A systemic study of dynamic morphology, cytology, and transcriptome reveals the cytological basis and fine-regulation by multiple factors, especially phytohormones, of rice lamina joint.
Abstract
Rice (Oryza sativa) leaf angle is determined by lamina joint and is an important agricultural trait determining leaf erectness and, hence, the photosynthesis efficiency and grain yield. Genetic studies reveal a complex regulatory network of lamina joint development; however, the morphological changes, cytological transitions, and underlying transcriptional programming remain to be elucidated. A systemic morphological and cytological study reveals a dynamic developmental process and suggests a common but distinct regulation of the lamina joint. Successive and sequential cell division and expansion, cell wall thickening, and programmed cell death at the adaxial or abaxial sides form the cytological basis of the lamina joint, and the increased leaf angle results from the asymmetric cell proliferation and elongation. Analysis of the gene expression profiles at four distinct developmental stages ranging from initiation to senescence showed that genes related to cell division and growth, hormone synthesis and signaling, transcription (transcription factors), and protein phosphorylation (protein kinases) exhibit distinct spatiotemporal patterns during lamina joint development. Phytohormones play crucial roles by promoting cell differentiation and growth at early stages or regulating the maturation and senescence at later stages, which is consistent with the quantitative analysis of hormones at different stages. Further comparison with the gene expression profile of leaf inclination1, a mutant with decreased auxin and increased leaf angle, indicates the coordinated effects of hormones in regulating lamina joint. These results reveal a dynamic cytology of rice lamina joint that is fine-regulated by multiple factors, providing informative clues for illustrating the regulatory mechanisms of leaf angle and plant architecture.
As an important agronomic trait determined by multiple factors, architecture is crucial for crop growth and yields. Leaf erectness, one of the key traits of rice (Oryza sativa) architecture and ideotype (Donald, 1968), will help dense planting, enhance light capture, and improve photosynthesis (Sinclair and Sheehy, 1999; Sakamoto et al., 2006). Leaf angle is determined by the shape of the lamina joint, the portion connecting the leaf blade and sheath (Hoshikawa, 1989). Lamina joint provides a unique path for transportation between the leaf blade and sheath and, additionally, a supporting foundation for leaf erectness and expansion as a mechanical tissue.
The well-ordered cell organization provides the structural basis of lamina joint and abnormal cell constitutions, and division and elongation lead to the altered leaf inclination. Deficiency of lamina joint structure (Lee et al., 2007), suppressed longitudinal elongation of adaxial parenchyma cells, and increased division of abaxial sclerenchyma cells of the lamina joint (Zhang et al., 2009a; Sun et al., 2015) result in leaf erectness. Conversely, increased expansion or proliferation of adaxial parenchyma cells leads to increased leaf inclination (Zhao et al., 2010, 2013; Zhang et al., 2015). Abnormal vascular bundle formation and cell wall composition also result in changed leaf angle (Ning et al., 2011).
Genetic and functional studies indicate the involvement of various factors in the regulation of lamina joint development and leaf inclination, especially phytohormones. Mutants with deficiencies of brassinosteroid (BR) biosynthetic genes, dwarf4-1 (Sakamoto et al., 2006), ebisu dwarf (d2; Hong et al., 2003), and chromosome segment deleted dwarf1 (Li et al., 2013), show reduced leaf inclination, which also is observed in the BR signaling-defective mutant d61 (Yamamuro et al., 2000) or transgenic rice plants with suppressed expression of OsBAK1 (Li et al., 2009) and OsBZR1 (Bai et al., 2007). On the other hand, transgenic rice plants overexpressing sterol C-22 hydroxylase (an important enzyme of BR biosynthesis; Wu et al., 2008), OsDim/dwf1 (which participates in late steps of sterol biosynthesis; Hong et al., 2005), and OsBAK1 (Li et al., 2009) present increased leaf inclination, which is consistent with BR promoting cell elongation at the adaxial side of the lamina joint (Cao and Chen, 1995). A recent study showed that BR signaling inhibits the proliferation of sclerenchyma cells at the abaxial side of the lamina joint by coordinately regulating the U-type cyclin CYCU4;1 (Sun et al., 2015).
Auxin homeostasis and signaling regulate leaf inclination, and gain-of-function mutants or transgenic plants overexpressing GH3 family members, including OsGH3-1, OsGH3-2, OsGH3-5, and OsGH3-13, present reduced auxin levels and enlarged leaf angles (Zhang et al., 2009b, 2015; Du et al., 2012; Zhao et al., 2013). Overexpression of miR393a/b, which suppresses the expression of OsAFB2 or OsTIR1, leads to the enlarged inclination of flag leaf (Bian et al., 2012). Transgenic rice overexpressing OsARF19 shows enlarged leaf inclination due to the increased expression level of GH3s and the decreased free IAA content (Zhang et al., 2015), and overexpression of OsIAA1 results in the increased leaf angle (Song et al., 2009), suggesting the negative effects of auxin in leaf inclination. Interestingly, most auxin-related mutants present changed BR sensitivity (Song et al., 2009; Zhao et al., 2013; Zhang et al., 2015), and the inclination by treatment with brassinolide alone is suppressed by an inhibitor of IAA transport (Cao and Chen, 1995) or promoted by additional IAA (Han et al., 1997), suggesting a complex regulation of phytohormones in lamina joint development. In addition, BR-induced leaf inclination is accompanied by ethylene production and inhibited by an inhibitor of 1-aminocyclopropane-1-carboxylic acid oxidase (Cao and Chen, 1995) or suppressed by treatment with abscisic acid (ABA) and gibberellin (GA) (Han et al., 1997; Tong et al., 2014), further indicating that phytohormones synergistically and antagonistically regulate lamina joint development and leaf inclination.
Transcriptional regulation by transcription factors (TFs) and protein modification by protein kinases are involved in leaf inclination regulation. OsLIGULELESS1 (OsLG1) encodes an SBP domain-containing TF, and mutant oslg1 abolishes the ligule, auricle, and lamina joint (Lee et al., 2007). LAX PANICLE (LAX) encodes a basic helix-loop-helix TF the ectopic expression of which caused excessive bending of rice lamina joint (Komatsu et al., 2003). INCREASED LEAF ANGLE1 (ILA1) encodes a Raf-like MAPKKK of group C, and the ila1 mutant presents increased leaf angle by abnormal vascular bundle formation and cell wall composition in the lamina joint (Ning et al., 2011). In addition, LEAF INCLINATION2 (LC2), a VIN3-like protein, controls leaf inclination by inhibiting adaxial cell division (Zhao et al., 2010). Suppression of small RNA-producing genes, including RNA-dependent RNA polymerase2, RNase III-class Dicer-like3, and OsAGO4a and OsAGO4b (members of the Argonaute family), results in the increased bending of the lamina joint (Wei et al., 2014), while increased expression of OsAGO7 leads to the leaf erectness (Shi et al., 2007).
Although studies have demonstrated the involvement of multiple factors in lamina joint development, the underlying cellular and molecular mechanisms are rarely studied. Previous reports showed that when the leaf blade differentiates from the leaf primordium and grows to less than 1 cm, a hollow space appears at the base of the leaf blade and differentiation and development of the lamina joint initiate (Hoshikawa, 1989). Lamina joint develops along with the leaf blade and sheath, and fully developed lamina joint supports the leaf with an appropriate inclination for light capture and high-efficient photosynthesis. To illustrate the relevant regulatory mechanisms, a systematic study of lamina joint development, including the morphology, cytology, and global transcriptome analyses, was performed, and the results indicate that lamina joint development is well ordered and progressive, with a distinct cytological basis and dynamic processes. Genes associated with transcriptional regulation, signal transduction, and metabolic pathways, especially the biosynthesis and signaling of hormones, are involved in lamina joint development through regulating multiple biological processes, including elongation/division of parenchyma cells, sclerenchyma cell differentiation, vascular development, and programmed cell death (PCD). These results will provide informative clues illustrating the regulatory mechanism of rice lamina joint development and, hence, plant architecture formation.
RESULTS
Cytological Observation Reveals the Distinct Characteristics and Dynamic Development of Rice Lamina Joint
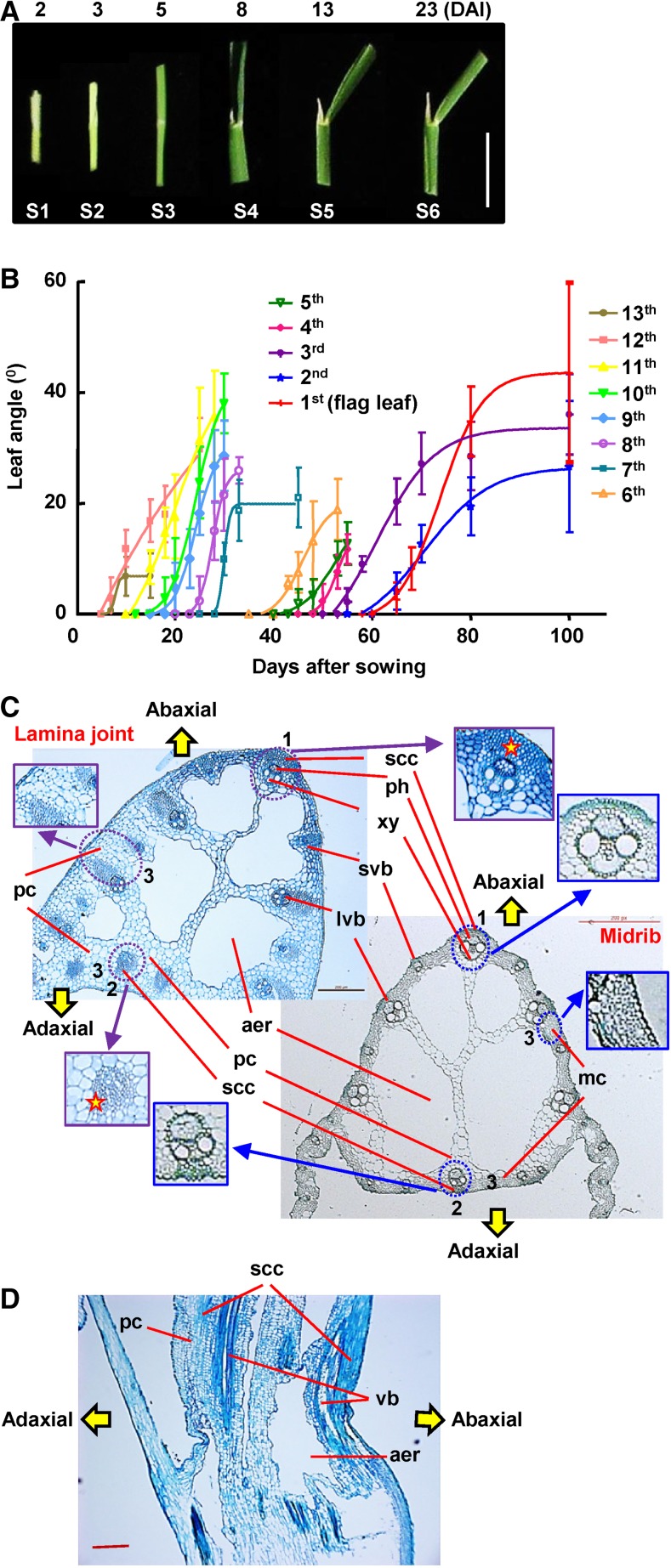
Leaf angle varies among different rice cultivars, and cv Zhonghua11 (ZH11) with intermediate leaf inclination was selected for these studies. Leaf angle increases after the lamina joint emerges from the prior leaf sheath, and daily observation of the dynamic growth shows the six distinct development stages of the lamina joint, from initiation to senescence, that encompass complex developmental and morphological changes (Table I; Fig. 1A). Interestingly, recording the angles of all 13 full leaves showed that, although the final inclination degrees, increasing periods, and maximum slopes of curves vary with each other, a similar and common S pattern, with the maximum increasing rates at stage 5, is observed (Fig. 1B), suggesting the uniformity and distinctness of lamina joint development of different leaves.
Table I. Developmental and cytological changes during lamina joint (LJ) growth.
| Stage | Morphology | Cytology |
|---|---|---|
| S1 (Initiation) | A ligule and a pair of auricles differentiate at the LJ initiation site | No aerenchyma in LJ, which is full with parenchyma cells, except vascular bundles arranged at the abaxial side |
| LJ is hollow and transparent | Small parenchyma cell clusters at the adaxial side and abaxial region between developing vascular bundles | |
| The periphery of abaxial vascular bundles protrudes to form ridges | ||
| S2 (Young) | LJ, leaf blade, and sheath are white or creamy yellow | LJ size increases mainly due to the extensive expansion of parenchyma cells |
| LJ protrudes and becomes larger | PCD of mature large parenchyma cells occurs to form small holes | |
| Longitudinal holes connect and fuse to form intact aerenchyma | ||
| S3 (Young) | Leaf continues to elongate, and leaf blade expands | Rapid cell division between the vascular bundles and epidermis cells at the abaxial domain |
| LJ is still enclosed by the prior leaf sheath | Small parenchyma cell clusters are formed at the adaxial side | |
| Leaf blade and sheath become green, while LJ is white (distinguishable from each other) | Parenchyma cells continue expanding | |
| A pair of horn-shaped auricles and a tongue-like ligule can be observed | Large and small vascular bundles locating at the abaxial side develop continuously | |
| S4 (Maturation) | Leaf blade and sheath fully develop | Thickening of cell walls of small cell clusters results in the formation of sclerenchyma cell clusters |
| LJ emerges from prior leaf sheath and is exposed in the air | Newly formed sclerenchyma cells at both sides perforate through longitudinally to form sclerenchyma fibers | |
| LJ is firm and plump | Vascular bundles and nearby wall-thickened small cell clusters at the adaxial side form ridges along the longitudinal direction | |
| LJ starts to bend | Parenchyma cells stop proliferation | |
| The periphery of whole LJ becomes smooth and the transverse size of the LJ achieves its maximum | ||
| S5 (Postmaturation) | LJ adaxial side elongates and bends to the abaxial side to form a curvature, resulting in the increasing leaf angle | Ongoing thickening of cell walls of small cell clusters on both sides |
| Parenchyma cells at the adaxial side undergo longitudinal elongation | ||
| Asymmetric cell growth between the adaxial and abaxial sides leads to increased leaf inclination | ||
| S6 (Senescence) | LJ approaches the maximum angle and begins to wither due to the loss of water | Larger aerenchyma and fewer layers of parenchyma cells |
| LJ is dehydrated and distorted |
Figure 1.
Morphological and cytological observation of the rice lamina joint. A, Morphology of the lamina joint of rice flag leaf at 2, 3, 5, 8, 13, and 23 d (corresponding to stages 1–6) after lamina joint initiation (DAI). Representative images are shown. Bar = 1 cm. B, Statistics of lamina joint inclination of rice leaves. Angles (°) of each lamina joint at different days after sowing (DAS) were measured and statistically analyzed. Rice leaves from the bottom leaf (13th leaf, the first full leaf) to the flag leaf (first leaf) were analyzed. Experiments were biologically repeated three times, and data are shown as means ± sd (n > 10). C, Comparison of cytology between the lamina joint and midrib of mature leaf (the eighth leaf was used as representative) by cross-section analysis. Highlighted and magnified differences include layers and cell numbers of abaxial sclerenchyma cells (1) and adaxial sclerenchyma cells (2); the mesophyll cells in the midrib were replaced by multiple layers of large parenchyma cells in the lamina joint (3). aer, Aerenchyma; lvb, large vascular bundle; mc, mesophyll cell; pc, parenchyma cell; ph, phloem; scc, sclerenchyma cluster; svb, small vascular bundle; xy, xylem. Bar = 200 μm. D, Longitudinal section of the lamina joint of a mature flag leaf. vb, Vascular bundle. Bar = 200 μm.
Observation of the cytology composition and structure of the lamina joint by cross sections showed that the parenchyma cells constitute the basic structure and abundant sclerenchyma tissues provide the mechanical support of the lamina joint. The ordered vascular bundles, together with large aerenchymas, provide channels for water/nutrient transport and air circulation (Fig. 1C). Structurally, vascular bundles are regularly spaced at both adaxial and abaxial sides and sclerenchyma cells are located at the fringe of vascular bundles. There are several aeration cavities (aerenchymas) inside the lamina joint, and the rest is filled with the parenchyma cells that connect the abaxial large vascular bundles with the adaxial side. Observation of the longitudinal sections shows the oppressed cells at the abaxial side, and sclerenchyma cells form the fibers from top to bottom (Fig. 1D).
Lamina joint connects the leaf blade and sheath and presents a similar anatomic structure but significantly different cell composition from the leaf midrib. Comparison reveals that there are no mesophyll cells but numerous layers of large parenchyma cells instead, and more sclerenchyma cells are seen at both abaxial and adaxial sides of the lamina joint (Fig. 1C), indicating that a distinct cell composition may result in the leaf inclination at the lamina joint.
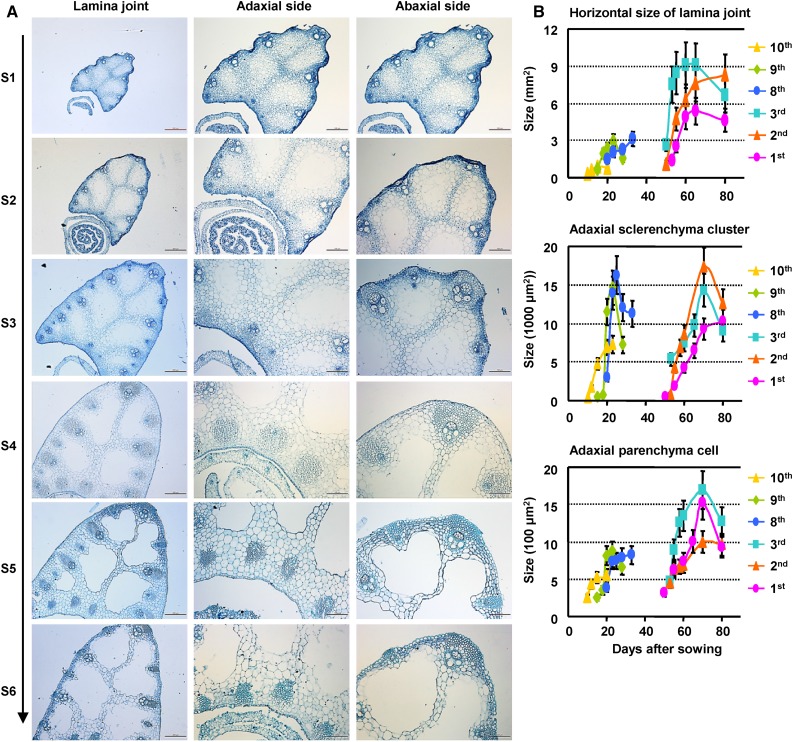
Systemic observation of the lamina joint at different developmental stages using paraffin sections reveals similar progressive and distinct cytological changes according to the morphological classification (stages 1–6; a cross section of the eighth leaf is shown as representative in Fig. 2A; a longitudinal section of a flag leaf is shown as representative in Supplemental Fig. S1). As described in Table I, the fine-controlled division and expansion of parenchyma cells, differentiation of sclerenchyma cells, cell wall thickening, and PCD are crucial for lamina joint development and final leaf inclination. The PCD proceeds continuously in the formation of aerenchyma and vascular bundles and the development of sclerenchyma tissues.
Figure 2.
Cytological dynamics of lamina joint development. A, Cytological dynamics of the lamina joint from initiation to maturation (left; bars = 200 μm), which can be divided into six stages (S1–S6). The lamina joint of the eighth leaf was observed by cross section, and the adaxial side (middle; bars = 100 μm) and the abaxial side (right; bars = 100 μm) were enlarged. B, Statistics of cytological changes of the lamina joint at different developmental stages. Lamina joints of first to third and eighth to 10th leaves were cross sectioned at different DAS, and sizes of horizontal area, adaxial sclerenchyma cluster, and adaxial parenchyma cells were measured and statistically analyzed. Data are shown as means ± sd (n = 3).
Detailed calculation reveals an active proliferation and expansion of cells in the horizontal direction. Different from the S curves of leaf inclination, the sizes of the lamina joint, adaxial sclerenchyma cluster, and parenchyma cells present an increase-decrease tendency (Fig. 2B). Sclerenchyma cells differentiate and develop synchronously at both adaxial and abaxial sides. At stage 5, the abaxial cells stop growing, while the remaining parenchyma cells at the adaxial side undergo vertical elongation (Supplemental Fig. S1), resulting in asymmetric growth between adaxial and abaxial sides and, hence, the increased leaf angle.
Global Transcriptome Analysis Reveals the Involvement of Multiple Biological Processes during Lamina Joint Development
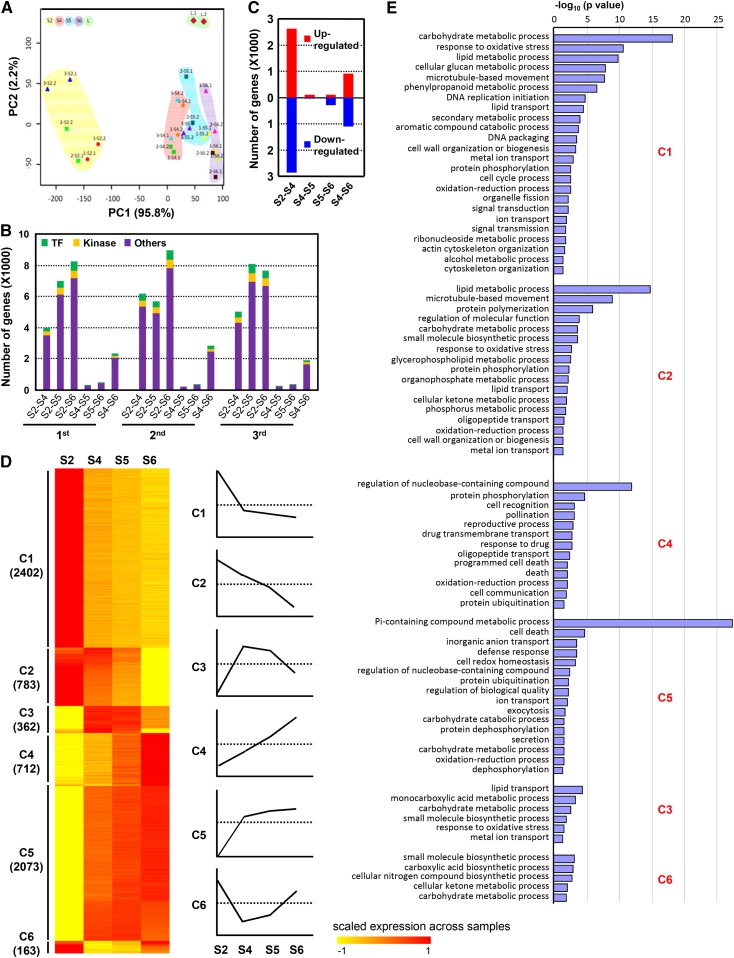
The lamina joints of the last three leaves develop across vegetative and reproductive growth and show typical leaf angle curves (Fig. 1B), and global transcriptome profiles were studied to investigate the underlying molecular mechanisms regulating lamina joint development and leaf inclination. Stages 2, 4, 5, and 6, representing distinct developmental phases designated early initiation, mature, inclination-increasing, and senescence of the lamina joint (Supplemental Fig. S2A), were analyzed and compared with the adjacent leaf parts of the last third lamina joint at stage 4 (defined as leaf) when the lamina joint is fully developed (Supplemental Fig. S2B). Pairwise Pearson correlation coefficient (PCC) analysis reveals the high correlation of gene expression profiles of different lamina joints at the same developmental stage (Supplemental Fig. S2C). The contiguous leaves and developmental stages tend to be more correlated. Stages 4 to 6 are highly correlated (PCC > 0.96), while stage 2 is distinct from the others, consistent with the cytological changes during lamina joint development. As expected, there is low correlation between the lamina joint and the adjacent part (0.88 < PCC < 0.96).
Based on the first two principal components (98% of variation captured), principal component analysis (PCA) additionally showed that expression profiles of the different developmental stages are well ordered and separated by principal component 1 (temporal factor; Fig. 3A), suggesting similar transcriptional regulation of different lamina joints at the same developmental stages. In addition, the adjacent leaf parts were separated from the lamina joint by principal component 2 (spatial factor), which may be due to the tissue specificity.
Figure 3.
Genome-wide gene expression profiles of the rice lamina joint. A, PCA reveals the spatial and temporal features of the examined samples (with repetition). Samples were named as follows: 1-S2.1, first repetition of the lamina joint of the first leaf at stage 2; 3-S4.2, second repetition of the lamina joint of the third leaf at stage 4; L, adjacent leaf parts. PC1 and PC2, Principal components 1 and 2. B, Number of genes showing differential expression between every two developmental stages (S2–S4, S2–S5, S2–S6, S4–S5, S5–S6, or S4–S6) of lamina joints from flag (first leaf), last second, and third leaves. Numbers of genes encoding TFs, kinases, or others are shown. C, Numbers of DEGs between two continuous stages of lamina joints of the last three leaves. D, Heat map of DEGs (identified in B), which can be classified into six distinct patterns (clusters 1–6) by the K-means clustering method. Expression values were scaled per gene across samples and are shown as the scaled expression. Diagrams of the expression patterns are shown (right), and gene numbers of each cluster are indicated. E, Enriched GO process terms for clusters 1 to 6 (from C), which were summarized using REVIGO. Column length indicates the overrepresentation significance of a group of GO terms.
The identification of organ developmental status could be defined by gene expression profiles, the differentially expressed genes (DEGs) were identified by pairwise comparison of the analyzed stages using Limma packages (Wettenhall and Smyth, 2004). The DEG numbers increase along with lamina joint development (Fig. 3B), and those at stages 2 to 4 are much more numerous than those between other contiguous stages, suggesting the complex regulation at stage 2, which is consistent with the cytological characteristics. DEG numbers at stages 4 and 5 or 5 and 6 are considerably low, indicating the similar gene expression profile at these three stages, and stage 5 is a transition period during lamina joint development. In sum, 5,842 (stages 2–4), 161 (stages 4 and 5), 420 (stages 5 and 6), and 2,180 (stages 4–6) DEGs of three lamina joints (based on similar gene expression profiles at the same developmental stage) were identified (Fig. 3C).
Preferential expression of previously reported genes regulating leaf angle, including LG1 (Lee et al., 2007), BU1 (Tanaka et al., 2009), LPA1 (Wu et al., 2013), and CYCU4:1 (Sun et al., 2015), at the lamina joint (Supplemental Fig. S2D) confirms the accuracy of the transcriptome analysis. Genes GRAS19 (Chen et al., 2013), LAZY1 (Li et al., 2007), CYP51G3 (Xia et al., 2015), and GSR1 (Wang et al., 2009) present high expression levels at the lamina joint, validating their roles in lamina joint development. In addition, genes involved in the biosynthesis and signaling of BR and auxin, which have been implicated in important roles in the leaf angle, present diverse expression patterns, including decreased expression (CYP51G3 and CYCU4:1), increased expression (DLT [Tong et al., 2009] and GRAS19 and GH3-2 [Tong et al., 2012]), constant high expression levels (LIC [Wang et al., 2008] and LAZY1 [Zhang et al., 2012]), and constant low expression levels (ILI1 [Zhang et al., 2009a] and REL1 [Chen et al., 2015a]), suggesting the complex regulation of leaf angles via different pathways.
Analysis of DEGs identifies six clusters with specific expression patterns: highest expression at stage 2 and reduced from stage 4 to stage 6 (C1); continued reduction from stage 2 to stage 6 (C2); relatively high expression at middle stages (C3); clusters C4, C5, and C6 showed opposite expression patterns to C2, C1, and C3 (Fig. 3D). Furthermore, enriched Gene Ontology (GO) analysis identifies stage-specific biological processes that are consistent with the cytological and functional features of each stage (Fig. 3E). Genes preferentially expressed at stage 2 (C1) participate in metabolism, cell cycle, signal transduction, and cell wall organization, which is consistent with cells at stage 2 being active and having partial primordium activity, enabling cells to divide and differentiate. Signal transduction and metabolism at all analyzed stages may play roles in determining cell fate. Genes that are gradually reduced (C2) are involved in microtubule-based processes in addition to primary metabolism, suggesting suppressed cell growth along with lamina joint development, which is consistent with the continuous and decreased cell expansion. Genes related to protein phosphorylation, PCD, and transport are enriched in C4 and C5, which are closely related to the cytological changes (vascular development, sclerenchyma fiber formation, and aerenchyma enlargement) at later stages and functions of the mature lamina joint (mechanical support and transportation).
The lamina joint connects the leaf blade and shares structural similarities with the nearby leaf midrib; however, the cell composition and functions differ significantly. Considering that the developmental processes of the three examined lamina joints are similar (Fig. 3, A and B; Supplemental Fig. S2C), the adjacent part (0.5 cm) of the lamina joint of the last third leaf at stage 4 was collected and used as a representative to study the difference between the lamina joint and the adjacent part. The last third leaf presents a typical structure of leaf blade and lamina joint, and the leaf is fully developed (with mature structures) at stage 4. Comparison of the transcriptomes between the lamina joint and the adjacent part reveals ∼1,000 up-regulated and ∼1,500 down-regulated genes in the lamina joint (fold change [FC] > 2, false discovery rate [FDR] < 0.05; Supplemental Fig. S3, A and B). GO analysis showed that cell wall organization and cell growth-related progress are enriched among up-regulated genes, which is consistent with the structural difference (more sclerenchyma cells featuring thickened cell walls and many more large parenchyma cells owing to additional cell division and growth) between the lamina joint and the adjacent leaf part (Supplemental Fig. S3C; Fig. 1C). The photosynthesis-related processes are enriched in down-regulated genes, which is consistent with the deficiency of chloroplast in the lamina joint (due to the lack of mesophyll cells; Fig. 1C). The difference in biological processes lays the foundation for leaf angle formation at the lamina joint rather than the leaf blade.
Expression Profile of TFs and Protein Kinases in the Lamina Joint
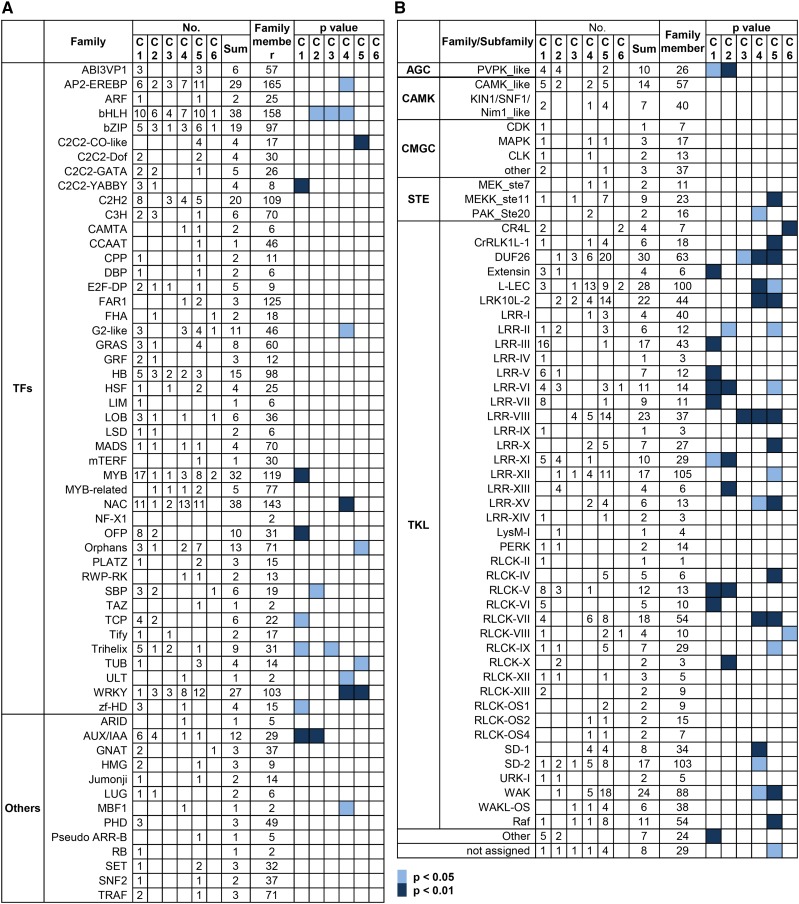
With the focus to identify the candidate regulators of lamina joint development, the expression profile of TFs was analyzed in detail. A total of 373 TFs belonging to 45 families and 37 transcription regulators belonging to 13 families were differentially expressed with distinct patterns (Fig. 4A), suggesting their functions at specific developmental stages of the lamina joint.
Figure 4.
Expression profiles of DEGs encoding TFs and kinases during lamina joint development. Numbers of DEGs and enrichment of a given gene family encoding TFs (A) or kinases (B) are shown. Gene members presenting expression patterns (C1–C6, as described in Fig. 3D) or members of a dedicated gene family are indicated. Enrichment of the gene family is indicated by calculating the P values by Fisher’s exact test.
Four C2C2-YABBY genes, DROOPING LEAF (Os03g11600), OsYABBY3 (Os04g45330), OsYABBY4 (Os02g42950), and OsYABBY1 (Os07g06620), which are reported to regulate midrib formation, lateral organ development, vascular development, and leaf angle (Dai et al., 2007; Liu et al., 2007; Tanaka et al., 2012; Yamaguchi et al., 2004), display rapid down-regulation from stage 2 to stage 4 (C1 and C2 patterns) and may regulate early lamina joint development and organogenesis. MYB members show dramatic changes from stage 2 to stage 4 (17 genes with the C1 pattern and eight genes with the C5 pattern), suggesting their potential roles in secondary cell wall synthesis in early lamina joint development and stress tolerance at later stages (Yang et al., 2014; Zhu et al., 2015). Most of the differentially expressed IAA genes show C1 and C2 patterns, suggesting the suppressed auxin signaling during lamina joint development (see below). WRKY genes exhibit an opposite pattern to IAAs and are enriched in C4 and C5 patterns, suggesting their roles at later stages of the lamina joint by regulating cell wall composition and hormonal signals (salicylic acid [SA] and ABA; Wang et al., 2007b; Zhang et al., 2009c). In addition, differentially expressed OFP, SBP, TCP, and Trihelix members mainly exhibit C1 and C2 patterns and AP2-EREBP, C2C2-CO-like, G2-like, NAC, and Orphans members are enriched in C4 and C5 patterns.
Analysis of the differentially expressed TFs between the lamina joint and adjacent leaf parts showed that 58 and 44 TFs are higher or lower expressed in the lamina joint, respectively (Supplemental Fig. S4, A and B). Genes belonging to growth-regulating factor, homeobox domain-containing, HRT domain-containing, and lateral organ boundaries families, which are involved in the regulation of cell expansion or differentiation, hormone response, or organ development (Raventós et al., 1998; Kappen, 2000; Kim et al., 2003; Husbands et al., 2007), are consistently enriched in higher expressed TFs, as well as LG1, which presents specific expression in the lamina joint (Supplemental Fig. S4B). Further analysis of the cis-elements regulated by lamina joint preferential TFs revealed the enrichment of ABA, auxin, and MYB-related elements, coinciding with the finding that ARF and MYB members are involved in lamina joint identification (Supplemental Fig. S4C). Unexpectedly, GA-related elements are enriched in down-regulated TF genes.
Protein modifications including phosphorylation, glycosylation, and ubiquitination are important in regulating various biological processes, and protein phosphorylation is highly enriched with C4 and C5 patterns (Fig. 3E), indicating the significance of protein kinase during lamina joint development. A total of 423 kinases belonging to 54 subfamilies are differentially expressed, and many of them present specific expression patterns (Fig. 4B). PVPK_like, Extensin, LRR-III/V/VI/VII/XI/XIII, and RLCK-V/VI subfamilies are enriched with C1 and C2 patterns, while MEKK, CrRLK1L, DUF26, l-LEC, LRR-VIII/X/XII/XV, RLCK-IV/VII, SD-1/2, WAK, and Raf subfamilies are enriched with C4 and C5 patterns. l-LEC and MEK kinases, the key regulators in biotic and abiotic stresses, are enriched in the C5 pattern, which is consistent with the senescence and enhanced stress tolerance at later stages. In addition, the similar expression pattern of the same family members suggests functional redundancy, which may be the reason why few kinase genes were reported by genetic studies. There are few reports on the roles of protein kinase in the lamina joint, and these results may provide clues for further studies, especially regarding the functions of protein kinases in organ development.
Drastic Expression Changes of Cell Division/Expansion-Related Genes at Early Stages
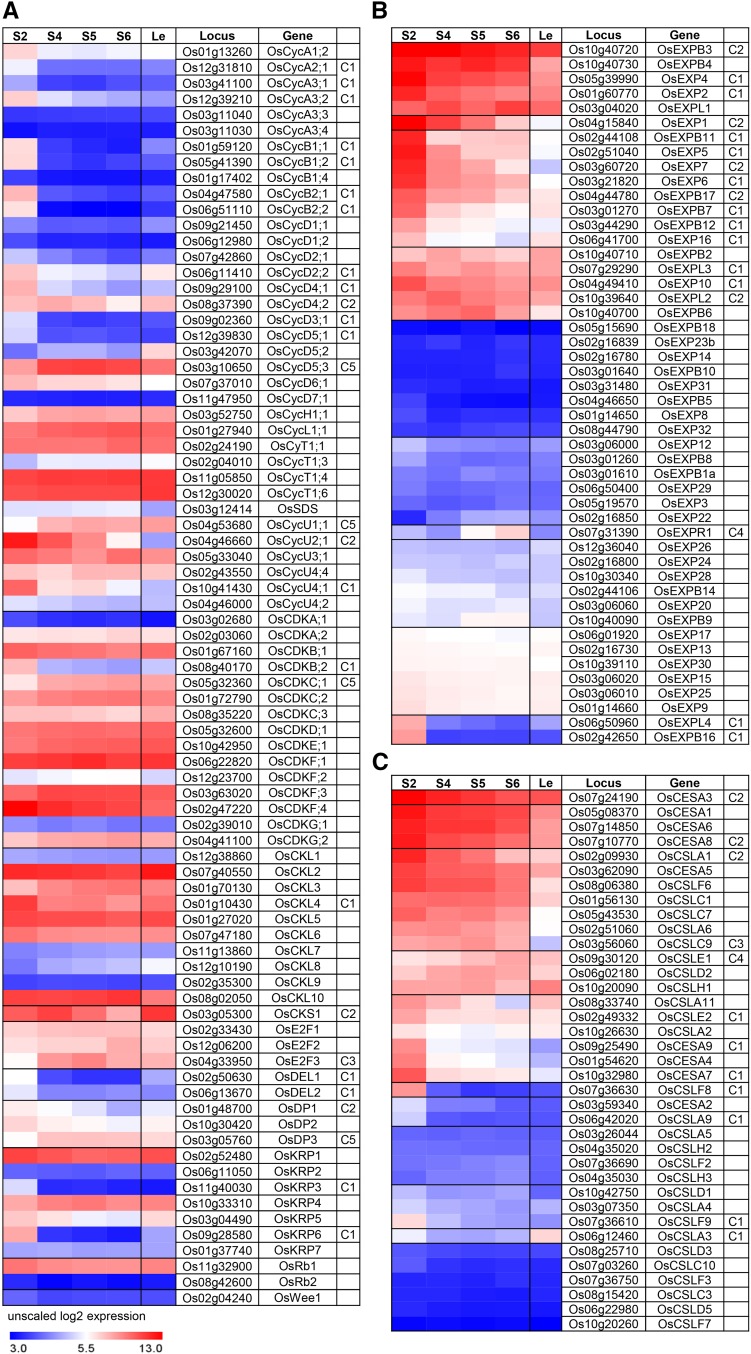
Cell division is a predominant process when the lamina joint initially develops, and along with cell division, cell expansion starts and lasts longer at the adaxial side, leading to asymmetric growth and, hence, leaf inclination (cell wall thickening provides mechanical support). Analysis of the expression profile of genes involving in cell cycle and expansion showed that cyclins, cyclin-dependent kinases (CDKs), and other core cell cycle regulators, including CDK inhibitors (KRPs), retinoblastoma (Rb/E2F/DP/DP-LIKE) pathway members, CDK subunit (CKS), and wee1-like protein kinases (Inzé and De Veylder, 2006), display a distinct stage- or tissue-specific expression (Fig. 5A). Consistent with the finding that OsCycU4;1 is involved in cell division at the abaxial side during lamina joint development (Sun et al., 2015), OsCycU2;1 and OsCycU4;1 are highly expressed in the lamina joint with C1 and C2 patterns, while OsCycD5;2 is lowly expressed, indicating a distinct regulatory mechanism. Interactions with cyclins and protein phosphorylation are important for CDK activity. Most cyclins (mainly in classes A–D) are highly expressed at early stages (especially stage 2) with C1 or C2 patterns, while the majority of CDK members are constantly highly expressed, indicating the significance of transcriptional regulation of cyclins in cell cycle determination.
Figure 5.
Expression profiles of cell cycle-, cell wall synthesis-, and cell expansion-related genes during lamina joint development. The heat maps show the expression profiles of genes related to cell cycle (cyclins, CDKs, and other core cell cycle regulators; A), cell expansion (EXPs; B), and cell wall synthesis (CESAs and CSLs; C) at four developmental stages of the lamina joint and adjacent leaf parts, which are shown as unscaled log2 expression levels. Gene expression patterns are indicated (C1–C6, as described in Fig. 3D).
Cell wall loosening and cell wall synthesis are involved in cell expansion (Shi et al., 2014), and analysis showed that highly expressed expansins (EXPB3, EXP7, and EXPL2) display C1 and C2 patterns as well (Fig. 5B). Further analysis of the cell wall synthases (CESAs; which are responsible for cellulose synthesis; Wang et al., 2010) showed that, unlike expansins, lamina joint preferential CESAs (CESA1, CESA5, CSLA6, and CSLC7) display constant expression during lamina joint development (Fig. 5C). However, most of the differentially expressed expansins and CESAs show C1 and C2 patterns, similar to cell cycle-related genes. High expression of cyclins, expansin, and CESAs leads to enhanced cell proliferation, expansion, elongation, and cell wall thickening and is consistent with the intensive cell division and growth at early stages of the lamina joint.
Vital Roles of Phytohormones in Lamina Joint Development
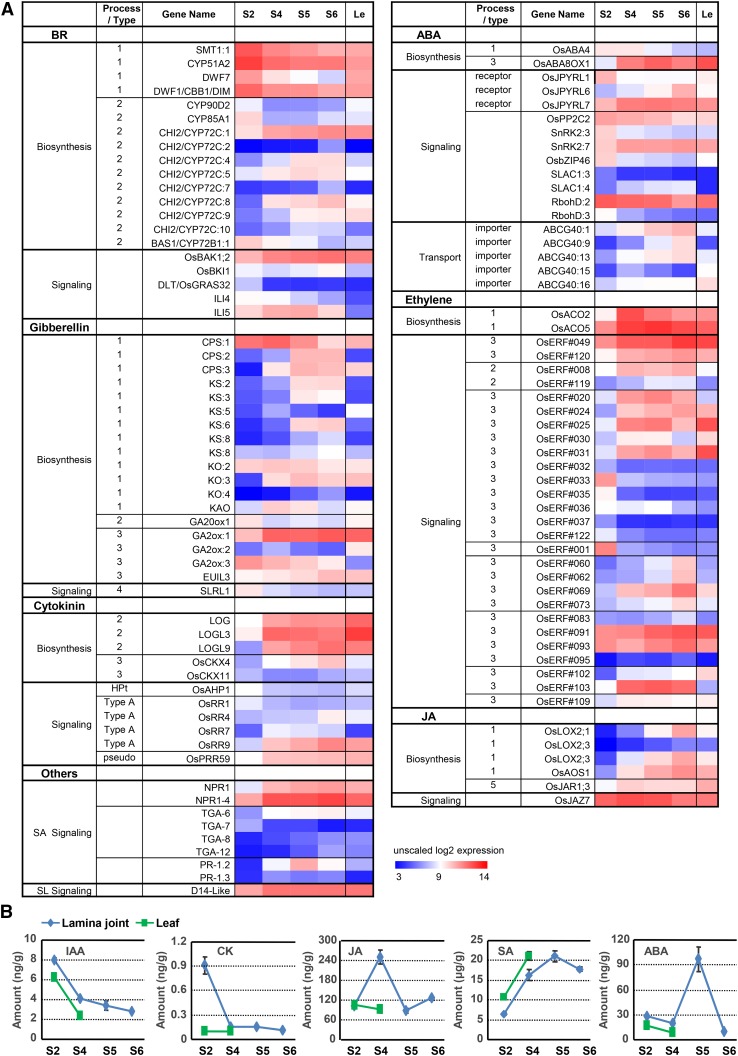
Detailed analysis of hormone biosynthesis- and signaling-related genes according to a previous definition (Supplemental Fig. S5; Chen et al., 2015b) showed that BR biosynthesis- and signaling-related genes display nonuniform expression patterns (Fig. 6A). Genes related to BR biosynthesis (D11 and D2) and signaling (BRI1, BAK1, and BU1) present a decreased tendency (Supplemental Fig. S2D), suggesting decreased BR biosynthesis and signaling during lamina joint development, which is consistent with the finding that enhanced BR signaling results in enlarged leaf inclination.
Figure 6.
Expression profiles of hormone-related genes during lamina joint development. A, Expression patterns of differentially expressed hormone-related genes at four developmental stages of the lamina joint and adjacent leaf parts, which are shown as unscaled log2 expression levels. Processes (types) of genes involved in hormone biosynthesis and signaling are indicated in Supplemental Figure S5. B, Amounts of various hormones at four developmental stages of the lamina joint and adjacent leaf parts (stages S2 and S4). For each sample, ∼30 to 80 lamina joints were collected for assay. Measurement was biologically repeated three times, and data are shown as means ± sd (n = 3).
The rate-limiting enzyme of GA biosynthesis, GA20ox1, shows decreased expression, while GA metabolic genes (GA2ox:1, GA2ox:2, and EUIL3) display the opposite tendency, which results in a decreased GA level at later stages. The expression synchronicity of GA- and BR-related genes may be due to the fact that they both promote cell elongation and to the complicated cross talk between them in rice (Tong et al., 2014). Similar to GA, decreased expression of an ABA biosynthesis gene (ABA4) and increased expression of a metabolic gene (ABA8OX1) may lead to a decreased active ABA level at later stages. In contrast to BR and GA, the expression of most ethylene biosynthesis and signaling, jasmonic acid (JA) biosynthesis, and SA signaling-related genes increases along with lamina joint development (Fig. 6A). In addition, compared with adjacent leaf parts, the expression of genes involved in auxin biosynthesis, signaling, and transport, BR signaling, and ethylene biosynthesis and signaling is much higher in the lamina joint (Supplemental Fig. S6).
Due to the complexity of hormone biosynthesis and signaling, especially the feedback and posttranscriptional regulation of hormone-related genes, it will be hard to predict the exact hormone biosynthesis and signaling based on nonuniform gene expression. The exact levels of various hormones were thus quantified at different stages (Fig. 6B; BR content is not measured due to the lack of an assay system). Consistent with the transcriptome analysis, levels of active auxin (IAA) and cytokinin (zeatin) decrease from stage 2 to stage 6, further confirming their roles in stimulating cell division at early stages. The content of free JA increases from stage 2 to stage 4, which is consistent with the elevated OsLOX and OsAOS1 expression (Supplemental Fig. S6), and decreases from stage 4 to stage 5, which may be due to the massive transformation from JA to the bioactive derivative JA-Ile (modified by OsJAR1; Fonseca et al., 2009; Sheard et al., 2010; Yan et al., 2016), suggesting the increased amount of bioactive JA on the whole. In addition, the amounts of SA increase along with lamina joint development, which is in accordance with their possible roles in improving biotic and abiotic stress tolerance and enriched cell death, protein phosphorylation, and defense responses at later stages (patterns C4 and C5; Fig. 3E). ABA amounts show an increase-decrease tendency with a maximum at stage 5, suggesting a complex regulation and function of ABA during lamina joint development, especially the senescence process at late stages.
Interestingly, amounts of auxin, cytokinin, and ABA in the lamina joint are higher, while SA is relatively lower, than those of adjacent leaf part, suggesting that auxin, cytokinin, and BR are possible regulators in promoting lamina joint organogenesis and development.
Auxin Regulates Lamina Joint Development through Distinct Processes
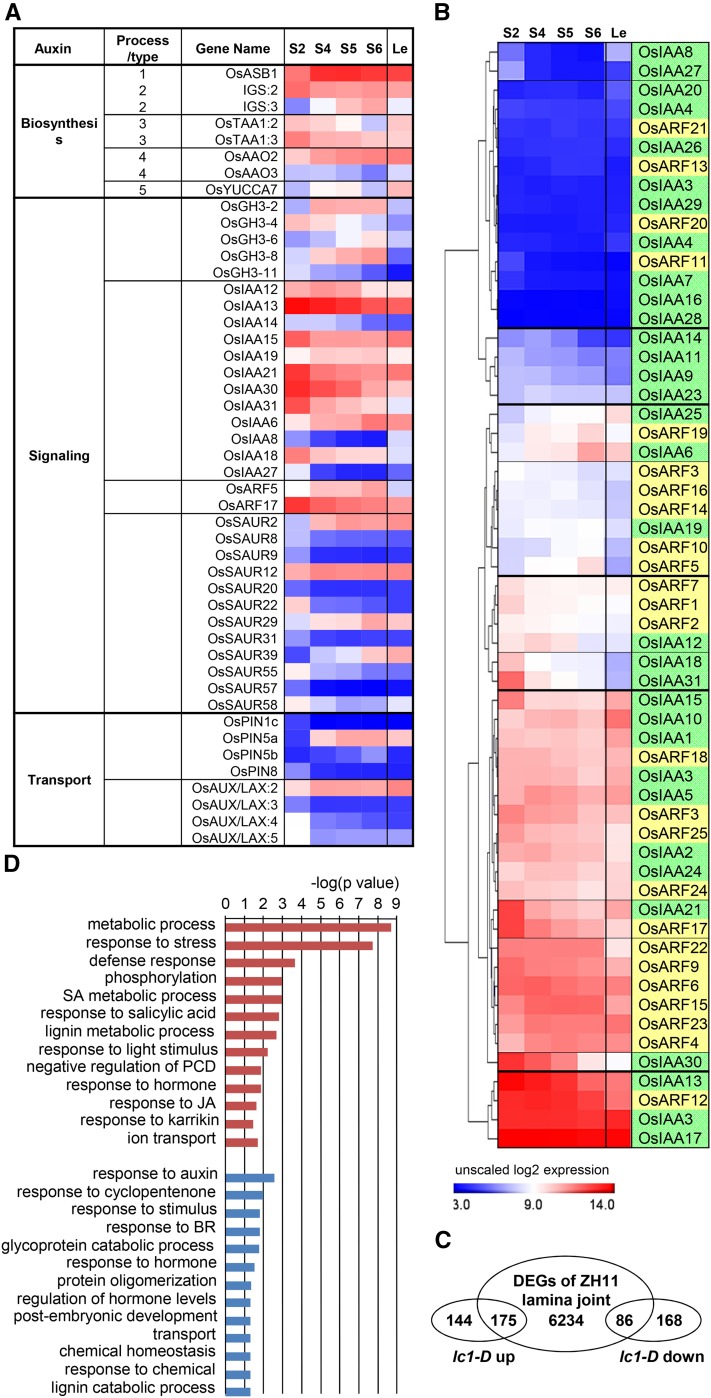
Based on the crucial roles of auxin in lamina joint development, auxin-related genes were particularly analyzed. The expression of auxin biosynthesis-, transport-, and signaling-related genes changes dynamically during lamina joint development (Fig. 7A). Auxin signaling and transport present a gradually decreasing tendency. Although auxin synthesis-related genes show unobvious changes, TAA1s (key genes involved in auxin biosynthesis) decrease and metabolic genes (GH3 family genes GH3-2, GH3-6, and GH3-8) increase, suggesting a decreased auxin level during lamina joint development, especially at the early stage, which is consistent with the measurement of free IAA content (Fig. 6B). In addition, auxin efflux carriers (OsPIN1C, OsPIN5a, OsPIN5b, and OsPIN8) and influx carriers (OsLAX:2, OsLAX:3, OsLAX:4, and OsLAX:5) exhibit obvious stage specificity, which may cause the asymmetrical auxin distribution to control cell division and elongation at the adaxial or abaxial side.
Figure 7.
Expression profiles of auxin-related genes and altered genes in the lc1-D mutant during lamina joint development. A, The heat map shows the expression patterns of differentially expressed auxin-related genes involving in biosynthesis, signaling, and transport at four developmental stages of the lamina joint and adjacent leaf parts, which are shown as unscaled log2 expression levels. Processes (types) of genes involved in auxin biosynthesis are indicated in Supplemental Figure S5. B, Hierarchical clustering analysis of all AUX/IAA and ARF genes. Genes with similar expression patterns are grouped by horizontal lines. IAAs and ARFs are distinguished by green and yellow backgrounds. C, Venn diagram of overlapping genes between DEGs in the lc1-D lamina joint and during lamina joint development of cv ZH11. The lc1-D microarray data sets can be found at the Gene Expression Omnibus database (accession no. GSE16640). D, GO enrichment analysis of auxin-regulating genes during lamina joint development. Genes were obtained from C, and GO terms of auxin up- or down-regulated genes are shown in blue or red respectively. P values were calculated by Fisher’s exact test.
Most differentially expressed IAA genes (except IAA6 and IAA19) show a decreased expression tendency. Considering that IAA functions by suppressing ARFs through complex interaction networks of IAAs-ARFs, clustering analysis of all 31 IAAs (Jain et al., 2006) and 25 ARFs (Wang et al., 2007a) based on the expression patterns provide further clues of the IAA-ARF interactions in regulating lamina joint development. IAA6/25-ARF19, IAA12/18/31-ARF1/2/7, and IAA21-ARF17 may cooperatively regulate lamina joint development at different stages (Fig. 7B). Interestingly, many ARFs and IAAs are constitutively expressed in the lamina joint (ARF3/4/6/9/12/15/18/22/23/24/25 and IAA1/2/3/5/10/17/24), and further analysis combined with interaction analysis will help to illustrate the distinct function of IAA-ARF in lamina joint development.
To further study the mechanism of auxin effects in regulating lamina joint development, a gain-of-function mutant, lc1-D (leaf inclination 1; which encodes OsGH3-1, an IAA amino synthetase), which was identified from the Shanghai T-DNA Insertion Population (Fu et al., 2009) and which presents a reduced auxin level and enlarged leaf angles due to the increased cell elongation in the adaxial surface of the lamina joint (Zhao et al., 2013), was used. Comparison of transcriptomes of the lamina joint of lc1-D and its corresponding original cultivar control, cv ZH11, identified 319 up-regulated and 254 down-regulated genes in lc1-D. Interestingly, 55% of the up-regulated genes (175) and 34% of the down-regulated genes (86) in lc1-D are detected in DEGs along with lamina joint development (Fig. 7C; Supplemental Fig. S7). Further GO analysis revealed the enrichment of cell division and elongation, stress, defense, and lignin metabolic-related processes (Fig. 7D), which is consistent with the cytological change and reduced level of auxin at later stages in lamina joint development.
In addition, JA and SA signaling-related genes are up-regulated and BR-related genes (mainly BR biosynthesis genes) are enriched in down-regulated genes of lc1-D (Fig. 7D; Supplemental Fig. S7), suggesting that auxin may suppress JA and SA signaling, or stimulate BR biosynthesis, to coordinately regulate lamina joint development.
Identification of the lc3 Mutant with Enlarged Leaf Angle
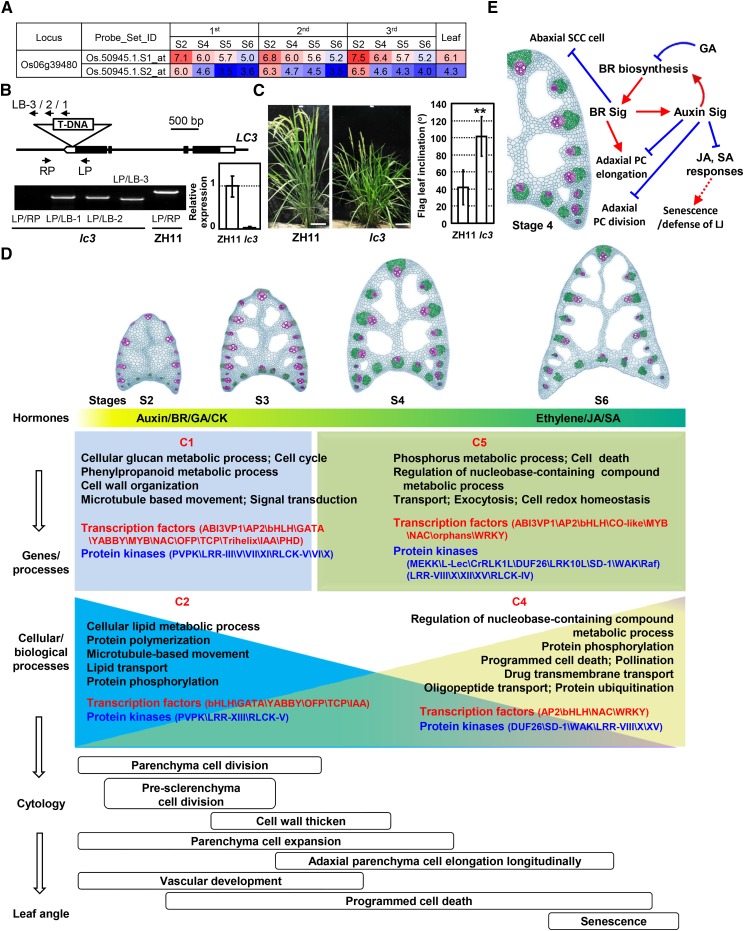
The cytology and transcriptome analyses provide informative clues to identify novel genes regulating leaf angles. By analyzing the DEGs and screening the mutants with altered leaf angles from the Shanghai T-DNA Insertion Population, a mutant designated as lc3 was identified. LC3 (Os06g39480) encodes a novel SPOC domain-containing protein and presents a down-regulated expression from stage 2 to stage 6 (Fig. 8A). T-DNA is located at the 3′ untranslated region and results in a significantly reduced expression of LC3 (Fig. 8B). Detailed observation and measurement showed that lc3 presents much enlarged leaf angles at the heading stage (Fig. 8C), indicating LC3 as a novel regulator of leaf inclination and confirming the value of the transcriptome studies. Detailed mechanisms will be investigated in the future.
Figure 8.
Identification and phenotypic observation of the rice lc3 mutant and schematic model of rice lamina joint development. A, Expression of the Os06g39480 (LC3) gene during lamina joint development at four developmental stages analyzed by Affymetrix chips. Expression levels are shown as log2 values. B, Schematic and confirmation of the lc3 mutant. The T-DNA insertion position is indicated (top: exons, black boxes; introns and intergenic regions, lines; untranslated regions, white boxes). Primers LP and RP, together with LB-1/2/3, were used to confirm the T-DNA insertion in lc3 and to identify the homozygous lines by PCR amplification using genomic DNA as a template (gel at bottom left). Quantitative real-time (qRT)-PCR analysis confirmed the significantly reduced expression of LC3 in the mutant (graph at bottom right). Transcript levels were normalized with Actin, and relative expression was shown by setting the LC3 expression of the wild-type flag leaf as 1. Experiments were biologically repeated three times, and data are shown as means ± se (n = 3). C, Phenotypic observation (left; bars = 10 cm) and measurement (right) showed the enlarged leaf inclination of the lc3 mutant. Flag leaf angles of the wild type and lc3 were measured at 10 d after flowering and statistically analyzed by Student’s t test. Data are shown as means ± sd (n > 20; **, P < 0.01). D, Rice lamina joint development contains distinct developmental stages and is fine-controlled by multiple factors. At the early initiation stage, auxin, BR, GA, and cytokinin stimulate cell division and expansion, while ethylene, JA, and SA control PCD and senescence at later stages. Crucial biological processes are enriched (corresponding genes with distinct expression patterns C1, C2, C4, and C5, as described in Fig. 4C), and specific TF and kinase families present certain expression patterns to modulate the corresponding cellular biological processes, leading to specific cytological processes and the formation of the leaf angle. In the cartoons showing lamina joint anatomies, vascular bundles, sclerenchyma cells, and parenchyma cells are highlighted with purple, green, and blue colors, respectively. E, A schematic model shows how phytohormones synergistically regulate rice lamina joint development.
DISCUSSION
Appropriate leaf inclination determines leaf erectness, photosynthesis efficiency, and, hence, crop yields. Considering the complex regulation and cytological changes (cell division and elongation, cell wall thickening, and PCD) and tissue development (vascular bundles, sclerenchyma, parenchyma, and aerenchyma are common in various organs), the lamina joint also is an ideal model in which to study the regulatory mechanism of cytology and tissue development as well as the cross talk of phytohormones. Through systemic cytology and transcriptome studies, it is shown that cell differentiation, cell division and elongation, cell wall thickening, PCD, and senescence are predominantly processes during lamina joint development. Consistently, most of the cyclin-CDKs (the master factor controlling cell division), expansin genes, and CESAs (the factors regulating cell expansion and cell wall thickening) are expressed preferentially at early stages and then decline, which may be regulated by upstream phytohormone signals (auxin, BR, and GA). Our studies reveal the distinct characteristics and cellular processes of rice lamina joint development, which is fine-controlled and cooperatively regulated by multiple factors, especially hormones (Fig. 8D), providing informative clues for understanding the regulatory network of lamina joint development and contributing to genetic breeding for the ideal architecture of crops.
Cytology during Lamina Joint Development
Cytological studies reveal that regularly occurring cell activities are closely related to specific processes at distinct stages during lamina joint development. At early/initiation stage (stage 2), when the organ pattern is formed, cell division and expansion, cell wall thickening, and vascular development occur. Longitudinal elongation of adaxial parenchyma cells after maturation stage (stage 4) leads to leaf inclination, and the degree of leaf angle is determined mainly at stage 4. The most significant change at the senescence stage (stages 5 and 6) is the distortion of the lamina joint. Interestingly, PCD happens throughout the process of lamina joint development (Fig. 8D).
In addition to cell proliferation and elongation, sclerenchyma cell properties and parenchyma cell numbers or size regulate leaf inclination as well. The parenchyma cells are living cells with plasticity, and the number of them at the adaxial side is the main factor determining leaf inclination. Indeed, altered parenchyma cell numbers or size lead to the altered leaf angle (Zhao et al., 2013; Zhang et al., 2015).
Sclerenchyma clusters are mechanical tissues to support the lamina joint, and change of their size and cell wall thickness results in altered mechanical strength and, hence, leaf angle, although there are only a few relevant reports (Ning et al., 2011; Sun et al., 2015).
Although PCD is a significant process throughout lamina joint development, there is no report on the effects of PCD-related genes in the regulation of leaf angle, which may be due to the fact that PCD happens not exclusively in the lamina joint but also in the leaf blade and sheath. Consistently, the PCD-related GO term is not enriched in the lamina joint compared with adjacent leaf parts. In addition, the simultaneous occurrence of PCD at both adaxial and abaxial sides indicates that PCD may not directly regulate leaf inclination.
Multilevel Regulation of Lamina Joint Development
Although the lamina joint starts to bend at stage 4 and constantly enlarges, molecular and cytological changes during the period from stage 2 (initiation) to stage 4 (maturation) are crucial and provide the foundation for leaf angle formation, which is consistent with the transcriptome analysis showing significant changes of gene expression at early stages and moderate but considerable changes at later stages.
Many TFs and kinases present distinct expression patterns, resulting in specifically enriched biological processes. At early/initiation stage, primary metabolism, cell cycle, cell wall organization, and microtubule based-movement are enriched, which may be regulated by organogenesis-related TFs (GATAs, YABBYs, OFPs, and IAAs). At maturation and the following senescence stage, NACs and WRKYs may regulate cell death, protein phosphorylation, and transport to achieve enhanced stress tolerance, function of the mature lamina joint, and ongoing PCD processes. In addition to transcriptional regulation, posttranslational modifications such as phosphorylation regulate lamina joint development by modulating the downstream signaling pathways, including the cell expansion or stress responses by extensins/WAKs, cell death, or defense responses by RLKs/MAPKs.
Although the inclination angle of each leaf differs one from another, lamina joints of all leaves display a consistent developmental process: the changing trajectories of leaf angles tend to display a similar S curve with varied maximum angles resulting from the differential cell properties, layers, and expansion, suggesting the existence of an intrinsic mechanism to determine the consistency and divergence of the lamina joints. The intrinsic factors, including spatial (leaf order from bottom to top) and temporal (leaf order from front to back) effects, and external factors, such as light, temperature, gravity, nutrition, and planting density, may synergistically regulate the cytology and final determination of leaf inclination by coordinating gene expression.
Crucial Roles of Phytohormones in Lamina Joint Development
Measurement and transcriptome analysis suggest that, at early stages, high amounts of auxin, BR, GA, and cytokinin promote cell division and expansion, while at later stages, ethylene, SA, and JA may regulate senescence and the stress response, of the lamina joint (Fig. 8D). BR-deficient or -insensitive mutants display erect leaves, and increased BR biosynthesis or signaling results in enlarged leaf angles. Endogenous BR regulates leaf angle either by enhancing parenchyma cell elongation at the adaxial side (similar to that caused by BR treatment) or inhibiting the proliferation of sclerenchyma cells at the abaxial side. The cytological and transcriptome analyses confirm the importance of these two cell types in lamina joint development and leaf inclination.
A reduced free auxin level causes enhanced leaf angle bending due to the stimulated elongation of parenchyma cells at the adaxial side (Zhao et al., 2013), suggesting the suppression effect of auxin on leaf inclination. Indeed, a high auxin level is detected at early stages and is reduced along with the developmental progress, which is consistent with the increased expression of GH3s. Combined with the cytological observations, it is suggested that, when the lamina joint initiates at the early stage, auxin is synthesized and polarly transported, resulting in a high local auxin concentration and stimulated cell division but suppressed elongation; meanwhile, along with the lamina joint development, the auxin level decreases, resulting in the promoted elongation of parenchyma cells. However, the increased auxin level does not lead to smaller leaf angles, implying the presence of a compensatory pathway in these cells that stimulates cell elongation. The fact that the expression levels of BR biosynthesis genes are increased in the lc1-D mutant suggests the possibly compensatory effects of BR and that auxin and BR cooperatively regulate lamina joint development.
In addition, various hormones may regulate lamina joint development synergistically and antagonistically. Auxin may function with BR to promote cell growth at early stages, and decreased auxin not only stimulates cell elongation but also relieves the suppression of JA and SA signals to regulate the stress response or senescence at later stages (Fig. 8E).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa) japonica cv ZH11 plants and the lc3 mutant for leaf angle recording and paraffin section were grown in a paddy field in Shanghai. For microarray analysis, cv ZH11 was cultivated in a greenhouse with a 12-h-light (28°C)/12-h-dark (22°C) cycle. The lamina joints were collected (those of the last third leaf at 50, 55, 60, and 70 DAS; those of the last second leaf at 55, 60, 65, and 75 DAS; and those of the last leaf at 60, 65, 70, and 80 DAS).
The adjacent part (0.5 cm) of the lamina joint of the last third leaf at stage 4 (55 DAS) was collected (defined as leaf) and used as a representative to study the difference between the lamina joint and the adjacent part. Thirteen samples in total were collected, and each sample had two biological replicates.
Leaf Angle Measurement
Leaf lamina joints with 1-cm sections of leaf and sheath on the main tiller were cut and photographed. Leaf angle was calculated by 180° minus the angle between sheath and leaf, which was measured with the aid of the ImageJ program (http://rsb.info.nih.gov/ij/). At least 10 leaf angles of individual plants were measured at each time point.
Paraffin Sections and Cytological Analysis
Rice lamina joints and leaves were fixed in FAA solution (50% ethanol, 5% acetic acid, and 10% formaldehyde in water), dehydrated in a graded ethanol series, and embedded in paraffin (Sigma). Microtome sections (8 mm) were applied onto microscope slides. The sections were deparaffinized in xylene, dehydrated through a graded ethanol series, and stained with Toluidine Blue. Sections were observed and photographed to measure the size of the lamina joint or calculate the cell number and length using ImageJ software.
RNA Extraction, Microarray Hybridization, and Data Normalization
Total RNA from lamina joints and adjacent leaf parts was extracted using Trizol reagent (15596-018; Life Technologies) and purified by the RNeasy Micro Kit (Qiagen) and RNase-Free DNase Set (Qiagen). The resultant total RNA was amplified, labeled, and purified using the GeneChip 3′IVT Express Kit (901229; Affymetrix) following the manufacturer’s instructions to obtain the biotin-labeled cRNA. Array hybridization and washing were performed using the GeneChip Hybridization, Wash, and Stain Kit (900720; Affymetrix) in Hybridization Oven 645 and Fluidics Station 450 (Affymetrix) following the manufacturer’s instructions. GeneChips were scanned using the GeneChip Scanner 3000 (Affymetrix). The obtained data of this study can be found at the Gene Expression Omnibus under accession number GSE92895.
Analyses were performed using the R software package (http://www.r-project.org) and packages by Bioconductor (Gentleman et al., 2004; http://www.bioconductor.org) if not specified. The raw data were evaluated (Gentleman et al., 2006; Wilson and Miller, 2005) to generate the QC reports. After comparing the performances of popular methods on our data, the RMA (Irizarry et al., 2003) method was used to obtain summary expression values for each probe set. Gene expression intensities were analyzed on a logarithmic scale. The data were separately normalized using the MAS 5.0 algorithm to generate present/absent calls. Probe sets showing absent (A) or marginal (M) in all samples were excluded for further analysis. PCCs were calculated from RMA-generated logarithmic scale expression values by Microsoft Excel 2010 to compare the relationships between samples. PCA was performed in the R environment.
Identification of DEGs and Clustering Analysis
The linear statistical model in the Limma package (Wettenhall and Smyth, 2004) from Bioconductor was used to identify the DEGs between two groups (lamina joint sample pairwise comparison or comparison between the lamina joint and adjacent leaf parts) by calculating FC and applying an FDR correction based on the method of Benjamini and Hochberg (1995). Only genes expressed in at least one sample were used for differential expression analysis (MAS 5.0 normalization). For pairwise comparison among lamina joint samples of three leaves (Fig. 4A), analyses were based on the two replicates; the threshold was set as FDR-adjusted P < 0.05 and FC > 2. For comparison between developmental stages of the lamina joint (Fig. 4B), samples at the same stage of three leaves were used as replicates. For comparison between lamina joint and adjacent leaf parts (Supplemental Fig. S3A), all lamina joint samples were used as replicates and two samples of adjacent leaf parts were used as leaf replicates. Different criteria were tested to identify DEGs, and adjusted P < 0.05 and FC > 2 were finally used for analysis. For comparison between the lc1-D mutant and the corresponding control cv ZH11 lamina joint (Fig. 7C), P < 0.05 and FC > 2 were used.
Identified DEGs were gathered after discarding the duplicate ones. The remaining gene average expression values of the replicates were normalized across all developmental stages using MultiExperiment Viewer 4.9 (Saeed et al., 2003; http://www.tm4.org/). K-means clustering with Euclidean distance was used to group genes based on merit analysis. Heat maps of gene expression values were generated in MultiExperiment Viewer 4.8 or Microsoft Excel. The hierarchical clustering used average linkage clustering and Pearson correlation within MultiExperiment Viewer 4.8.
GO Analysis
To annotate the functions of identified gene sets, the gene ontology information from AgriGO (Du et al., 2010; http://bioinfo.cau.edu.cn/agriGO/analysis.php), Rice Genome Annotation (Kawahara et al., 2013; http://rice.plantbiology.msu.edu/), and Arabidopsis (Arabidopsis thaliana) annotation (ftp://ftp.arabidopsis.org/home/tair/Ontologies/Gene_Ontology/) was used. GO analysis was performed using BiNGO software (Maere et al., 2005), which works as a Cytoscape plugin (Shannon et al., 2003). GO enrichment was derived with Fisher’s exact test and a cutoff FDR < 0.05. Only biological process GO terms were summarized using REVIGO (Supek et al., 2011) and shown.
Gene Family Enrichment Analysis
Rice TFs and other transcriptional regulators were referred to the plant TF databases PlnTFDB (Pérez-Rodríguez et al., 2010; http://plntfdb.bio.uni-potsdam.de/v3.0/) and PlantTFDB (Jin et al., 2014; http://planttfdb.cbi.pku.edu.cn/). TFs and other transcriptional regulators were gathered from the two Web sites after discarding the duplicates. Kinase genes were referred to the Rice Kinase Database (Jung et al., 2010; http://ricephylogenomics.ucdavis.edu/kinase/). Differentially expressed TFs or kinases were extracted from Figure 4C. Hypergeometric distribution analysis was performed to identify TF or kinase families enriched in cluster-grouped data sets.
cis-Elements Enrichment Analysis
The upstream sequence of each gene (up to 1,000 nucleotides from ATG) was extracted from the genome, and known cis-elements were mapped onto the promoters using the Plant cis-acting Regulatory DNA Elements database (Higo et al., 1999; http://www.dna.affrc.go.jp/htdocs/PLACE/). The ratio of each cis-element in a particular group was compared with that in the whole genome followed by performing hypergeometric distribution analysis. Raw P values were adjusted for multiple comparisons by the procedure of Benjamini and Hochberg (1995), which controls the FDR. If a known cis-element is enriched with a low adjusted P < 0.05, it will be annotated as enrichment in the promoters in a certain group.
Quantification of Various Hormones
The lamina joints of the cv ZH11 last leaf at 60, 65, 70, and 80 DAS (corresponding to stages 2, 4, 5, and 6) and the 0.5-cm adjacent part of the lamina joint of the last leaf blade at 60 and 65 DAS (corresponding to stages 2 and 4, as leaf samples) were collected for quantification assays of various hormones (Liu et al., 2010).
Auxin (IAA), ABA, SA, JA, and zeatin (cytokinin) contents were measured by liquid chromatography-tandem mass spectrometry (8030 plus; Shimadzu) as follows. A 200-mg fresh sample was frozen by liquid nitrogen and well homogenized using a TissueLyser homogenizer (Qiagen). Following the addition of 1 mL of 80% methanol, homogenates were well mixed in an ultrasonic bath and kept overnight (4°C). After centrifugation at 15,200g for 10 min, the supernatant was collected and vacuumed to dryness in a Jouan RCT-60 concentrator. Dried extract was dissolved in 200 µL of sodium phosphate solution (0.1 mol L−1, pH 7.8) and later passed through a Sep-Pak C18 cartridge (Waters) eluted with 1.5 mL of 80% methanol. The eluate was vacuumed to dryness again and dissolved in 10 mL of 10% methanol; 5 µL of solution was then injected into the liquid chromatography-tandem mass spectrometry system.
Liquid chromatography was performed using a 2-mm i.d. × 75-mm Shim-pack XR ODS I column (2.2 μm; Shimadzu) under a column temperature of 40°C. The mobile phase comprising solvent A (0.02% [v/v] aqueous acetic acid) and solvent B (100% [v/v] methanol) was employed in a gradient mode (time/A concentration/B concentration [min/%/%]: 0/90/10, 5/10/90, 6/10/90, and 6.1/80/20) at an eluent flow rate of 0.3 mL min−1.
The mass system was set to multiple reaction monitoring mode using electrospray ionization for different hormones. For IAA, ABA, JA, and SA, negative ion mode was used. For zeatin, positive ion mode was used. Operation conditions, including nebulizing gas flow, drying gas flow, desolvation temperature, and heat block temperature, were optimized for different hormones using standards. Deuterium-labeled hormones (Olchemim) were used as internal standards. Collision energy of −16 eV and mass-to-charge ratio (m/z) of 174.2 for IAA, collision energy of 11 eV and m/z of 263.2 for ABA, collision energy of 15 eV and m/z of 209.2 for JA, collision energy of 28 eV and m/z of 137 for SA, and collision energy of 32 eV and m/z of 218 for zeatin were employed.
Identification of the lc3 Mutant and qRT-PCR Analysis
T-DNA insertion was confirmed by PCR using primers LP (5′-GCAAACTCTTCCTCTTTCCGAC-3′) and LB-3 (5′-CCTAAAACCAAAATCCAG-3′), LB-2 (5′-GTGTTGAGCATATAAGAAACCCTTAG-3′), or LB-1 (5′-TAGGGTTCCTATAGGGTTTCGCTCA-3′). Identified homozygous lines were used for analysis.
qRT-PCR analysis was performed to examine the expression level of LC3 in the wild type and the lc3 mutant. Total RNAs were extracted from flag leaf (10 d after heading) with Trizol solution and reverse transcribed. qRT-PCR analyses were carried out on the Bio-Rad CFX Connect Real-Time PCR Detection System with a SYBR Green probe (SYBR Premix Ex Taq System; TaKaRa) using primers 5′-GGTGAAAAGACATCCACAAACGA-3′ and 5′-TATGAGTCAATGGTCCGCAAGA-3′. The rice Actin gene (03g50890) was amplified (primers 5′-CCTTCAACACCCCTGCTATG-3′ and 5′-TGAGTAACCACGCTCCGTCA-3′) and used as an internal reference control.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cytological dynamics of the lamina joint of rice flag leaf by longitudinal section analysis.
Supplemental Figure S2. Expression patterns of previously reported genes regulating lamina joint development.
Supplemental Figure S3. DEGs between the lamina joint and adjacent leaf parts.
Supplemental Figure S4. TFs showing differential expression between the lamina joint and adjacent leaf parts.
Supplemental Figure S5. Schematic diagram of hormonal biosynthesis and signaling pathways.
Supplemental Figure S6. Differentially expressed hormone-related genes between the lamina joint and adjacent leaf parts.
Supplemental Figure S7. Auxin-regulated genes involved in lamina joint development.
Acknowledgments
We thank Jian-Hua Tong at Hunan Agricultural University for the measurement of various phytohormones.
Glossary
- BR
brassinosteroid
- ABA
abscisic acid
- TF
transcription factor
- ZH11
Zhonghua11
- PCD
programmed cell death
- PCC
Pearson correlation coefficient
- PCA
principal component analysis
- DEG
differentially expressed gene
- GO
Gene Ontology
- FDR
false discovery rate
- FC
fold change
- JA
jasmonic acid
- SA
salicylic acid
- DAS
days after sowing
- qRT
quantitative real-time
Footnotes
This work was supported by the National Natural Science Foundation of China (NSFC, grant nos. 91535201 and 31570372) and Ministry of Science and Technology of China (MOST, grant nos. 2012CB944804 and 2013CBA01402).
Articles can be viewed without a subscription.
References
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196: 149–161 [DOI] [PubMed] [Google Scholar]
- Cao HP, Chen SK (1995) Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul 16: 189–196 [Google Scholar]
- Chen L, Xiong G, Cui X, Yan M, Xu T, Qian Q, Xue Y, Li J, Wang Y (2013) OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol Plant 6: 988–991 [DOI] [PubMed] [Google Scholar]
- Chen Q, Xie Q, Gao J, Wang W, Sun B, Liu B, Zhu H, Peng H, Zhao H, Liu C, et al. (2015a) Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. J Exp Bot 66: 6047–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shen LP, Wang DH, Wang FG, Zeng HY, Chen ZS, Peng YB, Lin YN, Tang X, Deng MH, et al. (2015b) A gene expression profiling of early rice stamen development that reveals inhibition of photosynthetic genes by OsMADS58. Mol Plant 8: 1069–1089 [DOI] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald CM. (1968) The breeding of crop ideotypes. Euphytica 17: 385–403 [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L (2012) A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63: 6467–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-Iso-jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Fu FF, Ye R, Xu SP, Xue HW (2009) Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res 19: 380–391 [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (2006) Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer Science, New York [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KS, Ko KW, Nam SJ, Park SH, Kim SK (1997) Optimization of a rice lamina inclination assay for detection of brassinosteroids. I. Effect of phytohormones on the inclination activity. J Plant Biol 40: 240–244 [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17: 2243–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa K. (1989) The Growing Rice Plant: An Anatomical Monograph. Nobunkyo, Japan. [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HM, Springer PS (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6: 47–59 [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 30: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Cao P, Seo YS, Dardick C, Ronald PC (2010) The Rice Kinase Phylogenomics Database: a guide for systematic analysis of the rice kinase super-family. Trends Plant Sci 15: 595–599 [DOI] [PubMed] [Google Scholar]
- Kappen C. (2000) The homeodomain: an ancient evolutionary motif in animals and plants. Comput Chem 24: 95–103 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Park JJ, Kim SL, Yim J, An G (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65: 487–499 [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li H, Jiang L, Youn JH, Sun W, Cheng Z, Jin T, Ma X, Guo X, Wang J, Zhang X, et al. (2013) A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytol 200: 1076–1088 [DOI] [PubMed] [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17: 402–410 [DOI] [PubMed] [Google Scholar]
- Liu HL, Xu YY, Xu ZH, Chong K (2007) A rice YABBY gene, OsYABBY4, preferentially expresses in developing vascular tissue. Dev Genes Evol 217: 629–637 [DOI] [PubMed] [Google Scholar]
- Liu X, Yang YL, Lin WH, Tong JH, Huang ZG, Xiao LT (2010) Determination of both jasmonic acid and methyl jasmonate in plant samples by liquid chromatography tandem mass spectrometry. Chin Sci Bull 55: 2231–2235 [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L (2011) Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. Plant Cell 23: 4334–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273: 23313–23320 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Xu X, Li H, Xu Q, Xu J (2014) Bioinformatics analysis of the expansin gene family in rice. [In Chinese.] Yi Chuan 36: 809–820 [DOI] [PubMed] [Google Scholar]
- Shi Z, Wang J, Wan X, Shen G, Wang X, Zhang J (2007) Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226: 99–108 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283: 1455 [Google Scholar]
- Song Y, You J, Xiong L (2009) Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70: 297–309 [DOI] [PubMed] [Google Scholar]
- Sun S, Chen D, Li X, Qiao S, Shi C, Li C, Shen H, Wang X (2015) Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev Cell 34: 220–228 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, et al. (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y (2007a) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007b) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65: 799–815 [DOI] [PubMed] [Google Scholar]
- Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, Xu Z, Wang Z, Chong K (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57: 498–510 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Y, Zhang C, Ma Q, Joo SH, Kim SK, Xu Z, Chong K (2008) OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, et al. (2014) Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA 111: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706 [DOI] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ (2005) Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics 21: 3683–3685 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Ou X, Tang H, Wang R, Wu P, Jia Y, Wei X, Xu X, Kang SH, Kim SK, et al. (2015) Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol 208: 790–802 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Li S, Gu M, Yao R, Li Y, Chen J, Yang M, Tong J, Xiao L, Nan F, et al. (2016) Endogenous bioactive jasmonate is composed of a set of (+)-7-iso-JA-amino acid conjugates. Plant Physiol 172: 2154–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li D, Liu X, Ji C, Hao L, Zhao X, Li X, Chen C, Cheng Z, Zhu L (2014) OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol 14: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu Y, Guo S, Zhu J, Huan Q, Liu H, Wang L, Luo G, Wang X, Chong K (2012) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009a) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, Geisler M, Jiang A, Qi Y (2015) The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ 38: 638–654 [DOI] [PubMed] [Google Scholar]
- Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y (2009b) Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol 151: 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Shin M, Zou X, Huang J, Ho TH, Shen QJ (2009c) A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol Biol 70: 139–151 [DOI] [PubMed] [Google Scholar]
- Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20: 935–947 [DOI] [PubMed] [Google Scholar]
- Zhao SQ, Xiang JJ, Xue HW (2013) Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6: 174–187 [DOI] [PubMed] [Google Scholar]
- Zhu N, Cheng S, Liu X, Du H, Dai M, Zhou DX, Yang W, Zhao Y (2015) The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci 236: 146–156 [DOI] [PubMed] [Google Scholar]