Abstract
Targeted gene modification mediated by single-stranded oligonucleotides (SSOs) holds great potential for widespread use in a number of biological and biomedical fields, including functional genomics and gene therapy. By using this approach, specific genetic changes have been created in a number of prokaryotic and eukaryotic systems. In mammalian cells, the precise mechanism of SSO-mediated chromosome alteration remains to be established, and there have been problems in obtaining reproducible targeting efficiencies. It has previously been suggested that the chromatin structure, which changes throughout the cell cycle, may be a key factor underlying these variations in efficiency. This hypothesis prompted us to systematically investigate SSO-mediated gene repair at various phases of the cell cycle in a mammalian cell line. We found that the efficiency of SSO-mediated gene repair was elevated by ≈10-fold in thymidine-treated S-phase cells. The increase in repair frequency correlated positively with the duration of SSO/thymidine coincubation with host cells after transfection. We supply evidence suggesting that these increased repair frequencies arise from a thymidine-induced slowdown of replication fork progression. Our studies provide fresh insight into the mechanism of SSO-mediated gene repair in mammalian cells and demonstrate how its efficiency may be reliably and substantially increased.
Keywords: single-stranded oligonucleotide, targeted gene repair, thymidine
Developed over the past few years, targeted genetic modification mediated by oligonucleotides promises to be an invaluable technique for use in functional genomic studies and may even form the basis of future gene therapy procedures (1–3). This approach is applicable for the specific alteration of short stretches of DNA to create deletions, short insertions, and point mutations (4, 5). Therefore, it is ideally suited for the study of single-nucleotide polymorphisms (SNPs) in model organisms such as mice and ultimately may be used for the treatment of diseases caused by small DNA alterations (6, 7).
Studies have shown that both chimeric RNA/DNA oligonucleotides (RDOs) and single-stranded DNA oligonucleotides (SSOs), which may contain a number of unnatural or “modified” linkages or bases, can introduce targeted genetic alterations in bacteria, fungi, and mammalian cells (8–13). For RDO-mediated repair, in which it is the DNA moiety that directs the genetic conversion event (14), there have been large variations in targeting efficiencies reported and problems with reproducibility (15). Compared with RDOs, SSOs appear to have the greater potential for development, because they have better stability, and are more easily synthesized, modified, and purified. SSOs also exhibit reliably higher in vivo targeting efficiencies than RDOs (16, 17).
To expedite the optimization of conditions for SSO-mediated repair, it is first vital to establish the exact mechanism(s). Analogous to the situation for RDOs, a two-step mechanism has been proposed, which involves the following steps: (i) DNA pairing/annealing, followed by (ii) DNA repair/recombination (18–20). First, the SSO (or the DNA component of the RDO) anneals with its complementary strand in the double-stranded DNA to form a D-loop structure, in a process mediated by cellular protein factors (13, 21–23). The formation of the D-loop, containing a centrally located mismatched base pair (in the case of a point mutation), results in a structural perturbation that most likely stimulates the endogenous protein machinery to initiate the second step in the repair process: site-specific modification of the chromosomal sequence. It has been suggested previously that this repair process also involved two steps (24). The annealed SSO/chromosome D-loop structure first directs a site-specific base conversion on one strand of the target gene, generating a single mismatched base pair in the original targeted helix, after dissociation of the SSO. This newly introduced mismatch then induces the DNA repair machinery to perform a second repair event, leading to the final revision of the target site (2). Some evidence suggests that mismatch repair may be responsible for the second step of the SSO-mediated repair process, but it appears inconsistent with the observation that the repair frequency is increased in cells that are defective for mismatch repair (14, 25, 26).
Significant variation in reproducibility reflects the complexity of oligonucleotide-mediated gene repair in higher organisms (27, 28). This complexity, in turn, indicates that many biological processes may affect the repair process. These processes may include transcription, recombination, DNA-damage repair, and cell-cycle-related events such as DNA replication and chromosome condensation. Indeed, transcriptional activity (25, 29) and DNA-damaging agents (30, 31) have been shown to have a positive influence on the efficiency of oligonucleotide-mediated gene alteration. DNA replication also was found to affect the frequency of gene repair (25, 32).
Taking into consideration the fact that DNA synthesis occurs predominantly in the S phase, and that chromatin structure and the expression of the DNA repair proteins varies considerably throughout the cell cycle (33), we investigated the efficiency of SSO-mediated gene repair during each phase of the cell cycle, by using synchronized cell populations. We found that the repair frequency was significantly elevated in thymidine-treated S phase cells, but not in cells treated with other similar inhibitors. Our results provide fresh insight into the mechanism of SSO-mediated gene repair and reveal a previously undescribed approach with which to increase its efficiency within mammalian cells.
Methods
Construction of Plasmid pmEGFP-C1 Containing the Mutant EGFP (mEGFP) Gene. By using a two-step, PCR-based strategy, an A613T alteration (to inactivate the ATG initiation codon of the EGFP gene) and an AgeI (ACCGGT) to EcoRI (GAATTC) restriction site change (nucleotides 600–605) were introduced into plasmid pEGFP-C1 (BD Biosciences Clontech, Palo Alto, CA) to generate plasmid pmEGFP-C1.
Screening Cell Clones for Stably Integrated mEGFP Gene. Plasmid pmEGFP-C1 was lineared by ApaLI and introduced into HeLa cells by electroporation. Briefly, 5 × 106 HeLa cells were transferred to an ice-cold 0.4-cm cuvette containing pmEGFP-C1 (10 μg). The mixture was electroporated (300 V, 975 μF, Gene Pulser, Bio-Rad), then immediately placed on ice for 10 min. The cells were cultured for 24 h before being diluted to a concentration of 100 cells per well in a 96-well plate and then cultured in selective medium (supplemented with 600 μg/ml G418) for 2 weeks. Resistant clones were analyzed by PCR and Southern blotting for successful mEGFP gene integration.
Transfection with SSO. Cells were seeded at 8 × 105 per 60-mm dishes and grown for 24 h to reach 60–80% confluence. SSO (3 μg) and Lipofectamine2000 (3 μl, 1 mg/ml, Invitrogen) were mixed, diluted with 100 μl of OPTIMEM (Invitrogen), and incubated at room temperature for 20 min. The SSO mixture was added to the cells without changing the culture medium (DMEM supplemented with 10% FBS, unless otherwise stated), which was continuously cultured for the time specified, before harvesting.
Cell Synchronization. Cell synchronization was performed as described in ref. 34 (see also Supporting Methods, which is published as supporting information on the PNAS web site).
FACS Analysis. Propidium iodide (PI) was used to stain fixed cells for cell cycle analysis as described in ref. 35 (see also Supporting Methods). Flow cytometry was used to detect fluorescing cells resulting from SSO-mediated mEGFP repair. Cells were harvested after trypsin digestion, washed with PBS, and then filtered (35 μm-pore size nylon mesh) to remove clumps and ensure a single-cell suspension.
Detection of Repaired Genomic DNA. Genomic DNA was extracted from SSO-E6/thymidine-treated cells. A nested PCR was used to amplify specific regions contained within the target locus first by using the primers 5′-TCAATGGGCGTGGATAGCG-3′ and 5′-GCTCAGGTAGTGGGTGTCG-3′, then the primers 5′-TCAATGGGCGTGGATAGCG-3′ and 5′-CGTTGTGGCTGTGTAGTTG-3′. PCR conditions for each round involved 30 cycles of 94°C for 40 s, 58°C for 40 s, and 72°C for 40 s. The second-round PCR products were subcloned into T-easy vectors (Promega) according to the manufacturer's instructions, with ligation mixtures transformed into competent Escherichia coli DH5α cells. Clones were transferred to a nitrocellulose membrane for hybridization with oligonucleotide probes. The probe 5′-GCTCACCATGGTGGCGG-3′ was used to detect the repaired gene, and the probe 5′-GCTCACCAAGGTGGCGG-3′ was used to detect the unrepaired gene copies. Conditions for hybridization were as described in ref. 5. Positive clones, which hybridized with the repaired EGFP gene probe, were selected for plasmid purification. Inserted fragments were analyzed by DNA sequencing.
Results
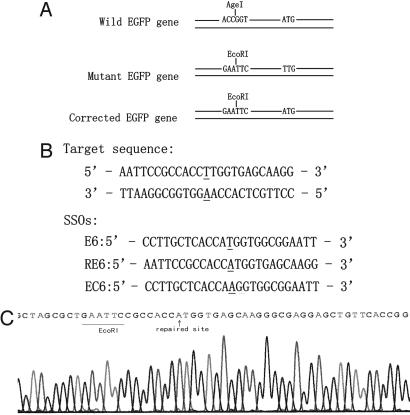
SSO-Mediated Repair of Chromosomal DNA. To detect SSO-mediated chromosomal DNA repair events, we established a mEGFP reporter cell line by stably integrating the pmEGFP plasmid into the genome of HeLa cells. The mEGFP gene contains two modifications compared with the wild-type (WT) EGFP: an A → T mutation that inactivates the initiation codon of the EGFP gene (ATG → TTG), and an ACCGGT → GAATTC change that converts an AgeI restriction site to EcoRI. EGFP expression was restored after specific correction mediated by the transfected SSO, which can be readily detected by fluorescence microscopy. The EcoRI restriction site was introduced to distinguish the repaired gene from WT EGFP contamination (Fig. 1A).
Fig. 1.
Illustration for SSO-mediated gene repair in reporter system. (A) In the WT EGFP reporter gene, the initiation codon of ATG contributes to the proper translation. The conversion from “A” in the WT to “T” in the mutant type of EGFP gene inactivates the initiation codon (ATG). Translation of the EGFP gene can occur only if the initiation codon is restored. An upstream ACCGGT in WT to GAATTC in mutant type converts an AgeI restriction site to an EcoRI. This alteration is included to identify the repaired EGFP gene from the WT EGFP. (B) SSO sequences. E6 is designed to be complementary to the untranscribed strand except a”T–T“mismatch, and RE6 is designed to be complementary to the transcribed strand except an”A–A“mismatch. EC6 is a control SSO that has the totally complementary sequence to the untranscribed strand of the mEGFP gene. The underlined residues indicate the target site. All SSOs have six phosphorothioate modifications at both terminals. (C) Sequence analysis of the EGFP gene isolated from the repaired clones, revealing a targeted gene correction (”A“in the initiation codon) and a new ATG-EcoRI haplotype, which indicates a successful repair event. WT contamination would have an ATG–AgeI combination.
After introduction of the pmEGFP plasmid into HeLa cells, we selected a stably integrated cell clone (F5) that did not fluoresce under UV light. We designed two “correction” SSOs, E6 and RE6, each 25 nt in length (Fig. 1B), to repair the inactivated initiation codon of mEGFP. The sequences of E6 and RE6 are identical to those of the transcribed and nontranscribed strands of the mEGFP gene, respectively, except for one centrally located nucleotide. SSO-EC6, whose sequence is identical to the transcribed strand of the mEGFP gene, was designed to act as a negative control. All three SSOs contained phosphorothioate linkages between each of the six terminal residues at both the 5′ and 3′ ends, which could increase the resistance of the oligonucleotide to intracellular exonucleases and improve its stability in cells.
A relatively low, but reproducible, repair frequency of 0.17% (defined as the percentage of green fluorescent cells per total cells surviving transfection with SSO) was measured 2 days after transfecting the F5 cells with SSO-E6. The mean repair frequency measured for SSO-RE6 (0.05%) was significantly lower than that of SSO-E6 (P < 0.05, n = 5). The repair frequency for the negative control, SSO-EC6, was essentially identical to background (spontaneous) repair levels of 0.01% (n = 5). This result indicated that the SSO-directed gene repair was site-specific and exhibited a strand bias of ≈3.4:1, favoring the SSO that annealed to the nontranscribed strand (W.-X.Y., X.-S.W., G. Liu, Z.-H. Li, J.-D.H., K.S.E.C., D.-P.L., and C.-C.L., unpublished data).
To confirm that the SSO-encoded mutation had been accurately introduced into the chromosome, genomic DNA was extracted from F5 cells treated with SSO-E6, and the target loci were specifically amplified by using a nested PCR procedure. The PCR products were ligated into T-easy vectors and screened by colony hybridization after transformation into competent E. coli cells. A 17-mer oligonucleotide probe was used to detect the single nucleotide difference between the repaired and nonrepaired mEGFP gene copies (5). Positive colonies were cultured, and plasmid DNA was extracted. Sequence analysis revealed that all of the fragments inserted in these plasmids contained a repaired ATG initiation codon together with an upstream EcoRI site (Fig. 1C). Because the EcoRI-ATG haplotype could only be present after SSO-mediated genomic DNA repair, contamination with WT EGFP DNA could be ruled out. Consequently, the F5 HeLa cell line and the E6 SSO were used in all further experiments, unless otherwise stated.
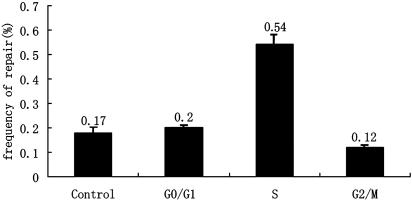
Repair Frequency During Different Phases of the Cell Cycle. To assess SSO-mediated repair frequencies during the different phases of the cell cycle, populations of F5 cells were synchronized at the G1, S, and M phases through treatment with l-mimosine, thymidine, and nocodazole, respectively (34). Synchronization rates were determined to be 87.0%, 96.7%, and 76.9% for G1, S, and M phases, respectively (Fig. 7, which is published as supporting information on the PNAS web site), by flow cytometry by using propidium iodide (PI) staining (35). Equal amounts of SSO-E6 were then introduced into the phase-synchronized cells, by using identical transfection conditions. After transfection, cells were cultured and sustained for at least 8 h in medium containing the respective phase-specific inhibitors, before they were released by changing the culture medium. After 2 days of culture, the fraction of the cells exhibiting fluorescence was quantified to determine the phase-specific gene repair frequencies.
Results indicated that the mEGFP gene repair frequencies varied considerably between the various phase-synchronized cell cultures (Fig. 2). The average repair frequency in the S phase (0.54%) was significantly higher than that in the G1 (0.20%) or M (0.12%) phases. Repair efficiencies in cell cultures synchronized in both the G1 and M phases were essentially the same as those for cells that had not been synchronized (0.17%). Even though the cell synchronization was not perfect, our data indicated that conditions for SSO repair were more favorable during the S phase than the other phases.
Fig. 2.
Evaluation of SSO-mediated repair frequency during different phases of the cell cycle. The cells were sustained in specific phases by corresponding inhibitors for 8 h after SSO transfection, then released by changing medium and detected in 2 days. The x-axis indicates the different phases in which cells were synchronized. Unsynchronized cells were treated at the same time as control (n = 3).
Effect of Thymidine on SSO-Mediated Gene Repair. We reasoned that the observed increase in repair efficiencies during the S phase may be attributable to the following two different factors: (i) the intracellular environment during the S phase, or (ii) the coincubation of the SSO with thymidine. If i is true, simply increasing the proportion of cells in the S phase should increase the repair efficiency. However, if ii is true, then the presence of thymidine is essential, even if the cells are already in the S phase. To distinguish between these two possible scenarios, cells first were arrested at the G1/S border by using a double-thymidine synchronization procedure (34). The synchronized cells then were released from the G1/S border by replacing the thymidinecontaining culture medium with fresh medium without thymidine, immediately before transfection with SSO. By monitoring the cell-cycle progression, we determined that the majority of the cells passed through the S phase in 9 h (Fig. 8, which is published as supporting information on the PNAS web site). However, the gene repair frequencies were found to be very low (0.10%) in these cells (Fig. 8B). This finding was in sharp contrast to the cells that were kept arrested in the S phase, by culturing in medium containing thymidine for 8 h after transfection with SSO (Fig. 2). This result demonstrated that simply increasing the proportion of cells that progressed through the S phase did not enhance the repair frequency, and it was the presence of thymidine (after transfection) that increased the levels of SSO-mediated gene repair.
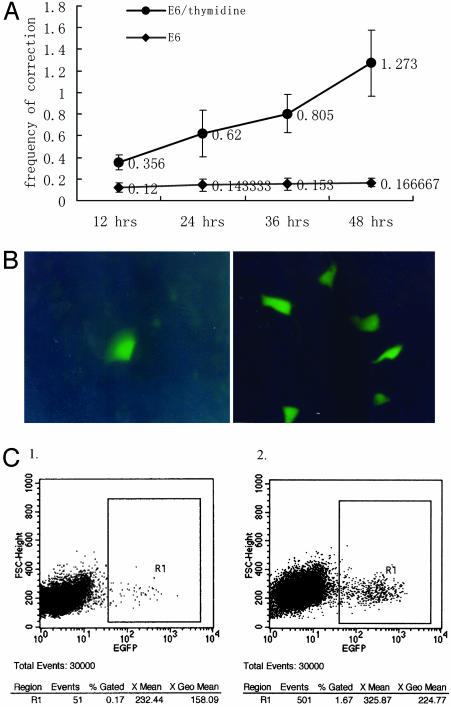
We then investigated the relationship between the duration of SSO/thymidine coincubation after transfection and the gene repair efficiency. Cells were coincubated with thymidine (2 mM)/SSO for a range of time periods before replacing the medium for the remainder of the incubation up to a total of 48 h, after which the repair frequency was determined (Fig. 3A). We found that the repair frequency rose steadily with the duration of thymidine/SSO-E6 coincubation, reaching ≈1.3% after 48 h (n = 5). When thymidine was omitted, there was only a very slight increase in repair frequency over the same time period. By using fluorescence microscopy, significantly more green cells were visible in the thymidine-treatment samples than in those without thymidine coincubation (Fig. 3B). In one experiment set, the E6-mediated mEGFP repair frequency after 48-h coincubation with thymidine was determined to be almost 10 times of that of the control without thymidine (see “events,” Fig. 3C).
Fig. 3.
Repair efficiency of E6 in the conditions with or without thymidine. (A) We incubated 2 mM of thymidine in medium 2 h before and 48 h after SSO transfection. The x-axis indicates the SSO/thymidine incubation time. The y-axis indicates the frequency of repaired cells. Compared with the control graph, where no thymidine was added before or after SSO transfection, the thymidine-administered group represents obviously higher frequency (P < 0.01, n = 5). (B) Repaired cells detected under UV light. (Left) Without thymidine, there is only one green cell in several eye fields. (Right) With thymidine there are about six green cells in almost every eye field. (C) FACS results comparing frequencies of SSO-mediated repair 48 h after transfection by E6 without (1) or with (2) thymidine incubation.
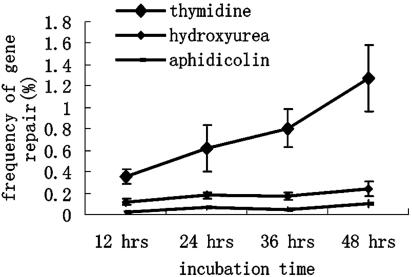
Replication Fork Leakage and SSO-Mediated Gene Repair. Because thymidine is known to synchronize cells by reducing the rate of DNA replication (36), we hypothesized that thymidine coincubation after transfection slowed the progression of the replication fork, creating a greater opportunity for the SSO to anneal to its homologous sequence. To investigate the relationship between the replication rate and the efficiency of SSO-mediated repair, we measured the repair frequencies in cells treated with three different S-phase inhibitors: thymidine (2 mM), hydroxyurea (1 mM), and aphidicolin (6 μM), which have been demonstrated to inhibit replication to varying extents. The drug concentrations used were identical to those previously reported to be effective for replication-inhibitory effects (37). Thymidine exerts its effect by inhibiting the formation of dCTP, one of the essential precursors for cellular DNA synthesis (36). Hydroxyurea has been shown to reduce the rate of DNA synthesis by indirectly depleting the pool of available deoxynucleotides (37, 38). Aphidicolin inhibits the replicative α-polymerase protein, while allowing cell-cycle progression in G2, M, and G1 cells, leading to synchronization and cell accumulation at the G1/S border (38, 39).
Flow cytometry data showed that after 24 h of continuous inhibition, all three drugs enriched the cells at the G1/S border and in the S phase. Cell-cycle progression was observed for both the thymidine- and hydroxyurea-inhibited cells, as indicated by monitoring changes in cellular DNA content. The cell-cycle progression was seen to be faster in the presence of thymidine than with hydroxyurea. After 24 h of incubation with 2 mM thymidine, the majority of the cells (77%) were at the G1/S border, and only a small fraction (23%) were in the S phase. After 36 h, 100% of the cells had entered the S phase, and after 48 h, more than half of the cells had left the S phase and had entered the G2/M phase (Fig. 9, which is published as supporting information on the PNAS web site). Our observations were consistent with previous findings that even when the concentration was raised to 10 mM, thymidine could not completely prevent de novo DNA synthesis and arrest the replication fork (36). After 36 h of continuous inhibition, only 67% of the hydroxyurea-treated cells had progressed into the S phase, and after 48 h, all of the hydroxyurea-inhibited cells still remained within the S phase, with none entering the G2/M phase (Fig. 9B). In the presence of aphidicolin, the most potent replication inhibitor, cells were totally stalled at the G1/S border and S phase. No cell-cycle progression was observed during inhibition time from 24 to 48 h (Fig. 9C).
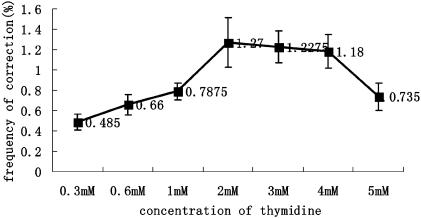
We then measured the efficiency of E6-mediated gene repair in cells undergoing DNA replication at different rates, as indicated by the speed of cell-cycle progression. As shown in Fig. 4, the thymidine-inhibited cells exhibited the highest repair frequencies, whereas inhibition with hydroxyurea led to more modest increases in the repair levels. Treatment with aphidicolin led to a slight decrease in the gene repair frequencies when compared with controls without inhibitor (compare Figs. 4 and 3A). The inability of hydroxyurea and aphidicolin to significantly increase the efficiency of the SSO-mediated repair process suggests that DNA replication leakage is required for elevated repair levels. We hypothesized that there may be an optimal rate of replication fork progression for maximum levels of SSO-mediated gene repair. Accordingly, neither a fast-moving replication fork (reduced opportunity for annealing) nor a stopped fork (lack of opportunity for annealing or lack of DNA synthesis) is conducive to high repair efficiencies. We further tested this hypothesis by investigating the relationship between thymidine concentration and the repair efficiency. We reasoned that varying the concentration of thymidine should alter the rate of replication fork progression, which should be reflected in the gene repair efficiencies measured. Our results suggested that this hypothesis was indeed the case, with a concentration of 2–4 mM of thymidine appearing to be optimal for SSO repair activity. The correction efficiencies were found to gradually decrease with higher thymidine concentrations (Fig. 5).
Fig. 4.
Effects of different types of S-phase inhibitors on the repair efficiency. Three different inhibitors were kept in the medium 2 h before and 48 h after transfection with E6. The x-axis indicates the SSO/inhibitor incubation time after transfection; the y-axis indicates the frequency of repaired cells (n = 3).
Fig. 5.
Efficiencies of E6-SSO in different concentration of thymidine. Thymidine was kept in the medium 2 h before and 48 h after transfection with E6. The x-axis indicates the thymidine concentration used; the y-axis indicates the frequency of repaired cells (n = 3).
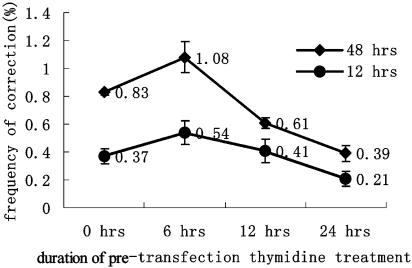
Thymidine-Induced Alteration of the Cellular Proteome. In addition to its ability to retard the progression of the replication fork, we investigated whether thymidine enhanced the efficiency of gene correction by creating a more favorable intracellular proteomic environment. We reasoned that by treating the cells with thymidine for a certain period before transfection with SSO, its putative beneficial effects on the proteome would be long-lived enough to cause an increase in the repair efficiency. To test this possibility, we preincubated the F5 cells with 2 mM thymidine for up to 24 h before transfection with E6. After transfection, incubation with thymidine was maintained for an additional 12 or 48 h, before determining the gene repair frequencies. Results indicated that the mEGFP gene correction efficiencies dropped markedly, because the preinhibition time was extended from 6 to 24 h (Fig. 6). In combination with the results shown in Fig. 8B, it can be seen that when thymidine was used for cell synchronization but withdrawn just before transfection, very low correction frequencies (0.10%) were obtained. These results suggest that thymidine enhances the repair efficiency through modulating the rate of replication fork progression and is not achieved by means of some other alteration of the cellular proteome.
Fig. 6.
Effect of preinhibition time on the SSO-mediated repair efficiency. Preinhibition time refers to the duration of thymidine incubation time for cells before transfection. The x-axis indicates the duration of pretransfection thymidine coincubation. Cells continued to incubate with thymidine/E6 for 12 h after transfection in one group and for 48 h for the other group. Results showed that frequencies of both groups decreased gradually as the duration of preinhibition increased from 6 to 24 h (n = 3).
Discussion
To maximize the efficiency by which oligonucleotides mediate genomic alterations, most research over the past few years has focused on oligonucleotide design and modification. This research has been conducted primarily to prolong its intracellular lifetime and maximize its chances of productively hybridizing with the target locus (27, 40, 41). We have found that SSOs 25 nt in length containing six phosphorothioate linkages at each terminus exhibit the optimal balance among the size, purity, and stability in vivo, for maximum repair activity (W.-X.Y., X.-S.W., G. Liu, Z.-H. Li, J.-D.H., K.S.E.C., D.-P.L., and C.-C.L., unpublished data).
When the SSO is introduced into the cell by transfection procedures, there are many factors impeding its intracellular survival and access to the chromosome. These include the plasma and nuclear membranes and various host nucleases. The SSO also may have difficulty in accessing its homologous double-stranded DNA target due to the tight packing of chromosomal DNA and associated proteins. Occasionally, the chromatin “opens up,” e.g., during transcription, DNA damage repair, replication, etc., which provides opportunities for the SSO to access its chromosomal target. It has been reported that transcriptional activity has a positive influence on the efficiency of SSO-mediated gene repair (29, 42). Recently, Parekh-Olmedo et al. (43) showed that treatment with trichostatin A, which could increase the acetylation level of histones, improved the SSO-mediated repair frequency by inducing a more open chromatin structure.
To explore the relationship between the efficiency of SSO-mediated gene repair and chromatin structure in mammalian cells, we established a chromosome-based fluorescence reporter system in a HeLa cell line. A mutant EGFP gene containing a disrupted initiation codon (ATG → TTG) was stably integrated into the chromosome. SSOs were transfected into the cells to correct the mutant initiation codon, restoring EGFP gene expression and creating a fluorescent phenotype that could be accurately quantified. We reasoned that the “correction” SSO would be able to anneal to its genomic target more efficiently during specific periods of the cell cycle, due to a more favorable chromatin arrangement. Through transfecting SSO-E6 into cells synchronized at different phases in the cell cycle, we discovered that the SSO-mediated repair efficiencies were highest in thymidine-enriched S-phase cells. This finding prompted us to speculate that the SSOs had a greater probability of accessing the target locus during the S phase, because of the increased presence of regions of single-stranded DNA exposed at the replication fork. It presumably provided a “window of opportunity” during the S phase, within which the SSOs could productively anneal with their homologous targets.
Further experiments demonstrated that it was not maintaining the cells in the S phase per se, but it was the presence of thymidine that was the key to higher efficiencies. Because it has previously been shown that thymidine reduces the rate of DNA replication (36), we analyzed the cell-cycle progression by flow cytometry, under the conditions analogous to those used for the thymidine-SSO repair experiments. It showed that the cells passed very slowly from the G1/S border to the G2/M phase in the presence of 2 mM thymidine. It took ≈48 h for all of the cells to pass through S phase. This finding correlated well with the results from the repair experiments, in which the repair efficiencies rose gradually over this period, peaking ≈48 h after transfection.
Our findings also indicated that DNA replication only should be inhibited to a certain level, to achieve optimal repair frequencies (Fig. 5). The complete inhibition of replication appears to be detrimental to the SSO-mediated repair process, suggesting the involvement of “replication fork leakage,” or de novo DNA synthesis during inhibition. This rationale is supported by the observation that the addition of hydroxyurea, which is a more potent (S phase) inhibitor of DNA replication than thymidine, leads to only a small increase in repair efficiencies compared with controls (Fig. 4). Furthermore, the addition of aphidicolin, which totally inhibits DNA replication, actually decreased the gene repair frequency.
Consistent with these results and a number of previous models (1, 17, 44), we put forward a replication fork leakage model for increased SSO-mediated gene repair levels. We propose that thymidine exerts its influence during the first step of the SSO-mediated gene repair process: namely that it improves the efficiency by which the SSO anneals to its homologous target DNA strand. This process is achieved by slowing down or retarding the progression of the replication fork, lengthening the period the homologous target DNA exists in single-stranded form, increasing the likelihood of hybridization with the SSO. Thymidine treatment thus provides the SSO with a greater window of opportunity within which it may initiate the gene repair process. In the absence of thymidine, the rate of replication fork progression may be too rapid, allowing little chance for the SSO to access and hybridize with its homologous single-stranded region. In the other extreme, a stalled or grossly inhibited replication fork may be detrimental to the repair process because of a lack of de novo DNA synthesis, or possibly due to a reduction in the amount of accessible single-stranded target DNA. Preincubation with thymidine before transfection with SSO also lowered the repair efficiency. This finding suggests that thymidine does not exert its effects through inducing a sustainable change in protein expression patterns. Rather, it appears to have a very short-lived or immediate mode of action.
After the annealing of the SSO to its homologous region, the second step in SSO-mediated gene repair is the alteration of chromosomal DNA sequence as encoded by the SSO. Several mechanisms including mismatch repair (2), homologous recombination (31), and direct incorporation of the SSO through DNA synthesis (13) have been proposed to occur during this step. Our results are consistent with the operation of all three of these possible mechanisms. However, we think that the direct incorporation model fits our data best: where the SSO first anneals with its homologous single-stranded target, most likely at the replication fork, and is subsequently integrated into the chromosome (rather than merely acting as a template for DNA alteration). Adherence to this model requires fresh DNA synthesis at the target locus (extension of the annealed SSO) and predicts a strand bias associated with the correction process. Indeed, we observed a distinct preferred strand in our gene targeting experiments (data not shown).
Nevertheless, the possibility that thymidine also can enhance or influence the operation of the other putative mechanisms remains intriguing. A slowed replication fork may trigger DNA damage repair by providing greater opportunity for the cellular surveillance systems to detect the SSO-chromosome DNA mismatch. The fact that SSO-mediated repair appears to occur during all cell phases, even in nonreplicating cells (45), indicates that there may be more than one mechanistic pathway in operation. It is possible that alternate pathways for gene repair are preferred under different intracellular conditions.
In conclusion, we have demonstrated that coincubation with thymidine can significantly increase the frequency of SSO-mediated chromosomal DNA repair in HeLa cells, by putatively retarding the progression of the replication fork. Because thymidine is a well-established and “mild” S phase inhibitor, we speculate that this approach has great potential for further adaptation and utilization in this and other gene targeting systems. By systematically modulating other factors that alter the chromosomal environment such as replication, transcription, mismatch repair, and the structure of chromatin, combined with varying oligonucleotide design, modification, and delivery, there is extensive scope for establishing optimal conditions for efficient and reproducible SSO-mediated genetic repair.
Supplementary Material
Acknowledgments
We thank Dr. K. M. Yao (University of Hong Kong) for critically reading the manuscript. This work was supported by National Basic Research Program of China Grant 2004CB518803, Hi-Tech Research and Development Program of China Grant 2003AA216020, the National Natural Science Foundation of China, and Research Grant Council of Hong Kong Grants 30218005 and N_HKU712/02.
Author contributions: X.-S.W., L.X., K.S.E.C., D.-P.L., and C.-C.L. designed research; X.-S.W., L.X., W.-X.Y., X.-Y.S., and L.L. performed research; and X.-S.W., L.X., R.M.W., and J.-D.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SSO, single-stranded oligonucleotide; RDO, chimeric RNA/DNA oligonucleotide.
References
- 1.Rice, M. C., Czymmek, K. & Kmiec, E. B. (2001) Nat. Biotechnol. 19, 321-326. [DOI] [PubMed] [Google Scholar]
- 2.Kmiec, E. B. (2003) J. Clin. Invest. 112, 632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullenger, B. A. (2003) J. Clin. Invest. 112, 310-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole-Strauss, A., Yoon, K., Xiang, Y., Byrne, B. C., Rice, M. C., Gryn, J., Holloman, W. K. & Kmiec, E. B. (1996) Science 273, 1386-1389. [DOI] [PubMed] [Google Scholar]
- 5.Yoon, K., Cole-Strauss, A. & Kmiec, E. B. (1996) Proc. Natl. Acad. Sci. USA 93, 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, L., Parekh-Olmedo, H. & Kmiec, E. B. (2003) Nat. Rev. Genet. 4, 679-689. [DOI] [PubMed] [Google Scholar]
- 7.Brachman, E. E. & Kmiec, E. B. (2002) Curr. Opin. Mol. Ther. 4, 171-176. [PubMed] [Google Scholar]
- 8.Tran, N. D., Liu, X., Yan, Z., Abbote, D., Jiang, Q., Kmiec, E. B., Sigmund, C. D. & Engelhardt, J. F. (2003) Mol. Ther. 7, 248-253. [DOI] [PubMed] [Google Scholar]
- 9.Parekh-Olmedo, H., Czymmek, K. & Kmiec, E. B. (March 13, 2001) Sci. STKE, 10.1126/stke.2001.73.pl1. [DOI] [PubMed]
- 10.Bennett, M. & Schaack, J. (2003) J. Gene Med. 5, 723-732. [DOI] [PubMed] [Google Scholar]
- 11.Alexeev, V. & Yoon, K. (1998) Nat. Biotechnol. 16, 1343-1346. [DOI] [PubMed] [Google Scholar]
- 12.Li, Z. H., Liu, D. P., Yin, W. X., Guo, Z. C. & Liang, C. C. (2001) Blood Cells Mol. Dis. 27, 530-538. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, H. M., Yu, D., DiTizio, T. & Court, D. L. (2001) Proc. Natl. Acad. Sci. USA 98, 6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamper, H. B., Parekh, H., Rice, M. C., Bruner, M., Youkey, H. & Kmiec, E. B. (2000) Nucleic Acids Res. 28, 4332-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taubes, G. (2002) Science 298, 2116-2120. [DOI] [PubMed] [Google Scholar]
- 16.Pierce, E. A., Liu, Q., Igoucheva, O., Omarrudin, R., Ma, H., Diamond, S. L. & Yoon, K. (2003) Gene Ther. 10, 24-33. [DOI] [PubMed] [Google Scholar]
- 17.Liu, L., Rice, M. C. & Kmiec, E. B. (2001) Nucleic Acids Res. 29, 4238-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen, M. S., Sorensen, C. B., Bolund, L. & Jensen, T. G. (2002) J. Mol. Med. 80, 770-781. [DOI] [PubMed] [Google Scholar]
- 19.Gamper, H. B., Hou, Y. M. & Kmiec, E. B. (2000) Biochemistry 39, 15272-15281. [DOI] [PubMed] [Google Scholar]
- 20.Court, D. L., Sawitzke, J. A. & Thomason, L. C. (2002) Annu. Rev. Genet. 36, 361-388. [DOI] [PubMed] [Google Scholar]
- 21.Drury, M. D. & Kmiec, E. B. (2003) Nucleic Acids Res. 31, 899-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, L., Cheng, S., van Brabant, A. J. & Kmiec, E. B. (2002) Nucleic Acids Res. 30, 2742-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, L., Usher, M., Hu, Y. & Kmiec, E. B. (2003) Appl. Environ. Microbiol. 69, 6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parekh-Olmedo, H., Krainc, D. & Kmiec, E. B. (2002) Neuron 33, 495-498. [DOI] [PubMed] [Google Scholar]
- 25.Li, X. T., Costantino, N., Lu, L. Y., Liu, D. P., Watt, R. M., Cheah, K. S., Court, D. L. & Huang, J. D. (2003) Nucleic Acids Res. 31, 6674-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costantino, N. & Court, D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Steege, G., Schuilenga-Hut, P. H., Buys, C. H., Scheffer, H., Pas, H. H. & Jonkman, M. F. (2001) Nat. Biotechnol. 19, 305-306. [DOI] [PubMed] [Google Scholar]
- 28.Graham, I. R., Manzano, A., Tagalakis, A. D., Mohri, Z., Sperber, G., Hill, V., Beattie, S., Schepelmann, S., Dickson, G. & Owen, J. S. (2001) Nat. Biotechnol. 19, 507-508. [DOI] [PubMed] [Google Scholar]
- 29.Liu, L., Rice, M. C., Drury, M., Cheng, S., Gamper, H. & Kmiec, E. B. (2002) Mol. Cell. Biol. 22, 3852-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrara, L., Parekh-Olmedo, H. & Kmiec, E. B. (2004) Exp. Cell Res. 300, 170-179. [DOI] [PubMed] [Google Scholar]
- 31.Ferrara, L. & Kmiec, E. B. (2004) Nucleic Acids Res. 32, 5239-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brachman, E. E. & Kmiec, E. B. (2004) J. Cell Sci. 117, 3867-3874. [DOI] [PubMed] [Google Scholar]
- 33.Kohn, K. W. (1999) Mol. Biol. Cell 10, 2703-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galgano, P. (1998) Cell Synchronization (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 35.Rodgers, L. (1998) Flow Cytometry (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 36.Bjursell, G. & Reichard, P. (1973) J. Biol. Chem. 248, 3904-3909. [PubMed] [Google Scholar]
- 37.Saintigny, Y., Delacote, F., Vares, G., Petitot, F., Lambert, S., Averbeck, D. & Lopez, B. S. (2001) EMBO J. 20, 3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pichierri, P., Franchitto, A., Mosesso, P. & Palitti, F. (2001) Mol. Biol. Cell 12, 2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedrali-Noy, G. & Spadari, S. (1979) Biochem. Biophys. Res. Commun. 88, 1194-1202. [DOI] [PubMed] [Google Scholar]
- 40.Parekh-Olmedo, H., Drury, M. & Kmiec, E. B. (2002) Chem. Biol. 9, 1073-1084. [DOI] [PubMed] [Google Scholar]
- 41.Alexeev, V., Igoucheva, O. & Yoon, K. (2002) Gene Ther. 9, 1667-1675. [DOI] [PubMed] [Google Scholar]
- 42.Igoucheva, O., Alexeev, V., Pryce, M. & Yoon, K. (2003) Nucleic Acids Res. 31, 2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parekh-Olmedo, H., Engstrom, J. U. & Kmiec, E. B. (2003) Ann. NY Acad. Sci. 1002, 43-55. [DOI] [PubMed] [Google Scholar]
- 44.Cole-Strauss, A., Gamper, H., Holloman, W. K., Munoz, M., Cheng, N. & Kmiec, E. B. (1999) Nucleic Acids Res. 27, 1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kren, B. T., Chen, Z., Felsheim, R., Roy Chowdury, N., Roy Chowdury, J. & Steer, C. J. (2002) Gene Ther. 9, 686-690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.