Abstract
The class and effector functions of antibodies are modulated through the process of Ig heavy chain class switch recombination (CSR). CSR occurs between switch (S) regions that lie upstream of the various Ig heavy chain constant region exons. Molecular analyses of S-region functions have been hampered by their large size and repetitive nature. To test potential relationships between S-region size and efficiency of CSR, we generated normal B lymphocytes in which the 12-kb S region flanking the Cγ1 exons (Sγ1) was replaced with synthetic or endogenous S regions of various lengths. Replacement of Sγ1 with 1- and 2-kb synthetic sequences representing the Sγ1 core repeats or a 4-kb portion of the core endogenous Sγ1 region supported CSR frequencies that directly correlated with S-region length. These findings indicate that S-region size is an important factor in determining endogenous CSR efficiency. Moreover, these results also will allow the development of a systematic system to test the function of various S-region motifs by replacing endogenous S regions with synthetic S regions controlled for size effects.
Keywords: IgH switch region, activation-induced deaminase, synthetic switch region, IgG1
Immunoglobulin (Ig) genes undergo several genetic alterations during the course of B lymphocyte development and activation. During early stages of mammalian B cell development, Ig heavy (IgH) and light (IgL) chain variable region exons are generated from germ-line V, D, and J gene segments via V(D)J recombination (1). When B cells are activated in the context of an immune response, the IgH and IgL variable region exons acquire point mutations through a process known as somatic hypermutation (SHM). SHM allows selection of mutated B cell clones that produce higher-affinity antibodies (2). In addition, the constant region (CH) of the IgH chain can be changed through the process of IgH class switch recombination (CSR) (3). CSR is a deletional recombination reaction that allows the Cμ, the first CH“gene” expressed during development, to be replaced with one of a number of downstream CH genes. In mouse, individual CH genes are organized in the order 5′-VDJ-Cμ-Cδ-Cγ3-Cγ1-Cγ2b-Cγ2a-Cε-Cα-3′. Each germ-line CH gene that undergoes CSR is preceded by a long, repetitive target sequence for CSR termed a switch (S) region. CSR results from recombination between the most 5′ S region (Sμ), which lies upstream of the Cμ gene, and one of the downstream S regions (e.g., Sγ3, Sγ1, etc.).
Individual CH genes are organized into germ-line transcription units that consist of a transcriptional promoter, a noncoding I exon, the S region, and the CH coding exons. S regions span most of the intron between I exons and the corresponding CH gene. Individual S regions range in size from ≈10 kb (Sγ1) to only 1 kb (Sε), and are characterized by highly repetitive (“core”) sequences. The core repeats of Sμ (4.0 kb) (4), Sε (1.0 kb) (5), and Sα (4.2 kb) (6) are homologous to each other and rich in GGGGT, GAGCT motifs. The core S regions of Sγ3 (2.5 kb) (7), Sγ1 (8 kb) (8), Sγ2b (5.0 kb) (9), and Sγ2a (2.5kb) (5) contain a characteristic 49-bp repeat that shares some of the repetitive elements (e.g., AGCT motifs) found in the Sμ, Sε, and Sα sequences. Although the four Sγ regions are related by overall homology of consensus repeats, there also are consistent differences among individual S regions (8) that have been suggested to provide some specificity in directing CSR (10). Intronic sequences external to the core repeats also contain similar repetitive elements interspersed with other sequences. Gene-targeted mutational studies have demonstrated that S regions are specialized targets for CSR. Thus, deletion of the 5′ donor Sμ greatly diminishes CSR (11, 12), whereas deletion of Sγ1, which is a 3′ acceptor, essentially abolishes CSR to Cγ1 (13). Moreover, replacement of Sγ1 with a 4-kb Xenopus Sμ region, which contains a region of AGCT repeats similar to those found in mammalian S regions, allowed substantial CSR. In contrast, replacement of Sγ1 with a 4-kb intronic sequence from Xpf gene, which lacks abundant AGCT motifs, did not support high-level CSR (14).
CSR absolutely requires the activation-induced deaminase (AID), a single-stranded-DNA-specific cytidine deaminase with no activity on double-stranded DNA (15, 16). AID initiates both CSR and SHM by deaminating cytidine residues in target S region or variable region DNA (3). Germ-line transcription through S regions also is required for CSR (17). Such germ-line CH gene transcription runs through the S region and is terminated downstream of the corresponding CH exons. Germ-line CH transcription of a given CH gene directly correlates with CSR potential of that CH gene and, thereby, serves to direct the CSR process. Transcription through S regions may contribute to generating a DNA substrate for AID in two manners. First, mammalian S regions are GC-rich and G-rich on their nontemplate strand, a property that allows them to generate single-stranded DNA in the form of R loops when transcribed (18, 19). Such R loops are effective AID substrates in vitro (20, 21). Second, in activated B cells, AID is phosphorylated, allowing it to interact with replication protein A (RPA) (22). In vitro, AID/RPA complex preferentially binds to and deaminates sequences, such as variable regions and S regions, which are rich in AGCT and related motifs. Together, these two properties, the ability to form R-loops and a high density of AGCT and related motifs, may help render transcribed S regions effective targets for AID in activated B cells.
Despite recent advances in our understanding of the role of AID and transcription in CSR, there are still many unknown issues related to potential S-region functions. For example, as mentioned above, S regions vary substantially in size, raising the possibility that size may increase the potential for AID targeting and contribute to effectiveness as a CSR substrate. In this regard, a synthetic 1-kb, G/C-rich, R-loop-forming sequence that lacks repetitive S-region motifs, provided ≈10% of the activity of Sγ1 when inserted in place of the 12-kb Sγ1 region in normal B cells (13). In these experiments, it was not clear whether reduced CSR activity of this synthetic sequence reflected either the lack of important elements in Sγ1 or the short length of the synthetic sequence (or both). With respect to S region length, it is notable that mouse IgG1, which derives from CSR to the very long Sγ1 region, constitutes the highest level of IgG subtypes in the serum (23). In addition, CSR to IgG1 predominates in B cells activated in vitro in the presence of IL-4, even though both Cγ1 and Cε germ-line transcription is induced by treatments involving IL-4 (24). Although the high level of Cγ1 CSR might be attributed to various causes, it is possible that the large size of Sγ1 region may be an important factor. Consistent with this possibility, increasing the number of Sγ3 consensus repeats from 1 to 6 in a transient CSR reporter substrate resulted in increased levels of recombination (25).
To further elucidate potential roles of S-region size with respect to targeting CSR and to develop a system to systematically assess the role of various S-region motifs in CSR, we now have replaced the endogenous Sγ1 regions with Sγ1 core sequences of various lengths and assayed their ability to support CSR in vivo. We find that, at least up to 4 kb, the length of S region directly influences CSR efficiency in vivo. We discuss the implications of this finding for the mechanism of CSR and also with respect to implications for the development of a systematic approach to assess functions of S-region motifs in vivo by using synthetic S regions that are controlled for size effects.
Materials and Methods
Generation of Synthetic Sγ1 Regions. Sγ1 consensus (8) oligonucleotides 5′-GATCC GGG AGC CAG GAC AGG TGG AAG TGT GGT GAC CCA GGC AGA GCA GCT CCA GG-3′ and 5′-GATCT CTG GAG CTG CTG CTC TGC CTG GGT CAG GAC ACT TCC ACC TGT CCT GGC TCC CG-3′ were phosphorylated by T4 polynucleotide kinase (New England Biolabs). Restriction sites outside of the Sγ1 repeat consensus are italicized. The phosphorylated oligonucleotides were annealed and ligated into BamHI site of S85 plasmid. Subsequent ligations of the same oligonucleotide resulted in the unidirectional cloning of desired length confirmed by sequencing analyses. A 6-kb StyI fragment containing the core Sγ1 repeat was cloned into the SmaI site of S85 vector. The 5′ end of this insert corresponds to StyI site at position 4,130 of the genomic sequence under GenBank accession no. D78344. To obtain a 4-kb fragment, this fragment was subject to exonuclease treatment from the 3′ end.
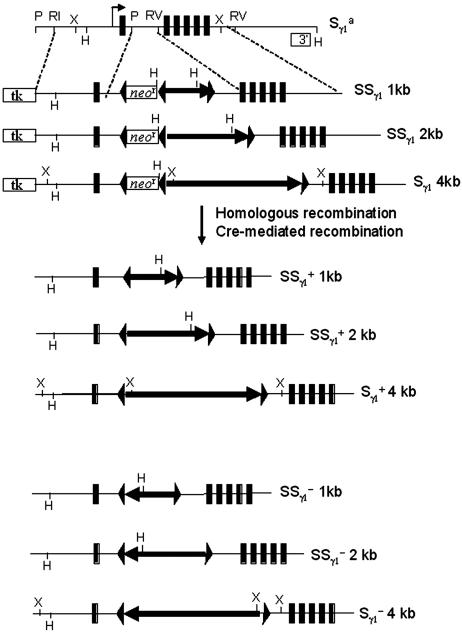
Generation of Targeting Vectors and Gene Targeting. Synthetic (1- or 2-kb) and 4-kb WT Sγ1 repeat regions were cloned into NotI and SalI sites of the targeting vector reported in ref. 14 (Fig. 2). These constructs were transfected into ΔSγ1 embryonic stem (ES) cells in which the Sγ1a region was deleted (13). Correctly targeted clones were identified by Southern analyses. The configurations of the targeted alleles were further confirmed by PstI digest with internal Iγ1 probe. The deletion of the neor gene and subsequent inversion of the inserts were achieved by using adenovirus Cre-expressing vector. The ES cells were injected into RAG-2-deficient blastocysts to obtain chimeric mice with mature lymphocytes. Animal work was approved by the Institutional Animal Care and Use Committee of Children's Hospital.
Fig. 2.
Targeting of the Sγ1a allele. Genomic organization of Δ Sγ1 is shown at the top. The design of targeting constructs is illustrated underneath. The structure of the targeted allele after Cre/loxP recombination is illustrated at the bottom. Iγ1 and Cγ1 are shown as rectangular black boxes. Other symbols are as follows: tk, gene encoding thymidine kinase; arrows, WT or targeted S regions with arrow direction indicating the physiological transcriptional orientation; triangles, loxP site; R1, EcoRI; RV, EcoRV; H, HindIII; P, PstI; X, XbaI; 1-SSγ1, 1-kb synthetic Sγ1; 2-SSγ1, 2-kb synthetic Sγ1; 4-Sγ1, 4-kb endogenous Sγ1; 8-Sγ1, 8-kb endogenous Sγ1; +, physiologic orientation; -, inverted transcriptional orientation.
Assays for Class Switching. All assays are described in detail in refs. 13 and 14. Briefly, splenocytes were stimulated with anti-CD40 and IL-4 for 4 days to generate hybridomas or for 6 days to perform ELISA. The monoclonal anti-IgG1a (Igh-4a, Pharmingen) was used to detect IgG1a (mutated alleles). Total IgG1 was measured by polyclonal anti-IgG1 (Southern Biotechnology Associates).
Results
Generation of Synthetic Sγ1 Sequences. To elucidate S-region functions, we developed a cloning strategy to generate synthetic S regions bearing repeat motifs similar to those of endogenous regions. In BALB/c mice, the core Sγ1 sequence in the intron between Iγ1 and Cγ1 consists of 120 tandem 49-bp repeats and is, thereby, ≈6 kb in length (8). In 129/Sv mice, the core Sγ1 repeat region is 2 kb larger (8 kb) than the core BALB/c Sγ1 (data not shown). The longest tandem repeat in the central part of Sγ1 consists of 79 repeat elements that show extensive (93.4%) homology to the consensus Sγ1 49-mer (8). To reproduce this structure, we generated 20-mer or 40-mer repetitive synthetic S regions by concatemerizing a consensus Sγ1 oligonucleotide that was flanked by BamHI and BglII restriction enzyme sites. Based on alignment of 540 bp of the synthetic Sγ1 with the endogenous Sγ1, the synthetic Sγ1 closely resembles the organization of endogenous Sγ1 repeats, both with respect to overall nucleotide content and to repeat structure and organization (Fig. 1). Also, similar to the endogenous S regions, all synthetic repeats are unidirectional. The only difference between synthetic and endogenous Sγ1 repeats is the presence of a 6-bp linker (AGATCC) between the 49-bp repeat units.
Fig. 1.
Sequence alignment of the synthetic Sγ1 and endogenous Sγ1. The location of a single consensus 49-bp repeat and the linker are indicated on top. The result of blast search on 540 bp (10 repeats) of synthetic Sγ1 (bold letters) is shown. The numbers on the left represent the beginning of synthetic Sγ1 or endogenous Sγ1 (GenBank accession no. M12389). Dashed lines represent gaps.
Replacement of Endogenous Sγ1 with Core Sγ1 Repeats of Different Length. To determine whether S-region length influences CSR frequency, we used gene-targeted mutation methods to replace the endogenous Sγ1a allele in an F1 ES cell line with 1 or 2 kb of synthetic Sγ1 repeats, as well as 4-kb Sγ1 of the endogenous Sγ1 core repeat region. The F1 ES cell was generated from the 129Sv-C57BL/6 mice in which the two Igh alleles represent the IgHa (from 129/Sv) or IgHb (from C57BL/6) allotypes, respectively (13, 26). The presence of allotypic markers and sequence polymorphism facilitates analyses of mutant mice. We have previously shown that the deletion of Sγ1 region abolishes CSR to Cγ1 (13, 14). We used our previous strategy (14) to target the different substitute S regions into the IgHa allele of an F1 derivative in which the Sγ1 region was deleted (referred to ΔSγ1 allele) (Fig. 2). Subsequently, the inserted neor gene was removed by Cre-mediated recombination through the flanking loxP sites. To analyze the effect of transcriptional orientation, the two loxP sites flanking the insert were placed in inverted orientation, allowing Cre-mediated recombination to invert the intervening sequence (Fig. 2). The size and integrity of the inserted synthetic S regions were confirmed by Southern blot analyses with a synthetic Sγ1 probe (Fig. 6A, which is published as supporting information on the PNAS web site). The strong sequence homology between synthetic and endogenous Sγ1 was evident by Southern blot, because under high stringency wash conditions, the synthetic Sγ1 probe remains bound to the endogenous Sγ1 (Fig. 6B).
Generation of B Lymphocytes with Sγ1 Core Sequences of Various Lengths. The Sγ1-replaced mutant F1 ES cells were injected into recombination activating gene 2 (RAG2)-deficient blastocysts to generate chimeric mice to obtain mature B lymphocytes harboring the targeted mutation (27). Splenocytes from mutant or control animals were activated with antibody against CD40 (anti-CD40) plus IL-4, a treatment that induces germ-line transcription of the Cγ1 gene and targets CSR to the Sγ1 region. Based on ELISA of culture supernatant after 6 days of treatment, we found overall IgG1 secretion in the various splenocyte cultures to all fall within the expected range with cells harboring the ΔSγ1 allele falling on the lowest end of the range (data not shown); we interpret this finding to indicate that all cultures were properly activated for CSR.
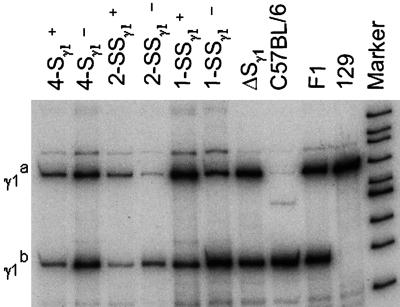
Because germ-line transcription is an important component of CSR, we assessed whether the targeted alleles were normally transcribed in B cells after anti-CD40 plus IL-4 treatment. For this purpose, we measured the relative levels of the steady-state germ-line transcripts from the WT γ1b and targeted γ1a alleles by reverse-transcription-mediated PCR (RT-PCR) (Fig. 3) (13, 14). Because the cultures were all similarly activated, the WT γ1b locus should be transcribed at normal levels and serve as the internal control for RT-PCR analysis. All of the mutant alleles in which the replacement Sγ1 sequences were inserted in the normal physiological orientation generated levels of germ-line transcripts comparable to those from the WT γ1b allele (Fig. 3). Whereas the “inverted” 1- and 4-kb Sγ1 alleles also generated levels of transcripts similar to those of the WT allele, the inverted 2-kb synthetic Sγ1 allele generated substantially lower levels of steady-state transcripts than the WT allele; it is not clear whether this represents decreased transcription or decreased stability of the transcripts (Fig. 3).
Fig. 3.
RT-PCR of germ-line γ1 transcripts. Germ-line transcripts were amplified by RT-PCR with Iγ1 and Cγ1 primers and subsequently subjected to primer extension. Representative data from a minimum of two separate stimulations employing cells from independent chimeras are shown.
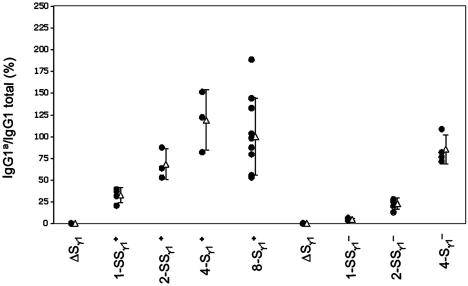
CSR Efficiency Correlates with Length of the Sγ1 Core Repeat Region. After activation of the various types of mutant B cells, we first assessed CSR by measuring the levels of secreted IgG1 from the various cultures via ELISA with allotype-specific antibodies against IgG1a. The level of IgG1b serves as an internal control for variations between experiments. Because antibody to IgG1b (WT) is not available, we measured the IgG1a/IgG1 ratio as an index for CSR. We arbitrarily defined the IgG1a/IgG1 ratio for WT F1 ES cells as 100% CSR efficiency for the WT IgG1a allele. Notably, the 1-kb synthetic sequence supported IgG1a secretion to ≈30% the level of the WT IgG1b allele, whereas 2-kb synthetic sequence supported IgG1a secretion to levels ≈60% those of the WT allele (Fig. 4). Moreover, the 4-kb endogenous Sγ1 core repeats supported IgG1a secretion at levels similar to those of WT (Fig. 4). The inverted forms of each of these alleles also supported CSR but at reduced levels compared with the “physiological” orientation (Fig. 4). These findings indicate that all of the replacement Sγ1 sequences can support a substantial level of IgH Class switching to IgG1.
Fig. 4.
Class-switching in mutant B cells. The ratio of IgG1a/IgG1 total of the WT mice is set arbitrarily as 100% for the IgHa allele. Each dot represents the average of at least two measurements of one mouse. Error bars represent standard deviation of the mean (open triangles). We have presented the endogenous Sγ1 region as 8 kb (8-Sγ1+), which reflects the core repetitive segment of the intronic region.
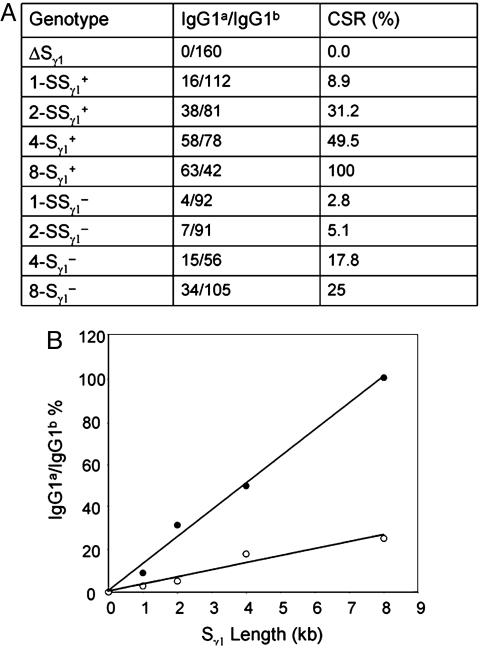
As described in refs. 13 and 14, hybridoma analyses provide a better means to quantitatively compare the level of CSR between different alleles, because this analysis measures the frequency of CSR based on IgG1a production at a single-cell level. Therefore, we generated a panel of IgG1-producing splenic B cell hybridomas from the various types of mutant B cells and assayed via ELISA whether they produced IgG1a. Those IgG1-producing hybridomas that do not produce IgG1a were considered to be IgG1b producers. Based on the hybridoma analyses (Fig. 5A), the CSR frequency was 9%, 31%, and 50% for 1-, 2-, or 4-kb Sγ1 replacement alleles as compared with WT allele. When each of these S regions was inverted, a similar trend was seen; however, in agreement with the results from the culture supernatants, the CSR frequency was reduced substantially relative to that of the corresponding constructs in the physiological orientation. The finding of reduced CSR by the inverted Sγ1 alleles is consistent with our previous finding that inverted endogenous Sγ1 allele supports CSR at reduced frequency (25%), as compared with endogenous Sγ1 allele in normal orientation. Although not directly demonstrated, based on our earlier studies, we presume that this decreased ability to support CSR might be due primarily to decreased ability to form R-loops. However, we do note that the inverted 2-kb Sγ1 sequence showed an even greater level of decrease as compared with the 2-kb Sγ1 sequence in the physiological orientation, which might arise, in part, from lower level of germ-line transcription of 2-kb synthetic allele in this configuration (Fig. 3). In this regard, a quantitative relationship between germ-line transcription and CSR remains to be fully elucidated, although a recent study found little impact of a 6-fold reduction in germ-line transcription of γ1 locus on CSR (28).
Fig. 5.
Linear correlation of S-region length and CSR frequency. (A) Ratio of IgG1a/IgG1b-producing hybridomas. The numbers of IgG1a- or IgG1b-producing hybridomas for each genotype are indicated. Relative CSR frequency is defined by the ratio of IgG1a- to IgG1b-producing hybridomas and is arbitrarily set as 100% for F1 cells. WT Sγ1 is presented as 8-Sγ1+ to illustrate size comparison. The data for inverted WT 8-Sγ1- were adopted from our previous study. (B) Linear correlation of S-region length and CSR frequency. The data for inverted WT Sγ1 (8-Sγ1) were incorporated from our previous study (13). Data from hybridoma analyses (A) of 1, 2, and 4 kb and WT (8 kb) were used for statistical analyses. R2 = 0.992 for physiologic orientation, and R2 = 0.9451 for inverted orientation. Filled circles, physiologic orientation; open circles, inverted orientation.
Discussion
In this study, we have demonstrated the feasibility of replacing endogenous S regions with synthetic sequences to assess, in vivo, parameters associated with S-region function. Based on this approach, we have, for the first time, assessed the effect of the size of an S region on CSR efficiency in vivo. By replacing the endogenous Sγ1 sequence with “core” synthetic or endogenous S-region sequences of various sizes, we have found a clear relationship between the size of the core S region at the endogenous Cγ1 locus and efficiency of CSR. To directly measure the significance of the apparent relationship between core Sγ1 size and CSR efficiency, we performed a statistical analysis of CSR with respect to the CSR efficiency of the 1-, 2-, and 4-kb core replacement alleles and the normal Sγ1 S region. For this comparison, we considered the endogenous Sγ1a allele as 8 kb in length, because this is the size of its core 49-bp repeat region within the 12-kb Iγ1-Cγ1 intron. Our statistical analysis demonstrated that, at least for Sγ1, there is a direct correlation between the length of the region of core Sγ1 49-bp repeats and the level of CSR that it supports (Fig. 5B). Therefore, the size of the core repeat Sγ1 region directly influences the ability to support CSR. In light of this correlation, the robust CSR to IgG1 and high serum levels of IgG1 in mice may, at least in part, be attributed to the exceptionally long endogenous Sγ1.
Core S region length may play several different roles in influencing CSR. In this regard, S regions may target AID activity and CSR through several mechanisms. One apparent function of mammalian S regions is ability to form R loops when transcribed in physiological orientation because of the G-rich composition of their nontemplate strands. The length of R-loops formed within endogenous S regions has been shown to exceed 1 kb (19); therefore, longer S regions conceivably could contribute to increased CSR by generating more extensive R-loops. However, we also see a direct correlation of CSR frequency with length of inverted Sγ1 regions, which do not detectably form R-loops and likely target CSR by a non-R-loop mechanism (13, 14). We have previously argued that the ability of the 4-kb Xenopus Sμ, which does not generate detectable R-loops, to support CSR in murine B cells is associated with its high number and density of AGCT motifs (14). Likewise, the presence of substantial numbers of AGCT and related motifs might account for the ability of the inverted endogenous Sγ1 sequence to promote CSR. In this context, increasing the number of such motifs by increasing the length of S-region core sequence might increase CSR potential by providing more substrates for the AID/replication protein A complex.
Although the present study was directed primarily at assessing the role of S-region length in CSR, it provides additional insights that may help interpret some unexplained differences in levels of CSR with respect to test sequences used in our prior replacement experiments (13, 14). A 1-kb random G-rich sequence, which forms R-loops but is devoid of standard S-region repeats and motifs, was able to support CSR to a comparable level to the 1-kb synthetic S region (7% vs. 9%) analyzed in this study (13). This finding is consistent with the notion that R-loop formation can contribute substantially to CSR activity of S regions. On the other hand, in inverted orientation, the CSR efficiency of the random G-rich sequence was reduced to background levels (13); whereas the inverted 1-kb S region still supported reduced, but readily detectable, levels of CSR. As the nucleotide composition of the random G-rich and synthetic Sγ1 sequences are comparable, the most obvious, major difference between the two is that the synthetic Sγ1 region contains a much higher number of AGCT motifs. Therefore, we would argue that the high density of AGCT motifs may be responsible the CSR activity of the inverted S region. Similarly, the inverted 4-kb Sγ1 shows comparable CSR activity to the 4-kb Xenopus S region analyzed in ref. 14. Because the Xenopus S region, like the inverted endogenous Sγ1 region, does not make substantial levels of R-loops, the comparable activity of these two sequences might again be attributed to the abundance of AGCT motifs. Therefore, our current findings are consistent with the possibility that both R-loop formation and density of AGCT and related motifs contribute to CSR by providing targets for AID.
Our development of a system to analyze synthetic Sγ1 regions lays the groundwork for more defined analyses of the function of S regions and their component motifs. Previous analyses of S regions in CSR have relied on CSR recombination substrates, which do not always appear to reflect the normal mechanism and control of endogenous CSR. On the other hand, previous studies of endogenous S regions have necessarily viewed S regions as large DNA fragments, with no ability to readily dissect function of their component motifs. While R-loop-forming ability and the density of AGCT and related motifs may be major contributors to targeting CSR, Ig isotype-specific elements and structures [inverted repeats and stem–loop structures (29)], G4 quartets (30), and other factors (e.g., transcription levels, location, etc.) also have been argued to contribute to differential CSR targeting (25). By using synthetic S regions, controlled for size effects, we can now begin to systematically investigate the contribution of these different elements with respect to the function of individual S regions in vivo.
Supplementary Material
Acknowledgments
We are indebted to Michael Lieber, Amy Kenter, and David Schatz for thoughtful comments and suggestions in the context of reviewing the manuscript. We thank Y. Fujiwara, N. Stokes, and A. Williams for mouse work and J. Chaudhuri, J. Manis, and S. Ranganath for helpful discussions. This work was supported by National Institutes of Health Grant AI31541 (to F.W.A.). F.W.A. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: A.A.Z., M.T., and F.W.A. designed research; A.A.Z., M.T., J.W., and T.B. performed research; J.W. analyzed data; and A.A.Z. and F.W.A. wrote the paper.
Abbreviations: AID, activation-induced deaminase; CSR, class switch recombination; ES, embryonic stem; IgH, Ig heavy chain; S, switch.
References
- 1.Bassing, C. H., Swat, W. & Alt, F. W. (2002) Cell 109, Suppl., S45-S55. [DOI] [PubMed] [Google Scholar]
- 2.Li, Z., Woo, C. J., Iglesias-Ussel, M. D., Ronai, D. & Scharff, M. D. (2004) Genes Dev. 18, 1-11. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhuri, J. & Alt, F. W. (2004) Nat. Rev. Immunol. 4, 541-552. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido, T., Nakai, S. & Honjo, T. (1981) Nature 292, 845-848. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido, T., Yamawaki-Kataoka, Y. & Honjo, T. (1982) J. Biol. Chem. 257, 7322-7329. [PubMed] [Google Scholar]
- 6.Arakawa, H., Iwasato, T., Hayashida, H., Shimizu, A., Honjo, T. & Yamagishi, H. (1993) J. Biol. Chem. 268, 4651-4655. [PubMed] [Google Scholar]
- 7.Szurek, P., Petrini, J. & Dunnick, W. (1985) J. Immunol. 135, 620-626. [PubMed] [Google Scholar]
- 8.Mowatt, M. R. & Dunnick, W. A. (1986) J. Immunol. 136, 2674-2683. [PubMed] [Google Scholar]
- 9.Kataoka, T., Miyata, T. & Honjo, T. (1981) Cell 23, 357-368. [DOI] [PubMed] [Google Scholar]
- 10.Shanmugam, A., Shi, M. J., Yauch, L., Stavnezer, J. & Kenter, A. L. (2000) J. Exp. Med. 191, 1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luby, T. M., Schrader, C. E., Stavnezer, J. & Selsing, E. (2001) J. Exp. Med. 193, 159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khamlichi, A. A., Glaudet, F., Oruc, Z., Denis, V., Le Bert, M. & Cogne, M. (2004) Blood 103, 3828-3836. [DOI] [PubMed] [Google Scholar]
- 13.Shinkura, R., Tian, M., Smith, M., Chua, K., Fujiwara, Y. & Alt, F. W. (2003) Nat. Immunol. 4, 435-441. [DOI] [PubMed] [Google Scholar]
- 14.Zarrin, A. A., Alt, F. W., Chaudhuri, J., Stokes, N., Kaushal, D., Du Pasquier, L. & Tian, M. (2004) Nat. Immunol. 5, 1275-1281. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 16.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102, 565-575. [DOI] [PubMed] [Google Scholar]
- 17.Manis, J. P., Tian, M. & Alt, F. W. (2002) Trends Immunol. 23, 31-39. [DOI] [PubMed] [Google Scholar]
- 18.Tian, M. & Alt, F. W. (2000) J. Biol. Chem. 275, 24163-24172. [DOI] [PubMed] [Google Scholar]
- 19.Yu, K., Chedin, F., Hsieh, C. L., Wilson, T. E. & Lieber, M. R. (2003) Nat. Immunol. 4, 442-451. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422, 726-730. [DOI] [PubMed] [Google Scholar]
- 21.Yu, K., Roy, D., Bayramyan, M., Haworth, I. S. & Lieber, M. R., Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 22.Chaudhuri, J., Khuong, C. & Alt, F. W. (2004) Nature 430, 992-998. [DOI] [PubMed] [Google Scholar]
- 23.Stavnezer, J. (1996) Adv. Immunol. 61, 79-146. [DOI] [PubMed] [Google Scholar]
- 24.Snapper, C. M., Finkelman, F. D., Stefany, D., Conrad, D. H. & Paul, W. E. (1988) J. Immunol. 141, 489-498. [PubMed] [Google Scholar]
- 25.Kenter, A. L., Wuerffel, R., Dominguez, C., Shanmugam, A. & Zhang, H. (2004) J. Exp. Med. 199, 617-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai, E., Bottaro, A., Davidson, L., Sleckman, B. P. & Alt, F. W. (1999) Proc. Natl. Acad. Sci. USA 96, 1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, J., Shinkai, Y., Young, F. & Alt, F. W. (1994) Curr. Opin. Immunol. 6, 313-319. [DOI] [PubMed] [Google Scholar]
- 28.Dunnick, W. A., Shi, J., Graves, K. A. & Collins, J. T. (2004) J. Immunol. 173, 5531-5539. [DOI] [PubMed] [Google Scholar]
- 29.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20, 165-196. [DOI] [PubMed] [Google Scholar]
- 30.Dempsey, L. A., Sun, H., Hanakahi, L. A. & Maizels, N. (1999) J. Biol. Chem. 274, 1066-1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.