Abstract
Transgenic mice that allow mutant cells to be visualized in situ were used to study variation in tumors. These mice carry the G11 placental alkaline phosphatase (PLAP) transgene, a mutant allele rendered incapable of producing its enzyme product by a frameshift caused by insertion of a tract of G:C base pairs in a coding region. Spontaneous deletion of one G:C base pair from this tract restores gene function, and cells with PLAP activity can be detected histochemically. To study tumors, the G11 PLAP transgene was introduced into the polyoma virus middle T antigen mammary tumor model. Tumors in these mice exhibited up to 300 times more PLAP+ cells than normal tissues. PLAP+ cells were located throughout each tumor. Many of the PLAP+ cells were singlets, but clusters also were common, with one cluster containing >30,000 cells. Comparison of these data to simulations produced by computer models suggested that multiple factors were involved in generating mutant cells in tumors. Although genetic instability appeared to have occurred in most tumors, large clusters were much more common than expected based on instability alone.
Keywords: microsatellites, mutation, instability, mouse
Cells in tumors tend to have multiple mutations (1–3). The presence of multiple changes may be due to genomic instability, where each cell in the tumor exhibits an increased rate of mutation (4). Alternatively, proliferation could be involved. For example, were a cell to acquire a mutation that causes it and its progeny to proliferate more, this would increase the chance of a second mutation in one of the progeny. Repeated cycles of this process would allow multiple mutations to accumulate in a single genome, even though the rate of mutation per cell per generation is normal (1).
The relative contributions of genomic instability and hyperproliferation to the mutations present in cells of a tumor cannot be resolved simply by counting mutant cells, because both processes can produce large numbers of these. However, the two processes would be expected to generate tumors with different phenotypes with respect to the positions of mutant cells. If genomic instability predominates, then many independent mutant cells would arise in the tumor. Most of these mutants would tend to reside at locations separate from other mutant cells. By contrast, if hyperproliferation predominates, then mutant cells would tend to be in a few large clusters.
The positions of mutant cells can be studied by using a transgenic mouse [G11 placental alkaline phosphatase (PLAP)] that allows mutant cells to be visualized in situ in tissue sections (5–8). G11 PLAP mice carry a mutant allele of a human PLAP transgene (G11). This mutant allele does not produce enzyme activity because of the presence of a tract of 11 G:C base pairs that shifts ribosomes into the wrong translational reading frame. Studies on cultured cells carrying this allele showed that deletion of one G:C base pair from this tract restores enzyme activity and produces a cell that stains histochemically (5–8).
The G11 tract in the PLAP transgene mimics mononucleotide repeats in the genome, which, considering their propensity to suffer mutation via deletion or insertion of base pairs during DNA replication, are surprisingly common in coding regions (9). More than 20 human genes have been reported to have a mononucleotide repeat (of at least 8 bp) in a coding region, and many of these have been observed to suffer mutation. The list of genes carrying a coding-mononucleotide repeat includes genes that perform functions critical for preventing cancer, such as DNA repair (RAD50, DNA-PKcs, hMSH3, hMSH6, and XPG) and regulation of cell proliferation, apoptosis, and tumor growth (TGFβRII, IGFIIR, CHK-1, BAX, ICE, caspase-5, FLASH, Apaf-1, E2F-4, TCF-4, Apc, c-myb, MYCL, CtIP, and MLSN1) (10–20). Many more human genes are at risk of suffering mutation due to mononucleotide repeat instability. A search of ≈33,000 human coding sequences identified mononucleotide repeats with 9 or more bp in 365 entries (21). Another study found 336 messenger RNAs containing either G7 or C7 and 4,382 containing either A7 or T7 (22).
Previous studies on the G11 PLAP mouse showed that PLAP+ cells were easily detectable in cells in the four organs examined in detail (brain, heart, kidney, and liver) (8). The average frequency of PLAP+ cells in normal tissues was ≈1.5 × 10-5, far lower than can be detected by DNA amplification but consistent with results of selection experiments, which have shown that mononucleotide tracts act as hot spots for frameshift mutations in vivo in lymphocytes (23).
It is well known that cells defective for mismatch repair exhibit a high-microsatellite-instability (MSI-H) phenotype, whereby changes in simple sequence tracts occur so frequently that populations of cells tend to be heterogeneous with respect to the length of any given tract (24). Cells proficient in repair do not exhibit MSI-H, but evidence is accumulating suggesting that low-level instability can occur (24). A so-called low-microsatellite-instability phenotype has been reported in cells from tissues that are either inflamed or hyperplastic and in carcinogen-induced rat mammary tumors (10, 14, 25, 26). In addition, changes were seen in somatic cell microsatellites of offspring of individuals exposed to radiation from the Chernobyl disaster (27). The PLAP+ cells present in mismatch repair-proficient mice may be formed by the same process that causes low microsatellite instability, and the G11 allele may respond to the same kinds of environmental insults.
To study mutation in tumors in situ, the G11 PLAP gene was introduced into a mammary tumor mouse model (28). These mice carry a transgene that causes expression of the polyoma virus middle T antigen in mammary cells. Middle T antigen is located at the cell membrane, where it binds and activates kinases that ultimately activate the Ras signal transduction pathway (29, 30). It was reasonable to anticipate that cells in the polyoma virus middle T antigen tumor model might exhibit reduced genetic stability, because Ras pathway activation has been linked to genetic instability (31–33). In addition, mouse mammary cancers induced by chronic c-erbB2 overexpression, which also disrupts cell cycle regulation, have been shown to be genetically unstable (34).
Tumors in polyoma virus middle T antigen mice contained up to 300 times more PLAP+ cells than normal tissues. PLAP+ cells were located throughout each tumor, and many were situated as singlets, suggesting that genetic instability occurred. However, comparison of the tumor data with simulations produced by computer models suggested that additional factors were involved in generating mutant cells in these tumors.
Methods
Mice. Transgenic FVB/N mice carrying the G11 PLAP frameshift reporter gene were generated as described (5, 7). Transgenic mice that develop mammary tumors with high frequency (strain FVB/N-TgN(MMTVPyVT)634Mul) were acquired from The Jackson Laboratory. Heterozygous MMTVPyVT mice were mated with G11 PLAP mice from line 100A (5, 7, 8). The PLAP gene was detected by amplification of tail DNA as described (8). The MMTVPyVT transgene was detected in tail DNA by using the PCR protocol provided by The Jackson Laboratory (http://jaxmice.jax.org). Mice were observed until tumors became apparent, which typically happened by the age of 6 mo regardless of sex. Tumors were taken from 20 mice (4 males and 16 females). Ten of these mice (3 males and 7 females) carried the G11 PLAP transgene. When multiple tumors occurred in a mouse, they were labeled with the mouse-identifier number followed by a letter, such as 125a, 125b, etc. Tumors in one location were often composed of more than one mass. It was not possible to determine whether such tumors were from a single source. Tumors varied in size, shape, and solidity. The largest was ≈6 cm3 in volume. Most, however, were between 1 and 3 cm3. There was no relationship between tumor size, shape, or solidity and PLAP phenotype.
Preparation and Staining of Tissues. Organs and tumors were removed and either frozen in OCT embedding compound on dry ice or snap frozen in liquid N2-cooled isopentane. Cryosections were cut 10 μm thick, mounted on slides, and fixed in 2% formaldehyde/0.2% glutaraldehyde in phosphate buffer (PB) (0.15 M NaCl/2.7 mM KCl/1.47 mM KH2PO4/4.86 mM Na2HPO4, pH 7.4) for 10 min at room temperature. Endogenous phosphatases in fixed tissues were inactivated by incubation of mounted sections at 65°C for 1 hr in PB. To detect PLAP activity, sections were incubated in 1 mg/ml 5-bromo-4-chloro-3-indolyl phosphate and nitro-BT in 0.1 M Tris, pH 10, for 30–90 min at 37°C. Sections were then washed in PB and stained for 5 min in Nuclear Fast Red (Vector Laboratories). Stained sections were dehydrated by sequential washes with aqueous in ethanol starting at 10% ethanol and ending with 90%. Dehydrated slides were dried in air, and sections were placed under cover slips by using Permount Mounting Medium (Fisher Scientific).
Enumeration of PLAP+ Cells. Images were captured with a Spot Jr digital camera (Diagnostic Instruments, Sterling Heights, MI). The number of cells examined per tissue sample was estimated as follows. The cells in a field of view (FOV) at ×400 were counted, and the area occupied by these cells was determined. The area of each section was measured from a digital image at ×20. Corrections were made to account for holes in sections. The number of cells in the section was computed from the ratio of section and ×400 FOV areas. The total number of tumor cells examined exceeded 1 × 108. The total number of PLAP+ cells observed in tumors exceeded 1 × 105. The number of PLAP+ cells in each tumor section varied, but at least 60 PLAP+ cells were counted in each tumor. This quota was easy to reach, because most tumors had >100 PLAP+ cells per section, and on the order of 1,000 PLAP+ cells were scored in most tumors. Normal tissues exhibited far fewer PLAP+ cells. Counting was pursued until at least 20 PLAP+ cells had been seen in each tissue sample examined.
Scoring Individual and Clusters of PLAP+ Cells. A PLAP+ cell was scored as a singlet if there were no other PLAP+ cells within an area defined by 10 cell diameters and preceding and succeeding serial sections lacked a PLAP+ cell in the location of interest. Clusters of PLAP+ cells were scored on individual sections, which defined them in two dimensions (x and y). The z dimension was examined by viewing serial sections, which showed that, in general, clusters extended into the z dimension, and that the larger the cluster, the more sections it occupied. These data confirmed expectations that clusters would occupy 3D space. However, data from the x-y dimension were sufficient for determining the relationship between cluster frequency and size, and ignoring the z dimension simplified the analysis. Therefore, only data from individual sections (x-y data) were used to define these relationships.
Computer Models. The genesis and proliferation of mutant cells in a tumor were simulated by an algorithm that did the following: Each simulation started with a single PLAP-negative cell, which produced two cells, which produced four cells, etc., until 1 million cells were produced. At each doubling, the number of mutant cells produced was determined by multiplying the number of cells present by the probability of mutation. Simulations were performed with the probability of mutation set at either 1 × 10-4,1 × 10-5,or1 × 10-6, and 100 simulations were performed for each probability. Spatial associations of PLAP+ cells originating from simulated spontaneous random mutations were represented in 1D space. Clusters of PLAP+ cells were defined as adjacent PLAP+ cells in the string of simulated cells. To assess the effects of hyperproliferation of mutant cells, the algorithm was modified to cause each mutant cell to double twice for each doubling of nonmutant cells.
Probability Analysis. If mutations are generated during DNA replication and the probability of mutation is p events per cell per replicative cycle, then there will be p(n/2) single-cell mutants present in a population of n cells because these mutants were generated during the previous round of cell division. If the population increases uniformly by doubling each cell in it at each generation, then the number of clusters containing two cells will be p(n/4), because these pairs of cells came from single cells in a previous generation when the population size was n/2. This relationship extends to clusters of all sizes and the number of clusters containing 2k cells will be p(n/2k+1). The proportions calculated for different ks were compared with observed proportions by using Fisher's exact test for Table 1.
Table 1. 2×2 table of values compared by the Fisher exact test.
| No. of single mutants | (n/2) - (no. of single mutants) |
| No. of clusters containing 2k mutants | (n/2k + 1) - (no. of clusters containing 2k mutants) |
Results
Tumors Exhibited an Elevated Number of Mutant Cells. To assess the instability of the PLAP-negative phenotype in situ, mice were killed, and tumors and major organs were removed and frozen. Sections were cut from frozen tissues and PLAP+ cells were detected by histochemical staining. PLAP+ cells were scored in 18 tumors from 10 G11 mice. Normal breast, brain, heart, kidney, and liver were also analyzed. MMTVPyVT mice that lacked the PLAP gene provided 11 control tumors. No PLAP+ cells were observed in such mice.
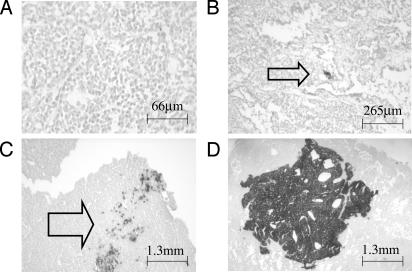
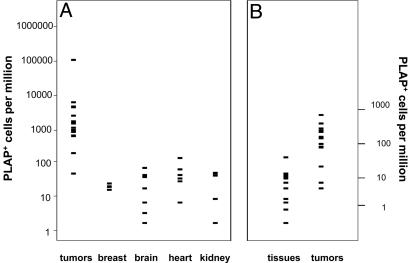
PLAP+ cells were common in sections cut from tumors. Fig. 1 shows some examples of the different section phenotypes observed. Tumors exhibited many more PLAP+ cells than normal tissues (Fig. 2A). The arithmetic mean frequency in tumors was 100-fold greater than that in the heart, which was the normal tissue that exhibited the most PLAP+ cells (Fig. 2 A). The frequency varied greatly among the tumors, and one tumor (714) exhibited a very large number of PLAP+ cells (108,513), which inflated the mean. However, 16 of the other 17 tumors exhibited at least 10 times more PLAP+ cells than normal breast tissue. The mean frequency in these 17 tumors was 2,036, which is 85-fold greater than in the normal breast. The median frequencies were 1,378 and 24 in tumors and normal breast tissues, respectively. Normal breast tissue from mice carrying the PyMT transgene did not exhibit more PLAP+ cells than breast tissue from mice that lacked the PyMT transgene. Immunohistochemical analysis of frozen sections showed that the viral oncoprotein was present in most, if not all, mammary cells of MMTVPyVT mice (data not shown). The MMTV promoter is estrogen-dependent and is not very active in the other tissues analyzed (brain, heart, kidney, and liver). Therefore, it was expected that the mean frequencies of PLAP+ cells in tissues of MMTVPyVT mice would not differ from those previously observed in mice lacking the PyMT transgene. Comparison of the MMTVPyVT with those previously obtained showed this to be the case (8). The frequency of PLAP+ cells in tumors was not correlated with frequency in the normal tissues of the same animal. None of the animals studied exhibited a generalized high level of mutation. Therefore, the phenotypes of the tumors were a feature of tumor cells.
Fig. 1.
Representative images of mammary tumor sections stained for PLAP activity. Cells with PLAP activity are dark. (A) Tumor from a mouse that did not carry the PLAP gene. Line indicates 66 microns. (B) Arrow indicates a single PLAP+ cell. Line indicates 265 microns. (C) Arrow indicates a group of PLAP+ cells. Line indicates 1,300 microns. (D) A very large group of PLAP+ cells. Line indicates 1,300 microns.
Fig. 2.
Frequencies of PLAP+ cells in tumors and normal tissues. (A) Total PLAP+ cells. (B) Single PLAP+ cells. Tissue data were from tissues listed in A.
The increased number of PLAP+ cells in tumors was not due to more extensive expression of the PLAP gene in tumors. Expression was assessed by examining tumors and tissues from mice that carried a revertant allele of the G11 PLAP transgene. In studies described elsewhere (35), mice carrying a reverted G11 PLAP gene were obtained by passing this gene through female mice that lacked a functional copy of the mismatch repair gene Pms2. Some of the offspring from these females were found to express PLAP in tail tissue. These putative germ-line revertant mice were crossed to wild-type mice to verify that the PLAP+ phenotype was transmitted as a dominant trait. Approximately half of the cells in tumors and tissues from mice carrying the germ-line revertant allele were positive for PLAP activity. Therefore, the PLAP gene was transcribed and translated in the same fraction of cells in tumors and normal tissues.
Prevalence of Single Mutant Cells Suggests Genetic Instability. PLAP+ cells were situated both alone and in clusters. Single PLAP+ cells were of particular interest, because their frequency should not be affected by proliferation of PLAP+ cells. Hence, the frequency of single PLAP+ cells should be indicative of the rate of mutation in a given tissue. Compared with normal tissues, tumors tended to exhibit many more single PLAP+ cells. Fig. 2B shows that tissues exhibited approximately four single PLAP+ cells per million cells (median, 3.5). This value was similar to that reported previously (8). By contrast, the median frequency of single PLAP+ cells in tumors was close to 50 per million. In addition, there were more single PLAP+ cells in all but 3 of 13 tumors. In large populations, mutation rates can be approximated from the frequency of mutants (36). Therefore, the rate of mutation in tumors appeared to be at least 10-fold higher than in normal cells.
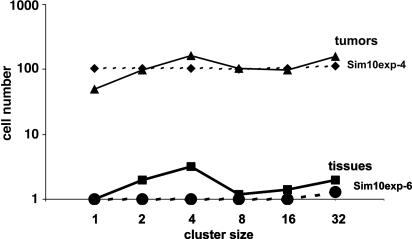
Clusters of Mutant Cells. If mutants arise at a constant rate at random times and proliferate normally, the sizes and frequencies of clusters are predictable. Under these conditions, the points on graphs that plot the product of cluster size and frequency vs. cluster size will fall on a horizontal line. Fig. 3 shows that normal tissues conformed to this expectation. The line produced from tissue data was similar to that produced by a computer simulation that used random mutation at a rate of 1 × 10-6 events per cell.
Fig. 3.
Median frequencies of clusters of PLAP+ cells in tissues and tumors conformed to expectation based on random mutation followed by normal proliferation in an exponential growth model. The values on the y axis are the product of cluster size and cluster frequency. Dotted lines labeled sim10exp-4 and sim10exp-6 show median values obtained from three simulations performed with the probability of mutation set at 1 × 10-4 and 1 × 10-6, respectively.
When considered together, tumors exhibited cluster phenotypes that also tended to conform to expectations based on stochastic mutation followed by uniform proliferation. Fig. 3 shows that the median values obtained from tumors produced a line quite similar to a computer simulation that used a random mutation at a rate of 1 × 10-4 events per cell. The median value line ends at 32 cells, because less than half of the tumors exhibited larger clusters. However, larger clusters were observed in some tumors (see Fig. 1 and below).
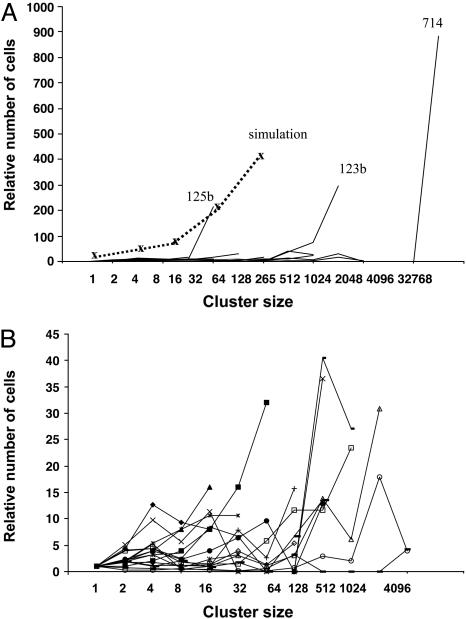
Although in the aggregate, tumors tended to produce clusters in numbers consistent with a simple stochastic model, individual tumors exhibited different cluster phenotypes, many of which were dramatically different from the predictions of simple models. Fig. 4 shows the relationship between cluster size and the product of cluster size and frequency for each of the tumors. To facilitate plotting all of the data on the same scale, cell-number values were normalized by dividing them by the number of single PLAP+ cells observed in a given tumor. When all of the data were plotted (Fig. 4A), three tumors (125b, 123b, and 714) stood out. Tumor 714 had a very large region of contiguous PLAP+ cells (see Fig. 1). All sections from this tumor had a very large patch of PLAP+ cells, which contained ≈30,000 cells. Similarly, most sections from tumor 123b contained a patch with ≈1,000 PLAP+ cells. Tumor 125b had a different phenotype. One section from tumor 125b exhibited 37 small clusters, each containing ≈30 cells. The shapes of the lines plotted for tumors 714, 123b, and 125b indicated they all had an overabundance of large clusters compared with what would be expected from the number of single PLAP+ cells. The sizes of the overabundant clusters varied in the three tumors. Tumor 125b was at one extreme, with an apparent excess of clusters larger than 16 cells. Tumor 714 was at the other extreme, with a very large cluster containing tens of thousands of cells. Tumor 123b exhibited an intermediate phenotype, with a large number of clusters larger than 128 cells.
Fig. 4.
Variation in frequencies of PLAP+ cell clusters in tumors. For each tumor, the relative number of clusters of a given size was calculated by dividing the number of clusters with more than one PLAP+ cell by the number of single PLAP+ cells. The values on the y axis are the products of these numbers and cluster sizes. (A) Data from all tumors. The simulation allowed mutant cells to double twice as fast as normal cells. (B) Data from all tumors except 123b, 125b, and 714.
It seemed possible that these large clusters formed via hyperproliferation of PLAP+ cells. To examine this possibility, a computer model that caused mutant cells to proliferate twice as fast as normal cells was developed. Although this model produced results that resembled the data from tumor 125b, the model data did not fit data from tumors 123b and 714 (Fig. 4A). These results suggest that PLAP activity did not directly cause hyperproliferation. Instead, it appears that the excess of larger clusters was formed by hyperproliferation of a subset of the PLAP+ cells in the tumor.
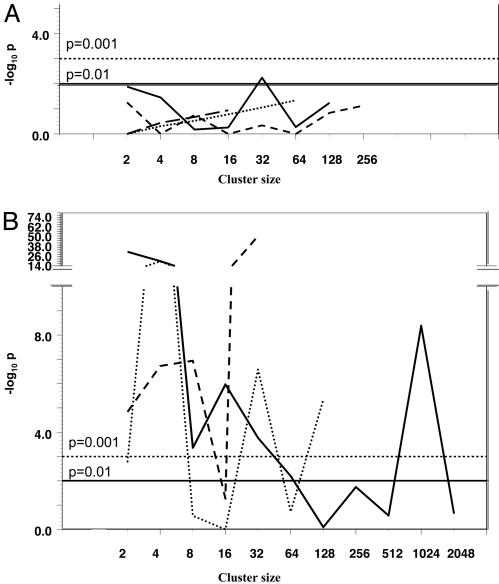
The large numbers of PLAP+ clusters in tumors 714, 123b, and 125b imposed a graphical scale that obscured details in the data from the other tumors. Therefore, a second graph, Fig. 4B, which did not include data from these three tumors, was constructed. Fig. 4B shows that tumors varied greatly with respect to PLAP+ cell cluster frequencies. To assess the significance of this variation, cluster counts from each tumor were subjected to probability analysis. The frequency of single PLAP+ cells in tumors was used to calculate the probabilities that larger clusters would form at the frequencies observed, assuming random mutation and exponential proliferation of all cells. For example, if the frequency of single PLAP+ cells in a tumor were 100, then random mutation and exponential proliferation would be expected to produce ≈50 two-cell clusters, 25 four-cell clusters, etc. Using these expected values, the probability of the observed frequency of each cluster (P) can be estimated via Fisher's exact test. Fig. 5 shows some of the results of the calculations performed on 11 tumors. Fig. 5 A and B display data from tumors selected and grouped according to cluster phenotypes. Fig. 5A shows data from the four tumors that exhibited cluster frequencies that were not extremely improbable. The P values for these tumors tended to exceed 0.05, although several values were as low as 0.01. Fig. 5B shows the opposite extreme in tumor phenotypes. These three tumors deviated markedly from expectations. Many of the P values were in the range of 10-6, and some were many orders of magnitude lower. The other four tumors subjected to probability analysis were more similar to those in Fig. 5B than to those in Fig. 5A. Therefore, formation of clusters of PLAP+ cells in most tumors was not governed by normal proliferation of randomly mutated cells. Instead, other factors, which presumably varied among tumors and within them, produced the highly idiosyncratic phenotypes observed.
Fig. 5.
Probabilities of observed cluster frequencies in tumors. For each tumor, the expected numbers of clusters of various sizes were calculated based on the frequency of single cells and assuming random mutation and exponential growth of all cells. The probabilities of the observed cluster frequencies are plotted on the y axis. (A) Data from the four tumors that deviated least from expectations. (B) Data from the three tumors that deviated most from expectations.
Discussion
In principle, both genetic instability and hyperproliferation can contribute to the number of genes suffering from point mutation in cells in tumors. Although a primary role for genetic instability has been established in cancers such as hereditary nonpolyposis colon carcinoma, which form from cells deficient in mismatch repair (37, 38), it is not clear that tumor evolution is generally driven by an increased mutation rate per cell. Mathematical models have demonstrated that selection could cause multiple mutations to arise in a tumor even when the mutation rate per cell remains normal (39). In addition, recent studies on cells from mismatch repair-proficient human colorectal neoplasias maintained in nude mice produced no evidence of an increased mutation rate (40). However, other studies have shown that cells in human tumors can have very high numbers of mutations (41). In addition, it has been pointed out that the failure to detect genetic instability in tumor cells does not necessarily indicate its absence, because the methods typically used cannot detect random mutations that occur in <10% of tumor cells (42). By contrast, studies in genetically engineered mice have the potential to detect mutations that occur in rare cells. The results of studies on transgenic mice have been mixed. The first study of this kind found no increase in the number of mutant lacI transgenes rescued from thymic tumors (43). However, two of three subsequent transgene rescue studies detected increased mutation (34, 44, 45). For example, a recent study reported that 5-fold more mutant lacZ transgenes were rescued from pristane-induced mouse plasmacytomas than from normal splenocytes (45). The results from plasmacytomas suggest a mutator phenotype, but because the mutant lacZ alleles were not sequenced, it is also possible that proliferation of mutant cells contributed to the increase observed. In a different study, where lymphomas and sarcomas in Trp-53-deficient mice were analyzed, two tumors of 17 exhibited a dramatic increase in the frequency of mutant lacI transgenes rescued (46). Analysis of the mutations showed that both of these increases were due to proliferation of cells with a mutant gene rather than to an increased mutation rate (46).
The studies on G11-PLAP mice described herein suggest that both genetic instability and proliferation were involved in generating the PLAP+ cells observed in the MMTVPyMT tumor model. A role for genetic instability is suggested by the higher numbers of single PLAP+ cells in tumors. Alternatively, some of the isolated PLAP+ cells may have been produced by other events, such as cell migration, differentiation, and apoptosis. However, studies on genetic heterogeneity of human tumors suggest that cell migration is limited (47). More importantly, migration, differentiation, and apoptosis would not be expected to produce the observed tendency of median tumor cluster counts to conform to expectations based on random mutation and exponential expansion of both mutant and wild-type cells. In other words, when tumors were analyzed as a group, the number of single PLAP+ cells was congruent with the number of PLAP+ doublets, quartets, etc., which is most simply explained as the result of random mutation followed by exponential proliferation. Therefore, models that incorporate factors other than mutation and proliferation into account are not needed to explain the frequency and distribution of PLAP+ cells when all tumors are considered as a group.
Whereas genetic instability is a plausible explanation for the numerous single PLAP+ cells in tumors, the frequencies and sizes of clusters of PLAP+ cells in individual tumors suggest that abnormal proliferation was sometimes a factor. However, simple hyperproliferation of PLAP+ cells does not explain the tumor phenotypes, because this phenomenon would produce more clusters of every size. In fact, cluster counts in tumors were extremely variable. In addition, studies on mouse 3T3 cells expressing PLAP showed that PLAP had no effect on proliferation in culture (J.R.S., unpublished observation). Furthermore, expression of ectopic alkaline phosphatase did not cause human cells to become oncogenic in nude mice (48).
Multiple factors appear to have been involved in generating the phenotypes of most tumors studied. One possibility is that different tumors, and/or different segments of a given tumor mass, contained cells that behaved differently, either with respect to mutation, proliferation, or both. Phenotypic and genotypic heterogeneity is common in human tumors (49–56). The evolution of heterogeneous tumors can be simulated by mathematical models that include selection in addition to mutation and proliferation (57). It seems probable that all three forces shaped the tumors in the MMTVPyMT model, and that the PLAP+ phenotype served as a neutral indicator of tumor heterogeneity.
Acknowledgments
We thank T. Doetschman and the Mouse Models of Human Cancers Consortium for help with support for mouse colonies; C. Tindal, M. Yin, A. Whitaker, C. York, S. Pawlowski, and M. Bender for technical assistance with mouse colonies; Yvette Doty and Sherri Crabtree for help with sectioning; and Peter J. Stambrook for comments on the manuscript. This work was supported by Grant P42 ES04908 from the National Institute of Environmental Health Sciences.
Author contributions: J.R.S., S.L.S., and M.N.H. designed research; S.L.S., J.S.L., J.M.F., and M.N.H. performed research; J.M.F., M.M., and G.P.B. contributed new reagents/analytic tools; J.R.S., S.L.S., M.M., J.S.L., J.M.F., and G.P.B. analyzed data; and J.R.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PLAP, placental alkaline phosphatase.
References
- 1.Tomlinson, I., Sasieni, P. & Bodmer, W. (2002) Am. J. Pathol. 160, 755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623-627. [DOI] [PubMed] [Google Scholar]
- 3.Sieber, O. M., Heinimann, K. & Tomlinson, I. P. (2003) Nat. Rev. Cancer 3, 701-708. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, A. L. & Loeb, L. A. (1998) Genetics 148, 1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePrimo, S. E., Cao, J., Hersh, M. N. & Stringer, J. R. (1998) Methods 16, 49-61. [DOI] [PubMed] [Google Scholar]
- 6.Cao, J., DePrimo, S. E., Hersh, M. N. & Stringer, J. R. (1998) Mutat. Res. 421, 163-178. [DOI] [PubMed] [Google Scholar]
- 7.DePrimo, S. E., Stambrook, P. J. & Stringer, J. R. (1996) Transgenic Res. 5, 459-466. [DOI] [PubMed] [Google Scholar]
- 8.Hersh, M. N., Stambrook, P. J. & Stringer, J. R. (2002) Mutat. Res. 505, 51-62. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty, R., Kimmel, M., Stivers, D. N., Davison, L. J. & Deka, R. (1997) Proc. Natl. Acad. Sci. USA 94, 1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iino, H., Jass, J. R., Simms, L. A., Young, J., Leggett, B., Ajioka, Y. & Watanabe, H. (1999) J. Clin. Pathol. 52, 5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikenoue, T., Togo, G., Nagai, K., Ijichi, H., Kato, J., Yamaji, Y., Okamoto, M., Kato, N., Kawabe, T., Tanaka, A., et al. (2001) Jpn. J. Cancer Res. 92, 587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz, S., Jr., Yamamoto, H., Navarro, M., Maestro, M., Reventos, J. & Perucho, M. (1999) Cancer Res. 59, 2995-3002. [PubMed] [Google Scholar]
- 13.Duval, A., Gayet, J., Zhou, X. P., Iacopetta, B., Thomas, G. & Hamelin, R. (1999) Cancer Res. 59, 4213-4215. [PubMed] [Google Scholar]
- 14.Codegoni, A. M., Bertoni, F., Colella, G., Caspani, G., Grassi, L., D'Incalci, M. & Broggini, M. (1999) Oncol. Res. 11, 297-301. [PubMed] [Google Scholar]
- 15.Yamamoto, H., Sawai, H. & Perucho, M. (1997) Cancer Res. 57, 4420-4426. [PubMed] [Google Scholar]
- 16.Laken, S. J., Petersen, G. M., Gruber, S. B., Oddoux, C., Ostrer, H., Giardiello, F. M., Hamilton, S. R., Hampel, H., Markowitz, A., Klimstra, D., et al. (1997) Nat. Genet. 17, 79-83. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz, S., Wang, J., Myeroff, L., Parsons, R., Sun, L., Lutterbaugh, J., Fan, R. S., Zborowska, E., Kinzler, K. W., Vogelstein, B., et al. (1995) Science 268, 1336-1338. [DOI] [PubMed] [Google Scholar]
- 18.Rampino, N., Yamamoto, H., Ionov, Y., Li, Y., Sawai, H., Reed, J. C. & Perucho, M. (1997) Science 275, 967-969. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J., Sun, L., Myeroff, L., Wang, X., Gentry, L. E., Yang, J., Liang, J., Zborowska, E., Markowitz, S. & Willson, J. K. (1995) J. Biol. Chem. 270, 22044-22049. [DOI] [PubMed] [Google Scholar]
- 20.Planck, M., Wenngren, E., Borg, A., Olsson, H. & Nilbert, M. (2000) Genes Chromosomes Cancer 29, 33-39. [DOI] [PubMed] [Google Scholar]
- 21.Woerner, S. M., Gebert, J., Yuan, Y. P., Sutter, C., Ridder, R., Bork, P. & von Knebel, D. M. (2001) Int. J. Cancer 93, 12-19. [DOI] [PubMed] [Google Scholar]
- 22.Olivero, M., Ruggiero, T., Coltella, N., Maffe', A., Calogero, R., Medico, E. & Di Renzo, M. F. (2003) Nucleic Acids Res. 31, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monroe, J. J., Kort, K. L., Miller, J. E., Marino, D. R. & Skopek, T. R. (1998) Mutat. Res. 421, 121-136. [DOI] [PubMed] [Google Scholar]
- 24.Duval, A. & Hamelin, R. (2002) Cancer Res. 62, 2447-2454. [PubMed] [Google Scholar]
- 25.Dahiya, R., Lee, C., Zhu, Z. & Thompson, H. J. (1998) Int. J. Oncol. 13, 23-28. [DOI] [PubMed] [Google Scholar]
- 26.Fleisher, A. S., Esteller, M., Harpaz, N., Leytin, A., Rashid, A., Xu, Y., Liang, J., Stine, O. C., Yin, J., Zou, T. T., et al. (2000) Cancer Res. 60, 4864-4868. [PubMed] [Google Scholar]
- 27.Weinberg, H. S., Korol, A. B., Kirzhner, V. M., Avivi, A., Fahima, T., Nevo, E., Shapiro, S., Rennert, G., Piatak, O., Stepanova, E. I., et al. (2001) Proc. R. Soc. London B Biol. Sci. 268, 1001-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy, C. T., Cardiff, R. D. & Muller, W. J. (1992) Mol. Cell. Biol. 12, 954-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dilworth, S. M., Hansson, H. A., Darnfors, C., Bjursell, G., Streuli, C. H. & Griffin, B. E. (1986) EMBO J. 5, 491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichaso, N. & Dilworth, S. M. (2001) Oncogene 20, 7908-7916. [DOI] [PubMed] [Google Scholar]
- 31.Wani, M. A., Denko, N. C. & Stambrook, P. J. (1997) Somat. Cell Mol. Genet. 23, 123-133. [DOI] [PubMed] [Google Scholar]
- 32.Denko, N., Stringer, J., Wani, M. & Stambrook, P. (1995) Somat. Cell Mol. Genet. 21, 241-253. [DOI] [PubMed] [Google Scholar]
- 33.Denko, N. C., Giaccia, A. J., Stringer, J. R. & Stambrook, P. J. (1994) Proc. Natl. Acad. Sci. USA 91, 5124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, S., Liu, W., Jakubczak, J. L., Erexson, G. L., Tindall, K. R., Chan, R., Muller, W. J., Adhya, S., Garges, S. & Merlino, G. (2002) Proc. Natl. Acad. Sci. USA 99, 3770-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson, J. S., Stringer, S. L. & Stringer, J. R. (2004) Mutat. Res. 556, 45-53. [DOI] [PubMed] [Google Scholar]
- 36.Hayes, W. (1968) The Genetics of Bacteria and Their Viruses (Wiley, New York).
- 37.Malkhosyan, S., McCarty, A., Sawai, H. & Perucho, M. (1996) Mutat. Res. 316, 249-259. [DOI] [PubMed] [Google Scholar]
- 38.Umar, A., Boyer, J. C., Thomas, D. C., Nguyen, D. C., Risinger, J. I., Boyd, J., Ionov, Y., Perucho, M. & Kunkel, T. A. (1994) J. Biol. Chem. 269, 14367-14370. [PubMed] [Google Scholar]
- 39.Tomlinson, I. P., Novelli, M. R. & Bodmer, W. F. (1996) Proc. Natl. Acad. Sci. USA 93, 14800-14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, T. L., Rago, C., Silliman, N., Ptak, J., Markowitz, S., Willson, J. K., Parmigiani, G., Kinzler, K. W., Vogelstein, B. & Velculescu, V. E. (2002) Proc. Natl. Acad. Sci. USA 99, 3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoler, D. L., Chen, N., Basik, M., Kahlenberg, M. S., Rodriguez-Bigas, M. A., Petrelli, N. J. & Anderson, G. R. (1999) Proc. Natl. Acad. Sci. USA 96, 15121-15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loeb, L. A., Loeb, K. R. & Anderson, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sands, A. T., Suraokar, M. B., Sanchez, A., Marth, J. E., Donehower, L. A. & Bradley, A. (1995) Proc. Natl. Acad. Sci. USA 92, 8517-8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakubczak, J. L., Merlino, G., French, J. E., Muller, W. J., Paul, B., Adhya, S. & Garges, S. (1996) Proc. Natl. Acad. Sci. USA 93, 9073-9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felix, K., Polack, A., Pretsch, W., Jackson, S. H., Feigenbaum, L., Bornkamm, G. W. & Janz, S. (2004) Cancer Res. 64, 530-537. [DOI] [PubMed] [Google Scholar]
- 46.Giese, H., Snyder, W. K., Van Oostrom, C., van Steeg, H., Dolle, M. E. & Vijg, J. (2002) Mutat. Res. 514, 153-163. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez-Garcia, I., Sole, R. V. & Costa, J. (2002) Proc. Natl. Acad. Sci. USA 99, 13085-13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latham, K. M. & Stanbridge, E. J. (1992) Cancer Res. 52, 616-622. [PubMed] [Google Scholar]
- 49.Fujii, H., Yoshida, M., Gong, Z. X., Matsumoto, T., Hamano, Y., Fukunaga, M., Hruban, R. H., Gabrielson, E. & Shirai, T. (2000) Cancer Res. 60, 114-120. [PubMed] [Google Scholar]
- 50.Barnetson, R., Jass, J., Tse, R., Eckstein, R., Robinson, B. & Schnitzler, M. (2000) Genes Chromosomes. Cancer 29, 130-136. [DOI] [PubMed] [Google Scholar]
- 51.Mora, J., Cheung, N. K. & Gerald, W. L. (2001) Br. J. Cancer 85, 182-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baisse, B., Bouzourene, H., Saraga, E. P., Bosman, F. T. & Benhattar, J. (2001) Int. J. Cancer 93, 346-352. [DOI] [PubMed] [Google Scholar]
- 53.Samowitz, W. S. & Slattery, M. L. (1999) Genes Chromosomes Cancer 26, 106-114. [PubMed] [Google Scholar]
- 54.Heimann, R., Ferguson, D., Recant, W. M. & Hellman, S. (1997) Cancer J. Sci. Am. 3, 224-229. [PubMed] [Google Scholar]
- 55.Heimann, R. & Hellman, S. (1998) J. Clin. Oncol. 16, 2686-2692. [DOI] [PubMed] [Google Scholar]
- 56.Hellman, S. & Heimann, R. (2000) Cancer J. 6, Suppl. 2, S131-S133. [PubMed] [Google Scholar]
- 57.Gatenby, R. A. & Vincent, T. L. (2003) Cancer Res. 63, 6212-6220. [PubMed] [Google Scholar]