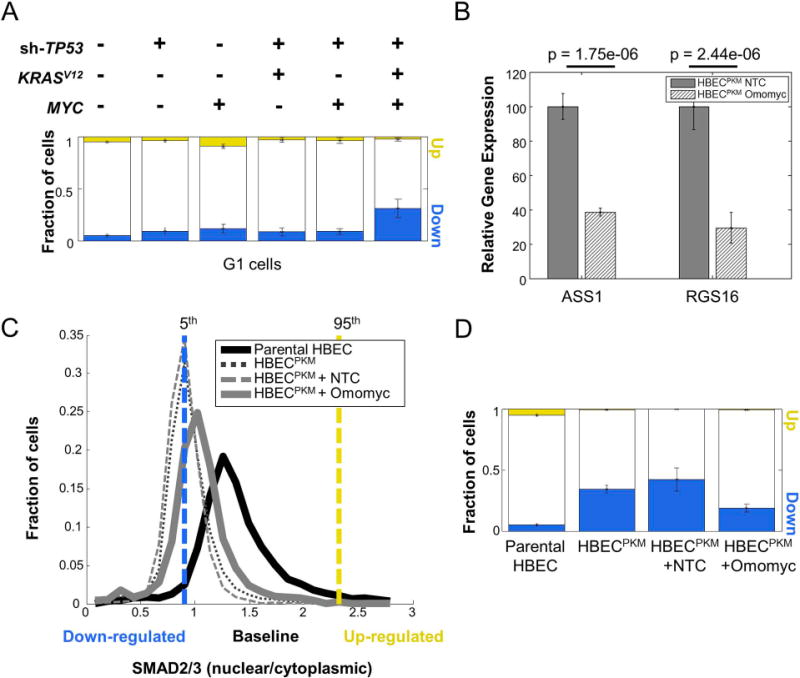
A. Single-cell quantification of altered fraction of cells across parental and oncogenically manipulated HBEC cell lines in G1 phase of cell cycle. Cells were computationally classified into various cell cycle phases based on their DNA intensity (5). For each step in the oncogenic progression, only cells belonging to the G1 phase were considered. SMAD2/3 signaling alteration was calculated as in Fig. 1C (n = 8 technical replicates).
B. qRT-PCR analysis of well-known MYC target genes (ASS1, RGS16) in HBECPKM cells with Non-target control (NTC) and Omomyc construct. The expression of each gene in HBECPKM cells with Omomyc construct is normalized to its expression in HBECPKM cells with NTC. Error bars represent standard deviation (n = 6 technical replicates); p-values are computed using a two-sided t-test.
C. Effect of MYC knockdown on SMAD2/3 signaling. Shown are the single-cell distributions of SMAD2/3 signaling (as in Fig. 1B) for HBECPKM (black dotted line), HBECPKM with non-target control vector (grey dashed line), HBECPKM with Omomyc vector (solid grey line), compared to parental HBEC (solid black line). The vertical lines denote 5th (blue) and 95th (yellow) percentiles of the parental HBEC distribution. Cells below and above these lines are considered down-regulated and up-regulated respectively. Results are obtained from pooling wells (n = 6 technical replicates).
D. Quantification of SMAD2/3 signaling heterogeneity showing fraction of upregulated (yellow) and downregulated (blue) cells, in HBECPKM, with non-target control and Omomyc construct. Error bars as in Fig. 1C (n = 6 technical replicates).
We next tested whether the oncogene-induced alteration in SMAD2/3 signaling is reversible. In particular, we wondered whether inhibition of MYC function in HBECPKM would reverse the observed signaling alteration. We stably introduced a MYC dominant negative construct, Omomyc (28), which allowed us to inducibly inhibit MYC’s transcriptional activity in HBECPKM. Upon induction of Omomyc, we observed a significant reduction in the expression of downstream target genes of MYC (RGS16 and ASS1, (29)), confirming functionality of the Omomyc construct (Fig. 3B). When assayed for SMAD2/3 signaling, the fraction of down-regulated HBECPKM cells decreased from ~40% to ~20% (Fig. 3C and 3D). This partial reversion suggests that MYC overexpression plays a role in the observed SMAD2/3 signaling alteration, and that this alteration is not simply due to permanent, genomic changes of our transformed HBECPKM cell line.
Alterations in SMAD2/3 downstream genes can reveal targetable vulnerabilities in HBECPKM
To target the subpopulation of cells with down-regulated SMAD2/3 signaling, we searched for genes with two properties. First, we looked for targets that are SMAD2/3-related. To this end, we compiled a list of 50 downstream target genes of TGFβ signaling from the literature (Methods). Second, we looked for targets that are up-regulated in HBECPKM cells primarily due to oncogenic changes rather than culture conditions. To this end, we made use of available microarray mRNA expression data to compare expression of genes in HBECPKM cells+/− serum vs. all the other HBECs in normal, serum-free growth conditions. We found 10 differentially expressed genes with significant p-values (Fig. 4A). Among these genes were MYC (which was expected and served as a positive control), BCL, MMP, FOXK and SOX family members, which were all confirmed by qRT-PCR (Fig. 4B). However, while the expression for all of these genes were strongly altered between HBECs and HBECPKM cells, BCL6 provided the most constant level of expression in HBECPKM cells (among the up-regulated alterations) independent of whether cells were cultured in short-term (40 minutes) or long-term (2 weeks) serum treatment conditions.
Figure 4. Alterations in SMAD2/3 downstream genes may reveal novel targetable vulnerabilities.
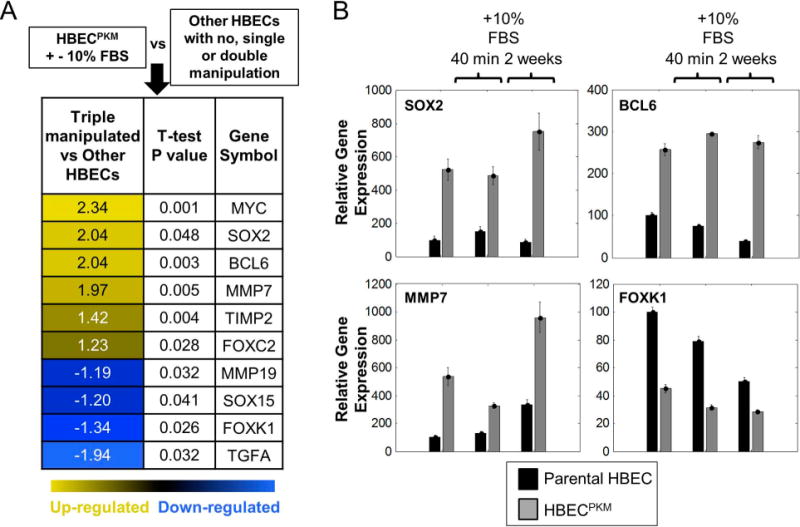
A. mRNA expression analysis of TGFβ downstream genes using microarray. HBECPKM cells grown in presence and absence of 10% FBS is in one group. Other HBECs (with no, single or double manipulations) are in the second group. Only differentially and significantly altered (two-sided t-test, p-value < 0.05) genes are shown. Numbers in the first column represent fold change (Log2) between the median level of gene expression of HBECPKM and other HBECs.
B. mRNA expression analysis of top 3 upregulated genes (SOX2, BCL6, MMP7) in HBECPKM cells using qRT-PCR. FOXK1 is used as an example of a downregulated gene in HBECPKM. Parental HBEC (black) or HBECPKM (grey) were grown in defined, serum-free growth condition, in 10% FBS treatment for 40 minutes or in 10% FBS treatment for 2 weeks. y-axis: gene expression relative to human reference (Methods). Error bars represent standard error (n = 6 technical replicates).
Next, we observed that MYC and BCL6 gene expressions are strongly correlated (Pearson's correlation coefficient 0.83) across 20 single cell clones of HBECPKM (Fig. 5A). Such correlation of BCL6 expression with the expression of other genetic manipulations (TP53 and KRAS) were relatively lower. Further, we found that inhibition of MYC function with Omomyc reduced BCL6 level by 2-fold (Fig. 5B). This low level of BCL6 expression after MYC inhibition is comparable to the level of BCL6 in the parental HBEC. Taken together, BCL6, a downstream gene of SMAD2/3 signaling (30–32), is significantly upregulated in a serum-independent manner, and strongly correlated to MYC expression in HBECPKM cells.
Figure 5. BCL6 is a targetable vulnerability in HBECPKM cells.
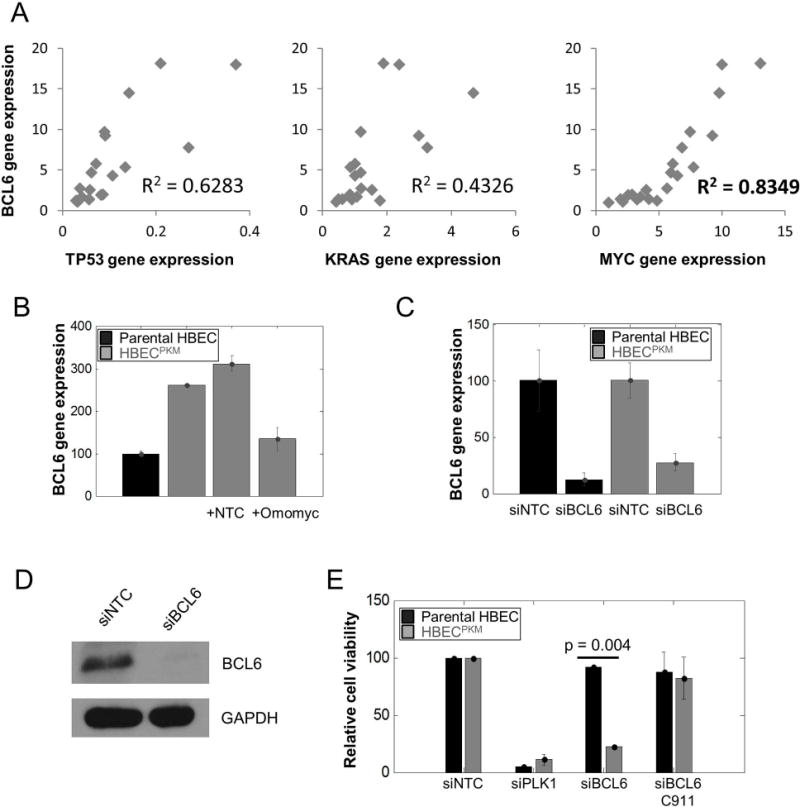
A. Correlation of relative BCL6 gene expression and TP53, K-RAS and MYC gene expression in 20 single-cell clonal populations of HBECPKM using qRT-PCR. The coefficients of determination R2 (0.6283, 0.4326 and 0.8466 respectively) for simple linear regression as shown.
B. qRT-PCR of BCL6 gene expression for (left to right): parental HBEC (black), HBECPKM, HBECPKM transfected with non-target control (NTC) and HBECPKM transfected with Omomyc construct. Error bars are as in Fig. 3B (n = 2 technical replicates); normalization is with respect to the parental HBEC.
C. Confirmation of BCL6 knockdown based on gene expression. Relative BCL6 gene expression using qRT-PCR for (left to right): parental HBEC transfected with non- target control, parental HBEC transfected with siRNA against BCL6, HBECPKM transfected with non-target control and HBECPKM transfected with siRNA against BCL6. Error bars are as in Fig. 3B (n = 6 technical replicates); the expression of BCL6 in each cell line with siBCL6 is normalized to BCL6 expression with siNTC.
D. Confirmation of BCL6 knockdown based on protein expression. Western blot showing BCL6 protein expression in HBECPKM transfected with non-target control and siRNA against BCL6
E. Effect of BCL6 knockdown on cell viability for parental HBEC and HBECPKM transfected with (left to right): non-target control, siRNA against PLK1 (positive toxic control), siRNA against BCL6, and C911 control for siBCL6. Error bars represent standard error (n = 2 technical replicates) and p-values are calculated using a two sided t-test.
BCL6 is a targetable vulnerability in HBECPKM cells
To test if BCL6 is a targetable vulnerability in HBECPKM, we made use of siRNA-mediated knockdown of BCL6 and searched for a differential change in viability of HBECPKM cells vs HBEC cells. First, we confirmed that our siRNA knocked down both BCL6 mRNA and protein expression (Fig. 5C, 5D, Suppl. Fig. 9). Next, we found that the BCL6 knockdown resulted in significantly higher cell death (4 fold) in HBECPKM cells than the parental HBECs (Fig. 5E). Using a C911 BCL6-siRNA construct (7) we showed that the siRNA knockdown was an “on target” effect. Thus, we found BCL6 to be a potential targetable vulnerability in the HBECPKM cells.
BCL6 can be a targetable vulnerability in a subset of non-small cell lung cancer lines
To examine the generality of BCL6 as a vulnerability, we tested 5 NSCLC cell lines: H1693, H1819, H1993, HCC827 and H2009. We compared SMAD2/3 signaling in these cells to parental HBECs (Fig. 6A). We utilized parental HBEC cells treated with 10% serum for 40 minutes as a positive control for SMAD2/3 up-regulation. Our analysis showed that the majority of cells (>50%) in H1693, H1993 and HCC827 show SMAD2/3 down-regulation compared with the parental HBEC. In contrast, H2009 and H1819 showed a smaller fraction of cells with SMAD2/3 down-regulation. Further, we found that H1693 and H1993 have significantly higher levels of BCL6 gene expression compared to the parental HBEC in our qRT-PCR and western blot assays (Fig. 6B, 6C). The high levels of BCL6 decreased significantly after MYC target gene knockdown using the Omomyc construct similar to the result we observed in the HBECPKM cells (Fig. 3C, D). After siRNA-mediated knockdown of BCL6 we observed that H1693 and H1993 showed significantly higher cell death than H2009 (Fig. 6D). Hence, our data suggests that BCL6 can be a targetable vulnerability in a subset of NSCLC cell lines that exhibit SMAD2/3 down-regulation coupled with increased BCL6 expression.
Figure 6. BCL6 in a targetable vulnerability in a subset of NSCLC cell lines.
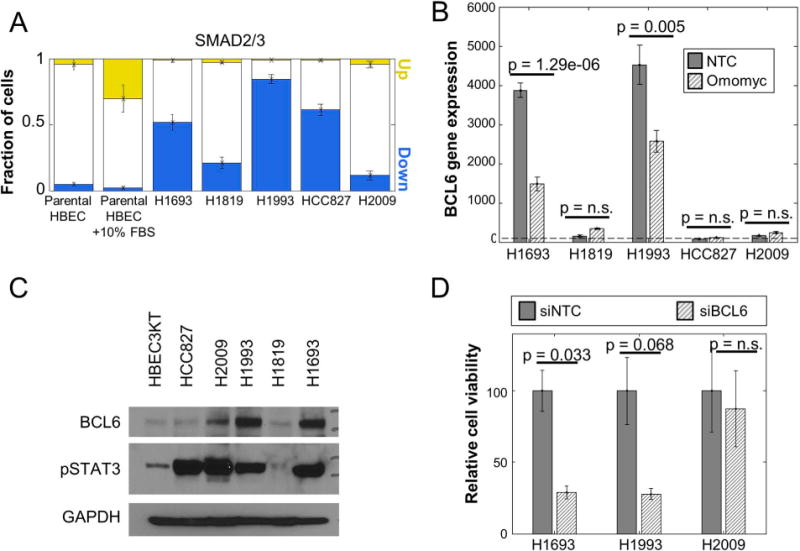
A. Quantitation of the fraction of cells with altered SMAD2/3 signaling in NSCLC cell lines H1693, H1819, H1993, HCC827 and H2009 compared to parental HBEC and parental HBEC treated with 10% FBS for 40 minutes (positive control) based on immunofluorescence microscopy. Error bars (n = 7 technical replicates) as in Fig. 1C.
B. Relative BCL6 gene expression from qRT-PCR assay of NSCLC cell lines transfected with non-target control and Omomyc construct compared to the level in parental HBEC (horizontal dotted line). Error bars and p-values are as in Fig. 3B (n = 6 technical replicates).
C. Protein expression of BCL6 and phospho-STAT3 in NSCLC cell lines H1693, H1819, H1993, HCC827 and H2009 compared to parental HBEC as measured by western blot. GAPDH is used as a loading control.
D. Relative viability of NSCLC cell lines H1693 and H1993 compared to H2009 after siRNA mediated BCL6 knockdown. Error bars and p-values are as in Fig. 3B (n = 2 technical replicates).
Combined treatment with BCL6 inhibitor and STAT3 inhibitor can achieve better response than treatment with each inhibitor alone in non-small cell lung cancer lines
After identifying BCL6 and phospho-STAT3 as targetable vulnerabilities, we tested the effect of small molecule inhibitors of these components alone and in combination using colony formation assays in characterized NSCLC lines. To pharmacologically block the transcriptional function of BCL6, we utilized FX-1 (12). Among our panel of lung cancer cell lines, H1693 was highly sensitive to FX-1 (Fig 7A, B). By contrast, HCC827 and H2009 were relatively resistant while H1993 had an intermediate response. To block the transcriptional function of STAT3, we utilized the highly potent inhibitor BBI-608 (10,11) that is known to down-regulate expression of STAT3 target genes. Unlike our previously used inhibitor (stattic), BBI-608 can be used in a more clinically relevant setting. Among our panel of lung cancer cell lines, HCC827 was highly sensitive to BBI-608 (Fig 7A, B). By contrast, H1693 and H2009 were relatively resistant. H1993 showed an intermediate response. We observed a correspondence between higher sensitivities to FX-1 with higher levels of BCL6 protein expression but no such correspondence of sensitivity to BBI-608 with phospho-STAT3 levels (Fig. 6C). Taken together, the combination treatment eliminated more cancer cells in vitro than the treatment with each inhibitor alone.
Figure 7. In vitro or in vivo combination treatment with BCL6 and STAT3 inhibitors eliminates more cancer cells than single agent treatments.
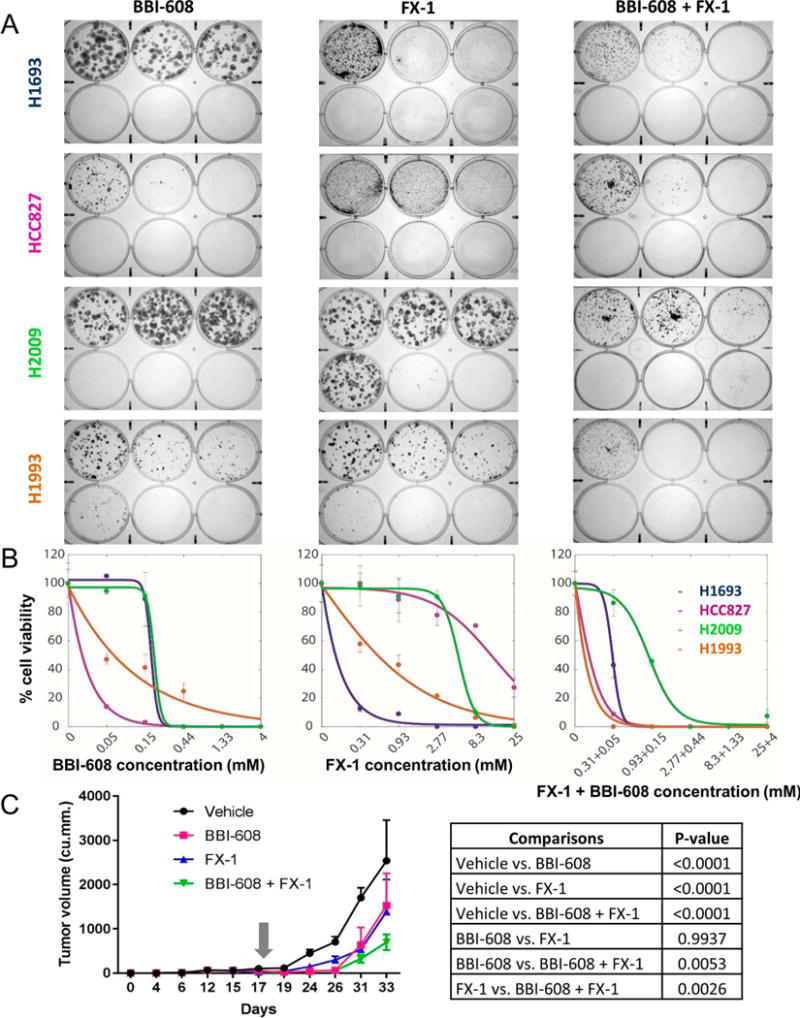
A. Liquid colony formation assay of four NSCLC cell lines H1693, HCC827, H2009 and H1993 with their responses to STAT3 inhibitor, BBI-608 alone (left), FX-1 alone (middle) and the combination treatment (right). Image shows crystal violet stained colonies after 2 weeks of drug treatment. For both single agent and combination treatments, BBI-608 concentration (mM) ranges in the six well plates are 0, 0.05, and 0.15 (top row left to right), and 0.44, 1.33 and 4 (bottom row left to right). FX-1 concentration (mM) ranges in the six well plates are 0, 0.31, and 0.93 (top row left to right), and 2.77, 8.3 and 25 (bottom row left to right).
B. Quantification of liquid colony formation assay of four NSCLC cell lines H1693, HCC827, H2009 and H1993 to BBI-608 alone (left), FX-1 alone (middle) and the combination treatment (right). The number of colonies in each experimental condition were normalized to vehicle controls. Error bars represent standard deviation (n=8 technical replicates). Solid curves were constructed using a sigmoidal curve fit.
C.Volume of H1993 cell line derived subcutaneous, xenografted tumors measured (1/2*length*width*width) and averaged in each of the 4 treatment groups (vehicle, BBI-608 alone, FX-1 alone and BBI-608+FX-1 combination) over time. BBI-608 was administered at 20 mg/kg. FX-1 was administered at 25 mg/kg. Gray arrow represents the start day of treatment. P-values are calculated by two way Anova with Tukey’s multiple comparison test. Error bars represent standard deviation (n = 10 technical replicates).
Combination of BBI-608 (STAT3 inhibitor) and FX-1 (BCL6 inhibitor) increased H1993 cell line derived xenografts’ response to therapy
We wondered whether this observed benefit of combined treatment would also hold in vivo for H1993-derived subcutaneous xenografts (see Supplementary Table 3 for doses and schedules). As with the in vitro results, we found that both single-agent therapies significantly reduced (P<0.0001) the growth in tumor volume as compared to the vehicle treated controls (Fig. 7C, Suppl. Table 3). More importantly, the combination therapy significantly increased (P<0.005) the response of the xenografted tumors compared to tumors from the single agent therapies. Taken together, the growth rate of the tumor supported the hypothesis that combined treatment was more effective than either treatment alone.
Discussion
We investigated whether oncogene-induced signaling alterations can provide insight into vulnerabilities of cancer cell populations and if these changes are homogeneous or heterogeneous in a defined, genetically manipulated, human bronchial epithelial cell preclinical model. Our study utilized HBECs, a simplified cellular model of defined oncogenesis (with TP53 knockdown, KRASV12 and MYC overexpression) and focused on signaling pathways commonly affected in lung cancer. We found that SMAD2/3 signaling becomes down-regulated and phospho-STAT3 signaling becomes up-regulated after all three oncogenic manipulations. Interestingly, in HBECPKM cells, SMAD2/3 down-regulation and phospho-STAT3 up-regulation occur in two mutually exclusive subpopulations. Thus, heterogeneity in signaling states is present, even within a defined oncogenotype. We found that the drug stattic, designed to target STAT3, eliminated cells with STAT3 up-regulation, but left a large surviving subpopulation of cells with down-regulated SMAD2/3 signaling. There is currently no obvious druggable target for such SMAD2/3 downregulated cells. To target the subpopulation with down-regulated SMAD2/3 signaling, we searched for downstream genes of SMAD2/3 that are correspondingly up-regulated in a serum-independent manner. This strategy identified the transcription factor BCL6 as such a SMAD2/3 downstream target making it a potential genetic vulnerability. This vulnerability was validated both in our HBECPKM model as well as NSCLC cell line models exhibited dysregulated SMAD2/3 signaling. Our ability to knock down only a sub-population of cells in the HBEC series with the STAT3 drug, led us to design a combination therapy with small molecule inhibitors targeting both STAT3 and BCL6 which provided increased response in vitro and in vivo in subcutaneous xenografted tumors.
BCL6 has recently been characterized as a vulnerability in a subset of breast cancers but to our knowledge has not been described as a therapeutic target in lung cancer (33). BCL6 was also identified as a target of miR-187-3p in NSCLC cell lines A549 and SPC-A-1 (34). Of course classically, BCL6 has been widely studied in the context of B-cell lymphoma (35) and its connection to TGFβ resistance (36). In particular, for a subgroup of lymphoma cases, known as “double-hit lymphoma”, BCL6 and MYC aberrations together are considered as prognostic factors (37). Interestingly, we find the analogous result that MYC regulates BCL6 expression in a subset of NSCLC cell lines. Our results point to a possible new strategy to target MYC- related BCL6 over-expressing lung cancers. Further clinical translation of such phospho-STAT3, BCL6 targeted therapy will require both development of small molecule inhibitors with appropriate characteristics for therapeutic delivery and information if and how lung cancers develop resistance to such combined phospho-STAT3 and BCL6 targeted therapy.
The paucity of “clinically actionable” targets for the majority of driver oncogenes and tumor suppressor genes in lung cancer indicates the need to find ways to therapeutically target these potential vulnerabilities. In the present work, we show that single-cell analysis of oncogene-induced signaling alterations can reveal targetable vulnerabilities for multiple cellular subpopulations, each having potentially distinct signatures of signaling alteration. Heterogeneity in cancer cell populations has been traditionally viewed as an impediment to effective diagnosis and treatment. Our work suggests that analysis of heterogeneity may, in fact, help identify molecular vulnerabilities and suggest rational, effective therapeutic combination strategies.
Supplementary Material
Acknowledgments
We thank Dr. Laura Soucek for kindly providing us with the Omomyc construct, Dr. Luc Girard for his help with the microarray data analysis, and Dr. Ari Melnick for advice and drug for the BCL6 studies. We also thank all the members of Altschuler lab, Wu lab, and Minna lab for useful discussions.
Financial support: University of Texas Southwestern Medical Center SPORE P50CA70907 (J. D. Minna), CPRIT RP110708 (J. D. Minna), U01 CA176284 (J. D. Minna), National Health and Medical Research Council of Australia Overseas-Based Biomedical Training Fellowship (494511) and TSANZ/Allen & Hanburys Respiratory Research Fellowship (J. E. Larsen), NCI-NIH RO1 CA133253 (S. J. Altschuler), NSF PHY-1545915 (S. J. Altschuler), SU2C/MSKCC (S. J. Altschuler), NCI-NIH RO1 CA185404 (L. F. Wu) and the Institute of Computational Health Sciences (ICHS) at UCSF (S. J. Altschuler and L. F. Wu).
Footnotes
Conflict of interest: The authors disclose no potential conflicts of interest.
References
- 1.Lauro S, Onesti CE, Righini R, Marchetti P. The use of bevacizumab in non-small cell lung cancer: an update. Anticancer research. 2014;34:1537–45. [PubMed] [Google Scholar]
- 2.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clinics in chest medicine. 2011;32:703–40. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. The Lancet Oncology. 2015;16:e342–51. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 4.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155:552–66. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh DK, Ku CJ, Wichaidit C, Steininger RJ, 3rd, Wu LF, Altschuler SJ. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Molecular systems biology. 2010;6:369. doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11:638–50. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buehler E, Chen YC, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PLoS One. 2012;7:e51942. doi: 10.1371/journal.pone.0051942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasband WS. ImageJ. National Institutes of Health B; Maryland, USA: 1997–2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 9.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–53. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1839–44. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Jin Z, Zhou H, Ou X, Xu Y, Li H, et al. Suppression of prostate cancer progression by cancer cell stemness inhibitor napabucasin. Cancer Med. 2016;5:1251–8. doi: 10.1002/cam4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, et al. Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J Clin Invest. 2016;126:3351–62. doi: 10.1172/JCI85795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science (New York, NY) 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science (New York, NY) 2014;346:256–9. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harbor perspectives in biology. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. The Journal of clinical investigation. 2009;119:1429–37. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science (New York, NY) 2004;306:1194–8. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 21.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer cell. 2006;10:459–72. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J, et al. Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PloS one. 2008;3:e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong A, Yu W, Liu Y, Sanders BG, Kline K. Elimination of ALDH+ breast tumor initiating cells by docosahexanoic acid and/or gamma tocotrienol through SHP-1 inhibition of Stat3 signaling. Molecular carcinogenesis. 2015 doi: 10.1002/mc.22291. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9623–8. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendell JC, Hong DS, Burris HA, 3rd, Naing A, Jones SF, Falchook G, et al. Phase 1, open-label, dose-escalation, and pharmacokinetic study of STAT3 inhibitor OPB-31121 in subjects with advanced solid tumors. Cancer chemotherapy and pharmacology. 2014;74:125–30. doi: 10.1007/s00280-014-2480-2. [DOI] [PubMed] [Google Scholar]
- 26.Kaowinn S, Kim J, Lee J, Shin DH, Kang CD, Kim DK, et al. Cancer upregulated gene 2 induces epithelial-mesenchymal transition of human lung cancer cells via TGF-beta signaling. Oncotarget. 2016 doi: 10.18632/oncotarget.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, He Z, Gao Y, Zheng R, Zhang X, Zhao L, et al. Induced Pluripotent Stem Cells Inhibit Bleomycin-Induced Pulmonary Fibrosis in Mice through Suppressing TGF-beta1/Smad-Mediated Epithelial to Mesenchymal Transition. Frontiers in pharmacology. 2016;7:430. doi: 10.3389/fphar.2016.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soucek L, Helmer-Citterich M, Sacco A, Jucker R, Cesareni G, Nasi S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–72. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 29.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and cellular biology. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huret J-L, Ahmad M, Arsaban M, Bernheim A, Cigna J, Desangles F, et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013;41:D920–4. doi: 10.1093/nar/gks1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–76. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Lin X, Cai Z, Zhang Z, Han C, Jia S, et al. Global identification of SMAD2 target genes reveals a role for multiple co-regulatory factors in zebrafish early gastrulas. J Biol Chem. 2011;286:28520–32. doi: 10.1074/jbc.M111.236307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker SR, Liu S, Xiang M, Nicolais M, Hatzi K, Giannopoulou E, et al. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene. 2015;34:1073–82. doi: 10.1038/onc.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F, et al. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem Biophys Res Commun. 2016;471:82–8. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
- 35.Lindsley RC, LaCasce AS. Biology of double-hit B-cell lymphomas. Current opinion in hematology. 2012;19:299–304. doi: 10.1097/MOH.0b013e328353bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Long J, Dai F, Liang M, Feng X-H, Lin X. BCL6 represses Smad signaling in transforming growth factor-beta resistance. Cancer research. 2008;68:783–9. doi: 10.1158/0008-5472.CAN-07-0008. [DOI] [PubMed] [Google Scholar]
- 37.Cinar M, Rosenfelt F, Rokhsar S, Lopategui J, Pillai R, Cervania M, et al. Concurrent inhibition of MYC and BCL2 is a potentially effective treatment strategy for double hit and triple hit B-cell lymphomas. Leukemia research. 2015 doi: 10.1016/j.leukres.2015.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data discussed in this publication have been made available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) public repository <http://www.ncbi.nlm.nih.gov/geo/> and are accessible through GEO Series accession number GSE40828. Raw images of a subset or complete dataset are available upon request.


