Abstract
Chemo-resistance, which is the main obstacle in cancer therapy, is caused by the onset of drug-resistant cells in the heterogeneous cell population in cancer tissues. MicroRNAs regulate gene expression at the post-transcriptional level, and they are involved in many different biological processes, including cell proliferation, differentiation, metabolism, stress response, and apoptosis. The aberrant expression of microRNAs plays a major pathogenic role from the early stages of the carcinogenesis process. Recently, microRNAs have been reported to play an important role in inducing resistance to anti-cancer drugs. Specific microRNA alterations occur selectively in cancer cells, rendering these cells resistant to various chemotherapeutic agents. For example, resistance to 5-fluorouracil is mediated by alterations in miR-21, miR-27a/b, and miR-155; the sensitivity to Docetaxel is influenced by miR-98, miR-192, miR-194, miR-200b, miR-212, and miR-424; and the resistance to Cisplatin is mediated by miR-let-7, miR-15, miR-16 miR-21 and miR-214. Chemo-resistant cancer cells are characterized by altered functions in enzymes that are involved in microRNA maturation, primarily including Dicer, as demonstrated in ovarian cancer, oral squamous cell carcinoma, breast cancer and cervical cancer. Based on the evidence reviewed in this paper, various strategies have been developed to artificially re-establish microRNA expression in resistant cells, thus restoring chemo-sensitivity. These strategies employ synthetic analogs, anti-microRNA oligonucleotides, locked nucleic acid, microRNA sponges, drugs that inhibit DNA methylation or histone deacetylation, and the introduction of microRNA mimics. The ability to modulate microRNA expression is a promising strategy for overcoming the problem of drug resistance in cancer treatment.
Keywords: MicroRNA, chemo-resistance, cancer chemotherapy
Introduction
Chemo-resistance is the main obstacle to the effectiveness of cancer therapies as it allows the cancer cells to survive the treatment and proliferate uncontrollably, leading to cells with a more aggressive phenotype that are more able to metastasize to other organs than naïve cancer cells [1]. Currently, no therapy has an efficacy of 100% [2] because drug resistance limits the potency of both conventional chemotherapeutic and novel biological agents [3].
Tumors can be intrinsically resistant to chemotherapy or the resistance can be acquired as a result of the pharmacological treatment of cancer cells that were previously sensitive to the drug [4]. Indeed, tumors can be characterized by molecular heterogeneity [5], and this heterogeneity can contribute to the onset of drug resistance through a therapy-induced selection of a resistant cell subpopulation that is present in the original tumor. Cancer tissues generally consist of a mixed population of malignant cells; some of these cells are sensitive to cancer drugs, while others are drug resistant. According to the Cancer Stem Cells theory, tumor tissues are composed of a sub-population of tumorigenic stem cells that undergo differentiation, giving rise to cancer cells. These cancer stem cells are chemo-resistant and are involved in the formation of metastases [6]. Meads et al. reported a chemo-resistance mechanism named environment-mediated drug resistance, which is based on communication between the tumor cells and their microenvironment. Signaling events originating in this microenvironment allow these cells to escape apoptosis induced by chemotherapeutic agents, leading to the creation of surviving foci of residual cells [7].
Chemotherapy kills drug-sensitive cells, but resistant cells survive and become more aggressive and prone to metastasis due to the hypoxic conditions established by the therapy in the neoplastic mass [8-10].
During the acquisition of resistance, cancer cells can become cross-resistant to a range of chemotherapies, which may ultimately lead to treatment failure [11].
This general situation is enhanced in lung cancer, which is highly prone to develop chemo-resistance since its early onset. Indeed, lung cancer typically occurs in smokers, and thus, it is composed of cells that have adapted for decades and are to resist the toxic environment established by the cigarette smoke. Lung cancer develops from cells that are targeted by multiple cigarette smoke-induced genetic and epigenetic alterations that escape the apoptotic pathway [12]. These cells grow well in the presence of highly genotoxic cigarette smoke condensate [13] due to the existence of effective mechanisms that counteract the cigarette smoke genotoxicity. These mechanisms mainly include the activation of glutathione conjugation-based phase II detoxification reactions and the up-regulation of multidrug resistance protein 1 (MDR1). Indeed, we demonstrated that in p53 mutant mice undergoing 4 weeks of exposure to cigarette smoke, phase I and II metabolic reactions are strongly induced, which is a feature that is paralleled by MDR1 up-regulation [14]. MDR1 is modulated by changes in microRNA expression and is a sensitive target of cigarette smoke-induced molecular alterations [15]. In particular, cigarette smoke alters the expression of miR-30c, miR-138, and miR-378, which play a pivotal role in activating the expression of the MDR1 protein that is involved in the extracellular extrusion of the glucurono-conjugated genotoxic metabolites of cancer chemotherapeutic drugs. This finding explains the mechanism by which lung cancer cells, particularly in smokers and ex-smokers, become highly chemo-resistant. To overcome drug resistance, the molecular mechanisms underlying drug resistance must be identified and understood with the aim of discovering new drugs that are able to interfere with chemo-resistance [1].
Etiology and mechanisms of chemo-resistance
Drug resistance is a complex phenomenon that can occur at different levels. One of the most common mechanisms is the action of a specific group of trans-membrane proteins, whose task is to remove cytotoxic molecules from the cell. These proteins belong to the class of ATP-binding cassette proteins, which are involved in the regulation of the absorption and excretion of many different toxic compounds. One of these proteins, P-glycoprotein (Pgp), is mainly responsible for drug resistance targeted at a wide range of chemotherapeutic agents with different mechanisms of action. Pgp is physiologically expressed in almost all tissues at low levels, but its expression is increased in the epithelial cells that are implicated in excretion, such as those located in the small intestine, pancreatic ductules and kidney proximal tubules [16]. In many cancer tissues, Pgp overexpression determines intrinsic drug resistance. However, chemotherapy can also enhance the expression of proteins, causing acquired resistance [17]. The overexpression of the Pgp protein in the membranes of cancer cell leads to an increased drug efflux and reduces the accumulation of the therapeutic effective dose in the cytoplasm, thus rendering the drugs ineffective for cancer treatment [18].
MicroRNAs play a major role in regulating Pgp expression and activity [19], as demonstrated by miR-145 in intestinal cells [20] and miR-130 in Cisplatin-resistant ovarian cancer cells. Accordingly, the modulation of these microRNAs may be a possible therapeutic strategy for overcoming multidrug resistance in cancer cells [21] and limiting the efficacy of hydrophobic and amphipathic molecules, such as vinca alkaloids, mitomycin C, anthracyclines, and taxanes [22].
Another mechanism underlying resistance is the lack of drug activation at the metabolic level. Indeed, many compounds must be converted to their active metabolites before they exert their cytotoxic effects. For example, the antimetabolite 5-Fluorouracil requires different enzymes, such as uridine phosphorylase, to become activated, and its therapeutic efficacy is associated with the levels of these enzymes in the liver [23,24]. Nevertheless, this drug is inactivated in the liver by dihydropyrimidine dehydrogenase before it reaches the tumor site, thus decreasing the amount of the drug that can reach the target site [25]. MicroRNAs play a major role in regulating the balance between the activation and detoxification of 5-Fluorouracil. For example, miR-27a/b are pivotal regulators of dihydropyrimidine dehydrogenase [26]. Regarding uridine phosphorylase, 2 bioinformatics databases [27] predict that this enzyme is targeted by 9 different microRNAs. These microRNAs, according to their decreasing target score in miRDB, are as follows: miR-581, miR-5011, miR-4446, miR-578, miR-221, miR-8073, miR-5002, miR-4635 and miR-5197.
Regarding the effectiveness of cancer therapies, drug targets must not undergo alterations in their expression level. Indeed, when the enzyme thymidylate synthase (the target of the 5-Fluorouracil metabolite fluorodeoxyuridine monophosphate) [28] is overexpressed, drug resistance occurs. The same situation occurs when the thymidylate synthase gene is amplified [29]. This amplification likely results from a loss of function of the molecule.
Modification of DNA-methylation also leads to drug resistance. In lung cancer, DNA methylation contributes to the down-regulation of the Dickkopf-related protein 3 (DKK3) gene, which is a tumor suppressing gene. This down-regulation makes the cells resistant to Docetaxel by promoting the proliferative capacity and inhibiting of apoptosis [30]. The same resistance to Docetaxel-based chemotherapy is also observed in human lung adenocarcinoma cells in which the gene RUNX3 is down-regulated because of its methylation [31]. MicroRNAs are deeply involved in the regulation of DNA methylation; particularly, miR-29 modulates DNA methyl-transferase expression, thus, in turn, regulating the epigenetic silencing of multiple genes that are involved in cancer growth and chemo/radio-resistance [32]. In glioma cells, the role of DNA-methylation is further supported by the action of Temozolomide on DNA methyltransferase through miR-29 [33].
Many drugs that are used in cancer chemotherapy act by targeting DNA directly or indirectly, causing DNA damage and leading to a cell cycle arrest as follows: if the damage can be repaired, the cell survives; otherwise, the cell dies. Platinum agents induce the formation of DNA adducts that are removed by the nucleotide excision repair (NER) pathway via a complex process that involves many different proteins, including the excision repair cross-complementary 1 (ERCC1) protein [34]. A high expression level of ERCC1 is associated with platinum chemo-resistance both in ovarian and gastric tumors. A positive correlation has been observed between the low expression of ERCC1 and the prolonged survival of patients with non-small cell lung cancer (NSCLC), who are treated with Cisplatin plus Gemcitabine [35]. MicroRNAs play a fundamental role in this type of chemo-resistance because ERCC1 is regulated by miR-139 and miR-199 as demonstrated in leukemic cells [36]. miR-199a is also involved in the progression of osteosarcoma, and its expression is down-regulated compared with that in hea-lthy osteoblasts. When miR-199a-3p is transfected into U-2OS osteosarcoma cells, it enhances the Doxorubicin sensitivity via CD44 inhibition [37]. miR-199a is also down-regulated in ovarian cancer, and its transfection into cancer-initiating cells leads to an increased sensitivity in these cells to Cisplatin, Paclitaxel, and Adriamycin. Moreover, in the transfected cells, CD44 expression is reduced, suggesting that miR-199a plays an inhibitory role against this protein [38].
Another mechanism underlying the resistance that is associated with DNA-damaging agents is represented by mutations in the DNA mismatch repair (MMR) genes. The loss of this repair mechanism leads to drug resistance, rendering the cell unable to recognize the damage and activate apoptosis. Furthermore, indirect damage caused by the loss of MMR includes genome instability that occurs as result of the increased mutation rate [39]. MMR undergoes microRNA regulation, particularly by miR-21 and miR-155 [40]. Indeed, Cisplatin inhibits the growth of aggressive A549 lung cancer cells by up-regulating MSH2 expression through miR-21 down-regulation [41].
RAD18 is a DNA damage-activated E3 ubiquitin ligase that is involved in DNA damage repair in cancer cells. Liu et al. showed that this protein is overexpressed in 5-Fluorouracil-resistant colon cancer, while miR-145 is down-regulated. Using luciferase reporter assays, the authors demonstrated that RAD18 is targeted by miR-145, and when the expression of this protein is decreased, 5-Fluorouracil chemo-resistance is overcome [42].
In cancer chemo-resistant cells, the mechanisms underlying apoptosis are often inhibited, including both intrinsic and extrinsic pathways, due to the overexpression of anti-apoptotic proteins, such as Bcl-2, or the decreased expression of pro-apoptotic proteins, such as Fas [43]. Regarding the Bcl-2 gene, its expression is increased in chemo-resistant acute myeloid leukemia [44] and breast cancer [45].
The Bcl-2 pathway in chemo-resistant cells is under microRNA control as demonstrated by miR-138 in glioblastoma [46] and miR-192 and miR-98 in lung cancer [47]. Xiang et al. transfected the Cisplatin-resistant A549 cell line with miR-98 and found that the expression of the anti-apoptotic proteins Bcl-2 and Bcl-XL is decreased, leading to spontaneous apoptosis [47]. In colorectal cancer, miR-139-5p is involved in the regulation of apoptosis by targeting Bcl-2. The lower expression of miR-139-5p in cancer cell lines and tumor specimens leads to decreased apoptosis. When colorectal cancer cells are transfected with miR-139-5p, Bcl-2 is reduced, and apoptosis is enhanced [48]. Liu et al. confirmed that miR-139-5p is down-regulated in colorectal cancer cells that are treated with 5-Fluorouracil and 5-Fluorouracil-resistant cancer cells. Moreover, these authors found that NOTCH-1 is targeted by miR-139-5p, and when this microRNA expression is enhanced, the chemotherapeutic effects of 5-Fluorouracil are increased [49]. In breast cancer, miR-139-5p is down-regulated, and cells transfected with this miRNA mimic are characterized by a decreased cell growth. Regarding Docetaxel-resistant breast cancer, the miR-139-5p down-regulation increases the chemo-sensitivity via the NOTCH-1 pathway [50].
In A549 Cisplatin-resistant lung cancer cell lines, the inhibition of apoptosis is due to the down-regulation of Bim as determined by high expression of miR-192 in these cells that directly targets Bim [51].
The so-called guardian of the human genome, P53, is also a pivotal regulator of apoptosis. The lack of functional P53 due to mutations leads to drug resistance, which is ultimately due to the lack of apoptosis activation after genotoxic drugs-induced DNA damage as demonstrated in 5-fluorouracil [52]. However, the link between p53 and chemo-resistance remains controversial. In fact, there are some studies that did not find any correlation between P53 expression and therapy failure. For example, Paradiso and colleagues performed a retrospective study on advanced colorectal cancer patients to detect the ability of the P53 status to predict the clinical response to chemotherapy. In this study, the authors could not show any association between the protein P53 positivity in the samples and the clinical response to chemotherapy [53]. This finding is likely related to the high heterogeneity in p53 mutations, and only a few of these mutations have an impact on cancer progression and chemo-resistance [54]. Overall, p53 mutations, if well characterized from a functional standpoint, are important predictors of cancer outcome [55]. The P53 status and function are modulated by microRNAs, particularly miR-34, which is recognized as a pivotal p53 effector [56,57].
MicroRNA and chemo-resistance
MicroRNAs are small non-coding RNAs consisting of 20-25 nucleotides. MicroRNAs are evolutionarily conserved, and their pivotal role is the regulation of gene expression at the post-transcriptional level [58]. MicroRNAs are involved in many different biological processes, including cell proliferation, differentiation, metabolism, stress response and apoptosis. Thus, their aberrant expression is associated with pathologies, particularly cancer [59]. Moreover, microRNA deregulation can influence the outcome of cancer therapies and allows tumors to acquire a chemo-resistant phenotype. This outcome was first demonstrated by a study that reported a difference in the microRNA expression profiles of Tamoxifen-sensitive and resistant MCF-7 breast cancer cells [60]. Similarly, Sarkar et al. noted that the expression levels of miR-194, miR-200b, and miR-212 are down-regulated in Docetaxel-resistant non-small cell lung cancer cells compared to those levels in sensitive cells, while miR-98, miR-192, and miR-424 are up-regulated [61,62]. Regarding lung cancer, the expression of miR-98 is decreased by 3-fold in the Cisplatin-resistant A549 cell line compared to that in the parental line; when miR-98 expression is restored, the cells become sensitive to Cisplatin partially through the miR-98-mediated regulation of high mobility group A2 (HMGA2) oncogene regulation [47]. In chronic myeloid leukemia, miR-424 expression is lower compared to that in healthy samples. When this microRNA is transfected into K562 chronic myeloid leukemia cells, its expression leads to a reduced proliferation and increased sensitivity to Imatinib, leading to apoptosis [63]. In non-small cell lung cancer, cell proliferation is influenced by miR-194 expression. The expression of this microRNA is down-regulated in cancerous-lung tissues compared with that in normal tissues. When exogenous miR-194 is introduced into lung cancer cells, proliferation, migration and invasion are reduced while apoptosis is increased. These results are also observed in the Cisplatin-resistant lung cancer cell line, in which miR-194 is down-regulated, and its ectopic expression leads to an increased Cisplatin sensitivity [64].
MicroRNAs influence the response to cancer treatments by regulating the expression of genes that are involved in the cell-cycle control, apoptosis, and DNA repair [1]. Regarding the latter, miR-155 and miR-21 are responsible for 5-Fluorouracil resistance in colorectal cancer by interfering with the DNA-repair system [65,66]. miR-125b, which targets p53, inhibits the apoptosis that leads to Doxorubicin, Vincristine, Etoposide and Mafosfamide resistance in Ewing sarcoma/primitive neuroectodermal tumors. High levels of this microRNA are observed in Doxorubicin-resistant Ewing sarcoma cells compared to those in sensitive cells [67]. In lung cancer, the resistance to apoptosis is mediated by miR-128-2, which renders the cells resistant to Cisplatin, Doxorubicin and 5-Fluorouracil. The overexpression of this microRNA determines chemo-resistance by targeting the transcriptional repressor E2F5, which leads to the accumulation of the p21 protein in the cytoplasm and the inhibition of apoptosis [68].
In contrast, certain microRNAs promote drug sensitivity. Li and co-workers reported that in colon cancers characterized by p53 mutations, miR-22, which is a positive regulator of the tumor suppressor gene phosphatase and tensin homolog (PTEN), enhances the sensitivity to Paclitaxel. This enhancement occurs through the promotion of PTEN expression and the consequent inhibition of Akt phosphorylation [69]. Another example is provided by miR-214, which increases the chemo-sensitivity to Cisplatin in cervical cancer by counteracting the expression of the anti-apoptotic protein Bcl-2 [70].
MicroRNAs are also involved in other mechanisms underlying resistance, such as the control of drug uptake and drug metabolism. Bao et al., analyzes the microRNA expression profiles of Doxorubicin-sensitive and Doxorubicin-resistant breast cancers and found that miR-298 is down-regulated in the resistant cells, which is associated with the increased expression of P-glycoprotein and the increased drug efflux [71]. This effect is also observed when miR-451 and miR-27a are up-regulated as determined by the overexpression of p-glycoprotein in multidrug-resistant ovarian and cervical cancer cell lines [72].
Regarding drug metabolism, the cytochrome p450 family enzymes play a pivotal role in this mechanism. The expression of many of these enzymes is under microRNA control. miR-27b is implicated in the regulation of the CYP1B1 protein, and when this microRNA is down-regulated, CYP1B1 is overexpressed in tumor tissues [73]. The miR-27b down-regulation is also evident in Tamoxifen-resistant breast cancer cells in which it targets HMGB3. When this microRNA is re-expressed, their cells become chemo-sensitive, their invasion is reduced and the epithelial-mesenchymal transition (EMT)-like properties are reversed [74].
Regarding melanoma, Fattore et al. found that miR-579-3p expression is significantly reduced in melanoma cell lines that are resistant to vemurafenib compared to that in wild-type cells. Moreover, the inverse correlation between the RNA levels of miR-579-3p and BRAF in resistant cells shows that miR-579-3p controls BRAF expression. The ectopic expression of this microRNA in vemurafenib-resistant melanoma cells reduces drug resistance [75].
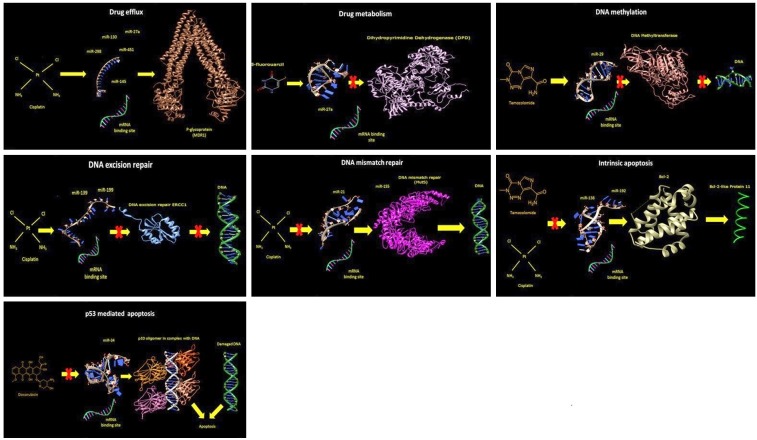
A schematic overview of the chemo-resistance mechanisms that are induced by alterations in different microRNAs is shown in Figure 1.
Figure 1.
A schematic view of the chemo-resistance mechanisms induced by miRNAs. Drug efflux: Expression of P-glycoprotein (MDR1) is regulated through the binding of microRNAs (miR27a, miR-130, miR145, miR-298, miR-451) to their target mRNA. The microRNA (miR-145) three-dimensional mature structure was modeled using the Web Server RNAComposer [Popenda et al., 2012]. The atomic structure of the human mitochondrial ABC transporter ABCB10 [Shintre et al., 2013; PDB code 3ZDQ] is represented in ribbons using the program Chimera [Pettersen et al., 2004]. Drug metabolism: Using the same software described above, the action of miR-27a on dihydropyrimidine dehydrogenase [Dobritzsch et al., 2001; PDB code 1H7X], a major determinant of the pharmacokinetics of the anti-cancer drug 5-Fluorouracil, is reported. DNA methylation: Gene methylation by DNA methyltransferases is regulated by miR-29. In this panel, the atomic structure of the protein structure of mouse DNMT1 and a fragment of hemi-methylated CpG DNA [Song et al., 2012; PDB code 4DA4]. DNA excision repair: The figure shows the 3D structure of the central domain of human ERCC1, a DNA excision repair protein [Tsodikov, 2005; PDB code 2A1I] that is regulated by miR-139 and miR-199. DNA mismatch repair: miR-21 and miR-155 are involved in the regulation of the DNA mismatch repair protein MutS [Oblomova et al., 2000; PDB code 1EWR]. Intrinsic apoptosis: Bcl-2 three-dimensional structure [Checco et al., 2015; PDB code 5AGW] and the regulators miR-138 and miR-192 are shown. p53-mediated apoptosis: The quaternary assembly of the p53 protein core domains bound to a naturally occurring response element (RE) located at the promoter of the Bcl-2-associated X protein (BAX) gene [Chen et al., 2013; PDB code 4HJE].
Most frequently involved microRNAs in cancer chemo-resistance
miR-let-7
Let-7, which is one of the best-known microRNA families, was first identified in Caenorhabditis elegans, and it is responsible for developmental timing [76]. It is evolutionarily conserved, and in many organisms, such as rats, mice and humans, it constitutes a family with multiple members, including eleven members in humans [77]. Let-7 is a tumor suppressor microRNA that targets the oncogene ras. In lung cancer, its expression is lower compared to that in normal lung tissues, while the expression of the RAS protein is enhanced in lung cancer [78]. The early let-7 down-regulation is a fundamental step in cigarette smoke-induced lung carcinogenesis [79] and is accompanied by the overexpression of multidrug resistance mechanisms [12].
Yang et al. performed a microRNA microarray analysis in both ovarian cancer samples and ovarian and breast cancer cell lines to identify the microRNAs that are involved in the resistance to Cisplatin. These authors found that the let-7i expression was reduced in the chemo-resistant specimens [80]. Another study showed that let-7e is down-regulated in the ovarian cancer cell line A2780CIS, which is resistant to Cisplatin. This result is also observed in the ovarian cancer cells A2780TC1 and A2780TC3, which are resistant to Paclitaxel [81]. Using the ovarian cancer Cisplatin-resistant cell line A2780/CP, Cai et al. found that the expression of let-7e is reduced compared to that in the parental cell line A2780. When let-7e is transfected into these cells, its consequent overexpression leads to a re-sensitization of A2780/CP, while the inhibition of this microRNA in the parental cell line determines the acquisition of Cisplatin-resistance [82].
Considering gastric cancer, Yang et al. reported that in the chemotherapy-resistant SGC7901/DDP and SGC7901/VCR gastric cancer cell lines, the expression of let-7 is lower compared to that in the chemotherapy-sensitive cells [83]. In the breast cancer Tamoxifen-resistant cell line MCF-7 Tam, the level of let-7b/i is significantly reduced compared to that in the parental cells, indicating that this microRNA in involved in chemo-resistance. Indeed, the transfection of let-7 mimics enhances the sensitivity of MCF7 cells to Tamoxifen [84].
The expression of let-7 is also decreased in esophageal squamous cell carcinoma. Sugimura et al. measured the microRNA expression in pretreated biopsies from 98 patients with esophageal cancer and found that let-7c is down-regulated, and this down-regulation is associated with a poor response to Cisplatin and a poor prognosis. However, when the authors induced the expression of let-7c in the esophageal squamous cell carcinoma cell lines, the chemo-sensitivity was restored [85].
miR-21
miR-21 is an oncomiR whose expression is frequently up-regulated in different tumors, including tongue and prostate carcinomas and non-small cell lung cancer [86-88]. When miR-21 is inhibited in breast and glioma cell lines, cell proliferation is reduced, and apoptosis is increased [89,90]. The miR-21 inhibition-induced apoptosis is found in Cisplatin-resistant ovarian cells. Chan et al. performed microRNA microarrays and discovered that miR-21 is overexpressed in the Cisplatin-resistant ovarian cell line A2780. When this microRNA is down-regulated by anti-microRNA inhibitors, the cells become more sensitive to the drug [91].
In pancreatic cancer, the overexpression of miR-21 is strongly associated with chemo-resistance. Paik et al. used the human pancreatic cancer cell line Panc-1 and noted that the suppression of miR-21 by indole-3-carbinol enhances the sensitivity of the cells to the drug Gemcitabine [92]. Indole-3-carbinol was recognized as a miR-21 modulator in mice [93].
The overexpression of miR-21 is associated with Cisplatin-resistance in osteosarcoma cells. When the cells are transfected with anti-miR-21, they show a higher sensitivity to Cisplatin because the knockdown of miR-21 leads to a down-regulation of the bcl-2 pro-apoptotic protein [94].
In the glioblastoma cells U373 MG, miR-21 is up-regulated, and this up-regulation causes a resistance to the drug VM-26 (Teniposide), which is a topoisomerase II inhibitor. Using specific antisense oligonucleotides in glioblastoma cells, the cytotoxicity of the drug is increased [95].
In non-small cell lung cancer, miR-21 expression is up-regulated in PC-9 gefitinib-resistant cells compared to that in the parental cells, while the silencing of miR-21 restored the chemo-sensitivity to this monoclonal antibody, inhibiting the epidermal growth factor receptor pathway [96]. Dong et al. discovered that miR-21 silencing reverses the lung cancer cell multidrug resistance. In the lung cancer cell line A549/DDP, which is resistant to Cisplatin, miR-21 silencing reduces the expression of P-glycoprotein and increases apoptosis. Furthermore, cell proliferation is inhibited, and cells are blocked in the G0/G1 phase of the cell cycle [97]. In the MCF-7 breast cancer cell line, miR-21 silencing provides another example of reversing multidrug resistance. Indeed, after this procedure, the MCF-7 cells become sensitive to the drug topotecan, which is a topoisomerase inhibitor [98].
miR-15/miR-16
miR-15 and miR-16 are members of a family that includes miR-15a/b, miR-16, miR-195, miR-497 and miR-322, is involved in cell cycle control by targeting CDK1, CDK2, and CDK6 as well as cyclins D1, D3, and E1, and, consequently, is responsible for cell cycle arrest at the G1 phase [99]. miR-15b is implicated in the modulation of the stress response by inducing p53 phosphorylation, DNA repair and apoptosis after irradiation of the human bronchial epithelial cell line HBEC [100].
Calin et al. reported that miR-15a and miR-16-1 under normal conditions negatively regulate the anti-apoptotic protein Bcl-2, leading to apoptosis; however, when these microRNAs are down-regulated, the cells can escape apoptosis, which contributes to malignant transformations [101,102].
In the multidrug resistant gastric cancer cell line SGC7901/VCR, which is resistant to Vincristine, the expression levels miR-15b and miR-16 are down-regulated compared to those in the parental cell line. This finding is confirmed by a drug sensitivity assay, which shows that the overexpression of these microRNAs renders the cells sensitive to anticancer drugs (Vincristine, Doxorubicin, Etoposide and Cisplatin). Moreover, the down-regulation of these microRNAs results in the up-regulation of the Bcl-2 protein, leading to apoptosis inhibition [103].
Blower et al. used the lung cancer A549 cell line and found that the inhibition of miR-16 is responsible for the decreased sensitivity of these cells to the drug Plumbagin (AKT inhibitor) [104]. Conversely, miR-15b overexpression determines resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in non-small cell lung cancer cells [105,106]. Zhao et al. showed that in Cisplatin-resistant lung adenocarcinoma A549/CDDP cells, miR-15 expression is enhanced compared to that in the parental cell line. This increased expression is also found in vivo in tissues sampled from lung adenocarcinoma patients, who received Cisplatin-based chemotherapy. Even in this situation, the overexpression of miR-15 is associated with Cisplatin resistance and a poor prognosis [107].
Regarding breast cancer, chemo-resistant MCF-7/HER2delta16 cells become sensitive to Tamoxifen when miR-15a/16 are re-introduced into these cells [108]. This finding was further confirmed by Chu et al. in Tamoxifen-resistant ER-positive breast cancer cell lines in which the introduction of miR-15a/16 leads to chemo-sensitivity through the inhibition of Cyclin E1 and Bcl-2 [109].
In the chemo-resistant ovarian cancer A2780-CP20 cell line, Dwivedi et al. reported that the ectopic expression of miR-15a and miR-16 is involved in the inhibition of chemo-resistance mechanisms and tumor progression, leading to sensitization to Cisplatin. This finding is confirmed in vivo in a mouse model of chemo-resistant ovarian cancer in which the combined delivery of these two microRNAs reduced the tumor burden [110].
In the Temozolomide-resistant U251MG/TR cell line, miR-16 expression is down-regulated compared to that in the parental cells. Treating the resistant cells with miR-16 mimics enhances chemo-resistance, while the use of miR-16 inhibitors increases Temozolomide sensitivity [111].
miR-34a
miR-34a, which is a pivotal actor in the p53 network, is involved in the inhibition of genes that are implicated in the cell cycle control and apoptosis by targeting the mRNA of these proteins, such as CDK4, CDK6, Cyclin D1, Cyclin E2, E2F3, and BCL2. miR-34a is frequently down-regulated in many tumors. Fujita et al. determined that the expression of miR-34a is significantly reduced in the prostate cancer p53-null cell line PC3 and the p53-mutated DU145 cell line compared to that in the p53 wild-type cells. The transfection of the miR-34a precursor into PC3 cells leads to the inhibition of cell growth. Moreover, this transfection can enhance the sensitivity to the drug Camptothecin through apoptosis activation [112]. In gastric cancer, Cao et al. found that in the Cisplatin-resistant cancer cell line SGC-7901/DDP, miR-34a is down-regulated, indicating that the overexpression of this microRNA increases the sensitivity of gastric cancer cells to Cisplatin [113]. The expression of miR-34a is reduced in breast cancer MCF-7 cells that are resistant to Adriamycin compared to that in sensitive cells [114]. Conversely, another study performed in breast cancer cells showed an up-regulation of miR-34a. Indeed, Kastl et al. found that in the MCF-7 Docetaxel-resistant cell line, the expression of miR-34a is increased, and the response to this drug could be modified through the modulation of miR-34a. Indeed, in the MCF-7 Docetaxel-resistant cells, the inhibition of this microRNA leads to chemo-sensitivity, while its overexpression is responsible for the onset of drug resistance in MCF-7 Docetaxel-sensitive cells [115].
miR-221/222
miR-221 and miR-222 are implicated in the regulation of endothelial inflammation and angiogenesis [116]. These microRNAs are overexpressed in many different cancer types, including breast cancer. miR-221 and miR-222 are involved in the resistance to Tamoxifen in breast cancer. To clarify the underlying mechanism, Miller et al. performed a microRNA microarray analysis using MCF-7 cell lines that are either sensitive or resistant to Tamoxifen. The expression of miR-221 and miR-222 was significantly up-regulated in the Tamoxifen-resistant cells compared to that in sensitive cells. Furthermore, the ectopic expression of these microRNAs renders the sensitive MCF-7 cells resistant to the drug. The expression of miR-221 and miR-222 is also enhanced in HER2/neu-positive primary human breast cancer tissues that are frequently resistant to endocrine therapy [117].
Zhao et al. found the same association between miR-221/miR-222 and the resistance to Tamoxifen in breast cancer [118].
Further evidence of the involvement of miR-221 in drug resistance is provided by Ye et al., who observed that in HER2-positive breast cancer, this microRNA is implicated in the inhibition of apoptosis and the induction of Trastuzumab resistance by targeting PTEN [119]. In the osteosarcoma SOSP-9607 and MG63 cell lines, the expression of miR-221 is increased compared to that in naive osteoblasts. This up-regulation is significantly associated with the Cisplatin-resistance and reduced apoptosis [120].
The specific relationships between the microRNAs and chemo-resistance in different cancer types are summarized in Table 1.
Table 1.
MicroRNAs altered in chemo-resistant cancer cells
| microRNA | Alteration trend | Main target | Drug resistance | Cancer | Experimental condition | References |
|---|---|---|---|---|---|---|
| miR-let-7b | ↓ | c-Myc | Cisplatin; Vincristine | Gastric cancer | Cell culture | [83] |
| miR-let-7b/i | ↓ | Estrogen receptor alpha | Tamoxifen | Breast cancer | Cell culture | [84] |
| miR-let-7c | ↓ | IL-6/STAT3 pathway | Cisplatin | Esophageal cancer | Human biopsy | [85] |
| miR-let-7e | ↓ | TGFBR1, OSMR, IL6, MMP11* | Cisplatin | Ovarian cancer | Cell culture | [81] |
| ↓ | TGFBR1, OSMR, IL6, MMP11* | Paclitaxel | Ovarian cancer | Cell culture | [81] | |
| ↓ | EZH2 CCND1 | Cisplatin | Ovarian cancer | Cell culture | [82] | |
| miR-let-7i | ↓ | H-RAS HMGA2 | Cisplatin | Ovarian and breast cancer | Cell culture, human biopsy | [80] |
| miR-15/miR-16 | ↓ | Bcl-2 | Vincristine | Gastric cancer | Cell culture | [103] |
| miR-15a/miR-16 | ↓ | Bcl-2 and cyclin E1 | Tamoxifen | Breast cancer | Cell culture | [109] |
| miR-16 | ↓ | Bcl-2 K-RAS-2A | Plumbagin (AKT inhibitor) | Lung cancer | Cell culture | [104] |
| ↓ | Bcl-2 | Temozolomide | Glioma | Cell culture | [111] | |
| miR-21 | ↓ | MSH2 | Cisplatin | Lung cancer | Cell culture | [41] |
| ↑ | DNA mismatch repair genes | 5-Fluorouracil | Colon cancer | Cell culture | [65,66] | |
| ↑ | PDCD4 c-IAP2 | Cisplatin | Ovarian cancer | Cell culture, human biopsy | [91] | |
| ↑ | PDCD4 | Gemcitabine | Pancreatic cancer | Cell culture | [92] | |
| ↑ | Bcl-2 | Cisplatin | Osteosarcoma | Cell culture | [94] | |
| ↑ | LRRFIP1 | VM-26 (Teniposide) | Glioblastoma | Cell culture | [95] | |
| ↑ | PTEN | Gefitinib | Lung cancer | Cell culture | [96] | |
| ↑ | MDR-related gene expression and inhibition of the AKT signaling pathway | Cisplatin | Lung cancer | Cell culture | [97] | |
| ↑ | Bcl-2 | Topotecan | Breast cancer | Cell culture | [98] | |
| miR-22 | ↑ | PTEN | Paclitaxel | Colon cancer | Cell culture | [69] |
| miR-27a | ↑ | MDR1/P-glycoprotein | Doxorubicin; Vinblastine | Ovarian and cervical cancer | Cell culture | [72] |
| ↑ | Dihydropyrimidine dehydrogenase | 5-Fluorouracil | Colon cancer | Cell culture | [26] | |
| miR-27b | ↓ | HMGB3 | Tamoxifen | Breast cancer | Cell culture | [74] |
| miR-29 | ↓ | O6-methylguanine-DNA methyltransferase | Temozolomide | Glioma | Cell culture | [33] |
| miR-34a | ↓ | SIRT1 | Camptothecin | Prostate cancer | Cell culture | [112] |
| ↓ | PI3K/AKT/survivin signaling pathway | Cisplatin | Gastric cancer | Cell culture | [113] | |
| ↓ | NOTCH1 | Adriamycin | Breast cancer | Cell culture | [114] | |
| ↑ | BCL-2 and cyclin D1 | Docetaxel | Breast cancer | Cell culture | [115] | |
| miR-98 | ↑ | HMGA2, Bcl-XL and Bcl-2 | Cisplatin | Lung cancer | Cell culture | [47] |
| miR-145 | ↓ | RAD18 | 5-Fluorouracil | Colon cancer | Cell culture | [42] |
| miR-125b | ↑ | P53 | Doxorubicin | Ewing sarcoma | Cell culture | [67] |
| miR-130 | ↑ | PTEN | Cisplatin | Ovarian cancer | Cell culture | [21] |
| miR-138 | ↑ | Bcl-2 pathway | Temozolomide | Glioma | Cell culture | [46] |
| miR-139 | ↓ | Bcl-2 | 5-Fluorouracil; Oxaliplatin | Colon cancer | Cell culture, mice model, human biopsy | [48] |
| ↓ | NOTCH-1 | 5-Fluorouracil | Colorectal cancer | Cell culture, human biopsy | [49] | |
| ↓ | NOTCH-1 | Docetaxel | Breast cancer | Cell culture | [50] | |
| miR-155 | ↑ | DNA mismatch repair (MMR) genes | 5-Fluorouracil | Colorectal cancer | Human biopsy | [40,65,66] |
| miR-192 | ↑ | Bim | Cisplatin | Lung cancer | Cell culture | [51] |
| miR-194 | ↓ | FOXA1 | Cisplatin | Lung cancer | Cell culture, human biopsy | [64] |
| miR-199a | ↓ | CD44 | Doxorubicin | Osteosarcoma | Cell culture, human biopsy | [37] |
| ↓ | CD44 | Cisplatin; Paclitaxel; Adriamycin | Ovarian cancer | Human biopsy | [38] | |
| miR-200b | ↓ | Bcl-2 | Docetaxel | Lung cancer | Cell culture | [61] |
| miR-212 | ↓ | BAG1, ABCC3, DYRK2, RNF8* | Docetaxel | Lung cancer | Cell culture | [61] |
| miR-214 | ↓ | Bcl-2 | Cisplatin | Cervical cancer | Lung cancer | [70] |
| miR-221/222 | ↑ | p27 | Tamoxifen | Breast cancer | Cell culture, human biopsy | [117] |
| ↑ | Estrogen receptor alpha | Tamoxifen | Breast cancer | Cell culture | [118] | |
| miR-221 | ↑ | PTEN | Trastuzumab | Breast cancer | Human biopsy | [119] |
| ↑ | PTEN | Cisplatin | Osteosarcoma | Cell culture | [120] | |
| miR-298 | ↓ | P-glycoprotein | Doxorubicin | Breast cancer | Cell culture | [154] |
| miR-424 | ↑ | LNA | Docetaxel | Lung cancer | Cell culture | [61,62] |
| ↓ | BCR-ABL | Imatinib | Chronic myeloid leukemia | Cell culture, human blood specimens | [63] | |
| miR-451 | ↑ | P-glycoprotein | Doxorubicin Vinblastine | Ovarian and Cervical cancer | Cell culture | [72] |
| miR-579 | ↓ | BRAF | Vemurafenib | Melanoma | Cell culture | [75] |
Putative targets identified by bioinformatics in silico models only.
Dicer
The mechanism that links alterations in microRNAs and chemo-resistance is, at least partially, related to molecular damage that hampers the function of enzymes involved in the microRNA maturation process.
RNase III ribonuclease Dicer (EC: 3.1.26.3) is a large multi-domain protein that plays a central role in microRNA biogenesis by cleaving the double stranded microRNA (dsRNA) precursor into segments, leading to fragments of approximately 25 nucleotides in length. Structurally, this enzyme includes a PAZ domain, which binds to the end of the dsRNA that is flanked by two catalytic ribonuclease III domains on the enzyme C-terminal domain [121]. The distance between the PAZ domain and the two RNase III domains is approximately 65 Å, which matches the length of a 25 base pair RNA [122].
Biochemical, structural and molecular modeling studies [123] initially indicated that the PAZ domain binds one end of the dsRNA, while a positively charged flat area between the PAZ and the RNAse III domains orients the double helix. More recently, the three-dimensional structures of the PAZ domain in human Dicer in complex with different siRNA were solved [124]. A visual analysis of these structures confirmed that the 2 nucleotide overhangs at the 3’ end of the siRNA are located in the PAZ domain in which they are tightly bound in place by hydrogen bonds. Furthermore, this study highlighted that the 5’-phosphate is located near the phosphate-binding site, which was identified in the platform domain, and it is connected to the side chains of Arg778, Arg780, Arg811 and His982. The phosphate-binding cleft is separated from the 3’-binding pocket by the Dicer-specific helix, which is an alpha-helical segment that extends from the protein in a knob-like manner. In a cleavage-competent complex model, this helix is disrupted by directing the RNA duplex towards the protein. The pre-miRNA 2-nucleotide 3’-overhang is positioned in the Dicer 3’ pocket, and the terminal pair mismatch is disrupted. In this case, the 5’-phosphate of pre-miRNA can occupy the phosphate-binding site. When pre-miRNA cleavage occurs, the helix Dicer-specific helix folds into a knob-like structure and orients the 5’-RNA away from the protein [124]. These data are summarized in diagrams shown in Figure 2.
Figure 2.
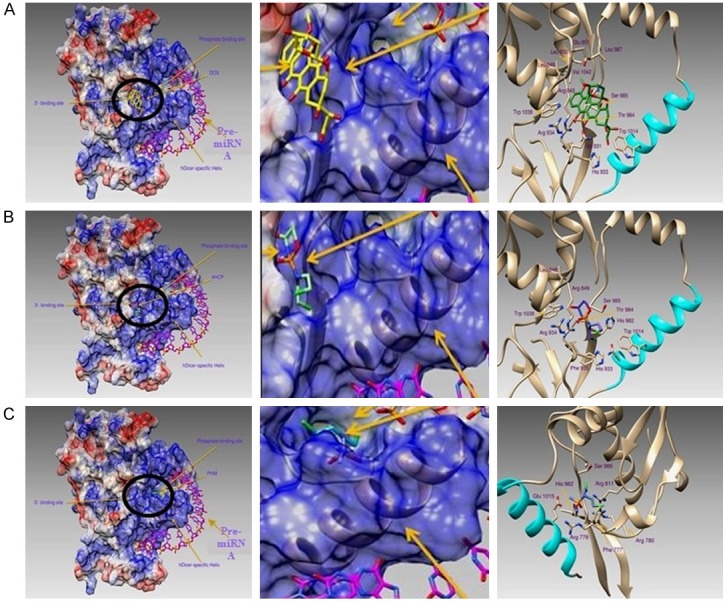
Binding modes of different drug active metabolites within the Dicer PAZ domain. A: Doxorubicinol-binding mode. Left panel: Protein surface is colored according to its electrostatic potential (red negative, blue positive) and is reported with the location of the 3’- and phosphate (5’-)-binding sites. Doxorubicinol, which is the doxorubicin active metabolite, is circled in black. Pre-miRNA is represented by a pink color. Central panel: Enlarged detailed interaction between the Paz domain and Doxorubicinol. Right panel: Binding modes of the active metabolites of Doxorubicinol. Residues involved in the metabolite binding are drawn in sticks and are labeled. The hDicer-specific helix is colored in cyan. B: 4-hydroxycyclophosphamide-binding mode. Left panel: Protein surface is colored according to its electrostatic potential (red negative, blue positive) and is reported with the location of the 3’- and phosphate (5’-)-binding sites. 4-hydroxycyclophosphamide (4HCP), which is one of the active metabolites of cyclophosphamide, is circled in black. Pre-miRNA is represented by a pink color. Central panel: Enlarged detail of the interaction between the Paz domain and 4-hydroxycyclophosphamide. Right panel: Binding modes of the active metabolites 4-hydroxycyclophosphamide. Residues involved in metabolite binding are drawn in sticks and are labeled. The hDicer-specific helix is colored in cyan. C: Phosphoramide mustard binding mode. Left panel: Protein surface is colored according to its electrostatic potential (red negative, blue positive) and is reported with the location of the 3’- and phosphate (5’-)-binding sites. Phosphoramide mustard (PHM), which is one of the active metabolites of cyclophosphamide, is circled in black. Pre-miRNA is represented by a pink color. Central panel: Enlarged detailed interaction between the Paz domain and phosphoramide mustard. Right panel: Binding modes of the active metabolites of phosphoramide mustard. Residues involved in metabolite binding are drawn in sticks and are labeled. The hDicer-specific helix is colored in cyan.
These structural and functional data are relevant because Dicer expression is closely related to chemo-resistance. Chen et al. performed a Western blot analysis and found that Dicer expression is increased in a gefitinib-resistant (PC9/GR) non-small cell lung cancer (NSCLC) cell line compared to that in the gefitinib-sensitive cell line. Moreover, specimens from lung cancer patients who were treated with gefitinib were analyzed by quantitative reverse transcriptase PCR to determine the Dicer mRNA expression level. In the chemotherapy responder group, the Dicer mRNA expression is lower than that in the non-responder group [125]. The opposite result was reported by Kuang et al., who compared Dicer expression in the ovarian cancer A2780 cell line and the Cisplatin-resistant A2780/DDP cell line. Both the Western blot analysis and quantitative reverse transcriptase PCR revealed that Dicer expression is decreased in resistant cells compared to that in the parental cells, indicating that the enzyme is involved in the resistance to Cisplatin [126].
In oral squamous cell carcinoma, an immunohistochemical analysis was performed using specimens obtained from patients who were treated with 5-FU chemoradiotherapy and showed a decrease in Dicer expression, which was also associated with a shorter overall survival [127]. Regarding breast adenocarcinoma, Kovalchuk et al. found that the expression of Dicer is decreased at the protein level in the Doxorubicin-resistant MCF-7 cell line compared to that in the parental cell line [128]. Conversely, Selever et al. performed an immunoblot analysis on Tamoxifen-resistant and -sensitive MCF-7 cell lines and found that the Dicer levels are elevated by 6-fold compared to those in the resistant cells. This result was further confirmed by a quantitative RT-PCR comparison of Tamoxifen-resistant metastatic tumors and Tamoxifen-sensitive tumors [129]. Moreover, when Dicer is knocked-down by siRNA in the MCF-7 cell line, the cell cycle is blocked in the G1 phase, and an increased sensitivity to Cisplatin occurs [130]. Even in glioblastoma cells, a Dicer-knockdown leads to an increased sensitivity to Temozolomide [131]. In cervical cancer, Cai et al. performed a Western blot analysis and found that Dicer expression is decreased in a Cisplatin-resistant HeLa/DDP cell line compared to that in the parental cell line [132]. Kim et al. studies amphiphilic peptides as potential anticancer drugs and discovered that LK-L1C/K6W/L8C enhances Dicer activity by 1.5-fold during the processing of pre-miR-29b into mature miR-29b. This result is indicative of an interaction between LK-L1C/K6W/L8C and the terminal loop region of pre-miR29b, which increases its ability to bind Dicer [133].
The data describing Dicer and the chemo-resistance of cancer cells and primary tumors are summarized in Table 2.
Table 2.
Dicer alteration in drug-resistant cancers
| Dicer alteration trend | Performed assay | Drug resistance | Cancer | Experimental condition | References |
|---|---|---|---|---|---|
| ↑ | Western blot; Quantitative reverse transcriptase PCR | Gefitinib | Lung cancer | Cell culture and human biopsy | [125] |
| ↓ | Western blot; Quantitative reverse transcriptase PCR | Cisplatin | Ovarian cancer | Cell culture | [126] |
| ↓ | Immunohistochemical analysis | 5-Fluorouracil | Oral squamous carcinoma | Human biopsy | [127] |
| ↓ | Western blot | Doxorubicin | Breast cancer | Cell culture | [128] |
| ↓ | Immunoblot analysis; Quantitative reverse transcriptase PCR | Tamoxifen | Breast cancer | Cell culture | [129] |
| ↓ | siRNA silencing | Cisplatin | Breast cancer | Cell culture | [130] |
| ↓ | siRNA silencing | Temozolomide | Glioblastoma | Cell culture | [131] |
| ↓ | Western blot | Cisplatin | Cervical cancer | Cell culture | [132] |
| ↑* | Gel mobility-shift assay | Amphiphilic peptide LK-L1C/K6W/L8C | Cervical and breast cancer | Recombinant human Dicer | [133] |
Functional analysis of Dicer activity.
Evaluation of the interaction between Dicer and cancer chemotherapeutic-drugs by computational analysis
To better understand the effects exerted by anticancer drugs on Dicer activity, we performed molecular docking simulations (Figure 2) using a three-dimensional structure of the Platform-PAZ-connector helix cassette of Human Dicer in complex with a 17-mer siRNA [124]. The residues that were missing in the X-ray diffraction data analysis were modeled using the program MODELLER (Biovia, San Diego, USA) [134] to yield a complete structure of the protein domain. The following three anticancer drugs were docked to the protein: (1) phosphoramide mustard (PHM); (2) 4-hydroxycyclophosphamides (4HCP), which are active metabolites of Cyclophosphamide; and (3) doxorubicinol (DOX), which is an active metabolite of Adriamycin (Doxorubicin).
The docking simulations were performed using the program GOLD v. 5.2.2 [135] and further validated using the program Autodock. Simulations with GOLD were executed using the standard defaults as follows: for all tested ligands, the number of islands was set to 5; the population size was set to 100 (algorithm-software parameters); the number of operations was set to 100,000; the niche size was set to 2; and the selective pressure was 1.1; ChemPLP scoring was used. The binding site center was positioned as the NE1 atom of Trp 1038, and the volume encompassed by 20 Å of that atom was considered the possible binding zone. Three-dimensional protein structures were drawn using the program Chimera [136], and the microRNA structures were modeled using the web server RNAcomposer [137].
The docking simulations demonstrated that 4HCP and DOX bind to the 3’-binding site, while PHM is located within the phosphate-binding pocket. All these metabolites, therefore, interfere with the correct activities of Dicer. In particular, DOX was bound in the 3’-anchor site through hydrogen bonds in a network that involved the Dicer residues Arg849, Glu851, Gln931, His933, Arg934, Thr984, and Ser985. This binding mode was further stabilized by van der Waals interactions with residues Leu848, Leu850, Leu897, Trp1014, Trp1038, and Val1042. The same binding site also hosts 4HCP, which is hydrogen bonded to the residues Arg849, His933, Arg934, Thr984, Ser985, and His982, forming hydrophobic interactions with Leu848, Phe935, Trp1014, and Trp 1038. The second active metabolite of Cyclophosphamide, PHM, is in the phosphate-binding site on the other side of Dicer-helix. PHM is hydrogen bonded to Arg778, Arg780, Arg811, His982, and Glu1015, forming a halogen bond between the Cl-atom and the -OH of Ser985, and is further stabilized by a hydrophobic interaction with the aromatic ring of Phe777.
The binding positions determined by GOLD were confirmed by the simulations performed using Autodock.
These computational findings indicate that Dicer is a major target of the pharmacodynamic effects of anticancer compounds on the microRNA machinery.
MicroRNA modulation as a therapeutic strategy to overcome chemo-resistance
MicroRNA alterations play a pivotal role in establishing chemo-resistance to anti-cancer drugs. Accordingly, a reasonable strategy to overcome chemo-resistance is to re-establish microRNA expression. The recent employment of synthetic analogs of small RNA molecules termed “antagomirs” has shown that particular microRNAs can be specifically targeted [138]. Various methods are available to artificially modulate microRNA expression using synthetic analogs, including anti-microRNA oligonucleotides, locked nucleic acid (LNA), and the microRNA “sponge”.
The introduction of anti-microRNA oligonucleotides that are fully complementary to the pri-microRNA, pre-microRNA or the mature microRNA can inhibit the microRNA activity [139].
Locked nucleic acids (LNA), also called “inaccessible RNAs”, are sequences of nucleotides in which sugars are chemically modified (protected) through a “bridge” that connects the 2’-oxygen with the 4’ carbon. LNA oligonucleotides display an unprecedented hybridization affinity toward complementary single-stranded RNA. Structural studies have demonstrated that LNA oligonucleotides cause A-type (RNA-like) duplex conformations [140].
The microRNA “sponge” method creates a continuous microRNA loss of function in cell culture lines and transgenic organisms. MicroRNA-sponges have complementary binding sites to specific microRNAs and are produced by transfecting cells with vectors containing multiple microRNA-binding sites into the 3’ UTR of a reporter gene. The sponge-binding sites are specific to the microRNA seed region. By binding to this seed region, a whole microRNA family can be blocked, thus inhibiting the expression of all controlled genes. This transgenic approach has been shown to be a promising tool for controlling microRNA functions in various experimental systems [141,142]. Sponges with microRNA-binding sites, which maintain an internal mismatch in the middle part to create a bulge, demonstrate an increased potential in suppressing the activity of intracellular microRNAs than those that are perfectly complementary to the microRNA of interest. This phenomenon is due to the decreased turnover of sponges hampering their endonucleolytic cleavage. Since microRNA sponges can escape destruction, they can sequester their target microRNA for a prolonged time [143].
There are two possible options for re-establishing normal levels of microRNAs that are silenced in cancer cells. The first option is the use of epigenetic drugs, such as inhibitors of DNA methylation (5-azacytidine, 5-aza-2’-deoxycytidine, 5-fluoro-2’-deoxycytidine) [144] or histone deacetylation (SAHA, Romidepsin) [145], that can inhibit tumor growth by restoring, among other mechanisms, the expression of epigenetically silenced microRNAs [146]. For example, miR-34a has a very low expression in different tumors, such as breast, lung, colon, kidney, pancreatic, and melanoma, because of the aberrant CpG methylation in its promoter region. The re-expression of miR-34a in the cancer cell line leads to senescence and a cell cycle block [147].
The second option is the direct introduction of microRNA mimics into the cancer cells; these mimics are synthetic small RNAs that consist of the exact sequence of the endogenous microRNAs [146]. Since miR-34a and let-7 are down-regulated in lung cancer, Trang et al. used the mimics of these two microRNAs in association with a neutral lipid emulsion as a delivery system and observed a tumor reduction in mice [148].
In April 2013, MiRNA Therapeutics [149] started a phase I study (NCT01829971) to investigate the safety, pharmacokinetics and pharmacodynamics of MRX34, which is a microRNA mimic of miR-34a. The main requirement for patient recruitment (in progress up to December 2016) is the presence of primary liver cancer, other selected solid tumors or hematologic malignancies characterized by a decreased expression of miR-34a [150].
Overall analysis of the role of microRNA in chemo-resistance to anti-cancer drugs and problems limiting their clinical applicability
Chemotherapy is one of the main strategies in cancer treatment; however, resistance to drugs due to mutations, changes in gene expression and drug-induced karyotypic alterations limits its effectiveness [151]. The development of high-throughput techniques allows for the identification of molecular signatures and genotypes that help predict the response to drugs. Moreover, these strategies are useful for identifying novel therapeutic targets in to overcome chemo-resistance [152]. In addition to the traditional mechanisms underlying resistance, such as an increased drug efflux, decreased drug activation, DNA methylation, DNA damage repair, and evasion of apoptosis, it has been recently discovered that microRNAs are fundamental players in drug resistance. Indeed, there are dramatic differences in the microRNA expression between cancerous and healthy tissues, particularly in drug-resistant cancer compared with that in normal tissue [153,154]. Thus, microRNAs have attracted increasing interest as potential agents to overcome chemo-resistance. The rationale for this application is based on the following two criteria: (a) in cancer tissue, the expression of microRNAs is dysregulated compared to that in normal tissues; (b) in cancer cells that are resistant to chemotherapies, the expression of microRNAs is dysregulated compared to that in sensitive cancer cells; and (c) the cancer phenotype can be modified by targeting microRNA expression. This microRNA-based adjuvant therapy has many advantages, such as the possibility of targeting multiple genes, most of which belong to the same pathway. However, the use of microRNAs as therapeutic agents is a challenge that includes issues regarding delivery, safety, and potential off-target effects. One of the main problems is the low stability of microRNAs in vivo that have a very short half-life in extracellular and intracellular compartments because of degradation by RNAses. Another obstacle is represented by the poor cellular uptake of oligonucleotides, which is mainly due to their negative charge and size, that could hinder their ability to cross the cell membrane [155]. To limit microRNA degradation, chemical modifications have been made to generate LNA nucleotides [156], peptide nucleic acid (PNA) [157], and phosphorothionate-containing oligonucleotides [158].
In addition to the problem of degradation, microRNAs must overcome other obstacles before reaching their final destination in the target tissue. For example, microRNAs must cross physiological barriers, such as the capillary endothelium or the blood-brain barrier in the therapy of brain tumors. To help these drugs reach their target, several strategies have been proposed. MicroRNAs can be conjugated to a cholesterol moiety to facilitate cell entry [159] or they can be inserted into liposome nanoparticles [160]. The main disadvantage of liposomes is that they could induce an immune response [161]. In particular, cationic liposomes could be characterized by toxicity because they are prone to bind serum proteins, thus forming complexes that activate the immune system [162].
An addition hurdle in the use of microRNA therapeutics is that one microRNA can target many different mRNAs, leading to off-target effects. This obstacle can be overcome by binding cancer-specific ligands to the nanoparticles to specifically reach the target tissue. For this purpose, hyaluronic acid, which binds the cancer stem cell marker CD44 or specific cancer cell receptors, such as EGFR, can be adopted [163,164].
Safety concerns regarding the use of microRNAs as therapeutic agents are also related to their pro-inflammatory activity. In particular, GC-rich oligonucleotides are potent activators of the TLR3/7 receptors, which trigger an interferon alpha-related inflammatory response. Indeed, a microRNA endogenous overload results in interferon alpha production and lymphocytosis, leading to neurological damage in the brain [165].
However, this immunological activation may have therapeutic benefits in the immune response of cancer patients. Regarding this issue, Warren et al. showed that GC dinucleotides enhanced the efficacy of antitumor monoclonal antibody therapy in mice with 38C13 murine B-cell lymphoma, confirming the immunostimulatory role of these dinucleotides [166].
Similarly, Kranz et al. used RNA-lipoplexes to stimulate the T-cell response in cancer immunotherapy by targeting antigen-presenting cells [167].
Conclusions
MicroRNAs could represent an effective therapeutic strategy for overcoming the obstacle of chemo-resistance to anti-cancer drugs. However, there are still many challenges, particularly their stability in body fluids and tissues, their ability to reach the target tissue, and their limited cellular uptake. These problems require further study before microRNAs can be effectively used in humans.
Acknowledgements
This study was supported by Italian Ministry of Health, current research project 2016, IRCCS AOU San Martino IST, Genoa, Italy.
Disclosure of conflict of interest
None.
Abbreviations
- Pgp
P-glycoprotein
- MDR1
multidrug resistance protein 1
- ERCC1
excision repair cross-complementary 1
- NSCLC
non-small cell lung cancer
- bcl-2
B-cell lymphoma 2
- PTEN
phosphatase and tensin homolog
- CDK
cyclin-dependent kinase
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- PAZ
Piwi, Argonaut and Zwille
- PHM
phosphoramide mustard
- 4HCP
4-hydroxycyclophosphamide
- DOX
doxorubicinol
- LNA
locked nucleic acid
References
- 1.Donzelli S, Mori F, Biagioni F, Bellissimo T, Pulito C, Muti P, Strano S, Blandino G. MicroRNAs: short non-coding players in cancer chemoresistance. Mol Cell Ther. 2014;2:16. doi: 10.1186/2052-8426-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen KE, Weiss GJ. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther. 2010;9:3126–36. doi: 10.1158/1535-7163.MCT-10-0397. [DOI] [PubMed] [Google Scholar]
- 3.Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat. 2009;12:114–26. doi: 10.1016/j.drup.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Kerbel RS, Kobayashi H, Graham CH. Intrinsic or acquired drug resistance and metastasis: are they linked phenotypes? J Cell Biochem. 1994;56:37–47. doi: 10.1002/jcb.240560108. [DOI] [PubMed] [Google Scholar]
- 5.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–82. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2010;29:4625–4635. doi: 10.1038/onc.2010.207. [DOI] [PubMed] [Google Scholar]
- 7.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16:5928–35. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9:1623–36. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science. 2016;352:175–80. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–92. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 12.Izzotti A, Pulliero A. Molecular damages and lung tumors in cigarette smoke exposed mice. Ann N Y Acad Sci. 2015;1340:75–83. doi: 10.1111/nyas.12697. [DOI] [PubMed] [Google Scholar]
- 13.Izzotti A. Molecular medicine and development of cancer chemopreventive agents. Ann N Y Acad Sci. 2012;1259:26–32. doi: 10.1111/j.1749-6632.2012.06646.x. [DOI] [PubMed] [Google Scholar]
- 14.Izzotti A, Cartiglia C, Longobardi M, Bagnasco M, Merello A, You M, Lubet RA, De Flora S. Gene expression in the lung of p53 mutant mice exposed to cigarette smoke. Cancer Res. 2004;64:8566–72. doi: 10.1158/0008-5472.CAN-04-1420. [DOI] [PubMed] [Google Scholar]
- 15.Izzotti A, Larghero P, Balansky R, Pfeffer U, Steele VE, De Flora S. Interplay between histopathological alterations, cigarette smoke and cancer chemopreventive agents in defining microRNA profiles in mouse lung. Mutat Res. 2011;717:17–24. doi: 10.1016/j.mrfmmm.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Ann Rev Pharmacol Toxicol. 1999;39:361–98. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 17.Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control. 2003;10:159–65. doi: 10.1177/107327480301000207. [DOI] [PubMed] [Google Scholar]
- 18.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 19.Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikemura K, Yamamoto M, Miyazaki S, Mizutani H, Iwamoto T, Okuda M. MicroRNA-145 posttranscriptionally regulates the expression and function of P-glycoprotein in intestinal epithelial cells. Mol Pharmacol. 2013;83:399–405. doi: 10.1124/mol.112.081844. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Li N, Wang H, Jia X, Wang X, Luo J. Altered microRNA expression in cisplatin-resistant ovarian cancer cells and upregulation of miR-130a associated with MDR1/P-glycoprotein-mediated drug resistance. Oncol Rep. 2012;28:592–600. doi: 10.3892/or.2012.1823. [DOI] [PubMed] [Google Scholar]
- 22.Krishna R, Mayer LD. Multidrug resistance (MDR) in cancer. Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 2000;11:265–83. doi: 10.1016/s0928-0987(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz PM, Moir RD, Hyde CM, Turek PJ, Handschumacher RE. Role of uridine phosphorylase in the anabolism of 5-fluorouracil. Biochem Pharmacol. 1985;34:3585–9. doi: 10.1016/0006-2952(85)90737-3. [DOI] [PubMed] [Google Scholar]
- 24.Houghton JA, Houghton PJ. Elucidation of pathways of 5-fluorouracil metabolism in xenografts of human colorectal adenocarcinoma. Eur J Cancer Clin Oncol. 1983;19:807–15. doi: 10.1016/0277-5379(83)90013-5. [DOI] [PubMed] [Google Scholar]
- 25.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clin Pharmacokinet. 1989;16:215–37. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 26.Offer SM, Butterfield GL, Jerde CR, Fossum CC, Wegner NJ, Diasio RB. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther. 2014;13:742–51. doi: 10.1158/1535-7163.MCT-13-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.miRDB-MicroRNA Target Prediction And Functional Study Database. 2016. http://mirdb.org/miRDB.
- 28.Peters GJ, Kohne CH. Antifolate drugs in cancer therapy. Humana Press; 1999. Fluoropyrimidines as antifolate drugs; pp. 101–145. [Google Scholar]
- 29.Copur S, Aiba K, Drake JC, Allegra CJ, Chu E. Thymidylate synthase gene amplification in human colon cancer cell lines resistant to 5-fluorouracil. Biochem Pharmaco. 1995;49:1419–26. doi: 10.1016/0006-2952(95)00067-a. [DOI] [PubMed] [Google Scholar]
- 30.Tao L, Huang G, Chen Y, Chen L. DNA methylation of DKK3 modulates docetaxel chemoresistance in human nonsmall cell lung cancer cell. Cancer Biother Radiopharm. 2015;30:100–6. doi: 10.1089/cbr.2014.1797. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Wang R, Song HZ, Pan BZ, Zhang YW, Chen LB. Epigenetic downregulation of RUNX3 by DNA methylation induces docetaxel chemoresistance in human lung adenocarcinoma cells by activation of the AKT pathway. Int J Biochem Cell Biol. 2013;45:2369–78. doi: 10.1016/j.biocel.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Sun J, Shi C, Sun C, Yu L, Wen Y, Zhao S, Liu J, Xu J, Li H, An T, Zhou X, Ren L, Wang Q, Yu S. miR-29s inhibit the malignant behavior of U87MG glioblastoma cell line by targeting DNMT3A and 3B. Neurosci Lett. 2015;590:40–6. doi: 10.1016/j.neulet.2015.01.060. [DOI] [PubMed] [Google Scholar]
- 33.Xiao S, Yang Z, Qiu X, Lv R, Liu J, Wu M, Liao Y, Liu Q. miR-29c contribute to glioma cells temozolomide sensitivity by targeting O6-methylguanine-DNA methyltransferases indirectely. Oncotarget. 2016;7:50229–38. doi: 10.18632/oncotarget.10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson TR, Longley DB, Johnston PG. Chemoresistance in solid tumours. Ann Oncol. 2006;17:315–24. doi: 10.1093/annonc/mdl280. [DOI] [PubMed] [Google Scholar]
- 35.Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sánchez JM, Gumerlock PH, Tarón M, Sánchez JJ, Danenberg KD, Danenberg PV, Rosell R. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–91. [PubMed] [Google Scholar]
- 36.Alemdehy MF, Haanstra JR, de Looper HW, van Strien PM, Verhagen-Oldenampsen J, Caljouw Y, Sanders MA, Hoogenboezem R, de Ru AH, Janssen GM, Smetsers SE, Bierings MB, van Veelen PA, von Lindern M, Touw IP, Erkeland SJ. Interstrand cross-link induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood. 2015;125:3937–48. doi: 10.1182/blood-2014-11-612507. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Feng Y, Shen JK, Lin M, Choy E, Cote GM, Harmon DC, Mankin HJ, Hornicek FJ, Duan Z. CD44 is a direct target of miR-199a-3p and contributes to aggressive progression in osteosarcoma. Sci Rep. 2015;5:11365. doi: 10.1038/srep11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–59. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 39.Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 40.Svrcek M, El-Murr N, Wanherdrick K, Dumont S, Beaugerie L, Cosnes J, Colombel JF, Tiret E, Fléjou JF, Lesuffleur T, Duval A. Overexpression of microRNAs-155 and 21 targeting mismatch repair proteins in inflammatory bowel diseases. Carcinogenesis. 2013;34:828–34. doi: 10.1093/carcin/bgs408. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YX, Yue Z, Wang PY, Li YJ, Xin JX, Pang M, Zheng QY, Xie SY. Cisplatin upregulates MSH2 expression by reducing miR-21 to inhibit A549 cell growth. Biomed Pharmacother. 2013;67:97–102. doi: 10.1016/j.biopha.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu RL, Dong Y, Deng YZ, Wang WJ, Li WD. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour Biol. 2015;36:5011–19. doi: 10.1007/s13277-015-3152-5. [DOI] [PubMed] [Google Scholar]
- 43.Bunz F. Cell death and cancer therapy. Curr Opin Pharmacol. 2001;1:337–41. doi: 10.1016/s1471-4892(01)00059-5. [DOI] [PubMed] [Google Scholar]
- 44.Campos L, Rouault JP, Sabido O, Oriol P, Roubi N, Vasselon C, Archimbaud E, Magaud JP, Guyotat D. High expression of bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Blood. 1993;81:3091–6. [PubMed] [Google Scholar]
- 45.Bonetti A, Zaninelli M, Leone R, Cetto GL, Pelosi G, Biolo S, Menghi A, Manfrin E, Bonetti F, Piubello Q. bcl-2 but not p53 expression is associated with resistance to chemotherapy in advanced breast cancer. Clin Cancer Res. 1998;4:2331–6. [PubMed] [Google Scholar]
- 46.Stojcheva N, Schechtmann G, Sass S, Roth P, Florea AM, Stefanski A, Stühler K, Wolter M, Müller NS, Theis FJ, Weller M, Reifenberger G, Happold C. MicroRNA-138 promotes acquired alkylator resistance in glioblastoma by targeting the Bcl-2-interacting mediator BIM. Oncotarget. 2016;7:12937–50. doi: 10.18632/oncotarget.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang Q, Tang H, Yu J, Yin J, Yang X, Lei X. MicroRNA-98 sensitizes cisplatin-resistant human lung adenocarcinoma cells by up-regulation of HMGA2. Pharmazie. 2013;68:274–281. [PubMed] [Google Scholar]
- 48.Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, Wang Z, Liu J, Cai G. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep. 2016;6:27157. doi: 10.1038/srep27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Prac. 2016;212:643–49. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 50.Zhang HD, Sun DW, Mao L, Zhang J, Jiang LH, Li J, Wu Y, Ji H, Chen W, Wang J, Ma R, Cao HX, Wu JZ, Tang JH. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun. 2015;465:702–13. doi: 10.1016/j.bbrc.2015.08.053. [DOI] [PubMed] [Google Scholar]
- 51.Zhang F, Li Y, Wu H, Qi K, You J, Li X, Zu L, Pan Z, Wang Y, Li Y, Li Y, Wang M, Shen W, Zhou Q. MiR-192 confers cisplatin resistance by targeting Bim in lung cancer. Zhongguo Fei Ai Za Zhi. 2014;17:384–90. doi: 10.3779/j.issn.1009-3419.2014.05.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paradiso A, Simone G, Petroni S, Leone B, Vallejo C, Lacava J, Romero A, Machiavelli M, De Lena M, Allegra CJ, Johnston PG. Thymidilate synthase and p53 primary tumour expression as predictive factors for advanced colorectal cancer patients. Br J Cancer. 2000;82:560–7. doi: 10.1054/bjoc.1999.0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie C, Wu B, Chen B, Shi Q, Guo J, Fan Z, Huang Y. Histone deacetylase inhibitor sodium butyrate suppresses proliferation and promotes apoptosis in osteosarcoma cells by regulation of the MDM2-p53 signaling. Onco Targets Ther. 2016;9:4005–13. doi: 10.2147/OTT.S105418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherwood J, Dearden S, Ratcliffe M, Walker J. Mutation status concordance between primary lesions and metastatic sites of advanced nonsmall-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res. 2015;34:92. doi: 10.1186/s13046-015-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle. 2012;11:1273–81. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 57.Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, Bu P, Vogel H, Jablons DM, Keller AC, Wilkinson JE, He B, Speed TP, He L. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–50. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 59.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rui W, Bing F, Hai-Zhu S, Wei D, Long-Bang C. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1) J Cell Mol Med. 2010;14:206–14. doi: 10.1111/j.1582-4934.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13:57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, Shpilberg O, Granot G. Restoration of miR-424 suppresses BCR-ABL activity and sensitizes CML cells to imatinib treatment. Cancer Lett. 2015;360:245–256. doi: 10.1016/j.canlet.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 64.Zhu X, Li D, Yu F, Jia C, Xie J, Ma Y, Fan S, Cai H, Luo Q, Lv Z, Fan L. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget. 2016;7:13139–52. doi: 10.18632/oncotarget.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, Costinean S, Sandhu SK, Nuovo GJ, Alder H, Gafa R, Calore F, Ferracin M, Lanza G, Volinia S, Negrini M, McIlhatton MA, Amadori D, Fishel R, Croce CM. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A. 2010;107:6982–7. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, Nuovo GJ, Fishel R, Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iida K, Fukushi J, Matsumoto Y, Oda Y, Takahashi Y, Fujiwara T, Fujiwara-Okada Y, Hatano M, Nabashima A, Kamura S, Iwamoto Y. miR-125b develops chemoresistance in Ewing sarcoma/primitive neuroectodermal tumor. Cancer Cell Int. 2013;13:21. doi: 10.1186/1475-2867-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donzelli S, Fontemaggi G, Fazi F, Di Agostino S, Padula F, Biagioni F, Muti P, Strano S, Blandino G. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19:1038–48. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Zhang Y, Zhao J, Kong F, Chen Y. Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signalling in p53-mutated colon cancer cells. Mol Cell Biochem. 2011;357:31–8. doi: 10.1007/s11010-011-0872-8. [DOI] [PubMed] [Google Scholar]
- 70.Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587:488–95. doi: 10.1016/j.febslet.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol. 2012;180:2490–503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–8. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–8. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 74.Li X, Wu Y, Liu A, Tang X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477:768–73. doi: 10.1016/j.bbrc.2016.06.133. [DOI] [PubMed] [Google Scholar]
- 75.Fattore L, Mancini R, Acunzo M, Romano G, Laganà A, Pisanu ME, Malpicci D, Madonna G, Mallardo D, Capone M, Fulciniti F, Mazzucchelli L, Botti G, Croce CM, Ascierto PA, Ciliberto G. miR-579-3p controls melanoma progression and resistance to target therapy. Proc Natl Acad Sci U S A. 2016;113:E5005–13. doi: 10.1073/pnas.1607753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 77.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 78.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 79.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber BL, Bützow R, Croce CM, Coukos G, Zhang L. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–14. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;11:478–86. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 82.Cai J, Yang C, Yang Q, Ding H, Jia J, Guo J, Wang J, Wang Z. Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis. 2013;2:e75. doi: 10.1038/oncsis.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X, Cai H, Liang Y, Chen L, Wang X, Si R, Qu K, Jiang Z, Ma B, Miao C, Li J, Wang B, Gao P. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncol Rep. 2015;33:1723–30. doi: 10.3892/or.2015.3757. [DOI] [PubMed] [Google Scholar]
- 84.Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M, Recker RR, Gatalica Z, Wang Z, Xiao GG. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol Med. 2011;17:1233–41. doi: 10.2119/molmed.2010.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugimura K, Miyata H, Tanaka K, Hamano R, Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori M, Doki Y. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res. 2012;18:5144–53. doi: 10.1158/1078-0432.CCR-12-0701. [DOI] [PubMed] [Google Scholar]
- 86.Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S, Salvioni R, Valdagni R, Daidone MG, Zaffaroni N. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 88.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 89.Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang GX, Jia Z, Pu PY, Kang CS, Yao Z. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010;9:77–86. doi: 10.1177/153303461000900109. [DOI] [PubMed] [Google Scholar]
- 90.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 91.Chan JK, Blansit K, Kiet T, Sherman A, Wong G, Earle C, Bourguignon LY. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol. 2014;132:739–44. doi: 10.1016/j.ygyno.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 92.Paik WH, Kim HR, Park JK, Song BJ, Lee SH, Hwang JH. Chemosensitivity induced by downregulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. 2013;33:1473–81. [PubMed] [Google Scholar]
- 93.Izzotti A, Cartiglia C, Steele VE, De Flora S. MicroRNAs as targets for dietary and pharmacological inhibitors of mutagenesis and carcinogenesis. Mutat Res. 2012;751:287–303. doi: 10.1016/j.mrrev.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ziyan W, Yang L. MicroRNA-21 regulates the sensitivity to cisplatin in a humanosteosarcoma cell line. Ir J Med Sci. 2016;185:85–91. doi: 10.1007/s11845-014-1225-x. [DOI] [PubMed] [Google Scholar]
- 95.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–8. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 96.Shen H, Zhu F, Liu J, Xu T, Pei D, Wang R, Qian Y, Li Q, Wang L, Shi Z, Zheng J, Chen Q, Jiang B, Shu Y. Alteration in Mir-21/PTEN expression modulates gefitinib resistance in non-small cell lung cancer. PLoS One. 2014;9:e103305. doi: 10.1371/journal.pone.0103305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dong Z, Ren L, Lin L, Li J, Huang Y, Li J. Effect of microRNA-21 on multidrug resistance reversal in A549/DDP human lung cancer cells. Mol Med Rep. 2015;11:682–90. doi: 10.3892/mmr.2014.2662. [DOI] [PubMed] [Google Scholar]
- 98.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 99.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 100.Rahman M, Lovat F, Romano G, Calore F, Acunzo M, Bell EH, Nana-Sinkam P. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J Biol Chem. 2014;289:26406–16. doi: 10.1074/jbc.M114.573592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 104.Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, Weinstein JN, Sadee W. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 105.Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–55. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 106.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–31. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 107.Zhao Z, Zhang L, Yao Q, Tao Z. miR-15b regulates cisplatin resistance and metastasis by targeting PEBP4 in human lung adenocarcinoma cells. Cancer Gene Ther. 2015;22:108–14. doi: 10.1038/cgt.2014.73. [DOI] [PubMed] [Google Scholar]
- 108.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–57. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chu J, Zhu Y, Liu Y, Sun L, Lv X, Wu Y, Hu P, Su F, Gong C, Song E, Liu B, Liu Q. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget. 2015;6:31944–57. doi: 10.18632/oncotarget.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dwivedi SK, Mustafi SB, Mangala LS, Jiang D, Pradeep S, Rodriguez-Aguayo C, Ling H, Ivan C, Mukherjee P, Calin GA, Lopez-Berestein G, Sood AK, Bhattacharya R. Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget. 2016;7:15093–104. doi: 10.18632/oncotarget.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han J, Chen Q. MiR-16 modulate temozolomide resistance by regulating BCL-2 in human glioma cells. Int J Clin Exp Pathol. 2015;8:12698–707. [PMC free article] [PubMed] [Google Scholar]
- 112.Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, Deguchi T, Ito M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–9. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 113.Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, Jin Y, Wu Y. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35:1287–95. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- 114.Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, Song E, Park JH. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573–82. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 115.Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat. 2012;131:445–54. doi: 10.1007/s10549-011-1424-3. [DOI] [PubMed] [Google Scholar]
- 116.Qin B, Cao Y, Yang H, Xiao B, Lu Z. MicroRNA-221/222 regulate ox-LDL-induced endothelial apoptosis via Ets-1/p21 inhibition. Mol Cell Biochem. 2015;405:115–24. doi: 10.1007/s11010-015-2403-5. [DOI] [PubMed] [Google Scholar]
- 117.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang L, Yang A, Zhao J, Jia L. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47:268–73. doi: 10.5483/BMBRep.2014.47.5.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao G, Cai C, Yang T, Qiu X, Liao B, Li W, Ji Z, Zhao J, Zhao H, Guo M, Ma Q, Xiao C, Fan Q, Ma B. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8:e53906. doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ligorio M, Izzotti A, Pulliero A, Arrigo P. Mutagens interfere with microRNA maturation by inhibiting DICER. An in silico biology analysis. Mutat Res. 2011;717:116–28. doi: 10.1016/j.mrfmmm.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 122.MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–98. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 123.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 124.Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell. 2014;53:606–16. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen JC, Su YH, Chiu CF, Chang YW, Yu YH, Tseng CF, Chen HA, Su JL. Suppression of Dicer increases sensitivity to gefitinib in human lung cancer cells. Ann Surg Oncol. 2014;21:555–63. doi: 10.1245/s10434-014-3673-y. [DOI] [PubMed] [Google Scholar]
- 126.Kuang Y, Cai J, Li D, Han Q, Cao J, Wang Z. Repression of Dicer is associated with invasive phenotype and chemoresistance in ovarian cancer. Oncol Lett. 2013;5:1149–54. doi: 10.3892/ol.2013.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kawahara K, Nakayama H, Nagata M, Yoshida R, Hirosue A, Tanaka T, Nakagawa Y, Matsuoka Y, Kojima T, Takamune Y, Yoshitake Y, Hiraki A, Shinohara M. A low Dicer expression is associated with resistance to 5-FU-based chemoradiotherapy and a shorter overall survival in patients with oral squamous cell carcinoma. J Oral Pathol Med. 2014;43:350–6. doi: 10.1111/jop.12140. [DOI] [PubMed] [Google Scholar]
- 128.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 129.Selever J, Gu G, Lewis MT, Beyer A, Herynk MH, Covington KR, Tsimelzon A, Dontu G, Provost P, Di Pietro A, Boumendjel A, Albain K, Miele L, Weiss H, Barone I, Ando S, Fuqua SA. Dicermediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clin Cancer Res. 2011;17:6510–21. doi: 10.1158/1078-0432.CCR-11-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bu Y, Lu C, Bian C, Wang J, Li J, Zhang B, Li Z, Brewer G, Zhao RC. Knockdown of Dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncol Rep. 2009;21:13–7. [PubMed] [Google Scholar]
- 131.Munoz JL, Rodriguez-Cruz V, Ramkissoon SH, Ligon KL, Greco SJ, Rameshwar P. Temozolomide resistance in glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic hedgehog level. Oncotarget. 2015;6:1190–201. doi: 10.18632/oncotarget.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cai L, Wang Z, Liu D. Interference with endogenous EZH2 reverses thechemotherapy drug resistance in cervical cancer cells partly by upregulating Dicer expression. Tumour Biol. 2016;37:6359–69. doi: 10.1007/s13277-015-4416-9. [DOI] [PubMed] [Google Scholar]
- 133.Kim S, Lee JH, Kang I, Hyun S, Yu J, Shin C. An amphiphilic peptide induces apoptosis through the miR29b-p53 pathway in cancer cells. Mol Ther Nucleic Acids. 2016;5:e330. doi: 10.1038/mtna.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 135.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 136.Pettersen EF, Goddard TD, Huagn CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 137.Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mattes J, Yang M, Foster PS. Regulation of microRNA by antagomirs: a new class of pharmacological antagonists for the specific regulation of gene function? Am J Respir Cell Mol Biol. 2007;36:8–12. doi: 10.1165/rcmb.2006-0227TR. [DOI] [PubMed] [Google Scholar]
- 139.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 140.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–41. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 141.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–27. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 144.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–37. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 145.Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 2011;3:166–79. [PMC free article] [PubMed] [Google Scholar]
- 146.Saito Y, Nakaoka T, Saito H. microRNA-34a as a therapeutic agent against human cancer. J Clin Med. 2015;4:1951–59. doi: 10.3390/jcm4111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 148.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. MiRNA Therapeutics website, http://www.mirnatherapeutics.com.
- 150.Registry and results database of publicly and privately supported clinical studies of human participants conducted around the world. U.S. National Institutes of Health, https://www.clinicaltrials.gov.
- 151.Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. MicroRNAs in cancer treatment and prognosis. Am J Cancer Res. 2012;2:414–33. [PMC free article] [PubMed] [Google Scholar]
- 152.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 153.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol. 2012;180:2490–503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin Drug Deliv. 2009;6:673–86. doi: 10.1517/17425240902992894. [DOI] [PubMed] [Google Scholar]
- 156.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hökfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A. 2000;97:5633–8. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 158.Crooke ST, Graham MJ, Zuckerman JE, Brooks D, Conklin BS, Cummins LL, Greig MJ, Guinosso CJ, Kornbrust D, Manoharan M, Sasmor HM, Schleich T, Tivel KL, Griffey RH. Pharmacokinetic properties of several novel oligonucleotide analogs in mice. J Pharmacol Exp Ther. 1996;277:923–37. [PubMed] [Google Scholar]
- 159.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–6. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–6. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–9. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 162.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41:2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Choi KY, Yoon HY, Kim JH, Bae SM, Park RW, Kang YM, Kim IS, Kwon IC, Choi K, Jeong SY, Kim K, Park JH. Smart nanocarrier based on PEGylated hyaluronic acid for cancer therapy. ACS Nano. 2011;5:8591–9. doi: 10.1021/nn202070n. [DOI] [PubMed] [Google Scholar]
- 164.Orian-Rousseau V. CD44, a therapeutic target for metastasising tumours. Eur J Cancer. 2010;46:1271–7. doi: 10.1016/j.ejca.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 165.Pulliero A, Fazzi E, Cartiglia C, Orcesi S, Balottin U, Uggetti C, La Piana R, Olivieri I, Galli J, Izzotti A. The aicardi-goutières syndrome. Molecular and clinical features of RNAse deficiency and microRNA overload. Mutat Res. 2011;717:99–108. doi: 10.1016/j.mrfmmm.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 166.Warren TL, Dahle CE, Weiner GJ. CpG oligodeoxynucleotides enhance monoclonal antibody therapy of a murine lymphoma. Clin Lymphoma. 2000;1:57–61. doi: 10.3816/clm.2000.n.005. [DOI] [PubMed] [Google Scholar]
- 167.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, Meng M, Fritz D, Vascotto F, Hefesha H, Grunwitz C, Vormehr M, Hüsemann Y, Selmi A, Kuhn AN, Buck J, Derhovanessian E, Rae R, Attig S, Diekmann J, Jabulowsky RA, Heesch S, Hassel J, Langguth P, Grabbe S, Huber C, Türeci Ö, Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]