Abstract
Egr2 is a transcription factor required for peripheral nerve myelination in rodents, and mutations in Egr2 are associated with congenital hypomyelinating neuropathy (CHN) in humans. To further study its role in myelination, we generated mice harboring a hypomorphic Egr2 allele (Egr2Lo) that survive for up to 3 weeks postnatally, a period of active myelination in rodents. These Egr2Lo/Lo mice provided the opportunity to study the molecular effects of Egr2 deficiency on Schwann cell biology, an analysis that was not possible previously, because of the perinatal lethality of Egr2-null mice. Egr2Lo/Lo mice phenocopy CHN, as evidenced by the severe hypomyelination and increased numbers of proliferating Schwann cells of the peripheral nerves. Comparison of sciatic nerve gene expression profiles during development and after crush injury with those of Egr2Lo/Lo Schwann cells revealed that they are developmentally arrested, with down-regulation of myelination-related genes and up-regulation of genes associated with immature and promyelinating Schwann cells. One of the abnormally elevated genes in Egr2Lo/Lo Schwann cells, Sox2, encodes a transcription factor that is crucial for maintenance of neural stem cell pluripotency. Wild-type Schwann cells infected with Sox2 adenovirus or lentivirus inhibited expression of myelination-associated genes (e.g., myelin protein zero; Mpz), and failed to myelinate axons in vitro, but had an enhanced proliferative response to β-neuregulin. The characterization of a mouse model of CHN has provided insight into Schwann cell differentiation and allowed the identification of Sox2 as a negative regulator of myelination.
Deficits in Schwann cell function are associated with a number of human diseases, from debilitating conditions like Charcot–Marie–Tooth disease (CMT) and diabetic neuropathy, to frequently lethal diseases, such as congenital hypomyelinating neuropathy (CHN) (1). Developmentally, Schwann cells progress from an immature state, expressing markers such as L1, Ncam, and Cyclin D1, to the nonmyelinating or the promyelinating state. Whereas nonmyelinating Schwann cells continue to express L1 and Ncam and are associated with multiple axons, promyelinating Schwann cells establish a one-to-one relationship with axons and are marked by expression of the POU domain transcription factor suppressed cAMP-inducible POU (SCIP) (2). Shortly after birth, promyelinating Schwann cells exit from the cell cycle and differentiate into myelinating Schwann cells, a process that involves the expression of genes such as Egr2, Mpz, Pmp22, Prx, and Gjb1 and the formation of myelin. A similar process occurs after axonal injury, in which Schwann cells must revert to a proliferative immature state and later redifferentiate and myelinate regenerating axons (3).
Studies of Egr2-null mice led to the discovery that this transcription factor is a prime regulator of Schwann cell myelination. Although most Egr2 null mice die at birth, a few Egr2-null mice survive for up to 2 weeks. Nerves from these rare survivors are hypomyelinated and are populated with Schwann cells that fail to exit the cell cycle (4). Enforced expression of Egr2 in cultured Schwann cells activates expression of numerous genes associated with myelination, including crucial myelin structural proteins and enzymes involved in lipid synthesis (5), further supporting the importance of Egr2 in regulating myelination.
More importantly, mutations in Egr2 are found in patients with CHN, CMT, or Dejerine–Sottas syndrome (6–9). Most Egr2 mutations in CHN patients occur in the zinc-finger DNA-binding domain. These DNA-binding mutants act in a dominant-negative fashion to inhibit wild-type Egr2 activity (5). Additionally, a recessive form of CHN is caused by a mutation in the R1 domain, a site of interaction for the Nab proteins that modulate Egr2 activity (6). While these data indicate that alterations in Egr2 activity result in Schwann cell dysfunction and aberrant myelination, the molecular alterations that occur in these cells are unknown.
To understand the molecular consequences of Egr2-deficiency in the context of CHN, we have studied mice homozygous for an Egr2 hypomorphic allele (Egr2Lo/Lo) that consistently survive into the third postnatal week. These mice display characteristics of CHN, including severe hypomyelination and an absence of Schwann cell differentiation. Global gene expression analysis of CHN Schwann cells from these Egr2Lo/Lo mice identified numerous proteins that are likely to play important roles in Schwann cell development and may directly contribute to Schwann cell dysfunction in CHN. Further examination of one of these candidates, Sox2, indicates a role for this transcription factor in maintaining the undifferentiated Schwann cell state.
Methods
Microarray Analysis. We compared microarray data from three different paradigms in this study. Two of the data sets, nerve crush injury (10) and development (11), were previously generated. The third data set was generated from two independent samples of sciatic nerve total RNA (10 μg) for each genotype, Egr2Lo/Lo and wild type. Each sample contained nerves from 10 animals. RNA extraction, probe generation, and chip hybridization to MU74A V2 microarrays were described (10), and chips were scaled to 1,500 (mas 5.0, Affymetrix).
For each paradigm, pairwise comparisons were performed (a total of nine analyses). The baseline points, wild type (for Egr2Lo/Lo), uninjured (for nerve crush), or embryonic day (E)17 (for development), were compared with mutant (Egr2Lo/Lo), or to each monitored time point in the nerve crush or development paradigms, respectively. For each comparison, genes called absent in all chips were excluded. These filtered data sets were subjected to significance analysis of microarrays by using the pairwise comparison option with the following parameters: significantly altered >2-fold, and a false-discovery rate of 10% (12). The list of differentially regulated genes in injury or development was obtained by taking the union of significantly altered genes in each pairwise comparison within each paradigm. A summary of these analyses is in Table 1, which is published as supporting information on the PNAS web site. The array data are available upon request.
Supporting Methods. Generation of the Egr2Lo targeting construct, sciatic nerve histology and ultrastructure, immunohistochemistry, western analysis, quantitative RT-PCR, sciatic nerve crush injury, primary Schwann cell cultures, proliferation assays, adenoviral infection, lentiviral infection, in vitro myelination assay, and K-means clustering of microarray profiles, are all detailed in Supporting Methods, which is published as supporting information on the PNAS web site.
Results
Generation of Egr2Lo/Lo Mice. Because most Egr2-null mice die at birth, a model of CHN with a longer lifespan would enhance the identification of important Schwann cell deficits in this disease. In attempting to introduce specific point mutants into the Egr2 locus, we found that insertion of a PGK-Neo cassette into the first intron of Egr2 generated a hypomorphic Egr2 allele (Egr2Lo). The decreased expression from this mutant allele is presumably due to the introduction of the PGK promoter, as has been observed at other loci (13). Mice heterozygous (Egr2+/Lo) for the hypomorphic allele were overtly normal, and homozygous mice (Egr2Lo/Lo) were produced at expected Mendelian ratios. Southern analysis confirmed homologous recombination at the Egr2 locus (Fig. 6, which is published as supporting information on the PNAS web site).
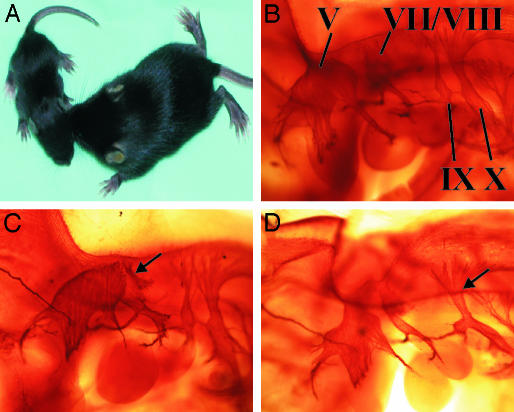
The Egr2Lo/Lo mice were smaller and displayed severe tremors and impaired coordination (Fig. 1A and Movie 1, which is published as supporting information on the PNAS web site). Although some Egr2Lo/Lo animals died within the first postnatal week, 60% of them survived into the third week of life, a time when peripheral nerve myelination in rodents is well underway. The early neonatal lethality observed in some Egr2Lo/Lo mice was reminiscent to that observed for Egr2-null mice, where the lethality is thought to be due to a hindbrain segmentation defect. Egr2 is expressed in rhombomeres 3 and 5, which are lost in Egr2-null mice, resulting in a notable fusion of cranial nerve V with VII and VIII and IX with X, respectively (14, 15). Analysis of the hindbrain of Egr2Lo/Lo animals at E10.5 demonstrated a segmentation defect that showed variable expressivity (Fig. 1 B–D). Whereas 14% of Egr2Lo/Lo embryos had a loss of both rhombomeres 3 and 5, 57% had a fusion of CN V with VII and VIII, and 29% had a fusion of CN IX and X (Fig. 1 C and D). The reduced severity of the hindbrain defect in Egr2Lo/Lo mice, compared with that observed in Egr2-null mice, is the presumptive cause of their extended lifespan.
Fig. 1.
Egr2Lo/Lo mice have a variably expressed hindbrain segmentation defect. (A) P14 mice homozygous for the hypomorphic allele, Egr2Lo/Lo, were runted and displayed severe trembling. (B–D) In contrast to wild-type E10.5 embryos (B), Egr2Lo/Lo embryos showed a range of hindbrain segmentation defects that include fusion of cranial nerves V with VII and VIII (C, arrow) or fusion of cranial nerves IX and X (D, arrow).
Peripheral Nerves in Egr2Lo/Lo Mice Are Severely Hypomyelinated. The tremors and uncoordinated behavior of Egr2Lo/Lo mice suggested a deficit in Schwann cell function, resulting from insufficient Egr2 activity. We compared expression of Egr2 in postnatal day (P)14 nerves from wild type and Egr2Lo/Lo by immunohistochemistry, immunoblotting, and quantitative RT-PCR, and found decreased levels of Egr2 protein (2.9-fold) and mRNA (3.1-fold) in Egr2Lo/Lo nerves (Fig.7, which is published as supporting information on the PNAS web site).
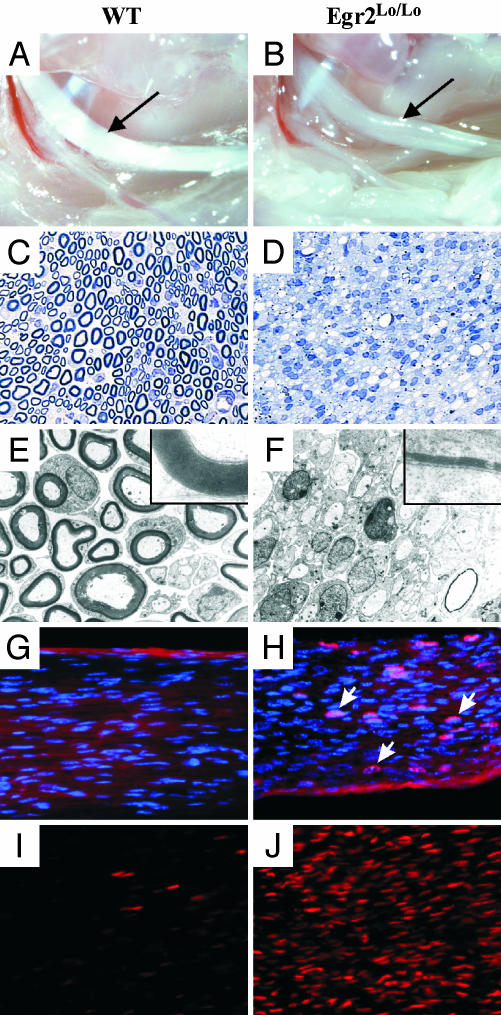
The sciatic nerves of P14 Egr2Lo/Lo mice were thin and translucent compared with those of wild-type mice (Fig. 2 A and B). Congruently, examination of plastic-embedded sciatic nerve sections confirmed that, whereas P14 wild-type nerves contained numerous thickly myelinated large and small caliber axons (Fig. 2C), Egr2Lo/Lo nerves showed an absence of myelination, and instead, contained an abundance of undifferentiated Schwann cells (Fig. 2D). No evidence of an inflammatory process or necrosis was observed, and no onion bulb formations, reflecting a demyelination/remyelination scenario seen in later-onset CMT diseases, were present. Electron microscopic examination revealed normal numbers of large- and medium-caliber axons in the Egr2Lo/Lo nerve, but the majority of these were unmyelinated (Fig. 2 E and F). Many Schwann cells were associated with individual axons, suggesting that they could progress to the promyelinating stage, but not beyond. Rare hypomyelinating figures had very thin myelin sheaths compared with wild-type myelinating Schwann cells (Fig. 2 E and F Insets). Consistent with the predominance of undifferentiated Schwann cells in Egr2Lo/Lo nerves, BrdUrd incorporation assays revealed increased numbers of proliferating cells in P14 nerve from Egr2Lo/Lo mice (wild type = 0.69 ± 0.5% vs. Egr2Lo/Lo = 11.3 ± 1.9%) (Fig. 2 G and H). Furthermore, Egr2Lo/Lo nerves had a substantial increase in the number of SCIP-immunoreactive nuclei, reflecting the large number of promyelinating Schwann cells that were unable to terminally differentiate (Fig. 2 I and J).
Fig. 2.
Egr2Lo/Lo sciatic nerves are congenitally hypomyelinated. Gross examination revealed that at P14, wild-type (WT) nerves (A, arrow) were opaque white because of abundant myelination, whereas P14 Egr2Lo/Lo nerves (B, arrow) were transparent because of defective myelination. Toluidine blue-stained sections (C and D) and electron micrographs (E and F) illustrate that WT nerves (C and E Inset) have thick and abundant myelination of medium- and large-caliber axons, whereas Egr2Lo/Lo (D and F Inset) nerves have densely packed Schwann cells that show minimal myelination of a few axons. BrdUrd incorporation (red nuclei) showed that P14 nerves from Egr2Lo/Lo mice (H) have higher numbers of proliferating Schwann cells than do wild-type nerves (G). Immunohistochemical analysis with antibodies against the promyelinating Schwann cell marker SCIP demonstrated that P14 wild-type nerve has few SCIP-immunoreactive cells (red nuclei) (I), whereas Egr2Lo/Lo nerve has abundant SCIP-positive cells (J).
Expression Profiling of Egr2Lo/Lo CHN Nerves Identifies Genes Involved in Schwann Cell Differentiation. The extended lifespan of Egr2Lo/Lo mice presented a unique opportunity to explore the Schwann cell molecular defects that underlie the hypomyelinating phenotype of this CHN model. Microarray expression profiling was performed on sciatic nerves from P14 Egr2Lo/Lo and wild-type mice, and significance analysis of microarrays analysis was performed to compare the Egr2Lo/Lo and wild-type data sets (see Methods). We identified 528 probe sets (henceforth labeled as genes) that were significantly changed. There were 262 genes expressed at higher levels in Egr2Lo/Lo nerves (INC set) and 266 genes expressed at lower levels in Egr2Lo/Lo nerves (DEC set).
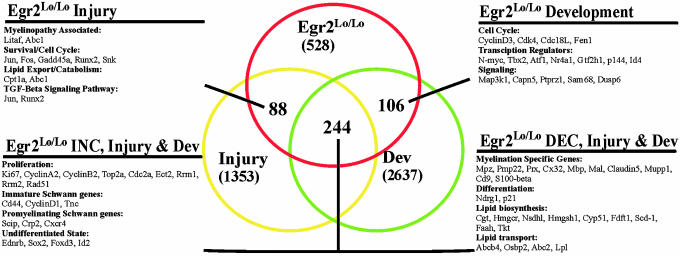
To further evaluate the importance of these genes in Schwann cell function, these data were integrated with data sets derived from expression profiles obtained by using paradigms where dynamic alterations in Schwann cell phenotype occur, such as during peripheral nerve development and after nerve crush injury. The developmental data, including E17, P0, P2, P4, P10, and P56 nerves, was made available by Verheijen et al. (11). The sciatic nerve crush data were generated as described (10), and included samples harvested at 0, 4, 7, and 14 days after injury. When these data sets were analyzed (see Methods), we found that 123 genes in the DEC set and 121 genes in the INC set were also significantly altered in the nerve development and injury data sets. These genes were designated the DEC ALL and INC ALL sets, respectively (Fig. 3 and Table 2, which is published as supporting information on the PNAS web site).
Fig. 3.
Intersection of genes altered in Egr2Lo/Lo CHN nerve with those altered during development or after sciatic nerve injury. Using significance analysis of microarrays analysis (see Methods), we identified 528 genes that were altered in Egr2Lo/Lo CHN sciatic nerves (red set), 1,353 genes that were altered after nerve crush injury, and 2,637 genes that were altered during development. Of the 528 genes associated with CHN, 438 genes were also altered during development (green set) or after nerve crush injury (yellow set). Whereas 244 genes in this subset were altered in all three paradigms, 106 genes were altered in development alone, and 88 genes were altered only after injury. Representative functional gene categories are included for each subset.
Myelination-Associated Genes Are Enriched in the Egr2Lo/Lo DEC ALL Subset. The congenital hypomyelination of Egr2Lo/Lo nerves indicate a halt in the maturation of these Schwann cells. In accordance, the subset of genes with decreased expression (DEC ALL) included genes associated with myelination, some of which are frequently mutated in patients with inherited peripheral neuropathies, including Mpz, Pmp22, Prx, Gjb1, and Ndrg1.
Classification of the DEC ALL gene subset by gene ontology by using dchip software (Table 3, which is published as supporting information on the PNAS web site) revealed an enrichment for proteins involved in lipid metabolism (e.g., high-mobility group-CoA reductase and CGT), which is consistent with the high lipid content in myelin and the coregulation of lipid biosynthesis with myelin protein expression (10, 11). Additionally, myelin sheath structural proteins, such as Claudin 5 and Mupp1 (16), which interact with each other in the Schmidt-Lanterman incisures, were present in the DEC ALL data set. These results encouraged us to perform quantitative RT-PCR analysis to examine the expression of other DEC ALL genes, including EphB6, p21, Pea15, Nr4a2, and the potassium ion channel KcnK1 in immature cultured Schwann cells, and in developing and injured nerves. We found a strong association between their expression levels and the differentiated Schwann cell phenotype (Fig. 8A, which is published as supporting information on the PNAS web site). These results confirm that the DEC ALL subset is enriched for genes involved in establishing and/or maintaining the myelinating phenotype, in accordance with the inability of Egr2Lo/Lo CHN Schwann cells to terminally differentiate.
Identification of Immature and Promyelinating Schwann Cell Markers by Analysis of the Egr2Lo/Lo INC ALL Subset. The failure of Egr2Lo/Lo CHN Schwann cells to exit the cell cycle and differentiate results in nerves containing increased numbers of both immature and promyelinating Schwann cells. To identify genes selective for these Schwann cell stages, we performed K-means clustering on the INC ALL subset by using both the nerve development and sciatic nerve crush injury data sets (Fig. 9 A and B and Table 4, which are published as supporting information on the PNAS web site). The resulting clusters were then categorized by inspection for well known marker genes (e.g., Cyclin D1 for immature Schwann cells, SCIP for promyelinating Schwann cells, and Ki67 for proliferating Schwann cells).
Through this approach, clusters of proliferation-associated genes, in accord with the high proliferative index of Egr2Lo/Lo Schwann cells, were identified in both the injury (cluster 2) and development (clusters 1 and 2) data sets. A promyelinating Schwann cell cluster, including SCIP and its putative transcriptional targets, Crp2 and Cxcr4 (17, 18), was also found by this analysis. Furthermore, clusters containing genes associated with immature Schwann cells were identified in both the developing (clusters 1 and 2) and injured (cluster 4) nerve data sets. By inference, other genes in these clusters, such as Pde8A, FoxD3, and Sox2 (see below), are also likely to be involved in Schwann cell maturation.
Sox2 Is a Marker of Immature Schwann Cells. The delineation of differentially expressed genes in the CHN Schwann cells of Egr2Lo/Lo nerves offers the opportunity to identify genes that play important roles in normal Schwann cell differentiation and in CHN Schwann cell dysfunction. Whereas Egr2 is associated with the differentiated state, and SCIP expression is correlated with the promyelinating state, transcriptional regulators for the immature undifferentiated state have not yet been defined. From our K-means analysis of INC ALL genes, we found that Sox2, FoxD3, and Ets1 had expression patterns similar to markers of immature Schwann cells (e.g., Cyclin D1 and Cd44).
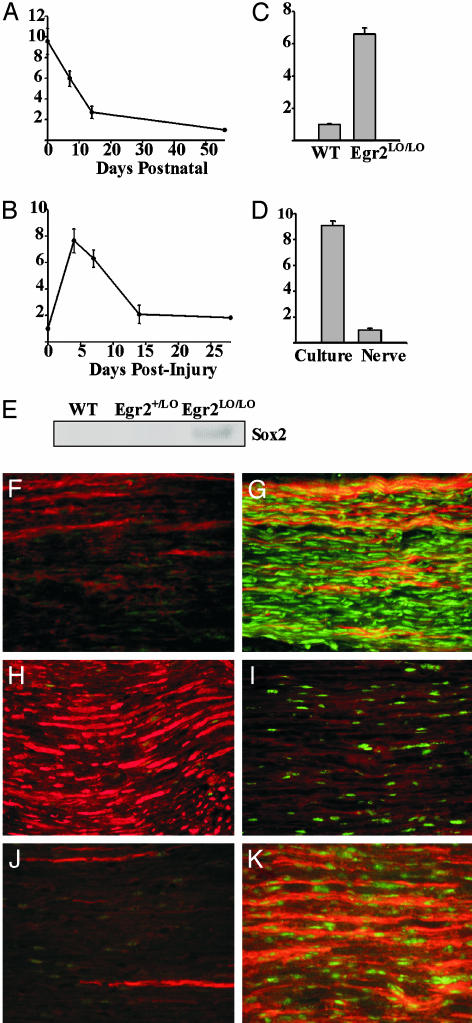
Sox2 is particularly interesting in this regard, because it is expressed in multipotent undifferentiated cells from a variety of tissues, including embryonic epiblasts and neuronal stem cells. In each case, Sox2 expression is down-regulated as these progenitors differentiate, suggesting that it is important for maintaining the undifferentiated state (19–21). From these studies, we hypothesized that Sox2 might also be important for maintaining Schwann cells in the undifferentiated state. Indeed, a recent study of avian neural crest (22) found that Sox2 expression is modulated during development, with down-regulation of Sox2 associated with neuronal commitment and continued Sox2 expression in early immature Schwann cells. We performed quantitative RT-PCR analysis and confirmed that Sox2 has an expression profile associated with the immature Schwann cell state in all nerve paradigms examined (Fig. 4 A–D). Sox2 protein levels were also high in P14 Egr2Lo/Lo nerves compared with age-matched wild-type nerves, which is consistent with the undifferentiated state of the Schwann cells in these CHN nerves (Fig. 4E). Immunohistochemical studies using antibodies to Sox2 and L1, a marker of undifferentiated Schwann cells, further demonstrated that the increased Sox2 expression in Schwann cells of Egr2Lo/Lo CHN sciatic nerve correlated with increased L1 expression (Fig. 4 F and G). Likewise, after crush injury and axonal Wallerian degeneration, as seen by loss of neurofilament staining, Sox2 was also significantly up-regulated in dedifferentiated Schwann cells (Fig. 4 H and I). The increased Sox2 expression after injury again paralleled the induction of L1 (Fig. 4 J and K), thus leading us to conclude that Sox2 is an important marker for immature Schwann cells.
Fig. 4.
Sox2 is preferentially expressed in undifferentiated Schwann cells. Analysis of Sox2 expression by quantitative RT-PCR during Schwann cell development (A), after nerve crush injury (B), in P14 Egr2Lo/Lo CHN Schwann cells (C), and in immature nonmyelinating cultured Schwann cells (D) indicate elevated Sox2 expression in undifferentiated Schwann cells. Sox2 protein (E) is present at higher levels in P14 Egr2Lo/Lo CHN nerves than in the age-matched myelinated wild-type or Egr2+/Lo nerves. In contrast to wild-type nerves (F), Egr2Lo/Lo nerves (G) have increased Sox2 staining (green nuclei) along with increased L1 staining (red), a marker of undifferentiated Schwann cells. In comparison to uninjured adult P56 nerve (H), nerve 4 days after crush (I) demonstrated diminished neurofilament staining (red), and an increased number of Sox2-positive nuclei (green). Additionally, the intact nerve (J) showed minimal L1 (red) and Sox2 (green) staining, whereas nerve 4 days after crush (K) displayed increased L1 (red) and Sox2 (green) staining.
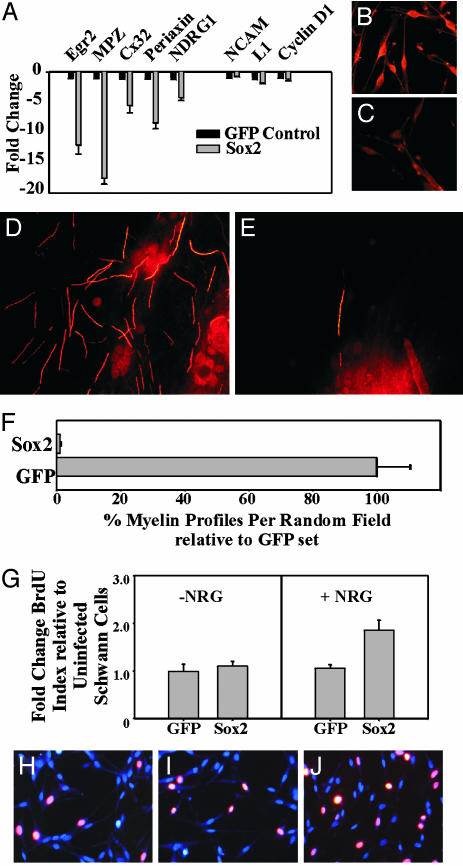
Sox2 Is a Critical Regulator of Schwann Cell Differentiation. Schwann cell progression from the undifferentiated state to the myelinating state requires the down-regulation of immature Schwann cell markers with concomitant up-regulation of genes associated with differentiated Schwann cells. To determine whether Sox2 can influence the expression of genes associated with myelination, we infected Schwann cells cultured in the presence of forskolin (6 μM), a treatment that induces expression of myelination-associated genes (23), with adenovirus-expressing Sox2. Using quantitative RT-PCR, we found that Sox2 suppressed the expression of several myelination-related genes, including Egr2, Mpz, Gjb1, Prx, and Ndrg1 (Fig. 5A). In contrast, genes expressed in undifferentiated Schwann cells, such as L1, Cyclin D1, and Ncam, were not affected by Sox2 overexpression. Additionally, we used Mpz immunostaining to confirm that forskolin-treated Schwann cells infected with adenovirus-expressing GFP have elevated Mpz levels, whereas those infected with Sox2-expressing adenovirus have low levels of Mpz (Fig. 5 B and C).
Fig. 5.
Sox2 inhibits myelination and enhances Schwann cell proliferation. (A) Quantitative RT-PCR after infection of Schwann cells, in the presence of forskolin (6 μM), with GFP or Sox2 adenovirus demonstrated the suppression of differentiation genes (left group), whereas immature marker genes (right group) were not affected. Immunostaining for Mpz in Schwann cells infected with GFP control (B) or Sox2 (C) adenovirus illustrated the decrease in Mpz levels in Sox2-expressing Schwann cells. Myelination in vitro using either control GFP or Sox2 lentivirus-infected Schwann cells showed many myelin basic protein-positive myelin profiles in cocultures containing GFP-expressing Schwann cells (D), but not in cocultures containing Sox2-expressing Schwann cells (E). Quantification of the myelin profiles from three independent in vitro myelination experiments is summarized in F. BrdUrd incorporation assays performed on uninfected, or GFP or Sox2 lentivirus-infected Schwann cells demonstrate similar proliferation rates in the absence of β-neuregulin (G Left). In the presence of β-neuregulin (G Right), uninfected Schwann cells (H) and GFP-expressing Schwann cells (I) showed similar proliferation rates, whereas Sox2-expressing Schwann cells (J) showed a 1.8-fold greater BrdUrd incorporation rate than did GFP-expressing Schwann cells (P = 0.035, Student's t test, data represent three independent experiments). NRG, neuregulin.
Enforced Sox2 expression inhibited the induction of several crucial Schwann cell differentiation genes. We therefore assessed whether Schwann cells constitutively expressing Sox2 could myelinate axons. For these experiments, we produced lentiviruses expressing either Sox2 or enhanced GFP (EGFP) and performed myelination assays in vitro with lentivirus-infected Schwann cells. Schwann cells infected with the EGFP lentivirus effectively myelinated DRG axons, as observed by the presence of many myelin basic protein-immunoreactive myelin profiles (Fig. 5 D–F). However, we rarely detected myelin profiles in cultures containing Schwann cells infected with the Sox2 lentivirus, indicating that they were essentially incapable of myelinating DRG axons. Thus, consistent with its ability to suppress expression of crucial myelination genes such as Egr2 and Mpz, continuous expression of Sox2 in Schwann cells maintained them in the undifferentiated state and prevented them from myelinating axons.
Although Schwann cells infected with the Sox2 lentivirus were unable to myelinate axons, they rapidly populated the cocultures. To determine whether Sox2 affected Schwann cell growth, the proliferative capacity of these Schwann cells was assessed by BrdUrd incorporation (Fig. 5 G–I). When cultured in serum-free defined media, Schwann cells infected with either EGFP or Sox2 lentivirus proliferated at a rate indistinguishable from uninfected Schwann cells (BrdUrd index: uninfected, 7.7 ± 2.1%; GFP-infected, 8.6 ± 3.0%; Sox2-infected, 7.1 ± 1.5%). However, in response to β-neuregulin, a potent mitogenic factor for Schwann cells, Sox2-expressing Schwann cells proliferated twice as fast as did uninfected or EGFP-expressing Schwann cells (BrdUrd index: uninfected, 20.8 ± 4.5%; EGFP-infected, 22.3 ± 4.6%; Sox2-infected, 40.0 ± 6.7%). Thus, enforced expression of Sox2 enhanced the proliferative response of Schwann cells to β-neuregulin stimulation, supporting its role in the maintenance of the immature Schwann cell state.
Discussion
Inherited peripheral neuropathies are common genetic diseases that occur in ≈1 of 2,500 people (24). Mutations in a number of genes, including Egr2, have been associated with these conditions; however, little is known of the molecular alterations underlying these neuropathies. Using a hypomorphic allele of Egr2 to circumvent the neonatal lethality of traditional Egr2-null mice, we generated a longer-lived mouse model of CHN, the most severe form of myelinopathy. Histologically, Egr2Lo/Lo sciatic nerves were very similar to those of CHN patients, with a marked deficiency in myelination of axons from birth and an absence of axonopathy and onion bulb formations, hallmarks that distinguish CHN from other peripheral neuropathies. The molecular alterations in dysfunctional CHN Schwann cells from the Egr2Lo/Lo mice largely reflected their developmental arrest and the role of Egr2 in promoting Schwann cell differentiation (25).
Curiously, however, a number of genes were altered in Egr2Lo/Lo nerves and after nerve injury, but were not differentially regulated during development (Table 5, which is published as supporting information on the PNAS web site). This expression pattern was present, even in the absence of the inflammatory processes that normally occur after nerve injury, perhaps reflecting a general stress-related response in Egr2Lo/Lo Schwann cells that results from a failure to properly enter the differentiation pathway. Significance classification of this subset of genes based on ontology identified a group of downstream effectors of TGFβ1 activation, e.g., Runx2 and Jun (26) (Table 6, which is published as supporting information on the PNAS web site, and Fig. 8B). Indeed, TGFβ1 has demonstrated to be important modulator of Schwann cell phenotype and an activator of Jun (27, 28). A group of genes involved in lipid catabolism, including Abc1 and Cpt1a, was also identified in this subset. Interestingly, mutations in ABC1 result in Tangiers disease, which commonly presents with a peripheral demyelinating neuropathy (29). Likewise, LITAF, which is mutated in patients with demyelinating neuropathy variant CMT1C (30), was also present in this injury-associated set (Fig. 8B). Thus, we identified several injury-related genes in CHN nerves of Egr2Lo/Lo mice; the importance of these genes may be exemplified by the finding that mutations in some of them (e.g., LITAF and ABC1) are found in patients with demyelinating neuropathies.
The molecular events underlying the reversion of Schwann cells to the immature state after crush injury, followed by the subsequent maturation and remyelination of regenerating axons, is commonly believed to recapitulate events that occur during nerve development. However, we found a subset of genes whose expression was altered in Egr2Lo/Lo and developing nerves, but was not modulated after nerve crush injury (Table 7, which is published as supporting information on the PNAS web site, and Fig. 8C). This finding implies the existence of distinct molecular differences between developing Schwann cells and differentiated Schwann cells forced to repeat the maturation process after injury. Interestingly, gene ontology significance classification of these genes (Table 8, which is published as supporting information on the PNAS web site) revealed a category of cyclin and cyclin-dependent protein kinases that included Cyclin D3 and Cdk4. Compensation by Cyclin D3 during development but not after nerve injury may explain why proliferation in Cyclin D1-deficient Schwann cells is impaired after injury, but is normal during development (31, 32). The differential regulation of this subset of development-related genes in Egr2Lo/Lo nerves supports the finding that CHN Schwann cells are halted in maturation, and reflects the molecular disparity between Schwann cells undergoing development versus redifferentiation after injury.
Through the molecular analysis of Egr2Lo/Lo nerves, we identified a number of genes that are potentially crucial for normal Schwann cell function and may be involved in the Schwann cell deficits that characterize CHN. Several of these genes encode transcription factors, such as FoxD3, Ets1, Sox2, Id2, and HMG 1C; thus, they can potentially regulate entire genetic programs. We focused on Sox2 because it was primarily expressed in immature Schwann cells. The Sox (SRY-related high-mobility group box) family of transcription factors are characterized by their DNA-binding high-mobility group domain and are involved in the developmental regulation of numerous cell types (33), including Schwann cells, as for Sox10 (34).
Similar to the role of Sox2 in other systems, where it is involved in maintaining pluripotency (19, 20, 22), Sox2 is preferentially expressed in immature Schwann cells. Its expression is evident early in postmigratory avian neural crest stem cells, and its misexpression in chick neural crest appears to inhibit glial differentiation (22). We have demonstrated that enforced expression of Sox2 inhibits Schwann cell differentiation, promoting increased responsiveness to proliferative stimuli, and preventing myelin gene expression, and thus, the ability to myelinate axons. The mechanism by which Sox2 suppresses expression of genes associated with myelination is still being explored. However, it is possible that Sox2 directly inhibits Egr2 expression, and thereby, suppresses subsequent expression of myelin genes (e.g., Mpz, Gjb1, or Prx). It is also possible that Egr2 is responsible for suppressing Sox2 expression to promote proper differentiation of Schwann cells. For example, in neuronal stem cell differentiation, Sox2 expression is inhibited by prodifferentiation basic helix–loop–helix transcription factors (19).
Whereas Sox2 inhibits differentiation of myelinating Schwann cells, its role in nonmyelinating Schwann cell differentiation is presently unclear. For instance, Sox2 overexpression in cultured Schwann cells did not inhibit expression of nonmyelinating markers, such as NCAM and L1. However, Sox2 immunostaining of adult sciatic nerve did reveal a faint signal in some nuclei that may indicate expression in nonmyelinating Schwann cells, although this staining was not always associated with L1 staining. Further examination of the role of Sox2 in nonmyelinating Schwann cells, and in Schwann cell development in general, must await the generation of mice with Schwann cell-specific Sox2 deletion, because Sox2-null animals are lethal at periimplantation (21).
In this study, we have characterized a mouse model of CHN and have identified a number of potential regulators of Schwann cell development, including Sox2. Understanding the genetic programs they control will provide important insights into normal Schwann cell functions and help identify and characterize abnormalities associated with myelinopathies.
Supplementary Material
Acknowledgments
We thank Tatiana Gorodinsky, Nina Panchenko, and Amber Nielson for technical assistance and members of the Milbrandt laboratory for comments on the manuscript.We thank members of the Siteman Cancer Center Bioinformatics Core for assistance in microarray analysis. This work was supported by National Institutes of Health Grants NS4074 (to J.M.), R37 DK19645 (to R.E.S.), and R01 AG10299 (to R.E.S.).
Author contributions: N.L., R.N., and J.M. designed research; N.L., R.N., J.Y.T.W., and R.E.S. performed research; N.L., R.N., and J.M. contributed new reagents/analytic tools; N.L., R.N., and R.E.S. analyzed data; and N.L., R.N., T.A., R.E.S., and J.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHN, congenital hypomyelinating neuropathy; CMT, Charcot–Marie–Tooth disease; En, embryonic day n;Pn, postnatal day n; EGFP, enhanced GFP; Mpz, myelin protein zero; SCIP, suppressed cAMP-inducible POU.
References
- 1.Warner, L. E., Garcia, C. A. & Lupski, J. R. (1999) Annu. Rev. Med. 50, 263-275. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo, E. J., Bermingham, J. R., Jr., Rosenfeld, M. G. & Scherer, S. S. (1998) J. Neurosci. 18, 7891-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toews, A. D., Barrett, C. & Morell, P. (1998) J. Neurosci. Res. 53, 260-267. [DOI] [PubMed] [Google Scholar]
- 4.Topilko, P., Schneider-Maunoury, S., Levi, G., Baron-Van Evercooren, A., Chennoufi, A. B. Y., Seitanidou, T., Babinet, C. & Charnay, P. (1994) Nature 371, 796-799. [DOI] [PubMed] [Google Scholar]
- 5.Nagarajan, R., Svaren, J., Le, N., Araki, T., Watson, M. & Milbrandt, J. (2001) Neuron 30, 355-368. [DOI] [PubMed] [Google Scholar]
- 6.Warner, L. E., Mancias, P., Butler, I. J., McDonald, C. M., Keppen, L., Koob, K. G. & Lupski, J. R. (1998) Nat. Genet. 18, 382-384. [DOI] [PubMed] [Google Scholar]
- 7.Bellone, E., Di Maria, E., Soriani, S., Varese, A., Doria, L. L., Ajmar, F. & Mandich, P. (1999) Hum. Mutat. 14, 353-354. [DOI] [PubMed] [Google Scholar]
- 8.Botti, S., Pareyson, D., Sghirlanzoni, A., Nemni, R., Riva, D. & Taroni, F. (1998) Am. J. Hum. Genet. 63, A352. [Google Scholar]
- 9.Timmerman, V., De Jonghe, P., Ceuterick, C., De Vriendt, E., Lofgren, A., Nelis, E., Warner, L. E., Lupski, J. R., Martin, J. J. & Van Broeckhoven, C. (1999) Neurology 52, 1827-1832. [DOI] [PubMed] [Google Scholar]
- 10.Nagarajan, R., Le, N., Mahoney, H., Araki, T. & Milbrandt, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8998-9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verheijen, M. H., Chrast, R., Burrola, P. & Lemke, G. (2003) Genes Dev. 17, 2450-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pham, C. T., MacIvor, D. M., Hug, B. A., Heusel, J. W. & Ley, T. J. (1996) Proc. Natl. Acad. Sci. USA 93, 13090-13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider-Maunoury, S., Topilko, P., Seitanidou, T., Levi, G., Cohen-Tannoudji, M., Pournin, S., Babinet, C. & Charnay, P. (1993) Cell 75, 1199-1214. [DOI] [PubMed] [Google Scholar]
- 15.Swiatek, P. J. & Gridley, T. (1993) Genes Dev. 7, 2071-2084. [DOI] [PubMed] [Google Scholar]
- 16.Poliak, S., Matlis, S., Ullmer, C., Scherer, S. S. & Peles, E. (2002) J. Cell Biol. 159, 361-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bermingham, J. R., Jr., Shumas, S., Whisenhunt, T., Sirkowski, E. E., O'Connell, S., Scherer, S. S. & Rosenfeld, M. G. (2002) J. Neurosci. 22, 10217-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kury, P., Koller, H., Hamacher, M., Cornely, C., Hasse, B. & Muller, H. W. (2003) Mol. Cell. Neurosci. 24, 1-9. [DOI] [PubMed] [Google Scholar]
- 19.Bylund, M., Andersson, E., Novitch, B. G. & Muhr, J. (2003) Nat. Neurosci. 6, 1162-1168. [DOI] [PubMed] [Google Scholar]
- 20.Graham, V., Khudyakov, J., Ellis, P. & Pevny, L. (2003) Neuron 39, 749-765. [DOI] [PubMed] [Google Scholar]
- 21.Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N. & Lovell-Badge, R. (2003) Genes Dev. 17, 126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakamatsu, Y., Endo, Y., Osumi, N. & Weston, J. A. (2004) Dev. Dyn. 229, 74-86. [DOI] [PubMed] [Google Scholar]
- 23.Morgan, L., Jessen, K. R. & Mirsky, R. (1991) J. Cell Biol. 112, 457-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupski, J. R. (1997) Hosp. Pract. 32, 83-84, 89-91, 94-95. [DOI] [PubMed] [Google Scholar]
- 25.Zorick, T. S., Syroid, D. E., Brown, A., Gridley, T. & Lemke, G. (1999) Development (Cambridge, U.K.) 126, 1397-1406. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. S., Kim, H. J., Li, Q. L., Chi, X. Z., Ueta, C., Komori, T., Wozney, J. M., Kim, E. G., Choi, J. Y., Ryoo, H. M. & Bae, S. C. (2000) Mol. Cell. Biol. 20, 8783-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awatramani, R., Shumas, S., Kamholz, J. & Scherer, S. S. (2002) Mo. Cell. Neurosci. 19, 307-319. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson, D. B., Dong, Z., Bunting, H., Whitfield, J., Meier, C., Marie, H., Mirsky, R. & Jessen, K. R. (2001) J. Neurosci. 21, 8572-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remaley, A. T., Rust, S., Rosier, M., Knapper, C., Naudin, L., Broccardo, C., Peterson, K. M., Koch, C., Arnould, I., Prades, C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 12685-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Street, V. A., Bennett, C. L., Goldy, J. D., Shirk, A. J., Kleopa, K. A., Tempel, B. L., Lipe, H. P., Scherer, S. S., Bird, T. D. & Chance, P. F. (2003) Neurology 60, 22-26. [DOI] [PubMed] [Google Scholar]
- 31.Atanasoski, S., Shumas, S., Dickson, C., Scherer, S. S. & Suter, U. (2001) Mol. Cell. Neurosci. 18, 581-592. [DOI] [PubMed] [Google Scholar]
- 32.Kim, H. A., Pomeroy, S. L., Whoriskey, W., Pawlitzky, I., Benowitz, L. I., Sicinski, P., Stiles, C. D. & Roberts, T. M. (2000) Neuron 26, 405-416. [DOI] [PubMed] [Google Scholar]
- 33.Kamachi, Y., Uchikawa, M. & Kondoh, H. (2000) Trends Genet. 16, 182-187. [DOI] [PubMed] [Google Scholar]
- 34.Britsch, S., Goerich, D. E., Riethmacher, D., Peirano, R. I., Rossner, M., Nave, K. A., Birchmeier, C. & Wegner, M. (2001) Genes Dev. 15, 66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.