Abstract
Lung cancers are the leading cause of cancer-related deaths worldwide, with small cell lung cancer (SCLC) being the most aggressive type. At the time of diagnosis, SCLC has usually already metastasized, and an astonishing 95% of patients eventually succumb to the disease. This highlights the need for more effective SCLC screening and treatment options. Interestingly, the earliest and most frequent genetic alteration associated with lung cancers involves a lesion in the region to which the RNA binding protein RBM5 maps. We have recently shown that a decrease in RBM5 expression may be a key step in SCLC development, as RBM5 regulated many transformation-associated processes in SCLC cells. RBM5 is structurally and functionally similar to another RNA binding protein, RBM10. Both proteins have tumor-suppressor properties in a variety of cancer cell lines, and it has been suggested that RBM5 expression can influence RBM10. Due to their similarities, and the recent evidence that RBM10 is mutated in up to 21% of lung cancers, we hypothesized that RBM10 would share RBM5’s tumor-suppressor properties in SCLC. Using transcriptome analysis and functional assays, we show, however, that RBM10’s function was opposite to what we hypothesized; in the endogenously RBM5-null GLC20 SCLC cell line, RBM10 actually promoted cell proliferation and other transformation-associated processes. Using RNA immunoprecipitation followed by next generation sequencing (RIP-Seq) and Western blotting, we demonstrate that RBM5 post-transcriptionally regulated RBM10 expression via direct interaction with specific RBM10 splice variants. We propose a working model describing the impact of this interaction on cellular processes. Our results provide evidence that RBM10 expression, in RBM5-null tumors, may contribute to tumor growth and metastasis. Measurement of both RBM10 and RBM5 expression in clinical samples may therefore hold prognostic and/or potentially predictive value.
Introduction
According to the World Health Organization, cancer is one of the main causes of morbidity and mortality worldwide. Lung cancers are not only one of the most diagnosed cancers, but also the main cause of these cancer-related deaths, claiming over 1.6 million lives in 2012 [1]. The most aggressive form of lung cancer is small cell lung cancer (SCLC) [2]. In 90% of SCLC cases, the disease has already metastasized when the patient is diagnosed, which limits treatment options [3]. An astonishing 95% of SCLC patients eventually succumb to the disease, highlighting the need for more effective detection and treatment options [4]. Interestingly, the earliest and most frequent genetic alteration that occurs in lung cancer involves deletion within the 3p21.3 region [5–7]. The RNA binding protein, RBM5, resides within this region and is significantly downregulated [8], but not deleted [6], in the majority of lung cancers. Recently, our group demonstrated that this decline in RBM5 expression may be a key step in the establishment of the transformed state of lung cells; RBM5 is responsible for slowing the cell cycle, promoting apoptosis, and downregulating transformation-associated processes such as angiogenesis in SCLC cells [9]. RBM5 may therefore be an important marker for SCLC risk, and could guide the development of more effective screening and/or treatment options.
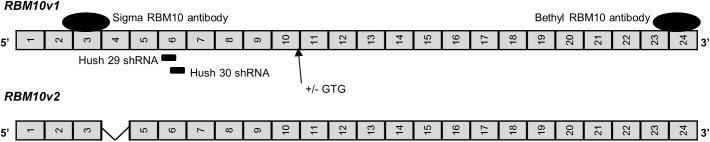
RBM5 is structurally similar to another RNA binding protein, RBM10. RBM10 has two main alternative splice variants termed RBM10 variant 1 (RBM10v1) and RBM10 variant 2 (RBM10v2) [10–12]. Each main RBM10 splice variant also codes for alternative isoforms with or without the addition of one valine residue (Fig 1) [13]. Structurally, RBM10v2 and RBM5 share 53% homology at the amino acid level [12].
Fig 1. Schematic representation of RBM10v1/v2 exons.
Exons are represented by grey boxes outlined in black. Box sizes are not representative of actual exon size. Black ovals represent approximate location of the epitopes recognized by the antibodies used in this study. Solid black lines represent the approximate location of shRNA RBM10 targets, which were not variant specific. Corresponding names of antibodies and shRNA are listed next to their approximate location. Arrow indicates position of the GTG codon coding for the valine residue that can be differentially spliced from RBM10 transcripts (last amino acid coded by exon 10).
RBM5 and RBM10 also share functional similarities, and both have tumor-suppressor properties in various cancer cell lines [10, 12, 14]. Functional data for RBM10 are limited, but in certain studies, RBM10 promoted apoptosis and/or decreased cell proliferation; (1) RBM10 expression correlated with decreased cell proliferation and increased apoptosis in primary chondrocytes induced to hypertrophy [15], (2) RBM10v1 expression in breast cancer specimens correlated with the expression of proapoptotic BAX and the tumor suppressor gene p53 [16], (3) stable RBM10 knockdown (KD) in ovarian cancer cells (HeLa) correlated with a significant increase in colony formation by clonogenic assays [17], (4) in lymphoblastic leukemia (Jurkat) and breast cancer (MCF-7) cells, transient RBM10 overexpression correlated with increased levels of apoptosis, whereas stable RBM10KD correlated with lower TNF-α protein levels, as well as decreased TNF-α mediated apoptosis [10], and (5) in a mouse xenograft tumor model with HeLa cells, RBM10KD enhanced tumor growth [14]. In contrast, other studies suggest an anti-apoptotic function for RBM10; (1) RBM10KD in SHSY5Y human neuronal cells augmented proapoptotic caspase activity after staurosporine exposure [18], (2) expression of both RBM10 mRNA variants in breast cancer cells positively correlated with VEGF mRNA, a promoter of new blood vessel growth [16], and (3) in patients with metastatic melanoma, high RBM10 expression correlated with increased disease aggressiveness [19]. Considered as a whole, these results suggest that the regulation of cell growth is an important aspect of RBM10 function, but that the mechanism regulating this function requires elucidation. Limited evidence suggests that this regulation depends, at least in part, on the related RNA binding protein RBM5 [20].
Considering the similarities between RBM5 and RBM10, and the recent finding that RBM10 is mutated in up to 21% of invasive lung adenocarcinomas [21], we hypothesized that RBM10 shares RBM5’s tumor-suppressor properties in SCLC. Consequently, RBM10 may also hold prognostic potential for assessing SCLC risk and provide important information regarding the development and/or progression of this particularly aggressive form of lung cancer. To this end, we set out to determine RBM10’s role in SCLC, and compare it to that of RBM5.
Materials and methods
Cell culture & subline establishment
SCLC GLC20 cells were gifted by the late Dr. Charles Buys from the University of Groningen (Groningen, Netherlands) and cultured as previously described [9]. Stable RBM5-expressing T2 and C4 GLC20 sublines were established as previously described [9].
SCLC GLC20 RBM10 stable KD sublines were generated from passage 7 GLC20 wild type cells. Several different dilutions of GLC20 cells were plated in 24-well flat-bottom plates (Sarstedt AG & Co., Nümbrecht, Germany) and incubated at 37°C with 5% CO2 in a humidified chamber for 24 hours. Half of the cells were transfected with a control shRNA vector “Hush 300” (TR30003, Origene Technologies, Inc, Rockville, U.S.A) and the other half with a 50:50 shRNA mix of “Hush 29” (TI308329, Origene, 5’-GCCTTCGTCGAGTTTAGTCACTTGCAGGA) and “Hush 30” (TI308330, Origene, 5’-AGTCACTTGCAGGACGCTACACGATGGAT). Hush 29 and 30 both target exon 6 of RBM10 and are not variant 1 or variant 2 specific (Fig 1). Cells were transfected using Lipofectamine 2000 (L2000) (Invitrogen, Life Technologies, Burlington, Canada). Transfected cells were selected using 0.1 μg/mL puromycin (Invitrogen) starting at 48 hours post-transfection. After seven days of selection, cells were treated with a Histopaque solution (Sigma-Aldrich) to separate live from dead cells. The stable negative control shRNA subline was named G300.3, and three surviving RBM10KD sublines were named G29/30.1, G29/30.3 and G29/30.4.
RNA sequencing and analysis
RNA samples were isolated using Tri-Reagent (BioCan Scientific, Mississauga, Canada). All sequencing was performed by the Donnelly Sequencing Centre (Toronto, Canada) on the Illumina HiSeq 2500 platform. RNA-Seq library preparation and sequencing specifications were as previously described [9]. All specimens, therefore, had (1) quality score distributions over all sequences above 37 (on a phred 33 quality scale), as determined by FastQC (Babraham Bioinformatics, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and (2) low quality ends trimmed during adapter trimming (quality cut-off of 26), as previous recommended [22]. Specifications for the GLC20 stable scramble control and RBM10KD (G29/30.4) were as follows; (1) RNA was not DNAse treated prior to sequencing, (2) reads were 125 bp long, and (3) both samples were multiplexed together and run in one lane. Sequencing data has been deposited to the NCBI Sequence Read Archive (SRA) database, under the SRA accession number (https://www.ncbi.nlm.nih.gov/Traces/study/) SRP106846, Bioproject PRJNA386251.
Sequencing results were analyzed and pathway analysis carried out as previously described [9].
Western blotting
Performed as previously described [9]. Two rabbit anti-human RBM10 primary antibodies were also used: Sigma-Aldrich, HPA034972 and Bethyl Laboratories, A301-006A-1.
RNA immunoprecipitation followed by next generation sequencing (RIP-Seq)
The basic RIP technique was carried out as previously described [23]. RNA was sequenced by the Donnelly Sequencing Centre on the Illumina HiSeq 2500 platform. Libraries were prepared using the Illumina TruSeq Stranded Total RNA library preparation kit with Ribozero Gold depletion and random priming. Associated samples were multiplexed together for sequencing, and each run was 125 bp and pair-ended. The RBM10 antibody used for RBM10 immunoprecipitation was from Sigma-Aldrich (catalogue number: HPA034972), and the non-specific IgG Control was from Cell Signaling (catalogue number: 2729). Sequencing data has been deposited to the NCBI Sequence Read Archive (SRA) database, under the SRA accession number (https://www.ncbi.nlm.nih.gov/Traces/study/) SRP106846, Bioproject PRJNA386251.
The FPKM and log2-fold change expression values generated by Cuffdiff were used to determine protein targets. The potential target had to meet the following inclusion criteria in the RBM5 or RBM10 RIP sample, as previously described [9]: (1) an FPKM value above one, and (2) a log2-fold change above one (and necessarily positive, indicating it was more highly expressed than in the Control RIP).
Cell growth assay
GLC20 cells and RBM10-knockdowns were plated, as previously described [9], and monitored every day for five days by MTT assays. At the indicated time intervals, cells were treated with 10 μL of 5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) Reagent (Life Technologies) in PBS (Gibco) for 2 hours and 45 min at 37°C, as previously described [24]. Following incubation, cells were transferred to a 96-well Vee-bottom plate (Sarstedt) and subjected to centrifugation. Supernatant was discarded and the blue formazan precipitate was dissolved in DMSO (BDH Chemicals, VWR). Dissolved crystals were then transferred to a 96-well flat-bottom plate and, after a ten minute incubation, absorbances were read at 540 nm using a BioTek Synergy S4 Spectrophotometer (BioTek Instruments, Inc). Three biological replicates, using cells with different passage numbers, were performed.
Absorbances of each biological replicate were normalized to their respective day 0 absorbance value, and the averages of three or four biological replicates for each day are presented. A Two-way ANOVA statistical analysis was performed with the Bonferroni post-hoc test, comparing all growth to the pcDNA3 subline for the RBM5 sublines or G300.3 for the RBM10KD sublines, using Graphpad Prism 5 (Graphpad Software, Inc., San Diego, U.S.A).
Results
RBM10 RNA targets in GLC20 cells that express RBM5 cDNA
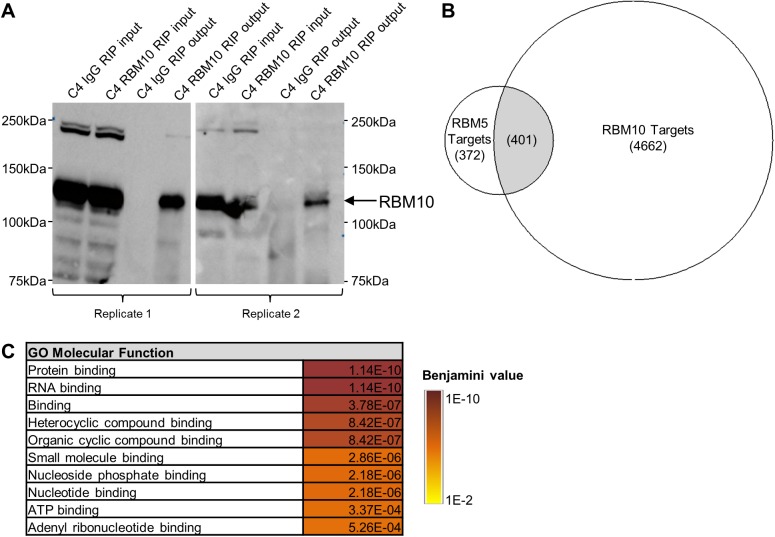
We first set out to compare RBM5 and RBM10 targets in SCLC. As RBM10 is an RNA binding protein, and was previously shown to be a part of prespliceosomal complexes A and B [25, 26], we determined that RNA Immunoprecipitation followed by next generation sequencing (RIP-Seq) was the most suitable method for identification of RBM10 targets. RIP-Seq involves antibody-driven immunoprecipitation of targeted endogenous protein without breakdown of protein-protein interactions, thus any RNA that is either directly bound to RBM10, and/or bound to RBM10 via another RBM10-bound protein would be captured. We anticipated that, having homology and similar functions, RBM10 targets would overlap with those of RBM5. Our group recently identified RBM5 targets using an endogenously RBM5-null SCLC cell line (S1 Table), GLC20 [24], from which three stable sublines—an empty vector control and two RBM5 expressing (T2 and C4)–had been generated [9]. The “T2” subline was derived from a pool of transfected cells, and had RBM5 expression levels comparable to non-tumor lung tissue, while the “C4” subline was derived from a single transfected cell, and had 6-fold higher levels of RBM5 than the T2 subline [9]. As RBM5 targets were previously identified in the C4 GLC20 subline, RBM10 RIP-Seq experiments were also performed in this subline in order to directly compare RBM5 and RBM10 targets. Specifically, RBM10 RIP-Seq was performed in biological duplicate in C4 cells using a non-specific IgG as a Control IP and an anti-RBM10 antibody for RBM10-specific IP. Following confirmation of successful IP (Fig 2A), the associated RNA was sequenced (S1 Table).
Fig 2. RBM10 RIP-Seq results and comparison to identified RBM5 targets.
(A) Successful immunoprecipitation of RBM10 from C4 cells, demonstrated by Western blot of input and output protein samples. Blots probed for RBM10, using the same RBM10 antibody used in the immunoprecipitation experiment (Sigma-Aldrich). Control immunoprecipitation performed, using a non-specific rabbit IgG antibody. (B) Overlap between RBM5 and RBM10 targets in C4 GLC20 subline. (C) GO Molecular Function gene sets enriched in common RBM5 and RBM10 RIP-Seq targets, as determined by FIDEA. Values indicated are Benjamini values.
Using moderate binding criteria, as previously described [9], we identified 5063 RBM10 targets. In our previous study, using the same RIP-Seq procedure and binding criteria, we had identified 773 RBM5 targets in this GLC20 subline [9]. RBM10, therefore, had many more targets than RBM5, suggesting the scope of RBM10’s influence in SCLC may be greater than that of RBM5.
Interestingly, more than half of RBM5’s targets were shared by RBM10 (Fig 2B), confirming that they do have overlapping targets. Furthermore, this suggests that RBM10 plays a key role in RBM5 regulated processes but not visa versa. To determine the functional relevance of these common targets, we performed pathway analyses using the Functional Interpretation of Differential Expression Analysis (FIDEA) Program, a program that can be used to analyze various types of differential expression data [27]. Two different databases were used with the FIDEA Program; (1) the Kyoto Encyclopedia of Genes and Genomes (KEGG) Database, which groups genes based on established molecular interactions [28, 29], and (2) the Gene Ontology’s (GO) Molecular Function Database, which groups genes based on cellular functions [30, 31]. Using a Benjamini value cut off of 0.005, no KEGG pathways were found to be enriched, a not unexpected result since multiple targets within a single signaling pathway would not necessarily be required for an effect. Using the same Benjamini value parameter, ten GO Molecular Function gene sets were enriched (Fig 2C). Interestingly, all enriched gene sets involved binding, including to ATP, nucleotides, protein, RNA and small molecules. Thus RBM5 and RBM10’s common targets may regulate a plethora of cellular processes, which may also explain why no individual KEGG pathways were enriched.
Since RBM10 had so many RNA targets that were not shared by RBM5 (Fig 2B), we went on to determine the functional importance of these RBM10-specific genes (will be referred to as RBM10-only targets). Using the FIDEA Program with the KEGG Database, we identified eight significantly enriched pathways (Table 1). The most enriched pathway was ‘Metabolic pathways’, consistent with the top RBM10 targets, which are all involved in different aspects of cell metabolism: NDUFA6 (oxidoreductase activity), MDP1 (phosphatase activity) and ATP6VIF (ion transfer). We were particularly interested to see that the ‘Oxidative phosphorylation’ pathway was the second most-enriched pathway. Notably, changes in oxidative phosphorylation are not only linked to metabolism (the top enriched pathway identified), but also the three disease states enriched in RBM10-only targets: Parkinson’s [32, 33], Huntington’s [34] and Alzheimer’s [35, 36]. The fact that five of the eight enriched KEGG pathways are intimately linked to oxidative phosphorylation strongly suggests that RBM10 plays a particularly important role in this process.
Table 1. Pathways and gene sets enriched in RBM10-only RIP-Seq targets.
| Pathways & gene sets | Benjamini value |
|---|---|
| KEGG | |
| Metabolic pathways | 1.78E-8 |
| Oxidative phosphorylation | 9.34E-5 |
| Base excision pair | 1.56E-4 |
| Huntington’s disease | 4.37E-4 |
| Alzheimer’s disease | 1.77E-3 |
| Neurotrophin signaling pathway | 1.79E-3 |
| RNA transport | 1.79E-3 |
| Parkinson’s disease | 2.46E-3 |
| GO Molecular Function | |
| Protein binding | 3.62E-34 |
| Binding | 6.15E-21 |
| RNA binding | 1.07E-13 |
| Structural constituent of ribosome | 1.63E-7 |
| Catalytic activity | 5.31E-7 |
| Transcription cofactor activity | 2.07E-7 |
| Heterocycylic compound binding | 2.07E-6 |
| Organic cyclic compound binding | 2.07E-6 |
| Transcription factor binding transcription factor activity | 4.98E-6 |
| Transferase activity | 3.29E-6 |
| Protein binding transcription factor activity | 4.34E-6 |
| Enzyme binding | 2.30E-6 |
| Nucleic acid binding | 2.30E-5 |
| Kinase binding | 2.30E-5 |
| Transcription corepressor activity | 3.61E-4 |
| Protein kinase binding | 1.19E-4 |
| Mitogen-activated protein kinase binding | 2.13E-3 |
Significantly enriched gene sets with Benjamini values below 0.005 are listed.
To determine if these RBM10-only targets shared similar functionality, we used the GO Molecular Function Database with the FIDEA Program. Enriched gene sets are listed in Table 1. Many of the enriched gene sets are still showing RBM10 targets involved in a plethora of binding activities, but a new functional activity involving transcription regulation emerged, suggesting that RBM10 has distinct RBM5-independent functions.
Taken together our RIP-Seq results show that although RBM5 and RBM10 share many common targets, both have distinct targets, with RBM10 having over six times more than RBM5. RBM5 and RBM10 targets, themselves, bind various cellular components, suggesting that RBM5 and RBM10 work together to regulate a variety of cellular processes in SCLC. On the other hand, targets bound by RBM10, but not RBM5, regulated oxidative phosphorylation and numerous aspects of gene expression and protein activity, suggesting RBM10 has distinct mechanisms of action that could be RBM5-independent.
Effect of RBM10 inhibition on dehydrogenase activity in GLC20 cells
Our RIP-Seq results showed that RBM10 directly influenced metabolism, most specifically oxidative phosphorylation. Work in HeLa cells and primary chondrocytes had previously correlated RBM10 expression with decreased cell proliferation [15, 17]. Taken together, these observations suggested that RBM10 expression would result in decreased SCLC cell proliferation and/or metabolic activity. We therefore chose to use the MTT assay to examine the functional significance of RBM10 expression to SCLC.
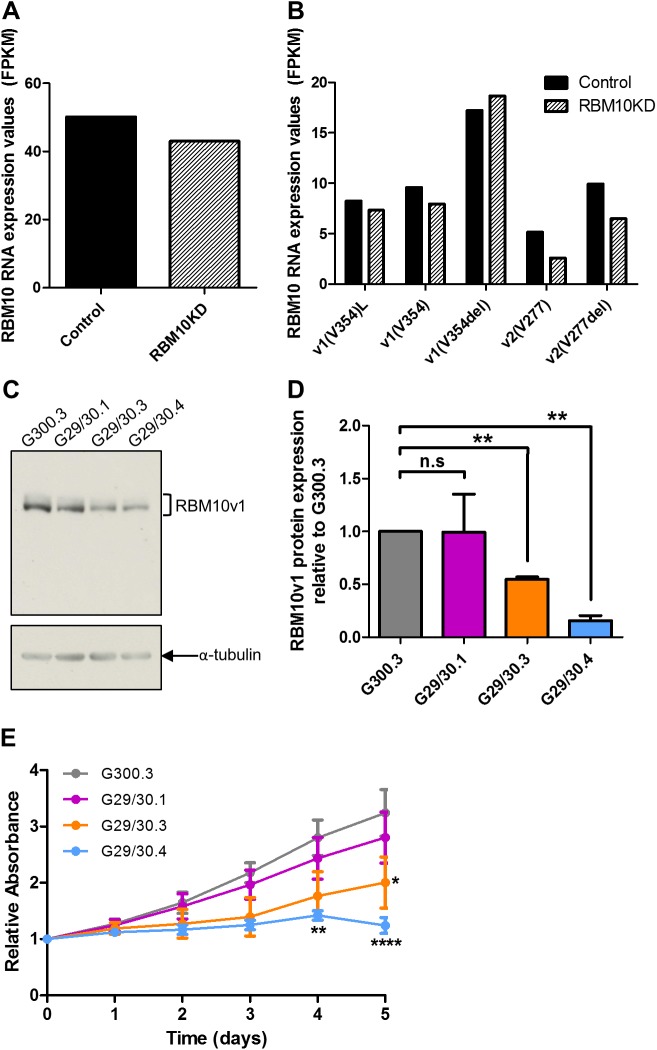
As a first step, we generated stable RBM10 knockdowns (KDs) in the parental RBM5-null GLC20 cells. It is important to note that this GLC20 SCLC cell line is a particularly appropriate model for aggressive SCLC, as RBM5 is downregulated in the majority of lung cancers [8]. RBM10KD was not performed in the T2 or C4 sublines since all RBM5 expressed in these sublines results from exogenously introduced RBM5 cDNA. These sublines, therefore, do not express any of the various RBM5 splice variants that are present in endogenously RBM5-expressing systems, rendering results from RBM10KD experiments in these sublines non-physiological. As noted in Materials and Methods, GLC20 cells were subjected to transfection with two RBM10KD shRNA plasmids, together termed 29/30, that targeted an exon common to both RBM10v1 and RBM10v2. Thus, knockdown of RBM10v1 was not specifically targeted in this study, but due to the almost exclusive expression of RBM10v1 in the GLC20 cells at the protein level (Fig 3C), all decreases in RBM10 protein were reported as RBM10v1. Three stable sublines derived from three distinct pooled RBM10KD populations were produced, termed G29/30.1, G29/30.3 and G29/30.4, as well as one control subline, termed G300.3. Somewhat surprisingly, but in line with one of the known mechanisms of shRNA regulation [37, 38], RNA levels of RBM10 were not diminished in any of the KDs (Fig 3A and 3B). At the protein level, although the G29/30.1 subline demonstrated great variance in protein expression, with no significant decrease in RBM10v1 expression relative to Control (99% expression, p = 0.9836), both G29/30.3 and G29/30.4 had significantly lower RBM10v1 protein levels relative to G300/3, with 55% (p = 0.0026) and 16% (p = 0.0033) the level of Control cells, respectively (Fig 3C and 3D).
Fig 3. RBM10 expression and dehydrogenase activity in control and stable RBM10KD GLC20 sublines.
(A & B) RBM10 mRNA levels, determined by RNA-Seq, for all RBM10 variants combined (A) or for individual splice variants (B). ‘Control’ represents values from parental GLC20 cells and the control G300.3 GLC20 subline. RBM10KD sample used was the G29/30.4 subline, as it showed greatest decrease in RBM10 protein expression. (C & D) RBM10v1 protein levels were monitored in the G300.3, G29/30.1, G29/30.3 and G29/30.4 sublines using the Bethyl RBM10 antibody. (C) One representative Western blot result is presented for RBM10v1 and α-tubulin (loading control). (D) Densitometric results of the average RBM10v1 protein levels from three biological replicates performed in duplicate. Analysis was performed using the AlphaEaseFC, ‘1D-Multi’ analysis tool. Values of RBM10v1 were normalized to the α-tubulin of each biological replicate, and then made relative to the G300.3 control subline. Standard error is presented. Subline expression levels were compared using the Student’s unpaired t-test, between sublines. (E) G300.3, G29/30.1, G29/30.3 and G29/30.4 were grown for five days and dehydrogenase activity was monitored daily using an MTT assay. Absorbance was plotted relative to day 0. The average of three biological replicates performed in eight technical replicates with standard error is presented. A Two-way ANOVA was performed between the G300.3 and other sublines, with Bonferroni post-hoc analysis. * p < 0.05, ** p < 0.01, and **** p < 0.0001.
The effect of RBM10KD on dehydrogenase activity was analyzed by MTT assay over a five day period (Fig 3E). The G300.3 and G29/30.1 sublines had similar dehydrogenase activity, a result that was anticipated as both sublines had nearly identical levels of RBM10v1 protein expression (Fig 3D). The G29/30.3 and G29/30.4 sublines, however, both displayed significantly reduced activity, relative to the G300.3 control. G29/30.4 had the greatest reduction in dehydrogenase activity, with significant differences on day four and day five (p < 0.01 and p < 0.0001, respectively), compared to G300.3.
These results indicate that the effect of RBM10 was level-dependent. More importantly, however, is the observation that inhibition of RBM10 expression correlated with dehydrogenase activity that was decreased. Decreased activity could have resulted from either a decrease in cell number and/or a decrease in the metabolic rate of existing cells. Since this finding in an RBM5-null background is in contradiction to the previous work in RBM5-retaining HeLa cells [14, 17], our results suggest that RBM10 function can not only be altered, but actually reversed by RBM5.
Effect of RBM10 inhibition on signaling pathways in GLC20 cells
To gain a more comprehensive understanding of the extent of RBM10’s influence in SCLC, we carried out RNA-Seq, because the MTT assay used above identifies only one functional consequence resulting from inhibition of RBM10. For these experiments, the stable RBM10KD G29/30.4 subline was used, as it had the greatest RBM10KD (referred to simply as RBM10KD in the following figures and the Discussion). As our Control, we sequenced and combined data for the stable pcDNA3 transfected GLC20 cells [9] and the stable scramble shRNA control G300.3. Sequencing and analysis were performed as previously described [9]. Specific parameters are listed in Materials and Methods. Using the Cufflinks suite, we identified 1157 significantly differentially expressed genes, representing approximately 4.45% of the transcriptome (S2 Table).
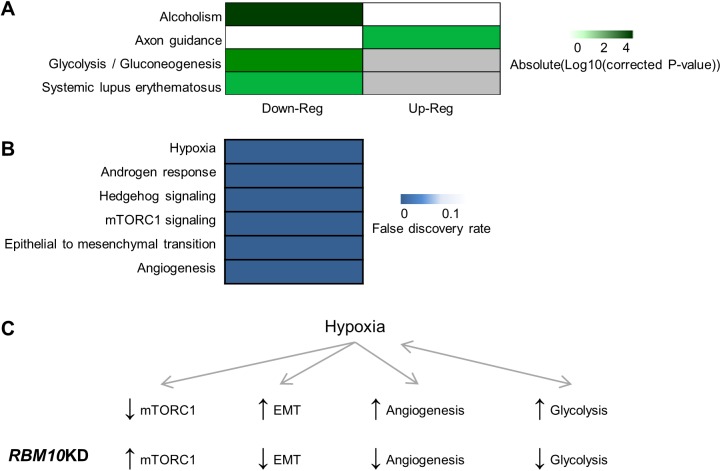
To determine the functional relevance of the differentially expressed genes, we performed pathway analyses. To ensure the validity of our results, as for the RIP-Seq data analyses, two different pathway analysis programs were used, each with their own database. We began with the FIDEA Program and the KEGG Database. With these tools, that group genes based on molecular interactions, we identified four enriched pathways (Fig 4A). The only upregulated pathway upon RBM10KD was ‘Axon guidance’. Genes involved in axon guidance are deleted or the subject of promoter hypermethylation in many cancers [39]. As RBM10 expression would correlate with decreased expression of these ‘Axon guidance’ factors, this may be one way by which RBM10 could promote the transformed state of lung cells. On the other hand, all three pathways that were significantly downregulated upon RBM10KD are positively linked with cancer; (1) ‘Alcoholism’-related pathways have been shown to promote the transformed state, in part via increased EMT [40], (2) ‘Glycolysis’ is upregulated in cancers and is the main pathway for ATP production, even in the presence of oxygen [41], and (3) ‘Systemic lupus erythematosus’, an autoimmune disease, is linked with an increased risk of cancer, especially cancers of hematologic origin, as well as lung and hepatobiliary cancers [42, 43]. These results suggest that RBM10 could promote the transformed state in a wide variety of ways, including modulation of the immune system and various aspects of cell metabolism.
Fig 4. Pathway analysis of genes differentially expressed upon RBM10KD in GLC20 cells.
(A) KEGG pathways significantly enriched upon RBM10KD in GLC20 cells. Analysis performed using subline RNA-Seq expression data and the FIDEA pathways analysis program. (B) MSigDB Hallmark gene sets enriched at a false discovery rate of below 5% upon RBM10KD in GLC20 cells. Analysis performed using subline RNA-Seq expression data and the GSAASeqSP pathway analysis program. (C) Relationship between select RBM10-altered pathways in GLC20 cells. Hypoxia is associated with decreased levels of mTORC1 signaling, and promotion of EMT and angiogenesis. In addition, hypoxia is associated with increased levels of glycolysis, and likewise, survival in hypoxic conditions is promoted by increased levels of glycolysis. The influence of RBM10KD on these pathways, as determined by RNA-Seq is indicated.
To complement our FIDEA and KEGG pathway analysis, we used the Gene Set Association Analysis with Sequence Permutation (GSAASeqSP) Program, which is specific for RNA-Seq data [44], with the Broad Institute’s Molecular Signatures Hallmark Database (MSigDB), which groups genes based on known functions [45]. As shown in Fig 4B, six gene sets with false discovery rates (FDR) below 5% were enriched upon RBM10KD in GLC20 cells. The most enriched gene set was ‘Hypoxia’, which coordinates well with the KEGG pathway analysis results described above; hypoxic conditions increase rates of glycolysis, and likewise, increased glycolysis supports tumor growth in hypoxic conditions [46]. Interestingly, three of the other five gene sets significantly enriched upon RBM10KD are also influenced by hypoxia; ‘mTORC1 signaling’, ‘Epithelial to Mesenchymal Transition’ (EMT) and ‘Angiogenesis’. Of note, the ‘mTORC1 signaling’ pathway, which regulates cell growth, is downregulated during hypoxia [47]. Conversely, EMT and angiogenesis can be promoted during hypoxia [48, 49] (Fig 4C). Although our GSAASeqSP analysis showed that these three hypoxia-related gene sets were enriched upon RBM10KD, we went on to determine if RBM10KD specifically promoted or inhibited these processes.
Firstly, in hypoxic conditions, downregulation of mTORC1 signaling is accomplished, in part, by ATM protein kinase, REDD1 and BNIP3 [47, 50, 51]. Interestingly, in our system, RBM10KD correlated with decreased levels of all three of these genes: ATM levels decreased from 27.98 fragments per kilobase of transcript per million mapped read (FPKM) (Control) to 20.188 FPKM (RBM10KD), REDD1 levels decreased from 71.65 FPKM (Control) to 30.96 FPKM (RBM10KD), and BNIP3 levels decreased from 58.02 FPKM (Control) to 19.70 FPKM (RBM10KD). These results suggest that mTORC1 signaling is promoted upon RBM10KD (Fig 4C). Secondly, we examined the impact of RBM10KD on the expression of previously published EMT-associated genes [52]. Notably, all genes listed as downregulated during EMT in the referenced review were significantly upregulated upon RBM10KD in GLC20 cells, or showed no significant change in expression. The significantly upregulated genes included CDH1, TJP1, PATJ, JUP and DSC2. On the other hand, all genes listed as upregulated during EMT, except fibronectin, were either significantly downregulated upon RBM10KD in GLC20 cells or showed no significant change expression. The significantly downregulated genes included VIM, ID1 and ZEB1. Our results therefore show that RBM10KD is associated with decreased EMT (Fig 4C). This is in line with our KEGG pathway analysis, which showed that RBM10KD decreased the EMT-promoting ‘Alcoholism’ pathway.
Finally, we examined the expression of angiogenesis markers [49] in our RBM10KD samples: RBM10KD significantly decreased the expression of three by at least nine FPKM (VEGF from 20 to 9 FPKM; FGF10 from 10 to 1 FPKM; PLGF from 75 to 33 FPKM), and increased the expression of two others, but only by 1–2 FPKM (THBS1 from 1.75 to 3.8 FPKM; ANGPT1 from 0.89 to 1.8 FPKM). These results show that, in general, RBM10KD correlates with decreased angiogenic marker expression (Fig 4C).
In summary, our GSAASeqSP pathway analysis showed that RBM10KD correlated with increased mTORC1 signaling, and decreased EMT and angiogenesis. Therefore, RBM10 expression would be expected to downregulate mTORC1 signaling and promote EMT and angiogenesis, thereby influencing these processes in a similar way as hypoxia, and consequently promoting the transformed state. The large overlap in the results obtained using the two different pathway analysis programs (FIDEA and GSAASeqSP), each with their own database (KEGG and MSigDB), strongly supports our conclusion that RBM10 influences various hypoxia-related pathways to promote the transformed state. Together, our pathway analysis results suggest that, unlike RBM5, RBM10 promotes the transformed state. RBM10 does this specifically by (1) increasing glycolysis, (2) reducing mTORC1 signaling, which is associated with deregulated cell growth and could contribute to the higher viable cell metabolism activity observed in RBM10 expressing GLC20 cells by MTT (Fig 3E), (3) promoting EMT, thereby contributing to the establishment and progression of the transformed state, and (4) promoting angiogenesis. This wide scope of influence is supported by the large number of RBM10-only targets identified by RIP-Seq.
Differential gene expression in GLC20 RBM10KD cells compared to RBM5 expressing GLC20 cells
Having determined that RBM5 and RBM10 share similar targets in the C4 subline, and affect similar processes in SCLC, but with opposing functional consequences, we decided to identify and compare the genes influenced by both proteins. To this end, we examined differential gene expression following either RBM10KD or RBM5 expression in GLC20 cells. We anticipated that many of the same genes would be influenced, but in an opposite manner, by each protein.
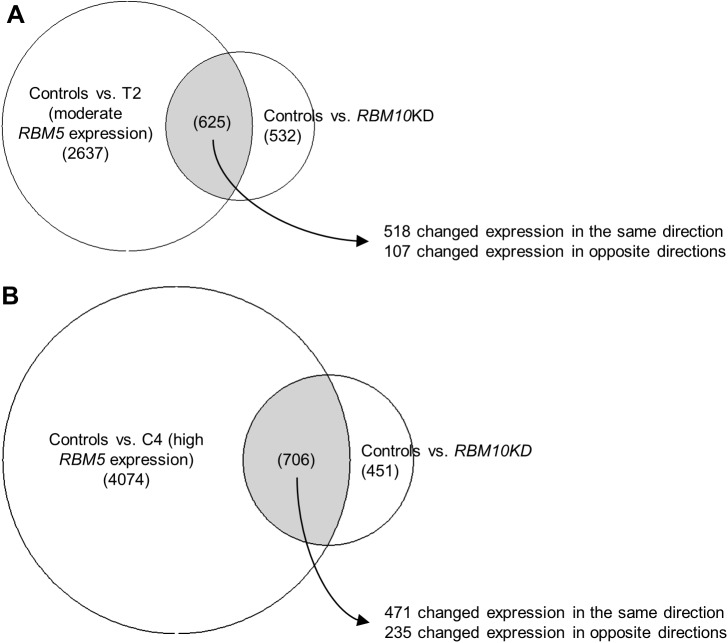
Comparing the RNA-Seq data from our G29/30.4 GLC20 RBM10KD subline with the RNA-Seq data from the stable RBM5-expressing GLC20 sublines T2 and C4, we determined that many of the same genes were in fact differentially expressed following either RBM10KD or RBM5 expression; of the significantly differentially expressed genes in RBM10KD, 53.8% were also significantly differentially expressed between Control and T2, and 60.7% were also differentially expressed between Control and C4 (Fig 5). As expected, well over half of these common differentially expressed genes (428 genes) were the same whether RBM10KD was compared to T2 or C4. Therefore, RBM5 and RBM10 do affect the expression of many of the same genes.
Fig 5. Comparison of genes differentially expressed upon RBM10KD in GLC20 cells or RBM5 expression in GLC20 sublines.
Differentially expressed genes, based on RNA-Seq results, in GLC20 cells upon RBM10KD, and moderate (A) or high (B) expression levels of RBM5 expression.
We then went on to determine how RBM5 and RBM10 levels affected the expression of these commonly differentially expressed genes. Between RBM10KD and T2 RBM5 expression, 82% of the common significantly differentially expressed genes changed expression in the same direction (Fig 5A). As for RBM10KD and C4 RBM5 expression, 67% of the differentially expressed genes changed expression in the same direction (Fig 5B). Next, we wanted to determine if this similarity would extend to genes specifically associated with SCLC. In June 2014, the National Cancer Institute compiled a list of ‘Genes of Interest in SCLC’, as part of their report entitled Scientific Framework for Small Cell Lung Cancer. We recently demonstrated that RBM5 expression in C4 significantly altered the expression of 11 of these SCLC-associated genes in a way which would suppress tumor growth [9]. Here we now show that RBM10KD altered the expression of nine of these 11 genes in the same direction as RBM5 expression; BLC2, CCNE1, CREBBP, EPHA7, MED12L, MYCL, SLIT2, SMO and SOX2 (Table 2). This high level of similarity between the effect of RBM5 expression and RBM10KD on gene expression, especially SCLC-associated genes, suggests opposing roles for RBM5 and RBM10 in aggressive SCLC, and supports our MTT and RNA-Seq findings.
Table 2. Expression of select National Cancer Institute (NCI) ‘Genes of Interest in SCLC’ in Control and RBM10KD samples, as determined by RNA-Seq.
| Gene name | Control | RBM10KD | p-value |
|---|---|---|---|
| BCL2 | 1.81928 | 1.26218 | 0.29525 |
| CCNE1 | 18.6741 | 15.4965 | 0.2551 |
| COBL † | 0.774161 | 1.82919 | 0.00095 |
| CREBBP ‡ | 5.54439 | 9.8752 | 0.0001 |
| EPHA7 | 16.5578 | 21.467 | 0.09965 |
| MED12L † | 4.10916 | 2.75174 | 0.0181 |
| MYCL | 11.3944 | 8.92857 | 0.17185 |
| RAB37 | 0.737284 | 0.803781 | 0.758 |
| SLIT2 ‡ | 5.30312 | 10.1706 | 0.00005 |
| SMO † | 5.8898 | 4.02038 | 0.04565 |
| SOX2 † | 13.317 | 8.46985 | 0.0124 |
† Genes significantly downregulated upon RBM10KD (p < 0.05).
‡ Genes significantly upregulated upon RBM10KD (p < 0.05).
Effect of RBM5 on RBM10 expression
Previous studies have shown tumor-suppressor properties for both RBM5 and RBM10. In the results presented herein, however, using an aggressive RBM5-null SCLC model, we show an opposite effect for RBM10, correlating its expression with promotion of various transformation-associated processes. These results suggested to us that RBM5 may be a regulator of RBM10 function, a relationship previously supported by our work in a rat myoblast system [20]. To investigate if and how RBM5 might influence RBM10, we examined RBM10 mRNA and protein expression in our stable RBM5-expressing GLC20 sublines T2 and C4.
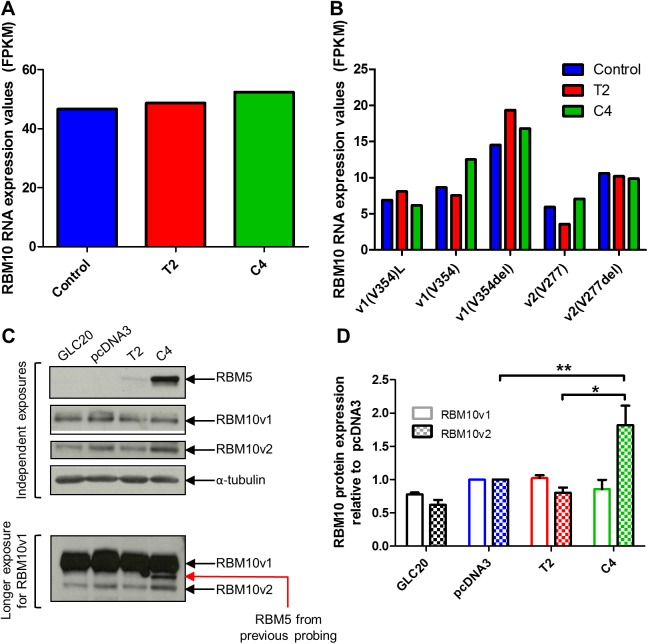
RBM10 mRNA levels in T2 and C4 were determined from RNA-Seq data [9]. Since RBM10 splice variants differ, in some cases, by only one amino acid, this level of resolution was required to precisely quantify the expression of each variant (Fig 1). Differential expression analysis of RNA-Seq results showed no significant change in RBM10 mRNA expression levels following RBM5 expression in GLC20 cells (Fig 6A). There was also no significant change in the expression of any RBM10 splice variant when RBM5 levels were altered (Fig 6B). RBM5 did not, therefore, influence RBM10 transcription or alternative splicing.
Fig 6. RBM10 expression in the parental GLC20 cell line and stable RBM5-expressing sublines.
(A) Expression of all RBM10 mRNA variants, as determined by RNA-Seq. (B) mRNA expression of specific RBM10 splice variants, as determined by RNA-Seq. ‘Control’ in A and B refers to the GLC20 parental cell line and stable pcDNA3-transfected GLC20 subline. (C) Representative Western blot for RBM5 and RBM10 (Sigma antibody) protein expression levels. Alpha-tubulin was used as loading control. (D) Densitometric results of the average RBM10v1/v2 expression of three biological replicates each performed in technical duplicate. Analysis was performed using AlphaEaseFC, ‘1D-Multi’ analysis tool. Values of RBM10v1 and RBM10v2 were normalized to the α-tubulin of each biological technical duplicate, and then made relative to the pcDNA3 controls. Standard error is presented. One-way ANOVA was performed with Tukey post-hoc analysis, between sublines. * p < 0.05 and ** p < 0.01.
To determine if RBM5 is capable of influencing RBM10 post-transcriptionally, RBM10 protein levels were investigated via Western blot. To note, only the two major isoforms of RBM10 (v1 and v2) are distinguishable on Western blots. Interestingly, RBM10v1 levels were not altered between sublines, whereas RBM10v2 levels were significantly increased in C4 compared to either T2 or the Control (Fig 6C and 6D). These results demonstrated that RBM5 was capable of influencing RBM10 expression, that this influence occurred post-transcriptionally, and that the effect was restricted to RBM10v2.
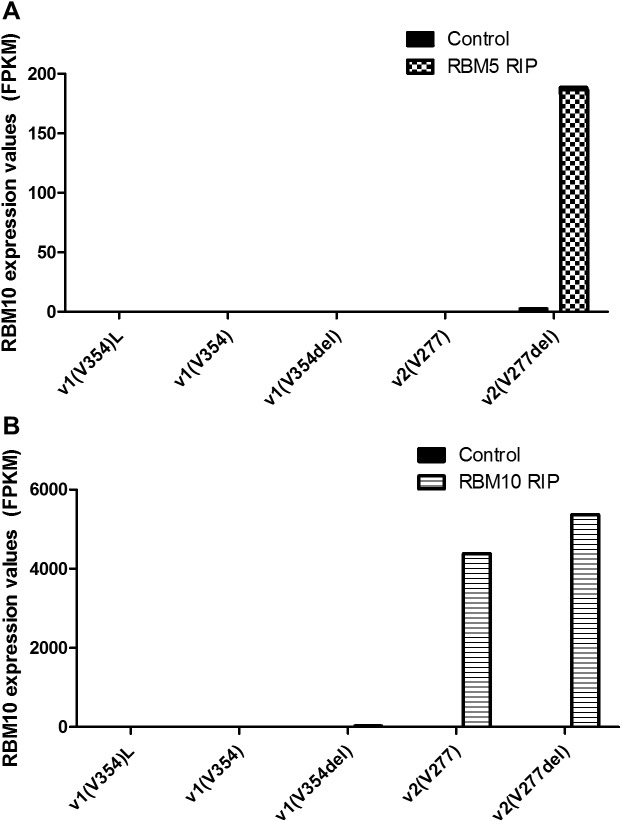
This increase in RBM10v2 levels could potentially have resulted from a targeted increase of RBM10v2 translation, or from stabilization of the RBM10v2 isoform, by high levels of RBM5. Increased RBM10v2 translation would likely involve binding of RBM5 protein to RBM10v2 transcript, whereas stabilization of the RBM10v2 isoform by RBM5 would involve a protein/protein interaction. As a next step in our investigations, we decided to examine the ability of RBM5 protein to interact with RBM10 transcripts. Towards this end, we re-examined our previous RBM5 RIP-Seq data (from C4 cells) [9]. As RIP-Seq provides a single nucleotide resolution of all RNA targets, we would be able to determine if RBM5 interacted with specific RBM10 splice variants. As shown in Fig 7A, RBM5 bound only the one specific RBM10 splice variant, RBM10v2(V277del). RBM5 bound 186.357 FPKM of RBM10v2(V277del), compared to 2.62 FPKM of IgG bound RBM10v2(V277del) in the Control RIP. All other RBM10 splice variants had counts of 0.1 FPKM or below, in the RBM5 RIP sample, a result clearly unrelated to variant expression levels in C4 (Fig 6B). This level of specificity, in regards to RBM5’s binding of only one RBM10 splice variant, is likely due to structural differences between RBM10 alternative splice transcripts, which is supported by our preliminary RNA structure analysis (S1 Fig) [53]. Interestingly, in an RBM5-null environment, we found that RBM10 was also capable of binding self; specifically, using our RIP-Seq data, we found that just like RBM5, RBM10 bound RBM10v2(V277del) mRNA. Unlike RBM5, which was only able to bind the one RBM10v2 variant, RBM10 was able to bind not only both variants of RBM10v2, at high levels (Fig 7B), but also RBM10v1(V354del), to a lesser extent (28.1 FPKM). These results show that RBM10 protein can in fact bind RBM10 mRNA and thus it may, like RBM5, be involved in regulating RBM10 isoform expression.
Fig 7. RBM5 and RBM10 RIP-Seq results for RBM10 splice variants.
Graph representing the expression of the various RBM10 splice variants in RBM5 (A) and RBM10 (B) RIP-Seq experiments, which were carried out in C4 cells.
Discussion
Functional RBM10 is important for normal cellular processes. For instance, mutations in RBM10 are the cause of TARP syndrome, which results in many developmental abnormalities and often lethality before or soon after birth [11, 54, 55]. Furthermore, RBM10 is mutated in pancreatic intraductal papillary mucinous neoplasms [56], 7% of lung adenocarcinomas [57], and 21% of invasive lung adenocarcinomas [21]. RBM10 has also been suggested to be important to rat myoblast differentiation [20]. Therefore, understanding the regulation of RBM10, as well as the range of its downstream effects, is of interest to various areas of cell, developmental and cancer biology.
Previous studies relating to RBM10, which have been performed in RBM5-expressing systems, have primarily linked RBM10 expression with reduced cell proliferation and promotion of apoptosis [10, 14–17]. The study reported here explores, for the first time, the function of RBM10 in an RBM5-null system, and the regulation of RBM10 by RBM5. As RBM5 is downregulated in many cancers [8, 58–62], this work is particularly meaningful. Here we show that RBM10 actually promotes transformation-associated processes in an RBM5-null environment. Supporting our findings, RBM10 has been associated with increased expression of VEGF, a promoter of new blood vessel growth, in breast cancer samples [16]. In addition, in patients with metastatic melanoma, high RBM10 expression was correlated with increased disease aggression [19]. Of note, the RBM5 promoter was identified as a “Signature of accelerated somatic evolution” in melanomas, with mutations in this region correlating with significantly lower survival rates and a higher incidence of metastasis [63]. Therefore, altered RBM5 levels in melanoma may result in decreased translational regulation of RBM10, and thus the pro-transformation characteristics associated with RBM10 in melanomas, and here in our RBM5-null system. In fact, in solid tumors in general, RBM5 is one of nine genes, within a 17 gene signature associated with metastasis, that are downregulated in various humans [64] and mice [65]. Thus, the increased metastatic characteristics of tumors with decreased RBM5 levels could in part involve decreased translational regulation of RBM10 by RBM5. This is particularly relevant as we show that RBM10 upregulates processes such as EMT and angiogenesis in an RBM5-null system. The majority of functional studies relating to RBM10 have focused on its role in modulation of alternative splicing [14, 17, 66–69], likely due to the early findings that RBM10 is a core component of spliceosomal A and B complexes [25, 26, 70]. Our RNA-Seq results show, however, no significant shifts in alternative splicing upon RBM10KD in GLC20 cells (no genes had a significant increase in the expression of one alternative splice variant and a significant decrease in the expression of another) (S2 Table), a finding we postulate may be dependent on RBM5 expression. Of note, upon RBM5 expression in GLC20 cells, using the same techniques and analysis parameters, we detected multiple alternative splicing changes. Our transcriptomic analyses, which show no effect of RBM10 downregulation on alternative splicing in GLC20 cells, are supported by (1) our RBM10-only targets (Table 1), which are involved in various aspects of gene expression regulation, excluding alternative splicing, (2) RBM10’s mRNA stabilization effect on its only identified direct RNA target, the AT1 receptor [71], and (3) RBM10’s interaction with the 2A-DUB deubiquitinase protein complex [72] and the Rac-specific GTPase-activitating protein FilGAP [73]. These findings are not unanticipated considering RNA binding proteins are known to participate in multiple aspects of RNA metabolism, including RNA editing, polyadenylation, export, localization, translation and stability [74]. Future studies regarding RBM10 should therefore attempt to capture its influence on these other aspects of RNA biology, and not be restricted to alternative splicing. Falling to do so would result in an incomplete view of the scope of RBM10’s influence on the cell.
More important than the role of RBM10 in the regulation of alternative splicing might be the role of alternative splicing of RBM10 itself. The single valine residue that differentiates the two RBM10v1 variants, as well as the two RBM10v2 variants, occurs within the second RNA Recognition Motif (RRM) of RBM10. The presence or absence of this valine residue can influence the α-helical structure of this RRM domain, and could thus influence RBM10’s binding targets [13, 14]. Although the functional implications of this modification have yet to be tested, the level of binding specificity exhibited by RBM5 suggests that (1) the presence or absence of this valine residue modifies RBM10’s structure sufficiently for proteins (at least RBM5) to be able to distinguish between variants, and (2) RBM10 splice variants have specific, potentially opposing, roles in the cell, and thus require specific regulation.
Based on our results and those of previous RBM10-related studies, we propose a working model describing RBM5’s influence on RBM10 and yet to be confirmed functional interactions between RBM5 and RBM10 (S2 Fig). This is an extension of a previous model we developed in a non-transformed rat myoblast system, which described how RBM10 might influence RBM5 [20]. We hypothesize that RBM10’s binding to RBM10v2 mRNA results in a downregulation of RBM10v2 protein expression levels. This would explain the much lower levels of RBM10v2 protein, compared to RBM10v1, that we observed in all GLC20 sublines (Fig 6C), even though mRNA levels for both variants were very similar (Fig 6A and 6B). Furthermore, we suggest that RBM5’s binding to RBM10v2(V277del) promotes this transcript’s translation or prevents its degradation, resulting in the higher levels of RBM10v2 protein seen in the C4 population. The lack of significantly higher RBM10v2 protein expression in T2 (Fig 6C), compared to Control, is not unexpected since this subline has substantially less RBM5 expressed than C4. We postulate that RBM10v2 is an essential component, or participates in the formation, of certain RBM5 protein complexes, which would explain why (1) RBM5 would upregulate RBM10v2 protein levels, (2) such a large proportion of RBM5 targets were also shared by RBM10 in our RIP-Seq experiments, and (3) RBM10 would have so many RBM10-only targets; these targets would represent RBM10v1 targets, which would be independent from those targeted by the RBM5-RBM10v2 complex.
We propose that the RBM5-RBM10v2 complex regulates cell cycle progression and promotes apoptosis, functions with which RBM5 has been previously associated. We suggest that most, but not all, RBM5’s tumor-suppressor properties occur via this RBM5-RBM10v2 complex. The RBM5-RBM10v2 complex could also be responsible for the alternative splicing events previously associated with RBM10. This would explain why, in an RBM5-null system, these alternative splicing events do not appear to be influenced by RBM10 (e.g., there would be no RBM5-RBM10v2 complex). On the other hand, we suggest that RBM10v1 promotes transformation-associated processes, including glycolysis, EMT and angiogenesis. Some of the downstream RBM5-RBM10v2 and RBM10v1 targets overlap, with both complexes affecting the expression of these genes in an opposing manner. This would explain the similarity of differentially expressed genes upon RBM5 expression and RBM10KD in GLC20 cells, but why RBM5 and RBM10 affected the expression of these genes in contrasting ways (Fig 5). We also propose that the RBM5-RBM10v2 complex has a more significant influence on transformation-associated pathways than RBM10v1, explaining why cells expressing physiological levels of both RBM5 and RBM10 have regulated cell growth (as seen in the T2 subline) [9].
According to our working model, upon RBM5 expression, an RBM5-RBM10v2 complex would be formed, resulting in higher levels of apoptosis and cell cycle arrest (S3A Fig). This is in line with what we have previously shown experimentally upon RBM5 expression in GLC20 cells [9]. Inversely, upon RBM5KD, less RBM5-RBM10v2 complex would be formed, resulting in decreased apoptosis and deregulated cell growth, as well as less competition for RBM10v1 regarding their common targets (S3B Fig). This would all contribute to promotion of a transformation-like phenotype. Indeed, this is what we observed previously in parental GLC20 cells, compared to the RBM5-expressing sublines [9]. Upon RBM10KD in an RBM5-expressing system, we would expect almost no RBM5-RBM10v2 complex to be formed, especially as RBM10v2 levels are already so much lower than RBM10v1 in GLC20 cells (S3C Fig). This would result in a significant decrease in apoptosis, as well as reduced regulation of cell cycle progression. Although knockdown of RBM10 would also significantly decrease RBM10v1 levels, since RBM10 would not be completely knocked out in this situation, some RBM10v1 would remain in the cell. This small amount of RBM10v1 could still promote transformation-associated processes to a certain point, especially with much less competition from the RBM5-RBM10v2 complex. In this case, we would expect a slight increase in cell growth upon RBM10KD in RBM5-expressing systems. This hypothesis is in line with previous functional work performed for RBM10 in endogenously RBM5-expressing HeLa cells [17]. Finally, upon RBM10KD in an RBM5-null system, since there would be no influence from the RBM5-RBM10v2 complex, RBM10KD would only significantly reduce levels of RBM10v1 (S3D Fig). This would result in diminished promotion of transformation-associated processes, and thus lower levels of cell growth and/or metabolism. This proposition is in line with the MTT and RNA-Seq results presented herein regarding RBM10KD in endogenously RBM5-null GLC20 cells (Figs 3E and 4).
Interestingly, RBM10 mutations have recently been associated with lung adenocarcinoma pathogenesis [66]. Loss-of-function RBM10 mutations would result in no functional RBM10 in the cell and, based on our working model, no RBM5-RBM10v2 complex. This would have two opposing consequences; (1) loss of RBM10v1 would result in reduced promotion of transformation-associated processes, and (2) loss of RBM5-RBM10v2 complex would result in reduced regulation of the cell cycle and decreased apoptosis, and thus promotion of transformation. Since, as we proposed above, the influence of RBM5-RBM10v2 is greater than RBM10v1, we would expect the overall effect of loss-of-function RBM10 mutations to be pro-transformatory, similar to the effects of RBM5KD. This is indeed the result seen in clinical samples; loss-of-function RBM10 mutations were associated with increased pathogenesis of lung adenocarcinomas [21]. These findings further support our working model.
A very recent study in non-transformed mouse cells not only supports our working model, but extends its relevance beyond cancer-based systems [75]. In that study, Rodor et al. knocked out RBM10 in mouse cells and examined a number of processes, including cell proliferation. According to our working model, since RBM10 would be completely depleted in this scenario, RBM10v1 could no longer promote transformation-associated processes and RBM5-RBM10v2 complexes would be completely eliminated. Since RBM10 would no longer influence cellular processes, RBM5 could exercise its RBM10v2-independent tumor-suppressor properties. We would thus predict that RBM10 knockout would result in a decrease in cell proliferation. This is indeed what the authors found. Although Rodor et al. strongly associated RBM10 with the regulation of alternative splicing in that study [75], as mouse cells in general express RBM5 [76], this could be an RBM5-dependant function of RBM10, as we propose above.
Considered together, our results suggest that the RBM10 reversal-of-function associated with downregulation of RBM5 is one means by which cells transition to a cancerous state, largely through processes that involve regulation of a hypoxic state. Our results shed light on the relationship between RBM5 and RBM10, a particularly important aspect to remember when studying either of these genes individually. We not only determine, for the first time, the role of RBM10 in an RBM5-null system, but also propose a working model to explain how RBM10 is influenced by RBM5 expression. We also identify all RBM10 targets and explore the extent of RBM10’s influence on cellular processes. In addition, we show that RBM5 directly binds RBM10 to regulate its expression post-transcriptionally, and that RBM5 and RBM10 share similar direct (RIP-Seq) and downstream (RNA-Seq) targets. These results lay the groundwork for additional studies to examine the role(s) of specific RBM10 isoforms, and the relationship between RBM5 and RBM10 in different systems and disease states.
Supporting information
Most probable RNA structure for (A) RBM10v1(V354), (B) RBM10v1(V354del), (C) RBM10v2(V277) and (D) RBM10v2(V277del), as determined by RNAstructure MaxExpect using standard settings. Structure generated is composed of highly probable base pairs. Black arrow indicates position of the ‘GTG’ codon alternatively spliced from both RBM10v1 and RBM10v2, which results in their respective +/- valine isoforms.
(PPTX)
Rectangles represent mRNA, whereas ovals represent protein. Blue ovals are unspecified proteins.
(PPTX)
Using our working model, we predict the effects of (A) RBM5 overexpression, (B) RBM5KD, (C) RBM10KD in an RBM5-expressing system, and (D) RBM10KD in an RBM5-null system. Open green, blue and purple arrows indicate effect, and thickness of said arrows indicates activity levels (dotted arrows indicate least activity). Closed red arrows indicate direction of expression change only. For detailed description of model, refer to text.
(PPTX)
RNA expression levels of all genes examined in RBM5 and RBM10 RIP-Seq experiments. Gene expression quantification was performed using Cuffdiff v2.2.1; however, targets were identified using the inclusion criteria elaborated in the Materials and Methods section. Genes listed based on log2(fold-change) in expression, and then their value in the experimental RIP sample.
(XLSX)
RNA expression levels for all genes examined, as well as their specific splice variants. Genes listed based on their q-value, and log2(fold-change) in expression. Ctrl refers to combined gene expression values for parental GLC20 cells and the stable pcDNA3 transfected GLC20 subline. Differential expression testing was performed using Cuffdiff v2.2.1. The third sheet contains the information required to identify a particular gene’s alternative splice variant(s) (based on their TCONS assigned number from Cufflinks).
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequencing data files are available from the NCBI Sequence Read Archive (SRA) database (accession number SRP106846): https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP106846&go=go.
Funding Statement
This work was supported by the Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) (JJL) (http://www.nserc-crsng.gc.ca/Students-Etudiants/PG-CS/BellandPostgrad-BelletSuperieures_eng.asp). Discovery Grant #9043429 from the Natural Sciences and Engineering Research Council of Canada (NSERC) (LCS) (http://www.nserc-crsng.gc.ca/Professors-Professeurs/Grants-Subs/DGIGP-PSIGP_eng.asp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E86. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366(9494):1385–96. doi: 10.1016/S0140-6736(05)67569-1 [DOI] [PubMed] [Google Scholar]
- 3.Johnson DH, Schiller JH, Bunn PA Jr., Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32(10):973–82. doi: 10.1200/JCO.2013.53.1228 [DOI] [PubMed] [Google Scholar]
- 4.Chute JP, Chen T, Feigal E, Simon R, Johnson BE. Twenty years of phase III trials for patients with extensive-stage small-cell lung cancer: perceptible progress. J Clin Oncol. 1999;17(6):1794–801. doi: 10.1200/JCO.1999.17.6.1794 [DOI] [PubMed] [Google Scholar]
- 5.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60(21):6116–33. [PubMed] [Google Scholar]
- 6.Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, et al. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60(7):1949–60. [PubMed] [Google Scholar]
- 7.Hung J, Kishimoto Y, Sugio K, Virmani A, McIntire DD, Minna JD, et al. Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. JAMA. 1995;273(24):1908 . [DOI] [PubMed] [Google Scholar]
- 8.Oh JJ, West AR, Fishbein MC, Slamon DJ. A candidate tumor suppressor gene, H37, from the human lung cancer tumor suppressor locus 3p21.3. Cancer Res. 2002;62(11):3207–13. [PubMed] [Google Scholar]
- 9.Loiselle JJ, Roy JG, Sutherland LC. RBM5 reduces small cell lung cancer growth, increases cisplatin sensitivity and regulates key transformation-associated pathways. Heliyon. 2016;2(11):e00204 doi: 10.1016/j.heliyon.2016.e00204 ; PubMed Central PMCID: PMCPMC5133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Bacon ML, Tessier JJ, Rintala-Maki ND, Tang V, Sutherland LC. RBM10 Modulates Apoptosis and Influences TNA-α Gene Expression. Journal of Cell Death. 2012;5:1–19. doi: 10.4137/JCD.S9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, Chong K, et al. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86(5):743–8. doi: 10.1016/j.ajhg.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94(1):5–24. doi: 10.1002/jcb.20204 [DOI] [PubMed] [Google Scholar]
- 13.Tessier SJ, Loiselle JJ, McBain A, Pullen C, Koenderink BW, Roy JG, et al. Insight into the role of alternative splicing within the RBM10v1 exon 10 tandem donor site. BMC Res Notes. 2015;8:46 doi: 10.1186/s13104-015-0983-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13(4):466–72. doi: 10.1080/15476286.2016.1144004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James CG, Ulici V, Tuckermann J, Underhill TM, Beier F. Expression profiling of Dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development. BMC Genomics. 2007;8:205 doi: 10.1186/1471-2164-8-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Arribas F, Agudo D, Pollan M, Gomez-Esquer F, az-Gil G, Lucas R, et al. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97(6):1275–82. doi: 10.1002/jcb.20725 [DOI] [PubMed] [Google Scholar]
- 17.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 Differentially Regulate NUMB Alternative Splicing to Control Cancer Cell Proliferation. Mol Cell. 2013;52(5):720–33. doi: 10.1016/j.molcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 18.Jackson TC, Du L, Janesko-Feldman K, Vagni VA, Dezfulian C, Poloyac SM, et al. The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J Cereb Blood Flow Metab. 2015;35(4):655–66. doi: 10.1038/jcbfm.2014.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrisi VM, Strippoli S, De SS, Pinto R, Perrone A, Guida G, et al. Proteomic profile and in silico analysis in metastatic melanoma with and without BRAF mutation. PLoS One. 2014;9(12):e112025 doi: 10.1371/journal.pone.0112025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loiselle JJ, Tessier SJ, Sutherland LC. Post-transcriptional regulation of Rbm5 expression in undifferentiated H9c2 myoblasts. In Vitro Cell Dev Biol Anim. 2015. doi: 10.1007/s11626-015-9976-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinayanuwattikun C, Le Calvez-Kelm F, Abedi-Ardekani B, Zaridze D, Mukeria A, Voegele C, et al. Elucidating Genomic Characteristics of Lung Cancer Progression from In Situ to Invasive Adenocarcinoma. Sci Rep. 2016;6:31628 doi: 10.1038/srep31628 ; PubMed Central PMCID: PMCPMC4992872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One. 2013;8(12):e85024 doi: 10.1371/journal.pone.0085024 ; PubMed Central PMCID: PMCPMC3871669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain R, Devine T, George AD, Chittur SV, Baroni TE, Penalva LO, et al. RIP-Chip Analysis: RNA-Binding Protein Immunoprecipitation-Microarray (Chip) Profiling In: Nielsen H, editor. RNA. Methods in Molecular Biology: Humana Press; 2011. p. 247–63. [DOI] [PubMed] [Google Scholar]
- 24.Smit EF, de Vries EG, Timmer-Bosscha H, de Leij LF, Oosterhuis JW, Scheper RJ, et al. In vitro response of human small-cell lung-cancer cell lines to chemotherapeutic drugs; no correlation with clinical data. Int J Cancer. 1992;51(1):72–8. [DOI] [PubMed] [Google Scholar]
- 25.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26(6):1737–48. doi: 10.1038/sj.emboj.7601631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26(14):5528–43. doi: 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Andrea D, Grassi L, Mazzapioda M, Tramontano A. FIDEA: a server for the functional interpretation of differential expression analysis. Nucleic Acids Res. 2013;41(Web Server issue):W84–8. doi: 10.1093/nar/gkt516 ; PubMed Central PMCID: PMCPMC3692084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. ; PubMed Central PMCID: PMCPMC102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070 ; PubMed Central PMCID: PMCPMC4702792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556 ; PubMed Central PMCID: PMCPMC3037419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56. doi: 10.1093/nar/gku1179 ; PubMed Central PMCID: PMCPMC4383973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finsterer J, Jarius C, Baumgartner M. Parkinson's disease associated with impaired oxidative phosphorylation. Neuroradiology. 2001;43(11):997–1000. . [DOI] [PubMed] [Google Scholar]
- 33.Shoffner JM, Watts RL, Juncos JL, Torroni A, Wallace DC. Mitochondrial oxidative phosphorylation defects in Parkinson's disease. Ann Neurol. 1991;30(3):332–9. doi: 10.1002/ana.410300304 . [DOI] [PubMed] [Google Scholar]
- 34.Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington's disease. Biochim Biophys Acta. 2010;1802(1):52–61. doi: 10.1016/j.bbadis.2009.07.012 . [DOI] [PubMed] [Google Scholar]
- 35.Biffi A, Sabuncu MR, Desikan RS, Schmansky N, Salat DH, Rosand J, et al. Genetic variation of oxidative phosphorylation genes in stroke and Alzheimer's disease. Neurobiol Aging. 2014;35(8):1956 e1–8. doi: 10.1016/j.neurobiolaging.2014.01.141 ; PubMed Central PMCID: PMCPMC4329419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer's disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5(2):147–62. doi: 10.1385/NMM:5:2:147 . [DOI] [PubMed] [Google Scholar]
- 37.Gu S, Kay MA. How do miRNAs mediate translational repression? Silence. 2010;1(1):11 doi: 10.1186/1758-907X-1-11 ; PubMed Central PMCID: PMCPMC2881910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. doi: 10.1101/gad.1399806 . [DOI] [PubMed] [Google Scholar]
- 39.Chedotal A, Kerjan G, Moreau-Fauvarque C. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ. 2005;12(8):1044–56. doi: 10.1038/sj.cdd.4401707 . [DOI] [PubMed] [Google Scholar]
- 40.Forsyth CB, Tang Y, Shaikh M, Zhang L, Keshavarzian A. Alcohol stimulates activation of Snail, epidermal growth factor receptor signaling, and biomarkers of epithelial-mesenchymal transition in colon and breast cancer cells. Alcohol Clin Exp Res. 2010;34(1):19–31. doi: 10.1111/j.1530-0277.2009.01061.x ; PubMed Central PMCID: PMCPMC3689303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. . [DOI] [PubMed] [Google Scholar]
- 42.Dey D, Kenu E, Isenberg DA. Cancer complicating systemic lupus erythematosus—a dichotomy emerging from a nested case-control study. Lupus. 2013;22(9):919–27. doi: 10.1177/0961203313497118 ; PubMed Central PMCID: PMCPMC4107831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernatsky S, Boivin JF, Joseph L, Rajan R, Zoma A, Manzi S, et al. An international cohort study of cancer in systemic lupus erythematosus. Arthritis Rheum. 2005;52(5):1481–90. doi: 10.1002/art.21029 . [DOI] [PubMed] [Google Scholar]
- 44.Xiong Q, Mukherjee S, Furey TS. GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Sci Rep. 2014;4:6347 doi: 10.1038/srep06347 ; PubMed Central PMCID: PMCPMC4161965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. doi: 10.1016/j.cels.2015.12.004 ; PubMed Central PMCID: PMCPMC4707969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robin ED, Murphy BJ, Theodore J. Coordinate regulation of glycolysis by hypoxia in mammalian cells. J Cell Physiol. 1984;118(3):287–90. doi: 10.1002/jcp.1041180311 . [DOI] [PubMed] [Google Scholar]
- 47.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell. 2010;40(4):509–20. doi: 10.1016/j.molcel.2010.10.030 ; PubMed Central PMCID: PMCPMC3004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10(3):295–305. doi: 10.1038/ncb1691 . [DOI] [PubMed] [Google Scholar]
- 49.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2(12):1117–33. doi: 10.1177/1947601911423654 ; PubMed Central PMCID: PMCPMC3411127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007;282(49):35803–13. doi: 10.1074/jbc.M705231200 . [DOI] [PubMed] [Google Scholar]
- 51.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–904. doi: 10.1101/gad.1256804 ; PubMed Central PMCID: PMCPMC534650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96. doi: 10.1038/nrm3758 ; PubMed Central PMCID: PMCPMC4240281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129 doi: 10.1186/1471-2105-11-129 ; PubMed Central PMCID: PMCPMC2984261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorlin RJ, Cervenka J, Anderson RC, Sauk JJ, Bevis WD. Robin's syndrome. A probably X-linked recessive subvariety exhibiting persistence of left superior vena cava and atrial septal defect. Am J Dis Child. 1970;119(2):176–8. . [PubMed] [Google Scholar]
- 55.Kurpinski KT, Magyari PA, Gorlin RJ, Ng D, Biesecker LG. Designation of the TARP syndrome and linkage to Xp11.23-q13.3 without samples from affected patients. Am J Med Genet A. 2003;120A(1):1–4. doi: 10.1002/ajmg.a.10201 . [DOI] [PubMed] [Google Scholar]
- 56.Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, Yamamoto M, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161 doi: 10.1038/srep00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welling DB, Lasak JM, Akhmametyeva E, Ghaheri B, Chang LS. cDNA microarray analysis of vestibular schwannomas. Otol Neurotol. 2002;23(5):736–48. [DOI] [PubMed] [Google Scholar]
- 59.Zhao L, Li R, Shao C, Li P, Liu J, Wang K. 3p21.3 tumor suppressor gene RBM5 inhibits growth of human prostate cancer PC-3 cells through apoptosis. World J Surg Oncol. 2012;10:247 doi: 10.1186/1477-7819-10-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller G, Socci ND, Dhall D, D'Angelica M, DeMatteo RP, Allen PJ, et al. Genome wide analysis and clinical correlation of chromosomal and transcriptional mutations in cancers of the biliary tract. J Exp Clin Cancer Res. 2009;28:62 doi: 10.1186/1756-9966-28-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng J, Valeshabad AK, Li Q, Wang Y. Differential expression of RBM5 and KRAS in pancreatic ductal adenocarcinoma and their association with clinicopathological features. Oncol Lett. 2013;5(3):1000–4. doi: 10.3892/ol.2012.1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim YS, Hwan JD, Bae S, Bae DH, Shick WA. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010;10:576 doi: 10.1186/1471-2407-10-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith KS, Yadav VK, Pedersen BS, Shaknovich R, Geraci MW, Pollard KS, et al. Signatures of accelerated somatic evolution in gene promoters in multiple cancer types. Nucleic Acids Res. 2015;43(11):5307–17. doi: 10.1093/nar/gkv419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060 [DOI] [PubMed] [Google Scholar]
- 65.Qiu TH, Chandramouli GV, Hunter KW, Alkharouf NW, Green JE, Liu ET. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res. 2004;64(17):5973–81. doi: 10.1158/0008-5472.CAN-04-0242 [DOI] [PubMed] [Google Scholar]
- 66.Zhao J, Sun Y, Huang Y, Song F, Huang Z, Bao Y, et al. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci Rep. 2017;7:40488 doi: 10.1038/srep40488 ; PubMed Central PMCID: PMCPMC5238425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Gogol-Doring A, Hu H, Frohler S, Ma Y, Jens M, et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol Med. 2013;5(9):1431–42. doi: 10.1002/emmm.201302663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng S, Damoiseaux R, Chen L, Black DL. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res. 2013;23(6):998–1007. doi: 10.1101/gr.147546.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inoue A, Yamamoto N, Kimura M, Nishio K, Yamane H, Nakajima K. RBM10 regulates alternative splicing. FEBS Lett. 2014;588(6):942–7. doi: 10.1016/j.febslet.2014.01.052 [DOI] [PubMed] [Google Scholar]
- 70.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12(8):1231–45. doi: 10.1101/gr.473902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller CF, Berger A, Zimmer S, Tiyerili V, Nickenig G. The heterogenous nuclear riboprotein S1-1 regulates AT1 receptor gene expression via transcriptional and posttranscriptional mechanisms. Arch Biochem Biophys. 2009;488(1):76–82. doi: 10.1016/j.abb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 72.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27(4):609–21. doi: 10.1016/j.molcel.2007.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamada H, Tsutsumi K, Nakazawa Y, Shibagaki Y, Hattori S, Ohta Y. Src Family Tyrosine Kinase Signaling Regulates FilGAP through Association with RBM10. PLoS One. 2016;11(1):e0146593 doi: 10.1371/journal.pone.0146593 ; PubMed Central PMCID: PMCPMC4709192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–86. doi: 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodor J, FitzPatrick DR, Eyras E, Caceres JF. The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 2017;14(1):45–57. doi: 10.1080/15476286.2016.1247148 ; PubMed Central PMCID: PMCPMC5270529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozuemba B, Masilamani TJ, Loiselle JJ, Koenderink B, Vanderbeck KA, Knee J, et al. Co- and post-transcriptional regulation of Rbm5 and Rbm10 in mouse cells as evidenced by tissue-specific, developmental and disease-associated variation of splice variant and protein expression levels. Gene. 2016;580(1):26–36. doi: 10.1016/j.gene.2015.12.070 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Most probable RNA structure for (A) RBM10v1(V354), (B) RBM10v1(V354del), (C) RBM10v2(V277) and (D) RBM10v2(V277del), as determined by RNAstructure MaxExpect using standard settings. Structure generated is composed of highly probable base pairs. Black arrow indicates position of the ‘GTG’ codon alternatively spliced from both RBM10v1 and RBM10v2, which results in their respective +/- valine isoforms.
(PPTX)
Rectangles represent mRNA, whereas ovals represent protein. Blue ovals are unspecified proteins.
(PPTX)
Using our working model, we predict the effects of (A) RBM5 overexpression, (B) RBM5KD, (C) RBM10KD in an RBM5-expressing system, and (D) RBM10KD in an RBM5-null system. Open green, blue and purple arrows indicate effect, and thickness of said arrows indicates activity levels (dotted arrows indicate least activity). Closed red arrows indicate direction of expression change only. For detailed description of model, refer to text.
(PPTX)
RNA expression levels of all genes examined in RBM5 and RBM10 RIP-Seq experiments. Gene expression quantification was performed using Cuffdiff v2.2.1; however, targets were identified using the inclusion criteria elaborated in the Materials and Methods section. Genes listed based on log2(fold-change) in expression, and then their value in the experimental RIP sample.
(XLSX)
RNA expression levels for all genes examined, as well as their specific splice variants. Genes listed based on their q-value, and log2(fold-change) in expression. Ctrl refers to combined gene expression values for parental GLC20 cells and the stable pcDNA3 transfected GLC20 subline. Differential expression testing was performed using Cuffdiff v2.2.1. The third sheet contains the information required to identify a particular gene’s alternative splice variant(s) (based on their TCONS assigned number from Cufflinks).
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequencing data files are available from the NCBI Sequence Read Archive (SRA) database (accession number SRP106846): https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP106846&go=go.