Abstract
Establishment of a successful symbiosis between rhizobia and legumes results from an elaborate molecular dialogue between both partners. Bacterial nodulation (Nod) factors are indispensable for initiating plant responses, whereas bacterial surface polysaccharides are important for infection progression and nodule development. The mutant ORS571-oac2 of Azorhizobium caulinodans, affected in its surface polysaccharides, provokes a defective interaction with its host Sesbania rostrata. ORS571-oac2 induced structures with retarded development and continued generation of infection centers and organ primordia, leading to multilobed ineffective nodules. Bacterial development throughout the interaction occurred without major defects. A functional bidirectional complementation was obtained upon coinfection of ORS571-oac2 and a Nod factor-deficient mutant, indicating that the Fix- phenotype of ORS571-oac2-induced nodules resulted from the absence of a positive signal from ORS571-oac2. Indeed, the Fix- phenotype could be complemented by coinoculation of ORS571-oac2 with lipopolysaccharides (LPSs) purified from A. caulinodans. Our data show that Nod factors and LPSs are consecutive signals in symbiosis. Nod factors act first to trigger the onset of the nodulation and invasion program; LPSs inform the plant to proceed with the symbiotic interaction and to develop a functional fixation zone.
Keywords: nodulation, plant–bacterium interaction, polysaccharides
Under conditions of nitrogen limitation, symbiotic rhizobia are able to induce the formation of new organs, the nodules, on roots of leguminous plants. Nodule ontogenesis and bacterial invasion are highly coordinated multistep processes that result from a molecular dialogue that begins with the recognition by the bacteria of plant flavonoids (1). These compounds induce the synthesis of bacterial nodulation (Nod) factors that trigger the onset of plant responses (2, 3).
Nod factors are not the only bacterial signals required for a successful symbiosis. As in other interactions between bacteria and plants or animals (4), bacterial surface polysaccharides (SPSs), especially exopolysaccharides (EPSs) and lipopolysaccharides (LPSs), are believed to play an important role in symbiosis either as structural components or as signaling molecules (1, 5–9).
The water-tolerant tropical legume Sesbania rostrata (10) exhibits versatile nodulation features upon interaction with the microsymbiont Azorhizobium caulinodans (11–13). Nodules can form at the bases of lateral roots of hydroponically grown plants and at bases of adventitious root primordia that are located on the stems. During crack entry invasion, bacteria proliferate in the epidermal fissures at the lateral root bases, and colonize intercellularly the first outer cortical cell layers by inducing local cell death for infection pocket formation (14). Subsequently, infection threads guide the bacteria toward a globular nodule primordium formed upon de novo induced cell divisions in the inner cortex. The central tissue develops into an open basket-shaped structure that is zonated with an apical meristem, an infection zone, and a fixation zone. In the infection zone, bacteria are released and develop into nitrogen-fixing bacteroids that are individually packed in symbiosomes. This transient indeterminate phase of nodule development ends when the meristematic activity ceases. Further growth of the typical globular shape of a determinate nodule is accomplished by cell enlargement (15). Both primordium formation and invasion of the host tissues are Nod factor-dependent processes (2).
The importance of SPSs in the Azorhizobium–Sesbania interaction has been illustrated by the analysis of two A. caulinodans mutant strains, ORS571-X15 and ORS571-oac2 (16, 17), that have truncated LPSs with an altered O-antigen structure. Moreover, strain ORS571-X15 is completely deficient in the production of EPSs, whereas ORS571-oac2 produces EPSs in smaller amounts than the wild type (9, 16–18). After inoculation of adventitious rootlets on S. rostrata stems, ORS571-X15 bacteria form infection pockets in the outer cortex and trigger cortical cell division, but are unable to induce the formation of infection threads and to invade the plant tissues. Nodule development is arrested at the globular primordium stage and the bacteria are restricted to superficial plant cell layers. Coinfection of S. rostrata with ORS571-X15 and a Nod factor-deficient strain ORS571-V44 (19) leads to a functional, but unidirectional, complementation with the formation of Fix+ nodules inhabited solely by ORS571-V44 bacteria (18). Upon inoculation of S. rostrata with ORS571-oac2, Fix- nodules are formed. Nodule development and bacterial invasion are greatly retarded, and a functional central tissue never develops (17).
We report on the fate of the ORS571-oac2 bacteria during the later stages of the interaction with S. rostrata. Our data support the hypothesis that the aberrant nodule phenotype is a consequence of a defective communication between both partners that blocks nodule development. This defect can be rescued by coinoculation with ORS571-V44 and also with purified azorhizobial LPSs, showing that LPSs function as “progression” signals informing the plant to continue the symbiotic process.
Materials and Methods
Bacterial Strains, Growth Conditions, and Plasmids. Bacterial strains and plasmids are listed in Table 1, which is published as supporting information on the PNAS web site. A. caulinodans strains were grown overnight at 37°C in yeast extract broth medium (20).
To localize ORS571-oac2 upon coinoculation and to evaluate its nitrogen-fixing potential, plasmids pXLGD4 (carrying a constitutively expressed lacZ gene) (21) and pRS2002 (carrying a nifH-lacZ fusion) (22) were introduced into ORS571-oac2 by triparental mating with pRK2073 (23) as helper plasmid.
Plant Growth Conditions and Inoculations. S. rostrata Brem seeds were surface sterilized and grown as described (24). After 4 weeks, ≈100 adventitious rootlets were inoculated by painting the stem with a washed bacterial culture (5 × 108 cells per ml) (25). Typically, upon infection with ORS571, ORS571-oac2, and a mixture of ORS571-oac2 and ORS571-V44, all inoculated primordia were reactive. Infection with ORS571-V44 did not trigger any plant response (18). For root inoculations, plants were grown and treated as described (11). For each inoculation, six plants were used per strain, and every experiment was repeated at least once.
Histochemical Stains and Microscopy. Infected root primordia and developing nodules were harvested at different time points for microscopic analysis. Histochemical β-glucuronidase (GUS) staining was performed (18) with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid or magenta-β-d-glucuronic acid as substrates and LacZ staining as described (26). For the double staining of coinoculated tissues, the GUS reaction was done first, after which the material was prefixed overnight, followed by the LacZ reaction. All staining reactions were performed for 5 h.
For light and electron microscopy analyses, the material was embedded in Technovit 7100 (Kulzer Histo-Technik, Wehrheim, Germany), and LR White hard grade (London Resin, Basingstoke, U.K.), respectively. The material was processed as described (18).
LPS Preparation and Coinoculation. LPSs of A. caulinodans were prepared with the hot-phenol water method, purified by size exclusion chromatography, and analyzed by deoxycholate PAGE combined with oxidation and silver staining to visualize the LPS, and with Alcian blue followed by silver staining, but without the oxidation step to address purity as described (27). For coinoculations, 109 bacteria of either ORS571 or ORS571-oac2 were resuspended in LPS solutions of 100 or 10 μg/ml in 2.5 mg/ml deoxycholic acid. Without a preincubation period, per condition, 10-μl spots of these mixtures were locally applied to 50 adventitious root primordia on three individual S. rostrata stems. The experiment was repeated once. Control inoculations were performed on stems with similar mixtures in which either bacteria or LPS were omitted.
For the root inoculations (11), 5 × 107 bacteria were resuspended in 100 μl of a concentrated LPS solution in 2.5 mg/ml deoxycholic acid that was diluted to a final concentration of 100 μg/ml LPS. For both strains, on average, 10 nodules per plant were obtained, of which all were Fix+.
Results
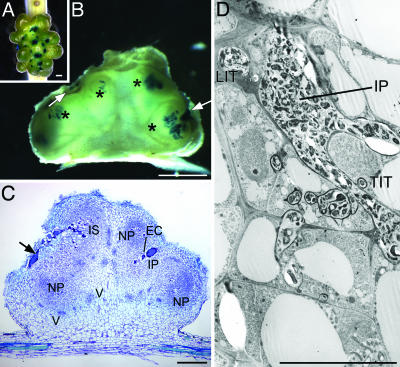
ORS571-oac2 Induces the Formation of Fix- Multilobed Nodules by Reiteration of Early Infection Steps. Mature wild-type nodules on S. rostrata typically retain a globular shape (Fig. 6 A and B, which is published as supporting information on the PNAS web site), whereas ORS571-oac2-induced nodules continued to develop primordia, resulting in large multilobed structures (Fig. 1A). The interaction remained Fix-, as indicated by the absence of leghemoglobin and severe symptoms of nitrogen deficiency (17). To track the bacteria, plants were inoculated with GUS-marked strains. Histochemical staining of wild-type nodules 30 days postinfection (dpi), revealed hardly any bacteria at the surface of the nodules, but a densely colonized fixation zone (Fig. 6A). ORS571-oac2-induced nodules at 60 dpi had multiple blue spots on the surface (Fig. 1 A) that corresponded with colonization of the outermost cortical cells and a limited deeper tissue invasion (Fig. 1B).
Fig. 1.
Multilobed nodules induced by A. caulinodans strain ORS571-oac2 on S. rostrata stems. (A) GUS-stained multiple infection sites (blue dots) in an ORS571-oac2-induced multilobed stem nodule (60 dpi). (B) Hand-cut section through a nodule (30 dpi) with superficial (arrows) and deeper (asterisks) colonization and the absence of a fixation zone. (C) Semithin section through a nodule (21 dpi) showing multiple nodule primordia (NP), an infection pocket (IP) that is surrounded by enlarged cells (EC), a large infection structure (IS) that reaches from the surface deep into the nodule tissues, the nodular vascular bundles (V), and the colonized fissure (arrow). (D) TEM image of an infection pocket (IP) with ramifying thin (TIT) and large (LIT) infection threads. (Bars: 1 mm in A–C and 10 μm D.)
Morphological analysis throughout the development of the ORS571-oac2-induced nodules showed that the initial steps of the interaction followed a delayed, but otherwise normal, pattern of primordium initiation and formation of infection pockets and infection threads (Fig. 6C). Multiple large infection pockets were observed, surrounded by unusually enlarged cells (Figs. 1C and 6 D and E). Eventually, the stage of a young developing indeterminate nodule was reached (at 21 dpi vs. 4 dpi for ORS571) with nodular parenchyma, vascular bundles, and central tissue differentiation to an open basket structure with a nodule meristem and infection zone, but without functional fixation zone. The activity of the nodule meristem was arrested, and the final outcome was a determinate nodule (Fig. 1C). The initial stages were repeatedly reinitiated, resulting in the multilobed shape (Fig. 1 A). Aberrant structures were observed, such as secondary infection pockets formed from enlarged infection threads that originated from primary infection pockets. Occasionally, huge infection structures extended from the surface deep into the nodule tissue (Figs. 1 C and D and 6 D–F). The spatial and temporal occurrence of two early nodulins of S. rostrata analyzed by immunolocalization confirmed the reiteration of early events (Fig. 7, which is published as supporting information on the PNAS web site).
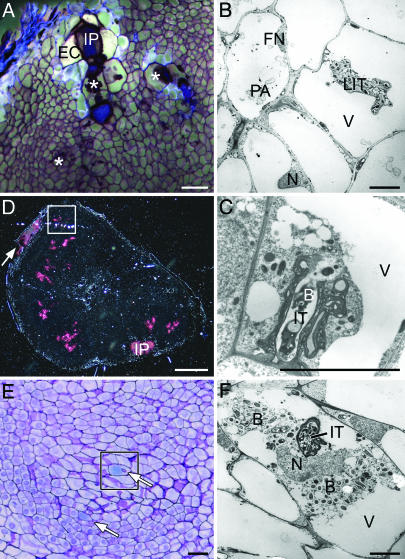
Plant Defense Fails to Restrict ORS571-oac2 Progression. Analysis of sections under UV light revealed unusually large autofluorescing cells surrounding primary and secondary infection pockets and infection threads. Nevertheless, infection centers were not always associated with autofluorescence, because large cells flanking thin infection threads and newly formed secondary infection pockets did not autofluoresce (Fig. 2A and Fig. 8A, which is published as supporting information on the PNAS web site). Moreover, no autofluorescence was seen in plant cells that harbored intracellular infection threads or other infection structures (Figs. 2 A and 8A).
Fig. 2.
Inability of plant defense responses to block infection thread progression and bacterial release. (A) Autofluorescence in enlarged cells that surround a primary infection pocket. Plant cells that harbor infection structures (asterisks) do not autofluoresce (61 dpi). (B) TEM image showing signs of decay, such as protein aggregates (PA) in the vacuole and flattened nuclei (FN) in plant cells that surround cells containing a large infection thread. (C) Release of bacteria from an infection thread in a healthy looking plant cell (46 dpi). (D) Semithin section through a nodule with several infection centers surrounded by diffuse pink-colored GUS staining viewed in dark-field optics (21 dpi); the arrow indicates the colonized fissure. (E) Detail of D showing cyan blue GUS staining in the plant cell cytoplasm (arrows). (F) TEM image of a plant cell similar to the squared cell in E harboring an infection thread and released bacteria. B, bacterium; EC, enlarged cell; IP, infection pocket; IT, infection thread; LIT, large infection thread; N, nucleus; V, vacuole. (Bars: 100 μm in A and E; 5 μm in B, C, and F; and 1 mm in D.)
Autofluorescence in plant cells is often indicative for cell death (28). Indeed, transmission electron microscopy (TEM) showed the occurrence of plant cells with flattened and more granular nuclei and with large vacuoles containing aggregates, one cell layer away from or adjacent to infection structures (Figs. 2B and 8 B and C). This location corresponded to the autofluorescing cells.
Another sign of defense, enlarged infection threads with thickened cell walls and many bacteria, was commonly observed; however, a similar number of infection threads had thin walls and contained a single file of bacteria (Fig. 1D). Often, the infection threads were extremely ramified and budded (Figs. 1D, 6G, and 8 B and C).
Bacteria could be released from infection threads, which was not a rare event (Fig. 2 C–F). Upon nodule induction with a GUS-marked ORS571-oac2 (Fig. 2D), plant cells of the infection zone had cyan blue precipitates in the cytoplasm (Fig. 2E). TEM confirmed the presence of bacteria in plant cells that, atypically, still contained several vacuoles (Fig. 2F).
Signs of defense could be associated with an attempt by the plant to limit bacterial multiplication. Therefore, ORS571 and ORS571-oac2 bacteria were counted at different time points after infection. The kinetics of recovery of viable bacteria differed for the two strains: compared with ORS571, much fewer ORS571-oac2 bacteria could be recovered 4 h after infection, but at later time points the bacterial counts increased steadily. Nevertheless, the highest number of ORS571-oac2 bacteria remained ≈100-fold lower than that of the final wild-type population (Table 2, which is published as supporting information on the PNAS web site). No exponential growth could be observed for ORS571-oac2, which is in agreement with the absence of fixation zones.
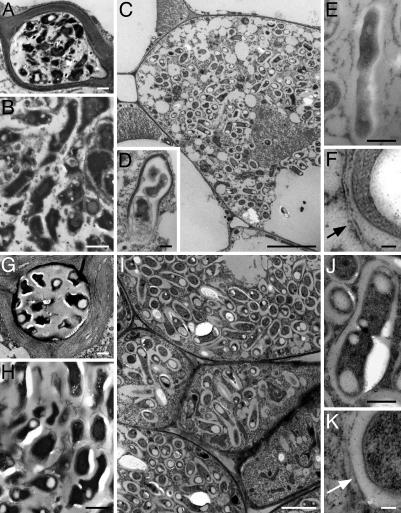
ORS571-oac2 Expresses nifH upon Release but Cannot Completely Fill the Plant Cell. Comparison of the morphology of ORS571-oac2 and ORS571 by TEM showed, for both strains, electron-dense and irregularly shaped bacteria in infection threads and pockets (Fig. 3 A, B, G, and H). During plant cell invasion, both strains recovered a rod shape (Fig. 3 C and I), which was accompanied by decondensation of the electron-dense material (Fig. 3D).
Fig. 3.
Bacteria and bacteroids in ORS571- and ORS571-oac2-induced nodules on S. rostrata stems. (A–F) ORS571-oac2 (46 dpi). (G–K) ORS571 (6 dpi). Bacteria in intercellular infection threads (A and G) and infection pockets (B and H) are not rod shaped. (C and I) Bacterial release in plant cells leading to individually packed symbiosomes. (D) Recovery of the bacterial rod shape upon release in the plant cell. (E and J) Bacterial cell division in infected plant cells. (F and K) Peribacteroid membrane (arrow) bordering the symbiosomes. (Bars: 1 μm in A, B, D, E, G, H, and J; 5 μm in C and I; and 100 μm in F and K.)
Similar to wild-type bacteria, released ORS571-oac2 bacteria were surrounded by a peribacteroid membrane (Fig. 3 F and K) in individually packed symbiosomes (Fig. 3 C and I) and, occasionally, division was observed within the plants cells (Fig. 3 E and J). However, the invaded plant cells were never completely filled with ORS571-oac2 bacteria (Fig. 3C).
The presence of a nifH–lacZ fusion in ORS571-oac2 showed no nifH expression in infection pockets and infection threads. However, dotted staining could be observed in the cytoplasm of plant cells of the infection zone (see below).
Wild-Type LPS Is Required for Nodule Progression. To test the hypothesis that a defective communication between both symbiotic partners is caused by the absence of a signal from the SPS-impaired ORS571-oac2 bacteria, a coinoculation experiment was performed by combining the marked strains ORS571-oac2[pXLGD4] and ORS571-V44[pRG960SD-32], expressing the lacZ and GUS reporter genes, respectively, in a 1:1 ratio. The latter bacteria have a wild-type envelope but are unable to produce Nod factors and, therefore, cannot induce nodule formation (Nod-) (18).
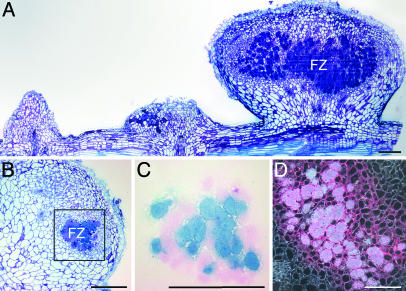
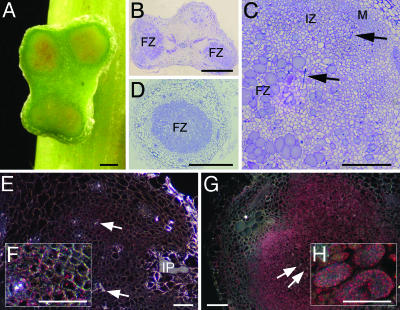
Inoculation of S. rostrata roots or stems with either ORS571-oac2 or ORS571-V44 alone resulted in poorly growing nitrogen-deprived chlorotic plants. Within the same time frame of wild-type infections, coinoculations consistently resulted in a Fix+ phenotype, exemplified by dark-green plants growing vigorously. For 32–40% of the nodules, a Fix+ phenotype was suggested by the presence of leghemoglobin. In most Fix+ nodules, both strains were equally represented (Table 3, which is published as supporting information on the PNAS web site). A fixation zone comparable to that of wild type developed in the Fix+ nodules, whereas in Fix- nodules, typical mutant morphologies were observed (Fig. 4A). The two strains in the cells of the fixation zone were not distributed randomly, as each infected plant cell predominantly contained one of the two azorhizobial mutants (Fig. 4 B and C). Thus, coinoculation of the two mutants on S. rostrata resulted in a bidirectional complementation of the symbiotic defects (Nod factor synthesis and SPS composition).
Fig. 4.
Coinoculation on S. rostrata stems of ORS571-oac2 with ORS571-V44. (A) Three consecutive, infected adventitious root primordia (14 dpi) showing, from right to left, a functional nodule with a clear fixation zone, a nodule with typical features of an ORS571-oac2 phenotype, and a root primordium without obvious signs of development, typical for a ORS571-V44 infection. (B and C) Section through a fixation zone stained with toluidine blue and stained for bacterial reporter gene expression (lacZ ORS571-oac2 in blue and gus ORS571-V44 in pink), respectively, illustrating the presence and distribution of both strains. (D) Section through a Fix+ nodule viewed under dark-field optics with nifH expression of ORS571-oac2 in the fixation zone. FZ, fixation zone. (Bars, 1 mm.)
The coinfection was repeated with an ORS571-oac2 strain carrying a nifH–lacZ fusion. Semithin sections through the Fix+ nodules showed a patchy staining in the fixation zone (Fig. 4D).
Finally, we tested whether the nodulation defect of ORS571-oac2 could be rescued by coinoculation with LPS purified from free-living A. caulinodans. Control inoculations without bacteria or LPSs showed that the deoxycholic acid, in which the LPS was dissolved, had some deleterious effect on the primordia, explaining the low reactivity of the plants. For the wild-type strain, on average, 50% of the inoculated primordia reacted and consistently developed into large Fix+ nodules (data not shown). For ORS571-oac2, only with 100 μg/ml LPS, an average of nine nodules per plant was obtained (18% reactivity), of which 20% were partially Fix+. As shown in Fig. 5A, nodules with a light-red central tissue, indicative for leghemoglobin synthesis, were obtained. The development of these nodules was delayed compared to that of wild type. Morphologically, these nodules differed from ORS571-oac2-induced nodules because no exaggerated infection structures, such as secondary infection pockets and enlarged infection threads, were observed. Large cells completely filled with bacteria reminiscent of cells of a fixation zone were present (Fig. 5 B and C). Bacterial counts from such nodules revealed a colonization level intermediate between that of wild type and ORS571-oac2 (Table 3).
Fig. 5.
Coinoculation on S. rostrata with ORS571-oac2 and purified LPS (100 μg/ml). (A) Pale-red central tissue indicative for the presence of leghemoglobin in a partial Fix+ stem nodule (17 dpi). (B) Semithin section of a stem nodule with a wild-type-like fixation zone. (C) Detail of B showing the zonation of the nodule. (D) Root nodule with wild-type fixation zone. (E) ORS571-oac2[pRS2002] inoculation without LPS (19 dpi); dotted staining from nifH expression visible in bacteria located in the plant cell cytoplasm and not in bacteria present in infection pockets and threads. (F) Detail of E. (G and H) ORS571-oac2[pRS2002] inoculation with LPS (19 dpi); extensive staining from nifH expression in bacteria throughout the infection zone (G) and nifH-expressing bacteria densely colonizing infected plant cells (H). FZ, fixation zone; IP, infection pocket; IZ, infection zone; M, nodule meristem; arrows, infection threads. (Bars: 1 mm in A, B, and D; and 250 μm in C and E–H.)
The LPS coinoculation was repeated with a nifH–lacZ-marked strain. Similar nodules developed that contained plant cells completely filled with bacteria expressing nifH. Overall, nifH expression was much more pronounced than after inoculation with ORS571-oac2 alone (Fig. 5 E–H). The functionality of these nodules was confirmed by coinoculation of roots with LPS and ORS571-oac2. Under this condition, nodule development was delayed compared to that of wild type, but, for both LPS concentrations, the interaction consistently resulted in Fix+ plants with dark-green leaves and root nodules with a dark-red central tissue and a wild-type fixation zone (Fig. 5D). Bacterial counts from such nodules showed a colonization that was comparable to that from the wild type (Table 3). The enhanced complementation level in root vs. stem nodules probably reflects differences in the inoculation conditions.
Discussion
The relative importance of different SPSs in nodule invasion and development depends on the nodule type. For indeterminate nodules, EPSs are essential for infection thread formation, whereas LPSs are required for persistent intracellular colonization. The SPS requirements for determinate nodule formation are just the opposite (5, 6). The mutation in the A. caulinodans strain ORS571-oac2 affects the synthesis of SPSs that are indispensable for the establishment of a functional symbiosis (17). The interaction between ORS571-oac2 and S. rostrata suffers from a strongly delayed progression and is arrested before a fixation zone develops. Developing nodules have a transiently active nodule meristem and an invasion zone where bacterial release occurs from intracellular infection threads. The released bacteria are surrounded by a peribacteroid membrane and express the nifH promoter, but apparently no leghemoglobin is produced in the infected plant cells, which are never completely filled with symbiosomes. The early steps are continuously repeated, an observation corroborated by the continuous expression of two early nodulins, resulting in large multilobed Fix- nodules. The reiteration is also obvious at the level of infection structures.
Based on these observations, we hypothesize that, in the ORS571-oac2–S. rostrata interaction, the absence of a bacterial signal fails to trigger progression of the symbiotic program of the host. As a result, bacteria are released in plant cells that are not ready to support symbiosis. A possible sign of unpreparedness is the absence of leghemoglobin production and the inability of the infected cells to develop into a fixation zone. In support of our hypothesis, a functional symbiosis is established upon mixed infection with two nodulation defective mutants, ORS571-V44 (Nod factor deficient) and ORS571-oac2 (SPS deficient). The successful outcome of the extracellular complementation allowing both mutant strains to inhabit the fixation zone demonstrates that the altered SPSs in ORS571-oac2 are not dominant negative signals that trigger enhanced plant defense. This result also demonstrates that Nod factors as well as components of the bacterial envelope act as consecutive signals that first initiate the nodule developmental program (Nod factor), and then inform the host cells to proceed with that program (SPS). The patchy distribution of both bacterial strains in the cells of the fixation zone suggests that every individual infection thread that releases bacteria contains siblings of only one or a few bacteria. A clonal origin of the bacterial population in invaded cells has been proposed (29), but in the context of the signaling model, it infers that SPSs act as signals “at a distance.” Indeed, plant cells that are predominantly colonized by the ORS571-oac2 bacteria in the coinoculated Fix+ nodules appear like wild-type-infected cells, and the released bacteria express nifH. Hence, the plant cells are prepared to sustain the mutant bacteria, suggesting that the development of a functional fixation zone occurs in a non-cell autonomous fashion. Bidirectional complementation has been reported for the coinoculation of Nod- and EPS- mutants of Sinorhizobium meliloti on alfalfa (5, 30, 31). Similar to our observations, the resulting Fix+ nodules contained a 1:1 ratio of both strains and the spatial distribution differed for every infected cell with sometimes only a single strain present. Thus, EPS would be permanently required during invasion, and a close contact between both mutant strains would be necessary for infection thread formation and progression, but not during subsequent steps of nodule development (31). The contrasting inability of another SPS mutant of A. caulinodans, ORS571-X15, which completely lacks EPS, to invade the nodule tissue upon coinfection with ORS571-V44 most probably originates from the poor fitness of that strain and its extreme sensitivity toward H2O2 (9).
Knocking out the oac2 gene in A. caulinodans decreases EPS production, but more importantly, alters the LPSs. No elaborate structural data are available for the azorhizobial LPSs, but the O-antigen has a lower polymerization level and its sugar composition is changed with a decreased ratio of glucose and rhamnose vs. 3-deoxy-d-manno-2-octulosonic acid. The ORS571-oac2 LPSs partition in the water phase during the hot-phenol extraction procedure, and the bacteria exhibit a higher hydrophobicity (17). The Fix+ phenotype of stem and root nodules induced upon coinoculation of ORS571-oac2 and purified wild-type LPSs proves that LPSs are the signal molecules that are dysfunctional in ORS571-oac2, leading to the miscommunication between both partners. Nodulation is still delayed, but the induction of aberrant structures is completely suppressed in the presence of LPS, and, upon release, the nifH-expressing bacteria can divide efficiently, completely fill the infected cells, and provide the plant with nitrogen. Although effects of purified LPSs on root hairs and the efficiency of wild-type nodulation have been reported (32), our data demonstrate a functional extracellular complementation of an LPS mutant. A signaling function in nodule development has been proposed for LPS, mainly as a suppressor of plant defense (6, 32). Striking similarities exist between the ORS571-oac2 nodules and the indeterminate nodule phenotypes induced by LPS- mutants of S. meliloti (33, 34) and Rhizobium leguminosarum bv. viciae (35) on their hosts: nodulation is delayed, the overall nodule morphology appears normal, bacterial release occurs but without effective colonization of the infected cells, and the outcome of the interaction is a Fix- nodule. However, bacteroid development is disturbed in these interactions, which is not evident for ORS571-oac2.
Inoculations with LPS mutants often enhance plant defense responses resulting in autofluorescence around infection structures (35), necrosis in the central tissue (33), or even killing of the released bacteria (34). Upon infection of S. rostrata with ORS571-oac2, there are apparent defense responses of the plant (17). Autofluorescence is commonly observed around ORS571-oac2-induced infection structures. However, autofluorescence also occurs during wild-type nodulation (data not shown), suggesting that it is a transient feature in normal nodule development. Defense responses do not seem to effectively contain or kill the bacteria in infection structures. Viable ORS571-oac2 cells could be isolated even 4 months after inoculation, and novel nodule primordia formed continuously, a feature that requires bacterial Nod factors produced by healthy bacteria. Instead of being a true sign of plant defense, the autofluorescence associated with ORS571-oac2 might reflect the ongoing formation of infection structures. This finding is supported by a lack of induction of typical defense-related genes, such as chalcone synthase and the pathogenesis-related gene PR1 (unpublished data).
Another sign of defense, the thickening of infection thread cell walls, is thought to affect ramification and efficient penetration of the developing nodule or to physically block the bacteria within the threads (8, 17, 33, 35, 36). However, this response of the plant to ORS571-oac2 does not prevent the progression of the mutant into the nodule primordium and the release of bacteria in plant cells.
We propose that, in the interaction of A. caulinodans ORS571-oac2 with S. rostrata, the following sequence of events occurs. After the host is perceived, Nod factors are produced that are essential for the onset of nodule primordium and infection pocket formation (2). For the successful continuation of invasion and infection thread development, EPSs are essential (9, 18). Because ORS571-oac2 has fewer EPSs, the initiation of infection pocket and thread formation is considerably delayed. In the infection pockets and outgrowing infection threads, EPSs protect the bacteria against H2O2 abundantly produced by the plant in these early steps of nodulation (9, 14). The low recovery of ORS571-oac2 bacteria at early time points and their increasing numbers at later stages of the interaction support the role of EPS in bacterial survival and explain the delayed nodulation. Finally, the bacteria reach the infection zone and are released into the plant cells, indicating that neither of the SPS defects prevent endocytosis. The recovery of a regular rod shape suggests that the intracellular environment is less restrictive and does not require a thick protective EPS layer. In Rhizobium, expression of genes involved in the production of EPS ceases upon release in the host cells (37). During progression into the nodule tissue, LPSs are required as a positive signal toward the plant cells. Therefore, upon release of ORS571-oac2 and activation of nifH expression, no efficient bacterial division occurs and no functional nitrogen fixation can be established, not because of a structural defect of the bacteria, but of the unpreparedness of the plant to continue the symbiotic process. This signaling event is probably related to activation of plant functions, such as leghemoglobin and peribacteroid proteins, that alter the cells of the invasion zone to balance their energy household and allow efficient bacteroid proliferation.
Supplementary Material
Acknowledgments
We thank Koen Goethals and Sofie Goormachtig for critical reading of the manuscript, the referees for their useful comments, R. W. Carlson (Complex Carbohydrate Research Center, University of Georgia, Athens) for help with the preparation and purification of A. caulinodans LPSs, Willem Van De Velde for providing chitinase antibodies, Christa Verplancke for growing plants, and Martine De Cock and Karel Spruyt for help in preparing the paper and the figures, respectively. This work was supported by Interuniversity Poles of Attraction Program-Belgian Science Policy Grant P5/13 and European Commission Research Training Networks Grant HPRN-CT-2000-000984. W.D. was indebted to the European Molecular Biology Organization for a long-term postdoctoral fellowship.
Abbreviations: SPS, surface polysaccharide; EPS, exopolysaccharide; LPS, lipopolysaccharide; GUS, β-glucuronidase; TEM, transmission electron microscopy.
References
- 1.Perret, X., Staehelin, C. & Broughton, W. J. (2000) Microbiol. Mol. Biol. Rev. 64, 180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Haeze, W. & Holsters, M. (2002) Glycobiology 12, 79R-105R. [DOI] [PubMed] [Google Scholar]
- 3.Geurts, R. & Bisseling, T. (2002) Plant Cell 14, Suppl., S239-S249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, H., Baldini, R. L. & Rahme, L. G. (2001) Annu. Rev. Phytopathol. 39, 259-284. [DOI] [PubMed] [Google Scholar]
- 5.Becker, A. & Pühler, A. (1998) in The Rhizobiaceae: Molecular Biology of Model Plant-Associated Bacteria, eds. Spaink, H. P., Kondorosi, A. & Hooykaas, P. J. J. (Kluwer Academic, Dordrecht, The Netherlands), pp. 97-118.
- 6.Kannenberg, E. L., Reuhs, B. L., Forsberg, L. S. & Carlson, R. W. (1998) in The Rhizobiaceae: Molecular Biology of Model Plant-Associated Bacteria, eds. Spaink, H. P., Kondorosi, A. & Hooykaas, P. J. J. (Kluwer Academic, Dordrecht, The Netherlands), pp. 119-154.
- 7.Spaink, H. P. (2000) Annu. Rev. Microbiol. 54, 257-288. [DOI] [PubMed] [Google Scholar]
- 8.Fraysse, N., Couderc, F. & Poinsot, V. (2003) Eur. J. Biochem. 270, 1365-1380. [DOI] [PubMed] [Google Scholar]
- 9.D'Haeze, W., Glushka, J., De Rycke, R., Holsters, M. & Carlson, R. W. (2004) Mol. Microbiol. 52, 485-500. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfus, B., Garcia, J. L. & Gillis, M. (1988) Int. J. Syst. Bacteriol. 38, 89-98. [Google Scholar]
- 11.Fernández-López, M., Goormachtig, S., Gao, M., D'Haeze, W., Van Montagu, M. & Holsters, M. (1998) Proc. Natl. Acad. Sci. USA 95, 12724-12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goormachtig, S., Capoen, W., James, E. K. & Holsters, M. (2004) Proc. Natl. Acad. Sci. USA 101, 6303-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goormachtig, S., Capoen, W. & Holsters, M. (2004) Trends Plant Sci. 9, 518-522. [DOI] [PubMed] [Google Scholar]
- 14.D'Haeze, W., De Rycke, R., Mathis, R., Goormachtig, S., Pagnotta, S., Verplancke, C., Capoen, W. & Holsters, M. (2003) Proc. Natl. Acad. Sci. USA 100, 11789-11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndoye, I., de Billy, F., Vasse, J., Dreyfus, B. & Truchet, G. (1994) J. Bacteriol. 176, 1060-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goethals, K., Leyman, B., Van den Eede, G., Van Montagu, M. & Holsters, M. (1994) J. Bacteriol. 176, 92-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, M., D'Haeze, W., De Rycke, R., Wolucka, B. & Holsters, M. (2001) Mol. Plant-Microbe Interact. 14, 857-866. [DOI] [PubMed] [Google Scholar]
- 18.D'Haeze, W., Gao, M., De Rycke, R., Van Montagu, M., Engler, G. & Holsters, M. (1998) Mol. Plant-Microbe Interact. 11, 999-1008. [Google Scholar]
- 19.Van den Eede, G., Dreyfus, B., Goethals, K., Van Montagu, M. & Holsters, M. (1987) Mol. Gen. Genet. 206, 291-299. [Google Scholar]
- 20.Geremia, R. A., Mergaert, P., Geelen, D., Van Montagu, M. & Holsters, M. (1994) Proc. Natl. Acad. Sci. USA 91, 2669-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leong, S. A., Williams, P. H. & Ditta, G. S. (1985) Nucleic Acids Res. 13, 5965-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminski, P. A., Mandon, K., Arigoni, F., Desnoues, N. & Elmerich, C. (1991) Mol. Microbiol. 5, 1983-1991. [DOI] [PubMed] [Google Scholar]
- 23.Ditta, G., Stanfield, S., Corbin, D. & Helinski, D. R. (1980) Proc. Natl. Acad. Sci. USA 77, 7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goethals, K., Gao, M., Tomekpe, K., Van Montagu, M. & Holsters, M. (1989) Mol. Gen. Genet. 219, 289-298. [DOI] [PubMed] [Google Scholar]
- 25.Goormachtig, S., Valerio-Lepiniec, M., Szczyglowski, K., Van Montagu, M., Holsters, M. & de Bruijn, F. J. (1995) Mol. Plant–Microbe Interact. 8, 816-824. [DOI] [PubMed] [Google Scholar]
- 26.Etchebar, C., Trigalet-Demery, D., van Gijsegem, F., Vasse, J. & Trigalet, A. (1998) Mol. Plant-Microbe Interact. 11, 869-877. [Google Scholar]
- 27.Gudlavaletti, S. K. & Forsberg, L. S. (2003) J. Biol. Chem. 278, 3957-3968. [DOI] [PubMed] [Google Scholar]
- 28.Parrott, D. L., Anderson, A. J. & Carman, J. G. (2002) Physiol. Mol. Plant Pathol. 60, 59-69. [Google Scholar]
- 29.Gage, D. J. & Margolin, W. (2000) Curr. Opin. Microbiol. 3, 613-617. [DOI] [PubMed] [Google Scholar]
- 30.Müller, P., Hynes, M., Kapp, D., Niehaus, K. & Pühler, A. (1988) Mol. Gen. Genet. 211, 17-26. [Google Scholar]
- 31.Kapp, D., Niehaus, K., Quandt, J., Müller, P. & Pühler, A. (1990) Plant Cell 2, 139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dazzo, F. B., Truchet, G. L., Hollingsworth, R. I., Hrabak, E. M., Pankratz, H. S., Philip-Hollingsworth, S., Salzwedel, J. L., Chapman, K., Appenzeller, L., Squartini, A., et al. (1991) J. Bacteriol. 173, 5371-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niehaus, K., Lagares, A. & Pühler, A. (1998) Mol. Plant-Microbe Interact. 11, 906-914. [Google Scholar]
- 34.Campbell, G. R. O., Reuhs, B. L. & Walker, G. C. (2002) Proc. Natl. Acad. Sci. USA 99, 3938-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perotto, S., Brewin, N. J. & Kannenberg, E. L. (1994) Mol. Plant-Microbe Interact. 7, 99-112. [Google Scholar]
- 36.Keating, D. H., Willits, M. G. & Long, S. R. (2002) J. Bacteriol. 184, 6681-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latchford, J. W., Borthakur, D. & Johnston, A. W. B. (1991) Mol. Microbiol. 5, 2107-2114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.