Abstract
To help advance the species definition for prokaryotes, we have compared the gene content of 70 closely related and fully sequenced bacterial genomes to identify whether species boundaries exist, and to determine the role of the organism's ecology on its shared gene content. We found the average nucleotide identity (ANI) of the shared genes between two strains to be a robust means to compare genetic relatedness among strains, and that ANI values of ≈94% corresponded to the traditional 70% DNA–DNA reassociation standard of the current species definition. At the 94% ANI cutoff, current species includes only moderately homogeneous strains, e.g., most of the >4-Mb genomes share only 65–90% of their genes, apparently as a result of the strains having evolved in different ecological settings. Furthermore, diagnostic genetic signatures (boundaries) are evident between groups of strains of the same species, and the intergroup genetic similarity can be as high as 98–99% ANI, indicating that justifiable species might be found even among organisms that are nearly identical at the nucleotide level. Notably, a large fraction, e.g., up to 65%, of the differences in gene content within species is associated with bacteriophage and transposase elements, revealing an important role of these elements during bacterial speciation. Our findings are consistent with a definition for species that would include a more homogeneous set of strains than provided by the current definition and one that considers the ecology of the strains in addition to their evolutionary distance.
Keywords: prokaryotic diversity, species concept, nucleotide identity, comparative genomics, evolution
Abacterial species is essentially considered to be a collection of strains that are characterized by at least one diagnostic phenotypic trait and whose purified DNA molecules show 70% or higher reassociation values, following the recommendations in the classical paper by Wayne et al. (1). This species definition, while pragmatic and universally applicable within the prokaryotic world (2–4), has been criticized for being difficult to implement because of technological limitations in identifying diagnostic traits and in performing the pairwise DNA–DNA reassociation experiments, and for being often not adequately predictive of phenotype (5–7). Furthermore, this definition is much broader and is not encompassed by any of the eukaryotic species definitions (8). Indeed, applying this standard to eukaryotic species would lead to the inclusion of members of many taxonomic tribes in the same species, e.g., all of the primates should then belong to the same species (8–10). Last, several strains that show >70% DNA–DNA reassociation values are classified into different species, even different genera, usually on the basis of pathogenicity or host range, such as strains of Escherichia coli and Shigella spp. (11), making the current prokaryotic classification somehow inconsistent.
To gain insight into these issues, we performed pairwise, whole-genome comparisons between all related (i.e., showing >94% 16S rRNA gene identity) sequenced bacterial strains to determine both the conserved predicted protein-coding genes between the pair of strains and the strain-specific genes. We then studied how these parameters correlate with the evolutionary distance between the strains and the strain assignment to species. This analysis is most informative, with respect to the species definition, because it concerns genes that largely determine the organism's phenotype. Further, our strain set represents several major bacterial lineages, including α- and β-proteobacteria, low GC Gram-positive bacilli, streptococci, and staphylococci, and high GC Gram-positive mycobacteria, allowing for robust interpretations. We found that strains of the same species can vary up to 30% in gene content, raising questions as to whether they should belong to the same species, and that these intraspecies differences are presumably driven by differences in the ecology of the strains, lending support for a more ecological and stringent definition for prokaryotic species.
Materials and Methods
Seventy fully sequenced and closely related genomes were used in this study (Table 1, which is published as supporting information on the PNAS web site). The genomic sequences and sequence annotation for 63 of the 70 closed genomes that were published at the time of this study (August 2004) were obtained from National Center for Biotechnology Information's ftp site, which can be accessed at ftp://ftp.ncbi.nih.gov. The remaining seven genomes were closed at the time of this study; however, their annotation was not completed (denoted by NA in Table 1). These seven strains were: Salmonella bognori 12419, Yersinia enterocolitica, Neisseria meningitidis FAM 18, produced by the Sanger Center and obtained through the Sanger ftp site at ftp://ftp.sanger.ac.uk/pub; and Mycobacterium avium, Staphylococcus epidermitidis RP62A, and Clostridium perfrigens (ATCC 13124), produced by The Institute for Genomic Research and obtained through their web site at www.tigr.org. Neisseria gonorrhoeae FA1090 was produced at the Advanced Center for Genome Technology at the University of Oklahoma (Norman; which can be accessed at www.genome.ou.edu/gono.html).
Determination of Conserved Genes and Evolutionary Relatedness. The conserved genes between a pair of genomes were determined by whole-genome sequence comparisons using the blast algorithm, release 2.2.5 (12). For these pairwise comparisons, all predicted protein-coding sequences (CDSs) from one genome (hereafter known as the query genome) were searched against the genomic sequence of a closely related genome (hereafter known as the reference genome). CDSs from the query genome were considered conserved when they had a blast match of at least 60% overall sequence identity (recalculated to an identity along the entire sequence) and an alignable region >70% of their length (nucleotide level) in the reference genome, whereas CDSs that had no match or a match below this cutoff were considered genome-specific in the query genome. The blast search was run with the following settings: x = 150 (drop-off value for gapped alignment), q = -1 (penalty for nucleotide mismatch), and F = F (filter for repeated sequences), and the rest of the parameters were at default settings. These settings give better sensitivity with more distantly related genomes, compared with default settings, because the default settings target more highly identical sequences. The genomes that were used as query genomes, the genome sizes and total number of CDSs for all genomes used is this study, and the raw data from the pair-wise comparisons are summarized in Table 1.
Searching at the amino acid level predicted more conserved genes than the nucleotide level search only when the evaluated strains show less than ≈97% 16S rRNA sequence identity. For this case, there was only a slight upward shift of the left end of the regression line in Fig. 4A. Furthermore, the use of less stringent cutoffs for the determination of conserved sequences did not significantly alter our final conclusions (data not shown). Last, the use of a cutoff for match length and identity without manual inspection of the alignments proved highly accurate for the prediction of conserved sequences. For instance, Parkhill et al. (13) have identified 4,297 and 3,394 CDSs of Bordetella bronchiseptica RB50 to have orthologs in Bordetella parapertussis and Bordetella pertussis, respectively, whereas our approach predicted 4,261 and 3,382 CDSs for the same comparison, respectively.
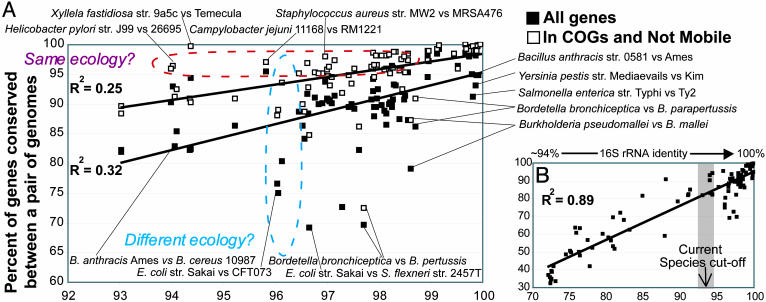
Fig. 4.
Correlation between conserved genes and evolutionary distance for bacterial species. Each data point represents the percent of conserved genes between two strains plotted against their evolutionary distance, measured as ANI of all conserved genes between the strains. Black squares represent all genes and white squares represent the fraction of all genes that are well characterized genes (see Materials and Methods). (A) Only pairs of strains that should belong in the same species, according to the current species definition standard. (B) Pairs of more distantly related strains are also included.
The evolutionary distance between a pair of strains was measured by the average nucleotide identity (ANI) of all conserved genes between the strains as computed by the blast algorithm. Duplicated genes within a genome were defined as the genes that had a better match within their genome than in the reference genome during a pairwise whole-genome comparison, using, in all cases, a minimum cutoff for a match of 60% identity over at least 70% of the length of the query gene. Despite the use of the rather stringent cut-off in these comparisons, cases of independent acquisition of very similar genes (instead of gene duplication) cannot be excluded.
Determination of DNA–DNA Reassociation, 16S rRNA Gene Sequence Identity, and Sequence Divergence. DNA–DNA reassociation values between species were obtained from the literature (11, 14–18). When the sequenced strains were the same as the ones used in the DNA–DNA reassociation experiments, we directly compared the DNA–DNA reassociation values with the ANI of the sequenced genomes. When the strains were different (the majority of cases), we used the average DNA–DNA reassociation and ANI values for several strains of the same species for the comparisons. The 16S rRNA sequence identity between strains was determined as the average identity between all copies of the 16S rRNA gene the strains possess. The 16S rRNA sequence identity was determined by using the phylip package with the Kimura two-parameter method, from the Ribosomal Database Project, which can be accessed at http://rdp.cme.msu.edu/cgis/phylip.cgi (19). Sequence divergence at synonymous (Ks) and nonsynonymous (Ka) sites was calculated with diverge software from the gcg package, which uses the method of Li (20).
CDS Functional Annotation and Intergenic Regions. We obtained more high-level annotation (compared with the annotation found in the GenBank files) of the CDSs in the query genome by using the 20 functional categories in the recently updated Cluster of Ortholgous Genes (COG) database (21). Each COG functional category represents a major cellular process, like transcription, signal transduction, etc. For genomes not incorporated into the COG database, we performed our own CDS assignment to the COG database as described (22). Our assignments were >99% consistent with those already in the COG database when tested for those strains.
CDSs that were assignable to the COG database and were not associated with phage or transposase elements were denoted as well characterized genes. Hypothetical genes were defined in this study as the genes that were not assignable to the COG database and were annotated as hypothetical or unknown functions in the primary annotation (GenBank files), including hypothetical genes carried by phages. This category included the majority (>60%) of the genes not assignable to the COG database and consisted of 10–20% of the total number of annotated genes in a genome. Genes that were annotated as hypothetical in the primary annotation and were assignable to the COG database conserved hypothetical category or other category were considered conserved hypothetical (and well-characterized) and denoted as such in the article.
perl scripts were used to edit CDSs assignments where necessary, extracting sequences from GenBank files, formatting databases for blast searches, and automatically parsing blast outputs.
Results and Discussion
ANI to Measure Genetic Distance. We needed a more precise measure of the genetic relatedness between any two strains. The main limitation to a universal measure for all prokaryotic taxa is the lack of genes that are widely distributed in all taxa, e.g., recent estimates suggest that there are <100 such genes (23, 24). Even these widely distributed genes, however, frequently show conflicting values of genetic relatedness because of their varied evolutionary histories (mutation rate and selection pressures) and the as-yet-unclear and not quantifiable effect of horizontal gene transfer (HGT). For these reasons, and to maximize the robustness of our approach, we used the ANI of all conserved genes between two strains to measure their genetic relatedness. There are several strengths in using ANI for these purposes. First, ANI is a simple, useful, overall descriptor of genetic relatedness. Second, it is derived from lineage-specific genes, in addition to the widely distributed genes (typically >1,000 genes in total), increasing the robustness and resolution of the phylogenetic signal extracted. Furthermore, because of the large number of genes used in the calculations, ANI should be superior to a single gene, such as the 16S rRNA gene, for measuring relatedness, and should not be prone to varied evolutionary rates or HGT events of single genes, or a few genes. Even if genes with different evolutionary histories represent a large fraction of the genome, their effect on AAI is minimized when some evolve faster but others evolve slower than the average of the genome, and hence should not be problematic for ANI. Consistent with this statement, we obtained the same phylogenetic relationship between strains of the same species (for E. coli, nine strains were used, see below) when we used all genes in the genome, and when the analysis was restricted only to genes conserved in all strains, excluding many auxiliary, strain-specific genes (analytical data not shown).
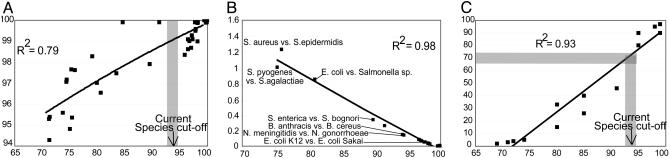
We also found that ANI strongly correlates (R2 = 0.79 for logarithmic correlation) with the 16S rRNA gene sequence identity and can resolve areas where the 16S rRNA gene is inadequate, such as the species level, because a 0–5% 16S rRNA sequence mispairing is spread between 0% and 30% average nucleotide mispairing (Fig. 1A). Furthermore, the average rate of synonymous substitutions shows a tight correspondence to ANI, suggesting that ANI may also be a useful descriptor of the evolutionary distance, in addition to genetic distance, between two organisms (Fig. 1B). Finally, ANI shows a strong linear correlation to DNA–DNA reassociation values, and the 70% DNA–DNA reassociation standard corresponds to ≈93–94% ANI (Fig. 1C), which agrees approximately with previous experimental evidence (reviewed in ref. 25). Therefore, strains that show >94% ANI should belong to the same species, according to the DNA–DNA reassociation standard. This finding was also confirmed by the fact that almost all strains in our set that reside in the same species showed >94% ANI.
Fig. 1.
Relationships between ANI, 16S rRNA, mutation rate, and DNA–DNA reassociation. Each black square represents the ANI of all conserved genes between two strains (x axes) plotted against (y axes) the 16S rRNA sequence identity (A), the average rate of synonymous nucleotide substitutions (B), and the DNA–DNA reassociation values (C) of the two strains. The shaded bar represents 93–94% ANI, which approximately corresponds to 70% DNA–DNA reassociation value, i.e., the species cutoff for prokaryotic species, according to the regression analysis in C. Shown in B is the average rate of synonymous substitutions for all genes in the genome. Comparable results were also obtained when analysis was restricted to genes with no apparent codon biases, or to only fourfold-degenerated sites, as opposed to all sites in a gene (data not shown).
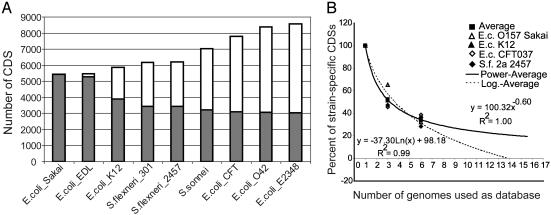
Conserved Gene Core and Genetic Diversity Within a Species. Using the 94% ANI criterion for strain assignment to species, we first attempted to evaluate the extent of genetic diversity within a single bacterial species. Our results for E. coli, the best-sampled species with genomic sequences, show that the number of unique genes in all nine available genomes together exceeds 8,000, whereas the conserved gene core for all nine genomes is only 3,050 genes, which is 55–60% of the genes that most strains of the species possess (Fig. 2A). Results from seven genomes of Salmonella enterica and the five genomes of the Gram-positive Staphylococcus aureus indicate that other species may show extensive genetic diversity as well (Fig. 3). Our analysis of E. coli species also indicates that the number of novel genes in a strain declines with greater coverage of the species with genomic sequences, although the number of available genomic sequences is still too limited to predict how many strains would need to be sequenced to discover most of the gene diversity within the species (Fig. 2B). Nonetheless, extrapolation from the current genomic sequences suggests that when ≈12–15 strains of E. coli are sequenced, the amount of new genes in the next sequenced strain would be <5% of the total CDSs in the genome. However, this prediction might be biased because almost all evaluated strains are pathogens of animal or human hosts, i.e., they have similar ecological niches, and some E. coli are known to colonize water and soil.
Fig. 2.
Conserved gene core vs. genetic diversity within E. coli species. (A) Starting with the 5,447 CDSs in the genome of E. coli O157 strain Sakai, the next bar to the right represents how many unique CDSs in total are found in strain EDL and Sakai together (white bars), and how many of the 5,447 CDSs are conserved in strain EDL (gray bars), etc. Hence, white bars represent the total genetic diversity within species and gray bars represent the conserved gene core for species. (B) All CDSs in a strain (graph label) were searched against a database of an increasing number of genomes. The number of strain-specific CDSs, expressed as a percentage of the strain-specific CDSs when only one genome was used as the database, is plotted against the number of genomes used as the database. The almost identical genomes of E. coli O157 and Shigella flexneri 2a lineages were pooled together so that the seven genomes finally compared showed similar ANIs between each other. The logarithmic and power correlations shown are not statistically different from each other.
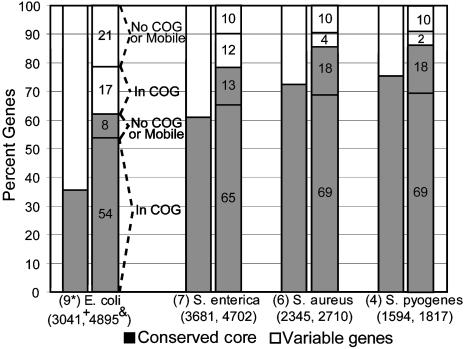
Fig. 3.
Conserved gene core vs. genetic diversity of species. The first column for each species (x axis) shows what fraction of all unique (nonredundant) genes found in all genomes of the species belongs to the species' conserved core (gray part) and what fraction is variable, i.e., not in the core (white part). The second column shows the same distribution for the average strain of the species. The functional annotation of the genes in the average strain of the species is also shown, as exemplified for E. coli. E. coli shows the greatest and Streptococcus pyogenes shows the lowest genetic diversity; note, however, that E. coli genomes are generally more distantly related between each other compared with genomes of the other species, based on ANI measurements (ANI between E. coli genomes is ≈96–97% vs. >98% for the others). *, number of genes used; +, number of genes in the core; &, number of genes in the average strain.
Despite the extensive genetic diversity revealed among closely related bacteria, species-specific diagnostic genetic signatures appear to be identifiable, thus, a species definition for prokaryotes appears meaningful. For example, by comparing the nine E. coli-Shigella spp. genomes against the seven genomes of Salmonella spp. (a close relative of E. coli; the ANI between E. coli and Salmonella spp. genomes is ≈80%), we identified ≈300 genes, i.e., ≈6% of the total genes, in any E. coli-Shigella spp. strain that are not conserved in any Salmonella spp. genome, whereas the reverse comparison revealed in ≈12% of the genes to be Salmonella spp.-specific. Approximately one-half of the genes in these signatures are related to traits that are known to differentiate E. coli-Shigella spp. from Salmonella spp. species; for instance, the E. coli-Shigella contain ≈80 genes involved in transport and metabolism of sugars, amino acids, and oligopeptides, which is consistent with this species' growth on sucrose and production of indole from tryptophan, whereas Salmonella spp. can do neither (11). Likewise, the Salmonella spp. signature-included genes for growth on hydrogen sulfide, which is not used by E. coli-Shigella spp. (11). The other half of the genetic signatures involves genes not assignable to COGs or of general function prediction only, which may yield even more distinguishing phenotypic traits.
The Current Species Definition Appears to Be Too Liberal. The combined data for all 70 compared strains reveal that two-thirds of the strains with >94% ANI differ in at least 5%, and up to 35%, of their total genes, revealing an extensive genetic diversity within a species (Fig. 4A). When a reciprocal best match approach was used to determine the orthologous fraction of the conserved genes in an effort for a more conservative estimation of functional similarity, then the gene differences were even greater (but generally, not considerably greater) by an average of 1.12% (SD = 1.15, MAX = 6.78%). To extend the comparison with higher organisms, only ≈25% of the human genes do not have homologs in the distantly related fish genome, Fugu rubripes (26), whereas the ANI between humans and chimpanzees is 98.7% (27), i.e., much higher than the current standard for prokaryotic species. Therefore, the genetic differences we find among several strains of the same bacterial species are extensive when viewed from a eukaryotic perspective as well. Furthermore, a significant fraction, up to 50%, of the genes that vary within a prokaryotic species is well characterized, as opposed to hypothetical, phage- or transposase-related genes (see Materials and Methods), whose significance on the cell phenotype remains largely unexplored (Fig. 4A). These results, and results from individual species presented previously (Fig. 3), indicate that current species includes only moderately homogeneous strains, perhaps not homogeneous enough for species to be predictive of the phenotype and ecological niche of the strains it encompasses. Several additional lines of evidence support that the genetic differences revealed among strains of the same species are often large enough to justify the description of the strains as different species.
First, the ecology of a species appears to be a distinguishing factor for some species. Pairs of strains that presumably have an overlapping ecological niche, like Xyllela fastidiosa and Helicobacter pylori strains that cause the same disease in closely related plant species and humans, respectively (28, 29), have more genes conserved relative to the mean. On the other hand, pairs of strains that show comparable evolutionary relatedness but presumably have nonoverlapping ecological niches, as with E. coli strains that cause different diseases in humans (enterohemorrhagic vs. uropathogenic) (30), have much fewer genes conserved relative to the mean (Fig. 4A, contrasting dashed ovals). The former cases typically involve obligatory pathogens with small genome sizes, whereas the latter involve free-living or opportunistic pathogens with large genomes. Species with larger genomes are thought to be more ecologically versatile (22); therefore, it is more likely for strains of such species to have evolved within different ecological niches, further supporting the previous interpretations. Furthermore, sexual isolation is more pronounced for obligatory pathogens because of restrictions in their dispersion, as is documented by H. pylori biogeography (28), which may explain why strains of these species show substantial nucleotide divergence, while sharing a very identical gene content.
Second, genetic signatures, like the ones described previously between E. coli-Shigella spp. and Salmonella spp. genomes, are identifiable among some groups of strains that show between 94% and 99% ANI. For example, the two pathogenic genomes of the S. enterica pathovar Typhi share ≈325 genes that are not conserved in any of the three pathovar Typhimurium, strain PT2 and Salmonella gallinarum strain 287/91 genomes (ANI between the Typhi genomes is >99% between Typhi genomes and the ANI for others is 97–98.5%) (see Fig. 6A, which is published as supporting information on the PNAS web site). Many of the Typhi-specific genes are potential pathogenicity factors, such as fimbrial and exported polysaccharide gene clusters, further supporting the ecological importance of this genetic signature. These extensive gene differences may also indicate that Typhi strains do not directly compete with the other S. enterica strains in situ (i.e., they exploit a different ecological niche); otherwise, the genetic differences should have been purged by natural selection. The lack of competition between two populations is considered strong evidence toward describing the populations as different species by several microbiologists (5, 7). A similar comparison revealed ≈4% of the genes to be Typhimurium-specific, whereas comparable results were obtained for other groups with several sequenced representatives, such as the Listeria monocytogenes and Neisseria spp. Importantly, the E. coli-Shigella spp. and Salmonella spp. genomes compared previously are much more distantly related (i.e., ≈80% ANI) than the genomes compared here; nonetheless, the genetic signatures revealed are comparable in size.
Moreover, in at least two cases in our data set, we could not identify species-specific genetic signatures when applying the current definition. For instance, there are two strains of Bacillus cereus fully sequenced, strains ATCC 10987 and ATCC 14579, with the former showing ≈94% ANI to the Bacillus anthracis strains (thus, albeit marginally, strain 10987 should belong to the same species with B. anthracis, according to current standard) and the latter showing only ≈91% (ANI between the two B. cereus genomes is 91.2%). Strain 14579 however, has more genes conserved with the B. anthracis genomes than does strain 10987, and no genetic signature is identifiable for the B. anthracis strain 10987 group (see Fig. 6B). Such instances prove that the current standard is rather arbitrary and suggest that any species definition (like the DNA–DNA reassociation) that does not consider the ecology of the strains, in addition to their evolutionary distance, is problematic with regard to conserved gene content and phenotypic potential. This finding is also evident by the low correlation observed between conserved gene content and genetic distance over a short evolutionary scale (Fig. 4A). Finally, gene expression, which is another important determinant of organism's phenotype apart from gene presence (27, 31), is likely to be different between strains that show a substantial number of nucleotide substitutions, as between strains that show 94–97% ANI. Notably, approximately one-half of the nucleotide substitutions between such strains cause nonsynonymous amino acids substitutions in our data set.
How Should Ecotypes and Species Be Differentiated? If a more stringent definition for species is to eventually be implemented, as this analysis supports, and others have suggested, based on different approaches (5–7), then it remains unclear what may constitute an ecotype. In our view, an ecotype is a population that has acquired a small number of extra genetic elements or mutations, enabling the population to exploit a slightly different ecological niche but preserving the genetic signature and the full ecological potential that characterizes its species. Our genomic comparisons indicate that such ecotypes appear to exist even among strains that show higher than 99% ANI, i.e., among (almost) indistinguishable strains by conventional means. For example, several B. anthracis or S. enterica pathovar Typhi strains that show >99.6% ANI have significant gene differences, primarily involving plasmids and secondarily phage- and transposase-related genes (Fig. 4A). These plasmids have been connected to a strain's ability to cause increased disease symptoms (see, for instance, ref. 32), i.e., they enable the strains to exploit a slightly different but highly overlapping ecological niche compared with their species. Such genetic differences borne as plasmids or mobile elements may not be viewed as genetic signatures that justify a description as a new species because they are not stable properties of the genome. Moreover, the H. pylori and X. fastidiosa strains mentioned previously can also be viewed as ecotypes of the same species that show substantial evolutionary divergence presumably because of their spatial isolation. Notably, an evolutionary species definition, e.g., a multilocus sequence typing-based definition (5, 33), would have probably considered such cases to be different species. This definition, however, may not be realistic from a practical, human-oriented, perspective given that these strains presumably share the same ecological niche and almost identical gene content, whereas it may be realistic from the bacterial perspective because this example could be regarded an allopatric speciation event.
There are a few more complicated cases with respect to speciation in our data set, which can be exemplified by the three pathogenic Bordetella spp. genomes. These organisms that are colonizers of the respiratory tracts of mammals, show 97.8–98.7% ANI between each other's genomes and it appears that B. pertussis and B. parapertussis have evolved by a (considerable) genome reduction from an B. bronchiseptica-like ancestor; presumably as a result of population bottlenecks or ecological specialization because these genomes show increased host specificity compared with Bordetella bronchoseptica (13) (see Fig. 4A). However, no clear and ecologically meaningful genetic signature is identifiable for B. pertussis or B. parapertussis to support their description as separate species because the genes specific to these two genomes are limited or are of hypothetical and/or transposase function. Viewing these genomes as ecotypes of B. bronchiseptica would deviate from the rule that an ecotype should preserve the full potential of its species because B. bronchiseptica has at least 600 additional genes compared with B. pertussis or B. parapertussis. One possibility is that the latter genomes represent snapshots of an active speciation process that might have not yet reached the stage of a diagnosable species-specific genetic signature. Alternatively, such instances indicate that some species are likely to show a continuum/gradient of genetic diversity, rather than defined boundaries diagnosable by species-specific genetic signatures, or that one should look for species-specific signatures at a different level, e.g., the gene expression level or deletion (instead of acquisition) of specific pathways to achieve ecological specialization. Examples like that of Bordetella also indicate that species might be found, even among strains that show >99% ANI if the populations have undergone major ecological constraints.
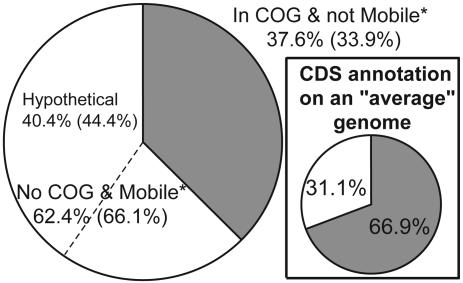
Functional Biases in the Genome-Specific Genes. The functional annotation of the genes that constitute the genome-specific genes in all of the pairwise comparisons between the 70 strains used in this study was also evaluated to provide insights into the factors that might foster speciation. We found that hypothetical, phage-, and transposase-associated genes comprise 62.4% of the genome-specific genes, with the hypothetical genes comprising the majority, 40.4%; the former percentage becomes even larger, 66.1%, when the analysis is restricted to strains of the same species (Fig. 5). The former results contrast with an average of 31.1% of hypothetical, phage-, and transposase-related genes in a typical genome, indicating that hypothetical, phage-, and transposase-related genes might play a more important role in the speciation process than expected, based on the frequency at which these genes are encountered in the genome. These genes are, however, largely species- or genome-specific (see also Fig. 3), revealing a weak positive selection for these functions. Collectively, this information is congruent with phage and mobile elements being ephemeral intruders of the genome and have little, if any, value for the cell, but that occasionally might be important, e.g., when carrying ecologically important genes, and lead to speciation (for examples, see ref. 34).
Fig. 5.
Functional distribution of genome-specific CDSs from 90 pairwise, whole-genome comparisons. Results using only strains showing >94 ANI are shown in parentheses. (Inset) Mean functional distribution of annotated CDSs for the 64 genomes deposited in GenBank as of October 2003. *Mobile, phage- or transposase-associated genes.
The fraction of the genome-specific genes that is well characterized is, on average, 37.6%, which contrast with an average of 69.9% of such genes in a typical genome (Fig. 5). Restriction of the analysis to orthologous genes (i.e., reciprocal best match approach vs. one-way match approach) did not significantly affect these results. Finally, gene duplication appears to play a significant but not major role in the genetic diversity within species. The occurrence of duplicated genes among the genome-specific genes during comparisons of strains of the same species ranged from <1% to 30%, and this variation appeared to be species-dependent.
Perspective
While the current data set is rather limited compared with the total breadth of prokaryotic diversity and is biased toward pathogenic species, it is large enough to reveal some major trends in, and impressions about, the prokaryotic species definition. Most importantly, it reveals that a more stringent and natural definition for prokaryotic species that should be flexible to accommodate the ecological distinctiveness of the organisms, is more appropriate than the current definition because such a definition would be more reasonably predictive of the phenotype and ecological potential of species. Although a larger data set should be analyzed to more knowledgeably guide standards for a new species definition, the current data set indicates that these standards could be as stringent as including only strains that show a >99% ANI or are less identical at the nucleotide level, but share an overlapping ecological niche, should belong to the same species because such strains show minimum gene differences, e.g., <5% of their well characterized genes differ. This standard is closer to eukaryotic standards as well. It would, however, be impractical to implement because it would instantaneously increase the number of existing species probably by a factor of 10 (2), and cause considerable confusion in the diagnostic and regulatory (legal) fields. Furthermore, our analysis does show that the existing classification system is congruent with the current genomic information in that almost all strains that should belong to the same species, according to the current DNA–DNA reassociation standard, share at least 80% of their well characterized genes (Fig. 4A). Hence, we feel that the existing system is serviceable while the necessary new information is obtained for new standards.
Our analysis also reveals several issues that must be addressed before more robust interpretations are possible. To obtain a large enough data set, we had to pool comparisons between strains of different genera, resulting in discontinuities in the results. Therefore, a better sampling of species with genomic sequences is still needed to reject, for instance, the hypothesis that there is a continuum of genetic diversity, as opposed to species-specific genetic signatures, which would not be supportive of a prokaryotic species definition. Finally, there is inadequate knowledge on the population sizes and activities in the natural environments of most (even the sequenced!) species, and hence, the quantification of the effect of ecology on the conserved gene content is not currently feasible. Studying natural populations at the genomic level and over time will also allow us to more fully evaluate the importance of mobile elements in the process of bacterial speciation. Nonetheless, with the rapid growth in genome information and the emerging evolutionary and ecological insight, the necessary data for a more appropriate prokaryotic species definition should be possible in the near future.
Supplementary Material
Acknowledgments
We thank The Institute for Genomic Research and the Sanger Center for permission to use preliminary sequence data and several reviewers for helpful comments. This work was supported by the Bouyoukos Fellowship Program (K.T.K.), the Department of Energy's Microbial Genome Program, the Ribosomal Database Project (supported by the Department of Energy, the National Science Foundation, and the National Institutes of Health), and the Center for Microbial Ecology.
Abbreviations: ANI, average nucleotide identity; CDS, protein-coding sequence; COG, Cluster of Ortholgous Genes.
References
- 1.Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., Moore, L. H., Moore, W. E. C., Murray, R. G. E., Stackebrandt, E., et al. (1987) Int. J. Syst. Bacteriol. 37, 463-464. [Google Scholar]
- 2.Brenner, D., Staley, J. & Krieg, N. (2000) Bergey's Manual of Systematic Bacteriology (Springer, New York).
- 3.Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kampfer, P., Maiden, M. C. J., Nesme, X., Rossello-Mora, R., Swings, J., Truper, H. G., et al. (2002) Int. J. Syst. Evol. Microbiol. 52, 1043-1047. [DOI] [PubMed] [Google Scholar]
- 4.Rossello-Mora, R. & Amann, R. (2001) FEMS Microbiol. Rev. 25, 39-67. [DOI] [PubMed] [Google Scholar]
- 5.Cohan, F. M. (2002) Annu. Rev. Microbiol. 56, 457-487. [DOI] [PubMed] [Google Scholar]
- 6.Staley, J. T. (2004) Microbial Diversity and Bioprospecting (Am. Soc. Microbiol., Washington, DC).
- 7.Ward, D. M. (1998) Curr. Opin. Microbiol. 1, 271-277. [DOI] [PubMed] [Google Scholar]
- 8.Staley, J. T. (1997) Curr. Opin. Biotechnol. 8, 340-345. [DOI] [PubMed] [Google Scholar]
- 9.Sibley, C. G. & Ahlquist, J. E. (1987) J. Mol. Evol. 26, 99-121. [DOI] [PubMed] [Google Scholar]
- 10.Sibley, C. G., Comstock, J. A. & Ahlquist, J. E. (1990) J. Mol. Evol. 30, 202-236. [DOI] [PubMed] [Google Scholar]
- 11.Brenner, D. (1984) Bergey's Manual of Systematic Bacteriology (Williams & Wilkins, Baltimore).
- 12.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids. Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkhill, J., Sebaihia, M., Preston, A., Murphy, L. D., Thomson, N., Harris, D. E., Holden, M. T., Churcher, C. M., Bentley, S. D., Mungall, K. L., et al. (2003) Nat. Genet. 35, 32-40. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura, Y., Hou, X. G., Sultana, F., Miura, H. & Ezaki, T. (1995) Int. J. Syst. Bacteriol. 45, 406-408. [DOI] [PubMed] [Google Scholar]
- 15.Tonjum, T., Welty, D. B., Jantzen, E. & Small, P. L. (1998) J. Clin. Microbiol. 36, 918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vauterin, L., Hoste, B., Kersters, K. & Swings, J. (1995) Int. J. Syst. Bacteriol. 45, 472-489. [Google Scholar]
- 17.Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., Ezaki, T. & Arakawa, M. (1992) Microbiol. Immunol. 36, 1251-1275. [DOI] [PubMed] [Google Scholar]
- 18.Imaeda, T. (1985) Int. J. Syst. Bacteriol. 35, 147-150. [Google Scholar]
- 19.Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., Chandra, S., McGarrell, D. M., Schmidt, T. M., Garrity, G. M., et al. (2003) Nucleic Acids Res. 31, 442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, W. H. (1993) J. Mol. Evol. 36, 96-99. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov, R., Fedorova, N., Jackson, J., Jacobs, A., Kiryutin, B., Koonin, E., Krylov, D., Mazumder, R., Mekhedov, S., Nikolskaya, A., et al. (2003) BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstantinidis, K. T. & Tiedje, J. M. (2004) Proc. Natl. Acad. Sci. USA 101, 3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. (2003) Nat. Rev. Microbiol. 1, 127-136. [DOI] [PubMed] [Google Scholar]
- 24.Santos, S. R. & Ochman, H. (2004) Environ. Microbiol. 6, 754-759. [DOI] [PubMed] [Google Scholar]
- 25.Goodfellow, M. & O'Donnell, A. (1993) Handbook of New Bacterial Systematics (Academic, San Diego).
- 26.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.-M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301-1310. [DOI] [PubMed] [Google Scholar]
- 27.Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. (2002) Science 296, 340-343. [DOI] [PubMed] [Google Scholar]
- 28.Falush, D., Wirth, T., Linz, B., Pritchard, J. K., Stephens, M., Kidd, M., Blaser, M. J., Graham, D. Y., Vacher, S., Perez-Perez, G. I., et al. (2003) Science 299, 1582-1585. [DOI] [PubMed] [Google Scholar]
- 29.Van Sluys, M. A., de Oliveira, M. C., Monteiro-Vitorello, C. B., Miyaki, C. Y., Furlan, L. R., Camargo, L. E. A., da Silva, A. C. R., Moon, D. H., Takita, M. A., Lemos, E. G. M., et al. (2003) J. Bacteriol. 185, 1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch, R. A., Burland, V., Plunkett, G., III, Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S. R., Boutin, A., Hackett, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oleksiak, M. F., Churchill, G. A. & Crawford, D. L. (2002) Nat. Genet. 32, 261-266. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmaster, A. R., Ravel, J., Rasko, D. A., Chapman, G. D., Chute, M. D., Marston, C. K., De, B. K., Sacchi, C. T., Fitzgerald, C., Mayer, L. W., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feil, E. J. (2004) Nat. Rev. Microbiol. 2, 483-495. [DOI] [PubMed] [Google Scholar]
- 34.Boyd, E. F. & Brussow, H. (2002) Trends Microbiol. 10, 521-529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.