Abstract
Much of the flowering time variation in wild strains of Arabidopsis thaliana is due to allelic variation at two epistatically acting loci, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FLC encodes a MADS (MCM1/AGAMOUS/DEFICIENS/SRF1) domain transcription factor that directly represses a series of flowering-promoting genes. FRI and FLC, however, do not explain all of the observed variation, especially when plants are grown in short days. To identify loci that act in addition to FRI and FLC in controlling flowering of natural accessions, we have analyzed a recombinant inbred line population derived from crosses of accession Niederzenz (Nd) to Columbia, both of which contain natural FRI lesions. Quantitative trait locus mapping and genomic DNA analysis by microarray hybridization were used to identify candidate genes affecting variation in flowering behavior. In both long and short days, the quantitative trait locus of largest effect, termed FLOWERING 1 (FLW1), was found to be associated with a Nd-specific deletion of FLOWERING LOCUS M (FLM), which encodes a floral repressor closely related to FLC. Analysis of near isogenic lines and quantitative transgenic complementation experiments confirmed that the FLM deletion is, in large part, responsible for the early flowering of the Nd accession.
A variety of environmental inputs, both biotic and abiotic, affect the onset of the reproductive phase in plants. Principal among these factors are temperature and light, which vary with geographic location (e.g., latitude) and undergo daily and seasonal cycles. Given their consistent annual patterns, light and temperature are also effective cues for deducing the time of year and hence are key stimuli for the initiation of reproductive development. Indeed, the flowering of many species is accelerated significantly by the longer days that accompany the more favorable growing conditions of spring and summer. Natural variation within these timing mechanisms is expected to provide the necessary phenotypic variation on which selection can act in response to a changing environment.
In creating an effective reproductive program, perhaps equally important as the ability to recognize and respond to a favorable situation is the capacity to prevent flowering in anticipation of unfavorable conditions. One demonstration of such behavior is the distinction between summer and winter annual growth habits seen in many species, including Arabidopsis thaliana. For plants that germinate in late summer or early fall, day-length and temperature may be sufficiently similar to spring to encourage flowering. However, in colder regions, flowering shortly before winter could be futile and would be strongly selected against. Winter annual varieties maintain a molecular brake on floral initiation that is only relieved after winter has passed.
In A. thaliana, inappropriate flowering is prevented primarily through regulation of the potent floral inhibitor FLC. In winter annual accessions of A. thaliana, functional alleles at FRI and its (distant) relatives FRIGIDA-LIKE 1 (FRL1) and FRL2 cause strong expression of FLC (1–3). The MADS domain protein FLC, in turn, represses genes that promote flowering (4). Long periods of low temperature, such as those experienced in winter, lead to epigenetic modifications of the FLC locus that render it insensitive to the positive effects of FRI and related activators, thereby allowing flowering to occur (5–7).
Natural variation at the FRI and FLC loci is widespread within the growing collection of A. thaliana accessions sampled from around the world. Numerous mutations that inactivate FRI and FLC, and thereby cause early flowering, have been identified (8–10). The prevalence of FRI lesions and the characteristics of nucleotide variation across the FRI locus have suggested that FRI may be a target of natural selection (10).
There is also allelic variation at FLC, with weak alleles providing an alternate route to summer annualism. The limited ability of the FLC allele from the accession Landsberg erecta (Ler) to respond to FRI activity or to loss of the so-called autonomous pathway of floral regulators is well known (11, 12). Recently, the attenuated nature of the Ler FLC allele was shown to be caused by an insertion of a transposon-related sequence within the first intron, a large intron that is required for proper transcriptional regulation (13, 14). FLC alleles showing similar attenuation were found in accessions Shahdara and Da(1)-12 (13).
Although the flowering time effects of the FRI/FLC system are dramatic, there is substantial variation apart from the winter versus summer annual distinction (15, 16). We have mapped genomic regions that contribute to differences in flowering time between two A. thaliana accessions that both lack functional FRI alleles. Using a combination of quantitative trait locus (QTL) mapping and hybridization of genomic DNA to microarrays, we have identified a natural deletion of FLOWERING LOCUS M (FLM) as being associated with early flowering of the Nd-1 (Nd, Niederzenz) compared with the Col-3 and Col-5 strains (Col, Columbia). This effect is consistent with knockout and overexpression studies of FLM (17), also called MADS AFFECTING FLOWERING 1 (MAF1) (18), a floral inhibitor that is closely related to FLC.
Materials and Methods
Plant Materials. The complete set of 98 recombinant inbred lines (RILs) derived from crosses of Nd-1 with Col-3 and Col-5 was obtained from the Arabidopsis Biological Resource Center as stock set CS1696 (19, 20). The Col-5 parent carries a glabrous 1 (gl1) mutation and lacks trichomes, whereas Col-3 and Nd-1 both have trichomes. Thus, approximately half of the RILs derived from Col-5 are expected to be glabrous. Four lines (CS1728, CS1765, CS1788, and CS1792) were found to yield both glabrous and nonglabrous plants. Seed and tissue were collected from single plants for all lines, including glabrous and nonglabrous segregants for these four lines, resulting in 102 lines total. After genotyping, six lines were later found to be redundant with other lines (with adjacent stock center numbers) and were consequently merged for further analysis (see below).
Genotyping. All lines from the stock center set [including the glabrous (G) and nonglabrous (T) isolates] were genotyped for a set of 29 PCR-based markers spanning the genome. Using genotypes from 24 of these markers, it was discovered that the following lines had identical genotypes: CS1723 = CS1769, CS1729 = CS1728-G, CS1737 = CS1782, CS1765-G = CS1766, CS1772 = CS1774, and CS1788-G = CS1788-T. These lines also had similar flowering times and morphological phenotypes. As a result of this analysis, the RIL set was found to actually include only 96 different lines.
Ninety-three lines (excluding CS1698, CS1792-G, and CS1796) were selected for single-nucleotide polymorphism (SNP) genotyping with the Sequenom MassARRAY system (21), performed by Genaissance Pharmaceuticals. SNP discovery data were provided by Magnus Nordborg (http://walnut.usc.edu/2010/2010.html). Marker data for all markers are provided in the supporting information, which is published on the PNAS web site. Additional information is available from the authors upon request.
Additional PCR markers for genotyping the FLM region were F22K20 (amplified with oligonucleotides 5′-TTTTTGGTGAGATTTTAAGCCC and 5′-ATATCTCCATCGCTGCAACC), FLM (see supporting information), and the CAPS (22) marker T14N5-2 (amplified with oligonucleotides 5′-TGGGAGATGTGCT T T TAGT and 5′-T TCCAGAGAGAGAAATTCC, followed by digest with restriction enzyme DdeI).
Genetic Map. Genotype data for the 96 different RILs generated by PCR or MassARRAY were analyzed together with mapmaker/exp 3.0 (23). All marker orders were as expected, given the known locations of the markers on the pseudochromosomes of the Col reference sequence (24). Map distances were then obtained from mapmaker by using the Kosambi map function.
Growth Conditions. Seeds were imbibed in 0.1% Phytagar (Invitrogen), sown directly onto soil, and grown at 22°C in growth rooms. Long-day conditions consisted of 16 h of light provided by a 3:1 mixture of Cool White and Gro-Lux (Sylvania) fluorescent bulbs, followed by 8 h of darkness. Short days (SD) were 9 h of light.
Two pots of six plants for each RIL and four pots of six plants for the parental lines were grown in each condition. To minimize environmental variation, flats were rotated across and between shelves on a daily basis and all pots were randomized across all flats several times over the course of the experiment.
Phenotyping. Flowering time was measured both as total leaf number (TLN) and days to flowering (DTF). Rosette leaf number (RLN) and cauline leaf number (CLN) on the main shoot were determined independently and added to yield the TLN before formation of the first flower. DTF was recorded as the number of days from sowing until the macroscopic appearance of the first floral buds. For fine mapping and analysis of transgenic plants, only DTF was recorded, to simplify data collection and allow better seed set for later analysis. Best linear unbiased estimates (BLUPs) were obtained for DTF by accounting for pot effect in both environments. Growth rate (leaves per day) was estimated by subtracting log(TLN) from log(DTF) for each plant and calculating BLUPs. Leaf ratio (CLN/RLN) was estimated by subtracting log CLN from log RLN and calculating BLUPs (see supporting information).
QTL Mapping. All QTL analyses were performed by using standard interval mapping in bqtl (http://hacuna.ucsd.edu/bqtl). Significance thresholds for DTF in long days (LD) and SD were obtained from 1,000 permutations of each data set (25). For both environments, the 5% logarithm of odds threshold of 2.64 was applied. All results presented were performed with untransformed data. Similar results were obtained when analyzing log-transformed data. The following markers were selected as suggestive QTL for further analysis: FLM, M2.36, gen7327, and gen7750. A two-dimensional genome scan was performed, testing models with and without epistatic terms (26) in both LD and SD and with thresholds set by permutations. In LD, weak evidence for epistasis was identified between markers at 30 and 50 cM on chromosome 1 [≈20% false discovery rate (FDR)], but this was not included in the final QTL model (see supporting information).
Construction of Near Isogenic Lines (NILs) and Fine Mapping. An NIL containing the Nd-1 FLW1 region introgressed into a Col-5 background was created by backcrossing RIL CS1705 twice to Col-5. BC2 plants (BC, backcross) were genotyped for several markers around FLW1 as well as microsatellite markers on chromosomes 2–5. A line that was heterozygous around FLM and homozygous for the Col alleles at the other markers was backcrossed two additional times to Col-5, selecting for heterozygosity at the FLW1 region. One BC4 plant was selfed to obtain the final NIL. Progeny homozygous for the FLW1 region were used for transformation with a genomic FLM fragment from Col.
Likewise, for fine mapping, an NIL heterozygous for the FLW1 region from Col introgressed into the Nd-1 background was constructed. RIL CS1712 was backcrossed twice to Nd-1. Microsatellite markers were used to identify plants homozygous for Nd-1 alleles at as many loci as possible outside the FLW1 region. An appropriate line was backcrossed two more times, again selecting for heterozygosity in the FLW1 region. One BC4 plant was selfed, and the progeny was genotyped for many markers in the FLW1 region, ultimately leading to the establishment of lines 88 and 820. Progeny from these plants was grown in SD, phenotyped, and genotyped at F22K20, FLM, and T14N5-2.
Transgenic Experiments. The FLM genomic region was isolated from BAC F22K20 as an 8.8-kb XhoI/Asp718 fragment and subcloned into pBluescript (Stratagene). A BamHI/Asp718 fragment was cloned into a derivative of pPZP212 (27), pJHA212, a binary vector lacking viral promoter and enhancer sequences surrounding the multiple-cloning site (kind gift of Ji Hoon Ahn, Korea University, Seoul). Transformants were selected on 0.7% Phytagar plates supplemented with 0.5× Murashige and Skoog Minimal Organics Medium without sucrose (Invitrogen) and 50 μg/ml kanamycin. After 10 days, transgenic plants and unselected control plants were transplanted to soil.
Results
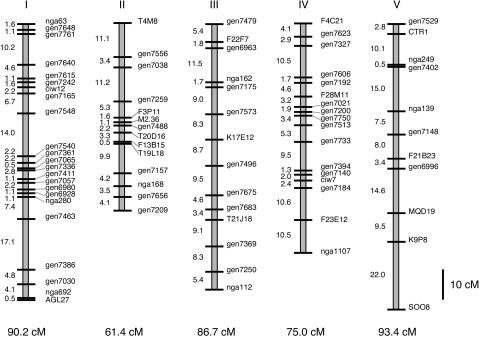
Genetic Map. In total, the 96 Nd/Col RILs were genotyped at 79 loci (Fig. 1). The average distance between markers is 5.5 cM, and the largest distance between markers is a 22-cM gap at the bottom of chromosome 5, with the total genetic distance covered being 408 cM. The relative genetic lengths of chromosomes 1, 2, and 4 are similar to that seen for the Col/Ler and Cvi/Ler RIL genetic maps (28, 29). Chromosome 3 is relatively longer, and chromosome 4 relatively shorter, than in the other RIL sets.
Fig. 1.
Genetic map of for Nd/Col RIL population. The 79 molecular markers are described in the supporting information.
The population shows severe segregation distortion for several regions of the genome, most dramatically at the bottom of chromosome 1, where nearly three times as many lines are homozygous for Nd-1 alleles compared with homozygotes for Col alleles (see supporting information). Other regions with departures from the expected frequencies are the top of chromosomes 2, the top of chromosome 4, and chromosome 5 at marker MQD19 (P < 0.05). All combinations of markers that were not physically linked were tested for linkage. No evidence for linkage disequilibrium was found, as may be expected if synthetic lethality was the cause of the pairwise segregation distortion.
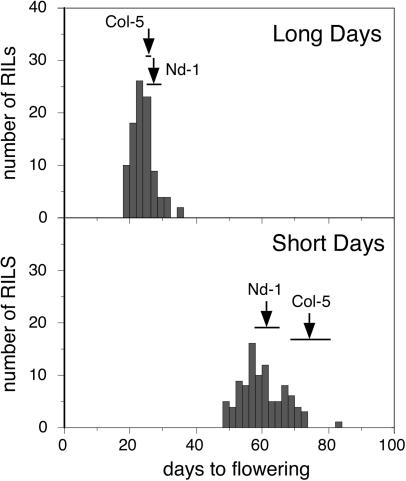
Flowering-Time Variation in Nd/Col RILs. The flowering times of 96 RILs along with Nd-1 and Col-5 parents were measured in LD and SD (Fig. 2). In LD, the parents flowered at about the same time, with Col-5 being slightly earlier than Nd-1. The RILs, however, showed substantial transgression, with many lines having flowering times more extreme than the parents. In SD, however, Nd-1 flowered much earlier than Col-5. Furthermore, although there were many lines with flowering times significantly earlier than the early parent Nd-1, only one line, CS1710, flowered later than the late parent Col-5.
Fig. 2.
Distribution of flowering times for Nd/Col RILs. Flowering time was measured for the 96 lines as DTF in LD (Upper) and SD (Lower). Arrowheads and horizontal bars indicate parental means and two standard deviations, respectively.
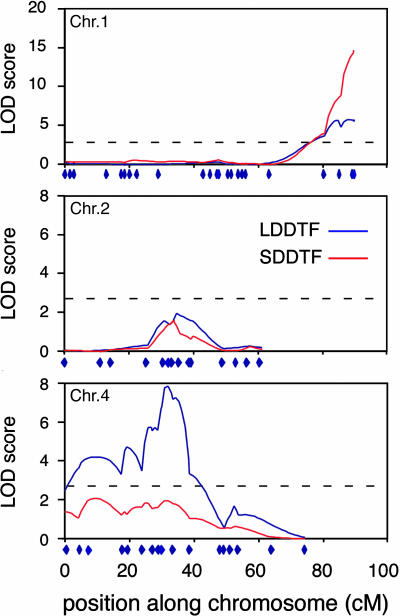
Identification of Flowering-Time QTL. Phenotype and genotype data were used to generate QTL maps for different flowering-time-associated traits (Fig. 3) in LD and SD (see Materials and Methods for details). Using a significance threshold (P < 0.05) of 2.64, established by permutation, we detected four distinct QTL on chromosomes 1, 2, and 4 (Table 1). The QTL with the largest effect was found on the bottom of chromosome 1 and named FLOWERING 1 (FLW1). Homozygous Col alleles at this locus delayed flowering by 3 days in LD and 10 days in SD relative to homozygous Nd alleles (Table 1). Col alleles at a minor QTL (35 cM on chromosome 2) also delayed flowering, with a larger effect in SD. In contrast, Col alleles at two other minor QTL, which are on chromosome 4 and linked, promoted flowering, with a larger effect in the LD environment. In summary, this pattern of positive and negative QTL effects is consistent with the flowering times of the parents. Significant QTL were also found for growth rate (ratio of TLN to DTF) and partitioning of TLN into rosette and cauline leaves and are presented as supporting information.
Fig. 3.
QTL maps of chromosomes 1, 2, and 4 for DTF in LD (blue) or SD (red). Results from interval mapping using bqtl are shown with logarithm of odds scores on the y axis versus position on the chromosomes on the x axis. Marker positions are indicated at the bottom of each chromosome. The dotted gray line corresponds to a logarithm of odds score threshold of 2.64, which represents a P = 0.05 genome wide threshold set by permutations.
Table 1. QTL effects on flowering time.
| Long-day effects
|
Short-day effects
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Chr. | cM | 2a* | %ft† | %var‡ | 2a* | %ft† | %var‡ |
| FLM | 1 | 98 | -3.1 | -13.2 | 26.6 | -9.9 | -16.5 | 62.3 |
| M2.36 | 2 | 36 | -0.9 | -3.9 | 2.4 | -2.7 | -4.4 | 4.4 |
| gen7327 | 4 | 7 | 1.4 | 5.7 | 4.9 | 2.0 | 3.3 | 2.5 |
| gen7750 | 4 | 32 | 2.4 | 10.0 | 15.2 | 1.4 | 2.3 | 1.2 |
2a represents the estimated phenotypic effect associated with the replacement of 2 Col alleles by 2 Nd alleles at the QTL
Allele effect on flowering time (2a) divided by the RIL mean
Variance explained by each QTL
Identification of an FLM Deletion in Nd-1 by Comparative DNA Hybridization. To identify candidate genes for flowering-time QTL, we turned to comparative hybridization of genomic DNA to oligonucleotide arrays designed for expression studies (30). Genomic DNA from both parents was labeled by random-priming and hybridized to Affymetrix ATH1 arrays. Approximately 10,000 (FDR < 5%) of the >200,000 features showed stronger hybridization in Col than in Nd-1 and were designated as single-feature polymorphisms.
We decided to focus on genes that showed an excess of single-feature polymorphisms, as candidates for genes that are either particularly polymorphic or partially or even completely deleted in Nd-1. (Because the ATH1 array was designed based on the Col reference sequence, the reverse approach was not possible.) A linear-clustering algorithm was used to find strings of consecutive polymorphic features in individual genes (30). The resulting list of 210 potential deletions or highly polymorphic genes was then compared with the positions of flowering-time QTL.
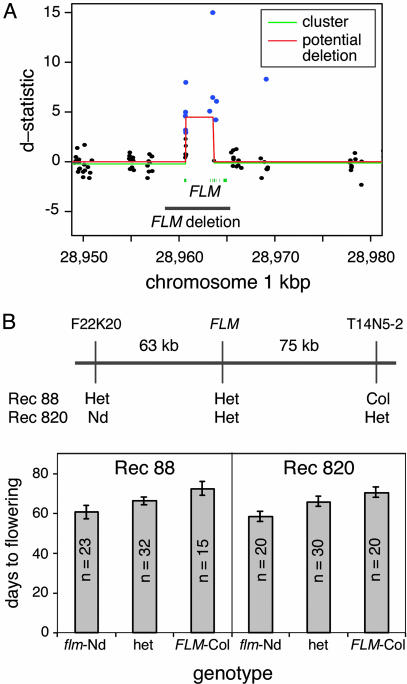
A strong candidate gene for the major FLW1 QTL was revealed by an accumulation of single-feature polymorphisms in FLM (Fig. 4A). FLM encodes a close homolog of FLC, for which natural variation was already known (1, 2, 8, 13). In addition, overexpression and knockout studies in several strains have shown that FLM inhibits flowering in both LD and SD (17, 18). The long- and short-day responses observed for the flm-1 and flm-2 knockout alleles in the Ws background (17) are very similar to those estimated for FLW1 (Table 1).
Fig. 4.
Deletion of the FLM locus in Nd. (A) Results of linear-clustering algorithm based on hybridization of labeled genomic DNA from Nd-1 and Col to ATH1 expression arrays. Closed circles represent d-statistics for each feature; single-feature polymorphisms are shown in blue, and nonsignificant features are shown in black. The lines correspond to clusters of features (green) that qualify as potential deletions (red) (see ref 31). The FLM deletion in Nd-1, verified by sequencing, is indicated by a black line. FLM exons are shown in blue. The deleted sequence is flanked by parallel repeats of GTATAAT. (B) Fine mapping of FLW1 QTL. Genotypes at markers F22K20, FLM, and T14N5-2 are shown for two plants with recombination events flanking FLM (see Materials and Methods). Flowering time means were derived for progeny of recombinants grown in SD and genotyped at FLM. Error bars denote 95% confidence intervals. Regression of DTF on the number of Col alleles showed the additive allele effect (2a) to be 12.1 days (P < 10-7) in Rec820 and 11.5 days (P < 10-7) in Rec88; dominance effects could not be rejected.
We sequenced various PCR products of the FLM genomic region from Nd-1, showing that the sequences corresponding to the FLM probe set on the ATH1 array were deleted. Compared with Col, Nd-1 harbors a 6,817-bp deletion that removes the entire transcribed region of FLM (Fig. 4A). The results of DNA gel-blot analysis confirmed this deletion and showed no evidence for other copies of the FLM coding region in the Nd-1 genome (data not shown). As would be expected, no FLM transcript is detectable in Nd-1 by RT-PCR, whereas it can be readily amplified from Col (data not shown). Considering the similar effects of the FLW1 QTL and flm mutations on flowering time, FLM is a very strong candidate for the underlying cause of the FLW1 phenotype. Interestingly, the sequence deleted in Nd-1 is flanked by direct GTATAAT repeats in Col, of which only one remains in Nd-1, suggesting that illicit recombination could have played a role in the deletion event. Another possibility is that the original FLM insertion was created by a transposon insertion that created a 7-bp target-site duplication. Transposon-mediated gene evolution has recently been shown to be an important mechanism for creating novel gene function in plants (31).
Cosegregation of FLM and the FLW1 QTL. To support the conclusion that the FLM deletion is causal for the FLW1 QTL, we first tested for tight linkage of the deletion with flowering time. In the process of creating a FLW1-Col NIL, we identified two plants having recombination events within a 138-kb region centered on FLM (Fig. 4B). When grown in SD, progeny from both recombinants segregated for different flowering times. Genotyping showed that plants homozygous for the FLM deletion flowered ≈12 days earlier than those that were homozygous for the Col allele. Heterozygotes had an intermediate behavior, although dominance could not be rejected (Fig. 4B). In both lines, the observed effects of FLM on flowering time were very similar to the FLW1 effect predicted from the QTL studies. In addition, the effects were very similar to those reported for the flm-1 and flm-2 alleles in the Ws background (17).
The Col FLM Allele Delays Flowering. To demonstrate that the Col FLM allele is functional and inhibits flowering, we created transgenic plants in which a genomic fragment of the Col FLM locus had been transformed into the background of a NIL that carried the FLW1-Nd allele in a Col-5 background (Fig. 5). In SD, the mean flowering time for both untransformed Nd-1 plants and the FLW1-Nd NIL was on average 70 days, whereas Col-5 flowered ≈17 days later. As expected for a gene that delays flowering in a dosage-dependent manner, the 74 FLM-Col T1 lines showed wide variation in flowering time. Fifty-five lines flowered within the 6 months of the experiment, with a median flowering time of 98 days (mean = 99 days), which is significantly later than that observed for the untransformed background and the Nd parent (P < 0.0017). These results confirm that the Col FLM is active.
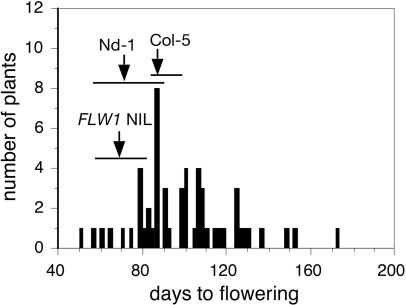
Fig. 5.
Flowering time of 55 T1 lines transformed with the FLM-Col locus grown in SD. Fifty-five of the 74 T1 lines in the FLW1 NIL flowered within the 6 months of the experiment. Flowering time ranges and means of untransformed FLW1 NIL, Nd-1, and Col-5 are indicated by the corresponding horizontal lines and arrows.
The Nd FLM Deletion Is Unique. In contrast to the common FRI deletions found in Col and Ler (9), specific dysfunctional FLC alleles are rare (8, 13, 32). To determine the prevalence of the FLM deletion identified in Nd-1, we analyzed 144 accessions by PCR. None of them shared the Nd allele of FLM, except Nd-0, which is also from Niederzenz. This pattern is similar to that of the EDI allele of CRY2, another large-effect flowering time allele that appears to be limited to accessions originating from the Cape Verde Islands (33).
Discussion
To map loci responsible for flowering time differences between the natural accessions Nd and Col, we genotyped 96 Nd × Col (NdC) RILs for 79 markers. Segregation distortion is not uncommon in A. thaliana RIL populations (28, 29). In the Nd/Col RIL set, we found substantial segregation distortion for several regions of the genome, with the most extreme distortion being at the bottom of chromosome 1, which also comprises the flowering-time QTL with the strongest effect, FLW1. The Nd allele at this QTL causes early flowering, and there was a large excess of plants with Nd alleles in this region. Early-flowering alleles may be expected to increase in frequency during reiterative selfing if there is an unintended bias for the earliest plants. An alternative explanation would be that these regions directly decrease fertility or viability. The absence of interchromosomal linkage disequilibrium, however, indicates that there are no synthetic, viability-decreasing interactions between loci in different regions of the genome.
In LD, Nd-1 and Col-5 flower at about the same time, as measured by both DTF and TLN. However, when grown in SD, Col-5 is much later than Nd-1. Thus, Col-5 shows a more pronounced response to photoperiod than Nd-1. The photoperiod-specific effects of the majority of the QTL mapped in the Nd/Col RIL set, including a large difference in the effect of FLW1, provide a genetic explanation for these differences. Transgression was observed for all combinations of flowering-time-associated traits and environments, indicating that each parent carries loci that positively and negatively influence these traits in both conditions.
We mapped FLW1 to a small interval of 138 kb, spanning 38 annotated genes including FLM on chromosome 1. NILs carrying this region from Nd-1 in an otherwise Col background flowered similarly to Nd-1, confirming that this region is causal for the FLW1 QTL. Additionally, a large percentage of transgenic plants carrying the genomic FLM region from Col flowered significantly later than the untransformed NIL background. Taken together, these results strongly suggest that the FLW1 phenotype is due, at least in large part, to the absence of FLM in Nd-1.
Surveying a large collection of accessions for the Nd-1 FLM deletion identified only one additional accession, also from Nd, that shared the deletion. However, in the process of characterizing the Nd-1 FLM deletion, we identified several other accessions, including Lz-0 and Shahdara, with possible rearrangements of genomic sequences near FLM, as deduced from DNA blot analyses (J.D.W. and D.W., unpublished data). Further analysis revealed the insertion of three different transposable elements upstream of the FLM coding regions in these accessions. That the FLM transcript can be detected in both accessions indicates that the effect of the transposon insertions on FLM activity, if any, is more subtle than that of the Nd deletion. However, a QTL affecting flowering time has been mapped to the same region of chromosome 1 in a cross between Bay-0 and Shahdara (34). In SD, the Bay-0 allele delays flowering relative to the Shahdara allele by ≈6 days, consistent with the Shahdara allele being due to reduced FLM activity.
One other report suggests that FLM polymorphisms may deserve additional attention. While exploring the flowering time roles of the five FLC paralogs (FLM/MAF1, MAF2, MAF3, MAF4, and MAF5), Ratcliffe and colleagues (35) found differences in FLM/MAF1 expression levels among three accessions. These differences may affect the flowering time of these accessions, although it is not known whether the variation is due to cis- or trans-regulatory effects. Recently, the Paf1 complex was identified as a common regulator of the FLC/MAF clade of MADS box genes (36, 37). Because this group of genes contains both activators and repressors of flowering, natural variation in this complex may have complex effects on flowering time.
Supplementary Material
Acknowledgments
We thank the National Science Foundation-supported Arabidopsis Biological Resource Center at Ohio State University and R. Amasino for seed stocks, J. H. Ahn for pJHA212, Christopher Tan and Tsegaye Dabi for technical assistance, and Genaissance Pharmaceuticals for performing genotyping assays. Support for the joint program in quantitative genetics in the laboratories of D.W. and J.C. has come from a National Science Foundation Predoctoral Fellowship (to J.D.W.), funds from the Howard Hughes Medial Institute (HHMI) (to J.C.), National Institutes of Health Grant GM62932 (to J.C. and D.W.), and a core grant from the Max Planck Society (to D.W.). J.C. is an Investigator of the HHMI, and D.W. is a Director of the Max Planck Institute.
Author contributions: J.D.W., J.O.B., N.W., J.C., and D.W. designed research; J.D.W., G.T.T., and N.W. generated data; and J.D.W., J.O.B., J.R.E., J.C., and D.W. analyzed data and wrote the paper.
Abbreviations: Col, Columbia; DTF, days to flowering; LD, long days; Nd, Niederzenz; NIL, near isogenic line; QTL, quantitative trait locus; RIL, recombinant inbred line; SD, short days; TLN, total leaf number.
References
- 1.Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J. & Dennis, E. S. (1999) Plant Cell 11, 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaels, S. D. & Amasino, R. M. (1999) Plant Cell 11, 949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaels, S. D., Bezerra, I. C. & Amasino, R. M. (2004) Proc. Natl. Acad. Sci. USA 101, 3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson, I. R., Shindo, C. & Dean, C. (2003) Annu. Rev. Genet. 37, 371-392. [DOI] [PubMed] [Google Scholar]
- 5.Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A. & Dean, C. (2004) Nature 427, 164-167. [DOI] [PubMed] [Google Scholar]
- 6.Sung, S. & Amasino, R. M. (2004) Nature 427, 159-164. [DOI] [PubMed] [Google Scholar]
- 7.He, Y., Michaels, S. D. & Amasino, R. M. (2003) Science 302, 1751-1754. [DOI] [PubMed] [Google Scholar]
- 8.Gazzani, S., Gendall, A. R., Lister, C. & Dean, C. (2003) Plant Physiol. 132, 1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. & Dean, C. (2000) Science 290, 344-347. [DOI] [PubMed] [Google Scholar]
- 10.Le Corre, V., Roux, F. & Reboud, X. (2002) Mol. Biol. Evol. 19, 1261-1271. [DOI] [PubMed] [Google Scholar]
- 11.Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W. & Peeters, T. (1994) Plant J. 6, 911-919. [Google Scholar]
- 12.Lee, I., Michaels, S. D., Masshardt, A. S. & Amasino, R. M. (1994) Plant J. 6, 903-909. [Google Scholar]
- 13.Michaels, S. D., He, Y., Scortecci, K. C. & Amasino, R. M. (2003) Proc. Natl. Acad. Sci. USA 100, 10102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheldon, C. C., Conn, A. B., Dennis, E. S. & Peacock, W. J. (2002) Plant Cell 14, 2527-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordborg, M. & Bergelson, J. (1999) Am. J. Bot. 86, 470-475. [PubMed] [Google Scholar]
- 16.Hagenblad, J., Tang, C., Molitor, J., Werner, J., Zheng, H., Marjoram, P., Weigel, D. & Nordborg, M. (2004) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 17.Scortecci, K. C., Michaels, S. D. & Amasino, R. M. (2001) Plant J. 26, 229-236. [DOI] [PubMed] [Google Scholar]
- 18.Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L. & Riechmann, J. L. (2001) Plant Physiol. 126, 122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holub, E. B. & Beynon, J. L. (1997) Adv. Bot. Res. 24, 227-273. [Google Scholar]
- 20.Deslandes, L., Pileur, F., Liaubet, L., Camut, S., Can, C., Williams, K., Holub, E., Beynon, J., Arlat, M. & Marco, Y. (1998) Mol. Plant Microbe Interact. 11, 659-667. [DOI] [PubMed] [Google Scholar]
- 21.Tang, K., Fu, D. J., Julien, D., Braun, A., Cantor, C. R. & Köster, H. (1999) Proc. Natl. Acad. Sci. USA 96, 10016-10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konieczny, A. & Ausubel, F. M. (1993) Plant J. 4, 403-410. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Day, M. J., Lincoln, S. E. & Newberg, L. (1987) Genomics 1, 174-181. [DOI] [PubMed] [Google Scholar]
- 24.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 25.Doerge, R. W. & Churchill, G. A. (1996) Genetics 142, 285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borevitz, J. O., Maloof, J. N., Lutes, J., Dabi, T., Redfern, J. L., Trainer, G. T., Werner, J. D., Asami, T., Berry, C. C., Weigel, D. & Chory, J. (2002) Genetics 160, 683-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajdukiewicz, P., Svab, Z. & Maliga, P. (1994) Plant Mol. Biol. 25, 989-994. [DOI] [PubMed] [Google Scholar]
- 28.Lister, C. & Dean, C. (1993) Plant J. 4, 745-750. [Google Scholar]
- 29.Alonso-Blanco, C., Peeters, A. J., Koornneef, M., Lister, C., Dean, C., van den Bosch, N., Pot, J. & Kuiper, M. T. (1998) Plant J. 14, 259-271. [DOI] [PubMed] [Google Scholar]
- 30.Borevitz, J. O., Liang, D., Plouffe, D., Chang, H.-S., Zhu, T., Weigel, D., Berry, C. C., Winzeler, E. & Chory, J. (2003) Genome Res. 13, 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang, N., Bao, Z., Zhang, X., Eddy, S. R. & Wessler, S. R. (2004) Nature 431, 569-573. [DOI] [PubMed] [Google Scholar]
- 32.Caicedo, A. L., Stinchcombe, J. R., Olsen, K. M., Schmitt, J. & Purugganan, M. D. (2004) Proc. Natl. Acad. Sci. USA 101, 15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Assal, S. E.-D., Alonso-Blanco, C., Peeters, A. J., Raz, V. & Koornneef, M. (2001) Nat. Genet. 29, 435-440. [DOI] [PubMed] [Google Scholar]
- 34.Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D. & Daniel-Vedele, F. (2002) Theor. Appl. Genet. 104, 1173-1184. [DOI] [PubMed] [Google Scholar]
- 35.Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J. & Riechmann, J. L. (2003) Plant Cell 15, 1159-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He, Y., Doyle, M. R. & Amasino, R. M. (2004) Genes Dev. 18, 2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh, S., Zhang, H., Ludwig, P. & van Nocker, S. (2004) Plant Cell 16, 2940-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.